In this study, Li et al. conducted comprehensive gain- and loss-of-function screens using a human DUB cDNA library of 65 genes and an siRNA library of 98 genes, and identified USP20 as a deubiquitinase (DUB) that regulates a metastasis-promoting transcription factor, SNAI2, ubiquitination, and stability.
Keywords: deubiquitinase, USP20, SNAI2, SLUG, migration, invasion, metastasis
Abstract
SNAI2/SLUG, a metastasis-promoting transcription factor, is a labile protein that is degraded through the ubiquitin proteasome degradation system. Here, we conducted comprehensive gain- and loss-of-function screens using a human DUB cDNA library of 65 genes and an siRNA library of 98 genes, and identified USP20 as a deubiquitinase (DUB) that regulates SNAI2 ubiquitination and stability. Further investigation of USP20 demonstrated its function in promoting migration, invasion, and metastasis of breast cancer. USP20 positively correlates with SNAI2 protein level in breast tumor samples, and higher USP20 expression is associated with poor prognosis in ER− breast cancer patients.
SNAI2 (also known as SLUG), one of the three members in the SNAIL family transcription factors, has been reported to promote cancer metastasis by enhancing cell mobility and invasion (Shih and Yang 2011), promoting the survival of metastatic cells (Kim et al. 2014), and enhancing the activities of breast cancer stem cells (Guo et al. 2012). Elevated expression of SNAI2 has been observed in many cancer types (Alves et al. 2009) and correlates with increased risks of metastasis and postoperative relapse, as well as a shorter survival period for patients with a variety of cancers (Shih et al. 2005; Shioiri et al. 2006; Alves et al. 2009).
SNAI2 level can be regulated both transcriptionally and post-translationally. Multiple signaling pathways and specific molecules have been identified to induce SNAI2 transcription, such as TGF-β, Wnt, and Notch signaling (Shih and Yang 2011). SNAI2 protein level is also controlled by its ubiquitination and degradation through the ubiquitin-proteasome system (Vernon and LaBonne 2006; Wang et al. 2009; Xu et al. 2015; Lin et al. 2017; Ouchida et al. 2018). In the ubiquitin-proteasome pathway, proteins designated for degradation are first covalently conjugated with polyubiquitin chain, which are then recognized and degraded by the 26S proteasome complex (Hochstrasser 1996). Countering the action of ubiquitination enzymes, deubiquitinases (DUBs) have emerged as pivotal regulators of the ubiquitin–proteasome pathway by removing ubiquitin from their specific protein substrates and thus increasing the stability of those proteins (Komander et al. 2009).
Despite SNAI2's critical role in regulating breast cancer metastasis, so far there is no effective way to directly target SNAI2 pharmaceutically. Since DUBs are enzymes that are more druggable than the transcription factor SNAI2 itself, DUBs that recognize and stabilize SNAI2 may serve as novel drug targets to reduce SNAI2 level and inhibit tumor progression. Here, we performed comprehensive screenings of siRNA and cDNA libraries and identified USP20 as the bona fide SNAI2-stabilizing DUB that promotes breast cancer cell migration, invasion, and metastasis.
Results and Discussion
Identification of SNAI2-regulating DUBs
To identify the DUBs that can potentially deubiquitinate and stabilize SNAI2, we screened a human DUB siRNA library consisting of siRNAs targeting each of the 98 DUB genes. Previous studies have demonstrated that estrogen receptor α (ERα) represses SNAI2 expression (Ye et al. 2010), and SNAI2 correlates inversely with ERα expression in human breast cancer tissues and cell lines (Bai et al. 2017). Thus, we performed the siRNA screening in a triple-negative breast cancer cell line MDA-MB-231, which has relatively high endogenous SNAI2 expression. Pooled siRNAs (4 siRNAs per gene) were transfected into MDA-MB-231 cells, and protein lysates were collected 48 h later. SNAI2 knockdown was included as a positive control to confirm the consistency of siRNA transfection efficiency. Western blot results for SNAI2 from the DUB knockdown cells were quantified and normalized to β-actin level before comparing with that from the control knockdown cells (Supplemental Fig. S1A). The top 20 knockdowns of DUB genes that reduced SNAI2 level most significantly were selected for a second-round screening (Fig. 1A; Supplemental Table S1), during which each of the 20 DUB genes was targeted by four individual siRNAs in MDA-MB-231 cells. The lowest SNAI2 level achieved among the four siRNAs was used for comparison. Among these 20 candidates, the knockdown of two genes, USP20 and USP52, resulted in a reduction of SNAI2 protein level to less than half compared with control (Fig. 1B; Supplemental Fig. S1B).
Figure 1.
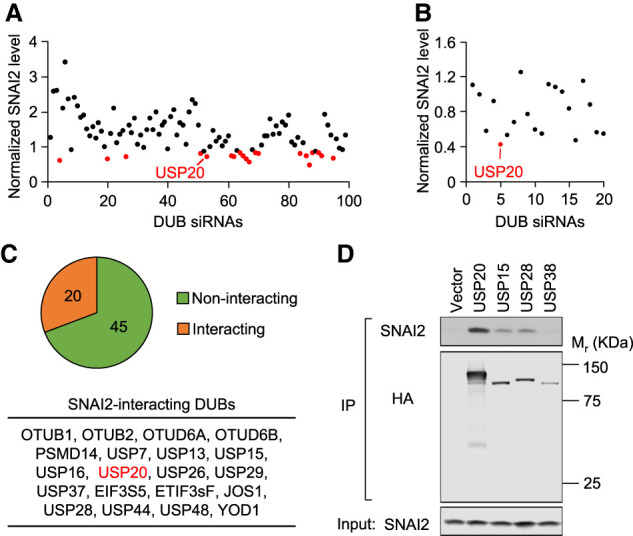
Identification of candidate DUBs of SNAI2. (A) Each of the 98 human DUB genes was knocked down in MDA-MB-231 cells with pooled siRNAs. The 20 genes with the lowest SNAI2 levels when knocked down were marked in red and selected for the second round of screening. (B) The second-round screening was conducted in MDA-MB-231 cells to evaluate the top 20 candidates. (C) Summary of the coimmunoprecipitation (co-IP) screening conducted in HEK293T cells to search for DUBs that interact with SNAI2 when coexpressed. (D) Representative Western blots for one of the co-IP experiments that include USP20.
The siRNA library allowed us to screen for knockdowns of DUBs that reduce endogenous SNAI2 level, and thus the candidates are more likely to be physiologically relevant in maintaining high SNAI2 level in breast cancer cells. However, this approach is limited by the expression level of DUBs in the specific cell line used for screening and the knockdown efficiency of pooled siRNAs, which could also explain some of the discrepancies in results obtained in different cell lines. For example, two studies reported that DUB3 and USP10 stabilize SNAI2 in different cell lines (Lin et al. 2017; Ouchida et al. 2018), which did not pass the criteria for positive hits in our siRNA screen, and thus may have limited effects in breast cancer cells. To complement the siRNA library screening, we also screened a DUB cDNA library by coimmunoprecipitation to examine the interaction of each DUB with SNAI2. Most of the DUB cDNA plasmids were constructed by molecular cloning to express the DUB protein with an HA tag. Each of those DUBs was then co-overexpressed together with SNAI2 in 293T cells. Coimmunoprecipitation (co-IP) experiments were conducted using HA antibody to pull down DUBs, and SNAI2 was probed in the co-IP samples (Supplemental Fig. S1C; Supplemental Table S2). While the co-overexpression of SNAI2 and DUBs may increase the chance of getting false positive hits, this approach allowed us to directly screen a DUB cDNA library for interactors of SNAI2 in a simplified yet robust manner. Sixty-five DUBs were successfully pulled down by HA antibody, and among them, 20 DUBs, including USP20, showed interaction with SNAI2 (Fig. 1C,D; Supplemental Fig. S1C).
The present study focuses on USP20, the most promising candidate based on the overlap of these two screenings. However, since both approaches have their own advantages and limitations, unique candidates from each of the screenings could also be critical regulators of SNAI2 degradation in different contexts. These hits need to be carefully evaluated by further functional assays to rule out false positives and confirm that the identified DUB does have deubiquitinating activity and stabilizing effect on SNAI2 protein.
USP20 inhibits SNAI2 degradation by deubiquitinating SNAI2
Since knockdown of USP20 significantly reduces SNAI2 protein level and there is a strong interaction between USP20 and SNAI2, we considered USP20 as the most promising candidate and further investigated its role in regulating SNAI2 stability. First, we confirmed that the decrease of SNAI2 protein level is not due to the down-regulation of SNAI2 mRNAs (Fig. 2A). Next, we tested whether USP20 can indeed deubiquitinate SNAI2. USP20 was knocked down by siRNAs in MDA-MB-231 cells. Cells were treated with MG132 for 6 h before protein sample collection to inhibit proteasomal degradation, so that polyubiquitinated SNAI2 can be accumulated and detected later. Endogenous SNAI2 protein was pulled down, and polyubiquitinated SNAI2 was detected using anti-ubiquitin antibody. When USP20 was knocked down, higher polyubiquitination level was observed for SNAI2 (Fig. 2B). Similar ubiquitination assay was also performed in MDA-MB-231 cells stably overexpressing SNAI2. Only the overexpression of wild-type USP20 but not the catalytically dead mutant USP20-C154S (Zhu et al. 2014) reduced SNAI2 polyubiquitination (Fig. 2C). Taken together, these ubiquitination assays indicate that USP20 is the bona fide deubiquitinating enzyme for SNAI2.
Figure 2.
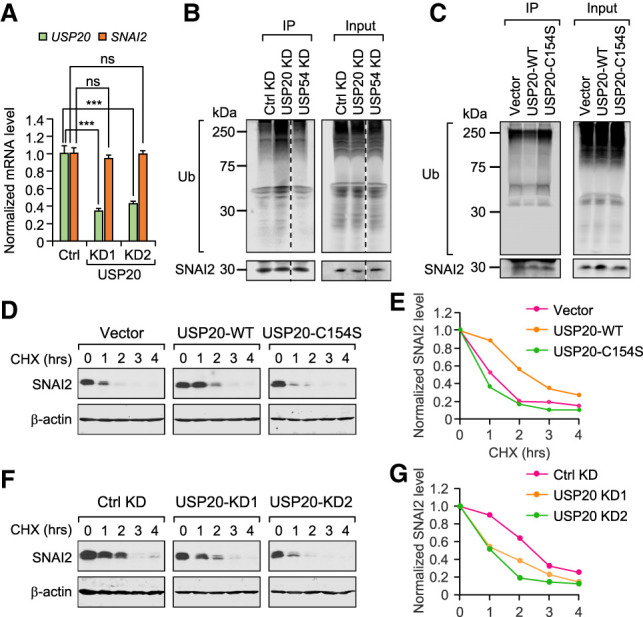
USP20 protects SNAI2 from degradation by deubiquitinating SNAI2. (A) USP20 knockdown in LM2 cells did not affect SNAI2 mRNA level. Data represent mean ± standard deviation. (ns) Nonsignificant, (***) P < 0.001 by one-tailed Student's t-test. (B) USP20 knockdown but not USP54 knockdown (negative control) increased accumulation of polyubiquitinated SNAI2 when treated with MG132. SNAI2 was pulled down and anti-ubiquitin antibody was used to detect polyubiquitinated SNAI2. (C) Overexpression of wild-type USP20 (USP20-WT) but not the catalytically inactive USP20 mutant (USP20-C154S) reduced polyubiquitinated SNAI2. (D,E) Overexpression of wild-type USP20 (USP20-WT) but not the catalytically inactive USP20 mutant (USP20-C154S) stabilized SNAI2. Cells were treated with cycloheximide (CHX) for 0–4 h as indicated before being lysed for Western blot. (F,G) USP20 knockdown using two different siRNAs in LM2 cells resulted in accelerated degradation of SNAI2. Cells were treated with cycloheximide (CHX) for 0–4 h as indicated before being lysed for Western blot.
Since polyubiquitinated SNAI2 is targeted by proteasome for degradation, we tested the effect of USP20 knockdown on SNAI2 degradation rate by cycloheximide (CHX) pulse-chase assay. Cells were treated with or without CHX (50 μg/mL) for 1, 2, 3, and 4 h to inhibit new protein synthesis, and the remaining SNAI2 level after CHX treatment was subsequently determined by Western blot. Consistent with the ubiquitination assay results, the overexpression of wild type but not the catalytically inactive mutant USP20 stabilized SNAI2 (Fig. 2D,E), while USP20 depletion resulted in accelerated SNAI2 degradation (Fig. 2F,G). Taken together, these results indicate that USP20 stabilizes SNAI2 by deubiquitinating SNAI2.
USP20 knockdown inhibits migration and invasion via reducing SNAI2
We knocked down USP20 in LM2 and SCP28 cells, two metastatic sublines derived from the MDA-MB-231 breast cancer cells (Kang et al. 2003; Minn et al. 2005), as well as SUM159-M1a cells, a metastatic subline derived from the SUM159 breast cancer line (Esposito et al. 2019). As expected, SNAI2 protein level was reduced after USP20 was knocked down (Fig. 3A–C; Supplemental Fig. S2A,B). Such an effect is specific to SNAI2, as there was no significant difference in the protein levels of c-MYC, SNAI1, and another reported USP20 substrate β-catenin (Wu et al. 2018). On the other hand, the expression of wild-type USP20 but not the catalytically dead mutant USP20-C154S increased SNAI2 level in LM2 cells overexpressing SNAI2 (Fig. 3D).
Figure 3.
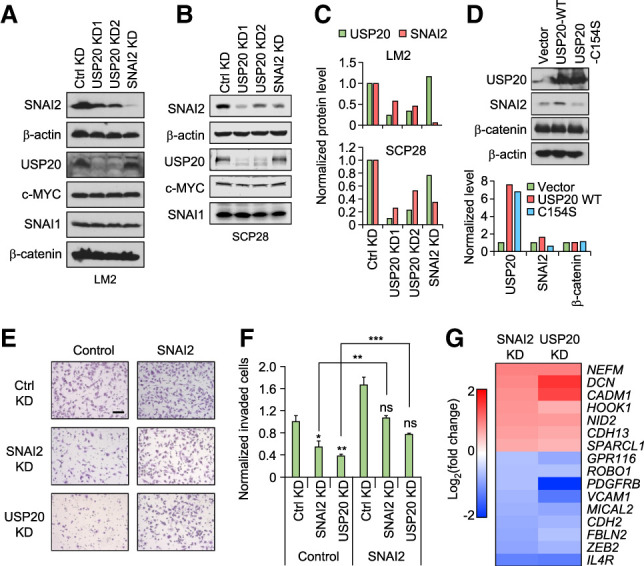
USP20 knockdown inhibits cell invasion via reducing SNAI2. (A) USP20 knockdown reduced SNAI2 level in LM2 cells. (B) USP20 knockdown reduced SNAI2 level in SCP28 cells. (C) Quantification of A (top) and B (bottom). (D) Wild-type but not mutant USP20 increased SNAI2 level. (E,F) Invasion assays with USP20 and SNAI2 knockdown in control or SNAI2-overexpressing LM2 cells. (E) Representative microscope images. Scale bar, 100 μm. (F) Quantified results. Data represent mean ± SEM. (ns) Nonsignificant, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 by one-tailed Student's t-test. P-values marked above the SNAI2 and USP20 KD bars represent their comparison with the control KD bar in the control group. (G) Heat map presentation of microarray data demonstrating the expression changes of several migration-related genes upon SNAI2 or USP20 KD.
SNAI2 has been well known to promote cell migration and invasion (Shih and Yang 2011). To test whether USP20 may regulate migration and invasion through targeting SNAI2, we performed Transwell migration and invasion assays. When USP20 or SNAI2 was knocked down in SCP28 cells using siRNAs, the number of migrated cells was significantly reduced when compared with the cells treated with control siRNA (Supplemental Fig. S2C,D). Similar results were observed for invasion assay using the highly metastatic LM2 cells. Similar to SNAI2-knockdown cells, USP20-depleted cells demonstrated significantly less invasive capabilities compared with control cells. Importantly, the effect of USP20 or SNAI2 knockdown on invasion was fully rescued by SNAI2 overexpression (Fig. 3E,F; Supplemental Fig. S2E), suggesting that the influence of USP20 on migration and invasion is mediated by SNAI2. Microarray analysis of control, USP20, and SNAI2 knockdown LM2 cells revealed that many migration/invasion-related genes showed consistent changes in USP20 and SNAI2 knockdown cells, indicating that USP20 and SNAI2 share similar downstream effectors in regulating migration and invasion (Fig. 3G).
USP20 knockdown inhibits lung colonization by breast cancer cells
Given the important role of SNAI2 in metastasis, we next investigated the effect of USP20 on lung metastasis of breast cancer cells. First, we generated cell lines with stable USP20 knockdown using lentiviruses containing USP20-targeting shRNAs. The protein level of SNAI2 was decreased when the cell lines were first generated. However, the SNAI2 protein level in USP20 knockdown cells recovered after several passages, possibly because SNAI2 is essential for LM2 cells and the cells developed compensating mechanisms to regain SNAI2 expression over time. Thus, we took an alternative approach of knocking down USP20 using two different siRNAs, and the siRNA knockdown effect was confirmed to last for at least 10 d, which is sufficient for in vivo experimental lung colonization studies (Supplemental Fig. S3A,B). Firefly luciferase-labeled siRNA transfected SUM159-M1a and LM2 cells were injected intravenously into NSG mice, and bioluminescent imaging (BLI) was used to monitor their metastatic seeding and growth in the lung. Five days after injection, there was already a significantly lower number of cancer cells seeded in the lung for USP20 or SNAI2 knockdown cells compared with the control cells (Fig. 4A; Supplemental Fig. S4A). This effect was maintained for at least 3 wk for both siRNAs of USP20 after injection of LM2 cells, and then the two siRNAs of USP20 diverged at week 4, possibly due to different knockdown efficiencies in vivo (Fig. 4B). Mice were sacrificed 4 wk later and metastasis nodules on the lungs were counted. Significantly less lung metastasis nodules were observed for USP20 and SNAI2 knockdown groups (Fig. 4C,D), which could result from both reduced lung seeding as well as a modest decrease in proliferation (Supplemental Fig. S4B). These results suggest that USP20 knockdown suppresses lung colonization by breast cancer cells, which leads to reduced formation of metastasis nodules. Such effect can be partially rescued by SNAI2 overexpression (Supplemental Fig. S4C), suggesting that there might be other substrates of USP20 also contributing to its effect on metastasis.
Figure 4.
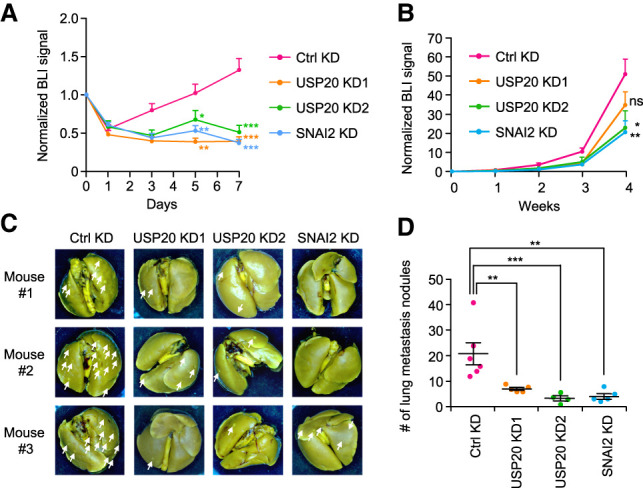
USP20 knockdown inhibits breast cancer lung metastasis. (A) The seeding of SUM159-M1a cells in the lung was monitored by bioluminescence imaging (BLI) every other day and was normalized to day 0. Data represent mean + SEM. (B) The growth of metastasis was monitored by BLI weekly. Data represent mean ± SEM. (C) Representative images of lungs with metastasis nodules (pointed by white arrows) at 28 d after injection. (D) Quantified results of lung metastasis nodules in each group. Each dot represents data from one individual mouse. Lines represent mean ± SEM. (ns) Nonsignificant, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 by one-tailed Student's t test.
USP20 positively correlates with SNAI2 in clinical breast cancer samples
Since in ER− breast cancer patients higher SNAI2 mRNA level correlates with poorer prognosis (Bai et al. 2017), we focused our clinical study on ER− breast cancer patients. We first tested the prognostic value of USP20 in a large public clinical microarray database using an online Kaplan–Meier plotter tool (Györffy et al. 2010). Higher expression of USP20 mRNA is associated with worse metastasis-free survival (Fig. 5A), consistent with our observations in mice that USP20 promotes metastasis. We tested the correlation between USP20 and SNAI2 at the mRNA level in ER− breast cancer patients using the TCGA-BRCA sample set (The Cancer Genome Atlas Network 2012; Ciriello et al. 2015). No significant correlation was observed between USP20 and SNAI2 mRNA expression (Fig. 5B), similar to the lack of regulation of SNAI2 transcription by USP20 as we observed in cell culture (Fig. 2A). To determine whether USP20 is correlated with SNAI2 at the protein level, we next collected breast cancer samples from 84 ER− breast cancer patients treated at Fudan University Shanghai Cancer Center, and examined SNAI2 and USP20 protein levels using immunohistochemistry (IHC) staining. Supporting our hypothesis that USP20 stabilizes SNAI2, these two proteins’ expression patterns showed a significant positive correlation (Fig. 5C,D). Importantly, as expected from their prometastatic functions, higher protein levels of USP20 and SNAI2 in breast cancer tissues correlate with worse relapse-free survival and metastasis-free survival of breast cancer patients (Fig. 5E,F).
Figure 5.
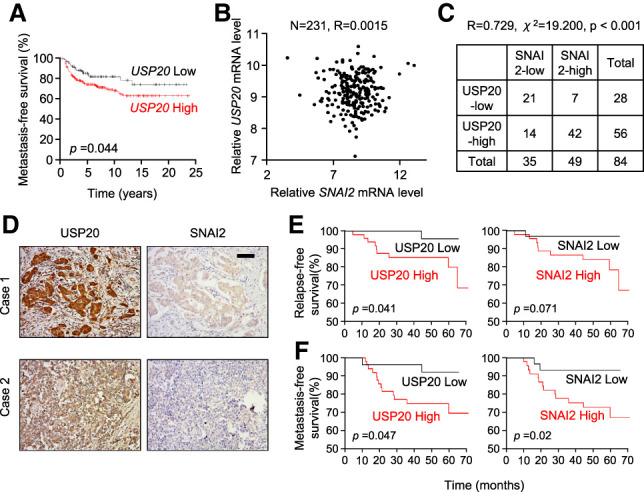
USP20 positively correlates with SNAI2 protein level and predicts poor prognosis in ER− breast cancer patients. (A) Kaplan–Meier plot of metastasis-free survival in ER− breast cancer patients, stratified by USP20 mRNA level. Data were obtained from the Kaplan–Meier plotter database. (B) USP20 mRNA does not correlate with SNAI2 mRNAs in 231 ER− breast cancer patients. Data were obtained from the TCGA database. R = 0.0015 by Pearson analysis. (C,D) USP20 positively correlates with SNAI2 at the protein level. Tissues were obtained from 84 ER− breast cancer patients and were subjected to IHC staining of USP20 and SNAI2 proteins. (C) Correlation between USP20 and SNAI2 proteins. R = 0.729 by Pearson analysis. χ2 = 19.200, P < 0.001 by χ2 test. (D) Representative IHC images of USP20 and SNAI2 in breast tumors. (E,F) Kaplan–Meier plots of relapse-free survival (E) and metastasis-free survival (F) in 84 ER− breast cancer patients, stratified by protein expression of USP20 or SNAI2. Log rank analysis was conducted to determine significance.
Our investigations on USP20 and SNAI2 in breast cancer patient samples demonstrated a promising clinical relevance of USP20. Thus, our discovery provided a new potential therapeutic option to reduce SNAI2 level in breast cancer patients by indirectly inhibiting the protease USP20 that is protecting it from proteasomal degradation. Selective inhibitors against USP20 can be developed through structure-guided design and screening, similar to those for USP4 (Nguyen et al. 2019) and USP7 (Lamberto et al. 2017). Compared with the proteasome inhibitor bortezomib, which has been approved by the Food and Drug Adminstration for the treatment of relapsed and/or refractory multiple myeloma (Manasanch and Orlowski 2017), such DUB-selective inhibitors can target more specific pathways and potentially achieve better efficacy without deleterious side effects.
Materials and methods
Antibodies and reagents
Primary antibodies for Western blotting and immunohistochemical staining included anti-SNAI2 (Cell Signaling Technology 9585), anti-USP20 (Proteintech 17491-1-AP), anti-β-actin (Abcam ab6276, clone AC-15), anti-HA (Roche 11867423001), anti-ubiquitin (Santa Cruz Biotechnology sc-271289), anti-SNAI1 (Cell Signaling Technology 3895S), anti-MYC (9E10) (Santa Cruz Biotechnology), and anti-β-catenin (Cell Signaling Technology 9585).
The negative control siRNA (D-001810-01-05), SNAI2 siRNA (J-017386-06-0002), and the human DUB siRNA library GU-104705) were purchased from GE Healthcare Dharmacon. Individual USP20 siRNAs were synthesized or purchased from Sigma (siRNA#1 sequence: GGACAAUGAUGCUCACCUA, and siRNA#2 siRNA ID: SASI_Hs02_00324906).
Cell culture and stable cell lines
HEK293T, MDA-MB-231, LM2, and SCP28 cell lines were maintained in DMEM medium (Sigma) supplemented with fetal bovine serum (10%; Gemini Bio Products), penicillin/streptomycin (1%; Corning), and amphotericin (0.2%; Corning). SUM159-M1a cells were grown in F12 medium supplemented with 10% FBS, 10 μg/mL insulin, and 20 ng/mL EGF. Plasmids or siRNAs were transfected into cells using Lipofectamine 2000 or RNAiMax following the manufacturer's manual (Life Technology).
To generate lentiviruses for USP20 overexpression, pLEX empty vector, pLEX-USP20, or pLEX-USP20-C154S was cotransfected with VSV-G and p8.9 into HEK293T cells. Media containing viruses were collected 2 and 3 d after transfection. Recipient LM2 cells were incubated with virus-containing media supplemented with 2 μg/mL polybrene for 24 h. To generate stable cell lines, puromycin was used to select the infected cells.
Molecular cloning and plasmids
Most of the human DUB expression constructs were generated by regular PCR from human reference cDNA and molecular cloning into pLEX, pcDNA3.1 or pKH3 plasmids. Primer sequences are available on request. FLAG-HA-DUB3, FLAG-HA-OTUD1, FLAG-HA-OTUD4, FLAG-HA-OTUD5, FLAG-HA-OTUD7B, FLAG-HA-PSMD7, FLAG-HA-STAMBPL1, FLAG-HA-USP1, FLAG-HA-USP14, FLAG-HA-USP18, FLAG-HA-USP2, FLAG-HA-USP21, FLAG-HA-USP26, FLAG-HA-USP29, FLAG-HA-USP43, FLAG-HA-USP50, FLAG-HA-USP52, and FLAG-HA-VCPIP were gifts from Dr. Wade Harper (Addgene plasmids) (Sowa et al. 2009).
Coimmunoprecipitation assay
Two days after cotransfection of SNAI2-FLAG plasmid and DUB-HA plasmids, HEK293T cells were lysed with 500 μL of IP lysis buffer (50 mM Tris-Cl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) with protease inhibitors (Roche 11836170001). Five percent input samples were saved and loaded for Western blot. After incubation with mouse HA antibody (Santa Cruz Biotechnology sc-7392) overnight at 4°C, 30 μL of prewashed protein A/G beads (Santa Cruz Biotechnology sc-2003) was added and incubated for 1 h. After four washes, beads were pelleted and boiled with SDS loading buffer for Western blot analysis.
Quantitative real-time qPCR
Primer sequences used were as follows: SNAI2 (forward: 5′-CGAACTGGACACACATACAGTG-3′; reverse: 5′-CTGAGGATCTCTGGTTGTGGT-3′), USP20 (forward: 5′-GGTTGCCTGCCCCTATGTT-3′; reverse: 5′-GGTTCACGGTCAAGTTGTGC-3′), and GAPDH (internal control, forward: 5′-TGCACCACCAACTGCTTAGC-3′; reverse: 5′-GGCATGGACTGTGGTCATGAG-3′).
Ubiquitination assay
Two days after siRNA transfection or 3 d after infection with lentiviruses containing control, USP20 wild-type, or mutant cDNAs, proteosomal degradation was blocked by treating the cells with 20 μM MG132 for 6 h. Cells were then lysed with 150 μL of denaturing lysis buffer (50 mM Tris-Cl at pH 6.8, 1.5% SDS), and protein samples were collected by scraping followed by boiling for 15 min. Ninety microliters of the denatured protein samples was added to 1.2 mL of EBC/BSA buffer (50 mM Tris-Cl at pH 6.8, 180 mM NaCl, 0.5% NP40, 0.5% BSA), and incubated with anti-SNAI2 antibody or anti-FLAG antibody overnight and with protein A/G beads for 1 h at 4°C. Ubiquitin antibody was then used to detect polyubiquitinated SNAI2 in the IP samples.
Cycloheximide pulse-chase assay
Cells were seeded on 12-well plates at 1 × 105 to 2 × 105 cells/well and cultured overnight before adding cycloheximide (CHX). Cells were treated with 50 μg/mL CHX for 1–4 h as indicated prior to Western blot analysis.
Migration and invasion assays
For migration assay, cells were seeded at 5 × 104 cells per well in cell culture inserts (Falcon 08-771-21) placed in 24-well companion plates (Falcon 353504). The cells were incubated in serum-free medium in the inserts, while regular DMEM containing 10% FBS was added to the wells to induce cell migration for 12 h. Cells were then fixed with 4% paraformaldehyde (PFA) and stained with crystal violet (0.05% [w/v]). The cells inside the inserts were removed by cotton swabs, and images of the migrated cells were taken under the bright field of a microscope.
Invasion assays were conducted following similar procedure. Cells (1 × 105) were seeded on Matrigel (final concentration of 1–2 mg/mL; BD Biosciences) precoated inserts in serum-free medium for 16 h before they were fixed using 4% PFA.
Microarray
Forty-eight hours after LM2 cells were transfected with control nontargeting siRNA, USP20 siRNA#1, USP20 siRNA#2, and SNAI2 siRNA, cells were lysed for RNA preparation. To profile the gene expression of USP20 and SNAI2 knockdown LM2 cells, microarray was conducted using Agilent human SurePrint G3 GE 8x60K microarray (Agilent G4851B, design ID 039494) following the manufacturer's instructions. Two independent biological repeats of each condition were used for the microarray. The microarray data have been submitted to NCBI Gene Expression Omnibus (GEO) with accession number GSE112017.
Animal studies
All procedures and protocols involving mice were approved by the Institutional Animal Care and Use Committee of Princeton University. LM2 cells were trypsinized and resuspended in phosphate-buffered saline (PBS) 48 h after transfection with nontargeting control, USP20, or SNAI2 siRNA at 2 × 104 cells/mL. One-hundred microliters of cell suspension was injected through the tail vein of NSG mice. SUM159-M1a cells and LM2 cells stably overexpressing control vector or SNAI2 were transfected and prepared similarly and were injected at 8 × 104 and 3 × 104 cells per injection, respectively. To monitor the seeding and growth of cancer cells in the lung, bioluminescent imaging was conducted by retro-orbital injection of 100 μL of luciferin solution (15 mg/mL) into anesthetized mice. Luciferase activity was measured by scanning the mice with a Xenogen IVIS 200 imaging system.
Clinical analysis of public data sets
The Kaplan–Meier plotter was used to evaluate the prognosis value of USP20 mRNA expression (Györffy et al. 2010). The data from a total of 351 ER− breast cancer patients were used for analyzing the distant metastasis free survival, and the patients were split based on an automatically selected cutoff for USP20.
The TCGA (The Cancer Genome Atlas) BRCA (breast invasive carcinoma) exon expression data were downloaded from the UCSC genomic center. The mRNA expression levels of USP20 and SNAI2 in a total of 231 ER− breast cancer patients were used for the Pearson correlation analysis.
Immunohistochemistry staining and scoring
A cohort of 84 patients was assembled from patients with primary ER− breast cancer treated at Fudan University Shanghai Cancer Center in 2011. Anonymized paraffin-embedded formalin-fixed breast cancer tissues were deparaffinized using xylene and rehydrated in a graded alcohol series. For immunorecognition, tissue sections mounted on slides were heated in 0.01 M citric buffer (pH 6.0) for 15 min with autoclaving. After cooling down to room temperature, sections were treated with 3% H2O2 in PBS for 30 min at room temperature. Slides were washed, blocked, and then incubated with the primary antibodies overnight at 4°C, followed by incubation with corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies. HRP enzyme activity was detected using a labeled streptavidin–biotin (LSAB) system with 3,3-diaminobenzidine tetrachloride. The immunostained sections were counterstained using hematoxylin and were dehydrated and mounted.
Three pictures of each slide were generated and evaluated based on staining intensity (0–3) and area (0–100) by a pathologist. The score of each slide was indicated by the average of intensity multiplied by area. Slides scored ≥180 were classified into high expression group while the remaining were classified as low expression group.
Statistical analysis
The statistical methods used for data analysis are specified in the figure legends.
Supplementary Material
Acknowledgments
This research was supported by a Cancer Institute of New Jersey Research Development Award, the Brewster Foundation, and grants from the Breast Cancer Research Foundation, American Cancer Society, and Susan G. Komen Foundation to Y.K.
Author contributions: W.L. designed and performed the experiments, analyzed data, and wrote the manuscript. M.S. assisted with cloning of DUB genes and mouse experiments. R.Z. assisted with experiments. H.Z. assisted with cloning of DUB genes and provided advice for ubiquitination assay. Y.-Z.J. and Z.-M.S. provided patient samples and performed patient sample-related experiments. Y.W. assisted with microarray experiment and data analysis. Y.K. supervised the overall study, designed experiments, analyzed data, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339804.120.
References
- Alves CC, Carneiro F, Hoefler H, Becker KF. 2009. Role of the epithelial–mesenchymal transition regulator Slug in primary human cancers. Front Biosci 14: 3035–3050. 10.2741/3433 [DOI] [PubMed] [Google Scholar]
- Bai JW, Chen MN, Wei XL, Li YC, Lin HY, Chen M, Li JW, Du CW, Man K, Zhang GJ. 2017. The zinc-finger transcriptional factor Slug transcriptionally downregulates ERα by recruiting lysine-specific demethylase 1 in human breast cancer. Oncogenesis 6: e330 10.1038/oncsis.2017.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. 2015. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163: 506–519. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M, Mondal N, Greco TM, Wei Y, Spadazzi C, Lin SC, Zheng H, Cheung C, Magnani JL, Lin SH, et al. 2019. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat Cell Biol 21: 627–639. 10.1038/s41556-019-0309-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Hardi U, Bell G, et al. 2012. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148: 1015–1028. 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123: 725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. 1996. Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439. 10.1146/annurev.genet.30.1.405 [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549. 10.1016/S1535-6108(03)00132-6 [DOI] [PubMed] [Google Scholar]
- Kim S, Yao J, Suyama K, Qian X, Qian BZ, Bandyopadhyay S, Loudig O, De Leon-Rodriguez C, Zhou ZN, Segall J, et al. 2014. Slug promotes survival during metastasis through suppression of Puma-mediated apoptosis. Cancer Res 74: 3695–3706. 10.1158/0008-5472.CAN-13-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563. 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- Lamberto I, Liu X, Seo HS, Schauer NJ, Iacob RE, Hu W, Das D, Mikhailova T, Weisberg EL, Engen JR, et al. 2017. Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem Biol 24: 1490–1500.e11. 10.1016/j.chembiol.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wang Y, Shi Q, Yu Q, Liu C, Feng J, Deng J, Evers BM, Zhou BP, Wu Y. 2017. Stabilization of the transcription factors slug and twist by the deubiquitinase dub3 is a key requirement for tumor metastasis. Oncotarget 8: 75127–75140. 10.18632/oncotarget.20561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manasanch EE, Orlowski RZ. 2017. Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol 14: 417–433. 10.1038/nrclinonc.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. 2005. Genes that mediate breast cancer metastasis to lung. Nature 436: 518–524. 10.1038/nature03799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HH, Kim T, Nguyen T, Hahn MJ, Yun SI, Kim KK. 2019. A selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating β-catenin signaling. Cell Physiol Biochem 53: 157–171. 10.33594/000000127 [DOI] [PubMed] [Google Scholar]
- Ouchida AT, Kacal M, Zheng A, Ambroise G, Zhang B, Norberg E, Vakifahmetoglu-Norberg H. 2018. USP10 regulates the stability of the EMT-transcription factor Slug/SNAI2. Biochem Biophys Res Commun 502: 429–434. 10.1016/j.bbrc.2018.05.156 [DOI] [PubMed] [Google Scholar]
- Shih JY, Yang PC. 2011. The EMT regulator slug and lung carcinogenesis. Carcinogenesis 32: 1299–1304. 10.1093/carcin/bgr110 [DOI] [PubMed] [Google Scholar]
- Shih JY, Tsai MF, Chang TH, Chang YL, Yuan A, Yu CJ, Lin SB, Liou GY, Lee ML, Chen JJ, et al. 2005. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res 11: 8070–8078. 10.1158/1078-0432.CCR-05-0687 [DOI] [PubMed] [Google Scholar]
- Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. 2006. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer 94: 1816–1822. 10.1038/sj.bjc.6603193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. 2006. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development 133: 3359–3370. 10.1242/dev.02504 [DOI] [PubMed] [Google Scholar]
- Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, et al. 2009. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 11: 694–704. 10.1038/ncb1875 [DOI] [PubMed] [Google Scholar]
- Wu C, Luo K, Zhao F, Yin P, Song Y, Deng M, Huang J, Chen Y, Li L, Lee S, et al. 2018. USP20 positively regulates tumorigenesis and chemoresistance through β-catenin stabilization. Cell Death Differ 25: 1855–1869. 10.1038/s41418-018-0138-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhu C, Zhao X, Chen C, Zhang H, Yuan H, Deng R, Dou J, Wang Y, Huang J, et al. 2015. Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget 6: 979–994. 10.18632/oncotarget.2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX, Shetuni B, Barsky SH. 2010. ERα signaling through slug regulates E-cadherin and EMT. Oncogene 29: 1451–1462. 10.1038/onc.2009.433 [DOI] [PubMed] [Google Scholar]
- Zhu M, Zhao H, Liao J, Xu X. 2014. HERC2/USP20 coordinates CHK1 activation by modulating CLASPIN stability. Nucleic Acids Res 42: 13074–13081. 10.1093/nar/gku978 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


