Abstract
The MLE DExH helicase and the roX lncRNAs are essential components of the chromatin modifying Dosage Compensation Complex (DCC) in Drosophila. To explore the mechanism of ribonucleoprotein complex assembly, we developed vitRIP, an unbiased, transcriptome-wide in vitro assay that reveals RNA binding specificity. We found that MLE has intrinsic specificity for U-/A-rich sequences and tandem stem-loop structures and binds many RNAs beyond roX in vitro. The selectivity of the helicase for physiological substrates is further enhanced by the core DCC. Unwinding of roX2 by MLE induces a highly selective RNA binding surface in the unstructured C-terminus of the MSL2 subunit and triggers-specific association of MLE and roX2 with the core DCC. The exquisite selectivity of roX2 incorporation into the DCC thus originates from intimate cooperation between the helicase and the core DCC involving two distinct RNA selection principles and their mutual refinement.
INTRODUCTION
The assembly of lncRNAs with chromatin modifying complexes into ribonucleoprotein (RNP) regulators represents an increasingly important mechanism of gene regulation (1). Prominent examples where such a mechanism has been unequivocally established are PAPAS RNA, which recruits epigenetic repressors to rDNA loci (2), XIST, which tethers repressive activities to one of the female X chromosomes in mammals for inactivation (3,4), and roX RNAs, which tether activating activities to the single male X chromosome in Drosophilids (5).
Although in some cases the critical RNA determinants for complex formation have been identified, the rules according to which chromatin proteins bind lncRNAs specifically are only beginning to emerge (6). Some RNA binding modes are predicted because the proteins involved contain RNA-binding domains (RBDs) of known structure, such as the canonical RNA-recognition motif (RRM), the oligosaccharide binding (OB)-fold domain or catalytic structures such as ribonuclease or helicase domains (7). These domains bind many RNAs with a range of affinities, indicating a degenerate intrinsic specificity that may be tuned by context (6). However, RNA binding was also mapped to chromodomains and bromodomains, which are better known for their interactions with modified peptides, or to intrinsically disordered regions (IDRs) (8–10), which complicates the determination of intrinsic RNA binding propensity.
While RBDs read out nucleotide sequence in unfolded RNA, RNA secondary structure, such as stem-loops (SL) or G quartets (G4) may contribute to defining protein binding sites (6). Such structures may be substrates of RNA helicases, which utilize adenosine triphosphate (ATP) to disrupt intramolecular base pairing. The existence of numerous RNA helicases testifies to the importance of their activities for a variety of cellular processes (11,12). Many helicases of the Superfamily 2 (SF2), in particular members of the DEAD-box and DExH subfamilies, display modular RBD structures. Because they work on various substrates in diverse cellular processes, they are suggested to lack intrinsic substrate specificity and to interact primarily with the ribose-phosphate backbone (11–13). Instead, function and properties of individual DExH proteins may be regulated by auxiliary domains or protein cofactors (14).
Further insight into the mechanism of DExH helicases may come from the helicase MLE (maleless) in Drosophila melanogaster. This helicase is thought to incorporate at least one of two long, non-coding roX RNAs into the male-specific lethal dosage compensation complex (MSL-DCC or DCC), which further consists of three MSL proteins (MSL1-3) and the acetyltransferase MOF (15–17) (Supplementary Figure S1). The DCC is essential for compensating the sex chromosome monosomy in male flies (genotype XY) by enhancing the transcriptional output of X chromosomal genes through acetylation of lysine 16 on histone H4 (H4K16ac) (5).
The DCC contains a DNA-binding module (MSL1-MSL2), which recognizes X chromosomal ‘High Affinity Sites’ (HAS), and an epigenetic ‘reader/writer’ module (MSL1-MSL3-MOF) that recognizes transcribed, H3K36-methylated chromatin and acetylates it (Supplementary Figure S1). The presence of roX RNA is required for full functioning of the complex. Indeed, if either the roX RNAs or MLE are missing, partial complexes accumulate at HAS and fail to contact target genes (18–22).
The two roX RNAs are remarkably different in size and sequence but share in their 3′ region SL structures with short conserved uridine-rich sequence elements, termed roX boxes. MLE remodels these roX box-containing SLs in an ATP-dependent manner. The resulting alternative secondary structures are supposed to provide a platform for the assembly of a functional DCC (23–28). The structure of MLE’s active core in complex with a single-stranded poly-uridine model substrate revealed roles of auxiliary OB fold and helicase-associated domains in imparting unexpected substrate specificity (29).
Other studies suggest that MLE may also function in processes beyond dosage compensation (30–32). MLE has remarkable structural similarity to spliceosomal helicases (29,33), it was identified in a screen for mRNA-binding proteins in early Drosophila embryos (34) and it was hypothesized to resolve adenosine-to-inosine edited RNA structures (32). However, how MLE recognizes its RNA substrates in general and how MLE and roX connect to the DCC remains elusive.
A deeper understanding of the physiological function of MLE would be facilitated by the characterization of the intrinsic RNA binding specificities of MLE and its functional context, the subunits of the DCC. Since RNA binding in vivo is generally modulated by cooperating factors, intrinsic specificity can only be determined in vitro. Several high-throughput techniques, such as RNAcompete (35), RNA Bind-n-Seq (36,37) and in vitro iCLIP (38), have been used to determine the RNA binding profiles of individual proteins. In these studies, RNA oligonucleotide libraries or pools of in vitro-transcribed model RNAs served as substrates. Despite the high complexity of such RNA libraries, they only represent a subset of the cellular RNA pool and may not contain complex secondary structures.
Here, we present vitRIP, an unbiased transcriptome-wide in vitro assay that aims to unravel intrinsic RNA-binding specificities of isolated proteins in context of a complex pool of transcripts. RNA–protein complexes are retrieved under native, i.e. non-crosslinked, conditions by simple and quick one-step purification and bound RNAs are identified by deep sequencing. Applying the procedure to MLE we identified roX1 and roX2 and several coding and non-coding RNAs as MLE substrates in vitro. In addition, we derived the intrinsic affinity of MLE for poly-uridine and poly-adenosine sequences and for defined secondary structure elements.
Applying a similar RNA immunoprecipitation strategy to endogenous MLE demonstrates that MLE binds almost exclusively roX in vivo. This suggests that dosage compensation is the main function of the helicase. In search for the missing determinant of specificity, we explored the intrinsic RNA binding of the reconstituted DCCcore composed of the subunits MSL1, MSL2, MSL3 and MOF. In the absence of MLE, the complex did not show any specific RNA binding. However, the complex modulated the substrate selectivity of the helicase, resulting in an increased specificity of MLE for physiological targets. Furthermore, we demonstrated an almost exclusive enrichment of roX2 with the DCCcore as a consequence of an active MLE helicase. The helicase selectively transferred roX2 to the DCC subunits MSL1 and MSL2, which acquired specific binding. The data suggest a synergistic mechanism for exclusive roX2 incorporation and assembly of a functional DCC.
MATERIALS AND METHODS
Cell lines and culture conditions
Drosophila melanogaster cell lines were used for in vitro and in vivo RNA immunoprecipitation (RIP) experiments. Male S2 cells were a gift from Philipp Zamore, the S2 subclone L2-4 was a gift from Patrick Heun. Male clone eight cells were obtained from the Drosophila Genomics Resource Center. All cell lines were tested negative for mycoplasma. S2 were cultured in Schneider's Drosophila medium (Gibco) supplemented with 10% fetal calf serum and penicillin-streptomycin at 26°C. Clone eight cells were cultured in Shields and Sang M3 insect medium (Sigma-Aldrich) supplemented with 2.5% fly extract, 2% fetal calf serum, 5 μg/ml insulin (Sigma-Aldrich) and penicillin-streptomycin at 26°C. Spodoptera frugiperda 21 (SF21) cells (Gibco) were used for amplification of recombinant baculoviruses and baculovirus-driven expression of recombinant proteins. SF21 cells were cultured at 26°C in Sf-900 II medium (Gibco) supplemented with 10% fetal calf serum and gentamycin.
Drosophila strain
Drosophila melanogaster of the w1118 Oregon R genotype was used to extract total RNA for in vitro RNA immunoprecipitation experiments. Flies were reared under standard conditions.
Antibodies
Rat monoclonal anti-MLE 6E11, rabbit anti-MSL1, rabbit anti-MSL2 and rat monoclonal anti-MSL3 1C9 antibodies were previously described in (39–42). Mouse anti-FLAG M2 affinity gel and mouse monoclonal anti-FLAG M2 antibody were from Sigma-Aldrich. Mouse monoclonal anti-Lamin antibody (T40) was provided by Prof. H. Saumweber. Mouse monoclonal anti-GFP antibody was from Roche.
Cloning, protein expression and purification
Full-length D. melanogaster MLE in pFastbac-1 was described in (25). Mutations K413E (MLEGET) and H1032E-K1033E (MLEHKE), respectively, in pFastbac-MLE-FLAG were introduced by site-directed mutagenesis and were described in (29,39). Full-length human DHX9 cDNA was a gift from Frank Grosse (FSU Jena) and was cloned with a C-terminal FLAG tag in pFastbac-1 using restriction enzymes BamHI and NotI. For hsp70 promoter-driven expression of C-terminally GFP-tagged DHX9 in Drosophila S2 cells, full-length DHX9 cDNA was cloned into plasmid pHsp70-eGFP (41). Vectors for baculovirus-driven expression of full-length MSL1-FLAG and MSL2-FLAG wild-type and MSL2 mutants (ΔRING (MSL2Δ1-131), ΔCXC (MSL2Δ523-565), ΔCTD (MSL2Δ650-778)) were described in (43). MSL1 mutants (ΔN (MSL1Δ1-191), central (MSL1192-864), ΔC (MSL1Δ865-1039)) were cloned with a C-terminal FLAG tag in pFastbac-1 using restriction enzymes StuI and XbaI.
Drosophila melanogaster Dosage Compensation Complex (DCC) sub-complexes (Supplementary Figure S1) were cloned and expressed using the Multibac technology (44). Full-length cDNAs encoding DCC subunits were cloned into pFBDM expression cassettes and combined using the multiplication module to yield a single expression vector for each sub-complex. For the DCCcore complex, MSL2 with a C-terminal FLAG tag, MSL1, MOF and MSL3 with an N-terminal hexahistidine tag were combined. The chromatin-binding module is composed of MSL1 with a C-terminal FLAG tag, MOF, and MSL3 with an N-terminal hexa-histidine tag. The DNA-binding module consists of MSL2 with a C-terminal FLAG tag and MSL1 lacking the C-terminal PEHE domain (MSL1Δ865-1039). Bacmids were recombined in Escherichia coli Multibac cells (Geneva Biotech). Baculoviruses were amplified in SF21 insect cells in two successive rounds. For protein expression, SF21 insect cells were infected with 1/1000 (v/v) baculovirus, cultured for 72 h at 26°C and collected by centrifugation.
MLE-FLAG (wild-type and mutant) and DHX9-FLAG, respectively, were purified by FLAG-affinity chromatography as described in (25). Single MSL1-FLAG and MSL2-FLAG wild-type and mutant proteins were purified by FLAG-affinity chromatography as described in (43).
DCC sub-complexes were purified from isolated SF21 nuclei according to (45) with modifications. For nuclei extraction, pellets of 2.5 × 108 baculovirus-infected SF21 cells were resuspended in 10 ml cold buffer A (10 mM HEPES-KOH pH 7.9, 10 mM KCl, 1 mM dithiothreitol (DTT), 0.1 mM ethylenediaminetetraacetic acid (EDTA), 0.1 mM ethylene glycol-bis N,N,N',N'-tetraacetic acid (EGTA), 0.5 mM phenylmethylsulfonyl flouride (PMSF), 1× Complete EDTA-free protease inhibitor (Roche)) and incubated on ice for 15 min. Following addition of 0.625 ml 10% (v/v) NP40 and brief vortexing, nuclei were spun down for 30 s at 17 000 × g. The supernatant was discarded and the nuclei were washed with another 10 ml buffer A and pelleted by centrifugation for 30 s at 17 000 × g. For protein solubilization, the nuclear pellet was resuspended in 3 ml cold buffer B (20 mM HEPES-KOH pH 7.9, 400 mM KCl, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, 0.5 mM PMSF, 1× Complete EDTA-free protease inhibitor (Roche)) and incubated at 4°C on a rotating wheel. The soluble nuclear fraction containing recombinant complexes was obtained after pelleting the nuclear debris at 4°C for 10 min at 17 000 × g. The nuclear extract was diluted 2-fold with buffer BC-0 (20 mM HEPES-KOH pH 7.9, 1 mM DTT, 10% glycerol, 0.2 mM PMSF) to reach a final salt concentration of less than 200 mM KCl. Precipitates were removed by centrifugation at 4°C for 10 min at 5000 × g. For FLAG affinity chromatography, 750 μl anti-FLAG M2 agarose slurry (Sigma-Aldrich) was washed three times with BC-100 buffer (BC-0 with 100 mM KCl). Diluted nuclear extract was treated with 25 μg RNase A and incubated with equilibrated FLAG beads for 1 h at 4°C on a rotating wheel. Beads were washed extensively with 10 ml of buffer: twice with BC-100, once with BC-200 (BC-0 with 200 mM KCl) and once with BC-100. Protein complexes were eluted in three successive rounds with one bead volume elution buffer each (BC-100 supplemented with 0.5 mg/ml FLAG peptide (Sigma-Aldrich) and 1× Complete EDTA-free protease inhibitor (Roche)) for 20 min at 4°C on a rotating wheel. Elution fractions were pooled, not further concentrated, flash-frozen in liquid nitrogen and stored at −80°C.
Quality of all purified proteins was assessed by sodium dodecyl sulphate-polyacrylamide gelelectrophoresis (SDS-PAGE) and Coomassie staining and concentration was determined in Image Lab 6.0 (Bio-Rad) using bovine serum albumin (BSA) standards (Thermo Fisher) as reference.
Extraction of total RNA
Total RNA was extracted from 5 × 106 cells (S2, cl.8) using the RNeasy Mini kit (Qiagen) according to the manufacturers instructions. Total RNA from was isolated from 30–40 heads of sex-sorted flies using QIAzol Lysis Reagent (Qiagen) followed by ethanol precipitation. Extracted RNA was photometrically quantified using a DeNovix Spectrophotometer.
In vitro RNA immunoprecipitation (vitRIP)
The described native (i.e. without crosslinking) vitRIP protocol is common to the different experimental setups detailed in Supplementary Table S1. vitRIP was performed with highly purified recombinant protein and 2 μg of total RNA, extracted from Drosophila cell lines or Drosophila heads. The concentration of total RNA was kept constant throughout all experiments regardless of the RNA source.
Two microgram of total RNA was diluted with vitRIP-100 buffer (25 mM HEPES-NaOH pH 7.6, 100 mM NaCl, 0.05% NP40, 3 mM MgCl2) to a final volume of 10 μl. Ten percent of this RNA mix (1 μl) served as input sample and was kept on ice. The remaining 90% (9 μl) of the RNA mix were incubated with purified protein (protein concentrations are given below) in a total volume of 100 μl in vitRIP-100 buffer supplemented with 10 μg BSA (New England Biolabs), 0.1 U/μl RNase-free recombinant DNase I (Roche) and 0.8 U/μl RNasin recombinant RNase inhibitor (Promega). For vitRIP experiments with DCC sub-complexes 10 μM ZnCl2 was included in the buffer. If applicable, 1 mM fresh ATP was added to the reaction. All in vitro mixtures were incubated for 30 min at 25°C.
Antibodies were pre-bound to sepharose beads for 3 h at 4°C in vitRIP-100 buffer. Subsequently, antibody-conjugated sepharose beads and anti-FLAG M2 affinity gel (Sigma-Aldrich) were blocked for 1 h with 2% (w/v) BSA and 0.1 mg/ml yeast tRNA (Sigma-Aldrich) and washed once with vitRIP-100 buffer. The following antibodies were used: vitRIP of FLAG-tagged MLE alone was performed using 15 μl anti-FLAG M2 affinity gel; vitRIP of FLAG-tagged MLE in absence or presence of recombinant DCCcore was performed using 50 μl of monoclonal rat anti-MLE 6E11 hybridoma supernatant and 15 μl protein G sepharose; vitRIP of DCC sub-complexes in absence or presence of MLE was performed using 3 μl of rabbit anti-MSL1 antibody and 15 μl protein A+G sepharose mix.
Protein–RNA complexes were retrieved in 45 min at room temperature (22°C) on a rotating wheel. Beads were washed three times with vitRIP-100 buffer, vitRIP-250 buffer (vitRIP-100 buffer with 250 mM NaCl) and vitRIP-100 buffer, respectively. Of 80% of the bead material, RNA was extracted using Proteinase K (100 μg in vitRIP-100 buffer with 0.5% SDS; 55°C for 45 min), phenol–chloroform extraction and ethanol precipitation in presence of 20 μg glycogen (Roche). The RNA pellet was resuspended in 20 μl RNase-free water. Input material (10%) was treated equally. RNA input and vitRIP samples were quantified using the Qubit RNA High Sensitivity Assay Kit (Invitrogen). RNA-seq libraries were prepared with 30–40 ng of RNA material. If applicable, 25% of RNA input and IP material was analyzed by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) using the SuperScript III First-Strand Synthesis System (Thermo Fisher) and SYBR green dye (Applied Biosystems) with primers specific for roX2 and 7SK (roX2-SL7.fw 5′-GACGTGTAAAATGTTGCAAATTAAG; roX2-SL7.rv 5′-TGACTGGTTAAGGCGCGTA; 7SK.fw 5′-GATAACCCGTCGTCATCCAG; 7SK.rv 5′-AGTAATTCTGCCTGGCGTTG). The fraction of bound RNA relative to input was calculated. Highly expressed 7SK RNA served as an unbound control in each experiment. Statistical analysis of qPCR results was performed by linear regression models on the log2-transformed values and is given in Supplementary Table S3. Western blot analysis of 20% bead material was performed with antibodies against the FLAG tag, MLE, MSL1 and MSL3.
The following protein concentrations were used in vitRIP experiments: MLE-FLAG titration experiments with S2 total RNA: [High] = 50 nM MLE, [Med] = 20 nM MLE and [Low] = 5 nM MLE; anti-FLAG vitRIP with S2, cl.8 or male/female Drosophila head RNA: [High] = 50 nM wild-type and mutant MLE; anti-FLAG-vitRIP with DHX9 and S2 RNA: [High] = 50 nM DHX9; anti-MLE vitRIP with S2 RNA: [Low] = 5 nM MLE in absence or presence of 20 nM dimeric DCCcore; anti-MSL1 vitRIP with S2 RNA: 40 nM dimeric DCCcore in absence or presence of 10 nM wild-type and mutant MLE; anti-MSL1 vitRIP with cl.8 RNA: 40 nM dimeric DCCcore in absence or presence of 10 nM MLE; anti-MSL1 vitRIP with S2 RNA: around 40 nM DNA-binding module or reader-writer module (balancing the levels of MSL1 between the different complexes) in absence or presence of 10 nM MLE.
In vivo RNA immunoprecipitation (RIP)
RIP of endogenous MLE (eMLE) was essentially performed as described in (46) with modifications. For each replicate of specific (anti-MLE) and mock (bead control) RIP, 1 × 108 exponentially grown S2 cells were collected, washed once with phosphate-buffered saline (PBS) and flash frozen in liquid nitrogen. Cell pellets were thawed on ice and resuspended in 1 ml of cold lysis buffer (20 mM HEPES-NaOH pH 7.6, 125 mM NaCl, 0.05% SDS, 0.25% sodium deoxycholate, 0.5% NP40, 1.5 mM MgCl2, 0.25 mM DTT) supplemented with 0.05 U/μl RNase-free recombinant DNase I (Roche), 0.4 U/μl RNasin recombinant RNase inhibitor (Promega) and 1× Complete EDTA-free protease inhibitor (Roche). The lysate was incubated for 15 min on ice with 5 s vortexing every 5 min. Following centrifugation for 30 min at 14 000 rpm at 4°C, the cleared lysate was transferred to a fresh low-binding tube. 2.5% of the lysate was kept as input material for RNA extraction. The lysate was mixed with 40 μl protein G beads (mock treated or coupled with 100 μl monoclonal rat anti-MLE 6E11 hybridoma supernatant) and incubated at 4°C for 2 h on a rotating wheel. Beads were washed three times with RIP-100, RIP-250 and RIP-100 buffer (25 mM HEPES-NaOH pH 7.6, 0.05% NP40, 3 mM MgCl2 with 100 mM NaCl and 250 mM NaCl, respectively). Of 80% of the bead material, RNA was extracted using Proteinase K (100 μg in RIP-100 buffer with 0.5% SDS; 55°C for 45 min), phenol–chloroform extraction and ethanol precipitation in presence of 20 μg glycogen (Roche). Input material (2.5%) was treated equally. MLE RIP efficiency was controlled using anti-MLE western blot analysis of 20% bead material. RNA samples (Input and mock and MLE RIP, respectively) were quantified using the Qubit RNA High Sensitivity Assay Kit (Invitrogen). Of each sample, 40 ng RNA was used to prepare rRNA-depleted RNA-seq libraries.
Library preparation and sequencing
The amount of starting material for depletion of ribosomal RNA and following RNA-seq library preparation varied between different sets of experiments and is given in the respective method section. Ribosomal RNA was depleted using the NEBNext rRNA Depletion Kit (Human/Mouse/Rat; New England Biolabs) and samples were analyzed on a Bioanalyzer using the RNA 6000 Pico Kit (Agilent). Of the rRNA-depleted samples directional libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the recommended protocol. The quality of the libraries was assessed on a Bioanalyzer using the DNA 1000 or DNA High Sensitivity Kit (Agilent). Libraries were sequenced on an Illumina HiSeq1500 instrument in paired-end mode.
Data analysis
A total of 50 bp paired-end reads were aligned to the D. melanogaster reference genome (release 6) using STAR aligner (version 2.5.3a) with providing GTF annotation (dmel-all-r6.17.gtf). Reads with multiple alignments were filtered by setting outFilterMultimapNmax parameter to 1. Reads were counted per gene with parameter –quantMode GeneCounts. BAM files were converted to normalized bedgraph coverages using genomeCoverageBed command (bedtools version 2.27.1) with -scale parameter set to divide by the total number of reads and multiplied by a million. Bedgraph files were converted to tdf files (igvtools version 2.3.98) to visualize in the IGV browser.
Count tables (read counts per gene) were read into R and low count genes were filtered out (at least 1 read per gene in 75% of the samples analyzed together). Differential expression analysis was performed by DESeq2 package (version 1.24) by adding replicate information as batch variable. Samples that were directly compared to each other were fitted in the same DESeq2 model (groups and batches are given in Supplementary Table S1). Log2FoldChange estimates and adjusted P-values were obtained by the results function (DESeq2) and adjusted P-value cutoff was set 0.01 (Supplementary Table S1). For principal component analysis (PCA) analysis batch effect was corrected by the ComBat function (sva package version 3.32) on the normalized read counts.
Nucleotide frequencies for each gene were calculated by oligonucleotideFrequency function (Biostrings package version 2.52) by setting width to four, five or six, step to 1 and as.prob to TRUE. Nucleotide frequencies for genes significantly enriched (adjusted P-value < 0.01 and log2FC > 0) were visualized by boxplots, where kmer sequences were sorted by their median frequency. The association between RNA-seq log2FC (e.g. IP—Input) and nucleotide frequencies were determined by Spearman's correlation.
Annotations for exons, introns, 5′ and 3′ UTRs as well as for snRNAs, snoRNAs and tRNAs were extracted from the GTF annotation (dmel-all-r6.17.gtf). Transcripts with long 3′ UTRs were selected as the top 5% transcripts (n = 654) with a 3′ UTR longer than 2027 bp. In cases, when a gene was annotated with 3′ UTR isoforms, the isoform with the longest 3′ UTR was taken. Annotation of snoRNA classes (C/D or H/ACA box) were taken from the SnOPY database (snoRNA orthological gene database) (47).
The list of edited RNAs were obtained from (48) (nsmb.2675-S2.xlsx).
The enrichment of selected classes of RNAs was analyzed by Fisher exact test (Supplementary Table S2).
Plots were generated using R graphics.
Data and code availability
Sequencing data were deposited to GEO with accession number GSE143455. Analysis code is available upon request.
Genomic DNA preparation
Pellet from 6 × 107 S2 cells was suspended in 1.2 ml of lysis buffer (10 mM Tris pH 8, 100 mM NaCl, 25 mM EDTA pH 8, 0.5% SDS, 0.15 mg/ml of proteinase K) and incubated at 56°C overnight. After addition of sodium acetate to a final concentration of 0.3 M, the nucleic acids were extracted with phenol–chloroform and precipitated with an equal volume of isopropanol at −20°C for 1 h. Precipitated nucleic acids were centrifuged and washed with 70% ethanol. Dried pellets were resuspended in TE buffer and sonicated with Covaris AFA S220 (microTUBEs, Peak Incident Power 175 W, Duty Factor 10%, Cycles per Burst 200, 430 s) to generate 200 bp fragments. After RNAse digestion (0.1 mg/ml, 1 h at 37°C), DNA was purified with the GenElute kit (Sigma-Aldrich).
DNA immunoprecipitation (DIP)
DNA immunoprecipitation (DIP) experiments were performed as in (22) with few modifications. Briefly, 400 ng of genomic DNA (gDNA) was incubated with 2800 ng DCCcore ( = 40 nM dimeric DCCcore) at 26°C for 30 min in 100 μl of binding buffer (100 mM KCl, 2 mM MgCl2, 2 mM Tris–HCl pH 7.5, 10% Glycerol, 10 μM ZnCl2). Ten percent of the reaction was taken as input material and subjected to quantitative PCR. DNA–protein complexes were immunoprecipitated using 30 μl of beads pre-incubated with an anti-MSL2 antibody (30 μl of A/G sepharose beads, 2 μl of polyclonal guinea pig anti-MSL2 antibody, 400 μl of 2% BSA for 3 h at 4°C) for 20 min at room temperature and washed twice with 100 μl of binding buffer to eliminate unbound DNA. After proteinase K digestion (0.5 mg/ml, 1 h at 56°C), DNA was purified with AMPure beads (Beckman Coulter). Input and IP samples were subjected to quantitative PCR with primers specific for hiw and CG8097 PionX sites and for the spt4 control locus (hiw.fw 5′-TCATCAGATTGGCACTGCAC; hiw.rv 5′-AACCGTGTTCTTCCATCTCG; CG8097.fw 5′-CGACAAGCTCTCGGAG; CG8097.rv 5′-CCATCAGCTCGTGCTG; spt4.fw 5′-GCTCCGATTCATAAGCCCAG; spt4.rv 5′-GCCTCTTTCGGAGCAGCTTT). Enrichment of PionX sites and control region is displayed as percentage of input.
RNA interference and immunostaining
Drosophila male S2 cells (subclone L2-4) stably expressing DHX9-GFP were established by co-transfection of 500 ng pHsp70-DHX9-GFP plasmid and 25 ng of a plasmid encoding a blasticidin resistance gene using the Effectene transfection reagent (Qiagen). Stable clones were selected for 2 weeks in complete medium containing 25 ng/ml blasticidin, followed by recovery for 1 week in complete medium without blasticidin.
RNA interference of target genes was essentially performed as described (49). Double-stranded RNA fragments (dsRNA) were generated using the MEGAscript T7 transcription kit (Thermo Fisher) from DNA templates generated by PCR with the following oligonucleotides: mle RNAi: 5′-TTAATACGACTCACTATAGGGAGAATGGATATAAAATCTTTTTTGTACCAATTTTG, 5′-TTAATACGACTCACTATAGGGAGAACAGGGCGCATGACTTGCT; gst RNAi: 5′-TTAATACGACTCACTATAGGGAGAAT, 5′-GTCCCCTATACTAGGTTA (amplified from pGEX-6P-1 (GE Healthcare)). Ten micrograms of dsRNA were added to 1.5 × 106 DHX9-GFP expressing S2 cells and non-transfected S2 cells, respectively, seeded in 6-well plates. Cells were incubated with dsRNA for 7 days at 26°C and then collected for analysis. Western blot analysis of 1 × 106 cells was performed with primary antibodies against MLE, GFP and Lamin. Immunostaining with mouse anti-GFP, rat anti-MLE and rabbit anti-MSL2 primary antibodies was performed according to (50). RNAi-treated cells were settled and fixed with PBS/3.7% paraformaldehyde for 10 min at room temperature, permeabilized with PBS/0.25% Triton X-100 for 6 min on ice and blocked with image iT FX signal enhancer (Invitrogen). Cells on coverslips were incubated over night at 4°C with primary antibodies. Following two washes with PBS/0.1% Triton X-100, fluorophore-coupled secondary antibodies were added for 1 h at room temperature. DNA was counterstained with DAPI. After PBS/0.1% Triton X-100 and PBS washes, cells were mounted in VECTASHIELD (Vector Laboratories).
Fluorescence microscopy
Fluorescence images were recorded with a Leica Thunder Imager 3D based on a DMi8 stand, equipped with a Leica DFC900 GTC sCMOS camera and a LUMENCOR Spectra X as fluorescence excitation source with individually switchable LEDs for specific excitation. DAPI, GFP/Alexa Fluor 488, Cy3 and Alexa647 signals were recorded with a Quad-Band filter cube and an additional emission filter wheel in the emission beam path to avoid channel crosstalk. Image stacks of 13 planes with a step size of 0.42 μm were recorded with a 1.3 NA 63× Glycerol immersion objective at a pixel size of 103 nm. All stacks were deconvolved with Huygens Professional in batch deconvolution mode with standard settings and a SNR of 25.
Image processing was done in Fiji (51). Images shown are maximum intensity projections of deconvolved stacks. Images of individual cells shown in Supplementary Figure S8G and I were additionally resized by a factor of four without interpolation followed by gaussian filtering with a radius of two pixels.
RNA oligos and secondary structure prediction
The modules of roX2 stem loop (SL) seven mimicking alternative secondary structures were synthesized with a 3′ biotin moiety at Biomers. The sequences were as follows: SL7-5′: 5′-AAGACGUGUAAAAUGUUGCAAA; SL7-Loop: 5′-AUUAAGCAAAUAUAUAUGCAUAUAU; SL7-3′: 5′-UGGGUAACGUUUUACGCGCCUU. Secondary structure prediction and drawing was performed using RNA structure and Structure Editor (52).
Sequence alignment and prediction of a common secondary structure of five selected snoRNA was performed using RNAalifold of the LocARNA package (53). Sequences of the snoRNAs Psi28S-3186, Psi28S-2622, Psi28S-3327b, Psi18S-525b, Psi18S-920 were retrieved from the snoRNA Orthological Gene Database snOPY (47).
In vitro UV crosslinking experiments
Fifty picomoles of 3′ biotinylated roX2 fragments were mixed with 5 pmol of recombinant single proteins (MLE, MSL1, MSL2 (wild-type or mutant)) and 2.8 pmol of recombinant DCCcore, respectively, in absence or presence of 10–50 pmol PionX CG8097 dsDNA. Mixtures were incubated in a total volume of 20 μl in XL buffer (20 mM HEPES-NaOH pH 7.6, 100 mM NaCl, 3 mM MgCl2) for 30 min at 25°C. Samples were transferred to a 384-well plate and crosslinked for 5 min on ice at 254 nm wavelength using a UV-Stratalinker 1800 (Stratagene). Crosslinked samples were resolved by 8% SDS-PAGE and transferred to a nitrocellulose membrane. Protein-crosslinked biotinylated roX2 fragments were detected using Streptavidin-HRP (Pierce; 1:20000 in TBS-T). FLAG-tagged proteins MLE, MSL1 and MSL2, respectively, were detected by anti-FLAG western blot. The DCCcore was visualized in SDS-PAGE using the stain-free technology (Biorad). Crosslinking of roX2 to MSL1 variants was visualized using IRDye 800 Streptavidin on a LI-COR Odyssey imager (green channel), FLAG-tagged MSL1 variants were detected by anti-FLAG western blot (red channel).
RESULTS
vitRIP provides a comprehensive in vitro transcriptome binding profile of the DExH helicase MLE
Drosophila MLE is best characterized for its ability to bind and unwind roX RNA in the context of dosage compensation in male flies. This highly specific function appears surprising since MLE as a DExH-type helicase might well act on other RNA substrates than roX (32,34) and is also present in female flies, which lack roX RNA (15). Therefore, we set out to comprehensively determine the intrinsic RNA binding properties of MLE itself and of the remaining DCC subunits. Toward this end we developed vitRIP (Figure 1A), a native in vitro RNA immunoprecipitation approach. In vitRIP, recombinant, highly purified proteins are incubated with total RNA extracted from Drosophila cells or tissues in a native, i.e. non-crosslinked, form to avoid UV crosslinking bias to pyrimidine bases (54). RNA–protein complexes are retrieved on beads using a protein-fused affinity tag or protein-specific antibodies and RNA is identified by deep sequencing. On average, we obtained about 30 million sequencing reads per RNA input and immunoprecipitate (IP) sample and detected comparable numbers of genes per million sequencing reads in input samples from multiple RNA sources (Supplementary Figure S2A).
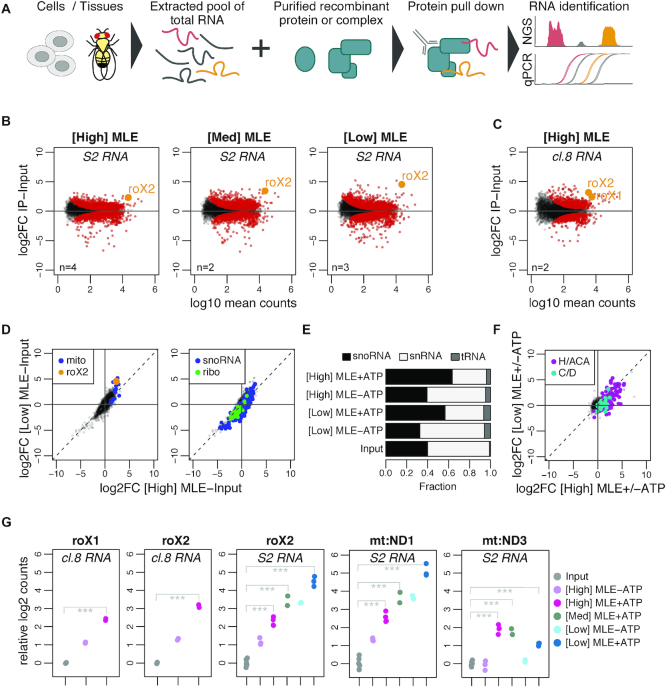
Figure 1.
vitRIP provides a comprehensive in vitro transcriptome binding profile of the DExH helicase MLE. (A) Schematic representation of the vitRIP strategy. (B) Log2 fold change (log2FC) of S2 RNA in MLE vitRIP relative to input in relation to log10 mean RNA-seq read counts. vitRIP was performed with a constant concentration of total S2 RNA and an MLE protein concentration series of [High] = 50 nM, [Med] = 20 nM and [Low] = 5 nM. Reactions contained ATP. Red: significant RNAs (P < 0.01); orange: selected significant RNA; n: number of biological replicates. Differential analysis of the individual comparisons is given in Supplementary Table S1. (C) vitRIP analysis as in (B), but with [High] = 50 nM MLE and total cl.8 RNA. (D) Scatter plots comparing the log2FC of S2 RNA in vitRIP with [High] MLE and [Low] MLE relative to input. Reactions contained ATP. roX2 and mitochondrial RNAs (mito) are highlighted on the left plot, small nucleolar RNAs (snoRNA) and ribosomal protein-coding RNAs (ribo) on the right plot. Enrichment analysis of RNA classes is given in Supplementary Table S2. (E) Bar plot displaying the fraction of snoRNA, snRNA and tRNA RNA-seq read counts in input and MLE vitRIP with S2 RNA in absence or presence of ATP. The sum of snoRNA, snRNA and tRNA read counts constitute on average 1% of all mapped RNA-seq reads. (F) Scatter plot comparing vitRIP of [High] MLE and [Low] MLE with S2-RNA in presence of ATP relative to absence of ATP. Box H/ACA and box C/D snoRNAs are highlighted. Enrichment analysis of RNA classes is given in Supplementary Table S2. (G) Dot plot representing enrichment of roX1, roX2, mt:ND1 and mt:ND3 in MLE vitRIP with RNA derived from cl.8 or S2 cells, in absence or presence of ATP. Log2 counts relative to input are displayed; ***P < 0.001. Differential analysis of individual comparisons is given in Supplementary Table S1.
To determine the in vitro binding landscape of MLE, we performed vitRIP with decreasing concentration of FLAG-tagged MLE and total RNA derived from Drosophila S2 cells, a male cell line that serves widely as a model to study dosage compensation (Figure 1B; Supplementary Figure S2B and Table S1). S2 cells express predominantly roX2 and only very low amounts of roX1. The MLE concentrations for vitRIP were determined empirically and ranged from 50 nM (henceforth labeled [High]) to 5 nM [Low], the lowest concentration for reliable immunoprecipitation (Supplementary Figure S2C). To distinguish sole binding and unwinding functions, MLE vitRIP was performed in absence and presence of ATP. PCA confirmed the reproducibility of the individual replicates in the vitRIP titration series (Supplementary Figure S2B). In presence of ATP, MLE significantly enriched roX2 from the S2 transcriptome in vitro, a proof-of-concept suggesting that vitRIP can recapitulate known binding events (Figure 1B and G). Strikingly, we detected a number of additional coding and non-coding RNAs that were significantly enriched by MLE in presence of ATP across the tested protein concentration range (Supplementary Table S1), suggesting a broad spectrum of potential substrates in vitro. To test the reproducibility and robustness of the approach, we expanded MLE vitRIP to RNA pools isolated from another male Drosophila cell line (clone 8; cl.8) or from male and female head tissue, respectively. Clone eight cells and adult male flies express both roX RNAs at high levels, allowing the analysis of roX1 binding by MLE and the DCC. We detected a number of RNAs significantly enriched by MLE from the transcriptomes of cl.8 cells or fly heads in vitro, including roX1 and roX2 (Figure 1C and G; Supplementary Figure S2D and Table S1).
Notably, female fly heads express low levels of both roX species, which are retrieved by MLE in vitRIP, illustrating the sensitivity of the method (Supplementary Figure S2D). According to modENCODE expression data, MLE is highly expressed in the nervous system and brain of adult flies, arguing for a physiological role in these tissues. Indeed, the mlenapts allele was previously linked to a splicing catastrophe of the para transcript and associated neurophysiological defects (32). Para is one of over thousand transcripts in Drosophila, many of them neuronal, which undergo ADAR-dependent adenosine-to-inosine editing (48,55). It was hypothesized that MLE resolves dsRNA structures around edited sites in para to enable correct splicing (32). Intriguingly, we identified not only para as an MLE target in vitRIP with RNA from fly heads, but also other transcripts, which have been described to be edited in vivo (48) (Supplementary Figure S2E and Table S1). The data suggest that the MLE helicase may act during the process of editing independent of its main function in dosage compensation.
Analysis of the S2 transcriptome binding profiles revealed clusters of transcripts that were enriched by MLE in a dose-dependent and/or ATP-dependent manner (Supplementary Figures S2F and G). Among the top 100 targets, we identified a near-complete pool of RNAs encoded in the mitochondrial genome, which are bound with qualitative and quantitative differences depending on the MLE concentration or availability of ATP (Figure 1D; Supplementary Figure S2F and Table S2). By contrast, mRNAs encoding ribosomal proteins are generally not bound by MLE (Figure 1D and Supplementary Table S2). Furthermore, we found a striking ATP-dependent enrichment of small nucleolar (sno) RNAs, but not of small nuclear RNAs or transfer RNAs, across all Drosophila transcriptomes analyzed in MLE vitRIP (Figure 1D-E and Supplementary Figure S2H). In particular, H/ACA-box snoRNAs were enriched, unlike C/D box snoRNAs (Figure 1F and Supplementary Table S2). The clear ATP-dependence of binding suggests that MLE recognizes snoRNAs as substrates for unwinding.
The titration series further allowed concluding on the relative binding affinity of MLE to its targets. Targets that enrich best at [Low] MLE concentration classify as high-affinity targets, as exemplified by roX2 and the mitochondrial ND1. On the other hand, low-affinity targets, such as mitochondrial ND3, are characterized by enrichment upon increase of MLE concentration (Figure 1G).
In summary, the vitRIP data provide an overview about potential substrates for MLE beyond its main target roX, including edited neuronal mRNAs, mitochondrial RNAs and H/ACA-box snoRNAs.
vitRIP uncovers intrinsic RNA binding specificity of MLE
To identify the intrinsic RNA binding specificity of MLE, we dissected the sequence composition of its targets. In an unbiased approach, we calculated tetra-, penta- and hexa-nucleotide frequencies of S2-derived transcripts, which were significantly enriched with [High] and [Low] MLE in vitRIP relative to input (adjusted P-value < 0.01 and log2FC > 0). We observed that MLE-enriched transcripts exhibit high frequencies of poly-uridine and poly-adenosine motifs (Figure 2A and Supplementary Figure S3). This is in good agreement with our previous structural work, in which we had identified base-specific contacts of four uridines in the ssRNA-binding channel of MLE (29). Since the unbiased kmer analysis revealed homo-polymeric stretches in enriched transcripts as the most frequent motifs, we compared the frequency of tetra-nucleotide motifs in all RNAs with their relative enrichment by MLE (i.e. log2FC IP-Input). We observed that the enrichment correlated the best with tetra-uridine (U4) motifs, both at [High] and [Low] MLE concentration and across different pools of Drosophila total RNA (Figure 2B and Supplementary Figure S4). Consistently, most mitochondrial RNAs exhibit an increased frequency of U4 stretches, in direct correlation with the high selectivity of MLE for these RNAs in vitro. Also tetra-adenosine (A4) motifs were enriched in MLE targets to a similar extent and in agreement with the unbiased kmer analysis, which is readily explained by the U-A base pairing in RNA secondary structures. By contrast, C4 and G4 motifs were not enriched to substantial degrees in MLE targets, confirming a clear selectivity of MLE for U-/A-rich motifs (Figure 2A-B and Supplementary Figure S4). In the absence of ATP the U4/A4 specificity was less pronounced (compare Figure 2B; Supplementary Figures S4A and S5A-B), which may reflect the requirement for unwinding of secondary structure to accommodate the single-stranded U4 sequence in the RNA binding channel, as we had observed in the MLE:U10RNA:ADPAlF4 structure (29).
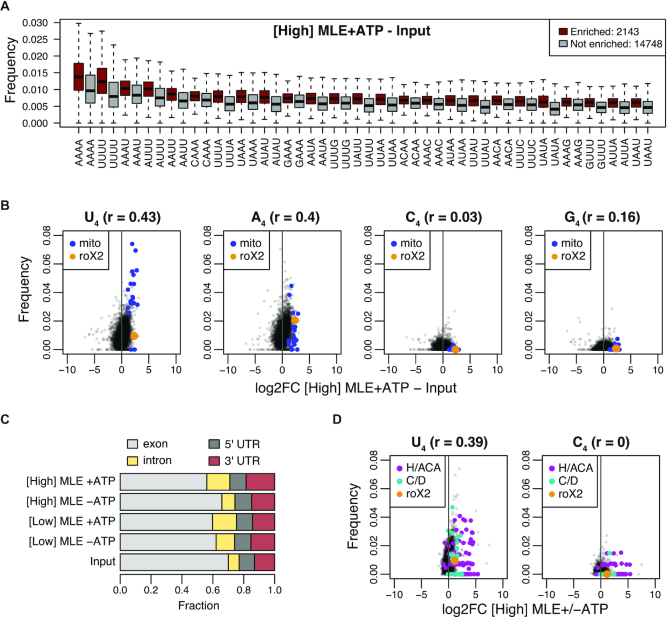
Figure 2.
vitRIP uncovers intrinsic RNA binding specificity of MLE. (A) Boxplot of tetra-nucleotide frequencies for transcripts significantly enriched by [High] MLE in presence of ATP relative to input (red) and for non-significant transcripts (gray). Frequencies were ordered by the median in the enriched class. Top 25 are shown. (B) Comparison of tetranucleotide frequency U, A, C or G and log2 fold change (log2FC) of S2 RNA in vitRIP with [High] MLE relative to input. Reactions contained ATP. roX2 and mitochondrial (mito) RNAs are highlighted. Spearman's correlation (r) was calculated for all datapoints. (C) Bar plot displaying the fraction of RNA-seq read counts mapping to the indicated pre-mRNA features in input and MLE vitRIP with S2 RNA in absence or presence of ATP. The sum of pre-mRNA read counts including exonic, intronic and UTR elements constitute on average 99% of all mapped RNA-seq reads. (D) Comparison of U4 and C4 frequency and log2FC of S2 RNA in vitRIP with [High] MLE depending on the presence of ATP. roX2 and snoRNA classes box H/ACA and box C/D are highlighted. Spearman's correlation (r) was calculated for all datapoints.
About 99% of all uniquely mapped sequencing reads in MLE vitRIP originate from pre-mRNAs. We observed a subtle binding preference of MLE to introns and 3′ untranslated regions (3′ UTR), independent of the MLE concentration and variably affected by ATP (Figure 2C). MLE enriched in particular transcripts containing long 3′ UTRs (>2027 bases, constituting the top 5%, Supplementary Table S2), which were reported to occur in neuronal tissue during Drosophila development (56). U4 motifs were enriched in MLE-bound long 3′ UTRs, in line with our earlier finding (Supplementary Figure S5C and D).
RoX2 exhibits only three U4 motifs, one of them within the essential roX box (GUUUUACG) in SL7 (Supplementary Figure S9B), and several imperfect U4 motifs, often accompanied by adenosines. The highly selective retrieval of roX2 by MLE suggests that additional features contribute to roX2 recognition. Indeed, structural integrity of SL7 in roX2 is essential for MLE in vitro binding and male fly viability (24–25,27).
Thus, we wondered if the tentative rules for MLE-roX recognition may be applied to other in vitro targets. Since MLE-bound H/ACA-box snoRNAs did not feature a particular N4 motif prevalence, which would explain robust MLE interaction (Figure 2D and Supplementary Figure S6A-B), we used RNAalifold of the LocARNA package (53) to predict the common secondary structure of five snoRNAs, which are among the most highly enriched snoRNAs (Supplementary Figure S6C). The analysis revealed a common fold into two adjacent SL structures, which confirms the canonical structure of H/ACA-box snoRNAs (57) and is reminiscent of the conserved tandem SL structures in the 3′ region of roX RNAs (24,25).
The DCCcore modulates the intrinsic RNA binding specificity of MLE
Our study so far highlighted the intrinsic binding selectivity of MLE for U-/A-rich sequences and tandem stem-loop structures in vitro, which in combination is perfectly represented in roX. However, according to these loose rules, many other cellular RNAs qualify as potential MLE targets. Previously, CLIP-based methods had revealed roX1 and roX2 as the main substrates for MLE in cells (23,24). One of these studies (23) reported binding of an ATPase-deficient mutant (MLEGET) to introns and 3′ UTRs in S2 pre-mRNAs. This mutant binds roX2 less well in vivo leading to the suggestion that MLE redistributes to other targets, i.e. 3′ UTRs, if the main target roX2 is not accessible, due to lack of SL unwinding activity. Interestingly, our vitRIP titration experiments did reveal MLE binding to introns and 3′ UTRs in addition to significant roX2 binding, suggesting that under physiological conditions more transient RNA interactions may be missed due to dominating effects by stable roX2 binding.
We recapitulated MLE binding in vivo under conditions comparable to vitRIP (avoiding crosslinking) by immunoprecipitation of endogenous MLE (eMLE) from S2 cell extracts. Among the few RNAs that were significantly enriched relative to input were roX2, roX1 (which is present in only few copies in S2 cells) and RpS29, which had been identified by UV-CLAP before (Figure 3A and B; Supplementary Figure S7A-B and Table S1) (23). In strong contrast to vitRIP and in line with the earlier studies, we neither observed distinct U-rich motifs in MLE targets (Figure 3C) nor binding to intronic regions, 3′ UTRs or snoRNAs (Supplementary Figure S7C and D). We concluded that the intrinsic binding properties of MLE are modulated by extrinsic factors in vivo, of which the MSL proteins of the DCC are the best candidates.
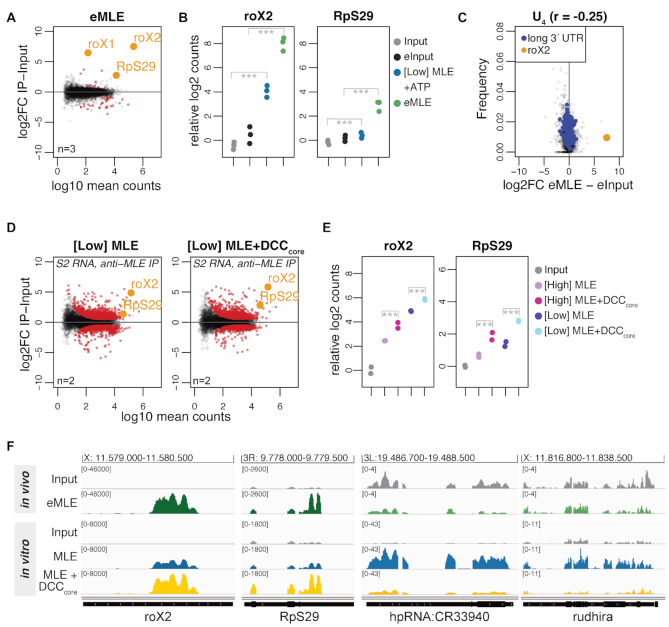
Figure 3.
The DCCcore modulates the intrinsic RNA binding specificity of MLE. (A) Log2 fold change (log2FC) of RNA recovered from S2 extracts in RNA immunoprecipitation (RIP) with endogenous MLE (eMLE) relative to input in relation to log10 mean counts. Red: significant RNAs (P < 0.01); orange: selected significant RNAs; n: number of biological replicates. (B) Dot plot representing roX2 and RpS29 enrichment in eMLE RIP from S2 extracts and in [Low] MLE vitRIP with S2 RNA, respectively. Log2 counts relative to input are displayed; ***P < 0.001. Differential analysis of the individual comparisons is given in Supplementary Table S1. (C) Comparison of U4 frequency and log2FC of RNA in eMLE RIP from S2 cells relative to input. roX2 and pre-mRNAs with a 3′ UTR longer than 2027 bp (top 5%) are highlighted. Spearman's correlation (r) was calculated for all datapoints. (D) Log2FC of S2 RNAs in vitRIP with [Low] MLE relative to input in absence (left) or presence (right) of recombinant DCCcore, in relation to log10 mean counts. MLE vitRIP was performed with a monoclonal anti-MLE antibody. The DCCcore comprises the subunits MSL1, MSL2, MSL3 and MOF. Red: significant RNAs (P < 0.01); orange: selected significant RNAs; n: number of biological replicates. (E) Dot plot representing roX2 and RpS29 enrichment with [High] or [Low] MLE in vitRIP with S2 RNA in absence or presence of recombinant DCCcore. Log2 counts relative to input are displayed; ***P < 0.001. Differential analysis of the individual comparisons is given in Supplementary Table S1. (F) Genome browser views of representative in vivo and in vitro binding profiles of MLE on roX2, RpS29, hpRNA:CR33940 and rudhira in comparison to input. Genomic coordinates for each region are given above the graph.
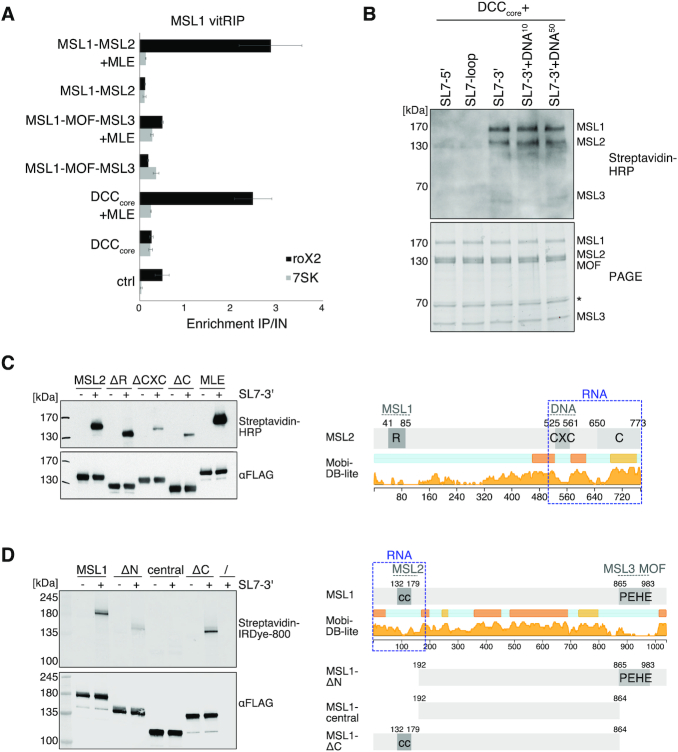
Since manipulation of DCC subunit levels in cells results in disassembly of the complex and destabilization of roX, we analyzed the impact of the DCC on the RNA binding specificity of MLE in vitro. To this end, we purified the recombinant DCC core complex (DCCcore) comprising MSL1, MSL2, MSL3 and MOF with good stoichiometry from baculovirus-infected insect cells (Supplementary Figure S1). As a proxy for functionality, we assessed the DNA binding capability of the recombinant DCCcore in a DIP assay using sheared genomic DNA from Drosophila S2 cells (22). The DCCcore enriched various PionX high affinity sites relative to control regions, indicating that MSL2 as part of the recombinant DCCcore can recognize specific X-chromosomal binding sites (Supplementary Figure S7E).
We then performed vitRIP with [Low] or [High] MLE and total S2 RNA in absence or presence of recombinant DCCcore. Since both MLE and MSL2 within the DCCcore comprise a FLAG-tag for purification, MLE vitRIP was done using an MLE-specific antibody, which correlated well with the FLAG vitRIP shown in Figure 1 (Supplementary Figure S7G and H). Overall, MLE-bound targets did not change much in presence of the DCCcore (Figure 3D and Supplementary Table S1). However, we observed an intriguing stimulating effect of the DCCcore on the enrichment of roX2 by MLE (Figure 3D–F and Supplementary Figure S7F). Remarkably, enrichment of the physiological target RpS29 with MLE increased in the presence of the DCCcore (Figure 3D–F). The DCCcore not only promoted the selectivity of MLE binding to physiological targets, but also reduced the helicase affinity to non-physiological targets, like hpRNA:CR33940 and rudhira, two RNAs that we had only identified as MLE targets in vitro and not in vivo (Figure 3F and Supplementary Figure S7F). The data suggest that in cells the DCC acts as an extrinsic modulator of MLE’s intrinsic RNA binding preference and thus plays an important role in the selective target identification.
Specific roX2 binding by the DCC is achieved by two distinct selection principles
How does the DCCcore influence target selectivity of MLE? The most likely explanation is that the DCCcore acts directly on MLE and/or the RNA substrate, but whether MLE and roX are integral components of the DCC is an unresolved question. With the notable exception of MSL1, the core proteins MSL2, MSL3 and MOF were shown to bind RNA without apparent specificity in vitro (25,43,58–59), and in case of MSL2 also to roX in vivo (24). However, RNA binding of the complete DCC was never explored in absence of extrinsic factors. Thus, we first needed to determine the intrinsic RNA binding specificity of the DCC. We incubated the reconstituted DCCcore with total S2 or cl.8 RNA in absence or presence of sub-stoichiometric amounts of MLE and ATP and retrieved the complex with an MSL1-specific antibody. In absence of MLE, we observed low-level promiscuous RNA binding by the DCCcore, but no significant enrichment of specific transcripts (Figure 4A; Supplementary Figure S8A-B and Table S1). In presence of MLE, however, the DCCcore specifically enriched roX2 and a small subset of H/ACA-box snoRNAs and mitochondrial RNAs from the S2 transcriptome, which we had identified before as specific MLE targets (Figure 4A–C; Supplementary Figure S8A-B and Table S2). Notably, from the cl.8 transcriptome only a single RNA, roX2, was retrieved by the DCC (Figure 4A; Supplementary Figure S8C and Table S1). Interestingly, despite expressed at high levels in cl.8 cells, roX1 did not significantly enrich with the DCCcore in presence of MLE, which hints at a substantially different binding mode of roX1 (Figure 4A; Supplementary Figure S8C and Table S1). We propose that MLE uses its intrinsic RNA binding specificity to select roX2 and other RNAs from the transcriptome but mediates primarily the association of roX2 with the DCCcore in a mechanism involving a second level of selectivity intrinsic to the DCCcore (Figure 3D–F).
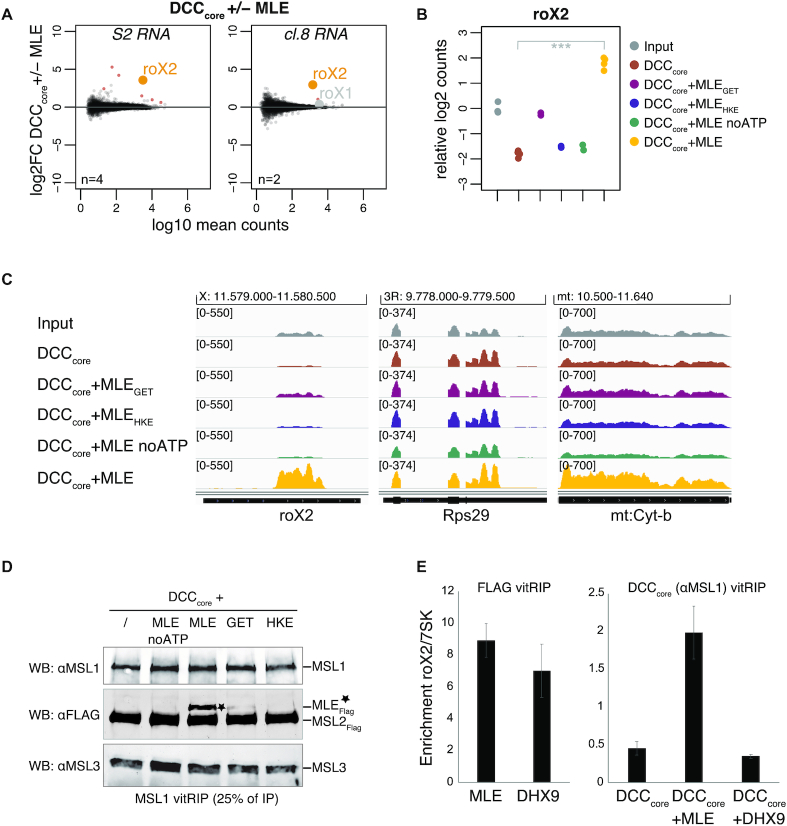
Figure 4.
Specific roX2 association with the DCC depends on Drosophila MLE. (A) Log2 fold change (log2FC) of RNA from S2 (left) or cl.8 cells (right) in vitRIP with the DCCcore in presence or absence of [Low] MLE in relation to log10 mean counts. Reactions contained ATP. vitRIP of the DCCcore employed an anti-MSL1 antibody. Red: significant RNAs (P < 0.01); orange: selected significant RNA; gray: selected non-significant RNA; n: number of biological replicates. (B) Dot plot showing log2 counts of roX2 relative to input in DCCcore vitRIP with S2 RNA in absence or presence of MLE variants and ATP unless indicated. vitRIP of the DCCcore utilized an MSL1 antibody. Analyzed variants are MLEGET (MLEK413E) and MLEHKE (MLEH1032E-K1033E). ***P < 0.001. Differential analysis of the individual comparisons is given in Supplementary Table S1. (C) Genome browser views of representative in vitro binding profiles of the DCC shown in (B) on roX2, RpS29 and mt:Cyt-b in comparison to S2 input. Genomic coordinates for each region are given above the graph. (D) Western blot analysis of the integrity of the DCC in MSL1 vitRIP samples shown in (B) and (C). 25% of each immunoprecipitate was analyzed using MSL1, MSL3 and FLAG antibodies. The anti-FLAG antibody detects FLAG-tagged MSL2 and MLE variants (indicated by asterisk). (E) Left: FLAG vitRIP of FLAG-tagged [High] MLE and [High] DHX9, respectively, with S2 RNA. Right: DCCcore vitRIP using an anti-MSL1 antibody with S2 RNA in absence or presence of [Low] MLE and [Low] DHX9, respectively. Enrichment of roX2 relative to 7SK was analyzed by RT-qPCR. Error bars represent standard deviation for two independent replicates. Statistical analysis of RT-qPCR is given in Supplementary Table S3.
MLE had previously been described to unwind roX in an ATP-dependent manner (25). RoX2 binding by the DCC may thus depend on the RNA unwinding ability of the helicase. We performed vitRIP with the DCCcore, total RNA from S2 cells and MLE derivatives, either mutated in the ATP-binding site (MLEGET) (60) or mutated in the OB-fold domain that contacts the unwound RNA substrate in the helicase channel (MLEHKE) (29). Neither MLE mutant was able to faithfully mediate roX2 association with the DCCcore, suggesting that an active helicase with single-stranded substrate binding properties is required (Figure 4B and C; Supplementary Figure S8D and Table S1). The same effect was observed when vitRIP was performed with wild-type MLE in absence of ATP. Of note, and in contrast to a published study (23), the MLEGET mutant still enriched roX2 in vitRIP (Supplementary Figure S8E and Table S3), which explains the subtle roX2 enrichment with the DCCcore in presence of this mutant (Figure 4B and C). Western blot analysis of the DCC vitRIP samples confirmed the integrity of the DCCcore upon MSL1 pull down and further showed that MLE associates faithfully with the complex only in its wild-type form in the presence of ATP (Figure 4D). In the other conditions tested (lack of ATP, mutated MLE), the unwinding properties of MLE are impaired, but general RNA binding through the dsRDB2 is unaffected (29,39). Thus, we propose that roX2 unwinding by MLE generates binding sites on the RNA and on the helicase itself, allowing specific interaction with the DCCcore.
Harnessing MLE for a Drosophila-specific function
So far, the data suggest that the exquisite selectivity of roX2 RNA binding by the DCC originates from an intimate functional relationship between MLE and the DCCcore, where the intrinsic RNA binding properties of the latter serves as a second selector applied to a helicase preselection of RNAs. Conceivably, such a specific functional relationship may have evolved specifically to coordinate the dosage compensation process. To experimentally test this hypothesis, we searched the databases for close MLE homologs but could not identify any other DExH helicase in D. melanogaster resembling the domain arrangement of MLE. Therefore, we focused on the mammalian helicase DHX9, [a.k.a RNA helicase A (61)], the closest ortholog of MLE with identical domain arrangement and 51% sequence identity. The functions of DHX9 in various cellular processes are well studied (62). Recombinant, FLAG-tagged DHX9 (Supplementary Figure S8F) was subjected to vitRIP with total S2 RNA and roX2 binding was quantified by RT-qPCR. DHX9 was able to enrich roX2 relative to the 7SK control to an extent that was comparable to MLE (Figure 4E and Supplementary Table S3). This suggests that both helicases have a similar intrinsic RNA binding specificity. To test if DHX9 could catalyse the enrichment of roX2 with the DCC, we performed vitRIP with the DCCcore in absence or presence of MLE or DHX9 and analyzed roX2 recovery by RT-qPCR. Surprisingly, despite its ability to bind roX2, DHX9 could not mediate roX2 association with the DCC (Figure 4E and Supplementary Table S3). To validate this finding in vivo, we monitored the functional association of the helicase with the dosage-compensated X chromosome, as we did previously for Drosophila MLE (29,46). We generated an S2 cell line stably expressing GFP-tagged DHX9 and monitored the localization of the ectopic helicase by immunostaining. With reference to endogenous MSL2 and MLE, DHX9-GFP did not localize to the X chromosomal territory but appeared as speckles in the nucleoplasm (Supplementary Figure S8G). DHX9-GFP also failed to substitute for endogenous MLE in maintaining the X territory, if the MLE helicase was depleted by RNA interference (Supplementary Figure S8H and I). We conclude that the ability of the DExH helicase MLE to unwind roX2 RNA and to interact with the DCCcore subunits constitutes a species-specific mechanism ensuring Drosophila dosage compensation.
The MSL1-MSL2 module of the DCC recruits MLE and incorporates remodeled roX2
The results suggest that MLE and roX2 RNA are integrated into the DCC through a mechanism, which combines stringent RNA selection using the intrinsic RNA binding specificities of MLE and DCCcore, and active unwinding of the RNA substrate. In the context of the complex, the substrate specificity of MLE is modulated (Figure 3E and F). But which subunits of the DCCcore contribute to it? The DCCcore might form a hetero-octamer (5) and can functionally be divided into two modules. Dimeric MSL1 constitutes a scaffold and interacts through its N-terminal coiled-coil domain with MSL2 (DNA-binding module) and through its C-terminal PEHE domain with MSL3 and MOF (epigenetic reader-writer module) (Supplementary Figure S1). Thus, we reconstituted the DNA-binding module (MSL1-MSL2) and the reader-writer module (MSL1-MSL3-MOF) with good stoichiometry from baculovirus-infected insect cells (Supplementary Figure S1) and compared their ability to cooperate with MLE to select roX2 from the transcriptome in vitro. We performed vitRIP with S2 RNA in absence or presence of MLE and quantified roX2 enrichment by RT-qPCR. In the absence of MLE, none of the tested complexes enriched roX2 compared to the 7SK control RNA. In presence of MLE and ATP, however, the DNA-binding module and the DCCcore showed substantial roX2 enrichment, while the reader-writer module did not (Figure 5A and Supplementary Table S3). In essence, whenever MSL2 was present, MLE was more efficiently recruited (Supplementary Figure S9A) and roX2 was specifically associated with the complex. We propose that the DNA-binding module, in particular MSL2, is necessary and sufficient to recruit MLE and remodeled roX2, presumably via direct interaction with MSL2.
Figure 5.
The MSL1-MSL2 module of the DCC recruits MLE and incorporates remodeled roX2. (A) vitRIP of recombinant DCC DNA-binding module (MSL1-MSL2), reader-writer module (MSL1-MOF-MSL3) and DCCcore (MSL1-MSL2-MOF-MSL3) with S2 RNA in absence or presence of [Low] MLE and ATP. vitRIP utilized an MSL1 antibody. Enrichment of roX2 and 7SK was analyzed by RT-qPCR and is displayed relative to input. Ctrl indicates an RNA-only MSL1 vitRIP sample lacking recombinant proteins. Error bars represent standard deviation for two or three independent replicates. Statistical analysis of RT-qPCR is given in Supplementary Table S3. (B) UV crosslinking assay showing direct binding of the DCCcore to biotinylated roX2 RNA fragments mimicking alternative secondary structures. Unwinding of stem loop 7 (SL7) in the 3′ region of roX2 by MLE generates alternative stem loops as predicted in Supplementary Figure 9B. Annotation of roX2 secondary structure is based on (25). The presence of 10 pmol or 50 pmol PionX CG8097 dsDNA in addition to 50 pmol RNA in the reaction is indicated by DNA10 and DNA50, respectively. Protein-crosslinked biotinylated roX2 fragments were detected by Streptavidin-HRP (upper panel). The DCCcore proteins were visualized in a stain-free image of the SDS-PAGE (lower panel). The asterisk indicates a co-purifying contaminant. (C) Left: UV crosslinking assay showing direct binding of 10 pmol MLE and MSL2 to 50 pmol biotinylated remodeled roX2 SL7-3′ RNA. Of MSL2, wild-type and the mutants ΔRING (MSL2Δ1-131), ΔCXC (MSL2Δ523-565) and ΔC (MSL2Δ650-778) were analyzed. Upper panel shows the protein-bound biotinylated SL7-3′ RNA detected by Streptavidin-HRP. Lower panel shows an anti-FLAG western blot for detection of FLAG-tagged MLE and MSL2 proteins, respectively. Right: Domain structure of MSL2. The RING (R), CXC, and C-terminal domain (C) are labeled. Intrinsic disorder probability was predicted by MobiDB 3.0 (72). Dashed gray lines indicate known DCC interfaces. Dashed blue box indicates the putative non-canonical RNA binding region in MSL2. (D) UV crosslinking assay probing 10 pmol MSL1 for direct binding to 50 pmol biotinylated remodeled roX2 SL7-3′ RNA. Of MSL1, wild-type and the mutants ΔN (MSL1Δ1-191), central (MSL1192-864) and ΔC (MSL1Δ865-1039) were analyzed. Protein-bound biotinylated SL7-3′ RNA was detected using IRDye 800 Streptavidin. FLAG-tagged MSL1 variants were detected by anti-FLAG western blot. Right: Domain structure of MSL1. The coiled-coil region (cc) and the PEHE domain are indicated. Intrinsic disorder probability was predicted by MobiDB 3.0 (72). Dashed gray lines indicate known DCC interfaces. Dashed blue box indicates the putative non-canonical RNA binding regions in MSL1.
To identify if subunits of the DCCcore bind roX2 directly or only indirectly through interaction with MLE, we employed an in vitro UV crosslinking (XL) assay. Biotinylated roX2 substrates were crosslinked to the DCCcore, fixed RNA–protein complexes were transferred to a membrane and roX2-bound proteins were detected using Streptavidin-HRP. The roX2 substrates used in the assay mimic the alternative secondary structures of stem loop 7 (SL7), which are the result of MLE-dependent remodeling (Supplementary Figure S9B) (25). Thus, putative direct RNA binding would not require the presence of MLE or MSL2-dependent recruitment of roX2-loaded MLE. We subjected the DCCcore to the UV-XL assay with different roX2 substrates and identified MSL2 and, to our surprise, also MSL1 as direct roX2 interactors. Intriguingly, MSL1 and MSL2 specifically crosslinked to the SL7-3′ alternative secondary structure, which constitutes the 3′ half of SL7 and includes the conserved roX-box consensus, but not to the original loop region (Figure 5B and Supplementary Figure S9B). Addition of dsDNA resembling a PionX HAS (22) did not interfere with crosslinking to SL7-3′ (Figure 5B). We confirmed the direct binding of MSL1 and MSL2 to remodeled roX2 in an UV-XL assay with individual proteins (Figure 5C-D and Supplementary Figure S9C). Deletion of the CXC DNA-binding domain or the basic C-terminal domain (C) severely diminished crosslinking of MSL2 to SL7-3′ of roX2 (Figure 5C), suggesting that roX2 RNA binding is integrated with DNA binding (43) in the C-terminus of MSL2. Following the same strategy with MSL1, we observed compromised SL7-3′ binding upon deletion of an N-terminal region spanning 191 amino acids (Figure 5D). In contrast, the intrinsically disordered central region and the C-terminal MSL3-/MOF-interacting region did not contribute to SL7-3′ binding in the UV-XL assay. The data suggest that the putative non-canonical RNA-binding region resides in the N-terminus of MSL1 close to the MSL2-interaction coiled-coil domain.
We propose that roX2 remodeling by MLE generates binding sites, which are specifically recognized through the C-terminal region in MSL2 and through a hitherto undescribed RNA-binding function in the N-terminus of MSL1.
DISCUSSION
vitRIP, a versatile tool to comprehensively characterize intrinsic RNA binding specificities
Generally, RNA binding specificity of a given protein can be analyzed in vitro on pools of in vitro-transcribed or chemically synthesized RNAs, or in a competitive cellular environment using state-of-the-art CLIP technologies on natural transcripts. Here, we characterized the intrinsic RNA binding specificities of recombinant, highly purified subunits of the Drosophila DCC using a simple and versatile in vitro transcriptome binding assay, which aims to bridge the gap between in vitro and in vivo approaches. vitRIP takes advantage of working in a controllable biochemical setting, yet using the complex cellular transcriptome as substrate. Interestingly, a high throughput Bind-n-Seq assay using natural RNAs of 109 nt length illustrated the importance of secondary structure as a key determinant for RNA motif binding specificity (63). In vitRIP the RNAs may be much longer and folded by longer-range base pairing, further increasing the complexity of the target pool of binding determinants. Of note, the experimental design of vitRIP (i.e. protein and RNA concentrations) does not intend to recapitulate physiological conditions. Rather, vitRIP informs on the intrinsic binding specificity of an individual protein outside of its physiological context and, by employing protein titration series, on binding affinities. In addition, the controlled experimental setup allows to evaluate the impact of defined extrinsic factors on binding selectivity. Critical to the success of vitRIP, however, is the availability of highly pure, homogeneous protein preparations. Titrating the total RNA pool instead, may circumvent the need for high protein quantities.
We designed vitRIP as a native technology avoiding the well-known pyrimidine bias induced by UV-based crosslinking (64). However, we also recognize the potential to combine vitRIP with crosslinking to identify the exact protein-binding motifs on RNA substrates. While this study was in preparation, an in vitro RNA pulldown approach similar to ours was applied to analyze binding of purified BAF60 and BRG1 to Xist RNA using RNA extracted from fibroblast cells (65). This study did not include next-generation sequencing, which is a crucial aspect of vitRIP as it enables a comprehensive survey of target RNAs.
The intrinsic RNA selectivity of MLE suggests potential substrates beyond roX RNAs
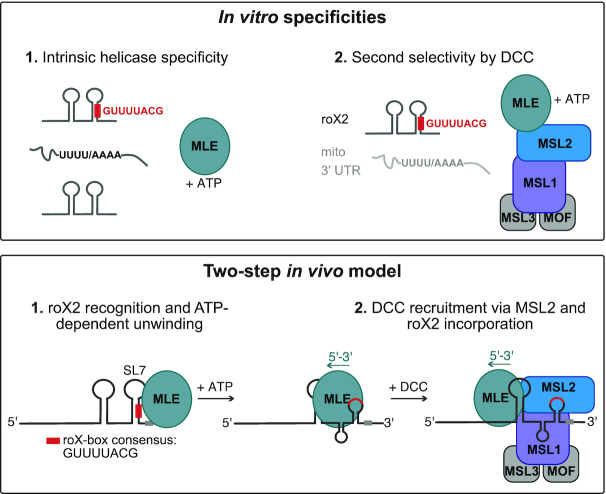
Central to our study was the determination of intrinsic RNA binding specificity of the DExH helicase MLE itself and the modulation by extrinsic factors, for example the functional interaction with the male-specific lethal DCC. MLE binds a broad spectrum of RNAs in vitro with remarkable selectivity toward uridine- and adenosine-rich sequences in male and female transcriptomes (Figure 6), confirming our previous observation of a base-specific readout of the unwound RNA substrate in the helicase channel (29). Base-specific binding to uridines was recently also reported for the related mammalian DExH helicase DHX37 (66), suggesting that substrate selectivity among DExH RNA helicases is more prevalent than expected (11). Among the most enriched substrates of MLE, we identified H/ACA-box snoRNAs, which fold into a defined tandem stem loop structure (57). Poly-uridine or poly-adenosine motifs are not a specific feature of H/ACA-box snoRNAs, suggesting that MLE has affinity for stem loop structures arranged in tandem (Figure 6). Both sequence and structural binding determinants are combined in the critical functional parts of roX RNAs (24,25), explaining the high affinity of MLE for these lncRNAs in vitro and in vivo (Figure 6). This intrinsic specificity classifies MLE as a helicase with a broad, but defined target spectrum, similarly to the close structural homolog Prp43p (11). Indeed, we identified U-rich, long 3′ UTR regions and edited RNAs as in vitro substrates of MLE, arguing for a physiological role in unwinding these transcripts in the context of pre-mRNA processing or RNA editing, respectively. A role of MLE in the process of adenosine-to-inosine editing has long been hypothesized (32). In concordance with a recently proposed function of the mammalian MLE ortholog DHX9 on edited Alu repeats (67), we suggest that MLE acts as a resolvase of double-stranded editing intermediates.
Figure 6.
Two-step model for selective roX2 integration into the DCC. Upper panel: In vitro, the helicase MLE exhibits intrinsic specificity for U-/A-rich linear sequence motifs and tandem stem-loop structures. These features are combined in roX1/2 RNA. Here the stems of RNA secondary structures contain stretches of U:A base pairing. ATP-dependent unwinding thus exposes linear U-/A-rich sequences. The DCCcore provides additional selectivity, which increases the affinity to the physiological substrate roX2 and decreases the affinity to other U-/A-rich transcripts. Lower panel: In the first step of roX2 integration into the DCC, MLE uses its intrinsic specificity to bind and unwind roX2 with 3′–5′ directionality. The unwinding process exposes binding sites on the RNA (the conserved roX-box consensus sequence). In the second step, additional selectivity provided by the DCCcore prevents unspecific binding of other MLE substrates and allows specific interaction of MLE and DCCcore via MSL2. Within the DCC, the MSL1-MSL2 module contributes to roX2 integration by direct binding to exposed roX-box sequences.
It is likely that the intrinsic RNA specificity of MLE is modulated by diverse molecular factors in cells, reflecting its different functions. This is nicely illustrated by our finding that DCCcore enhances the affinity of MLE to roX RNAs. In male cells, the dosage compensation system appears the dominant modulator of MLE substrate selectivity. In genetic terms, this dominance is explained by the lack of functional redundancy with other helicases. In molecular terms, we consider that MLE may have evolved to be more stably associated with the DCC, involving roX RNA as a molecular tether. We speculate that the association of MLE with the DCC may also involve protein interactions that have not been characterized so far.
Interactions of MLE with other RNAs in vivo may be more transient in the context of RNA processing events. In support of this hypothesis, a helicase-deficient MLE mutant was reported to bind 3′ UTRs in male cells (23), in agreement with our in vitro observation. Further insights into the physiological role of MLE may come from in vitro approaches, such as vitRIP, and from studies in female flies, which lack roX RNA and a functional dosage compensation complex.
De novo assembly of roX2-containing Drosophila DCC
The Drosophila DCC requires MLE and roX for its function, but if and how RNA and helicase are connected to the complex was mysterious. We ultimately reconstituted the assembly of the complete roX2-comprising DCC using vitRIP and found that the DCCcore incorporates roX2 from male transcriptomes with high specificity in presence of an active MLE helicase. We propose that MLE uses its intrinsic specificity to select roX2 from the transcriptome, possibly at the site of roX2 transcription, and upon unwinding, associates with the DCCcore (Figure 6), in agreement with our previous hypothesis (25). This mechanism appears to be specific to MLE, since in D. melanogaster no other DExH helicase exists with similar domain architecture. Even the closest mammalian ortholog DHX9 could not replace the function of MLE in vitro and in vivo, which suggests that the male-specific lethality of mle loss is directly linked to its role in selecting and incorporating roX2 into the DCC. In vitro, DHX9 was able to select roX2 from the transcriptome and it is possible that it can unwind roX2 in vivo due to almost complete conservation of helicase-relevant motifs (29). The failure to complement an MLE deficiency thus likely resides in MLE-specific surfaces that we predict to interact with MSL2, bringing MSL2 in close proximity to remodeled roX2. Surprisingly, we did not observe MLE-dependent incorporation of roX1, although the main recognition principles (i.e. roX boxes, tandem stem loop structures) apply to both roX1 and roX2 and both RNAs are contacted by MLE and MSL2 in cl.8 cells (68). We speculate that roX1 may integrate via a different mechanism or via an unknown co-factor. Alternatively, roX1 may have a different role during the process of dosage compensation early in development as suggested by a recent study (69).
Our data suggest that the DCCcore adds a second selectivity step to the roX2 incorporation to prevent unspecific binding of other MLE substrates. We found that the DNA-binding module of the DCC, composed of MSL1 and MSL2, recruits the roX2-loaded helicase and binds directly to the remodeled roX2 substrate (Figure 6). While MSL2 had been proposed to bind roX2 in vitro and in vivo (24–25,43), our study identified MSL1 as a novel RNA binding protein. Intriguingly, neither MSL1 nor MSL2 possess canonical RNA binding domains and may use intrinsically disordered regions (IDRs) for RNA binding (7). Both proteins comprise extended regions with high disorder probability (Figure 5C and D), rendering IDRs as potential RNA-binding domains in MSL1 and MSL2. Notably, we identified the largely disordered C-terminal region in MSL2 as the roX2-interacting domain, thus confirming a long-proposed model of interaction of the MSL2 C-terminus with MLE and roX2 (70). Within MSL1 we identified the N-terminus as a putative non-canonical RNA binding region. This region is largely disordered with the exception of the well-structured MSL2 interaction domain (Figure 5D) (71), emphasizing the importance of a functional unit composed of MSL1 and MSL2 for MLE and roX2 integration into the DCC.
Our biochemical approach now paves the way for a dissection of the molecular interactions that define the pathway for the assembly of the dosage compensation complex, a vital ribonucleoprotein chromatin regulator. Beyond dosage compensation, our study outlines a strategy to determine intrinsic target selectivity of diverse RNA-binding proteins and the modulation of specificity through extrinsic factors.
DATA AVAILABILITY
Sequencing data were deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE143455. Analysis code is available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Frank Grosse for kindly providing DHX9 cDNA, Nadia Prayitno for sharing clone eight cells and all members of the Becker laboratory for discussion. We thank Helmut Blum and Stefan Krebs for the next-generation sequencing service.
Authors’ contributions: M.M. and P.B.B. conceived experiments; M.M., S.K., R.V. and A.W.T. performed experiments; M.M., T.S. and A.W.T. analyzed data; M.M. and P.B.B. wrote the manuscript; P.B.B. secured funding.
Contributor Information
Marisa Müller, Molecular Biology Division, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany.
Tamas Schauer, Molecular Biology Division, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany; Bioinformatics Unit, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany.
Silke Krause, Molecular Biology Division, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany.
Raffaella Villa, Molecular Biology Division, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany.
Andreas W Thomae, Molecular Biology Division, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany; Core Facility Bioimaging at the Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany.
Peter B Becker, Molecular Biology Division, Biomedical Center, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Be1140/8-1, Be1140/9-1]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rutenberg-Schoenberg M., Sexton A.N., Simon M.D.. The properties of long noncoding RNAs that regulate chromatin. Annu. Rev. Genomics Hum. Genet. 2016; 17:69–94. [DOI] [PubMed] [Google Scholar]
- 2. Zhao Z., Senturk N., Song C., Grummt I.. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018; 32:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. da Rocha S.T., Heard E.. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 2017; 24:197–204. [DOI] [PubMed] [Google Scholar]
- 4. Moindrot B., Brockdorff N.. RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin. Cell Dev. Biol. 2016; 56:58–70. [DOI] [PubMed] [Google Scholar]
- 5. Samata M., Akhtar A.. Dosage compensation of the X chromosome: a complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu. Rev. Biochem. 2018; 87:323–350. [DOI] [PubMed] [Google Scholar]
- 6. Jankowsky E., Harris M.E.. Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015; 16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 8. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.M., Foehr S., Curk T., Krijgsveld J., Hentze M.W.. Comprehensive identification of RNA-Binding domains in human cells. Mol. Cell. 2016; 63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He C., Sidoli S., Warneford-Thomson R., Tatomer D.C., Wilusz J.E., Garcia B.A., Bonasio R.. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell. 2016; 64:416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Panhale A., Richter F.M., Ramirez F., Shvedunova M., Manke T., Mittler G., Akhtar A.. CAPRI enables comparison of evolutionarily conserved RNA interacting regions. Nat. Commun. 2019; 10:2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jarmoskaite I., Russell R.. RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 2014; 83:697–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ozgur S., Buchwald G., Falk S., Chakrabarti S., Prabu J.R., Conti E.. The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 2015; 282:850–863. [DOI] [PubMed] [Google Scholar]
- 13. Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008; 37:317–336. [DOI] [PubMed] [Google Scholar]
- 14. Sloan K.E., Bohnsack M.T.. Unravelling the mechanisms of RNA helicase regulation. Trends Biochem. Sci. 2018; 43:237–250. [DOI] [PubMed] [Google Scholar]
- 15. Amrein H., Axel R.. Genes expressed in neurons of adult male Drosophila. Cell. 1997; 88:459–469. [DOI] [PubMed] [Google Scholar]
- 16. Kelley R.L., Solovyeva I., Lyman L.M., Richman R., Solovyev V., Kuroda M.I.. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995; 81:867–877. [DOI] [PubMed] [Google Scholar]
- 17. Maenner S., Muller M., Becker P.B.. Roles of long, non-coding RNA in chromosome-wide transcription regulation: lessons from two dosage compensation systems. Biochimie. 2012; 94:1490–1498. [DOI] [PubMed] [Google Scholar]
- 18. Figueiredo M.L., Kim M., Philip P., Allgardsson A., Stenberg P., Larsson J.. Non-coding roX RNAs prevent the binding of the MSL-complex to heterochromatic regions. PLos Genet. 2014; 10:e1004865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franke A., Baker B.S.. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell. 1999; 4:117–122. [DOI] [PubMed] [Google Scholar]
- 20. Gu W., Wei X., Pannuti A., Lucchesi J.C.. Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J. 2000; 19:5202–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meller V.H., Rattner B.P.. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002; 21:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Villa R., Schauer T., Smialowski P., Straub T., Becker P.B.. PionX sites mark the X chromosome for dosage compensation. Nature. 2016; 537:244–248. [DOI] [PubMed] [Google Scholar]
- 23. Ilik I.A., Maticzka D., Georgiev P., Gutierrez N.M., Backofen R., Akhtar A.. A mutually exclusive stem-loop arrangement in roX2 RNA is essential for X-chromosome regulation in Drosophila. Genes Dev. 2017; 31:1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ilik I.A., Quinn J.J., Georgiev P., Tavares-Cadete F., Maticzka D., Toscano S., Wan Y., Spitale R.C., Luscombe N., Backofen R. et al.. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell. 2013; 51:156–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maenner S., Muller M., Frohlich J., Langer D., Becker P.B.. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol. Cell. 2013; 51:174–184. [DOI] [PubMed] [Google Scholar]
- 26. Park S.W., Kang Y., Sypula J.G., Choi J., Oh H., Park Y.. An evolutionarily conserved domain of roX2 RNA is sufficient for induction of H4-Lys16 acetylation on the Drosophila X chromosome. Genetics. 2007; 177:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park S.W., Kuroda M.I., Park Y.. Regulation of histone H4 Lys16 acetylation by predicted alternative secondary structures in roX noncoding RNAs. Mol. Cell. Biol. 2008; 28:4952–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stuckenholz C., Meller V.H., Kuroda M.I.. Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics. 2003; 164:1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prabu J.R., Muller M., Thomae A.W., Schussler S., Bonneau F., Becker P.B., Conti E.. Structure of the RNA helicase MLE reveals the molecular mechanisms for uridine specificity and RNA-ATP coupling. Mol. Cell. 2015; 60:487–499. [DOI] [PubMed] [Google Scholar]
- 30. Cugusi S., Kallappagoudar S., Ling H., Lucchesi J.C.. The Drosophila helicase maleless (MLE) is implicated in functions distinct from its role in dosage compensation. Mol. Cell. Proteomics. 2015; 14:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cugusi S., Li Y., Jin P., Lucchesi J.C.. The Drosophila helicase MLE targets hairpin structures in genomic transcripts. PLos Genet. 2016; 12:e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reenan R.A., Hanrahan C.J., Ganetzky B.. The mle(napts) RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000; 25:139–149. [DOI] [PubMed] [Google Scholar]
- 33. Tauchert M.J., Fourmann J.B., Luhrmann R., Ficner R.. Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. Elife. 2017; 6:e21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wessels H.H., Imami K., Baltz A.G., Kolinski M., Beldovskaya A., Selbach M., Small S., Ohler U., Landthaler M.. The mRNA-bound proteome of the early fly embryo. Genome Res. 2016; 26:1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al.. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dominguez D., Freese P., Alexis M.S., Su A., Hochman M., Palden T., Bazile C., Lambert N.J., Van Nostrand E.L., Pratt G.A. et al.. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell. 2018; 70:854–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sutandy F.X.R., Ebersberger S., Huang L., Busch A., Bach M., Kang H.S., Fallmann J., Maticzka D., Backofen R., Stadler P.F. et al.. In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res. 2018; 28:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izzo A., Regnard C., Morales V., Kremmer E., Becker P.B.. Structure-function analysis of the RNA helicase maleless. Nucleic Acids Res. 2008; 36:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Straub T., Grimaud C., Gilfillan G.D., Mitterweger A., Becker P.B.. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLos Genet. 2008; 4:e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Straub T., Neumann M.F., Prestel M., Kremmer E., Kaether C., Haass C., Becker P.B.. Stable chromosomal association of MSL2 defines a dosage-compensated nuclear compartment. Chromosoma. 2005; 114:352–364. [DOI] [PubMed] [Google Scholar]
- 42. Vogt S., Schneider G., Steuernagel A., Lucchesi J., Schulze E., Rudolph D., Schmahl G.. X-ray microscopic studies of the Drosophila dosage compensation complex. J. Struct. Biol. 2000; 132:123–132. [DOI] [PubMed] [Google Scholar]
- 43. Fauth T., Muller-Planitz F., Konig C., Straub T., Becker P.B.. The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome. Nucleic Acids Res. 2010; 38:3209–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berger I., Fitzgerald D.J., Richmond T.J.. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004; 22:1583–1587. [DOI] [PubMed] [Google Scholar]
- 45. Dann G.P., Liszczak G.P., Bagert J.D., Muller M.M., Nguyen U.T.T., Wojcik F., Brown Z.Z., Bos J., Panchenko T., Pihl R. et al.. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature. 2017; 548:607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ankush Jagtap P.K., Muller M., Masiewicz P., von Bulow S., Hollmann N.M., Chen P.C., Simon B., Thomae A.W., Becker P.B., Hennig J.. Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless. Nucleic Acids Res. 2019; 47:4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoshihama M., Nakao A., Kenmochi N.. snOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes. 2013; 6:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. St Laurent G., Tackett M.R., Nechkin S., Shtokalo D., Antonets D., Savva Y.A., Maloney R., Kapranov P., Lawrence C.E., Reenan R.A.. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat. Struct. Mol. Biol. 2013; 20:1333–1339. [DOI] [PubMed] [Google Scholar]
- 49. Maiato H., Sunkel C.E., Earnshaw W.C.. Dissecting mitosis by RNAi in Drosophila tissue culture cells. Biol. Proced. Online. 2003; 5:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomae A.W., Schade G.O., Padeken J., Borath M., Vetter I., Kremmer E., Heun P., Imhof A.. A pair of centromeric proteins mediates reproductive isolation in Drosophila species. Dev. Cell. 2013; 27:412–424. [DOI] [PubMed] [Google Scholar]
- 51. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H.. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013; 41:W471–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Raden M., Ali S.M., Alkhnbashi O.S., Busch A., Costa F., Davis J.A., Eggenhofer F., Gelhausen R., Georg J., Heyne S. et al.. Freiburg RNA tools: a central online resource for RNA-focused research and teaching. Nucleic Acids Res. 2018; 46:W25–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.L., Hentze M.W., Kohlbacher O., Urlaub H.. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 2014; 11:1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yablonovitch A.L., Deng P., Jacobson D., Li J.B.. The evolution and adaptation of A-to-I RNA editing. PLos Genet. 2017; 13:e1007064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hilgers V., Perry M.W., Hendrix D., Stark A., Levine M., Haley B.. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:15864–15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 58. Akhtar A., Becker P.B.. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell. 2000; 5:367–375. [DOI] [PubMed] [Google Scholar]
- 59. Morales V., Straub T., Neumann M.F., Mengus G., Akhtar A., Becker P.B.. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 2004; 23:2258–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee C.G., Chang K.A., Kuroda M.I., Hurwitz J.. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997; 16:2671–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee C.G., Hurwitz J.. Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem. 1993; 268:16822–16830. [PubMed] [Google Scholar]
- 62. Fuller-Pace F.V. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006; 34:4206–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Taliaferro J.M., Lambert N.J., Sudmant P.H., Dominguez D., Merkin J.J., Alexis M.S., Bazile C., Burge C.B.. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol. Cell. 2016; 64:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wheeler E.C., Van Nostrand E.L., Yeo G.W.. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip. Rev. RNA. 2018; 9:e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jegu T., Blum R., Cochrane J.C., Yang L., Wang C.Y., Gilles M.E., Colognori D., Szanto A., Marr S.K., Kingston R.E. et al.. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 2019; 26:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boneberg F.M., Brandmann T., Kobel L., van den Heuvel J., Bargsten K., Bammert L., Kutay U., Jinek M.. Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis. RNA. 2019; 25:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aktas T., Avsar Ilik I., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A.. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017; 544:115–119. [DOI] [PubMed] [Google Scholar]
- 68. Quinn J.J., Ilik I.A., Qu K., Georgiev P., Chu C., Akhtar A., Chang H.Y.. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 2014; 32:933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prayitno K., Schauer T., Regnard C., Becker P.B.. Progressive dosage compensation during Drosophila embryogenesis is reflected by gene arrangement. EMBO Rep. 2019; 20:e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li F., Schiemann A.H., Scott M.J.. Incorporation of the noncoding roX RNAs alters the chromatin-binding specificity of the Drosophila MSL1/MSL2 complex. Mol. Cell. Biol. 2008; 28:1252–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hallacli E., Lipp M., Georgiev P., Spielman C., Cusack S., Akhtar A., Kadlec J.. Msl1-mediated dimerization of the dosage compensation complex is essential for male X-chromosome regulation in Drosophila. Mol. Cell. 2012; 48:587–600. [DOI] [PubMed] [Google Scholar]
- 72. Piovesan D., Tabaro F., Paladin L., Necci M., Micetic I., Camilloni C., Davey N., Dosztanyi Z., Meszaros B., Monzon A.M. et al.. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018; 46:D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited to GEO with accession number GSE143455. Analysis code is available upon request.
Sequencing data were deposited in NCBI’s Gene Expression Omnibus (GEO) with accession number GSE143455. Analysis code is available upon request.