Abstract
Liver and intestinal flukes of the family Fasciolidae cause zoonotic food–borne infections that impact both agriculture and human health throughout the world. Their evolutionary history and the genetic basis underlying their phenotypic and ecological diversity are not well understood. To close that knowledge gap, we compared the whole genomes of Fasciola hepatica, Fasciola gigantica, and Fasciolopsis buski and determined that the split between Fasciolopsis and Fasciola took place ∼90 Ma in the late Cretaceous period, and that between 65 and 50 Ma an intermediate host switch and a shift from intestinal to hepatic habitats occurred in the Fasciola lineage. The rapid climatic and ecological changes occurring during this period may have contributed to the adaptive radiation of these flukes. Expansion of cathepsins, fatty-acid-binding proteins, protein disulfide-isomerases, and molecular chaperones in the genus Fasciola highlights the significance of excretory–secretory proteins in these liver-dwelling flukes. Fasciola hepatica and Fasciola gigantica diverged ∼5 Ma near the Miocene–Pliocene boundary that coincides with reduced faunal exchange between Africa and Eurasia. Severe decrease in the effective population size ∼10 ka in Fasciola is consistent with a founder effect associated with its recent global spread through ruminant domestication. G-protein-coupled receptors may have key roles in adaptation of physiology and behavior to new ecological niches. This study has provided novel insights about the genome evolution of these important pathogens, has generated genomic resources to enable development of improved interventions and diagnosis, and has laid a solid foundation for genomic epidemiology to trace drug resistance and to aid surveillance.
Keywords: food–borne flukes, Fasciola hepatica, Fasciola gigantica, Fasciolopsis buski, genome evolution, adaptive radiation
Introduction
Digenetic trematodes (flukes) are a major group of helminth parasites of humans and animals. Among them, Fasciolopsis buski (Fb. buski), Fasciola gigantica (Fa. gigantica), and Fasciola hepatica (Fa. hepatica), the intestinal and liver flukes of the family Fasciolidae, cause zoonotic food–borne infections that have a substantial impact on both agriculture (3 billion USD per year) and human health (∼90,000 disability-adjusted life years) throughout the world (Torgerson et al. 2015; Cwiklinski et al. 2016). Fasciolopsis buski (subfamily Fasciolopsinae) is a large fluke (up to 7.5 cm long, 2.5 cm wide) that infects the small intestine of humans and pigs in East and Southeast Asia, causing diarrhea, abdominal pain, fever, ascites, and bowel obstruction. Fasciola hepatica and Fa. gigantica (subfamily Fasciolinae) cause liver disease in ruminants and humans in Europe, the Americas and Australasia (where only Fa. hepatica is transmitted) and in Africa and Asia (where the two species overlap). When present, clinical symptoms include fever, malaise, abdominal pain, eosinophilia, and hepatomegaly during the acute phase, whereas biliary tree obstruction symptoms predominate in chronic disease. Fasciolid flukes have a heteroxenous life cycle, which involves a definitive vertebrate host (where the adult worms live, mate, and produce eggs), an intermediate molluscan host (where the larval stages develop and multiply), and a carrier (suitable aquatic plants). A previous phylogenetic study of the family Fasciolidae (Lotfy et al. 2008) indicates that Fp. buski (subfamily Fasciolopsinae) is descended from a relatively early-diverging lineage, whereas Fa. gigantica and Fa. hepatica are derived sister species. Somewhere along the line leading to Fasciola species and other members of the Fasciolinae, two events of great importance occurred: a host switch from planorbid snails to lymnaeid snails, and a habitat switch by adults from intestinal sites to the liver. Here, we present the genomes of Fp. buski and Fa. gigantica, making the genomes of all three human-infecting fasciolid flukes available and via comparisons provide a better understanding of their evolutionary history and diversification, and the genetic bases underlying their phenotypic and ecological divergence and adaptation to different host species and habitats.
Results and Discussion
Genome Features of the Intestinal and Liver Flukes in the Family Fasciolidae
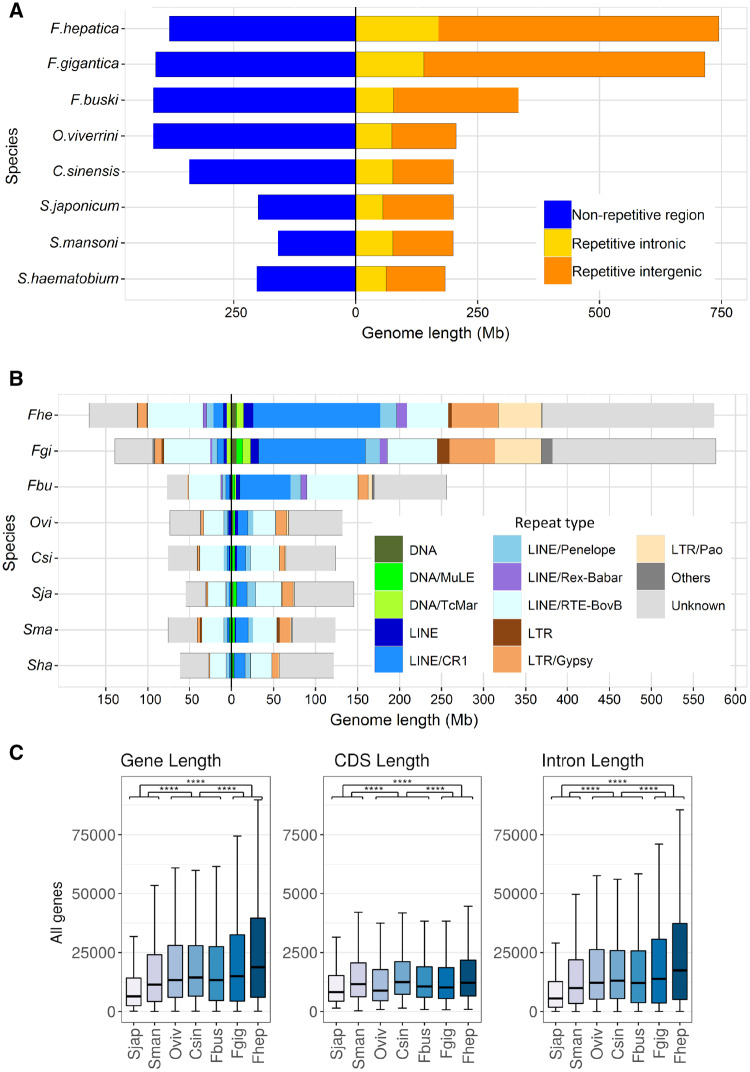
The nuclear and mitochondrial genomes of Fb. buski and Fa. gigantica were sequenced, assembled, and annotated (table 1 and supplementary table 1, Supplementary Material online). To facilitate more robust interspecies comparisons, our previously published Fa. hepatica genome (GenBank accession number: GCA_002763495) (McNulty et al. 2017) was reannotated using an improved methodology and updated RNA-seq and protein homology databases. The total assembly lengths of Fp. buski and Fa. gigantica draft genomes were 748 Mb and 1.13 Gb, respectively. Although the former is comparable to outgroup species in the family Opisthorchiidae, the latter with its expanded genome size is similar to Fa. hepatica (1.14 Gb), suggesting that the increased genome size is a derived trait that emerged in the lineage leading to Fasciola. Despite the genome size differences, the total numbers of protein-coding genes annotated in Fp. buski and the two Fasciola species were similar, ranging from 11,218 to 12,647 and representing 91.5% to 93.0% BUSCO completeness. These numbers were also comparable to other distantly related digenean taxa (table 1), suggesting that the relatively larger Fasciola genomes did not evolve through whole-genome duplications. Interestingly, the patterns of variation in transposable element (TE) contents of these and related genomes indicate that lineage-specific differential accumulation of TE families may have played a central role in genome size evolution in Fasciolidae (fig. 1A). The nonrepeat genome sizes are similar in Fp. buski (400 Mb), Fa. hepatica (372 Mb), Fa. gigantica (409 Mb), and the Opisthorchiidae, while smaller in the Schistosomatidae (200 Mb). However, the genomic regions containing interspersed repetitive elements are more than twice as long in Fasciola spp. (658–707 Mb) than in Fp. buski (318 Mb), and longer in fasciolids than in other trematodes. Most of the enrichment in repetitive elements is due to intergenic elements, although intronic elements are twice as long in Fasciola spp. than in Fp. buski. Notably, Fasciola genomes carry about 3–4 times more DNA transposons (e.g., Tc1/mariner) and about seven times more long-terminal-repeat retrotransposons (e.g., Gypsy, Pao, Copia) as compared with the Fp. buski genome (fig. 1B and supplementary table 2, Supplementary Material online). These are strongly enriched in intergenic regions, but other abundant long interspersed nuclear elements (LINEs) such as RTE-BovB are equally distributed between intronic and intergenic locations (fig. 1B). Gene length has also increased in Fasciolinae, due to longer introns, consistent with the increased presence of TEs within them (fig. 1C). As repeat elements degenerate through time, the sequence similarity measured as per-copy distances to consensus provides reasonable evidence that TE activity is currently low in the fasciolid genomes (supplementary fig. 1, Supplementary Material online).
Table 1.
Assembly and Annotation Statistics of Analyzed Genomes.
| Fasciolopsis buski | Fasciola gigantica | Fasciola hepatica after Reannotation | Fasciola hepatica Original Annotation | Clonorchis sinensis | Opisthorchis viverrini | Schistosoma mansoni | |
|---|---|---|---|---|---|---|---|
| GenBank assembly accession | LUCM00000000 | SUNJ00000000 | JXXN00000000 | GCA_002763495 | GCA_000236345 | GCA_000715545 | GCA_000237925 |
| Total genome length (Mb) | 748.5 | 1,128 | 1,138.3 | 1,138.3 | 547.3 | 620.5 | 364.5 |
| N50 scaffold length (kb) | 190.8 | 181.8 | 161.1 | 161.1 | 417.5 | 1,324 | 32,115.3 |
| L50 scaffold count | 1,104 | 1,805 | 2,036 | 2,036 | 408 | 138 | 4 |
| Protein-coding genes | 11,747 | 12,647 | 11,218 | 14,642 | 13,634 | 16,356 | 11,940 |
| %BUSCO (complete) | 73.9 | 77.2 | 78.6 | 43.9 | 77.5 | 79.5 | 89.4 |
| %BUSCO (fragmented) | 19.1 | 14.9 | 12.9 | 35.3 | 13.5 | 11.9 | 7.6 |
| Mean CDS length | 1,420 | 1,376 | 1,633 | 837 | 1,591 | 1,301 | 1,433 |
| Mean intron length | 3,708 | 3,982 | 4,170 | 2,902 | 2,757 | 3,550 | 2,409 |
| Mean exons per mRNA | 5.2 | 5.9 | 7.5 | 3.2 | 6.9 | 5.7 | 5.9 |
| % of genome covered by CDS | 2.2 | 1.5 | 1.6 | 1.1 | 4 | 3.4 | 4.6 |
Fig. 1.
Trematode genome nonrepetitive and repetitive contents. (A) Relative genome sizes of different trematodes, with nonrepetitive fraction (blue) represented to the left of the central axis, and repetitive fractions to the right indicating intronic (yellow) and intergenic (orange) repeats, respectively. (B) Classification of repeated elements present in intronic locations (to the left of the central axis) and intergenic repeats (to the right of the axis). Repeats classes are color coded. (C) Variation in gene, CDS, and intron lengths for the different trematode species. Statistically significant differences are indicated.
The influence of TEs on animal genome size variation is widely accepted, and it is increasingly recognized that changes in TE activity may have a major effect on adaptation of populations and species facing novel habitats and large environmental perturbations (Chenais et al. 2012). TEs are potent sources of mutation that can rapidly create genetic variance, especially following genetic bottlenecks and severe environmental changes, providing bursts of allelic and phenotypic diversity upon which selection can act (Stapley et al. 2015; Schrader and Schmitz 2018). Thus, our data lead to the hypothesis that TE-mediated genomic changes likely have contributed to the increased adaptive capacity of Fasciola spp. to new habitats and host species after their divergence from Fasciolopsis. Because TEs are highly mutagenic, either directly (e.g., insertions in coding or regulatory regions) or indirectly (e.g., chromosomal rearrangements), molecular countermeasures such as chromatin modifications suppress their activity and TE-derived transcripts are targeted for cleavage by npc silencing and piwi-interacting RNAs (Slotkin and Martienssen 2007). Interestingly the main silencing mechanism, the Piwi pathway, is incomplete in all parasitic flatworms including the Fasciolidae here analyzed and alternative silencing mechanisms based on conserved duplicated flatworm-specific Argonaute proteins (FLAgos) have been suggested (Skinner et al. 2014; Fontenla et al. 2017) (supplementary fig. 2, Supplementary Material online). Environmentally induced physiological or genomic stress can modulate TE activity by activating transposition or by inhibiting genomic silencing mechanisms (Rey et al. 2016), thus facilitating adaptive responses in species experiencing changed or diverse environments, as faced by invasive, pathogenic or parasitic species during the developmental cycle (Schrader and Schmitz 2018).
Lineage Diversification, Trait Evolution, and Historical Demography
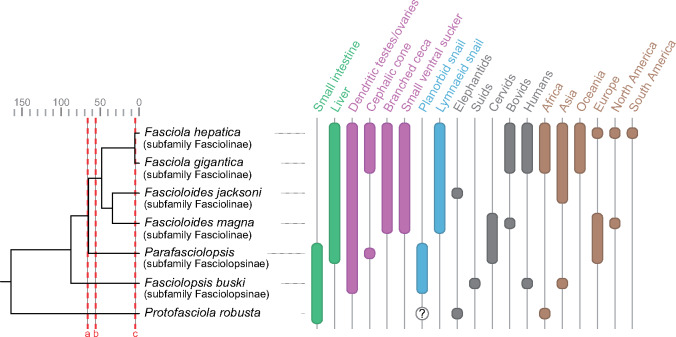
Despite their public health and veterinary significance, the evolutionary history of fasciolid flukes remains understudied. The Fasciolidae may have originated in African proboscideans and later radiated in Eurasian herbivores (Lotfy et al. 2008). As the family diversified, host shifts occurred in both molluscan and mammalian hosts. There was also a switch in habitat within the definitive host from the small intestine to the liver (fig. 2). Morphological, ecological, and molecular phylogenetic data support relatively basal divergence for the lineage leading to Fp. buski and a derived position for the species of the genus Fasciola (Lotfy et al. 2008). Although the former genus has a planorbid snail as intermediate host, the latter genus exploits the Lymnaeidae, indicating a host-switch at some point in the lineage leading to Fasciola. The intestinal fluke Fp. buski has a large ventral sucker and simple bifurcated digestive ceca, whereas branched digestive caeca and a reduced ventral sucker are characteristics of the liver flukes. Accordingly, the comparative analysis of their genomes offers the opportunity to gain insights into the evolution of key processes of parasitism such as host selection, tissue tropism, and morphological adaptations. To investigate prospective correlations between biogeographical events and lineage diversification in the Fasciolidae, we constructed a dated phylogeny using a Bayesian multilocus coalescent method with 30 nuclear protein-coding genes (supplementary table 3, Supplementary Material online) and a node height prior taken from the age of Protostomia estimated in a previous fossil-calibrated eukaryote phylogeny (Parfrey et al. 2011). The molecular dating revealed that the split between the genera Fasciolopsis and Fasciola took place around 88.1 Ma (73.0–102.9, 95% highest posterior density [HPD]) in the late Cretaceous period, and the divergence between Fa. hepatica and Fa. gigantica occurred around 5.3 Ma (3.4–7.2, 95% HPD) near the Miocene–Pliocene boundary (supplementary fig. 3, Supplementary Material online). The estimated date for the divergence of Fasciolopsis and Fasciola seems rather ancient, given what we know about the evolution of mammals. Monotreme mammals were already established at 112–121 Ma (Rowe et al. 2008). Meredith et al. (Meredith et al. 2011) have placentals as arising around 100 Ma, mammals as a group arising much earlier than this, and ungulates a bit later. It is possible that fasciolids originated early in another group of mammals and switched into their current host groups later. Lotfy et al. (2008) suggested that fasciolids emerged in proboscideans, a hypothesis supported by the fact that the extant more basal fasciolid (Protofasciola robusta) lives in the small intestine of African elephants. The proboscideans evolved in Africa and radiated to Eurasia, and the host transition to ungulates might have occurred during this dispersion. To narrow down the time-window during which the most distinctive apomorphic traits of the Fasciolinae (genera Fasciola and Fascioloides) (i.e., lymnaeid snail hosts, hepatic habitats, branched intestinal caeca, dendritic testes and ovaries) originated, the ages of the stem node (the last common ancestor of Fasciolinae and Fasciolopsinae) and the crown node (the last common ancestor of all living members of Fasciolinae) were estimated using a published phylogeny of Fasciolidae that included Parafasciolopsis fasciolaemorpha (Fasciolopsinae) (Lotfy et al. 2008) and a whole-genome mitochondrial phylogeny including Fascioloides magna (Fasciolinae) (fig. 2 and supplementary fig. 4, Supplementary Material online). The data suggested that the intermediate host switch and shift from intestinal to hepatic habitats occurred between 65 Ma (stem node; 43.2–85.7, 95% HPD) and 55.9 Ma (crown node; 42.0–70.8, HPD) in the lineage leading to Fasciolinae. The profound climatic and ecological changes that occurred during this period (e.g., Cretaceous–Paleogene mass extinction and Paleocene–Eocene Thermal Maximum) may have contributed to the adaptive radiation of these flukes to new niches. The divergence time estimate of 5.3 Ma between Fa. hepatica and Fa. gigantica is substantially more recent than the previously suggested date of 19 Ma based on cathepsin L-like cysteine proteases (Irving et al. 2003). Most notably, our speciation time estimate coincides with the Miocene–Pliocene boundary that was characterized by a reduced faunal exchange between Africa and Eurasia (Bibi 2011), which may have contributed to the speciation process through an increased and sustained disruption of gene flow, resulting in two locally adapted sister taxa, that is, Fa. gigantica in Africa and Fa. hepatica in Eurasia. Based on comparative data on infectivity, life span, egg shedding and immunity among modern hosts, it was proposed that Fa. hepatica emerged in Eurasian ovicaprines, whereas Fa. gigantica originated in an African ruminant phylogenetically close to present-day bovines (Mas-Coma et al. 2009). These sister taxa still hybridize producing an intermediate form in regions where they currently occur sympatrically in Africa and Asia (Saijuntha et al. 2018). This has important implications for epidemiology such as the potential for crossover of anthelmintic resistance between the two species or emergence of more pathogenic variants.
Fig. 2.
Dated phylogeny and trait distribution in the Fasciolidae. Cladogram and parasite traits are based on Lotfy et al. (2008). Node ages in Ma are based on nuclear and mitochondrial molecular dating analyses: primary host body habitat (green), morphology (purple), snail host (blue), vertebrate host (gray), and geographic distribution (brown). Cretaceous–Paleogene mass extinction (a), Paleocene–Eocene Thermal Maximum (b), reduced faunal exchange between Africa and Eurasia (c).
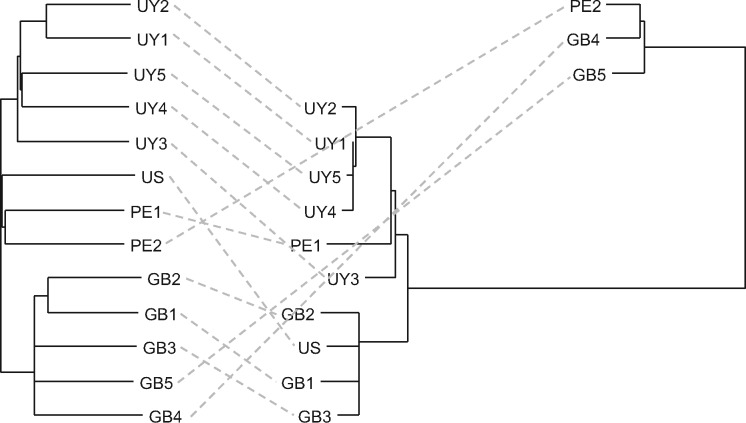
Using genome-wide variation data of 13 samples from the United States, Uruguay, Peru, and the United Kingdom (supplementary table 1, Supplementary Material online), genetic structuring within Fa. hepatica was assessed. A moderate to low-level, geographic population structure, reflecting the country of origin, was observed in the nuclear genome (fig. 3) (e.g., FST between Uruguay and the United Kingdom: 0.094). Mitochondrial genome variation, however, revealed a striking pattern of discordance in which two deeply diverged clades were apparent. These haplogroups correspond to the two previously reported mitochondrial lineages in Fa. hepatica (defined based on a ∼1.4-kb region that overlaps with cytochrome oxidase subunit III gene, tRNA-His gene, and cytochrome b gene) in geographically diverse European and Australian populations (Teofanova et al. 2011; Walker et al. 2011, 2012) (supplementary fig. 5, Supplementary Material online). Interestingly, both mitochondrial lineages were observed in Peru, suggesting that both haplogroups have been introduced to the New World, although their precise frequency and distribution in the Americas will need to be determined. Our molecular dating analysis indicated that these haplogroups originated around 1.1 Ma (0.07–2.7, 95% HPD) in the Pleistocene period (supplementary fig. 4, Supplementary Material online). Domesticated sheep (Ovis aries) fall into five mitochondrial haplogroups, whose radiation has been dated to be 0.92 ± 0.19 Ma, substantially predating the domestication event (∼8–11 ka) (Meadows et al. 2011). It is thus tempting to hypothesize that a process of host–parasite codiversification has played a central role in the development of these haplogroups in Fa. hepatica where haplogroup formation in the host led to genetic structuring in the parasite. The observed mito-nuclear discordance is consistent also with high levels of nuclear gene flow and mixing of alleles within each metapopulation (Beesley et al. 2017) in contrast to the patterns of (nonrecombining) mitochondrial allelic diversity where ancestral haplotypes can persist in a population alongside derived forms.
Fig. 3.
Nuclear (left) and mitochondrial (right) phylogeny of Fasciola hepatica based on genome-wide SNPs showing mito-nuclear discordance. US, USA; UY, Uruguay; PE, Peru; GB, United Kingdom.
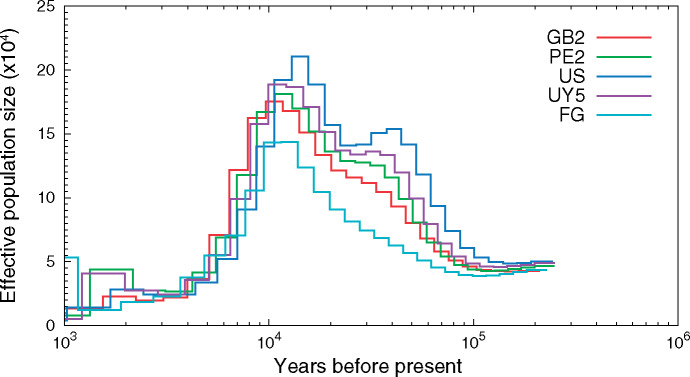
We reconstructed the historical demography of Fasciola and found evidence of a rapid decline in its effective population size (i.e., a signature of founder effect) ∼10–11 ka, which is consistent with a recent global spread associated with the ruminant domestication (fig. 4). An evolutionarily very recent spread of Fasciola spp. from their origin in the Eurasian Near East area (Fa. hepatica) and East Africa (Fa. gigantica) has been proposed based on the ribosomal DNA sequence diversity, and the spread of both species in postdomestication times likely has led to their present overlap in Africa and Asia where lymnaeid snails are suitable for the development of both species (Mas-Coma et al. 2009). Abnormal ploidy and aspermic parthenogenesis in hybrids between the two species also suggest their near-complete genetic isolation and separate evolution in predomestication times (Mas-Coma et al. 2009).
Fig. 4.
Fasciola historical demography. The PSMC model was used to characterize historical demography by examining heterozygosity densities across the genome. US, USA; UY, Uruguay; PE, Peru; GB, United Kingdom; FG, Fasciola gigantica.
Gene Family Dynamics in Medically Important Trematodes
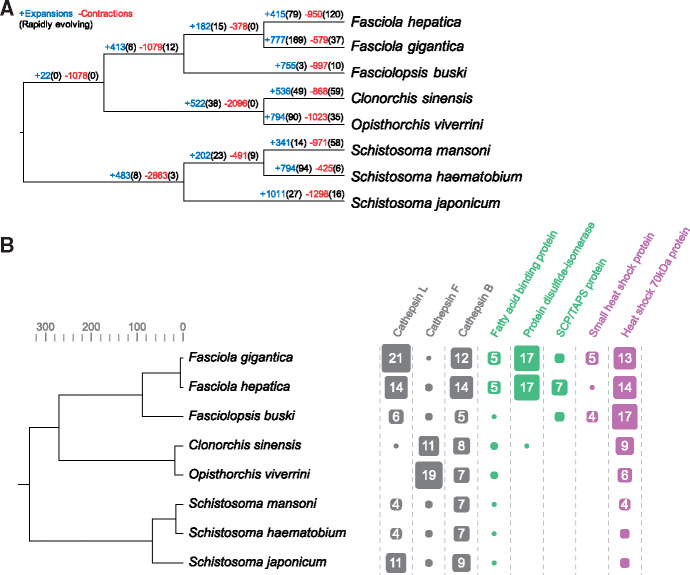
To understand the genomic changes underlying phenotypic variation within fasciolid species and between families of digenetic trematodes of medical importance (Schistosomatidae, Opisthorchiidae, and Fasciolidae), we investigated large-scale differences in gene complements among lineages. Using orthologous groups (OGs) of genes identified across eight digenean species with sequenced genomes (supplementary table 4, Supplementary Material online), we modeled gene gain and loss while accounting for the species’ phylogenetic history (Han et al. 2013) (fig. 5A). Based on the birth–death (λ) parameter estimate (0.00575), statistical significance of the observed family size differences among taxa was assessed. Gene families of interest that displayed most pronounced differential expansions or contractions included the cathepsin cysteine proteases (L, F, and B families), fatty-acid-binding proteins, protein disulfide-isomerases and molecular chaperones, highlighting the significance of excretory–secretory proteins in lineage-specific adaptation (fig. 5B). Although several of these protein families were highlighted as relevant in adaptation by Fa. hepatica (McNulty et al. 2017), the present study provides evidence that their amplification occurs at or after the split between Fasciolinae and Fasciolopsinae.
Fig. 5.
Gene family dynamics in trematodes of medical importance. (A) Gene family gains/losses were modeled by estimating birth–death (λ) parameters while accounting for the species’ phylogenetic history: when P < 0.01 they were considered rapidly evolving. (B) Gene families of interest that showed differential expansion/contraction in trematodes of medical importance. Gene family size and count are indicated by scaled boxes.
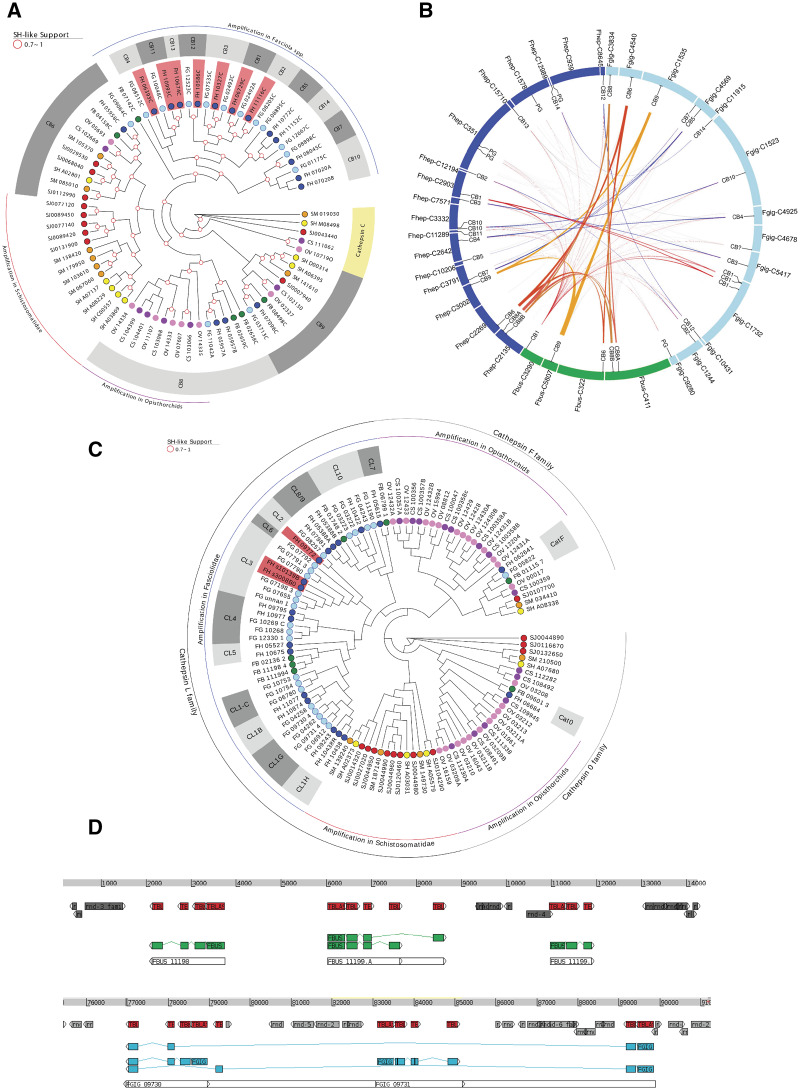
The cathepsin superfamily encompasses several cysteine protease genes present in diverse flatworms with differential expression according to the parasite stage and multiple overlapping functions (Caffrey et al. 2018). Cathepsins constitute a substantial fraction of the excretory–secretory products and endow Fasciola with the ability to migrate through tissue and digest matrix, to break down proteins including hemoglobin for nutrition, and to modulate the immune response through digestion of immunoglobulins (Cancela et al. 2008; McGonigle et al. 2008; Robinson et al. 2008). In Fa. hepatica, a particular cathepsin L (FhCL3) with an unusual collagenolytic activity and several cathepsin Bs have been implicated in the early stages of invasion through the intestinal wall (Corvo et al. 2009; Cancela et al. 2010; Robinson et al. 2011; Meemon and Sobhon 2015). Five cathepsin B genes are annotated in Fp. buski, whereas 16 genes are annotated in Fa. hepatica and Fa. gigantica. Comparative analysis within cathepsins Bs (OG0000035) shows four conserved enzymes present in the three species as well as other trematodes, namely CatB9, CatB6, and a tandem duplication of CatB8 (fig. 6A and B). The remaining single gene in Fp. buski is at the base of an expansion of cathepsin Bs in the Fasciola spp. resulting in more than ten discrete genes. Given that almost all of these novel genes are shared between Fa. hepatica and Fa. gigantica, the genomic event that conferred this gene gain predated the separation of these species. A subclade of the novel cathepsin B genes occurring in Fasciola spp. corresponds to those that are differentially expressed during the intestine invasive stage in Fa. hepatica (fig. 6A and B). Similarly, within the cathepsin Ls (OG0000050), although some members are conserved (particularly CatL0, expressed in eggs in Fa. hepatica), an expansion and diversification process has taken place in the Fasciolinae lineage after it diverged from the Fasciolopsinae (fig. 6C). A tandem array of at least three cathepsin L genes with repeated exons that might produce diverse transcripts by alternative splicing is present in the Fp. buski genome (fig. 6D). Similar complex structures occur repeatedly in diverse contigs within the genomes of Fa. gigantica and Fa. hepatica, giving rise to more than a dozen cathepsin Ls (fig. 6C). It is noteworthy that the clade including the collagenolytic juvenile-specific cathepsin L3 with a suggested role in early invasion and the clade containing the mature cathepsin L1 members involved in immune evasion both seem to be related to the same Fp. buski cluster (fig. 6C). Although the still fragmentary status of the three assemblies does not allow to trace precisely the possible duplication events, it is plausible to consider that a region similar to the Fp. buski CatL cluster might have been the origin of the amplifications in the Fasciolinae lineage. In addition to amplifications of cathepsins B and L in genomes of the Fasciolinae, other independent amplifications of these gene families are observed in the Opisthorchiidae and Schistosomatidae. By contrast, cathepsin F genes (OG0000076) are amplified only in the Opisthorchiidae (Kang et al. 2010; Sripa et al. 2010) (fig. 6C). Notably, the exopeptidase cathepsin C (OG0007199), which is implicated in terminal processing of peptides in schistosomes (Hola-Jamriska et al. 1998; Caffrey et al. 2018) and Clonorchis (Liang et al. 2014), was absent from the three species of the Fasciolidae studied here. Cathepsin C is conserved in the Schistosomatidae and Opisthorchiidae (fig. 6A).
Fig. 6.
Cathepsins in trematodes of medical importance. (A) Phylogenetic tree of cathepsins B and C from the family Fasciolidae and other trematodes. (B) Circos representation of the cathepsin B-containing contigs in Fasciolopsis buski (green), Fasciola hepatica (blue), Fasciola gigantica (light blue). The relative position of the diverse cathepsin B genes is indicated. Conservation between Fp. buski and the Fasciolinae cathepsin Bs is highlighted in black and those between the Fasciolinae cathepsin Bs in red. (C) Phylogenetic tree of cathepsins L and F from the family Fasciolidae and other trematodes. (D) Visualization of the gene structure of the cathepsins L cluster present in Fp. buski contig 4155 (top) and Fa. gigantica contig 4972 (bottom). Conserved regions (as detected by BLAST) are indicated in red, gene CDS are indicated in green (Fp. buski) and sky blue (Fa. gigantica). Repetitive sequences are indicated in gray. Note the presence of repeated exons that allow alternative transcripts from the same gene.
Asparaginyl endopeptidases, better known as legumains have been implicated in the maturation of cathepsin proenzymes (Robinson et al. 2009). We observed that Fa. gigantica shares the amplification of legumains (OG0000019) already described for Fa. hepatica (McNulty et al. 2017). By contrast, a restricted set of only three legumain genes is present in Fp. buski (supplementary fig. 6A, Supplementary Material online). The coincident amplification of cathepsins and legumains in the genus Fasciola might reflect the likely diversification of regulatory legumains involved in the maturation of the diverse cathepsin proenzymes. Other regulatory proteins of cysteine protease activity, such as the cystatin family of cysteine-protease inhibitors occurs broadly and similarly across the Fasciolidae (supplementary fig. 6B, Supplementary Material online).
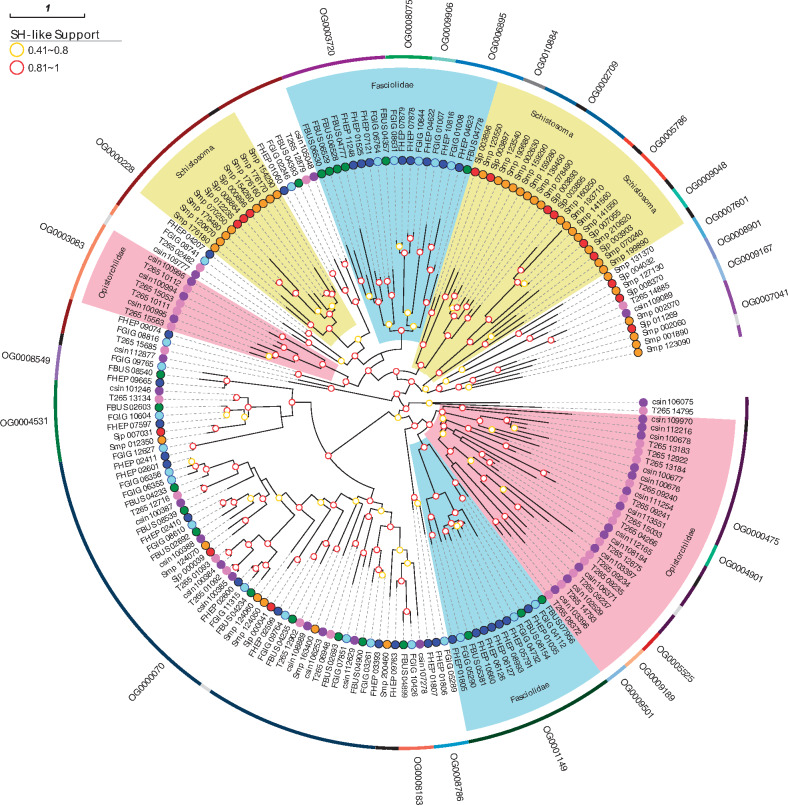
Analysis of gene gain and loss showed that an orthogroup corresponding to CAP domain-containing proteins (OG0001149) was extensively amplified in the Fasciolidae compared with other trematodes. Proteins with this domain, which also is known as the SCP/TAPS domain, appear to be involved in helminth parasite–mammalian host interactions. These proteins are excreted and secreted, and are differentially expressed in the parasitic stages of hookworms (Datu et al. 2008). In platyhelminths, most studies have been undertaken in Schistosoma mansoni, in which different superfamily members are expressed in different developmental stages. Some CAP domain-containing proteins are specifically expressed in the intrasnail stages or in the intramammal stages (Chalmers et al. 2008; Rofatto et al. 2012). Several specific duplications have been reported for this superfamily among diverse helminth taxa (Tang et al. 2014; Hunt et al. 2016; Costabile et al. 2018; International Helminth Genomes Consortium 2019). In particular, these duplications are lineage specific, implying that each lineage has duplicated and maintained particular superfamily members, influenced by the biological differences in hosts and/or life cycle stages. To explore this further, all the orthogroups with genes annotated as CAP domain-containing proteins from trematodes were retrieved and analyzed. The phylogenetic analysis (fig. 7) reveals that although several orthogroups are lineage specific, most of them are phylogenetically related to other lineage-specific orthogroups. The Fasciolidae-specific OG0001149 is phylogenetically related to five Opisthorchiidae-specific orthogroups that diverged after the Fasciolidae/Opisthorchiidae split. Several of their members are preferentially expressed in the invasive stage. Four other small OGs occur exclusively in the Fasciolidae, expressed across diverse life stages such as those presented in the phylogenetically conserved OG0000070. As reported for other helminths, different lineages have duplicated specific groups of genes, and this may be related to the differences in their life cycles and host immune-evasion mechanisms.
Fig. 7.
CAP domain-containing proteins of the Fasciolidae and other trematodes. Phylogeny of CAP domain-containing proteins from trematodes is depicted. The outer ring indicates the diverse orthogroups containing CAP domain proteins. Lineage-specific amplifications are highlighted (pink: Opisthorchiidae, light blue: Fasciolidae, yellow: Schistosomatidae).
Other excretory–secretory product components like the fatty-acid-binding proteins have potent immunomodulatory effects inhibiting activation of TLR4 and macrophages, allowing the parasite to establish a long-lasting infection (Figueroa-Santiago and Espino 2014; Martin et al. 2015) (supplementary fig. 7A, Supplementary Material online). Lipid transporters, such as the NPC2 protein, were also amplified in the Fasciolinae, confirming observations on Fa. hepatica (McNulty et al. 2017). This may represent a metabolic adaptation to the rich lipid conditions characteristic of the biliary tract, the predilection site for the adult stage of Fa. gigantica and Fa. hepatica (supplementary fig. 7B, Supplementary Material online). Heat-shock proteins expanded in fasciolid flukes (supplementary fig. 8, Supplementary Material online). Flatworm heat-shock proteins perform crucial roles in protein homeostasis and protection from stress-induced damage. They also participate in development and modulation of immune responses through their immunogenic and immunomodulative properties (Yang et al. 2012; Chung et al. 2017). The protein disulfide isomerases described in Fasciola may contribute to survival of the parasite in an environment like the bile ducts, with high oxidative stress and protein damage (Salazar-Calderon et al. 2003).
Genome-Wide Signatures of Adaptive Evolution in Fasciola
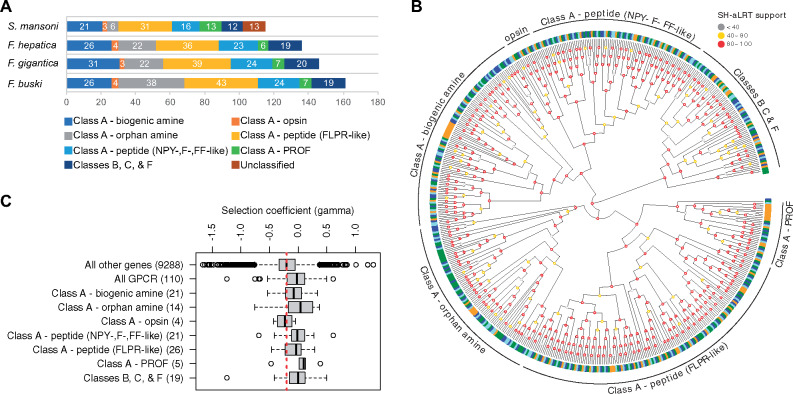
We examined genome-wide signatures of selection from patterns of genetic polymorphism (within Fa. hepatica) and divergence (between Fa. hepatica and Fa. gigantica) using SnIPRE, a Bayesian implementation of the McDonald and Kreitman (MK) test developed for genome-wide analysis (Eilertson et al. 2012). The Kolmogorov–Smirnov test was performed to identify enriched gene ontology (GO) terms among genes with high selection coefficient γ (indicating positive selection) (table 2). A marked enrichment was observed in GO terms related to G-protein-coupled receptors (GPCRs), indicating that GPCRs are more likely to be under positive and/or relaxed purifying selection than other genes, suggesting their involvement in adaptive evolution of these flukes. Using S. mansoni GPCRs (n = 115) as the reference (Hahnel et al. 2018), putative Fa. hepatica (117), Fa. gigantica (126), and Fp. buski (142) GPCRs were assigned into classes A, B, C, and F (fig. 8A and 8B). Class A GPCRs were further classified into aminergic receptors (including orphan amines, biogenic amines, and opsins), peptidergic receptors (including neuropeptide Y, neuropeptide F, and neuropeptide FF, and FMRFamide-like peptide), and the platyhelminth-specific rhodopsin-like orphan-family. Phylogenetic analysis indicated Fasciolidae-specific expansions among biogenic and orphan amine receptors (fig. 8B). The selection coefficient (γ) of GPCRs, on average, was higher than those of all other genes (Kruskal–Wallis test, P = 1.7 × 10−10), and this pattern was observed across all classes of GPCRs except for opsins (fig. 8C). GPCRs translate sensory inputs into cellular responses and are thus crucial for tuning physiology and behavior in response to the environment. Expansion of GPCR odorant receptors (ORs), for example, increases the repertoire of odorant signals that species detect, allowing them to occupy new ecological niches (e.g., terrestrial vs. aquatic vertebrates) (Kishida 2008). As a comparison, genomes of great apes contain about 1,000 OR genes, of which one-third appear to be functional, as acquisition of trichromatic color vision in primates caused the parallel pseudogenization of OR genes (the “vision-priority” hypothesis) (Gilad et al. 2004). Hence, it is reasonable to hypothesize that differential gene family expansion and positive selection of GPCRs, along with parallel pseudogenization of genes under relaxed purifying selection, represent one mechanism by which species of Fasciola adapted to a new ecological niche.
Table 2.
Enriched GO Terms among Fasciola Genes with Signatures of Adaptive Evolution (i.e., high gamma values indicating positive selection).
| GO Term | Kolmogorov–Smirnov Test P-Value | |
|---|---|---|
| Biological process | GPCR signaling pathway | 9.3E-09 |
| Potassium ion transport | 3.0E-04 | |
| Transmembrane transport | 3.6E-03 | |
| Molecular function | GPCR activity | 1.7E-11 |
| Ionotropic glutamate receptor activity | 3.4E-04 | |
| Extracellular-glutamate-gated ion channel activity | 3.8E-04 | |
| Calcium ion binding | 7.3E-04 | |
| Potassium channel activity | 8.9E-04 | |
| G-protein-coupled peptide receptor activity | 9.4E-03 | |
| Cellular component | Integral component of membrane | 2.6E-11 |
| Membrane | 4.9E-08 | |
Fig. 8.
G-protein-coupled receptor (GPCR). (A) total GPCR gene counts in each genome. (B) phylogeny of GPCRs and classification into different classes using Schistosoma mansoni GPCRs as the reference. Fasciolopsis buski (green), Fasciola hepatica (blue), Fasciola gigantica (light blue), S. mansoni (yellow) (C) selection coefficient of GPCRs showing signatures of positive and/or relaxed purifying selection. Positive gamma values indicate positive selection and negative gamma values indicate purifying selection.
Conclusions
This comparative analysis provides novel insight into the biology and evolution of Fasciolidae and other fluke families of medical importance. Rapid climatic and ecological changes may have contributed to the adaptive radiation of fasciolids, which were accompanied by lineage-specific gene family expansions and differential rates of molecular evolution among different gene families. The genomic resources that these studies have provided should enhance development of novel interventions and diagnosis, and underpin genomic epidemiologic investigation of new disease outbreaks, virulence and drug resistance.
Materials and Methods
Parasite Specimens
Genomes of Fa. gigantica and Fp. buski were assembled de novo using specimens from Uganda (cattle liver) and Vietnam (pig intestine), respectively. Adult Fa. gigantica were isolated from the livers of Ankole cattle at the abattoir of Fort Portal in the Western Region of Uganda in 1993. The samples of Fp. buski belong to the HT strain and were collected in Ha Tay town, near Hanoi, Vietnam. Parasites were stored in 80% ethanol at −20 °C until isolation of nucleic acids. Whole-genome resequencing data for Fa. hepatica were generated using specimens collected from Uruguay (Montevideo) and Peru (Cusco), which were analyzed together with published samples from the United States (McNulty et al. 2017) and the United Kingdom (Cwiklinski et al. 2015).
DNA/RNA Isolation, Genome Sequencing, and Assembly
Nucleic acids were extracted using QIAGEN DNeasy (DNA) and RNeasy (RNA) mini kits and cleaned up by ethanol precipitation for Fa. gigantica. For Fp. buski, ethanol-preserved adult worms were chopped up using a scalpel blade, and genomic DNA (gDNA) was extracted and purified using the kit E.Z.N.A and SQ Tissue DNA Kit (Omega Bio-tek), and the yield and purity were assessed by Bio-Analyzer as described (McNulty et al. 2017). DNA small-insert (fragment) and mate-pair (jump) libraries were constructed using gDNA extracted from an individual adult worm and sequenced on Illumina HiSeq platform (2X100bp) as described (McNulty et al. 2017). Pacific Biosciences sequencing (PacBio RS II P5-C3/P6-C4, 20-kb library) was performed to complement the Illumina data and improve scaffolding and gap-filling. ALLPATHS-LG (release 44837) (Gnerre et al. 2011) was used to assemble the Illumina reads after adapter sequences were removed with Trimmomatic v0.36 (Bolger et al. 2014). To scaffold the assembled contigs, SSPACE-standard v3.0 (Boetzer et al. 2011) and SSPACE-longread v1.1 (Boetzer and Pirovano 2014) were sequentially run using the Illumina and PacBio reads, respectively. Gapfiller v1.10 (Boetzer and Pirovano 2012) and PBJelly v15.8.24 (English et al. 2012) were used to close gaps, and the resulting assembly was error-corrected using Pilon v1.20 (Walker et al. 2014) and screened for contaminants using blobtools v0.9.19. Mitochondrial genomes (mtDNA) were assembled with NOVOPlasty v2.6.3 using the Illumina fragment reads (Dierckxsens et al. 2017).
Genome Annotation
The nuclear genomes of Fa. gigantica and Fp. buski were annotated using the MAKER pipeline v2.31.8 (Holt and Yandell 2011). The published Fa. hepatica genome assembly (GenBank accession number: GCA_002763495.1) was reannotated using the same pipeline. Repetitive elements were soft-masked with RepeatMasker v4.0.6 using a species-specific repeat library created by RepeatModeler v1.0.8, RepBase repeat libraries (Bao et al. 2015), and a list of known TEs provided by MAKER (Holt and Yandell 2011). From the NCBI Sequence Read Archive, Fa. hepatica (SRR2038730, SRR2038734, SRR2038743, SRR2039050, SRR2039051, ERS524681-3, ERS524685-91, ERS524693, ERS524696), Fa. gigantica (SRR094761), and Fp. buski (SRR941773, SRR5929441) RNA-seq data were obtained. Additional RNA-seq was performed for Fa. gigantica in biological duplicates (adult stage) on the Illumina HiSeq platform (2X100bp TruSeq Stranded mRNA library) to support the genome annotation. After adapter trimming using Trimmomatic v0.36 (Bolger et al. 2014), RNA-seq reads were aligned to their respective genome assemblies using HISAT2 v2.0.5 (Kim et al. 2015) with the –dta option and subsequently assembled using StringTie v1.2.4 (Pertea et al. 2015). The resulting alignment and transcript assembly were used by BRAKER (Hoff et al. 2016) and MAKER pipelines, respectively, as extrinsic evidence data. In addition, mRNA and EST sequences for each species were retrieved from NCBI and passed to MAKER as transcript evidence. Protein sequences from UniRef100 (UniProt Consortium 2017) (Trematoda-specific, n = 205,161) and WormBase ParaSite WBPS7 (Howe et al. 2017) (Clonorchis sinensis PRJDA72781, Opisthorchis viverrini PRJNA222628, S. mansoni PRJEA36577) were provided to MAKER as protein homology evidence. Ab initio gene predictions from BRAKER v1.9 (Hoff et al. 2016) and AUGUSTUS v3.2.2 (trained by BRAKER and run within MAKER) were refined using the transcript and protein evidence. Previously unpredicted exons and untranslated regions were added, and split models were merged. The best-supported gene models were chosen based on Annotation Edit Distance (Eilbeck et al. 2009). To reduce false positives, gene predictions without supporting evidence were excluded during building the final annotation, with the exception of those encoding Pfam domains, as detected by InterProScan v5.19 (Jones et al. 2014). These Pfam domain sequences were rescued to improve the overall annotation accuracy by balancing sensitivity and specificity (Holt and Yandell 2011; Campbell et al. 2014). Unfiltered set of gene models are available upon request. PANNZER2 (Koskinen et al. 2015) and sma3s v2 (Casimiro-Soriguer et al. 2017) were employed to name gene products. The completeness of annotated gene sets was assessed using BUSCO v3.0 (Waterhouse et al. 2017). GO and KEGG annotations were performed using InterProScan v5.19 (Jones et al. 2014) and BlastKOALA (Kanehisa et al. 2016), respectively. rRNA and tRNA were annotated using RNAmmer v1.2.1 (Lagesen et al. 2007) and tRNAscan-SE v1.23 (Lowe and Eddy 1997), respectively. Mitochondrial genomes were annotated using MITOS2 (Bernt et al. 2013).
Repeat Analysis
RepeatModeler v1.0.8 (with WU-BLAST as its search engine) was used to build, refine, and classify consensus models of putative interspersed repeats for each species. With the resulting repeat libraries, genomic sequences were screened using RepeatMasker v4.0.6 in “slow search” mode to generate a detailed annotation of the interspersed and simple repeats. Per-copy distances to consensus were calculated (Kimura two-parameter model, excluding CpG sites) and were plotted as repeat landscapes where divergence distribution reflected the activity of TEs on a relative time scale per genome using the calcDivergenceFromAlign.pl and createRepeatLandscape.pl scripts included in the RepeatMasker package. Intergenic and intragenic repeats were identified by comparing the genic and repeat annotation coordinates. The distribution of gene lengths, coding, and intronic sequences for different species were calculated, and the statistical significance of the observed size differences among taxa was assessed.
Molecular Divergence Dating Analysis
Diversification timeframe for Fasciolidae was estimated using StarBEAST2, a multiindividual, multilocus coalescent method implemented in *BEAST v2.4.7 (Ogilvie et al. 2017). To infer times to the most-recent common ancestor for Fasciolopsis–Fasciola and Fa. hepatica–Fa. gigantica, 30 single-copy protein-coding nuclear loci were selected randomly from OGs of genes identified across 11 protostome taxa using OrthoFinder v1.1.4 (Emms and Kelly 2015). In S. mansoni, which is currently the only trematode for which a chromosome-level assembly is available, all 30 loci are located on autosomes and at least 500 kb apart from each other, suggesting that these genes are unlinked and evolve independently. For each orthologous gene group, PRANK (Loytynoja and Goldman 2005) was used within the framework of GUIDANCE2 (Sela et al. 2015) to generate codon-based multiple sequence alignments with removal of unreliable columns (below the default cutoff of 0.93). A relaxed molecular clock analysis was run with the Calibrated Yule model (Heled and Drummond 2012) and bModelTest (Bouckaert and Drummond 2017) as the tree prior and the site model, respectively. A most-recent common ancestor prior was set on the root height for the species tree, taken from the age of Protostomia estimated in a previous fossil-calibrated eukaryote phylogeny (Parfrey et al. 2011)—a normal prior with mean = 632 Ma, SD = 29.3 Ma. Twenty independent Markov chain Monte Carlo chains were run, each for 2 × 109 generations, sampling every 105 states. The topology was held constant when estimating other parameters, including divergence times. Convergence, mixing, and ESS values for each parameter were assessed using Tracer v1.6 (Rambaut et al. 2018). The last 2 × 104 trees sampled from the stationary posterior distribution of each of the 20 Markov chain Monte Carlo runs were combined using LogCombiner v1.8.4, and a maximum clade credibility tree was generated using TreeAnnotator v2.4.7. An earlier fasciolid phylogeny (based on 28S, ITS1, ITS2, NAD1) (Lotfy et al. 2008) and a whole-genome mitochondrial phylogeny were dated, using the same methods, but adjusting gene ploidy for mitochondrial loci, and using the resulting divergence time estimates (Fasciolopsis–Fasciola: mean = 88.1 Ma, SD = 7.7 Ma; Fa. hepatica–Fa. gigantica: mean = 5.29 Ma, SD = 0.9 Ma) as node age priors.
Gene Family Evolution
OGs of genes were inferred with OrthoFinder v1.1.4 (Emms and Kelly 2015) using the longest isoform for each gene. The CAFE method (Han et al. 2013) was employed to model gene gain and loss while accounting for the species’ phylogenetic history based on an ultrametric species tree, generated as described above for molecular dating, and the number of gene copies found in each species for each gene family. Birth–death (λ) parameters were estimated, and the statistical significance of the observed family size differences among taxa was assessed. Gene trees from selected Fasciolidae-enriched families were generated by aligning with MAFFT (Katoh and Standley 2014) and PHYML tree building (Guindon et al. 2010), with models predicted by Model Generator (Keane et al. 2006). Trees were visualized with Evolview (He et al. 2016). Cryptic or partial copies of gene family members were captured by tBLASTn on the genomes with coding sequence (CDS) of OG members as queries, and visualized and inspected manually with Artemis (Carver et al. 2012).
Genome Variation Analysis
Individual worm gDNA reads from Illumina small-insert libraries were aligned to the corresponding genome assemblies using BWA-MEM v0.7.15 (Li and Durbin 2009). Polymerase chain reaction and optical duplicates were removed using picard tools v2.8.3 (http://broadinstitute.github.io/picard; last accessed February 9, 2017). Reads that aligned on the edges of indels were realigned to achieve the most consistent placement. Single-nucleotide variants were called via local de novo assembly of haplotypes using GATK v3.7 (McKenna et al. 2010) and quality-filtered as previously described (Van der Auwera et al. 2013; McNulty et al. 2017). For mitochondrial loci, sample ploidy was set to 1. Full mitochondrial haplotype sequences were reconstructed based on single nucleotide polymorphisms (SNPs) for individual Fa. hepatica samples and were used to build maximum-likelihood phylogenetic trees with RAxML v8.2.9 under GTRCAT model and with autoMRE bootstrapping (Stamatakis 2014). The pairwise genetic distance between samples (1-ibs, identity by state) was computed using PLINK v1.90 after excluding loci with missing genotypes in any of the samples for both the nuclear and mitochondrial genomes. The computed distances were subsequently used to generate a tanglegram based on neighbor-joining trees in Dendroscope v3.5.9. To correlate our mitochondrial genome sequences with the previously published ∼1.4-kb partial mitochondrial sequences overlapping with cytochrome oxidase subunit III (cox3) gene, tRNA-His gene, and cytochrome b (cytb) gene, the corresponding regions were extracted from the genomes and subjected to phylogenetic analysis using MAFFT and RAxML (Katoh and Standley 2014; Stamatakis 2014). Variants were annotated according to their genomic locations and predicted coding effects using SnpEff (Cingolani et al. 2012).
Historical Demography
The Pairwise Sequentially Markovian Coalescent (PSMC) model (Li and Durbin 2011) was used to characterize historical demography by examining heterozygosity densities in 100-bp sliding windows across the genome. Consensus genomic sequence data (contig length > 50 kb) were generated for each diploid individual worm using SAMtools/BCFtools (mapping quality > 20; base quality > 20; median × 0.33 < depth of coverage < median × 2) (Li et al. 2009) based on the deduplicated and indel-realigned alignments. Because PSMC is sensitive to variation in coverage depth, it was run twice for each individual using parameters -N25 -t15 -r5 -p “4+25*2+4 + 6.” First, it was run utilizing all mapped sequence data and then utilizing data down-sampled to 10× coverage using the DownsampleSam tool (picard). Results were scaled by a mutation rate estimated based on genome size (1.6 × 10−8 per base pair per generation for Fasciola spp.) (Crellen et al. 2016) and a generation time of 0.25 years (Phalee et al. 2015), resulting in distributions of Ne through time. Subsequently, 100 PSMC bootstrap replicates were performed for both full-coverage and down-sampled data to confirm consistent distributional patterns.
Genome-Wide Analysis of Signatures of Adaptive Evolution
To identify genome-wide signatures of selection from patterns of genetic polymorphism (within Fa. hepatica) and divergence (between Fa. hepatica and Fa. gigantica), we performed the MK test within a Bayesian framework using SnIPRE (Eilertson et al. 2012). The MK table of fixed or polymorphic replacement and silent substitutions was prepared using PopGenome (Pfeifer et al. 2014) based on exonic SNPs (identified using the GATK pipeline as described above) in 13 Fa. hepatica and 1 Fa. gigantica samples. Gene loci with >8× sequencing coverage over >70% CDS length in all samples were included in the analysis (n = 9,398). The Kolmogorov–Smirnov test was performed to identify enriched GO terms among gene with high gamma (positive selection).
GPCR Annotation and Analyses
Based on available high-confidence GPCR sequences in S. mansoni (Hahnel et al. 2018) and Fa. hepatica (McVeigh et al. 2018), orthologous GPCR sequences were identified in our fasciolid genomes by Inparanoid v4.1 (Sonnhammer and Ostlund 2015) and Reciprocal Best Hits methods. These sequences were supplemented with those annotated with the GO term GO:0004930 (GPCR activity) by InterProScan v5.19. False-positive GPCR sequences were removed through manual curation involving an iterative process of inspecting multiple sequence alignments, building phylogenetic trees, and identifying anomalous phylogenetic placements using the S. mansoni phylogeny (Hahnel et al. 2018) as the reference. Multiple sequence alignments were generated using TM-Aligner (Bhat et al. 2017), and phylogenetic trees were inferred by maximum likelihood using IQ-TREE (Nguyen et al. 2015) with the best-fit model automatically selected (Kalyaanamoorthy et al. 2017) and the SH-aLRT test (Guindon et al. 2010) performed with 10,000 replicates.
Command lines used to perform the analyses are made available in supplementary text 1, Supplementary Material online.
Supplementary Material
Acknowledgments
Sequencing of the genomes was supported by the “Sequencing the etiological agents of the Food–Borne Trematodiases” project (National Institutes of Health—National Human Genome Research Institute award number U54HG003079). Comparative genome analysis was funded by grants National Institutes of Health—National Institute of Allergy and Infectious Diseases AI081803 and National Institutes of Health—National Institute of General Medical Sciences GM097435 to M.M.
Author Contributions
Conceptualization: M.M., P.J.B.; formal analysis: Y.-J.C., J.F.T., S.F., A.C.; funding acquisition: P.J.B., J.F.T., M.M.; methodology: J.F.T., M.M.C., P.U.F., T.H.L., D.B., M.M.; resources: M.M.C., T.H.L., P.U.F., M.M.; visualization: Y.-J.C., J.F.T., S.F., A.C.; writing—original draft: Y.-J.C., J.F.T., M.M.; writing—review & editing: M.M., D.B., P.J.B., P.U.F., J.F.T., M.M.C.
References
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley NJ, Williams DJ, Paterson S, Hodgkinson J.. 2017. Fasciola hepatica demonstrates high levels of genetic diversity, a lack of population structure and high gene flow: possible implications for drug resistance. Int J Parasitol. 47(1):11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Bhat B, Ganai NA, Andrabi SM, Shah RA, Singh A.. 2017. TM-Aligner: multiple sequence alignment tool for transmembrane proteins with reduced time and improved accuracy. Sci Rep. 7(1):12543.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi F. 2011. Mio-Pliocene faunal exchanges and African biogeography: the record of fossil bovids. PLoS One 6(2):e16688.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W.. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13(6):R56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W.. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15:211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR, Drummond AJ.. 2017. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 17(1):42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey CR, Goupil L, Rebello KM, Dalton JP, Smith D.. 2018. Cysteine proteases as digestive enzymes in parasitic helminths. PLoS Negl Trop Dis. 12(8):e0005840.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, Hufnagel DE, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, et al. 2014. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164(2):513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancela M, Acosta D, Rinaldi G, Silva E, Duran R, Roche L, Zaha A, Carmona C, Tort JF.. 2008. A distinctive repertoire of cathepsins is expressed by juvenile invasive Fasciola hepatica. Biochimie 90(10):1461–1475. [DOI] [PubMed] [Google Scholar]
- Cancela M, Ruetalo N, Dell’Oca N, da Silva E, Smircich P, Rinaldi G, Roche L, Carmona C, Alvarez-Valin F, Zaha A, et al. 2010. Survey of transcripts expressed by the invasive juvenile stage of the liver fluke Fasciola hepatica. BMC Genomics 11(1):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA.. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28(4):464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro-Soriguer CS, Munoz-Merida A, Perez-Pulido AJ.. 2017. Sma3s: a universal tool for easy functional annotation of proteomes and transcriptomes. Proteomics 17(12):1700071.. [DOI] [PubMed] [Google Scholar]
- Chalmers IW, McArdle AJ, Coulson RM, Wagner MA, Schmid R, Hirai H, Hoffmann KF.. 2008. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics 9:89.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenais B, Caruso A, Hiard S, Casse N.. 2012. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509(1):7–15. [DOI] [PubMed] [Google Scholar]
- Chung EJ, Jeong YI, Lee MR, Kim YJ, Lee SE, Cho SH, Lee WJ, Park MY, Ju JW.. 2017. Heat shock proteins 70 and 90 from Clonorchis sinensis induce Th1 response and stimulate antibody production. Parasit Vectors. 10(1):118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvo I, Cancela M, Cappetta M, Pi-Denis N, Tort JF, Roche L.. 2009. The major cathepsin L secreted by the invasive juvenile Fasciola hepatica prefers proline in the S2 subsite and can cleave collagen. Mol Biochem Parasitol. 167(1):41–47. [DOI] [PubMed] [Google Scholar]
- Costabile A, Koziol U, Tort JF, Iriarte A, Castillo E.. 2018. Expansion of cap superfamily proteins in the genome of Mesocestoides corti: an extreme case of a general bilaterian trend. Gene Rep. 110–120. [Google Scholar]
- Crellen T, Allan F, David S, Durrant C, Huckvale T, Holroyd N, Emery AM, Rollinson D, Aanensen DM, Berriman M, et al. 2016. Whole genome resequencing of the human parasite Schistosoma mansoni reveals population history and effects of selection. Sci Rep. 6:20954.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwiklinski K, Dalton JP, Dufresne PJ, La Course J, Williams DJ, Hodgkinson J, Paterson S.. 2015. The Fasciola hepatica genome: gene duplication and polymorphism reveals adaptation to the host environment and the capacity for rapid evolution. Genome Biol. 16:71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwiklinski K, O’Neill SM, Donnelly S, Dalton JP.. 2016. A prospective view of animal and human Fasciolosis. Parasite Immunol. 38(9):558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datu BJ, Gasser RB, Nagaraj SH, Ong EK, O’Donoghue P, McInnes R, Ranganathan S, Loukas A.. 2008. Transcriptional changes in the hookworm, Ancylostoma caninum, during the transition from a free-living to a parasitic larva. PLoS Negl Trop Dis. 2(1):e130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Moore B, Holt C, Yandell M.. 2009. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10:67.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilertson KE, Booth JG, Bustamante CD.. 2012. SnIPRE: selection inference using a Poisson random effects model. PLoS Comput Biol. 8(12):e1002806.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7(11):e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Santiago O, Espino AM.. 2014. Fasciola hepatica fatty acid binding protein induces the alternative activation of human macrophages. Infect Immun. 82(12):5005–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenla S, Rinaldi G, Smircich P, Tort JF.. 2017. Conservation and diversification of small RNA pathways within flatworms. BMC Evol Biol. 17(1):215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S.. 2004. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2(1):E5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hahnel S, Wheeler N, Lu Z, Wangwiwatsin A, McVeigh P, Maule A, Berriman M, Day T, Ribeiro P, Grevelding CG.. 2018. Tissue-specific transcriptome analyses provide new insights into GPCR signalling in adult Schistosoma mansoni. PLoS Pathog. 14(1):e1006718.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- He Z, Zhang H, Gao S, Lercher MJ, Chen WH, Hu S.. 2016. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44(W1):W236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ.. 2012. Calibrated tree priors for relaxed phylogenetics and divergence time estimation. Syst Biol. 61(1):138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32(5):767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hola-Jamriska L, Tort JF, Dalton JP, Day SR, Fan J, Aaskov J, Brindley PJ.. 1998. Cathepsin C from Schistosoma japonicum–cDNA encoding the preproenzyme and its phylogenetic relationships. Eur J Biochem. 255(3):527–534. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M.. 2017. WormBase ParaSite—a comprehensive resource for helminth genomics. Mol Biochem Parasitol. 215:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt VL, Tsai IJ, Coghlan A, Reid AJ, Holroyd N, Foth BJ, Tracey A, Cotton JA, Stanley EJ, Beasley H, et al. 2016. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet. 48(3):299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Helminth Genomes Consortium. 2019. Comparative genomics of the major parasitic worms. Nat Genet. 51:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving JA, Spithill TW, Pike RN, Whisstock JC, Smooker PM.. 2003. The evolution of enzyme specificity in Fasciola spp. J Mol Evol. 57(1):1–15. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K.. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 428(4):726–731. [DOI] [PubMed] [Google Scholar]
- Kang JM, Bahk YY, Cho PY, Hong SJ, Kim TS, Sohn WM, Na BK.. 2010. A family of cathepsin F cysteine proteases of Clonorchis sinensis is the major secreted proteins that are expressed in the intestine of the parasite. Mol Biochem Parasitol. 170(1):7–16. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2014. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 1079:131–146. [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO.. 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 6:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T. 2008. Pattern of the divergence of olfactory receptor genes during tetrapod evolution. PLoS One 3(6):e2385.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen P, Toronen P, Nokso-Koivisto J, Holm L.. 2015. PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics 31(10):1544–1552. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW.. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.; 1000 Genome Project Data Processing Group. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, He L, Xu Y, Chen X, Huang Y, Ren M, Liang C, Li X, Xu J, Lu G, et al. 2014. Identification, immunolocalization, and characterization analyses of an exopeptidase of papain superfamily, (cathepsin C) from Clonorchis sinensis. Parasitol Res. 113(10):3621–3629. [DOI] [PubMed] [Google Scholar]
- Lotfy WM, Brant SV, DeJong RJ, Le TH, Demiaszkiewicz A, Rajapakse RP, Perera VB, Laursen JR, Loker ES.. 2008. Evolutionary origins, diversification, and biogeography of liver flukes (Digenea, Fasciolidae). Am J Trop Med Hyg. 79(2):248–255. [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102(30):10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Caban-Hernandez K, Figueroa-Santiago O, Espino AM.. 2015. Fasciola hepatica fatty acid binding protein inhibits TLR4 activation and suppresses the inflammatory cytokines induced by lipopolysaccharide in vitro and in vivo. J Immunol. 194(8):3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas-Coma S, Valero MA, Bargues MD.. 2009. Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol. 69:41–146. [DOI] [PubMed] [Google Scholar]
- McGonigle L, Mousley A, Marks NJ, Brennan GP, Dalton JP, Spithill TW, Day TA, Maule AG.. 2008. The silencing of cysteine proteases in Fasciola hepatica newly excysted juveniles using RNA interference reduces gut penetration. Int J Parasitol. 38(2):149–155. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SN, Tort JF, Rinaldi G, Fischer K, Rosa BA, Smircich P, Fontenla S, Choi YJ, Tyagi R, Hallsworth-Pepin K, et al. 2017. Genomes of Fasciola hepatica from the Americas reveal colonization with Neorickettsia endobacteria related to the agents of Potomac horse and human sennetsu fevers. PLoS Genet. 13(1):e1006537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVeigh P, McCammick E, McCusker P, Wells D, Hodgkinson J, Paterson S, Mousley A, Marks NJ, Maule AG.. 2018. Profiling G protein-coupled receptors of Fasciola hepatica identifies orphan rhodopsins unique to phylum Platyhelminthes. Int J Parasitol Drugs Drug Resist. 8(1):87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JR, Hiendleder S, Kijas JW.. 2011. Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel. Heredity (Edinb). 106(4):700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meemon K, Sobhon P.. 2015. Juvenile-specific cathepsin proteases in Fasciola spp.: their characteristics and vaccine efficacies. Parasitol Res. 114(8):2807–2813. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334(6055):521–524. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie HA, Bouckaert RR, Drummond AJ.. 2017. StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol Biol Evol. 34(8):2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA.. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 108(33):13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsburger U, Ramos-Onsins SE, Lercher MJ.. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31(7):1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalee A, Wongsawad C, Rojanapaibul A, Chai JY.. 2015. Experimental life history and biological characteristics of Fasciola gigantica (Digenea: Fasciolidae). Korean J Parasitol. 53(1):59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey O, Danchin E, Mirouze M, Loot C, Blanchet S.. 2016. Adaptation to global change: a transposable element-epigenetics perspective. Trends Ecol Evol (Amst). 31(7):514–526. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Corvo I, Jones PM, George AM, Padula MP, To J, Cancela M, Rinaldi G, Tort JF, Roche L, et al. 2011. Collagenolytic activities of the major secreted cathepsin L peptidases involved in the virulence of the helminth pathogen, Fasciola hepatica. PLoS Negl Trop Dis. 5(4):e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S.. 2009. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics. 8(8):1891–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MW, Tort JF, Lowther J, Donnelly SM, Wong E, Xu W, Stack CM, Padula M, Herbert B, Dalton JP.. 2008. Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol Cell Proteomics. 7(6):1111–1123. [DOI] [PubMed] [Google Scholar]
- Rofatto HK, Parker-Manuel SJ, Barbosa TC, Tararam CA, Alan Wilson R, Leite LC, Farias LP.. 2012. Tissue expression patterns of Schistosoma mansoni Venom Allergen-Like proteins 6 and 7. Int J Parasitol. 42(7):613–620. [DOI] [PubMed] [Google Scholar]
- Rowe T, Rich TH, Vickers-Rich P, Springer M, Woodburne MO.. 2008. The oldest platypus and its bearing on divergence timing of the platypus and echidna clades. Proc Natl Acad Sci U S A. 105(4):1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijuntha W, Tantrawatpan C, Agatsuma T, Wang C, Intapan PM, Maleewong W, Petney TN.. 2018. Revealing genetic hybridization and DNA recombination of Fasciola hepatica and Fasciola gigantica in nuclear introns of the hybrid Fasciola flukes. Mol Biochem Parasitol. 223:31–36. [DOI] [PubMed] [Google Scholar]
- Salazar-Calderon M, Martin-Alonso JM, Castro AM, Parra F.. 2003. Cloning, heterologous expression in Escherichia coli and characterization of a protein disulfide isomerase from Fasciola hepatica. Mol Biochem Parasitol. 126(1):15–23. [DOI] [PubMed] [Google Scholar]
- Schrader L, Schmitz J.. 2018. The impact of transposable elements in adaptive evolution. Mol Ecol. 28(6):1537–1549. [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T.. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DE, Rinaldi G, Koziol U, Brehm K, Brindley PJ.. 2014. How might flukes and tapeworms maintain genome integrity without a canonical piRNA pathway? Trends Parasitol. 30(3):123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R.. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 8(4):272–285. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Ostlund G.. 2015. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43(Database issue):D234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripa J, Laha T, To J, Brindley PJ, Sripa B, Kaewkes S, Dalton JP, Robinson MW.. 2010. Secreted cysteine proteases of the carcinogenic liver fluke, Opisthorchis viverrini: regulation of cathepsin F activation by autocatalysis and trans-processing by cathepsin B. Cell Microbiol. 12(6):781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Santure AW, Dennis SR.. 2015. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol. 24(9):2241–2252. [DOI] [PubMed] [Google Scholar]
- Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, Tyagi R, Heizer E, Zhang X, Bhonagiri-Palsikar V, et al. 2014. Genome of the human hookworm Necator americanus. Nat Genet. 46(3):261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teofanova D, Kantzoura V, Walker S, Radoslavov G, Hristov P, Theodoropoulos G, Bankov I, Trudgett A.. 2011. Genetic diversity of liver flukes (Fasciola hepatica) from Eastern Europe. Infect Genet Evol. 11(1):109–115. [DOI] [PubMed] [Google Scholar]
- Torgerson PR, Devleesschauwer B, Praet N, Speybroeck N, Willingham AL, Kasuga F, Rokni MB, Zhou XN, Fevre EM, Sripa B, et al. 2015. World Health Organization estimates of the global and regional disease burden of 11 foodborne parasitic diseases, 2010: a data synthesis. PLoS Med. 12(12):e1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 43:11.10.1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SM, Johnston C, Hoey EM, Fairweather I, Borgsteede F, Gaasenbeek C, Prodohl PA, Trudgett A.. 2011. Population dynamics of the liver fluke, Fasciola hepatica: the effect of time and spatial separation on the genetic diversity of fluke populations in the Netherlands. Parasitology 138(2):215–223. [DOI] [PubMed] [Google Scholar]
- Walker SM, Prodohl PA, Hoey EM, Fairweather I, Hanna RE, Brennan G, Trudgett A.. 2012. Substantial genetic divergence between morphologically indistinguishable populations of Fasciola suggests the possibility of cryptic speciation. Int J Parasitol. 42(13–14):1193–1199. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM.. 2017. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yang L, Lv Z, Wang J, Zhang Q, Zheng H, Wu Z.. 2012. Molecular cloning and characterization of a HSP70 gene from Schistosoma japonicum. Parasitol Res. 110(5):1785–1793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.