In this review, Soffers and Workman discuss the initial discovery of the canonical SAGA complex, the subsequent studies that have shaped our view on the internal organization of its subunits into modules, and the latest structural work that visualizes the modules and provides insights into their function.
Keywords: DUB, HAT, SAGA, activators, adaptor complex, adaptors, chromatin, coactivator, structure, transcription
Abstract
There are many large protein complexes involved in transcription in a chromatin context. However, recent studies on the SAGA coactivator complex are generating new paradigms for how the components of these complexes function, both independently and in concert. This review highlights the initial discovery of the canonical SAGA complex 23 years ago, our evolving understanding of its modular structure and the relevance of its modular nature for its coactivator function in gene regulation.
Chromatin-modifying and remodeling complexes are important for epigenetic regulation of gene expression. Studies into their exact molecular mode of action have inspired different research areas. Chromatin and structural biology work has helped us understand how these complexes interact with chromatin and their enzymatic substrates. Translational research has profiled epigenetic landscapes in homeostatic, differentiation, and pathological conditions to better understand how chromatin modifications regulate gene expression. However, a detailed understanding of the molecular basis of the gene regulatory actions of chromatin modifiers ultimately starts with a profound understanding of subunit functions, mode of assembly, and complex structure, which together dictate their capacity to alter chromatin structure. This review focuses on one of the first identified chromatin-modifying complexes, the Spt/Ada/Gcn5 acetyltransferase (SAGA) complex. We review its initial discovery, studies that have shaped our view on the internal organization of its subunits into modules, and the latest structural work that visualizes the modules and provides insights into their function. Finally, we discuss how modules also function as noncanonical complexes.
Discovery of the yeast SAGA complex
Studies designed to investigate the molecular mechanisms of eukaryotic gene regulation in yeast led to the discovery of the first chromatin-modifying and remodeling complexes. One set of genetic experiments took advantage of the Ty transposable elements. Ty element insertion mutations interfere with transcription because of Ty-encoded transcription signals. Suppressor of Ty (Spt) mutations restore transcription as they disrupt the genes encoding factors that recognize the Ty transcription signals (Winston et al. 1987; Fassler and Winston 1988). As such, Spt mutations uncovered proteins that promote eukaryotic gene expression and identified the TATA-binding protein (TBP)-related set of Spt proteins that includes Spt3 (Eisenmann et al. 1992; Laprade et al. 2007), Spt7 (Gansheroff et al. 1995), Spt8 (Eisenmann et al. 1994), and Spt20 (Marcus et al. 1996; Roberts and Winston 1996), also (for review, see Winston and Carlson 1992).
Another set of genetic experiments identified proteins that interacted with transcription activator proteins. Activators bind a specific promoter DNA sequence, recruit components of the transcription machinery and stimulate transcription (Hall and Struhl 2002; Carpenter et al. 2005; Ferreira et al. 2005). Natural activators in yeast are Gal4 and Gcn4. An unusually potent artificial hybrid Gal4-VP16 activator is formed by the DNA-binding portion of Gal4 fused to a highly acidic portion of the herpes simplex virus protein VP16 that interacts with DNA-binding proteins (Sadowski et al. 1988). Gal4-VP16 overexpression is lethal and may be due to sequestration of general transcription factors away from productive transcription complexes, perhaps via concentration-dependent liquid phase separation of abundant Gal4-VP16 and its interactors (Guarente 1995; Bolognesi et al. 2016; Boija et al. 2018). Like the Spt mutations, mutations that suppress Gal4-VP16 toxicity identified proteins that promote gene expression. These proteins were termed transcriptional adaptor (Ada) proteins, because they are thought to form a physical bridge between the upstream DNA-bound activators and the transcriptional machinery at a promoter (Guarente 1995). Ada1–5 interact with Gcn4 and Gal4-VP16 in yeast (Marcus et al. 1994). Ada5 is identical to Spt20 (Marcus et al. 1996; Roberts and Winston 1996). Ada4 is currently better known as Gcn5, a nuclear histone acetyltransferase (HAT) enzyme (Brownell et al. 1996; Wang et al. 1997). Ada2, Ada3, and Gcn5 not only interact with activators, but also with each other, and Ada2 also interacts indirectly with TBP (Georgakopoulos and Thireos 1992; Marcus et al. 1994; Silverman et al. 1994; Horiuchi et al. 1995; Candau and Berger 1996; Saleh et al. 1997). These data suggested that Ada proteins are part of activator complexes that mediate interactions between acidic activators and TBP.
The roles for the Spt and Ada proteins in transcription regulation converged with the discovery of the Spt/Ada/Gcn5 acetyltransferase (SAGA) complex. A collaboration of biochemists and geneticists discovered that the adaptor Gcn5 functions as a catalytic subunit in a high-molecular-mass native HAT complex that acetylated nucleosomal histones at H3 and H2B (Grant et al. 1997). This complex contained Ada2, Ada3, Ada5/Spt20, as well as Spt3, Spt7, and Spt8. The function of Gcn5 as a histone acetyltransferase within the SAGA adaptor complex indicated the importance of histone acetylation during steps in transcription activation, mediated by interactions with transcription activators and general transcription factors. Concomitantly with its discovery, Roberts and Winston (1997) demonstrated that SAGA genetically interacted with the Switch/ Sucrose nonfermentable (Swi/Snf) chromatin remodeling and Mediator complexes. This formed the first indication that SAGA was important for additional aspects of chromatin remodeling and transcription regulation.
A comprehensive overview of SAGA subunits identified in yeast and higher eukaryotes
Much of the early work described above was performed using yeast as a model organism, but the SAGA complex is also conserved in higher eukaryotes. To date, 18 conserved subunits have been identified, as well as some species-specific associated factors (Table 1). Concomitantly, additional coactivator functions of the SAGA complex emerged. This section summarizes the discovery of these additional subunits and the implications for gene regulation.
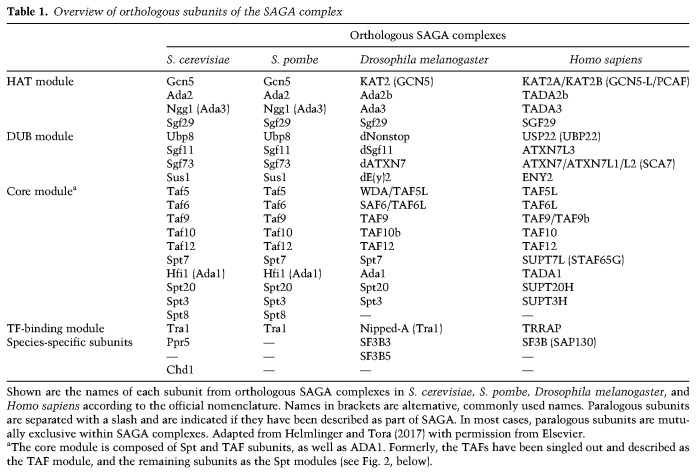
Table 1.
Overview of orthologous subunits of the SAGA complex
Gcn5-associated adaptor proteins
The adaptor proteins are conserved in higher eukaryotes. In Drosophila and humans, two Ada2 paralogs exist: Ada2a and Ada2b (Barlev et al. 2003; Kusch et al. 2003; Guelman et al. 2006a). Ada2a incorporates into the Drosophila Ada2a-containing ATAC complex, also a Gcn5-type HAT complex (Guelman et al. 2006a; Suganuma et al. 2008). Ada2b is part of Drosophila and human SAGA (Martinez et al. 1998; Muratoglu et al. 2003; Qi et al. 2004; Gamper et al. 2009). In fact, two Ada2b isoforms exist in Drosophila; SAGA contains the long isoform of Ada2b (Kusch et al. 2003), and so does another Gcn5-type HAT complex named ADA (Soffers et al. 2019). The Drosophila Chiffon Gcn5-type HAT incorporates the shorter Ada2b isoform (Torres-Zelada et al. 2019).
The Gcn5 family member proteins are PCAF (p300/CBP-associated factor, also known as KAT2B) and two isoforms of GCN5 (Gcn5-L and Gcn5-S). The PCAF protein shares extensive similarity with yeast Gcn5, but it has an N-terminal extension that interacts with the transcriptional coactivator p300/CBP (Yang et al. 1996; Smith et al. 1998). The less abundant Gcn5-S protein is similar in length to yeast, mouse, and Drosophila Gcn5p (Candau and Berger 1996; Yang et al. 1996; Smith et al. 1998; Xu et al. 1998), whereas the predominantly expressed Gcn5-L protein contains an extended N-terminal domain akin to PCAF (Smith et al. 1998). The human Ada2 and Gcn5-S proteins correspond to functional homologs of the yeast adaptors (Candau and Berger 1996). Gcn5-L was identified as part of the human SAGA complex (Smith et al. 1998).
SAGA factor 29 (Sgf29) is a conserved SAGA subunit (Sanders et al. 2002; Vermeulen et al. 2010; Lee et al. 2011). It is a bone fide Ada factor that rescues Gal4-VP16-mediated toxicity, just like the other Ada proteins that had been identified previously (Lee et al. 2011). This subunit contains a tandem tudor domain that recognizes H3K4me3 (Li et al. 2010; Bian et al. 2011).
The SAGA complex contains TBP-associated factors
Transcription initiation requires the assembly of the preinitiation complex (PIC), formed by polymerase II (Pol II) along with the general transcription factors, which include TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. PIC formation typically starts with the recognition of the core promoter by TFIID, which consists of TBP and up to 14 other proteins called TBP-associated factors (TAFs) (Thomas and Chiang 2006). In agreement with its widespread role in transcription initiation, the SAGA complex also contains TAF subunits. In fact, human SAGA was initially named STAGA because it contained TAF9 (TAFII31) (Ogryzko et al. 1998; Martinez et al. 2001).
SAGA-related complexes also contain TAFs. One such complex is the TBP-free TAF-containing complex (TFTC) (Wieczorek et al. 1998). TFTC acetylates similar lysine residues to SAGA, and is in fact very similar in structure and subunit composition to mammalian SAGA (Brand et al. 1999a,b; Cavusoglu et al. 2003). Furthermore, PCAF also associates with multiple TAFs (Kotani et al. 1998).
The TAFs that form an integral part of yeast SAGA are Taf4, Taf6, Taf9, Taf10, and Taf12 (Grant et al. 1998a). Although both yeast and metazoan incorporate TAFs, metazoans have evolved several paralogs not found in yeast. Whereas the TAFs that are part of yeast SAGA are all shared with TFIID, in Drosophila SAF6 is the SAGA-specific paralog of TAF6, TAF10b is the paralog of TAF10 (Georgieva et al. 2000; Weake et al. 2009), and WDA is the paralog of TAF5 (Guelman et al. 2006b). Similarly, human SAGA incorporates TAF5L, TAF6L, and TAF9B, whereas TAF5, TAF6, and TAF9 are found in TFIID (Helmlinger and Tora 2017).
The SAGA complex contains the activator-interacting protein Tra1
Yeast Tra1 is related to the ataxia telangiectasia mutated (ATM) family of phosphatidylinositol 3 (PI 3) kinases (PIKKs) and is a conserved SAGA subunit (Grant et al. 1998b; McMahon et al. 1998; Saleh et al. 1998; Kusch et al. 2003). PIKKs contain numerous conserved domains, including HEAT (huntingtin, elongation factor 3, protein phosphatase 2A, and lipid kinase TOR) and FAT (focal adhesion translocase) domains (Knutson and Hahn 2011; Cheung and Díaz-Santín 2019). The Tra1 pseudokinase domain is required for nuclear localization and incorporation into SAGA and other coactivator complexes (Berg et al. 2018; Elías-Villalobos et al. 2019). The best characterized function of Tra1 in SAGA is its ability to associate with activators. VP16 interacts with the C-terminal half of Tra1 (Brown et al. 2001), whereas the N-terminal HEAT domain binds yeast Gal4 (Lin et al. 2012), Gcn4 and Rap1 (Han et al. 2014), human p53 (Li et al. 2004), and c-Myc (Park et al. 2001). The multiple activator interaction domains may provide a simple mechanism to integrate signaling from multiple activators; for example, by different activators binding Tra1 simultaneously and/or competitively (Cheung and Díaz-Santín 2019). In addition, the presence of multiple activator binding sites may serve to increase avidity by binding single activator with multiple contact points as described between activator Gcn4 and Mediator subunit Med15 (Tuttle et al. 2018; Cheung and Díaz-Santín 2019).
Tra1 is not the only subunit of SAGA that interacts with activators. Gcn5 interacts with Myc (Liu et al. 2003; Zhang et al. 2014) and Ada1, Taf6, Taf9, and Taf12 interact with Gcn4 and/or VP16 (Goodrich et al. 1993; Uesugi et al. 1997; Natarajan et al. 1998; Klein et al. 2003; Fishburn et al. 2005; Reeves and Hahn 2005). These multiple interactions facilitate the recruitment of SAGA coactivation activities to an activator-bound promoter.
The SAGA complex not only contains histone acetylation, but also ubiquitin protease activity
The next major discovery was that yeast SAGA contains not only HAT activity, but also histone deubiquitinase (DUB) activity. H2B lysine 123 undergoes ubiquitination, which serves as a transient signal that orchestrates a diverse set of events during transcription (Shilatifard 2006; Fleming et al. 2008; Cheon et al. 2020). Four subunits have been identified in yeast SAGA that are required to cleave monoubiquitin from H2B. The ubiquitin protease 8 (Ubp8) is the catalytic protease subunit (Sanders et al. 2002; Henry et al. 2003; Daniel et al. 2004). SAGA-associated factor 73 (Sgf73) (McMahon et al. 2005) anchors the DUB subunits to the SAGA complex (Rodríguez-Navarro et al. 2004; McMahon et al. 2005; Köhler et al. 2008) and zinc finger protein SAGA-associated factor 11 (Sgf11) and SI gene upstream of ySa1 (Sus1) are required for protease activity (Powell et al. 2004; Ingvarsdottir et al. 2005; Lee et al. 2005; Köhler et al. 2006). In humans, more variety exists in the subunits that encode DUB activity (Köhler et al. 2010; Atanassov et al. 2016). Furthermore, ATAXIN-7, the homolog of yeast Sgf73, exists as three paralogous proteins in humans (Helmlinger and Tora 2017).
Sus1 is also a functional component of the nuclear pore-associated mRNA export machinery TREX-2 (Rodríguez-Navarro et al. 2004; García-Oliver et al. 2012). A physical interaction between SAGA and TREX-2 (Ellisdon et al. 2010) is essential for targeting the transcriptional machinery to the periphery of the nuclear pore complex and contributes to transcription-coupled mRNA export. Moreover, a functional interaction is required for maintaining the ubiquitination status linked to genome stability, since SAGA deubiquitination activity depends on TREX-2 integrity (Evangelista et al. 2018).
Species-specific subunits
The SAGA complex incorporates subunits that are important for mRNA splicing. These are associated with the U2 small nuclear ribonucleoprotein particle (U2 snRNP) that forms a core constituent of the spliceosome (Zhang et al. 2020.). Human SAGA contains spliceosome-associated protein 130 (SAP130), a subunit of splicing factor SF3B found in the U2 snRNP (Martinez et al. 2001). This finding suggested a function for SAGA in transcription-coupled pre-mRNA splicing. Drosophila SAGA contains SF3B3 and SF3B5. Although SF3B5 does not appear to function in SAGA's histone-modifying activities, it is still required for expression of a subset of SAGA-regulated genes (Stegeman et al. 2016). Both factors associated with SAGA independent of RNA and interacted with Sgf29 and Spt7 (Stegeman et al. 2016). S. cerevisiae SAGA interacts with Ppr5 via Spt3 and Spt7 in Shao et al. (2020). Prp5p is a spliceosomal RNA-dependent ATPase required for stable binding of U2 snRNP to the pre mRNA and subsequent prespliceosome assembly (Ruby et al. 1993; Xu and Query 2007). Shao et al. (2020) proposed that the interaction with SAGA mediates a balance between transcription initiation/elongation and prespliceosome assembly.
Finally, the chromatin remodeling protein chromo-ATPase/helicase DNA-binding domain 1 (Chd1) associates with SAGA and SAGA-like complex (SLIK) in S. cerevisiae (Pray-Grant et al. 2005). Chd1 has functions in chromatin remodeling, transcription regulation, and transcription elongation (Tran et al. 2000; Simic et al. 2003). One of the two chromodomains of Chd1 specifically interacts with the H3K4me3, a mark that is associated with transcriptional activity (Pray-Grant et al. 2005). Furthermore, the SLIK complex shows enhanced acetylation of a methylated substrate, and this activity was dependent on a functional methyl-binding chromodomain (Pray-Grant et al. 2005). This suggest that there can be crosstalk between Gcn5-mediated acetylation and the recognition of promoter methylation marks.
SAGA structure meets function
SAGA subunits coactivate gene expression at multiple points during the process of transcription. How the subunits organize and how this potentiates transcription is discussed next.
Genetic and biochemical experiments identify critical core subunits
The first genetic and biochemical studies grouped SAGA subunits based on their mutant phenotypes and effects on the composition of the complex (Grant et al. 1998c). Complete deletion of the adaptor proteins Gcn5, Ada2, or Ada3 cause the Ada− growth phenotype, which is characterized by the inability to grow on minimal medium and the inability to repress Gal4-VP16-mediated toxicity (Berger et al. 1992; Piña et al. 1993; Marcus et al. 1994; Eberharter et al. 1999; Lee et al. 2011). Deletion of Spt3 or Spt8 causes the Spt− phenotype, which is characterized by the abnormal initiation of certain transcripts (Winston and Carlson 1992). Deletion of the gene Spt7 or Spt20 causes both Ada− and Spt− phenotypes (Marcus et al. 1996; Roberts and Winston 1996) and their deletion is synthetically lethal in combination with mutations in genes that encode members of the Swi/Snf or Mediator complexes (Roberts and Winston 1997). In contrast, null mutations in the SAGA genes Gcn5, Spt3, and Spt8 do not cause lethality in combination with mutations in these other transcriptional regulators, which sets Spt20 and Spt7 apart (Roberts and Winston 1997). In addition, Spt20, Spt7, and also Ada1 deletion disrupts SAGA complex formation as well as function (Grant et al. 1997; Horiuchi et al. 1997; Roberts and Winston 1997; Sterner et al. 1999; Wu and Winston 2002).Together, these studies demonstrate that Spt7, Spt20 and Ada1 are critical structural core components of the S. cerevisiae SAGA complex.
Architecture of the SAGA complex
The first structure of the SAGA complex was derived from electron microscopy and mapped subunit positions with different labeling techniques and mutant yeast strains (Wu et al. 2004). This divided SAGA into five domains (Fig. 1A). Domains I and V are involved in transcriptional regulatory functions as domain I consists mainly of the Tra1 protein, required for interaction with activators, and V contains the TBP-binding protein Spt3. Domain III contains the HAT Gcn5, and central domains II, III, and IV contain the TAF subunits and the structural components Spt7, Spt20, and Ada1. Later studies roughly mapped the DUB subunits a region corresponding to domain III, close to TAF subunits Ada1 and Spt7 (Fig. 1B; Setiaputra et al. 2015). These models showed that the Spt subunits form a TBP-binding surface that is adjacent to Tra1 on one side and both enzymatic modules at the other lateral side of the TBP-binding surface (Wu et al. 2004; Setiaputra et al. 2015).
Figure 1.
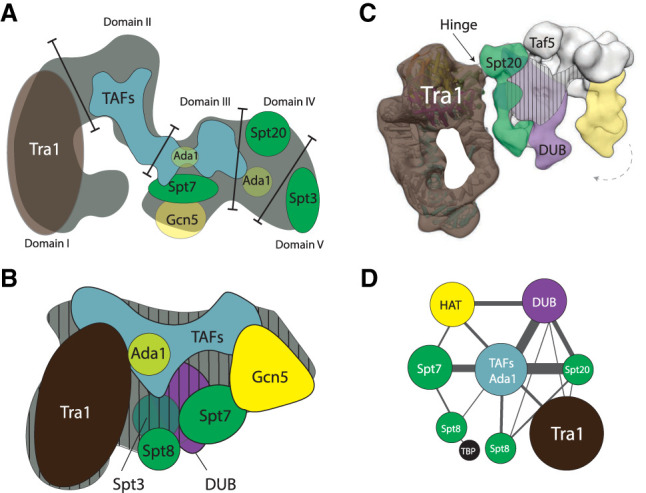
Schematic overview of the architecture of the SAGA complex based on electron microscopy, immunolabeling, and cross-linking mass spectrometry techniques. (A) The first S. cerevisiae SAGA structure divides it in five domains (adapted from Wu et al. 2004, with permission from Elsevier). Tra1 occupies domain I, and domains II–IV contain TAFs, core (Ada1, Spt7, and Spt20) and HAT subunits (Gcn5). Domain V is flexible and contains Spt3. (B) The subsequent SAGA structure locates the DUB module as bulging density in close proximity to TAF and Spt subunits. (Adapted from research originally published in Setiaputra et al. 2015. © the American Society for Biochemistry and Molecular Biology). This structure highly resembles the previous structure (Wu et al. 2004), when domain V is flipped toward domain I. The Spt subunits form a TBP-binding surface adjacent to Tra1, and the DUB and HAT modules locate to the right. Vertical dashes indicate the midplane. Subunit Spt3 and the DUB module are located on the backside. (C) Cryo-EM structure showing the narrow hinge region that forms the peripheral connection between Tra1 and the core modules (arrow; Tra1 cross-links to Spt20 and Taf12). The DUB (purple) and HAT module (yellow) associate with the structural core (green/white). The vertical stripes indicate the midplane. The DUB module anchors via the backside. Both enzymatic modules form a clamp that engages nucleosomes, and the HAT module can swirl ∼15° toward the DUB module (dashed arrow) (adapted from Cheung and Díaz-Santín 2019). (D) The architectural map determined by cross-linking mass spectrometry (adapted with permission from Han et al. 2014; © 2014 The Authors) reveals the close proximity of Tra1 and the Spt subunits, and the peripheral connections of Tra1 to the rest of the complex. The TAFs with Ada1 assume a central position and are interlinked with Spt subunits. The HAT and DUB module link to the TAF/Ada1 center, and the DUB module cross-links also to Spt20.
Tra1 was initially thought to be a critical structural core subunit, mainly because its large size (∼430 kDa) would allow it to act as a scaffolding protein. However, the Ada1 and Spt20 mutants that lack Tra1 are still able to form partial SAGA complexes; thus, Tra1 cannot be a critical subunit that nucleates SAGA assembly (Wu and Winston 2002; Lee et al. 2011). In fact, Tra1 assumes a peripheral position and is not located centrally in the complex at all, nor does it extensively cross-link to different subunits (Figs. 1C,D, 3) (Han et al. 2014; Diaz-Santin et al. 2017; Sharov et al. 2017). High-resolution cryogenic electron microscopy (cryo-EM) studies show that Tra1 connects via a narrow hinge region to SAGA (Fig. 1C; Diaz-Santin et al. 2017; Sharov et al. 2017; Cheung and Díaz-Santín 2019). The main connection forms between the Tra1 FAT domain and several regions of Spt20 and Taf12 (Fig. 3A,B; Han et al. 2014; Elías-Villalobos et al. 2019; Papai et al. 2020; Wang et al. 2020)
Figure 3.
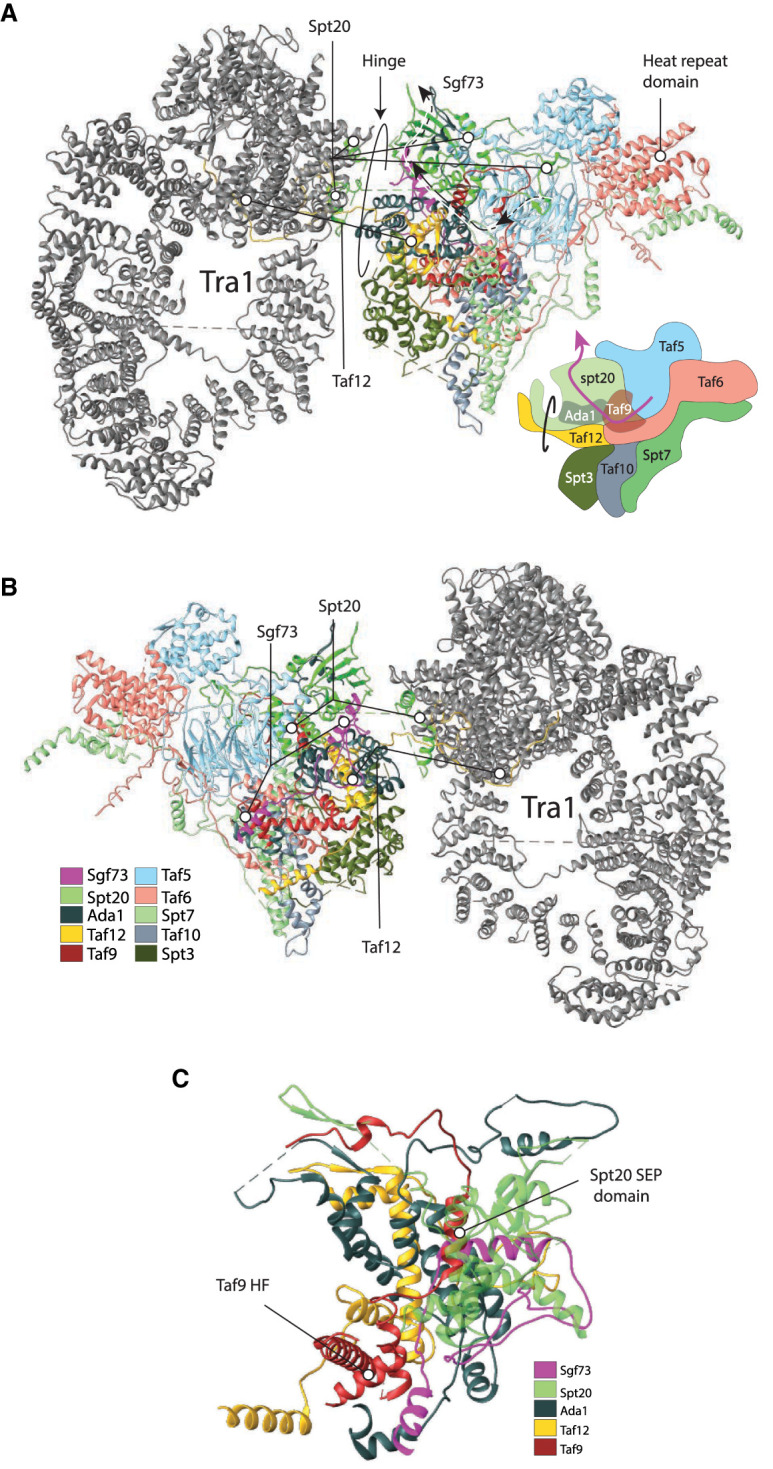
Anchor points for the enzymatic modules of the SAGA complex (A) Front view. PDB ID 6T9I and stylized cartoon. (B) Back view. (C) Back view close-up showing the insertion of Sgf73. (PDB ID 6T9I) (Wang et al. 2020). The DUB module connects via the insertion the C-terminal part of Sgf73, which enters the core proximal to the Taf9 histone fold, traverses an elongated domain of Ada1, and exits next to the Spt20 SEP domain (Papai et al. 2020; Wang et al. 2020). Spt20 is critical for the association of the DUB module. The HAT module docks to SAGA at subunit Taf6, where two helical domains—attributable to Ada3—lie at the surface of the Taf6 HEAT repeat domain. Taf5, Ta6, and Taf9 likely stabilize the configuration of Taf6 as well as the overall structure of the core module. The conformations of the enzymatic modules are dynamic in vivo, and this mobility allows the modifications on a stretch of nucleosomal histone tails up along the promoter region.
Further cross-linking mass spectrometry experiments corroborated these data and show that TAFs and Ada1 assume a central position and are interlinked with Spt subunits, and link the HAT and DUB module to the TAF/Ada1 center, whereas Tra1 does not form many interprotein connections (Fig. 1D; Han et al. 2014).
Modular divisions of the SAGA complex
A yeast combinatorial depletion study generated a comprehensive interaction matrix for all the SAGA subunits (Lee et al. 2011). The modular organization of the complex as well as the interrelationships between its subunits were determined with computational methods after the systematic purification of several tagged SAGA subunits in genetic deletion strains (Lee et al. 2011). This model divided the SAGA subunits in four modules (Fig. 2A): the enzymatic HAT module (Gcn5, Ada3, Ada2, and Sgf29); the DUB module (Ubp8, Sgf73, Sgf11, and Sus1); the TAF module, composed of all of the SAGA TAF proteins (Taf6, Taf5, Taf12, Taf9, and Taf10); and the SPT module, consisting of all of SAGA's SPT proteins (Spt7, Spt8, Spt3, and Spt20) together with Tra1 and Ada1.
Figure 2.
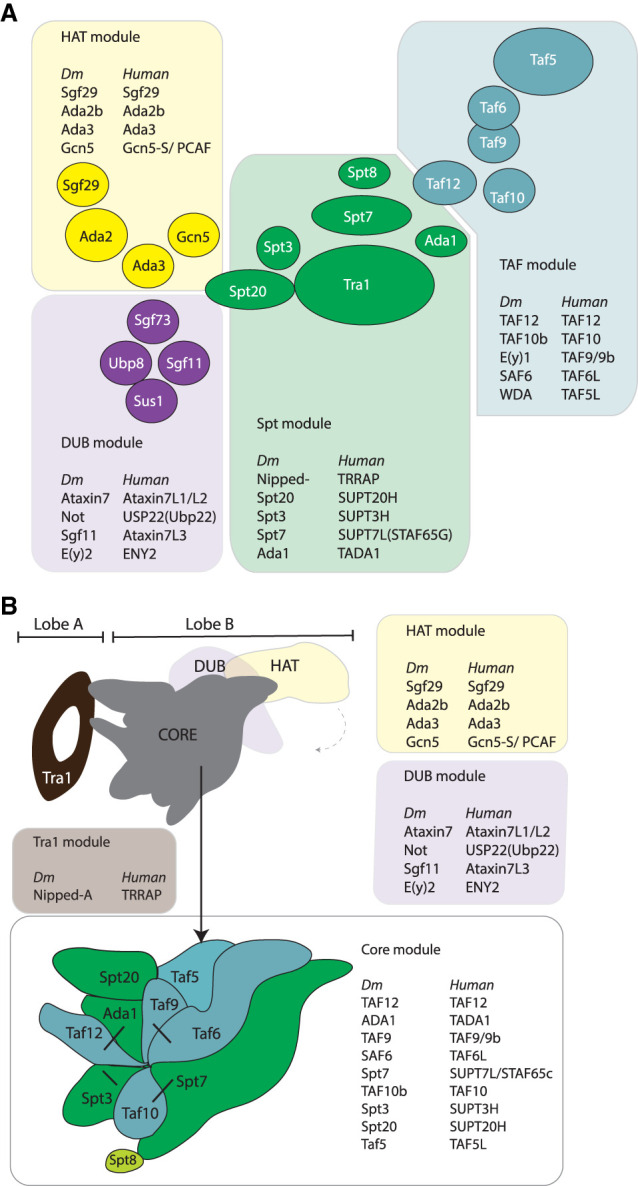
Modular division of the SAGA complex. (A) combinatorial depletion study in S. cerevisiae reveals the interprotein relationships of the complete SAGA complex. SAGA is divided into four modules that are conserved in Drosophila and humans. S. cerevisiae names are listed in the cartoon, and Drosophila and human paralogs are listed in the expanded boxes that are color-coded after their corresponding module. The four modules are the HAT module, the DUB module, TAF module, and Spt module. Note that Spt8 is not conserved in Drosophila and humans. (B) New modular division. The latest cryo-EM structure of SAGA in S. cerevisiae and P. pastori form a framework to further understand the relationship between subunits and the function of these subunits (Papai et al. 2020; Wang et al. 2020). The large, peripheral protein Tra1 is now considered its own module. The core module is composed of the former Spt and TAF module joint together. TAF and Spt proteins form an asymmetric octamer via histone fold dimer pairs (indicated with black lines). Spt20 is interwoven through the core and connects peripherally with Tra1. Spt20 and Taf12 anchor Tra1. Taf5 organizes the structure of the core and regulates the interactions of the Taf6/Taf9 dimer pair. The structure of the SAGA core allows for a flexible insertion of the two enzymatic modules.
Thus, the critical core subunits (Ada1, Spt7, and Spt20) that had been mapped to a central location within the complex (Wu et al. 2004; Han et al. 2014; Setiaputra et al. 2015) also reside together in the Spt module (Lee et al. 2011). However, the exact positions of Ada1, Spt7, and Spt20 raised the question of how they could form a central structural core. Spt7 and Spt20 cross-linked to different sets of subunits and not to each other (Fig. 1C; Han et al. 2014). Specifically, Spt7 cross-linked to Spt8, primarily between the C terminus of Spt7 and WD40 repeats at the center of Spt8 (Han et al. 2014). Spt7 also cross-linked to Gcn5 and Ada3, showing close positioning of Spt7 to the HAT module, in agreement with the models by Wu et al. (2004) and Setiaputra et al. (2015) In contrast, Spt20 cross-linked to neither Spt7, Spt8, nor the HAT module. Instead, Spt20 cross-linked to TAFs, the DUB module, and Spt3 (Han et al. 2014).
The latest high-resolution cryo-EM structures of SAGA from S. cerevisiae and P. pastori corroborate these results and show how Ada1, Spt20, and Spt7 together can be part of one and the same structural core (Fig. 2B; Papai et al. 2020; Wang et al. 2020). These structures reveal that the TAFs, Ada1, and Spt subunits together form the structural core of the SAGA complex. Spt20 and Spt7 are found at opposing lateral sides and Ada1 at a central position (Figs. 2B, 3A). Moreover, they resolve questions on the subunit stoichiometry. Both SAGA and TFIID contain Taf5, Taf6, Taf9, Taf10, and Taf12, but TFIID contains two copies of these TAFs (Leurent et al. 2002). The first structures could not yet conclusively resolve their stoichiometry (Wu et al. 2004; Setiaputra et al. 2015). However, these cryo-EM studies reveal that each subunit of the core module is present as one copy (Fig. 2B). Finally, these studies categorize the SAGA modules as follows: Tra1 module, core module (Spt, TAFs, Ada1, and Spt20), HAT module (Gcn5, Ada2, Ada3, and Sgf29), and DUB (Sgf73, Sgf11 Sus1, and Ubp8) (Fig. 2B; Papai et al. 2020; Wang et al. 2020).
The histone fold gives rise to a histone-like octamer structure within SAGA's core module
All of the SAGA core module subunits, except for Taf5 and Spt20, have a histone fold (HF) motif (Gangloff et al. 2001a,b). The HF is a protein–protein interaction motif that was originally described in the heterodimerization of histones H4 and H3, and also H2A and H2B and their assembly from a central H3-H4-heterotetramer into the histone octamer as it is found in nucleosomes (Luger et al. 1997). The motif is composed of three helices separated by two short strap loops and it assembles into heterodimers by interleaving the helices into the “handshake motif” and juxtaposing the strap loops into short parallel β-bridges (Arents et al. 1991).
TAFs similar to H3, H4, and H2B were identified in Drosophila, (Goodrich et al. 1993; Weinzierl et al. 1993; Kokubo et al. 1994; Mengus et al. 1995), humans (Weinzierl et al. 1993; Hisatake et al. 1995; Mengus et al. 1995; Hoffmann et al. 1996; Ogryzko et al. 1998), and yeast (Poon et al. 1995; Reese et al. 2000) (also for review, see Burley and Roeder 1996). Soon, biochemical studies emerged that showed that the TAFs in TFIID could form a histone octamer-like structure, like histones do as part of the nucleosome (Hoffmann et al. 1996). The Drosophila H3–H4-resembling TAFs were found to dimerize via HF domains (Nakatani et al. 1996) and Xie et al. (1996) demonstrated with X-ray diffraction studies that a Drosophila [Taf6-9]x2 tetramer forms, and postulated that four copies of Taf12 will complete the octamer. In 2001, Selleck et al. (2001) crystalized an octamer composed of two copies of the tetramer formed by Taf6, Taf9, Taf4, and Taf12, and other studies show that this octamer is complemented by a hexamer of remaining TAF subunits in TFIID (Kolesnikova et al. 2018) in Pichia pastoris, which is in agreement with findings in insect cells (Bieniossek et al. 2013).
Whether histone-like TAFs arranged into an octamer core in SAGA remained largely unclear until the stoichiometry and structure of the SAGA core subunits were resolved. The SAGA core contains an octamer of TAF subunits formed by the following histone fold dimer pairs: Taf12–Ada1, Taf9–Taf6, Taf10–Spt7, and Spt3 (Fig. 2B; Papai et al. 2020; Wang et al. 2020). The Spt3 dimer pair is not formed by two proteins, as one copy of Spt3 contains both halves of an interacting histone fold pair located at the N-terminal and C-terminal ends (Birck et al. 1998). This octamer configuration explains why previous EM studies located Spt7 in proximity to the TAFs and especially Taf10, and mapped Ada1 in proximity of Taf12 (Fig. 2B; Wu et al. 2004; Han et al. 2014).
In contrast to the canonical symmetric histone octamer that contains two copies of each histone, the octamer in SAGA is asymmetric and formed by one copy of each HF protein (Papai et al. 2020; Wang et al. 2020). In addition, it is not built up from a central histone fold domain, but from one end through interactions with Taf5 (Papai et al. 2020; Wang et al. 2020). Taf5 lacks a HF domain but contains WD40 domain repeats (Durso et al. 2001). WD40 repeat proteins act as scaffold proteins because of their β propeller structure (Xu and Min 2011). In SAGA, the Taf5 WD40 propeller docks to two HF pairs (Taf6-9 and Taf10-Spt7) and binds Spt20 (Wang et al. 2020). Spt20 also contains an extended loop that forms a wedge between the N-terminal and C-terminal Taf5 domains, thereby stabilizing them in a defined orientation The N-terminal helical domain of Taf5 binds the HEAT repeat region of Taf6 (Papai et al. 2020; Wang et al. 2020). This interaction is conserved, and influences the dimerization of human TAF6 and TAF9 (Scheer et al. 2012). Conversely, Taf6 contributes one β strand to the Taf5 propeller, making them obligate heterodimers (Papai et al. 2020; Wang et al. 2020). Work on human cells further demonstrated that TAF6's HF dimer partner TAF9 also interacts with the WD40 domain of TAF5 in a discrete TAF5–TAF6–TAF9 subcomplex, which further emphasizes the elaborate interactions between Taf5, Taf6, and Taf9 (Antonova et al. 2018). Taken together, both HF and non-HF core subunits interact and define the architecture of the core.
The core structure is important for TBP binding
Recent work has addressed how TBP is loaded onto prompters by TFIID (Patel et al. 2020), but how SAGA mechanistically contributed to TBP loading had remained largely unclear. Now that the structure of the SAGA core is resolved, several studies converge into a model where SAGA tethers TBP to Spt subunits and loads it onto DNA when certain conformational changes are induced, as discussed next.
Genetic and biochemical studies have implicated all SAGA Spt subunits in SAGA–TBP association, and Spt3 and Spt8 bind TBP directly (Eisenmann et al. 1992, 1994; Marcus et al. 1996; Roberts and Winston 1996; Sterner et al. 1999; Belotserkovskaya et al. 2000; Larschan and Winston 2001; Warfield et al. 2004; Sermwittayawong and Tan 2006; Laprade et al. 2007). Sermwittayawong and Tan (2006) proposed a two-step model in which SAGA, via Spt8, binds TBP to facilitate its subsequent transfer to a TATA box. However, the role of Spt8 has remained controversial. No Spt8 homologs have been described in higher eukaryotes (Helmlinger and Tora 2017), suggesting that other subunits must sustain TBP binding. Even in yeast, two SAGA-like complexes that lack Spt8 are able to engage TBP and coactivate gene expression, indicating that Spt8 is not necessarily always required (Saleh et al. 1997; Grant et al. 1998b). In agreement with a more modulatory role for Spt8 in TBP binding, impairment of Spt8 barely impacts mRNA synthesis, whereas Spt3 is absolutely required (Baptista et al. 2017). In the model proposed by Ben-Shem and colleagues (Papai et al. 2020), Spt8 engages TBP, and introduction of TFIIA leads to transfer of TBP from Spt8 to Spt3. Spt8 binds the N-terminal region of TBP (Wu and Winston 2002; Han et al. 2014; Papai et al. 2020). The cryo-EM structures show how multiple Spt3 helices bind the C-terminal stirrup of TBP (Papai et al. 2020). In SAGA, the Spt3 HFs are tilted compared with its analogous H2A–H2B histone pair in the nucleosome. This tilt prevents the participation of Taf10 α3 helix in the interaction, such that Spt3 is barely linked to Taf10 and able to engage TBP (Papai et al. 2020). Spt20 may also aid the Spt3–TBP binding. Spt20 cross-links to the N-terminal side of TBP near the Spt8 interaction site, but also to both ends of the Spt3 C-terminal HF (Han et al. 2014), and Spt20 deletion leads to a severe reduction in TBP binding at the GAL 1 locus (Dudley et al. 1999). TBP–Spt3 binding induces a conformational change in Spt3 that allows DNA to pass between Tra1 and the remainder of the SAGA complex, which facilitates TBP loading onto promoters (Papai et al. 2020). Since Spt3 and TFIIB mutually exclusively bind TBP (Warfield et al. 2004), the actual release of TBP from SAGA is important for further PIC assembly (Papai et al. 2020), but how this is regulated is not currently understood. Biochemical studies suggest that TBP/TFIIA binding directs the dissociation of the TATA box from the surface of the histone octamer (Godde et al. 1995). Histone acetylation also weakens the interaction between nucleosomal DNA and histone tails, since lysine acetylation affects the higher-order folding of chromatin fibers and loosens—but does not totally abolish—the contacts between the DNA and the nucleosomes (Li et al. 2007). Indeed, in vitro binding of the TFIIA–TBP complex is stimulated by Gcn5 HAT activity (Biswas et al. 2004). Thus, SAGA HAT activity might support the role of TFIIA and help to facilitate TBP release by promoting its binding to the TATA box.
The core structure allows a flexible link of the enzymatic modules
The yeast DUB module forms two functional lobes organized around the catalytic subunit. N-terminal residues of Sgf73 are located between the two Ubp8 domains and potentiate Ubp8 activity (Köhler et al. 2010; Samara et al. 2010). Ubp8, the subunit containing DUB activity, is inactive unless associated with the three other subunits of the DUB module (Köhler et al. 2008; Lang et al. 2011). In contrast to yeast, in humans and Drosophila the anchoring subunit Ataxin-7 is completely dispensable for DUB module enzymatic activity (Mohan et al. 2014). The DUB module joins the SAGA core module close to the Tra1 hinge region and subunit Sgf73 forms the anchor (Köhler et al. 2010; Samara et al. 2010; Morgan et al. 2016; Wang et al. 2020). The C-terminal part of Sgf73 is required for insertion into the core module (Lee et al. 2009; Kamata et al. 2013; Durand et al. 2014; Han et al. 2014) and enters the core proximal to the Taf9 histone fold, traverses an elongated domain of Ada1, and exits next to the Spt20 SEP (shp1, eyc, and p47) domain (Fig. 3A–C; Papai et al. 2020; Wang et al. 2020). Tra1 and Spt20 are critical for the association of the DUB module in S. pombe (Elías-Villalobos et al. 2019). Likely, Tra1 stabilizes the configuration of Spt20 in such a way that the DUB module can anchor. This finding further stresses that non-HF-containing core subunits are also important for SAGA structure and explains why Ada1 and Spt20 are critical structural subunits, required for the integration of the DUB activity.
Several studies investigated how the enzymatic modules contact the nucleosome, but exact interaction sites remain hard to identify (Durand et al. 2014; Papai et al. 2020; Wang et al. 2020). The structure of the DUB module bound to an ubiquitinated nucleosome superimposed onto the S. cerevisiae SAGA structure shows that one nucleosome fits between the HAT and DUB module (Wang et al. 2020). The HAT module maps close to the DUB module, and is the most mobile region of the complex (Setiaputra et al. 2015; Papai et al. 2020; Wang et al. 2020). It adopts multiple conformations and orientations. Accordingly, the bulk of this module is visible only as an elongated featureless density. In contrast to the DUB module, no parts of the HAT module are embedded in the SAGA core. The HAT module docks to SAGA at subunit Taf6, where two helical domains—attributable to Ada3—lie at the surface of the Taf6 HEAT repeat domain (Papai et al. 2020; Wang et al. 2020), which agrees with earlier work that demonstrated that Ada3 links the HAT module to the SAGA complex (Han et al. 2014).
It is interesting that the Taf6 HEAT repeat domain is important for anchoring the HAT module. As eluded to earlier, this domain is also important for the modulation of TAF9–TAF6 dimerization by TAF5 (Scheer et al. 2012; Antonova et al. 2018). Deletion of the Taf6 HF domain does not affect the stability of the yeast SAGA complex, even though it does disrupt the H3/H4-like Taf6/9 dimer pair (Dahiya and Natarajan 2018), which agrees with the notion that other interactions stabilize Taf6 and Taf9. Indeed, in the human TAF5/TAF6/TAF9 subcomplex, the TAF9 C-terminal domain wraps around the TAF5 WD40 repeat domain like a clamp with three major anchor points (Antonova et al. 2018). In TAF9 mutants where the TAF9/TAF5 interaction surface is completely disrupted, SAGA fails to form completely. The only interaction that this mutant TAF9 can maintain is that with TAF6. TFIID is less stable, and interference with one of the three major anchor points is already sufficient to affect complex stability (Antonova et al. 2018). This indicates that TAF5/TAF6 have different stabilizing protein contacts in each complex also (for review, see Timmers 2020). In TFIID, Taf5 docks to the octamer-like fold, where it is stabilized by Taf4 (Wang et al. 2020). In SAGA, which lacks Taf4, Ada1 assumes the position of Taf4 and stabilizes Taf5, and the Taf5 N-terminal tail occupies a rather distant position where it is further stabilized by Spt20 and contacts Taf6 (Wang et al. 2020). Thus, in the SAGA complex, Taf5, Spt20, and Ada1 are important stabilizers of Taf6 and Taf9, which is in agreement with all the genetic and biochemical studies that have uncovered since the very early discovery of the SAGA complex that Spt20 and Ada1 are critical core components for structural integrity and SAGA function.
The composition of the HAT module facilitates its function
The SAGA HAT module contains four Ada subunits that are critical for the regulation of the enzymatic lysine acetyltransferase activity as well as specific lysine residue recognition (Piña et al. 1993; Horiuchi et al. 1995; Syntichaki and Thireos 1998; Grant et al. 1999; Balasubramanian et al. 2002; Ringel et al. 2015). Recombinant Gcn5 acetylates free histones (Kuo et al. 1996), but the Gcn5-associated proteins enhance the ability of Gcn5 to acetylate nucleosomal histones and confer upon Gcn5 the ability to acetylate an expanded set of lysine residues (Grant et al. 1997, 1999) on both histone and nonhistone substrates (Kim et al. 2010).
The particular requirement for Ada2 became evident when Gcn5 with an active HAT domain but missing Ada2-interacting portion could not sustain yeast growth (Candau et al. 1997). Recent structural work reveals the molecular explanation. Ada2 contains the SANT histone-binding domain (found in Swi3, ADA2, N-Cor, and TFIIIB, hence named SANT) (Sterner et al. 2002), a ZZ-type zinc finger (ZZ) domain involved in histone tail binding (Zhang et al. 2019) and a Swi3p, Rsc8p, and Moira (SWIRM) domain, a eukaryotic domain found in proteins implicated in chromatin remodeling and gene expression (Boyer et al. 2002; Sterner et al. 2002; Qian et al. 2005; Da et al. 2006). The SANT domain does not appear to act as a histone tail-binding module, because it is positioned away from the Gcn5 peptide-binding pocket. Instead, the structure of Gcn5 and Ada2 associated with acetyl-CoA substrate suggests that the SANT domain enhances binding of the acetyl-CoA substrate (Sun et al. 2018). Gcn5, Ada2, and Ada3 form a heterotrimeric complex when expressed and combined in vitro. They form an oblong complex where Ada2 as the linchpin holds Ada3 and Gcn5 together (Horiuchi et al. 1995; Balasubramanian et al. 2002). Ada3 links the HAT module to SAGA, but deleting the most C-terminal portion of Ada3 does not compromise SAGA stability, nor does it lead to disassociation of Gcn5 from the SAGA complex (Han et al. 2014). However, it still causes the classical Ada− growth phenotype, and to an extent equivalent to complete deletion of the ADA3 gene (Han et al. 2014). Therefore, the C-terminal domain of Ada3 is required for full Gcn5 activity in vivo.
In vitro, the trimeric complex of Ada3, Ada2, and Gcn5 is sufficient for nucleosomal H3K9 and H3K14 acetylation (Balasubramanian et al. 2002). In vivo, Sgf29 is likely required for promoter recruitment and H3K9 acetylation at these regions is impaired in Sgf29 mutants (Bian et al. 2011). Sgf29 contains a tandem tudor domain that binds histone H3 methylated on lysine 4 (Bian et al. 2011). H3K4me3 recognition allows SAGA to dock to pre-existing trimethyl marks at promoter regions and promotes subsequent processive acetylation (Ringel et al. 2015). The C-terminal Ada3 mutant likely cannot bind Sgf29 and processive acetylation is disturbed such that the Ada− growth phenotype presents itself.
Distinct requirements of different modules during gene regulation
Although SAGA holistically integrates multiple coactivator functions, there is a distinct genetic requirement for each module during gene regulation. For example, even though Tra1 is important for recruitment of SAGA to promoter regions, S. pombe SAGA complexes lacking Tra1 display only very limited defects in gene regulation and show no global decrease in gene expression (Helmlinger et al. 2011). In addition, gene expression profiles of spt3Δ or spt8Δ mutants are distinct from those of ubp8Δ or sgf11Δ, indicating that loss of TBP-binding ability affects gene expression to a different degree than loss of DUB activity (Ingvarsdottir et al. 2005). Deletion strains that interfered with HAT function (gcn5Δ, ada2Δ, or ada3Δ mutants) behaved comparably, but affected only a moderate number of genes (178), and only a subset of these were affected by loss of Sgf29 (Helmlinger et al. 2011). Together, these data suggest that SAGA's coactivator function depends on its structural integrity, ability to engage TBP, anchor the DUB module, and HAT activity. However, not all genes require all of these activities.
One striking observation is that loss of Spt20 causes severe defects in gene regulation in S. pombe and S. cerevisiae (Lee et al. 2000; Helmlinger et al. 2011; Bonnet et al. 2014; Warfield et al. 2017), which suggests that its deletion disrupts multiple coactivation functions simultaneously, which is in keeping with its role as a key structural component. However, Spt20, Ada1, and Spt7 mutants do not behave identically, suggesting that even though these are all critical for the structure of the SAGA complex, they do not govern the same coactivating functions equally (Helmlinger et al. 2011). Spt20 and Spt8 mutants grossly affect the same genes. This indicates that a portion of the genes that require Spt20 rely on TBP interactions. However, the number of genes deregulated in Spt20 mutants exceeded that of Ada1, Spt7, and also exceeded the number of genes that are misexpressed in the Sgf73 and Ubp8 mutants. Therefore, the additional defects in Spt20 mutants compared with Ada1 and Spt7 can be explained by the inability to anchor the complete DUB module to SAGA, as has been observed in multiple studies (Lee et al. 2011; Elías-Villalobos et al. 2019). A disassociated submodule of the four DUB subunits in yeast is likely still catalytically active (Köhler et al. 2010; Samara et al. 2010; Lim et al. 2013). This suggests that not associating the active DUB module to SAGA leads to more severe consequences than a catalytically dead module and raises intriguing questions about how DUB function is regulated.
Different studies have addressed how DUB activity is regulated at the promoter and gene body. In S. cerevisiae, global ubH2B levels increase upon deletion of Ubp8 or the anchoring subunit Sgf73, indicating that the DUB is inactive when not anchored to SAGA (Bonnet et al. 2014). However, in humans, loss of the anchoring subunit leads to a decrease in global ubH2B levels and increased ubH2B levels in gene bodies of expressed genes (Bonnet et al. 2014). Also, in Drosophila, a global decrease in ubH2B is observed upon deletion of the anchoring subunit (Mohan et al. 2014). In Drosophila, DUB module activity is regulated slightly differently than in yeast, in that Sgf11, e(y)2, and Nonstop without Sgf73 form a catalytically active complex (Mohan et al. 2014; Cloud et al. 2019). In fact, even in wild-type conditions, the DUB module (Sgf11 and Nonstop) associates with ∼600 genomic regions that are not targeted by the canonical SAGA complex. Approximately half of these map near TSS and ∼10% require ubiquitin protease activity, indicating that the Nonstop regulates gene expression independent of canonical SAGA (Li et al. 2017).
Other studies in S. cerevisiae suggest that Spt20 is also important for integration of the SAGA HAT module. spt20Δ mutants fail to integrate the HAT module, yet the HAT module exists as stable subcomplex (Lee et al. 2011). Spt20 and Ada2 assemble cotranslationally (Kassem et al. 2017), which raises the possibility that the SAGA core module already engages the HAT module in the cytoplasm. In humans, recombinant expression of all four HAT module subunits leads to the formation of a stable HAT subcomplex (Nguyen-Huynh et al. 2015). The same components form the Drosophila ADA complex (Soffers et al. 2019), which is similar to the yeast ADA complex, which preferentially acetylates H3K14, H3K18, and H2B (Eberharter et al. 1999; Grant et al. 1999) but lacks the yeast-specific Ada HAT components Ahc1 and Ahc2. The Drosophila nuclear complex does not associate with Spt20; thus, it is not the imported form of a Spt20–Ada2b cotranslational assembly complex (Soffers et al. 2019). The ADA complex localizes to sites that canonical SAGA does not occupy, and it has HAT activity not only on recombinant histones and core histones, but also polynucleosome substrates, suggesting that it can also acetylate chromatin in a cellular context. Loss of Drosophila Ada2b influences the expression of ∼600 genes in larvae and ∼900 genes in pupa (Zsindely et al. 2009; Pankotai et al. 2013). These observations raise questions about which genes depend on the metazoan ADA complex and which are regulated by SAGA, and also what advantage such redundant complexes may confer.
Conclusion
Combined biochemical, genetic, and structural studies have painted a picture of the versatile and diverse functions of the SAGA complex during gene regulation, starting from its role in transcription initiation to functions in elongation and RNA processing and export. Our understanding of its molecular mode of action keeps evolving with the identification of new subunits that account for additional functions, substrates, and knowledge about the roles of individual modules. Current questions are how the conformational flexibility directs function in vivo, and we are eagerly awaiting further structures of nucleosome-bound SAGA. These will help understand whether SAGA clamps one nucleosome at a time or can modify histone tails of adjacent nucleosomes. From here, further questions arise. Are pre-existing methylation marks directing levels of acetylation and ubiquitination at the same nucleosome and vice versa? Do multiple SAGA copies bind at different regions of a gene and exert different activities, and does this explain why some SAGA-associated marks are prevalent upstream of the transcription start site, yet others in the gene body? Can we account for these differences by enzymatic activities of isolated SAGA modules? Indeed, 23 yr after its discovery, the SAGA histone acetyltransferase complex keeps generating new paradigms for how the components of chromatin-modifying complexes function together and independently.
Acknowledgments
We thank the Workman laboratory members, Dr. Randal Halfmann, Dr. Ryan D. Mohan, Professor Patrick Cramer, and their group members for critical reading of the manuscript, discussion, and suggestions. These studies were supported by funds from the Stowers Institute and the National Institutes of General Medical Sciences grant R35GM118068 (to J.L.W.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.341156.120.
References
- Antonova SV, Haffke M, Corradini E, Mikuciunas M, Low TY, Signor L, van Es RM, Gupta K, Scheer E, Vos HR, et al. 2018. Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly. Nat Struct Mol Biol 25: 1119–1127. 10.1038/s41594-018-0156-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. 1991. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci 88: 10148–10152. 10.1073/pnas.88.22.10148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, McIvor E, Li W, Zhang Y, Florens L, et al. 2016. ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell 62: 558–571. 10.1016/j.molcel.2016.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem 277: 7989–7995. 10.1074/jbc.M110849200 [DOI] [PubMed] [Google Scholar]
- Baptista T, Grunberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, Tora L. 2017. SAGA is a general cofactor for RNA polymerase II transcription. Mol Cell 68: 130–143.e5. 10.1016/j.molcel.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Emelyanov AV, Castagnino P, Zegerman P, Bannister AJ, Sepulveda MA, Robert F, Tora L, Kouzarides T, Birshtein BK, et al. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol 23: 6944–6957. 10.1128/MCB.23.19.6944-6957.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol 20: 634–647. 10.1128/MCB.20.2.634-647.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, Genereaux J, Karagiannis J, Brandl CJ. 2018. The pseudokinase domain of Saccharomyces cerevisiae Tra1 is required for nuclear localization and incorporation into the SAGA and NuA4 complexes. G3 (Bethesda) 8: 1943–1957. 10.1534/g3.118.200288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Piña B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70: 251–265. 10.1016/0092-8674(92)90100-Q [DOI] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 30: 2829–2842. 10.1038/emboj.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. 2013. The architecture of human general transcription factor TFIID core complex. Nature 493: 699–702. 10.1038/nature11791 [DOI] [PubMed] [Google Scholar]
- Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, Davidson I, Moras D. 1998. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94: 239–249. 10.1016/S0092-8674(00)81423-3 [DOI] [PubMed] [Google Scholar]
- Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. 2004. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein–TFIIA–DNA complex. Mol Cell Biol 24: 8312–8321. 10.1128/MCB.24.18.8312-8321.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175: 1842–1855.e16. 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi B, Lorenzo Gotor N, Dhar R, Cirillo D, Baldrighi M, Tartaglia GG, Lehner B. 2016. A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep 16: 222–231. 10.1016/j.celrep.2016.05.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28: 1999–2012. 10.1101/gad.250225.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10: 935–942. 10.1016/S1097-2765(02)00634-2 [DOI] [PubMed] [Google Scholar]
- Brand M, Leurent C, Mallouh V, Tora L, Schultz P. 1999a. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286: 2151–2153. 10.1126/science.286.5447.2151 [DOI] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L. 1999b. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem 274: 18285–18289. 10.1074/jbc.274.26.18285 [DOI] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337. 10.1126/science.1060214 [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851. 10.1016/S0092-8674(00)81063-6 [DOI] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65: 769–799. 10.1146/annurev.bi.65.070196.004005 [DOI] [PubMed] [Google Scholar]
- Candau R, Berger SL. 1996. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem 271: 5237–5245. 10.1074/jbc.271.9.5237 [DOI] [PubMed] [Google Scholar]
- Candau R, Zhou JX, Allis CD, Berger SL. 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J 16: 555–565. 10.1093/emboj/16.3.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Memedula S, Plutz MJ, Belmont AS. 2005. Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol 25: 958–968. 10.1128/MCB.25.3.958-968.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavusoglu N, Brand M, Tora L, Van Dorsselaer A. 2003. Novel subunits of the TATA binding protein free TAFII-containing transcription complex identified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry following one-dimensional gel electrophoresis. Proteomics 3: 217–223. 10.1002/pmic.200390030 [DOI] [PubMed] [Google Scholar]
- Cheon Y, Kim H, Park K, Kim M, Lee D. 2020. Dynamic modules of the coactivator SAGA in eukaryotic transcription. Exp Mol Med 52: 991–1003. 10.1038/s12276-020-0463-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ACM, Díaz-Santín LM. 2019. Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes. Transcription 10: 37–43. 10.1080/21541264.2018.1530936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V, Thapa A, Morales-Sosa P, Miller TM, Miller SA, Holsapple D, Gerhart PM, Momtahan E, Jack JL, Leiva E, et al. 2019. Ataxin-7 and Non-stop coordinate SCAR protein levels, subcellular localization, and actin cytoskeleton organization. Elife 8: e49677 10.7554/eLife.49677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. 2006. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci 103: 2057–2062. 10.1073/pnas.0510949103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya R, Natarajan K. 2018. Mutational analysis of TAF6 revealed the essential requirement of the histone-fold domain and the HEAT repeat domain for transcriptional activation. FEBS J 285: 1491–1510. 10.1111/febs.14423 [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR III, Grant PA. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867-1871. 10.1074/jbc.C300494200 [DOI] [PubMed] [Google Scholar]
- Diaz-Santin LM, Lukoyanova N, Aciyan E, Cheung AC. 2017. Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 at 3.7 angstrom resolution. Elife 6: e28384 10.7554/eLife.28384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev 13: 2940–2945. 10.1101/gad.13.22.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand A, Bonnet J, Fournier M, Chavant V, Schultz P. 2014. Mapping the deubiquitination module within the SAGA complex. Structure 22: 1553–1559. 10.1016/j.str.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Durso RJ, Fisher AK, Albright-Frey TJ, Reese JC. 2001. Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol Cell Biol 21: 7331–7344. 10.1128/MCB.21.21.7331-7344.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR III, Berger SL, Workman JL. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol 19: 6621–6631. 10.1128/MCB.19.10.6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6: 1319–1331. 10.1101/gad.6.7.1319 [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elías-Villalobos A, Fort P, Helmlinger D. 2019. New insights into the evolutionary conservation of the sole PIKK pseudokinase Tra1/TRRAP. Biochem Soc Trans 47: 1597–1608. 10.1042/BST20180496 [DOI] [PubMed] [Google Scholar]
- Elías-Villalobos A, Toullec D, Faux C, Séveno M, Helmlinger D. 2019. Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription co-activator complexes in yeast. Nat Commun 10: 5237 10.1038/s41467-019-13243-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisdon AM, Jani D, Köhler A, Hurt E, Stewart M. 2010. Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1. J Biol Chem 285: 3850–3856. 10.1074/jbc.M109.070839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista FM, Maglott-Roth A, Stierle M, Brino L, Soutoglou E, Tora L. 2018. Transcription and mRNA export machineries SAGA and TREX-2 maintain monoubiquitinated H2B balance required for DNA repair. J Cell Biol 217: 3382–3397. 10.1083/jcb.201803074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler JS, Winston F. 1988. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics 118: 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira ME, Hermann S, Prochasson P, Workman JL, Berndt KD, Wright AP. 2005. Mechanism of transcription factor recruitment by acidic activators. J Biol Chem 280: 21779–21784. 10.1074/jbc.M502627200 [DOI] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, Hahn S. 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell 18: 369–378. 10.1016/j.molcel.2005.03.029 [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66. 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- Gamper AM, Kim J, Roeder RG. 2009. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol 29: 266–280. 10.1128/MCB.00315-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. 2001a. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci 26: 250–257. 10.1016/S0968-0004(00)01741-2 [DOI] [PubMed] [Google Scholar]
- Gangloff YG, Sanders SL, Romier C, Kirschner D, Weil PA, Tora L, Davidson I. 2001b. Histone folds mediate selective heterodimerization of yeast TAF(II25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol Cell Biol 21: 1841–1853. 10.1128/MCB.21.5.1841-1853.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansheroff LJ, Dollard C, Tan P, Winston F. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Oliver E, García-Molinero V, Rodríguez-Navarro S. 2012. mRNA export and gene expression: the SAGA–TREX-2 connection. Biochim Biophys Acta 1819: 555–565. 10.1016/j.bbagrm.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Georgakopoulos T, Thireos G. 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J 11: 4145–4152. 10.1002/j.1460-2075.1992.tb05507.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S, Kirschner DB, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B, et al. 2000. Two novel Drosophila TAFIIs have homology with human TAFII30 and are differentially regulated during development. Mol Cell Biol 20: 1639–1648. 10.1128/MCB.20.5.1639-1648.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde JS, Nakatani Y, Wolffe AP. 1995. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res 23: 4557–4564. 10.1093/nar/23.22.4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R. 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75: 519–530. 10.1016/0092-8674(93)90386-5 [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650. 10.1101/gad.11.13.1640 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR III, Workman JL. 1998a. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53. 10.1016/S0092-8674(00)81220-9 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR III, Workman JL. 1998b. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell 2: 863–867. 10.1016/S1097-2765(00)80300-7 [DOI] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. 1998c. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol 8: 193–197. 10.1016/S0962-8924(98)01263-X [DOI] [PubMed] [Google Scholar]
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem 274: 5895–5900. 10.1074/jbc.274.9.5895 [DOI] [PubMed] [Google Scholar]
- Guarente L. 1995. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci 20: 517–521. 10.1016/S0968-0004(00)89120-3 [DOI] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR III, Washburn MP, Abmayr SM, et al. 2006a. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol 26: 871–882. 10.1128/MCB.26.3.871-882.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Weake V, Swanson SK, Washburn MP, Abmayr SM, Workman JL. 2006b. The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol Cell Biol 26: 7178–7189. 10.1128/MCB.00130-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DB, Struhl K. 2002. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem 277: 46043–46050. 10.1074/jbc.M208911200 [DOI] [PubMed] [Google Scholar]
- Han Y, Luo J, Ranish J, Hahn S. 2014. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J 33: 2534–2546. 10.15252/embj.201488638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Tora L. 2017. Sharing the SAGA. Trends Biochem Sci 42: 850–861. 10.1016/j.tibs.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villén J, Swaney DL, Gygi SP, Bähler J, Winston F. 2011. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J 30: 2843–2852. 10.1038/emboj.2011.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663. 10.1101/gad.1144003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatake K, Ohta T, Takada R, Guermah M, Horikoshi M, Nakatani Y, Roeder RG. 1995. Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors. Proc Natl Acad Sci 92: 8195–8199. 10.1073/pnas.92.18.8195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Chiang CM, Oelgeschläger T, Xie X, Burley SK, Nakatani Y, Roeder RG. 1996. A histone octamer-like structure within TFIID. Nature 380: 356–359. 10.1038/380356a0 [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Marcus GA, Guarente L. 1995. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol 15: 1203–1209. 10.1128/MCB.15.3.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Piña B, Marcus GA, Guarente L. 1997. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol 17: 3220–3228. 10.1128/MCB.17.6.3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol 25: 1162–1172. 10.1128/MCB.25.3.1162-1172.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata K, Hatanaka A, Goswami G, Shinmyozu K, Nakayama J, Urano T, Hatashita M, Uchida H, Oki M. 2013. C-terminus of the Sgf73 subunit of SAGA and SLIK is important for retention in the larger complex and for heterochromatin boundary function. Genes Cells 18: 823–837. 10.1111/gtc.12075 [DOI] [PubMed] [Google Scholar]
- Kassem S, Villanyi Z, Collart MA. 2017. Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res 45: 1186–1199. 10.1093/nar/gkw1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Saraf A, Florens L, Washburn M, Workman JL. 2010. Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes Dev 24: 2766–2771. 10.1101/gad.1979710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Nolden M, Sanders SL, Kirchner J, Weil PA, Melcher K. 2003. Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA. J Biol Chem 278: 6779–6786. 10.1074/jbc.M212514200 [DOI] [PubMed] [Google Scholar]
- Knutson BA, Hahn S. 2011. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol Cell Biol 31: 818–831. 10.1128/MCB.00687-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Pascual-García P, Llopis A, Zapater M, Posas F, Hurt E, Rodríguez-Navarro S. 2006. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell 17: 4228–4236. 10.1091/mbc.e06-02-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. 2008. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10: 707–715. 10.1038/ncb1733 [DOI] [PubMed] [Google Scholar]
- Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617. 10.1016/j.cell.2010.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo T, Gong DW, Wootton JC, Horikoshi M, Roeder RG, Nakatani Y. 1994. Molecular cloning of Drosophila TFIID subunits. Nature 367: 484–487. 10.1038/367484a0 [DOI] [PubMed] [Google Scholar]
- Kolesnikova O, Ben-Shem A, Luo J, Ranish J, Schultz P, Papai G. 2018. Molecular structure of promoter-bound yeast TFIID. Nat Commun 9: 4666 10.1038/s41467-018-07096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Zhang X, Schiltz RL, Ogryzko VV, Howard T, Swanson MJ, Vassilev A, Zhang H, Yamauchi J, Howard BH, et al. 1998. TBP-associated factors in the PCAF histone acetylase complex. Cold Spring Harb Symp Quant Biol 63: 493–500. 10.1101/sqb.1998.63.493 [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272. 10.1038/383269a0 [DOI] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol 23: 3305–3319. 10.1128/MCB.23.9.3305-3319.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31: 3734–3744. 10.1128/MCB.05231-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade L, Rose D, Winston F. 2007. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3–TBP allele-specific interactions and bypass of Spt8. Genetics 177: 2007–2017. 10.1534/genetics.107.081976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Winston F. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15: 1946–1956. 10.1101/gad.911501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. 10.1038/35015104 [DOI] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol 25: 1173–1182. 10.1128/MCB.25.3.1173-1182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. 2009. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin 2: 2 10.1186/1756-8935-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. 2011. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol 7: 503 10.1038/msb.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J 21: 3424–3433. 10.1093/emboj/cdf342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Cuenin C, Murr R, Wang ZQ, Herceg Z. 2004. HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J 23: 4824–4834. 10.1038/sj.emboj.7600479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128: 707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Li J, Xue X, Ruan J, Wu M, Zhu Z, Zang J. 2010. Cloning, purification, crystallization and preliminary crystallographic analysis of the tandem tudor domain of Sgf29 from Saccharomyces cerevisiae. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 902–904. 10.1107/S1744309110016726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Seidel CW, Szerszen LT, Lange JJ, Workman JL, Abmayr SM. 2017. Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev 31: 1588–1600. 10.1101/gad.300988.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kwak J, Kim M, Lee D. 2013. Separation of a functional deubiquitylating module from the SAGA complex by the proteasome regulatory particle. Nat Commun 4: 2641 10.1038/ncomms3641 [DOI] [PubMed] [Google Scholar]
- Lin L, Chamberlain L, Zhu LJ, Green MR. 2012. Analysis of Gal4-directed transcription activation using Tra1 mutants selectively defective for interaction with Gal4. Proc Natl Acad Sci 109: 1997–2002. 10.1073/pnas.1116340109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. 2003. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem 278: 20405–20412. 10.1074/jbc.M211795200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J 13: 4807–4815. 10.1002/j.1460-2075.1994.tb06806.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Horiuchi J, Silverman N, Guarente L. 1996. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol 16: 3197–3205. 10.1128/MCB.16.6.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Kundu TK, Fu J, Roeder RG. 1998. A human SPT3–TAFII31–GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem 273: 23781–23785. 10.1074/jbc.273.37.23781 [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol 21: 6782–6795. 10.1128/MCB.21.20.6782-6795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374. 10.1016/S0092-8674(00)81479-8 [DOI] [PubMed] [Google Scholar]
- McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR III, Grant PA. 2005. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci 102: 8478–8482. 10.1073/pnas.0503493102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. 1995. Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J 14: 1520–1531. 10.1002/j.1460-2075.1995.tb07138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan RD, Dialynas G, Weake VM, Liu J, Martin-Brown S, Florens L, Washburn MP, Workman JL, Abmayr SM. 2014. Loss of Drosophila Ataxin-7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev 28: 259–272. 10.1101/gad.225151.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. 2016. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351: 725–728. 10.1126/science.aac5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratoglu S, Georgieva S, Pápai G, Scheer E, En ünl ü I, Komonyi O, Cserpán I, Lebedeva L, Nabirochkina E, Udvardy A, et al. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol 23: 306–321. 10.1128/MCB.23.1.306-321.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Bagby S, Ikura M. 1996. The histone folds in transcription factor TFIID. J Biol Chem 271: 6575–6578. 10.1074/jbc.271.12.6575 [DOI] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. 1998. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell 2: 683–692. 10.1016/S1097-2765(00)80166-5 [DOI] [PubMed] [Google Scholar]
- Nguyen-Huynh NT, Sharov G, Potel C, Fichter P, Trowitzsch S, Berger I, Lamour V, Schultz P, Potier N, Leize-Wagner E. 2015. Chemical cross-linking and mass spectrometry to determine the subunit interaction network in a recombinant human SAGA HAT subcomplex. Protein Sci 24: 1232–1246. 10.1002/pro.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94: 35–44. 10.1016/S0092-8674(00)81219-2 [DOI] [PubMed] [Google Scholar]
- Pankotai T, Zsindely N, Vamos EE, Komonyi O, Bodai L, Boros IM. 2013. Functional characterization and gene expression profiling of Drosophila melanogaster short dADA2b isoform-containing dSAGA complexes. BMC Genomics 14: 44 10.1186/1471-2164-14-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben-Shem A. 2020. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577: 711–716. 10.1038/s41586-020-1944-2 [DOI] [PubMed] [Google Scholar]
- Park J, Kunjibettu S, McMahon SB, Cole MD. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev 15: 1619–1624. 10.1101/gad.900101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AB, Greber BJ, Nogales E. 2020. Recent insights into the structure of TFIID, its assembly, and its binding to core promoter. Curr Opin Struct Biol 61: 17–24. 10.1016/j.sbi.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B, Berger S, Marcus GA, Silverman N, Agapite J, Guarente L. 1993. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol 13: 5981–5989. 10.1128/MCB.13.10.5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D, Bai Y, Campbell AM, Bjorklund S, Kim YJ, Zhou S, Kornberg RD, Weil PA. 1995. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci 92: 8224–8228. 10.1073/pnas.92.18.8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol 24: 7249–7259. 10.1128/MCB.24.16.7249-7259.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR III, Grant PA. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434–438. 10.1038/nature03242 [DOI] [PubMed] [Google Scholar]
- Qi D, Larsson J, Mannervik M. 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol 24: 8080–8089. 10.1128/MCB.24.18.8080-8089.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. 2005. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol 12: 1078–1085. 10.1038/nsmb1022 [DOI] [PubMed] [Google Scholar]
- Reese JC, Zhang Z, Kurpad H. 2000. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem 275: 17391–17398. 10.1074/jbc.M001635200 [DOI] [PubMed] [Google Scholar]
- Reeves WM, Hahn S. 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol 25: 9092–9102. 10.1128/MCB.25.20.9092-9102.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel AE, Cieniewicz AM, Taverna SD, Wolberger C. 2015. Nucleosome competition reveals processive acetylation by the SAGA HAT module. Proc Natl Acad Sci 112: E5461–E5470. 10.1073/pnas.1508449112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol 16: 3206–3213. 10.1128/MCB.16.6.3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S, Fischer T, Luo MJ, Antúnez O, Brettschneider S, Lechner J, Pérez-Ortín JE, Reed R, Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. 10.1016/S0092-8674(03)01025-0 [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. 1993. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev 7: 1909–1925. 10.1101/gad.7.10.1909 [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564. 10.1038/335563a0 [DOI] [PubMed] [Google Scholar]
- Saleh A, Lang V, Cook R, Brandl CJ. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem 272: 5571–5578. 10.1074/jbc.272.9.5571 [DOI] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR III, Lees-Miller SP, Cole MD, Brandl CJ. 1998. Tra1p is a component of the yeast Ada·Spt transcriptional regulatory complexes. J Biol Chem 273: 26559–26565. 10.1074/jbc.273.41.26559 [DOI] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029. 10.1126/science.1190049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol 22: 4723–4738. 10.1128/MCB.22.13.4723-4738.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer E, Delbac F, Tora L, Moras D, Romier C. 2012. TFIID TAF6–TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5 protein. J Biol Chem 287: 27580–27592. 10.1074/jbc.M112.379206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S. 2001. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol 8: 695–700. 10.1038/90408 [DOI] [PubMed] [Google Scholar]
- Sermwittayawong D, Tan S. 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J 25: 3791–3800. 10.1038/sj.emboj.7601265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK. 2015. Conformational flexibility and subunit arrangement of the modular yeast Spt–Ada–Gcn5 acetyltransferase complex. J Biol Chem 290: 10057–10070. 10.1074/jbc.M114.624684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Ding Z, Zheng ZZ, Shen JJ, Shen YX, Pu J, Fan YJ, Query CC, Xu YZ. 2020. Prp5−Spt8/Spt3 interaction mediates a reciprocal coupling between splicing and transcription. Nucleic Acids Res 48: 5799–5813. 10.1093/nar/gkaa311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov G, Voltz K, Durand A, Kolesnikova O, Papai G, Myasnikov AG, Dejaegere A, Ben Shem A, Schultz P. 2017. Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Nat Commun 8: 1556 10.1038/s41467-017-01564-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269. 10.1146/annurev.biochem.75.103004.142422 [DOI] [PubMed] [Google Scholar]
- Silverman N, Agapite J, Guarente L. 1994. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci 91: 11665–11668. 10.1073/pnas.91.24.11665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856. 10.1093/emboj/cdg179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Belote JM, Schiltz RL, Yang XJ, Moore PA, Berger SL, Nakatani Y, Allis CD. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res 26: 2948–2954. 10.1093/nar/26.12.2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffers JHM, Li X, Saraf A, Seidel CW, Florens L, Washburn MP, Abmayr SM, Workman JL. 2019. Characterization of a metazoan ADA acetyltransferase complex. Nucleic Acids Res 47: 3383–3394. 10.1093/nar/gkz042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman R, Spreacker PJ, Swanson SK, Stephenson R, Florens L, Washburn MP, Weake VM. 2016. The spliceosomal protein SF3B5 is a novel component of Drosophila SAGA that functions in gene expression independent of splicing. J Mol Biol 428: 3632–3649. 10.1016/j.jmb.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98. 10.1128/MCB.19.1.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J Biol Chem 277: 8178–8186. 10.1074/jbc.M108601200 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. 2008. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol 15: 364–372. 10.1038/nsmb.1397 [DOI] [PubMed] [Google Scholar]
- Sun J, Paduch M, Kim SA, Kramer RM, Barrios AF, Lu V, Luke J, Usatyuk S, Kossiakoff AA, Tan S. 2018. Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2. Proc Natl Acad Sci 115: 10010–10015. 10.1073/pnas.1805343115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Thireos G. 1998. The Gcn5·Ada complex potentiates the histone acetyltransferase activity of Gcn5. J Biol Chem 273: 24414–24419. 10.1074/jbc.273.38.24414 [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178. 10.1080/10409230600648736 [DOI] [PubMed] [Google Scholar]
- Timmers HTM. 2020. SAGA and TFIID: friends of TBP drifting apart. Biochim Biophys Acta Gene Regul Mech 10.1016/j.bbagrm.2020.194604 [DOI] [PubMed] [Google Scholar]
- Torres-Zelada EF, Stephenson RE, Alpsoy A, Anderson BD, Swanson SK, Florens L, Dykhuizen EC, Washburn MP, Weake VM. 2019. The Drosophila Dbf4 ortholog Chiffon forms a complex with Gcn5 that is necessary for histone acetylation and viability. J Cell Sci 132: jcs214072 10.1242/jcs.214072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HG, Steger DJ, Iyer VR, Johnson AD. 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J 19: 2323–2331. 10.1093/emboj/19.10.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle LM, Pacheco D, Warfield L, Luo J, Ranish J, Hahn S, Klevit RE. 2018. Gcn4-mediator specificity is mediated by a large and dynamic fuzzy protein–protein complex. Cell Rep 22: 3251–3264. 10.1016/j.celrep.2018.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. 1997. Induced α helix in the VP16 activation domain upon binding to a human TAF. Science 277: 1310–1313. 10.1126/science.277.5330.1310 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980. 10.1016/j.cell.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis CD, Berger SL. 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol 17: 519–527. 10.1128/MCB.17.1.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dienemann C, Stutzer A, Urlaub H, Cheung ACM, Cramer P. 2020. Structure of the transcription coactivator SAGA. Nature 577: 717–720. 10.1038/s41586-020-1933-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Ranish JA, Hahn S. 2004. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev 18: 1022–1034. 10.1101/gad.1192204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S. 2017. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol Cell 68: 118–129.e5. 10.1016/j.molcel.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, Workman JL. 2009. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev 23: 2818–2823. 10.1101/gad.1846409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl RO, Ruppert S, Dynlacht BD, Tanese N, Tjian R. 1993. Cloning and expression of Drosophila TAFII60 and human TAFII70 reveal conserved interactions with other subunits of TFIID. EMBO J 12: 5303–5309. 10.1002/j.1460-2075.1993.tb06226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek E, Brand M, Jacq X, Tora L. 1998. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature 393: 187–191. 10.1038/30283 [DOI] [PubMed] [Google Scholar]
- Winston F, Carlson M. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8: 387–391. 10.1016/0168-9525(92)90300-S [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Malone EA, Clare J, Kapakos JG, Farabaugh P, Minehart PL. 1987. Three genes are required for trans-activation of Ty transcription in yeast. Genetics 115: 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Winston F. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol 22: 5367–5379. 10.1128/MCB.22.15.5367-5379.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15: 199–208. 10.1016/j.molcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. 1996. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380: 316–322. 10.1038/380316a0 [DOI] [PubMed] [Google Scholar]
- Xu YZ, Query CC. 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28: 838–849. 10.1016/j.molcel.2007.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J. 2011. Structure and function of WD40 domain proteins. Protein Cell 2: 202–214. 10.1007/s13238-011-1018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Roth SY. 1998. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol 18: 5659–5669. 10.1128/MCB.18.10.5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382: 319–324. 10.1038/382319a0 [DOI] [PubMed] [Google Scholar]
- Zhang N, Ichikawa W, Faiola F, Lo SY, Liu X, Martinez E. 2014. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta 1839: 395–405. 10.1016/j.bbagrm.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mi W, Xue Y, Shi X, Kutateladze TG. 2019. The ZZ domain as a new epigenetic reader and a degradation signal sensor. Crit Rev Biochem Mol Biol 54: 1–10. 10.1080/10409238.2018.1564730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Will CL, Bertram K, Dybkov O, Hartmuth K, Agafonov DE, Hofele R, Urlaub H, Kastner B, Lührmann R, et al. 2020. Molecular architecture of the human 17S U2 snRNP. Nature 583: 310–313. 10.1038/s41586-020-2344-3 [DOI] [PubMed] [Google Scholar]
- Zsindely N, Pankotai T, Ujfaludi Z, Lakatos D, Komonyi O, Bodai L, Tora L, Boros IM. 2009. The loss of histone H3 lysine 9 acetylation due to dSAGA-specific dAda2b mutation influences the expression of only a small subset of genes. Nucleic Acids Res 37: 6665–6680. 10.1093/nar/gkp722 [DOI] [PMC free article] [PubMed] [Google Scholar]