Here, Van Nostrand et al. investigated the mechanisms of action of the biguanide drug metformin by using a new RaptorAA mouse model, in which AMPK phospho-serine sites Ser722 and Ser792 of RAPTOR were mutated to alanine. The hepatic transcriptional response in mice on a high-fat diet treated with metformin was largely ablated by AMPK deficiency under the conditions examined, indicating the essential role of this kinase and its targets in metformin action in vivo.
Keywords: AMPK, mTOR, RAPTOR, TSC2, metformin, STAT3
Abstract
Despite being the frontline therapy for type 2 diabetes, the mechanisms of action of the biguanide drug metformin are still being discovered. In particular, the detailed molecular interplays between the AMPK and the mTORC1 pathway in the hepatic benefits of metformin are still ill defined. Metformin-dependent activation of AMPK classically inhibits mTORC1 via TSC/RHEB, but several lines of evidence suggest additional mechanisms at play in metformin inhibition of mTORC1. Here we investigated the role of direct AMPK-mediated serine phosphorylation of RAPTOR in a new RaptorAA mouse model, in which AMPK phospho-serine sites Ser722 and Ser792 of RAPTOR were mutated to alanine. Metformin treatment of primary hepatocytes and intact murine liver requires AMPK regulation of both RAPTOR and TSC2 to fully inhibit mTORC1, and this regulation is critical for both the translational and transcriptional response to metformin. Transcriptionally, AMPK and mTORC1 were both important for regulation of anabolic metabolism and inflammatory programs triggered by metformin treatment. The hepatic transcriptional response in mice on high-fat diet treated with metformin was largely ablated by AMPK deficiency under the conditions examined, indicating the essential role of this kinase and its targets in metformin action in vivo.
Type 2 diabetes is a global health crisis affecting 8.3% of adults worldwide (https://www.diabetesatlas.org). The frontline treatment and most prescribed antidiabetic drug is the biguanide metformin, which prevents cardiovascular events, limits inflammation, and lowers blood glucose by decreasing hepatic glucose production, decreasing intestinal glucose absorption and increasing peripheral glucose utilization (Rena et al. 2017; Vial et al. 2019). However, the molecular mechanisms underlying these benefits of metformin are complex and still not fully understood.
Metformin has been shown to inhibit mitochondrial respiratory complex I, subsequently decreasing cellular respiration and cellular ATP levels (Vial et al. 2019). Metformin-induced energy depletion rapidly activates the AMP-activated protein kinase (AMPK), which is a serine–threonine kinase conserved in all eukaryotes that plays a critical role in sensing intracellular energy levels and suppressing cell growth under low-energy conditions (González et al. 2020). AMPK phosphorylates multiple downstream targets to restore energy balance by promoting ATP-producing catabolic processes and inhibiting ATP-consuming anabolic processes (Garcia and Shaw 2017; González et al. 2020).
One of the central downstream pathways inhibited by AMPK is the mechanistic target of rapamycin complex I (mTORC1) pathway, a major driver of anabolic metabolism that is inappropriately activated by obesity and implicated in diabetes (Khamzina et al. 2005; Laplante and Sabatini 2012). mTORC1 is a highly conserved nutrient-responsive regulator of cell growth composed of the mTOR serine-threonine kinase, the scaffold protein RAPTOR, and associated subunits (Liu and Sabatini 2020). mTORC1 directly phosphorylates downstream substrates including ribosomal S6 kinase-1 (S6K1), eukaryotic initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1), unc-51 like autophagy-activating kinase 1 (ULK1), and transcription factor binding to IGHM enhancer 3 (TFE3), to regulate protein synthesis and autophagy in order to promote cell proliferation and biogenesis of building blocks for cell replication (Liu and Sabatini 2020).
mTORC1 activity is regulated by nutrients, growth factors, and energy status through multiple upstream avenues. Growth factor- and insulin-stimulated signaling pathways signal through the small G protein RHEB, which activates mTORC1 when GTP-bound (Gomes and Blenis 2015). The GTP-status of RHEB is directly regulated by a protein complex composed of the tuberous sclerosis complex proteins (TSC1 and TSC2), which functions as a GTPase-activating protein (GAP) for RHEB thereby inhibiting mTORC1 (Huang and Manning 2008). As such, many signals that turn off mTORC1, such as insulin withdrawal, do so through the TSC complex. Consequently, loss of any component of the TSC complex leads to sustained mTORC1 activity under conditions that would normally inhibit mTORC1, such as during fasting in the liver (Sengupta et al. 2010). Energy levels signal to mTORC1 through AMPK via two mechanisms. AMPK phosphorylates TSC2 on Serine 1387 to activate TSC2 and promote inhibition of mTORC1 through the Rheb axis (Inoki et al. 2003; Shaw et al. 2004). AMPK can also phosphorylate RAPTOR on serine 722 and serine 792 to directly inhibit mTORC1 activity (Gwinn et al. 2008). Additional AMPK-independent mechanisms have also been suggested to inhibit mTORC1 upon energy stress, particularly in cell culture (Kalender et al. 2010; Ben Sahra et al. 2011). However, the relative importance of the various mechanisms by which metformin inhibits mTORC1 and the circumstances under which AMPK phosphorylation of RAPTOR and TSC2 inhibit mTORC1 are not clearly elucidated.
Previous analyses have shown that metformin is capable of inhibiting hepatic mTORC1 signaling via AMPK and partially via the TSC complex (Howell et al. 2017). However, the relative contribution of the AMPK-dependent phosphorylation of RAPTOR on metformin's inhibition of mTORC1 has not been assessed. Additionally, the role of mTORC1 in the action of metformin in the physiological context of the liver has been understudied, in part due to the lack of genetic tools to dissociate mTORC1 signaling from AMPK. Using novel genetic models, we found that metformin-induced inhibition of mTORC1 is dependent on AMPK, TSC2, and AMPK-dependent phosphorylation of RAPTOR. We found that metformin regulates the transcription of a subset of genes that is common to that observed with direct inhibition of mTORC1 by INK128, a portion of which is dependent on AMPK regulation of mTORC1. We also reveal that while global protein synthesis is AMPK-independent, inhibition of gene-specific translation is dependent on complete mTORC1 inhibition by AMPK via phosphorylation of RAPTOR and TSC2. Finally, in addition to established roles in metabolism, gene expression analysis reveals an important role for mTORC1 regulation and activity in the anti-inflammatory response to metformin.
Results
Metformin inhibits mTOR-induced transcriptional induction of anabolism
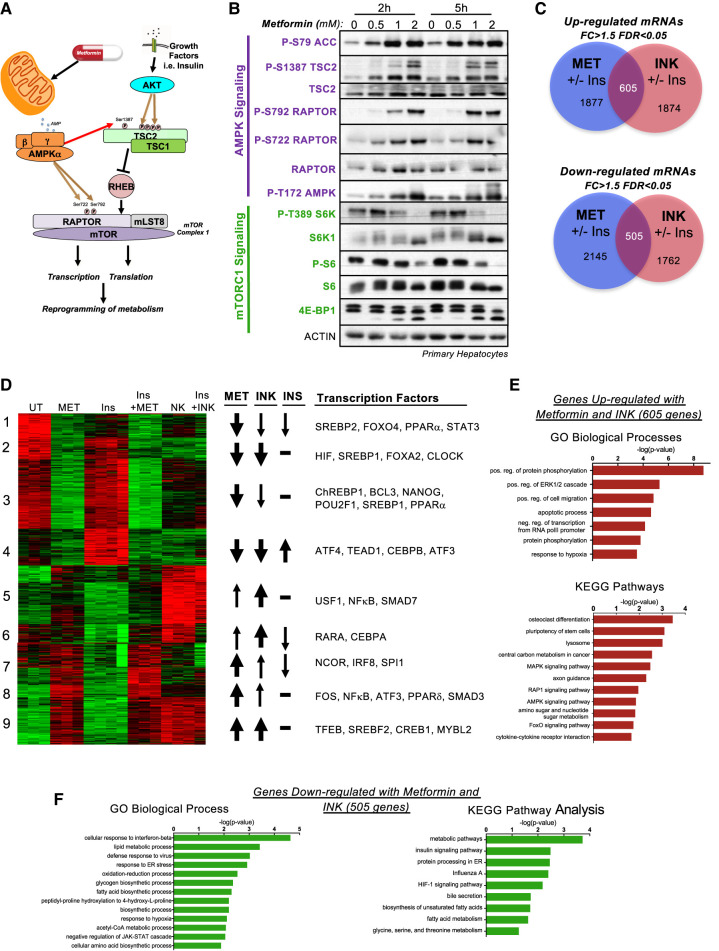
The kinetics of metformin action on AMPK-mTORC1 signaling was evaluated in primary hepatocytes. Activation of AMPK and inhibition of mTORC1 signaling through AMPK phosphorylation of RAPTOR Ser722 and Ser792 and TSC2 Ser1387 was observed after 2 and 5 h of metformin treatment in a dose dependent manner (Fig. 1A,B). Since metformin exerts part of its long-term effects through transcriptional regulation, we analyzed gene expression after 5 h of metformin treatment. To ascertain what transcriptional changes were due to mTOR inhibition, we also treated some primary hepatocytes with the direct mTOR catalytic kinase inhibitor INK128 (“INK”), also known as MLN0128 (Hsieh et al. 2012; Janes et al. 2013). In addition to its inhibition by AMPK, mTORC1 is also positively regulated by insulin through inhibition of the TSC2 protein. Therefore, to understand the consequences of metformin treatment under conditions of stimulated mTORC1 signaling, we also assessed the effect of metformin in the presence of insulin pretreatment. We observed 1324 genes or 2017 genes up-regulated by metformin in the absence or presence of insulin pretreatment, respectively. We also observed 1834 genes or 2051 genes down-regulated by metformin in the absence or presence of insulin pretreatment, respectively. In comparison, we observed 1210 genes and 1607 genes down-regulated by INK, with and without insulin, and 1010 genes and 2215 genes up-regulated by INK, with and without insulin, respectively (Supplemental Fig. S1A,B). Notably, many of the genes regulated with and without insulin by each treatment were overlapping, suggesting that insulin treatment did not affect the primary response to metformin and INK (Fig. 1C; for gene lists for all clusters and Venn diagrams, see Supplemental Table S1). From these differentially regulated genes, we observed a wide variety of interactions between metformin, INK, and insulin (Supplemental Fig. S1A). The metformin-specific subset of genes is responsive to only metformin and not INK (potentially mTOR-independent genes), whereas the mTOR-specific subset responds only to INK and not metformin (potentially regulated by mTORC2 rather than mTORC1). Nevertheless, a substantial fraction of genes are regulated in the same direction by both metformin and INK treatments, and are classified as metformin and mTOR-responsive genes (most likely mTORC1-dependent genes). We also observed a class of genes that respond to insulin; a subset of which was also responsive to INK and/or metformin. Gene set enrichment analysis of genes modulated by metformin identified the AKT–mTOR pathway, further validating the role of mTORC1 signaling in the function of metformin (Supplemental Fig. S1C).
Figure 1.
Metformin inhibits mTOR signaling. (A) Schematic of known AMPK and mTORC1 regulation. (B) Immunoblot analysis of primary serum-starved hepatocytes from wild-type mice treated with increasing doses of metformin (0, 0.5, 1, 2 mM) for 2 or 5 h. (C) Venn diagram of genes regulated in primary hepatocytes by 2 mM metformin (Met) and 0.1 μM INK-128 (INK) in the presence or absence of a 15 min pretreatment with 1 nM Insulin (Ins). Fold change >1.5, FDR <0.05. (D) Heat map of genes regulated similarly between 2 mM metformin (Met) and 0.1 μM INK-128 (INK) in the absence or presence of 1 nM insulin 15′ pretreatment (Ins) for 5 h. Clusters annotated for metformin (Met), INK-128 (INK), and insulin (Ins) responsiveness (arrow weights) and critical transcription factors driving gene signatures, according to Enrichr. (E,F) GO biological process and KEGG pathway analyses of genes up-regulated (E) or down-regulated (F) by metformin and INK as described in D.
We decided to focus on the 1110 genes regulated in a similar manner by both metformin and INK in the presence or absence of insulin pretreatment, expecting that these genes would be most informative in further understanding the role of AMPK regulation of mTORC1 signaling. Hierarchical clustering of this gene set revealed multiple clusters, including those that are dependent on insulin and those differentially dependent on metformin or INK (Fig. 1D). GO term and KEGG pathway enrichment analysis revealed that genes up-regulated by metformin and INK are involved in the lysosome, central carbon metabolism, cell migration, and stem cell pluripotency, while genes down-regulated are involved in fatty acid biosynthesis, anabolic metabolism, and the HIF pathway (Fig. 1E,F). A closer look at the clusters identified that are regulated by both metformin and INK uncovered numerous previously characterized signaling pathways shown to be responsive to mTORC1 such as HIF (cluster 2), SREBP1 (cluster 2), and TFEB (cluster 9) (Düvel et al. 2010; Young et al. 2016; Collodet et al. 2019). Notably, we also observed multiple understudied potential regulators of these clusters, such as STAT3 and CLOCK, providing possible novel avenues to investigate. In all, from these analyses, we compiled a list of genes regulated by metformin in an mTOR-dependent manner (Supplemental Table S1). We suspect that many of these metformin-regulated genes are AMPK-dependent; however, they could also be regulated through parallel pathways.
Generation of Raptor knock-in mouse model
Previous work has attempted to understand the mechanism of dependency of metformin on mTORC1 signaling (Dowling et al. 2007; Kalender et al. 2010; Ning and Clemmons 2010). Notably, in hepatocytes, loss of Tsc1 was found to only partially recapitulate the effects of AMPK loss on metformin action (Howell et al. 2017), illustrating the importance of other targets of AMPK in mTORC1 regulation. One possible alternate avenue for AMPK regulation of mTORC1 upon metformin treatment in hepatocytes is via the AMPK-dependent phosphorylation of RAPTOR, as has been previously demonstrated in fibroblasts (Gwinn et al. 2008). In order to address the importance of RAPTOR regulation by AMPK upon metformin treatment (and potential other stimuli), we used CRISPR in utero to generate a whole-body Raptor knock-in mouse model, in which both AMPK phosphorylation sites (Ser722/Ser792) in RAPTOR were mutated to alanine (Fig. 2A; Supplemental Fig. S2). Intercrossing of heterozygous RaptorAA/+ knock-in mice resulted in viable homozygous RaptorAA/AA (RaptorAA) mice born in Mendelian ratios. Immunoblot analysis of livers from homozygous RaptorAA mice on ad lib fed a high-fat diet revealed loss of both P-S722 and P-S792 with full retention of the RAPTOR protein (Fig. 2B). Mice were maintained on a high-fat diet to mimic metabolic syndrome in humans, including insulin resistance and obesity, for which metformin is most commonly prescribed (Garcia and Shaw 2017). We next assessed the role of RAPTOR phosphorylation upon fasting, a context under which mTORC1 is normally inhibited (Howell et al. 2017). Surprisingly, loss of AMPK-dependent phosphorylation of RAPTOR resulted in higher mTORC1 activity levels, as revealed by increased phosphorylation the S6K1 downstream target S6 and the mTOR site Ser757 in ULK1 (Fig. 2C). This suggests that AMPK activation is required at least in part for the inhibition of mTORC1 activity in the fasted state.
Figure 2.
Generation and validation of Raptor knock-in mouse model. (A) Targeting schematic to mutate serine 722 and serine 792 to alanines in Raptor in utero. (Red text) PAM (NGG) site, (blue text) gRNA sequence, (green text) Ser/Ala residue, (yellow highlight) wobble base pairs, (italics) recombinant enzyme sites added or deleted by recombination. (B) Immunoblot analysis of ad lib fed wild-type and RaptorAA homozygous mice following 20 wk on a high-fat diet (HFD). (C) Immunoblot analysis of livers from wild-type and RaptorAA homozygous mice fasted overnight (O/N) following 20 wk on a high-fat diet. (Bottom) Quantification of P-S6 and P-S6K relative to total protein levels (n = 6–12). SEM. Welch t-test, (*) P-value < 0.05 compared with wild type. (D) Immunoblot analysis of livers from wild-type and RaptorAA homozygous mice fasted overnight, refed for 1 h with HFD, and treated with saline or 250 mg/kg metformin for 2 h following 20 wk on HFD. (Bottom) Quantification of P-S6 and P-S6K relative to total protein levels and ratio of untreated (n = 6). SEM. Welch t-test, (*) P-value < 0.05 compared with wild type.
Further analysis of livers from mice fasted overnight, refed for 1 h to induce mTORC1 activity, and then treated with metformin for 2 h revealed induction of AMPK signaling. This fasting–refeeding model has been established as a method to consistently elevate mTORC1 activity to reveal the pharmacological effects of metformin on AMPK activation and mTORC1 inhibition in the liver. Additionally, dosing mice with 200–250 mg/kg metformin has been shown to result in plasma metformin concentrations similar to those seen in humans on metformin treatment (Howell et al. 2017). Following this metformin treatment scheme, RAPTOR was phosphorylated at Ser722 and Ser792, and mTORC1 activity was inhibited in wild-type livers, as measured by P-S6K1, P-S6, and P-4E-BP1 and their corresponding mobility shifts. Importantly, in the RaptorAA mice following metformin treatment, phosphorylation of RAPTOR was absent and the inhibition of mTORC1 was attenuated (red arrows, Fig. 2D). Therefore, phosphorylation of RAPTOR by AMPK is partially necessary for mTORC1 inhibition by metformin.
We next assessed the effect of RaptorAA mutation on metformin-induced mTORC1 inhibition in primary hepatocytes. Primary hepatocytes derived from wild-type or RaptorAA livers were treated with metformin and mTORC1 signaling was assessed 2 or 5 h later (Supplemental Fig. S2B). We observed that while mTORC1 was inhibited in wild-type cells, RaptorAA hepatocytes showed modest attenuation of mTORC1 inhibition at high doses, as measured by P-S6K1 and P-S6 (Supplemental Fig. S2B). We next assessed whether any of the transcriptional changes we observed in wild-type hepatocytes treated with metformin and INK were due to inhibition of mTORC1 via AMPK-dependent phosphorylation of RAPTOR. Comparison of genes regulated in wild-type primary hepatocytes upon metformin and INK treatment and genes regulated in RaptorAA primary hepatocytes upon metformin treatment revealed ∼30% were dependent on RAPTOR phosphorylation (Supplemental Fig. S2C). GO term and KEGG pathway analysis of RAPTORAA-dependent genes that change upon metformin and INK treatment identified ER stress, glucose response, and regulation of cell growth (Supplemental Fig. S2D,E). Therefore, Raptor phosphorylation by AMPK is partly necessary for mTORC1-dependent transcriptional changes upon metformin.
RAPTOR phosphorylation and TSC2 are both required for metformin-dependent inhibition of mTORC1 signaling
Since we observed that RAPTOR phosphorylation by AMPK is required for partial inhibition of mTORC1 by metformin, we questioned the relative contribution of RAPTOR and TSC2 phosphorylation by AMPK for complete mTORC1 inhibition. RaptorAA mutant mice were crossed to Liver-Tsc2 knockout mice (Tsc2-floxed [Tsc2fl] mice crossed with Albumin-Cre mice) or to iLiver-Tsc2KO mice (Tsc2-floxed mice crossed with inducible Albumin-CreERT2 mice in which liver deletion is induced upon tamoxifen treatment) (Supplemental Fig. S3A,B). Using RaptorAA mutant mice, or Tsc2fl;Alb-Cre mice, or mice combined for these two mutations (RaptorAA;Tsc2fl;Alb-Cre [AA;Tsc2]), we assessed the effect of metformin on mTORC1 signaling in the livers of mice fed a high-fat diet. Mice were fasted overnight, refed for 1 h, and treated with metformin for 2 h before analysis of liver signaling (Fig. 3A). In agreement with previous results, we observed induction of AMPK signaling upon metformin, as seen by increased P-AMPK and P-RAPTOR, and inhibition of mTORC1 signaling as seen by decreased P-S6K1, P-S6, and P-4E-BP1 in wild-type livers. In contrast, we saw an allelic dose dependency of the resistance to mTORC1 inhibition in RaptorAA, Tsc2-null, and RaptorAA;Tsc2-null livers, as determined by P-4E-BP1 and P-S6, with the strongest effect observed in RaptorAA;Tsc2-null livers. Thus, regulation of both RAPTOR and TSC2 by AMPK is required in vivo in the liver for the complete acute inhibition of mTORC1 signaling by metformin.
Figure 3.
RAPTOR phosphorylation and TSC2 are both required for metformin-induced inhibition of mTORC1. (A) Immunoblot analysis of wild-type (WT), RaptorAA (AA), Tsc2;Alb-Cre (TSC2), and RaptorAA;Tsc2;Alb-Cre (AA;TSC2) livers fasted overnight, refed for 1 h, and treated with saline or 250 mg/kg metformin for 2 h. (Bottom) Quantification of P-56K and P-S6 relative to total protein and ratio of saline treatment (n = 3–6). SEM. t-test, (*) P-value = 0.05 compared with wild type. (B) Immunoblot analysis of serum-starved primary hepatocytes derived from wild-type (WT), RaptorAA (AA), Tsc2;AlbCreER (TSC2), or Raptor;Tsc2;Alb-CreER (AA;T) mice treated with increasing doses of metformin (0, 1, or 2 mM) for 2 or 5 h. (Bottom) Quantification of P-S6K and P-S6 relative to total protein. (C,D) Immunoblot analysis of primary hepatocytes serum-starved or pretreated with 1 nM insulin for 15′ prior to treatment with 2 mM metformin for 2 h (C) or 5 h (D). Treatment with 0.1 μM INK-128 (INK) was used as a positive control for mTOR inhibition.
We next looked at the requirement for RAPTOR phosphorylation and TSC2 in primary hepatocytes treated with metformin. Primary hepatocytes were collected, serum starved, then treated with metformin for 2 or 5 h (Fig. 3B). With increasing doses of metformin, we observed concomitant induction of AMPK and inhibition of mTORC1 in wild-type cells. Similar to previous results, we observed an allelic dose-dependent attenuation of mTORC1 inhibition, with RaptorAA;Tsc2 double-mutant cells being the most resistant to metformin's effects.
We observed that the basal rate of mTORC1 signaling, as assessed by basal P-S6K and P-S6 levels, were significantly increased in Tsc2-null and RaptorAA;Tsc2-null hepatocytes. We hypothesized that this was due to the insulin-independent hyperactivation of mTORC1 signaling due to Tsc2 deficiency (Sengupta et al. 2010). Therefore, we examined the impact of metformin treatment in the context of a pretreatment of serum-starved cells with a low dose of insulin to physiologically potentiate mTORC1 signaling in the wild-type cells. Insulin pretreatment indeed induced basal mTORC1 signaling in wild-type and RaptorAA hepatocytes to levels comparable with Tsc2 loss (Fig. 3C,D). Furthermore, treatment with insulin partially rescued the mTORC1 inhibition due to metformin in both wild-type and RaptorAA cells after 2 and 5 h, suggesting that the inhibition of TSC2 by insulin signaling forces AMPK to rely more on RAPTOR regulation to inhibit mTORC1 (hepatocytes treated with INK-128 as a positive control for mTOR inhibition) (Fig. 3C,D; Supplemental Fig. S3C,D).
AMPK and AMPK phosphorylation of RAPTOR and TSC2 play critical roles in select, but not global, protein translation in response to metformin
mTORC1 plays a critical role in protein translation and metformin has been shown to inhibit global translation in part through the TSC complex (Howell et al. 2017). Therefore, using our genetic system in primary hepatocytes, we tested whether AMPK regulation of RAPTOR phosphorylation and TSC2 plays a role in decreased translation upon metformin treatment. In addition to RaptorAA;Tsc2-null cells, we also generated Ampk-null primary hepatocytes to test the AMPK dependency of metformin treatment. Ampk-null primary hepatocytes were generated from Ampkα1 and Ampkα2 double floxed mice harboring Albumin-CreER that were treated with tamoxifen at 8 wk of age to create adult liver-specific deletion of both isoforms of the AMPK catalytic subunits.
Global protein synthesis was assayed using the Sunset assay, which quantifies puromycin incorporation into newly synthesized proteins (Schmidt et al. 2009). Hepatocytes of each genotype were treated with metformin in the absence or presence of insulin pretreatment for 2 h and pulsed with puromycin in the last 10 min prior to collection (Supplemental Fig. S4A). In agreement with previous literature, we observed that metformin inhibited global protein synthesis and this was tempered by TSC2 loss, presumably due to hyperactivation of mTORC1 in Tsc2-deficient cells (Fig. 4A; Supplemental Fig. S4B). Surprisingly, neither RaptorAA nor Ampk deficiency rescued acute protein synthesis inhibition by metformin. However, insulin treatment did blunt the percent inhibition by metformin (Supplemental Fig. S4B). Thus, global translation inhibition by metformin is not mediated by AMPK but can be buffered by Tsc2 deficiency.
Figure 4.
RAPTOR phosphorylation and TSC2 regulate translational changes observed upon metformin. (A) Sunset assay on primary hepatocytes treated with 2 mM metformin (Met) or 0.1 μM INK-128 (INK) for 2 h. (Bottom) Puromycin incorporation was quantified with ImageJ. t-test, (**) P-value < 0.01. (B) mRNA 5′ cap-binding complexes were isolated with m7GTP-agarose from primary hepatocytes treated with 2 mM metformin or 0.1 μM INK-128 (INK) for 2 h and analyzed by immunoblotting. (C) Analysis of the fold change in translational efficiency between treated and untreated in genes with 1.5-fold down-regulation in translational efficiency upon metformin and INK treatment as described in A in wild-type cells. Kolmogorov–Smirnov test comparing with wild-type treated with metformin. n = 3.
Because mTORC1 plays a well-established role in promoting translation, we analyzed the ability of metformin to regulate cap-dependent translation initiation in hepatocytes of each genotype. When mTORC1 signaling is inhibited, 4E-BP1 becomes dephosphorylated and is able to associate with the translation initiation protein eIF4E at the 7-methylguanosine moiety (m7GTP) cap at the 5′ end of mRNA, thereby blocking translation initiation. Consistent with their suppression of mTORC1 signaling in hepatocytes, metformin and INK treatments in wild-type primary hepatocytes induced binding of 4E-BP1 to eIF4E on m7GTP-agrose beads, which mimic the mRNA 5′ cap structure (Fig. 4B). Similarly, induction of 4E-BP1 association with eIF4E was observed in RaptorAA and Tsc2-null cells treated with metformin. In contrast, the ability of metformin to induce 4E-BP1 association with eIF4E was greatly diminished in RaptorAA;Tsc2-null and Ampk-null cells, validating the mTORC1 and AMPK dependency of metformin effects on cap-dependent translation initiation.
We further explored which proteins were specifically regulated by metformin to assess the role of AMPK, RAPTOR phosphorylation, and TSC2 in directed translation. In order to focus on the direct translational effects of mTORC1 and avoid secondary effects, we performed polysome profiling on hepatocytes treated with metformin for 2 h in the absence or presence of insulin pretreatment (Supplemental Fig. S4C). In order to uncouple the transcriptional and translational effects of metformin, we calculated the translational efficiency of each mRNA. As previously described, due to the robust inhibition of global translation, which was in part genotype-dependent, we chose to focus on genes most down-regulated in the presence of metformin (Supplemental Fig. S4D; Thoreen et al. 2012). We identified the genes with down-regulated translation efficiency by at least a 1.5-fold upon metformin treatment between untreated and metformin treatment in three independent rounds of polysome profiling, two in the presence of insulin pretreatment and one in the absence of insulin. Using wild-type cells, we identified 82 genes that exhibited decreased translation efficiency upon metformin treatment in all three experiments and plotted the average translation efficiency (Fig. 4C). GO term and KEGG pathway analysis of these genes revealed enrichment in genes involved in translation and oxidative phosphorylation (Supplemental Fig. S4E). Comparing with INK-treated samples, 62 of the 82 genes were regulated by both metformin and INK. Using previously reported gene lists for genes down-regulated with the mTORC1 inhibitor Torin1 in MEFs or PP242 in prostrate cancer cells, we observed 23 genes with TOP motifs or PRTE motifs down-regulated upon metformin (Hsieh et al. 2012; Thoreen et al. 2012; Philippe et al. 2020). Thus, we were able to generate a list of 82 genes with decreased translation upon metformin treatment that correlates with mTORC1 inhibition (Supplemental Table S2). We next used this list of metformin-responsive genes to assess the effect of the various genotypes on translation. We observed an allelic dose dependency of metformin on translation inhibition with Ampk-null cells having the strongest effect on translation and RaptorAA;Tsc2-null hepatocytes closely mimicking Ampk-null cells, while RaptorAA and Tsc2-null hepatocytes had a partial response (Fig. 4C). Performing similar analyses of genes identified with reduced translational efficiency in wild-type cells upon INK treatment (Supplemental Fig. S4F,G) or genes previously identified as containing TOP motifs (Supplemental Fig. S4H) revealed similar allelic dose dependency of metformin on translation inhibition. Overall, we conclude that inhibition of select translation by metformin is regulated by mTORC1 in an AMPK-dependent manner.
Metformin requires RAPTOR phosphorylation and TSC2 to regulate transcriptional control of mTORC1-dependent pathways
With the observations that metformin regulates mTORC1 signaling and mTORC1-specific translation via TSC2 and phosphorylation of RAPTOR, we wanted to explore which of the transcriptional effects of metformin were also dependent on regulation of RAPTOR phosphorylation and TSC2. Therefore, we performed transcriptional analysis of primary hepatocytes treated with metformin for 5 h in the absence or presence of insulin pretreatment. We then assessed the effect of RaptorAA mutation, Tsc2 loss, combined RaptorAA;Tsc2 loss or Ampk loss on those genes identified as being regulated by metformin and INK in the same direction. Analysis of the various genotypes reveals multiple types of gene interactions, including genes that are independent of AMPK or are dependent on AMPK, RAPTOR phosphorylation, TSC2, or both RAPTOR phosphorylation and TSC2 (Supplemental Fig. S5, S6). Comparison of genes regulated in wild-type primary hepatocytes upon metformin and INK treatment and genes regulated in Tsc2-null and RaptorAA;Tsc2-null primary hepatocytes upon metformin treatment revealed about 80% and 50% of genes up-regulated and down-regulated, respectively, in a TSC2 and RAPTOR;TSC2-dependent manner (Supplemental Fig. S6A). Similarly, comparison of genes regulated in wild-type primary hepatocytes upon metformin and INK treatment and genes regulated in RaptorAA;Tsc2-null or Ampk-null primary hepatocytes upon metformin treatment revealed 70%–80% of genes up-regulated in wild-type hepatocytes were dependent on AMPK and RAPTOR;TSC2; in contrast, only 35%–50% of genes down-regulated in wild-type hepatocytes were found to be dependent on AMPK and RAPTOR;TSC2 (Supplemental Fig. S6B).
A more focused analysis of pathways known to be affected by mTOR, such as lysosome/TFEB, the HIF pathway, and lipogenesis, revealed dependence on RAPTOR phosphorylation and TSC2, and AMPK for metformin-induced expression changes as expected (Fig. 5A; Supplemental Fig. S7A–C). Notably, a number of key HIF and SREBP target genes are greater impacted in AA;Tsc2-null than in Tsc2-null alone, including Fasn, Acaca/Acc1, Pdk1, and Egln1. Additionally, the relative effect of INK and metformin appeared to vary among these signaling pathways, with INK having comparable strength in transcriptional modulation to metformin in HIF signaling but having a less robust effect on TFEB signaling.
Figure 5.
RAPTOR phosphorylation and TSC2 regulate transcriptional changes observed upon metformin. (A) Heat map of fold change for genes involved in HIF signaling and SREBP signaling upon 2 mM metformin or 0.1 μM INK-128 (INK) treatment for 5 h compared with untreated. (Right) Quantification of fold change of genes identified with fold change >1.5 in wild type. Box plot with bars denoting minimum to maximum. Kolmogorov–Smirnov test comparing wild-type treated with metformin. (B) GO term analysis of genes down-regulated in wild-type hepatocytes with metformin and INK as described in Figure 1D (FC >1.5, FDR <0.05) in an AA;TSC- and AMPK-dependent manner. (C) Heat map of fold change for genes involved in STAT signaling upon metformin and INK as described in Figure 1D. Genes with fold change >1.5 are denoted in black; >1.2 are denoted in blue. (Right) Quantification of fold change of genes identified with fold change >1.2 in wild type. Kolmogorov–Smirnov test comparing with wild type treated with metformin. (D) qRT-PCR analysis of STAT3-dependent genes in wild-type primary hepatocytes treated with 2 mM metformin or 50 μM S3I-201 (STAT3 inhibitor) for 5 h. n = 3, t-test, (*) P-value < 0.05, (**) P-value < 0.01. (E) Heat map of the average FPKM value of STAT3-dependent genes upon 2 mM metformin or 0.1 μM INK treatment for 5 h, as shown in Figure 4F, in primary hepatocytes of each genotype. (F) Immunoblot analysis of primary hepatocytes treated with 2 mM of metformin or 0.1 μM INK for 2 h. (Bottom) Quantification of P-S727 STAT3 (left) and P-Y705 STAT3 (right) relative to STAT3. n = 4–7. Mean ± SEM, t-test, (*) P value < 0.05.
To identify additional pathways that show dependency on AMPK and RAPTOR;TSC2, we performed GO term and KEGG pathway analysis on genes down-regulated by metformin and INK in an RaptorAA;Tsc2- and Ampk-dependent manner (Supplemental Fig. S6B). In addition to an expected signature for lipid metabolism and HIF1 signaling, we also observed signatures for genes responsive to interferon-β and viral defense (Fig. 5B; Supplemental Fig. S6B). We were intrigued by the presence of inflammatory gene signatures, considering metformin's purported role to inhibit inflammation in patients (Saisho 2015; Cameron et al. 2016), and decided to investigate whether this effect is AMPK- and mTORC1-dependent.
Interferon β signaling is modulated by STAT1, STAT2, and STAT3 transcription factors and mTOR has previously been associated with STAT1 and STAT3 activity. Of note, mTOR activation has been associated with decreased STAT1 activity, yet increased STAT3 activity suggesting that the signature we observed following metformin may be more closely related to changes in STAT3 activity (Fielhaber et al. 2009; Kim et al. 2009; Karonitsch et al. 2018). To query whether STATs are indeed regulated by mTORC1 in response to metformin, we first assessed gene expression of GSEA- and ChEA-annotated STAT3 pathway modulated genes and observed that many “STAT3 target” genes are down-regulated by metformin in wild-type hepatocytes, but this regulation is blunted in RaptorAA;Tsc2-null and Ampk-null hepatocytes (Fig. 5C; Supplemental Fig. S7D). To validate that regulation of these genes in primary hepatocytes is STAT3-dependent, we assessed gene expression of a subset of these genes following treatment with the specific small molecule STAT3 inhibitor S3I-201 in wild-type cells. Treatment with either metformin or S3I-201 was able to reduce expression of these annotated STAT3 genes, confirming these genes are STAT3-dependent in the context of primary hepatocytes (Fig. 5D). Intriguingly, closer analysis of a subset of these STAT3-dependent genes revealed down-regulation of the basal gene expression in conditions of mTORC1 hyperactivation caused by Tsc2 or Ampk loss, suggesting that dysregulation of some STAT3 target genes may be due to adaptation to mTORC1 hyperactivation, perhaps due to mRNA changes in cytokines or other regulators of STAT3 (Fig. 5E).
Previous reports have suggested that mTORC1 can phosphorylate both STAT1 and STAT3 at serine 727 (Yokogami et al. 2000; Fielhaber et al. 2009; Kim et al. 2009), so we decided to query whether this occurred in our system using our genetic models. Analysis of P-S727 of STAT1 in hepatocytes showed no reduction upon metformin treatments. However, analysis of P-S727 of STAT3 in hepatocytes treated with metformin revealed decreased P-STAT3 in wild-type cells, but not RaptorAA:Tsc2-null or Ampk-null cells (compare with P-S6K1) (Fig. 5F). Intriguingly, P-S727 STAT3 was unchanged upon INK treatment, suggesting that the regulation of this site is more likely to be caused by a secondary effect of chronic mTORC1 activity caused by Tsc2 and Ampk loss (Fig. 5F; Supplemental Fig. S7E). STAT3 can also be phosphorylated at Tyr705 by the JAK proteins and other RTKs, and this phosphorylation is required for STAT3 dimerization and nuclear translocation. Analysis of P-Y705 STAT3 revealed a similar decrease upon metformin in wild-type cells, which was partially, but significantly, rescued in RaptorAA:Tsc2-null or Ampk-null cells (Fig. 5F). Importantly, unlike STAT3 P-S727, INK treatment significantly reduced P-Y705 STAT3 levels. Furthermore, treatment of wild-type cells with AZD8055 (mTOR inhibitor), 991 (AMPK activator), and MK2206 (AKT inhibitor) reduced P-Y705 STAT3 levels but not P-S727 STAT3 levels (Supplemental Fig. S7E), confirming the acute mTORC1 dependency of P-Y705 STAT3 in primary hepatocytes. Thus, STAT3 can be modulated by mTORC1 via two distinct mechanisms: (1) chronic mTORC1 activation ameliorates the inhibition of P-S727 by metformin treatment, while acute mTOR inhibition in wild-type cells does not affect P-S727, and (2) acute mTOR inhibition suppresses P-Y705. Both of these mechanisms could contribute to the overall STAT3-dependent gene signature observed upon metformin treatment. Thus, we found that STAT3 is regulated by AMPK in an mTORC1-dependent manner in these cells and may elicit part of the transcriptional changes triggered by metformin that control its effects on inflammation.
Metformin inhibits inflammatory signature in vivo
We next wanted to extend our findings in hepatocytes to the intact mouse liver, a major target tissue for metformin action (Johnson et al. 1993; Rena et al. 2017). Therefore, we treated mice with metformin for 4 h following our fasting/refeeding protocol, and evaluated effects of the various genotypes on signaling. Similar to mice treated for 2 h (Fig. 3A), we observed that metformin activated AMPK signaling and inhibited mTORC1 signaling in wild-type livers. This inhibition was attenuated in the various genotypes, with the strongest attenuation occurring in the RaptorAA;Tsc2-null and Ampk-null livers (Fig. 6A). In addition to inhibition of P-S6K and P-TFE3 in wild-type livers, we also observed inhibition of P-S727 STAT3 that occurred in an mTORC1/AMPK-dependent manner. Thus, our observation in hepatocytes that metformin inhibition of STAT3 is dependent on mTORC1 was recapitulated in vivo.
Figure 6.
Metformin regulates STAT3 in vivo in mTORC1-dependent manner. (A) Immunoblot analysis of wild-type (WT), RaptorAA (AA), Tsc2;Alb-CreER (Tsc), RaptorAA;Tsc2;Alb-CreER (AA;T) and Ampk;Alb-CreER (Ampk) livers fasted overnight, refed for 1 h, and treated with 200 mg/kg metformin for 4 h. (Bottom) Quantification of P-T389 S6K and P-S727 STAT3 relative to total protein and ratio of saline treatment (n = 3–5). SEM. (*) P-value = 0.05, (**) P-value = 0.01 compared with wild type. (B) GO biological process analysis of AMPK-dependent genes up-regulated (left; red) or down-regulated (right; green) by at least 1.5-fold with an FDR <0.05 in wild-type livers upon metformin as described in A. (C) Heat map of fold change for genes involved in HIF signaling upon metformin treatment as described in A compared with saline treated. Genes with fold change >1.5 in wild-type livers are denoted in black; FC >1.2 is denoted in blue. Asterisks represent adjusted P-value in WT livers. (Right) Quantification of fold change for genes identified with fold change >1.2 in wild-type liver upon metformin treatment. Box plot with bars denoting minimum to maximum. Kolmogorov–Smirnov test compared with wild-type treated with metformin. (D) Heat map of fold change for genes involved in STAT signaling upon metformin treatment as described in A with a fold change of ≥1.5 (black) or 1.2 (blue) in wild-type livers. Asterisks represent adjusted P-value in WT livers. (Right) Quantification of fold change of genes identified with fold change >1.2 in wild type. Kolmogorov–Smirnov test compared with wild-type treated with metformin.
We next decided to explore the transcriptional effects of metformin in the liver. We performed RNA-seq on livers from the varying genotypes treated with saline or metformin for 4 h (Supplemental Fig. S8A). From this analysis, we observed 391 and 576 genes down-regulated or up-regulated, respectively, in wild-type livers. In contrast, we observed only 116 and 188 gene down-regulated or up-regulated, respectively, in RaptorAA livers and only three and 16 genes down-regulated or up-regulated, respectively, in Ampk-null livers, suggesting that the majority of metformin's function in vivo in the liver is through AMPK (Supplemental Fig. S8B). GO term analysis of metformin-responsive genes in wild-type livers revealed signatures for immune processes, extracellular matrix, and translation (Fig. 6B; Supplemental Fig. S8C). Analysis of genes involved in HIF signaling, SREBP signaling, and TFEB signaling revealed only minor changes with a fold change of >1.5, but a larger percent with a fold change of 1.2 or greater (Fig. 6C; Supplemental Fig. S8D).
Because we saw a STAT3 signature in hepatocytes and a genotype-dependent inhibition of P-STAT3 in the livers, we next performed a more directed evaluation of genes involved in STAT3 signaling. Analysis of STAT3-responsive genes indeed revealed that many were down-regulated in wild-type livers and this down-regulation was largely diminished in the RaptorAA;Tsc2-null and Ampk-null livers (Fig. 6D). Notably, more genes in the STAT3 signature were significantly regulated by AMPK and mTORC1 in the liver of mice on a high-fat diet than were the genes in the HIF, SREBP1, or TFEB signatures under these conditions. Since it has been established previously that metformin can reduce inflammation in patients (Saisho 2015; Cameron et al. 2016), we propose that AMPK-dependent suppression of mTORC1-activation of STAT3 may be mediating some of metformin's anti-inflammatory effects.
Discussion
Altogether, our data support the hypothesis that AMPK-dependent regulation of mTORC1 requires RAPTOR phosphorylation and TSC2. Furthermore, metformin induces gene expression changes and translational changes in part through regulation of mTORC1 downstream from AMPK. Finally, metformin regulates known transcriptional programs, such as those mediated by TFEB, HIF, and SREBP1, as well as less-appreciated pathways, such as STAT3 signaling, through inhibition of mTORC1 downstream from AMPK (Fig. 7). Metformin inhibition of mTORC1 requires both RAPTOR phosphorylation and TSC activity, and only with loss of both of these axes does metformin fail to inhibit mTORC1 signaling and the resulting repression of translation and transcription in a manner comparable with AMPK loss. Notably, this study highlights the importance of RAPTOR phosphorylation as a mechanism of mTORC1 regulation by AMPK and metformin, both alone and in combination with Tsc2 loss. Analysis of our panel of mutants revealed that even at higher doses of metformin, RaptorAA;Tsc2 double mutants largely maintain loss of mTORC1 regulation following both the acute and long-term treatment, unlike what we observed in the Tsc2-null state. A confounding factor here is that complete loss of Tsc2 causes basal hyperactivation of mTORC1 equivalent to the signal from chronic growth factor signaling from AKT that inactivates TSC2 biochemically (Hopkins et al. 2020). Thus, in the Tsc2-null state, basal mTORC1 is hyperactive, and the effects of Tsc2 deficiency on transcription appear more prominent and potentially obscures some of the metformin-induced transcriptional changes. In the future it will be important to generate mice with knock-in Ser-Ala mutations in the AMPK phosphorylation sites in TSC2 (e.g., Ser1387), as they will not bear constitutive basal hyperactivation of mTORC1 like the Tsc2-null cells do. Such Tsc2 AMPK-site Ser-Ala knock-in mice can be combined with RaptorAA mice to more clearly define the role of AMPK regulation of mTORC1. Regardless, the observation that RaptorAA;Tsc2-null primary hepatocytes and livers show additional activation of mTORC1 signaling and transcriptional effects beyond what is observed in Tsc2-null mice illustrates that RAPTOR phosphorylation is necessary for a full response of mTORC1 inhibition by AMPK.
Figure 7.

Model of metformin effects that are dependent on AMPK and mTORC1 via phosphorylation of RAPTOR and TSC2.
A major controversy in the metformin field has related to how important a role AMPK plays in different pharmacological outcomes of metformin treatment (An and He 2016; Rena et al. 2017). Issues surrounding dosage used; volumetric rodent:human comparisons and the potential for cell culture artifacts from an absence of physiological clearance from hepatocytes in the intact organism have led to a range of discrepancies and debates among researchers (He and Wondisford 2015; Chandel et al. 2016). Interestingly, under the conditions used here, at 4 h after a single dose of 200 mg/kg metformin treatment of refed C57/Bl6 mice on a high-fat diet for 12 wk, we observed that metformin-induced transcription changes were all nearly completely AMPK-dependent in vivo (Supplemental Fig. S8B). This is in comparison with the large number of genes that change in response to metformin in primary hepatocytes in AMPK-deficient hepatocytes (Supplemental Fig. S6B), suggesting that the AMPK-independent effects observed in hepatocytes are largely remediated in the liver. It has been reported in multiple other contexts that AMPK-deficient cells are unable to survive energetic stress (Shaw et al. 2004; Eichner et al. 2019), so one possibility is that AMPK-deficient hepatocytes in culture begin undergoing apoptosis after 5 h, unlike the AMPK-deficient livers in the intact mouse where ATP loss from metformin may be balanced by nutrients available in vivo.
Using RaptorAA;Tsc2-null double-mutant primary hepatocytes and livers, we show that mTORC1 is necessary for transcriptional changes in multiple signaling pathways including STAT3 signaling pathway, suggesting that an AMPK/mTORC1/STAT3 pathway play an essential role in the anti-inflammatory effects of metformin. Obesity is associated with low-grade inflammation, and patients on metformin have decreased levels of chronic inflammation markers, irrespective of diabetes status (Cameron et al. 2016; Jing et al. 2018). Additionally, mTOR has been associated with inhibition of NFκB and activation of STAT3, although this has largely been shown in macrophages and other immune cells as opposed to the liver (Yokogami et al. 2000; Weichhart et al. 2008; Kim et al. 2009; Li et al. 2014; Karonitsch et al. 2018). Notably, there has been little evidence of a direct role of mTORC1 downstream from metformin in the liver for STAT3 inhibition or its effect on inflammation. Thus, the finding that mTORC1 can regulate STAT3 phosphorylation and transcriptional activity in primary hepatocytes and the liver upon metformin treatment reveals a new mechanism for the action of metformin and validates the importance of the liver for metformin's immunosuppressive function. Notably, we observed both a chronic and acute role for mTORC1 regulation of STAT3 affecting phosphorylation of both S727 and Y705, respectively, suggesting that mTORC1's ability to inhibit STAT3-dependent transcription may be in part dependent on additional pathways activated by metformin. Additionally, the observation that AA;Tsc2-null cells are not able to completely rescue the inhibition of P-Y705 STAT3, suggests that metformin also uses a secondary mTORC1-independent mechanism to inhibit STAT3. In subsequent studies, it will be important explore the kinases or phosphatases downstream from mTORC1 that are activated or inhibited to modulate STAT3 phosphorylation both in the chronic and acute context. Additionally, it will be worth investigating the physiological consequences of metformin inhibition of mTORC1 activity on circulating cytokines and inflammation, as well as the impact of new selective AMPK agonists on STAT3 signaling and the inflammatory response (Myers et al. 2017). The discovery that metformin controls STAT3-dependent transcription in an AMPK- and mTORC1-dependent manner in the liver of mice on a high-fat diet suggests new therapeutic possibilities for metabolic diseases (including type 2 diabetes and NAFLD), as well as a range of diseases where inflammation is a prime contributor to pathology.
Materials and methods
Animals
RaptorS722A;S792A mice were generated using CRISPR in utero using gRNAs, donor dsDNA (IDT) and Cas9 mRNA (Thermo Fisher A25640) as shown in Figure 2A and as described previously (Yang et al. 2014). RaptorAA double-mutant mice were crossed to Tsc2 floxed mice (Jackson Laboratory 027458) (Hernandez et al. 2007) and Albumin-Cre (Jackson Laboratory 003574) or Albumin-CreER (Imai et al. 2000) mice on a predominately C57/Bl6 background. Ampkα1(Prkaa1)fl/fl and Ampkα2(Prkaa2)fl/fl mice on C57/Bl6 were previously described (Hasenour et al. 2014). The final cohorts were generated from the genotypes described (Supplemental Fig. S3A) and tamoxifen (1.5 mg) treatment every other day for a total of 3 i.p. injections (Supplemental Fig. S3B). Experiments or primary hepatocyte isolation was carried out ∼2–4 wk after tamoxifen injection. For fasting–refeeding studies, mice were fed a high-fat diet (45% kcal from fat) (Research Diets D12451) for at least 8 wk prior to the experiment. Mice were fasted overnight and either euthanized or refed a high-fat diet for 3–5 h before euthanasia. Vehicle (0.9% saline) or 250 mg/kg metformin (Sigma PHR1084) reconstituted in 0.9% saline was administered via intraperitoneal injection 2 or 4 h prior to euthanasia. All animal procedures were approved by the Salk Institute's Institutional Animal Care and Use Committee. Detailed methods are in the Supplemental Material.
Primary hepatocytes
Primary mouse hepatocytes were isolated as described previously (Garcia et al. 2019) from 8- to 16-wk-old mice. For global protein synthesis analysis, we used the sunset assay as described previously (Schmidt et al. 2009) following 2 h of treatment. For polysome profiling, primary hepatocytes were treated for 2 h prior to collection. Cells were lysed in polysome lysis buffer containing cycloheximide, centrifuged in a sucrose gradient, and fractions collected and pooled. RNA was purified and sequenced on the Illumina HiSeq 2500 platform at the Salk Next-Generation Sequencing Core. Detailed methods are provided in the Supplemental Material.
Protein extraction and immunoblotting
Upon euthanization, liver tissue was harvested immediately and flash-frozen in liquid nitrogen. Liver tissue lysates were generated from 50- to 100-mg pieces of frozen liver tissue homogenized in CST lysis buffer. Hepatocytes were lysed using CST lysis buffer or pull-down buffer. Detailed methods regarding cell and tissue lysis, m7GTP pull-down, immunoblotting, and antibodies used are provided in the Supplemental Material.
RNA isolation and sequencing
Primary hepatocytes and livers were treated as described. RNA was isolated using the RNeasy lipid tissue mini kit (Qiagen). mRNA-seq libraries were prepared using the TruSeq RNA library preparation kit (version 2), according to the manufacturer's instructions (Illumina) and sequenced by using the Illumina HiSeq 2500 or NovsSeq SP platform at the Salk Next-Generation Sequencing Core. Detailed methods are in the Supplemental Material. Gene lists for all clusters and Venn diagrams are in Supplemental Table S1. Data have been submitted to Gene Expression Omnibus.
Statistical analyses
Data are presented as mean ± SEM and statistical significance was determined using a Welsh's two-tailed t-test, with significance at P-values of <0.05 (*) and <0.01 (**), or Kolmogorov–Smirnov test, with significance at P-values of <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****).
Supplementary Material
Acknowledgments
We thank Benoit Viollet for Prkaa1 and Prkaa2 floxed models. We thank T.J. Rymoff, Debbie Ross, Sonja Brun, and Amanda Hutchins for mouse colony help and technical assistance with maintaining the Shaw laboratory mouse colony. We thank Amandine Chaix, Elijah Trefts, and Sonja Brun for critical reading of the manuscript. This work was supported in part by the National Institutes of Health (R01-DK080425, R35-CA220538, and P01-CA120964 to R.J.S.) Further support was provided by a fellowship from the Damon Runyan Cancer Research Foundation (DRG 2219-15 to J.L.V.N.).
Author contributions: J.L.V.N. designed and conducted experiments, analyzed results, and wrote the manuscript. K.H. designed and validated experiments. E.-C.L. performed polysome profiling, and E.L.V.N. analyzed polysome data in the laboratory of G.W.Y. A.D. supported mouse experiments. J.Y. and M.N.S. analyzed RNA-seq data. Y.D. assisted with generation of Raptor KI mouse model. R.J.S. designed experiments, analyzed results, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339895.120.
References
- An H, He L. 2016. Current understanding of metformin effect on the control of hyperglycemia in diabetes. J Endocrinol 228: R97–R106. 10.1530/JOE-15-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, Tanti J-F, Giorgetti-Peraldi S, Bost F. 2011. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res 71: 4366–4372. 10.1158/0008-5472.CAN-10-1769 [DOI] [PubMed] [Google Scholar]
- Cameron AR, Morrison VL, Levin D, Mohan M, Forteath C, Beall C, McNeilly AD, Balfour DJK, Savinko T, Wong AKF, et al. 2016. Anti-inflammatory effects of metformin irrespective of diabetes status. Circ Res 119: 652–665. 10.1161/CIRCRESAHA.116.308445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel Navdeep S, Avizonis D, Reczek Colleen R, Weinberg Samuel E, Menz S, Neuhaus R, Christian S, Haegebarth A, Algire C, Pollak M. 2016. Are metformin doses used in murine cancer models clinically relevant? Cell Metab 23: 569–570. 10.1016/j.cmet.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Collodet C, Foretz M, Deak M, Bultot L, Metairon S, Viollet B, Lefebvre G, Raymond F, Parisi A, Civiletto G, et al. 2019. AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR. FASEB J 33: 12374–12391. 10.1096/fj.201900841R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJO, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. 2007. Metformin inhibits mammalian target of rapamycin–dependent translation initiation in breast cancer cells. Cancer Res 67: 10804–10812. 10.1158/0008-5472.CAN-07-2310 [DOI] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183. 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner LJ, Brun SN, Herzig S, Young NP, Curtis SD, Shackelford DB, Shokhirev MN, Leblanc M, Vera LI, Hutchins A, et al. 2019. Genetic analysis reveals AMPK is required to support tumor growth in murine kras-dependent lung cancer models. Cell Metab 29: 285–302.e7. 10.1016/j.cmet.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielhaber JA, Han Y-S, Tan J, Xing S, Biggs CM, Joung K-B, Kristof AS. 2009. Inactivation of mammalian target of rapamycin increases STAT1 nuclear content and transcriptional activity in α4- and protein phosphatase 2A-dependent fashion. J Biol Chem 284: 24341–24353. 10.1074/jbc.M109.033530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Shaw RJ. 2017. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 66: 789–800. 10.1016/j.molcel.2017.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Hellberg K, Chaix A, Wallace M, Herzig S, Badur MG, Lin T, Shokhirev MN, Pinto AFM, Ross DS, et al. 2019. Genetic liver-specific AMPK activation protects against diet-induced obesity and NAFLD. Cell Rep 26: 192–208.e6. 10.1016/j.celrep.2018.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Blenis J. 2015. A nexus for cellular homeostasis: the interplay between metabolic and signal transduction pathways. Curr Opin Biotechnol 34: 110–117. 10.1016/j.copbio.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A, Hall MN, Lin S-C, Hardie DG. 2020. AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell Metab 31: 472–492. 10.1016/j.cmet.2020.01.015 [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226. 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenour CM, Ridley DE, Hughey CC, James FD, Donahue EP, Shearer J, Viollet B, Foretz M, Wasserman DH. 2014. 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) effect on glucose production, but not energy metabolism, is independent of hepatic AMPK in vivo. J Biol Chem 289: 5950–5959. 10.1074/jbc.M113.528232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wondisford Fredric E. 2015. Metformin action: concentrations matter. Cell Metab 21: 159–162. 10.1016/j.cmet.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Hernandez O, Way S, McKenna J III, Gambello MJ. 2007. Generation of a conditional disruption of the Tsc2 gene. Genesis 45: 101–106. 10.1002/dvg.20271 [DOI] [PubMed] [Google Scholar]
- Hopkins BD, Goncalves MD, Cantley LC. 2020. Insulin–PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat Rev Endocrinol 16: 276–283. 10.1038/s41574-020-0329-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, Hellberg K, Turner M, Talbott G, Kolar MJ, Ross DS, Hoxhaj G, Saghatelian A, Shaw RJ, Manning BD. 2017. Metformin inhibits hepatic mTORC1 signaling via dose-dependent mechanisms involving AMPK and the TSC complex. Cell Metab 25: 463–471. 10.1016/j.cmet.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61. 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning Brendan D. 2008. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 412: 179–190. 10.1042/BJ20080281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Chambon P, Metzger D. 2000. . Inducible site-specific somatic mutagenesis in mouse hepatocytes. Genesis 26: 147–148. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan K-L. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590. 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- Janes MR, Vu C, Mallya S, Shieh MP, Limon JJ, Li LS, Jessen KA, Martin MB, Ren P, Lilly MB, et al. 2013. Efficacy of the investigational mTOR kinase inhibitor MLN0128/INK128 in models of B-cell acute lymphoblastic leukemia. Leukemia 27: 586–594. 10.1038/leu.2012.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Wu F, Li D, Yang L, Li Q, Li R. 2018. Metformin improves obesity-associated inflammation by altering macrophages polarization. Mol and Cell Endocrinol 461: 256–264. 10.1016/j.mce.2017.09.025 [DOI] [PubMed] [Google Scholar]
- Johnson AB, Webster JM, Sum CF, Heseltine L, Argyraki M, Cooper BG, Taylor R. 1993. The impact of metformin therapy on hepatic glucose production and skeletal muscle glycogen synthase activity in overweight type II diabetic patients. Metab Clin Exp 42: 1217–1222. 10.1016/0026-0495(93)90284-U [DOI] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brûlé S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, et al. 2010. Metformin, independent of AMPK, inhibits mTORC1 in a Rag GTPase-dependent manner. Cell Metab 11: 390–401. 10.1016/j.cmet.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karonitsch T, Kandasamy RK, Kartnig F, Herdy B, Dalwigk K, Niederreiter B, Holinka J, Sevelda F, Windhager R, Bilban M, et al. 2018. mTOR senses environmental cues to shape the fibroblast-like synoviocyte response to inflammation. Cell Rep 23: 2157–2167. 10.1016/j.celrep.2018.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamzina L, Veilleux A, Bergeron S, Marette A. 2005. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology 146: 1473–1481. 10.1210/en.2004-0921 [DOI] [PubMed] [Google Scholar]
- Kim J-H, Yoon M-S, Chen J. 2009. Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem 284: 35425–35432. 10.1074/jbc.M109.051516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini David M. 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lee J, He C, Zou M-H, Xie Z. 2014. Suppression of the mTORC1/STAT3/Notch1 pathway by activated AMPK prevents hepatic insulin resistance induced by excess amino acids. Am J Physiol Endocrinol Metab 306: E197–E209. 10.1152/ajpendo.00202.2013 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu GY, Sabatini DM. 2020. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21: 183–203. 10.1038/s41580-019-0199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RW, Guan H-P, Ehrhart J, Petrov A, Prahalada S, Tozzo E, Yang X, Kurtz MM, Trujillo M, Gonzalez Trotter D, et al. 2017. Systemic pan-AMPK activator MK-8722 improves glucose homeostasis but induces cardiac hypertrophy. Science 357: 507–511. 10.1126/science.aah5582 [DOI] [PubMed] [Google Scholar]
- Ning J, Clemmons DR. 2010. AMP-activated protein kinase inhibits IGF-I signaling and protein synthesis in vascular smooth muscle cells via stimulation of insulin receptor substrate 1 S794 and tuberous sclerosis 2 S1345 phosphorylation. Mol Endocrinol 24: 1218–1229. 10.1210/me.2009-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe L, van den Elzen A, Watson M, Thoreen C. 2020. Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5′ TOP motifs. Proc Natl Acad Sci 117: 5319–5328. 10.1073/pnas.1912864117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Hardie DG, Pearson ER. 2017. The mechanisms of action of metformin. Diabetologia 60: 1577–1585. 10.1007/s00125-017-4342-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho Y. 2015. Metformin and inflammation: its potential beyond glucose-lowering effect. Endocr Metab Immune 15: 196–205. 10.2174/1871530315666150316124019 [DOI] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. 2010. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468: 1100–1104. 10.1038/nature09584 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA 101: 3329–3335. 10.1073/pnas.0308061100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial G, Detaille D, Guigas B. 2019. Role of mitochondria in the mechanism(s) of action of metformin. Front Endocrinol (Lausanne) 10: 294 10.3389/fendo.2019.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Hörl WH, Hengstschläger M, et al. 2008. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29: 565–577 10.1016/j.immuni.2008.08.012 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Jaenisch R. 2014. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9: 1956–1968. 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
- Yokogami K, Wakisaka S, Avruch J, Reeves SA. 2000. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol 10: 47–50. 10.1016/S0960-9822(99)00268-7 [DOI] [PubMed] [Google Scholar]
- Young NP, Kamireddy A, Van Nostrand JL, Eichner LJ, Shokhirev MN, Dayn Y, Shaw RJ. 2016. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev 30: 535–552. 10.1101/gad.274142.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.