Abstract
Identification of progenitor cells that generate differentiated cell types during development, regeneration, and disease states is central to understanding the mechanisms governing such transitions. For more than a century, different lineage-tracing strategies have been developed, which helped disentangle the complex relationship between progenitor cells and their progenies. In this review, we discuss how lineage-tracing analyses have evolved alongside technological advances, and how this approach has contributed to the identification of progenitor cells in different contexts of cell differentiation. We also highlight a few examples in which lineage-tracing experiments have been instrumental for resolving long-standing debates and for identifying unexpected cellular origins. This discussion emphasizes how this century-old quest to delineate cellular lineage relationships is still active, and new discoveries are being made with the development of newer methodologies.
Development of multicellular organisms from a single-cell zygote is a complex process involving highly synchronized events of cellular proliferation, differentiation, and patterning. For development to proceed in a regulated manner, mechanisms exist that properly regulate the generation of cell progenies from specific progenitor populations. These progenitor cells traverse a defined developmental trajectory involving progressive fate decision steps, each refining their identity, until they reach their functional end state (Weinreb et al. 2018). Progenitor cells also play important roles in the mature animal, by serving as reservoirs of new cell types in contexts of normal tissue turnover, damage, and regeneration (Pellettieri and Alvarado 2007). Concordantly, a major endeavor of developmental biology has been to identify the progenitors from which different cell types originate, thereby establishing lineage relationships between different cells. Over the last century, a large number of techniques and model systems have been developed for efficient and reliable “lineage tracing”—the identification of progenies generated from a progenitor cell type (reviewed by Kretzschmar and Watt 2012; Woodworth et al. 2017).
The importance of lineage tracing lies not only in revealing the identity of progenitor cells, but is also central to the quest for understanding how the different fate decisions are made and how different cell types can be produced from the same progenitors. The identification of cellular origins for differentiated cell types also have important functional implications, especially in cases where seemingly identical cell types are derived from ontologically distinct subsets. A growing number of studies have described how similar cell types, derived from ontologically distinct subsets, can show different properties (Topouzis and Majesky 1996; Cheung et al. 2012; Plein et al. 2018; Sande-Melón et al. 2019). Indeed, the importance of “lineage history” of a cell is underlined by the recent proposal that a full characterization of cellular identity requires understanding of three essential aspects: lineage, cellular function, and state (Morris 2019).
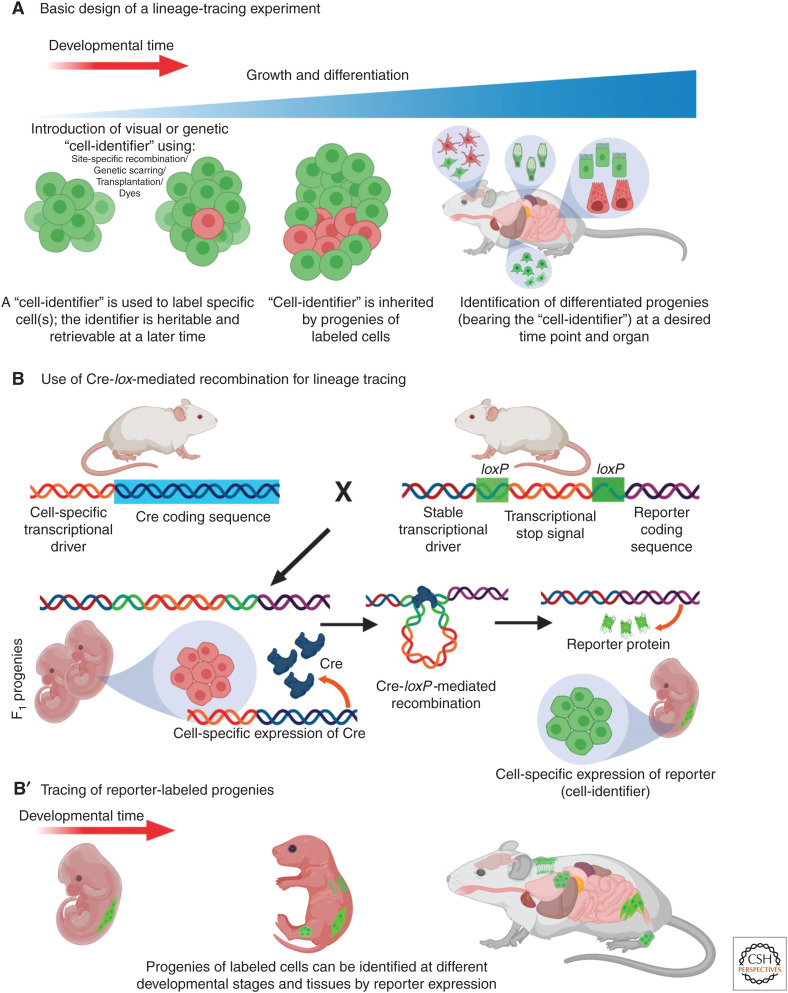
Identification of progenitors of differentiated cells is not a straightforward process. Because differentiation can make progenies very distinct from their progenitors, both in morphology and molecular composition, clonal relationships between cell types can only be discovered through well-strategized techniques. The basis of lineage-tracing experiments consists in selecting a reliable and specific identifier for a cell or a cell population that is stable over the experimental time and is inherited by all its progenies through the subsequent rounds of cell division and differentiation (Fig. 1A). At the experimental end point, the identifier should be readily detected in the cells that inherited it (the progenies), thus allowing to establish lineage relationships between them and the cells bearing the identifier at the beginning of the experiment. Typically, visual identifiers are used such as dyes or fluorescent proteins (FPs), but other identifiers like genetic barcodes, or heritable change in the genome have also been successfully used.
Figure 1.
Lineage tracing and usage of Cre-lox-mediated site-specific recombination. (A) Basic outline of lineage tracing. “Cell-identifier” of various kinds are introduced in selected cells (red), that are inherited by the daughter cells. “Cell-identifier”-tagged cells (red) at later stages of life are identified as the progenies of these initially tagged cell(s). (B,B′) Cre-lox recombination system is commonly used for lineage tracing. Cell-specific transcriptional drivers, inducing expression of Cre recombinase allows recombination between loxP sites, which in turn results in expression of a reporter gene. The progenies of these cells (in green), also expressing the reporter, can be identified at different stages of life.
Herein, we review different lineage-tracing techniques and discuss how they help uncover novel progeny–progenitor relationships among various cell types and in different contexts. We focus primarily on model systems that were most influential in establishing vertebrate cell lineages, including mouse, zebrafish, salamander, and quail-chick chimeras.
TOOLS FOR LINEAGE TRACING
A Historical Perspective
As advancements were made on light microscopy, the 17th century saw an increased interest in microscopic observation and description of living specimens. As a result, the cell theory, recognizing that living organisms are made of cells, and that cells derive from preexisting cells (for reviews, see Wolpert 1995; Mazzarello 1999), was formally formulated in 1838–1839. This new insight led to a fundamental question—“If all cells come from preexisting cells, then how is the whole array of spectacular cell diversity within an organism generated?” The quest for understanding the mechanisms underlying cell-type diversification, has led to an interesting path of discoveries that remains active to this day.
Since the beginning, one of the biggest challenges of lineage-tracing experiments has been finding ways to reliably track specific cells and their progenies within the entire population. Some of the earliest work on lineage tracing took advantage of invertebrate animal models, such as leech (Whitman 1887) and sea squirt (Conklin 1905), for direct microscopic observations of early development of the fertilized egg. Edwin Conklin, for instance, used a natural label—an orange-yellow spot—in the cytoplasm of the one-cell-stage sea-squirt egg, that was carried with the cells in subsequent divisions and could be followed up to the tadpole stage (Conklin 1905). This spot, called the “yellow crescent” is now known to contain lipid inclusions that migrate to the vegetal hemisphere of the egg immediately after sperm entry (Gilbert 2000). Conklin was able to trace the cells for up to eight rounds of cell division and found the yellow cytoplasmic cells to generate mesodermal structures. Unfortunately, natural labels are not readily available for most model systems, and, thus, highlighting specific cells in the rapidly dividing and differentiating embryo called for the establishment of more sophisticated methodologies.
The use of “vital dyes” that stain living cells without killing them became one such way to label and follow cells. The power of this approach was shown by Walter Vogt, who constructed a fate map of the 32-cell-stage Xenopus embryo (Vogt 1929; discussed in Anderson and Dietrich 2012). Small pieces of agar chips were saturated with vital dyes and applied to the different regions of the late-blastula embryo. Cells that absorbed the dye retained the label over days and passed it on to their daughter cells, thereby enabling tracing of cells from gastrulation to the early organs they formed. This approach was further improved by the use of lipid-soluble, but water-insoluble, carbocyanine dyes that reduce their diffusion to neighboring cells, thus increasing the reliability of the results (for review, see Hsu 2015). However, externally added dyes whose intracellular concentrations become diluted with every round of cell division, could not be used to trace cell progenies for long time periods. In recent years, the advent of transgenic techniques and genetically expressed FPs, has significantly expanded the term of lineage-tracing experiments (discussed in the section below on lineage tracing).
In addition to labeling cells to serve as identifiers, experiments involving transplantation of cells from one embryo to another have also yielded important cell lineage discoveries. Many important notions in developmental biology, such as the organizer concept, have emerged from transplantation experiments. This technique involves grafting a desired piece of tissue from one embryo (donor) to another (host), and then tracing the fate of the donor cells within the host embryo. The important requirement of this methodology is the ability to distinguish between donor and host cells in the host embryo. In the classic Spemann–Mangold organizer experiment, for instance, a salamander host embryo with a distinct pigmentation pattern was grafted with tissue from an unpigmented donor (Spemann and Mangold 1924). Hence, by tracing the unpigmented cells within the resulting chimera, the fate of the donor tissue could be revealed. Additional important progenitor cell discoveries were made using the quail-chick chimera system. Chimeras generated from chick and Japanese quail embryos have been useful because the cells from each animal could be identified by the differences in their interphase nuclei (Le Douarin 1973). This system has contributed extensively to the identification of the progenies of neural crest cells and the neural primordium (for reviews, see Le Douarin 1980, 2004). Mammalian chimeras have also been insightful for lineage tracing (for review, see Mascetti and Pedersen 2016). Using mouse chimeras, Gardner and Rossant (1979) identified the progenitors present in the late blastocyst that generate the entire fetus versus those that generate the extraembryonic tissues. Thus, transplantation-based lineage tracing has been used in different models for identifying new progenitor–progeny relationships. However, these experiments could also render misleading conclusions as the donor cells might behave differently in the host body. Moreover, the transplantation process involves wounding that can also change the behavior of the cells.
During the last decades, site- and temporal-specific genetic recombination has become the most extensively applied lineage-tracing method. The development of various FPs, microscopy, transgenesis techniques, and genome editing has further improved this endeavor.
Lineage Tracing through Site-Specific Recombination
The discovery of site-specific recombination systems in bacteriophages and yeasts, and the exploitation of these systems as tools for specific manipulation of the genome, have brought about major improvements in lineage tracing. One of the most commonly used site-specific recombination system in vertebrates is the Cre-lox system (Fig. 1B). LoxP sites are short DNA stretches from the coliphage P1, which under the catalyzing influence of the recombination enzyme Cre recombinase (Cre), undergo DNA cleavage and reciprocal strand exchange with another loxP site (Sternberg and Hamilton 1981; reviewed by Gierut et al. 2014). Because animals lack this system, it can only be engineered through transgenesis, allowing site-specific recombination at the genomic locations where loxP has been introduced. For lineage tracing, a stable transcriptional driver is engineered upstream to a reporter gene (usually coding for an FP or LacZ), and a loxP flanked stop signal (loxP-STOP-loxP) is placed between these two sequences (Fig. 1B). In the presence of Cre, recombination of the loxP sites removes the interfering loxP-STOP-loxP sequence and allows expression of the reporter gene. Cre, on the other hand, is expressed through a cell-specific transcriptional driver that can be specifically induced within a desired cell population. To achieve temporal control of the system, Cre is fused with a mutant ligand-binding domain of the human estrogen receptor (CreER) (Danielian et al. 1993; Metzger et al. 1995; Feil et al. 1996) that does not bind to the endogenous estradiol, but can be activated through experimental exposure of synthetic ligands such as tamoxifen or 4-hydroxytamoxifen (CreERT2). Exposure to tamoxifen leads to translocation of CreER into the nucleus, where it induces recombination of the DNA at the loxP sites. Because this change in DNA is heritable and irreversible, the progenies of these cells can be traced throughout life based on the expression of the reporter (Fig. 1B′).
Lineage tracing through site-specific recombination is not without drawbacks (discussed in Semo et al. 2016). Conclusions derived from these experiments largely depend on the accuracy of the transcriptional drivers used for expressing Cre. In certain cases, leakiness and promiscuity of Cre expression, may cause undesirable labeling, hampering the interpretation of the results. For example, Payne et al. (2018) have described how certain Cre lines commonly used in endothelial cell (EC) research also highlight non-EC populations.
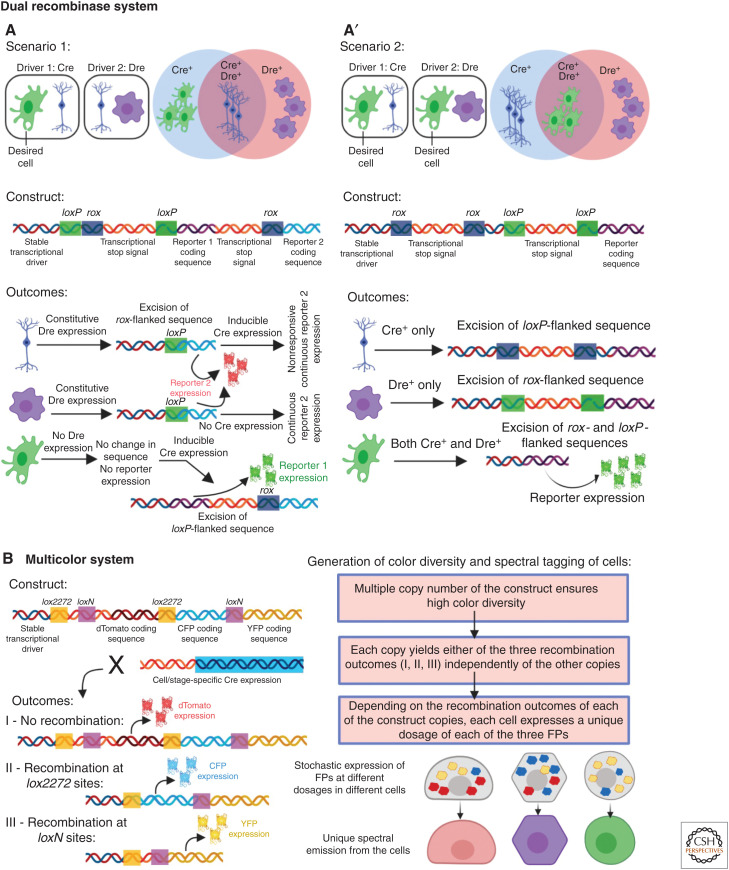
One way to tackle the problem of promiscuous Cre expression has been the use of a “dual recombinase system” (Fig. 2A,A′; He et al. 2017). This method uses the Dre-rox recombination system (Sauer and McDermott 2004), in which Dre induces DNA recombination at the rox sites. The method has been used when a Cre driver is available for labeling the desired cell type (“dCells”), but it also labels additional cells (“aCells”). This method has been used in two different ways: In one case (Fig. 2A), a transcriptional driver expressed in aCells but not in dCells, is engineered to express Dre (Li et al. 2018). By using a dual reporter line “driver-loxP-rox-Stop-loxp-reporter1-Stop-rox-reporter2,” it is ensured that Dre-rox recombination will excise reporter 1 sequence in the Dre+ aCells, rendering sole expression of reporter 2. Subsequent induction CreERT2 activity will result in exclusive expression of reporter 1 in cells where Dre-rox has not disrupted its sequence (i.e., dCells, which are CreERT2+, Dre–). The second strategy (Fig. 2A′; Liu et al. 2019) is useful when two different drivers are available for the dCells (Dre+, Cre+), but both of them also label additional sets of aCells (each either Cre+ or Dre+). In this case, the reporter line is programmed to allow simultaneous recombination of both loxP and rox sites (through a “driver-rox-Stop-rox-loxp-Stop-loxp-reporter”) in a given cell, thereby rendering exclusive expression of the reporter only in cells that are both Cre+ and Dre+. The dual recombinase system has been successfully used to analyze cardiomyocyte renewal (Li et al. 2018, 2019) (further discussed below in the section Assessing Progenitor Cells in Adult Health and Disease) as well as liver (Han et al. 2019) and lung regeneration (Liu et al. 2019).
Figure 2.
Different applications of site-specific recombination systems for precise lineage tracing. (A,A′) The dual recombination system uses Cre-lox and Dre-rox recombination systems together to increase the precision of lineage tracing by resolving the issues of promiscuous Cre expression. (A) In some studies (scenario 1), promiscuous expression of Cre is abolished by Dre-rox-mediated excision of a specific reporter gene (reporter 1) from the nonspecific cell types. Cells that express only Dre (violet) or both Dre and Cre (blue) can only express reporter 2 (red). The desired cell only expresses Cre and thus the Cre-lox-mediated recombination allows it to express reporter 1 (green). (A′) In certain cases (scenario 2), combinatorial expression of both Cre and Dre is required for the reported gene expression, restricting the reporter expression only in the overlapping cells. Cells that express only Cre (blue) or Dre (violet) cannot express the reporter, but combinatorial recombination in the desired cell (green) allows the expression of the reporter. (B) The multicolor system depends on stochastic and combinatorial expression of different fluorescent proteins (FPs) that allow unique spectral emission from different cells and, thus, uniquely labeling them. The construct allows expression of dTomato (outcome I) without any recombination. Cre mediates recombination either between the lox2272 or the loxN sites, resulting in the expression of CFP (outcome II) or YFP (outcome III). By inheriting multiple copies of the construct, and different outcomes from each copy, each cell independently acquires a unique combination of the FPs, resulting in unique spectral emission.
Another innovative application of the Cre-lox system for lineage tracing consists of the labeling of any given cell with a unique color resulting from the combinatorial expression of three or four FPs (Fig. 2B). This method, known as “Brainbow” as was first used to label neurons in the brain (Livet et al. 2007), is engineered such that induction of Cre at a particular time point would trigger stochastic recombination and therefore variable expression of FPs, rendering cells displaying distinctive spectral emissions that will be inherited by its progenies. Thus, this technique allows tracking of multiple cells in the same animal, each distinguished based on their unique spectral emission (Hampel et al. 2011; Gupta and Poss 2012; Pan et al. 2013; Currie et al. 2016). Simplified versions of this approach, including the confetti (Snippert et al. 2010) and the Rainbow systems (Red-Horse et al. 2010; Rinkevich et al. 2011), were created through modification of the Brainbow cassette (Snippert et al. 2010). These systems harbor one copy of the construct per haploid genome (reviewed by Weissman and Pan 2015), and thus Cre induction, instead of producing combinatorial expression of FPs, allows each cell to express only one of the three (Rainbow) or four (confetti) FPs. Although in these systems cells can acquire a limited number of colors, image acquisition and analysis are less complex than with the Brainbow system.
Identifying Progenitors through Live Imaging and Photoconvertible Proteins
Direct visualization of cellular genesis is one of the concrete methods for identifying progenitor cells. However, limited optical accessibility has traditionally restricted the widespread use of live imaging in most model organisms. In recent years, with advancements in imaging technologies, real-time observation of cellular events has become more achievable. The zebrafish is one of the most widely adopted vertebrates for live imaging because of its transparency during embryonic and larval stages (discussed in Nguyen and Currie 2018). In addition, the availability of pigmentation mutants (White et al. 2008), along with the advancements in anesthetic methods (Daetwyler et al. 2019), have enabled further improvement in deep tissue- and long-term imaging. In the case of murine models, creation of imaging windows exposing specific organs (intravital imaging) combined with the use of multiphoton microscopy have provided ample opportunities for recording progenitor cell behavior (for review, see Nguyen and Currie 2018). Such “imaging windows” have been successfully used to visualize cellular dynamics in the hair follicle (HF) (Rompolas et al. 2012), intestinal crypts (Ritsma et al. 2014), mammary glands (Zomer et al. 2013), hippocampus (Pilz et al. 2018), and many more (reviewed by Prunier et al. 2017). One great advantage of live imaging is that not only it reveals the progenitor–progeny relationships, but also the interaction between the progenitor and its microenvironment. This is particularly important because the microenvironment is known to play a major role in determining progenitor cell behavior and specification (reviewed by Batlle and Clevers 2017).
Another approach for lineage tracing involves usage of photoconvertible FPs. These proteins can be transformed from one fluorescent state to another, following activation by specific wavelengths. For instance, the photoconvertible protein Kaede, isolated from the stony coral Trachyphyllia geoffroyi, switches from green to red fluorescence emission after ultraviolet/violet light irradiation (Ando et al. 2002). This property allows the use of violet light to specifically highlight a single cell or a small group of cells within a large population of Kaede expressing cells. This is particularly useful in optically accessible tissues and in cases where it is not possible to genetically label small-cell populations because of the lack of specific markers. The tool is often combined with live imaging to enable precise tracking of the trajectory of a photoconverted cell, or can be used for pulse-chase experiments, in which photoconverted cells and their progenies are identified at later time points. The use of both these strategies in the transparent zebrafish embryo revealed, for instance, how specialized progenitor cells in the embryonic vein generate lymphatic vessels and blood vessels of various developing organs (Hen et al. 2015; Nicenboim et al. 2015). Over the years, a number of photoconvertible proteins have been discovered (Wiedenmann et al. 2004; Tsutsui et al. 2005), and variants have been created for increased efficiency (McKinney et al. 2009). Yet, it is important to keep in mind that photoconvertible proteins are suited for tracing cells for relatively short timescales, as their concentration is diluted with each round of cell division. Interestingly, recent work analyzing zebrafish neuromast regeneration has suggested that nuclear localized photoconvertible proteins are suited for longer tracing experiments by being protected from degrading components of the cytoplasm (Thomas and Raible 2019).
In the next section, we summarize some of the important discoveries of progenitor cells made through the use of lineage-tracing tools.
IDENTIFICATION OF NOVEL CELLULAR ORIGINS DURING EARLY DEVELOPMENT
Progenitors of Hematopoietic Stem Cells
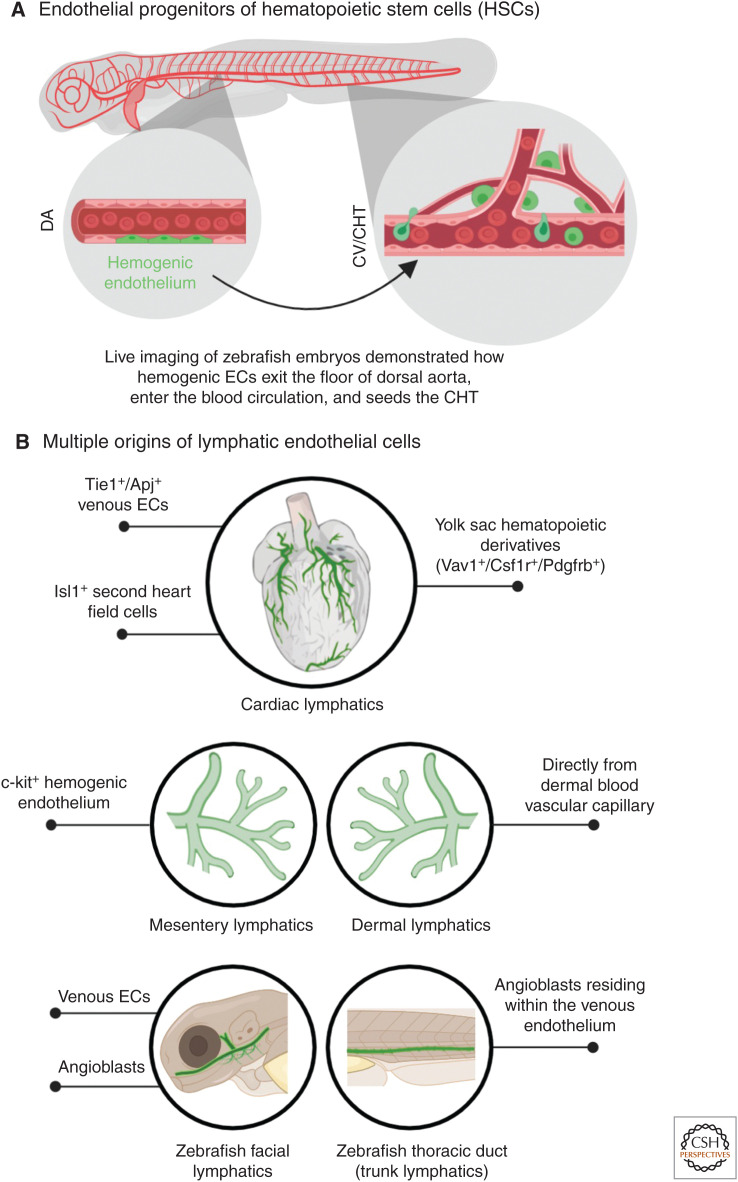
The origin of blood cells has been a focus of intense debate for most of the twentieth century. The identification of hematopoietic stem cells (HSCs) that can generate all blood cell lineages (for review, see Orkin 2000) called for the determination of their own progenitors. Lineage-tracing experiments performed in chick, mouse, and zebrafish provided strong evidence for the role of early ECs, called hemogenic endothelium (reviewed by Gritz and Hirschi 2016), in generating HSCs. In chick, ECs were originally labeled by injection of a nonreplicative spleen necrosis virus (SNV) vector harboring the lacZ reporter gene (Jaffredo et al. 2000). By visualizing the lacZ-labeled ECs at different time points, it was established that many of them express CD45, an important surface antigen of hematopoietic progenitors. ECs expressing CD45 were also detected in the floor of the aorta (Jaffredo et al. 2000). Later on, a more definite proof of the existence of the hemogenic endothelium came from lineage-tracing experiments in the mouse. Using VE-cadherin-CreERT2 transgenic mice (Monvoisin et al. 2006), which label both developing and mature endothelium, Zovein et al. (2008) labeled ECs at E9.5 and showed that the label was carried over at later stages by HSCs. These EC-derived progenies were also shown to populate the fetal liver and bone marrow, the sites of haematopoesis at different developmental stages. Definite confirmation for these results arrived from live-imaging experiments in mouse (Boisset et al. 2010) and zebrafish (Bertrand et al. 2010; Kissa and Herbomel 2010; Lam et al. 2010). In zebrafish, three separate studies used elegant live imaging of transgenic embryos bearing fluorescently labeled early hematopoietic progenitors to show that ECs from the floor of the dorsal aorta indeed bud off, migrate to the HSC niche, and seed the HSC population (Fig. 3A).
Figure 3.
Origins of hematopoietic stem cells (HSCs) and lymphatic endothelial cells (ECs). (A) Live imaging of zebrafish embryos revealed the existence of hemogenic ECs (green) in the dorsal aorta (DA), which migrate to the caudal end of the embryo and seeds in the cardinal vein (CV), also called caudal hematopoietic tissue (CHT). Here, these cells function as the HSCs. (B) Different progenitor populations that generate organ-specific lymphatic vessels (green) have been revealed through lineage-tracing experiments over the years.
The Origins of Blood Vasculature
Blood vessels are one of the early forming systems in the embryo. Sabin, in 1917, made visual observations in live chick embryos to describe the emergence of angioblasts from mesoderm, which serve as progenitors for ECs (Sabin 1917). Later on, the splanchnopleuric mesoderm was identified as the subset from which ECs emerge (reviewed by Risau and Flamme 1995). This origin of early ECs remained almost unchallenged for decades, with the exception of the liver, in which certain EC subsets were shown to be generated from endoderm (Goldman et al. 2014) and endocardium (Zhang et al. 2016b). Most recently, a lineage-tracing study in mouse revealed a new and surprising cell type contributing to blood vessels—the yolk sack–derived erythromyeloid progenitors (EMPs) (Plein et al. 2018). Analysis of an inducible Csf1r-Cre mice, which labels EMPs along with their derived microglia and other differentiated myeloid cells, also revealed sparse labeling of ECs. These EMP-derived ECs were detected in the adult animal and in multiple organs, including the liver, where ∼60% of its ECs derived from this population.
One of the vascular beds that has attracted a lot of interest because of its high clinical importance are the coronary vessels (for review, see Sharma et al. 2017). The origins of the coronary vasculature have a been a subject of extensive debate, propelled in large part by lineage-tracing experiments, each highlighting the contribution of different progenitor populations. Early anatomical examinations in mammals, such as pig (Bennett 1936), led to the idea that coronary arteries are derived from the aorta and veins from the sinus venosus (SV) (reviewed by Tian et al. 2015; Sharma et al. 2017). However, a number of lineage-tracing studies using chick and quail-chick chimeras identified the proepicardium, a transient extracardiac structure that gives rise to most of the epicardium as the major source for coronary vessels (Mikawa and Fischman 1992; Pérez-Pomares et al. 2002; reviewed by Majesky 2004). Cre-lox-based lineage tracing in mouse models, however, showed conflicting results, and did not support proepicardium or its epicardium derivatives as a major contributor of coronary vessels. Reporter-mediated labeling of different epicardium populations, such as Gata5+ (Merki et al. 2005), Tbx18+ (Cai et al. 2008), and Wt1+ (Zhou et al. 2008), did not show major labeling of coronary vessel endothelium. Later, two subsets of proepicardium (Sema3D+ and Scx+) were shown to contribute toward a fraction of the coronary endothelium (Katz et al. 2012). Yet, the main source of coronary vessels was found to be the SV (Red-Horse et al. 2010). By following coronary ECs specifically labeled by the apelin-nlacZ driver and clonal analysis of VE-cadherin-CreERT2-derived clones, the SV was identified as the major source of coronary ECs. These results were further confirmed by lineage tracing of SV endothelium-specific Apj-CreERT2 cells (Chen et al. 2014). Interestingly, the endocardium was also proposed as an additional source of coronary vessels (Chen et al. 2014). Recently, it has been reported that the SV-derived coronary veins give rise to coronary arteries as well through gradual fate switching (Red-Horse et al. 2010; Su et al. 2018). However, a minor fraction of coronary arteries was shown to be derived from the Npr3+ endocardial population (Zhang et al. 2016a).
Mixed Origins of Lymphatic Endothelial Cells
The origins of lymphatic endothelial cells (LECs) have been pondered over and debated since the early twentieth century (reviewed by Semo et al. 2016). Two main theories took shape—the centrifugal or venous theory—which argued that lymphatics originate from the cardinal vein (CV) and spread in the body from the center toward the periphery (Sabin 1902, 1904), and the centripetal or nonvenous theory, claiming that LECs derive from mesenchymal clusters that grow from the periphery toward the center (Huntington and McClure 1910). The controversy between these two models remained vivid until the past decade, when the use of lineage-tracing tools shed much light on this issue. By following the development of the vasculature through live imaging of zebrafish embryos, Yaniv and colleagues showed that posterior CV (PCV)-derived ECs generate the trunk lymphatics (Yaniv et al. 2006). In similar fashion, induction of lacZ reporter through Prox1-CreERT2, confirmed that LECs emerge from veins in the mouse as well (Srinivasan et al. 2007). Although these studies strongly supported the “venous theory,” later work in zebrafish and mouse revealed that lymphatic vessels have much more heterogeneous origins (reviewed by Ulvmar and Mäkinen 2016). A combination of live imaging and lineage-tracing approaches in zebrafish led to the surprising finding that although LEC precursors emerge from the PCV, they do so from a population of angioblasts residing within the PCV (Nicenboim et al. 2015). Indeed, these PCV-resident progenitors, which display certain features of undifferentiated ECs, could also generate veins and arteries for the neighboring organs (Hen et al. 2015). Also in the mouse, a recent study reinforces the idea that only specific cells in the CV have the potency to generate LECs (Stone and Stainier 2019). Though the large majority of CV-ECs are known to derive from the lateral plate mesoderm, Stone and Stainier, showed that Pax3-Cre, which traces the paraxial mesoderm (PXM), labeled a subset of cells in the CV. Interestingly, LECs of multiple organs and tissues were in fact derived from this specific subset. Furthermore, conditional deletion of Prox1 in the PXM led to severe defects in LEC differentiation. Many other studies pointed at a mixed origin for specific lymphatic populations (Fig. 3B). These included c-kit+ cells in the mouse mesentery (Stanczuk et al. 2015) and local blood capillary plexus in the skin (Pichol-Thievend et al. 2018). In the case of the heart, the majority of the LECs are thought to be of venous origin (Klotz et al. 2015) that emerge from the CV ∼E9.5–10.5 (Gancz et al. 2019), with additional contributions from nonvenous sources such as the yolk sac hematopoietic derivatives (Klotz et al. 2015) as well as Isl1+ second heart field cells (Maruyama et al. 2019). Similarly, live imaging of the facial lymphatics in the zebrafish revealed mixed venous and nonvenous origins (Okuda et al. 2012; Eng et al. 2019).
Multiple Progenitors of Vascular Smooth Muscle Cells
Vascular smooth muscle cells (VSMCs) are intimately associated with the endothelium, and their dysregulation is involved in various human diseases (for review, see Gaengel et al. 2009). Lineage-tracing experiments have revealed multiple origins for VSMCs, and differences in their ontogeny have been linked to functional differences. Neural crest cells were the first to be reported to give rise to VSMCs in the “quail-chick” chimera. When a piece of quail cranial neural tube was grafted into the chick, the donor generated progenies that populated the walls of the pharyngeal arch arteries, whereas the ECs of these vessels remained that of the host (Le Lièvre and Le Douarin 1975). More than two decades later, the same was seen in a mouse model when Cre-lox-mediated lacZ-labeled neural crest cells were shown to generate VSMCs (Jiang et al. 2000). Since then, several lineage-tracing studies have uncovered a wide variety of progenitor cell types generating VSMCs (for review, see Majesky 2007). These include second heart field cells (Waldo et al. 2005), different subsets of mesothelium (Mikawa and Gourdie 1996; Wilm et al. 2005), somites (Pouget et al. 2006), mesoangioblasts (Minasi et al. 2002), certain progenitors that reside in vessel walls (Hu et al. 2004), and ECs (Chen et al. 2016). Recent work in zebrafish used live imaging of photoconvertible Kaede-expressing cells and showed that the VSMCs recruited in the developing dorsal aorta derive from sclerotomal progenitors, whereas neural crest cells were not found to contribute (Stratman et al. 2017).
VSMCs, along with their multiple progenitors, form an important model to understand the potential link between lineage history and function. Indeed, many of these ontologically different subsets of VSMCs show anatomical segregation (Sinha et al. 2014; Sawada et al. 2017), and single-cell transcriptional profiling shows that the anatomical segregation often correlates with differences in gene expression (Dobnikar et al. 2018). In addition, distinct VSMC subtypes have been shown to respond differently to various signaling triggers (Topouzis and Majesky 1996; Cheung et al. 2012), a fact that is important in atherosclerosis (Cheung et al. 2012) and aneurysms (Lindsay and Dietz 2011). Accordingly, lineage-tracing tools have been applied to understand the relative contribution of different VSMCs to the formation of atherosclerotic plaques (for review, see Basatemur et al. 2019). Multicolor “confetti” labeling of SMCs in mice showed that only a few VSMC cells are involved in generating most of the plaques (Chappell et al. 2016; Jacobsen et al. 2017), raising the possibility of a link between the lineage history of these cells and disease susceptibility.
ASSESSING PROGENITOR CELLS IN ADULT HEALTH AND DISEASE
Adult Progenitor Cells and Regeneration
In addition to embryonic development, maintenance of tissues throughout the life span involves cellular turnover, proliferation, and differentiation (Pellettieri and Alvarado 2007). These processes require a source of new differentiated cells, and lineage-tracing tools have been instrumental in revealing the identities of these progenitor sources. In general, different organs have evolved different strategies for progenitor cell recruitment (reviewed by Clevers and Watt 2018). One of them is the use of a dedicated stem cell pool, as is the case in the mouse HF. Each HF harbors stem cell populations that can regenerate the follicle during hair cycle or injury (reviewed by Blanpain and Fuchs 2006). Using [3H]TdR-labeling experiments, a site within the HF called bulge was shown to harbor slow cycling cells, a feature of quiescent stem cells (Cotsarelis et al. 1990). These cells were later shown to generate other cells of the HF (Taylor et al. 2000) and also displayed distinctive gene expression patterns (Morris et al. 2004; Tumbar et al. 2004). Using adult bulge-specific, inducible Krt1-15-Cre, these cells were shown to generate all components of the cutaneous epithelium (Morris et al. 2004). A direct visualization of this process was performed through intravital microscopy that revealed that even within the cells in the bulge region, there are locations from where the stem cells are more likely to participate in the differentiation process (Rompolas et al. 2012), showing the presence of more active cells within the quiescent population of stem cells. Interestingly, it was also revealed that ablation of the bulge cells induce neighboring epithelial cells (that are not usually involved in hair growth) to replenish the stem cell compartment from which follicle growth resumes (Rompolas et al. 2012). This showed the importance of the microenvironment in inducing and maintaining stem cells pools.
Tissue-resident stem cells have also been well described in the adult intestinal epithelium, brain, and blood-generating cells (reviewed by Li and Clevers 2010; Bond et al. 2015). However, existence of progenitor pools is not a general mechanism. In the heart for instance, the dual recombinase system has been used to show that nonmyocytes do not generate myocytes in the neonatal or the adult mouse (Li et al. 2018, 2019), providing strong support for the absence of cardiac stem cells in the adult, an issue of intense controversy in the last two decades (discussed in Lee 2018; Maliken and Molkentin 2018). Existence of dedicated stem cells in liver has also been controversial. A study that labeled adult differentiated hepatocytes found that in steady-state these can replicate to produce additional daughter cells without dedifferentiation (Malato et al. 2011; for reviews, see Stanger 2015; Clevers and Watt 2018). Moreover, during regeneration, there seems to be minimal contribution from other cell types. This was in contrast to previous findings claiming the existence of a Sox9+ progenitor population in the liver (Furuyama et al. 2011; discussed in Stanger 2015). Recently, however, a new study has proposed that within the hepatocyte population some, which are Sox9+, can produce both hepatocytes and ductal cells, displaying bipotent characteristics (Han et al. 2019). Another study using single-cell RNA sequencing of human liver suggested the existence of bipotent progenitors expressing intermediate levels of TROP2 (Aizarani et al. 2019), although in vivo lineage-tracing experiments confirming these findings are lacking.
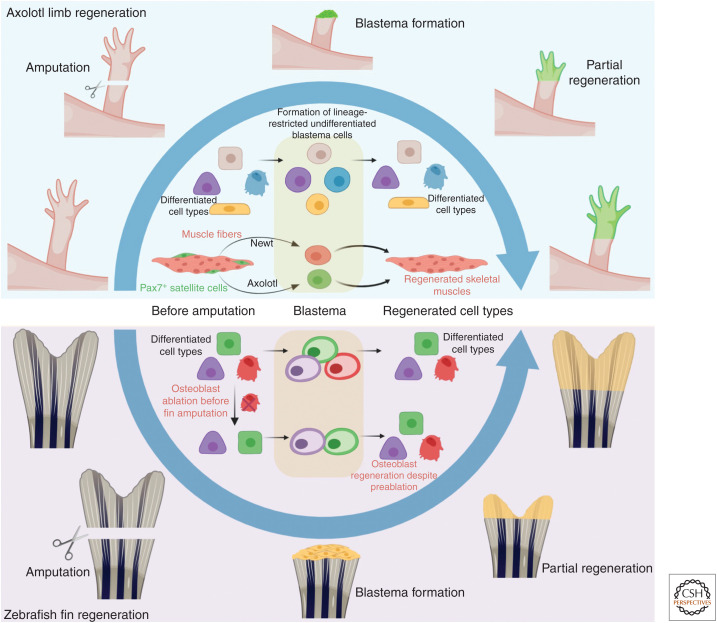
Adult mammals possess very limited regeneration capabilities. However, certain vertebrates, such as fishes and amphibians, show remarkable abilities to regenerate their organs. A description of salamander limb and tail regeneration came as early as 1768 (Spallanzani 1768; reviewed by Tsonis and Fox 2009), followed by fish fin regeneration in 1786 (Broussonet 1786). In both cases, the loss of appendage causes generation of a transient and heterogeneous mass of proliferating cells called blastema, which rapidly regenerates the lost structure (for reviews, see Tanaka 2016; Chen and Poss 2017). Lineage-tracing tools and live imaging have been instrumental in revealing the dynamics of progenitor to progeny differentiation taking place during the formation of the blastema and the subsequent regenerated structures (Fig. 4). Through a series of green fluorescent protein (GFP)-labeled transplants in unlabeled salamander, it was revealed that blastema cells are restricted progenitors that can only yield cells of the same lineage from which they are born (Kragl et al. 2009). Interestingly, Cre-lox-based lineage-tracing studies have uncovered multiple strategies used by tissues for progenitor recruitment during regeneration. For instance, the progenitor cells that generate skeletal muscles in the regenerating limb of the salamander species Notophthalmus viridescens, originate through dedifferentiation of existing muscle fibers. In contrast, in the salamander species Ambystoma mexicanum, the muscle-generating progenitors derive from pax7+ satellite cells (Fig. 4; Sandoval-Guzmán et al. 2014). Lineage-tracing analyses in zebrafish fin regeneration also supported a fate-restricted nature of blastema cells (Fig. 4; Tu and Johnson 2011). However, a subsequent study showed that, although normally lineage-restricted progenitors (osteoblasts in this case) give rise to the same cell types, on osteoblast ablation new osteoblasts can derive from alternative sources within the blastema (Singh et al. 2012). These results indicated that blastema cells may have more potency than previously appreciated and the microenvironment may play an important role in such contexts.
Figure 4.
Progenitors in appendage regeneration. Axolotl limb and zebrafish fin regenerate through the formation of undifferentiated blastema (depicted in green for axolotl and yellow for zebrafish), which generates the differentiated cell types of the regenerating structure. Blastema cells are fate-restricted, which yields differentiated progenies of the same lineage from which they are born. The exact progenitors might differ between species, as progenitors for regenerating muscles are derived from differentiated muscles in newts, but from the satellite cells in case of axolotl. Although lineage-restricted blastemal progenitors are also described in zebrafish fin regeneration, osteoblasts (red cells) ablated before the amputation can still yield regenerating osteoblasts. This indicates that other cell types in the blastema can also differentiate to osteoblasts when required.
The origins of the newly formed vasculature in the regenerating fin have also been investigated through live imaging of adult fish (Xu et al. 2015). Interestingly, the study revealed that the arteries of the regenerating fin are generated from venous-derived ECs. Further lineage-tracing work on development of these vessels would be required to fully understand whether such a mode of fin vasculature formation is seen only during regeneration or also during normal development.
Overall, the status of progenitor cells in different organs of the adult organism differs substantially, and many remain understudied. Future lineage-tracing and functional studies would be essential to shed light on the contribution of these progenitors to organ regeneration and repair.
Lineage Tracing in Cancer Research
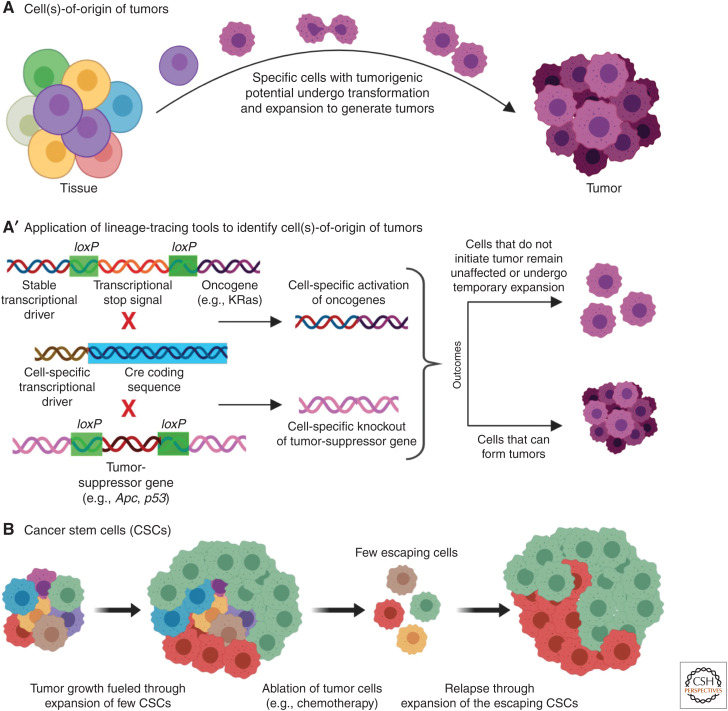
Although the last decades have witnessed enormous advances in our understanding of the genetic basis of cancer and the molecular events that lead to cancerous growth and malignancy, much less is known regarding specific cell subpopulations in which accumulation of mutations allows cancer development and progression (discussed in Blanpain 2013). Several studies have shown that certain cells within a tissue have higher tumorigenic potential, even when other cells accumulate equivalent mutations (Fig. 5A; reviewed by Visvader 2011; Blanpain 2013). Uncovering the identity of these “cell(s)-of-origin,” holds great promise for therapies aimed at preventing cancer progression at the initial phases of the disease (Visvader 2011). In the last decade, lineage-tracing tools have been increasingly used to identify the “cell-of-origin” for different cancer types. A common approach for such experiments has been the use of a Cre-lox system engineered to activate specific oncogenes or to knock out specific tumor-suppressor genes in restricted cell subsets (Fig. 5A′). Using this strategy, it was shown that knockout of the Apc tumor-suppressor gene in the Lgr5+ intestinal stem cells causes rapid transformation of these cells and results in formation of long-lived macroscopic adenomas within 3–5 weeks (Barker et al. 2009). In contrast, Apc loss in a subset of transit-amplifying cells, resulted in microadenomas, whose growth was rapidly arrested. A more detailed work has been performed on mouse models of skin cancer (for reviews, see Blanpain 2013; Sánchez-Danés and Blanpain 2018). In squamous cell carcinoma (SCC), for instance, it was found that activating oncogenic KRas expression, in combination with p53 deletion in K19+ or K15+ HF bulge cells and their immediate progenies or in the Inv-CreER-labeled interfolicular epidermis (IFE), led to SCC (Lapouge et al. 2011; White et al. 2011). However, Shh+ HF transit-amplifying cells with the same genetic alterations were not competent to initiate SCC. Although both HF bulge cells and IFE were competent to initate SCC, the bulge cells produced less differentiated and more invasive tumors, whereas IFE yielded well-differentiated tumors (Latil et al. 2017). This is because different cell types feature distinct transcriptional and chromatin landscapes and, thus, similar genetic alterations can result in different tumorigenic potential. In this case, the HF bulge cells showed epithelial-to-mesenchymal transition (EMT), whereas IFE did not (Latil et al. 2017). Thus, cell-of-origin for a tumor also determines the final phenotype and the cancerogenic potential.
Figure 5.
Lineage tracing in cancer. (A) Specific cells (depicted in violet) in a tissue have higher tumorigenic potential. (A′) These cell(s)-of-origin of tumors are identified using Cre-lox lineage-tracing strategies. Through Cre-lox-mediated excision, oncogenes can be expressed by cell-specific transcriptional drivers (gain-of-function of oncogenes), whereas tumor-suppressor genes can be excised out in specific cells (loss-of-function of tumor-suppressor genes). The ability of different cell populations to produce cancerous growth is documented on such manipulations. (B) The basic concept of cancer stem cells that are the prime drivers of tumor growth. Only a few cells in a tumor are responsible for most of the tumor expansion. During destruction of tumor cells, only a few escaping CSCs are enough to cause regrowth of the tumor, and this is understood to be the underlying reason for relapse of cancer after treatments such as chemotherapy.
Another intensively debated subject in cancer research is the existence of “cancer stem cells” (CSCs) (Fig. 5B; reviewed by Kreso and Dick 2014; Batlle and Clevers 2017; Scott et al. 2019). Akin to the stem cells that constitute a fraction of different adult organs, tumors have also been suggested to possess CSCs that can self-renew and generate cells for tumor growth and malignancy (Beck and Blanpain 2013). The presence of these CSCs is proposed to be one of the main reasons for relapse after chemotherapy, which while destroying most of the tumor cells, may miss a small fraction of CSCs that fuel new neoplastic growth. Thus, the identification of CSCs, along with the understanding of their unique properties and cellular behaviors, may hold a key to cancer therapy. In 2012, three studies (Chen et al. 2012; Driessens et al. 2012; Schepers et al. 2012) applying lineage-tracing tools provided strong evidence supporting the CSC hypothesis (discussed in Gilbertson and Graham 2012). Using a mouse model of papilloma, it was shown that only a small number of proliferating cells within a tumor generate the majority of the tumor cells (Driessens et al. 2012). In this study, the Cre-lox system was used to sparsely label basal cells (i.e., the proliferating cells) of a papilloma. Although within 7 days 80% of the labeled cells were lost, the remaining 20% produced enormous numbers of daughter cells whose progenies were capable of differentiating into all other cell types of the tumor. A similar conclusion was reached using the multicolor confetti system to trace the behavior of Lgr5+ stem cells in intestinal adenomas (Schepers et al. 2012). In this study, single Lgr5+ cells were found to give rise to a large number of different cell types in the growing tumor. Chen et al. (2012) further showed that eliminating a bulk of the tumor cells in a mouse glioblastoma model, triggers repopulation of the tumor through a few escaping quiescent CSCs. However, tumor growth could be completely arrested by additional elimination of the CSC population. In a similar fashion, a combination of confetti lineage tracing with intravital imaging of mammary gland adenoma showed how a few stem cells clonally expand, fuel tumor growth, and generate multiple cell types (Zomer et al. 2013).
The identification of CSCs had inspired newer strategies of cancer therapies aiming at their elimination. Nevertheless, growing evidences indicate that both CSC and non-CSC populations within a tumor might be plastic, and their tumorigenic potential may be altered by the tumor microenvironment (for reviews, see Batlle and Clevers 2017; Scott et al. 2019). In support of this idea, it has been shown that in intestinal tumors, epithelial non–stem cells can dedifferentiate into tumor-initiating cells (Schwitalla et al. 2013). Furthermore, lineage-tracing experiments in human colorectal cancer organoids showed that selective ablation of CSCs leads to the formation of new CSCs from differentiated non-CSCs (Shimokawa et al. 2017). Thus, differentiated tumor cells can act as progenitors for CSCs, leading to a fluidity in progeny–progenitor cell relationship in tumors.
In summary, lineage analyses of adult tissues are becoming increasingly possible and continue providing surprising, and in many cases, contradictory answers. This is partly because of the difficulty of accessing and analyzing adult tissues and partly because of the caveats of the techniques.
FUTURE OF PROGENITOR CELL DISCOVERY
As discussed so far, lineage-tracing approaches are majorly dependent on visual/microscopic methodologies. With the advent of next-generation sequencing technologies, new avenues become available, which bear the potential of revealing new progeny–progenitor relationships.
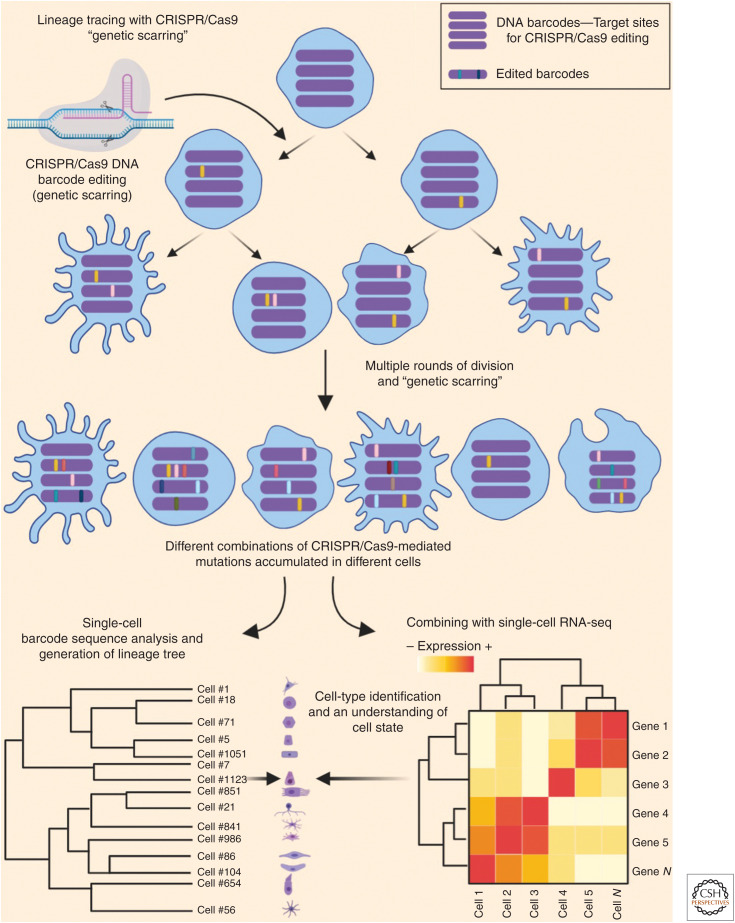
Apart from introducing visual labels, studies have used viral-mediated introduction of DNA barcodes as means for labeling specific cells, whose progenies can be later identified by DNA sequencing (Lu et al. 2011; Bhang et al. 2015; Eirew et al. 2015). Additional strategies combining the Cre-lox system with DNA barcodes have also been successfully used for lineage tracing. One such example is the polylox system (Pei et al. 2017), which makes use of an array of DNA barcodes interspersed by multiple loxP sites. In this system, Cre activation induces a number of different DNA barcode combinations in the specific cells of interest. With the advance of CRISPR/Cas9 genome editing, new ways of tagging cells have emerged (Fig. 6). In absence of a specific template for homologous repair, the Cas9 enzyme generates random “repaired” sequences (refered as “genetic scars”), making cells harbor a unique DNA barcode that is inherited by progenies (reviewed by McKenna and Gagnon 2019). This technique can be designed to induce the genetic scars in specific loci or within a transgene. Single-cell DNA sequencing of the targetted loci enables clustering of all cell types that have inherited the same genetic scars and construction of a lineage map. This method was recently applied in zebrafish through a technique called GESTALT (genome editing of synthetic target arrays for lineage tracing) (McKenna et al. 2016), where the genetic scarring was restricted to early embryogenesis by transient use of CRISPR reagents. Thus, through CRISPR activity, the early embryonic cells acquired unique barcodes and passed them onto their progenies. By harvesting different organs in the adult fish and revealing the mutated barcodes through single-cell sequencing, it was revealed that most organs in the fish use only a few progenitor cells to develop. In case of blood cells, only five different barcodes were found in >98% of the sequenced cells, whereas for most organs >50% of the cells harbored seven different barcodes (McKenna et al. 2016). A similar work, also on zebrafish, using the same “genetic scarring” strategy, extended the lineage tracing to fin regeneration and concluded that most clones that participated in the development of the fin were also used in the regeneration process (Junker et al. 2017). Lineage-analysis of blastema in axolotl limb regeneration, also accomplished through this method, largely supported the prevailing model (Fig. 5) that embryonic lineages are mostly maintained in the regenerating limbs (Flowers et al. 2017). The technique was later adapted to mice using a “homing CRISPR” technique (Kalhor et al. 2017), which uses an engineered guide RNA (gRNA) (homing gRNA [hgRNA]) to induce “genetic scars” in its own sequence, so that the hgRNA sequence itself acts as the barcode. Thus, barcode editing can be initiated by crossing a Cas9 expressing mouse with a transgenic mouse carrying hgRNA sequences (Kalhor et al. 2018).
Figure 6.
Use of CRISPR/Cas9-mediated DNA barcode editing for lineage tracing. CRISPR/Cas9 system is used for editing specific DNA sequences (barcode; shown as violet bars) that are inherited by their progenies. With multiple rounds of such editing (genetic scarring), different cells accumulate different combinations of the “genetic scars” (shown as different colors on the barcodes). Retrieval of the genetic scars from the barcodes of each cell allows to understand the progressive divergence from common progenitors and thus used for construction of a lineage tree. Simultaneous RNA-seq from the same cells is used for precise cell-type and cell-state identification.
The CRISPR/Cas9-based lineage tracing has now been further improved through simultaneous single-cell RNA (scRNA)-seq of these cells, as performed by three independent zebrafish studies (Alemany et al. 2018; Raj et al. 2018; Spanjaard et al. 2018). These studies reported the identification of cell types and states of the cells that had been “lineage-traced” through the CRISPR/Cas9 method (Fig. 6). Furthermore, combined with data on the complete transcriptome of each cell, this approach allows to gain an unprecedented amount of information on progenitor–progeny relationships, although high cost and technical complexity in these methodologies might restrict their widespread usage. The lack of spatial information is another big drawback of these techniques.
So far, we have discussed techniques that rely on introducing a heritable label—be it a visual label or a genetic tag. However, scRNA-seq analyses are now being increasingly used to predict lineage relationships in an unbiased manner, without prelabeling the cells. Single-cell transcriptomics is used for revealing the extent of trancriptional activity of all genes in thousands of individual cells. For most differentiation events, a cell's switch from a progenitor cell to a fully differentiated progeny is not a binary event, but rather a gradual change in the transcriptional landscape that involves progressive aquisition of the differentiated cell expression signature, while losing many of the transcriptional features of the progenitor cell type. Thus, for a given tissue, in which a number of differentiation events occur simultaneously, a single-cell transcriptomic experiment will capture the transcriptional states of all cells, including the progenitors, progenies, and intermediate cell states. Subsequently, the cells are ordered in a hypothetical trajectory (the pseudotime) based on the quantified gene expression similarity between the sampled cells (Tritschler et al. 2019). However, it is important to recognize that lineage reconstruction solely through scRNA-seq data is not enough to confirm progenitor–progeny relationship (discussed in Kester and van Oudenaarden 2018); thus, conventional lineage-tracing analyses are still required to confirm the lineage relationships predicted by this method. Nevertheless, for every identified progeny or differentiation intermediate, this technique allows us to have a peek at the entire transcriptome, opening up immense possibilities for understanding the biology behind such transitions.
CONCLUDING REMARKS
The discovery of progenitor cells remains an important pursuit in the understanding of cell differentiation in different contexts of health and disease. With the availibility of newer technologies, lineage-tracing experiments have improved in precision and efficiency, remaining an active and lively area of research. Imaging-based lineage tracing is still the most used methodology, but next-generation sequencing-based techniques are fast capturing interest. scRNA-seq, in combination with DNA barcode editing through CRISPR/Cas9, is taking lineage tracing beyond cell identification by providing in-depth information of gene expression associated with changes in cell states.
Another important change in lineage tracing has been the work on adult tissues. Earlier, because of experimental constraints, much of the work was restricted to the developing embryo. Now, technological advances facilitate the investigation of later events of organogenesis, maintenance, and regeneration. Many pathological states, including cancer, which depend on the behavior of progenitor cells, are also being investigated through lineage-tracing methods.
As we dive deeper in this pursuit, an increased appreciation of cellular lineage relationships in shaping functional outcomes is attained. The information from lineage tracing and the progenitor–progeny biology is deeply influencing the growing areas of tissue engineering and organoid systems. Thus, looking at the trend, it seems that lineage-tracing techniques will continue to thrive and even improve.
ACKNOWLEDGMENTS
We sincerely apologize to colleagues whose important work could not be cited owing to space limitations. We thank Dr. Kamalesh Kumari (Weizmann Institute of Science) for assisting with the illustrations. The illustrations were generated using Biorender (see biorender.com) and Adobe photoshop CS5.1. This work was supported, in part, by the European Research Council 818858 CoG (to K.Y.), Binational Science Foundation-2015289 (to K.Y.), Minerva Foundation-712610 (to K.Y.), the H&M Kimmel Institute for Stem Cell Research (to K.Y.), and the Estate of Emile Mimran (SABRA program) (to K.Y.). K.Y. is supported by the Daniel Shapiro Cardiovascular Fund. R.N.D. was supported by European Molecular Biology Organization (EMBO) long-term fellowship (ALTF 1532-2015), Edith and Edward F. Anixter Postdoctoral Fellowship and a senior postdoctoral fellowship from the Weizmann Institute of Science.
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, Pessaux P, Baumert TF, Grün D. 2019. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572: 199–204. 10.1038/s41586-019-1373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany A, Florescu M, Baron CS, Peterson-Maduro J, Van Oudenaarden A. 2018. Whole-organism clone tracing using single-cell sequencing. Nature 556: 108–112. 10.1038/nature25969 [DOI] [PubMed] [Google Scholar]
- Anderson N, Dietrich MR. 2012. The educated eye: visual culture and pedagogy in the life sciences. Dartmouth College Press, Hanover, NH. [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. 2002. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci 99: 12651–12656. 10.1073/pnas.202320599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, Van Es JH, Van De Wetering M, Begthel H, Van Den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611. 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z. 2019. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol 16: 727–744. 10.1038/s41569-019-0227-9 [DOI] [PubMed] [Google Scholar]
- Batlle E, Clevers H. 2017. Cancer stem cells revisited. Nat Med 23: 1124–1134. 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- Beck B, Blanpain C. 2013. Unravelling cancer stem cell potential. Nat Rev Cancer 13: 727–738. 10.1038/nrc3597 [DOI] [PubMed] [Google Scholar]
- Bennett HS. 1936. The development of the blood supply to the heart in the embryo pig. Am J Anat 60: 27–53. 10.1002/aja.1000600103 [DOI] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, Traver D. 2010. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464: 108–111. 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhang HEC, Ruddy DA, Radhakrishna VK, Caushi JX, Zhao R, Hims MM, Singh AP, Kao I, Rakiec D, Shaw P, et al. 2015. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 21: 440–448. 10.1038/nm.3841 [DOI] [PubMed] [Google Scholar]
- Blanpain C. 2013. Tracing the cellular origin of cancer. Nat Cell Biol 15: 126–134. 10.1038/ncb2657 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. 2006. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol 22: 339–373. 10.1146/annurev.cellbio.22.010305.104357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, Van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464: 116–120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Bond AM, Ming GL, Song H. 2015. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17: 385–395. 10.1016/j.stem.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussonet M. 1786. Observations sur la régénération de quelques parties du corps des poissons. Histoire de l'Académie Royale des Sciences 684–688. [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. 2008. A myocardial lineage derives from Tbx18 epicardial cells. Nature 454: 104–108. 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jørgensen HF. 2016. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res 119: 1313–1323. 10.1161/CIRCRESAHA.116.309799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Poss KD. 2017. Regeneration genetics. Annu Rev Genet 51: 63–82. 10.1146/annurev-genet-120116-024554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. 2012. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488: 522–526. 10.1038/nature11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HI, Sharma B, Akerberg BN, Numi HJ, Kivela R, Saharinen P, Aghajanian H, McKay AS, Bogard PE, Chang AH, et al. 2014. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development 141: 4500–4512. 10.1242/dev.113639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang H, Liu Y, Adams S, Eilken H, Stehling M, Corada M, Dejana E, Zhou B, Adams RH. 2016. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat Commun 7: 12422 10.1038/ncomms12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C, Bernardo AS, Trotter MWB, Pedersen RA, Sinha S. 2012. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat Biotechnol 30: 165–173. 10.1038/nbt.2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Watt FM. 2018. Defining adult stem cells by function, not by phenotype. Annu Rev Biochem 87: 1015–1027. 10.1146/annurev-biochem-062917-012341 [DOI] [PubMed] [Google Scholar]
- Conklin EG. 1905. The organization and cell lineage of the ascidian egg. J Acad Nat Sci Phila 12: 1–19. [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61: 1329–1337. 10.1016/0092-8674(90)90696-C [DOI] [PubMed] [Google Scholar]
- Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, Tanaka EM. 2016. Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev Cell 39: 411–423. 10.1016/j.devcel.2016.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler S, Günther U, Modes CD, Harrington K, Huisken J. 2019. Multi-sample SPIM image acquisition, processing and analysis of vascular growth in zebrafish. Development 146: dev173757 10.1242/dev.173757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, White R, Hoare SA, Fawell SE, Parker MG. 1993. Identification of residues in the estrogen receptor that confer differential sensitivity to estrogen and hydroxytamoxifen. Mol Endocrinol 7: 232–240. [DOI] [PubMed] [Google Scholar]
- Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jørgensen HF. 2018. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun 9: 4567 10.1038/s41467-018-06891-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. 2012. Defining the mode of tumour growth by clonal analysis. Nature 488: 527–530. 10.1038/nature11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. 2015. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518: 422–426. 10.1038/nature13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng TCY, Chen W, Okuda KS, Misa JP, Padberg Y, Crosier KE, Crosier PS, Hall CJ, Schulte-Merker S, Hogan BM, et al. 2019. Zebrafish facial lymphatics develop through sequential addition of venous and non-venous progenitors. EMBO Rep 20: e47079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. 1996. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci 93: 10887–10890. 10.1073/pnas.93.20.10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers GP, Sanor LD, Crews CM. 2017. Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. eLife 6: e25726 10.7554/eLife.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. 2011. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41. 10.1038/ng.722 [DOI] [PubMed] [Google Scholar]
- Gaengel K, Genové G, Armulik A, Betsholtz C. 2009. Endothelial-mural cell signaling in vascular development and angiogenesis. Arter Thromb Vasc Biol 29: 630–638. 10.1161/ATVBAHA.107.161521 [DOI] [PubMed] [Google Scholar]
- Gancz D, Raftrey BC, Perlmoter G, Marín-Juez R, Semo J, Matsuoka RL, Raviv H, Moshe M, Addadi Y, Golani O, et al. 2019. Distinct origins and molecular mechanisms contribute to lymphatic formation during cardiac growth and regeneration. eLife 8: e44153 10.7554/eLife.44153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RL, Rossant J. 1979. Investigation of the fate of 4–5 day post-coitum mouse inner cell mass cells by blastocyst injection. J Embryol Exp Morphol 52: 141–152. [PubMed] [Google Scholar]
- Gierut JJ, Jacks TE, Haigis KM. 2014. Strategies to achieve conditional gene mutation in mice. Cold Spring Harb Protoc 2014: 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. 2000. Developmental biology, 6th ed Sinauer, Sunderland, MA. [Google Scholar]
- Gilbertson RJ, Graham TA. 2012. Resolving the stem-cell debate. Nature 488: 462–463. 10.1038/nature11480 [DOI] [PubMed] [Google Scholar]
- Goldman O, Han S, Hamou W, De Villeroche VJ, Uzan G, Lickert H, Gouon-Evans V. 2014. Endoderm generates endothelial cells during liver development. Stem Cell Reports 3: 556–565. 10.1016/j.stemcr.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritz E, Hirschi KK. 2016. Specification and function of hemogenic endothelium during embryogenesis. Cell Mol Life Sci 73: 1547–1567. 10.1007/s00018-016-2134-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Poss KD. 2012. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 484: 479–484. 10.1038/nature11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. 2011. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat Methods 8: 253–259. 10.1038/nmeth.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang Y, Pu W, Huang X, Qiu L, Li Y, Yu W, Zhao H, Liu X, He L, et al. 2019. Lineage tracing reveals the bipotency of SOX9+ hepatocytes during liver regeneration. Stem Cell Rep 12: 624–638. 10.1016/j.stemcr.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Li Y, Li Y, Pu W, Huang X, Tian X, Wang Y, Zhang H, Liu Q, Zhang L, et al. 2017. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med 23: 1488–1498. 10.1038/nm.4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen G, Nicenboim J, Mayseless O, Asaf L, Shin M, Busolin G, Hofi R, Almog G, Tiso N, Lawson ND, et al. 2015. Venous-derived angioblasts generate organ-specific vessels during zebrafish embryonic development. Development 142: 4266–4278. 10.1242/dev.129247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC. 2015. Theory and practice of lineage tracing. Stem Cells 33: 3197–3204. 10.1002/stem.2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhang Z, Torsney E, Afzal AR, Davison F, Metzler B, Xu Q. 2004. Abundant progenitor cells in the adventitia contribute to atherosclerosis of vein grafts in ApoE-deficient mice. J Clin Invest 113: 1258–1265. 10.1172/JCI19628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington GS, McClure CFW. 1910. The anatomy and development of the jugular lymph sacs in the domestic cat (Felis domestica). Am J Anat 10: 177–312. 10.1002/aja.1000100108 [DOI] [Google Scholar]
- Jacobsen K, Lund MB, Shim J, Gunnersen S, Füchtbauer E-M, Kjolby M, Carramolino L, Bentzon JF. 2017. Diverse cellular architecture of atherosclerotic plaque derives from clonal expansion of a few medial SMCs. JCI Insight 2: 95890 10.1172/jci.insight.95890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo T, Gautier R, Brajeul V, Dieterlen-Lièvre F. 2000. Tracing the progeny of the aortic hemangioblast in the avian embryo. Dev Biol 224: 204–214. 10.1006/dbio.2000.9799 [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. 2000. Fate of the mammalian cardiac neural crest. Development 127: 1607–1616. [DOI] [PubMed] [Google Scholar]
- Junker JP, Spanjaard B, Peterson-Maduro J, Alemany A, Hu B, Florescu M, van Oudenaarden A. 2017. Massively parallel clonal analysis using CRISPR/Cas9 induced genetic scars. bioRxiv 56499. [Google Scholar]
- Kalhor R, Mali P, Church GM. 2017. Rapidly evolving homing CRISPR barcodes. Nat Methods 14: 195–200. 10.1038/nmeth.4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Kalhor K, Mejia L, Leeper K, Graveline A, Mali P, Church GM. 2018. Developmental barcoding of whole mouse via homing CRISPR. Science 361: eaat9804 10.1126/science.aat9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. 2012. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell 22: 639–650. 10.1016/j.devcel.2012.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester L, van Oudenaarden A. 2018. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23: 166–179. 10.1016/j.stem.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Klotz L, Norman S, Vieira JM, Masters M, Rohling M, Dubé KN, Bollini S, Matsuzaki F, Carr CA, Riley PR. 2015. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 522: 62–67. 10.1038/nature14483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. 2009. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460: 60–65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Kreso A, Dick JE. 2014. Evolution of the cancer stem cell model. Cell Stem Cell 14: 275–291. 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. 2012. Lineage tracing. Cell 148: 33–45. 10.1016/j.cell.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Lam EYN, Hall CJ, Crosier PS, Crosier KE, Flores MV. 2010. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 116: 909–914. 10.1182/blood-2010-01-264382 [DOI] [PubMed] [Google Scholar]
- Lapouge G, Youssef KK, Vokaer B, Achouri Y, Michaux C, Sotiropoulou PA, Blanpain C. 2011. Identifying the cellular origin of squamous skin tumors. Proc Natl Acad Sci 108: 7431–7436. 10.1073/pnas.1012720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil M, Nassar D, Beck B, Boumahdi S, Wang L, Brisebarre A, Dubois C, Nkusi E, Lenglez S, Checinska A, et al. 2017. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell 20: 191–204.e5. 10.1016/j.stem.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. 1973. A biological cell labeling technique and its use in experimental embryology. Dev Biol 30: 217–222. 10.1016/0012-1606(73)90061-4 [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. 1980. The ontogeny of the neural crest in avian embryo chimaeras. Nature 286: 663–669. 10.1038/286663a0 [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. 2004. The avian embryo as a model to study the development of the neural crest: a long and still ongoing story. Mech Dev 121: 1089–1102. 10.1016/j.mod.2004.06.003 [DOI] [PubMed] [Google Scholar]
- Lee RT. 2018. Adult cardiac stem cell concept and the process of science. Circulation 138: 2940–2942. 10.1161/CIRCULATIONAHA.118.036407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Lièvre CS, Le Douarin NM. 1975. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol 34: 125–154. [PubMed] [Google Scholar]
- Li N, Clevers H. 2010. Coexistence of quiescent and active adult stem cells in mammals. Science 327: 542–545. 10.1126/science.1180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, He L, Huang X, Bhaloo SI, Zhao H, Zhang S, Pu W, Tian X, Li Y, Liu Q, et al. 2018. Genetic lineage tracing of nonmyocyte population by dual recombinases. Circulation 138: 793–805. 10.1161/CIRCULATIONAHA.118.034250 [DOI] [PubMed] [Google Scholar]
- Li Y, Lv Z, He L, Huang X, Zhang S, Zhao H, Pu W, Li Y, Yu W, Zhang L, et al. 2019. Genetic tracing identifies early segregation of the cardiomyocyte and non-myocyte lineages. Circ Res 125: 343–355. 10.1161/CIRCRESAHA.119.315280 [DOI] [PubMed] [Google Scholar]
- Lindsay ME, Dietz HC. 2011. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature 473: 308–316. 10.1038/nature10145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, Qin Z, Li Y, Yang R, Pu W, et al. 2019. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet 51: 728–738. 10.1038/s41588-019-0346-6 [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Lu R, Neff NF, Quake SR, Weissman IL. 2011. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol 29: 928–933. 10.1038/nbt.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majesky MW. 2004. Development of coronary vessels. Curr Top Dev Biol 62: 225–259. 10.1016/S0070-2153(04)62008-4 [DOI] [PubMed] [Google Scholar]
- Majesky MW. 2007. Developmental basis of vascular smooth muscle diversity. Arter Thromb Vasc Biol 27: 1248–1258. 10.1161/ATVBAHA.107.141069 [DOI] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schürmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. 2011. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest 121: 4850–4860. 10.1172/JCI59261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliken BD, Molkentin JD. 2018. Undeniable evidence that the adult mammalian heart lacks an endogenous regenerative stem cell. Circulation 138: 806–808. 10.1161/CIRCULATIONAHA.118.035186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Miyagawa-Tomita S, Mizukami K, Matsuzaki F, Kurihara H. 2019. Isl1-expressing non-venous cell lineage contributes to cardiac lymphatic vessel development. Dev Biol 452: 134–143. 10.1016/j.ydbio.2019.05.002 [DOI] [PubMed] [Google Scholar]
- Mascetti VL, Pedersen RA. 2016. Contributions of mammalian chimeras to pluripotent stem cell research. Cell Stem Cell 19: 163–175. 10.1016/j.stem.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarello P. 1999. A unifying concept: the history of cell theory. Nat Cell Biol 1: E13–E15. 10.1038/8964 [DOI] [PubMed] [Google Scholar]
- McKenna A, Gagnon JA. 2019. Recording development with single cell dynamic lineage tracing. Development 146: dev169730 10.1242/dev.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J. 2016. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353: aaf7907 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. 2009. A bright and photostable photoconvertible fluorescent protein. Nat Methods 6: 131–133. 10.1038/nmeth.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JCI, et al. 2005. Epicardial retinoid X receptor α is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci 102: 18455–18460. 10.1073/pnas.0504343102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. 1995. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci 92: 6991–6995. 10.1073/pnas.92.15.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Fischman DA. 1992. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci 89: 9504–9508. 10.1073/pnas.89.20.9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. 1996. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol 174: 221–232. 10.1006/dbio.1996.0068 [DOI] [PubMed] [Google Scholar]
- Minasi MG, Riminucci M, De Angelis L, Borello U, Berarducci B, Innocenzi A, Caprioli A, Sirabella D, Baiocchi M, De Maria R, et al. 2002. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development 129: 2773–2783. [DOI] [PubMed] [Google Scholar]
- Monvoisin A, Alva JA, Hofmann JJ, Zovein AC, Lane TF, Iruela-Arispe ML. 2006. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev Dyn 235: 3413–3422. 10.1002/dvdy.20982 [DOI] [PubMed] [Google Scholar]
- Morris SA. 2019. The evolving concept of cell identity in the single cell era. Development 146: dev169748 10.1242/dev.169748 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. 2004. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22: 411–417. 10.1038/nbt950 [DOI] [PubMed] [Google Scholar]
- Nguyen PD, Currie PD. 2018. In vivo imaging: shining a light on stem cells in the living animal. Development 145: 150441 10.1242/dev.150441 [DOI] [PubMed] [Google Scholar]
- Nicenboim J, Malkinson G, Lupo T, Asaf L, Sela Y, Mayseless O, Gibbs-Bar L, Senderovich N, Hashimshony T, Shin M, et al. 2015. Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature 522: 56–61. 10.1038/nature14425 [DOI] [PubMed] [Google Scholar]
- Okuda KS, Astin JW, Misa JP, Flores MV, Crosier KE, Crosier PS. 2012. lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish. Development 139: 2381–2391. 10.1242/dev.077701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet 1: 57–64. 10.1038/35049577 [DOI] [PubMed] [Google Scholar]
- Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, Schier AF. 2013. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140: 2835–2846. 10.1242/dev.094631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S, De Val S, Neal A. 2018. Endothelial-specific Cre mouse models. Arter Thromb Vasc Biol 38: 2550–2561. 10.1161/ATVBAHA.118.309669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Feyerabend TB, Rössler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, et al. 2017. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548: 456–460. 10.1038/nature23653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Alvarado AS. 2007. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet 41: 83–105. 10.1146/annurev.genet.41.110306.130244 [DOI] [PubMed] [Google Scholar]
- Pérez-Pomares JM, Phelps A, Sedmerova M, Carmona R, González-Iriarte M, Muñoz-Chápuli R, Wessels A. 2002. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs). Dev Biol 247: 307–326. 10.1006/dbio.2002.0706 [DOI] [PubMed] [Google Scholar]
- Pichol-Thievend C, Betterman KL, Liu X, Ma W, Skoczylas R, Lesieur E, Bos FL, Schulte D, Schulte-Merker S, Hogan BM, et al. 2018. A blood capillary plexus-derived population of progenitor cells contributes to genesis of the dermal lymphatic vasculature during embryonic development. Development 145: dev160184 10.1242/dev.160184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz GA, Bottes S, Betizeau M, Jörg DJ, Carta S, Simons BD, Helmchen F, Jessberger S. 2018. Live imaging of neurogenesis in the adult mouse hippocampus. Science 359: 658–662. 10.1126/science.aao5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plein A, Fantin A, Denti L, Pollard JW, Ruhrberg C. 2018. Erythro-myeloid progenitors contribute endothelial cells to blood vessels. Nature 562: 223–228. 10.1038/s41586-018-0552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget C, Gautier R, Teillet MA, Jaffredo T. 2006. Somite-derived cells replace ventral aortic hemangioblasts and provide aortic smooth muscle cells of the trunk. Development 133: 1013–1022. 10.1242/dev.02269 [DOI] [PubMed] [Google Scholar]
- Prunier C, Chen N, Ritsma L, Vrisekoop N. 2017. Procedures and applications of long-term intravital microscopy. Methods 128: 52–64. 10.1016/j.ymeth.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF. 2018. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 36: 442–450. 10.1038/nbt.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman IL, Krasnow MA. 2010. Coronary arteries form by developmental reprogramming of venous cells. Nature 464: 549–553. 10.1038/nature08873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. 2011. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476: 409–413. 10.1038/nature10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risau W, Flamme I. 1995. Vasculogenesis. Annu Rev Cell Dev Biol 11: 73–91. 10.1146/annurev.cb.11.110195.000445 [DOI] [PubMed] [Google Scholar]
- Ritsma L, Ellenbroek SIJ, Zomer A, Snippert HJ, De Sauvage FJ, Simons BD, Clevers H, Van Rheenen J. 2014. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507: 362–365. 10.1038/nature12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, Greco V. 2012. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487: 496–499. 10.1038/nature11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin FR. 1902. On the origin of the lymphatic system from the veins and the development of the lymph hearts and thoracic duct in the pig. Am J Anat 1: 367–389. 10.1002/aja.1000010310 [DOI] [Google Scholar]
- Sabin FR. 1904. On the development of the superficial lymphatics in the skin of the pig. Am J Anat 3: 183–195. 10.1002/aja.1000030205 [DOI] [Google Scholar]
- Sabin FR. 1917. Preliminary note on the differentiation of angioblasts and the method by which they produce blood-vessels, blood-plasma and red blood-cells as seen in the living chick. Anat Rec 13: 199–204. 10.1002/ar.1090130403 [DOI] [PubMed] [Google Scholar]
- Sánchez-Danés A, Blanpain C. 2018. Deciphering the cells of origin of squamous cell carcinomas. Nat Rev Cancer 18: 549–561. 10.1038/s41568-018-0024-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande-Melón M, Marques IJ, Galardi-Castilla M, Langa X, Pérez-López M, Botos MA, Sánchez-Iranzo H, Guzmán-Martínez G, Ferreira Francisco DM, Pavlinic D, et al. 2019. Adult sox10+ cardiomyocytes contribute to myocardial regeneration in the zebrafish. Cell Rep 29: 1041–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Guzmán T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, Simon A. 2014. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell 14: 174–187. 10.1016/j.stem.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Sauer B, McDermott J. 2004. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res 32: 6086–6095. 10.1093/nar/gkh941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H, Rateri DL, Moorleghen JJ, Majesky MW, Daugherty A. 2017. Smooth muscle cells derived from second heart field and cardiac neural crest reside in spatially distinct domains in the media of the ascending aorta—brief report. Arter Thromb Vasc Biol 37: 1722–1726. 10.1161/ATVBAHA.117.309599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337: 730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. 2013. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152: 25–38. 10.1016/j.cell.2012.12.012 [DOI] [PubMed] [Google Scholar]
- Scott JG, Dhawan A, Hjelmeland A, Lathia J, Chumakova A, Hitomi M, Fletcher AG, Maini PK, Anderson ARA. 2019. Recasting the cancer stem cell hypothesis: unification using a continuum model of microenvironmental forces. Curr Stem Cell Reports 5: 22–30. 10.1007/s40778-019-0153-0 [DOI] [Google Scholar]
- Semo J, Nicenboim J, Yaniv K. 2016. Development of the lymphatic system: new questions and paradigms. Development 143: 924–935. 10.1242/dev.132431 [DOI] [PubMed] [Google Scholar]
- Sharma B, Chang A, Red-Horse K. 2017. Coronary artery development: progenitor cells and differentiation pathways. Annu Rev Physiol 79: 1–19. 10.1146/annurev-physiol-022516-033953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, Sato T. 2017. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545: 187–192. 10.1038/nature22081 [DOI] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. 2012. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell 22: 879–886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Iyer D, Granata A. 2014. Embryonic origins of human vascular smooth muscle cells: implications for in vitro modeling and clinical application. Cell Mol Life Sci 71: 2271–2288. 10.1007/s00018-013-1554-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. 2010. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Spallanzani L. 1768. Prodromo di un opera da imprimersi sopra la riproduzioni anamali. Giovanni Montanari, Modena, Italy. [Google Scholar]
- Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP. 2018. Simultaneous lineage tracing and cell-type identification using CrIsPr-Cas9-induced genetic scars. Nat Biotechnol 36: 469–473. 10.1038/nbt.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spemann H, Mangold H. 1924. Über Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Arch Mikrosk Anat Entwicklungsmech 100: 599–638. [Google Scholar]
- Srinivasan RS, Dillard ME, Lagutin OV, Lin FJ, Tsai S, Tsai MJ, Samokhvalov IM, Oliver G. 2007. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev 21: 2422–2432. 10.1101/gad.1588407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczuk L, Martinez-Corral I, Ulvmar MH, Zhang Y, Laviña B, Fruttiger M, Adams RH, Saur D, Betsholtz C, Ortega S, et al. 2015. cKit lineage hemogenic endothelium-derived cells contribute to mesenteric lymphatic vessels. Cell Rep 10: 1708–1721. 10.1016/j.celrep.2015.02.026 [DOI] [PubMed] [Google Scholar]
- Stanger BZ. 2015. Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol 77: 179–200. 10.1146/annurev-physiol-021113-170255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D. 1981. Bacteriophage P1 site-specific recombination. I: Recombination between loxP sites. J Mol Biol 150: 467–486. 10.1016/0022-2836(81)90375-2 [DOI] [PubMed] [Google Scholar]
- Stone OA, Stainier DYR. 2019. Paraxial mesoderm is the major source of lymphatic endothelium. Dev Cell 50: 247–255.e3. 10.1016/j.devcel.2019.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratman AN, Pezoa SA, Farrelly OM, Castranova D, Dye LE, Butler MG, Sidik H, Talbot WS, Weinstein BM. 2017. Interactions between mural cells and endothelial cells stabilize the developing zebrafish dorsal aorta. Development 144: 115–127. 10.1242/dev.143131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Stanley G, Sinha R, D'Amato G, Das S, Rhee S, Chang AH, Poduri A, Raftrey B, Dinh TT, et al. 2018. Single-cell analysis of early progenitor cells that build coronary arteries. Nature 559: 356–362. 10.1038/s41586-018-0288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM. 2016. The molecular and cellular choreography of appendage regeneration. Cell 165: 1598–1608. 10.1016/j.cell.2016.05.038 [DOI] [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. 2000. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102: 451–461. 10.1016/S0092-8674(00)00050-7 [DOI] [PubMed] [Google Scholar]
- Thomas ED, Raible DW. 2019. Distinct progenitor populations mediate regeneration in the zebrafish lateral line. eLife 8: e43736 10.7554/eLife.43736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Pu WT, Zhou B. 2015. Cellular origin and developmental program of coronary angiogenesis. Circ Res 116: 515–530. 10.1161/CIRCRESAHA.116.305097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topouzis S, Majesky MW. 1996. Smooth muscle lineage diversity in the chick embryo: two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-β. Dev Biol 178: 430–445. 10.1006/dbio.1996.0229 [DOI] [PubMed] [Google Scholar]
- Tritschler S, Büttner M, Fischer DS, Lange M, Bergen V, Lickert H, Theis FJ. 2019. Concepts and limitations for learning developmental trajectories from single cell genomics. Development 146: dev170506 10.1242/dev.170506 [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Fox TP. 2009. Regeneration according to spallanzani. Dev Dyn 238: 2357–2363. 10.1002/dvdy.22057 [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. 2005. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep 6: 233–238. 10.1038/sj.embor.7400361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Johnson SL. 2011. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 20: 725–732. 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. 2004. Defining the epithelial stem cell niche in skin. Science 303: 359–363. 10.1126/science.1092436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvmar MH, Mäkinen T. 2016. Heterogeneity in the lymphatic vascular system and its origin. Cardiovasc Res 111: 310–321. 10.1093/cvr/cvw175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. 2011. Cells of origin in cancer. Nature 469: 314–322. 10.1038/nature09781 [DOI] [PubMed] [Google Scholar]
- Vogt W. 1929. Gestaltungsanalyse am amphibienkeim mit Örtlicher Vitalfärbung. Wilhelm Roux Arch Entwickl Mech Org 120: 384–706. 10.1007/BF02109667 [DOI] [PubMed] [Google Scholar]
- Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML. 2005. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev Biol 281: 78–90. 10.1016/j.ydbio.2005.02.012 [DOI] [PubMed] [Google Scholar]
- Weinreb C, Rodriguez-Fraticelli AE, Camargo FD, Klein AM. 2018. Lineage tracing on transcriptional landscapes links state to fate during differentiation. bioRxiv 467886. 10.1101/467886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman TA, Pan YA. 2015. Brainbow: new resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics 199: 293–306. 10.1534/genetics.114.172510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, et al. 2008. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2: 183–189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Tran K, Khuu J, Dang C, Cui Y, Binder SW, Lowry WE. 2011. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc Natl Acad Sci 108: 7425–7430. 10.1073/pnas.1012670108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman CO. 1887. A contribution to the history of the germ-layers in Clepsine. J Morphol 1: 105–182. 10.1002/jmor.1050010107 [DOI] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. 2004. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci 101: 15905–15910. 10.1073/pnas.0403668101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B, Ipenberg A, Hastie ND, Burch JBE, Bader DM. 2005. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 132: 5317–5328. 10.1242/dev.02141 [DOI] [PubMed] [Google Scholar]
- Wolpert L. 1995. Evolution of the cell theory. Philos Trans R Soc B 349: 227–233. 10.1098/rstb.1995.0106 [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Girskis KM, Walsh CA. 2017. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat Rev Genet 18: 230–244. 10.1038/nrg.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Hasan SS, Schmidt I, Rocha SF, Pitulescu ME, Bussmann J, Meyen D, Raz E, Adams RH, Siekmann AF. 2015. Arteries are formed by vein-derived endothelial tip cells. Nat Commun 5: 5758 10.1038/ncomms6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K, Isogai S, Castranova D, Dye L, Hitomi J, Weinstein BM. 2006. Live imaging of lymphatic development in the zebrafish. Nat Med 12: 711–716. 10.1038/nm1427 [DOI] [PubMed] [Google Scholar]
- Zhang H, Pu W, Li G, Huang X, He L, Tian X, Liu Q, Zhang L, Wu SM, Sucov HM, et al. 2016a. Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ Res 118: 1880–1893. 10.1161/CIRCRESAHA.116.308749 [DOI] [PubMed] [Google Scholar]
- Zhang H, Pu W, Tian X, Huang X, He L, Liu Q, Li Y, Zhang L, He L, Liu K, et al. 2016b. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat Genet 48: 537–543. 10.1038/ng.3536 [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, Von Gise A, Ikeda S, Chien KR, et al. 2008. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454: 109–113. 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Ellenbroek SIJ, Ritsma L, Beerling E, Vrisekoop N, Van Rheenen J. 2013. Brief report: intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31: 602–606. 10.1002/stem.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3: 625–636. 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]