In this study, Parisian et al. introduced an inducible SMARCB1 loss-of-function system into human iPSCs that were subjected to either directed neuronal differentiation or differentiation into cerebral organoids to identify potential interactions between SMARCB1 loss and the process of neural development. Their findings provide insight into how SMARCB1 loss might interact with neural development in the process of ATRT tumorigenesis.
Keywords: SMARCB1, ATRT, iPSC, organoid, BAF complex, neural development, tumor modeling
Abstract
Atypical teratoid rhabdoid tumors (ATRTs) are challenging pediatric brain cancers that are predominantly associated with inactivation of the gene SMARCB1, a conserved subunit of the chromatin remodeling BAF complex, which has known contributions to developmental processes. To identify potential interactions between SMARCB1 loss and the process of neural development, we introduced an inducible SMARCB1 loss-of-function system into human induced pluripotent stem cells (iPSCs) that were subjected to either directed neuronal differentiation or differentiation into cerebral organoids. Using this system, we identified substantial differences in the downstream effects of SMARCB1 loss depending on differentiation state and identified an interaction between SMARCB1 loss and neural differentiation pressure that causes a resistance to terminal differentiation and a defect in maintenance of a normal cell state. Our results provide insight into how SMARCB1 loss might interact with neural development in the process of ATRT tumorigenesis.
The gene SMARCB1 encodes a subunit of the BAF (also known as SWI/SNF) chromatin remodeling complex. The BAF complex uses ATP hydrolysis to restructure chromatin through alterations of nucleosome positioning and occupancy (Cairns 2007), leading to downstream changes in chromatin accessibility (Tolstorukov et al. 2013; Kadoch et al. 2017) and enhancer activity (Nakayama et al. 2017; Wang et al. 2017). The BAF complex has important roles in development and cellular differentiation. Subunit composition has been shown to change as pluripotent cells differentiate (Lessard et al. 2007; Ho and Crabtree 2010), and a distinct version of the complex with defined subunit composition has been identified in stem cells (Ho et al. 2009). In addition, members of the complex have been identified as reprogramming factors to generate pluripotent cells from somatic cells (Singhal et al. 2010). Nucleosomal occupancy changes are an important aspect of the epigenetic alterations that undergo cellular differentiation (West et al. 2014), and the BAF complex in general along with SMARCB1 in particular have been shown to be important for the regulation of normal nucleosomal occupancy patterns (Tolstorukov et al. 2013; You et al. 2013), with downstream effects on transcription factor binding, enhancer activity, and gene expression.
In addition to their normal roles during development, many BAF complex genes have demonstrated roles as tumor suppressor genes. When taken together, the 20 BAF subunit genes have been shown to be mutated in 19% of all tumor types (Shain and Pollack 2013). This speaks to the important genome-wide role of this complex in maintenance of a stable epigenome. Genetic loss of function of SMARCB1 in particular has been shown to be both sufficient and necessary for tumorigenesis of atypical teratoid rhabdoid tumors (ATRTs) (Versteege et al. 1998; Reincke et al. 2003; Jackson et al. 2009), a highly aggressive and early onset pediatric brain tumor. The mutation rate in ATRTs is very low (Lee et al. 2012; Johann et al. 2016), with no other consistent recurrent mutations identified. This low number of mutations is consistent with an early age of onset, but also implies that SMARCB1 loss likely leads to tumorigenesis through initiation of epigenetic changes rather than through the combined effect of multiple genetic mutations. With a median age of onset of 11 months and a lethality rate of 80%–90% (Roberts and Orkin 2004), these tumors are responsible for a huge loss of potential life. In addition, very few effective therapies are available for the treatment of ATRTs and treatment is complicated by the negative cognitive effects of brain radiation in young children (Ginn and Gajjar 2012). Targeted therapeutics could provide a much-needed alternative to radiation, the development of which would be aided by a greater understanding of the mechanisms driving ATRT tumorigenesis and access to additional model systems with relevance to the human disease.
While transcriptomic and epigenomic analyses of ATRT samples (Johann et al. 2016; Torchia et al. 2016; Chun et al. 2019; Erkek et al. 2019) have characterized the epigenetic alterations that take place following SMARCB1 loss, the mechanisms by which SMARCB1 loss leads to these changes and the factors required for SMARCB1 loss to initiate cellular transformation are not well understood. Increased polycomb-repressive complex 2 (PRC2) binding (Wilson et al. 2010; Kadoch et al. 2017) and skewed SMARCB1-deficient BAF complex binding at superenhancers (Nakayama et al. 2017; Wang et al. 2017) have been suggested mechanisms of tumorigenesis due to SMARCB1 loss, but many questions still remain unanswered. The sufficiency of SMARCB1 deletion to drive pediatric tumor growth but lack of SMARCB1 mutation as an exclusive driver mutation in adult cancers along with the demonstrated role of the BAF complex in development and differentiation leads us to the hypothesis that the ability of SMARCB1 deletion to cause tumorigenesis may be dependent on the epigenetic environment of a particular stage in cellular differentiation. Engineering of induced pluripotent stem cells (iPSCs) with known tumorigenic alterations has been shown to be an effective technique for modeling of glioblastoma (Koga et al. 2020), leading us to apply an inducible system of SMARCB1 loss in iPSCs to address this question.
Results
SMARCB1 loss causes differential phenotypes in pluripotent and committed cell types
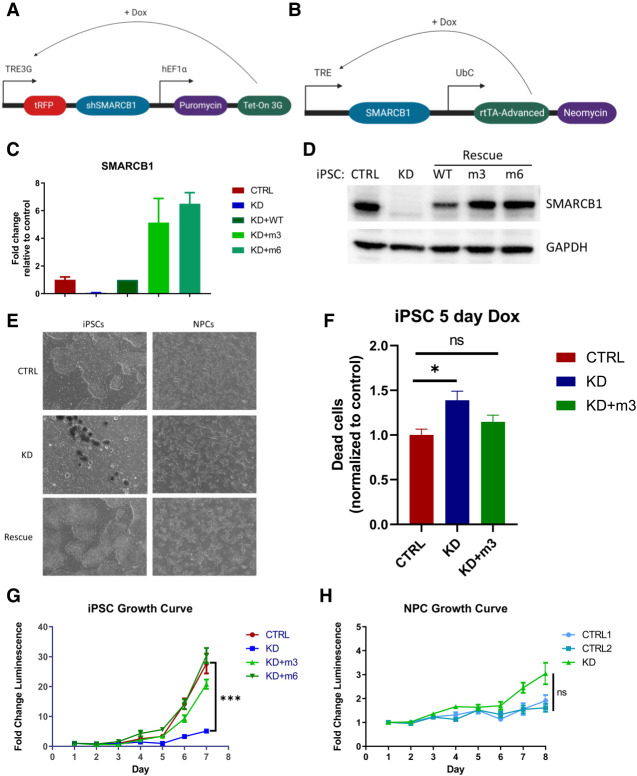
To interrogate possible interactions between SMARCB1 loss and cellular differentiation state, we generated a doxycycline-inducible SMARCB1 loss-of-function system in an iPSC line using an inducible shRNA construct targeting SMARCB1 (Fig. 1A; Supplemental Fig. S1A). To rule out the possibility that any observed effects could be due to shRNA off-target effects on genes other than SMARCB1, a doxycycline-inducible SMARCB1 re-expression vector was engineered with either three (m3) or six (m6) silent mutations in the shRNA target sequence (Fig. 1B; Supplemental Fig. S1A). Treatment of this cell line with doxycycline resulted in rapid reduction of SMARCB1 transcript and protein levels (Fig. 1C,D), both of which were successfully rescued in the presence of the re-expression vector. With this inducible system, SMARCB1 loss could be initiated at various stages of differentiation to observe the interplay between cell state and the effects of SMARCB1 loss. After initial doxycycline induction at the iPSC state, it was observed that prolonged induction of SMARCB1 loss resulted in a pronounced cell death phenotype in shSMARCB1 iPSCs (Fig. 1E,F; Supplemental Fig. S1B) but not in control iPSCs engineered with a nontargeting shRNA. Beginning 3 d after doxycycline induction, a pronounced decrease in growth rate was observed (Fig. 1G) along with an increase in cell death as measured by cell cycle assay, which showed an increase in sub-G-phase dead and dying cells (Fig. 1F; Supplemental Fig. S1B). This SMARCB1-induced cell death phenotype is consistent with the mouse model data showing embryonic lethality of SMARCB1 knockout mice (Roberts et al. 2000; Han et al. 2016), but has not been previously demonstrated in human cells. Cell death induced by SMARCB1 loss was replicated in a separate doxycycline-inducible SMARCB1 knockdown iPSC line using a CRISPR interference method of transcription repression (Supplemental Fig. S2A–E). However, this system proved to be less stable than the shRNA method and was subject to silencing during differentiation. For this reason, all differentiation experiments were conducted using the shRNA knockdown method with rescue vector. To investigate whether the effects of SMARCB1 loss might vary with differentiation state, iPSCs were differentiated into neural progenitor cells (NPCs) according to the protocol described by Reinhardt et al. (2013) (Supplemental Fig. S1C) prior to exposure to doxycycline. Cells were induced with doxycycline for 5 d and monitored for changes in morphology or growth rate. In contrast to the iPSCs, SMARCB1 knockdown NPCs tolerated the loss and displayed no changes in growth rate or morphology (Fig. 1E,H), even with extended doxycycline treatment (data not shown) and a similar level of knockdown as observed in the iPSCs (Supplemental Fig. S1D). SMARCB1 knockdown NPCs displayed changes in expression of BAF complex subunits similar to those observed in SMARCB1-deficient rhabdoid cell lines and reductions in BAF complex stability (Supplemental Fig. S1E) consistent with those observed in the literature with ATRT cell lines (Wang et al. 2017), suggesting that shRNA knockdown of SMARCB1 has a similar molecular effect to SMARCB1 loss occurring through genomic deletion.
Figure 1.
Development of an inducible SMARCB1 knockdown system reveals that SMARCB1 loss causes lethality in pluripotent cells but not neural progenitors. (A) Schematic representation of doxycycline-inducible SMARCB1 shRNA construct, which was stably transduced into induced pluripotent stem cells (iPSCs). (B) Schematic representation of doxycycline-inducible SMARCB1 rescue construct, which was stably transduced into shSMARCB1 iPSCs to rescue SMARCB1 knockdown. (C,D) Efficacy of shSMARCB1 and rescue vector was tested in iPSCs after 3 d of doxycycline induction using qRT-PCR to measure SMARCB1 transcript levels and standard deviation relative to control mean (C), or Western blot to measure SMARCB1 protein levels (D). (E) Phase contrast images at 4× magnification of control, SMARCB1 knockdown, and rescue iPSCs and NPCs after 5 d of doxycycline induction. (F) Percentage of dead cells based on cell cycle assay of control, rescue, and SMARCB1 knockdown iPSCs induced with doxycycline for 5 d. (G,H) Growth of iPSCs (G) and NPCs (H) was also assessed with doxycycline induction beginning at day 0 of assay.
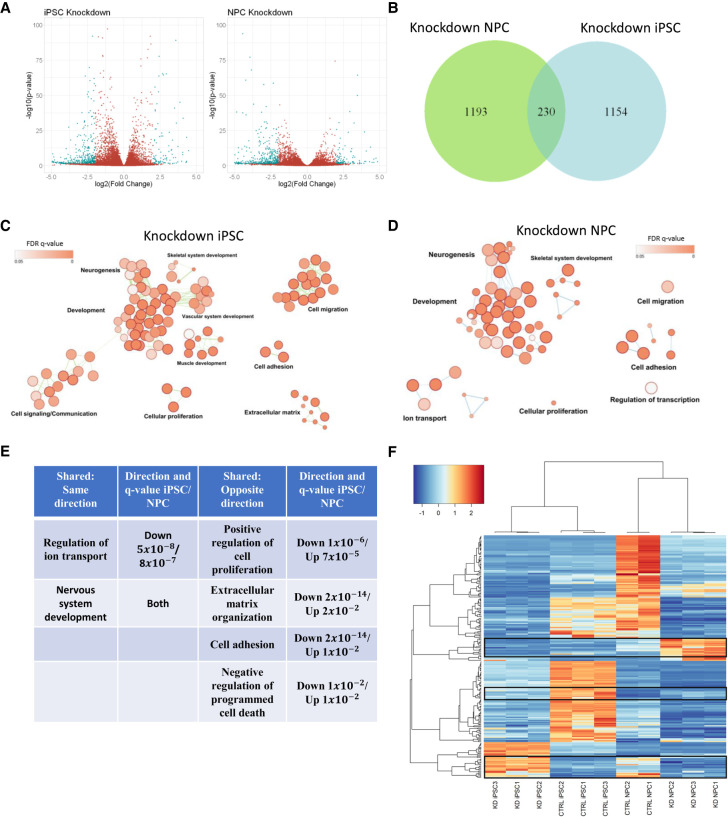
To identify transcriptional differences underlying these contrasting phenotypes, we conducted RNA sequencing on control and SMARCB1 knockdown iPSCs and NPCs. In both cell types, more down-regulated genes were observed in SMARCB1 knockdown cells than up-regulated genes (Fig. 2A; Supplemental Fig. S2D,E). This is consistent with the previously described mechanism for epigenetic and transcriptional changes underlying ATRTs, in which loss of SMARCB1 leads to a decrease in BAF complex activity and a corresponding decrease in H3K27Ac active histone marks, along with altered activity of the repressive PRC2 complex (Wilson et al. 2010; Kadoch et al. 2017; Nakayama et al. 2017; Erkek et al. 2019). Comparison of the genes differentially expressed by SMARCB1 loss in the two cell types revealed very little overlap between knockdown NPCs and iPSCs (Fig. 2B), suggesting that the downstream targets of SMARCB1 can vary substantially in different cell types. Gene ontology analysis of the dysregulated genes show similarities in the classes of genes altered by SMARCB1 loss in the two cell types, including genes associated with neural development, cellular proliferation, and cellular adhesion (Fig. 2C,D). However, many of these shared genes were altered in opposite directions in iPSCs and NPCs, both on the ontology level (Fig. 2E) and on the individual gene level (Fig. 2F). About a quarter of genes that were dysregulated in both iPSCs and NPCs were up-regulated in one cell type but down-regulated in the other (Fig. 2F). This unexpected result suggests that the transcriptional effects of SMARCB1 loss can vary dramatically in different cell types, even leading to opposite phenotypic and transcriptional effects, and explains the very different growth phenotypes observed in knockdown iPSCs and NPCs. These results, along with the established role of the BAF complex in developmental processes (Lessard et al. 2007; Ho et al. 2009; Ho and Crabtree 2010), lead us to believe that SMARCB1 loss might also have dramatic impacts on cellular differentiation processes, potentially highlighting an interplay between differentiation state and ATRT tumorigenesis.
Figure 2.
SMARCB1 loss leads to differing transcriptional effects at defined stages of differentiation. RNA sequencing was conducted on SMARCB1 knockdown and shControl iPSCs and NPCs. (A) Volcano plot of genes differentially expressed in SMARCB1 knockdown cells relative to controls in each cell type. (B) Overlap of genes differentially expressed in SMARCB1 knockdown iPSCs and NPCs relative to controls. (C,D) Gene ontology networks for differentially expressed genes in iPSCs (C) and NPCs (D). Dots represent statistically significant gene ontology terms, clustered based on overlap of the genes contained in each term. Dot size indicates the number of genes included in each term and darker color corresponds to smaller adjusted P-value. Labels indicate the main process making up each cluster. (E) Table comparing shared gene ontology results and direction of alteration between SMARCB1 knockdown iPSCs and NPCs. q-value was obtained using the Benjamini-Hochberg adjustment for multiple comparisons. (F) Heat map showing genes that are differentially expressed in both iPSCs and NPCs. Boxes indicate regions that are altered in opposite directions between iPSCs and NPCs. Overall, 62 of 230 overlapping genes (27%) were altered in opposite directions between the two cell types.
Neural development without SMARCB1 leads to defects in neuron formation in an organoid model
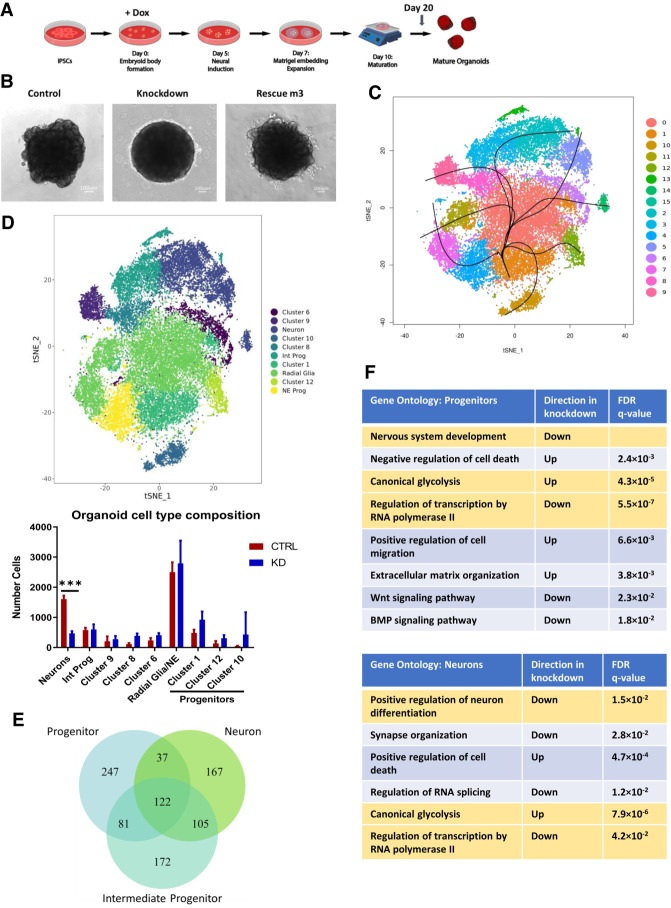
To assess the effect of SMARCB1 loss on neural differentiation, we used a cerebral organoid model of neural development (Fig. 3A; Lancaster and Knoblich 2014). Because this protocol results in the formation of multiple regional identities without selecting for specific neural cell types (Lancaster et al. 2013), the model allows a relatively unbiased assessment of the impact of SMARCB1 loss on the neural developmental process. shControl or shSMARCB1 iPSCs were induced to form cerebral organoids with doxycycline induction beginning at various time points through the protocol and assessed for changes in expression of various neural marker genes (Supplemental Fig. S3A). While no obvious changes were observed in markers of pluripotency (Nanog) or neural progenitor formation (Pax6), decreases were observed in markers of neuronal commitment and maturation (Dcx, visible trend in Map2), especially with earlier doxycycline induction. It was also observed that these early knockdown organoids demonstrated morphological differences relative to the control during expansion and early maturation phases of the protocol (Fig. 3B). Knockdown organoids were defective for the outward expansion of neuroepithelium (Lancaster et al. 2013) into the surrounding matrix typically observed after Matrigel embedding at day 7 of the differentiation protocol, suggestive of a defect in normal cell differentiation. Immunofluorescence images of day 20 organoids show a reduction in staining of neuron marker STMN2 at the periphery of knockdown organoids (Supplemental Fig. S3B), supporting this hypothesis. In addition, no visible differences in Ki67 or Caspase3 staining were observed in the knockdown organoids (Supplemental Fig. S3C,D), making it unlikely that these morphological changes were driven by increased cell death or a reduction in cellular proliferation. These results imply that there is a window early in development where cells are especially sensitive to the effects of SMARCB1 loss.
Figure 3.
SMARCB1 loss during cerebral organoid development leads to neural differentiation defects. Cerebral organoids were formed from shSMARCB1 and shControl iPSCs in the presence of doxycycline from day 0 of differentiation protocol. (A) Schematic showing stages of organoid generation from iPSCs. (B) At day 10 of the protocol, organoids were examined for morphology and presence of neuroepithelial expansion at 4× magnification in control, SMARCB1 knockdown, and rescue organoids. Knockdown organoids exhibit absence of neuroepithelial expansion. (C) Single-cell RNA sequencing was conducted on three day 20 control and SMARCB1 knockdown organoids using droplet-based scRNA-seq methodology. Canonical correlation analysis was conducted on combined single-cell data and displayed on a tSNE graph. Clustering and pseudotime analysis was conducted on the combined data to identify variability in cell types and lineages within the organoids. Fifteen clusters were identified, of which cluster 15 was excluded from analysis due to its small size. (D, top) Clusters were analyzed for expression of neural differentiation markers and grouped together by cell type where possible. Clusters not matched to a particular cell type were left unnamed. (Bottom) Mean with standard deviation of the number of each cell type in control and knockdown organoids. Statistical comparisons were conducted using two-way ANOVA with Sidak multiple comparisons test. (***) Adjusted P-value < 0.001. (E) Venn diagram of the overlap across cell types of genes differentially expressed in SMARCB1 knockdown organoids relative to control organoids. (F) Tables showing top gene ontology results from genes differentially expressed in SMARCB1 knockdown organoids relative to controls relative to a list of all expressed genes in progenitor cells (top) and neurons (bottom). Ontologies highlighted in gold are similarly altered in both cell types.
To better assess the impact of early-stage SMARCB1 loss on neural differentiation we conducted droplet-based single-cell RNA sequencing (scRNA-seq) on three control and three SMARCB1 knockdown organoids at day 20 of the differentiation protocol, when morphological differences were apparent. These six organoids (Supplemental Fig. S4A) were aligned and clustered using canonical correlation analysis (Butler et al. 2018) in order to compare numbers of neural cell types between control and knockdown organoids. Cluster analysis resulted in 15 distinct clusters (Fig. 3C; Supplemental Fig. S4B,C), one of which was excluded for containing <100 cells. All but one of these remaining clusters (cluster 10) contained a similar distribution of cells across replicate organoids (Supplemental Fig. S4A). Organoids had similar distributions of UMI counts and detected genes (Supplemental Fig. S4D), and SMARCB1 knockdown organoids displayed loss of SMARCB1 transcript in nearly all cells analyzed (Supplemental Fig. S4E). Clusters were analyzed for expression of several neural development marker genes to identify corresponding cell types (Supplemental Fig. S5A–F) and Slingshot pseudotime analysis (Street et al. 2018) was performed to identify differentiation trajectories across clusters (Fig. 3C). This analysis revealed a mix of clusters representing neural progenitors, positive for markers such as Sox2, Pax6, Hes1, and Hes5 (Supplemental Fig. S5A,C,D), and various stages of neuronal differentiation including intermediate progenitors positive for markers NHLH1 and EOMES (Supplemental Fig. S5E), immature neurons positive for DCX, and more mature neurons with high expression of markers such as STMN2, MAP2, and NCAM1 (Supplemental Fig. S5F,G). Within the progenitor clusters, some seemed to represent neuroepithelial-like cells positive for early neural markers Sox2 and Hes1 while others appeared more radial glia-like, with expression of markers such as VIM, Hes5, and Pax6 (Supplemental Fig. S5B,D), while others lacked consistent expression of these markers and may represent progenitors of a distinct lineage (clusters 1, 10, and 12). Other clusters were defined by aspects of cell state such as cell cycle stage or apoptosis (Supplemental Fig. S5B) rather than cell type. Grouping together the identifiable clusters representing neuroepithelial-like progenitors (cluster 4) and radial glia-like cells (clusters 0, 7, and 11), intermediate progenitors (clusters 3 and 13), and committed neurons (clusters 2, 5, and 14) (Fig. 3D), the number of cells in each group were quantified in both control and SMARCB1 knockdown organoids. The number of cells in neuron-associated clusters was substantially lower in SMARCB1 knockdown organoids (P < 0.001) than controls (Fig. 3D; Supplemental Fig. S4C) and the expression of individual neuronal markers was lower in knockdown organoids (Supplemental Fig. S5G), suggesting that the knockdown might be causing a differentiation block and preventing cells from achieving a neuronal cell fate. Although no differences were observed in the number of neuroepithelial or radial glial progenitors, some apparent increases (although not statistically significant to P < 0.05 with current number of replicates), spread across progenitor clusters 1, 12, and 10 (Fig. 3D; Supplemental Fig. S4C) suggest that SMARCB1 loss might lead to a shift in the lineage preference of cells during differentiation while contributing toward a preference for less differentiated cell types.
Differences in the gene expression changes caused by SMARCB1 loss across cell types within the organoids highlight the effects of SMARCB1 loss on differentiation as well as further demonstrating that the transcriptional effects of SMARCB1 loss vary with cell state. For differential expression analysis, related clusters were combined to form larger groups representing different stages of neural differentiation: neural progenitor cells (combining neuroepithelial progenitors, radial glia, and progenitor clusters 1 and 12), intermediate neuronal progenitors, and committed neurons. For each cell type, differential expression analysis was conducted comparing cells of that type in the control and knockdown organoids. A similar number of genes were significantly dysregulated in each cell type, but only about a quarter of these were dysregulated in all three cell types (Fig. 3E). In addition, the number of overlapping genes was greater between more closely related cell types (progenitors and intermediates or intermediates and neurons) than between the more distantly related progenitors and committed neurons. This suggests that there may be a spectrum of transcriptional changes occurring with SMARCB1 loss that varies throughout the developmental process and has different effects on cells at different stages of cellular differentiation. Gene ontology analysis of dysregulated genes in neural progenitors and neurons showed that different biological processes were affected in the two cell types (Fig. 3F). While canonical glycolysis was up-regulated in both cell types and genes associated with both neural development and transcriptional regulation were down-regulated, cell death processes were altered in opposite directions. In addition, neural progenitors had additional changes in pathways associated with cellular migration, extracellular matrix organization, Wnt signaling, and BMP signaling, which were not observed in the more differentiated cells. These differences in transcriptional state illustrate how SMARCB1 loss could lead to distinct cellular phenotypes depending on the stage of cellular differentiation.
SMARCB1 loss during neuronal differentiation leads to a lack of stability among neural progenitors that may contribute to tumorigenesis
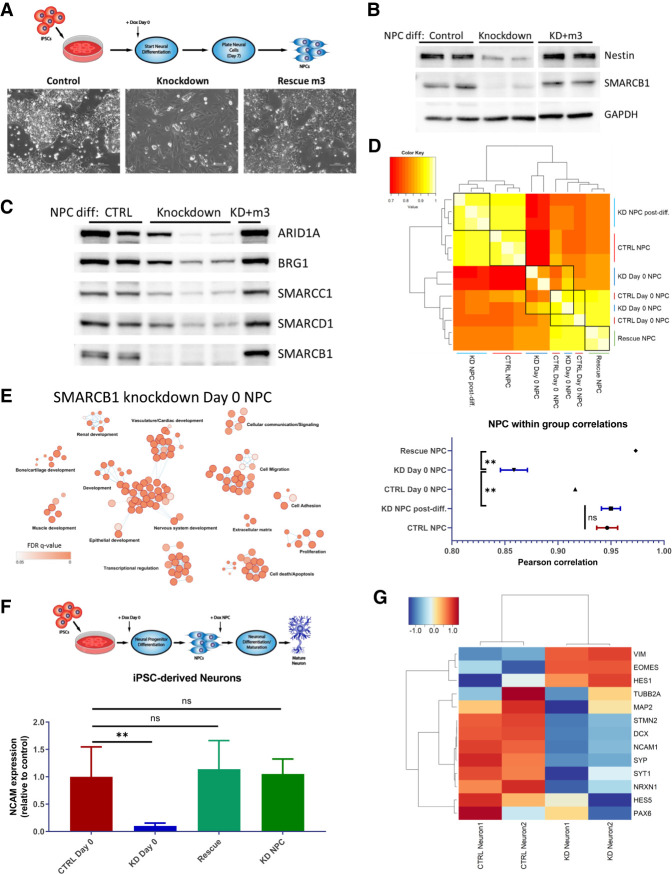
To validate that the differentiation defects observed in the organoid system are reproducible in other contexts, and to further investigate the effects of SMARCB1 loss on neural differentiation processes, iPSCs were induced with doxycycline and simultaneously differentiated into neural progenitor cells (Fig. 4A). Resulting progenitors were cultured for several passages postdifferentiation to assess their ability to maintain an NPC state, and it was observed that NPCs differentiated without SMARCB1 were prone to morphology changes two to five passages postdifferentiation (Fig. 4A), while control and SMARCB1 rescue NPCs maintained a consistent morphology for up to 10 passages (data not shown). These morphological alterations resulted in an increased cell surface area and tendency to spread across the tissue culture dish away from distinct groups of cells, and suggest a loss in ability to maintain a consistent differentiation state. NPCs differentiated without SMARCB1 were also subject to a low-frequency enhancement in growth rate (Supplemental Fig. S6A), another indication of a lack of stability in these cells relative to control or rescue NPCs. These cells demonstrated a reduction in levels of neural progenitor marker Nestin (Fig. 4B) that is prevented by SMARCB1 rescue, implying a defect in differentiation in the absence of SMARCB1, consistent with observed results using the organoid system. Analysis of BAF complex expression levels in NPCs differentiated without SMARCB1 revealed a decrease in levels of nuclear BAF complex subunits ARID1A, BRG1, SMARCC1, and SMARCD1 relative to control NPCs (Fig. 4C), consistent with what has been observed in SMARCB1 re-expression cell lines (Wang et al. 2017). However, the level of decrease varied substantially in different batches of differentiation for the same level of SMARCB1 knockdown (Fig. 4C), suggesting a stochasticity in the downstream effects of SMARCB1 loss after application of cellular differentiation pressures. RNA-seq of four NPC lines differentiated in the absence of SMARCB1 also revealed a higher transcriptomic variability than was observed in control or rescue cells differentiated with doxycycline or in NPCs subjected to SMARCB1 loss postdifferentiation (Fig. 4D). Correlations within replicates of NPCs differentiated without SMARCB1 were significantly lower than rescue NPCs or NPCs with SMARCB1 knockdown induced at the NPC state. A comparison in the genes dysregulated when SMARCB1 is absent throughout NPC differentiation and those altered with SMARCB1 loss at the NPC state (Fig. 4E; Supplemental Fig. S6B,C) revealed that SMARCB1 loss throughout the differentiation process leads to changes in a wide variety of differentiation-associated pathways ranging from renal development to ossification in addition to the expected neural development-associated genes. Changes in pathways associated with cell death, cellular proliferation, and TGF-β signaling are also observed in genes dysregulated by SMARCB1 loss when it occurs during NPC differentiation. A time course of doxycycline application throughout the NPC differentiation process (Supplemental Fig. S6D) verified that more deleterious effects on neural development are observed with earlier induction of SMARCB1 loss. Immunofluorescence analysis of neural progenitor marker Hes5 in NPCs differentiated without SMARCB1 (Supplemental Fig. S7A) reveals significant reduction of Hes5 levels consistent with an inability to differentiate that is not observed with postdifferentiation SMARCB1 knockdown.
Figure 4.
SMARCB1 loss throughout neural differentiation leads to aborted differentiation and a lack of stability in resulting neural progenitor cells. (A, top) Schematic of directed differentiation of iPSCs into neural progenitor cells, with doxycycline induction at day 0. (Bottom) Phase contrast images at 20× magnification of resulting NPC morphology at days 10–14 of protocol. (B) Western blot showing protein expression of SMARCB1, neural marker Nestin, and control GAPDH in control, SMARCB1 knockdown, or rescue NPCs differentiated in the presence of doxycycline. (C) Western blot of BAF complex subunit protein expression in nuclear lysates of NPCs differentiated in the presence of doxycycline. (D, top). Pearson correlation chart comparing transcriptome similarity between control, knockdown, or rescue NPCs differentiated with or without doxycycline. Black boxes indicate groups being compared. White corresponds to greater correlation and red to lower correlation. (Bottom) Mean within group correlation values with standard error of the mean for each group of NPCs. Within-group correlations were compared using one-way ANOVA with Tukey's multiple comparisons test. (**) Adjusted P-value < 0.01. (E) Gene ontology network of top 500 genes that were differentially expressed in NPCs differentiated with SMARCB1 knockdown relative to controls but not in NPCs with knockdown postdifferentiation relative to controls. Dots represent statistically significant gene ontology terms, clustered based on overlap of the genes contained in each term. Dot size indicates the number of genes included in each term and darker color corresponds to smaller adjusted P-value. Labels indicate the main process making up each cluster. (F, top) Schematic showing directed neuronal differentiation with doxycycline induction either at day 0 or at the NPC state. Control, SMARCB1 knockdown, and rescue neurons differentiated in this manner were assessed for neuronal maturation efficacy by FACS analysis for neuronal surface marker NCAM1. (Bottom) Mean and standard deviation percentages of NCAM-positive cells in postdifferentiation neurons. Comparisons between groups were conducted by one-way ANOVA with Tukey's multiple comparisons test. (**) P-value < 0.01. (G) Heat map of scaled transcript expression of neuronal differentiation markers in control or SMARCB1 knockdown neurons differentiated with day 0 doxycycline. VIM, HES1, HES5, PAX6, and EOMES are markers of less differentiated neural cells. All other genes are markers of committed neurons.
To validate results from the organoid model and further assess the interaction between SMARCB1 loss and differentiation, control and knockdown cells were subjected to in vitro directed neuronal differentiation (Reinhardt et al. 2013). Neuronal maturation efficacy was measured using FACS analysis for surface expression of NCAM, a marker of mature neurons (Fig. 4F). Cells subjected to SMARCB1 knockdown from day 0 of NPC differentiation had lower numbers of NCAM-positive cells after 25 d of neuronal differentiation and maturation than control cells, as well as when compared with cells subjected to knockdown beginning at the NPC state. RNA-seq analysis of control and knockdown neurons showed a reduction in the expression of neuronal markers in cells differentiated in the absence of SMARCB1, along with a retention of some markers of earlier stages of neural differentiation (Fig. 4G). Immunofluorescence of these neurons (Supplemental Fig. S7B) also shows a loss in neuronal marker expression in neurons differentiated entirely without SMARCB1. These data suggest that SMARCB1 loss during neuronal differentiation leads to a failure in maturation in multiple contexts and validate that cells are particularly vulnerable to SMARCB1 loss early in neural development. This window of vulnerability is consistent between organoid and directed neuronal differentiation experiments and demonstrates a similar trend to that previously observed in an inducible SMARCB1 knockout mouse model (Han et al. 2016).
Neural progenitors differentiated without SMARCB1 are transcriptionally similar to ATRTs, particularly the SHH subgroup
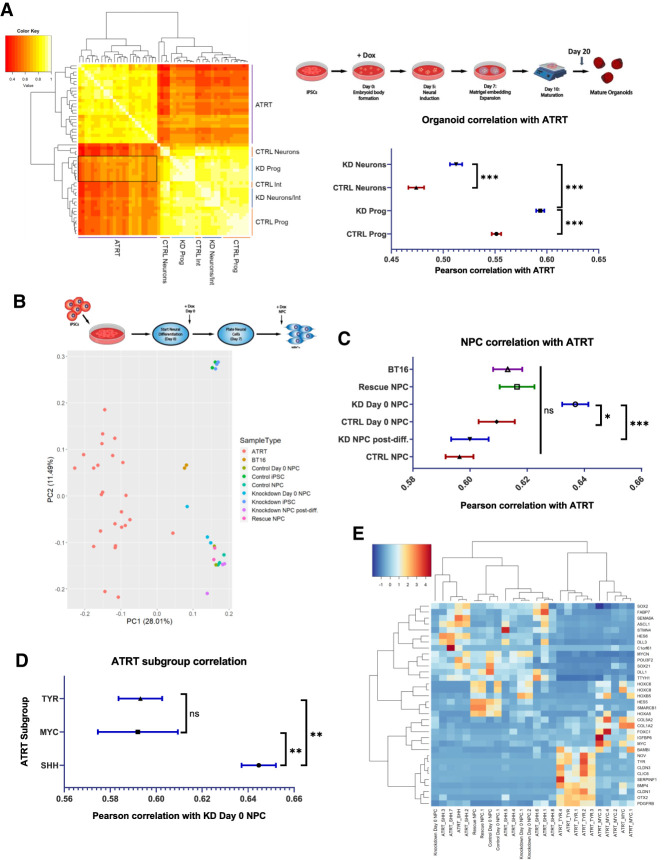
It seems probable that these observed interactions between SMARCB1 loss and neural differentiation could play a role in ATRT tumorigenesis. To investigate this, previously published bulk RNA-seq data generated from ATRT tumors (Johann et al. 2016) was obtained in order to determine the similarity of this cellular model to patient tumors and to identify cell types with the greatest similarity. To compare the tumor data with the organoid scRNA-seq data, averaged transcriptomic data for each organoid cluster was computed and correlated to the ATRT samples (Fig. 5A). While correlations were generally higher within the organoid or tumor groups, there was variability in the similarity of different organoid cell types to tumors. Neurons in the control organoids were the least similar to the tumors, while progenitor clusters in the SMARCB1 knockdown organoids were most similar (Fig. 5A). This is consistent with the concept of a SMARCB1-deficient early neural progenitor acting as the cell of origin for ATRTs. Progenitor clusters in the control organoids were generally less similar to the tumor samples than the same clusters in the knockdown organoids, with the least differentiated clusters (10 and 12) showing the greatest similarity to tumors (Fig. 5A). These clusters also demonstrate a possible (but not statistically significant with n = 3 organoids) expansion in knockdown organoids relative to controls (Supplemental Fig. S4C), and thus their development may be favored in the absence of SMARCB1 expression. SMARCB1 knockdown progenitors also show changes in genes associated with transcriptional regulation, nervous system development, and extracellular matrix organization (Fig. 3F), all pathways identified as being altered in ATRTs (Johann et al. 2016; Torchia et al. 2016). Comparison of ATRT transcriptomes with RNA-seq data from control and SMARCB1 knockdown iPSCs and NPCs (Fig. 5B,C; Supplemental Fig. S8A) revealed greater ATRT similarity to NPCs differentiated without SMARCB1 than either knockdown iPSCs or NPCs induced with SMARCB1 loss postdifferentiation. Previous transcriptomic and epigenomic analyses have identified three subgroups within ATRTs with differing epigenetic landscapes and gene expression profiles (Johann et al. 2014; Torchia et al. 2016). A comparison of both NPCs differentiated without SMARCB1 via small molecule-directed differentiation and progenitors within SMARCB1 knockdown organoids with ATRTs from each of the three subgroups revealed the greatest similarity with the SHH, or Neurogenic subgroup (Fig. 5D,E; Supplemental Fig. S8B–D). This suggests a possible mechanism of ATRT tumorigenesis, likely most relevant to the SHH subgroup, in which focal deletion of SMARCB1 occurs early in neural development, leading to unstable NPCs with tendencies toward differentiation defects, cellular transformation and tumorigenesis (Fig. 6).
Figure 5.
Neural progenitor cells differentiated without SMARCB1 are transcriptomically similar to atypical teratoid rhabdoid tumors. (A) RNA sequencing data from 25 ATRTs (Johann et al. 2014) was compared with averaged single-cell RNA sequencing data for each cluster in control and SMARCB1 knockdown cerebral organoids. (Left) Chart of Pearson correlation values between individual clusters and ATRT samples, clustered by similarity. White indicates highest correlation and red corresponds to lowest correlation. Labels indicate cell types corresponding to clusters. Box indicates region of highest similarity to ATRT samples. (Top right) Schematic of protocol used for organoid generation. (Bottom right) Mean and standard error of Pearson correlation values of progenitors and neurons from control and SMARCB1 knockdown organoids. Comparisons between groups conducted using one-way ANOVA with Tukey's multiple comparisons test. (***) Adjusted P-value < 0.001. (B, top) Schematic of protocol used for NPC differentiation either in the presence of doxycycline from day 0 (KD day 0) or at the NPC state (KD post-diff.). (Bottom) Principal component analysis of RNA sequencing results from 25 ATRT samples compared with directed differentiation of control or SMARCB1 knockdown iPSC-derived NPCs, along with BT16 ATRT cell line and undifferentiated iPSCs induced with doxycycline. (C) Mean and standard error of pearson correlation values of control and SMARCB1 knockdown NPCs and BT16 cell line with ATRT samples. Comparisons between groups conducted using one-way ANOVA with Tukey's multiple comparisons test. (*) Adjusted P-value < 0.05, (***) adjusted P-value < 0.001. (D) Mean and standard error of pearson correlation values of NPCs differentiated without SMARCB1 and samples from each of the three ATRT subgroups. Comparisons between groups conducted using one-way ANOVA with Tukey's multiple comparisons test. (**) Adjusted P-value < 0.01. (E) Heat map showing expression of ATRT subtype-specific genes across ATRT samples and day 0 doxycycline-induced NPCs, clustered by similarity of expression (displayed by dendrogram at the top).
Figure 6.
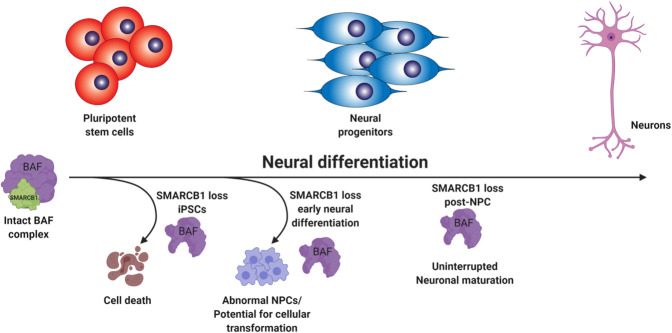
SMARCB1 loss interacts with developmental state to redirect cell fate. Schematic summarizing findings on the interaction between neural differentiation state and the effect of SMARCB1 loss. In pluripotent cells, SMARCB1 loss results in cell death. In the early stages of neural differentiation, SMARCB1 loss induces dedifferentiation, morphology changes, and lack of stability in resulting NPCs along with defects in capacity for further neuronal differentiation. With induction of knockdown in later stages of differentiation, little to no effect on differentiation capacity or cell growth was observed.
Discussion
SMARCB1 is an important chromatin remodeling subunit as well as a known tumor suppressor whose loss is the primary driver of pediatric rhabdoid tumors. In this study, we interrogated the interactions between SMARCB1 loss, cellular differentiation state, and transcriptional changes associated with tumorigenesis, while generating a cellular model that will have utility for future mechanistic studies as well as for identification of potential therapeutic vulnerabilities in SMARCB1-deficient cells. While other systems of SMARCB1 loss or reintroduction have been used to study the mechanisms underlying ATRTs in a controlled manner (Wilson et al. 2010; Han et al. 2016; Nakayama et al. 2017; Wang et al. 2017; Carugo et al. 2019; Langer et al. 2019), this complementary system has the benefit of using human cells, having the flexibility to take into account the effects of differentiation processes, and using SMARCB1 loss alone without additional oncogenic drivers, consistent with the human tumor phenotype (Lee et al. 2012). In addition, similar to a recent publication (Langer et al. 2019), our study provides an interrogation of the interactions between SMARCB1 loss and neural development; however, here we illustrate novel insight into the dramatic phenotypic differences that can occur with loss of SMARCB1 at different stages of differentiation, such as lethality in pluripotent cells and impairment of neuronal commitment and maturation. This is the first study to model the interaction between SMARCB1 loss and cellular differentiation state that likely contributes to ATRT tumorigenesis in human cells, and to monitor the accompanying gene expression and phenotypic changes.
Here, we demonstrated significant differences in the response of cells to SMARCB1 loss at differing stages of neural differentiation and identified a window early in neural commitment in which cells seem to be particularly vulnerable to SMARCB1 loss of function and in which SMARCB1 loss results in profound defects in the progression of differentiation. SMARCB1 loss during this period results in cells with greater similarity to ATRT tumors than loss at an earlier pluripotent or later committed neural progenitor state, along with a lack of stability resulting in a tendency toward stochastic alterations in cellular morphology and gene expression. This provides insight into a possible mechanism for ATRT tumorigenesis in which a loss of SMARCB1 during embryogenesis could result in cells that are primed for cellular transformation. This is consistent with both the early age of onset of this disease and heterogeneity of presentation, as well as with mouse data showing development of ATRT-like tumors with SMARCB1 loss during early mouse embryogenesis (Han et al. 2016). While it is clear that the neural progenitors differentiated without SMARCB1 in this study are most similar to the SHH/Neurogenic subgroup of ATRTs, more work is needed to determine the mechanism underlying this similarity. It is possible that the transcriptomic and epigenetic differences between the subgroups are driven by different cells or developmental stages of origin, and the origin of SHH/Neurogenic tumors more closely resembles the loss of function early in development that was applied in this study. Indeed, the SHH/Neurogenic subgroup of tumors does tend to occur in younger children, consistent with this hypothesis (Johann et al. 2016; Torchia et al. 2016). Another possibility is that the mechanism of SMARCB1 loss could play a role, with the larger chromosomal alterations observed more often in the other subgroups of ATRTs leading to additional effects on neighboring genes or regulatory regions not replicated with a knockdown model. Thus, the predominance of smaller focal or point mutations in the SHH subgroup might more closely resemble a SMARCB1 knockdown system.
In this study we focused on the interactions of SMARCB1 loss with neural development, but molecular heterogeneity and dysregulated developmental pathways observed in extra-cranial malignant rhabdoid tumors (Chun et al. 2016, 2019) suggest that a similar mechanism might take place in other types of rhabdoid tumors. Indeed, abnormal neural differentiation patterns has been implicated as playing a role in the tumorigenesis of a number of different pediatric and adult brain tumors (Ong et al. 2017; Jessa et al. 2019; Vladoiu et al. 2019), suggesting that the mechanisms observed in this study could have applications to a wider variety of tumor types. In all, we presented here an in-depth investigation into the stages of neural differentiation in which SMARCB1 loss has the greatest effects on cellular outcome, chronicled gene expression changes resulting from SMARCB1 loss at various stages of differentiation, and generated a novel platform on which to expand our understanding of the mechanisms and vulnerabilities underlying ATRT tumorigenesis.
Materials and methods
Pluripotent stem cell culture and neural differentiations
Human induced pluripotent stem cell (iPSC) line iPS12 was purchased from Cell Applications. This cell line is integration-free and was validated for pluripotency, viability, karyotype normality, and normal disease status by Cell Applications. CRISPRi Gen1C and WTC iPS lines were obtained from the Conklin laboratory at University of California at San Francisco. iPSCs were cultured using standard feeder-free conditions with mTESR1 or mTESR Plus medium on Matrigel-coated plates. iPSCs were induced to form neural progenitor cells using a small-molecule based differentiation protocol as described in Reinhardt et al. (2013), using combined small-molecule inhibition of BMP and TGFβ signaling along with WNT and SHH pathway stimulation. Neuron differentiations were also conducted as described in Reinhardt et al. (2013) for peripheral neurons, starting from NPCs of three to six passages in smNPC maintenance medium (N2B27 medium + CHIR + PMA). Neurons were harvested after 2 wk in neuronal maturation medium (N2B27 medium + dbcAMP + TGF-b3 + BDNF + GDNF). Both neural progenitor and neuron differentiations were conducted under 0.5 µg/mL puromycin to prevent loss of shRNA expression. Rescue cell lines were differentiated in the presence of 0.5 µg/mL puromycin and 100 µg/mL G418. When needed, doxycycline was applied at a 1 µg/mL concentration for all experiments.
Plasmids and cell line engineering
Doxycycline-inducible shRNA constructs against SMARCB1 and nontargeting controls were purchased from Dharmacon (SMARTvector Inducible Lentiviral shRNA) and transductions were conducted using Dharmacon trans-lentiviral packaging kit according to kit protocol. After selection with puromycin, individual clones were screened for SMARCB1 knockdown by quantitative real-time PCR and top clones were pooled to obtain highly efficient knockdown. Of three shRNA constructs tested, only one was capable of efficient SMARCB1 knockdown (sh905). Rescue vectors were engineered from pInducer20 plasmid backbone (Addgene 44012) to express SMARCB1 cDNA with either three or six silent mutations at the sh905 target site. See Supplemental Figure S1 for shRNA target and rescue sequences. For CRISPRi experiments, CRISPRi Gen1C cell line was transduced with several guide RNAs targeting the SMARCB1 locus and selected with blasticidin for guide RNA expression. Clones were then obtained, screened for SMARCB1 knockdown and top clones pooled to obtain a highly efficient level of knockdown in iPSCs.
Real-time PCR
RNA was extracted from cell pellets using Qiagen RNeasy Plus minikit and converted to cDNA using Takara RNA to cDNA EcoDry premix. Reactions were run in triplicate using cDNA converted from 10 ng of RNA. Primers used for real-time PCR analysis of SMARCB1 and neural developmental genes are included in Supplemental Table S1. Data were normalized to GAPDH expression.
Organoid development and culture
Organoids were generated using the STEMdiff cerebral organoid kit from Stem Cell Technologies (08570), which is based on the Lancaster and Knoblich (2014) protocol for cerebral organoid formation and development. Organoids were developed in the presence of 0.5 µg/mL puromycin and 1 µg/mL doxycycline. Organoids were matured for 10–50 d in maturation medium (day 10 of protocol). Organoids used for single-cell RNA-seq were matured for 10 d to look at early-stage commitment and development of neural progenitors.
Growth, cell cycle, and cell death assays
For growth assays, 1000–2000 cells/well with five to 10 replicates per cell line were plated on Matrigel-coated black 96-well plates in maintenance medium without antibiotic selection. First timepoint was read within 24 h of plating for baseline comparison and subsequent readings were performed every 24 h following. Medium changes were conducted as needed (every 2–3 d) throughout the assay. ATPlite 1step assay kit (PerkinElmer 6016731) was used to estimate cell number. For cell cycle assays, eight replicate doxycycline inductions of ∼1 × 106 cells were harvested and fixed overnight in 70% ethanol, washed three times with PBS and stained for 30 min with 0.5 mL of FxCycle PI/RNase staining solution (Life Technologies F10797) before quantification with a BD LSR II flow cytometer. Cell cycle percentages were calculated using FlowJo software and the Dean-Jett-Fox model. Doxycycline was added to shRNA iPSCs for 5 d before fixation and to CRISPRi iPSCs for 9 d before fixation.
Western blots and immunoprecipitations
For Western blots, cells were lysed in RIPA lysis buffer with proteinase and phosphatase inhibitors. Twenty micrograms of lysate was run on an SDS-PAGE gel, transferred at 350 mA over 1.5 h to EMD Millipore Immobilon-P PVDF membrane, blocked for 1 h in 5% BSA, and probed with primary antibody overnight. Membranes were washed with PBS + 0.1% Tween-20, probed for 1–2 h with secondary HRP-conjugated antibody and exposed using Thermo Scientific SuperSignal West Pico or Femto chemiluminescent substrate. For BAF complex immunoprecipitations, nuclear extractions were first performed using Thermo Scientific NE-PER nuclear and cytoplasmic extraction reagents (78833) according to kit instructions. For immunoprecipitations, 2 µg of BRG1 antibody (Santa Cruz Biotechnology sc-17796) was incubated for 1 h with 20 µL of Dynabeads Protein G magnetic beads (Thermo Fisher Scientific 10004D), washed, then incubated overnight at 4°C with 500 µg of nuclear lysate, washed, and prepared for SDS-PAGE. Washes were conducted with either citric or RIPA buffer, as specified. Western blots were run as described previously. Primary antibodies used for Western blots were SMARCB1/BAF47 (mouse, 1:500; BD Biosciences 612110), GAPDH (rabbit, 1:5000; 2118), HDAC1 (rabbit, 1:1000; Cell Signaling 2062), SMARCD1 (mouse, 1:1000; Santa Cruz Biotechnology sc-135843), SMARCC1 (rabbit, 1:1000; Santa Cruz Biotechnology sc-10756), BRG1 (mouse, 1:1000; Santa Cruz Biotechnology sc-17796), and Nestin (rabbit, 1:1000; EMD Millipore).
Bulk RNA sequencing preparation and analysis
RNA was extracted using Qiagen RNeasy Plus minikit and library preparation was conducted using Illumina NEBNext Ultra Directional library preparation kit. Samples were sequenced using an Illumina sequencer with a minimum of 20 million reads per sample. Transcriptome data were aligned using the STAR aligner to a reference human genome (hg19). Reads were counted using featureCounts with default settings, and differential expression analysis conducted using DESeq2 R package (Love et al. 2014). Significant genes were considered to be those with a Benjamini-Hochberg adjusted P-value of <0.05 and a fold change of >2. Gene ontology analysis for up-regulated and down-regulated genes was conducted using the GOrilla web-based tool (Eden et al. 2009), comparing a list of up to 500 most significant genes (based on adjusted P-value) to a background list of all expressed genes (rpkm > 4 across all samples). For gene ontology networks, top 500 significant genes were analyzed for GO biological process and Reactome biological pathway enrichment by gProfiler (Raudvere et al. 2019) and output file visualized using Cytoscape (Shannon et al. 2003) software with EnrichmentMap plugin (Merico et al. 2010).
Single-cell RNA sequencing preparation and analysis
Three replicate control and knockdown day 20 organoids (10-d maturation) were collected, washed, and incubated for 1 h on a 37°C shaker in Accutase + 50 µg/mL DNaseI to aid in generation of a single-cell suspension. Organoids were dissociated by gentle pipetting with a wide-bore pipette after 15 and 45 min of incubation. Clumps were removed by filtration through a cell strainer, resuspended in PBS + 0.04% BSA, counted, and resuspended to a 1.2 × 106 cells/mL concentration. Cell viabilities were 60%–80% with a total of 5 × 104 to 2 × 105 cells collected per organoid. Samples were prepared for single-cell RNA sequencing as detailed in the 10× Chromium Single-Cell 3′ reagent kits v2 user guide. Reagents used included the Chromium Single-Cell 3′ library and gel-bead kit v2 (PN-120237), Chromium Single-Cell A Chip kit (PN-120236) and Chromium i7 multiplex kit (PN-120262). The protocol was followed for a desired 10,000 cells per organoid using 10 cycles of cDNA amplification. Quality control was conducted after cDNA amplification and library construction on a BioAnalyzer TapeStation instrument. Sample libraries were pooled, and shallow sequencing conducted on an Illumina HiSeq4000 to estimate cell numbers and read counts for each sample and a new pool generated to obtain 50,000 reads/cell for each sample. Final sequencing was conducted on an Illumina NovaSeq 6000. Analysis of the resulting data was conducted using the Cell Ranger pipeline for counting and aggregation of sequencing reads. Additional analysis was then conducted using the Seurat R toolkit for single cell genomics (Butler et al. 2018). Cells with a mitochondrial DNA percentage above 0.1 were filtered out of subsequent analysis. Canonical correlation analysis was conducted using variable genes from the control and knockdown conditions, and a tSNE plot of the first 20 aligned subspaces used for visualization. Clusters were generated using a resolution value of 0.6 and the first 20 CCA subspaces, resulting in the identification of 15 clusters, of which cluster 15 was excluded from later analysis due to its small size. Identification of cluster markers, differential expression analysis, and cluster quantifications were all conducted using the Seurat toolkit. Gene ontology analysis of differential expression gene sets was conducted using the GOrilla (Eden et al. 2009) website in comparison with a background list of expressed genes.
Flow cytometry
For neuron FACS experiments, day 25 neurons were dissociated with Accutase + DNase I for 30 min, with occasional gentle pipetting to break up clumps. Cells were filtered through a cell strainer and resuspended in PBS + 1% FBS. 1 × 106 cells were stained with NCAM-1 antibody (CD56 anti-human Alexa fluor 700, 1:200 dilution; Fisher Scientific BDB557919) for 1 h, washed three times with PBS + 1% FBS, and analyzed on a Sony SH800 instrument.
Immunofluorescence
NPCs or neurons were plated on poly-d-lysine coated coverslips and allowed to attach and proliferate for 3–5 d. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, washed with PBS, and permeabilized 10 min at room temperature with 0.1% Triton X-100 in PBS. Coverslips were incubated 30 min to 1 h in 2% IgG-free BSA, then stained with a 1:100 dilution of primary antibody overnight at 4°C. Coverslips were washed, stained for 1-2 h in a 1:1000 dilution of fluorescent secondary antibody, washed with PBS, and mounted on slides using Fluoro-Gel II mounting medium (Electron Microscopy Sciences 1798550). Organoids were fixed 15 min in 4% paraformaldehyde, washed with PBS and incubated overnight at 4°C in 30% sucrose solution before embedding in Tissue-Tek O.C.T. compound, and frozen at −80°C for cryosectioning. Slides containing organoid sections were thawed for 30 min at room temperature, permeabilized 10 min with 0.1% Triton X-100 in PBS, and incubated 30 min to 1 h in 2% IgG-free BSA before staining with a 1:100 dilution of primary antibody overnight at 4°C in a humidity chamber. Coverslips were washed, stained for 1–2 h in a 1:1000 dilution of fluorescent secondary antibody, washed with PBS and mounted on slides using Fluoro-Gel II mounting medium (Electron Microscopy Sciences 1798550). Imaging was conducted using a Keyence microscope with 20× or 40× magnification. Organoid images were generated by stitching multiple 20× images using Keyence BZ-X software and processed with haze reduction (blur size 8, brightness 1.6, reduction rate 0.6) in order to remove blur from images. If needed for visualization, brightness was increased so that all images were at a similar brightness level.
Data accessibility
Data have been deposited at Gene Expression Omnibus. Bulk RNA-seq data are available under accession number GSE157012, and single-cell RNA-seq data are available under accession number GSE157525.
Supplementary Material
Acknowledgments
We thank Dr. Bruce Conklin for providing CRISPR interference cell lines and constructs, and Dr. Diana Hargreaves for provision of BAF complex antibodies. In addition, we thank Rachel Reed and Dr. Ciro Zanca, Dr. Jorge Benitez, Dr. Jianhui Ma, Dr. Amy Haseley Thorne, Dr. Nathan Jameson, and Dr. Tiffany Taylor for their technical contributions to this project, and Dr. Gene Yeo, Dr. Robert Wechsler-Reya, Dr. Rafael Bejar, Dr. Alysson Muotri, and Dr. Lorraine Pillus for their continued advice and mentorship. This work was supported by Padres Pedal the Cause/RADY grant (PTC2017), the National Institute of General Medical Sciences (T32GM008666; to A.D.P.), the National Institute of Neurological Diseases and Stroke (R01-NS080939; to F.B.F.), and a Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowship (S.M.). This publication includes data generated at the UC San Diego IGM Genomics Center using an Illumina NovaSeq 6000 that was purchased with funding from a National Institutes of Health SIG grant (S10 OD026929).
Author contributions: A.D.P. performed cell line generation, all analytical experiments and sample processing, and genomic analysis and played a primary role in drafting the manuscript. T.K. aided in cell line generation and maintenance and single-cell RNA sequencing sample preparation in addition to aiding in initial project conception and data interpretation. S.M. aided in cell culture experiments and provided guidance to the project. P.D.J. and M.K. collected and analyzed ATRT RNA sequencing data and provided them for use in this project. J.R.C. aided in obtaining funding for this project and provided advice. F.B.F. provided funding for the project, was responsible for the conception of the work, and contributed to data interpretation and project design. All authors contributed to revising the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339978.120.
References
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. 2007. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol 14: 989–996. 10.1038/nsmb1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo A, Minelli R, Sapio L, Soeung M, Carbone F, Robinson FS, Tepper J, Chen Z, Lovisa S, Svelto M, et al. 2019. p53 is a master regulator of proteostasis in SMARCB1-deficient malignant rhabdoid tumors. Cancer Cell 35: 204–220.e9. 10.1016/j.ccell.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun H-JE, Lim EL, Heravi-Moussavi A, Saberi S, Mungall KL, Bilenky M, Carles A, Tse K, Shlafman I, Zhu K, et al. 2016. Genome-wide profiles of extra-cranial malignant rhabdoid tumors reveal heterogeneity and dysregulated developmental pathways. Cancer Cell 29: 394–406. 10.1016/j.ccell.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun HE, Johann PD, Milne K, Zapatka M, Buellesbach A, Ishaque N, Iskar M, Erkek S, Wei L, Tessier-Cloutier B, et al. 2019. Identification and analyses of extra-cranial and cranial rhabdoid tumor molecular subgroups reveal tumors with cytotoxic T cell infiltration. Cell Rep 29: 2338–2354.e7. 10.1016/j.celrep.2019.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S, Johann PD, Finetti MA, Drosos Y, Chou H-C, Zapatka M, Sturm D, Jones DTW, Korshunov A, Rhyzova M, et al. 2019. Comprehensive analysis of chromatin states in atypical Teratoid/Rhabdoid tumor identifies diverging roles for SWI/SNF and polycomb in gene regulation. Cancer Cell 35: 95–110.e8. 10.1016/j.ccell.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn KF, Gajjar A. 2012. Atypical teratoid rhabdoid tumor: current therapy and future directions. Front Oncol 2: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZY, Richer W, Fréneaux P, Chauvin C, Lucchesi C, Guillemot D, Grison C, Lequin D, Pierron G, Masliah-Planchon J, et al. 2016. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat Commun 7: 10421 10.1038/ncomms10421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463: 474–484. 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci 106: 5181–5186. 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EM, Sievert AJ, Gai X, Hakonarson H, Judkins AR, Tooke L, Perin JC, Xie H, Shaikh TH, Biegel JA. 2009. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res 15: 1923–1930. 10.1158/1078-0432.CCR-08-2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, Poreau B, De Jay N, Hébert S, Monlong J, et al. 2019. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 51: 1702–1713. 10.1038/s41588-019-0531-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johann P, Korshunov A, Kerl K, Huang A, Jabado N, Hasselblatt M, Frühwald M, Pfister S, Kool M. 2014. CNS AT/RTs are a heterogeneous entity that comprises subgroups with distinct molecular profiles. Cancer Genet 207: 447 10.1016/j.cancergen.2014.09.011 [DOI] [Google Scholar]
- Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, Jones DT, Sturm D, Hermann C, Segura Wang M, et al. 2016. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 29: 379–393. 10.1016/j.ccell.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, Crabtree GR. 2017. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 49: 213–222. 10.1038/ng.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Chaim IA, Benitez JA, Markmiller S, Parisian AD, Hevner RF, Turner KM, Hessenauer FM, D'Antonio M, Nguyen N-p, et al. 2020. Longitudinal assessment of tumor development using cancer avatars derived from genetically engineered pluripotent stem cells. Nat Commun 11: 550 10.1038/s41467-020-14312-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 9: 2329–2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer LF, Ward JM, Archer TK. 2019. Tumor suppressor SMARCB1 suppresses super-enhancers to govern hESC lineage determination. Elife 8: e45672 10.7554/eLife.45672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, et al. 2012. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122: 2983–2988. 10.1172/JCI64400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, et al. 2017. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49: 1613–1623. 10.1038/ng.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DST, Hu B, Ho YW, Sauvé C-EG, Bristow CA, Wang Q, Multani AS, Chen P, Nezi L, Jiang S, et al. 2017. PAF promotes stemness and radioresistance of glioma stem cells. Proc Natl Acad Sci 114: E9086–E9095. 10.1073/pnas.1708122114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47: W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reincke BS, Rosson GB, Oswald BW, Wright CF. 2003. INI1 expression induces cell cycle arrest and markers of senescence in malignant rhabdoid tumor cells. J Cell Physiol 194: 303–313. 10.1002/jcp.10201 [DOI] [PubMed] [Google Scholar]
- Reinhardt P, Glatza M, Hemmer K, Tsytsyura Y, Thiel CS, Höing S, Moritz S, Parga JA, Wagner L, Bruder JM, et al. 2013. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One 8: e59252 10.1371/journal.pone.0059252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CWM, Orkin SH. 2004. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer 4: 133–142. 10.1038/nrc1273 [DOI] [PubMed] [Google Scholar]
- Roberts CWM, Galusha SA, McMenamin ME, Fletcher CDM, Orkin SH. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci 97: 13796–13800. 10.1073/pnas.250492697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8: e55119 10.1371/journal.pone.0055119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR. 2010. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–955. 10.1016/j.cell.2010.04.037 [DOI] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19: 477–477. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson BG, Park PJ, et al. 2013. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci USA 110: 10165–10170. 10.1073/pnas.1302209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J, Golbourn B, Feng S, Ho KC, Sin-Chan P, Vasiljevic A, Norman JD, Guilhamon P, Garzia L, Agamez NR, et al. 2016. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell 30: 891–908. 10.1016/j.ccell.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394: 203–206. 10.1038/28212 [DOI] [PubMed] [Google Scholar]
- Vladoiu MC, El-Hamamy I, Donovan LK, Farooq H, Holgado BL, Sundaravadanam Y, Ramaswamy V, Hendrikse LD, Kumar S, Mack SC, et al. 2019. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572: 67–73. 10.1038/s41586-019-1158-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, et al. 2017. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49: 289–295. 10.1038/ng.3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Cook A, Alver BH, Stadtfeld M, Deaton AM, Hochedlinger K, Park PJ, Tolstorukov MY, Kingston RE. 2014. Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat Commun 5: 4719 10.1038/ncomms5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho Y-J, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CWM. 2010. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18: 316–328. 10.1016/j.ccr.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, De Carvalho DD, Dai C, Liu M, Pandiyan K, Zhou XJ, Liang G, Jones PA. 2013. SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS Genet 9: e1003459 10.1371/journal.pgen.1003459 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited at Gene Expression Omnibus. Bulk RNA-seq data are available under accession number GSE157012, and single-cell RNA-seq data are available under accession number GSE157525.