In this study, King et al. identified additional effectors that enable cell growth dependent on the uptake of extracellular proteins, and used pancreatic ductal adenocarcinoma (PDA) cells selected for growth in medium where extracellular albumin was the obligate source of the essential amino acid leucine. Their results indicate that the Hippo pathway effectors Yap and Taz are important transcriptional regulators of endocytic nutrient uptake.
Keywords: Yap, cancer metabolism, macropinocytosis
Abstract
The uptake of macromolecules and cellular debris through macropinocytosis has emerged as an important nutrient acquisition strategy of cancer cells. Genetic alterations commonly found in human cancers (e.g. mutations in KRAS or loss of PTEN) have been shown to increase macropinocytosis. To identify additional effectors that enable cell growth dependent on the uptake of extracellular proteins, pancreatic ductal adenocarcinoma (PDA) cells were selected for growth in medium where extracellular albumin was the obligate source of the essential amino acid leucine. Analysis of global changes in chromatin availability and gene expression revealed that PDA cells selected under these conditions exhibited elevated activity of the transcriptional activators Yap/Taz. Knockout of Yap/Taz prevented growth of PDA cells in leucine-deficient medium, but not in complete medium. Furthermore, constitutively active forms of Yap or Taz were sufficient to stimulate macropinocytosis of extracellular protein. In addition to promoting the uptake of plasma proteins, Yap/Taz also promoted the scavenging of apoptotic cell bodies and necrotic debris by PDA cells. The Yap/Taz transcriptional target Axl was found to be essential for cell growth dependent on the uptake of dead cells and cell debris. Together, these studies suggest that the Hippo pathway effectors Yap and Taz are important transcriptional regulators of endocytic nutrient uptake.
Anabolic growth is the process by which cells convert sources of reduced carbon and nitrogen to higher order structures, such as lipids, proteins and nucleotides, to build the biomass that supports proliferation (DeBerardinis and Chandel 2016). In metazoans, the major sources of reduced carbon and nitrogen are glucose and amino acids, respectively, which are taken up through specialized cell surface transporters (Chantranupong et al. 2015). Yet, we and others have previously shown that mammalian cells can survive and, in many cases, grow in conditions where amino acids are scarce as long as the cells are provided with a sufficient supply of extracellular protein (Commisso et al. 2013; Palm et al. 2015; Wyant et al. 2017; Kim et al. 2018). Under these conditions, cells take up extracellular proteins and deliver them to the lysosome, where they can be hydrolyzed to the free amino acids needed to fuel cell growth.
Macropinocytosis, a bulk fluid phase type of endocytosis, is a major means by which cells take up macromolecules, such as proteins, cell debris, or viruses, from their environment (Kerr and Teasdale 2009). More recently, it has been demonstrated that macropinocytosis can function as a means of nutrient acquisition in mammalian cells, particularly in cancer (Finicle et al. 2018). Specifically, transformed cells expressing Ras oncogenes can grow in amino acid-depleted medium when albumin is supplemented at a concentration comparable to what is observed in human plasma (∼3%–5%) (Commisso et al. 2013; Palm et al. 2015). Oncogenic Ras has been shown to directly promote the scavenging of macromolecules, including extracellular proteins, through the activation of macropinocytosis (Bar-Sagi and Feramisco 1986; Commisso et al. 2013; Davidson et al. 2017). However, Ras is not essential for this process: Cells lacking all three major Ras genes (Kras, Hras, and Nras) can still engage in macropinocytosis (Palm et al. 2017). Thus, Ras appears to act by increasing the rate of macropinocytosis. In contrast, other signaling molecules including the PI3 and Pak family of kinases contribute to regulation of the essential machinery that executes the engulfment and internalization of extracellular macromolecules through macropinocytosis (Araki et al. 1996; Dharmawardhane et al. 2000).
The high prevalence of Kras mutations present in pancreatic ductal adenocarcinoma (PDA; ∼90%) as well as the observation that pancreatic tumors are relatively depleted of free amino acids (Kamphorst et al. 2015) has made pancreatic cancer a tractable model for the study of macropinocytosis. Multiple studies have demonstrated that the level of macropinocytosis in Kras mutant PDA cells can vary with changes in nutrient availability (Nofal et al. 2017; Lee et al. 2019). Therefore, the macropinocytic activity of PDA cells is not only dependent on underlying genetic alterations but also their metabolic context. We were inspired by the idea that changes in nutrient availability could provide a selective pressure that influences the evolution of cancer cell populations as tumors grow larger or metastasize (García-Jiménez and Goding 2019).
Here we found that when PDA cells are challenged to grow in conditions where extracellular proteins are the obligate source of essential amino acids, persistent cells exhibited elevated activity of Yap/Taz. Yap/Taz transcriptional activity was essential for growth of PDA cells in this context and was required for these cells to engage in macropinocytosis. Finally, we identified a target of Yap/Taz transcription, Axl, which can directly activate macropinocytosis in a PI3-kinase-dependent manner and is specifically required for the uptake of cell debris by PDA cells.
Results
Pancreatic cancer cells selected for extracellular protein-dependent growth exhibit increased macropinocytosis and a unique gene expression program
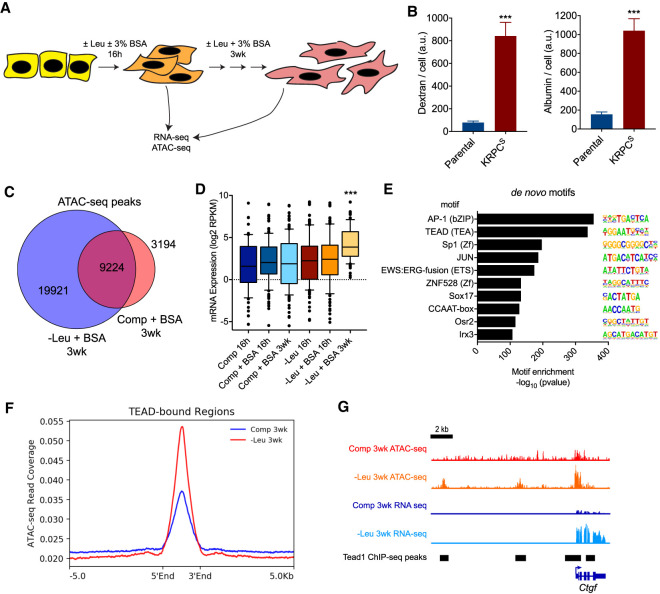
To identify epigenetic and transcriptional features that are preferentially enriched upon long-term, albumin-dependent growth, murine PDA cells were cultured for 21 d in leucine-free or complete medium supplemented with 3% bovine serum albumin (BSA) and subjected to ATAC-seq and RNA-seq analysis (Fig. 1A). The cells selected in long-term culture in leucine-free medium supplemented with BSA (KRPCS) exhibited an increased ability to scavenge proteins through macropinocytosis, as measured by the uptake of fluorescently labeled dextran or albumin, in comparison with parental cells (KRPC) (Fig. 1B).
Figure 1.
Pancreatic cancer cells selected for extracellular protein-dependent growth exhibit increased macropinocytosis and a unique gene expression program. (A) Schematic representation of experimental strategy to measure global changes in gene expression and chromatin accessibility after short-term (16 h) or long-term (3 wk) culture of PDA cells in complete (+Leu) or leucine-free (−Leu) medium with or without supplementation of 3% bovine serum albumin (BSA). (B) Uptake of fluorescent dextran (left panel) and albumin (right panel) by parental KRPC and KRPCS cells in complete medium as measured by fluorescence confocal microscopy. Bars represent mean ±SEM fluorescence intensity per cell, across 12 fields of view. (***) P < 0.0001, unpaired two-tailed t-test. (C) Venn diagram depicting total number of unique and shared called peaks from ATAC-seq of KRPC cells cultured for 3 wk in ±Leu + 3% BSA. (D) Box plot of mean log2 RPKM expression values of genes within 5 kb of unique ATAC-seq peaks identified in KRPCS cells. (***) P < 0.0001, one-way ANOVA. (E) Top de novo transcription factor motifs enriched within ATAC-seq peaks identified in KPRCS cells. (F) Average ATAC-seq read coverage across genomic regions bound by Tead1 ±5 kb in KRPC cells cultured for 3 wk in ±Leu + 3% BSA. (G) Tracks showing ATAC-seq and RNA-seq read coverage as well as called Tead1 ChIP-seq peaks at the Ctgf locus.
Chromatin analysis of the selected cells revealed that KRPCS cells had an overall increase in chromatin accessibility, as measured by ATAC-seq analysis, exhibiting a 2.3-fold increase in the total number of called peaks genome-wide compared with cells grown in complete medium (Fig. 1C). The majority of these accessible regions were at intergenic and intronic loci (Supplemental Fig. S1A). Importantly, gains in chromatin accessibility correlated with changes in gene expression: Genes associated with enriched accessible regions were significantly up-regulated in the KRPCS cells (Fig. 1D). Principal component analysis of the RNA-seq data delineated the selected KRPCS cells as a distinct cluster, while the cells cultured acutely in leucine-deficient medium or complete medium clustered independently (Supplemental Fig. S1B).
To identify potential transcription factors associated with this unique gene expression program in KRPCS cells, de novo motif enrichment analysis was performed across genomic regions that were found to be significantly enriched in accessibility. DNA motifs that closely resemble those bound by AP-1 or TEAD transcription factors were the most highly enriched in these regions (Fig. 1E). Using publicly available ChIP-seq data (Tremblay et al. 2014), we confirmed that the KRPCS cells had greater chromatin accessibility at genomic loci bound by Tead1 (Fig. 1F) and this was associated with increased expression of known Tead1 targets such as Ctgf (Fig. 1G). Cooperation between AP-1 and TEAD at active gene enhancers has been previously demonstrated to promote tumor growth across multiple cancer types (Stein et al. 2015; Zanconato et al. 2015; Koo et al. 2020), but a role of these transcription factors in nutrient scavenging has not been reported previously.
Enrichment of Yap/Taz activity in cells selected for ability to use extracellular proteins for growth
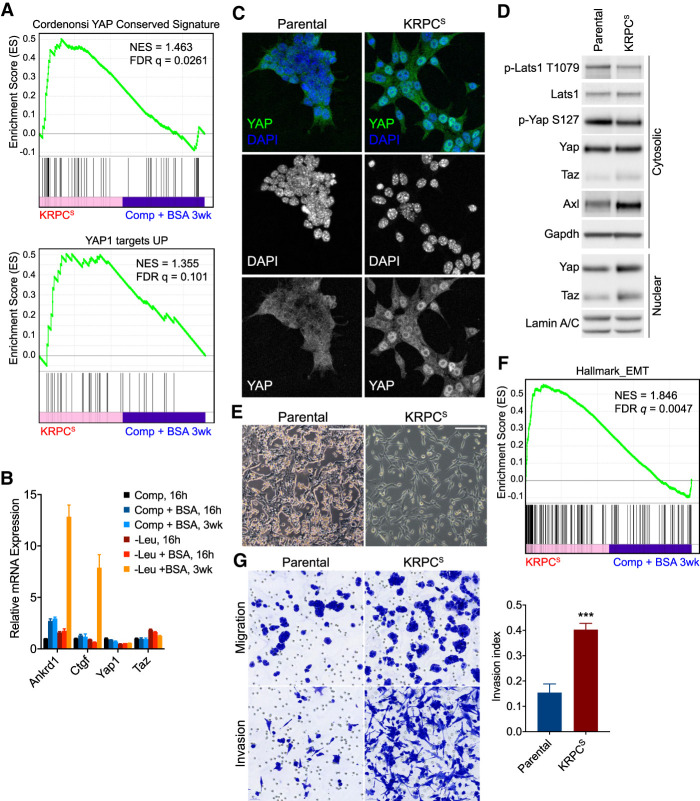
Two closely related proteins, Yap and Taz, are known to form complexes with TEAD transcription factors and activate target gene transcription as components of the Hippo pathway, which has been implicated in organ development and tissue regeneration (Zhao et al. 2008). Using gene set enrichment analysis, we observed that genes activated by Yap were significantly enriched in our RNA-seq profiling of KRPCS cells (Fig. 2A). Elevated expression of canonical Yap targets, including Ctgf and Ankrd1, was confirmed independently by qRT-PCR (Fig. 2B). However, expression of Yap target genes was not increased after 16 h of leucine deprivation, suggesting that Yap/Taz activity is not acutely responsive to essential amino acid deprivation. In the selected cells, expression of Yap or Taz themselves were not reproducibly increased at the mRNA level. Nuclear-cytoplasmic shuttling of Yap/Taz is known to be repressed by Hippo signaling in response to cell–cell contact and other signaling pathways including G protein coupled receptors (GPCRs) (Meng et al. 2016). Immunofluorescent imaging revealed that the selected cells had more prominent nuclear Yap than the parental line, where the majority of cells exhibited a diffuse, cytoplasmic staining pattern (Fig. 2C), suggesting that the increased Yap/Taz transcriptional activity observed in KRPCS cells results from the nuclear translocation of these factors, rather than their increased transcription or translation. Given that the subcellular localization of Yap/Taz is primarily regulated through their phosphorylation by Lats kinases (Lats1/2), Lats activity was compared in KRPCS cells relative to parental KRPC cells. KRPCS cells exhibited a decrease in phosphorylation of both Lats1 and Yap (Fig. 2D). Together with the data demonstrating increased Yap nuclear localization observed in cells selected in leucine-deficient, albumin substituted media (Fig. 2C), these findings suggest that one or more of the kinases that activate Lats1 is less active in cells selected for the use of extracellular proteins as a source of essential amino acids.
Figure 2.
Increased Yap/Taz transcriptional activity and EMT phenotype in cells selected for extracellular protein-dependent growth. (A) Gene set enrichment analysis (GSEA) plots showing enrichment in expression of Yap target genes, as measured by RNA sequencing of KRPCS cells, relative to expression in KRPC cells cultured for 3 wk in complete medium + 3% BSA. (B) Relative mRNA expression of Yap, Taz, and target genes as measured by qRT-PCR in KRPC cells cultured in ±Leu ±3% BSA for 16 h or 3 wk. Bars represent mean ± SD from three biological replicates. (C) Immunofluorescence micrographs showing localization of Yap (antibody also cross-reacts with Taz) in parental KRPC and KRPCS cells. (D) Immunoblot of cytosolic and nuclear extracts from parental KRPC and KRPCS cells. (E) Phase contrast micrographs of parental KRPC and KRPCS cells. Scale bars = 200 µm. (F) GSEA plot showing enrichment in expression of EMT genes in KRPCS cells, relative to expression in KRPC cells cultured for 3 wk in complete medium + 3% BSA. (G) Light micrographs showing migration of parental or KRPCS cells across control (top panels) or matrigel-coated (bottom panels) membranes. Bar plot represents mean ± SEM of invasion index (ratio of cells that migrated across matrigel membrane to control membrane) across three biological replicates (***) P = 0.0005, unpaired two-tailed t-test.
Elevated Yap/Taz activity has been associated with epithelial–mesenchymal transition (EMT) and invasiveness across multiple cancers (Overholtzer et al. 2006; Cordenonsi et al. 2011). Consistent with these prior results, we found that cells selected in leucine-deficient, albumin-substituted medium were larger, more spindle shaped, and formed fewer cell-cell contacts relative to parental KRPC cells (Fig. 2E). Unbiased analyses of gene expression signatures enriched in KRPCS cells also identified multiple gene sets associated with tumor cell invasion and epithelial–mesenchymal transition (EMT) (Fig. 2F). Moreover, in in vitro invasion assays, we observed that KRPCS cells were significantly more invasive than parental KRPC cells (Fig. 2G). These results suggested that the increased invasiveness observed in KRPCS cells could be a functional consequence of elevated Yap/Taz activity. To test whether constitutively nuclear Yap and/or Taz would be sufficient to induce an invasive phenotype in the parental cells, we used mutants of Yap or Taz that could not be phosphorylated by Hippo pathway kinases Lats1/2 (Zhao et al. 2007). Expression of either Yap5SA or TazS89A was sufficient to activate an invasive phenotype in the parental cells (Supplemental Fig. S2A,B).
Yap/Taz are essential for macropinocytosis and growth that requires scavenging of extracellular proteins
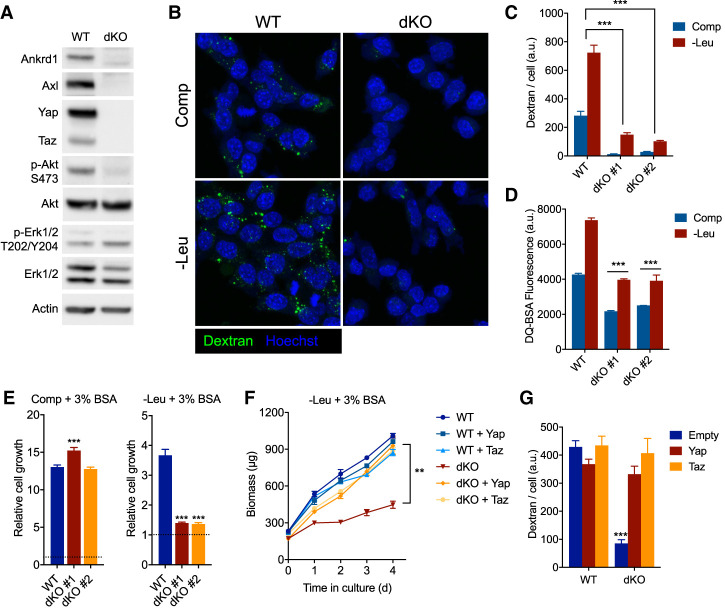
While invasiveness has been associated with the nuclear translocation of Yap and Taz, a role for these transcriptional activators in macropinocytosis has not been described. Therefore, we next investigated whether disrupting the function of these genes would impair this process. Knockout of either Yap or Taz alone did not significantly decrease macropinocytosis or albumin-dependent growth in unselected PDA cells (Supplemental Fig. S3A,B). However, we observed a compensatory increase in Taz when Yap was depleted from these cells, and vice versa (Supplemental Fig. S3C). Thus, we generated Yap/Taz-deficient (dKO) PDA clones by transfection with vectors expressing sgRNAs targeting both Yap and Taz together with Cas9. Yap/Taz dKO cells had undetectable levels of Axl and Ankrd1 protein (Fig. 3A). Dextran and albumin uptake were severely impaired in two independently derived Yap/Taz dKO clones (Fig. 3B–D). Furthermore, while Yap/Taz dKO PDA cells could grow at least as well as Yap/Taz wild-type (WT) cells in complete medium, Yap/Taz dKO PDA cells grew significantly slower in leucine-free medium supplemented with albumin (Fig. 3E). Importantly, complementation with either Yap or Taz cDNA in the dKO cells restored albumin-dependent growth (Fig. 3F) and macropinocytosis (Fig. 3G) in leucine-free medium, confirming that these phenotypes are due to a deficiency of Yap/Taz and not an artifact of clonal selection.
Figure 3.
Yap/Taz are essential for macropinocytosis and cell growth that is dependent on the catabolism of extracellular proteins. (A) Immunoblot of whole-cell lysates from Yap/Taz-deficient (dKO) or wild-type (WT) KRPC cells. (B) Confocal micrographs showing uptake of fluorescent dextran (green) and nuclei (Hoechst, blue) in Yap/Taz dKO or WT KRPC cells after 16 h of culture in complete (Comp) or leucine-free (−Leu) medium + 3% BSA. (C) Bar plot represents mean ± SEM dextran fluorescence intensity per cell, across 12 fields of view. (D) DQ-BSA fluorescence as measured by flow cytometry in Yap/Taz dKO and WT KRPC cells cultured for 24 h in complete or leucine-free medium + 3% BSA. DQ-BSA was added 4 h prior to analysis. Bars represent mean fluorescence intensity ± SD across three technical replicates. (E) Growth of Yap/Taz dKO or WT KRPC clones in complete (48 h) or leucine-free medium (96 h) + 3% BSA. (F) Growth of Yap/Taz dKO or WT KRPC cells transduced with retroviruses expressing a doxycycline-inducible Yap or Taz cDNA or empty vector in leucine-free medium + 3% BSA with 100 ng/mL doxycycline. (G) Dextran uptake in Yap/Taz dKO or WT KRPC transduced with retroviruses as in F, after culture for 16 h in leucine-free medium with 3% BSA and 100 ng/mL doxycycline. Bars represent mean fluorescence intensity per cell ±SEM across 12 fields of view. (***) P < 0.0001, (**) P < 0.005, one-way ANOVA, for C–G.
Activation of nuclear Yap/Taz promotes macropinocytosis in PDA cells
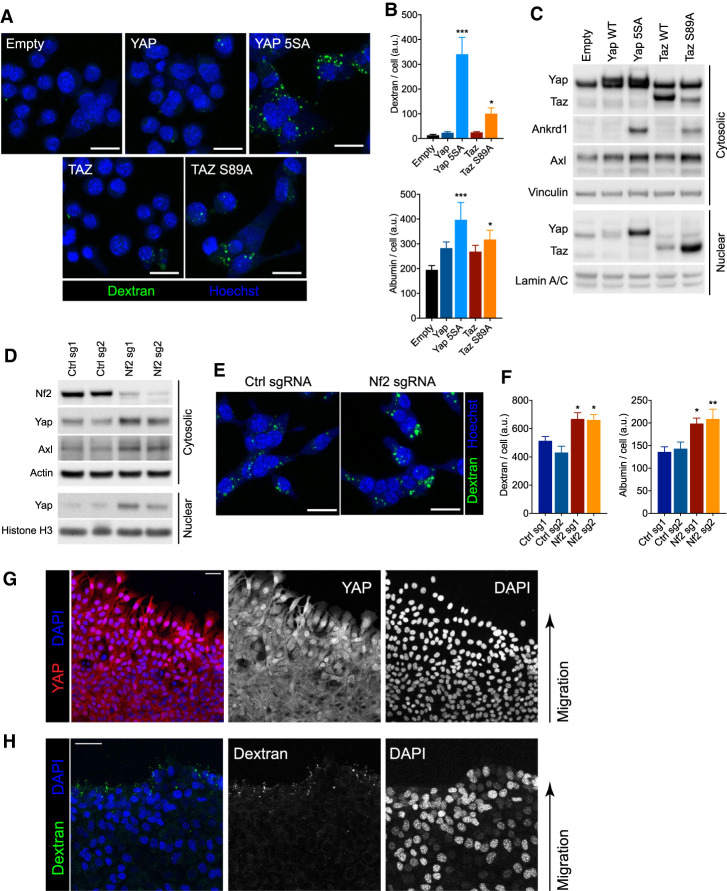
We next tested whether enforced expression of nuclear Yap/Taz in the absence of nutrient stress would result in increased macropinocytosis. Transient overexpression of Yap5SA or TazS89A mutants, but not wild-type Yap or Taz, led to a significant increase in macropinocytosis (Fig. 4A,B). Increased expression of transcriptional targets Axl and Ankrd1 was also observed (Fig. 4C). To determine whether activation of Yap or Taz could also stimulate macropinocytosis in cells without a Ras mutation, we introduced Yap5SA or TazS89A into KRAS wild-type BxPC-3 human PDA cells. Even in the absence of mutant Ras, constitutively nuclear Yap or Taz activated expression of Axl and Ankrd1 (Supplemental Fig. S4A) and significantly increased macropinocytosis and albumin uptake in these cells (Supplemental Fig. S4B), demonstrating that Yap/Taz can be sufficient to activate macropinocytosis.
Figure 4.
Activation of nuclear Yap/Taz increases macropinocytosis. (A,B) Representative confocal micrographs showing uptake of fluorescent dextran (green) and nuclei (Hoechst; blue) in KRPC cells upon ectopic expression of Yap and Taz in complete medium. Scale bars, 30 µm. (B) Bar plots represent mean ± SEM dextran (top) or albumin (bottom) fluorescence intensity per cell, across 12 fields of view. (C) Immunoblot of cytosolic and nuclear lysates from KRPC cells transduced with retroviruses expressing doxycycline-inducible Yap (wild-type or 5SA) or Taz (wild-type or S89A) cDNA, or empty vector, and cultured with 100 ng/mL doxycycline for 24 h. (D) Immunoblot of cytosolic and nuclear lysates from KRPC cells transduced with lentirviruses expressing Cas9 and an sgRNA targeting either the Nf2 gene or the Rosa26 locus (Ctrl). (E) Representative confocal micrographs showing uptake of fluorescent dextran (green) and nuclei (Hoechst, blue) in KRPC cells expressing Cas9 and sgRNAs as in D. Scale bars, 30 µm. (F) Bar plots represent mean ± SEM dextran (left) or albumin (right) fluorescence intensity per cell, across 12 fields of view. (G,H) Representative confocal micrographs showing Yap immunofluorescence (G; red), dextran uptake (H; green), and nuclei (DAPI; blue) in KPC (KrasLSL-G12D/+, Trp53LSL-R272H/+, Pdx1-Cre) cells at migrating edge of wound boundary 4 h after administration of scratch to a confluent monolayer. Scale bars, 30 μm. (***) P < 0.0001, (**) P < 0.005, (*) P < 0.05, one-way ANOVA, for B and F.
To independently confirm that endogenous Yap and Taz could increase macropinocytosis if redirected to the nucleus, we next tested whether loss of a negative regulator of Yap/Taz, specifically the tumor suppressor Nf2 (Zhang et al. 2010), would phenocopy the effect of overexpressing constitutively nuclear Yap/Taz. CRISPR/Cas9-mediated deletions in Nf2 led to an accumulation of nuclear Yap and increased expression of the Yap target gene Axl (Fig. 4D). Loss of Nf2 also led to a significant increase in macropinocytosis and albumin uptake (Fig. 4E,F).
As Nf2 and Yap/Taz have been shown to be positively and negatively regulated by cell contact inhibition (Shaw et al. 2001; Zhao et al. 2007), respectively, we also tested whether relief of contact inhibition could similarly activate macropinocytosis in PDA cells. To this end, wild-type PDA cells were grown to confluency and subjected to a scratch assay, whereby fluorescent dextrans could be added during wound closure. Cells at the invading edge of the wound were enriched in nuclear Yap (Fig. 4G) and this increase correlated with elevated levels of macropinocytosis near the scratch boundary (Fig. 4H).
Yap/Taz must bind TEAD proteins to activate macropinocytosis
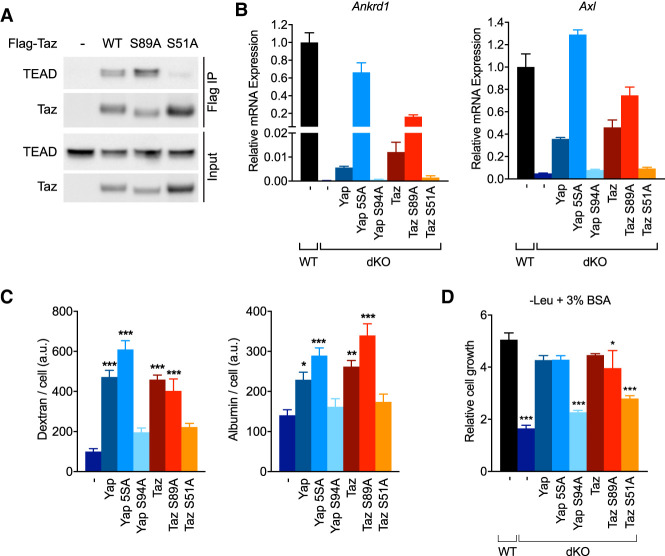
Since nontranscriptional functions for Yap/Taz have been described (Xu et al. 2018), we next tested whether their association with the DNA-binding TEAD factors was required to promote macropinocytosis. An S94A mutation in Yap that impairs its binding to TEAD proteins has been identified (Zhao et al. 2008), and we also generated a substitution in Taz at an analogous serine residue, S51A. We confirmed that TazS51A did not interact with TEAD proteins by coimmunoprecipitation, while we observed a robust association with wild-type Taz or TazS89A (Fig. 5A). Moreover, unlike their wild-type or constitutively nuclear counterparts, YapS94A and TazS51A failed to rescue expression of target genes Ankrd1 or Axl upon expression in Yap/Taz dKO PDA cells (Fig. 5B). Consequently, neither YapS94A nor TazS51A could restore macropinocytosis or albumin scavenging activity (Fig. 5C), and these cells failed to grow in leucine-free medium supplemented with albumin (Fig. 5D). Thus, the ability of Yap and Taz to transactivate TEAD target genes is required for the activation of macropinocytosis and albumin-dependent growth.
Figure 5.
Transcriptional activity of Yap/Taz is essential for macropinocytosis and cell growth requiring the catabolism of extracellular proteins. (A) Western blot of FLAG immunoprecipitates and input lysates from Yap/Taz dKO KRPC cells transduced with retroviruses containing a doxycycline-inducible, FLAG-tagged, Taz cDNA (either wild type, S89A, or S51A mutant) or empty vector. Cells were cultured for 24 h in 100 ng/mL doxycycline prior to harvest. (B) Relative expression of Ankrd1 and Axl mRNA in WT KRPC or Yap/Taz dKO cells transduced with retroviruses containing a doxycycline-inducible Yap or Taz cDNA (wild type or with the indicated mutation) after culture for 24 h in 100 ng/mL doxycycline. Bars represent mean ± SD of three biological replicates. (C) Uptake of fluorescent dextran (left panel) and albumin (right panel) in Yap/Taz dKO cells transduced with retroviruses containing a doxycycline-inducible Yap or Taz cDNA after culture for 24 h in 100 ng/mL doxycycline. Bars represents mean ± SEM fluorescence intensity per cell, across 12 fields of view. (D) Growth of WT KRPC or Yap/Taz dKO cells transduced with retroviruses containing a doxycycline-inducible Yap or Taz cDNA after 96 h in leucine-free medium + 3% BSA and 100 ng/mL doxycycline. Bars represent mean ± s.d. of three technical replicates. (***) P < 0.0001, (**) P < 0.005, (*) P < 0.05 one-way ANOVA, for C and D.
Uptake of necrotic cells by macropinocytosis is dependent on the Yap/Taz transcriptional target Axl
The above results demonstrate that the transcriptional activity of Yap/Taz contributes to the regulation of macropinocytosis. We next explored whether one or more Yap/Taz target genes could be key effectors of this process. In eukaryotic cells, macropinocytosis can be stimulated by growth factor or oncogene-induced activation of PI3-kinase (PI3K) (Araki et al. 1996; Amyere et al. 2000; Palm et al. 2017). Surprisingly, we found that when transcriptionally active forms of Yap/Taz were introduced into dKO PDA cells, Akt became phosphorylated (Fig. 6A), which suggested that PI3-kinase (PI3K) signaling could be activated by a factor downstream from Yap/Taz. The Yap/Taz target gene Axl is a receptor tyrosine kinase (RTK) known to activate PI3K and Akt (Goruppi et al. 1997). Axl, a member of the TAM (Tyro-Axl-Mer) family of RTKs, can be activated by its ligand Gas6 (Stitt et al. 1995); recombinant Gas6 increased phosphorylation of Akt in PDA cells, but not in Yap/Taz dKO cells (Supplemental Fig. S5A). Furthermore, expression of exogenous Axl in dKO cells increased Akt phosphorylation, either upon stimulation with Gas6 following serum starvation or in leucine-free medium supplemented with albumin (Supplemental Fig. S5B,C).
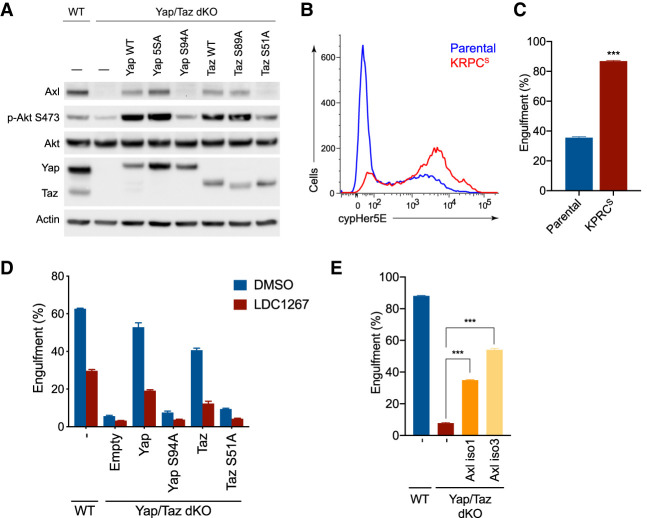
Figure 6.
Uptake of necrotic cells by macropinocytosis is dependent on the Yap/Taz transcriptional target Axl. (A) Immunoblot of whole cell lysates from WT KRPC or Yap/Taz dKO cells transduced with retroviruses containing a dox-inducible Yap or Taz cDNA after culture for 24 h in 100 ng/mL doxycycline. (B) Histogram showing engulfment of cypHer5E-stained necrotic FL5.12 cells by KRPCS cells or parental KRPC after 2 h in complete medium, as measured by flow cytometry. (C) Bar plot represents mean ± SD frequency of cypHer5E-positive cells from three technical replicates. (***) P < 0.0001, unpaired two-tailed Student's t-test. (D) Engulfment of cypHer5E-stained necrotic FL5.12 cells after 6 h by WT or Yap/Taz dKO KRPC cells. Cells were transduced with retroviruses containing a doxycycline-inducible Yap or Taz cDNA and necrotic cell engulfment was measured after 6 h in the presence of 100 nM LDC1267 or vehicle. Assay was performed following 18 h culture in 100 ng/mL doxycycline. Bars represent mean ± SD cypHer5E-positive cells, as measured by flow cytometry, from three technical replicates. (E) Engulfment of cypHer5E-stained necrotic FL5.12 cells after 6 h by WT KRPC or Yap/Taz dKO cells transduced with retroviruses containing a doxycycline-inducible mouse Axl cDNA (either isoform 1 or 3). Assay was performed following an 18-h culture in 100 ng/mL doxycycline. Bars represent mean ± SD cypHer5E-positive cells, as measured by flow cytometry, from three technical replicates. (***) P < 0.0001, one-way ANOVA.
Gas6 has been reported to function as a bridging molecule between phosphatidylserine on the surface of dead cells or cellular debris and TAM receptor-expressing phagocytes (Lemke and Rothlin 2008). Recently, it was demonstrated that necrotic cell debris are taken up through macropinocytosis and, similar to albumin, can serve as a source of amino acids in nutrient-deprived cells (Kim et al. 2018). In an approach similar to that of Kim et al. (2018), we used IL-3-deprived FL5.12 cells as a source of necrotic cells, which we then labeled with a pH-sensitive fluorescent ester, cypHer5E, prior to feeding to PDA cells. CypHer5E fluorescence is increased as the engulfed material is endocytosed and this gain in fluorescence is readily quantified by confocal microscopy or flow cytometry (Morioka et al. 2018). Using this technique, we compared the engulfment of cypHer5E-stained dead cells by KRPCS or parental KRPC. In agreement with their increased level of macropinocytosis, KRPCS cells were more effective at engulfing dead cells than parental KRPC (Fig. 6B,C). Conversely, in Yap/Taz dKO PDA cells, where macropinocytosis was severely reduced, dead cell engulfment was significantly lower (Fig. 6D). Introduction of either Yap or Taz in dKO cells rescued dead cell engulfment, but this rescue of macropinocytosis was lost when cells were treated with the TAM receptor inhibitor LDC1267, suggesting that Axl activity is required downstream from Yap/Taz for the macropinocytic uptake of necrotic cell debris.
Finally, although the above data demonstrate that Yap/Taz induction of Axl correlates with the ability to engulf dead cells, we asked whether the expression of Axl would be sufficient to induce the engulfment of dead cells in the absence of Yap and Taz. Ectopic expression of Axl in the Yap/Taz dKO was sufficient to rescue dead cell engulfment (Fig. 6E). Treatment with either a PI3-kinase or TAM receptor inhibitor similarly blocked dead cell engulfment by PDA cells (Supplemental Fig. S5D,E), indicating that PI3K is a critical downstream effector of Axl in this context.
The ability of Axl to increase macropinocytosis is evolutionarily conserved in mammals
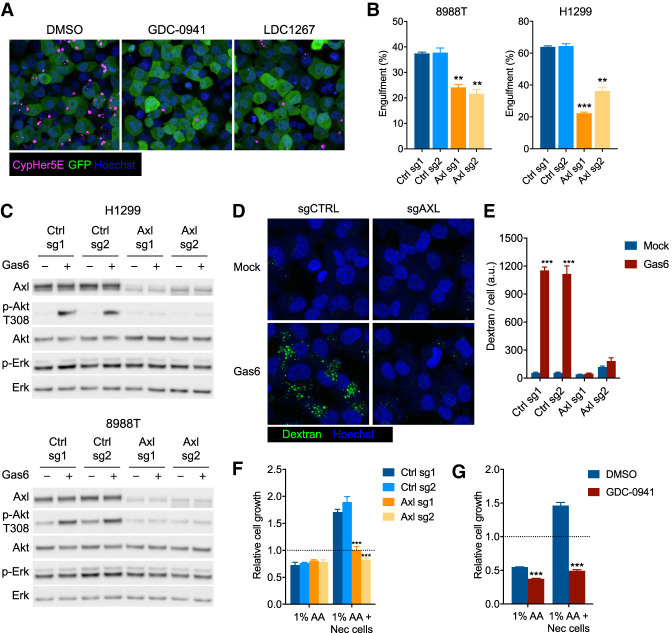
Since the preceding experiments were performed in mouse PDA cells, we next assessed whether these findings could be generalized to other cell types and/or species. To this end, we used two human KRAS mutant cell lines that highly expressed AXL: NCI-H1299 (lung adenocarcinoma) and PaTu 8988T (pancreatic ductal adenocarcinoma). Both cell lines actively engulfed necrotic FL5.12 cells labeled with cypHer5E (Fig. 7A,B). Either PI3-kinase or TAM receptor inhibition could block the engulfment of dead cells (Fig. 7A) and knockout of AXL alone was sufficient to significantly impair dead cell engulfment in either H1299 or 8988T cells (Fig. 7B). Gas6 stimulation increased phosphorylation of Akt in either cell line, but this activity was lost entirely upon depletion of AXL (Fig. 7C). Transient exposure to Gas6 was also sufficient to activate macropinocytosis in these cells, but not if AXL was deleted (Fig. 7D,E). Finally, the ability of these cells to grow in low amino acid (1% AA) medium was supported by supplementing the medium with necrotic FL5.12 cells. The necrotic cell-dependent growth was prevented following either deletion of AXL (Fig. 7F) or PI3K inhibition (Fig. 7G).
Figure 7.
Evolutionarily conserved role for Gas6/Axl/PI3K signaling in the activation of macropinocytosis. (A) Engulfment of cypHer5E-stained FL5.12 cells (magenta) by H1299 cells (green) in the presence of 1 µM GDC-0941, 100 nM LDC1267, or vehicle. Representative images taken by live confocal microscopy at 4 h. Nuclei were stained with Hoechst-33342 (blue). (B) Engulfment of cypHer5E-stained FL5.12 cells after 4 h by PaTu 8988t or H1299 cells transduced with lentiviruses expressing Cas9 and sgRNAs targeting AXL or control genomic loci. Bars represent mean ± SD cypHer5E-positive cells, as measured by flow cytometry, from three technical replicates. (**) P < 0.005, (***) P < 0.0001, one-way ANOVA. (C) Immunoblot of whole-cell lysates from H1299 or PaTu 8988t cells transduced with lentiviruses expressing Cas9 and sgRNAs targeting AXL or control genomic loci. Cells were serum-starved in 0.1% FBS for 8 h, followed by 15 min ±recombinant Gas6 (400 ng/mL) prior to harvest. (D) Uptake of fluorescently labeled dextran by H1299 cells transduced with lentiviruses expressing Cas9 and sgRNAs targeting either AXL or control genomic loci. Cells were serum-starved in 0.1% FBS for 8 h prior to addition of dextran ± recombinant human Gas6 (400 ng/mL) for 30 min. (E) Bar plot representing mean ± SEM fluorescence intensity per cell, across 12 fields of view. (***) P < 0.0001, one-way ANOVA. (F) Growth of PaTu 8988t cells transduced with lentiviruses expressing Cas9 and sgRNAs targeting either AXL or control genomic loci in medium containing 1% amino acids ±necrotic FL5.12 cells (25:1). (***) P < 0.0001, one-way ANOVA. (G) Growth of PaTu 8988t cells in medium containing 1% amino acids ± necrotic FL5.12 cells (25:1) with 1 µM GDC-0941 or vehicle. (***) P < 0.0001, two-tailed t-test.
Discussion
The above results suggest a previously uncharacterized role for Yap and Taz in supporting cell growth by enhancing macropinocytosis and the uptake of necrotic cells and cell debris through their transcriptional target, Axl. Since the discovery of the their ortholog yorkie (yki) in Drosophila, Yap and Taz have been implicated in the control of cell growth and tissue regeneration (Huang et al. 2005).
In mouse models of PDA, amplification of Yap has been observed in tumors that relapse upon inactivation of Kras (Kapoor et al. 2014; Shao et al. 2014). While the complete loss of Yap/Taz in PDA cells was well-tolerated in complete medium, we found that Yap/Taz were required for cell growth when protein or necrotic cells were the obligate source of amino acids. Pancreatic tumors have been reported to be vascularly compromised and often depend on alternative sources of nutrients to maintain viability and growth (Kamphorst et al. 2015). Furthermore, we observed that loss of a negative regulator of Yap/Taz, Nf2, also led to an increase in protein scavenging. These results are in agreement with studies that observed elevated macropinocytosis in Nf2-deficient cancer cells (Chiasson-Mackenzie et al. 2018). Nf2 is a known tumor suppressor gene that is frequently mutated or deleted in human cancer.
Our results implicate the TAM receptor Axl can function as a critical downstream mediator of Yap/Taz in the activation of macropinocytosis. Although the role of TAM receptors in the phagocytosis of apoptotic cells has been well studied in macrophages (Lemke and Rothlin 2008), our data demonstrate that tumor cells can also use these receptors to efficiently engulf dead cells and recycle their contents to fuel growth. Moreover, we observed that the ligand for Axl, Gas6, is a potent inducer of macropinocytosis and can directly activate PI3-kinase in PDA cells. Gas6 secretion by stromal cells was shown to be induced in response to oncogenic Kras activation in the pancreas (Tape et al. 2016). In this context, PI3-kinase activation by Kras was non-cell-autonomous and was dependent on stromal production of Gas6. Gas6 activity requires the vitamin K-dependent γ carboxylation of glutamate residues (Varnum et al. 1995), which means it can be also be inhibited by vitamin K antagonists such as warfarin. This could partially explain the intriguing trend that PDA patients on warfarin exhibit slightly improved outcomes (Kirane et al. 2015). Recently, blockade of Gas6 in mouse models of pancreatic cancer was also shown to be effective in reducing metastatic tumor burden (Ireland et al. 2020).
A major reason why PDA has such a dismal prognosis is the propensity of this cancer to metastasize (Hidalgo 2010). Our observation that PDA cells that persist after chronic leucine deprivation are more invasive and exhibit hallmarks of EMT implies that nutrient availability could influence clonal evolution as tumors grow larger or metastasize. Recent studies have demonstrated that cancer cells with greater metastatic potential are prone to use alternative metabolic fuels such as lactate or fatty acids (Pascual et al. 2017; Tasdogan et al. 2020). Thus, it has been theorized that the ability of cells to endure different tissue microenvironments with varying concentrations of nutrients would necessitate a higher degree of metabolic plasticity. Intriguingly, when cells were selected for their ability to scavenge an alternative metabolic fuel, namely extracellular protein, these selected cells were also more invasive and enriched in nuclear Yap/Taz. Although it remains unclear in this context which signals contribute to the decrease in Lats-mediated phosphorylation of Yap/Taz, increasing nuclear Yap or Taz alone was sufficient to enable PDA cells to scavenge extracellular proteins and promoted their invasion through basement membrane. Previous studies have shown Yap/Taz can activate EMT and promote metastasis (Overholtzer et al. 2006; Mueller et al. 2018). Yap/Taz are transcriptional activators and the overlap of Yap/Taz target genes required to induce EMT and those required to induce macropinocytosis remains to be determined. To date, other well-characterized activators of EMT, such as Snail, Slug, or Twist, have not been implicated in the induction of macropinocytosis. Nevertheless, the present data suggest that EMT and macropinocytosis can both be supported by Yap/Taz and could contribute to the ability of PDA cells to metastasize.
Materials and methods
Reagents
Antibodies to Akt (2920), Akt pSer473 (4060), Akt pThr308 (13038), S6K1 pThr389 (9234), S6K (2708), S6 pSer235/236 (4856), S6 (2317), Erk1/2 pThr202/Tyr204 (4377), Erk1/2 (9107), Lats1 pT1079 (8654), Lats1 (3477), Nf2 (6995), Lamin A/C (4777), Histone H3 (3638), pan-TEAD (13295), Taz (4883), and human Axl (8661) were purchased from Cell Signaling Technology. Antibodies to Yap (63.7) and Ankrd1 (G-2) were purchased from Santa Cruz Biotechnology. Antibodies to Vinculin (V9131), Actin (A5441), and Gapdh (G8795) were purchased from Sigma-Aldrich. Antibodies to mouse Axl (AF854) and recombinant human Gas6 (895-GSB-050) were from R&D Systems. Secondary antibodies were from GE Healthcare (HRP-linked anti-rabbit and HRP-linked anti-mouse) and Thermo Fisher Scientific (HRP-linked anti-goat).
Alexa fluor 647 BSA, Oregon Green dextran (70 kDA), DQ Green BSA, and Hoechst 33342 were from Life Technologies. DAPI was from Sigma-Alrich. Inhibitors were purchased from Cayman Chemical (GDC-0941), Tocris Bioscience (Torin1), and Selleck Chemicals (LDC1267). Amino acid starvation media were from Athena (DMEM/F12 0423) or prepared by the Memorial Sloan Kettering Cancer Center Media Core Facility. Dialyzed FBS was from Gemini Biosystems and BSA (A1470) was from Sigma.
Plasmids
LentiCRISPR v2 (Addgene plasmid 52961), PX458 (Addgene plasmid 48138), and pX459 (Addgene plasmid 62988) were a gift from Feng Zhang. pLenti-EF-FH-Taz-ires-blast (Addgene plasmid 52083) and pLenti-EF-FH-Taz S89A-ires-blast (Addgene plasmid 52084) were a gift from Yukata Hata. pQCXIH-Myc-Yap (Addgene plasmid 33091) and pQCXIH-Myc-Yap 5SA (Addgene plasmid 33093) were a gift from Kun-Liang Guan. Yap S94A and Taz S51A mutants were generated by PCR mutagenesis using the following primer pairs: YapS94AF (CGGCTTGAAGAAGGCGTCGGGCAGCTTCC) and YapS94AR (GGAAGCTGCCCGACGCCTTCTTCAAGCCG), and TazS51AF (GGCTCCTTAAAGAAAGCCTCCGGCAGGATCTTC) and Taz S51AR (GAAGATCCTGCCGGAGGCTTTCTTTAAGGAGCC). Yap and Taz cDNAs were PCR-amplified and subcloned into pTURN (pSIN-TREtight-UbiC-rtTA-ires-Hygro) vectors by Gibson Assembly (NEB). Mouse Axl isoforms 1 and 3 (NM_009465 and NM_001190975) were cloned from cDNA prepared from KRPCS cells using the following primer pair: mAxlF (TGTCTGGAGGATGGGCAG) and mAxlR (AGATTGTCTCAGGCTCCG).
CRISPR sgRNA sequences targeting genes of interest were generated with GuideScan (Perez et al. 2017). The following sgRNA sequences were cloned into LentiCRISPRv2 to generate knockout cell lines: Yap sg1 (ATCAGCGTCTGGGGCACCGC), Yap sg2 (CGGCTGTTGCGCGGGCTCCA), Taz sg1 (CGCCAGCTAGTCGAGGCCCC), Taz sg2 (GGCTAGTGGCCACGACTTGC), Nf2 sg1 (AGAGCACGGACGCCTCGGGA), Nf2 sg2 (TGTCGCTGTAGGAGATGTTT), AXL sg1 (CCGAAGCCAATGTACCTCGG), and AXL sg2 (CACCCCTTATCACATCCGCG). Control sgRNA sequences were as follows: Ctrl sg1 (mouse; GCTGGAGTTGCAGATCACGA), Ctrl sg2 (mouse; GAAGATGGGCGGGAGTCTTC), Ctrl sg1 (human; GGATACTTCTTCGAACGTTT), and Ctrl sg2 (human; GGGGCCACTAGGGACAGGAT). For generation of Yap/Taz dKO cells, Yap sg2 and Taz sg1 were each cloned into PX459 and transiently cotransfected into KRPC cells. Puromycin-resistant cells were then plated at clonal density and selected clones were screened for loss of Yap and Taz expression by Western blot.
Cell culture
KRPC (Lito et al. 2014) and KPC (KrasLSL-G12D Trp53LSL-R272H Pdx1-Cre) murine pancreatic ductal adenocarcima cell lines were kindly provided by Scott Lowe. FL5.12 cells were described previously (Vander Heiden et al. 1997). NCI-H1299 lung carcinoma, BxPC-3, and Pa-Tu-8988t pancreatic ductal adenocarcinoma cell lines were obtained from the ATCC and verified by the Memorial Sloan Kettering Cancer Center cell line distribution service. KRPC, KPC, NCI-H1299, and Pa-Tu-8988t cells were cultured at 37°C and 5% CO2 in DMEM/F12 with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine. FL5.12 and BxPC-3 cells were cultured in RPMI with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine. For FL5.12, 55 µM 2-mercaptoethanol (Gibco) and 1.4 ng/mL murine IL-3 (Peprotech 213-13) were also supplemented. To prepare necrotic FL5.12 cells, cells were pelleted, washed three times with PBS, and seeded in medium without IL-3 at high density (107/mL) for 72–96 h, as described previously (Kim et al. 2018). All cell lines were routinely tested for mycoplasma contamination.
Lentivirus (pLentiCRISPRv2) was prepared by cotransfection of HEK293T cells with lentiviral vector, psPAX2, and pMD2.G (4:3:1) complexed with PEI (linear MW 25,000; Polysciences, Inc.). Retrovirus (pTURN) was similarly prepared using pCG-gagpol in lieu of psPAX2. Viral supernatant was collected 48 h later and filtered through a 0.45-µm polyethersulfone membrane. Cells were infected in the presence of 4 µg/mL polybrene and selected 32 h later with 5 µg/mL puromycin or 200 µg/mL hygromycin.
Cell growth experiments were performed by washing cells once with PBS, dissociating cells with trypsin and then resuspending in leucine-free medium (Athena SILAC medium 0423; reconstituted with lysine, arginine, and methionine with 10% dialyzed FBS, 4 mM glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin). Cells (2.5 × 104 to 5 × 104) were seeded directly in complete or leucine-free medium with 3% BSA in 12-well plates in triplicate. For complete medium, free leucine was added back to leucine-free medium to a final concentration of 400 µM. Cells were counted 16 h after seeding to determine starting population (day 0) and every 24 h thereafter, up to 96 h later.
To measure cell growth dependent on the uptake of necrotic cells, 5 × 104 cells were first seeded in triplicate in complete DMEM/F12 in 12-well plates. Sixteen hours later, initial cell counts were determined; wells were washed once with PBS and medium was exchanged to 1% AA DMEM/F12 (amino acid-free DMEM/F12 mixed 99:1 with complete DMEM/F12 + 10% dialyzed FBS, 100 U/mL penicillin, 100 μg/mL streptomycin) with or without necrotic FL5.12 (25:1 necrotic cells:target cells). Necrotic FL5.12 cells were washed twice with PBS prior to resuspending in assay medium. No change in cell growth was observed upon addition of necrotic cells in complete medium.
Relative cell growth was calculated as the ratio of cells counted at the indicated experiment endpoint relative to Day 0. Biomass was calculated by multiplying cell number by mean cell volume and theoretical cell density. All cell counts and volume measurements were determined using a Beckman Coulter Counter Multisizer 3.
Western blots
Whole-cell lysates were prepared by washing cell monolayers once with ice-cold PBS and scraping cells in ice-cold RIPA buffer (10 mM Tris at pH 8, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 0.5 mM EGTA, 1× Halt protease, phosphatase inhibitor mixtures; Thermo Scientific). For instances when cells were grown in media with supplemented BSA, cells were first dissociated with 0.25% trypsin and resuspended in serum-containing media, pelleted at low speed (500×g for 5 min at 4°C), washed once with ice-cold PBS, and lysed in RIPA buffer. Lysis was allowed to proceed for 15 min on ice and soluble lysate fractions were isolated by centrifugation at 16,000g for 10 min.
Nuclear and cytoplasmic fractions were prepared by first resuspending cells in Harvest buffer (10 mM HEPES KOH at pH 7.9, 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA, 0.5% Triton X-100, 1× Halt protease, phosphatase inhibitors) for 5 min to solubilize the cytosol, followed by pelleting the remaining material at low speed (500×g for 5 min), washing once with buffer A (10 mM HEPES KOH at pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA), and solubilizing nuclei in buffer C (10 mM HEPES KOH at pH 7.9, 500 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% NP-40 substitute, 1× Halt protease, phosphatase inhibitors), followed by brief sonication.
For immunoprecipitation of FLAG-tagged proteins, cells were lysed in 1% Triton X-100, 20 mM Tris (pH 8), 140 mM NaCl, 10% glycerol, 2 mM EDTA with 1× Halt protease, and phosphatase inhibitors. FLAG-containing complexes from 1 mg of cell lysate were captured with 40 µL of anti-FLAG M2 magnetic beads (Sigma-Aldrich) for 1 h, followed by four washes with lysis buffer and elution in 30 µL of 2× Laemmli buffer for 5 min at 95°C. Equal volumes of eluate were run alongside 25 µg of input on an SDS–polyacrylamide gel.
Protein concentrations were determined with the Pierce BCA protein assay (Thermo Scientific) and equal amounts of proteins were separated by SDS-PAGE (NuPAGE 4%–12% Bis-Tris gels, Thermo Fisher) and transferred to nitrocellulose membrane (Bio-Rad). Membranes were blocked for 1 h in TBST with 5% nonfat dry milk and incubated with primary antibodies (diluted 1:500–5000 in TBST with 5% BSA) overnight (16–24 h). Following three 10-min washes in TBST, membranes were placed in secondary antibodies (diluted 1:5000 in TBST with 5% milk) for 1 h, washed three times for 10 min in TBST, briefly air-dried and developed with Pierce ECL (Thermo Fisher) or Super Digital ECL solution (Kindle Biosciences). Chemiluminescent exposures were captured with a ChemiDoc Touch imaging system (Bio-Rad) and images were processed with Image Lab software. When necessary, nitrocellulose membranes were stripped with Restore Stripping solution (Thermo Fisher) for 20 min at 37°C, blocked with 5% milk in TBST, and probed with additional antibodies.
Fluorescence microscopy
For quantification of dextran uptake, cells were seeded onto glass chamber slides in complete or leucine-free medium + 3% BSA and cultured for 16 h. Uptake of macromolecules was determined by addition of 0.1 mg/mL fluorescently labeled 70-kDa dextran and BSA for 30 min, followed by three washes with ice-cold PBS and fixation in 4% formaldehyde in PBS for 15 min. After fixation, cells were labeled for 5 min with 2 µg/mL Hoechst in PBS and washed once with PBS. Images were acquired with a Zeiss LSM 880 confocal microscope. Cellular uptake of fluorescently labeled dextran or albumin was quantified using the particle analyzer function of ImageJ. Mean fluorescence intensity was determined by calculating the integrated signal density per cell in randomly chosen fields of view.
Immunofluorescence staining for Yap was performed after culturing cells for 48 h on poly-L-lysine-coated coverslips. Cells were washed once with PBS, fixed for 20 min in 4% formaldehyde in PBS, washed twice more with PBS and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Coverslips were blocked in PBG (0.5% BSA/0.2% cold water fish skin gelatin in PBS) for 1 h, followed by addition of primary Yap antibody (63.7; 1:200; Santa Cruz Biotechnology) in PBG overnight at 4°C. Following three 5-min washes with 4% normal goat serum (Cell Signaling Technology) in PBG, secondary antibody solution (Alexa fluor 647 goat anti-mouse IgG; ThermoFisher, diluted 1:1000 in 4% normal goat serum in PBG) was added for 1 h at room temperature. After three final 5-min washes with PBS, coverslips were mounted onto glass slides in ProLong Gold Antifade medium with DAPI (Life Technologies). Images were acquired with a Leica SP5 confocal microscope.
Live imaging of uptake of cypHer5E-stained FL5.12 cells was performed by seeding H1299 cells transduced with pMSCV-puro-ires-GFP retrovirus into glass chamber slides. CypHer5E-stained necrotic FL5.12 cells (106/mL), Hoechst 33342 (0.2 µg/mL), and inhibitors were added to the medium and fields were imaged every 20 min on a Zeiss LSM880 confocal microscope equipped with a CO2 humid chamber for 6 h at 37°C.
ATAC-seq
ATAC-seq libraries were prepared from 5 × 104 cells per sample as described previously (Buenrostro et al. 2015) using the Nextera DNA library preparation kit (Illumina). Libraries were amplified by PCR with barcoded primers using NEBNext Hi-Fidelity 2× Mastermix (New England Biolabs) and purified with the Qiagen MinElute PCR purification kit. Sample libraries were quantified by qPCR using a KAPA library quantification kit, pooled and sequenced on an Illumina NextSeq 500. Paired-end, 50-bp reads from demultiplexed fastq files were trimmed using TrimGalore and trimmed reads were aligned to genome build mm9 with bowtie2 with a maximum fragment length (-X) of 2000 bp. Genome-wide sequence coverage was determined with DeepTools bamCoverage, using 1× RPGC normalization, and visualized with Integrated Genome Viewer (IGV2.0). ATAC-seq peaks were called using MACS2 and peaks unique to KRPCS were determined using compiled data from KRPC cells grown in complete medium as background. Enriched DNA motifs associated with transcription factors were determined within 200-bp window centered on peak summits from MACS2 (using BedTools) and de novo motif analysis was performed using Homer. ATAC-seq read coverage across TEAD-bound genomic loci was calculated with DeepTools using publically available Tead1 ChIP-seq data (GSE 55186). Raw and processed ATAC-seq data was deposited into the Gene Expression Omnibus (GSE155674).
RNA-seq
RNA was isolated from cells using Trizol and libraries were prepared using the TruSeq RNA sample preparation kit v2 (Illumina) in accordance with the manufacturer's instructions. Libraries were sequenced using an Illumina HiSeq 4000 generating 150-bp paired-end reads. Sequences from demultiplexed fastq files were trimmed using TrimGalore and trimmed reads were aligned to genome build mm9 using TopHat2 with default parameters. Aligned features were counted with htseq-count and differential expression was determined using the edgeR package in Bioconductor as described previously (Anders et al. 2013). Genes with > 1 aligned read per million in at least two replicates were included for further analysis. Principal component analysis was performed using the DESeq2 package in Bioconductor. Gene set enrichment analysis (Broad Institute) was performed using log2 ratio of classes as a ranking metric with 1000 permutations. Pairwise comparisons of RNA-seq data from each sample group were tested across all curated (c2) and hallmark gene sets available in the Molecular Signature Database. Gene sets related to Yap target genes (“Cordenonsi Yap conserved signature” and “YAP1 targets up”) were generated from previously published studies (Zhang et al. 2008; Cordenonsi et al. 2011). Raw and processed RNA-seq data was deposited into the Gene Expression Omnibus (GSE155650).
Flow cytometry
To measure catabolism of extracellular albumin, cells were directly seeded in complete or leucine-free medium supplemented with 3% BSA and cultured overnight (16 h). DQ-BSA Green (0.05 mg/mL) and LysoTracker Red (10 nM; Life Technologies) were added to the cells for 4 h, after which cells were washed once with PBS, dissociated with trypsin, resuspended in serum-containing medium, and pelleted at 400g for 5 min at 4°C. Pelleted cells were resuspended in FACS buffer (PBS, 2% FBS) and samples were analyzed immediately.
Determination of necrotic cell engulfment was achieved by first pelleting 5 × 107 necrotic FL5.12 cells at 700g for 5 min at room temperature and resuspending them in 10 mL of PBS with 1 µM cypHer5E NHS ester (GE Healthcare) for 30 min at 37°C. Stained cells were pelleted again and resuspended in assay medium at a concentration of 2 × 106/mL along with inhibitors or vehicle controls and added to the wells of a tissue culture plate containing target cells. Plates were spun in a swinging bucket rotor centrifuge twice at 500g for 1 min at room temperature, rotating the plate in between spins, and returned to the tissue culture incubator for 2–6 h. Cells were harvested in the same manner as for the DQ-BSA assay above. Samples were analyzed on a BD Fortessa or LSRII flow cytometer. Analysis of data and generation of histograms was performed with FlowJo (Treestar).
In vitro invasion assay
Control and matrigel invasion chambers (Corning, PET membrane, 8-µm pore size, 24-well) were prepared according to the manufacturer's instructions. Cells were dissociated with nonenzymatic cell dissociation reagent and resuspended in DMEM/F12 with 0.1% BSA at a concentration of 5 × 105/mL. One-hundred microliters of cell suspension was added to the upper chamber, followed by addition 650 µL of DMEM/F12 with 10% FBS to the outer/lower chamber. Invasion/migration was allowed to proceed overnight (16 h) in a tissue culture incubator. Nonmigrated cells were scrubbed away with a cotton-tipped applicator and membranes were fixed in 70% ethanol for 10 min, stained in 0.2% crystal violet for 5 min, washed in water, and air-dried. Membranes were excised with a scalpel and mounted on a cover-slipped glass slide with Cytoseal-70 (Thermo Fisher). Images were acquired with a Zeiss AxioCam microscope or Mirax Slide Scanner.
Statistical analyses
One-way ANOVA and Student's t-tests were performed with GraphPad Prism, using Tukey's method to correct for multiple comparisons. Graphs were generated using R or GraphPad Prism software.
Competing interest statement
C.B.T. is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He is also a former member of the Board of Directors and stockholder of Merck and Charles River Laboratories. He holds patents related to cellular metabolism.
Supplementary Material
Acknowledgments
We thank members of the C.B.T. laboratory for critical review of the data presented in this manuscript, Justin Perry for discussions and technical expertise related to cell engulfment assays, and the Memorial Sloan Kettering Cancer Center (MSKCC) Molecular Cytology Core Facility for assistance with microscopy. B.K. was a Berger Foundation Fellow of the Damon Runyon Cancer Research Foundation (DRG 2234-15). This work is supported by grants from the National Cancer Institute (to C.B.T.) and by the Cancer Center support grant (P30 CA008748) to MSKCC.
Author contributions: B.K. and C.B.T. conceived the study and wrote the manuscript. B.K. and J.A. performed experiments and analyzed the data. W.P. provided technical expertise and reagents. All authors participated in discussing and finalizing the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.340661.120.
References
- Amyere M, Payrastre B, Krause U, Smissen PVD, Veithen A, Courtoy PJ. 2000. Constitutive macropinocytosis in oncogene-transformed fibroblasts depends on sequential permanent activation of phosphoinositide 3-Kinase and phospholipase C. Mol Biol Cell 11: 3453–3467. 10.1091/mbc.11.10.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8: 1765–1786. 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- Araki N, Johnson MT, Swanson JA. 1996. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol 135: 1249–1260. 10.1083/jcb.135.5.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco J. 1986. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 233: 1061–1068. 10.1126/science.3090687 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson Rachel L, Sabatini David M. 2015. Nutrient-sensing mechanisms across evolution. Cell 161: 67–83. 10.1016/j.cell.2015.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson-Mackenzie C, Morris ZS, Liu C-H, Bradford WB, Koorman T, McClatchey AI. 2018. Merlin/ERM proteins regulate growth factor-induced macropinocytosis and receptor recycling by organizing the plasma membrane:cytoskeleton interface. Genes Dev 32: 1201–1214. 10.1101/gad.317354.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497: 633–637. 10.1038/nature12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti Anna R, Poletti A, et al. 2011. The hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147: 759–772. 10.1016/j.cell.2011.09.048 [DOI] [PubMed] [Google Scholar]
- Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, Wyckoff J, Del Rosario AM, Whitman M, Chin CR, et al. 2017. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med 23: 235–241. 10.1038/nm.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Chandel NS. 2016. Fundamentals of cancer metabolism. Sci Adv 2: e1600200 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhane S, Schürmann A, Sells MA, Chernoff J, Schmid SL, Bokoch GM. 2000. Regulation of macropinocytosis by p21-activated kinase-1. Mol Biol Cell 11: 3341–3352. 10.1091/mbc.11.10.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finicle BT, Jayashankar V, Edinger AL. 2018. Nutrient scavenging in cancer. Nat Rev Cancer 18: 619–633. 10.1038/s41568-018-0048-x [DOI] [PubMed] [Google Scholar]
- García-Jiménez C, Goding CR. 2019. Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metab 29: 254–267. 10.1016/j.cmet.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S, Ruaro E, Varnum B, Schneider C. 1997. Requirement of phosphatidylinositol 3-kinase-dependent pathway and Src for Gas6-Axl mitogenic and survival activities in NIH 3T3 fibroblasts. Mol Cell Biol 17: 4442–4453. 10.1128/MCB.17.8.4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo M. 2010. Pancreatic cancer. N Engl J Med 362: 1605–1617. 10.1056/NEJMra0901557 [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. 2005. The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating yorkie, the Drosophila homolog of YAP. Cell 122: 421–434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Ireland L, Luckett T, Schmid MC, Mielgo A. 2020. Blockade of stromal Gas6 alters cancer cell plasticity, activates NK cells, and inhibits pancreatic cancer metastasis. Front Immunol 11: 297 10.3389/fimmu.2020.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. 2015. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res 75: 544–553. 10.1158/0008-5472.CAN-14-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu C-J, Sadanandam A, Hu B, et al. 2014. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158: 185–197. 10.1016/j.cell.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MC, Teasdale RD. 2009. Defining macropinocytosis. Traffic 10: 364–371. 10.1111/j.1600-0854.2009.00878.x [DOI] [PubMed] [Google Scholar]
- Kim SM, Nguyen TT, Ravi A, Kubiniok P, Finicle BT, Jayashankar V, Malacrida L, Hou J, Robertson J, Gao D, et al. 2018. PTEN deficiency and AMPK activation promote nutrient scavenging and anabolism in prostate cancer cells. Cancer Discov 8: 866–883. 10.1158/2159-8290.CD-17-1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirane A, Ludwig KF, Sorrelle N, Haaland G, Sandal T, Ranaweera R, Toombs JE, Wang M, Dineen SP, Micklem D, et al. 2015. Warfarin blocks Gas6-mediated Axl activation required for pancreatic cancer epithelial plasticity and metastasis. Cancer Res 75: 3699–3705. 10.1158/0008-5472.CAN-14-2887-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JH, Plouffe SW, Meng Z, Lee D-H, Yang D, Lim D-S, Wang C-Y, Guan K-L. 2020. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes Dev 34: 72–86. 10.1101/gad.331546.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-W, Zhang Y, Jung M, Cruz N, Alas B, Commisso C. 2019. EGFR–Pak signaling selectively regulates glutamine deprivation-induced macropinocytosis. Dev Cell 50: 381–392.e5. 10.1016/j.devcel.2019.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke G, Rothlin CV. 2008. Immunobiology of the TAM receptors. Nat Rev Immunol 8: 327–336. 10.1038/nri2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, Saborowski M, Kastenhuber E, Fellmann C, Ohara K, et al. 2014. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan K-L. 2016. Mechanisms of Hippo pathway regulation. Genes Dev 30: 1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka S, Perry JSA, Raymond MH, Medina CB, Zhu Y, Zhao L, Serbulea V, Onengut-Gumuscu S, Leitinger N, Kucenas S, et al. 2018. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563: 714–718. 10.1038/s41586-018-0735-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, Konukiewitz B, Öllinger R, Zwiebel M, Strong A, et al. 2018. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 554: 62–68. 10.1038/nature25459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofal M, Zhang K, Han S, Rabinowitz JD. 2017. mTOR inhibition restores amino acid balance in cells dependent on catabolism of extracellular protein. Mol Cell 67: 936–946.e5. 10.1016/j.molcel.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci 103: 12405–12410. 10.1073/pnas.0605579103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. 2015. The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell 162: 259–270. 10.1016/j.cell.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Araki J, King B, DeMatteo RG, Thompson CB. 2017. Critical role for PI3-kinase in regulating the use of proteins as an amino acid source. Proc Natl Acad Sci 114: E8628–E8636. 10.1073/pnas.1712726114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS-O, Berenguer A, Prats N, Toll A, Hueto JA, et al. 2017. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541: 41–45. 10.1038/nature20791 [DOI] [PubMed] [Google Scholar]
- Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A. 2017. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol 35: 347–349. 10.1038/nbt.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. 2014. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158: 171–184. 10.1016/j.cell.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Paez JG, Curto M, Yaktine A, Pruitt WM, Saotome I, O'Bryan JP, Gupta V, Ratner N, Der CJ, et al. 2001. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev Cell 1: 63–72. 10.1016/S1534-5807(01)00009-0 [DOI] [PubMed] [Google Scholar]
- Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schübeler D, et al. 2015. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet 11: e1005465 10.1371/journal.pgen.1005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Conn G, Goret M, Lai C, Bruno J, Radzlejewski C, Mattsson K, Fisher J, Gies DR, Jones PF, et al. 1995. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell 80: 661–670. 10.1016/0092-8674(95)90520-0 [DOI] [PubMed] [Google Scholar]
- Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, Norrie IC, Miller CJ, Poulogiannis G, Lauffenburger DA, et al. 2016. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell 165: 910–920. 10.1016/j.cell.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, Murphy MM, Gu Z, Gu W, Martin M, et al. 2020. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577: 115–120. 10.1038/s41586-019-1847-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay AM, Missiaglia E, Galli GG, Hettmer S, Urcia R, Carrara M, Judson RN, Thway K, Nadal G, Selfe JL, et al. 2014. The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell 26: 273–287. 10.1016/j.ccr.2014.05.029 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Williamson EK, Schumacker PT, Thompson CB. 1997. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91: 627–637. 10.1016/S0092-8674(00)80450-X [DOI] [PubMed] [Google Scholar]
- Varnum BC, Young C, Elliott G, Garcia A, Bartley TD, Fridell Y-W, Hunt RW, Trail G, Clogston C, Toso RJ, et al. 1995. Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth-arrest-specific gene 6. Nature 373: 623–626. 10.1038/373623a0 [DOI] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, Sabatini DM. 2017. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 171: 642–654.e12. 10.1016/j.cell.2017.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Vanderzalm PJ, Ludwig M, Su T, Tokamov SA, Fehon RG. 2018. Yorkie functions at the cell cortex to promote myosin activation in a non-transcriptional manner. Dev Cell 46: 271–284.e5. 10.1016/j.devcel.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17: 1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA. 2008. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res 68: 2789–2794. 10.1158/0008-5472.CAN-07-6205 [DOI] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19: 27–38. 10.1016/j.devcel.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.