Here, Lasman et al. sought to understand the role of three homologous m6A-binding proteins (Ythdf1, Ythdf2, and Ythdf3) and systematically knocked out the Mettl3 writer, each of the Ythdf readers, and the three readers together (triple-KO). Their findings suggest a novel model for the Ythdf reader function, in which there is profound dosage-dependent redundancy when all three readers are equivalently coexpressed in the same cell types.
Keywords: RNA methylation, m6A, stem cells
Abstract
The N6-methyladenosine (m6A) modification is the most prevalent post-transcriptional mRNA modification, regulating mRNA decay and splicing. It plays a major role during normal development, differentiation, and disease progression. The modification is regulated by a set of writer, eraser, and reader proteins. The YTH domain family of proteins consists of three homologous m6A-binding proteins, Ythdf1, Ythdf2, and Ythdf3, which were suggested to have different cellular functions. However, their sequence similarity and their tendency to bind the same targets suggest that they may have overlapping roles. We systematically knocked out (KO) the Mettl3 writer, each of the Ythdf readers, and the three readers together (triple-KO). We then estimated the effect in vivo in mouse gametogenesis, postnatal viability, and in vitro in mouse embryonic stem cells (mESCs). In gametogenesis, Mettl3-KO severity is increased as the deletion occurs earlier in the process, and Ythdf2 has a dominant role that cannot be compensated by Ythdf1 or Ythdf3, due to differences in readers’ expression pattern across different cell types, both in quantity and in spatial location. Knocking out the three readers together and systematically testing viable offspring genotypes revealed a redundancy in the readers’ role during early development that is Ythdf1/2/3 gene dosage-dependent. Finally, in mESCs there is compensation between the three Ythdf reader proteins, since the resistance to differentiate and the significant effect on mRNA decay occur only in the triple-KO cells and not in the single KOs. Thus, we suggest a new model for the Ythdf readers function, in which there is profound dosage-dependent redundancy when all three readers are equivalently coexpressed in the same cell types.
RNA modifications are a layer of gene expression regulation, similar to DNA and protein modifications (Heck and Wilusz 2019). N6-methyladenosine, also known as m6A, is the most abundant mRNA modification (Heck and Wilusz 2019). It was first discovered in the 1970s, but major progress was done in recent years due to new approaches of mapping m6A sites (Dominissini et al. 2012; Meyer et al. 2012; Garcia-Campos et al. 2019). Its importance was shown in a wide range of organisms and processes, in yeast meiosis (Schwartz et al. 2013), sex determination in Drosophila (Kan et al. 2017), and in mammalian early development (Geula et al. 2015), neural development (Wang et al. 2018) and hematopoiesis (Lee et al. 2019).
m6A modification is regulated by writer, reader, and eraser proteins. Mettl3 forms a heterodimer with Mettl14 (Liu et al. 2014), and together with the supporting WTAP protein, catalyzes m6A with preference to 3′UTR, 5′UTR, long exons, and near stop codons (Heck and Wilusz 2019). Alkbh5 is a well-established eraser enzyme found in vertebrates (Jia et al. 2011; Zheng et al. 2013).
Multiple proteins were identified as m6A readers, with the YTH domain-containing proteins (Ythdf and Ythdc) stand out among them. Previous studies linked these proteins to specific functions in RNA metabolism; YTHDF1 and YTHDF3, but not YTHDF2, were reported to promote translation by recruiting translation initiation factors in HeLa cells (Wang et al. 2015; Li et al. 2017; Shi et al. 2017). YTHDF2, but not YTHDF1, was linked to mRNA degradation, by recruiting the CCR4–NOT deadenylase complex (Wang et al. 2014; Du et al. 2016).
However, it is not fully clear whether indeed each one of the Ythdf readers fulfills a distinct role. Their sequence similarity, and the fact that they are all localized in the cytoplasm (Wang et al. 2014, 2015; Shi et al. 2017) and share many of their targets (Patil et al. 2016, 2018; Li et al. 2017) indicate, at least, partial redundancy. However, knockout (KO) of Ythdf2 alone is sufficient to stop proper oocyte maturation (Ivanova et al. 2017), and a single KO of Ythdf1 or Ythdf2 causes neural defects (Li et al. 2018; Shi et al. 2018), suggesting that in certain systems, Ythdf readers cannot compensate for each other. However, this could be a result of differences in expression levels in the different tissues that has not been fully addressed until now. Comprehensive research into the redundancy between the three Ythdf proteins has not been conducted, and the effect of knocking out all three readers in vivo and in vitro in the same defined systems side by side, has not been described so far.
In this study, we used KOs of Mettl3, Ythdf1, Ythdf2, and Ythdf3, and show that both Mettl3 and Ythdf2 are essential for proper gametogenesis, and that mice lacking these proteins are either hypofertile or sterile. The severity of the phenotype is increased when the Mettl3 deletion is done earlier in the developmental process. We found that the Ythdf readers have different expression patterns during gametogenesis, which might explain their lack of compensation during this process. In addition, only in triple-KO embryos (but not single-KO embryos), we found impaired development as early as E7.5, and early embryonic lethality as previously described for Mettl3-KO embryos. By evaluating offspring viability of compound Ythdf1/2/3 mutants, we found that in early development there is compensation between the readers, which is dosage-dependent; that is, Ythdf2 hetrozygouse mice need to have at least one functional copy of another Ythdf1 or Ythdf3 readers to escape mortality. Furthermore, we used mouse embryonic stem cells (mESCs), which naturally express all three Ythdf proteins, to analyze the effect of ablating individual or multiple Ythdf readers. We found that only triple-KO mESCs are not able to differentiate properly, and present a prolonged mRNA half-life, similar to the effect shown in Mettl3-KO, while no significant effect is seen in the single-KOs or in rescue cell lines with any of the three known Ythdf reader proteins. This suggests that also in mouse ESCs, there is a redundancy between Ythdf readers, which enables adequate functional compensation.
Results
Mettl3 plays an essential role in oogenesis and spermatogenesis
Ythdf1, Ythdf2, and Ythdf3 share high-protein sequence similarity (67%–70%) (Supplemental Fig. S1A). In addition, they have coevolved during evolution. While Drosophila melanogaster has one copy of Ythdf protein (named Ythdf), vertebrates have three functional proteins (Fig. 1A), probably generated following duplication events (Pervaiz et al. 2019). This has provoked the interest to experimentally define the extent of their ability to compensate for each other by comparing their depletion, individually or collectively, with that of depleting Mettl3. We started by systematically testing the three readers in a specific system in vivo, focusing on spermatogenesis and oogenesis. m6A writers Mettl3 and Mettl14 and m6A eraser ALKBH5 were found to be essential for proper gametogenesis in mice. Their KO typically leads to defective maturation of sperm or ova, and hypofertility (Zheng et al. 2013; Lin et al. 2017; Xu et al. 2017; Tang et al. 2018; Lasman et al. 2020).
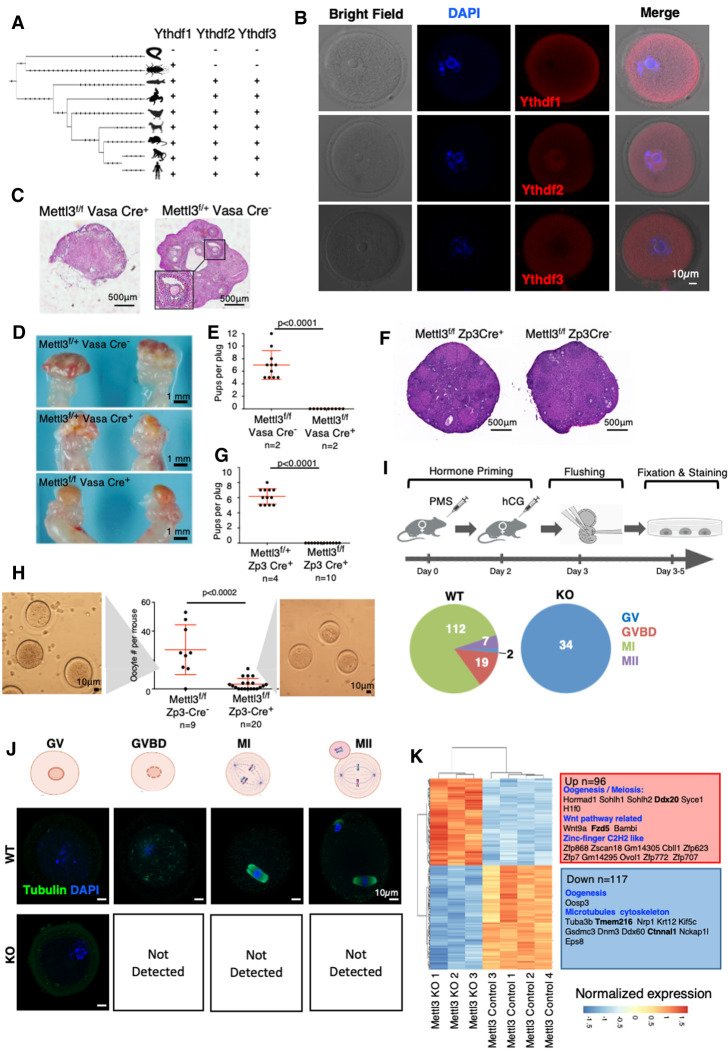
Figure 1.
Mettl3 is essential for female mice fertility. (A). Phylogenetic tree of the protein sequences of Ythdf1, Ythdf2 and Ythdf3, based on UCSC database. The three readers appeared together in vertebrates, possibly after whole genome duplication. (B) Immunostaining of Ythdf1, Ythdf2, and Ythdf3 in ICR wild-type (WT) oocytes. (BF) Bright field. (C) H&E staining of ovaries, showing normal follicle structure in Mettl3f/f VasaCre− control, and a severe abnormality in Mettl3f/f VasaCre+ females. (D) Gross morphology of Mettl3f/fVasaCre+ and control female ovaries. Cre+ females show a smooth shape that lacks the typical follicular morphology. (E) Number of pups per plug produced by mating Mettl3f/f Vasa Cre+ females, compared with Mettl3f/f Vasa Cre− control females. The fathers in both cases were WT. A significant difference between Cre+ and Cre− female fertility is observed (P < 0.0001, Mann–Whitney test). (F) H&E staining of ovaries, showing normal morphology both in Mettl3f/f Zp3 Cre− and Mettl3f/f Zp3 Cre+ ovaries. (G) Number of pups per plug produced by mating a Mettl3f/f Zp3 Cre+ female, compared with a Mettl3f/+ Zp3 Cre+ control female. The fathers in both cases were WT. A significant difference between f/f and f/+ female fertility is observed (P < 0.0001, Mann-Whitney test). (H) Number of oocytes per mouse flushed from Mettl3f/f Zp3 Cre+ females, compared with Mettl3f/f Zp3 Cre− control females. A significant difference between the number of oocytes of Mettl3f/f ZP3 Cre+ and Mettl3f/f ZP3 Cre− is observed (P < 0.0002, Mann–Whitney test). (I, top) Experimental design – Mett3f/f Zp3 Cre+ and Cre− as control underwent hormone priming, oocyte flushing, fixation, and staining for tubulin. (Bottom) Number of oocytes observed in the different stages of meiosis. In the control, most of the oocytes were in MI stage, in KO (Cre+) all of the observed oocytes were in the GV state. (J) Staining examples of oocytes in the different stages of meiosis as observed in KO (Cre+) and control (Cre−). (K) Differentially expressed genes between Mettl3f/f Zp3 Cre− (control) and Mettl3f/f Zp3 Cre+ (KO) oocytes, along with selected enriched categories. m6A methylated genes appear in bold. Ninety-six genes are up-regulated in the KO, and 117 are down-regulated in the KO.
As for m6A readers, both Ythdc1 and Ythdc2 have an essential role in gametogenesis. Their KO in spermatogenesis or oogenesis leads to a severe hypofertility phenotype (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018; Kasowitz et al. 2018). All Ythdf readers are expressed in developing oocytes (Fig. 1B). Knocking out Ythdf2 leads to normal ovulation but an inability to down-regulate maternal mRNA. Thus, Ythdf2-KO females are sterile (Ivanova et al. 2017). In contrast, Ythdf2-KO males show normal seminiferous tubule histology (Ivanova et al. 2017). Depletion of Ythdf2 in mouse spermatogonia leads to defective cell morphology and decreases cell proliferation (Huang et al. 2020). However, further examination of all Ythdf readers in spermatogenesis is still required. We assessed the importance of m6A in gametogenesis, both in males and females, using conditional KOs of the m6A writer Mettl3 (Supplemental Fig. S2). Mettl3flox/flox mice were crossed with mice carrying one of the following Cre constructs: Zp3-Cre, which is activated during oogenesis; Stra8-Cre and Prm1-Cre, which are activated during spermatogenesis; and Vasa-Cre, which is activated in the early stages of both. Thus, we could test the effect of Mettl3-KO systematically in different time points during gametogenesis, in both males and females.
Mettl3f/fVasaCre+ KO had a major effect on oocyte development. Dissection of Mettl3f/fVasaCre+ female mice showed abnormal ovary morphology (Fig. 1C,D), and the mice were sterile (Fig. 1E). Zp3 is expressed during a later stage of the oocyte maturation, prior to the completion of the first meiosis (Gao et al. 2017). Accordingly, Mettl3f/f Zp3Cre+ female mice showed a normal ovary morphology (Fig. 1F). However, the female mice were sterile (Fig. 1G). Flushing oocytes from the oviduct revealed an overall significant low number of oocytes (P-value < 0.002) (Fig. 1H). All the flushed oocytes of the KO were stalled at the germinal vesicle (GV) stage and did not reach the two-cell stage upon fertilization attempts (Fig. 1I; Supplemental Fig. S3A), meaning that they have not completed the first meiosis. Indeed, immunostaining of tubulin in KO and WT oocytes, showed that KO oocytes were arrested in the GV stage, and did not proceed for GV breakdown and completion of the first meiosis (Fig. 1J). The transcriptional profile of Mettl3-cKO and control oocytes revealed a major change in transcription (Supplemental Fig. S3B), including aberrant expression of genes related to oocyte development (Fig. 1K; Supplemental Fig. S3C; Supplemental Table S1).
Next, we tested the role of Mettl3 in spermatogenesis. Mettl3f/fVasaCre+ male mice, in which the KO was activated in primordial germ cells, showed a massive reduction in the testis volume (Supplemental Fig. S4A), severe degenerative defects (Supplemental Fig. S4B), and sterility (Supplemental Fig. S4C), as was reported elsewhere (Lin et al. 2017; Xu et al. 2017). Similarly, a dissection of Mettl3f/fStra8Cre+ male mice, in which the KO was activated during early-stage spermatogonia, showed a significantly reduced testis volume (Supplemental Fig. S4D), mild degenerative changes in seminiferous tubules (Supplemental Fig. S4E), and ∼75% reduction in sperm quantity observed in the cauda epididymis (Supplemental Fig. S4E). Mettl3f/fStra8Cre+ mice showed significant hypofertility compared with their counterpart control (Supplemental Fig. S4F), similar to what was previously reported (Lin et al. 2017). Interestingly, Mettl3f/fPrm1Cre+ male mice, in which the KO was activated in the spermatids (Supplemental Fig. S4G), showed normal fertility (Supplemental Fig. S4H) and typical seminiferous tubules morphology (Supplemental Fig. S4I), indicating that m6A machinery becomes dispensable once maturity is established in sperm as marked by protamine induction stage. In summary, our genetic dissection of Mettl3's role during gametogenesis shows the pivotal role of m6A modifications in both oogenesis and spermatogenesis. The severity of the phenotype was dependent on the stage in which Mettl3 was depleted.
Ythdf2 is the only Ythdf reader that is essential for gametogenesis, and has a different expression pattern than Ythdf1 and Ythdf3
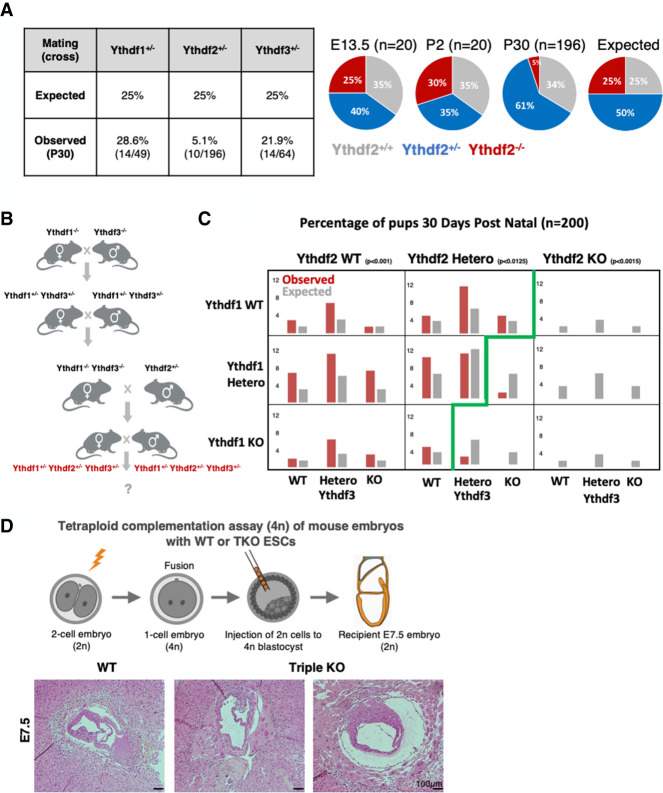
We next moved to test the effect of Ythdf1/2/3 KO, which might have a milder effect on the process. We knocked out each of the three readers using CRISPR–Cas9 (Supplemental Fig. S1B). Heterozygous mice were further crossed to give full KO for each of the Ythdf proteins. The mice that were born were validated by genotyping (Supplemental Fig. S1C). Ythdf1−/− and Ythdf3−/− mice were viable, were born in the expected Mendelian ratio (Fig. 3A, below), and showed no apparent gross defects. However, ∼80% of Ythdf2−/− pups died shortly after birth, leading to a sub-Mendelian ratio 30 d after birth (Fig. 3A, below).
Figure 3.
Characterization of compound Ythdf1/2/3 KO mice. (A, left) Statistics of KO offspring received from crossing of Heterozygous mice from each of the indicated strains (Ythdf1+/−, Ythdf2+/−, and Ythdf3+/−). (Right) Distribution of Ythdf2 WT, HET, and KO offspring at days E13.5, 2 d postnatal (DPN), and 30 DPN (compared with expected ratios). (B) Crossing strategy for generating triple-heterozygous mice that were further crossed, and their offspring statistics are presented in C. (C) Percentage of genotypes received by crossing triple-heterozygous mice, out of 200 pups tested 30 DPN. (Red) Observed percentage; (gray) expected under null assumption. No pups with Ythdf2-KO genotype survived 30 DPN. In Ythdf2+/− genotype, pups with KO in either Ythdf1 or Ythdf3 were born in a sub-Mendelian ratio. χ2 test P-values are indicated. (D) Using tetraploid complementation assay, triple-KO embryos were generated and examined on E7.5. (Bottom) H&E staining showing aberrant morphology of triple-KO E7.5 embryos, compared with WT control.
Ythdf1-KO and Ythdf3-KO mice were fertile as their control counterparts (Supplemental Fig. S5A–D) and did not show any histological defect in their reproductive organs (Supplemental Fig. S5E,F). As for Ythdf2, the few viable KO mice that survived were analyzed. While Ythdf2-KO female mice showed normal oocyte morphology (Supplemental Fig. S5G) and could be fertilized, they were still sterile (Supplemental Fig. S5H), with a defect that occurs at a later stage than that observed in Mettl3f/fZp3Cre+ oocytes when all m6A machinery is neutralized (Fig. 1I). Consistently, a recent work showed that Ythdf2-KO oocytes could be fertilized but do not develop beyond the eight-cell stage (Ivanova et al. 2017). Next, the oocytes were flushed after hormone priming and measured for RNA levels using SMART-seq. Although the morphology was indistinguishable from WT, on the molecular level, the cells of Ythdf2-KO were already distinctly clustered compared with WT (Supplemental Fig. S5I). Among the 311 genes that were down-regulated in the KO (Supplemental Fig. S5J; Supplemental Table S1), 72 are related to extracellular matrix (P < 1.35 × 10−06), which is crucial for oocyte competence. Interestingly, in sperm also, m6A regulation of metallopeptidase is mediated by Ythdf2 (Huang et al. 2020).
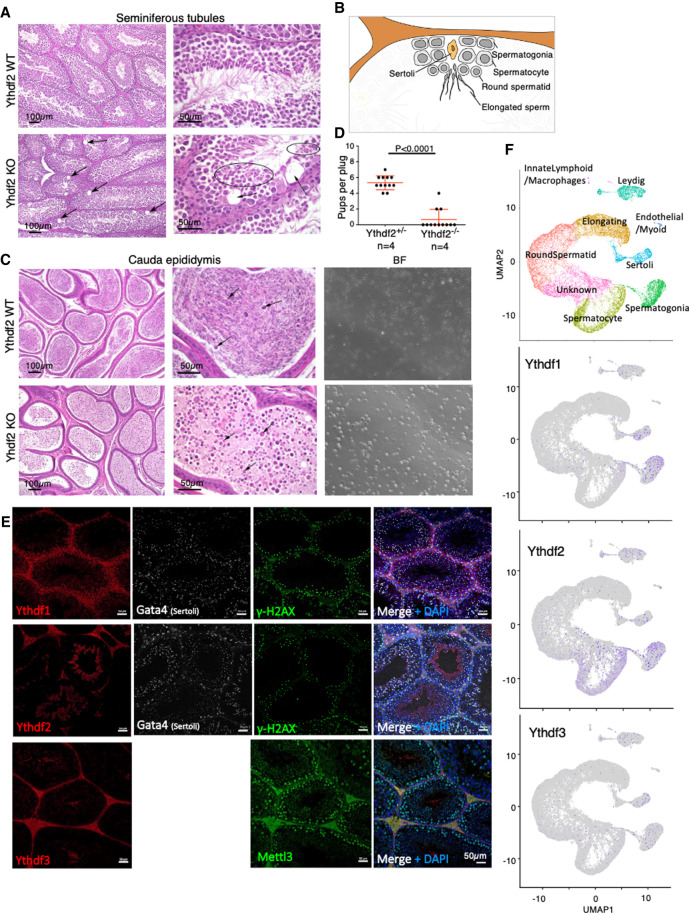
Ythdf2-KO male mice showed mild degenerative changes in the seminiferous tubules, including large scattered vacuoles in Sertoli cells (Fig. 2A,B), and severe loss of sperm in the cauda epididymis (Fig. 2C). Accordingly, these males were hypofertile (Fig. 2D). Measuring expression in WT and KO round spermatids, we observed changes in expression in 301 genes (Supplemental Fig. S5K; Supplemental Table S1), many of them associated with cytoskeleton, microtubules, and cilium functions, possibly explaining the impaired sperm maturation. In addition, some metallopeptidases were up-regulated in the KO (e.g. Adam4, Adamts3, and Cpxm1).
Figure 2.
Ythdf2 is essential for male mice fertility. (A) H&E staining showing mild degenerative changes, including scattered vacuoles marked by arrows in the seminiferous tubules in Ythdf2-KO males, compared with WT control. (B) Schematic representation of spermatogenesis inside seminiferous tubules. Differentiation is progressing from spermatogonia at the periphery, via spermatocytes and round spermatids, and ends with elongated mature sperm in the center of the tubule. (C, left) H&E staining of the cauda epididymis, showing severe loss of sperm in Ythdf2-KO compared with control. (Right) Bright field of sperm extracted from the cauda epididymis of KO and control, showing a severe reduction in normal sperm quantity in the KO sample, compared with control. (D) Number of pups per plug produced by mating Ythdf2−/− males, compared with Ythdf2+/− control males. The mothers in both cases were WT. A significant difference between the fertility of KO and heterozygous males is observed (P < 0.0001, Mann–Whitney test). (E) Immunostaining of Ythdf1, Ythdf2, Ythdf3, and Mettl3 in seminiferous tubules, showing that each of the Ythdf proteins is expressed at different stages of spermatogenesis. Costaining of Gata4 typically marks Sertoli cells, costaining of γ-H2AX marks different cells during early spermatogenesis. (F) Dimension reduction representation of single-cell RNA-seq (UMAP) measured in adult mouse testis, showing mild expression of Ythdf1 and Ythdf3 in spermatogonia and in Sertoli cells, and more substantial expression of Ythdf2 in spermatogonia and in spermatocytes.
Although Ythdf1 and Ythdf3 share high sequence homology with Ythdf2, they cannot fully compensate for its depletion during gametogenesis. One possible explanation for this observation is that the proteins are not expressed in the same spatial or temporal space during the process. Indeed, immunostaining of seminiferous tubules (Fig. 2E) showed that Ythdf1 is expressed in spermatogonia, while Ythdf2 is expressed in a later stage. Furthermore, analysis of single-cell RNA-seq data from adult mouse testis (Green et al. 2018) showed that Ythdf1 and, to a lesser extent, Ythdf3 are mainly expressed in spermatogonia, while Ythdf2 is expressed both in spermatogonia and in spermatocytes (Fig. 2F). Similarly, immunostaining of GV oocytes showed a different expression pattern for Ythdf2, as it is the only Ythdf reader that is located both in the cytoplasm and nucleus (Fig. 1B; Supplemental Fig. S6). Thus, the different expression pattern of the Ythdf readers in gametogenesis likely explains the lack of compensation for Ythdf2 depletion in this context.
Ythdf readers compensate for one another in a dosage-dependent manner during embryonic development and gestation
Next, we tried to generate Ythdf triple-KO mice for a more comprehensive understanding of the readers’ roles in vivo at the organismal systemic level. Ythdf1−/− and Ythdf3−/− mice were crossed, and the double-heterozygote offspring were further crossed with Ythdf2+/− mice to generate triple-heterozygote mice to all three readers (Ythdf1+/− Ythdf2+/−Ythdf3+/− or “triple-HET”) (Fig. 3B). Triple-HET mice were crossed, and all of their offspring (n = 200) were genotyped on postnatal day 30 (Fig. 3C). The ratio of offspring with Ythdf2-WT genotype was as or above the expected Mendelian ratio, while no offspring with Ythdf2-KO were detected (as was observed previously in pure background strains) (Ivanova et al. 2017). Interestingly, even when the mice were only heterozygous for Ythdf2 (Ythdf2+/−), the Mendelian ratio of offspring with another missing reader was below the expected ratio (Ythdf1−/−2+/−3+/−, threefold below expected; Ythdf1+/− 2+/−3−/−, fourfold below expected; P < 0.012). Moreover, we could not detect any offspring that were Ythdf2+/− and null in the two other readers (Ythdf1−/−Ythdf2+/− Ythdf3−/−). These results suggest that lack of Ythdf1 or Ythdf3 can be compensated for by the two other readers. However, the lack of Ythdf2 cannot be compensated by Ythdf1 or Ythdf3. In addition, the fact that partial expression of Ythdf2 in the heterozygous lineage requires the expression of at least one other Ythdf protein, suggests that the function of the readers is dosage-dependent, and that cumulative expression of Ythdf readers beyond a certain threshold is required for embryonic viability.
Further supporting functional redundancy between Ythdf reader proteins is the observation that while Mettl3−/− embryos die at E7.5 at the postimplantation stage, single-KO mice for Ythdf1/2/3 developed until later developmental stages (Fig. 3A–C). However, by using tetraploid (4N) complementation assay, triple-KO embryos did not develop after E7.5 (Fig. 3D) and recapitulated the previously described Mettl3-KO embryo lethality phenotype (Geula et al. 2015).
Ythdf triple-KO mESCs recapitulate the phonotypes observed in Mettl3-KO ESCs
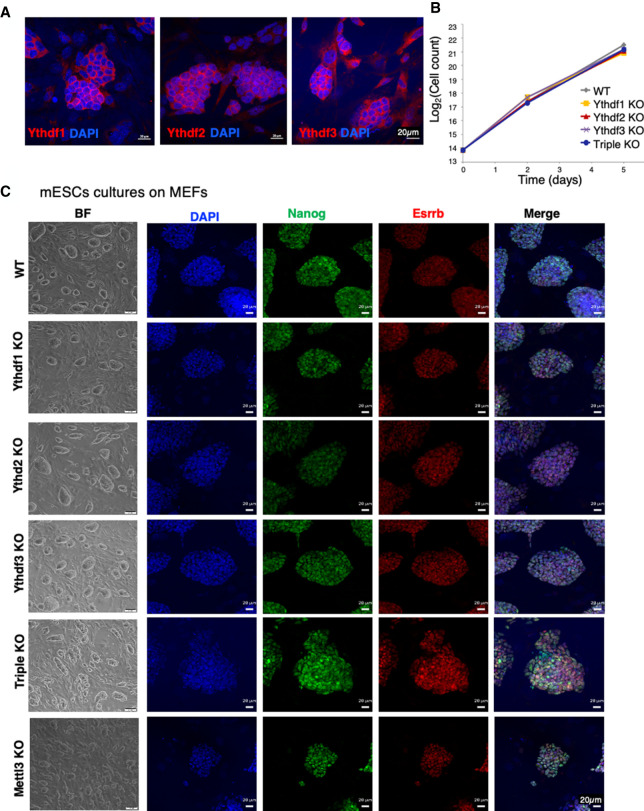
We found that the Ythdf proteins can compensate for one another in vivo when expressed in the same cells, and that this compensation is dosage-dependent. Next, we wanted to better understand the molecular mechanism through which the different Ythdfs process mRNA molecules and thus affect cell viability and differentiation potential. We reasoned that mESCs would be a good model for studying the molecular role of the readers in vitro, since this is a system in which we can systematically perturb the cells and test the stem cell activity outcomes (self-renewal and differentiation). In addition, in contrast to gametogenesis, all of the three Ythdf readers are expressed in mouse ESCs (Fig. 4A; Supplemental Fig. S7), thus enabling us to test different compensation mechanisms. We stained for the readers in mouse ESCs, and indeed all were found to be expressed in the cytosolic compartment of the cells (Fig. 4A). Next, we knocked out each of the Ythdfs in mouse ESCs using the CRISPR/Cas9 strategy (Supplemental Fig. S7A). In addition, we generated a triple-KO line, Ythdf1/2/3 KO, using sequential CRISPR KO. All KO cell lines were validated by immunostaining, AP staining, Western blot, DNA sequencing, RNA sequencing, and ribosomal footprint (Supplemental Figs. S7–S9).
Figure 4.
Ythdf1, Ythdf2, and Ythdf3 are redundant in mouse naïve ESC maintenance. (A) Immunostaining of Ythdf1, Ythdf2, and Ythdf3 in WT mESCs, showing a protein expression in the cytosolic compartment of the cell. (B) Cell growth curve of all KO lines and WT control. Cells were grown on mouse feeders, in serum/LIF conditions. (C) Bright field and immunostaining of Nanog (green), Esrrb (red), and DAPI (blue) in KO cells (single, triple, and Mettl3) and WT control, showing that all cell lines express Nanog and Esrrb.
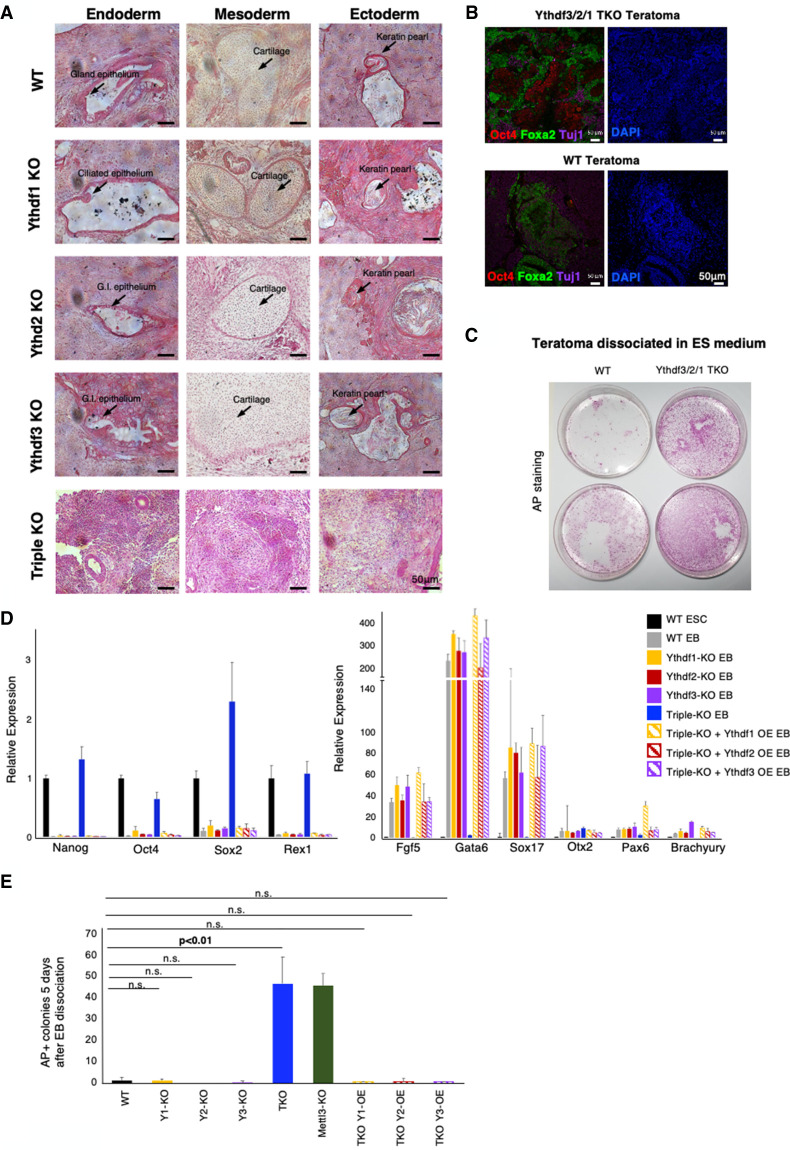
All ESC lines were viable and showed similar self-renewal ability as WT cells in mESC naïve growing conditions (Fig. 4B). All cell lines adequately expressed pluripotent markers (Fig. 4C; Supplemental Fig. S8). We next wanted to test their ability to undergo differentiation using in vivo and in vitro assays. First, we tested their ability to generate teratomas upon injection to immune-deficient mice. While WT and single-KO teratomas generated differentiated structures containing the three germ layers and stained for developmental markers such as Foxa2 and Tuj1 (Fig. 5A,B), triple-KO teratomas were poorly differentiated, and broadly stained for Oct4, a pluripotent marker, indicating their poor ability to differentiate.
Figure 5.
Ythdf1, Ythdf2, and Ythdf3 are functionally redundant in ESC differentiation. (A) Teratomas generated from KO cell line and from WT control. Single-KO teratomas show all germ layers, while triple-KO (TKO) teratomas are poorly differentiated. Selected differentiated structures are marked by arrows. (B) Immunostaining of triple-KO and WT control to Oct4 (red), Foxa2 (green), Tuj1(purple) and DAPI (blue). Triple-KO teratomas contain patches of Oct4 staining, unlike WT teratomas. (C) Alkaline phosphatase (AP) staining of disassociated teratomas from triple-KO and WT control samples, showing a greater AP staining in the Ythdf triple-KO. (D) RT-PCR of pluripotency genes (left) and differentiation genes (right), measured in WT mESCs, and mEBs from the following cell lines: WT control, Ythdf single-KO, Ythdf triple-KO, and Ythdf triple-KO + overexpression (OE) of Ythdf1/2/3 (rescued cell lines). Triple-KO EBs express pluripotent markers and repress differentiation markers similarly to mESCs. Rescued EBs express differentiation markers, similarly to single-KO EBs. (E) EBs were induced for 7 d from the indicated cell lines, and dissociated into single cells at day 7 and comparable amounts of cells were replated in ES medium. Undifferentiated AP+ colonies number per 1000 plated cells was evaluated 5 d later.
To further examine the differentiation ability of the cells, the teratomas were disaggregated and cells were replated in mouse ES medium for 5 d. Triple-KO cells generated significantly more pluripotent colonies, as shown by alkaline phosphatase staining (Fig. 5C). Next, embryoid bodies (EBs) were generated from all our cell lines (Supplemental Fig. S9A), followed by RNA extraction and RT-PCR. Once again, differentiation markers were not induced, and pluripotent markers remained highly expressed in the triple-KO EBs (Fig. 5D compared with WT and single-KO cell lines), indicating their poor differentiation tendency. This phenotype is highly similar to the “hyperpluripotency” phenotype that we observed previously in Mettl3-KO cells (Geula et al. 2015). Dissociating triple-KO day 7 EBs and replating the cells in ES medium yielded significantly higher undifferentiated AP+ ESC colonies when compared with WT EBs, while single-KO Ythdf EB-derived cells behaved like WT controls (Fig. 5E). Importantly, to test the specificity of the results, we attempted to rescue the phenotype by overexpressing each of the Ythdf readers in the triple-KO mESC line (Supplemental Fig. S9B). The EBs that were generated from the rescued cell lines, differentiated similarly to WT and single-KO EBs (Fig. 5D,E), showing that the expression of any of the three Ythdf readers alone is sufficient for the proper differentiation of mESCs and rescue of the triple-KO phenotype in ESCs.
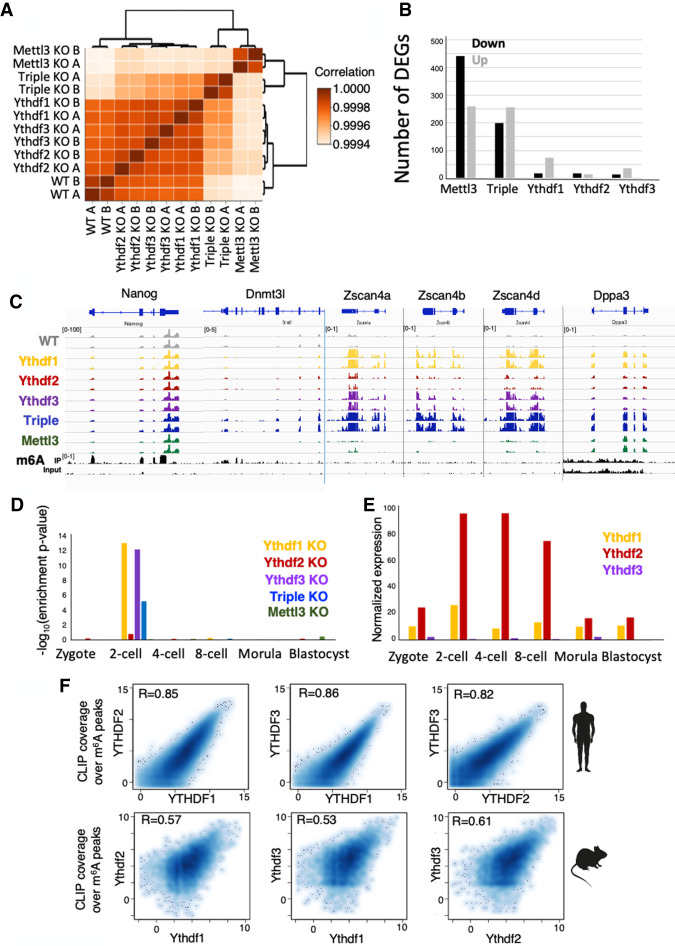
To dissect the molecular profile of single-KOs and the triple-KO, transcription profiles were measured using RNA-seq from each of the cell lines (Ythdf1−/−, Ythdf2−/−, Ythdf3−/−, and Ythdf1/2/3−/−). In addition, we had a WT control and a positive control consisting of Mettl3-KO cells, which lack m6A methylation and were previously shown to be hyperpluripotent (Geula et al. 2015). Clustering the samples based on their transcriptional profile showed that while single reader KO samples cluster together with WT samples, triple-KO samples cluster more closely to Mettl3−/− samples (Fig. 6A; Supplemental Fig. S10A), suggesting that single-KOs do not have a dramatic effect on the transcriptional profile of the cells, consistent with the existence of significant redundancy in this context.
Figure 6.
Ythdf triple-KO has a dramatic effect on gene expression. (A) Hierarchical clustering of samples based on Pearson correlation, showing that single-KO samples are highly similar to WT. (B) Number of differentially expressed genes in each of the KO cell lines, compared with WT. (Black) Down-regulated genes, (gray) up-regulated genes. (C) RNA-seq and m6A methylation landscape of selected genes. Normalized coverage is presented. Only Nanog and Dnmt3l are m6A methylated. Dnmt3l, Zscan4a, Zscan4b, Zscan4d, and Dppa3 are overexpressed in triple-KO. (D) Enrichment of up-regulated genes in each category, to early embryo genes (Gao et al. 2017). Genes that are up-regulated in KO of Ythdf1 and Ythdf3 are specifically enriched for two-cell embryo genes. (E) Normalized expression of Ythdf1, Ythdf2, and Ythdf3, as measured in early mouse embryo (Gao et al. 2017). (F) CLIP coverage over m6A peaks of Ythdf readers as measured in humans (n = 41,885) (Patil et al. 2016), and in mice (n = 9861), showing high correlation between coverage by different Ythdf readers.
When we analyzed the number of differentially expressed genes in each of the cell lines (Fig. 6B; Supplemental Table S2), we could see that the few genes that were up-regulated in the single-KOs (77 In Ythdf1, 16 in Ythdf2, and 37 in Ythdf3), greatly overlapped with the genes that were up-regulated in the triple KO and to a lesser extent in the Mettl3-KO (Supplemental Fig. S10B). Interestingly, several of the genes that were up-regulated in Ythdf1-KO, Ythdf3-KO, and triple KO, but not in Ythdf2-KO, were significantly enriched for two-cell stage embryo genes (genes that are expressed after the first division of the zygote), such as Zscan4a,c,d,f, Usp17a,b,c,e, Zfp352, Gm20767, and Tcstv1 (Fig. 6C–E; Supplemental Fig. S10B; Storm et al. 2009). This suggests that even though most of Ythdfs’ effects on expression are redundant, some Ythdf readers cannot fully and identically compensate for the absence of the others in all settings.
To further investigate the role of the Ythdfs, their RNA-binding target profile was measured in three different single-Ythdf FLAG-tagged mESC lines using the eCLIP method (Van Nostrand et al. 2016). The binding peaks had a high degree of overlap with previously published binding data, and with known m6A sites, and harbored RRACH motif in 18%–25% of the targets (Supplemental Fig. S11). Reassuringly, binding levels of Ythdf1,2&3, measured over known m6A sites, were highly correlated, both in previously published human data (Patil et al. 2016) and in mice (Fig. 6F).
When we analyzed the common peaks between the readers, we found that the peaks bound by Ythdf1 and Ythdf3 highly overlap with the peaks bound by Ythdf2 (72% and 49%, respectively) (Supplemental Fig. S11E; Supplemental Table S3), indicating again a possible redundancy between the readers’ binding sites. However, targets of Ythdf1 and Ythdf3 were not enriched for two-cell genes, which are typically not expressed in mESCs, but rather for blastocyte genes that are expressed in mESCs (Fig. 11F). To investigate the roles of Ythdf1&3 in the context of two-cell genes, binding profiling needs to be done in two-cell stage embryos, which is not feasible with the current technology (Van Nostrand et al. 2016).
Increase in m6A methylated mRNA half-life is achieved in ESCs only upon ablation of all Ythdf reader proteins
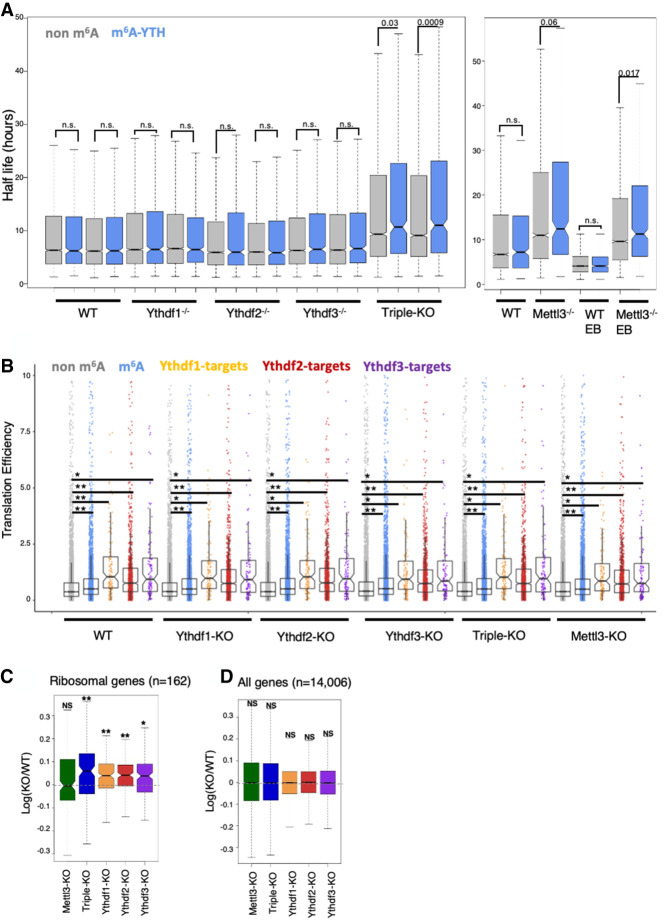
Previous studies proposed that m6A has a role in mRNA degradation (Geula et al. 2015; Du et al. 2016). Specifically, Wang et al. (2014) suggested that Ythdf2 binds methylated transcripts and directs them to mRNA decay sites. We therefore examined the decay rate in mouse ESCs, in each of the single-KO and triple-KO cell lines. We treated the cells with actinomycin-D, and harvested RNA at three time points (t = 0, 4 h, 8 h, with duplicates of 0 and 8). We estimated transcription levels using 3′ poly-A RNA-seq (Geula et al. 2015), and calculated the mRNA half-life (Supplemental Table S4). Overall, half-life is mildly shorter as the number of m6A in the transcript is higher (Supplemental Fig. S12). In single-KO cells, including Ythdf2-KO, m6A methylated mRNA was degraded at a similar rate to nonmethylated mRNA (Fig. 7A). Only in the triple-KO cells we observed a significant increase in the half-life of m6A methylated mRNA, compared with nonmethylated mRNA, similar to what was observed in the Mettl3-KO (Fig. 7A). The fact that in single-KOs there was no significant effect on degradation, suggests that all of the readers have similar roles in mRNA degradation and can compensate for each other. A previous study (Du et al. 2016) showed that Ythdf2 recruits the CCR4–NOT complex to mediate accelerated deadenylation and decay. Ythdf1 and Ythdf3 were also shown to promote deadenylation (Du et al. 2016), further supporting the hypothesis that the three readers contribute to mRNA decay, and may compensate in case of a partial loss.
Figure 7.
Loss of all Ythdf reader proteins is required to perturb mRNA degradation in mESCs. (A) mRNA half-life calculated in non-m6A genes (gray) and m6A genes (blue) in each of the KO cell lines and WT control. Only in the triple-KO and Mettl3-KO are there significant differences between the half-life of m6A and non-m6A genes. (B) Distribution of translation efficiencies of gene groups in the different samples. Graph shows that m6A methylated genes and Ythdf targets are translated in a higher efficiency, consistently across all samples (including WT cells), compared with nonmethylated genes. (**) P < 1×1015; (*) P < 1×106, Kolmogorov–Smirnov test. Mettl3 and Ythdf KO hardly affect translation efficiency. (C) Log ratio(KO/WT) distribution of ribosomal genes (n = 162), showing a significant increase in their expression in the single and triple KO. (*) P-value < 10−6; (**) P-value < 10−10, paired Wilcoxon test. (D) Log ratio(KO/WT) distribution of all genes (n = 14,006), showing a nonsignificant difference in their expression. P ≥ 0.01, paired Wilcoxon test.
Translation was also reported as a possible biological process that is affected directly by m6A methylation (Shi et al. 2017). We therefore set to measure translation in our cell lines, using a ribosomal footprint assay, which measures fragments of mRNA that are bound to a ribosome (Stern-Ginossar et al. 2012). To compare translation, the ribosomal footprint was normalized by mRNA levels, yielding a translation efficiency level for each gene in each cell line. We observed higher translation efficiency of m6A methylated genes, and of Ythdf targets, compared with nonmethylated genes (Fig. 7B). However, this difference in translation efficiency was not affected by any of the knockouts and was present in WT cells before introducing any of the perturbations. Interestingly, a mild but significant increase in the expression of ribosomal genes (Fig. 7C) was observed in single-KO and triple-KO cell lines, an increase that is not apparent in the global gene population (Fig. 7D). These results suggest that minor changes observed elsewhere in translation efficiency are not mediated directly by m6A methylation or the Ythdf proteins, but rather by the indirect effect on ribosomal gene transcript expression levels.
Discussion
Previous studies have suggested that each of the Ythdf readers has unique functions with limited redundancy (Wang et al. 2014, 2015; Du et al. 2016; Li et al. 2017; Shi et al. 2017). We propose a different model, according to which Ythdf readers have redundant functions to a profound extent, and directly influence mRNA stability via an m6A-dependent manner, rather than translation efficiency.
By analyzing Ythdf1/2/3 compound mutant mouse viability, we observed that there is compensation between the readers, and this compensation is dosage-dependent: Ythdf2 full KO or Ythdf2 heterozygotes that are also null in the two other readers are not viable. Ythdf2 heterozygote mice need at least one functional copy of another Ythdf reader to escape embryonic mortality (Fig. 3C). This provides strong genetic evidence for functional compensation between Ythdf1/2/3 at the organismal level. The fact that Ythdf2-KO has the most severe lethality phenotype, compared with Ythdf1 and Ythdf3 knockouts may be due to differences in protein expression patterns, as seen in oogenesis and spermatogenesis (Figs. 1B, 2E,F).
The strongest evidence for Ythdf redundancy was observed in mESCs, a system in which all Ythdf readers are expressed in the cytoplasmatic compartment, thus allowing examination of the redundancy hypothesis. Indeed, in mESCs, redundancy in the function of Ythdf readers was observed. Single-KO ESCs were viable, had normal self-renewal ability, expressed pluripotent markers, and differentiated normally. Only Ythdf triple-KO ESCs presented an impaired differentiation ability in vivo and in vitro, similar to the Mettl3-KO phenotype (Geula et al. 2015). In addition, only the triple-KO cells presented a significant decrease in the degradation rate of m6A methylated transcripts, while no change was observed in the single KOs. Redundancy is also supported by the observation that all Ythdf readers promote deadenylation (Du et al. 2016), although the question as to whether they do so using the same mechanism is still open, specifically whether Ythdf1 and Ythdf3, like Ythdf2, interact with CCR4–NOT complex.
Notably, while this work was under review, an independent work (Zaccara and Jaffrey 2020) reached a similar conclusion that the YTHDF readers are redundant and predominantly influence human cell behavior through RNA destabilizations, by using a series of siRNA knockdown in HeLa cell and leukemia cell line models. Structural examination showed that YTHDF1 and YTHDF2 have an identical RNA-binding surface in their RNA-binding domain, similar protein interaction networks, cellular localization, and RNA-binding preferences (Zaccara and Jaffrey 2020), thus presenting a strong support for a functional and binding redundancy model for YTHDF proteins in human cells.
The difference in expression patterns of Ythdf1/2/3 in vivo in several tissues tested herein hints that Ythdf readers are differentially regulated. Further experiments that induce the expression of Ythdf1/3 under the promoter of Ythdf2 can strongly support this hypothesis. Such a system can be examined in additional developmental processes such as neurogenesis or hematopoiesis. Last, the mechanisms that regulate Ythdf expression, such as transcription factors that bind the readers, promoters, and the readers' response to external signals, will be important to elucidate.
Materials and methods
Stem cell lines and cell culture
Maintenance of WT or mutant murine ESCs was done as described previously (Geula et al. 2015). Briefly, mESCs expansion was carried out in 500 mL of high-glucose DMEM (ThermoScientific), 15% USDA certified fetal bovine serum (FBS; Biological Industries), 1 mM L-glutamine (Biological Industries), 1% nonessential amino acids (Biological Industries), 0.1 mM β-mercaptoethanol (Sigma), 1% penicillin–streptomycin (Biological Industries), 1% sodium-pyruvate (Biological Industries), and 10 μg of recombinant human LIF (Peprotech). Cells were maintained in 20% O2 conditions on irradiation-inactivated mouse embryonic fibroblast (MEF) feeder cells and were passaged following 0.25% trypsinization. For RNA extraction, cells were grown on gelatin for three passages in FBS free N2B27-based medium (Gafni et al. 2013). Briefly, 500 mL of N2B27 media was produced by including: 250 mL of DMEM:F12 (ThermoScientific), 250 mL of neurobasal (ThermoScientific), 5 mL N2 supplement (Invitrogen 17502048 or in-house prepared), 5 mL of B27 supplement (Invitrogen 17504044), 1 mM L-glutamine (Biological Industries), 1% nonessential amino acids (Biological Industries), 0.1 mM β-mercaptoethanol (Sigma), and penicillin–streptomycin (Biological Industries). Naïve conditions for murine ESCs included 10 μg of recombinant human LIF (Peprotech), 3 μM small molecule inhibitors CHIR99021 (CH; Axon Medchem), and 1 μM PD0325901 (PD; Axon Medchem) termed 2i.
Generation of Ythdf1, Ythdf2, and Ythdf3 knockout murine ESC lines with CRISPR/Cas9
To knock out the Ythdf genes in mESCs, oligos for gRNAs were cloned into px335 vector (Addgene 42335) encoding SpCas9 nickase. A pair of unique gRNA sequences for each gene were chosen with the help of the Zhang laboratory website (http://www.genome-engineering.org/crispr) and a 20- to 30-bp offset was left between the CRISPR target sites. One-hundred micrograms of resulting constructs and 10 µg of GFP-expressing vector were electroporated into V6.5 mESCs. Three days later, GFP-expressing cells were sorted by FACS and seeded at low density. Nine days after seeding, colonies were picked and their DNA was analyzed by high-resolution melt assay (HRM) using MeltDoctor reagent (Life Technologies). The clones that showed reduced Tm for the targeted locus were expanded and sequenced to confirm mutations.
The following gRNAs were used for knocking out Ythdf genes in mES cells: mYTHDF1 gRNA1 (ATTTCCTTACTCCCTCAGCG; targeting site: 5′ region of Exon4), mYTHDF1 gRNA2 (GGATAGTAACTGGACAGGTA; targeting site: 5′ region of Exon4), mYTHDF2 gRNA1 (CTTACTTGAGCCCACAGGCA; targeting site Exon3), mYTHDF2 gRNA2 (ACAGAACCATTTTGTACTAG; targeting site: Exon3), mYTHDF3 gRNA1 (ATTGGATTTCCATATTCTCT; targeting site: 5′ region of Exon4), and mYTHDF3 gRNA2 (ATATATGGATCTGACATTGG; targeting site: 5′ region of Exon4).
Generation of Ythdf1, Ythdf2, and Ythdf3 knockout mouse strains via CRISPR/Cas9 in zygotes
The gRNA sequences were designed with the help of the Zhang laboratory website (http://www.genome-engineering.org/crispr). For Ythdf1 and Ythdf3 genes, single gRNAs were chosen targeting exon3. For the Ythdf2 gene, we designed a pair of gRNAs flanking exon4. The deletion of this exon creates an out-of-frame mutation in the coding sequence. Targeting Ythdf genes in mouse single-cell embryos (zygotes) was performed as described in Yang et al. (2014). Briefly, Cas9 and respective gRNA-coding sequences tagged with T7 promoter were transcribed using mMESSAGE mMACHINE T7 ULTRA kit and MEGA shortscript T7 kit, then purified with a MEGA clear kit (all the kits were from Thermo Fisher Scientific). CB6F1 (C57BL/6 × BALB/c) and ICR mouse strains were used as embryo donors and foster mothers, respectively. Superovulated CB6F1 mice (8–10 wk old) were mated to CB6F1 stud males, and fertilized embryos were collected from oviducts. Cas9 mRNAs and sgRNA (50 ng/μL) were injected into the cytoplasm of fertilized eggs with well-recognized pronuclei in M2 medium (Sigma). The injected zygotes were cultured in KSOM with amino acids (Sigma) at 37°C under 5% CO2 in air until blastocyst stage by 3.5 d. Thereafter, 15–25 blastocysts were transferred into the uterus of pseudopregnant ICR females at 2.5 d postcoitum (dpc). Mutated mice were screened for deletions by sequencing the targeted loci. Ythdf+/− mice were backcrossed with C57BL/6 mice for two generations before mating to generate Ythdf−/− knockout mice. Please note that no offspring with the Ythdf2-KO genotype were detected (Fig. 3C), as observed previously in pure background strains (Ivanova et al. 2017).
The following gRNAs were used for knocking out Ythdf genes in mouse zygotes: mYhdf1 CRISPR (ATTGGACTGTCCAGAAAGGT; targeting site: Exon3), mYhdf2 5′ CRISPR (GTAAATTTTAGGACTACGGT; targeting site: Intron34), mYhdf2 3′ CRISPR (GTAAATTTTAGGACTACGGT; targeting site: Intron45), and mYhdf3 CRISPR (TTTGTCTGGCTACTTAAGTA; targeting site: Exon3).
Generation of a Mettl3 conditional knockout mouse model
Stem cell lines and mice deficient for Mettl3 were generated by targeted disruption of the endogenous Mettl3 locus via homologous recombination. The targeting strategy and construct Knockout Mouse Project repository [Mettl3:tm1a(KOMP)Wtsi] introduced loxP sites spanning the fourth exon that would result in an out-of-frame and truncated product upon deletion and introduced a LacZ reporter cassette driven by the endogenous Mettl3 promoter. Fifty micrograms of DNA of the targeting construct was linearized and electroporated into a V6.5 ESC line that was then subjected to selection with 300 µg/mL G418. After 10 d of selection, resistant clones were analyzed for correct targeting. Mettl3f/f floxed ESCs were injected to BDF1 host blastocyst and chimeric mice were generated. Chimeric male mice were mated with C57BL/6 females. F1 offspring were screened for germline transmission by agouti coat color and validation via PCR of LacZ transgene reporter. To remove neomycin and lacz cassette F1 offspring were mated with Rosa26-FlpE mice (Jackson Laboratory 003946) and offspring pups were validated for the removal of LacZ transgene. The mice were crossed to C57BL/6 for three generations before being used for any experiment.
Generation of Mettl3flox/flox Cre+ knockout mice
Cre+ mice were crossed with Mettl3flox/flox mice to generate Mettl3flox/flox Cre+ mice, as detailed in Supplemental Figure S2B. The following Cre mice were used: Prm1-Cre (Jax 003328), Stra8-Cre (Jax 017490), Vasa-Cre (Jax 006954), and Zp3-Cre+ (Jax 003651).
Western blot analysis
Cells were harvested, and whole-cell protein was extracted by lysis buffer containing 150 mM NaCl, 150 mM Tris-Hcl (pH 7.4), 0.5% NP40, 1.5 mM MgCl2, and 10% glycerol. Protein concentration was determined by BCA kit (ThermoScientific). SDS/PAGE was performed according to Laemmli and transferred to nitrocellulose membranes for immunostaining. Membranes containing the transferred proteins were blocked with 5% (w/v) nonfat dried skimmed milk powder in PBST, and then incubated with the primary antibody in 5% BSA in PBST overnight at 4°C. Secondary antibodies used were peroxide-conjugated AffiniPure goat anti-rabbit (1:10,000; Jackson ImmunoResearch 111-035-003). Blots were developed using SuperSignal West Pico chemiluminescent substrate (ThermoScientific 34080). The following primary antibodies were used: Ythdf2 (Aviva Systems Biology ARP67917_P050), Ythdf3 (Santa Cruz Biotechnology SC-87503), Cnot1 (Proteintech 14276-1-A) and Hsp90 (Epitomics 1492-1).
Real-time (RT)-PCR analysis
Total RNA was extracted from the cells using Trizol. One microgram of RNA was then reverse transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was performed with 10 ng of cDNA, in triplicates, on Viia7 platform (Applied Biosystems), using Fast SYBR Master Mix (Applied Biosystems). Error bars indicate standard deviation of triplicate measurements for each measurement. The primers used for amplification are listed in Table 1.
Table 1.
Primers
Embryoid bodies and teratoma formation
For the embryoid body (EB) in vitro differentiation assay, 5 × 106 ESCs were disaggregated with trypsin and transferred to nonadherent suspension culture dishes and cultured in MEF medium (DMEM supplemented with 1% L-glutamine, 1% nonessential amino acids, 1% penicillin–streptomycin, 1% sodium-pyruvate, 15% FBS) for 8–10 d (time points were always matched with control cells). Medium replacement was carried out every 2 d.
For teratoma formation, 5 × 106 ESCs were injected subcutaneously to the flanks of immune-deficient NSG mice. After 4–6 wk, all injected mice were sacrificed, and the tumor mass extracted and fixed in 4% paraformaldehyde overnight. Slides were prepared from the paraffin embedded fixed tissues, which were next hematoxylin and eosin (H&E)-stained and inspected for representation of all three germ layers.
Histology
Ovaries and testis were fixed in 4% PFA overnight at 4°C. The fixed tissues were washed with 25%, 50%, and 70% ethanol, embedded in paraffin, and sectioned in 4-μm thickness.
Oocyte isolation and immunostaining
Female mice (5- to 8-wk-old ICR) were injected with 5 i.u. of pregnant mare serum gonadotropin (PMSG; Sigma), followed by an injection of 5 i.u. of human chorionic gonadotrophin (hCG; Sigma) 46 h later. Mouse oocytes were extracted from the oviduct by flushing the oviduct with M2 medium 24 h after hCG injection. Somatic cells were removed from the oocytes by gentle pipetting in M2 medium supplemented with hyaluronidase (Sigma). Oocytes were transferred to an embryological watch-glass and fixed with 4% PFA EM grade (Electron Microscopy Sciences) in PBS overnight at 4°C. Next, oocytes were washed three times in PBS (5 min each), permeabilized in PBS with 0.3% Triton X-100 for 30 min, blocked with 2% normal donkey serum/0.1% BSA/0.01% Tween-20 in PBS for 1 h at room temperature, and incubated overnight at 4°C with primary antibodies diluted in blocking solution. Oocytes were rinsed three times for 15 min each in blocking solution, incubated for 1 h at room temperature with secondary antibodies diluted 1:500 in blocking solution, counterstained with DAPI (1 µg/mL in PBS) for 5 min, and washed with PBS/0.01% Tween-20 for 5 min three times. Finally, oocytes were mounted in 96-well glass-bottom plates for confocal imaging. The following primary antibodies were used: mouse monoclonal antiacetylated α-Tubulin (Santa Cruz Biotechnology sc-23950), Ythdf1 (Proteintech 17479-1-AP), Ythdf2 (Aviva Systems Biology ARP67917_P050), and Ythdf3 (Santa Cruz Biotechnology SC-87503).
Flushing oocytes
For the collection of oocytes by flushing, female mice were injected with 5 i.u. of pregnant mare serum gonadotropin (PMSG), and after 46 h with 5 i.u. of human chorionic gonadotropin (hCG). Oocytes were extracted from the oviduct 24 h after the hCG injection. Next, somatic cells were removed from the oocytes by gentle pipetting in M2 medium supplemented with hyaluronidase.
Poking ovaries
For the collection of GV oocytes, female mice were injected with 5 i.u. of pregnant mare serum gonadotropin (PMSG). After 48 h, the ovaries were punctured in M2 medium using tweezers. Next, somatic cells were removed from the oocytes by gentle pipetting in M2 medium supplemented with hyaluronidase.
Immunofluorescence staining
Cells subjected to immunofluorescence staining were washed three times with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. Cells were then washed three times with PBS and blocked for 15 min with 5% FBS in PBS containing 0.1% Triton X-100. After incubation with primary antibodies overnight at 4°C in 5% FBS in PBS containing 0.1% Tween20, cells were washed three times with PBST (PBS containing 0.1% Tween20) and incubated for 1 h at room temperature with fluorophore-labeled appropriate secondary antibodies purchased from Jackson ImmunoResearch. Next, cells were washed and counterstained with 1 μg/mL DAPI (MP Biomedical 0215754), mounted with Shandon Immu-Mount (ThermoScientific 9990412), and imaged. All images were collected on LSM700 confocal microscope and processed with Zeiss ZenDesk and Adobe Photoshop CS4 (Adobe Systems).
Sections subjected to immunofluorescence staining were rehydrated, treated with antigen retrieval, rinsed in PBS for 5 min, permeabilized in 0.1% Triton X-100 in PBS, and then blocked in blocking solution (5% normal donkey serum in PBST) in a humidified chamber for 1 h at room temperature. Slides were then incubated in the appropriate primary antibodies diluted in blocking solution overnight at 4°C. Next, sections were washed three times in PBST, incubated with appropriate fluorochrome-conjugated secondary antibodies diluted in blocking solution for 1 h at room temperature in the dark, washed once in PBS, counterstained with DAPI for 10 min, rinsed twice in PBS, mounted with Shandon Immu-Mount (ThermoScientific 9990412), and imaged. All images were collected on LSM700 confocal microscope and processed with Zeiss ZenDesk and Adobe Photoshop CS4 (Adobe Systems). The following primary antibodies were used: Ythdf1 (Proteintech 17479-1-AP), Ythdf2 (Aviva Systems Biology ARP67917_P050), Ythdf3 (Santa Cruz Biotechnology SC-87503), Mettl3 (Proteintech Group 15073-1-AP), Nanog (Bethyl A300-397A or eBioscience 14-5761), Esrrb (R&D Systems PP-H6705-00), Oct4 (Santa Cruz Biotechnology SC9081 or SC5279), Foxa2 (Santa Cruz Biotechnology sc-6554), Tuj1 (BioLegend 801202), Tubulin (Santa Cruz Biotechnology sc-23950), Gata4 (Santa Cruz Biotechnology sc-1237), and H2AX (BD Biosciences 560443). Throughout the study, experimental and control samples were handled for staining, exposure, and analysis under identical conditions simultaneously to eliminate variability or bias.
Alkaline phosphatase (AP) staining
Alkaline phosphatase (AP) staining was performed with an AP kit (Millipore SCR004) according to the manufacturer's protocol. Briefly, cells were fixated using 4% PFA for 2 min and later washed with TBST. The reagents were then added to the wells, followed by an incubation of 10 min at room temperature.
Tetraploid complementation (4N) of mouse embryos
4N tetraploid complementation assay was performed by flushing BDF2 embryos at the two-cell stage, and subsequently allowing the embryos to develop until the blastocyst stage. At day 3.5, they were used for PSC microinjection of triple-KO cell line and its corresponding WT cell line. Embryos were recovered for analysis at E7.5 during development. Embryos were subjected to H&E staining and were observed for developmental defects. All animal studies were conducted according to the guideline and following approval by the Weizmann Institute Institutional Animal Care and Use Committee.
RNA stability assay
For the RNA stability assay, 5 × 105 cells of each cell type were plated on a gelatin-coated 6-cm plate. Forty-eight hours later, the medium was replaced with fresh medium containing 5 μM actinomycin-D for the inhibition of mRNA transcription. Cell samples were harvested at the indicated time points (0, 4, and 8 h) and total RNA was extracted using Trizol, followed by 3′poly(A) RNA-seq library preparation as described previously (Geula et al. 2015).
Male germ cell isolation
Male germ cell populations were isolated using FACS as described previously (Mahadevaiah et al. 2001; Bastos et al. 2005; DiGiacomo et al. 2013). Total RNA was isolated from FACS-sorted round spermatids, from WT control and Ythdf2-KO, using Trizol. The RNA was used for RNA-seq (described below).
RNA-seq library preparation
Total RNA was extracted from the indicated mESC cultures using Trizol and treated with DNase to avoid DNA contamination. Polyadenylated RNA was purified using the Dynabeads mRNA purification kit (Invitrogen 61006), followed by library preparation using ScriptSeq v2 RNA-seq library preparation kit (Illumina) according to the manufacturer's instructions.
Male germ cell populations were isolated using FACS as described previously (Mahadevaiah et al. 2001; Bastos et al. 2005; DiGiacomo et al. 2013). Total RNA was extracted using Trizol and purified using rRNA depletion (Ribo-Zero rRNA removal kit, Illumina), followed by library preparation using ScriptSeq V2 RNA-seq library preparation kit (Illumina) according to the manufacturer's instructions.
SMART-seq2 library preparation
The library was prepared according to the SMART-seq2 protocol as described previously (Picelli et al. 2014), with a few changes: Oocytes from each mouse were collected in 3 µL of M2 and added to 7.9 µL of lysis buffer. An additional 1.1 µL of DDW was added prior to the library preparation.
Ribosome profiling and analysis
Ribosome binding profiles in ESCs were measured in WT and KO conditions. For ribosome profiling, cells were treated with cycloheximide as described previously (Ingolia et al. 2009; McGlincy and Ingolia 2017). Cells were then lysed in lysis buffer (20 mM Tris 7.5, 150 mM NaCl, 15 mM MgCl2, 1 mM dithiothreitol) supplemented with 0.5% triton, 30 U/mL Turbo DNase (Ambion), and 100 μg/mL cycloheximide. Ribosome-protected fragments were then generated as described previously (McGlincy and Ingolia 2017).
Reads were preprocessed by trimming their linker (sequence CTGTAGGCACCATCAAT) and poly(A) removal with CutAdapt. Reads were aligned to mouse genome version mm10 with Bowtie aligner (parameters -v -m 16 -p 8 –max), where only uniquely aligned reads were used for further analyses. Per gene, for translation calculation, reads were counted in the coding region excluding 15 and 6 nt from the beginning and end of each coding sequence (CDS), respectively (Ingolia et al. 2009; McGlincy and Ingolia 2017). Translation efficiency was measured for each gene g and each condition i as log2(Ribogi/RNAgi). Normalized translation levels (RPKM) are available alongside the raw data at NCBI GEO series GSE148039.
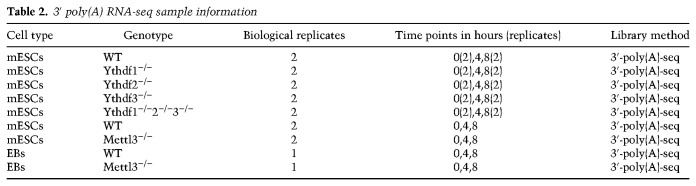
3′ poly(A) RNA sequencing analysis
3′-poly(A) RNA-seq was measured from WT and KO mESCs. KO cell lines used were as follows (Table 2): Ythdf1−/−, Ythdf2−/−, Ythdf3−/−, Ythdf1−/−Ythdf2−/−Ythdf3−/−, and Mettl3−/−. In each condition, two biological replicates were generated, and in each replicate, three time points were measured: 0, 4, and 8 h after actinomycin-D induction, with two replicates at time points 0 and 8. In addition, a similar 3′-poly(A) RNA-seq data set from a previous study (Geula et al. 2015), including samples from mESCs and mouse embryoid bodies (EBs), of Mettl3−/− and WT, measured in the same time points, was reanalyzed as described here.
Table 2.
3′ poly(A) RNA-seq sample information
Reads were aligned to mouse genome version mm10 with Bowtie2 software (Langmead and Salzberg 2012) using its default parameters. Gene expression levels were estimated using ESAT software (Derr et al. 2016), and normalized by library size of each sample (FPM [fragments per million reads]). To reduce noise, genes were filtered in each sample to include only genes with at least three positive FPM calls (two in Geula's data set), and at least one FPM call >3 (0.5 in Geula's data set), leaving 9000–12,000 genes in each sample.
The 3′ poly(A) RNA-seq values are available alongside the raw data at NCBI GEO series GSE148039.
mRNA half-life calculation
The half-life of all genes was calculated according to the following equation: ln(Ci/C0) = -kti, where k is degradation rate, Ci is the mRNA value at time i, and ti is the time interval in hours (Chen et al. 2008). Degradation rate k was estimated for each gene and each sample from its levels in time points 0, 4, and 8 h (as explained above) using linear regression lm() function in R, on the log transformed levels. Half-life t1/2 is ln(2)/k, where k is the degradation rate. Genes that had negative half-life due to slight experimental noise were ignored for the rest of the analysis.
Half-life distribution was calculated for each sample, for two groups of genes: non-m6A genes and m6A genes, which were also bound by either Ythdf1, Ythdf2, or Ythdf3 (m6A_YTH). P-value was calculated using Kolmogorov–Smirnov test.
RNA-seq analysis
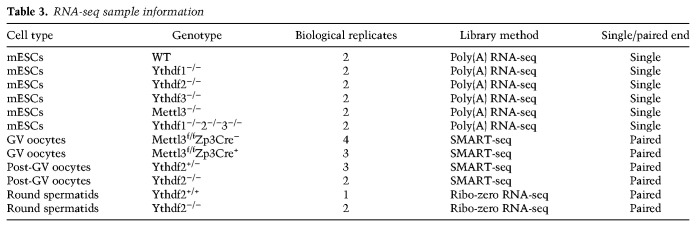
RNA sequencing was measured in mESCs, spermatids, and oocytes as detailed in Table 3.
Table 3.
RNA-seq sample information
Samples were analyzed using UTAP software (Kohen et al. 2019). Reads were trimmed using CutAdapt (Martin 2011) (parameters: -a ADAPTER1 -a “A{10}” -a “T{10}” -A “A{10}” -A “T{10}” –times 2 -q 20 -m 25). Reads were mapped to genome mm10 using STAR (Dobin et al. 2013) v2.4.2a (parameters: –alignEndsType EndToEnd, –outFilterMismatchNoverLmax 0.05, –twopassMode Basic). Sample counting was done using STAR, quantifying mm10 RefSeq annotated genes. Further analysis was done for genes having a minimum of five reads in at least one sample. Normalization of the counts and differential expression analysis was performed using DESeq2 (Love et al. 2014) with the parameters betaPrior = true, cooksCutoff = false, and independentFiltering = false. Raw P-values were adjusted for multiple testing using the procedure of Benjamini and Hochberg. Differentially expressed genes were selected with the following parameter: Padj ≤ 0.05, |log2 fold change| ≥ 1, baseMean ≥ 5. PCA and hierarchical clustering were generated in UTAP software. The normalized expression levels are available alongside the raw data at NCBI GEO series GSE148039.
Single-cell RNA-seq analysis
Single-cell spermatogenesis RNA-seq was taken from Green et al. (2018) and was analyzed using Seurat package version 3.0 (Butler et al. 2018). Low-expressing single cells (<500 expressing genes) and cells with extensive mitochondrial expression (>10% mitochondrial gene expression) were filtered out as performed in Green et al. (2018). All samples were then normalized and log-transformed using scale factor = 10,000. The top 5000 most variable genes were found by the variance stabilizing transformation method.
PCA was performed for dimensional examination using the “elbow” method. Fifteen dimensions showed the majority of data variability, and UMAP dimensional reduction was performed on aforementioned dimensions. Using Seurat's FindClusters, we adjusted the resolution parameter to correlate as much as possible to clusters found in Green et al. (2018), settling at 0.07. The log fold-change-threshold parameter used for identification of differentially expressed genes was 0.25. For cluster annotations, we used (Green et al. 2018) Differentially expressed genes (DEGs) found for each of their annotated clusters. These genes were compared with our DEGs. Clusters showing highest percentile overlap were selected. This method showed a unique identity to all but one cluster.
Enrichment analysis
Enrichment analysis was done either by using the GeneAnalytics tool (Fuchs et al. 2016), or by using Fisher's exact test.
Generating FLAG tag Ythdf proteins for eCLIP
The Ythdfs coding sequences were ordered from OriGene (MMCD00319825-mYthdf1, MR209085-mYthdf2, and MR209274-mYthdf3) and cloned with N-terminal FLAG extension into pBRY-CAG IRES-PURO plasmid using EcoRI sites. Fifty micrograms of DNA of the construct was linearized and electroporated into a V6.5 ESC line that was then subjected to selection with 2 μg/mL puromycin. After 10 d of selection, resistant clones were picked and analyzed for expression by Western blot.
eCLIP protocol and eCLIP analysis
Binding targets of Ythdf1, Ythdf2, and Ythdf3 were determined in mESCs using the eCLIP method, as described previously (Van Nostrand et al. 2016). We identified 291, 2061, and 306 peaks, mapped to 147, 1034, and 149 genes, respectively. The significance of overlap with previous targets and with m6A methylated genes was estimated using Fisher's exact test. Targets were mapped to human targets in order to test overlap, as previously published targets were measured in humans (Niu et al. 2013; Wang et al. 2015; Patil et al. 2016; Li et al. 2017; Shi et al. 2017). RRACT (R = G/A) motif was detected using MEME package in Ythdf targets, and compared with scrambled control. eCLIP coverage was estimated over 200-bp window around m6A peak centers. Mouse m6A peaks were taken from Garcia-Campos et al. (2019) (n = 9861). Human m6A peaks were taken from the WHISTLE data base (n = 41,885, high-confidence) (Chen et al. 2019), and human CLIP coverage was taken from Patil et al. (2016).
Generation of rescued Dox-inducible Ythdf1/2/3-KO cells
Ythdf expression was rescued or activated by the addition of 1 μg/mL doxycycline.
The coding sequences were amplified from FLAG-YTHDF's constructs (used for the eCLIP) and cloned in pBS31-Flp-in vector. Constructs were introduced separately to KH2-based triple-KO cells as described previously (Beard et al. 2006).
Competing interest statement
J.H.H. is an advisor to Biological Industries Ltd. G.W.Y. is cofounder, member of the Board of Directors, on the Scientific Advisory Board, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. G.W.Y.'s interest(s) have been reviewed and approved by the University of California at San Diego in accordance with its conflict of interest policies.
Supplementary Material
Acknowledgments
We thank Igor Ulitsky, Tsviya Olender, Eli Arama, and Benny Motro for insightful discussions and support. We thank the Weizmann Institute management and board for providing critical financial and infrastructural support. J.H.H. and N.N. are funded by the Nella and Leon Benoziyo Center for Neurological Diseases, the David and Fela Shapell Family Center for Genetic Disorders Research, the Kekst Family Institute for Medical Genetics, the Helen and Martin Kimmel Institute for Stem Cell Research, the Flight Attendant Medical Research Council (FAMRI), the Dr. Barry Sherman Center for Medicinal Chemistry, Pascal and Ilana Mantoux, the Dr. Beth Rom-Rymer Stem Cell Research Fund, the Edmond de Rothschild Foundations, the Zantker Charitable Foundation, the estate of Zvia Zeroni, the European Research Council (ERC-CoG), the Israel Science Foundation (ISF), Minerva, the Israel Cancer Research Fund (ICRF), and the U.S.-Israel Binational Science Foundation (BSF). This work was partly funded by National Institutes of Health grant HG004659 to G.W.Y.
Author contributions: L.L. and J.H.H. conceived the idea for this project and designed and conducted the experiments. L.L. and N.N. wrote the manuscript. N.N. supervised all bioinformatics analysis and analyzed the data. L.L. and V.K. conducted sequencing library preparation, immunostaining, and tissue culture. L.L., S.G., and V.K. engineered and generated cell lines and mouse strains under S.V.’s supervision. S.G. assisted in teratoma formation, immunostaining, and Western blots. M.Z. assisted in mouse dissection and oocyte flushing. N.M. and I.K. assisted in tissue culture and Western blots. L.L. and A.A.-C. conducted oocyte staining. J.B. and V.K. ran qPCR rescue experiments. S.T and. S.P. assisted in mouse genotyping and analysis. O.M. assisted in Ribo-seq library preparation, supervised by N.S.-G.; J.R.M., A.S., and S.A. conducted the eCLIP experiments. A.N. assisted in Ribo-seq analysis. S. Sathe analyzed the eCLIP data. G.W.Y. supervised the execution of the eCLIP experiments and analyses. T.S. conducted single-cell RNA-seq analysis, J.H.H. and N.N. supervised execution of experiments, adequate analysis of data, and presentation of conclusions made here.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.340695.120.
References
- Bailey AS, Batista PJ, Gold RS, Grace Chen Y, de Rooij DG, Chang HY, Fuller MT. 2017. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6: E26116 10.7554/eLife.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. 2005. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytom Part A 65A: 40–49. 10.1002/cyto.a.20129 [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28. 10.1002/gene.20180 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYA, Ezzeddine N, Bin SA. 2008. Messenger RNA half-life measurements in mammalian cells. Meth Enzymol 448: 335–357. 10.1016/S0076-6879(08)02617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wei Z, Zhang Q, Wu X, Rong R, Lu Z, Su J, De Magalhães JP, Rigden DJ, Meng J. 2019. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res 47: e41 10.1093/nar/gkz074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr A, Yang C, Zilionis R, Sergushichev A, Blodgett DM, Redick S, Bortell R, Luban J, Harlan DM, Kadener S, et al. 2016. End sequence analysis toolkit (ESAT) expands the extractable information from single-cell RNA-seq data. Genome Res 26: 1397–1410. 10.1101/gr.207902.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo M, Comazzetto S, Saini H, DeFazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608. 10.1016/j.molcel.2013.04.026 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. 2016. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat Commun 7: 12626 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SBA, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, Bogoch Y, Plaschkes I, Shitrit A, Rappaport N, et al. 2016. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray sata. Omi A J Integr Biol 20: 139–151. 10.1089/omi.2015.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- Gao LL, Zhou CX, Zhang XL, Liu P, Jin Z, Zhu GY, Ma Y, Li J, Yang ZX, Zhang D. 2017. ZP3 is required for germinal vesicle breakdown in mouse oocyte meiosis. Sci Rep 7: 41272 10.1038/srep41272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis A, et al. 2019. Deciphering the ‘m6A code’ via antibody-independent quantitative profiling. Cell 178: 731–747.e16. 10.1016/j.cell.2019.06.013 [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S, Moritz L, Sultan C, Gurczynski SJ, Moore BB, et al. 2018. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev Cell 46: 651–667.e10. 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck AM, Wilusz CJ. 2019. Small changes, big implications: the impact of m6A RNA methylation on gene expression in pluripotency and development. Biochim Biophys Acta-Gene Regul Mech 1862: 194402 10.1016/j.bbagrm.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. 2017. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27: 1115–1127. 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Liu Z, Zheng Y, Feng T, Gao Q, Zeng W. 2020. YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m6A/mRNA pathway. Cell Death Dis 11: 37 10.1038/s41419-020-2235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, O'Carroll D. 2017. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell 67: 1059–1067.e4. 10.1016/j.molcel.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, Anderson KV, Keeney S. 2018. Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife 7: E30919 10.7554/eLife.30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS, Huang YC, Joseph B, Lin CJ, Despic V, et al. 2017. The m6A pathway facilitates sex determination in Drosophila. Nat Commun 8: 15737 10.1038/ncomms15737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ. 2018. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 14: e1007412 10.1371/journal.pgen.1007412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohen R, Barlev J, Hornung G, Stelzer G, Feldmesser E, Kogan K, Safran M, Leshkowitz D. 2019. UTAP: user-friendly transcriptome analysis pipeline. BMC Bioinformatics 20: 154 10.1186/s12859-019-2728-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, Hanna JH, Novershtern N. 2020. Role of m6A in embryonic stem cell differentiation and in gametogenesis. Epigenomes 4: 5 10.3390/epigenomes4010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, Hanna JH, Ding L. 2019. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol 21: 700–709. 10.1038/s41556-019-0318-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al. 2017. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 27: 444–447. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, Perez SP, Suganthan R, He C, Bjørås M, et al. 2018. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol 19: 69 10.1186/s13059-018-1436-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al. 2017. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res 27: 1216–1230. 10.1038/cr.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JMA, Baudat F, Rogakou EP, De Boer P, Blanco-Rodríguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276. 10.1038/85830 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McGlincy NJ, Ingolia NT. 2017. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126: 112–129. 10.1016/j.ymeth.2017.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. 2013. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics, Proteomics Bioinforma 11: 8–17. 10.1016/j.gpb.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. M6 A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Pickering BF, Jaffrey SR. 2018. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol 28: 113–127. 10.1016/j.tcb.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervaiz N, Shakeel N, Qasim A, Zehra R, Anwar S, Rana N, Xue Y, Zhang Z, Bao Y, Abbasi AA. 2019. Evolutionary history of the human multigene families reveals widespread gene duplications throughout the history of animals. BMC Evol Biol 19: 128 10.1186/s12862-019-1441-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9: 171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. 2013. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. 10.1016/j.cell.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. 2017. YTHDF3 facilitates translation and decay of N 6-methyladenosine-modified RNA. Cell Res 27: 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, et al. 2018. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563: 249–253. 10.1038/s41586-018-0666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VTK, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. 2012. Decoding human cytomegalovirus. Science 338: 1088–1093. 10.1126/science.1227919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm MP, Kumpfmueller B, Thompson B, Kolde R, Vilo J, Hummel O, Schulz H, Welham MJ. 2009. Characterization of the phosphoinositide 3-kinase-dependent transcriptome in murine embryonic stem cells: identification of novel regulators of pluripotency. Stem Cells 27: 764–775. 10.1002/stem.3 [DOI] [PubMed] [Google Scholar]
- Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, Zheng H, Klungland A, Yan W. 2018. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc Natl Acad Sci 115: E325–E333. 10.1073/pnas.1717794115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, et al. 2018. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci 21: 195–206. 10.1038/s41593-017-0057-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. 2017. Regulation of m 6 A transcripts by the 3′→5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell 68: 374–387.e12. 10.1016/j.molcel.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, Chen YS, Zhang XX, Wang CX, Jiang LY, et al. 2017. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res 27: 1100–1114. 10.1038/cr.2017.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Jaenisch R. 2014. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9: 1956–1968. 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
- Zaccara S, Jaffrey SR. 2020. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181: 1582–1595. 10.1016/j.cell.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.