Abstract
Synthetic mRNA represents an exciting cancer vaccine technology for the implementation of effective cancer immunotherapy. However, inefficient in vivo mRNA delivery along with a requirement for immune co-stimulation present major hurdles to achieving anti-tumor therapeutic efficacy. Here, we demonstrate a proof-of-concept adjuvant-pulsed mRNA vaccine nanoparticle (NP) that is composed of an ovalbumin-coded mRNA and a palmitic acid-modified TLR7/8 agonist R848 (C16-R848), coated with a lipid-polyethylene glycol (lipid-PEG) shell. This mRNA vaccine NP formulation retained the adjuvant activity of encapsulated C16-R848 and markedly improved the transfection efficacy of the mRNA (>95%) and subsequent MHC class I presentation of OVA mRNA derived antigen in antigen-presenting cells. The C16-R848 adjuvant-pulsed mRNA vaccine NP approach induced an effective adaptive immune response by significantly improving the expansion of OVA-specific CD8+ T cells and infiltration of these cells into the tumor bed in vivo, relative to the mRNA vaccine NP without adjuvant. The approach led to an effective anti-tumor immunity against OVA expressing syngeneic allograft mouse models of lymphoma and prostate cancer, resulting in a significant prevention of tumor growth when the vaccine was given before tumor engraftment (84% reduction vs. control) and suppression of tumor growth when given post engraftment (60% reduction vs. control). Our findings indicate that C16-R848 adjuvant pulsation to mRNA vaccine NP is a rational design strategy to increase the effectiveness of synthetic mRNA vaccines for cancer immunotherapy.
Keywords: mRNA vaccine, Adjuvant pulsation, Nanoparticle, Cancer, Immunotherapy
1. Introduction
Adjuvant addition has been widely used for various prophylactic vaccines, such as the use of alum in the vaccine formulations for immunization against hepatitis A and B, diphtheria and tetanus, Haemophilus influenzae B, and others [1]. The use of immune stimulants as adjuvants for cancer treatment has also been reported, with the use of Bacillus Calmette-Guérin (BCG) for bladder cancer treatment [2] and interleukins for melanoma [3] as examples. In particular, there is significant interest in understanding how the concurrent stimulation of both the innate and adaptive immune responses can generate effective cytotoxic T cell responses to eliminate tumor cells. Utilizing cancer vaccine strategies is an alternative type of cancer immunotherapy [4]; Sipuleucel-T is an example of a dendritic cell (DC)-based vaccine that has been approved for the treatment of prostate cancer [5]. It is worth noting that a significant limitation of functional immunotherapy is the absence of an effective and durable CD8+ T cell-mediated cytotoxic response. Stimulation and expansion of antigen-specific CD8+ T cell populations are often not sufficient to mediate a persistent response and, even in the presence of antigen-specific CD8+ T cells, tumors can persist and grow [6]. To overcome this limitation, it is important to stimulate the innate immune response to unique tumor epitopes in order to generate CD8+ T cells with appropriate effector phenotypes is crucial which is associated with functional therapeutic efficacy and tumor-suppressive outcomes. Effective T cell-mediated immune response can be induced by the careful use of a combination of exposure to antigen along with the co-delivery of immune stimulating adjuvants [7].
mRNA-based therapeutics as well as vaccines hold incredible therapeutic potential and there are currently a number of mRNA-based systems that are progressing to clinical use, including mRNA vaccines for COVID-19 [[8], [9], [10], [11], [12], [13], [14], [15]]. Previous studies have demonstrated the clinical promise of mRNA for a range of therapeutic applications [8,[16], [17], [18], [19], [20], [21], [22], [23], [24]], proving the utility of mRNA as a powerful immuno-therapeutic platform that can achieve the necessary requirements for safety and scaled-up GMP production [25]. One method used to improve mRNA stability is the modification of mRNA transcripts with alternative naturally occurring nucleotides, such as pseudouridine (Ψ), 5-methylcytidine (5meC) and N1-methylpseudouridine (m1Ψ) [[26], [27], [28], [29], [30]]. However, with these modifications, mRNA can become ‘immunosilent’ and no longer trigger type I interferon (IFN) induction via recognition by toll-like receptor 3 (TLR3), TLR7, and TLR8 [31]. These modifications diminish RNA's self-adjuvant-effect, thus weakening DC activation and subsequent T-cell priming. We propose that incorporating adjuvant pulsation into an anti-tumor vaccine could restore innate immune activation in the context of these synthetic mRNAs and lead to greater immune stimulation [31].
TLR7 and TLR8 mediate type 1 IFN signaling in response to viral infection [[32], [33], [34]]. Synthetic small molecular TLR7/8 agonists from the imidazoquinoline family, including imiquimod and resiquimod (R848), have demonstrated potent anti-tumor immunity and are currently being evaluated in clinical trials [35,36]. Resiquimod (R848) has been shown to induce immune cell activation and improve therapeutic outcomes when used as an adjuvant in the context of cancer vaccination [37]. In recent work, R848 showed the polarization of tumor-associated macrophages from M2 to M1 phenotype to enhance cancer immunotherapy [38]. The objective of this study was to develop and evaluate a novel vaccine strategy that combines the use of antigen-encoding mRNA with a TLR7/8 agonist, thereby stimulating concurrent innate immune activation to enhance the magnitude and quality of the resulting antigen-specific T cell response.
Nanoparticle (NP)-based platforms offer a promising solution to not only effectively deliver mRNA but also other payloads such as adjuvants to improve therapeutic efficacy [[39], [40], [41], [42], [43]]. The design of such systems, however, is not without challenges. The particles need to be appropriately sized to condense mRNA and increase its stability while protecting against degradation and rapid clearance from the body and avoiding off-target interactions. These properties are also required for NP-mediated delivery of adjuvants (e.g., R848) capable of modulating and potentiating the antigen-specific immune responses induced by mRNA vaccines. Moreover, one of the essential features of NP-mediated delivery of R848 vs. free R848 is to avoid systemic cytokine response, thus reducing systemic toxicity [37]. Consequently, a NP co-delivery system consisting of both an mRNA vaccine and an adjuvant, such as R848, could provide concurrent stimulation of innate and adaptive immune responses with minimal toxic side-effects, resulting in potent protective or therapeutic anti-tumor immunity.
In this report, we demonstrate the promising potential of utilizing a co-stimulatory NP platform for the co-delivery of a synthetic mRNA antigen along with a chemically-modified R848 adjuvant to stimulate concurrent innate and adaptive immune responses. To overcome hydrophilicity and improve encapsulation efficacy, we modified R848 with palmitic acid to make lipophilic C16-R848. This co-delivery strategy demonstrated high transfection efficacy, robust expression of MHC class I antigen presentation in vitro in APCs, as well as an enhanced T cell response in vivo. We have further shown the efficacy of this approach in both prophylactic and therapeutic treatment settings in two murine syngeneic allograft tumor models. Together this work demonstrates the potential of NP delivery of synthetic mRNA pulsed with a chemically-modified TLR7/8 adjuvant to increase the anti-tumor efficacy of mRNA vaccines.
2. Materials and methods
2.1. Materials
EGFP mRNA and OVA mRNA (each modified with pseudouridine and 5-methylcytidine) were purchased from TriLink Biotechnologies. Cationic ethylenediamine core-poly(amidoamine) (PAMAM) generation 0 (G0) dendrimer, palmitic acid (C16–COOH), resiquimod (R848), anhydrous dichloromethane (DCM), N,N′-dicyclohexylcarbodiimide (DCC), anhydrous N,N-dimethylformamide (DMF), 4-dimethylaminopyridine (DMAP), hydrochloric acid (HCl), and magnesium sulfate were purchased from Sigma-Aldrich. Ceramide-PEG (N-palmitoyl-sphingosine-1-(succinyl{methoxy[polyethylene glycol]}) with PEG MW of 2000 was obtained from Avanti Polar Lipids. Lipofectamine 2000 (L2K) was purchased from Invitrogen.
2.2. Synthesis of C16-R848
C16-R848 was synthesized as we recently described [44]. R848 (50 mg) was dissolved in a co-solvent (3 ml of DCM plus 1 ml of DMF) and stirred at 1600 rpm under inert conditions using nitrogen gas. 100 mg of palmitic acid and DCC were then added to the stirring solution. The inert conditions were restored and the reaction mixture was stirred for ~10 min. Once the compounds were well dissolved, 21 mg of DMAP was added and the reaction mixture was allowed to stir overnight at 1600 rpm under nitrogen gas prior to vacuum filtered through a Büchner funnel to eliminate the precipitates made during the reaction. Then the mixer was washed with water for six times (three washes with pH 2.0 and three with pH 7.0) to remove hydrophilic reactants and other by-products, while the hydrophobic C16-R848 remained in the DCM/DMF mixture (the organic layer). Then to remove the remaining water, the organic layer was placed into a funnel containing magnesium sulfate desiccant. Further, to remove residual magnesium sulfate, this solution was then filtered into a pre-weighed 40 ml glass vial. A rotary evaporator at 300 mbar and 35 °C with 135 rpm was used to evaporate the residual solvent. After the evaporation, a wax-like residue was obtained. The product was then weighed and given to the Massachusetts Institute of Technology's Biopolymers and Proteomics Lab for purification. To purify, 2 mL of hexafluoro-2-propanol (HFIP) was used to dissolve C16-R848 and further diluted in 50 μl of dimethyl sulfoxide (DMSO). It was then purified using a Gilson GX-271 HPLC (Gilson, Middleton, Wisconsin, USA) with a Vydac 214TP101522 22 × 250 mm C4 column (Grace, Columbia, Maryland, USA). The method used was 10-10%B/15′-100%B/60’. The detection wavelengths were 254 and 320 nm. The identity and purity of the compound were verified using NMR and MALDI-TOF (Bruker, Billerica, Massachusetts, USA) and purity was analyzed using analytical HPLC.
2.3. Preparation and stability of G0-C14/mRNA complex
To assess mRNA complexation capacity by G0-C14 and evaluate stability of G0-C14/mRNA complex in organic solvent (DMF), naked EGFP-mRNA or EGFP-mRNA complexed G0-C14 (at different weight ratios from 1 to 20) were kept with or without DMF for 30 min. The samples were then run into a Novex® 10% Tris-glycine gel for 30 min at 50 V. UV light was used to image the gel and the bands were evaluated.
2.4. Preparation of mRNA vaccine NP
A self-assembly method was used to make the mRNA vaccine NP. In brief, 10 μg of C16-R848 (1 mg/ml in DMF) was mixed with 250 μg of G0-C14 (2.5 mg/ml in DMF) in a small glass vial in DMF. Then 15 μg of mRNA (1 mg/ml in aqueous solution) was mixed into the C16-R848/G0-C14 organic solution (the ratio of aqueous and organic solvent was 1:20) to form cationic lipid/mRNA/adjuvant nanocomplexes. This solution was then nanoprecipitated slowly into 10 ml of aqueous solution containing lipid-PEG (i.e. ceramide-PEG) at a concentration of 0.1 mg/ml in DNase/RNase-free Hypure water. NPs formed rapidly upon nanoprecipitation. The NPs were stirred at 600 rpm for 30 min at room temperature to stabilize. The ice-cold Hypure water was used to wash the NPs using Amicon tubes (MWCO 100 kDa; Millipore) to remove the organic solvent and other free compounds. The NPs were finally concentrated in PBS to make an mRNA concentration of 30 μg/ml for in vitro or further concentrated to make 300 μg/ml for in vivo studies and were either used fresh or kept at −80 °C to use later. Gel electrophoresis was employed to run the mRNA NPs to check for any free mRNA and/or mRNA leaching from the NPs. The encapsulation percentage of C16-R848 was analyzed by HPLC.
2.5. Physicochemical characterization of mRNA vaccine NP
The size, surface charge, and morphology were measured as the key parameters to characterize the mRNA vaccine NP. NanoSIGHT (Malvern, NS300) was used to measure the size at 20 °C and Nanoparticle Tracking Analysis (NTA) was used to analyze NP size. The dynamic light scattering (DLS) with 15-mW laser and an incident beam of 676 nm (Brookhaven Instrument Corporation) was used to measure surface charge of mRNA vaccine NP. To assess morphology and shape of NPs, a transmission electron microscope (TEM; Tecnai G2 Spirit BioTWIN microscope; FEI Company) was used. For TEM, the mRNA vaccine NPs were stained with 1% uranyl acetate and imaged at 80 kV.
2.6. Cell culture
Mouse dendritic cells (DC2.4), HEK-Blue™ mTLR7 cells (A HEK 293 engineered cell line), EG.7-OVA (lymphoma) cells, and RM1-OVA (prostate cancer) cells were used in various in vitro and in vivo studies. The EG.7-OVA cells were purchased from American Type Culture Collection (ATCC). The HEK-Blue™ mTLR7 cells were purchased from InvivoGen. The DC2.4 cell line was used from our in-house cell bank and RM1-OVA cell line was a kind gift from Dr. Xiang Yang Wang at Virginia Commonwealth University. Cells were maintained in F–12K (ATCC), Eagle's Minimum Essential Medium (EMEM; ATCC), or Roswell Park Memorial Institute (RPMI) 1640 (ATCC) cell-culture medium, according to the ATCC or cell source culture methods for each cell type. The cells were supplemented with high-glucose (4500 mg/L), 10% fetal bovine serum (FBS; Gibco®), and 1% penicillin/streptomycin antibiotic (Thermo-Fisher Scientific). For the RM1-OVA cells, 2% geneticin was used as an antibiotic (Thermo-Fisher Scientific), and for the HEK-Blue™ TLR cells Normocin, Blasticidin, and Zeocin (InvivoGen) were used as additional antibiotics. Blasticidin and Zeocin were not added to the media during early passages of HEK-Blue™ TLR cells, only when the cells were beginning to grow well. All biological experiments and cell culture were performed at 37 °C in 5% CO2 conditions in a cell-culture incubator. Cells authentication (service provided by DDC Biomedical) and mycoplasma contamination were checked before cell experiments in vitro and syngeneic allograft tumor model preparation in vivo.
2.7. In vitro cytotoxicity and transfection activity of mRNA vaccine NP
DC2.4 cells were seeded at a density of 5 × 104 cells per well in 24-well plates and grow until they reached ~80% confluence. Then the cells were transfected with mRNA vaccine NP at varying mRNA concentrations [from 0.100 to 0.700 or 0.300 to 0.500 μg/ml for the cytotoxicity study of NP (EGFP mRNA) or NP (EGFP mRNA + C16-R848), respectively and from 0.062 to 0.250 μg/ml for the transfection experiment with both NPs] for 16 h and then washed with fresh complete medium, followed by further incubation for 24 h to check cytotoxicity and transfection efficiency. An MTT assay was used for NP (EGFP mRNA) and an AlamarBlue assay was employed for NP (EGFP mRNA + C16-R848) to measure cytotoxicity according to the manufacturer's protocols and read using a microplate reader (TECAN, Infinite M200 Pro). To assess the transfection efficiency of the mRNA vaccine NP, cells were collected with trypsin (25% EDTA), washed (2X) and resuspended in PBS. The GFP expression was then measured using flow cytometry and the GFP-positive cell percentages and geometric mean fluorescence intensity (Geo MFI) were calculated using Kaluza Analysis Software and plotted using GraphPad Prism.
2.8. C16-R848 activity in mRNA vaccine NP by HEK-blue assay
To assess C16-R848 activity in the mRNA vaccine NPs, a HEK-blue assay was performed conferring to the company's protocol (InvivoGen). In this protocol, 20 μl of mRNA vaccine NP [without or with C16-R848 termed as NP (OVA mRNA) or NP (OVA mRNA + C16-R848), respectively] and free R848 at various R848 concentrations (10 to 2000 nM) were added to respective wells of a clear flat-bottom 96-well plate in triplicates. The HEK-Blue™ mTLR7 cells were harvested from their cultured flask by gently rinsing cells with pre-warmed 5 to 10 ml of PBS. 3 ml pre-warmed PBS was added again and the cells were kept at 37 °C for 1 to 2 min. The cells were then detached from the flask by gentle pipetting (without trypsinization) and made into a homogenize cell suspension for counting. A suspension of cells was prepared in HEK-Blue detection media with a concentration of 220,000 cells per ml and 180 μl of this cell suspension (~40,000 cells) was immediately added to each well containing the different groups of NP and free R848. The cells were then incubated at 37 °C in 5% CO2 for 24 h. The secreted embryonic alkaline phosphatase (SEAP) was observed and measured at 620 to 655 nm using a spectrophotometer. The optical density (OD) values of the NP and free R848 groups were normalized using the untreated control cells and the response ratio was presented graphically using GraphPad prism.
2.9. In vitro antigen presentation by MHC class-I of DC2.4 cells
DC2.4 cells (at a density of 5 × 104 cells per well) were seeded on 24-well plate and grown until ~80% confluence. Cells were then transfected with OVA mRNA NP (without or with C16-R848) at an mRNA concentration of 0.250 μg/ml for 24 h. L2K-OVA mRNA complex was used as a positive control, and untreated cells and naked OVA mRNA were used as negative controls. After 24 h of incubation at standard cell culture conditions, the cells were harvested and washed with FACS buffer (1х PBS with 5% FBS). The cells were then incubated with CD16/32 antibody (Bio Legend) (1.0 μg per sample with 5 × 104 cells in 100 μl volume) for 5 to 10 min on ice prior to immunostaining. Three μl of PE/Cy7 anti-mouse H-2Kb-SIINFEKL antibody was added to each sample and incubated on ice for 40 min in the dark followed by addition of 100 μl of fix medium A (Invitrogen) and further incubation for 15 min in the dark. 3 ml of FACS buffer was added to wash by centrifugation at 350 g for 5 min at 4 °C. Finally, 200 μl of FACS buffer containing 0.2% PFA was used to re-suspend the cell pellets and test using flow cytometry. The percentages of H-2Kb-SIINFEKL positive cells were analyzed by FlowJo software and graphed using GraphPad Prism.
2.10. Animals
Six-week-old C57/BL6 male wild-type black mice were used for testing in vivo CD8+ T cell response, and immunoprophylactic and immunotherapeutic studies with syngeneic allograft tumor models. The mice were obtained from Charles River Laboratories International, Inc. All animal experiments were performed under pathogen-free conditions and in accordance with the regulations of the animal facility of Brigham and Women's Hospital and the animal care guidelines of the National Institute of Health. The animals had sterile water/food pellets and were kept in standard animal facility condition under a 12-h light/dark cycle with the temperature and relative humidity maintained at 23±2 °C and 50 ± 20%, respectively. The mice were kept for at least one week to acclimatize to the food and environment of the animal facility before any prior treatment. The animal protocol was approved by the Institutional Animal Care and Use Committees at Harvard Medical School.
2.11. OVA expressing lymphoma and prostate cancer syngeneic allograft mouse tumor model preparation
To prepare the EG.7-OVA and RM1-OVA syngeneic allograft tumor mice models, cells (in 100 μL of culture medium) were implanted subcutaneously on the right flank of six-week-old C57/BL6 male wild-type black mice after mixing with 100 μl of matrigel (BD Biosciences). Mice were shaved before tumor cell inoculation to be able to observe the implanted tumor clearly and monitor the tumor growth daily. Approximately 1 105 of EG.7-OVA and 5 × 104 of RM1-OVA cells were implanted for the immunoprophylactic and immunotherapeutic studies, respectively.
2.12. Immunization
Mice were immunized with 100 μl PBS containing 30 μg OVA mRNA per mouse as described previously [45]. We calculated that if we use 30 μg of OVA mRNA, mice also received R848 (encapsulated inside the same nanoparticle vaccine formulation) at a concentration of 5.6 μg per mouse, which was quite similar to our previous report [37].
2.13. In vivo CD8+ T cell and CTL response in PBMC, spleen and tumor-draining lymph nodes by flow cytometry
Mice were anesthetized to collect blood retro-orbitally and then euthanized to aseptically harvest the spleen and tumor-draining lymph nodes (TDLNs) to make single cell suspensions using 70 μm strainers (BD Biosciences). In brief, first cell strainers were wet by adding 1 ml of cold media to a 50 ml falcon tube followed by placing the spleen or TDLNs on the strainer. Then the organs were gently ground and single cell suspensions were made by passing through strainers. The strainer was then washed with 4 ml of cold media to collect the splenocytes. The cells were centrifuged at 250 g for 5 min at RT and the media was aspirated off. The cell pellet was then resuspended using 3 ml of media and the suspension was transferred to a 15 ml falcon tube. To collect peripheral blood mononuclear cells (PBMC) from blood samples, ≥200 μl of blood was collected in heparinized tubes (Fisher Scientific) and immediately diluted with 3 ml of PBS. 3 ml of Histopaque 1083 (Sigma-Aldrich) was then slowly added to the bottom of the cell suspensions (PBMC, splenocytes and LNs samples) and carefully centrifuged at 330 g for 30 min at RT to separate the splenocytes and remove the red blood cells. The purified cells were carefully collected from the top layer and transferred to a 15 ml falcon tube. To wash the cells, 5 ml of cold media was added and centrifuged at 250 g for 5 min at 4 °C, followed by aspiration of the media. The cells were then resuspended with 40 μl MACS Buffer (Miltenyi Biotec) and 10 μl of MACS Biotin-antibody cocktail and incubated on ice for 5 min. 30 μl of MACS Buffer with 20 μl of anti-Biotin microbeads were then added, mixed well, and incubated on ice for 10 min. 400 μl of MACS buffer was finally added to run the suspension through the LS column (Miltenyi Biotec). To run the LS column, the column was placed into the magnet and rinsed with 3 ml of MACS buffer. The cell suspension was then run through the column and the cells were collected in a clean 15 ml tube. These cells, purified CD8+ T cells, were kept on ice. The cells were then counted and resuspended in FACS buffer for antibody staining. For antibody staining, 2 × 106 cells were taken in FACS tubes and washed 1 time with 3 ml of FACS buffer (centrifuged at 350 g for 5 min at 4 °C). The cells were then stained with tetramer (SIINFEKL-H2Kb PE), CD8a-FITC, KLRG1-Percp Cy5.5 (BD Biosciences) for 20 min in 100 μl of FACS buffer in the dark at room temperature. The cells were then washed with 2 ml of FACS buffer and resuspended in 200 μl of fix reagent A (BD Biosciences) and incubated for an additional 15 min on ice. The cells were then washed with 1x PBS and finally resuspended with 0.2% PFA to use for flow cytometry. Single staining was done for each of the antibodies for appropriate flow cytometry compensation. The percentages of in vivo CD8+ T cell and CTL responses in PBMC, spleen, and TDLNs were analyzed using FlowJo and graphed using GraphPad Prism software.
2.14. In vivo immunoprophylactic efficacy of mRNA vaccine NP in EG.7-OVA syngeneic allograft tumor model
To test the in vivo immunoprophylactic efficacy of the mRNA vaccine NPs in the EG.7-OVA syngeneic allograft tumor model, mice were immunized at days −25, −11 and −4. On day 0, four days after the last immunization, 1 × 105 EG.7-OVA cancer cells were injected subcutaneously in the right flank of the C57BL/6 black mice (after a clean shave). The mice were monitored daily and tumor measurement was started as soon as the tumors were first palpable, on day 9. Tumor measurements were performed daily using calipers and the average tumor size was calculated as ½(length × width × height). The average mouse weight was also determined and calculated. The mice were imaged at day 19 and day 27 post tumor induction, and the image backgrounds were removed using Adobe Photoshop software. On day 19 [for PBS and NP (EGFP mRNA + C16-R848) groups] and day 27 [for NP (OVA mRNA) and NP (OVA mRNA + C16-R848)] groups, the mice were sacrificed to evaluate the CD8+ T cell and CTL responses in PBMC, spleen, and TDLNs.
2.15. In vivo immunotherapeutic efficacy of mRNA vaccine NP in RM1-OVA syngeneic allograft tumor model
To check the in vivo immunotherapeutic efficacy of the mRNA vaccine NPs in the RM1-OVA syngeneic allograft tumor model, 5 × 104 RM1-OVA mouse prostate cancer cells were injected subcutaneously in the right flank of C57BL/6 black mice (after a clean shave) on day 0. The mice were immunized as soon as the tumors were first palpated on day 7. Two more immunizations were performed on day 11 and 15. The mice were monitored, and the tumor volume was measured daily after the start of the treatment on day 7. The average tumor size was calculated as ½(length × width × height). For survival analysis, the mice with tumor volumes over 2200 mm3 were considered to be dead, and the average percentages of survival were calculated. The average mouse weight was also determined. The mice were imaged at day 15 post-tumor induction, and Adobe Photoshop software was used to remove the image backgrounds. The mice were sacrificed to evaluate CD8+ T cell and CTL response in spleen and TME.
2.16. Detection of in vivo CD8+ T cell infiltration in TME by flow cytometry and immunohistochemistry
Mice were anesthetized and then euthanized to aseptically harvest tumors. The tumors were kept in ice-cold complete media (RPMI with 10% FBS). The tumors were then bisected using a sharp scalpel. Part of the tumor was kept in 10% formalin (for 2–3 days) followed by storage in 70% ethanol prior to slide preparation for the immunofluorescence testing. Another part of the tumor was taken to isolate myeloid cells to check CD8+ T cell infiltration in TME by flow cytometry. Briefly, each tumor tissue was taken into 2 ml of dissociation buffer (100 U/ml of collagenase IV and 100 μg/ml DNase I in RPMI media with 10% FBS) and kept for 30–40 min at 37 °C. A single cell suspension was made with FACS buffer (PBS with 5% FBS) through 70 μm strainer. The resulting suspension was then centrifuged at 400×g for 10 min at 4 °C and then washed one time with FACS buffer. CD8+ T cells were then purified (described in the method section in detail) and counted to take 2 × 106 cells per FACS tube for the respective sample. The cells were stained with tetramer (SIINFEKL-H2Kb PE) and CD8a-FITC (BD Biosciences) to determine tetramer+ CD8+ T cells by flow cytometry assay. To perform immunohistochemistry assay to further detect CD8+ T cell infiltration in TME, paraffin-embedded sections were incubated in xylene for 10 min, following rehydration in 100% ethanol three times for 10 min each. They were subsequently placed in 95%, 70%, and 50% ethanol for 5 min. The rehydration process was then completed by a series of washes in deionized water. The slides were placed into Dako Target Retrieval Solution (Agilent, S1700) and incubated for 30 min in a water bath preheated to 98 °C. The slides were left to equilibrate for 10 min at room temperature and then were cooled down for 10 min at 4 °C in the Target Retrieval Solution (Agilent). The slides were washed with water and permeabilized in a solution of 0.1% Triton-X100 in 1X TBS for 15 min, followed by further water wash. The slides were then incubated with a protein blocking solution at room temperature for 30 min and then incubated with the primary antibodies (CD8) diluted at 1:200 in Dako Antibody Diluent (Agilent S3022) overnight at 4 °C. The next day, the slides were allowed to come to room temperature for 30 min followed by washing and incubation with a blocking solution for 10 min. They were then incubated for 45 min with the secondary antibodies [FITC conjugated Goat Anti-Mouse IgG H&L (Abcam, ab6785) and Alexa Flour 594 conjugated Goat anti-Rabbit IgG (H + L) (ThermoFisher, A11037)] at room temperature. The slides were washed with PBS before being mounted with Vectashield Antifade Mounting Medium with DAPI (Vectorlabs, H-1200). The slides were imaged using confocal microscopy (FV1000 Olympus) immediately or within 4–5 days, keeping the slides stored at 4 °C.
2.17. Statistical analysis
GraphPad Prism 7.0 software was used to prepare all graphs and perform statistical analysis, including one-way ANOVA. All experiments were performed in triplicate unless otherwise stated. Error bars indicate standard deviation (SD), unless specified to be standard error means (SEM). A P < 0.05 value is considered statistically significant, and all statistically significant values shown in figures are indicated as: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
3. Results
3.1. Development and characterization of R848-pulsed mRNA vaccine NP
In the first set of experiments, we synthesized low molecular weight palmitic acid-conjugated R848 (C16-R848) to enhance the encapsulation efficiency of R848, control its release, minimize systemic exposure, and provide persistent APC uptake and stimulation with sustained immune surveillance. C16-R848 was synthesized using the Steglich esterification process (Supplementary Fig. S1a). The progress of the reaction was followed using thin-layer chromatography (TLC), and after synthesis, crude was purified using column chromatography. The product was characterized using mass spectrometry (Supplementary Fig. S1b) and 1H NMR analysis (Supplementary Fig. S1c). Molecular weight in the MALDI-TOF profile along with the 1H NMR peaks around 1, 3 and 7–8 ppm confirms the successful conjugation of palmitic acid (C16) to R848.
Next, a robust self-assembly approach was used to formulate the mRNA NPs using C16-R848 and G0-C14, a cationic lipid-like compound [46], coated with a lipid-poly(ethylene glycol) (lipid-PEG) shell (Fig. 1 a). The G0-C14 was used for EGFP mRNA (used as a model mRNA) complexation. The G0-C14/EGFP mRNA complexes coated with ceramide-PEG are herein referred to as NP (EGFP mRNA). The addition of C16-R848 during the preparation of NP (EGFP mRNA) makes the NP (EGFP mRNA + C16-R848) formulation platform. The stability and integrity of EGFP mRNA, whether naked, complexed with G0-C14, or encapsulated in NPs had no adverse effects in the presence of organic solvent (N, N-dimethyl formamide) (Fig. 1b). G0-C14 effectively complexed EGFP mRNA at a weight ratio of 5 or above as shown in Fig. 1b. The NPs, prepared at a G0-C14/mRNA weight ratio of 15, showed no leaching of mRNA as analyzed by electrophoresis, indicating that most mRNA (~100%) was entrapped inside the NP. The NP (EGFP mRNA) was 110.7 ± 1.8 nm in size whereas the size of NP (EGFP mRNA + C16-R848) increased to 137 ± 2.8 nm after C16-R848 loading as characterized by NanoSIGHT. The polydispersity indexes for NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) were 0.303 and 0.317, respectively. Both the NPs were spherical as revealed by transmission electron microscopy (TEM) (Fig. 1c and d). The particle sizes measured in the liquid state by NanoSIGHT (Fig. 1e) were visually found to be little different from those observed by TEM which were measured in the dry state. At −80 °C storage, the size distribution and morphologies were also characterized using NanoSIGHT and TEM, respectively with no significant differences [NP (EGFP mRNA) was 112.1 ± 2.3 nm and NP (EGFP mRNA + C16-R848) was 144 ± 6.4 nm; and both NPs retained their morphological integrity], suggesting that they are stable in cryo-storage (Supplementary Fig. S2). With an outer lipid-PEG shell, the average surface charges were slightly positively charged; 8.2 ± 0.3 and 11.2 ± 0.21 mV for NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848), respectively, measured by dynamic light scattering (DLS) (Fig. 1e). We also quantitatively analyzed C16-R848 encapsulation efficacy by high-performance liquid chromatography (HPLC) and found about ~51% C16-R848 encapsulation with this NP formulation which was improved compared to our previous systems [37] as well as other systems reported recently, such as cyclodextrin NPs (~11%) [38], poly(l-histidine) NPs (23%) [47], and polylactic acid NPs (~36%) [48].
Fig. 1.
Preparation and characterization of adjuvant-pulsed mRNA nanovaccine particle (NP). (a) Schematic representation of mRNA NP structure. After self-assembly of cationic G0-C14 to anionic mRNA together with C16-R848, the formulated adjuvant-pulsed mRNA NP was coated with lipid-PEG. Here, EGFP-mRNA was used as a reporter mRNA. (b) mRNA stability in organic solvent, as naked or complexed with cationic G0-C14, at different weight ratios (from 0.1 to 20) determined by agarose gel electrophoresis assay. The formulated NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) were also run through gel to detect any mRNA leaching from the NPs. Naked EGFP mRNA and Naked EGFP mRNA in DMF were used as controls. About 0.125 μg of EGFP-mRNA was used for all groups in this assay. (c, d) The NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) were characterized with NanoSIGHT to check size distribution (n = 3 batches), and transmission electron microscope (TEM) to observe morphologies. (e) Size and zeta potential of NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) were measured by NanoSIGHT and dynamic light scattering. A weight ratio of 1:15 for mRNA: G0-C14 was used for the mRNA NP preparation.
3.2. mRNA vaccine NP exhibits high transfection efficiency in dendritic cells (DCs) in vitro
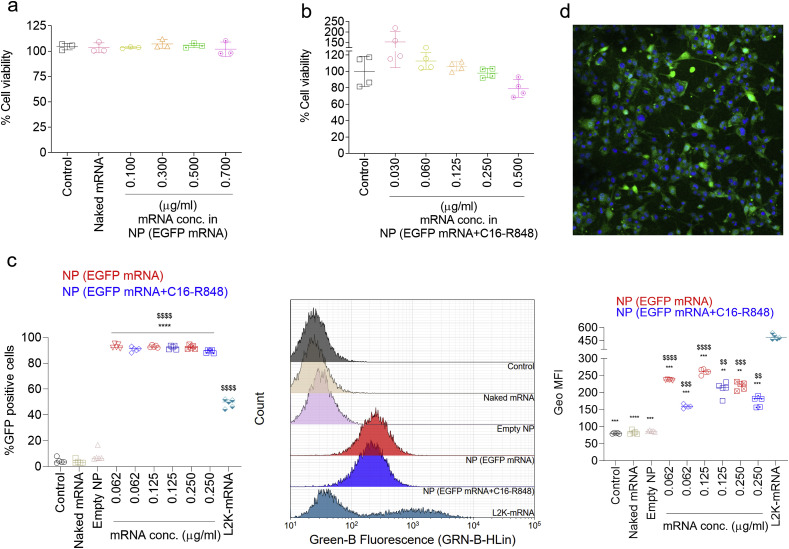
The initial priming of T cells to vaccine-derived antigens relies upon antigen processing and presentation by APCs. Here we investigated the safety and transfection activity of the NP (EGFP mRNA) in DCs (DC2.4 cells), the most crucial APC to determine if mRNA introduced by NP delivery resulted in protein expression in this APC cell type. Following 16 h transfection and additional 24 h incubation, nearly 100% of the cells were viable at different mRNA concentrations from 0.100 to 0.700 μg/ml (Fig. 2 a), indicating that the NP (EGFP mRNA) are not cytotoxic in these cell types. The cytotoxicity of NP (EGFP mRNA) with C16-R848 adjuvant was also evaluated under similar conditions as described above with a range of mRNA concentrations (from 0.030 to 0.500 μg/ml). We found minimal or no cytotoxic effect within the mRNA concentrations of 0.030 to 0.250 μg/ml and even at the highest mRNA concentration of 0.500 μg/ml, more than 80% cells were found viable (Fig. 2b). Notably, both NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) demonstrated a significantly higher transfection efficiency (~95%) in DC2.4 cells even at a lower mRNA dose of 0.062 μg/ml, as validated by the expression of EGFP mRNA and the detection of fluorescent protein expression by flow cytometry (Fig. 2c). On the other hand, the transfection agent Lipofectamine 2000 (L2K) showed only ~50% transfection capacity even at a high mRNA dose of 0.250 μg/ml (although it is worth noting that the geometric mean fluorescence intensity (Geo MFI) for L2K was significantly higher than all groups including the NP vaccines). This high transfection percentage of NP (EGFP mRNA + C16-R848) in DC2.4 cells suggested that the C16-R848 co-encapsulation did not compromise the bioactivity of the mRNA NP. The transfection efficiency of the mRNA NP was also examined by fluorescence microscopy, which showed enhanced cytosolic GFP expression in DC2.4 cells following treatment with the mRNA NP (Fig. 2d). Together, these results demonstrated that both NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) displayed high mRNA transfection efficiency with minimal cytotoxicity in DC2.4 cells.
Fig. 2.
Toxicity and transfection efficiency of mRNA NP in DC2.4 cells. Cells were treated with different mRNA concentrations (at 0.10, 0.30, 0.50, and 0.70 μg/mL) of NP (EGFP mRNA) for 16 h and further incubated in standard cell culture incubation conditions for 24 h. (a) Cytotoxicity assay of NP (EGFP mRNA) by MTT assay; cell viability was normalized with the untreated control group. Naked EGFP mRNA was used as control without NP formulation. Error bars represent the s.d. (n = 3). (b) Cytotoxicity assay of NP (EGFP mRNA + C16-R848) by AlamarBlue assay; cell viability was normalized with the untreated control group. Error bars represent the s.d. (n = 4). (c) Flow cytometry was used to determine transfection efficiency (% GFP positive cells) of NP (EGFP mRNA) and NP (EGFP mRNA + C16-R848) ($$$$P < 0.0001 compared to the controls (untreated control cells, Naked mRNA and empty NP) and ****P < 0.0001 compared to L2K-mRNA group). Error bars represent the s.d. (n = 5) and significance was determined using One-Way ANOVA test. Transfection efficiency was analyzed with the histograms and by analyzing geometric mean fluorescence intensity (Geo MFI) for the respective groups using Kaluza Analysis Software and (d) fluorescence microscopy images of DC2.4 cells captured after transfection with NP (EGFP mRNA) (magnification at 20 × ).
3.3. In vitro C16-R848 activity, MHC class-I presentation of OVA mRNA-derived peptides, and in vivo antigen-specific CD8+ T cell response by mRNA vaccine NP
As a surrogate for vaccine efficacy, we investigated the ability of the C16-R848 mRNA vaccine NP to activate dendritic cells and present OVA mRNA-derived peptides using MHC I, two critical events for facilitating a functional vaccine-mediated immune response. Stimulation of HEK293 TLR7 reporter cells with R848 or C16-R848 mRNA NP resulted in increased reporter cell activity in a dose-dependent manner. TLR7 activity of the NP (OVA mRNA + C16-R848) and free R848 treatment groups was significantly higher than the response to NP (OVA mRNA) control (without C16-R848) (Fig. 3 a). This result suggested that NP (OVA mRNA + C16-R848) retained TLR7 agonist activity and could provide a functional adjuvant activity via the encapsulated C16-R848.
Fig. 3.
Characterizing in vitro and in vivo immune surveillance by adjuvant-pulsed mRNA nanovaccine. (a) HEK-blue assay to determine R848 activity in NP (OVA mRNA + C16-R848). Method section explains the detail procedure of the assay. Free R848 and NP (OVA mRNA) without R848 were used a positive and negative controls, respectively. (b) In vitro antigen presentation by MHC class-I of DC2.4 cells. Cells were transfected with OVA mRNA NPs (without or with C16-R848) and other treatments at an mRNA concentration of 0.250 μg/ml for 24 h. Cells were then harvested and stained with anti-mouse H-2Kb-SIINFEKL antibody followed by flow cytometry assay. The percentages of H-2Kb-SIINFEKL positive cells were analyzed by FlowJo software. Untreated control cells and naked OVA mRNA were used as negative controls and L2K-mRNA was used as a positive control. (c) In vivo antigen-specific CD8+ T cell response by mRNA nanovaccine. Mice were immunized at day 0 and boost at day 14 and splenocytes were isolated 7 days-post boost. CD8+ T cells were purified (described in the method section in detail) and cells were stained with tetramer (SIINFEKL-H2Kb PE), and CD8a-FITC to determine SIINFEKL+ CD8+ T cells by flow cytometry assay. Naked OVA mRNA and NP (C16-R848) were used as negative controls. Error bars represent the s.d. (n = 3) and significance was determined using One-Way ANOVA test (*P < 0.05, **P < 0.01 and ***P < 0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To assess the ability of NPs to facilitate MHC class-I presentation of peptide epitopes in DCs in vitro, DC2.4 cells were incubated with NP (OVA mRNA) or NP (OVA mRNA + C16-R848) for 24 h. After treatment, the cells were collected and stained with H-2Kb SIINFEKL specific antibodies to assess the presence of presented antigen on the surface of the DCs. NP mediated delivery of mRNA resulted in significantly higher MHC class-I presentation of OVA derived SIINFEKL peptides compared with L2K-complexed or naked mRNA (Fig. 3b and analysis strategies with histograms were shown in Supplementary Fig. S3a). Within 24 h, >5.5% of DCs were expressing H-2Kb/SIINFEKL on their surface, indicating that NP (OVA mRNA) itself was sufficient to initiate antigen presentation on the surface of DCs. Pulsation of C16-R848 to NP (OVA mRNA) further improved H-2Kb/SIINFEKL expression (~7%) on dendritic cells, suggesting that the addition of C16-R848 did not negatively impact on the antigen-specific immunity but possibly increased the immune-stimulating activity of the OVA mRNA vaccine NP. Both NP (OVA mRNA) and NP (OVA mRNA + C16-R848) showed significantly better MHC-class I presentation compared to L2K-OVA mRNA presumably because of the superior transfection activity in DC2.4 cells by both the NPs found in Fig. 2c. This result also suggested that the NP (OVA mRNA) itself was sufficient to stimulate MHC-class I responses better than L2K, and C16-R848 pulsation could further improve its immune activity.
Following the demonstration that NP mediated delivery of mRNA and C16-R848 results in mRNA encoded antigen expression and stimulation of TLR7 in APCs; we next investigated the in vivo vaccine efficacy of our mRNA NP. Mice were subcutaneously (s.c., footpad) immunized on days 0 and 14 (100 μl containing 30 μg OVA mRNA with or without 5.6 μg C16-R848 per mice). On day 21, all CD8+ T cells were isolated from the single-cell splenocytes of the vaccinated mice and stained with H-2Kb/SIINFEKL specific TCR. The results showed that the percentage of antigen-specific CD8+ T cells in the total CD8+ T cell population was higher in both NP (OVA mRNA) and NP (OVA mRNA + C16-R848) groups compared to that of the negative controls, NP (C16-R848) and naked OVA mRNA showed little to no response (Fig. 3c, and Supplementary Fig. S3b). Particularly, the antigen-specific CD8+ T cells response by NP (OVA mRNA + C16-R848) was significantly higher compared to NP (OVA mRNA). These results suggest that this vaccination strategy can induce an antigen-specific T cell response and the addition of C16-R848 may further improve the potency of the mRNA vaccine NP system.
3.4. In vivo prophylactic efficacy of the mRNA vaccine NP in EG.7-OVA syngeneic allograft tumor model and the infiltration of CD8+ T cells to the tumor microenvironment (TME)
We next investigated the in vivo prophylactic efficacy of NP (OVA mRNA + C16-R848) in an EG.7-OVA-bearing syngeneic allograft tumor model. Mice were vaccinated 25, 11 and 4 days prior to tumor inoculation into the right flank of C57BL/6 immunocompetent mice (n = 4 or 5 mice) (Fig. 4 a). The tumor growth inhibitions observed in NP (OVA mRNA) and NP (OVA mRNA + C16-R848) treated mice were significantly greater than that of the controls, NP (EGFP mRNA + C16-R848) and PBS treated groups on day 19 post tumor induction (Fig. 4b–e). Specifically, the average tumor sizes rapidly increased in controls, reaching mean volumes of ~1044 mm3 and ~1280 mm3 at day 19 post tumor inoculation in NP (EGFP mRNA + C16-R848) and PBS groups, respectively. This is significantly higher than the mean volume of ~202 mm3 for NP (OVA mRNA) and only ~48 mm3 for NP (OVA mRNA + C16-R848) treatment groups measured at day 19 post tumor induction (Fig. 4c). After day 19 post tumor induction, tumor growth for NP (OVA mRNA) without adjuvant exponentially increased over the investigated period (reached to ~900 mm3), whereas the NP (OVA mRNA + C16-R848) group continued to suppress tumor growth significantly till day 27 (reached to ~208 mm3) (Fig. 4c and d). At day 27 post tumor induction, there were no differences in average tumor sizes among PBS, NP (EGFP mRNA + C16-R848) and NP (OVA mRNA). However the average tumor size for NP (OVA mRNA + C16-R848) was significantly lower compared to all other groups (Fig. 4d). The tumor size of the individual mice from each separate group was shown in Fig. 4e. Of note, the mice in the NP (EGFP mRNA + C16-R848) and PBS groups were sacrificed at day 19 as tumors were growing rapidly and exceeded the permissible size limit. On the terminal days, CD8+ T cells were isolated from splenocytes, PBMC, and tumor-draining lymph nodes of the vaccinated mice. The cells were then stained with tetramer and KLRG1 antibodies to check for KLRG1-specific CD8+ T cell expression. KLRG1 staining was used to further determine the immunophenotype tetramer + CD8+ T cells. Its inclusion in the staining panel is supported by previous work demonstrating that the percentage of KLRG1+ effector-memory CD8+ T cells can predict the degree of therapeutic efficacy of the cancer vaccines [49]. The results showed nearly 2-fold higher KLRG1-positive CD8+ T cells for NP (OVA mRNA + C16-R848) group in the spleen, PBMC, and tumor-draining lymph nodes (Supplementary Fig. S4a). These results show the promising prophylactic efficacy of the NP (OVA mRNA + C16-R848) vaccine platform in preventing tumor progression. NP (OVA mRNA) group also showed 2-fold higher KLRG1-positive CD8+ T cells in the spleen, PBMC, and tumor-draining lymph nodes at the terminal day 27, although we found an increase in tumor size for this group, suggesting the necessity to investigate CD8+ T cells infiltration within the tumor bed (Supplementary Fig. S4a). To check for CD8+ T cells infiltration in TME after immunization with NP (OVA mRNA + C16-R848) vs. PBS or NP (EGFP mRNA + C16-R848) or NP (OVA mRNA), tumor sections were analyzed after CD8 antibody staining. A significantly increased level of CD8+ T cell infiltration was observed for the NP (OVA mRNA + C16-R848) treatment group relative to that of the PBS or NP (EGFP mRNA + C16-R848) or NP (OVA mRNA) groups (Fig. 5 a and b), validating the mechanism of action by NP (OVA mRNA + C16-R848) vaccination in preventing tumor growth. Mice body weight did not undergo significant changes for any of the group, suggesting toxicity was not an issue (Supplementary Fig. S4b).
Fig. 4.
In vivo immunoprophylactic efficacy of mRNA NP in EG.7-OVA syngeneic allograft tumor model. (a) Scheme of immunizations (s.c., foot pad) with PBS (n = 4), NP (EGFP mRNA + C16-R848) (n = 4), NP (OVA mRNA) (n = 5) or NP (OVA mRNA + C16-R848) (n = 5) and EG.7-OVA tumour inoculation in male C57/BL6 mice. Mice were immunized at days −25, −11 and −4. On day 0, four days after the last immunization, 1 × 105 EG.7-OVA cancer cells were injected subcutaneously in right flank of the C57BL/6 black mice (after a clean shave). (b) The mice of all groups were imaged at day 19 post tumor induction. (c) The mice of NP (OVA mRNA) and NP (OVA mRNA + C16-R848) groups and their excised tumors were imaged at the terminal day 27 post tumor induction. The mice were monitored daily and tumor measurement was started as soon as the tumors were first palpable, on day 9. Tumor measurements were performed daily using calipers and the average tumor volume was calculated as ½(length × width × height). (d) Represents average tumor size and statistical size differences amongst the groups. (e) Tumor sizes for individual mouse separately for each group. In this prophylactic study, PBS and NP (EGFP mRNA + C16-R848) groups were used as controls. Error bars represent the s.e.m. and significance was determined using One-Way ANOVA test (*P < 0.05 and **P < 0.01).
Fig. 5.
Immunohistochemistry to detection in vivo CD8+ T cell infiltration in TME for the mice studied in immunoprophylactic settings (see scheme in Fig. 4a). Mean fluorescent intensity (MFI) was measured using ImageJ software and plotted with GraphPad prism. PBS and NP (EGFP mRNA + C16-R848) groups were used as negative controls. Error bars represent the s.d. and significance was determined using One-Way ANOVA test (*P < 0.05 and **P < 0.01).
3.5. In vivo therapeutic efficacy of the mRNA vaccine NP in RM1-OVA syngeneic allograft tumor model and analysis of the TME
To investigate the in vivo therapeutic efficacy of NP (OVA mRNA + C16-R848), RM1-OVA syngeneic allograft tumor models were prepared by s.c. injection of RM1-OVA mouse prostate cancer cells into the right flank of C57BL/6 immunocompetent mice (n = 4 or 5). The mice were immunized on day 7 upon the development of palpable tumors and immunizations were repeated at days 11 and 15 (Fig. 6 a). In therapeutic settings, tumor growth was much more aggressive. Similarly to the prophylactic model, the average tumor size increased to ~2115 mm3 and ~2482 mm3 for the NP (EGFP mRNA + C16-R848) and PBS controls, respectively, while the NP (OVA mRNA + C16-R848) treatment significantly limited the tumor growth to a mean size of ~965 mm3, which was also significantly lower compared to the mean size of ~1847 mm3 by NP (OVA mRNA) vaccinated group (Fig. 6b–d). Specifically, NP (OVA mRNA) failed to show any immunotherapeutic effect to suppress tumor growth, whereas NP (OVA mRNA + C16-R848) group exhibited tumor suppression capacity throughout the study period, indicating a robust therapeutic efficacy to the C16-R848 adjuvant-pulsed mRNA vaccine NP platform and its significant potential for initiating tumor-specific responses to neo-antigens in vivo. Tumor size of the individual mice from each separate group is shown in Fig. 6d. Moreover, we further monitored for CD8+ T cell infiltration in TME in this therapeutic tumor model after immunization with NP (OVA mRNA + C16-R848) vs. PBS or NP (EGFP mRNA + C16-R848) or NP (OVA mRNA) group. On day 15, two mice from PBS, NP (EGFP mRNA + C16-R848), and NP (OVA mRNA + C16-R848) groups and one mouse from NP (OVA mRNA) group were euthanized and tumors were collected to analyze CD8+ T cell infiltration in TME by flow cytometry. Tumors were collected and CD8+ T cells were isolated and then purified (described in the method section in detail). The cells were stained with tetramer SIINFEKL-H2Kb PE and CD8a-FITC (BD Biosciences) to determine tetramer+ CD8+ T cells by flow cytometry assay. Supplementary Fig. S5 showed a trend of an increased level of tetramer+ CD8+ T cells by NP (OVA mRNA + C16-R848) group compared to that by PBS, NP (EGFP mRNA + C16-R848) or NP (OVA mRNA) group. Moreover, CD8+ T cells in TME were further evaluated by immunohistochemistry at the terminal days. In this experiment, mice were sacrificed and tumor sections were analyzed after CD8 antibody staining. NP (OVA mRNA + C16-R848) treatment group showed a significantly increased level of CD8+ T infiltration compared to that of the PBS, NP (EGFP mRNA + C16-R848) or NP (OVA mRNA) groups (Fig. 7 a and b). Note that the mouse body weight had no significant change between the vaccinated groups (Supplementary Fig. S6).
Fig. 6.
In vivo immunotherapeutic efficacy of mRNA NP in RM1-OVA syngeneic allograft tumor model. (a) Scheme of RM1-OVA tumor inoculation (at day 0 after a clean shave) and immunization (s.c., foot pad) of PBS (n = 5), NP (EGFP mRNA + C16-R848) (n = 5), NP (OVA mRNA) (n = 4) or NP (OVA mRNA + C16-R848) (n = 5) in RM1-OVA tumour-bearing male C57/BL6 mice. Mice were immunized at day 7 when tumors were first palpated and further boosted on days 11 and 15. On day 19, mice were euthanized and tumors were collected and processed to analyze TME by immunohistochemistry. (b) The mice were imaged at day 15 post tumor induction. The mice were monitored daily and tumor measurement was started as soon as the tumors were first palpable, on day 7. Tumor measurements were performed daily using calipers and the average tumor volume was calculated as ½(length × width × height). (c) Represents average tumor size and statistical size differences amongst the groups. (d) Shows tumor sizes for individual mouse for each group separately. On day 15, two mice from PBS, NP (EGFP mRNA + C16-R848) and NP (OVA mRNA + C16-R848), and one mouse from NP (OVA mRNA) were randomly selected to harvest tumors and check tetramer+ CD8+ T cells infiltration in tumor bed (see Supplementary Fig. S5 for the result). In addition, note that one mouse died on day 15 from the PBS group. Error bars represent the s.e.m. and significance was determined using One-Way ANOVA test (*P < 0.05 and **P < 0.01).
Fig. 7.
Immunohistochemistry to further confirm in vivo CD8+ T cell infiltration in the TME for the mice investigated in immunotherapeutic settings (see scheme in Fig. 6a). Mean fluorescent intensity (MFI) was measured using ImageJ software and plotted with GraphPad prism. PBS and NP (EGFP mRNA + C16-R848) groups were used as negative controls. Error bars represent the s.d. and significance was determined using One-Way ANOVA test (**P < 0.01 and ***P < 0.001).
4. Discussion
The development of effective and durable anti-tumor immunity relies on the initial priming of the adaptive immune activity via the proliferation of antigen-specific cytotoxic T cells [[50], [51], [52]]. Current vaccination approaches to stimulating this response are complicated by the need for the adoptive transfer of ex vivo antigen-loaded dendritic cells [53,54]. The de novo expression of exogenous mRNA delivered via NPs allows antigen-presenting cells to be preferentially transfected and results in the increased presentation of tumor-associated neo-antigens. When these APCs are concomitantly stimulated through pathogen recognition receptors (PRRs) including TLR7/8, MHC-I antigen presentation is further increased and stimulates a more robust CD8+ T cell response [55]. Resiquimod (R848) is a TLR7/8 agonist that is used as an adjuvant, in part for its ability to increase antigen-specific CD8+ T cell expansion and impede tumor growth [56]. In this report, we present a stable NP platform for the co-delivery of synthetic OVA mRNA as a model antigen and C16-R848 as a modified TLR7/8 agonist (Supplementary Fig. S1 and Fig. 1). The delivery platform was conceptualized from a new-generation polymer-lipid hybrid NP system reported in our recent work [9] that demonstrated effective systemic delivery of tumor suppressor mRNA to various tumor models in vivo resulting in a significant anti-tumor effect. In the present study, we prepared a nanovaccine formulation using the lipid-like material G0-C14 to encapsulate both mRNA antigen and C16-R848 coated with lipid-PEG (i.e., ceramide-PEG) and to be efficiently taken up by the antigen-presenting cells (APCs). To accelerate the de-PEGylation kinetics and subsequent cellular uptake of the NPs, we chose to use ceramide-PEG [57,58]. This approach prevents agglomeration, enabling re-distribution after freezing and offering the baseline stability required for synthesis and storage, while also providing rapid dissociation kinetics in vivo. Once inside the cell, these NPs show proton sponge effect, releasing the cargo into the cytosol of the cell. Within the APC, the C16-R848 and OVA mRNA act synergistically to increase MHC I mediated antigen presentation. First, the OVA mRNA is translated into OVA protein where it is processed by the immunoproteasome, allowing efficient presentation of antigen by the APC to CD8+ T cells. The C16-R848 adjuvant supplements this immune response in two ways: first, the C16 (palmitic acid) stabilizes the OVA peptide on the APC during antigen presentation [59,60] and second, the C16-R848 activates TLR7/8 expressing APCs, activating a type 1 IFN response and upregulating the pathways responsible for antigen presentation.
mRNA delivery can be achieved via DC transfection ex vivo followed by re-engraftment in vivo [61] or via mRNA delivery in vivo, with or without the use of a delivery vehicle. Although ex vivo manipulation of DCs with mRNA provides increased control of the transfection process and cellular conditions, its potential as a clinical therapy is hindered by the sub-optimal efficacy of re-engraftment and the prohibitive resource requirement. Conversely, the administration of mRNA in vivo is simpler. However, it requires a vehicle to transfect immune cells and increase antigen presentation efficiently. Delivery methods involving gene guns [62], in vivo electroporation [63] and in vivo administration of naked mRNA have been evaluated previously [64], but are not effective in delivering mRNA to immune cells, leading to suboptimal immune responses [65]. Cationic lipid and polymer-based delivery systems are favorable for transfection, because their properties can be tuned using different formulation chemistries to modulate interaction behavior. Protamine-based mRNA vaccine systems are an example of a cationic peptide delivery platform that has demonstrated significant T cell and B cell stimulation resulting in enhanced levels of cellular immunity and anti-tumor effect in clinical trials [66]. For polymer-lipid delivery platforms, the two most prominent commercially available mRNA delivery vehicles are TransIT-mRNA (Mirus Bio LLC) which is a cationic polymer-lipid structure, and Lipofectamine (Invitrogen) which forms a cationic liposome structure. Although cationic liposomes demonstrate moderate transfection efficiency, 65% for TransIT-mRNA (Mirus, TransIT mRNA) and 40% for Lipofectamine in the DC2.4 cell line, they perform poorly in vivo and are highly toxic [65]. Recently, transfection efficiencies of ~85% have been achieved in DC2.4 cells using lipid-polymer nanoparticles made of the cationic lipid 1,2-di-O-octadecenyl-3-trimethylammonium propane-coated polylactide-co-glycolide (PLGA), which maintained a cell viability of 90%. Alternatively, a well-established delivery platform involving cationic polymer chitosan-coated PLGA NPs achieved a transfection efficiency of 5% [67]. The NP (EGFP mRNA) platform presented in our study demonstrated transfection efficiencies of more than 95% in DC2.4 cells either with or without C16-R848 pulsation while maintaining cell viability from 80%–100% depending on the mRNA NP concentrations (Fig. 2).
In addition to mRNA delivery, adjuvant delivery is a critical component of a vaccine formulation. Adjuvants can improve immune responses by activating APCs to increase antigen presentation (signal 1), promote the co-expression of costimulatory molecules (signal 2) and the production of specific cytokines (signal 3) that, when properly coordinated, enhance T cell responses [68]. Adjuvants currently approved for human use in Europe and the United States include aluminum salts, oil-in-water emulsions as well as TLR4 and TLR9 adjuvants [69]. Aluminum salts and oil-in-water emulsions are the most commonly used, however they are limited by their need for repeated administration in order to induce protection [55]. On the other hand, the most effective adjuvants for cancer vaccines are those that target and activate TLRs. Common adjuvants are detoxified congener endotoxins [70] and monophosphoryl lipid A (MPLA) [7] which stimulate TLR4 used by the immune system to recognize pathogen-associated molecular patterns, enabling for immune responses specific to particular stimuli. TLR9 agonists, oligonucleotides (ODN) containing unmethylated CpG sequences, have also proven successful in promoting Th1 versus Th2 effector T cell responses, and have been implemented as adjuvants in approved vaccines [71]. In this context, TLR7/8 agonists have been rapidly gaining traction due to their ability to stimulate strong T cell responses [72]. The TLR7/8 agonist R848 induces DC maturation, stimulates pro-inflammatory cytokines production and enhances humoral and cellular adaptive immunity [73]. In a recent study, R848-loaded β-cyclodextrin NPs inhibited tumor growth in multiple tumor models through the conversion of the immunosuppressive M2 macrophage phenotype to the M1 that is associated with positive clinical outcomes [38]. Additionally, co-delivery of lipopolysaccharide (LPS) with R848 induced the generation of a spectrum of DC maturation phenotypes, including less mature DCs that are required for the generation of an effective memory CD8+ T cell response [74]. Administration of R848 also showed tumor growth inhibition >50% when used in combination with taxol-based chemotherapy to treat primary human colorectal xenografts in mice [75].
In this study, we used a modified version of R848 called C16-R848 synthesized by conjugating palmitic acid (C16) to R848 to achieve more stable co-encapsulation of this adjuvant into the mRNA NP. To evaluate the effect of C16-R848 activity in vitro, a cell-based reporter measuring NF-kb and AP-1 activation was used following exposure to C16-R848 encapsulated in NPs or freely dissolved in media. In this system, the cell response to C16-R848 (co-encapsulated in mRNA NP) compared with free R848 was similar (Fig. 3a), suggesting that NP (OVA mRNA + C16-R848) retained R848 activity. To further test whether incorporation of C16-R848 into the NP (OVA mRNA + C16-R848) increases the expression of the OVA-derived peptides via MHC Class I molecules on the cell surface, we evaluated surface expression with an H-2Kb-SIINFEKL specific antibody. We found that OVA-mRNA with or without co-delivery of C16-R848 increased antigen expression by MHC class-I compared to Lipofectamine-mediated delivery of OVA-mRNA or free OVA mRNA alone, whereas MHC class-I expression was improved by C16-R848 adjuvant-pulsation to the NP (OVA-mRNA). Moreover, an increased expansion of H2kB-SIINFEKL specific CD8+ T cells was observed in vivo when NP (OVA mRNA) was pulsed with C16-R848 (Fig. 3b) compared to those of naked OVA mRNA, NP (C16-R848) and NP (OVA mRNA) alone, suggesting that the increased MHC class-I antigen presentation by DCs resulted in stronger CD8+ T cell activation and proliferation, which led to more enhanced CD8+ T cell activation in vivo by C16-R848-pulsed NP (OVA mRNA).
The co-delivery of mRNA and C16-R848 in a NP as a cancer vaccine strategy has not been previously reported, although attempts have been made previously to achieve similar results. In a recent report, a charge-altering releasable transporter (CART) NP delivery system was used to deliver OVA mRNA in combination with CpG (a synthetic DNA molecule which is short, single-stranded and contains a cytosine triphosphate deoxynucleotide followed by a guanine triphosphate deoxynucleotide) in an A20-OVA lymphoma syngeneic allograft model, resulting in complete regression of the tumor which extended survival beyond 75 days, indicating the establishment of a durable anti-tumor immune response [76]. In this CART-OVA mRNA study, the treatment increases cytotoxic T cell responses and the tumor-specific killing capacity. This also aligned with the results presented in our study where we found higher CD8+ T cell response in the NP (OVA-mRNA + C16-R848) treated mice compared to that of control groups, naked OVA mRNA, NP (C16-R848) as well as NP (OVA mRNA) alone (Fig. 3c). In another approach, a DOTAP-cholesterol lipoplex NP was utilized in a co-delivery system, loaded with monophosphoryl lipid A (MPLA, a TLR4 agonist which is an FDA approved vaccine adjuvant) and nucleoside-modified OVA mRNA [31]. In this study mRNA was modified with 5′-methylcytidine and pseudouridine to increase the translational potential of the mRNA lipoplexes, however this modification reduced the stimulation of type I IFN release and overall immunoactivity of the mRNA. To improve the efficacy of the vaccine platform, MPLA was co-delivered to compensate for the reduction in immune activity due to the modification of the mRNA. Although this study hypothesized that the MPLA-OVA mRNA co-delivery vaccine platform is a feasible system for safe vaccine platform, it did not demonstrate anti-tumor efficacy. In our study, however, we found a high CD8+ T cell response by NP (OVA mRNA + C16-R848) system provoked both prophylactic and therapeutic efficacy, resulting in an 84% reduction of tumor volume when used as a prophylactic treatment and a 60% reduction when administered therapeutically after establishment of palpable tumors (~100 mm3) in the mouse EG.7-OVA lymphoma and RM1-OVA prostate cancer syngeneic allograft models, respectively (Figs. 4 and 6).
In addition to the initial activation and expansion of an antigen-specific CD8+ T cell population, subsequent T cell infiltration into the tumor is essential for positive therapeutic outcomes, especially in the context of immune checkpoint therapy [[77], [78], [79]]. In our prophylactic study, treatment with the NP (OVA mRNA + C16-R848) significantly stimulated an antigen-specific CD8+ T cell response. This response was further differentiated by the subsequent antigen-specific CD8+ T cell infiltration within the tumor bed in mice by NP (OVA-mRNA + C16-R848) vaccine, compared with PBS, NP (EGFP-mRNA + C16-R848) or NP (OVA-mRNA) alone (Fig. 5). The CD8+ T cell infiltration correlated with a reduction in tumor size, suggesting that effective tumor control relies on both expansion and infiltration of the tumor antigen specific CD8+ T cell population. The therapeutic treatment following establishment of the tumor also indicates that NP (OVA mRNA + C16-R848) can control tumor progression to a greater extent than controls as well as NP (OVA-mRNA) without adjuvant pulsation and this tumor prevention was correlated with the increased infiltration of CD8+ T cells at the TME site (Fig. 7 and Supplementary Fig. S5). Overall, we found that the antigen-specific CD8+ T cell population in the NP (OVA mRNA + C16-R848) group infiltrated and proliferated at the TME site, likely due to improved effector function. The increased effector function was occurred presumably because they were well primed due to increased antigen-presentation at the APC surface and/or also present a co-stimulatory TLR7/8 mediated co-activation of the antigen presenting cell, wherein all these were deficient for the NP (OVA mRNA) group without the adjuvant pulsation. Together, these results demonstrate that the addition of C16-R848 adjuvant pulsation to the mRNA nanovaccine can enhance the efficacy of synthetic mRNA as a source of antigen to trigger a therapeutic CD8+ T cell-mediated anti-tumor response.
5. Conclusion
In summary, this study is a proof-of-principle for the development of a vaccine using a NP encapsulating a synthetic mRNA antigen and the chemically-modified TLR7/8 agonist C16-R848. The co-delivery of the C16-R848 adjuvant with OVA mRNA improved the vaccine's therapeutic and prophylactic efficacy by increasing the tumor-associated antigen (TAA) presentation, while also promoting CD8+ T cell recruitment into the tumor and enhancing the overall anti-tumor response. The NP strategy resulted in a high mRNA transfection efficiency which led to a robust antigen specific CD8+ T cell population and observable tumor infiltration. The efficiency of this vaccine platform could potentially be further improved through additional use of immune checkpoint inhibitors including anti-PD-1 that can downregulate inhibitory immune signaling in the TME, reducing tumor cell immune tolerance and increasing T cell-mediated anti-tumor activity [80]. The versatility of this mRNA nanovaccine platform allows for the encapsulation and delivery of other TAA-encoding mRNA payloads, having the potential to expand potential indications to a wide range of diseases. Recent progress in the field of tumor associated T-cell epitope prediction [81] makes this especially exciting, opening up the potential possibilities to easily facilitate the presentation of multiple tumor associated T cell epitopes on activated resident antigen-presenting cells, without the requirement for DC-meditated antigen cross-presentation. Robust peripheral expansion of antigen-specific effector CD8+ T cells has the potential to overcome some of the limitations of immune checkpoint therapy where a lack of T cell expansion and infiltration hinders the clinical efficacy.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.
Authorship contribution statement
Mohammad Ariful Islam: Conceptualization, Methodology, directed the project, led and performed all the in vitro and in vivo experiments as well as analyzed the data, Writing - original draft, revised along with J.R. according to the comments of J.S., O.C.F., and B.R.Z, .. Jamie Rice: helped in all the in vivo studies. Emma Reesor: helped in nanoparticle preparation and experimental assays under the supervision of M.A.I., provided technical support and corrections of the manuscript. Harshal Zope: conceived an idea of adjuvant modification, synthesized and characterized C16-R848. Wei Tao: provided technical support and corrections of the manuscript. Michael Lim: helped in nanoparticle preparation and experimental assays under the supervision of M.A.I., provided technical support and corrections of the manuscript. Jianxun Ding: provided technical support and corrections of the manuscript. Yunhan Chen: synthesized and characterized C16-R848. Dike Aduluso: helped in nanoparticle preparation and experimental assays under the supervision of . Bruce R. Zetter: Conceptualization, Methodology, directed the project. Omid C. Farokhzad, Conceptualization, Methodology, directed the project, conceived an idea of adjuvant modification. Omid C. Farokhzad: Conceptualization, Methodology, directed the project, conceived an idea of adjuvant modification.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Omid C. Farokhzad has financial interest in Selecta Biosciences, Tarveda Therapeutics, and Seer.
Acknowledgements
This work was supported in part by the Prostate Cancer Foundation (PCF) Young Investigator Award (J.S.), the David Koch-PCF Award in Nanotherapeutics (O.C.F.), and the US National Institutes of Health (NIH) grant CA200900 (J.S. and B.R.Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. O.C.F. has financial interests in Selecta Biosciences, Tarveda Therapeutics, and Seer.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2020.120431.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Baylor N.W., Egan W., Richman P. Aluminum salts in vaccines—US perspective. Vaccine. 2002;20:S18–S23. doi: 10.1016/s0264-410x(02)00166-4. [DOI] [PubMed] [Google Scholar]
- 2.Sylvester R.J., van der Meijden A.P., Lamm D.L. Intravesical bacillus Calmette-Guerin reduces the risk of progression in patients with superficial bladder cancer: a meta-analysis of the published results of randomized clinical trials. J. Urol. 2002;168:1964–1970. doi: 10.1016/S0022-5347(05)64273-5. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg S.A., Packard B.S., Aebersold P.M., Solomon D., Topalian S.L., Toy S.T. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. N. Engl. J. Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 4.Goldman B., DeFrancesco L. The cancer vaccine roller coaster. Nat. Biotehnol. 2009;27:129. doi: 10.1038/nbt0209-129. [DOI] [PubMed] [Google Scholar]
- 5.Kantoff P.W., Higano C.S., Shore N.D., Berger E.R., Small E.J., Penson D.F. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 6.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed S.G., Orr M.T., Fox C.B. Key roles of adjuvants in modern vaccines. Nat. Med. 2013;19:1597. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 8.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. [Google Scholar]
- 9.Islam M.A., Xu Y., Tao W., Ubellacker J.M., Lim M., Aum D. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018;2:850. doi: 10.1038/s41551-018-0284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong N., Tao W., Ling X., Wang J., Xiao Y., Shi S. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y.X., Wang Y., Blake S., Yu M., Mei L., Wang H. RNA nanotechnology-mediated cancer immunotherapy. Theranostics. 2020;10:281–299. doi: 10.7150/thno.35568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Son S., Nam J., Zenkov I., Ochyl L.J., Xu Y., Scheetz L. Sugar-nanocapsules imprinted with microbial molecular patterns for mRNA vaccination. Nano Lett. 2020;20:1499–1509. doi: 10.1021/acs.nanolett.9b03483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Q., Lee G.Y., Ding J., Li W., Shi J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018;11:5281–5309. doi: 10.1007/s12274-018-2146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebastian M., Papachristofilou A., Weiss C., Früh M., Cathomas R., Hilbe W. Phase Ib study evaluating a self-adjuvanted mRNA cancer vaccine (RNActive®) combined with local radiation as consolidation and maintenance treatment for patients with stage IV non-small cell lung cancer. BMC Cancer. 2014;14:748. doi: 10.1186/1471-2407-14-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mockey M., Bourseau E., Chandrashekhar V., Chaudhuri A., Lafosse S., Le Cam E. mRNA-based cancer vaccine: prevention of B16 melanoma progression and metastasis by systemic injection of MART1 mRNA histidylated lipopolyplexes. Cancer Gene Ther. 2007;14:802. doi: 10.1038/sj.cgt.7701072. [DOI] [PubMed] [Google Scholar]
- 18.Gilboa E., Vieweg J. Cancer immunotherapy with mRNA‐transfected dendritic cells. Immunol. Rev. 2004;199:251–263. doi: 10.1111/j.0105-2896.2004.00139.x. [DOI] [PubMed] [Google Scholar]
- 19.Su Z., Dannull J., Heiser A., Yancey D., Pruitt S., Madden J. Immunological and clinical responses in metastatic renal cancer patients vaccinated with tumor RNA-transfected dendritic cells. Cancer Res. 2003;63:2127–2133. [PubMed] [Google Scholar]
- 20.Su Z., Dannull J., Yang B.K., Dahm P., Coleman D., Yancey D. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J. Immunol. 2005;174:3798–3807. doi: 10.4049/jimmunol.174.6.3798. [DOI] [PubMed] [Google Scholar]
- 21.Vik-Mo E.O., Nyakas M., Mikkelsen B.V., Moe M.C., Due-Tønnesen P., Suso E.M.I. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol. Immunother. 2013;62:1499–1509. doi: 10.1007/s00262-013-1453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suso E.M.I., Dueland S., Rasmussen A.-M., Vetrhus T., Aamdal S., Kvalheim G. hTERT mRNA dendritic cell vaccination: complete response in a pancreatic cancer patient associated with response against several hTERT epitopes. Cancer Immunol. Immunother. 2011;60:809–818. doi: 10.1007/s00262-011-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte J., Mu L., Aamdal S., Kvalheim G., Dueland S., Hauser M. Phase I/II trial of melanoma therapy with dendritic cells transfected with autologous tumor-mRNA. Cancer Gene Ther. 2006;13:905. doi: 10.1038/sj.cgt.7700961. [DOI] [PubMed] [Google Scholar]
- 24.Caruso D.A., Orme L.M., Amor G.M., Neale A.M., Radcliff F.J., Downie P. Results of a phase I study utilizing monocyte‐derived dendritic cells pulsed with tumor RNA in children with stage 4 neuroblastoma. Cancer. 2005;103:1280–1291. doi: 10.1002/cncr.20911. [DOI] [PubMed] [Google Scholar]
- 25.Grunwitz C., Kranz L.M. Cancer Vaccines: Springer; 2017. mRNA Cancer Vaccines—Messages that Prevail; pp. 145–164. [DOI] [PubMed] [Google Scholar]
- 26.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Kariko K. Nucleoside modifications in RNA limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 28.Kariko K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 31.Verbeke R., Lentacker I., Wayteck L., Breckpot K., Van Bockstal M., Descamps B. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: restoring the immunogenicity of immunosilent mRNA. J. Control. Release. 2017;266:287–300. doi: 10.1016/j.jconrel.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 32.Yrlid U., Milling S.W., Miller J.L., Cartland S., Jenkins C.D., MacPherson G.G. Regulation of intestinal dendritic cell migration and activation by plasmacytoid dendritic cells, TNF-α and type 1 IFNs after feeding a TLR7/8 ligand. J. Immunol. 2006;176:5205–5212. doi: 10.4049/jimmunol.176.9.5205. [DOI] [PubMed] [Google Scholar]
- 33.Yrlid U., Cerovic V., Milling S., Jenkins C.D., Klavinskis L.S., MacPherson G.G. A distinct subset of intestinal dendritic cells responds selectively to oral TLR7/8 stimulation. Eur. J. Immunol. 2006;36:2639–2648. doi: 10.1002/eji.200636426. [DOI] [PubMed] [Google Scholar]
- 34.Xu S., Koldovsky U., Xu M., Wang D., Fitzpatrick E., Son G. High-avidity antitumor T-cell generation by toll receptor 8–primed, myeloid-derived dendritic cells is mediated by IL-12 production. Surgery. 2006;140:170–178. doi: 10.1016/j.surg.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Williams H.C., Bath-Hextall F., Ozolins M., Armstrong S.J., Colver G.B., Perkins W. Surgery versus 5% imiquimod for nodular and superficial basal cell carcinoma: 5-year results of the SINS randomized controlled trial. J. Invest. Dermatol. 2017;137:614–619. doi: 10.1016/j.jid.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 36.Block M.S., Nevala W.K., Pang Y.-P., Allred J.B., Strand C., Markovic S.N. A pilot clinical trial testing topical resiquimod and a xenopeptide as immune adjuvants for a melanoma vaccine targeting MART-1. Melanoma Res. 2019;29:420–427. doi: 10.1097/CMR.0000000000000556. [DOI] [PubMed] [Google Scholar]
- 37.Ilyinskii P.O., Roy C.J., O'Neil C.P., Browning E.A., Pittet L.A., Altreuter D.H. Adjuvant-carrying synthetic vaccine particles augment the immune response to encapsulated antigen and exhibit strong local immune activation without inducing systemic cytokine release. Vaccine. 2014;32:2882–2895. doi: 10.1016/j.vaccine.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodell C.B., Arlauckas S.P., Cuccarese M.F., Garris C.S., Li R., Ahmed M.S. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng. 2018;2:578–588. doi: 10.1038/s41551-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coolen A.L., Lacroix C., Mercier-Gouy P., Delaune E., Monge C., Exposito J.Y. Poly(lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation. Biomaterials. 2019;195:23–37. doi: 10.1016/j.biomaterials.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Fan Y.N., Li M., Luo Y.L., Chen Q., Wang L., Zhang H.B. Cationic lipid-assisted nanoparticles for delivery of mRNA cancer vaccine. Biomater. Sci. 2018;6:3009–3018. doi: 10.1039/c8bm00908b. [DOI] [PubMed] [Google Scholar]
- 41.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sayour E.J., Grippin A., De Leon G., Stover B., Rahman M., Karachi A. Personalized tumor RNA loaded lipid-nanoparticles prime the systemic and intratumoral milieu for response to cancer immunotherapy. Nano Lett. 2018;18:6195–6206. doi: 10.1021/acs.nanolett.8b02179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Zope H., Islam M.A., Rice J., Dodman S., Lipert K. Lipidation approaches potentiate adjuvant-pulsed immune surveillance: a design rationale for cancer nanovaccine. Front. Bioeng. Biotech. 2020;8:787. doi: 10.3389/fbioe.2020.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X., Xie K., Zhang X.Q., Pridgen E.M., Park G.Y., Cui D.S. Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18638–18643. doi: 10.1073/pnas.1303958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Qiao L., Zhang S., Wan G., Chen B., Zhou P. Dual pH-responsive multifunctional nanoparticles for targeted treatment of breast cancer by combining immunotherapy and chemotherapy. Acta Biomater. 2018;66:310–324. doi: 10.1016/j.actbio.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Thauvin C., Widmer J., Mottas I., Hocevar S., Allemann E., Bourquin C. Development of resiquimod-loaded modified PLA-based nanoparticles for cancer immunotherapy: a kinetic study. Eur. J. Pharm. Biopharm. 2019;139:253–261. doi: 10.1016/j.ejpb.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 49.van Duikeren S., Fransen M.F., Redeker A., Wieles B., Platenburg G., Krebber W.J. Vaccine-induced effector-memory CD8+ T cell responses predict therapeutic efficacy against tumors. J. Immunol. 2012;189:3397–3403. doi: 10.4049/jimmunol.1201540. [DOI] [PubMed] [Google Scholar]
- 50.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 51.Hu Z., Ott P.A., Wu C.J. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 2018;18:168. doi: 10.1038/nri.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gubin M.M., Artyomov M.N., Mardis E.R., Schreiber R.D. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J. Clin. Invest. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sahin U., Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 54.Tacken P.J., de Vries I.J.M., Torensma R., Figdor C.G. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat. Rev. Immunol. 2007;7:790. doi: 10.1038/nri2173. [DOI] [PubMed] [Google Scholar]
- 55.Dowling D.J. Recent advances in the discovery and delivery of TLR7/8 agonists as vaccine adjuvants. ImmunoHorizons. 2018;2:185–197. doi: 10.4049/immunohorizons.1700063. [DOI] [PubMed] [Google Scholar]
- 56.Sabado R.L., Pavlick A., Gnjatic S., Cruz C.M., Vengco I., Hasan F. Resiquimod as an immunologic adjuvant for NY-ESO-1 protein vaccination in patients with high-risk melanoma. Cancer Immunol. Res. 2015;3:278–287. doi: 10.1158/2326-6066.CIR-14-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu X., Tao W., Liu D., Wu J., Guo Z., Ji X. Surface De-PEGylation controls nanoparticle-mediated siRNA delivery in vitro and in vivo. Theranostics. 2017;7:1990–2002. doi: 10.7150/thno.18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu X., Xu Y., Solis L.M., Tao W., Wang L., Behrens C. Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7779–7784. doi: 10.1073/pnas.1505629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson J., Case M., Brooks C. Palmitic acid conjugation of a protein antigen enhances major histocompatibility complex class II-restricted presentation to T cells. Immunology. 1992;76:593. [PMC free article] [PubMed] [Google Scholar]
- 60.Zaman M., Toth I. Immunostimulation by synthetic lipopeptide-based vaccine candidates: structure-activity relationships. Front. Immunol. 2013;4:318. doi: 10.3389/fimmu.2013.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benteyn D., Heirman C., Bonehill A., Thielemans K., Breckpot K. mRNA-based dendritic cell vaccines. Expert Rev. Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 62.Steitz J., Britten C.M., Wölfel T., Tüting T. Effective induction of anti-melanoma immunity following genetic vaccination with synthetic mRNA coding for the fusion protein EGFP. TRP2. Cancer Immunol. Immunother. 2006;55:246–253. doi: 10.1007/s00262-005-0042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson D.X., Ljungberg K., Kakoulidou M., Liljeström P. Intradermal electroporation of naked replicon RNA elicits strong immune responses. PloS One. 2012;7 doi: 10.1371/journal.pone.0029732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phua K.K., Leong K.W., Nair S.K. Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. J. Control. Release. 2013;166:227–233. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Islam M.A., Reesor E.K., Xu Y., Zope H.R., Zetter B.R., Shi J. Biomaterials for mRNA delivery. Biomater. Sci. 2015;3:1519–1533. doi: 10.1039/c5bm00198f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yasar H., Biehl A., De Rossi C., Koch M., Murgia X., Loretz B. Kinetics of mRNA delivery and protein translation in dendritic cells using lipid-coated PLGA nanoparticles. J. Nanobiotechnol. 2018;16:72. doi: 10.1186/s12951-018-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lynn G.M., Laga R., Darrah P.A., Ishizuka A.S., Balaci A.J., Dulcey A.E. In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat. Biotechnol. 2015;33:1201. doi: 10.1038/nbt.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shetab Boushehri M., Lamprecht A. TLR4-Based immunotherapeutics in cancer: a review of the achievements and shortcomings. Mol. Pharm. 2018;15:4777–4800. doi: 10.1021/acs.molpharmaceut.8b00691. [DOI] [PubMed] [Google Scholar]
- 70.Duthie M.S., Windish H.P., Fox C.B., Reed S.G. Use of defined TLR ligands as adjuvants within human vaccines. Immunol. Rev. 2011;239:178–196. doi: 10.1111/j.1600-065X.2010.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackson S., Lentino J., Kopp J., Murray L., Ellison W., Rhee M. Immunogenicity of a two-dose investigational hepatitis B vaccine, HBsAg-1018, using a toll-like receptor 9 agonist adjuvant compared with a licensed hepatitis B vaccine in adults. Vaccine. 2018;36:668–674. doi: 10.1016/j.vaccine.2017.12.038. [DOI] [PubMed] [Google Scholar]
- 72.Kim H., Niu L., Larson P., Kucaba T.A., Murphy K.A., James B.R. Polymeric nanoparticles encapsulating novel TLR7/8 agonists as immunostimulatory adjuvants for enhanced cancer immunotherapy. Biomaterials. 2018;164:38–53. doi: 10.1016/j.biomaterials.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 73.Tomai M.A., Vasilakos J.P. TLR7/8 agonists as vaccine adjuvnats. In: Singh M., editor. Novel Immune Potentiators and Delivery Technologies for Next Generation Vaccines. Springer Science & Business Media; New York: 2013. pp. 3–18. [Google Scholar]
- 74.Pufnock J.S., Cigal M., Rolczynski L.S., Andersen-Nissen E., Wolfl M., McElrath M.J. Priming CD8+ T cells with dendritic cells matured using TLR4 and TLR7/8 ligands together enhances generation of CD8+ T cells retaining CD28. Blood. 2011;117:6542–6551. doi: 10.1182/blood-2010-11-317966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stier S., Maletzki C., Klier U., Linnebacher M. Combinations of TLR ligands: a promising approach in cancer immunotherapy. Clin. Dev. Immunol. 2013;2013 doi: 10.1155/2013/271246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haabeth O.A., Blake T.R., McKinlay C.J., Waymouth R.M., Wender P.A., Levy R. mRNA vaccination with charge-altering releasable transporters elicits human T cell responses and cures established tumors in mice. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E9153–E9161. doi: 10.1073/pnas.1810002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Azoury S.C., Straughan D.M., Shukla V. Immune checkpoint inhibitors for cancer therapy: clinical efficacy and safety. Curr. Cancer Drug Targets. 2015;15:452–462. doi: 10.2174/156800961506150805145120. [DOI] [PubMed] [Google Scholar]
- 78.Kingwell K. Anticancer drugs: translational target for checkpoint inhibitors. Nat. Rev. Drug Discov. 2018;17:863. doi: 10.1038/nrd.2018.204. [DOI] [PubMed] [Google Scholar]
- 79.Kingwell K. Translational target for checkpoint inhibitors. Nat. Rev. Immunol. 2018;18:728–729. doi: 10.1038/s41577-018-0091-6. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y., Zhang L., Xu Z., Miao L., Huang L. mRNA Vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol. Ther. 2018;26:420–434. doi: 10.1016/j.ymthe.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Backert L., Kohlbacher O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015;7:119. doi: 10.1186/s13073-015-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.