Abstract
Fast microbiological diagnostics (MDx) are needed to ensure early targeted antimicrobial treatment in sepsis. This systematic review focuses on the impact on antimicrobial management and patient outcomes of MDx for pathogen and resistance gene identification compared with blood cultures. PubMed was searched for clinical studies using either whole blood directly or after short-term incubation. Twenty-five articles were retrieved describing the outcomes of 8 different MDx. Three interventional studies showed a significant increase in appropriateness of antimicrobial therapy and a nonsignificant change in time to appropriate therapy. Impact on mortality was conflicting. Length of stay was significantly lower in 2 studies. A significant decrease in antimicrobial cost was demonstrated in 6 studies. The limitations of this systematic review include the low number and observed heterogeneity of clinical studies. In conclusion, potential benefits of MDx regarding antimicrobial management and some patient outcomes were reported. More rigorous intervention studies are needed focusing on the direct benefits for patients.
Keywords: antimicrobial therapy, bacteremia, molecular microbiological diagnostics, patient outcomes
Sepsis is defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection and can lead to septic shock, which increases mortality considerably [1]. The mortality rate of patients with sepsis is 10%–20%, which increases to 40%–80% in patients developing septic shock [2]. Additionally, hospital length of stay (LOS) increases in patients with bloodstream infection (BSI), especially due to antimicrobial-resistant pathogens [3, 4].
Antimicrobial treatment should start as soon as possible and certainly within the first hour of recognizing sepsis. This should be preceded by taking blood cultures, if this does not cause any significant delay (>45 minutes) [5]. Kumar et al. showed a 7.6% decrease in survival rate for each hour of delay in administering antimicrobial therapy after the start of hypotension in patients with septic shock [6].
Blood culture (BC) is the gold standard for identifying bacteria that cause sepsis [7]. A serious limitation is that culture-based diagnostics are time-consuming, and the time to positivity is pathogen dependent [8, 9].
Due to this long turnaround time (TAT) and the need for immediate antimicrobial treatment, physicians start treatment empirically with broad-spectrum antibiotics, which contributes to antibiotic resistance [10, 11]. Thus, faster microbiological diagnostics are necessary to ensure an early targeted treatment. Molecular microbiological diagnostic techniques (MDx) could provide relevant results within a few hours [11, 12].
Ideally, results should be available within a single workday, and the time-consuming culture step should be avoided. Starting directly from the clinical specimen or after a short-term (a few hours) incubation of blood is beneficial [13–15]. In 2016, Timbrook et al. performed a meta-analysis of 5920 patients to assess the impact of rapid MDx starting from positive blood cultures, such as Matrix-assisted laser desorption/ionisation time of flight (MALDI-TOF), on clinical outcomes. They found that these MDx were associated with significant decreases in time to effective antibiotic therapy, in mortality risk in the presence of an antimicrobial stewardship plan, and in LOS [16]. The goal of this systematic review was to identify the clinical impact of rapid MDx for the identification of pathogens directly in whole-blood samples or after short-term incubation in terms of antimicrobial therapy management and patient outcomes.
METHODS
Literature Search
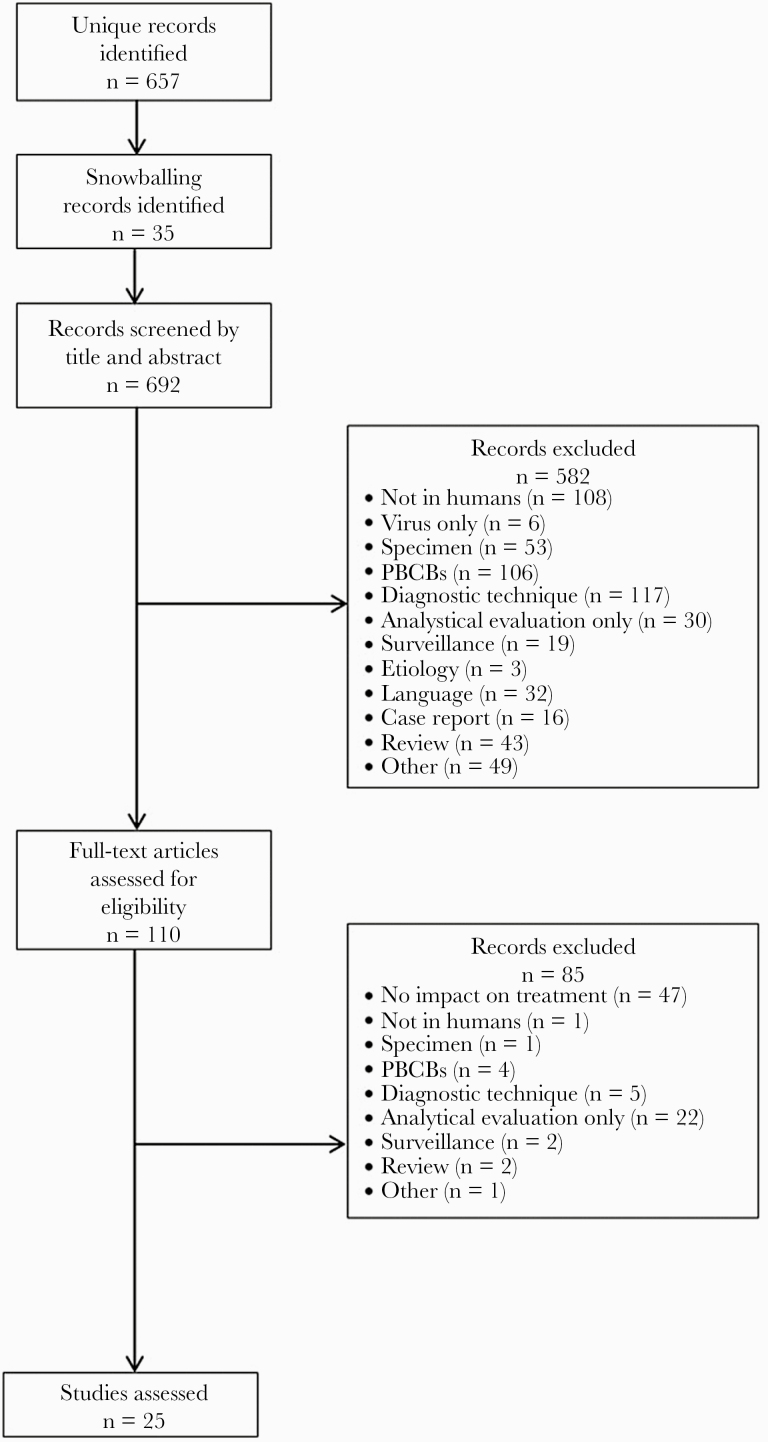
This systematic review was done according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines [17]. PubMed was searched through October 2019 for all types of studies evaluating MDx to identify pathogens and resistance genes either directly in whole-blood samples or after a short-term incubation step in patients with sepsis (cutoff date October 22, 2019). The search terms are shown in Supplementary Figure 1. No restrictions on the year of publication or language were set. Two researchers independently screened the corresponding studies by title and abstract. Studies were included in consensus and excluded according to criteria shown in Figure 1.
Figure 1.
Flow diagram of the selection process. In total, 657 records were retrieved from PubMed, and an additional 35 records were found after snowballing in references of included records. Title and abstract screening resulted in the inclusion of 110 articles. One hundred fifty-nine articles were excluded because MDx were not performed on whole blood or with a short-term incubation step. There was no or a nonmolecular technique evaluated in 117 records. No language restrictions were set, but 32 articles had to be excluded because of a language barrier. After full-text screening, 85 more records were excluded, 47 because they only evaluated analytical performance of MDx and did not contain data on impact. The final search result yielded 25 eligible articles. Abbreviation: PBCBs, positive blood culture bottles.
Study Selection Criteria
The objective was to identify articles that evaluated the clinical impact of rapid MDx on patients with sepsis or (suspected) BSI. Studies assessing the use of MDx to identify causative pathogens (bacteria and/or fungi) and/or genetic susceptibility directly from whole-blood specimens or after short-term incubation (shorter than BC incubation to positivity) in comparison with BCs were included. Both pathogen-specific and multiplex tests were eligible. The study population was any patients with sepsis, BSI, or suspected BSI of bacterial or fungal origin, from whom BCs were drawn. Studies concerned all settings, such as the emergency department, the intensive care unit (ICU), and the hospital. All study types (interventional, prospective, and retrospective), except case reports and reviews, were eligible.
Reasons for exclusion (Figure 1) were the use of other types of specimen than whole blood or the use of positive blood culture bottles. Nonmolecular techniques such as antigen detection or biomarkers were excluded as well. Studies on viral, protozoan, and parasitic infections were excluded. Studies using spiked samples or analyzing only performance characteristics were excluded.
Data Extraction
Data collected from included studies were study design, diagnosis at inclusion, population, number of patients included, type and (branded) name of MDx used, study location, funding source, number of antimicrobial therapy adjustments, number of patients on appropriate antimicrobial therapy, time to any antimicrobial therapy adjustment, time to appropriate antimicrobial therapy, LOS in-hospital and in-ICU, mortality, and costs or expenses. Additionally, test characteristics and test performances of MDx were extracted from studies that were found in the search but were excluded because they did not contain clinical data on antimicrobial therapy or patient outcomes.
Outcomes
The aims were to assess the impact of MDx on the antimicrobial management of patients with bacterial and fungal BSI or sepsis and the impact on all patient-related outcomes. Appropriateness of antimicrobial therapy regardless of definition, any treatment change (escalation, de-escalation, discontinuation), and the time to appropriateness or change of antimicrobial therapy was assessed. Eligible patient-related outcomes were mortality, LOS, ICU-LOS, sepsis severity, destination at discharge, and readmission. Lastly, costs associated with the use of MDx in the clinical setting were analyzed.
Risk of Bias
The risk of bias of randomized controlled trials (RCTs) was assessed by 1 reviewer using the Cochrane risk of bias tool for randomized trials (RoB2) [18]. This tool assesses 5 elements (sequence generation, allocation concealment, blinding, incomplete outcome data, and selective outcome reporting) of potential bias to estimate the effectiveness of an intervention. The risk of bias for prospective (nonrandomized) studies was also evaluated by 1 reviewer using the Cochrane risk of bias tool for nonrandomized studies of interventions (ROBINS-I) [19].
RESULTS
Study Design and Population
The search resulted in 692 records (Figure 1). The study characteristics of the included articles are presented in Table 1. In total, 25 eligible articles were retrieved. Three interventional studies (including 2 RCTs), 12 prospective studies, and 7 retrospective studies performed MDx directly on blood samples. Three prospective studies on different MDx used short-term BC incubation. MDx were compared with BC in all studies.
Table 1.
Characteristics of Studies Evaluating the Impact of MDx on Patient Outcomes, Antimicrobial Therapy, or Cost
| Author | Study Design | Diagnosis of Inclusion | Population | No. of Patients | Diagnostic Test | Study Location | Funding Source |
|---|---|---|---|---|---|---|---|
| Tafelski [20] | RCT | Suspected sepsis of abdominal or pulmonary origin | ICU patients | 41 intervention 37 control | SeptiFast, Roche | Germany | Roche Deutschland GmbH |
| Bhat [21] | RCT | Sepsis | Neonates | 183 intervention 185 control | Syndrome evaluation system | India | Council for Scientific and Industrial Research, New Millennium Indian Technological Leadership Initiative |
| Cambau [22] | Cluster-randomized interventional | Severe sepsis | In-hospital patients | 731 intervention 684 control | SeptiFast, Roche | France | French Ministry of Health |
| Septic shock | |||||||
| Infective endocarditis | |||||||
| Lodes [25] | Prospective intervention | SIRS | ICU patients | 104 intervention No control | SeptiFast, Roche | Germany | Not known |
| Mancini [23] | Prospective, pre-/postintervention | Suspected sepsis | Hematological patients | 137 intervention 138 control | SeptiFast, Roche | Italy | Roche Diagnostics |
| Bloos [42] | Prospective controlled observational | Severe sepsis | ICU patients | 142 prospective 63 control | SeptiFast, Roche | Germany, France | Roche Diagnostics |
| Septic shock | |||||||
| Wallet [43] | Prospective observational | Suspected BSI | ICU patients | 72 | SeptiFast, Roche | France | Roche Diagnostics provided materials |
| Bravo [26] | Prospective observational | Suspected BSI | Neutropenic and ICU patients | 86 neutropenic 53 ICU | SeptiFast, Roche | Spain | Not known |
| Maubon [27] | Prospective observational | Suspected sepsis | Patients with solid or hematological malignancies | 110 | SeptiFast, Roche | France | Roche Diagnostics, academic grants, Brahms Diagnostics, Gilead sciences, Merck, Pfizer |
| Bloos [32] | Prospective observational | Suspected sepsis | ICU patients | 245 | VYOO, SIRS-Lab | Germany | Thuringian Ministry of Education; the Thuringian Foundation for Technology, Innovation, and Research; STIFT, the German Sepsis Society; SIRS-Lab GmbH supplied the VYOO Kits and personnel |
| Tran [28] | Prospective observational | Suspected sepsis | Trauma, emergency, and burn surgery patients | 76 | SeptiFast, Roche | US | Roche Diagnostics, National Institute of Biomedical Imaging and Bioengineering |
| Vincent [34] | Prospective observational | Suspected/proven sepsis | ICU patients | 529 | PCR/ESI-MS | Belgium, France, Germany, UK, Switzerland, Poland | Ibis Biosciences, Abbott |
| O’Dwyer [40] | Prospective observational | Suspected/proven sepsis | ICU patients | 439 | PCR/ESI-MS | Belgium, France, Germany, UK, Switzerland, Poland | Ibis Biosciences, Abbott |
| Muñoz [38] | Prospective observational | Suspected invasive candidiasis | In-hospital | 54 | T2MR | Spain | European Regional Development Fund, T2 Biosystems |
| Muñoz [39] | Prospective observational | Candidemia | In-hospital | 44 | T2MR | Spain | European Regional Development Fund, T2 Biosystems |
| Dierkes [30] | Retrospective, pre-/postintervention | Sepsis | In-hospital patients | 77 post | SeptiFast, Roche | Germany | Not known |
| Alvarez [37] | Retrospective, pre-/postintervention | Sepsis | ICU patients | 48 post | SeptiFast, Roche | Spain | Not known |
| Severe sepsis | 54 pre | ||||||
| Septic shock | |||||||
| Lehmann [29] | Retrospective | Suspected sepsis | ICU and emergency patients | 436 | SeptiFast, Roche | Germany, Spain, Italy | Supported in part by Roche Diagnostics |
| Tschiedel [24] | Retrospective | Systemic infection | Pediatric ICU patients | 75 | SeptiFast, Roche | Germany | Not known |
| Herne [31] | Retrospective | Proven sepsis | In-hospital patients | 144 | SeptiFast, Roche | Estonia | Not known |
| Septic shock | |||||||
| Severe infection | |||||||
| Bloos [33] | Retrospective | Candidemia | ICU patients | 874 | VYOO, SIRS Lab | Germany | Pfizer Pharma GmbH |
| Patch [35] | Retrospective | Candidemia | In-hospital | 325 post | T2MR | US | No specific funding, part of a supplement sponsored by T2 Biosystems |
| 19 pre | |||||||
| McCann [44] | Prospective observational | Suspicion of BSI | ED + ICU | 918 | PCR/pyrosequencing | US | National Institute for Allergy and Infectious Diseases, NIH |
| Gebert [45] | Prospective observational | Suspected septicemia | In-hospital patients | 69 | Molysis + PCR, Molzym | Germany | German Ministry of Economy and Labour |
| Dryden [36] | Prospective observational | Clinical infection | In-hospital patients | 246 | ETGA Cognitor Minus, Momentum Bioscience | UK | No specific funding; Momentum Bioscience provided the Cognitor Minus kits for this study |
Abbreviations: BSI, bloodstream infection; ED, emergency department; ICU, intensive care unit; MDx, molecular microbiological diagnostics; RCT, randomized controlled trial; SIRS, systemic inflammatory response syndrome.
All patients in all studies had (suspected) bacterial or fungal BSI or (suspected) sepsis. However, inclusion criteria were not defined homogeneously. Suspected infection was the criterion for inclusion in 12 studies. However, criteria for suspicion were variable or not mentioned, and patients could be included based on BC draw or if the patient presented with fever or hypothermia. Ten studies concerned patients with proven sepsis, a subset of sepsis (severe sepsis, septic shock), or a systemic infection. Again, these terms were not always uniformly defined: Sepsis was diagnosed based on either the prevalence of systemic inflammatory response syndrome (SIRS) along with a suspicion of BSI or other definitions or national guidelines. All patients were sampled in-hospital in different types of wards. The ICU was represented in most studies (13/25), the emergency department in 3/25. Furthermore, 2/25 studies concerned neutropenic patients or patients with malignancies, and 2/25 concerned pediatric patients.
Antimicrobial Management
Antimicrobial management was the most frequently analyzed outcome parameter in 21/25 articles. However, the definition differed among the studies. A currently accepted general definition of appropriate antimicrobial therapy is the use of antimicrobials with in vitro activity against the causative microorganism. Data on antimicrobial management from included studies are summarized in Table 2. Various terms and definitions were used for appropriateness or effective treatment and for change and/or adjustments (escalation, de-escalation, discontinuation). These terms and synonyms were used interchangeably. Presence and tasks of antimicrobial stewardship programs during the study periods were not always described.
Table 2.
Data on Defined Antimicrobial Management Outcomes of Included Studies
| Author | Antimicrobial Treatment Adjustments | Appropriate Treatment | Time to Treatment Adjustment, h | Time to Appropriate Treatment, h |
|---|---|---|---|---|
| Tafelski [20] | Adjustments: 13.5% vs 9.8%a | - | 38.8 vs 18.8 (ns)a | - |
| Bhat [21] | Total adjustments: 15 vs 165 (P < .001)a; 5.8 vs 2.6 (P < .001)a antibiotics/patient | - | - | - |
| Cambau [22] | - | 23.6% vs 33.6% (P < .0001)a of patients | - | - |
| Lodes [25] | Adjustment: 25 (16.9%) | - | - | - |
| Mancini [23] | 61.67% vs 62.5% (P = 1) | - | - | - |
| Bloos [42] | - | - | - | - |
| Wallet [43] | Adjusted: 3 patients based on SF | - | - | - |
| Bravo [26] | Inappropriate addition: 2 neutropenic cases | Appropriate adjustment: 5 neutropenic cases | - | - |
| Appropriate adjustment: 10/13 ICU patients | ||||
| Maubon [27] | - | Adequate treatment: 4/32 | Earlier treatment: 4/32 BC+ | - |
| Adequate duration: 1/32 | Immediate prescription: 3 (2.8%) | |||
| Improved initial treatment: 11 (10%) | ||||
| Bloos [32] | - | 24.2% of PCR results suggest a de-escalation | - | - |
| Inadequate treatment: 34% of patients with PCR-positive result | ||||
| Tran [28] | - | Inadequate antibiotics: 29.2% PCR+ vs 18.7% BC+ patients | - | - |
| Vincent [34] | Adjustment recommendation: 41% | - | - | - |
| O’Dwyer [40] | - | - | - | - |
| Muñozb [38] | - | - | - | |
| Muñozb [39] | - | - | - | - |
| Dierkes [30] | Adjustment: 8/27 would be triggered | - | 3 of 8 adjustments would have been made earlier | - |
| Alvarez [37] | Reduced use of antibiotics: 5.1 vs 4.2 (P < .05)a | - | - | - |
| Lehmann [29] | Adjustment suggested by PCR result: 46 episodes | - | - | 22.8 days on early adequate treatment per 100 tests |
| Tschiedel [24] | Adjustment: 18%, 9 positive and 5 negative cases | - | - | - |
| Herne [31] | Adjustment: 38.9% of positive cases (7.4% de-escalation, 25.9% escalation) | - | - | - |
| Bloos [33] | - | - | - | 67.5 vs 31.0 (P < .01)a |
| Patch [35] | - | Antifungal treatment without positive blood cultures or positive T2MR: 42.8% and 41.6% | - | 34 h vs 6 h (P = .0013)a |
| McCann [44] | - | - | - | - |
| Gebert [45] | - | - | 5.7 h time difference between PCR result and positive signaling | - |
| Dryden [36] | - | Appropriate stewardship outcome: 73.6% | - | - |
| Inappropriate stewardship outcome: 23.9% |
Abbreviations: BC, blood culture; ICU, intensive care unit; ns, not significant; PCR, polymerase chain reaction.
aControl vs intervention.
bThese studies did not evaluate the impact on antimicrobial therapy. They were included because they assessed the impact on clinical severity; results are shown in Table 3.
In 2 randomized intervention studies by Tafelski et al. and Bhat et al., there were significantly fewer adjustments made when using SeptiFast (13.5% vs 9.8%) [20, 21] and another intervention study by Cambau et al. found a significant increase in the use of appropriate antimicrobials (23.6% vs 33.6%) [22]. In prospective studies by Mancini et al. and Tschiedel et al., the use of SeptiFast resulted in higher rates of antibiotic changes [23, 24]. Lodes et al. found appropriate changes in 16.9% of cases [25], and in the study conducted by Bravo et al. this was true in 77% of ICU cases [26]. Ten percent of patients would have an earlier or improved initial treatment, observed by Maubon et al. [27]. Contrarily, Tran et al. found an inappropriate use of antibiotics in 29.2% of polymerase chain reaction (PCR)–positive patients, while this was 18.7% in BC-positive patients [28]. In addition, Lehmann et al. calculated that there was a 57-day reduction of inadequate antibiotic therapy when SeptiFast was used and a 22.5-day reduction per 100 tests done [29]. Retrospectively, SeptiFast would have resulted in adjustment of antibiotic therapy in 29.6%–38.9% of cases [29–31]. One prospective study by Bloos et al. concluded that a de-escalation was suggested in 24.2% of cases based on the VYOO results [32]. A retrospective analysis by Bloos et al. found a shorter time to appropriate therapy when using VYOO (67.5 hours vs 31 hours) [33]. Based on the PCR-ESI-MS results, Vincent et al. concluded that there would have been a recommendation for a treatment change in 41% of the cases [34]. Bhat et al. used SES in a randomized controlled trial with neonates, which resulted in a significantly higher rate of adjustments of antibiotics (15 vs 165 changes) but an overall lesser use of antibiotics per patient (5.8 vs 2.6 antibiotics/patient) [21]. Retrospectively assessed by Patch et al., the use of T2MR was significantly associated with initiation of appropriate antifungal therapy 6 hours after blood draw, while this was 32 hours when using BCs [35]. Last, using ETGA for confirmation of negative samples after ≥12 hours of incubation (overnight) instead of 7 days, Dryden et al. found 73.6% appropriate stewardship outcomes [36].
Patient Outcomes
There were limited published data on the impact of MDx on clinical outcomes of patients. We were able to find data on mortality and/or LOS in-hospital or in-ICU in 11/25 articles (Table 3). Eleven studies described mortality differences when performing MDx directly on whole-blood samples. Mortality was assessed differently between studies and could be in-hospital or in-ICU, at 7 or 28 days. In the randomized intervention study by Cambau et al., there was no significant difference in mortality in the postintervention group [22], which was confirmed by the retrospective analysis by Alvarez et al. [37]. In the RCT by Bhat et al. using SES, in-hospital mortality was significantly lower when using SES [21]. Other differences in mortality were not significant. Interestingly, Lehmann et al. and Bloos et al. observed a higher mortality for PCR-positive patients compared with BC-positive patients [29, 32]. Additionally, the absence of Candida spp. DNA was found to be an independent predictor of survival [28], and a positive T2MR at baseline was an independent predictor of 7-day mortality [38].
Table 3.
Data on Defined Patient Outcomes (Hospital and ICU LOS, Mortality, and Costs) of Included Studies
| Author | Hospital LOS, d | ICU LOS, d | Mortality | Costs |
|---|---|---|---|---|
| Tafelski [20] | - | - | - | - |
| Bhat [21] | 19 vs 15 (P < .001)a | 11 vs 7 (P < .001)a | 17.8% vs 3.2% (P < .001)a in-hospital mortality | - |
| Cambau [22] | - | - | 15.8 vs 18.7% (P = .38)a 7-d mortality | Total cost per patient: €20 995 vs €19 329 (P = .09)a |
| Lodes [25] | - | - | 45/104 (43.2%) | - |
| Mancini [23] | - | - | - | Total cost per patient: €2010.53 vs €1579.80 |
| (P = .05)a | ||||
| Cost of antimicrobial therapy: | ||||
| €1384.87 vs €927.01 (P = .02)a | ||||
| Bloos [42] | - | - | In-hospital mortality: | - |
| PCR-/BC-: 27.8% | ||||
| PCR+: 36% | ||||
| PCR+/BC+: 34.5% | ||||
| Wallet [43] | - | - | - | - |
| Bravo [26] | - | - | - | - |
| Maubon [27] | - | - | - | - |
| Bloos [32] | - | - | - | - |
| Tran [28] | - | - | DNA absence is an independent predictor of survival (OR, 0.194; 95% CI, 0.045–0.840; P = .028) | - |
| Vincent [34] | - | - | - | - |
| O’Dwyer [40] | BC+ vs BC-: | BC+ vs BC-: | BC+ vs BC-: | - |
| 23 vs 23 (P = .8) | 7 vs 7 (P = .8) | 35% vs 32% (P = .7) 28-d mortality | ||
| PCR+ vs PCR-: | PCR+ vs PCR-: | PCR+ vs PCR-: | ||
| 22 vs 23 (P = .9) | 8 vs 7 (P = .8) | 42% vs 26% (P = .001) 28-d mortality | ||
| PCR+ was an independent predictor for 28-d mortality | ||||
| (OR, 1.7; 95% CI, 1.01–2.82; P = .04) | ||||
| Muñoz [38] | - | - | T2MR positivity is an independent predictor of poor outcome (7-d mortality; OR, 26.4; 95% CI, 2.1–327.3; P = .01) | - |
| Muñozb [39] | - | - | - | - |
| Dierkes [30] | - | - | - | - |
| Alvarez [37] | 21.3 vs 18.3 (P < .05)a | 31.0 vs 22.9 (P < .05)a | 24% vs 29%a 28-d mortality | Total cost per patient: €42 198 vs €32 228 (P < .05)a Costs of antibiotics: €3576 vs €2812 (P < .05)a |
| Lehmann [29] | - | - | 30-d mortality: PCR-/BC-: 14.1% | - |
| PCR+: 26.7% | ||||
| PCR+/BC+: 33.9% | ||||
| Tschiedel [24] | - | - | - | - |
| Herne [31] | - | - | - | - |
| Bloos [33] | - | 27.5 vs 19 (P = .386)a | 17 vs 3 (P = .138)a ICU mortality | - |
| Patch [35] | 20 vs 12 (P = .2041)a | 4 vs 6 (P = .7039)a | 9 vs 8 (P = .75)a in-hospital mortality | - |
| 9 vs 8 (P = .75)a 7-d mortality | ||||
| McCann [44] | - | - | - | - |
| Gebert [45] | - | - | - | - |
| Dryden [36] | - | - | - | - |
Abbreviations: BC, blood culture; ICU, intensive care unit; LOS, length of stay; PCR, polymerase chain reaction.
aControl vs intervention.
bThis study did not evaluate any of these outcomes but evaluated the impact on the severity of the disease course.
Bhat et al. reported a significantly lower in-hospital LOS in their RCT using SES [21]. Alvarez et al. retrospectively found a significantly decreased in-hospital LOS and ICU LOS when using SeptiFast compared with BC, respectively [37]. Other tests did not result in significant differences in any LOS. In the study by Muñoz et al., the role of T2MR in predicting complicated candidemia, which was defined as an episode involving metastatic spread to other organs or with attributable mortality, was assessed. Patients with complicated candidemia, thus having a more severe disease, were more likely to have positive T2MR results. Furthermore, a positive T2MR result early in the disease episode was an independent predictor of complicated candidemia [39]. Our search did not reveal MDx studies reporting data on the clinical outcomes transfer to ICU, destination at discharge, or readmissions.
Costs
Four of 25 studies reporting cost calculations are presented in Table 3. Mostly, cost identification studies that compared total costs and spending during hospitalizations [23, 37] were performed, rather than cost-effectiveness analyses. Only Cambau et al. calculated cost-effectiveness, together with cost identification, when using SeptiFast. The costs for SeptiFast, BCs, other diagnostic procedures, and anti-infective treatments were identified per patient and were adjusted for their LOS. They did not find a difference in costs in the complete study population between the control group and intervention group, nor for severe sepsis patients. Furthermore, the cost-effectiveness analysis did not show a significant increase in effectiveness, due to an ICU LOS that was not affected by an earlier microbiological identification [22].
In 2 other studies, cost calculations included diagnostic procedures and anti-infective treatment per patient during hospitalization [23, 37]. In both studies, it was concluded that, despite the high costs of tests, MDx were cost saving because of lower costs of antibiotic treatment. Contrary to Cambau et al., Mancini et al. found significant savings for patients with sepsis when using MDx [23]. The retrospective study by Alvarez et al. calculated that there was a 96.3% probability of cost savings when using SeptiFast (321 EUR/analysis) [37].
Molecular Diagnostics
Details on the MDx that were used in the 25 eligible articles assessing clinical impact, including performance characteristics, are shown in Supplementary Table 1. Eight different MDx, of which 5 commercial MDx used whole blood directly as sample material and 3 techniques used short-term incubation, were reported. None of the articles reported on in-house assays. One MDx, SeptiFast (Roche, Mannheim, Germany), was evaluated in 14/25 (63.6%) of the included studies. The VYOO system (SIRS-Lab, Jena, Germany) and PCR followed by electrospray ionization–mass spectrometry (PCR/ESI-MS; Plex-ID, Ibis/Abbott Inc., Abbott Park, IL, USA) were each evaluated in 2/25 (9.1%) studies, respectively. The T2 Candida magnetic resonance assay (T2MR, T2 Biosystems, Lexington, MA, USA) was studied in 3/25 articles, and the Syndrome Evaluation System (SES, Xcyton Diagnostics, Bangalore, India) was assessed in 1/25 (4.5%) study. Three are CE-IVD marked tests. Only T2MR has Food and Drug Administration approval. Of note, the most recent publications on SeptiFast dated from 2017, and the test was discontinued after the end of 2019. Additionally, VYOO was bought by Analytik Jena in 2013 and is not marketed anymore. Last, Ibis Biosciences, Abbott, has discontinued the PCR/ESI-MS system.
Three techniques used short-term incubation; PCR + pyrosequencing after 8 hours of incubation, Molysis + PCR with 2.6-hour and 6.3-hour incubation, and the Enzyme Template Generation and Amplification (ETGA) test (Cognitor Minus, Momentum Bioscience, Long Hanborough, UK) for the confirmation of negative samples after 12 hours of incubation.
Supplementary Table 1 also shows 2 CE-IVD marked tests, SepsiTest-UMD (Molzym, Bremen, Germany) and the MagicPlex Sepsis Test (Seegene, Seoul, South-Korea), which were identified by our search but excluded because of lack of reported clinical outcomes.
Risk of Bias
Using the Cochrane risk of bias tool, the 3 intervention studies were assessed for bias at the study level. The risk of bias assessment is summarized in Supplementary Table 2. The study by Cambau et al. scored best (low level of bias), while the 2 other studies showed some concerns in missing outcome data. The overall level of bias was considered to be moderate with some concerns. The overall risk of bias in the included nonrandomized studies was moderate to serious. The assessment is shown in Supplementary Table 3. There was serious risk of bias for the studies assessing therapeutic impact, as most studies used a nonblinded method of chart review, which is a subjective method. In addition, no information on confounders was given in most studies, and no information was provided on missing data, which are both factors that are likely to occur in prospective observational studies. Lastly, bias in the selection of the reported results is mostly moderate, as no protocol reviews have been done, which is needed to reach the level of certitude needed for a low risk of bias assessment in this domain.
DISCUSSION
The most striking finding of this systematic review was the scarcity of reports describing the impact on patient outcomes and antimicrobial management of rapid MDx either directly on whole blood or after short-term incubation. Our search yielded only 25 eligible articles on 8 commercially available tests, and no in-house test with data on clinical impact was identified. The heterogeneity of these 25 articles was too large and the number was too small to perform a meta-analysis.
The use of MDx could potentially result in better antimicrobial management before BC results become available, and thus in higher rates of appropriate antimicrobial therapy. However, in the few studies describing antimicrobial management, no uniform terminology was used, making comparisons very difficult. An earlier change in antimicrobial therapy has additional benefits for patients. However, time to change of antimicrobial therapy was only evaluated in 3 studies, of which only 1 found a significant reduction of inadequate antibiotic therapy [20, 29, 33]. Additionally, a faster detection of the causative agent of sepsis, but more importantly its susceptibility, could lead to an earlier de-escalation from broad-spectrum to narrow-spectrum, thereby reducing the selective pressure for antibiotic resistance.
The only patient outcomes studied for the use of MDx were LOS, ICU LOS, and mortality. Furthermore, few studies found improved patient outcomes, while others did not find a significant improvement or did not study these outcomes at all. Two studies found a significantly decreased hospital LOS and ICU LOS with the use of MDx. Again, a comparison between tests and studies is difficult, as different definitions for LOS were used. Additionally, conflicting results were reported on mortality. Interestingly, fungal DNA absence, determined via PCR, was found to be an independent predictor for survival, and a positive PCR result was found to be an independent predictor for mortality, which could be valuable in the fast identification of the most critical patients [28, 40]. Only 1 study evaluated disease severity [38]. Other important patient outcomes such as ICU transfer or 30-day readmission were not evaluated. Other outcomes relevant to the patient and society, such as destination upon discharge and return to former trajectory, need to be explored further. Studies show that inadequate antibiotic treatment is associated with higher mortality [41]. However, the lack of RCTs evaluating antibiotic therapy changes makes the added value of these MDx unknown at the patient level.
Lastly, MDx seemed to have an impact on cost. Even with expensive tests, 4 studies showed a significant decrease in costs. Of note: This decrease is not related to the test itself but mostly to reduced antimicrobial therapy. Although no cost-effectiveness studies were included here, 3 model calculations were identified in the search but were excluded as they were beyond the scope of our review.
All of the above findings, and especially the large gap of knowledge, could have led to a reluctancy of physicians and clinical microbiologists to implement MDx starting directly from whole blood in clinical practice. These tests are reported to be expensive, and the research and production as well, which may have led to the discontinuation of 3 out of the 5 systems.
This review on rapid MDx is, to our knowledge, the first to report on the added clinical value of MDx either directly from blood or after short-term incubation. The strengths of this analysis were the evaluation of outcomes that affect the individual patient directly (mortality and LOS) and indirectly (antimicrobial management). These outcomes also have an impact on society. However, this review has several limitations. We only searched 1 database and could only include 25 studies in this review. Only 3 of those were intervention studies. Most studies retrospectively evaluated hypothetical changes in antibiotic therapy and were not designed to find statistically significant differences. Even among this limited number of studies, the observed heterogeneity of studies was high. Studies used different terms, outcome measures, and analyses and reported their findings differently. This hampers comparisons and general conclusions. Furthermore, the target population, (suspected) BSI or sepsis, is also heterogenous, making comparison between studies even more challenging.
In conclusion, data on the clinical evaluation of rapid MDx in sepsis are limited. Only a handful of studies showed clear benefits in antimicrobial therapy management and patient outcomes. Commercially available MDx on whole blood have important shortcomings, such as low sensitivity, limited antibiotic resistance detection, and high cost. These are all probable reasons for discontinuation by companies. MDx combined with cultivation, for example, short-term incubation, seem more performant. In the future, more robust intervention studies should be performed on newly developed MDx, focusing on the added value of MDx in clinical practice and the possible benefits for critically ill patients.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This systematic review is part of the FAPIC project and has received funding from the European Union’s Horizon 2020 research and innovation program under GA 634137. This study is part of the Limburg Clinical Research Center (LCRC) UHasselt-ZOL-Jessa, supported by the foundation Limburg Sterk Merk (LSM), Hasselt University, Ziekenhuis Oost-Limburg, and Jessa Hospital.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This systematic review does not include factors necessitating patient consent or approval by a local ethical committee.
References
- 1. Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (SEPSIS-3). JAMA 2016; 315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin GS. Sepsis, severe sepsis and septic shock: changes in incidence, pathogens and outcomes. Expert Rev Anti Infect Ther 2012; 10:701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stewardson AJ, Allignol A, Beyersmann J, et al. The health and economic burden of bloodstream infections caused by antimicrobial-susceptible and non-susceptible Enterobacteriaceae and Staphylococcus aureus in European hospitals, 2010 and 2011: a multicentre retrospective cohort study. Euro Surveill 2016; 21:30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cassini A, Plachouras D, Eckmanns T, et al. Burden of six healthcare-associated infections on European population health: estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med 2016; 13:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 2017; 43:304–77. [DOI] [PubMed] [Google Scholar]
- 6. Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34:1589–96. [DOI] [PubMed] [Google Scholar]
- 7. Long B, Koyfman A. Best clinical practice: blood culture utility in the emergency department. J Emerg Med 2016; 51:529–39. [DOI] [PubMed] [Google Scholar]
- 8. Altindis M, Koroglu M, Demiray T, et al. A multicenter evaluation of blood culture practices, contamination rates, and the distribution of causative bacteria. Jundishapur J Microbiol 2016; 9:e29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ning Y, Hu R, Yao G, Bo S. Time to positivity of blood culture and its prognostic value in bloodstream infection. Eur J Clin Microbiol Infect Dis 2016; 35:619–24. [DOI] [PubMed] [Google Scholar]
- 10. Richter DC, Heininger A, Brenner T, et al. Bacterial sepsis: diagnostics and calculated antibiotic therapy. Anaesthesist 2017; 66:737–61. [DOI] [PubMed] [Google Scholar]
- 11. Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign Guidelines Committee including The Pediatric Subgroup Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med 2013; 39:165–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Medina E, Pieper DH. Tackling threats and future problems of multidrug-resistant bacteria. Curr Top Microbiol Immunol 2016; 398:3–33. [DOI] [PubMed] [Google Scholar]
- 13. Kozel TR, Burnham-Marusich AR. Point-of-care testing for infectious diseases: past, present, and future. J Clin Microbiol 2017; 55:2313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das S, Shibib DR, Vernon MO. The new frontier of diagnostics: molecular assays and their role in infection prevention and control. Am J Infect Control 2017; 45:158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pilecky M, Schildberger A, Orth-Höller D, Weber V. Pathogen enrichment from human whole blood for the diagnosis of bloodstream infection: prospects and limitations. Diagn Microbiol Infect Dis 2019; 94:7–14. [DOI] [PubMed] [Google Scholar]
- 16. Timbrook TT, Morton JB, McConeghy KW, et al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64:15–23. [DOI] [PubMed] [Google Scholar]
- 17. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med 2009; 6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sterne JAC, Savović J, Page MJ, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ 2019; 366:l4898. [DOI] [PubMed] [Google Scholar]
- 19. Sterne JA, Hernán MA, Reeves BC, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016; 355:i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tafelski S, Nachtigall I, Adam T, et al. Molecular Diagnostics of Sepsis Study Group Randomized controlled clinical trial evaluating multiplex polymerase chain reaction for pathogen identification and therapy adaptation in critical care patients with pulmonary or abdominal sepsis. J Int Med Res 2015; 43:364–77. [DOI] [PubMed] [Google Scholar]
- 21. Bhat BV, Prasad P, Ravi Kumar VB, et al. Syndrome evaluation system (SES) versus blood culture (BACTEC) in the diagnosis and management of neonatal sepsis—a randomized controlled trial Indian J Pediatr 2016; 83:370–9. [DOI] [PubMed] [Google Scholar]
- 22. Cambau E, Durand-Zaleski I, Bretagne S, et al. EVAMICA study team Performance and economic evaluation of the molecular detection of pathogens for patients with severe infections: the EVAMICA open-label, cluster-randomised, interventional crossover trial. Intensive Care Med 2017; 43:1613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mancini N, Sambri V, Corti C, et al. Cost-effectiveness of blood culture and a multiplex real-time PCR in hematological patients with suspected sepsis: an observational propensity score-matched study. Expert Rev Mol Diagn 2014; 14:623–32. [DOI] [PubMed] [Google Scholar]
- 24. Tschiedel E, Steinmann J, Buer J, et al. Results and relevance of molecular detection of pathogens by SeptiFast—a retrospective analysis in 75 critically ill children. Klin Padiatr 2012; 224:12–6. [DOI] [PubMed] [Google Scholar]
- 25. Lodes U, Bohmeier B, Lippert H, et al. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch Surg 2012; 397:447–55. [DOI] [PubMed] [Google Scholar]
- 26. Bravo D, Blanquer J, Tormo M, et al. Diagnostic accuracy and potential clinical value of the LightCycler SeptiFast assay in the management of bloodstream infections occurring in neutropenic and critically ill patients. Int J Infect Dis 2011; 15:e326–31. [DOI] [PubMed] [Google Scholar]
- 27. Maubon D, Hamidfar-Roy R, Courby S, et al. Therapeutic impact and diagnostic performance of multiplex PCR in patients with malignancies and suspected sepsis. J Infect 2010; 61:335–42. [DOI] [PubMed] [Google Scholar]
- 28. Tran NK, Wisner DH, Albertson TE, et al. Multiplex polymerase chain reaction pathogen detection in patients with suspected septicemia after trauma, emergency, and burn surgery. Surgery 2012; 151:456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lehmann LE, Alvarez J, Hunfeld KP, et al. Potential clinical utility of polymerase chain reaction in microbiological testing for sepsis. Crit Care Med 2009; 37:3085–90. [DOI] [PubMed] [Google Scholar]
- 30. Dierkes C, Ehrenstein B, Siebig S, et al. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect Dis 2009; 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herne V, Nelovkov A, Kütt M, Ivanova M. Diagnostic performance and therapeutic impact of LightCycler SeptiFast assay in patients with suspected sepsis. Eur J Microbiol Immunol (Bp) 2013; 3:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bloos F, Sachse S, Kortgen A, et al. Evaluation of a polymerase chain reaction assay for pathogen detection in septic patients under routine condition: an observational study. PLoS One 2012; 7:e46003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bloos F, Bayer O, Sachse S, et al. Attributable costs of patients with candidemia and potential implications of polymerase chain reaction-based pathogen detection on antifungal therapy in patients with sepsis. J Crit Care 2013; 28:2–8. [DOI] [PubMed] [Google Scholar]
- 34. Vincent JL, Brealey D, Libert N, et al. Rapid Diagnosis of Infections in the Critically Ill Team Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med 2015; 43:2283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patch ME, Weisz E, Cubillos A, et al. Impact of rapid, culture-independent diagnosis of candidaemia and invasive candidiasis in a community health system. J Antimicrob Chemother 2018; 73:iv27–30. [DOI] [PubMed] [Google Scholar]
- 36. Dryden M, Sitjar A, Gunning Z, et al. Can rapid negative exclusion of blood cultures by a molecular method, enzyme template generation and amplification technique (Cognitor® Minus), aid antimicrobial stewardship? Int J Pharm Pract 2018; 26:267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alvarez J, Mar J, Varela-Ledo E, et al. Cost analysis of real-time polymerase chain reaction microbiological diagnosis in patients with septic shock. Anaesth Intensive Care 2012; 40:958–63. [DOI] [PubMed] [Google Scholar]
- 38. Munoz P, Vena A, Machado M, et al. T2Candida MR as a predictor of outcome in patients with suspected invasive candidiasis starting empirical antifungal treatment: a prospective pilot study. J Antimicrob Chemother 2018; 73:iv6–12. [DOI] [PubMed] [Google Scholar]
- 39. Munoz P, Vena A, Machado M, et al. T2MR contributes to the very early diagnosis of complicated candidaemia. A prospective study. J Antimicrob Chemother 2018; 73:iv13–9. [DOI] [PubMed] [Google Scholar]
- 40. O’Dwyer MJ, Starczewska MH, Schrenzel J, et al. The detection of microbial DNA but not cultured bacteria is associated with increased mortality in patients with suspected sepsis—a prospective multi-centre European observational study. Clin Microbiol Infect 2017; 23:208.e1–6. [DOI] [PubMed] [Google Scholar]
- 41. Ibrahim EH, Sherman G, Ward S, et al. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000; 118:146–55. [DOI] [PubMed] [Google Scholar]
- 42. Bloos F, Hinder F, Becker K, et al. A multicenter trial to compare blood culture with polymerase chain reaction in severe human sepsis. Intensive Care Med 2010; 36:241–7. [DOI] [PubMed] [Google Scholar]
- 43. Wallet F, Nseir S, Baumann L, et al. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin Microbiol Infect 2010; 16:774–9. [DOI] [PubMed] [Google Scholar]
- 44. McCann CD, Moore MS, May LS, et al. Evaluation of real-time PCR and pyrosequencing for screening incubating blood culture bottles from adults with suspected bloodstream infection. Diagn Microbiol Infect Dis 2015; 81:158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gebert S, Siegel D, Wellinghausen N. Rapid detection of pathogens in blood culture bottles by real-time PCR in conjunction with the pre-analytic tool MolYsis. J Infect 2008; 57:307–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.