Abstract
In cells, proteins form macromolecular complexes to execute their own unique roles in biological processes. Conventional structural biology methods adopt a bottom-up approach starting from defined sets of proteins to investigate the structures and interactions of protein complexes. However, this approach does not reflect the diverse and complex landscape of endogenous molecular architectures. Here, we introduce a top-down approach called Electron Microscopy screening for endogenous Protein ArchitectureS (EMPAS) to investigate the diverse and complex landscape of endogenous macromolecular architectures in an unbiased manner. By applying EMPAS, we discovered a spiral architecture and identified it as AdhE. Furthermore, we performed screening to examine endogenous molecular architectures of human embryonic stem cells (hESCs), mouse brains, cyanobacteria and plant leaves, revealing their diverse repertoires of molecular architectures. This study suggests that EMPAS may serve as a tool to investigate the molecular
architectures of endogenous macromolecular proteins.
Keywords: microscopy, protein, screening, structure
INTRODUCTION
In cells, proteins interact with each other and form supramolecular complexes, which are responsible for biological processes such as homeostasis, metabolism, growth and reproduction (Han et al., 2004; Jeong et al., 2001). Specific sets of proteins are regulated to form diverse molecular complexes in a precise spatiotemporal manner during these processes (Ahn et al., 2004; Olayioye et al., 2019). To understand how proteins function as large molecular complexes by interacting with their binding partners, several tools including mass spectrometry-based proteomics and structural biology have successfully identified the interaction networks among proteins and elucidated their molecular architectures (Burley et al., 2017). Proteomics approaches have provided tremendous information regarding protein-protein interactions (PPIs) and their dynamic changes in various spatiotemporal statuses of cells (Collins et al., 2013; Pellegrini et al., 2004). However, it is extremely difficult to interpret the information regarding PPIs to distinguish whether the interactions observed occur in the same discrete complex. Structural studies mainly using X-ray crystallography have provided insights into the molecular architectures and functions of macromolecular complexes. In addition, structural genomics has aimed to determine all three-dimensional structures in proteomes provided wealthy information regarding the landscape of protein structures (Burley, 2000; Grabowski et al., 2016). However, these conventional structural approaches revealing the structures and interactions of protein complexes have limitations in that they start with defined sets of proteins based on PPI information and can only reveal molecular architectures whose compositions are already known. Therefore, it is likely that conventional structural approaches cover only a small portion of the diverse and complex landscape of endogenous molecular architectures. Recent advances in electron microscopy (EM) have revolutionized the way of examining the structures of supramolecular complexes (Kühlbrandt, 2014). Single-particle analysis using cryo-EM analysis enabled us to examine the structures of protein complexes in principle without size limitation issues. In addition, the structures of protein complexes in heterologous conformations could be resolved into each different state due to an advanced computational algorithm (Nguyen et al., 2016; Scheres, 2016). Furthermore, cryo-electron tomography (cryo-ET) has opened up a new possibility to determine high-resolution structures of protein complexes in cell, although there are several technical challenges to be overcome such as improving resolutions and identifying target proteins (Schaffer et al., 2019). Despite recent technical advances in examining the structures of macromolecular complexes, the diversity and complexity of the molecular architectures in cells are far from being understood.
Here, we introduce an integrative method combining biochemical fractionation, mass spectrometry and EM analysis to investigate diverse endogenous molecular architectures in cells. We called this method EMPAS (Electron Microscopy screening for endogenous Protein ArchitectureS). Through this approach, we identified a spiral architecture observed in Escherichia coli cell lysate to be the protein AdhE, which led us to determine the high-resolution cryo-EM structures (Kim et al., 2019; 2020). In addition, we screened various samples including human embryonic stem cells (hESCs), mouse brains, cyanobacteria and a plant leaf, revealing the diversity and complexity of molecular architectures in the samples. Our approach may provide an alternative tool to examine the diverse and complex landscape of macromolecular complexes.
MATERIALS AND METHODS
Sample preparation
E. coli
BL21 (DE3) RILP E. coli cells were grown to be saturated in Luria–Bertani broth media at 37°C. The cell pellet was collected by centrifugation at 4,000 rpm for 20 min and resuspended in buffer containing 50 mM Tris pH 8.0, 100 mM NaCl, 5% glycerol, and 1 mM PMSF. The cells were lysed with sonication, and the debris was cleared with centrifugation at 13,300 rpm for 10 min. The cell lysate was separated with a HiTrap Q column (GE Healthcare, USA), and the fractions having similar proteins were categorized. Each categorized sample was subjected to a SuperoseTM 6 Increase 10/300 column (GE Healthcare) equilibrated with 50 mM Tris pH 8.0 and 100 mM NaCl. Only high molecular weight fractions were pooled and concentrated before negative staining.
Mouse brain
To obtain mouse brain samples, seven mice were sacrificed, and perfusion was performed before snap freezing. The mouse brain sample was mildly homogenized with lysis buffer containing 50 mM Tris pH 8.0, 100 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol, 1 mM DTT, 4 mM PMSF, and cOmpleteTM protease inhibitor cocktail (Roche, Switzerland). Then, brief sonication and additional homogenization were performed using a Dounce homogenizer. The mouse brain sample lysate was clarified by centrifugation at 18,000 rpm for 90 min at 4°C. The supernatant was filtered by an MF-MillporeTM membrane filter with a 0.45 µm pore size before ion exchange and gel filtration. After ion exchange, the fractions having similar proteins were categorized, and each categorized sample was subjected to a SuperoseTM 6 Increase 10/300 column (GE Healthcare) equilibrated by 50 mM Tris pH 8.0 and 100 mM NaCl. Only high molecular weight fractions were pooled and concentrated before negative staining.
Cyanobacteria
Synechocystis sp. PCC 6803 cyanobacteria cells were collected by centrifugation at 4,000 rpm for 20 min and resuspended in a buffer containing 50 mM Tris pH 8.0, 100 mM NaCl, 5% glycerol, 0.1% NP-40, and 1 mM PMSF. The cells were lysed with 2 rounds of freezing and thawing followed by mild and brief sonication. The cell lysate was cleared with centrifugation at 18,000 rpm for 20 min. The cleared cell lysate was separated with a HiTrap Q column (GE Healthcare), and the fractions were categorized according to the colors. The category showing yellow was subjected to a SuperoseTM 6 Increase 10/300 column (GE Healthcare) equilibrated by 50 mM Tris pH 8.0 and 100 mM NaCl. Only high molecular weight fractions were pooled and concentrated before negative staining.
Embryonic stem cells
hESCs were cultured in 6-well plates and washed with serum-free media before harvest. Cell pellets were obtained by centrifugation at 4,000 rpm for 20 min and resuspended in buffer containing 50 mM Tris pH 8.0, 100 mM NaCl, 5% glycerol, and 6 mM PMSF. Brief and mild sonication was used for lysis. The hESC lysate was clarified by centrifugation at 13,300 rpm for 10 min at 4°C. A total of 500 μl of 6.36 mg/ml supernatant was subjected to a SuperposeTM 6 Increase 10/300 column (GE Healthcare) equilibrated by 50 mM Tris pH 8.0 and 100 mM NaCl. Only high molecular weight fractions were pooled and concentrated before negative staining.
Plant leaf
First, 0.5 g of tobacco leaf powder was dissolved in 50 mM Tris pH 8.0, 500 mM NaCl, 5% glycerol, and 60 mM PMSF. To remove insoluble materials, centrifugation was performed at 13,000 rpm for 15 min at 4°C. To remove invisible aggregates or cell debris, the supernatant was filtered with an MF-MilliporeTM membrane filter with a 0.22 µm pore size. Next, 500 µl of filtrated supernatant was further analyzed by size-exclusion chromatography. A SuperposeTM 6 Increase 10/300 column (GE Healthcare) was equilibrated by 50 mM Tris pH 8.0, 100 mM NaCl, and eluted high molecular weight fractions were concentrated before negative staining.
The initial concentration of each sample was approximately 5-10 mg/ml.
Negative EM
A PELCO easiGlowTM glow discharger was used to apply a negative charged to a Cu grid at 30 mA for 20 s. Then, 3 µl of 0.05 to 0.1 mg/ml fractionated cell lysate was applied to the glow-discharged carbon-coated Cu grid. The fractionated sample was incubated on the grid for 1 min, blotted, washed twice with distilled water, washed once with 1.5% (w/v) uranyl acetate and incubated on a 1.5% uranyl acetate drop for 1 min. The stained grid was blotted and air-dried before observation. The prepared grid was analyzed with a Tecnai F20 electron microscope (FEI) with a Gatan CCD camera at the KAIST Analysis Center for Research Advancement (KARA). A total of 46, 43, 11, and 16 micrographs were collected for Group 1, 2, 3, and 4 from the E. coli lysate. The collected micrographs were processed using EMAN 2.0. Particles were auto-picked in different box sizes, 200 Å, 300 Å, and 420 Å. Selected particles were subjected to 2D classification.
ID mass spectrometry
The fractionated sample was analyzed by 10% SDS-PAGE with fresh running buffer. After Coomassie blue staining, the visible band was cut with a clean razor blade. The cut gel was washed with 1 ml of distilled water three times and stored at 4°C before mass spectrometry analysis. The cut gel was sent to the Taplin Mass Spectrometry Facility at Harvard Medical School.
RESULTS
EMPAS
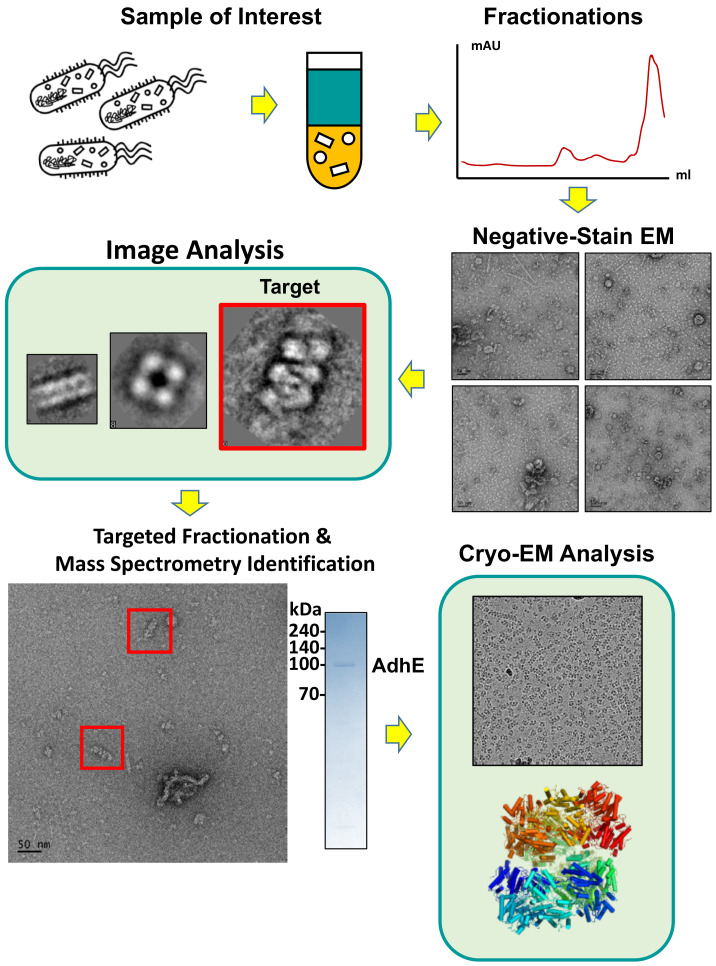
As shown in the scheme in Fig. 1, EMPAS starts by comprehensively examining the structural landscapes of endogenous macromolecules in cell lysates by using negative-stain EM. The cell lysate is then further separated by using additional biochemical tools, such as gradient ultracentrifugation, ion chromatography and size-exclusion chromatography. The separated fractions are collected into groups based on the similar protein composition assessed by SDS-PAGE. Each group from the sample is subsequently subjected to size-exclusion chromatography, and the fractions near the void volume are collected, which contain large molecular complexes suitable for further EM analysis. The fractionated samples are examined using negative-stain EM, and molecular architectures of interest are selected as targets for the second-round EMPAS. In the second-round EMPAS, the lysate is subjected to targeted fractionation. The fractions containing the targeted molecular architectures are evaluated by monitoring the presence of the targets by negative-stain EM until the fraction contains the targeted molecular architectures in near homogeneity. Proteins of the targets in the fraction are identified by mass spectrometry, the molecular architecture of proteins is reconstituted, and the high-resolution structure of the molecular architecture is resolved by cryo-EM. Using EMPAS, the molecular architectures of protein complexes in the endogenous environment can be examined.
Fig. 1. Schematic workflow of EMPAS: Fractionations, target selection, identification, and structural determination.
Fig. 1. Schematic workflow of EMPAS: Fractionations, target selection, identification, The lysate containing endogenous protein mixtures was fractionated and examined with negative-stain EM. From the collected micrographs, molecular architectures of interest are selected (indicated with red boxes). After target selection, targeted fractionation followed by mass spectrometry revealed the composition of the target molecular architectures. Further cryo-EM analysis determines the high-resolution structure of the targeted molecular architecture (Scale bar = 50 nm, bottom left panel).
Identification of endogenous molecular architectures in E. coli through EMPAS
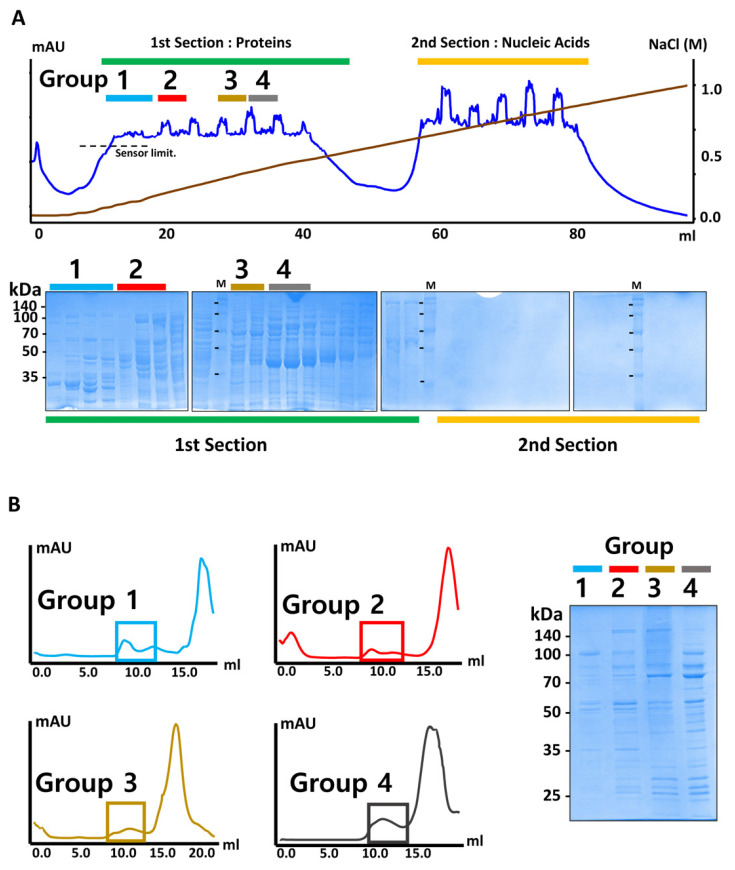
As a proof of principle for EMPAS, we aimed to screen and identify endogenous macromolecular architectures in E. coli lysate. E. coli cells in the stationary phase were collected and lysed. As we decided to focus on soluble protein, insoluble cell debris and the membrane fraction were cleared with centrifugation. The cell lysate was then separated with anion exchange chromatography, and the composition of proteins in the separated fractions was analyzed by SDS-PAGE. The fractions from anion exchange chromatography were divided into two large sections, and we analyzed all the fractions with SDS-PAGE to examine the protein components in the fractions (Fig. 2A). Interestingly, there was almost no protein in the second section, as assessed by SDS-PAGE. Consistent with this finding, the ratio of optical density at 260 to 280 nm (OD260 to OD280) was approximately 2, indicating that the second section contained mostly nucleic acids. We categorized the fractions containing proteins into four groups based on the protein band patterns, and each group was collected for the next step of EMPAS. As we aimed to examine large molecular architectures, we subjected the sample of each group to Superose 6 size-exclusion chromatography and collected fractions eluted in the high molecular weight range (Fig. 2B).
Fig. 2. E. coli sample preparation for EMPAS.
(A) An anion exchange chromatogram profile (top panel) and SDS-PAGE gel (bottom panel) of E. coli cell lysate. The fractionated samples are categorized into four groups based on the pattern of proteins analyzed by SDS-PAGE. (B) Gel filtration profile of each group of the samples (left panel). Proteins eluted at near void volume (boxed) were collected and analyzed by SDS-PAGE (right panel).
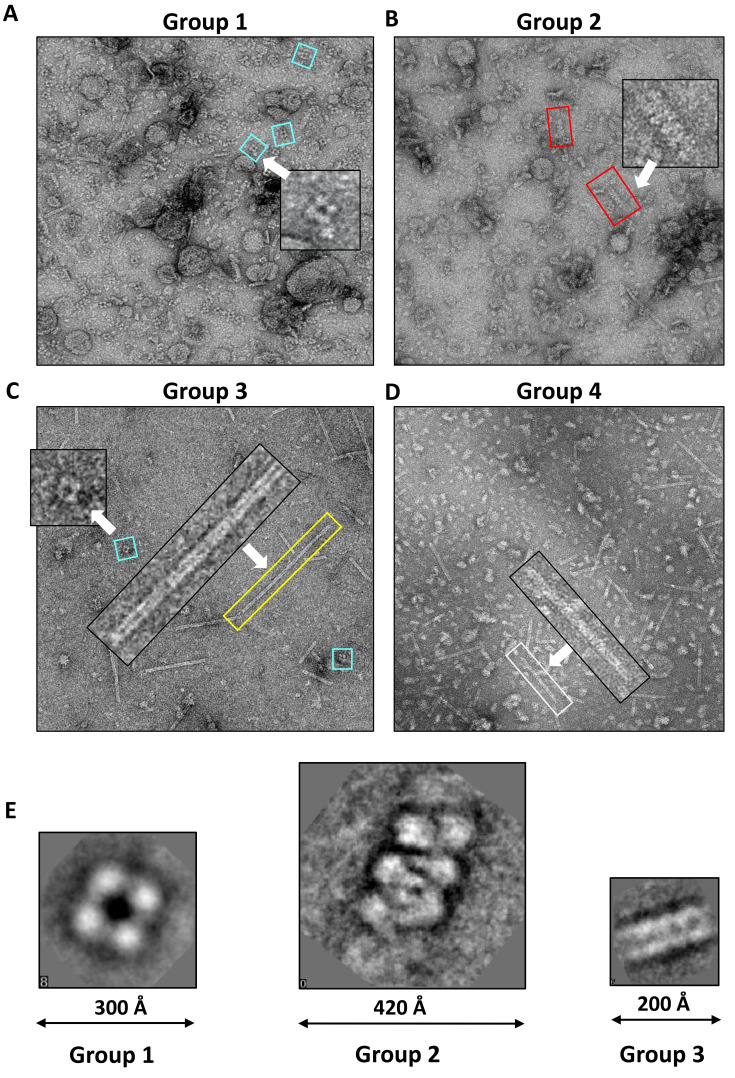
After preparing the fractionated samples containing endogenous macromolecular complexes, we moved to the next step of EMPAS, target searching and identification. We examined molecular architectures in each group by using negative-stain EM (Fig. 3A). The collected micrographs showed a vast diversity of molecular architectures. Each group contained different macromolecular architectures. Among these various architectures, some molecular structures showed prominent shapes due to their sizes or geometric features. In Group 1, we found large vacuole-like structures considered liposomes or micelles, which may originate from membrane debris (Fig. 3A). There were also square-shaped particles and particles with circular pores of various sizes (Fig. 3A). In Group 2, we found spiral structures composed of a repeating unit (Fig. 3B). These spiral architectures appeared to be formed by stacking subunits vertically in various lengths. The micrographs of Groups 3 and 4 show several tubular structures (Figs. 3C and 3D). To further characterize the structures of the prominent molecular architecture, we collected a number of micrographs from each group and performed 2D class averaging using EMAN2 (Tang et al., 2007) with random particle selection. The 2D class average analysis of the micrographs collected from Groups 1 and 3 shows a tetrameric structure and a long fibric structure, respectively (Fig. 3E). Interestingly, the 2D class average analysis of Group 2 reveals a spiral architecture approximately 100 Å in width and 70 Å in pitch (Fig. 3E). These data show that molecular architectures can be visualized through multiple fractionations followed by EM analysis.
Fig. 3. Representative negative-EM micrographs and 2D classes of endogenous macromolecular architectures.
Representative micrographs of Group 1 (A), Group 2 (B), Group 3 (C), and Group 4 (D). White arrows indicate proteins having prominent architectures, which are enlarged within the boxes. (E) Three representative 2D class averages of the distinct architectures from the micrographs collected using the samples in Groups 1, 2, and 3.
The spiral architecture is an aldehyde-alcohol dehydrogenase (AdhE)
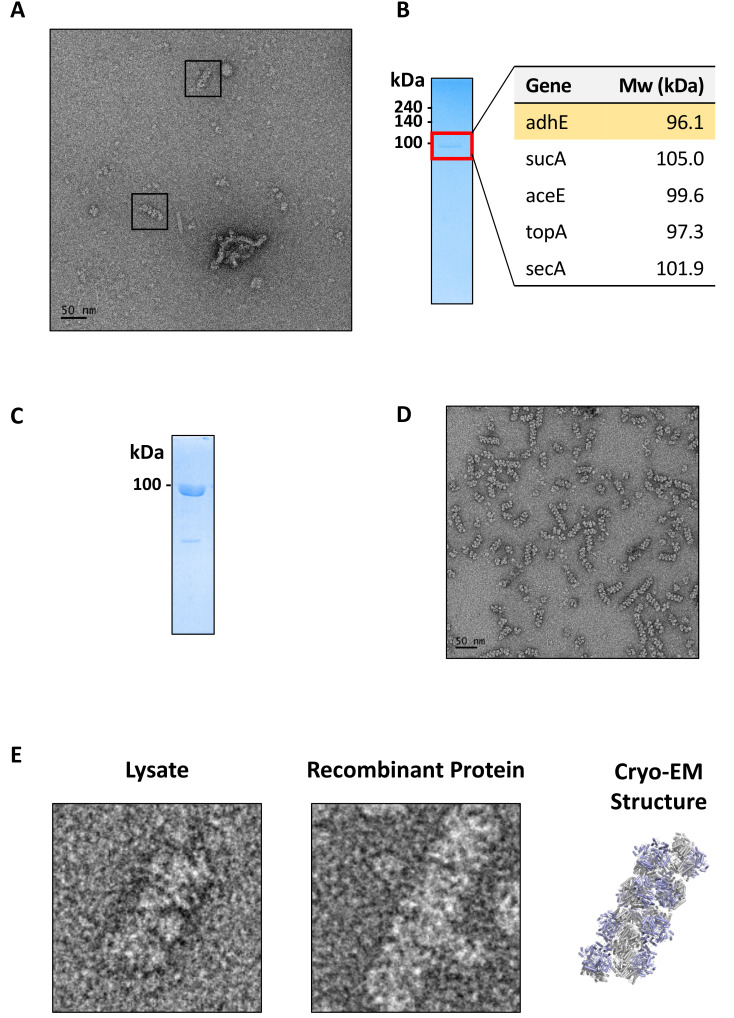
To determine the identity of the spiral architecture in Group 2, we performed targeted fractionations by following the presence of the spiral architecture examined by negative-stain EM (Fig. 4A). We prepared E. coli cell lysate and subjected it to anion exchange chromatography. We pooled the fractions containing the spiral architecture for further size-exclusion chromatography. One of the fractions from size-exclusion chromatography dominantly contained the spiral architecture. We concentrated the fraction and ran SDS-PAGE showing that there is a major band at approximately 100 kDa. We then identified the proteins by mass spectrometry (Fig. 4B). Among the five most proteins identified by mass spectrometry, we paid attention to AdhE, as it has been previously reported that AdhE forms oligomeric forms (Kessler et al., 1991). Therefore, to examine whether AdhE forms a spiral architecture, we cloned the adhE gene and purified AdhE to near homogeneity (Fig. 4C). Negative-stain EM analysis shows that recombinant AdhE indeed forms a spiral architecture, as observed in the cell lysate (Figs. 4D and 4E). Based on this finding, we reported the high-resolution cryo-EM structures of AdhE in two different conformations, showing that the formation of the spiral structure is important for AdhE enzymatic activity and that the structure dynamically changes, leading to a substrate channel (Kim et al., 2019; 2020).
Fig. 4. Identification and structural determination of aldehyde-alcohol dehydrogenase (AdhE).
(A) A negative-stain EM micrograph of the fraction containing the spiral architecture. Scale bar = 50 nm. (B) SDS-PAGE analysis of the fraction containing the spiral architecture (left panel). The top five abundant proteins from mass spectrometry identification are listed (right panel). (C) SDS-PAGE analysis of the purified recombinant E. coli AdhE protein. (D) A negative-stain EM micrograph of the recombinant E. coli AdhE protein showing spiral architectures. Scale bar = 50 nm. (E) Comparison among AdhE spiral architectures from negative-stain micrographs from cell lysates (left panel), recombinant protein (middle panel), and the cryo-EM structure (right panel; PDB ID: 6AHC).
EMPAS reveals the diverse and complex landscape of endogenous molecular architectures in various cells
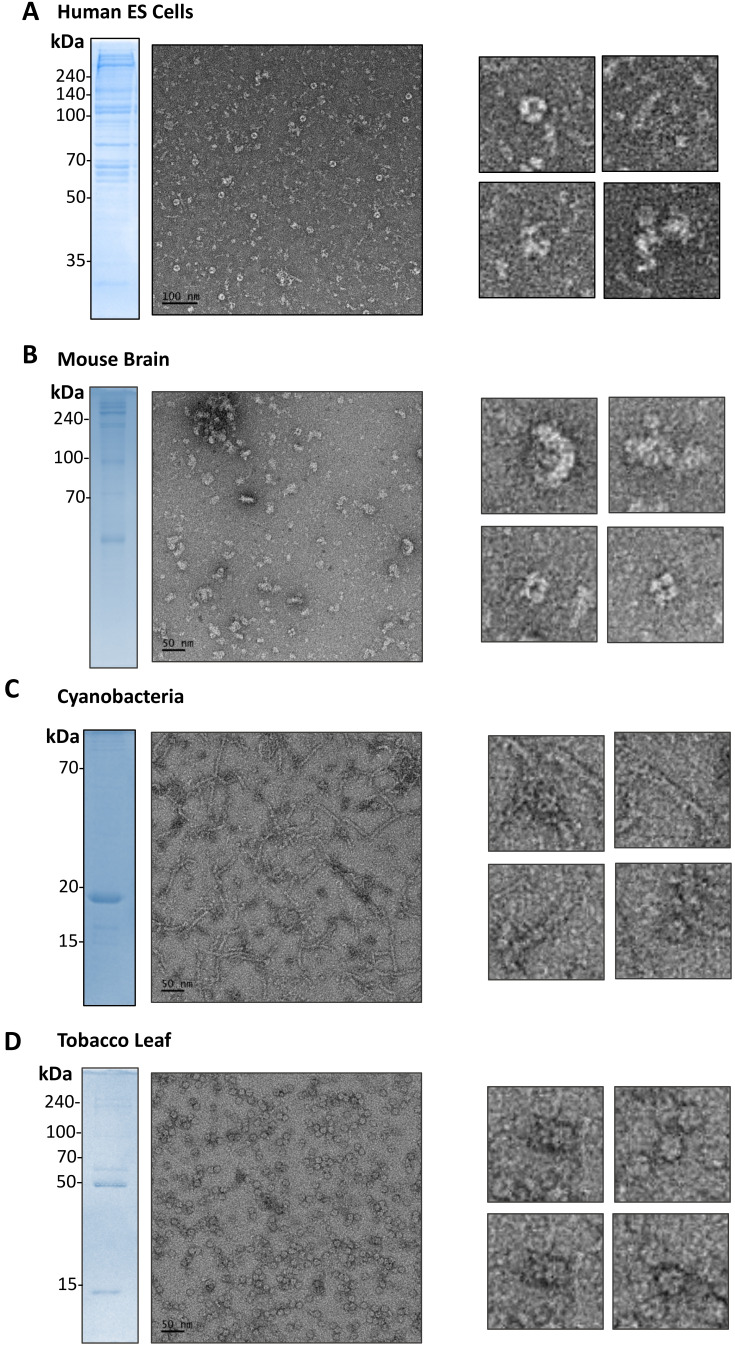
To test whether EMPAS can be applied to other cells to examine endogenous molecular architectures, we prepared lysates from different cells including hESCs, mouse brain cells, cyanobacteria and tobacco leaf cells. The lysates from the cells were prepared and fractionated using multiple chromatography, and high molecular weight fractions from size-exclusion chromatography were collected. We then examined the fractions using negative-stain EM. The micrograph from each sample shows various macromolecular architectures. Negative-stain EM micrographs from hESCs show molecular architectures exhibiting ring shapes of various sizes or cylindrical shapes (Fig. 5A). In addition, we were able to observe other molecular architectures with diverse geometries. The micrographs of mouse brains show very discrete and heterogeneous patterns, which might be lipid vesicles (Fig. 5B). In contrast to the presence of particles having diverse shapes in hESCs and mouse brains, the micrographs from cyanobacteria and tobacco leaves show dominantly tubular and spherical structures (Figs. 5C and 5D). The fractions from cyanobacteria and tobacco leaves contain rather homogeneous proteins. The spherical molecule from tobacco leaves is likely Rubisco, which is the most abundant protein in plants. Among these samples, we attempted to identify those molecular architectures from hESC cells and cyanobacteria by using mass spectrometry (Supplementary Fig. S1). However, the exact identity of these architectures needs further confirmation, as we have done with AdhE. Overall, our EMPAS analysis of several samples reveals the diverse landscape of endogenous molecular architectures, providing a potential tool to identify and investigate noble molecular architectures in cells.
Fig. 5. Representative micrographs of lysates from human stem cells, cyanobacteria, mouse brains and tobacco plant leaves showing the diverse and complex landscape of molecular architectures.
Representative negative-stain EM micrographs (left panels) from fractionated cell lysates of hESCs (A), mouse brains (B), cyanobacteria (C), and tobacco leaves (D) and SDS-PAGE analysis of the sample. A few interesting forms of molecular architectures are shown in the right panels. Scale bars = 50 nm (B-D) or 100 nm (A).
DISCUSSION
In this study, we have established a top-down approach methodology, EMPAS, which allows us to examine endogenous molecular architectures. Our approach to examine molecular architectures directly from cell lysates reveals the diverse endogenous molecular architectures of E. coli, hESCs, mouse brains, cyanobacteria and tobacco leaves, which could not be examined by conventional structural approaches investigating structures of reconstituted complexes with defined proteins.
EMPAS is composed of several steps. First, as direct visualization by EM is not possible due to the complexity of endogenous molecular complexes in cell lysates, the complexity is reduced by multiple fractionations followed by grouping the samples based on the compositions of proteins in fractions assessed by SDS-PAGE analysis. Among the molecular architectures visualized by negative-stain EM, a specific architecture of interest, a spiral structure in this case, is targeted. In this step, we should admit that a systematic approach is necessary to select targets from negative-stain EM micrographs, as we rely on manual inspection of every micrograph. The second-round fractionation is based on the presence of the targeted architecture. During the second-round fractionation, the targeted molecule will be enriched and identified by mass spectrometry. In our case, we targeted a spiral architecture and identified it as AdhE protein. We successfully applied EMPAS to endogenous protein complexes of E. coli to identify that AdhE forms a spiral structure, which led us to determine a high-resolution cryo-EM structure (Kim et al., 2019; 2020). This approach could be further applied to comparatively investigate the diversity and complexity of molecular architectures with different samples such as normal and cancerous cells, stem cells and differentiated cells and cells in different developmental stages. During our study, similar approaches have tried to identify large protein architectures and to determine the structures of protein architectures directly from the endogenous environment (Ho et al., 2020; Kastritis et al., 2017; Verbeke et al., 2018; Yi et al., 2019). In summary, our top-down methodology EMPAS could provide insights into endogenous protein complexes, which was not possible by conventional structural approaches.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
We thank the members of the Song lab as well as Yeon-Il Park, Unyoung Chae, and Gou Young Goh for providing samples. This work is partially supported by grants from KAIST (Grand Challenge 30 program) and from the National Research Foundation of Korea (2016K1A1A2912057). G.K. is a recipient of a Global Fellowship (NRF-2018H1A2A1061362).
Footnotes
AUTHOR CONTRIBUTIONS
G.K., S.J., and J.J.S. conceived idea. G.K., S.J., and E.L. performed the experiments. G.K., S.J., and J.J.S. wrote a manuscript. All examined data and J.J.S. supervised all process.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahn S., Shenoy S.K., Wei H., Lefkowitz R.J. Differential kinetic and spatial patterns of beta-arrestin and G protein-mediated ERK activation by the angiotensin II receptor. J. Biol. Chem. 2004;279:35518–35525. doi: 10.1074/jbc.M405878200. [DOI] [PubMed] [Google Scholar]
- Burley S.K. An overview of structural genomics. Nat. Struct. Biol. 2000;7(Suppl):932–934. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M., Kleywegt G.J., Markley J.L., Nakamura H., Velankar S. Protein Data Bank (PDB): the single global macromolecular structure archive. Methods Mol. Biol. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B.C., Gillet L.C., Rosenberger G., Röst H.L., Vichalkovski A., Gstaiger M., Aebersold R. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- Grabowski M., Niedzialkowska E., Zimmerman M.D., Minor W. The impact of structural genomics: the first quindecennial. J. Struct. Funct. Genomics. 2016;17:1–16. doi: 10.1007/s10969-016-9201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.D.J., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J., Cusick M.E., Roth F.P., et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Ho C.M., Li X., Lai M., Terwilliger T.C., Beck J.R., Wohlschlegel J., Goldberg D.E., Fitzpatrick A., Zhou Z.H. Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nat. Methods. 2020;17:79–85. doi: 10.1038/s41592-019-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Mason S., Barabási A., Oltvai Z. Lethality and centrality in protein networks. Nature. 2001;411:41. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Kastritis P.L., O'Reilly F.J., Bock T., Li Y., Rogon M.Z., Buczak K., Romanov N., Betts M.J., Bui K.H., Hagen W.J., et al. Capturing protein communities by structural proteomics in a thermophilic eukaryote. Mol. Syst. Biol. 2017;13:936. doi: 10.15252/msb.20167412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D., Leibrecht I., Knappe J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-A. [DOI] [PubMed] [Google Scholar]
- Kim G., Azmi L., Jang S., Jung T., Hebert H., Roe A.J., Byron O., Song J.J. Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity. Nat. Commun. 2019;10:4527. doi: 10.1038/s41467-019-12427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G., Yang J., Jang J., Choi J.S., Roe A.J., Byron O., Seok C., Song J.J. Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling. Commun. Biol. 2020;3:298. doi: 10.1038/s42003-020-1030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Nguyen T.H.D., Galej W.P., Bai X.C., Oubridge C., Newman A.J., Scheres S., Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye M.A., Noll B., Hausser A. Spatiotemporal control of intracellular membrane trafficking by Rho GTPases. Cells. 2019;8:1478. doi: 10.3390/cells8121478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M., Haynor D., Johnson J.M. Protein interaction networks. Expert Rev. Proteomics. 2004;1:239–249. doi: 10.1586/14789450.1.2.239. [DOI] [PubMed] [Google Scholar]
- Schaffer M., Pfeffer S., Mahamid J., Kleindiek S., Laugks T., Albert S., Engel B.D., Rummel A., Smith A.J., Baumeister W., et al. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat. Methods. 2019;16:757–762. doi: 10.1038/s41592-019-0497-5. [DOI] [PubMed] [Google Scholar]
- Scheres S.H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Verbeke E.J., Mallam A.L., Drew K., Marcotte E.M., Taylor D.W. Classification of single particles from human cell extract reveals distinct structures. Cell Rep. 2018;24:259–268.:e3. doi: 10.1016/j.celrep.2018.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Verbeke E.J., Chang Y., Dickinson D.J., Taylor D.W. Electron microscopy snapshots of single particles from single cells. J. Biol. Chem. 2019;294:1602–1608. doi: 10.1074/jbc.RA118.006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.