Abstract
Cellular delivery of DNA vectors for the expression of therapeutic proteins is a promising approach to treat monogenic disorders or cancer. Significant efforts in a preclinical and clinical setting have been made to develop potent nonviral gene delivery systems based on lipoplexes composed of permanently cationic lipids. However, transfection efficiency and tolerability of such systems are in most cases not satisfactory. Here, we present a one-pot combinatorial method based on double-reductive amination for the synthesis of short-chain aminolipids. These lipids can be used to maximize the DNA vector delivery when combined with the cationic lipid 1,2-dioleoyl-3-trimethylammonium propane (DOTAP). We incorporated various aminolipids into such lipoplexes to complex minicircle DNA and screened these systems in a human liver-derived cell line (HuH7) for gene expression and cytotoxicity. The lead aminolipid AL-A12 showed twofold enhanced gene delivery and reduced toxicity compared to the native DOTAP:cholesterol lipoplexes. Moreover, AL-A12-containing lipoplexes enabled enhanced transgene expression in vivo in the zebrafish embryo model.
Introduction
Delivery of genetic material to diseased cells is of growing clinical interest. Many inherited liver diseases including Crigler–Najjar syndrome or Gilbert’s syndrome are associated with genetic defects leading to loss of function of proteins or low levels of enzymatic activity.1−8 Gene therapy enables the introduction of genetic material (e.g., DNA vectors) encoding for a therapeutic protein, thereby restoring physiological functions. Alternatively, the expression of tumor suppressors, proapoptotic or oncotoxic proteins, offers an interesting option for the treatment of malignant disorders.9−16
In recent years, various nonviral delivery systems have been investigated in preclinical and clinical settings for the delivery of recombinant DNA.17 Electrostatic interaction of cationic lipids and negatively charged DNA vectors of virtually any size induces condensation. The resulting lipoplexes are formed spontaneously. Their preparation and handling are simple and cost-effective. Moreover, lipoplexes allow for a straightforward transfer “from bench to large-scale production” compared to viral vectors18 without the risk for insertion of genetic material into the host’s genome as shown for adeno-associated virus vectors.19−21 Although cationic lipids overcome some of the limitations described for viral vectors, they encompass some disadvantages: The permanent positive charge of cationic lipids is associated with cytotoxicity,22 and lipoplex systems generally lack efficiency compared to viral vectors.23,24 As a result, only a few types of cationic lipids have entered clinical trials. In 2017, lipofection (i.e., the introduction of exogenous genetic material by means of lipid-based transfection reagents) accounted for only 4.5% of all clinical trials involving gene therapy.25 As one of the exceptions, 1,2-dioleoyl-3-trimethylammonium propane and cholesterol (DOTAP:chol)-based lipoplexes have been clinically investigated for the treatment of various diseases including lung cancer, breast cancer, prostate cancer, or hepatocellular carcinoma.9,13,15,16,26,27 The cationic charge of DOTAP:chol lipoplexes enables facilitated cellular interactions and also accounts for decreased cell viability.24,28 Therefore, as observed for many lipoplexes, the use of DOTAP:chol systems is limited by poor transfection efficiency and cytotoxic effects.29−32
The aim of the present study was to maximize the potency of clinically tested DOTAP:chol lipoplexes while reducing toxicity. Based on our finding that short-chain (C8–C12) amide lipidoids are potential carriers for siRNA,33 a combination of short-chain aminolipids and DOTAP:chol-based lipoplexes was explored to enhance DNA vector delivery. It is noteworthy that enhanced gene silencing and reduced cytotoxic potential were demonstrated for polymeric nanoparticles after combination with natural polyphenols and polyphenol inspired polycatechols.34,35 In view of these results, a combinatorial library of different types of aminolipids with chain lengths of C10 or C12 and various headgroups, including aliphatic and heterocyclic functional groups, was synthesized by double-reductive amination. The designed aminolipids were incorporated into DOTAP:chol lipoplexes and screened in vitro for transfection efficiency and cellular toxicity. The most promising aminolipid-containing system was then compared to DOTAP:chol lipoplexes for in vivo transfection efficiency in the zebrafish embryo model.
Materials and Methods
Materials
Text adopted from Neuhaus et al.36 The starting compounds and solvents were purchased from Sigma-Aldrich (St. Louis, MO), Honeywell Fluka (Fisher Scientific AG, Reinach, Switzerland), ABCR (Zug, Switzerland), TCI Deutschland GmbH (Eschborn, Germany), or Acros Organics (Thermo Fisher Scientific, Geel, Belgium) and were used without further purification. For reactions under inert gas conditions, the solvent dichloromethane (DCM) was dried over molecular sieves 4 Å and degassed afterward. Column chromatography was carried out using 230–400 mesh, 60 Å silica gel (Chemie Brunschwig AG, Basel, Switzerland). TLC plates (Merck, Darmstadt, Germany, Silica gel 60 F254) were developed with KMnO4 solution. 1H and 13C NMR spectra were recorded (as indicated) on Bruker 300, 400, or 600 MHz spectrometer (Bruker, Billerica, MA) and are reported as chemical shifts in ppm relative to TMS, calibrated to the signal of the deuterated NMR solvent (300 and 400 MHz). Spin multiplicities are reported as singlet (s), doublet (d), triplet (t), quintet (qui), with coupling constants (J) given in hertz, or multiplet (m). Broad peaks are marked as br. NMR experiments (600 MHz) were performed on a Bruker Avance III NMR spectrometer operating at 600.13 MHz proton frequency. The instrument was equipped with a direct observe 5 mm BBFO smart probe. The experiments were performed at 298 K, and the temperature was calibrated using a methanol standard showing accuracy within ±0.2 K. HRESI-MS was performed on a QSTAR Pulsar (AB Sciex Switzerland GmbH, Baden, Switzerland) spectrometer and are reported as mass-per-charge ratio m/z. IR spectra were recorded on a PerkinElmer Spectrum One Fourier transform infrared (FT-IR) spectrometer (ATR, Golden Gate, PerkinElmer, Basel, Switzerland). 1,2-Dioleoyl-3-trimethylammonium-propane (DOTAP) was obtained from Corden Pharma Switzerland LLC (Liestal, Switzerland). Glucose was purchased from Roth AG, Switzerland. Dulbecco’s modified Eagle’s medium (DMEM) high glucose was obtained from Sigma-Aldrich Co. (St. Louis, MO) and supplemented with 10% FCS (BioConcept, Allschwil, Switzerland) and penicillin (100 units/mL)–streptomycin (100 μg/mL) (Sigma-Aldrich Co., St. Louis, MO). Trypsin/EDTA (0.25%) was purchased from Invitrogen, Life Technologies (Zug, Switzerland). FACS buffer, composed of D-PBS (Sigma-Aldrich Co, St. Louis, MO), was supplemented with 2% FCS and 0.1% NaN3 (Sigma-Aldrich Co., St. Louis, MO). The MTT reagent comprised a 1:10 dilution (v/v) of a 5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide stock solution in DMEM. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide was purchased from Carl Roth (Karlsruhe, Germany). SDS was purchased from Bio-Rad Laboratories, Cressier, Switzerland. HCl and isopropanol were obtained from Merck KGaA (Darmstadt, Germany).
The extrusion equipment consisted of 100 nm polycarbonate membranes (Whatman Nuclepore Track-Etched Membranes, GE Healthcare Life Sciences, Buckinghamshire, U.K.), filter supports (Whatman Drain Disc 10 mm PE, GE Healthcare Life Sciences, Buckinghamshire, U.K.), and a hand extruder (Avanti Mini Extruder, Avanti Polar Lipids, Inc., AL). Particle size and ζ-potential measurements were performed using a Delsa Nano C Particle Analyzer (Beckman Coulter, Inc., Indianapolis, IN). Transmission electron microscopy (TEM) was conducted using a CM-100 electron microscope (Philips, Eindhoven, the Netherlands). Transfection experiments and cytotoxicity experiments were conducted in 24-well plates (TPP Tissue Culture Testplate 24, TPP Techno Plastic Products AG, Trasadingen, Switzerland) and in 96-well plates (TPP Tissue Culture Testplate 96F, TPP Techno Plastic Products AG, Trasadingen, Switzerland), respectively. The minicircle DNA was kindly provided by the University Children’s Hospital Zürich, Switzerland. Statistical evaluation was carried out using OriginPro 2018 (64-bit) SR1 b9.5.1.195 (Academic) (OriginLab Corporation, Northampton, MA).
Synthesis of Aminolipids
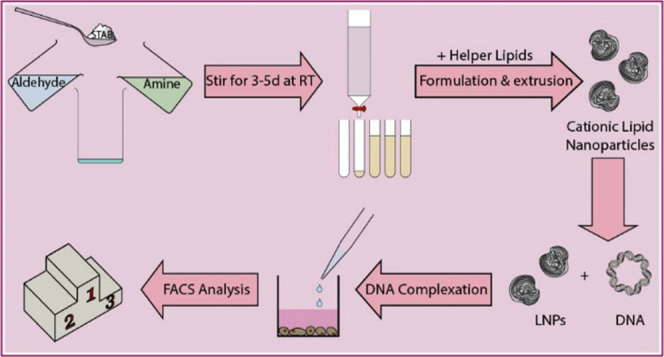
A small library of 12 multitailed aminolipids was obtained via reductive amination of decanal or dodecanal with different amines, as depicted in Figure 1. Briefly, the various amines representing the headgroups of the respective aminolipids were mixed with excess aldehyde (representing the tails) in dichloromethane (DCM). The mixture was stirred for 3 h at RT, followed by the addition of sodium triacetoxyborohydride (STAB) and further stirred for 2–5 days at RT. Afterward, the solvent was evaporated under reduced pressure. The obtained aminolipids were then redissolved in 30 mL of DCM. Brine (30 mL) was added, and the aminolipids were extracted three times with DCM. The crude product was then purified using column chromatography (silica) and a gradient of an appropriate solvent system, e.g., DCM/MeOH or EtOAc/MeOH (e.g., 98:2 to 90:10 (v/v)). The obtained aminolipid fractions were checked for the presence of aminolipid using thin-layer chromatography (TLC). Clean fractions containing the aminolipid (according to TLC) with the correct mass (determined by MS) were combined, and the solvent was evaporated under reduced pressure. For the final drying step, aminolipid samples were exposed to vacuum using an HV pump for at least 2 days. Detailed descriptions and NMR plots for each aminolipid can be found in the Supporting Information.
Figure 1.
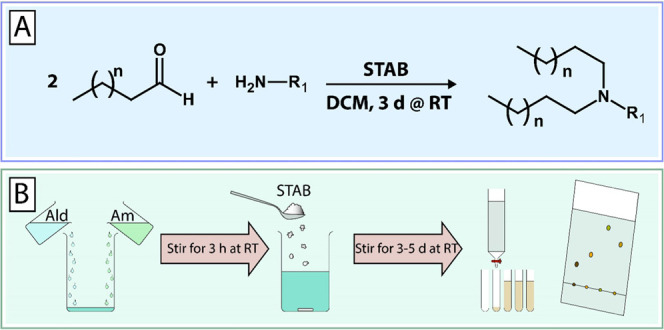
One-pot combinatorial synthesis of short-chain aminolipids via double-reductive amination. (A) Double-reductive amination of aldehydes with amines in dichloromethane (DCM) in the presence of sodium triacetoxyborohydride (STAB) as a reducing agent. (B) Briefly, amines (Am) are mixed with excess aldehyde (Ald) and stirred at RT for 3 h followed by the addition of STAB. The reaction continues under stirring at RT for 3–5 days. The obtained aminolipids are then extracted using DCM/brine and then purified by column chromatography (silica, DCM/MeOH 98:2 → 90:10). A detailed description of the chemical synthesis and experimental procedures is provided in the Supporting Information.
Preparation of Aminolipid-Based Systems
DNA Vector Delivery
Aminolipids in chloroform were combined with chloroform stock solutions of cholesterol, DOPE, or DOTAP (1:1 mol/mol) or a combination of either cholesterol and DOTAP, or DOPE and DOTAP (aminolipid:helper:DOTAP = 5:4:1 (mol/mol)) in a glass vial. The (amino-)lipid mixture in chloroform was subsequently dried under nitrogen flow overnight. On the next day, the components were rehydrated using 5% d(+)-glucose solution in Milli-Q ddH2O to obtain a final DOTAP (or aminolipid-) concentration of 8 mM. Afterward, the mixtures were stirred and vortexed to ensure complete rehydration of the (amino-)lipid components. Following this step, the rehydrated mixtures were subjected to five freeze–thaw cycles (i.e., frozen for 5 min using dry ice followed by thawing for 5 min in a water bath at 62 °C). After freeze–thawing, the (amino-)lipid mixtures were left at room temperature (RT) for 3 h. Subsequently, the mixtures were extruded 15 times through a Whatman Nuclepore Track-Etched Membrane with a pore size of 0.1 μm using an Avanti Mini Extruder.
Physicochemical Characterization of Aminolipid-Based Systems
Directly after extrusion, the size, polydispersity index (PDI), and ζ-potential of the aminolipid nanoparticles were measured using a Delsa Nano C Particle Analyzer at RT, as described previously.37,38 Particle size and PDI were determined using CONTIN data conversion after measurement of the extruded nanoparticle preparation (cDOTAP = 8 mM) using a 658 nm laser. Scattered light was detected at an angle of 165°. ζ-Potential data were converted using the Smoluchowski equation after measurement of nanoparticles diluted 1:50 (v/v) in 5% glucose solution at an angle of 15°. Transmission electron microscopy (TEM) images of both pure lipid nanoparticles (LNPs) and LNPs complexed with DNA were acquired on a CM-100 electron microscope (Philips, Eindhoven, the Netherlands). Thus, samples were loaded on a carbon-coated copper grid and counterstained using 2% uranylacetate solution. Excess uranylacetate was removed and the grids were dried at RT overnight prior to TEM imaging.
Cell Culture
HuH7 cells were cultured in high-glucose DMEM, supplemented with 10% fetal calf-serum (FCS), penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37 °C and 5% CO2. For transfection experiments, the cells were detached at 80–90% confluency using 0.25% trypsin/EDTA (Invitrogen, Life Technologies, Zug, Switzerland) and seeded in 24-well plates (TPP Tissue Culture Testplate 24, TPP Techno Plastic Products AG, Trasadingen, Switzerland) at a seeding density of 5 × 104 cells per well. For cytotoxicity experiments, the cells were detached as for transfection experiments and seeded in 96-well plates (TPP Tissue Culture Testplate 96F, TPP Techno Plastic Products AG, Trasadingen, Switzerland) at a seeding density of 104 cells per well.
Transfection Experiments
DNA Vector Delivery
Twenty-four hours after seeding, the cells were transfected with minicircle DNA (mcDNA) encoding for enhanced green fluorescent protein (eGFP 1) under the control of a liver-specific p3 promoter. Aminolipid–LNPs were combined with DNA at the indicated ratios and amounts of DNA. Briefly, aminolipid–LNPs and DNA were diluted in separate 1.5 mL Eppendorf tubes in 5% glucose solution. After gentle mixing, the contents of the tubes were combined, and the mixture was left for 30 min on the bench to allow for formation of complexes between aminolipid–LNPs and DNA. After incubation of the DNA with the LNPs, 50 μL of LNP/DNA nanoparticles was added to the cell culture medium (final volume: 1 mL). The ratios of the combination of DNA with LNPs are depicted as the amount of DNA in micrograms and the final concentration of LNPs (μM) in cell culture medium during transfection experiments. The reason for this depiction is that Lipofectamine 3000, whose composition is unknown, was included in the experiment according to the manufacturer’s protocol. The amount of Lipofectamine 3000 for transfection experiments was calculated based on an approach relying on the volume of Lipofectamine 3000 in microliters per weight of DNA in micrograms (v/w). This volume per weight approach was maintained throughout the whole study, resulting in a uniform depiction of all samples. Even though no concentration data is available for Lipofectamine 3000, and therefore, it is technically not correct to provide any concentrations for Lipofectamine 3000, we employed an arbitrary concentration for Lipofectamine 3000 assuming that 1 μL of Lipofectamine 3000—as used according to the manufacturer’s instructions—is equivalent to 1 μL of DOTAP:chol LNPs. N/P ratios are indicated where applicable.
Transgene Expression Analysis
Confocal Laser Scanning Microscopy
Live-cell images were acquired on an Olympus FV-1000 inverted confocal fluorescence microscope (Olympus Ltd., Tokyo, Japan) 24 and 48 h after transfection to qualitatively assess the transfection efficiency. GFP was excited at 488 nm, and emission was recorded at 516 nm using a UPlanSApo 10× objective (numerical aperture, 0.40).
Flow Cytometry
Transfection efficiency and transgene expression were both measured using an FACS Canto II flow cytometer (Becton Dickinson, San Jose, CA) 48 h after transfection. Briefly, the cells were detached using 0.25% trypsin/EDTA (m/V) for 7 min. Subsequently, trypsin digestion was stopped by the addition of DMEM cell culture medium and the cell suspension was transferred to a 1.5 mL Eppendorf tube. The cell suspension was centrifuged (Eppendorf 5424 R Centrifuge, Eppendorf AG, Hamburg, Germany) at 200g for 5 min at RT. The supernatant was aspirated, the cells were suspended in FACS buffer (D-PBS, 2% FCS, 0.1% NaN3), and %GFP-positive cells and mean fluorescence intensity (MFI) were measured after excitation at 488 nm. The fluorescence signal of cells expressing GFP was detected in fluorescence channel FL1 (505LP—530/30). Statistical evaluation of the obtained flow cytometry data was done using FlowJo Vx software (TreeStar, Ashland, OR). MFI values were calculated from GFP-positive cells.
Cytotoxicity Experiments
The HuH7 cells were treated with aminolipid–LNPs at various concentrations 24 h after seeding. Four hours after treatment, the medium containing LNPs was aspirated and replaced by a fresh medium. Twenty-four hours after treatment, the medium was aspirated and replaced by a medium containing 0.5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT). The 96-well plates were then incubated (37 °C, 5% CO2) for 2 h to allow formation of formazan crystals. Afterward, the medium was aspirated and the insoluble formazan crystals were solubilized using a mixture of 3% SDS in H2O and 40 mM HCl in isopropanol (1:6, v/v). The plates were then shaken for 2 h, followed by absorption measurement using a 96-well plate reader (SpectraMax M2e Microplate Reader, Molecular Devices LLC, San Jose, CA) at 570 (formazan signal) and 670 nm (background).
Statistical Evaluation
Statistical evaluation was done using OriginPro 2018 (64-bit) SR1 b9.5.1.195 (Academic) Software (OriginLab Corporation, Northampton, MA). A minimum of three independent measurements were used for statistical evaluation.
Zebrafish Embryo In Vivo Trials
Zebrafish culture and injection were carried out as described previously.39 Briefly, zebrafish embryos of the Tg(kdrl:EGFP)40 line were raised under standard conditions in accordance with Swiss animal welfare regulations. Fish were maintained at 28 °C in zebrafish culture media supplemented with 1-phenyl-2-thiourea (PTU). Calibrated volumes of 1 nL sample were injected into the Duct of Cuvier (80 pg DNA, 0.64 pmol DOTAP) using a micromanipulator (Wagner Instrumentenbau KG, Schöffengrund, Germany), a pneumatic PicoPump PV830 (WPI, Sarasota, Florida), and a Leica S8APO microscope (Leica, Wetzlar, Germany). Imaging was carried out 4 and 24 h post-injection using a LEICA POINT SCANNING CONFOCAL “SP5-II-MATRIX” (Leica, Wetzlar, Germany) equipped with a 25× HCX IRAPO L (NA 0.95) objective. Image analysis was carried out using Fiji ImageJ v. 1.52n.
Results and Discussion
Combinatorial Synthesis of Aminolipids
The aim of this study was the design, synthesis, and screening of novel types of short-chain aminolipids to maximize the DNA vector delivery of DOTAP:chol lipoplexes. To create a small lipid library for this proof-of-concept study, we developed a straightforward one-pot combinatorial synthesis based on a double-reductive amination strategy (Figure 1). A combination of various amines and aldehydes in the presence of the reducing agent sodium triacetoxyborohydride (STAB) resulted in the respective short-chain aminolipids with different headgroups and lipid tails.
An overview of selected building blocks as starting materials (i.e., amines and aldehydes) is given in Figure 2A–C. Based on previous findings by our group that short-chain (C8–C12) lipid-like materials result in improved nucleic acid delivery, decanal (C10) and dodecanal (C12) were selected as building blocks for lipid tails. In addition to aliphatic amines, heterocyclic amines were selected as headgroups based on previous reports that inclusion of aromatic or heterocyclic rings into transfection reagents can increase the transfection efficiency.41−45 To test whether these principles also apply to DNA vector delivery based on DOTAP:chol lipoplexes, different building blocks were selected and combined to afford the corresponding aminolipids (see the Supporting Information for synthesis details and aminolipid characterization). For example, aminolipid AL-A10 was synthesized using 3-methoxypropylamine (A) and decanal (C10).
Figure 2.
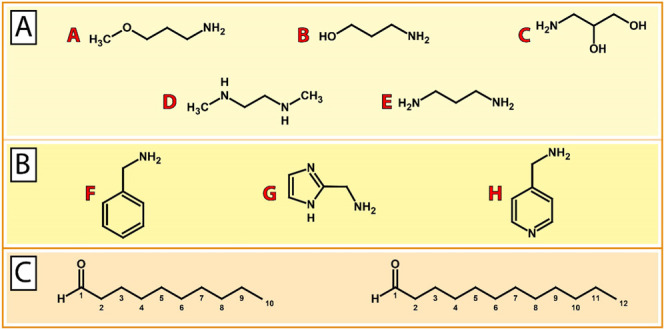
Building blocks for the combinatorial synthesis of aminolipids. (A) Aliphatic amine compounds including (A) 3-methoxypropylamine, (B) 3-aminopropan-1-ol, (C) 3-aminopropane-1,2-diol, (D) N,N′-dimethylethylenediamine, and (E) 1,3-diaminopropane. (B) Aromatic and heteroaromatic amine compounds including (F) benzylamine, (G) 1H-imidazol-2-ylmethan-amine, and (H) 4-(aminomethyl)pyridine. (C) Aldehydes decanal (C10) and dodecanal (C12) as building blocks for lipid tails.
In summary, 12 different short-chain aminolipids were successfully synthesized in high yield using a versatile two-step one-pot procedure. This combinatorial synthesis is easy to perform and fully scalable.
Physicochemical Characterization of Aminolipid-Containing Systems
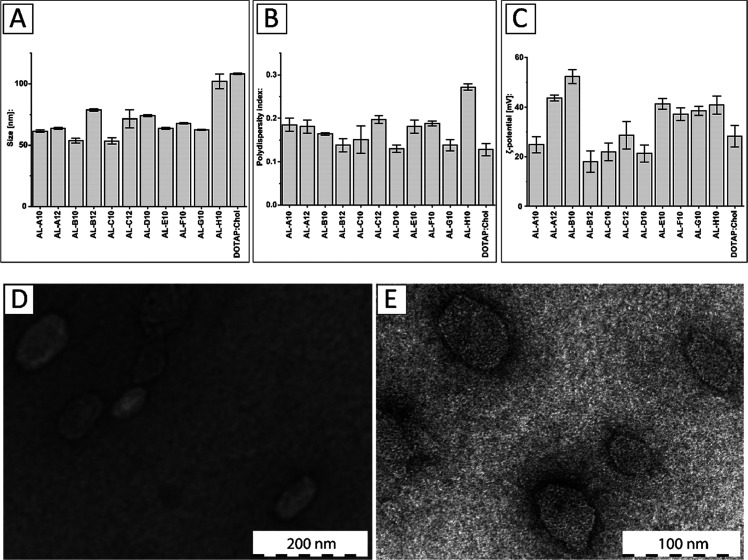
The obtained aminolipids were incorporated into DOTAP-based systems in a 1:1 ratio (mol/mol) using a lipid-film rehydration and extrusion method, as described in the Supporting Information. The results of the physicochemical characterization of aminolipid:DOTAP systems are shown in Figure 3. Most aminolipid:DOTAP systems showed hydrodynamic diameters in the range of 54–79 nm along with a monodisperse size distribution, as indicated by a polydispersity index (PDI) below 0.2 (Figure 3A,B). The largest diameter was measured for the conventional formulation based on DOTAP:chol (108.2 ± 0.6 nm) followed by the AL-H10-containing system (102.0 ± 5.9 nm). The smallest diameter was measured for the AL-C10 system (53.5 ± 2.5 nm) followed by the AL-B10 system (53.8 ± 2.0 nm). Both AL-A10 and AL-A12 systems showed hydrodynamic diameters of 61.5 ± 1.0 and 63.6 ± 0.8 nm and PDI values of 0.185 and 0.181, respectively. Interestingly, incorporation of C10-based aminolipids always resulted in smaller systems compared to their C12 counterparts. All measurements were carried out in triplicate.
Figure 3.
Physicochemical characterization of DOTAP systems combined with short-chain aminolipids. (A) Hydrodynamic diameter (nm), (B) polydispersity index (PDI), and (C) ζ-potential (mV) for aminolipid-containing DOTAP systems compared to the conventional DOTAP:cholesterol (chol) system at a 1:1 ratio (mol/mol). (D) Transmission electron microscopy (TEM) of DOTAP:chol and (E) AL-A12-containing systems.
The ζ-potential measurements for the obtained DOTAP-based systems revealed a positive surface charge in the range of 18–53 mV (Figure 3C). The highest ζ-potential was found for the formulation including AL-B10 with a highly positive value of 52.27 ± 2.80 mV followed by the AL-A12 system (43.66 ± 1.19 mV). The AL-B12 system showed the lowest ζ-potential with a value of 17.97 ± 4.28 mV followed by the AL-D10 system (21.29 ± 3.48 mV). The AL-A10 system showed a similar ζ-potential to DOTAP:chol (24.86 ± 3.26 vs 28.21 ± 4.31 mV). A highly positive ζ-potential (>30 mV) is indicative of a high colloidal stability resulting from electrostatic repulsive forces that prevent aggregation of particles.46,47 Good colloidal stability can still be assumed for systems with ζ-potentials in the range of ±30 mV. This in contrast to a range from 0 to ±5 mV, which is indicative of particle agglomeration and instability.48 In fact, during the time course of this study, no precipitation was observed for any formulation. All measurements were carried out in triplicate.
Representative TEM images of DOTAP:chol and the AL-A12-containing system are displayed in Figure 3D,E. Both systems resulted in spherical assemblies with diameters of 82.81 ± 24.64 nm (DOTAP:chol) and 46.13 ± 8.85 nm (AL-A12 containing system), which is slightly smaller than the diameters based on dynamic light scattering (DLS). In conclusion, short-chain aminolipids can be successfully incorporated into DOTAP-based systems, thereby influencing the size and charge of the resulting particles.
Assessment of DNA Vector Delivery In Vitro
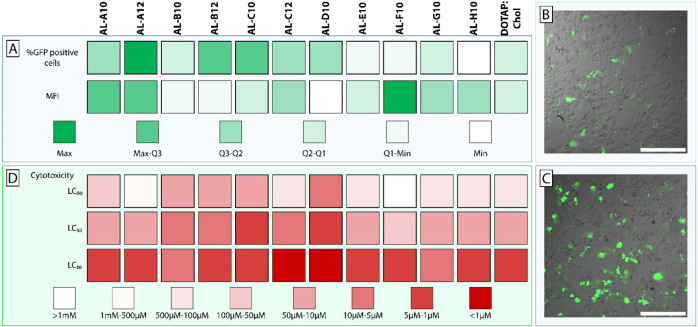
To assess the efficiency of DNA delivery, we selected an advanced DNA vector type, i.e., minicircle DNA, which has clear advantages over plasmid DNA. Several research groups have demonstrated that the use of minicircle DNA results in high transfection efficiencies and long-lasting transgene expression.49−51 Therefore, a minicircle DNA encoding the reporter gene for green fluorescent protein (GFP) was complexed using DOTAP:chol and aminolipid-containing systems. The lipoplexes were not PEGylated to reduce complexity, thereby allowing us to attribute observed effects directly to the formulation components. Resulting lipoplexes were used for transfection of human liver-derived HuH7 cells in vitro (Table S1). A color-coded summary of the transfection screening is shown in Figure 4A. DOTAP:chol lipoplexes (at 1 μg DNA and 8 μM lipid concentration) resulted in 5.7% GFP-positive cells. Compared to the conventional DOTAP:chol system, incorporation of various aminolipids based on aliphatic amines resulted in improved transfection efficiencies (i.e., AL-A to AL-D). For example, AL-B12 and AL-C12 systems resulted in 8.6 and 6.0% GFP-positive cells, respectively. The lipoplexes containing AL-A10 (7.7%) and AL-A12 (10.3%) showed a significantly increased transfection efficiency compared to DOTAP:chol (p < 0.001), which was even more distinct at 2 μg DNA and 16 μM lipid concentration with 28.7% (AL-A10) and 31.5% (AL-A12) compared to 13.2% (DOTAP:chol). Moreover, AL-A12 showed 72% of the transfection efficiency of Lipofectamine 3000 (47.5% GFP-positive cells). In contrast, DOTAP:chol was only 30% as efficient as Lipofectamine 3000. A statistically significant (p < 0.001) increase in transgene expression (i.e., assessed based on mean fluorescence intensity (MFI)) compared to DOTAP:chol was only observed for AL-A12. While DOTAP:chol only resulted in 10% of the gene expression achieved with Lipofectamine 3000, AL-A12-based lipoplexes exhibited 20% of the transgene expression of Lipofectamine 3000. The replacement of aliphatic amine headgroups with heterocyclic groups (i.e., AL-F10, AL-G10, and AL-H10) to improve the fusogenic properties (as previously described52) did not maximize the efficacy (i.e., %GFP-positive cells) of DOTAP-based lipoplexes (Figure 4A). However, AL-F10 (at 1 μg DNA and 8 μM lipid concentration) resulted in the highest transgene expression among all aminolipids of the combinatorial library. It is tempting to speculate that AL-F10 enhanced the endosomal escape in transfected cells, resulting in increased GFP expression. Due to the small number of investigated heterocyclic headgroups, further studies are warranted to clarify if such a strategy is applicable for DOTAP-based lipoplexes.
Figure 4.
In vitro assessment of transfection efficiency and cytotoxicity of lipoplex systems. (A) Heat map representing the transfection efficiency of aminolipid-containing DOTAP lipoplexes compared to conventional DOTAP:cholesterol (chol) composed lipoplexes. Transfection efficiency was assessed by means of percentage of GFP-positive cells and mean fluorescence intensity (MFI) of GFP expression using flow cytometry. The obtained results were allocated to different groups (color code) based on the quartiles of %GFP-positive cells and MFI. Representative confocal images of HuH7 cells transfected with (B) DOTAP:chol or (C) AL-A12-containing lipoplexes entrapping minicircle DNA coding for GFP (scale bar = 400 μm). (D) Cytotoxicity of aminolipid-containing DOTAP lipoplexes compared to conventional DOTAP:chol lipoplexes by means of lethal concentrations (LC 10/50/90) resulting in 90/50/10% cell viability, respectively.
To further improve the understanding of how the aminolipids’ structural parameters influence the transfection efficiency, specific compounds were selected for a structure–activity relationship (SAR) study (Figure S37). Aminolipids consisting of a bifunctional methoxy headgroup (AL-A10, AL-A12) showed significantly (p < 0.001) higher transfection efficiencies and transgene expression levels compared to aminolipids with a bifunctional hydroxy headgroup (AL-B10, AL-B12). SAR analysis of tail groups revealed significantly higher transfection efficiencies (p < 0.1) and transgene expression levels (p < 0.001) for aminolipids consisting of C12 tails compared to aminolipids consisting of C10 tails, as demonstrated by the superiority of AL-A12 over AL-A10. Due to the poor transfection efficiency of many of the tested aminolipids, an in-depth SAR analysis focusing on headgroups could not be performed. Further experiments with an expanded range of aminolipid tail lengths and alternative headgroups are needed to improve our understanding on how small changes in lipid tail length influence the delivery of DNA vectors.
In conclusion, the highest transfection efficiency was achieved using AL-A12 lipoplexes with a 2-fold increased number of GFP-positive cells as well as significantly higher transgene expression compared to DOTAP:chol lipoplexes. Representative confocal laser scanning microscopy images of HuH7 cells transfected with DOTAP:chol lipoplexes and AL-A12-containing lipoplexes are shown in Figure 4B,C, respectively. An increased efficiency results in a lower dose of the potentially cytotoxic transfection reagent that needs to be administered while maintaining the same effect and is consequently mitigating side effects of the gene therapy. Moreover, the known composition of these lipoplexes allows for surface modification (e.g., PEGylation), which is difficult to achieve for the proprietary formulation Lipofectamine 3000.
Cytotoxicity of Lipoplex Systems
Figure 4D displays the results of the cell viability assessment. Several aminolipids with aliphatic headgroups and improved transfection efficacy also resulted in enhanced cytotoxic effects (i.e., AL-B to AL-D). Generally, it was observed that aminolipids derived from C12 aldehydes are less toxic than aminolipids derived from C10 aldehydes (Tables S2 and S3). For example, AL-A10 systems showed a toxicity comparable to DOTAP:chol while at the same time having increased DNA delivery efficacy. Interestingly, the lead aminolipid AL-A12 demonstrated lower toxicity across the entire dose range than DOTAP:chol. Up to a concentration of 16 μM total lipid, AL-A12 systems resulted in cell viabilities of approximately 80%. At 64 μM total lipid, AL-A12 still resulted in cell viabilities of up to 50%. These results clearly indicate a lower cytotoxic potential for AL-A12-containing lipoplexes than for DOTAP:chol. Lipoplex systems composed of aminolipids with heterocyclic headgroups caused similar or reduced toxicity. The lowest cytotoxic effects were observed using AL-F10-based lipoplexes, thereby confirming that inclusion of aromatic or heterocyclic rings into a transfection reagent can mitigate cytotoxic effects.42−45
Conclusively, the lead structures from our transfection screening AL-A12 and AL-F10 (most GFP-positive cells and highest MFI) also outperformed the other aminolipids in the cytotoxicity screening assay. This confirms the generally accepted dogma that low cytotoxic side effects are a prerequisite to achieve high transfection rates.53
Assessment of DNA Vector Delivery In Vivo
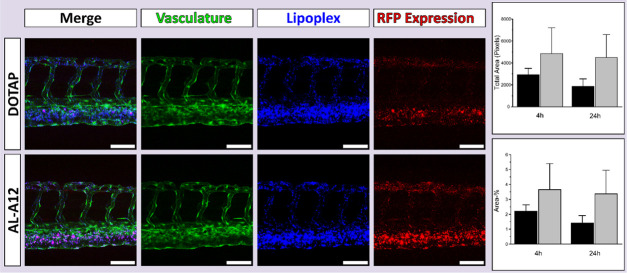
Due to its high transfection efficiency (% transfected cells) and low toxicity in vitro, AL-A12 was selected as lead aminolipid for the in vivo evaluation in the zebrafish embryo model, a validated in vivo tool to assess lipid-based delivery systems.39,54−56 To assess the systemic circulation of AL-A12 and DOTAP:chol lipoplexes, the fluorescent lipid 1,1′dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD) was incorporated as a tracer. To evaluate the utility of developed lipoplexes for in vivo gene delivery, lipoplexes entrapped DNA encoding for RFP. Both lipoplex systems were injected intravenously into transgenic zebrafish embryos expressing GFP in their vasculature. Biodistribution and reporter gene expression were analyzed 4 and 24 h post-injection using confocal microscopy (Figure 5).
Figure 5.
Assessment of in vivo transfection efficiency of the lead lipoplex system containing aminolipid AL-A12. Evaluation of lipoplex biodistribution and transfection efficiency in vivo in the zebrafish model. Confocal imaging (maximum intensity projections) of transgenic zebrafish embryos with GFP expressing vasculature endothelial cells was performed 4 and 24 h post-injection with fluorescent DOTAP:chol or AL-A12-based lipoplexes entrapping plasmid DNA encoding for RFP. Representative maximum intensity projections of a series of three images are shown (4 h post-injection). Displayed are the merged images, the vasculature expressing GFP (green), the DiD-labeled lipoplexes (blue), and the reporter gene expression (RFP, red). AL-A12 promoted a qualitatively higher gene expression than DOTAP:chol-based lipoplexes h. Scale bars = 80 μm. The right graphs show the quantitative image analysis using Fiji ImageJ of the total area in pixels (top) and the percentage of the area (bottom) of the RFP expression determined 4 and 24 h post-injection of DOTAP:chol (black) and AL-A12-based lipoplexes (gray). The results were not statistically significant at p < 0.05.
As expected for cationic systems, both lipoplexes associated with endothelial cells with preference for venous vasculature.57 The evaluation of reporter gene expression confirmed the fluorescence distribution patterns. AL-A12-based lipoplexes as well as DOTAP:chol resulted in RFP expression in endothelial cells 4 h post-injection with stronger expression in venous vasculature. Both qualitative confocal imaging and the image analysis confirmed a higher gene expression in vivo for AL-A12 compared to DOTAP-based lipoplexes. Further in vivo studies are needed to assess the potential to target actively growing tumor blood vessels as shown for other positively charged lipid systems.58
Conclusions
In recent decades, DOTAP:chol lipoplexes have been investigated in several clinical trials for the delivery of DNA vectors. However, none of these studies has advanced to a late clinical stage due to poor outcomes with respect to efficacy. The present study describes the design and one-pot synthesis of short-chain aminolipids enabling incorporation into DOTAP-based lipoplexes to maximize DNA vector delivery. The structure–activity relationship analysis revealed methoxy headgroups to be superior to hydroxy headgroups. Furthermore, C12 lipid tails promote higher transfection efficiencies than C10 tails. The DNA vector transfection ability in HuH7 cells in vitro was significantly increased using various aminolipids (e.g., AL-A12). In addition, incorporation of selected aminolipids clearly mitigated the cytotoxicity. In vivo studies in zebrafish embryos showed higher gene expression using the lead lipoplex system based on AL-A12 compared to conventional DOTAP:chol lipoplexes.
In conclusion, our presented one-pot combinatorial synthesis is a versatile approach to create lipid libraries for the development of DNA delivery systems. The combination of DOTAP with short-chain aminolipids promotes efficient DNA vector delivery in vitro and in vivo as well as reduced cellular toxicity.
Acknowledgments
D.M. and U.M. were supported by the Swiss National Science Foundation through a stipend professorship to A.Z. In addition, we thank Daniel Häussinger (Department of Chemistry, University of Basel) for NMR and MS analyses and Nicole Rimann (University Children’s Hospital Zurich) for supply with minicircle DNA. H.M.G.-C. acknowledges the support from Heidi Ras-Grant. Finally, the authors thank the Imaging Core facility (IMCF, University of Basel) and, in particular, Alexia Ferrand for the technical assistance provided on the confocal microscope for in vivo imaging.
Glossary
Abbreviations
- DCM
dichloromethane
- DLS
dynamic light scattering
- DOTAP:chol
DOTAP:cholesterol
- GFP
green fluorescent protein
- LC
lethal concentration
- MFI
mean fluorescence intensity
- PDI
polydispersity index
- SAR
structure–activity relationship
- STAB
sodium triacetoxyborohydride
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03303.
Details on materials and methods; results of the synthesis, transfection, and cytotoxicity of aminolipids; and statistical data (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. All authors contributed equally.
Swiss National Science Foundation (SNF grant no. 173057, Sinergia grant no. CRSII5_180257). Novartis Foundation for Medical-Biological Research (#19A004).
The authors declare no competing financial interest.
Supplementary Material
References
- Clayton P. T. Inborn Errors Presenting with Liver Dysfunction. Semin. Neonatol. 2002, 7, 49–63. 10.1053/siny.2001.0086. [DOI] [PubMed] [Google Scholar]
- Jansen P. L. M. Diagnosis and Management of Crigler-Najjar Syndrome. Eur. J. Pediatr. 1999, 158, S089–S094. 10.1007/PL00014330. [DOI] [PubMed] [Google Scholar]
- Marcuello E.; Altés A.; Menoyo A.; Rio E. del.; Gómez-Pardo M.; Baiget M. UGT1A1gene variations and irinotecan treatment in patients with metastatic colorectalcancer. Br. J. Cancer 2004, 91, 678–682. 10.1038/sj.bjc.6602042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadakol A.; Ghosh S. S.; Sappal B. S.; Sharma G.; Chowdhury J. R.; Chowdhury N. R. Genetic Lesions of Bilirubin Uridine-Diphosphoglucuronate Glucuronosyltransferase (UGT1A1) Causing Crigler-Najjar and Gilbert Syndromes: Correlation of Genotype to Phenotype. Hum. Mutat. 2000, 16, 297–306. . [DOI] [PubMed] [Google Scholar]
- Schuchman E. H.; Wasserstein M. P. Types A and B Niemann-Pick Disease. Best Pract. Res., Clin. Endocrinol. Metab. 2015, 29, 237–247. 10.1016/j.beem.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Marciniuk D. D.; Hernandez P.; Balter M.; Bourbeau J.; Chapman K. R.; Ford G. T.; Lauzon J. L.; Maltais F.; O’Donnell D. E.; Goodridge D.; Strange C.; Cave A. J.; Curren K.; Muthuri S. Canadian Thoracic Society COPD Clinical Assembly Alpha-1 Antitrypsin Deficiency Expert Working Group. Alpha-1 antitrypsin deficiency targeted testing and augmentation therapy: a Canadian Thoracic Society clinical practice guideline. Can. Respir. J. 2012, 19 (2), 272.Erratum in: Can Respir J. 2012 Jul-Aug; 19 (4) p 272 10.1155/2012/920918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruteau J.; Waddington S. N.; Alexander I. E.; Gissen P. Gene Therapy for Monogenic Liver Diseases: Clinical Successes, Current Challenges and Future Prospects. J. Inherited Metab. Dis. 2017, 40, 497–517. 10.1007/s10545-017-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudes P. F. Gene Therapy as a New Treatment Option for Inherited Monogenic Diseases. Eur. J. Intern. Med. 2014, 25, 31–36. 10.1016/j.ejim.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Lu C.; Stewart D. J.; Lee J. J.; Ji L.; Ramesh R.; Jayachandran G.; Nunez M. I.; Wistuba I. I.; Erasmus J. J.; Hicks M. E.; Grimm E. A.; Reuben J. M.; Baladandayuthapani V.; Templeton N. S.; McMannis J. D.; Roth J. A. Phase I Clinical Trial of Systemically Administered TUSC2(FUS1)-Nanoparticles Mediating Functional Gene Transfer in Humans. PLoS One 2012, 7, e34833 10.1371/journal.pone.0034833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R.; Saeki T.; Smyth Templeton N.; Ji L.; Stephens L. C.; Ito I.; Wilson D. R.; Wu Z.; Branch C. D.; Minna J. D.; Roth J. A. Successful Treatment of Primary and Disseminated Human Lung Cancers by Systemic Delivery of Tumor Suppressor Genes Using an Improved Liposome Vector. Mol. Ther. 2001, 3, 337–350. 10.1006/mthe.2001.0266. [DOI] [PubMed] [Google Scholar]
- Dai B.; Yan S.; Lara-Guerra H.; Kawashima H.; Sakai R.; Jayachandran G.; Majidi M.; Mehran R.; Wang J.; Bekele B. N.; Baladandayuthapani V.; Yoo S.-Y.; Wang Y.; Ying J.; Meng F.; Ji L.; Roth J. A. Exogenous Restoration of TUSC2 Expression Induces Responsiveness to Erlotinib in Wildtype Epidermal Growth Factor Receptor (EGFR) Lung Cancer Cells through Context Specific Pathways Resulting in Enhanced Therapeutic Efficacy. PLoS One 2015, 10, e0123967 10.1371/journal.pone.0123967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaobo C.; Majidi M.; Feng M.; Shao R.; Wang J.; Zhao Y.; Baladandayuthapani V.; Song J.; Fang B.; Ji L.; Mehran R.; Roth J. A. TUSC2(FUS1)-Erlotinib Induced Vulnerabilities in Epidermal Growth Factor Receptor(EGFR) Wildtype Non-Small Cell Lung Cancer(NSCLC) Targeted by the Repurposed Drug Auranofin. Sci. Rep. 2016, 6, 35741 10.1038/srep35741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimkus T.; Sirkisoon S.; Harrison A.; Lo H.-W. Tumor Suppressor Candidate 2 (TUSC2, FUS-1) and Human Cancers. Discovery Med. 2017, 23, 325–330. [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Xia W.; Li Z.; Kuo H.-P.; Liu Y.; Li Z.; Ding Q.; Zhang S.; Spohn B.; Yang Y.; Wei Y.; Lang J.-Y.; Evans D. B.; Chiao P. J.; Abbruzzese J. L.; Hung M.-C. Targeted Expression of BikDD Eradicates Pancreatic Tumors in Noninvasive Imaging Models. Cancer Cell 2007, 12, 52–65. 10.1016/j.ccr.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Li L.-Y.; Dai H.-Y.; Yeh F.-L.; Kan S.-F.; Lang J.; Hsu J. L.; Jeng L.-B.; Chen Y.-H.; Sher Y.-P.; Lin W.-C.; Hung M.-C. Targeted Hepatocellular Carcinoma Proapoptotic BikDD Gene Therapy. Oncogene 2011, 30, 1773–1783. 10.1038/onc.2010.558. [DOI] [PubMed] [Google Scholar]
- Xie X.; Li L.; Xiao X.; Guo J.; Kong Y.; Wu M.; Liu W.; Gao G.; Hsu J. L.; Wei W.; Hung M.-C.; Xie X. Targeted Expression of BikDD Eliminates Breast Cancer with Virtually No Toxicity in Noninvasive Imaging Models. Mol. Cancer Ther. 2012, 11, 1915–1924. 10.1158/1535-7163.MCT-12-0191. [DOI] [PubMed] [Google Scholar]
- Buck J.; Grossen P.; Cullis P. R.; Huwyler J.; Witzigmann D. Lipid-Based DNA Therapeutics: Hallmarks of Non-Viral Gene Delivery. ACS Nano 2019, 13, 3754–3782. 10.1021/acsnano.8b07858. [DOI] [PubMed] [Google Scholar]
- Tokunaga M.; Hazemoto N.; Yotsuyanagi T. Effect of Oligopeptides on Gene Expression: Comparison of DNA/Peptide and DNA/Peptide/Liposome Complexes. Int. J. Pharm. 2004, 269, 71–80. 10.1016/j.ijpharm.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Kaiser J. How Safe Is a Popular Gene Therapy Vector?. Science 2020, 367, 131. 10.1126/science.367.6474.131. [DOI] [PubMed] [Google Scholar]
- Nault J.-C.; Datta S.; Imbeaud S.; Franconi A.; Mallet M.; Couchy G.; Letouzé E.; Pilati C.; Verret B.; Blanc J.-F.; Balabaud C.; Calderaro J.; Laurent A.; Letexier M.; Bioulac-Sage P.; Calvo F.; Zucman-Rossi J. Recurrent AAV2-Related Insertional Mutagenesis in Human Hepatocellular Carcinomas. Nat. Genet. 2015, 47, 1187–1193. 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- Russell D. W.; Grompe M. Adeno-Associated Virus Finds Its Disease. Nat. Genet. 2015, 47, 1104–1105. 10.1038/ng.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S.; Wang Y.; Gong Y.; Lin X.; Zhao Y.; Zhi D.; Zhou Q.; Zhang S. Correlation of the Cytotoxic Effects of Cationic Lipids with Their Headgroups. Toxicol. Res. 2018, 7, 473–479. 10.1039/C8TX00005K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. J.; Lipps H. J.; Jans D. A. Towards Safe, Non-Viral Therapeutic Gene Expression in Humans. Nat. Rev. Genet. 2005, 6, 299–310. 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- Ramamoorth M.; Narvekar A. Non Viral Vectors in Gene Therapy- An Overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn S. L.; Amaya A. K.; Alexander I. E.; Edelstein M.; Abedi M. R. Gene Therapy Clinical Trials Worldwide to 2017: An Update. J. Gene Med. 2018, 20, e3015 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- Sher Y.-P.; Tzeng T.-F.; Kan S.-F.; Hsu J.; Xie X.; Han Z.; Lin W.-C.; Li L.-Y.; Hung M.-C. Cancer Targeted Gene Therapy of BikDD Inhibits Orthotopic Lung Cancer Growth and Improves Long-Term Survival. Oncogene 2009, 28, 3286–3295. 10.1038/onc.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Kong Y.; Tang H.; Yang L.; Hsu J. L.; Hung M.-C. Targeted BikDD Expression Kills Androgen-Dependent and Castration-Resistant Prostate Cancer Cells. Mol. Cancer Ther. 2014, 13, 1813–1825. 10.1158/1535-7163.MCT-13-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-S.; Liu F.; Huang L. Implications of Pharmacokinetic Behavior of Lipoplex for Its Inflammatory Toxicity. Adv. Drug Delivery Rev. 2005, 57, 689–698. 10.1016/j.addr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Lee C.-H.; Ni Y.-H.; Chen C.-C.; Chou C.-K.; Chang F.-H. Synergistic Effect of Polyethylenimine and Cationic Liposomes in Nucleic Acid Delivery to Human Cancer Cells. Biochim. Biophys. Acta, Biomembr. 2003, 1611, 55–62. 10.1016/S0005-2736(03)00027-0. [DOI] [PubMed] [Google Scholar]
- Li W.; Szoka F. C. Lipid-Based Nanoparticles for Nucleic Acid Delivery. Pharm. Res. 2007, 24, 438–449. 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- Yin H.; Kanasty R. L.; Eltoukhy A. A.; Vegas A. J.; Dorkin J. R.; Anderson D. G. Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15, 541–555. 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Anchordoquy T. J. The Role of Lipid Charge Density in the Serum Stability of Cationic Lipid/DNA Complexes. Biochim. Biophys. Acta, Biomembr. 2004, 1663, 143–157. 10.1016/j.bbamem.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Zumbuehl A.; Goldberg M.; Leshchiner E. S.; Busini V.; Hossain N.; Bacallado S. A.; Nguyen D. N.; Fuller J.; Alvarez R.; Borodovsky A.; Borland T.; Constien R.; de Fougerolles A.; Dorkin J. R.; Jayaprakash K. N.; Jayaraman M.; John M.; Koteliansky V.; Manoharan M.; Nechev L.; Qin J.; Racie T.; Raitcheva D.; Rajeev K. G.; Sah D. W. Y.; Soutschek J.; Toudjarska I.; Vornlocher H.-P.; Zimmermann T. S.; Langer R.; Anderson D. G. A Combinatorial Library of Lipid-like Materials for Delivery of RNAi Therapeutics. Nat. Biotechnol. 2008, 26, 561–569. 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.; Wang Q.; Shen Y.; Gao X.; Li L.; Yan Y.; Wang H.; Cheng Y. Green Tea Catechin Dramatically Promotes RNAi Mediated by Low-Molecular-Weight Polymers. ACS Cent. Sci. 2018, 4, 1326–1333. 10.1021/acscentsci.8b00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.; Wang R.; Fan Q.; Gao X.; Wang H.; Shen Y.; Li Y.; Cheng Y. Natural Polyphenol Inspired Polycatechols for Efficient SiRNA Delivery. CCS Chem. 2020, 2, 146–157. 10.31635/ccschem.020.201900084. [DOI] [Google Scholar]
- Neuhaus F.; Mueller D.; Tanasescu R.; Balog S.; Ishikawa T.; Brezesinski G.; Zumbuehl A. Vesicle Origami: Cuboid Phospholipid Vesicles Formed by Template-Free Self-Assembly. Angew. Chem., Int. Ed. 2017, 56, 6515–6518. 10.1002/anie.201701634. [DOI] [PubMed] [Google Scholar]
- Witzigmann D.; Wu D.; Schenk S. H.; Balasubramanian V.; Meier W.; Huwyler J. Biocompatible Polymer–Peptide Hybrid-Based DNA Nanoparticles for Gene Delivery. ACS Appl. Mater. Interfaces 2015, 7, 10446–10456. 10.1021/acsami.5b01684. [DOI] [PubMed] [Google Scholar]
- Witzigmann D.; Sieber S.; Porta F.; Grossen P.; Bieri A.; Strelnikova N.; Pfohl T.; Prescianotto-Baschong C.; Huwyler J. Formation of Lipid and Polymer Based Gold Nanohybrids Using a Nanoreactor Approach. RSC Adv. 2015, 5, 74320–74328. 10.1039/C5RA13967H. [DOI] [Google Scholar]
- Sieber S.; Grossen P.; Uhl P.; Detampel P.; Mier W.; Witzigmann D.; Huwyler J. Zebrafish as a Predictive Screening Model to Assess Macrophage Clearance of Liposomes in Vivo. Nanomedicine 2019, 17, 82–93. 10.1016/j.nano.2018.11.017. [DOI] [PubMed] [Google Scholar]
- Jin S.-W.; Beis D.; Mitchell T.; Chen J.-N.; Stainier D. Y. R. Cellular and Molecular Analyses of Vascular Tube and Lumen Formation in Zebrafish. Development 2005, 132, 5199–5209. 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Liu H.; Chang H.; Lv J.; Jiang C.; Li Z.; Wang F.; Wang H.; Wang M.; Liu C.; Wang X.; Shao N.; He B.; Shen W.; Zhang Q.; Cheng Y. Screening of Efficient SiRNA Carriers in a Library of Surface-Engineered Dendrimers. Sci. Rep. 2016, 6, 25069 10.1038/srep25069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. H.; Chen C.-K.; Ravikrishnan A.; Rane S.; Pfeifer B. A. Overcoming Nonviral Gene Delivery Barriers: Perspective and Future. Mol. Pharm. 2013, 10, 4082–4098. 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilies M. A.; Johnson B. H.; Makori F.; Miller A.; Seitz W. A.; Thompson E. B.; Balaban A. T. Pyridinium Cationic Lipids in Gene Delivery: An in Vitro and in Vivo Comparison of Transfection Efficiency versus a Tetraalkylammonium Congener. Arch. Biochem. Biophys. 2005, 435, 217–226. 10.1016/j.abb.2004.12.010. [DOI] [PubMed] [Google Scholar]
- van der Woude I.; Wagenaar A.; Meekel A. A.; ter Beest M. B.; Ruiters M. H.; Engberts J. B.; Hoekstra D. Novel Pyridinium Surfactants for Efficient, Nontoxic in Vitro Gene Delivery. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 1160–1165. 10.1073/pnas.94.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosjen A.; Šmisterová J.; Driessen C.; Anders J. T.; Wagenaar A.; Hoekstra D.; Hulst R.; Engberts J. B. F. N. Synthesis and Characteristics of Biodegradable Pyridinium Amphiphiles Used for in Vitro DNA Delivery. Eur. J. Org. Chem. 2002, 2002, 1271–1277. . [DOI] [Google Scholar]
- Faneca H.; Simões S.; Pedroso de Lima M. C. Evaluation of Lipid-Based Reagents to Mediate Intracellular Gene Delivery. Biochim. Biophys. Acta, Biomembr. 2002, 1567, 23–33. 10.1016/S0005-2736(02)00545-X. [DOI] [PubMed] [Google Scholar]
- Birchall J. C.; Kellaway I. W.; Mills S. N. Physico-Chemical Characterisation and Transfection Efficiency of Lipid-Based Gene Delivery Complexes. Int. J. Pharm. 1999, 183, 195–207. 10.1016/S0378-5173(99)00117-9. [DOI] [PubMed] [Google Scholar]
- Honary S.; Zahir F. Effect of Zeta Potential on the Properties of Nano-Drug Delivery Systems A Review (Part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. 10.4314/tjpr.v12i2.20. [DOI] [Google Scholar]
- Chen Z.-Y.; Riu E.; He C.-Y.; Xu H.; Kay M. A. Silencing of Episomal Transgene Expression in Liver by Plasmid Bacterial Backbone DNA Is Independent of CpG Methylation. Mol. Ther. 2008, 16, 548–556. 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- Viecelli H. M.; Harbottle R. P.; Wong S. P.; Schlegel A.; Chuah M. K.; VandenDriessche T.; Harding C. O.; Thöny B. Treatment of Phenylketonuria Using Minicircle-Based Naked-DNA Gene Transfer to Murine Liver. Hepatology 2014, 60, 1035–1043. 10.1002/hep.27104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay M. A. State-of-the-Art Gene-Based Therapies: The Road Ahead. Nat. Rev. Genet. 2011, 12, 316–328. 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- Csiszár A.; Hersch N.; Dieluweit S.; Biehl R.; Merkel R.; Hoffmann B. NovelFusogenic Liposomes for Fluorescent Cell Labeling and Membrane Modification. Bioconjugate Chem. 2010, 21, 537–543. 10.1021/bc900470y. [DOI] [PubMed] [Google Scholar]
- Durán M. C.; Willenbrock S.; Barchanski A.; Müller J.-M. V.; Maiolini A.; Soller J. T.; Barcikowski S.; Nolte I.; Feige K.; Murua Escobar H. Comparison of Nanoparticle-Mediated Transfection Methods for DNA Expression Plasmids: Efficiency and Cytotoxicity. J. Nanobiotechnol. 2011, 9, 47 10.1186/1477-3155-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S.; Grossen P.; Bussmann J.; Campbell F.; Kros A.; Witzigmann D.; Huwyler J. Zebrafish as a Preclinical in Vivo Screening Model for Nanomedicines. Adv. Drug Delivery Rev. 2019, 151–152, 152–168. 10.1016/j.addr.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Sieber S.; Grossen P.; Detampel P.; Siegfried S.; Witzigmann D.; Huwyler J. Zebrafish as an Early Stage Screening Tool to Study the Systemic Circulation of Nanoparticulate Drug Delivery Systems in Vivo. J. Controlled Release 2017, 264, 180–191. 10.1016/j.jconrel.2017.08.023. [DOI] [PubMed] [Google Scholar]
- Witzigmann D.; Uhl P.; Sieber S.; Kaufman C.; Einfalt T.; Schöneweis K.; Grossen P.; Buck J.; Ni Y.; Schenk S. H.; Hussner J.; Meyer zu Schwabedissen H. E.; Québatte G.; Mier W.; Urban S.; Huwyler J. Optimization-by-Design of Hepatotropic Lipid Nanoparticles Targeting the Sodium-Taurocholate Cotransporting Polypeptide. eLife 2019, 8, e42276 10.7554/eLife.42276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F.; Bos F. L.; Sieber S.; Arias-Alpizar G.; Koch B. E.; Huwyler J.; Kros A.; Bussmann J. Directing Nanoparticle Biodistribution through Evasion and Exploitation of Stab2-Dependent Nanoparticle Uptake. ACS Nano 2018, 12, 2138–2150. 10.1021/acsnano.7b06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt-Sody M.; Strieth S.; Krasnici S.; Sauer B.; Schulze B.; Teifel M.; Michaelis U.; Naujoks K.; Dellian M. Neovascular Targeting Therapy: Paclitaxel Encapsulated in Cationic Liposomes Improves Antitumoral Efficacy. Clin. Cancer Res. 2003, 9, 2335–2341. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.