In this study, Fan et al. studied the post-translational regulation of transcription factor SNAI2, which plays key roles during development and promotes metastasis by inducing invasive phenotype and tumor-initiating activity of cancer cells. They performed a dual-luciferase-based, genome-wide E3 ligase siRNA library screen and identified ASB13 as an E3 ubiquitin ligase that targets SNAI2 for ubiquitination and degradation, thereby establishing ASB13 as a suppressor of breast cancer metastasis.
Keywords: breast cancer, ASB13, SNAI2, ubiquitin proteasome system, Hippo–YAP pathway, migration, metastasis
Abstract
Transcription factor SNAI2 plays key roles during development and has also been known to promote metastasis by inducing invasive phenotype and tumor-initiating activity of cancer cells. However, the post-translational regulation of SNAI2 is less well studied. We performed a dual-luciferase-based, genome-wide E3 ligase siRNA library screen and identified ASB13 as an E3 ubiquitin ligase that targets SNAI2 for ubiquitination and degradation. ASB13 knockout in breast cancer cells promoted cell migration and decreased F-actin polymerization, while overexpression of ASB13 suppressed lung metastasis. Furthermore, ASB13 knockout decreased YAP expression, and such regulation is dependent on an increased protein level of SNAI2, which in turn represses YAP transcription. YAP suppresses tumor progression in breast cancer, as YAP knockout increases tumorsphere formation, anchorage-independent colony formation, cell migration in vitro, and lung metastasis in vivo. Clinical data analysis reveals that ASB13 expression is positively correlated with improved overall survival in breast cancer patients. These findings establish ASB13 as a suppressor of breast cancer metastasis by promoting degradation of SNAI2 and relieving its transcriptional repression of YAP.
The majority of cancer-related deaths result from distant metastasis (Valastyan and Weinberg 2011; Wan et al. 2013). As the first step in metastasis, local and lymph node invasion is a strong poor prognosis marker for patient survival (Rakha et al. 2012). SNAI2 (SLUG) is a C2H2 zinc finger transcriptional repressor belonging to the three-member family of SNAIL protein (SNAIL, SNAI2, and Smuc) (Nieto 2002). Although best known for its role in orchestrating epithelial–mesenchymal transition program (EMT), during which tumor cells lose their epithelial properties and resemble fibroblast-like cell phenotypes with increased cell migration ability, more recent studies reveal that SNAI2 has a much broader effect on cancer progression, including many functions independent of its role in regulating EMT. SNAI2 has been linked to the induction of tumor initiation cell, cell cycle regulation, as well as invasion and metastasis (Mittal et al. 2011; Guo et al. 2012; Nieto and Cano 2012; Uygur et al. 2015). A better understanding of the regulatory mechanisms for SNAI2 will provide critical information on how to block cancer progression and metastasis.
The SNAI2 protein, like SNAIL, is rapidly turned over by the ubiquitin proteasome system (UPS) (Zheng and Kang 2014). Previous results suggested β-TrCP1/FBXW1, CHIP (C terminus of Hsc70-interacting protein), MDM2, and FBXL14 as E3 ligases for SNAI2 degradation. However, none of these E3s have been tested for their roles in regulating endogenous SNAI2, especially in the context of cancer cell migration and metastasis. It is still not clear which E3 ligases are the major mediators regulating endogenous SNAI2 ubiquitination and degradation in breast cancer (Vernon and LaBonne 2006; Wang et al. 2009; Wu et al. 2012; Kao et al. 2014). Identification of such E3 ligase(s) and relevant signaling pathways will provide a comprehensive understanding of the regulatory mechanisms for SNAI2 in cancer. To this end, we performed a dual-luciferase-based E3 siRNA screening similarly as in our previous study (Zheng et al. 2014) and tested functions of the identified E3 ligase ASB13 in breast cancer tumorigenesis, migration, and metastasis.
ASB13 belongs to the ankyrin repeat and suppressor of cytokine signaling (SOCS) box (Asb) E3 ligase protein family (Liu et al. 2019). It has been implicated to be involved in tumor progression, although its functional role in cancer has not been directly tested (Blenk et al. 2007; Chi et al. 2018). Through stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomic profiling, several potential interacting proteins for ASB13 have been identified, including POLR3A, TCEB2, SSBP1, CCT8, etc. (Andresen et al. 2014). However, none of these proteins have been biochemically validated as direct protein substrate for ASB13.
Hippo pathway is a major modulator for organ size by controlling cell proliferation, differentiation, and migration in developing adult tissues (Meng et al. 2016). This pathway is controlled by intrinsic cell machineries, such as cell polarity and actin cytoskeleton, as well as a wide range of paracrine signals, including cell–cell contact, cellular energy status, mechanical cues, and hormonal signals that act through G-protein-coupled receptors (Zhao et al. 2007; Dupont et al. 2011; Yu et al. 2012). As an important downstream mediator, yes-associated protein (YAP) was originally found to promote cell proliferation and transformation. Overexpression of YAP has been linked to tumor progression and worse patient survival in many cancer types (Xu et al. 2009; Wang et al. 2010; Song et al. 2012; Su et al. 2012; Liu et al. 2013). However, in breast cancer, YAP has been suggested as a tumor suppressor gene, as it is located in a chromosome region with frequent loss of heterozygosity (LOH) (Carter et al. 1994; Hampton et al. 1994; Tomlinson et al. 1995). Decreased expression of YAP is correlated with tumor progression and worse survival in breast cancer (Gudmundsson et al. 1995; Winqvist et al. 1995; Tufail et al. 2012). However, the exact role of YAP in breast cancer progression and metastasis and whether YAP can cross-talk with critical metastasis determinants like SNAI2 is still not clear. Understanding such cross-talk may reveal possible windows for therapeutic targeting of SNAI2 and Hippo pathway in cancer.
Results
A dual-luciferase system for a genome-wide screen to identify E3 ligases targeting SNAI2 protein
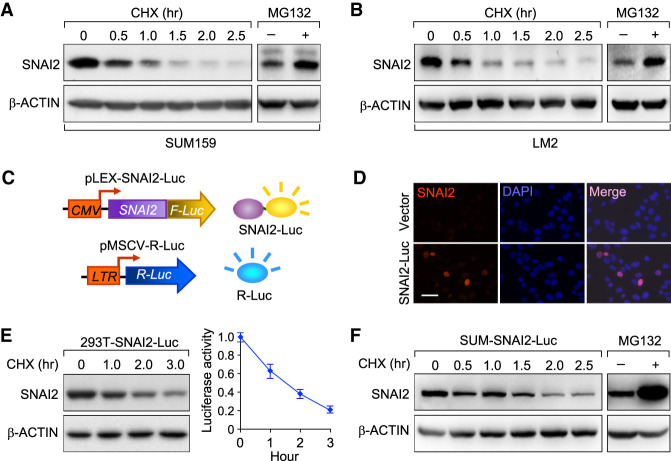
We first confirmed that the SNAI2 protein, like its family member SNAIL, is degraded through UPS (Zheng et al. 2014). The SUM159 breast cancer cell line (Forozan et al. 1999) culture was treated with cycloheximide (CHX) to block protein synthesis and the endogenous SNAI2 protein degradation was chased afterward (CHX chase assay). Indeed, the SNAI2 protein was degraded rapidly and kept at a low baseline expression level within 2.5 h of CHX treatment in SUM159 cells (Fig. 1A; Supplemental Fig. S1A,B). To confirm that SNAI2 degradation is mediated by the 26S proteasome, we blocked UPS with the proteasome inhibitor MG132, and the endogenous SNAI2 protein level was significantly increased (Fig. 1A; Supplemental Fig.S1A,B). Similarly, SNAI2 was also degraded rapidly in a proteasome-dependent manner in LM2 cell line, a highly lung-metastatic subline of the MDA-MB-231 breast cancer cell line (Fig. 1B; Supplemental Fig. S1C,D; Minn et al. 2005). To identify E3 ligase(s) responsible for SNAI2 ubiquitination and degradation, we generated a dual-luciferase-based siRNA library screening system that we developed in our previous study (Zheng et al. 2014). We fused the coding sequence of SNAI2 in-frame with the Firefly Luciferase coding sequence to produce lentiviruses containing this fusion gene (SNAI2-Luc). Breast cancer cell line SUM159 was then transduced with SNAI2-Luc lentiviruses and with retroviruses constitutively expressing renilla luciferase to generate a dual-luciferase reporter stable cell line. In this cell line, firefly luciferase activity reliably represents the expression level of the SNAI2-Luc protein, while renilla luciferase activity is used as internal control. This reporter cell line, denoted as “SUM-SNAI2-Luc/rLuc” to facilitate descriptions below, allowed us to monitor SNAI2 stable level and its degradation dynamics by measuring luciferase activities (Fig. 1C). When transfected and expressed in HEK293T cells, SNAI2-Luc mostly localized in the nucleus, similar to the nuclear localization of endogenous SNAI2 (Fig. 1D). In both HEK293T cells and SUM159, the SNAI2-Luc protein was degraded through UPS as demonstrated by CHX chase assay and proteasome inhibition assay (Fig. 1E,F; Supplemental Fig. S1E,F).
Figure 1.
A dual-luciferase system for the identification of SNAI2 targeting E3 ligase. (A) SUM159 cells were treated with 10 µM CHX for indicated time or MG132 for 6 h. The endogenous SNAI2 protein was detected by immunoblotting. β-ACTIN was used as internal loading control. (+) Treated with MG132, (−) no treatment. (B) LM2 cells were treated with 10 µM CHX for indicated time or MG132 for 6 h. The endogenous SNAI2 protein was detected by immunoblotting. β-ACTIN was used as internal loading control. (C) Illustration of the dual-luciferase reporter screening system for SNAI2 targeting E3 ligase. (CMV) Cytomegalovirus promoter, (LTR) long terminal repeat viral promoter. (D) Immunofluorescence staining of SNAI2-Luc in SUM159-SNAI2-Luc cell line with SNAI2 antibody. Scale bar, 50 µm. (E) In HEK293T cells stably expressing SNAI2-Luc, the turnover rate for SNAI2-Luc was determined by CHX pulse-chase assay and luciferase assay. Data are presented as mean ± standard error. (F) In SUM159 cells stably expressing SNAI2-Luc, the turnover rate for SNAI2-Luc was determined by CHX pulse-chase assays.
siRNA library screening identifies potential E3 ligases for targeting SNAI2 degradation
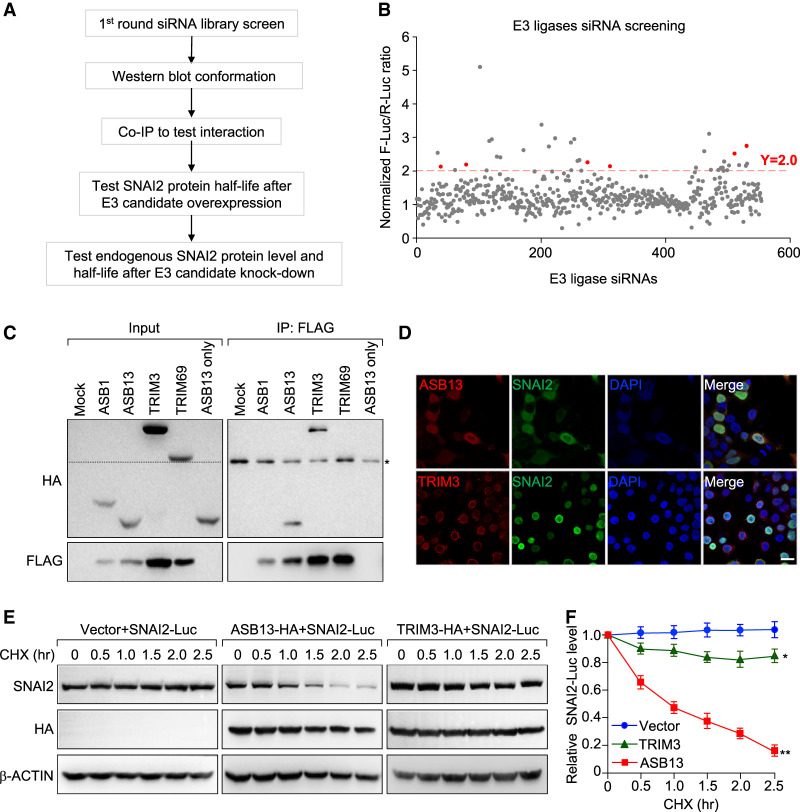
We followed the procedures outlined in Figure 2A to identify potential E3 ligase candidates for SNAI2 protein. Individual human E3 ligase was knocked down (KD) using pooled siRNAs (three siRNAs per gene) in SUM-SNAI2-Luc/R-Luc cells and those E3 ligase genes whose KD resulted in more than twofold increase in firefly luciferase/renilla-luciferase ratio were identified as candidate SNAI2 targeting E3s (Fig. 2B). Among those E3 ligases, there were several ASB (ankyrin repeat and suppressor of cytokine signaling box) family members and TRIM (tripartite motif) family members (pink dots on the graph). We next attempted to clone these E3 ligases and validate their interactions with SNAI2. Among the ones we were able to clone, ASB13 and TRIM3 were validated to interact with SNAI2 based on coimmunoprecipitation (co-IP) assay (Fig. 2C; Supplemental Fig. S2A). Immunofluorescence staining showed that ectopically expressed HA-ASB13 was mostly localized in the nucleus of HEK293T cells, and colocalized with SNAI2. In contrast, TRIM3 was not colocalized with SNAI2, as it mostly resided in the paranucleus region (Fig. 2D; Supplemental Fig. S2B). Interestingly, in SUM159 cells stably expressing HA-ASB13, immunofluorescent staining demonstrated that ASB13 was localized in both nucleus and cytoplasm (Supplemental Fig. S2C). Consistently, nuclear and cytoplasmic fractionation experiment demonstrated that ASB13 was partially localized in the nucleus and its expression reduced the SNAI2 protein stable level (Supplemental Fig. S2D). The interaction between ectopically expressed ASB13 and endogenous SNAI2 was also observed in SUM159 cells as demonstrated by co-IP experiment (Supplemental Fig. S2E). To functionally test the ability of ASB13 and TRIM3 to promote SNAI2 degradation, we cotransfected SNAI2-Luc with either ASB13 or TRIM3, and examined the degradation dynamics for SNAI2-Luc. ASB13 accelerated SNAI2-Luc protein degradation, whereas TRIM3 only had modest effect on SNAI2-Luc turnover (Fig. 2E,F). Taken together, we considered ASB13 as the primary E3 ligase candidate for SNAI2.
Figure 2.
A genome-wide siRNA library screen for E3 ubiquitin ligase(s) targeting the SNAI2 protein. (A) Experimental procedure flow chart for identification of E3 ubiquitin ligase(s) targeting SNAI2 protein. (B) Dual-luciferase-based siRNA library screen in HEK293T cells against human E3 ligases identified multiple E3 candidates. E3 ligase was considered to be a positive hit if its knockdown led to more than twofold increase in ff-luc/r-luc raito. (C) Lysates from HEK293T cells treated with 10 µM MG132 for 4 h were immunoprecipitated with FLAG beads, then immunoblotted with either HA antibody for E3 ligases or FLAG antibody for SNAI2 protein. The 50-kDa IgG bands in immunoprecipitated samples are labeled with an asterisk. Dashed line in input image indicates that the band for TRIM69 is above the 50-kDa IgG band. (D) HEK293 cells were transiently transfected with plasmids expressing HA-tagged ASB13 or TRIM3, and FLAG-tagged SNAI2 and stained with antibodies against HA or FLAG tag to visualize the cellular localizations of these proteins by immunofluorescence. Scale bar, 20 µm. (E) Two E3 ligase candidates or a control pLEX-vector were cotransfected with SNAI2-Luc plasmid into 293T cells, and a CHX pulse-chase assay was performed 48 h later. (F) Quantification of SNAI2 protein levels presented in E using ImageJ software. Vector versus TRIM3, P = 0.009; vector versus ASB13, P = 2.8 × 10−5.
ASB13 targets SNAI2 for ubiquitination and degradation
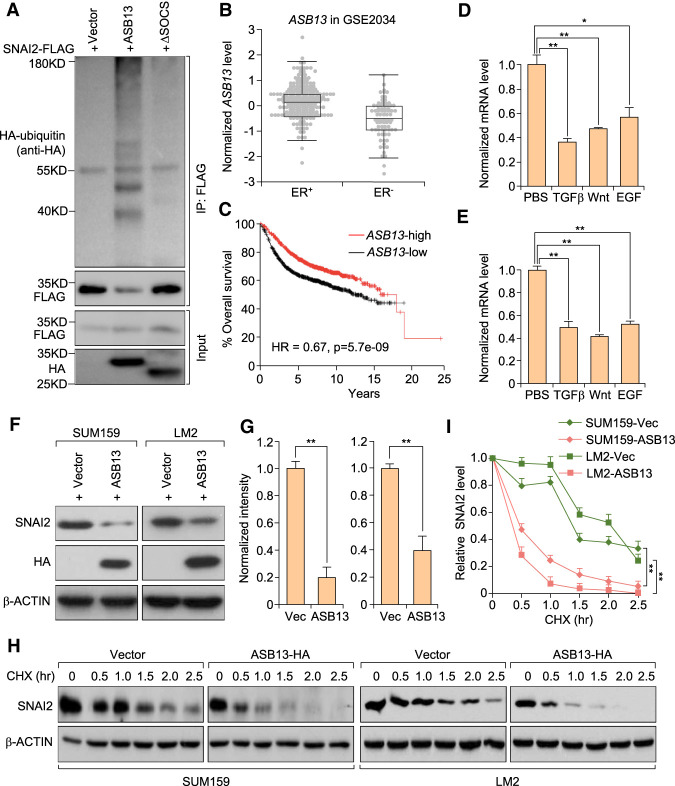
To investigate whether ASB13 functions as a bona fide E3 ligase that ubiquitinates SNAI2 protein, we cotransfected HEK293T cells with FLAG-tagged SNAI2, HA-tagged ASB13, and HA-tagged ubiquitin, and treated cells with MG132 for 6 h to prevent protein degradation before performing the ubiquitination assay. A significant increase of polyubiquitinated SNAI2 protein was observed in ASB13-transfected cells, whereas the SOCS domain (ElonginB/C–Culin5–SOCS complex formation, interacting domain for E3 complex formation) deletion mutant of ASB13 (ASB13-ΔSOCS) was not able to promote SNAI2 ubiquitination (Fig. 3A), confirming ASB13 as a bona fide E3 ubiquitin ligase for SNAI2. Domain deletion mutants of ASB13 were generated and their interactions with SNAI2 were tested with co-IP experiment. Deletion of either ANK3-4 or ANK5-6 almost completely diminished ASB13's interaction with SNAI2 (Supplemental Fig. S3A). SNAI2 protein has been reported as a marker for poor prognosis in cancer (Hajra et al. 2002; Uchikado et al. 2005, 2011), with its high expression correlated with worse patient outcome, and SNAI2 is also expressed at a higher level in estrogen receptor-negative (ER−) patients than ER+ patients in breast cancer (Chakrabarti et al. 2012). Thus, we hypothesized that E3 ligase(s) targeting SNAI2 could be a good prognosis marker in breast cancer and might be expressed at a lower level in ER− patients, based on its negative regulation of SNAI2. Indeed, analyses of clinical breast cancer data sets confirmed that ER− breast cancer patients have lower ASB13 level in ER− breast cancer, and higher ASB13 expression correlates with longer overall patient survival (Fig. 3B,C).
Figure 3.
ASB13 targets SNAI2 protein for ubiquitination and degradation. (A) HEK293T cells were cotransfected with plasmids expressing HA-Ub, SNAI2-FLAG together with either a vector control, ASB13, or the ASB13-ΔSOCS plasmid. Cells were treated with MG132 for 6 h before IP using a denature IP protocol to pull down the SNAI2 protein, and the polyubiquitinated SNAI2 protein was detected by an anti-HA antibody. (B) Box plot showing normalized ASB13 mRNA levels in ER+ and ER− breast cancer patients. Data are from the Wang et al. (2005) data set (GSE2034), and patients were free of lymph node invasion at the time of diagnosis. P = 0.0001 with unpaired two-tailed t-test. The middle line represents medium, the box represents 25%–75% values, while the error bar represents minimum and maximum without outlier. (C) Kaplan–Meier plot of overall survival of breast cancer patients stratified by the expression of ASB13 gene. Data were obtained from KMplot.com. (D,E) Endogenous ASB13 expression level was repressed during EMT inducer treatments like TGF-β, Wnt, and EGF signaling activation in MCF10A and EpRas cells. Cells were treated with TGF-β, LiCl (inhibitor of GSK3β kinase, activator of Wnt signaling pathway), and recombinant EGF protein, respectively. Data are presented as mean ± SEM. (*) P < 0.05; (**) P < 0.01 by Student's t-test. (F) ASB13 (HA-tagged) was stably expressed in SUM159 and LM2 cells by lentivirus transduction, cell lysates were collected, and the SNAI2 protein level was detected by immunoblotting. (G) Quantification of SNAI2 protein levels presented in F using ImageJ software. Data are presented as mean ± SEM. (**) P < 0.01 by Student's t-test. (H) Pulse-chase experiments in ASB13 overexpressing SUM159 cells and LM2 cells. (I) Quantification of SNAI2 degradation dynamics presented in H using ImageJ software. For LM2 cells, vector versus ASB13, P = 0.006; for SUM159 cells, vector versus ASB13, P = 2.3 × 10−5.
Many signaling pathways like TGF-β, Wnt, and EGF pathways can regulate EMT and cancer metastasis (Ciruna and Rossant 2001; Kim et al. 2002; Lu et al. 2003; Gotzmann et al. 2004; Thuault et al. 2006; Yook et al. 2006; Leong et al. 2007). We found that ASB13 level was reduced after treatment by these EMT inducers in MCF10A breast epithelial cells and EpRas breast cancer cells (Fig. 3D,E), suggesting that ASB13 might work downstream from these EMT-inducing signals to stabilize endogenous SNAI2 protein. To directly test the effect of ASB13 on endogenous SNAI2 degradation, we generated ASB13-overexpressing stable cell lines in SUM159 and LM2 cells. In both cell lines, we observed a dramatic decrease of the endogenous SNAI2 protein level and the acceleration of its degradation when ASB13 was overexpressed (Fig. 3F–I). To further test the effect of reducing endogenous ASB13 expression on SNAI2 degradation, we used siRNA to knock down ASB13 expression in SUM159 cells. SNAI2 protein was significantly stabilized after ASB13 KD in a pulse-chase assay (Supplemental Fig. S3B,C). Likewise, SNAI2 protein was significantly stabilized after ASB13 knockout (KO) by CRISPR-gRNA technology (Supplemental Fig. S3D,E).
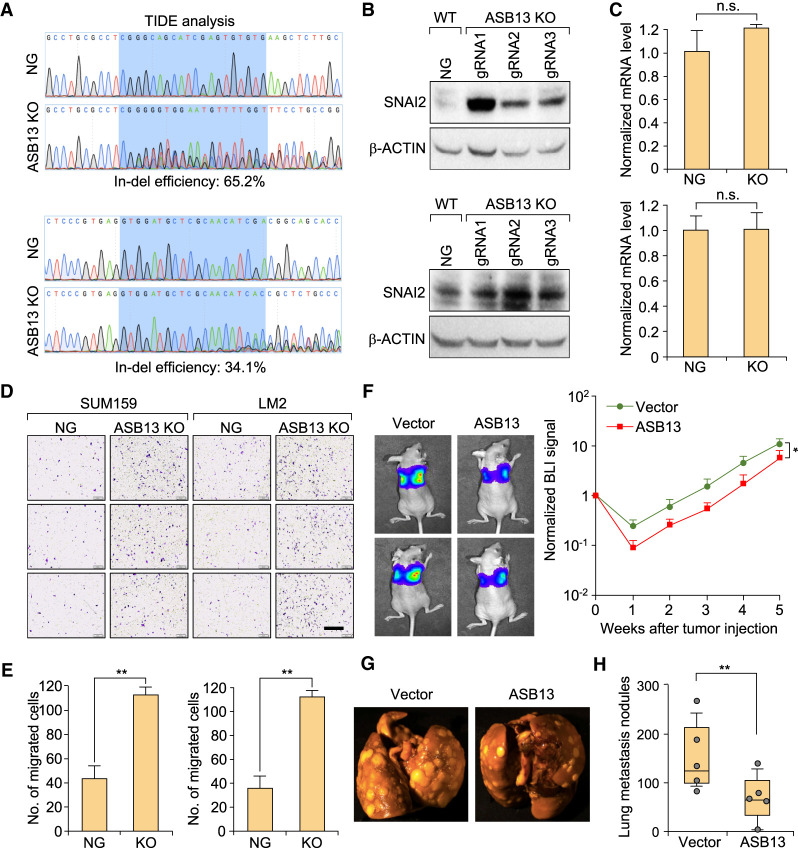
Consistently, the stable level of SNAI2 protein was found to be increased after ASB13 KO in both SUM159 and LM2 cells by three independent gRNA-mediated KOs (Fig. 4A,B; Supplemental Fig.S4A,B), while SNAI2 mRNA level was not changed after ASB13 KO (Fig. 4C). Interestingly, when we KO other previously reported E3s for SNAI2 such as BTRC1 and FBXO11 (Vernon and LaBonne 2006; Wang et al. 2009; Wu et al. 2012; Kao et al. 2014) in SUM159 or LM2 cells, the endogenous SNAI2 level was not changed significantly (Supplemental Fig. S4C,D). Taken together, our results suggest that ASB13 is a bona fide E3 ubiquitin ligase targeting endogenous SNAI2 protein for ubiquitination and degradation.
Figure 4.
ASB13 inhibits breast cancer migration and metastasis through promoting SNAI2 degradation. (A) ASB13 was KO by CRISPR–Cas9 system in SUM159 and LM2 cells. Genomic DNA was purified and the targeted locus was amplified by PCR. Representative sanger sequencing and TIDE analysis was used to confirm the ASB13 KO efficiency. (Top panel) gRNA1 in SUM159 cells. (Bottom panel) gRNA2 in LM2 cells. (NG) Nontargeting gRNA. (B) The expression level of the SNAI2 protein was determined by immunoblotting after ASB13 KO by CRISPR-Cas9 in SUM159 and LM2 cells. β-ACTIN was used as internal loading control. (C) The SNAI2 mRNA level was determined by real time qPCR in ASB13 KO cells compared with that of NG cells. (B,C, top panels) SUM159 cells (gRNA1). (Bottom panels) LM2 cells (gRNA2). Data are presented as mean ± SEM. n = 3, not significant (n.s.) by Student's t-test. (D) Representative images of Boyden chamber migration assay for NG cells and ASB13 KO cells using SUM159 and LM2 cell line. Scale bar, 400 µm. (E) Quantification of migrated cells from experiment in D. Data are presented as mean ± SEM. SUM159 on the left and LM2 at the right. n = 3. (**) P < 0.01 by Student's t-test. (F) We intravenously injected 105 vector control or ASB13-overexpressing LM2 cells into 6- to 8-wk-old female athymic nude mice to generate lung metastasis. Lung metastasis burden in mice were quantified weekly by bioluminescence imaging (BLI). (Left panel) Representative BLI images of each group at experimental endpoint. (Right panel) Normalized BLI signals of lung metastasis. Data are presented as mean ± SEM. n = 10. (*) P < 0.05 by Student's t-test. (G) Representative lung metastasis nodule images are presented from experiment in F. (H) Numbers of lung metastasis lesions of mice injected with the indicated LM2 cell lines. n = 10, (**) P < 0.01 by Mann–Whitney U-test.
ASB13 inhibits breast cancer metastasis through promoting SNAI2 degradation
SNAI2 has been reported to induce cell migration in cancer cells (Kurrey et al. 2005; Martin et al. 2005; Shih et al. 2005; Peinado et al. 2007). As ASB13 KO increased the SNAI2 protein level, we speculated that ASB13 KO may increase cell migration ability. Indeed, in both SUM159 and LM2 cells, there were significantly more migrated cells in ASB13 KO compared with control cells in the transwell migration assay (Fig. 4D,E). To further investigate the functional impact of ASB13 on SNAI2-induced breast cancer metastasis, we intravenously injected 6-wk-old female athymic nude mice with vector control or ASB13-overexpressed LM2 cells (labeled with luciferase for in vivo tracking). Overexpression of ASB13 significantly reduced lung metastasis progression as compared with that of control group by bioluminescence imaging (BLI) (Fig. 4F). There were also significantly less lung metastasis nodules in ASB13 overexpression group (Fig. 4G,H). In summary, these results suggest that ASB13 inhibits migration and metastasis by promoting the ubiquitination and degradation of the SNAI2 protein.
SNAI2 promotes cell migration through negatively regulating YAP expression
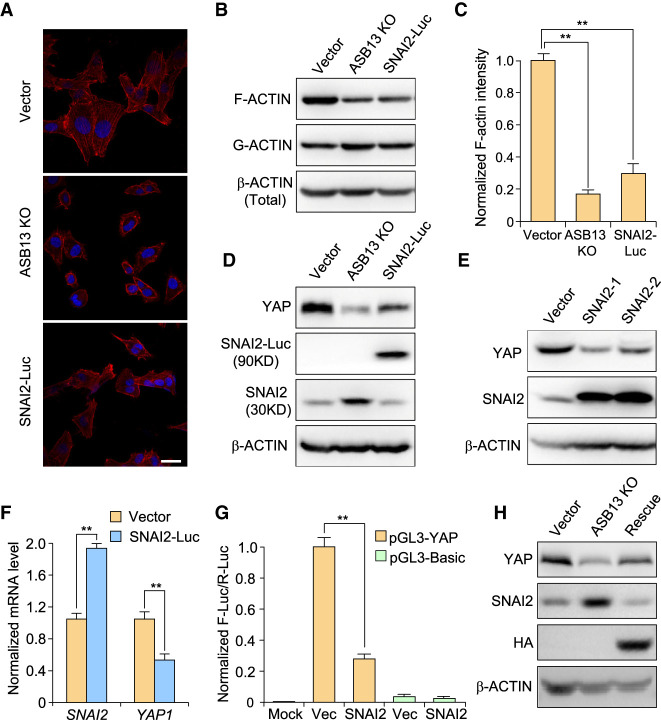
ASB13 KO or SNAI2-overexpressing cells displayed decreased cell spreading (Fig. 5A), promoting us to examine the F-actin polymerization status. Indeed, we noticed a significant decrease of F-actin fiber staining in ASB13 KO cells; this was also phenocopied in cells overexpressing SNAI2-Luc (Fig. 5A). Similarly, immunoblotting confirmed ASB13 KO or SNAI2-Luc expression significantly reduced F-actin polymerization compared with that in control cells, while there was slight increase of in G-actin level (Fig. 5B,C). These results thus suggest ASB13 KO induces F-actin depolymerization through up-regulating SNAI2 protein expression.
Figure 5.
ASB13 inhibits cell migration through SNAI2 degradation and inhibiting Hippo–YAP pathway. (A) F-actin was stained with phalloidin–rhodamine dye in WT, ASB13 KO, and SNAI2-Luc overexpression SUM159 cell lines. Scale bar, 30 µm. (B) F-actin and G-actin from the indicated SUM159 cells expressing different constructs were segmented by ultra-speed centrifugation and analyzed by immunoblotting using corresponding antibodies. (C) Quantification of endogenous F-actin levels presented in B using ImageJ software. Data are presented as mean ± SEM. n = 3. (**) P < 0.01 by Student's t-test. (D) The YAP protein level was detected by immunoblotting in ASB13 KO cells and SNAI2-Luc-overexpressing cells, and compared with the vector control of SUM159 cells. (E) Two independent SNAI2 stable cell lines were generated by transducing SNAI2-expressing lentivirus into SUM159 cells. YAP and SNAI2 expression in these cells was determined by immunoblotting. β-ACTIN was used as internal loading control. (F) The mRNA level of YAP and SNAI2 was determined by real-time qPCR in SNAI2-Luc overexpressing SUM159 cells compared with that of vector control cells. Data are presented as mean ± SEM. n = 3, (**) P < 0.01 by Student's t-test. (G) Indicated plasmids were cotransfected into 293T cells. Firefly luciferase reporter activity was measured and normalized to renilla luciferase internal control. Data are presented as mean ± SEM. n = 3; P = 0.001; (**) P < 0.01 by Student's t-test. (H) Lentivirus encoding gRNA1-resistant ASB13 was transduced into ASB13 KO SUM159 cell line to re-express ASB13. SNAI2 and YAP protein levels were determined by immunoblotting. β-ACTIN was used as loading control.
Previous studies have shown that F-actin cytoskeleton reorganization and cell detachment can be mechanical upstream signals of the Hippo–YAP pathway, which is a critical regulator for organ size, tumorigenesis, and metastasis (Gaspar and Tapon 2014; Yu et al. 2015). It is expected that decrease of cytoskeleton and F-actin would lead to decreased YAP protein level through post-translational modification and protein degradation (Aragona et al. 2013). To this end, we tested YAP protein level after ASB13 KO or SNAI2 overexpression in SUM159 cells. Indeed, YAP level was significantly reduced in ASB13 KO and SNAI2-Luc overexpressing cells (Fig. 5D; Supplemental Fig. S5A). Likewise, overexpression of SNAI2 in SUM159 cell line led to down-regulated YAP protein level (Fig. 5E; Supplemental Fig. S5B). Interestingly, the decrease of YAP protein level was also reflected at its mRNA level when we knocked out ASB13 or overexpressed SNAI2-Luc in SUM159 and LM2 cells (Fig. 5F; Supplemental Fig. S5C,D). Our results thus suggest that YAP expression is regulated at least in part at transcriptional level, likely mediated by transcriptional repression by SNAI2 protein. To further assess this hypothesis, we used a reporter system consisting of a 2-kb YAP promoter region containing five E-box sequences (Supplemental Fig. S5E). In the absence of SNAI2, pGL3-YAP displayed relative high luciferase activity. However, when SNAI2 was present, the reporter luciferase activity was strongly repressed (Fig. 5G). Furthermore, chromatin immunoprecipitation assay also confirms that SNAI2 directly binds to the YAP promoter through its first E-box region (Supplemental Fig. S5F). The reduction of YAP expression after ASB13 KO could also be rescued by exogenous expression of gRNA-resistant ASB13, suggesting that YAP was indeed a downstream gene of the ASB13-SNAI2 regulatory circuit (Fig. 5H). Since ASB13 gene expression was decreased (Fig. 3D,E) and SNAI2 was well known to be up-regulated by EMT inducing pathways, we tested whether YAP was also regulated during EMT. Indeed, in EpRas cells undergoing EMT after TGF-β treatment, there is a significant decrease in YAP protein expression level (Supplemental Fig. S5G,H).
YAP functions as a suppressor of breast cancer metastasis
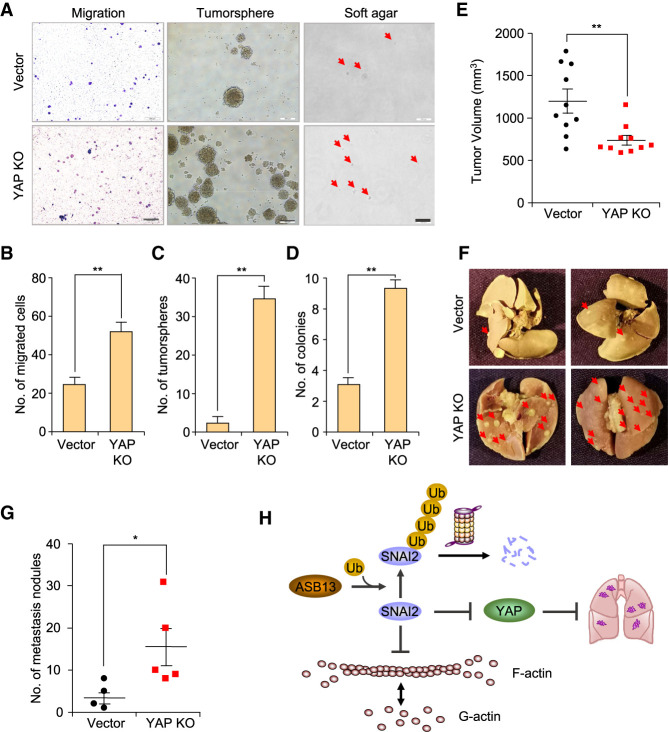
YAP is located on chromosome 11q22-23, a site of frequent loss of heterozygosity in breast cancer. Rather than being considered as an oncogene in many other cancer types, YAP was suggested to be a tumor suppressor in breast cancer with limited reports (Yuan et al. 2008). We sought to assess the functional impact of decreased YAP expression in breast cancer progression. We constructed YAP KO cells by CRISPR–Cas9 and examined its impact on cell migration (Supplemental Fig. S6A). Similar as ASB13 KO cells, YAP KO cells also increased cell migration significantly (Fig. 6A,B). Tumorsphere formation assay and soft agar colony formation assay suggested that YAP KO led to significantly more sphere formation and colony formation (Fig. 6A,C,D). We further investigated the impact of YAP loss on tumor progression and especially spontaneous metastasis in vivo. YAP KO and the vector control SUM159 cells were injected orthotopically into NSG mice, and tumor growth and lung metastasis were recorded. Interestingly, YAP KO cells formed smaller tumors than vector control cells, consistent with many previous reports of the tumor-promoting role of YAP (Fig. 6E; Lee et al. 2010; Wu et al. 2018). However, loss of YAP expression significantly increased spontaneous lung metastasis in vivo, indicating YAP might inhibit spontaneous lung metastasis (Fig. 6F,G). Finally, to test whether ASB13's tumor-suppressive function is mediated through its targeting of SNAI2, we knocked out both ASB13 and SNAI2 simultaneously in SUM159 cells (Supplemental Fig. S6B,C) and found that the migration ability of the double KO cells was dramatically decreased compared with ASB13 KO alone, and reduced to nearly baseline level (Supplemental Fig. S6D). Furthermore, the ability of tumorsphere formation was also significantly reduced in the ASB13/SNAI2 double KO cells (Supplemental Fig. S6E,F). Furthermore, when we ectopically expressed YAP in ASB13 knockout cells, the rescue of YAP expression significantly reduced the cell migration (Supplemental Fig. S6G,H). Collectively, these findings demonstrate that YAP potentially functions as a metastasis suppressor and reveal a new connection of the ASB13–SNAI2–YAP axis in the regulation of breast cancer metastasis (Fig. 6H).
Figure 6.
YAP functions as a tumor suppressor gene in breast cancer. (A) YAP KO cell lines generated in Supplemental Figure S6A were used to perform Boyden chamber migration assay, tumorsphere assay, and soft agar colony formation assay. Representative images are displayed. Scale bars, 500 µm. (B) Quantification of the number of migrated cells from Boyden chamber migration assay in A. Data are presented as mean ± SEM. n = 3, (**) P < 0.01 by Student's t-test. (C) Quantification of tumorsphere formation assay in A. Data are presented as mean ± SEM. n = 3, (**) P < 0.01 by Student's t-test. (D) Quantification of soft agar colony formation assay in A. Data are presented as mean ± SEM. n = 3, (**) P < 0.01 by Student's t-test. (E) A total of 2 × 106 SUM159 or SUM159 YAP KO cells was mixed 1:1 by volume with Matrigel (BD Biosciences) per injection. Mice were injected orthotopically in both flanks. Tumor volume was measured and calculated as volume = (length × width2)/2. Data are presented as mean ± SEM. n = 10, (**) P < 0.01 by Student's t-test. (F) Representative lung metastasis nodule images are presented from experimental end point in E. (G) Number of lung metastasis lesions from experiment in E were quantified. n = 5, (*) P < 0.05 by Student's t-test. (H) Schematic of ASB13-SNAI2-YAP regulation mechanism in breast cancer progression. SNAI2 inhibits F-actin polymerization and YAP mRNA expression to promote cancer cell migration and metastasis. SNAI2 protein is recognized and ubiquitinated by ASB13 for UPS-mediated protein degradation.
Discussion
SNAI2 has a broad effect on cancer progression, including cell migration, cell cycle regulation, and apoptosis as well as drug resistance (Wu et al. 2005; Haslehurst et al. 2012; Ye et al. 2015). In this study, we focused on identifying endogenous E3 ligases for SNAI2 degradation. Conventionally, identifying such E3 ligases is a difficult process. We previously developed a dual-luciferase-based screening method to streamline the E3-substrate pair discovery process (Zheng et al. 2014). Through the unbiased siRNA library screening and by additional confirmation steps, we discovered ASB13 as a bona fide E3 ubiquitin ligase for SNAI2. Functionally, knockdown of ASB13 stabilized the endogenous SNAI2 protein and increased the migration potential for SUM159 and LM2 cells, while overexpression of ASB13 promoted SNAI2 degradation and inhibited lung metastasis. Consistent with these results, higher ASB13 expression level correlates with longer survival time in breast cancer patients. Notably, ASB13 KO cells have much higher stable SNAI2 protein level compared with that of β-TrCP1/FBWX1 or FBXO11 KO cells, suggesting that ASB13 may be a major E3 ligase for endogenous SNAI2 in breast cancer.
Best known for its role in inducing EMT program, the mechanisms and downstream genes by which SNAI2 regulating tumorigenesis, migration, and metastasis independent of EMT are much less appreciated (Guo et al. 2012; Ye et al. 2015). We observed that there was a moderate increase in mesenchymal phenotype after ASB13 KO in SUM159 cells (data not shown); while no significant changes were observed after ASB13 overexpression in SUM159 and LM2 breast cancer cells, possibly because these cells were already locked in a relatively mesenchymal-like state. Instead, we noticed two important changes after ASB13 KO (and SNAI2 up-regulation). Firstly, F-actin polymerization was significantly decreased; and secondly, the YAP protein level was decreased after ASB13 KO or SNAI2 overexpression. The inhibition of YAP expression likely happened at the transcriptional level, as the mRNA level of YAP was also significantly reduced after ASB13 KO or SNAI2 overexpression, and a YAP promoter region containing multiple E-boxes were conferred transcriptional repression of the luciferase reporter by SNAI2. Changes in F-actin polymerization might additionally affect YAP protein level at the post-translational level as previously reported (Aragona et al. 2013).
SNAI2 possibly inhibits the transcription of YAP to execute its function in cell migration. Indeed, KO of YAP protein in SUM159 directly promoted cell migration. As a well-characterized core member of Hippo pathway, the role of YAP in cancer development is context-dependent. YAP was originally found to promote cell proliferation and transformation. Its expression was amplified in human hepatocellular carcinoma (HCC) and many other malignancies (Zender et al. 2006) and functional studies also confirmed its oncogenic role in tumorigenesis in many types of cancer (Lee et al. 2010; Wu et al. 2018). On the other hand, YAP has also been suggested to be a tumor suppressor in some cancer types. For example, YAP induces apoptosis in response to DNA damage in collaboration with p73 and promyelocytic leukemia (PML) (Strano and Blandino 2007; Lapi et al. 2008; Bertini et al. 2009). YAP is located on chromosome 11q22-23, a locus where frequent loss of heterozygosity (LOH) occurs in breast cancer. Reduced expression of YAP was significantly associated with ER and/or PR negativity in invasive carcinoma, suggesting that YAP acts as a tumor suppressor in invasive breast carcinomas (Tufail et al. 2012). However, most of the functional studies of YAP only focus on primary tumor growth but not metastasis. In our study, we noticed that ASB13 KO led to increased SNAI2 expression and decreased YAP expression, which was accompanied by increased cell migration. Depletion of YAP in breast cancer cells also led to increased cell migration. Interestingly, our in vivo primary tumor growth and spontaneous metastasis experiments revealed that YAP KO led to decreased primary tumor growth but significantly more lung metastasis. This result thus suggests that YAP pathway might functions similarly as TGF-β pathway in cancer progression, display dichotomous functions in primary tumor growth, and metastasis of breast cancer. Our findings may help explain the seemingly conflicting observation of YAP being a good prognosis marker in breast cancer (Gudmundsson et al. 1995; Winqvist et al. 1995; Tufail et al. 2012) despite its well-established function in promoting growth (Meng et al. 2016). However, the detailed molecular mechanisms mediating the metastasis-suppressing role of YAP in breast cancer need further investigation. Nevertheless, our study calls for caution and careful design in targeting YAP pathway for cancer therapy in breast cancer.
Materials and methods
Animal studies
All procedures involving mice and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Princeton University or Tsinghua University. For experimental lung metastasis assay, female athymic nude mice at 6–8 wk old were anaesthetized and tail vein-injected with 1×105 indicated tumor cells. Tumor cells were suspended in 100 µL of cold PBS before injection. Successful injections were confirmed by BLI imaging immediately after injection and lung metastasis progression was monitored by weekly BLI. By the experimental endpoint, mice were euthanized and lung tissues were dissected out and fixed for metastatic nodule counting. For primary tumor growth and spontaneous lung metastasis experiments using SUM159 cells in NSG mice, five female NSG mice aged 6–8 wk were used in each group. A total of 2 × 106 vector control cells or YAP KO cells was mixed 1:1 by volume with Matrigel (BD Biosciences) per injection. Each mouse was injected orthotopically in both flanks. Primary tumor growth was measured and tumor volume was calculated as volume = (length × width2)/2. At the experimental endpoint, mice were sacrificed and lung samples were removed and fixed for counting of lung metastasis nodules.
Actin segmentation by ultracentrifugation
Actin segmentation assay was performed following the protocol described in previous studies (Qiao et al. 2017). Cells were lysed directly in dishes using actin stabilization buffer (50 mM PIPES at pH 6.9, 50 mM NaCl, 5 mM MgCl2, 5 mM EGTA, 2 mM ATP, 5% glycerol, 0.1% Nonidet P-40, 0.1% Triton X-100, 0.1% Tween 20, 0.1% β-mercaptoethanol, 1:100 protease inhibitor mixture, 1:100 phosphatase inhibitor mixture) for 10 min at 37°C and collected into Eppendorf tubes by scraping, followed by centrifugation at 300g at room temperature to remove insoluble particles. An aliquot of the diluted cell lysates (10% of the volume) was kept separately as “total protein inputs,” to be analyzed by immunoblotting. The F-actin and G-actin pools of the diluted cell lysates were separated by ultracentrifugation at 100,000g for 1 h at 37°C. After centrifugation, F-actin precipitated at the bottom of the tube while G-actin remains in the supernatant. The supernatant containing the G-actin pool was removed to a fresh tube, while the pellet containing F-actin was resuspended in cold distilled water with 1 mM cytochalasin D (Sigma-Aldrich) and kept on ice for 45 min to dissolve F-actin. F-actin and G-actin fragments were used for immunoblotting by antibody against β-actin.
Anchorage-independent growth
Cells (5 × 103) were added to 1.5 mL of growth medium with 0.3% agar and layered onto 1.5 mL of 0.5% agar base in six-well plates. Cells were fed with 500 µL of medium every 3 d for 3 wk, after which colonies were photographed and counted. Assays were conducted in triplicate in three independent experiments.
Cell culture
HEK293T, EpRas, and LM2 cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gemini), penicillin/streptomycin (Corning). SUM159 cells were cultured in DMEM/F12 (1:1) medium added with 5% FBS, 5 µg/mL insulin, and 10 ng/mL EGF. MCF10A was cultured in DMEM/F12 (1:1) with 5% FBS, 20 ng/mL EGF, 0.5 µg/mL hydrocortisone and 0.1 µg/mL cholera toxin. The medium for H29 (packaged for retrovirus) added 0.5 µg/mL doxycline, 2 µg/mL puromycin, and 0.25 g/mL G418 based on the medium of HEK293T.
Chromatin immunoprecipitation assay
Four dishes of confluent SUM159 cells (10-cm dish) were fixed with 0.9% formaldehyde for 10 min and quenched in 0.125 M glycine for 5 min at 37°C. After centrifugation, cell pellets were resuspended in lysis buffer (1% SDS, 50 mM Tris⋅HCl at pH 8.1, 10 mM EDTA) with freshly prepared 1 mM DTT, 1 mM PMSF, and protease inhibitor. Chromatin DNA was fragmented by sonication to an average length of 500–1000 bp. A small aliquot (20 µL) was taken to reverse crosslinks and be saved as input control. Lysates were then incubated with 5 µg of control IgG antibody (Santa Cruz Biotechnology sc-166902X) or SNAI2 antibody (Santa Cruz Biotechnology sc-166902X) overnight at 4°C and then for 1 h with 30 µL of Dynabead protein G (Invitrogen). Beads were washed with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris at pH 7.9, 500 mM NaCl) for seven times. Samples were eluted by IP elution buffer (1%SDS, 50 mM Tris at pH 8.1, 1 mM EDTA, 0.1 M NaHCO3, 0.25 M NaCl and Proteinase K) twice for 20 min at 65°C. Cross-linking was reversed overnight at 65°C. DNA fragments were purified using Isopropyl alcohol precipitation. qPCRs were performed using 1 µL of DNA as template and the promoter fragment enrichment was calculated relative to input DNA. The primer sequences used were P1 (−2493 to −2257), ACAGAACATAGTCACTCCAAAG (forward) and CAGCCTGGGCAACACAGCGAGA (reverse); P2 (−2280 to −1950), TCAAGTGATTCTCATGCCTCAGC (forward) and ACTATGCATTAAAAACTTTACCT (reverse); P3 (−1006 to −791), TGTACATCAGGTGCCTGGGATG (forward) and AGTACTCGAATTTGCAATAGT (reverse); P4 (−786 to −579), AAGTTGATGTGATTTCAC (forward) and CCGGCATTGATGTTAATAAA (reverse); and P5 (−498 to −283), ACGCGAGGAGGAGGAGGTGGGT (forward) and CGCGAACCGCAAACGATGGG (reverse).
Clinical data set analysis
Gene expression data from the Wang et al. (2005) data set (GSE2034) was used for ASB13 expression level analysis in subtypes of breast cancer patients. To analyze prognosis value of ASB13 gene expression in breast cancer patients, patients were stratified by expression of ASB13. The association of ASB13 expression with overall patient survival was presented as Kaplan–Meier plots and tested for significance using online KMplot database (http://www.kmplot.com) (Györffy et al. 2010).
Coimmunoprecipitation assay
The SNAI2-FLAG (wild-type or mutant) plasmids were cotransfected with HA-tagged ASB13 or other E3 ligase candidate plasmids into HEK293T cells in six-well plates using PEI with the ratio 1:3. Two days after transfection, cells were treated with 10 µM MG132 for 4 h. Four-hundred microliters of cell lysates were then collected and immunoprecipitated with 20 µL of magnetic anti-FLAG-beads (Sigma) overnight at 4°C. After extensive washing (five times) with lysis buffer, the beads were spun down and resuspended with 50 µL of loading buffer. After boiling for 10 min, protein samples were run on SDS-PAGE along with 10% input sample and transferred to PVDF membrane to be detected with appropriate antibodies as described in each figure.
CHX pulse-chase assay
Cells were seeded on 12-well plates at a density of 2 × 105 cells per well. After culturing overnight, the cells were transfected with plasmids as desired. Two days after transfection, cells were treated with 50 µg/mL CHX dissolved in DMSO, total protein lysates were collected at different time points and subjected to immunoblotting for SNAI2 and related protein loading controls.
Generation of stable cell lines
Stable overexpression of all genes was achieved using the pLEX lentiviral plasmids or pMSCV retroviral plasmids. Lentiviruses were packaged in HEK293T cells, while retroviruses were packaged by using H29 cell line. After 48 h, viruses were collected, filtered, and used to infect target cells in the presence of 8 µg/mL polybrene. The infected cells were selected with puromycin or hygromycin for 5–7 d.
Immunofluorescence (IF) staining
Cells were seeded onto sterile glass coverslip for experiments. Cells were washed with cold PBS, fixed with 4% formaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 5 min, blocked with 5% BSA in PBS for 1 h, and incubated with proper primary antibodies overnight at 4°C and then with secondary antibodies for 1 h at 4°C in the dark. The antibodies used in the IF staining were anti-HA (1:100; Roche 11867423001), anti-SNAI2 (1:100; Santa Cruz Biotechnology A1317). Secondary antibodies used in the experiments were goat-anti-mouse, 488 and rabbit anti-rat, TRINC (Thermo).
Luciferase reporter assay-based siRNA library screening
SUM1315-SNAI2-Luc/R-Luc cells (1 × 105) were seeded in 24-well plates. On the following day, 10 ng/mL total pooled siRNAs (three siRNAs/gene) were transfected using RNAiMAX reagent (Life Technology) into cells. Cell lysates were collected 2 d after transfection and assayed for luciferase activity using the Glomax 96 microplate luminometer (Promega). The remaining cell lysates were used for immunoblotting to confirm SNAIL-Luc protein level changes indicated by luciferase assay.
YAP promoter luciferase assay
For the luciferase reporter assay, 2 × 105 293T cells were seeded in 12-well plates. pGL3-YAP or pGL3-Basic, pLEX-SNAI2 or pLEX-MCS, and pCMV-Renilla plasmids were cotransfected. Two days after transfection, cells were lysed and luciferase activity were assayed using a multiple microplate luminometer (EnVision). All luciferase activities were normalized to pGL3-YAP promoter activity, Renilla-luciferase activity was used as internal control. Around 2 kb YAP promoter region (−2493 to −436 bp relative to the transcriptional start site) was cloned from SUM159 genome and inserted into pGL3-Basic. The primers used for cloning were CGCGGTACCacagaacatagtcactcc (forward primer, KpnI site) and CCGCTCGAGaaagggccaaggccgaaa (reverse primer, XhoI site).
Tumorsphere formation assay
Cells to be used for generation of tumorspheres are trypsinized to generate a single cell suspension, plated to a final volume of 1 mL of serum free medium (DMEM/F12 [1:1] with B27 [Invitrogen], 20 ng/mL bFGF [Invitrogen], 20 ng/mL EGF [Invitrogen], penicillin/streptomycin [Corning]) in one well of a 12-well pretreated low attachment plate (coated with poly-HEMA, diluted in 95% ethanol and dried out before usage). A total of 5 × 103 cells was used per well. Fresh medium was added against the sides of the well every 3 d. The tumorspheres were cultured for 7–10 d before counting.
Molecular cloning and plasmids
The pLEX plasmids containing cDNAs for SNAI2-Luc, wild-type SNAI2, and E3 ligase candidates including ASB13 were generated by PCR from SUM159 cDNA with a FLAG tag or HA tag at their C-terminal and inserted into pLEX plasmid using BamH1–Xho1 restriction enzymes. ASB13-ΔSOCS-HA mutant was generated by PCR to delete amino acids 229–278 from full-length ASB13 DNA construct. HA-Ubiquitin expressing construct was generated as described (Zheng et al. 2014). ASB13 gRNA1 resistant plasmid was generated by two-fragment PCR to mutate the gRNA1 targeted DNA sequence and inserted into pMSCV-hygro plasmids using BgII–Xho1 restriction enzymes. The pGL3-YAP promoter plasmid containing YAP promoter region was generated by PCR from SUM159 genome sequence. Primer sequences are available upon request. ASB13, SNAI2, and YAP knockout plasmids are constructed based on Lenti-CRISPR V2. The gRNA sequences used were as follows: hASB13(g1) (forward: 5′-CGGGCAGCATCGAGTGTGTG-3′; reverse: 5′-CACACACTCGATGCTGCCCG-3′), hASB13(g2) (forward: 5′-GTGGATGCTCGCAACATCGA-3′; reverse: 5′-TCGATGTTGCGAGCATCCAC-3′), hASB13(g3) (forward: 5′-TATTGACGTCGGGGCCAATC-3′; reverse: 5′-GATTGGCCCCGACGTCAATA-3′), hSNAI2 (forward: 5′-ATCTCTGGTTGTGGTATGAC-3′; reverse: 5′-GTCATACCACAACCAGAGAT-3′), hYAP (forward: 5′-CCAAGGCTTGACCCTCGTTT-3′; reverse: 5′-AAACGAGGGTCAAGCCTTGG-3′), and NG (forward: 5′-ACGGAGGCTAAGCGTCGCAA-3′; reverse: 5′-TTGCGACGCTTAGCCTCCGT-3′).
Reverse transcription and qPCR analysis
Total RNAs were isolated from cells using TaKaRa kit following the manufacturer's instructions. RNAs were reverse-transcribed into cDNAs by using Reverse Transcription kit (Promega). Real-time quantitative PCR (qPCR) was performed using the SYBR Green PCR mix (DSBIO) with Bio-Rad CFX96 PCR machine. The gene-specific primer sets were used at a final concentration of 0.5 µM. All real-time RT-PCR assays were performed in triplicate in at least two independent experiments. Relative expression values of each target gene were normalized to GAPDH mRNA level. Sequences used were as follows: hGAPDH (forward: 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse: 5′-GAAGATGGTGATGGGATTTC-3′), hSNAI2 (forward: 5′-CTGGGCGCCCTGAACATGCAT-3′; reverse: 5′-GCTTCTCCCCCGTGTGAGTTCTA-3′), hASB13 (forward: 5′-CATGAGCGGGAGTTCCGAAT-3′; reverse: 5′-GACACAGTCCAGATGCTCCC-3′), and hYAP (forward: 5′-TGACCCTCGTTTTGCCATGA-3′; reverse: 5′-GTTGCTGCTGGTTGGAGTTG-3′).
TIDE analysis
Genomic DNA was purified from the indicated population, the targeted loci were amplified by PCR. The primers designed for PCR are ∼150 bp upstream of and 250 bp downstream from the targeted loci. KO efficiency was analyzed by the indel efficiency on the at https://tide.deskgen.com. Sequences used were as follows: hASB13(g1)3 (forward: 5′-CAAAGCAAACCGGTGCTTCTAA-3′; reverse: 5′-GCTTAGAATCTCTGCAGCTGAG-3′), hASB13(g2) (forward: 5′-ACTTCTCTACTCAGGAGGCCGTCG-3′; reverse: 5′-CCGAGTGAACCCGAAACGATGT-3′), and hSNAI2 (forward: 5′-TAAGAAAATCACGTGTGTGTTGCTA-3′; reverse: 5′-TGAAACTTTTCAGCTTCAATGGCA-3′).
Boyden chamber migration assay
Migration assays were performed in 12-well plate cell culture insert (8 μm). Cells were seeded at 1 × 104 cells per well in serum-free medium. The cells were induced to migrate toward bottom medium containing 10% FBS for 24 h in cell culture incubator. Migrated cell at the bottom chamber counted under a light microscope after staining with crystal violet (0.02% in PBS) staining.
Ubiquitination assay
Ubiquitination assay was done following a denaturing IP Protocal. HEK293T cells were transfected with HA-Ub plasmid, SNAI2-FLAG plasmid, and ASB13-HA or ΔSOCS mutant. Two days after transfection, cells were treated with 20 µM MG132 for 6 h to block proteasomal degradation of SNAI2 before being lysed with denature lysis buffer (denatured IP buffer: 50 mM Tris-Cl at pH 6.8, 1.5% SDS). Protein samples were collected by scraping hard and then boiling for 15 min. Seventy microliters of each sample was added to 1.2 mL of EBC/BSA buffer (50 mM Tris-Cl at pH 6.8, 180 mM NaCl, 0.5% NP40, 0.5% BSA) for IP with magnetic anti-FLAG M2 beads (Sigma) to specifically pull down FLAG-SNAI2 protein. Pull-down samples were subjected to immunoblotting with anti-HA (ubiquitin) to visualize polyubiquitinated SNAI2 protein bands.
Immunoblot analysis
For immunoblot analysis, whole cell lysate samples were collected using cell lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% NP-40). Lysates were heated to denature the proteins, loaded to SDS-PAGE gel for electrophoresis and subsequently transferred to PVDF membranes (Millipore). Membranes were blocked in 5% milk for 1 h at room temperature prior to overnight incubation with primary antibody. Primary antibodies used were anti-β-ACTIN (1:5000; Abcam ab6276), anti-SNAI2 (1:1000; sc-166476), anti-HA (1:5000; Roche 11867423001), anti-YAP1 (1:1000; Cell Signaling Biotechnology). Membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse, rabbit, or rat secondary antibody (1:10000; EASYbio) for 1 h at room temperature and chemiluminescence signals were detected by ECL substrate at ratio 1:1 (Thermo) on Champchemi digital image acquisition machine (Sagecreation). Image quantifications were performed using ImageJ (NIH).
Statistical analysis
Results were reported as mean ± SD (standard deviation) or mean ± SEM (standard error of the mean), as indicated in the figure legend. Statistical comparisons were performed using unpaired two-sided Student's t-test with unequal variance assumption. Statistical comparison for LM2 lung metastasis assay was performed using Mann–Whitney U-test. All the experiments with representative images (including Western blot and immunofluorescence) were repeated at least twice and representative images are shown.
Supplementary Material
Acknowledgments
We thank J. Meng, L. Xuan, and other laboratory members for the helpful discussions, and M. Yuan, J. Chen, X. Zhan, and T. Zhang for technical assistance. This research was supported by the National Science Foundation of China (81772981 and 81972462 to H.Z.), Rutgers Cancer Institute of New Jersey (RCINJ) Research Development Award and the Brewster Foundation (to Y.K.), postdoctoral fellowships from Susan G. Komen (PDF17332118 to M.S. and KG111164 to H.Z.), and from New Jersey Commission on Cancer Research (DFHS15PPCO21 to M.S.). This research was also supported by the Flow Cytometry Shared Resources of the RCINJ (P30CA072720).
Author contributions: H.Z., W.L., and M.S. performed the siRNA screen and ASB13 identification. H.F. designed and performed studies to elucidated mechanism of ASB13-SNAI2-YAP regulation and biological impact, analyzed data, and wrote the manuscript. X.W. provided technical help for experiments and helped data analysis. Y.K. and H.Z. developed the concept, designed the experiment, supervised the overall study, analyzed data, wrote and revised the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339796.120.
References
- Andresen CA, Smedegaard S, Sylvestersen KB, Svensson C, Iglesias-Gato D, Cazzamali G, Nielsen TK, Nielsen ML, Flores-Morales A. 2014. Protein interaction screening for the ankyrin repeats and suppressor of cytokine signaling (SOCS) box (ASB) family identify Asb11 as a novel endoplasmic reticulum resident ubiquitin ligase. J Biol Chem 289: 2043–2054. 10.1074/jbc.M113.534602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Bertini E, Oka T, Sudol M, Strano S, Blandino G. 2009. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle 8: 49–57. 10.4161/cc.8.1.7259 [DOI] [PubMed] [Google Scholar]
- Blenk S, Engelmann J, Weniger M, Schultz J, Dittrich M, Rosenwald A, Müller-Hermelink HK, Müller T, Dandekar T. 2007. Germinal center B cell-like (GCB) and activated B cell-like (ABC) type of diffuse large B cell lymphoma (DLBCL): analysis of molecular predictors, signatures, cell cycle state and patient survival. Cancer Inform 3: 399–420. 10.1177/117693510700300004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Negrini M, Baffa R, Gillum DR, Rosenberg AL, Schwartz GF, Croce CM. 1994. Loss of heterozygosity at 11q22-q23 in breast cancer. Cancer Res 54: 6270–6274. [PubMed] [Google Scholar]
- Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukačišin M, Romano RA, Smalley K, Liu S, Yang Q, Ibrahim T, et al. 2012. Elf5 inhibits the epithelial–mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol 14: 1212–1222. 10.1038/ncb2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Murphy LC, Hu P. 2018. Recurrent copy number alterations in young women with breast cancer. Oncotarget 9: 11541–11558. 10.18632/oncotarget.24336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. 2001. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell 1: 37–49. 10.1016/S1534-5807(01)00017-X [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. 1999. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer 81: 1328–1334. 10.1038/sj.bjc.6695007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Tapon N. 2014. Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol 31: 74–83. 10.1016/j.ceb.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Gotzmann J, Mikula M, Eger A, Schulte-Hermann R, Foisner R, Beug H, Mikulits W. 2004. Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat Res 566: 9–20. 10.1016/S1383-5742(03)00033-4 [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Barkardottir RB, Eiriksdottir G, Baldursson T, Arason A, Egilsson V, Ingvarsson S. 1995. Loss of heterozygosity at chromosome 11 in breast cancer: association of prognostic factors with genetic alterations. Br J Cancer 72: 696–701. 10.1038/bjc.1995.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, et al. 2012. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148: 1015–1028. 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123: 725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER. 2002. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res 62: 1613–1618. [PubMed] [Google Scholar]
- Hampton GM, Mannermaa A, Winqvist R, Alavaikko M, Blanco G, Taskinen PJ, Kiviniemi H, Newsham I, Cavenee WK, Evans GA. 1994. Loss of heterozygosity in sporadic human breast carcinoma: a common region between 11q22 and 11q23.3. Cancer Res 54: 4586–4589. [PubMed] [Google Scholar]
- Haslehurst AM, Koti M, Dharsee M, Nuin P, Evans K, Geraci J, Childs T, Chen J, Li J, Weberpals J, et al. 2012. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 12: 91 10.1186/1471-2407-12-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SH, Wang WL, Chen CY, Chang YL, Wu YY, Wang YT, Wang SP, Nesvizhskii AI, Chen YJ, Hong TM, et al. 2014. GSK3β controls epithelial–mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene 33: 3172–3182. 10.1038/onc.2013.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lu Z, Hay ED. 2002. Direct evidence for a role of β-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol Int 26: 463–476. 10.1006/cbir.2002.0901 [DOI] [PubMed] [Google Scholar]
- Kurrey NK, Amit K, Bapat SA. 2005. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol Oncol 97: 155–165. 10.1016/j.ygyno.2004.12.043 [DOI] [PubMed] [Google Scholar]
- Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, Pandolfi PP, Givol D, Strano S, Lu X, et al. 2008. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell 32: 803–814. 10.1016/j.molcel.2008.11.019 [DOI] [PubMed] [Google Scholar]
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, et al. 2010. The Hippo–Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci 107: 8248–8253. 10.1073/pnas.0912203107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. 2007. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med 204: 2935–2948. 10.1084/jem.20071082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Liu Y, Gao H, Meng F, Yang S, Lou G. 2013. Clinical significance of yes-associated protein overexpression in cervical carcinoma: the differential effects based on histotypes. Int J Gynecol Cancer 23: 735–742. 10.1097/IGC.0b013e31828c8619 [DOI] [PubMed] [Google Scholar]
- Liu P, Verhaar AP, Peppelenbosch MP. 2019. Signaling size: ankyrin and SOCS box-containing ASB E3 ligases in action. Trends Biochem Sci 44: 64–74. 10.1016/j.tibs.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Lu Z, Ghosh S, Wang Z, Hunter T. 2003. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell 4: 499–515. 10.1016/S1535-6108(03)00304-0 [DOI] [PubMed] [Google Scholar]
- Martin TA, Goyal A, Watkins G, Jiang WG. 2005. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol 12: 488–496. 10.1245/ASO.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. 2016. Mechanisms of Hippo pathway regulation. Genes Dev 30: 1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. 2005. Genes that mediate breast cancer metastasis to lung. Nature 436: 518–524. 10.1038/nature03799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal MK, Singh K, Misra S, Chaudhuri G. 2011. SLUG-induced elevation of D1 cyclin in breast cancer cells through the inhibition of its ubiquitination. J Biol Chem 286: 469–479. 10.1074/jbc.M110.164384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. 2002. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 3: 155–166. 10.1038/nrm757 [DOI] [PubMed] [Google Scholar]
- Nieto MA, Cano A. 2012. The epithelial–mesenchymal transition under control: global programs to regulate epithelial plasticity. Semin Cancer Biol 22: 361–368. 10.1016/j.semcancer.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428. 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Chen J, Lim YB, Finch-Edmondson ML, Seshachalam VP, Qin L, Jiang T, Low BC, Singh H, Lim CT, et al. 2017. YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep 19: 1495–1502. 10.1016/j.celrep.2017.04.075 [DOI] [PubMed] [Google Scholar]
- Rakha EA, Martin S, Lee AH, Morgan D, Pharoah PD, Hodi Z, Macmillan D, Ellis IO. 2012. The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer 118: 3670–3680. 10.1002/cncr.26711 [DOI] [PubMed] [Google Scholar]
- Shih JY, Tsai MF, Chang TH, Chang YL, Yuan A, Yu CJ, Lin SB, Liou GY, Lee ML, Chen JJ, et al. 2005. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res 11: 8070–8078. 10.1158/1078-0432.CCR-05-0687 [DOI] [PubMed] [Google Scholar]
- Song M, Cheong J-H, Kim H, Noh SH, Kim H. 2012. Nuclear expression of Yes-associated protein 1 correlates with poor prognosis in intestinal type gastric cancer. Anticancer Res 32: 3827–3834. [PubMed] [Google Scholar]
- Strano S, Blandino G. 2007. YAP1 meets tumor suppression. Mol Cell 27: 863–864. 10.1016/j.molcel.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Su LL, Ma WX, Yuan JF, Shao Y, Xiao W, Jiang SJ. 2012. Expression of Yes-associated protein in non-small cell lung cancer and its relationship with clinical pathological factors. Chin Med J (Engl) 125: 4003–4008. [PubMed] [Google Scholar]
- Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. 2006. Transforming growth factor-β employs HMGA2 to elicit epithelial–mesenchymal transition. J Cell Biol 174: 175–183. 10.1083/jcb.200512110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Strickland JE, Lee AS, Bromley L, Evans MF, Morton J, McGee JO. 1995. Loss of heterozygosity on chromosome 11 q in breast cancer. J Clin Pathol 48: 424–428. 10.1136/jcp.48.5.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufail R, Jorda M, Zhao W, Reis I, Nawaz Z. 2012. Loss of Yes-associated protein (YAP) expression is associated with estrogen and progesterone receptors negativity in invasive breast carcinomas. Breast Cancer Res Treat 131: 743–750. 10.1007/s10549-011-1435-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, Aikou T. 2005. Slug expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res 11: 1174–1180. [PubMed] [Google Scholar]
- Uchikado Y, Okumura H, Ishigami S, Setoyama T, Matsumoto M, Owaki T, Kita Y, Natsugoe S. 2011. Increased Slug and decreased E-cadherin expression is related to poor prognosis in patients with gastric cancer. Gastric Cancer 14: 41–49. 10.1007/s10120-011-0004-x [DOI] [PubMed] [Google Scholar]
- Uygur B, Abramo K, Leikina E, Vary C, Liaw L, Wu WS. 2015. SLUG is a direct transcriptional repressor of PTEN tumor suppressor. Prostate 75: 907–916. 10.1002/pros.22974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. 2011. Tumor metastasis: molecular insights and evolving paradigms. Cell 147: 275–292. 10.1016/j.cell.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. 2006. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development 133: 3359–3370. 10.1242/dev.02504 [DOI] [PubMed] [Google Scholar]
- Wan L, Pantel K, Kang Y. 2013. Tumor metastasis: moving new biological insights into the clinic. Nat Med 19: 1450–1464. 10.1038/nm.3391 [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365: 671–679. 10.1016/S0140-6736(05)17947-1 [DOI] [PubMed] [Google Scholar]
- Wang S-P, Wang W-L, Chang Y-L, Wu C-T, Chao Y-C, Kao S-H, Yuan A, Lin C-W, Yang S-C, Chan W-K, et al. 2009. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 11: 914–914. 10.1038/ncb0709-914 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. 2010. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci 101: 1279–1285. 10.1111/j.1349-7006.2010.01511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winqvist R, Hampton GM, Mannermaa A, Blanco G, Alavaikko M, Kiviniemi H, Taskinen PJ, Evans GA, Wright FA, Newsham I et al. 1995. Loss of heterozygosity for chromosome 11 in primary human breast tumors is associated with poor survival after metastasis. Cancer Res 55: 2660–2664. [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123: 641–653. 10.1016/j.cell.2005.09.029 [DOI] [PubMed] [Google Scholar]
- Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. 2012. Canonical Wnt signaling regulates Slug activity and links epithelial–mesenchymal transition with epigenetic breast cancer 1, early onset (BRCA1) repression. Proc Natl Acad Sci U S A 109: 16654–16659. 10.1073/pnas.1205822109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LMN, Deng Y, Wang J, Zhao C, Wang J, Rao R, Xu L, Zhou W, Choi K, Rizvi TA, et al. 2018. Programming of Schwann cells by Lats1/2–TAZ/YAP signaling drives malignant peripheral nerve sheath tumorigenesis. Cancer Cell 33: 292–308.e7. 10.1016/j.ccell.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MZ, Yao T-J, Lee NPY, Ng IOL, Chan Y-T, Zender L, Lowe SW, Poon RTP, Luk JM. 2009. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer 115: 4576–4585. 10.1002/cncr.24495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA. 2015. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525: 256–260. 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, et al. 2006. A Wnt–Axin2–GSK3β cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 8: 1398–1406. 10.1038/ncb1508 [DOI] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo–YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA, et al. 2008. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ 15: 1752–1759. 10.1038/cdd.2008.108 [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. 2006. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125: 1253–1267. 10.1016/j.cell.2006.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kang Y. 2014. Multilayer control of the EMT master regulators. Oncogene 33: 1755–1763. 10.1038/onc.2013.128 [DOI] [PubMed] [Google Scholar]
- Zheng H, Shen M, Zha YL, Li W, Wei Y, Blanco MA, Ren G, Zhou T, Storz P, Wang HY, et al. 2014. PKD1 phosphorylation-dependent degradation of SNAIL by SCF–FBXO11 regulates epithelial–mesenchymal transition and metastasis. Cancer Cell 26: 358–373. 10.1016/j.ccr.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.