Abstract
Currently the world is being challenged by a public health emergency caused by the coronavirus pandemic (COVID-19). Extensive efforts in testing for coronavirus infection, combined with isolating infected cases and quarantining those in contact, have proven successful in bringing the epidemic under control. Rapid and facile screening of this disease is in high demand. This review summarises recent advances in strategies reported by international researchers and engineers concerning how to tackle COVID-19 via rapid testing, mainly through nucleic acid- and antibody- testing. The roles of biosensors as powerful analytical tools are emphasized for the detection of viral RNAs, surface antigens, whole viral particles, antibodies and other potential biomarkers in human specimen. We critically review in depth newly developed biosensing methods especially for in-field and point-of-care detection of SARS-CoV-2. Additionally, this review describes possible future strategies for virus rapid detection. It helps researchers working on novel sensor technologies to tailor their technologies in a way to address the challenge for effective detection of COVID-19.
Keywords: SARS-CoV-2, COVID-19, Rapid detection, Point-of-care testing, Coronavirus, Biosensor
Highlights
-
•
Recent achievements in rapid COVID-19 detection have been comprehensively reviewed.
-
•
Applications of various biosensing strategies for the diagnosis of SARS-CoV-2 are discussed.
-
•
Commercial products are highlighted in this review.
-
•
New trends and future challenges are outlined.
1. Introduction
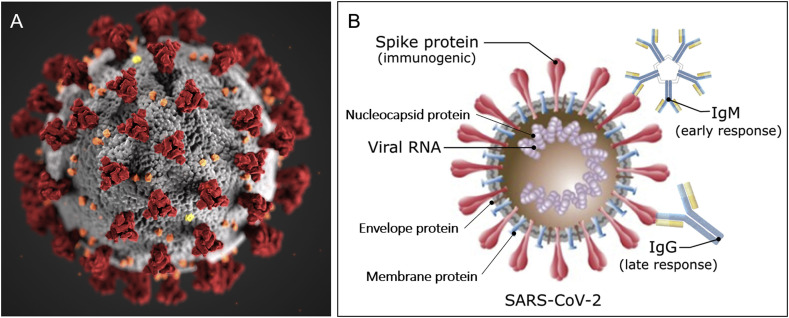
Since the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with the disease referred to as novel coronavirus disease (COVID-19) first reported in early January 2020 in Wuhan, China (Zhu et al., 2020a; Wang et al., 2020), the growing trend of infected cases is not yet under control (Chan et al., 2020; Huang et al., 2020a). COVID-19 was officially announced as a pandemic by the World Health Organisation (WHO) on March 11, 2020. So far there have been more than 7,823,289 cases confirmed globally, with 431,541 deaths from more than 210 countries and territories as of 15th June 2020 (World Health Organization, 2020a). SARS-CoV-2 is the virus strain that causes the respiratory illness COVID-19. SARS-CoV-2 is believed to have zoonotic origins and has close genetic similarity to bat coronaviruses (Chen et al., 2020a, Chen et al., 2020b). It is a positive-sense single-stranded RNA virus with approximately 50–200 nm in diameter (Fig. 1 A) (Xu et al., 2020). Similar to other coronaviruses, SARS-CoV-2 mainly has four structural proteins, namely, the spike (S), membrane (M), envelop (E), and nucleocapsid (N) proteins, respectively (Wrapp et al., 2020). The virus makes full use of S protein to bind to angiotensin-converting enzyme 2 (ACE2) on the human cell surface, to gain entry into a host cell. The N protein holds the viral genome but also involves in the host cellular response to viral infection. S, E, and M proteins together create the viral outer protecting membrane (Fig. 1B) (Wrapp et al., 2020). Both proteins (e.g. S protein) and viral RNA can be used as targets for COVID-19 detection. Alternatively, antibodies such as IgM and IgG from patient samples could also be detected for understanding the infection history. SARS-CoV-2 RNA is detectable 2–3 days before onset of symptoms and can remain up to 25–50 days afterwards, depending on disease severity (He et al., 2020). Many studies show IgM antibodies start to be detectable around 5–10 days after onset of symptoms and rise rapidly, followed by IgG antibody response closely (Peeling et al., 2020). These seroconversions are typically within the first 3 weeks with the mean time of 9–11 days after onset of symptoms for total antibodies (10–12 days for IgM and 12–14 days for IgG). RNA level can remain high despite high concentrations of IgM and IgG antibodies in patient blood (Zhao et al., 2020). These viral infection and immune response studies highlight the detection window for SARS-CoV-2 diagnosis and more importantly, guide the strategic implementation of appropriate types of testing at different infection stages. For example, immune testing can play a big role in tracing symptomatic cases at the middle/late stage of the infection (e.g. 5–10 days after symptom onset). IgM positive result in symptomatic patients fulfilling the COVID-19 case definition is strongly suggestive of SARS-CoV-2 infection. However, RNA testing is still recommended for confirming the case.
Fig. 1.
Schematic diagram of (A) 3D model of the SARS-CoV-2 virion. Reprint from CDC Public Health Image Library (ID 23312: Alissa Eckert and Dan Higgins). (B) Related targeting sites (biomolecules) for COVID-19 detection. Not to scale. Partially reprinted from (Morales-Narváez and Dincer, 2020).
A key message from the WHO in early March is: ‘test, test, test’ (World Health Organization, 2020b). Testing especially rapid detection is extremely critical and a powerful way to monitor and manage the pandemic before vaccines or effective drugs become available. Effective detection helps to confirm infected cases with symptoms shown or even when still within the virus incubation period (diagnostic testing), thus allowing treatment on-time. Millions of RNA-based tests have been carried out around the world looking for the presence of viral genes in a nose or throat swab as a sign of active infection. Additionally, blood tests for antibodies to SARS-CoV-2 can indirectly indicate an active infection or post-infection immunity/infection history (surveillance testing). Prompt testing also helps efficient allocation of medical resources in hospitals and saves time for frontline health workers. Particularly for low-income countries, fast, affordable, in-field and point-of-care testing can have substantial effects in controlling the spread of the disease where health systems may be weak and access to medical treatment limited. Furthermore, valuable information about the local distribution of infections enhances the accuracy of epidemiological prediction and facilitates corresponding policymaking. Therefore, rapid, facile, cost-effective and accessible detections for large-scale screening, in-field testing and point-of-care diagnosis of the disease are of great importance and urgency for quickly controlling the highly contagious and rapid spread of COVID-19. In this work, we first review the recent advance in rapid testing for COVID-19. We then highlight the roles of biosensors for rapid and facile detection of SARS-CoV-2 covering viral RNAs, surface antigens/whole viruses, antibodies and potential biomarkers detection. Finally, future developments of novel virus biosensors are discussed.
2. State-of-the-art of rapid testing approaches for SARS-CoV-2
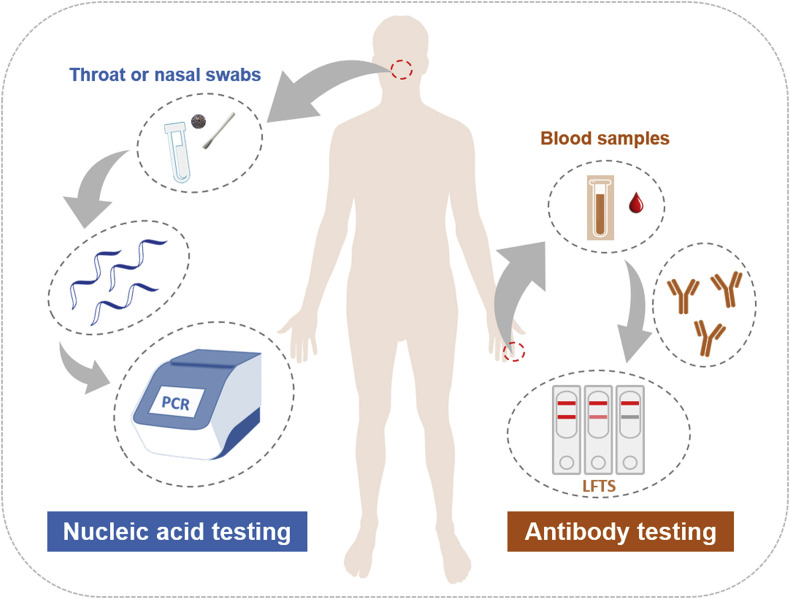
Currently, CT imaging, haematology tests and molecular methods based on viral genetic measurements are the primary tools used for clinical diagnosis of COVID-19, together with the identification of clinical symptoms to confirm infection (Jin et al., 2020b). These laboratory tests are essential to control the burgeoning of the disease. An RNA-based metagenomic next-generation sequencing (mNGS) approach was used to identify the sequence of SARS-CoV-2 immediately after the initial outbreak (Chen et al., 2020a, Chen et al., 2020b). mNGS is a sensitive technique, but it is restricted by throughput, turnaround time, high cost and a requirement for high technical expertise. Rapid detection approaches could usher in an era of point-of-care testing (POCT) or in-field screening of viruses. Fig. 2 shows the two main testing approaches that are currently being used for COVID-19 globally: nucleic acid testing and antibody testing.
Fig. 2.
Current main testing approaches for COVID-19: nucleic acid testing and antibody testing.
2.1. Nucleic acid (RNA) testing
Many laboratory-based molecular diagnostic kits have been developed by disease control organizations, research institutes and private companies and used for testing patients’ specimens since the beginning of COVID-19 pandemic (Chinese Center for Disease Control and Prevention, 2020; CDC, 2020; Corman et al., 2020; Roche Ltd, 2020a). Polymerase chain reaction (PCR)-based nucleic acid testing looks for viral RNAs in upper respiratory specimens (throat and/or nasal swabs) from an individual. Table 1 summarises recent viral nucleic acid-based detection methods for SARS-CoV-2 testing. The quantitative reverse transcription PCR (qRT-PCR) has gradually become the current gold standard for the diagnosis of SARS-CoV-2 infection. SARS-CoV-2 genes such as ORF1ab (open reading frame), RdRp (RNA-dependent RNA polymerase gene), E (envelope protein gene), and N (nucleocapsid protein gene) can be targeted for diagnosis. The general protocol of qRT-PCR is based on the extraction of RNA from respiratory swabs dissolved in viral transport media (VTM), and subsequent one-step reverse transcription and real-time qRT-PCR targeting one or several gene sequences from SARS-CoV-2 (Zou et al., 2020). Researchers have tried to simplify this current protocol by avoiding the RNA extraction step based on direct nasopharyngeal swab VTM heating before the qRT-PCR, which may provide viable options to overcome any supply chain issues and help to increase the testing throughput (Alcoba-Florez et al., 2020). Other new RNA-based methods for SARS-CoV-2 detection (Table 1) have also been developed to tackle this crisis, such as Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) technique (Lamb et al., 2020). Yan et al. evaluated a RT-LAMP assay for the SARS-COV-2 within 30 min using primers targeting ORF1ab and S (spike) genes, with a LOD of 2 × 101 copies and 2 × 102 copies of RNA per reaction, respectively (Yan et al., 2020). No cross-reactivity was found with another 60 respiratory pathogens. The sensitivity (true positive rate) for clinical specimen diagnosis (n = 130) was close to 100% (95% CI 92.3%–100%), as was its specificity (95% CI 93.7%–100%). Compared to qRT-PCR, RT-LAMP is faster and does not require prior RNA isolation from the samples. The reagents for RT-LAMP are relatively cheap and stable at room temperature, and therefore this technique holds promise for use outside of a central laboratory (in-field detection) by staff without special training and without need of advanced equipment (Park et al., 2020). A colorimetric-LAMP method was reported using pH-sensitive dyes to visualise the LAMP amplification via the change in pH resulting from proton accumulation due to the incorporation of deoxynucleoside triphosphates (dNTPs) (Zhang et al., 2020c). A sensitivity/LOD as low as 4.8 copies/μL was achieved in testing RNA samples purified from patient respiratory swabs, and the results were in 100% agreement with those from the qRT-PCR method. This effort expands the toolbox of molecular tests beyond sophisticated diagnostic laboratories in aiming to combat and monitor the growing public health threat. Additionally, digital PCR (dPCR) has been reported to improve the LOD to at least 10-fold lower than that of RT-PCR, and overall accuracy in the clinical detection of 109 samples was reported to be 96.3%, suggesting the potential of dPCR for the detection of asymptomatic and suspect patients (Lu et al., 2020).
Table 1.
Representative commercial test kits with POCT potential and other nucleic acid-based tests for screening of COVID-19.
| Sample volumea | Detection target | Detection method | Sensitivityb |
Specificityc (True negative rate) | Assay detection time | Turnaround time | Commercial products/registration status | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Limit of detection | True positive rate | ||||||||
| 5 μL | RNA (RdRp, E, N genes) | Real-time qRT-PCR | 3.9 copy/reaction (E gene); 3.6 copy/reaction (RdRp gene) | 100% (n = 297) | 100% (n = 297) | ~2 h | >4 h | Developed by academic and public laboratories in national and European research networks | Corman et al. (2020) |
| 5 μL | RNA | Real-time qRT-PCR | 3.2 copy/μL | / | / | ~2 h | >4 h | The CDC Flu SC2 Multiplex Assay; | CDC (2020) |
| FDA-EUA | |||||||||
| / | RNA (ORF-1a, E gene regions) | Real-time qRT-PCR | / | / | / | ~3–8 h | ~1 day | Roche Cobas® SARS-CoV-2 Test (cobas® 6800/8800 Systems); | Roche Ltd (2020a) |
| FDA-EUA + CE-IVD mark | |||||||||
| / | RNA | LAMP | / | / | / | 5 min (positive); 13 min (negative) | <30 min | Abbott ID NOW platform; | Abbott (2020) |
| FDA-EUA | |||||||||
| / | RNA | PCR with lateral flow assay | / | / | / | <30 min | <1 h | Mesa Biotech Accula SARS-CoV-2 Test; | Mesa Biotech (2020) |
| FDA-EUA | |||||||||
| / | RNA | Real-time qRT-PCR | / | / | / | <45 min | <1 h | Cepheid Xpert® Xpress SARS-CoV-2; | Cepheid (2020) |
| FDA-EUA | |||||||||
| / | RNA (N gene) | Isothermal DNA amplification | / | 95.0% (n = 20) | 100% (n = 30) | <30 min | <1 h | Cue Health, Cue COVID-19 Test; | Cue Health (2020) |
| FDA-EUA | |||||||||
| / | RNA | Molecular method | / | 98.7% (n = 102) | 100% (n = 102) | <90 min | <90 min | DRW SAMBA II machines | University of Cambridge (2020) |
| 20 μL | RNA | LAMP with colorimetric readout | 4.8 copy/μL | / | / | ~30 min | <1 h | Tested swab samples; in clinical validation stages | Zhang et al. (2020c) |
| <10 μL | RNA (E, N genes) | CRISPR-based LAMP with lateral flow assay | 10 copy/μL | 95% (n = 40) | 100% (n = 42) | <45 min | <1 h | Tested swab samples; in clinical validation stages | Broughton et al. (2020) |
| 14 μL | RNA | Digital PCR | >1 copy/μL | / | / | <45 min | <1 h | Tested swab samples; in clinical validation stages | Lu et al. (2020) |
| 25 μL | RNA (ORF1ab, S genes) | Reverse transcription-LAMP | 20 copy/reaction | 100% (n = 58) | 100% (n = 72) | <30 min | <1 h | Tested swab samples; in clinical validation stages | Yan et al. (2020) |
| 25 μL | RNA | Reverse transcription-LAMP | 1.02 fg | / | <30 min | <1 h | Only detects simulated patient samples | Lamb et al. (2020) | |
| 15 μL | RNA | Reverse transcription-LAMP | 100 copy/reaction | / | / | <30 min | <1 h | No clinical samples tested | Park et al. (2020) |
| / | Synthetic complementary DNA (RdRp) | RCA with magnetic nanoparticles | sub-femtomolar | / | / | ~100 min | <2 h | No clinical samples tested | Tian et al. (2020) |
Note: qRT-PCR (quantitative reverse transcription-polymerase chain reaction); LAMP (loop-mediated isothermal amplification); CRISPR (clustered regularly interspaced short palindromic repeats); FDA-EUA (Food and Drug Administration-Emergency Use Authorization); CE-IVD (CE marking-In Vitro Diagnostic); RCA (rolling circle amplification).
The sample volume is part of the viral transport medium (VTM) for transport of specimens collected by respiratory swabs (e.g. nasopharyngeal or oropharyngeal).
The sensitivity of an analytical method usually means the change of measured signal corresponding to the change of the concentration of analyte, and/or refers to a method's limit of detection (detection limit), which is the smallest amount of analyte that we can determine with confidence (Harvey, 2010); the sensitivity of a clinical test refers to the ability to correctly identify those patients with the disease (also called the true positive rate) (Lalkhen and McCluskey, 2008).
The specificity of a clinical test refers to the ability to correctly identify those patients without the disease (also called true negative rate) (Lalkhen and McCluskey, 2008).
Although qRT-PCR tests are sensitive and widely used as current diagnostic tools for SARS-CoV-2, they can only be pursued in certified laboratories with expensive equipment and trained technicians but not in places such as airports, borders, big shopping centres, etc., where testing facilities are not easily accessible. Considering the delivery time of samples to centralised labs, qRT-PCR testing could take over 24 h or longer from sampling to results (turnaround time), despite the assay itself taking only a few hours. Additionally, high false negative rates might be an issue for the qRT-PCR testing of COVID-19 due to errors in sampling and testing (Corman et al., 2020; Xie et al., 2020; Tang Xiao et al., 2020). Sometimes the identification of target viral gene sequences cannot confirm the presence of active virus particles, because a positive detection may be due to residues of viral RNA. As summarised in Table 1, many attempts have indeed been made to realise reliable and faster molecular diagnosis of COVID-19, such as LAMP as mentioned above. Many have been available on the market while some still need clinical validation using patient samples. Their commercialisation statuses are highlighted in Table 1 as FDA-EUA (The U.S. Food and Drug Administration-Emergency Use Authorization) or CE-IVD (European CE marking-In Vitro Diagnostic). As of Aug. 21, 2020, the FDA had issued more than 176 molecular tests to diagnose infections with the SARS-CoV-2 virus, including 17 “Diagnostics-Molecular-Home Collection” kits for collecting specimens at home and then sending them to the authorised lab for testing (US FDA, 2020). Some molecular diagnostic tests require a highly trained operator to manually perform the test (e.g., an RNA extraction step using specific extraction platforms and kits). For example, the Influenza SARS-CoV-2 (Flu SC2) Multiplex Assay developed by the Centres for Disease Control and Prevention (CDC) based on qRT-PCR can only be run in high complexity labs, as it can detect and differentiate SARS-CoV-2, influenza A, and/or influenza B in upper and lower respiratory specimens and rely on specific instruments (CDC, 2020). While others are automated and require only limited training to perform. Among those, the isothermal nucleic acid amplification approach with shorter sampling-to-result time and a simpler protocol shows promise in being able to overcome the drawbacks associated with conventional qRT-PCR. As a successful example, the ‘ID NOW Rapid Isothermal System’ was launched for the qualitative detection of COVID-19 with isothermal nucleic acid amplification technology on March 28, 2020 by one of the leading biomedical companies (Abbott (2020). This device is able to give positive results in 5 min and negative results in 13 min. Although it is only available for professional use at the moment, this portable coronavirus testing kit takes molecular testing to the frontlines and has substantially enhanced testing capacity in the USA from early April 2020 after approval by the U.S. FDA under EUA. However, clinical test performance results for this product have not been reported yet. Many other products for nucleic acid detection have also emerged for the point-of-care testing of COVID-19 which were recently approved by authorities for emergency use, such as the Accula SARS-CoV-2 Test from Mesa Biotech (Mesa Biotech, 2020), Xpert® Xpress SARS-CoV-2 from Cepheid Xpert (Cepheid, 2020), and SAMBA II machines (University of Cambridge, 2020) among others.
2.2. Antibody testing
Testing for antibodies in the patient's blood is another modality for COVID-19 detection. Antibody test kits are usually designed for the qualitative detection of IgM and/or IgG antibodies to SARS-CoV-2 in a given serum, plasma (EDTA, citrate) or venipuncture whole blood specimen from a patient. The lateral flow test strip (LFTS) or lateral flow immunoassay (LFIA) is widely used for this purpose. This is a simple cellulose-based device employing chromatographic lateral flow which is intended to detect the presence of a target analyte (antibody to SARS-CoV-2) in a liquid sample (blood/serum/plasma, etc.) without the need for specialized and costly equipment – although lab-based equipment can be used to achieve higher sensitivity (Posthuma-Trumpie et al., 2009). It usually contains a sample pad, a conjugate pad, a nitrocellulose membrane and an absorbent pad. The sample pad is exposed to the sample (a mixture of blood cells, vesicles, cell debris, antibodies, small molecules, etc.) and acts as a filter to promote lateral aid flow. The sample rehydrates the pre-immobilised gold-conjugated recombinant antigen (i.e. spike protein or its receptor binding domain (RBD)) on the conjugate pad and the antibodies bind with their matching antigens. Due to capillary force, the sample continues to flow along the nitrocellulose membrane to reach the test line and the control line. The absorbent pad will absorb excess sample fluid. Colloidal gold nanoparticles are often used for colorimetric visualisation, but coloured latex nanoparticles, fluorophores, etc., can also be used (Sajid et al., 2015). Table 2 summarises representative SARS-CoV-2 antibody-based test strips which are fast and at relatively low cost. For example, a rapid and point-of-care lateral flow immunoassay has been developed for the simultaneous detection of IgM and IgG antibodies against SARS-CoV-2 virus in blood within 15 min (Li et al., 2020b). A chemiluminescence-immunoassay has also been reported for the detection of SARS-CoV-2 infections and surveillance of changing antibody patterns based on the recombinant nucleocapsid antigen and magnetic beads (Lin et al., 2020). Clinical IgG testing identified 65 SARS-CoV-2 infections from 79 confirmed patients and only two false-positive cases from the control group (n = 80) with sensitivity and specificity values reaching 82.3% and 97.5% respectively. In addition, a colloidal gold-based immunochromatographic (ICG) strip test detecting viral IgM or IgG was carried out with 134 samples from 105 patients, and a sensitivity of 11.1% was achieved at the early stage (1–7 days after onset), 92.9% at intermediate stage (8–14 days after onset) and 96.8% at late stage (more than 15 days) (Pan et al., 2020). However, the specificity was not evaluated for this ICG assay. Nonetheless, according to a multi-centre cross-sectional study, the positive rate for IgG from antibody testing could reach 100% at around 20 days after symptom onset (Long et al., 2020), confirming the strong ability of antibody testing kits for use in late-stage infections. The positive rate of serum IgG single testing has been reported to be higher than that of IgM alone in COVID-19 detection, but the detection of both IgG and IgM was shown to be more accurate (Li et al., 2020b; Jin et al., 2020a).
Table 2.
Representative commercial POCT kits and reported antibody tests for screening of COVID-19.
| Sample volumea | Detection target | Detection method | Sensitivity (True positive rate)b | Specificity (True negative rate)c | Assay detection time | Turn-around time | Commercial products/registration status | Ref. |
|---|---|---|---|---|---|---|---|---|
| / | IgM and IgG | LFIA | / | / | <15 min | <30 min | National Bio Green Sciences, NBGS′ Novel Coronavirus (2019-nCoV) IgM/IgG Antibody Rapid Test Kits; | National Bio Green Sciences LLC (2020) |
| FDA-EUA | ||||||||
| / | IgM and IgG | LFIA (colloidal gold) | / | / | <15 min | <30 min | Cellex, qSARS-CoV-2 IgG/IgM Rapid Test; | Cellex (2020) |
| FDA-EUA | ||||||||
| / | IgG and IgM | LFIA | 99.0% (n = 128) | 99.0% (n = 312) | <15 min | <30 min | Autobio Diagnostics, Anti-SARS-CoV-2 Rapid Test; | Autobio (2020) |
| FDA-EUA | ||||||||
| / | Total antibody against N protein | Electrochemical- luminescence immunoassay | 100% (n = 29) | 99.8% (n = 5272) | ~18 min | <30 min | Roche Diagnostics, Elecsys Anti-SARS-CoV-2; | Roche Ltd (2020b) |
| FDA-EUA | ||||||||
| / | Total antibody against RBD of S1 protein | Chemi-luminescent microparticle immunoassay | 100% (n = 42) | 99.8% (n = 1091) | ~10 min | <20 min | Siemens Healthcare, Atellica IM SARS-CoV-2 Total (COV2T); | Siemens (2020) |
| FDA-EUA | ||||||||
| 10–15 μL | IgM and IgG | LFIA | 88.7% (n = 397) | 90.6% (n = 128) | <15 min | <30 min | Medomics Medical Technologies | Li et al. (2020b) |
| / | IgM and IgG | LFIA | 97.8% (IgM) and 99.6% (IgG) | / | <10 min | <25 min | SureScreen Diagnosis, COVID-19 Coronavirus Rapid Test Cassette | SureScreen Diagnostics (2020) |
| 50 μL | IgM and IgG (recombinant nucleocapsid) | Chemi-luminescence immunoassay | 82.3% (n = 79) | 97.5% (n = 80) | <30 min | <45 min | Tianshen Tech, A chemical immuno-luminescence analyzer ACCRE6 | Lin et al. (2020) |
| 10 μL (serum/plasma), 20 μL (whole blood) |
IgM or IgG | Colloidal gold-based immune-chromatographic (ICG) strip | 11.1% (early stage, 1–7 days after onset), 92.9% (inter-mediate stage, 8–14 days after onset) and 96.8% (late stage, >15 days) (n = 134) | / | <15 min | <30 min | Tested blood sample; in clinical validation stages | Pan et al. (2020) |
| / | Antibodies | Graphene field effect transistor (Gr-FET) | / | / | ~2 min | / | Only tested recombinant spike protein | Zhang et al. (2020b) |
| <1 μl (serum) | Antibody | Immune-precipitation and parallel DNA sequencing | 90–97% | ~97% | At least hours | ~1–2 week | / | Xu et al. (2015) |
Note: IgG (Immunoglobulin G); IgM (Immunoglobulin M); LFIA (lateral flow immunoassay); FDA-EUA (Food and Drug Administration-Emergency Use Authorization).
The blood sample is usually in small amount collected from fingertip by “finger-prick”.
The sensitivity of a clinical test refers to the ability to correctly identify those patient samples (also called the true positive rate) (Lalkhen and McCluskey, 2008).
The specificity of a clinical test refers to the ability to correctly identify those non-patient samples (also called true negative rate) (Lalkhen and McCluskey, 2008).
Antibody test kits are not yet available for home testing but do allow testing in laboratories or by healthcare workers at a point-of-care. Antibody testing cannot confirm the presence of the virus. Positive results mean acquired immunity against COVID-19 infection, which might be ascribed to past or present infections with non-SARS-CoV-2 strains such as coronavirus HKU1. In contrast, negative results do not rule out SARS-CoV-2 infection, particularly for those who have been in contact with virus carriers. IgM was found to be detectable in patient's blood after 3–6 days post-infection, with IgG detectable after 8 days (Xie et al., 2020; Long et al., 2020). Hence antibody testing is useful at the intermediate or late stages rather than the early stage of infection (Pan et al., 2020). In a word, this rapid screening tool is more suitable as a complementary method to nucleic acid testing (especially for negative results) by providing important immunological evidence for physicians to make diagnostic and pre-treatment decisions, but not as a sole basis for the diagnosis or exclusion of COVID-19 infection (Zhang et al., 2020a). Notably, once a vaccine for COVID-19 is available and people become immunised by vaccination, antibody testing may not be able to differentiate those who acquire immunity from those infected ones. Industries have been active in developing antibody test kits (mostly immunoassays) (Table 2), such as Novel Coronavirus (2019-nCoV) IgM/IgG Antibody Rapid Test Kits from National Bio Green Sciences LLC (FDA-EUA approved) (National Bio Green Sciences LLC, 2020), qSARS-CoV-2 IgG/IgM Rapid Test from Cellex (FDA-EUA approved) (Cellex, 2020), COVID-19 Coronavirus Rapid Test Cassette from SureScreen Diagnostics (SureScreen Diagnostics, 2020), etc. As of Aug. 21, 2020, the FDA had issued more than 39 serological (antibody) tests for SARS-CoV-2, as well as 3 antigen tests (US FDA, 2020). A technical report from the European Centre for Disease Prevention and Control released that there are over 60 CE-marked rapid SARS-CoV-2 antibody tests as of 1st April 2020 on the market (European Centre for Disease Prevention and Control, 2020). These tests for the qualitative detection of antibodies to SARS-CoV-2 in blood, serum, and/or plasma are intended for use as an aid in identifying individuals with an adaptive immune response to SARS-CoV-2, indicating recent or prior infection. Clinical validation for COVID-19 should be carried out by comparison with a gold standard test for a sufficiently large number of target samples before authorising them as stand-alone diagnostic tests (European Centre for Disease Prevention and Control, 2020). For instance, six commercial POCT lateral flow tests were evaluated for COVID-19 antibodies (Lassaunière et al., 2020) and their overall performance was ranked based on the detection sensitivity and specificity. These findings facilitate the selection of serological assays for the detection of SARS-CoV-2 specific antibodies for diagnostic purposes as well as sero-epidemiological and vaccine development studies. The pandemic is spreading around the world and many countries are facing a second wave of COVID-19. Therefore, more sensitive, low-cost, specific and fast antibody analytical methods are still in great demand to screen for immunity among populations and to help track the progress of the epidemic.
3. Biosensors for rapid and facile detection of SARS-CoV-2
3.1. Detection of nucleic acids
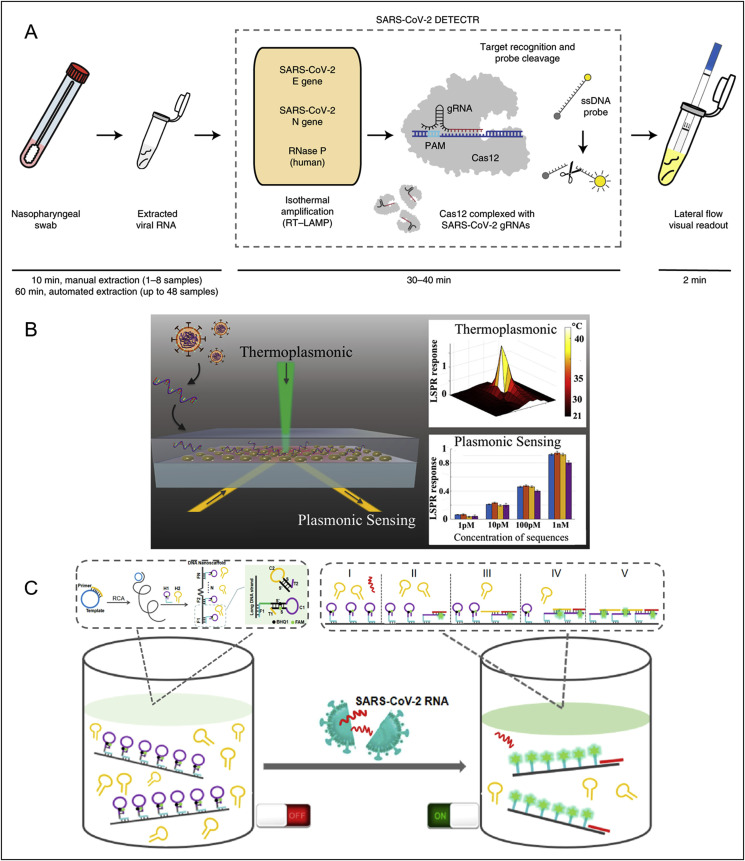
Biosensors as smart analytical devices combing specific recognition of target and sensitive readout of signals can facilitate rapid, facile and cost-effective detection of COVID-19 in field and at a point-of-care. The lateral flow technology can serve as a facile biosensing platform to couple with nucleic acid testing approaches for viral RNA detection. For example, a promising multiplex RT-LAMP coupled with a nanoparticle-based lateral flow biosensor (mRT-LAMP-LFB) was developed for diagnosing COVID-19 with LOD of 12 copies per reaction in 1 h (Zhu et al., 2020b). The sensitivity of SARS-CoV-2 test was 100% (33/33 oropharynx swab patient samples), and the specificity was also 100% (96/96 oropharynx swab non-patient samples). A CRISPR (clustered regularly interspaced short palindromic repeats)/Cas12-based biosensor combined with a lateral flow assay was recently developed for SARS-CoV-2 enabling a test result in around 30 min (Fig. 3 A) (Broughton et al., 2020). After extraction of RNAs from patients samples, the so-called DETECTR performs simultaneous reverse transcription and isothermal amplification using loop-mediated amplification (RT-LAMP) at 62 °C for 20 min, followed by the Cas12 detection of predefined coronavirus sequences at 37 °C for 10 min, after which the cleavage of a reporter molecule confirms detection of the virus as visualized on a lateral flow strip. The compatibility with the lateral strip also enables this CRISPR-based technology to be used for tests at the point-of-care away from the clinical diagnostics laboratory. Similarly, Huang and colleagues reported a rapid CRISPR-Cas12a fluorescent reporter assay coupled with one-step isothermal recombinase polymerase amplification (RPA) methods for amplifying target regions from extracted viral RNAs, with a sample-to-answer time of ~50 min, and a LOD of 2 copies per sample (Huang et al., 2020b). This assay can be readily performed in 96-well microtiter plates and is currently under investigation of the potential to integrate onto a microfluidic chip and smart phone reading system for point-of-care settings. Other automatic integrated gene detection systems are in clinical validation stage for COVID-19, for example, CoVIDNudge (Gibani et al., 2020), AIGS (Li et al., 2020a). In another example, a dual-functional plasmonic biosensor combining the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction has been reported to provide an alternative for clinical COVID-19 detection (Fig. 3B) (Qiu et al., 2020). This dual-functional LSPR biosensor exhibits a high sensitivity toward viral sequences including RdRp, ORF1ab, and E genes from SARS-CoV-2 with a lower LOD down to the concentration of 0.22 pM and allows precise detection of the specific target in a multigene mixture. This biosensor offers a reliable and easy-to-implement diagnosis platform to improve the diagnostic accuracy in clinical tests and relieve the pressure on PCR-based tests. Besides, Jiao et al. developed a fluorescence biosensor based on a DNA nanoscaffold hybrid chain reaction (DNHCR) for rapid detection of SARS-CoV-2 RNA (Fig. 3C) (Jiao et al., 2020). In this biosensor, the DNA nanoscaffolds constructed by the self-assembly of long DNA strands and self-quenching probes (H1) act as the sensing element. The target RNAs initiate the hybridization of H1 and free H2 DNA probes along the nanoscaffold to illuminate the DNA nanostring, which reflects the virus concentration. This DNHCR biosensor can detect SARS-CoV-2 within 10 min and under mild condition (15–35 °C), showing great potential in routine clinical diagnosis. In addition, researchers have conceived new concepts to largely improve the detection capacity of qRT-PCR testing, by taking a pooling approach to enable simultaneous detection of dozens of samples. A study shows that the group testing can identify a positive sample among 64 different samples with enough sensitivity (Yelin et al., 2020). Therefore, if scaled up appropriately, such pooling methods could facilitate mass and large-scale testing with less use of resources and quicker time.
Fig. 3.
Biosensors reported for viral RNA detection of SARS-CoV-2. (A) Schematic of SARS-CoV-2 DETECTR workflow. Conventional RNA extraction can be used as an input to DETECTR (LAMP preamplification and Cas12-based detection for E gene, N gene and RNase P), which is visualized by a fluorescent reader or lateral flow strip. Reprint from (Broughton et al., 2020). (B) A dual-functional plasmonic biosensor combining the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction for the clinical COVID-19 diagnosis. Reprint from (Qiu et al., 2020). (C) A DNA nano scaffold hybrid chain reaction (DNHCR)-based biosensor for the detection of SARS-CoV-2 RNA. Reprinted from (Jiao et al., 2020).
3.2. Direct detection of surface antigens and/or whole viruses
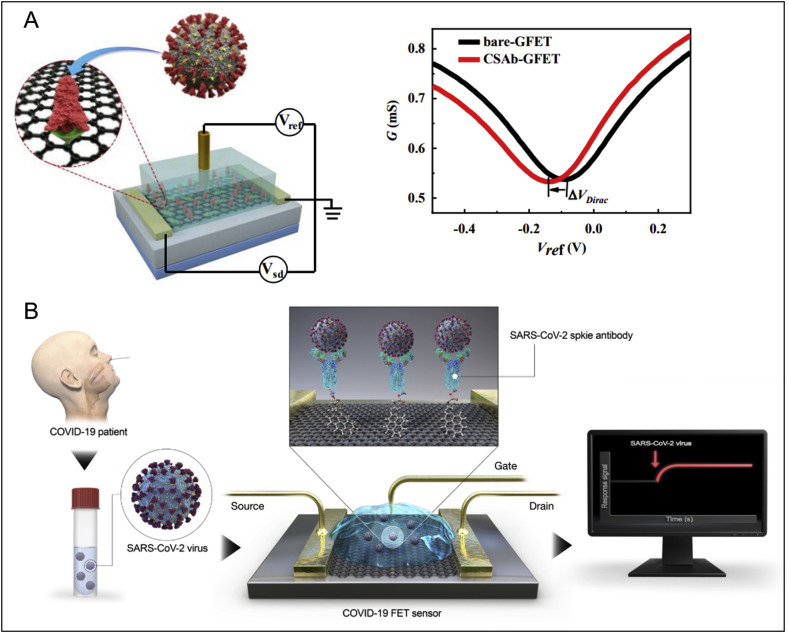
In addition to rapid detection of viral genetic components in swabs using biosensors, it would be attractive to implement the direct and facile sensing of the whole virus particle or its corresponding surface antigen epitope. This strategy could be employed for the development of a diagnostic or screening tool for COVID-19. There have been some relevant research studies reported so far. For example, an ultrasensitive graphene field-effect transistor (Gr-FET) immunosensor was reported recently as an effort towards simple and rapid screening for COVID-19 (Fig. 4 A) (Zhang et al., 2020b). The graphene surface was functionalized with SARS-CoV-2 spike S1 subunit protein antibody (CSAb) or ACE2 receptor. The hybridization of the slightly positively charged S1 protein (which contains a receptor binding domain, RBD) with the immobilised CSAb/ACE2 receptors alters its conductance/resistance via field effect, which can be electrically read out in a sensitive way. This Gr-FET immunosensor can rapidly identify (in about 2 min) and accurately capture the COVID-19 spike protein S1 at a LOD down to 0.2 pM, in a real-time and label-free manner. Although whole coronavirus particles instead of pure antigen proteins need to be tested for assay validation, and clinical trials are then required, this work represents an early proof-of-concept study and demonstrated the potential of Gr-FET technology for sensitive and rapid detection of coronaviruses. Interestingly, another field-effect transistor (FET)-based graphene biosensing device coated with a specific antibody against SARS-CoV-2 spike protein has been reported for direct detection of SARS-CoV-2 (Fig. 4B) (Seo et al., 2020). The introduced virus particles onto the antibody coated graphene surface generated readable electric changes. This COVID-19 FET sensor not only detects SARS-CoV-2 antigen protein transport medium for swab samples, but also detects cultured viruses and viruses in clinical samples. The LOD for clinical sample detection (n = 19 patients and normal subjects) reached 2.42 × 102 copies/mL. A larger clinical sample size would be needed to further validate its clinical potential for virus detection. Nevertheless, given the complexity of clinical samples, the development of novel materials for FET sensors which could overcome problems of non-specific interactions and screening effects associated with clinical samples would be necessary to provide more accurate detection. One of the key issues here is high assay sensitivity with a minimum of false alarms, which needs to be achieved in order to facilitate practical applications. Our team has been developing graphene sensors and FET technology for the detection of cells, exosomes, and biomolecules in biofluids over the past few years (Kwong Hong Tsang et al., 2019; Delle et al., 2018; Hanham et al., 2015). One of our recent work on Gr-FET biosensors for exosome detection can achieve LOD down to single particle level within 10 min, enhanced by surface nano decoration of specific carbon dots (Ramadan et al., 2020). Our carbon dot approach has boosted the detection limit for exosomes, and we expect a similar improvement for COVID-19 detection: exosomes exhibit many similar properties to SARS-CoV-2, including equivalent particle size (50–200 nm), abundant surface antigen molecules, shell-core spherical structure, etc. therefore it is theoretically reasonable to repurpose this ultrasensitive technology for COVID-19 detection. Current work focuses on wafer scalable fabrication of Gr-FET sensors towards sensitive, rapid, in-field and cost-effective screening of SARS-CoV-2, by working together with a world-leading graphene foundry (Graphenea, 2010).
Fig. 4.
Biosensors reported for direct viral antigen or viral particle detection of SARS-CoV-2. (A) A graphene field-effect transistor for electrical probing of SARS-CoV-2 surface antigen (spike protein Si subunit, or its receptor binding domain (RBD)). Reprint from (X. Zhang et al., 2020b). (B) A field-effect transistor-based biosensor for rapid detection of SARS-CoV-2 virus in human nasopharyngeal swab specimens. Reprint from (Seo et al., 2020).
It is important to understand the clinical infectious dose of SARS-CoV-2 in order to develop suitable biosensors for screening or diagnosis. The majority of viral RNA concentrations in upper respiratory tract samples are between 102 - 108 RNA copies (or virus particles) per swab, with the highest to be 7.11 × 108 copy/swab at day 4, first week of symptom onset, and in most cases viral doses in sputum and stool samples are above 102 copies per swab, as suggested by a recent virological analysis of 9 patients (Wölfel et al., 2020). Similar conclusions on viral load were drawn from another clinical study of 17 patients with COVID-19 (Zou et al., 2020). These data indicate that a biosensor capable of accurately detecting 100 virus particles or more (e.g. LOD ≤ 102 RNA/swab) would be potential as a screening tool, and even as a diagnostic tool. It might also be useful for hospital discharge management, since patients beyond day 10 of symptoms with less than 100 viral RNA copies per μL of sputum could be considered for early discharge and ensuing home isolation (Wölfel et al., 2020). Obviously it is easier to detect severe COVID-19 cases than mild ones, as the former tend to express higher viral load and longer virus-shedding periods (Liu et al., 2020). Hence, desired antigen testing methods are those sensitive enough to detect the low end of clinically relevant viral loads, typically around 103 viral nucleic acids per mL (one virion per μL), which is as low as a single particle within one sample, in order to compete with the current qRT-PCR diagnostic method whose LOD is as low as 3 copy/μL of input sample (equal to 3 viral particles) (CDC, 2020; Zou et al., 2020). Another important issue is the specificity, especially for surface antigen/whole virus detection. Positive results from antigen tests can be highly accurate, but there is a higher chance of false negatives, so negative results do not rule out infection, which may cause severe contagion. Other coronaviruses such as Middle East respiratory syndrome coronavirus (MERS-CoV) and Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) are likely to cause interference in the detection of SARS-CoV-2 due to their similar size, viral structure, and infective properties. Influenza virus A or B may also affect the selectivity of detection. Therefore, the biorecognition element, usually antibodies or aptamers, should be carefully selected in the design of biosensors and their specific binding with target virus should be well assessed against those potential interferable viruses.
With advance in functional materials, novel sensing mechanisms and nanotechnology, single-virus analysis would be possible, though still challenging in clinical settings (Schmidt and Hawkins, 2016). Meanwhile, emerging technical progress would help to overcome other challenging issues such as assay stability and reproducibility for biosensors in antigen/whole virus detection, thus facilitating accurate screening or diagnosis. SARS-CoV-2 is a spherical particle around 100 nm in diameter, with many antigens expressed on the surface (Bar-On et al., 2020). Such properties of the SARS-CoV-2 virus are similar to other viruses such as influenza viruses which researchers are more familiar with and more detection approaches are available. Therefore ideas to improve COVID-19 detection toward rapid, facile and reliable applications can be inspired by previous research (Schmidt and Hawkins, 2016; Wang and Li, 2016). For example, by working closely with industry partners and clinicians, our team is currently developing a portable graphene sensor for both viral antigen and whole virus detection, adapting from an established point-of-care platform for exosome analysis (Kwong Hong Tsang et al., 2019; Ramadan et al., 2020). Many other biosensing platforms developed for rapid, in-field, and portable detection of various viruses by our team over the past decades are promising to be repurposed for SARS-CoV-2. For instance, a target-responsive hydrogel aptasensor embedded with quantum dot fluorescent reporters could be used for rapid, one-step and in-field detection of virus in 30 min (Xu et al., 2016). A smart bio-nanogate controlled enzymatic biosensor could enhance the sensitivity of virus detection to almost single particle level (Wang et al., 2015). Other rapid and in-field technologies including an impedance immunosensor based on low-cost microelectrodes (Lin et al., 2015; Wang et al., 2011), an impedance biosensor with gold nanoparticles for signal amplification (Karash et al., 2016), a facile quartz crystal microbalance biosensor (Wang and Li, 2013), etc. have demonstrated outstanding capability to sensitively and specifically detect viruses in swab samples and these approaches are highly promising to be developed as portable COVID-19 detection. Aptamers are single-stranded RNA or DNA oligonucleotides capable of selective and sensitive binding to target antigens via hydrogen bonding, electrostatic, and hydrophobic interactions (Wang et al., 2013). They represent an alternative to antibodies as virus recognition agents in the design of such novel biosensors. Therefore, testing based on antigen/whole virus holds promise for rapid and facile screening in-field or at point-of-care.
3.3. Detection of antibodies
Apart from the reported and commercially available lateral flow test kits for detecting generated antibodies in blood, researchers around the world are intensifying the investigation of new and emerging biosensing technologies for antibody detection. For example, the Gr-FET sensing platform is possible to be developed for screening potential antibody candidates to SARS-CoV-2, if surface antigens like spike proteins are initially coated onto graphene for the sensor design. Zhang and the co-workers have demonstrated the principle of spike protein S1 antigen functionalized Gr-FETs for fast analysis and screening of neutralizing antibodies, which can block coronaviruses from attaching and infecting the health cell (Zhang et al., 2020b). Nevertheless, apart from the current lateral flow-based immunoassays which are based on mature industrial platforms, we believe more rapid detection approaches will be developed with the help of nanomaterials, microfluidics, and 3D printing, etc. for effective screening of COVID-19.
3.4. Detection of other biomarkers
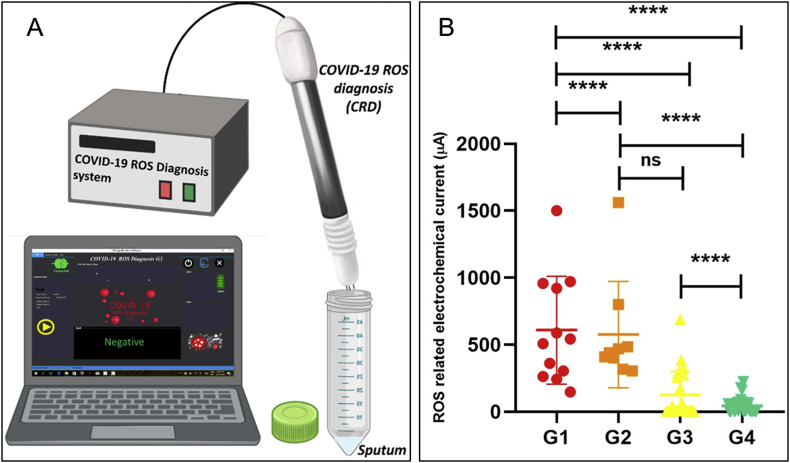
Apart from the detection of viral RNAs, surface antigens, whole virus particles, and the corresponding antibodies, to detect other novel biomarkers may present an interesting testing strategy. In a recent exploratory study to investigate a diagnostic mechanism based on early traces of mitochondrial reactive oxygen species (ROS) overproduction as lung cells’ dysfunctions induced by SARS-CoV-2, Miripour and coworkers developed a rapid, portable and simple electrochemical sensor for ROS measurement in the sputum sample (with a volume of <500 μL) (Miripour et al., 2020). This ROS detector system consists of a disposable sensor as the main diagnostic part of the system, an integrated portable automatic electrochemical readout board and a sample holding unit (Fig. 5 A). The sensor was fabricated by multi-wall carbon nanotubes (MWCNTs) on the tip of steel needles in the conformation of three electrodes (Working (WE), Counter (CE), and Reference (RE)) with a triangular distance of 3 mm from each other and can sensitively measure the current signal of the sample (Fig. 5B) under sweeping potential ranging from −0.8 to 0.8 V with a scan rate of 100 mV/s. Comparing to clinical diagnostics (n > 140 patient samples), more than 97% of true positive patients were detected while the ROS sensor declares the diagnosis in less than 30 s. The clinical analysis specificity would need to be verified. Nevertheless, this type of compact and portable sensing systems is attractive as a powerful assistant in the fast screening of the patients who need further medical examination during the pandemic. The analysis of recently published studies highlights the role of systemic vasculitis and cytokine mediated coagulation disorders as the principal actors of multi organ failure in severe COVID-19 patients and many potential biomarkers have been identified with homocysteine and angiotensin II, in particular, could play a significant role (Ponti et al., 2020). In addition, recent studies show individuals with severe COVID-19 may be at risk for cytokine storm syndrome (Mehta et al., 2020), therefore, point-of-care methods and biosensors that are capable for monitoring cytokine (e.g. IL-6) levels would be needed and beneficial for patients suffering from the severe viral inflammation (Russell et al., 2020).
Fig. 5.
The COVID-19 ROS diagnosis system consists of three needle electrodes coated by functionalized multi-wall carbon nanotubes (A) and is capable of current measurement for differentiating patient samples (B). G1: hospitalized in ICU (n = 25); G2: hospitalized without need to ICU care (n = 36); G3: PCR positive non-hospitalized (n = 45); G4: PCR negative healthy controls (n = 36). Reprinted from (Miripour et al., 2020).
3.5. “REASSURED” biosensors for virus detection
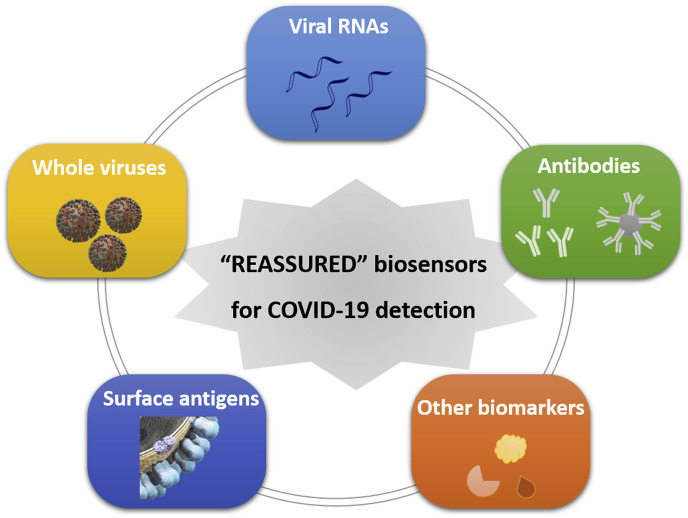
To mitigate the global spread of the COVID-19 pandemic, low-cost, fast, reliable, and sensitive detection methods are still in great demand to screen for the disease in field and at point-of-care and for the immunity among large populations. It is of primary importance to diagnose those already showing symptoms (suspects). It is highly desirable to enable the diagnosis of those without any symptoms (asymptomatic carriers), as in those cases the infected viral dose is usually low and they probably represent a sizeable percentage (more than one-third) of total infections (Qiu, 2020; Nishiura et al., 2020). Beyond the screening technologies discussed in this review, a promising and futuristic (albeit not unrealistic) type of fast detection kit for the SARS-CoV-2 and other viral infections can be visualized to be, for example, a small cartridge or chip with a handheld or portable device that would directly detect viral particles in a swab, or even a breath or saliva sample, within a short period of time without pre-treatment or enrichment. Alternatively, target molecules such as the RNA of the virus could be detected directly, followed by the simple breakage of the outer viral membrane to release the RNA. As one realistic possibility, single immobilised viral particles could be detected through light scattering, given that their relatively high refractive index resembles that of DNA, and commercial CMOS imaging sensors could be employed to detect single virus particles. To summarize, the criteria now known by the acronym ASSURED (Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, Delivered), as coined by the WHO in 2004, should represent the guidelines to be followed in building a strong health care system (Kettler et al., 2004). With rapid advances in digital and mobile health technology, so-called REASSURED (Real-time connectivity, Ease of specimen collection, Affordable, Sensitive, Specific, User-friendly, Rapid and Robust, Equipment-free or simple and Environmentally friendly, Deliverable to end-users) diagnostic systems could be established to strengthen health care systems and improve patient outcomes (Fig. 6 ) (Land et al., 2019). Particularly in low-income and middle-income countries (LMICs) and resource-limited areas without sufficient access to clinical laboratories, the use of point-of-care molecular assays and rapid immunodiagnostic tests (serology testing) should be recommended without any hesitation for SARS-CoV-2 detection (Peeling et al., 2020). Both are now commercially available, scalable, and affordable to enable rapid community-based testing for COVID-19 in these LMICs and areas. Global cooperation and international solidarity could empower their capacity and enhance the goods/reagents supply to combat this pandemic. The “REASSURED” biosensors for virus detection as discussed in this review can help triage symptomatic individuals in community settings, test contacts of confirmed cases, and assist in situational analysis and surveillance, providing the methods highly specific for the disease.
Fig. 6.
Schematic diagram of “RESSURED” biosensors for the detection of SARS-CoV-2 including approaches based on targeting viral RNAs, surface antigens, whole viruses, antibodies and other biomarkers in human specimens.
4. Conclusions and future perspectives
We do not doubt that the COVID-19 pandemic will be fought effectively in the near future. Lessons to be learned from the shocking number of deaths and huge economic crisis involved should alert us to the need to be well-prepared for any viral or other pathogenic microbial outbreaks in the future. In this context, rapid detection strategies are key to the prevention and management of potential future epidemics. Thus it is even more important and essential to develop lab-independent, hospital-decentralised, personalised, and point-of-care diagnostic approaches with cheap, fast, high-throughput and portable screening. Novel sensors based on functional materials, nanotechnologies and creative sensing mechanisms hold promise in terms of unprecedented sensitivity, minimal size and low cost. Progress in molecular and synthetic biology, the discovery of novel binding agents such as nanobodies and aptamers, and bioengineering may facilitate greater specificity in detection. New smart sensing approaches that combine the ultrahigh sensitivity of biosensors with advances in artificial intelligence and the Internet of Things can help to provide better control of any potential spread of diseases (Jeong et al., 2020). The present pandemic will clearly contribute to the definition of goals for agendas in interdisciplinary science in the near future.
CRediT authorship contribution statement
Lizhou Xu: Conceptualization, Writing - review & editing, contributed to the conception of the work, researching of data for article, discussion of content, writing, reviewing/editing of manuscript. Danyang Li: Conceptualization, Writing - review & editing, contributed to the conception of the work, researching of data for article, discussion of content, writing, reviewing/editing of manuscript. Sami Ramadan: Writing - review & editing, contributed to the discussion of content, literature searching, editing of manuscript/proofreading. Yanbin Li: Writing - review & editing, contributed to the conception of the work, interpretation of data, substantive review. Norbert Klein: Writing - review & editing, contributed to the conception, interpretation of data, and, substantively revised the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
L. Xu, S. Ramadan and N. Klein have received funding from the UK Engineering and Physical Science Research Council (EPSRC) through the active grants EP/P02985X/1 and EP/M020398/1.
References
- Abbott Detect COVID-19 in as little as 5 minutes | abbott newsroom. 2020. https://www.abbott.com/corpnewsroom/product-and-innovation/detect-covid-19-in-as-little-as-5-minutes.html [WWW Document]. Abbott.Com.
- Alcoba-Florez J., Gonzalez-Montelongo R., Inigo-Campos A., Artola D.G.-M. de, Gil-Campesino H., Team T.M.T.S., Ciuffreda L., Valenzuela-Fernandez A., Flores C. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int. J. Infect. Dis. 2020;2020 doi: 10.1016/j.ijid.2020.05.099. 04.08.20058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autobio Autobio's two antibody detection kits for SARS-CoV-2. 2020. http://www.autobio.com.cn/en/index.php?m=content&c=index&a=show&catid=29&id=54 [WWW Document] accessed 8.23.20.
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biotech Mesa. Accula SARS-CoV-2 diagnostic test for COVID-19 — Mesa Biotech. 2020. https://www.mesabiotech.com/coronavirus [WWW Document] accessed 4.13.20.
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;1–5 doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC CDC's diagnostic multiplex assay for flu and COVID-19 and supplies | CDC. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html [WWW Document] accessed 8.22.20.
- Cellex COVID-19, lgG/lgM rapid test, antibody detection, blood test, serological test, coronavirus disease. lateral flow immunoassay. 2020 https://cellexcovid.com/ [WWW Document] accessed 4.14.20. [Google Scholar]
- Cepheid Xpert® xpress SARS-CoV-2 has received FDA emergency use authorization. 2020. https://www.cepheid.com/coronavirus [WWW Document] accessed 4.13.20.
- Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H., Hui C.K.M., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B., Mei Y., Zhang W., Chen Y., Li Y., Shi M., Lan K., Liu Y. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microb. Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Center for Disease Control and Prevention Laboratory testing for COVID-19. 2020. http://www.chinacdc.cn/en/COVID19/202003/P020200323390321297894.pdf [WWW Document] [DOI] [PMC free article] [PubMed]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue Health Cue — COVID-19. 2020. https://www.cuehealth.com/covid-19 [WWW Document] accessed 8.23.20.
- Delle L.E., Pachauri V., Sharma S., Shaforost O., Ma H., Adabi M., Lilischkis R., Wagner P., Thoelen R., Klein N., O'Kennedy R., Ingebrandt S. ScFv-modified graphene-coated IDE-arrays for ‘label-free’ screening of cardiovascular disease biomarkers in physiological saline. Biosens. Bioelectron. 2018;102:574–581. doi: 10.1016/j.bios.2017.12.005. [DOI] [PubMed] [Google Scholar]
- Diagnostics SureScreen. COVID-19 coronavirus rapid test cassette | SureScreen Diagnostics. 2020. https://www.surescreen.com/products/covid-19-coronavirus-rapid-test-cassette [WWW Document] accessed 4.14.20.
- European Centre for Disease Prevention and Control . EU/EEA; Stockholm: 2020. An Overview of the Rapid Test Situation for COVID-19 Diagnosis in the. [Google Scholar]
- Gibani M.M., Toumazou C., Sohbati M., Sahoo R., Karvela M., Hon T.-K., De Mateo S., Burdett A., Felice Leung K.Y., Barnett J., Orbeladze A., Luan S., Pournias S., Sun J., Flower B., Bedzo-Nutakor J., Amran M., Quinlan R., Skolimowska K., Klaber R., Davies G., Muir D., Randell P., Crook D., Taylor G.P., Barclay W., Mughal N., P Moore L.S., Jeffery K., Cooke G.S., Graham Cooke P. CovidNudge: diagnostic accuracy of a novel lab-free point-of-care diagnostic for SARS-CoV-2 1. AUTHORS. medRxiv. 2020 doi: 10.1101/2020.08.13.20174193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graphenea High quality graphene producer – Graphenea. 2010. https://www.graphenea.com/ [WWW Document] accessed 4.16.20.
- Hanham S.M., Watts C., Otter W.J., Lucyszyn S., Klein N. Dielectric measurements of nanoliter liquids with a photonic crystal resonator at terahertz frequencies. Appl. Phys. Lett. 2015;107 doi: 10.1063/1.4927242. [DOI] [Google Scholar]
- Harvey D. Analytical Chemistry. vol. 2. 2010. Chapter 3: the vocabulary of analytical chemistry. 0. [DOI] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Adv S., Jeong Hyoyoung, Rogers J.A., Xu S. Continuous on-body sensing for the COVID-19 pandemic : gaps and opportunities. Sci. Adv. 2020;4794 doi: 10.1126/sciadv.abd4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z., Wang Y., Cui H., Pan K., Xu A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P., Fang C., Huang D., Huang L.Q., Huang Q., Han Y., Hu B., Hu F., Li B.H., Li Y.R., Liang K., Lin L.K., Luo L.S., Ma J., Ma L.L., Peng Z.Y., Pan Y.B., Pan Z.Y., Ren X.Q., Sun H.M., Wang Y., Wang Yun Yun, Weng H., Wei C.J., Wu D.F., Xia J., Xiong Y., Xu H.B., Yao X.M., Yuan Y.F., Ye T.S., Zhang X.C., Zhang Y.W., Zhang Y.G., Zhang H.M., Zhao Y., Zhao M.J., Zi H., Zeng X.T., Wang Yong Yan, Wang X.H. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020 doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karash S., Wang R., Kelso L., Lu H., Huang T.J., Li Y. Rapid detection of avian influenza virus H5N1 in chicken tracheal samples using an impedance aptasensor with gold nanoparticles for signal amplification. J. Virol. Methods. 2016;236:147–156. doi: 10.1016/j.jviromet.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Kettler H., White K., Hawkes S. UNICEF/UNDP/World Bank/WHO; 2004. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommandations. [Google Scholar]
- Kwong Hong Tsang D., Lieberthal T.J., Watts C., Dunlop I.E., Ramadan S., del Rio Hernandez A.E., Klein N. Chemically functionalised graphene FET biosensor for the label-free sensing of exosomes. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-50412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalkhen A.G., McCluskey A. Clinical tests: sensitivity and specificity. Cont. Educ. Anaesth. Crit. Care Pain. 2008;8:221–223. doi: 10.1093/bjaceaccp/mkn041. [DOI] [Google Scholar]
- Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel Coronavirus (COVID-19) by reverse transcription-loop-mediated isothermal amplification. medRxiv. 2020 doi: 10.1101/2020.02.19.20025155. 2020.02.19.20025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land K.J., Boeras D.I., Chen X.S., Ramsay A.R., Peeling R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019;4:46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C., Fomsgaard A., Krogfelt K.A., Jørgensen C.S. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.09.20056325. 2020.04.09.20056325. [DOI] [Google Scholar]
- Li Y., Li J., Zhang Y., Dai L., Li L., Liu J., Zhang S., Wu X., Hu Y., Qin C., Jiang T., Kang X. Development of an automatic integrated gene detection system for novel severe acute respiratory syndrome-related coronavirus (SARS-CoV2) Emerg. Microb. Infect. 2020;9:1489–1496. doi: 10.1080/22221751.2020.1782774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. jmv. 2020;25727 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Wang R., Jiao P., Li Yuntao, Li Yanbin, Liao M., Yu Y., Wang M. An impedance immunosensor based on low-cost microelectrodes and specific monoclonal antibodies for rapid detection of avian influenza virus H5N1 in chicken swabs. Biosens. Bioelectron. 2015;67:546–552. doi: 10.1016/j.bios.2014.09.037. [DOI] [PubMed] [Google Scholar]
- Lin D., Liu L., Zhang M., Hu Y., Yang Q., Guo J., Dai Y., Xu Y., Cai Y., Chen X., Huang K., Zhang Z. Evaluations of serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. medRxiv. 2020;27:20045153. doi: 10.1101/2020.03.27.20045153. 2020.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., Deng H., Chen J., Hu J., Liu B., Liao P., Lin Y., Yu L., Mo Z., Xu Y., Gong F., Wu G., Zhang Xian-xiang, Chen Y., Li Z., Wang K., Zhang Xiao-li, Tian W., Niu C., Yang Q., Xiang J., Du H., Liu H., Lang C., Luo X., Wu S., Cui X., Zhou Z., Wang J., Xue C., Li X., Wang L., Tang X., Zhang Y., Qiu J., Liu X., Li J., Zhang D., Zhang F., Cai X., Wang D., Hu Y., Ren J., Tang N., Liu P., Li Q., Huang A. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020 doi: 10.1101/2020.03.18.20038018. 2020.03.18.20038018. [DOI] [Google Scholar]
- Lu R., Wang J., Li M., Wang Y., Dong J., Cai W. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge. medRxiv. 2020 doi: 10.1101/2020.03.24.20042689. 2020.03.24.20042689. [DOI] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020 doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Bio Green Sciences LLC 15 minute COVID-19 rapid test kit announced - jems. 2020. https://www.jems.com/2020/04/01/15-minute-covid-19-rapid-test-kit-announced/ [WWW Document] accessed 4.14.20.
- Nishiura H., Kobayashi T., Suzuki A., Jung S.-M., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R., Linton N.M., Miyama T. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., Long X., Guo S., Zhao Z., Liu Y., Hu H., Xue H., Li Y. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 2020;81:28–32. doi: 10.1101/2020.03.13.20035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.-S., Ku K., Beak S.-H., Kim S.J., Kim S. Il, Kim B.-T., Maeng J.-S. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J. Mol. Diagnostics. 2020 doi: 10.1101/2020.03.09.983064. 2020.03.09.983064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Sall A., Tanuri A., Heymann D.L. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020 doi: 10.1016/s1473-3099(20)30517-x. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020 doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma-Trumpie G.A., Korf J., Van Amerongen A. Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009;393:569–582. doi: 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- Qiu J. Covert coronavirus infections could be seeding new outbreaks. Nature. 2020 doi: 10.1038/d41586-020-00822-x. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Ramadan S., Lobo R., Zhang Y., Xu L., Shaforost O., Tsang D.K.H., Yin T., Qiao M., Rajeshirke A., Jiao L.R., Petrov P.K., Dunlop I.E., Titirici M.-M., Klein N. Carbon dot (CDs) enhanced graphene field effect transistors for ultrasensitive detection of exosomes. Unpublished results. 2020 doi: 10.1021/acsami.0c18293. [DOI] [PubMed] [Google Scholar]
- Roche Ltd cobas® SARS-CoV-2 Test. 2020. https://diagnostics.roche.com/global/en/products/params/cobas-sars-cov-2-test.html [WWW Document] accessed 4.15.20.
- Roche Ltd Elecsys® anti-SARS-CoV-2. 2020. https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html [WWW Document] accessed 8.23.20.
- Russell S.M., Alba-Patiño A., Baron E., Borges M., González-Freire M., de la Rica R. Biosensors for managing the COVID-19 cytokine storm: challenges ahead. ACS Sens. 2020 doi: 10.1021/acssensors.0c00979. 2020. [DOI] [PubMed] [Google Scholar]
- Sajid M., Kawde A.N., Daud M. Designs, formats and applications of lateral flow assay: a literature review. J. Saudi Chem. Soc. 2015;19:689–705. doi: 10.1016/j.jscs.2014.09.001. [DOI] [Google Scholar]
- Schmidt H., Hawkins A.R. Single-virus analysis through chip-based optical detection. Bioanalysis. 2016 doi: 10.4155/bio-2016-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S. Il. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Siemens SARS-CoV-2 total assay - siemens healthineers USA. 2020. https://www.siemens-healthineers.com/en-us/laboratory-diagnostics/assays-by-diseases-conditions/infectious-disease-assays/cov2t-assay [WWW Document] accessed 8.23.20.
- Tang Xiao A., Xin Tong Y., Gao C., Zhu L., Jie Zhang Y., Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J. Clin. Virol. 2020:104346. doi: 10.2139/ssrn.3548769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- University of Cambridge Rapid COVID-19 diagnostic test developed by Cambridge team to be deployed in hospitals. 2020. https://www.cam.ac.uk/research/news/rapid-covid-19-diagnostic-test-developed-by-cambridge-team-to-be-deployed-in-hospitals [WWW Document]. Online. accessed 4.13.20.
- US F.D.A. FDA combating COVID-19 with medical devices. 2020. https://www.hsdl.org/?view&did=838993
- Wang R., Li Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013;42:148–155. doi: 10.1016/j.bios.2012.10.038. [DOI] [PubMed] [Google Scholar]
- Wang R., Li Y. Steps Forwards in Diagnosing and Controlling Influenza. InTech; 2016. Biosensors for rapid detection of avian influenza. [DOI] [Google Scholar]
- Wang R., Lin J., Lassiter K., Srinivasan B., Lin L., Lu H., Tung S., Hargis B., Bottje W., Berghman L., Li Y. Evaluation study of a portable impedance biosensor for detection of avian influenza virus. J. Virol. Methods. 2011;178:52–58. doi: 10.1016/j.jviromet.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhao J., Jiang T., Kwon Y.M., Lu H., Jiao P., Liao M., Li Y. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J. Virol. Methods. 2013;189:362–369. doi: 10.1016/j.jviromet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Wang R., Xu L., Li Y. Bio-nanogate controlled enzymatic reaction for virus sensing. Biosens. Bioelectron. 2015;67:400–407. doi: 10.1016/j.bios.2014.08.071. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease (COVID-2019) Situation Reports - 147. [Google Scholar]
- World Health Organization WHO Director-General’s opening remarks at the media briefing on COVID-19 - 16 March 2020. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-16-march-2020 [WWW Document]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 84. 2020;367:1260–1263. doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. 200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung’u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T., O'Connor K.C., Walker B., Larman H.B., Elledge S.J. Comprehensive serological profiling of human populations using a synthetic human virome. Science 84. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wang R., Kelso L.C., Ying Y., Li Y. A target-responsive and size-dependent hydrogel aptasensor embedded with QD fluorescent reporters for rapid detection of avian influenza virus H5N1. Sensor. Actuator. B Chem. 2016;234:98–108. doi: 10.1016/j.snb.2016.04.156. [DOI] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin I., Aharony N., Tamar E.S., Argoetti A., Messer E., Berenbaum D., Shafran E., Kuzli A., Gandali N., Shkedi O., Hashimshony T., Mandel-Gutfreund Y., Halberthal M., Geffen Y., Szwarcwort-Cohen M., Kishony R. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X. Lou, Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Qi Q., Jing Q., Ao S., Zhang Z., Ding M. 2020. Electrical probing of COVID-19 spike protein receptor binding domain via a graphene field-effect transistor. arXiv 1–20. [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020;2 doi: 10.1101/2020.02.26.20028373. 2020.02.26.20028373. [DOI] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]