Abstract
Purpose:
The objective was to develop a natural language processing (NLP) algorithm to identify vaccine-related anaphylaxis from plain-text clinical notes, and to implement the algorithm at five health care systems in the Vaccine Safety Datalink.
Methods:
The NLP algorithm was developed using an internal NLP tool and training dataset of 311 potential anaphylaxis cases from Kaiser Permanente Southern California (KPSC). We applied the algorithm to the notes of another 731 potential cases (423 from KPSC; 308 from other sites) with relevant codes (ICD-9-CM diagnosis codes for anaphylaxis, vaccine adverse reactions, and allergic reactions; Healthcare Common Procedure Coding System codes for epinephrine administration). NLP results were compared against a reference standard of chart reviewed and adjudicated cases. The algorithm was then separately applied to the notes of 6 427 359 KPSC vaccination visits (9 402 194 vaccine doses) without relevant codes.
Results:
At KPSC, NLP identified 12 of 16 true vaccine-related cases and achieved a sensitivity of 75.0%, specificity of 98.5%, positive predictive value (PPV) of 66.7%, and negative predictive value of 99.0% when applied to notes of patients with relevant diagnosis codes. NLP did not identify the five true cases at other sites. When NLP was applied to the notes of KPSC patients without relevant codes, it captured eight additional true cases confirmed by chart review and adjudication.
Conclusions:
The current study demonstrated the potential to apply rule-based NLP algorithms to clinical notes to identify anaphylaxis cases. Increasing the size of training data, including clinical notes from all participating study sites in the training data, and preprocessing the clinical notes to handle special characters could improve the performance of the NLP algorithms. We recommend adding an NLP process followed by manual chart review in future vaccine safety studies to improve sensitivity and efficiency.
Keywords: anaphylaxis, allergic reaction, clinical notes, natural language processing, vaccine safety
1 |. INTRODUCTION
Anaphylaxis is a rare but serious allergic reaction that is rapid in onset and may cause death.1,2 Anaphylaxis could cause multiple symptoms3 and it could be caused by various triggers. Vaccine components including antigens, adjuvants, excipients used in the manufacturing process (eg, gelatin, neomycin), or a latex stopper on the vial could each trigger an anaphylactic response.4
Anaphylaxis is a very difficult condition to identify using diagnosis codes and medications in vaccine safety studies because some anaphylaxis cases may be coded as general allergic reactions (eg, urticaria and allergy) and the temporal relatedness to a vaccination versus another potential exposure may be unclear. Possible cases are typically identified through diagnosis codes followed by manual chart review and expert adjudication to confirm case status, a labor-intensive process. In addition, information on sensitivity is unknown (ie, researchers do not have information to assess how many true cases are missed). One example is the recent study evaluating the risk of anaphylaxis due to vaccination in the Vaccine Safety Datalink (VSD) by McNeil et al, in which over a thousand possible cases were identified based on diagnosis codes and only 33 anaphylaxis cases due to vaccination were manually confirmed by medical chart review and adjudication.5
Natural Language Processing (NLP) techniques have been used to identify adverse events in electronic health records in several studies.6–8 Botsis et al developed an NLP algorithm to identify anaphylaxis cases.9 The NLP algorithm was essentially a rule-based classifier combined with machine learning techniques and was applied to detect reports of anaphylaxis in the Vaccine Adverse Event Reporting System database following H1N1 vaccine. In a later study, Ball et al reevaluated the algorithm against the unstructured data derived from electronic medical records (EMR).10 Although the algorithm demonstrated the ability to identify potential anaphylaxis cases, the algorithm did not detect timing, severity, or cause of symptoms. In this study, we developed an NLP algorithm with temporal detection, symptom severity detection and causality assessment to detect anaphylaxis cases caused by vaccination using unstructured data from five VSD sites.
2 |. METHODS
2.1 |. Study population
The study was conducted at five health care organizations within the VSD that were included in the study by McNeil et al, which analyzed data from 2009 through 2011.11 Each participating VSD site routinely creates structured datasets containing demographic and medical information (eg, vaccinations, diagnoses) on its members. For this study, participating sites also created text datasets of clinical notes from the organizations’ EMR according to a standard data dictionary. The text datasets included progress notes, discharge notes, and nursing notes from all care settings (outpatient, emergency department, and inpatient) and telephone notes from 0 to 8 days following the date of vaccination. Kaiser Permanente Southern California (KPSC) served as the lead study site responsible for development of the NLP system and creation of the algorithm using KPSC data. The other participating sites included Kaiser Permanente Colorado, Kaiser Permanente Northwest, Kaiser Permanente Washington, and Marshfield Clinic Research Institute. All five sites participated in the McNeil et al study. The Institutional Review Board of each participating organization approved this study.
The study population included patients who had a vaccination in 2008 or 2012 at KPSC (used to develop the training dataset), and patients who had a vaccination between 2009 and 2011 at KPSC and the other four participating sites mentioned above (used to develop the validation dataset). Potential anaphylaxis cases were identified by using the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes used in the McNeil et al study.5 These ICD-9-CM codes included anaphylaxis codes, vaccine adverse reaction E codes, and selected allergic reaction codes. The anaphylaxis codes were searched within the 0 to 2 days following vaccination. The E codes were searched only on day 0 (same day as vaccination date). Allergic reaction codes were identified only on day 0 and were without a prior diagnosis in the preceding 42 days. The potential allergic reaction cases were further restricted to those who received epinephrine (based on Healthcare Common Procedure Coding System codes) within 24 hours of vaccination, which was the same approach implemented in the McNeil et al study. Receipt of epinephrine was also used as a major criterion to identify anaphylaxis cases by Ball et al10
2.2 |. Training dataset
A total of 311 potential cases were identified in 2008 and 2012 at KPSC. The charts of these potential cases were manually reviewed by KPSC abstractors and adjudicated by a KPSC physician. The results were then used to develop the NLP algorithm. Out of the 311 potential cases, 15 (4.8%) were confirmed as anaphylaxis cases, of which 5 (33.3% of confirmed cases) were related to vaccination. Notes from sites other than KPSC were not included in the training dataset to avoid the transfer to KPSC of other sites’ protected health information in clinical notes text data which would have been needed for training purposes.
2.3 |. Validation dataset
The 731 potential cases following 13 939 925 vaccine doses administered during 2009 to 2011 at all five sites were used to generate the validation dataset. These cases were identified in the previous McNeil et al study.5 Chart reviews were performed by abstractors at each participating site, and adjudication was performed centrally by two physicians. Out of the 731 potential cases, 46 (6.3%) were confirmed as anaphylaxis cases, of which 21 (45.7% of confirmed cases) were related to vaccination. Of those 21, 16 (76.2%) were identified at KPSC and 5 (23.8%) were identified across the other four sites.
The NLP algorithm developed based on KPSC training data was pushed to each participating site to be run on each site’s validation dataset locally, with the NLP output sent back to KPSC for analysis. NLP developers were blinded to all validation datasets at the time of algorithm development.
2.4 |. NLP algorithm
We leveraged KPSC Research & Evaluation (RE) NLP software to develop the algorithm using the training dataset. The software was previously used in multiple KPSC RE studies12,13 to identify cases and retrieve clinical information. It was developed based on open source application programming interfaces (APIs)/libraries including the Natural Language Toolkit (NLTK),14 NegEx/pyConText,15 and Stanford Core NLP.16 The RE NLP software was created using various elements from these open source packages.
The NLP algorithm was developed in multiple steps which are described below. An example is presented in Figure 1.
Step 1. Clinical notes preprocessing. The extracted clinical notes were preprocessed through section boundary detection, sentence separation and word tokenization. Three paragraph separators (new line, carriage return, and paragraph sign) were used to identify the sections. The sentence boundary detection algorithm in NLTK and an additional customized sentence boundary detection algorithm were used to separate sentences. The algorithm was customized to handle some special cases with a nonterminating period. For example, in the sentence “Dr. xxx recommended …,” the period right after “Dr” was not considered as a sentence terminator.
Step 2. Anaphylaxis signs and symptoms list creation. A list of signs or symptoms that were indicative of anaphylaxis was built according to the Brighton Collaboration case definition of anaphylaxis which is used in the McNeil et al study5 and ontologies in the Unified Medical Language System.17 This was further enriched by linguistic variations identified in the training data.
Step 3. Symptom name entity identification. Pattern matching with predefined pattern strings was used to identify the signs and symptoms in the clinical notes. A dictionary was created to map regular expression patterns to the symptoms described as major or minor criteria defined in the Brighton Collaboration case definition of anaphylaxis.
Step 4. Negation detection. A negation algorithm based on NegEx/ pyConText was applied to the identified symptoms in Step 3. Negated symptoms were excluded.
Step 5. Relationship detection. A distance-based relationship detection algorithm was applied to relate the identified signs or symptoms to certain body sites as needed. For example, in the statement “Patient has swelling in his eyes,” the algorithm related “swelling” to “eyes” if the allowable distance (number of words) between the words was equal to or less than 5.
Step 6. Temporal relationship (timing) detection. Temporal relationship detection was utilized to identify and exclude any signs or symptoms prior to vaccination. For example, if the symptom “skin rash” happened before vaccination day, it was excluded. The process was based on pyConText, an algorithm for determining temporal relationship from clinical notes.
Step 7. Case classification. All remaining signs and symptoms identified from the above steps were grouped into the following categories as defined in the Brighton Collaboration case definition: Major Dermatology, Major Cardiovascular, Major Respiratory, Minor Dermatology, Minor Cardiovascular, Minor Respiratory, Minor Gastrointestinal, and Minor Laboratory. The grouped results were processed by the Brighton Collaboration case definition algorithm and produced the final outputs to determine the level of anaphylaxis diagnostic certainty for each clinical note. NLP note-level results were then combined into patient-level results, such that any positive note-level result was translated into a positive patient-level classification. The results of this step were also compared to the chart review and adjudication results from the training dataset. If there was a mismatch, the NLP algorithm was tuned until the NLP results completely matched with the results of chart review and adjudication.
Step 8. Causality assessment for cases meeting Brighton Collaboration criteria. Anaphylaxis cases were classified as vaccine-related based on the following three criteria: onset of the symptom(s) occurred after vaccination, vaccination was mentioned and related to anaphylaxis in the notes, and the anaphylaxis case was not caused by something other than vaccination. The algorithms in the above steps were combined in the final NLP algorithm.
FIGURE 1.
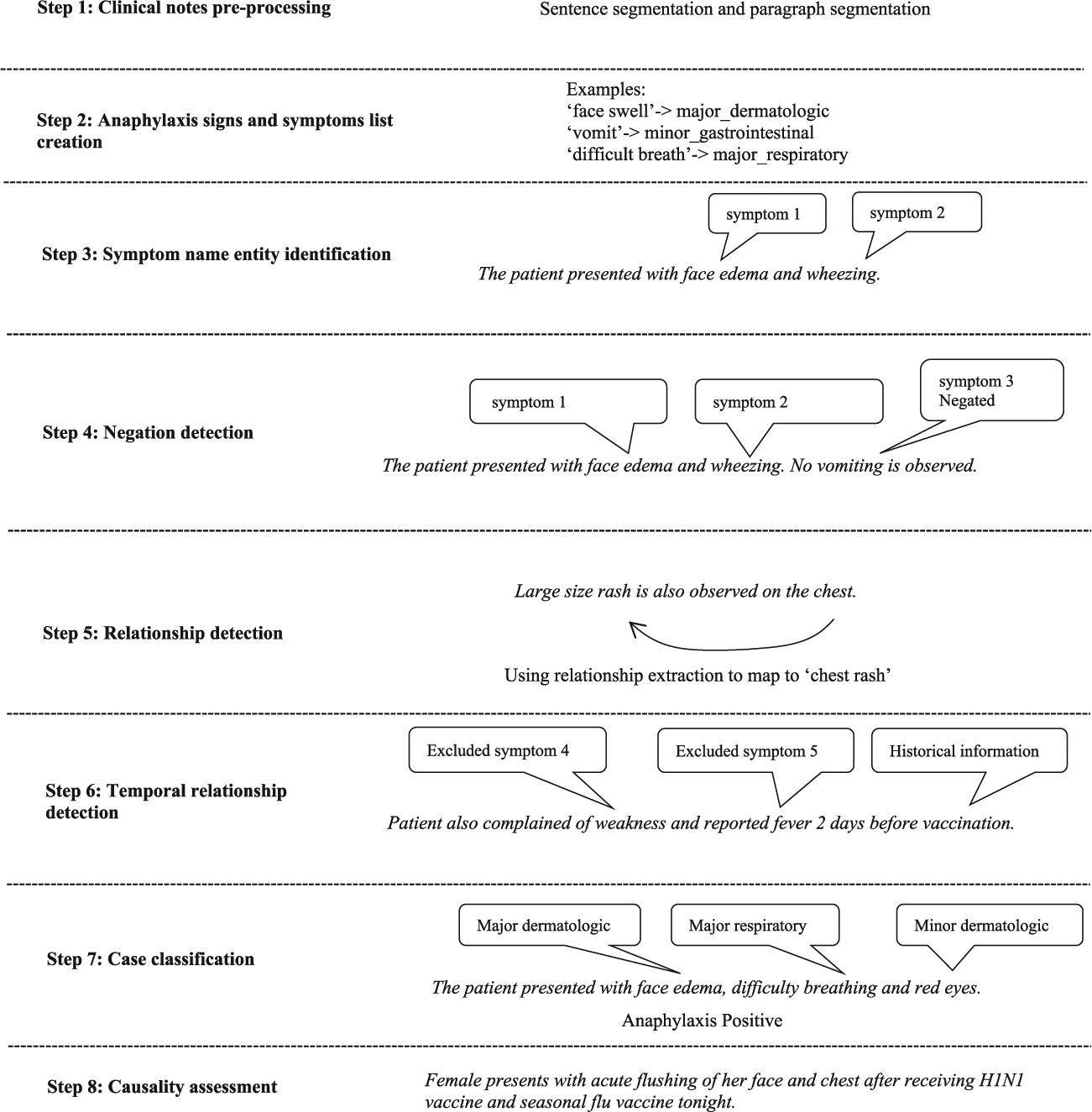
Flow of NLP process
2.5 |. Analysis
The results generated from the NLP algorithm were evaluated against the chart-reviewed and adjudicated reference standard validation dataset. Sensitivity (referred to as “recall” in the informatics literature), specificity, positive predictive value (PPV) (ie, precision), negative predictive value (NPV), and F-score (ie, harmonic mean of recall and precision) were estimated.18 The F-score can range from 0 to 1 with larger values indicating better accuracy. Confidence intervals (CIs) for sensitivity, specificity, PPV, NPV, and F-score were also estimated.
Using the validation dataset, we evaluated the performance of the NLP algorithm in identifying vaccine-related anaphylaxis for KPSC and for other sites. Two sets of performance measures were reported. The first set included patients with or without clinical notes, and the second set only included patients with clinical notes. Electronic text notes may not be available for treatment outside of the health care system, and therefore the set including only patients with clinical notes represents more reasonably the merit of the NLP algorithm. In addition, we evaluated the performance of the NLP algorithm in identifying anaphylaxis cases regardless of cause among all the study subjects (ie, regardless of the availability of clinical notes).
To identify anaphylaxis cases that would have been missed using diagnosis codes, we applied the NLP algorithm to the clinical notes of 6 427 359 KPSC vaccination visits from 2009 to 2011 without depending on the relevant codes (ie, diagnosis, medication). We then performed chart review and adjudication on the potential anaphylaxis cases identified by NLP.
3 |. RESULTS
The NLP algorithm was able to identify 12 of the 16 confirmed vaccine-related anaphylaxis cases at KPSC (sensitivity 75.0%). The NLP algorithm achieved a specificity of 98.5%, PPV of 66.7%, NPV of 99.0%, and F-score of 0.74 (Table 1). In addition, the level of Brighton Collaboration diagnostic certainty assigned by the NLP algorithm matched the manually validated Brighton levels for all 12 true positive cases.
TABLE 1.
Accuracy measurements of the NLP system in identifying vaccine-related anaphylaxis including cases without notes
| Site | Reference standard (n/N) | TP | TN | FN | FP | Sensitivity CI (%) | Specificity CI (%) | PPV CI (%) | NPV CI (%) | F-score CI |
|---|---|---|---|---|---|---|---|---|---|---|
| KPSC | 16/423 | 12 | 401 | 4 | 6 | 75.0 47.6–92.7 |
98.5 96.8–99.4 |
66.7 41.0–86.7 |
99.0 97.5–99.7 |
0.74 0.31–0.97 |
| Site 2 | 1/46 | 0 | 42 | 1 | 3 | 0.0 | 93.3 81.7–98.6 |
0.0 | 97.7 87.7–99.9 |
NA |
| Site 3 | 0/104 | 0 | 102 | 0 | 2 | NA | 98.1 93.2–99.8 |
0.0 | 100.0 97.1–100 |
NA |
| Site 4 | 1/83 | 0 | 80 | 1 | 2 | 0.0 | 97.6 91.5–99.7 |
0.0 | 98.8 93.3–99.9 |
0.00 |
| Site 5 | 3/75 | 0 | 68 | 3 | 4 | 0.0 | 94.4 86.4–98.5 |
0.0 | 95.8 88.1–99.1 |
0.00 |
Note: 95% confidence intervals are displayed beneath point estimates. Reference standard: 2009 to 2011 anaphylaxis cases abstracted and adjudicated as part of McNeil et al study.
Abbreviations: F-score: , with β = 3; FN, false negative—NLP negative, reference standard positive; FP, false positive—NLP positive, reference standard negative; n, number of positive vaccine-related anaphylaxis cases; N, number of cases reviewed; NPV, negative predictive value; TN, true negative—NLP negative, reference standard negative; TP, true positive—NLP positive, reference standard positive; PPV, positive predictive value.
Among the four false negative cases found at KPSC, one case was treated outside of the health care system and the clinical notes were not available. For two cases, the abstractor identified symptoms in a flowsheet which was not part of the clinical notes accessible to NLP. The remaining false negative case was due to symptom level misclassification in that the NLP algorithm failed to identify the cases with symptoms in multiple body locations. For example, “tingling on face and hands,” “itching on face, eyes and chest” were considered as generalized symptoms by manual chart review, but the NLP algorithm treated them as localized symptoms.
At other participating sites, the NLP algorithm failed to identify any of the five anaphylaxis cases. In one case, sentence seperation was missing. In two cases, clinical notes were not available. In another two cases, a generalized symptom was incorrectly recognized as a localized one. The site-specific sensitivity for all non-KPSC sites was either zero or not applicable.
When the patients without clinical notes were excluded, the NLP algorithm achieved a sensitivity of 85.7%, specificity of 98.5%, PPV of 66.7%, NPV of 99.5%, and F-score of 0.83 using KPSC data (Table 2).
TABLE 2.
Accuracy measurements of the NLP system in identifying vaccine-related anaphylaxis after removing cases without notes
| Site | Reference standard (n/N) | TP | TN | FN | FP | Sensitivity CI (%) | Specificity CI (%) | PPV CI (%) | NPV CI (%) | F-score CI |
|---|---|---|---|---|---|---|---|---|---|---|
| KPSC | 14/421 | 12 | 401 | 2 | 6 | 85.7 57.2–98.2 |
98.5 96.8–99.5 |
66.7 41.0–86.7 |
99.5 98.2–99.9 |
0.83 0.98–1.00 |
| Site 2 | 1/46 | 0 | 42 | 1 | 3 | 0.0 | 93.3 81.7–98.6 |
0.0 | 97.7 87.7–99.9 |
0.00 |
| Site 3 | 0/104 | 0 | 102 | 0 | 2 | NA | 98.1 93.2–99.8 |
0.0 | 100.0 97.1–100 |
NA |
| Site 4 | 1/75 | 0 | 72 | 1 | 2 | 0.0 | 97.3 90.6–99.7 |
0.0 | 98.6 92.6–99.9 |
0.00 |
| Site 5 | 1/67 | 0 | 62 | 1 | 4 | 0.0 | 93.9 85.2–98.3 |
0.0 | 98.4 91.5–99.9 |
0.00 |
Note: 95% confidence intervals are displayed beneath point estimates. Reference standard: 2009 to 2011 anaphylaxis cases abstracted and adjudicated as part of McNeil et al study.
Abbreviations: F-score: , with β = 3; FN, false negative—NLP negative, reference standard positive; FP, false positive—NLP positive, reference standard negative; n, number of positive vaccine-related anaphylaxis cases; N, number of cases reviewed; NPV, negative predictive value; TN, true negative—NLP negative, reference standard negative; TP, true positive—NLP positive, reference standard positive; PPV, positive predictive value.
The performance of the NLP algorithm in identifying anaphylaxis cases regardless of cause is summarized in Table 3. When cause was ignored, the number of confirmed cases at KPSC increased from 16 to 26. NLP continued to perform well at KPSC with a sensitivity of 84.6%; however, the sensitivity at non-KPSC sites ranged from 40.0% to 75.0%. At KPSC, the NLP algorithm achieved a sensitivity of 84.6%, specificity of 97.7%, PPV of 71%, NPV of 99%, and F-score of 0.83. Thirty out of 32 true positive cases across all sites matched the validation results for assignment of Brighton Collaboration criteria level. Some of the chart notes from non-KPSC sites were missing sentence-ending punctuations (eg, no period to separate consecutive sentences), so the NLP preprocessing step was not able to separate the text into individual sentences. This resulted in false negative cases at non-KPSC sites.
TABLE 3.
Accuracy measurements of the NLP system in identifying anaphylaxis regardless of cause, including cases without notes
| Site | Reference standard (n/N) | TP | TN | FN | FP | Sensitivity CI (%) | Specificity CI (%) | PPV CI (%) | NPV CI (%) | F-score CI |
|---|---|---|---|---|---|---|---|---|---|---|
| KPSC | 26/423 | 22 | 388 | 4 | 9 | 84.6 65.1–95.6 |
97.7 95.7–98.9 |
71.0 52.0–85.8 |
99.0 97.4–99.7 |
0.83 0.40–0.99 |
| Site 2 | 4/46 | 3 | 33 | 1 | 9 | 75.0 19.4–99.4 |
78.6 63.2–89.7 |
25.0 5.4–57.2 |
97.1 84.7–99.9 |
0.63 0.09–0.99 |
| Site 3 | 2/104 | 1 | 97 | 1 | 5 | 50.0 1.2–98.7 |
95.1 88.9–98.4 |
16.7 0.4–64.1 |
99.0 94.4–99.9 |
0.42 0.01–0.99 |
| Site 4 | 9/83 | 4 | 69 | 5 | 5 | 44.4 13.7–78.8 |
93.2 84.9–97.8 |
44.4 13.7–78.8 |
93.2 84.9–97.8 |
0.44 0.06–0.90 |
| Site 5 | 5/75 | 2 | 61 | 3 | 9 | 40.0 5.2–85.3 |
87.1 76.9–93.9 |
18.2 2.2–51.8 |
95.3 86.9–99.0 |
0.36 0.01–0.99 |
Note: 95% confidence intervals are displayed beneath point estimates. Reference standard: 2009 to 2011 anaphylaxis cases abstracted and adjudicated as part of McNeil et al study.
Abbreviations: F-score: , with β = 3; FN, false negative—NLP negative, reference standard positive; FP, false positive—NLP positive, reference standard negative; n, number of positive vaccine-related anaphylaxis cases; N, number of cases reviewed; NPV, negative predictive value; TN, true negative—NLP negative, reference standard negative; TP, true positive—NLP positive, reference standard positive; PPV, positive predictive value.
After the final anaphylaxis NLP algorithm was applied to the notes of 6 427 359 KPSC vaccination visits between 2009 and 2011 without relevant codes, 45 potential cases were identified by NLP as vaccine-related anaphylaxis cases (Table 4). Among them, 12 cases (26.7%) were confirmed by chart review and adjudication as anaphylaxis positive cases, and 8 out of the 12 (66.7%) were confirmed to be vaccine-related. Six of the eight vaccine-related cases (75.0%) had an allergic reaction code. With 9 402 194 vaccine doses given at KPSC during 2009 to 2011, the incidence rate of vaccine-related anaphylaxis using the traditional approach was 1.70 (95% CI, 0.98–2.75) per million doses, but after adding the cases found by NLP the incidence rate was 2.55 (95% CI, 1.64–3.77) per million doses.
TABLE 4.
Results of application of NLP algorithm to clinical notes of vaccinated subjects without relevant codes
| Possibly vaccine-related anaphylaxis positive cases identified by NLP | Anaphylaxis positive cases confirmed by chart review/adjudication n = 12 | |
|---|---|---|
| Chart review/adjudication: vaccine-related | Chart review/ adjudication: not vaccine-related | |
| 45 | 8 | 4 |
4 |. DISCUSSION
In this study, we demonstrated that NLP can be applied to identify anaphylaxis cases with reasonable accuracy at the site whose data were used for algorithm development. However, performance was lower at non-KPSC sites due to rarity of actual positives and inconsistency in note formatting between KPSC and non-KPSC sites. In order to improve performance at these sites, training data may be needed from each site to enhance the NLP algorithm.
Compared to Ball et al’s evaluation study,10 our study added temporal (timing) detection, severity detection and causality assessment to the NLP algorithm, which seemed to increase specificity but sacrifice sensitivity. However, the PPV and F-score from our study were similar to those of the Ball et al study. The most common reason for false negative cases (low sensitivity) was error in symptom severity detection (local vs generalized). Loosening or removal of the severity detection may yield higher sensitivity. Temporal detection and causality detection in the NLP algorithm helped to achieve high specificity.
This study demonstrated that NLP was able to identify cases directly from clinical notes that did not have specific anaphylaxis-related codes. In the McNeil et al study,5 the potential allergic reaction cases were further restricted to those with epinephrine receipt within 24 hours of vaccination, which markedly reduced the number of charts being reviewed. Using KPSC data, after applying the NLP algorithm to vaccinated subjects without the relevant diagnosis and medication codes and performing chart review and adjudication, we identified 12 additional confirmed anaphylaxis cases, 10 of which were coded as allergic reactions without documentation of epinephrine treatment. The remaining two cases only had diagnosis codes related to symptoms of interest (eg rash, wheezing, and urticaria).
When we included the anaphylaxis cases identified by NLP, the sensitivity based on the traditional approach (ie, codes with exclusions plus chart review and adjudication) was only 67% (=16/24) for vaccine-related anaphylaxis and 68% (=26/38) for anaphylaxis regardless of cause. The true sensitivity could be even lower, because NLP did not capture 100% of true medically-attended anaphylaxis cases in the validation data set. Moreover, we found that NLP performed better in identifying anaphylaxis cases regardless of cause. Without restricting to vaccine-related anaphylaxis, the total true positive anaphylaxis cases increased from 12 to 32 across all sites using the traditional approach.
While NLP alone was not able to identify all anaphylaxis cases, NLP identified additional cases that were missed by manual chart review. The addition of NLP with chart review and adjudication could be a supplemental approach to identify anaphylaxis cases. Initially, the resources and expertise to develop an NLP algorithm may be more costly than that required to conduct manual chart review; however, once an NLP algorithm is developed and achieves acceptable performance metrics, the subsequent manual chart review effort can be reduced.
This study also demonstrates that a distributed NLP system model may be utilized to avoid sharing clinical notes containing protected health information among multiple sites for safety surveillance. In this study, the NLP algorithm was transferred electronically to each site’s NLP server. Notes were processed locally at each respective site. The system was also able to handle large volumes of data associated with the vaccinated population. It processed nearly 12 million clinical notes on a single processor machine. This study demonstrated the feasibility of using NLP to reduce manual efforts in future vaccine safety studies by narrowing down the number of cases that would need to be manually chart reviewed and adjudicated.
Some limitations of this study may be noted. Due to the rarity of the condition, it is inevitable that the number of true cases available to train the NLP algorithm is limited. In the training dataset containing 311 potential anaphylaxis cases, there were only five confirmed vaccine-related anaphylaxis cases. Thus, rare symptoms were not identified in the training dataset, resulting in the failure of capturing some positive cases. For example, the symptom “prickle” was described as “Pt c/o feeling like pins and needles” in the validation dataset. Since there was no such a description in the training dataset, the NLP algorithm was not able to identify this symptom in the validation data. Another limitation is the use of data from 2008 to 2012 for the conduct of this study. There may have been secular changes since then, and ICD-10 codes were not incorporated into the analysis.
Without including all participating sites’ data in the training dataset, our ability to properly train the algorithm to achieve the best possible performance was limited. Although summary data (eg, average number of chart notes per patient, note types) were generated based on site-specific notes to examine data quality at a high level, we were unable to capture the details including the formatting of notes from participating sites. Differences in file structure and data format (eg different paragraph break symbol) between the lead site, where the training sample was generated, and the other participating sites also may have adversely affected NLP performance. Introducing a reformatting algorithm in a preprocessing step to harmonize clinical narratives from different sites could address this issue and improve overall NLP performance.
Lastly, the NLP process was limited to free text electronic chart notes, while medical record reviewers had access to all the available information stored in the EMR. For example, the medical record reviewers had access to scanned documentation or other non-EMR sources, such as insurance claims data, not available to NLP. In the future, complementary approaches using ICD codes, free text notes accessible to NLP, and other structured or semistructured data (eg, vital signs, medications, and so on) could be explored.
5 |. CONCLUSION
The current study demonstrated the potential to apply rule-based NLP algorithms to clinical notes to identify anaphylaxis cases. Increasing the size of training data, including clinical notes from all participating study sites in the training data, and preprocessing the clinical notes to handle special characters could improve the performance of the NLP algorithms. We recommend adding an NLP process followed by manual chart review in future vaccine safety studies to improve sensitivity and efficiency.
ACKNOWLEDGEMENTS
The authors thank Fernando Barreda, Theresa Im, Karen Schenk, Sean Anand, Kate Burniece, Jo Ann Shoup, Kris Wain, Mara Kalter, Stacy Harsh, Jennifer Covey, Erica Scotty, Rebecca Pilsner, and Tara Johnson for their assistance in obtaining data and conducting chart review for this study. The authors also thank Christine Taylor for her project management support and Dr. Bruno Lewin for his adjudication work on the training dataset. This study was funded through the Vaccine Safety Datalink under contract 200-2012-53580 from the Centers for Disease Control and Prevention.
Footnotes
CONFLICT OF INTEREST
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Sampson HA, Muñoz-Furlong A, Campbell RL, et al. Second symposium on the definition and management of anaphylaxis: Summary report—Second National Institute of Allergy and Infectious Disease/ Food Allergy and Anaphylaxis Network symposium. J Allergy Clin Immunol. 2006;117(2):391–397. [DOI] [PubMed] [Google Scholar]
- 2.Tintinalli JE, Stapczynski JS, Ma OJ, Yealy DM, Meckler GD, Cline DM. Tintinalli’s emergency medicine: a comprehensive study guide. New York, NY: McGraw-Hill; 2016. [Google Scholar]
- 3.Archived web page collected at the request of National Institute of Allergy and Infectious Diseases using Archive-It. This page was captured on Sep 06, 2016 and is part of the Public Website Archive. Archive collection 2016. https://wayback.archive-it.org/7761/20160906160831/http://www.niaid.nih.gov/topics/anaphylaxis/pages/default.aspx?wt.ac=bcNIAIDTopicsAnaphylaxis. Accessed February 4, 2016.
- 4.Madaan A, Maddox DE. Vaccine allergy: diagnosis and management. Immunol Allergy Clin. 2003;23(4):555–588. [DOI] [PubMed] [Google Scholar]
- 5.McNeil MM, Weintraub ES, Duffy J, et al. Risk of anaphylaxis after vaccination in children and adults. J Allergy Clin Immunol. 2016;137(3): 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazlehurst B, Naleway A, Mullooly J. Detecting possible vaccine adverse events in clinical notes of the electronic medical record. Vaccine. 2009;27(14):2077–2083. [DOI] [PubMed] [Google Scholar]
- 7.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005;12(4):448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murff HJ, Forster AJ, Peterson JF, Fiskio JM, Heiman HL, Bates DW. Electronically screening discharge summaries for adverse medical events. J Am Med Inform Assoc. 2003;10(4):339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botsis T, Nguyen MD, Woo EJ, Markatou M, Ball R. Text mining for the Vaccine Adverse Event Reporting System: medical text classification using informative feature selection. J Am Med Inform Assoc. 2011;18(5):631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ball R, Toh S, Nolan J, Haynes K, Forshee R, Botsis T. Evaluating automated approaches to anaphylaxis case classification using unstructured data from the FDA sentinel system. Pharmacoepidemiol Drug Saf. 2018;27(10):1077–1084. [DOI] [PubMed] [Google Scholar]
- 11.Baggs J, Gee J, Lewis E, et al. The Vaccine Safety Datalink: a model for monitoring immunization safety. Pediatrics. 2011;127:S45–S53. [DOI] [PubMed] [Google Scholar]
- 12.Zheng C, Yu W, Xie F, et al. The use of natural language processing to identify Tdap-related local reactions at five health care systems in the Vaccine Safety Datalink. Int J Med Inform. 2019;127:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang AH, Chow T, Mora-Marquez J, et al. Breastfeeding persistence at 6 months: Trends and disparities from 2008 to 2015. J Pediatr. 2019;208:169–175. e162 [DOI] [PubMed] [Google Scholar]
- 14.Loper E, Bird S. NLTK: The natural language toolkit. Paper presented at: Proceedings of the ACL-02 Workshop on Effective tools and methodologies for teaching natural language processing and computational linguistics; Vol 12002. [Google Scholar]
- 15.Chapman BE, Lee S, Kang HP, Chapman WW. Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform. 2011;44(5):728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning C, Surdeanu M, Bauer J, Finkel J, Bethard S, McClosky D. The Stanford CoreNLP natural language processing toolkit. Paper presented at: Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations; 2014. [Google Scholar]
- 17.Griffon N, Chebil W, Rollin L, et al. Performance evaluation of unified medical language system®’s synonyms expansion to query PubMed. BMC Med Inform Decis Mak. 2012;12(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altman DG, Bland JM. Statistics Notes: Diagnostic tests 2: predictive values. BMJ. 1994;309(6947):102. [DOI] [PMC free article] [PubMed] [Google Scholar]


