Abstract
The Preclinical Working Group of Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV), a public-private partnership spearheaded by the National Institutes of Health, has been charged with identifying, prioritizing, and communicating SARS-CoV-2 preclinical resources. Reviewing SARS-CoV-2 animal model data facilitates standardization and harmonization and informs knowledge gaps and prioritization of limited resources. To date, mouse, hamster, ferret, guinea pig, and non-human primates have been investigated. Several species are permissive for SARS-CoV-2 replication, often exhibiting mild disease with resolution, reflecting most human COVID-19 cases. More severe disease develops in a few models, some associated with advanced age, a risk factor for human disease. This review provides a snapshot that recommends the suitability of models for testing vaccines and therapeutics, which may evolve as our understanding of COVID-19 disease biology improves. COVID-19 is a complex disease, and individual models recapitulate certain aspects of disease; therefore, the coordination and assessment of animal models is imperative.
Keywords: animal models, COVID-19, SARS-CoV-2, mice, hamsters, macaques, therapeutics, vaccines
Animal models of COVID-19 disease have been rapidly developed. On behalf of the ACTIV public-private partnership led by NIH, Hewitt et al. compare these small and large models to human disease and make recommendations for preclinical testing of various classes of therapeutics and vaccines.
Introduction
COVID-19 took the world by surprise. While the disease spread around the globe, no vaccines or drugs were available to ameliorate its effects. As a result, the National Institutes of Health (NIH) and the Foundation for the NIH (FNIH) established the Accelerating COVID-19 Therapeutic Innovations and Vaccines (ACTIV) partnership in April 2020 comprised of experts in the fields of virology, public health, vaccine and drug development. This public-private partnership brings together industry, academic and government stakeholders in an unprecedented collaboration to share information and resources to impact the trajectory of the pandemic. There are four working groups: Preclinical, Clinical Therapeutics, Clinical Trial Capacity and Vaccines (Collins and Stoffels, 2020; NIH, 2020).
The Preclinical Working Group (WG) was charged “to standardize and share preclinical evaluation resources and methods and accelerate testing of candidate therapies and vaccines to support entry into clinical trials” (Collins and Stoffels, 2020). Under normal circumstances, the response to emerging pathogens is a methodical linear progression of in vitro, in vivo, and clinical studies, yet the urgency of the response to the current pandemic requires these activities to be carried out in parallel. Prior experience with other coronaviruses, both in vitro and in vivo models, provides a guiding foundation for current research and effectively enables interventions to the disease. ACTIV’s recommendations on accelerating preclinical in vitro studies are described in the accompanying manuscript (Grobler et al., 2020).
Animal models help us not only to understand the pathogenesis and mechanisms of SARS-CoV-2 disease biology but also to elucidate aspects of pharmacology, toxicology, and immunology of the therapeutic and vaccine strategies. These data may provide confidence that the products being developed can prevent or treat disease; however, no single animal model fully recapitulates human COVID-19 disease. While there is no substitute for randomized controlled clinical trials, there are great advantages to adjunctive research in animal models to explore in-depth mechanisms, immunology/pharmacology, and vaccine or therapeutic efficacy as well as to discover biomarkers that may be useful in the clinic and to ensure the safety of the candidates in humans. Understanding the pharmacokinetic/pharmacodynamic relationships such that effective and safe doses can be extrapolated from in vitro antiviral activity, preclinical pharmacokinetics, toxicology, and clinical modeling (FDA, 2020) can allow compounds to proceed into clinical testing without necessarily demonstrating efficacy in an animal model. Although animal model data may not be required to advance to the clinic, it can provide useful comparison data, which is especially informative in a pandemic.
The selection of appropriate animal models of infection, disease manifestation, and efficacy measurements is important for vaccines and therapeutics to be compared under ACTIV’s umbrella using Master Protocols with standardized endpoints and assay readouts. Models of SARS-CoV-2 infection include mice (ACE2 transgenic strains, mouse adapted virus, and AAV transduced ACE2 mice), hamsters, rats, ferrets, and non-human primates (NHPs).
The urgency of the need to coordinate efforts to reduce duplication and cycle times also necessitated an assessment of supply and demand. Specifically, resource optimization of all the models—but particularly that of the NHPs—and priority setting of experimental therapeutic and vaccine interventions was immediately warranted. The National Primate Research Centers (NPRCs), established 60 years ago with funding from the NIH, serve as a major resource in this effort to provide NHP expertise, models for human diseases, and NHP resources to NIH and other investigators (www.nprcresearch.org). Similarly, NIH-funded mouse repositories stand ready to supply mice to the broader scientific community for research and testing. The ACTIV Preclinical WG efforts are facilitating a coordinated effort of the seven NPRCs, which work together as a consortium to assure the highest degree of rigor and ethics and to manage the relative paucity of available primates and NPRC-ABLS3 facilities in which to perform SARS-CoV-2 in vivo research. The NPRCs are playing a major role in ACTIV, although the animal models in our assessment are not restricted to the NPRCs or those available through NIH-sponsored repositories.
Beyond ACTIV, Operation Warp Speed (OWS) is the US government’s effort to make 300 million doses of vaccine available by January 2021 (HHS, 2020). OWS is also making use of master protocols, and ACTIV and OWS are sharing information around protocols being developed. OWS plans to perform animal model testing at contract research organizations under contract to the Biodefense Advanced Research & Development Authority (BARDA) and NIAID as well as government labs in the Department of Defense. Coordinating protocols and plans between ACTIV and OWS for the supply of non-human primates is critical to the success of all pandemic-related efforts.
The ACTIV Preclinical WG aims to assess the status and applicability of all animal models that are under development and to share information in real time, including taking inventory of the resources and annotating the salient features of the models, establishing a data collection tool to assess the status and applicability, highlighting the gaps in effort and knowledge, and suggesting redirection to fill the gaps. This initial publication describes the current status of animal models as determined from the available data in the inventory. As features of the COVID-19 disease are described—for example, the rare but serious multisystem inflammatory syndrome in children (MIS-C) (Rowley, 2020) —new animal models can help address these complications, ideally in a coordinated fashion to accelerate our understanding. Additional animal model data are welcomed, particularly to address gaps in our knowledge, and we will describe how members of the scientific and medical community can submit data for inclusion.
Human Disease
It is appropriate to understand the course of human infections in order to model disease in animals; therefore, we present a summary of human disease first. Since the outbreak began in China, some of the earliest and most cited publications report numerous cases from China (Chauhan, 2020; Huang et al., 2020; Wu and McGoogan, 2020). COVID-19 is characterized by phases of increasing severity, generally described as asymptomatic, mild, moderate, or severe disease. Mild disease includes general symptoms such as fever, fatigue, cough, headache, diarrhea, sore throat, congestion, muscle or body aches, and/or vomiting; loss of sense of smell and/or taste and a dry, non-productive cough are more pathognomonic of COVID-19. Moderate disease is characterized by mild pneumonia, shortness of breath, and/or pressure in the chest. Severe symptoms include difficulty breathing, bluish lips, inability to stay awake, confusion, acute respiratory distress, septic shock, and/or multi-organ failure which may lead to death. The median time from symptom onset to pneumonia is about 5 days and to severe hypoxemia is 7 to 12 days (Chauhan, 2020). Progression to severe disease is associated with advanced age and/or comorbidities (Wang et al., 2020).
Siddiqi and Mehra (Siddiqi and Mehra, 2020) noted that the progression of disease was associated with transition from an antiviral response to an inflammatory response. García (2020) posits that responses at various immunological checkpoints determine the course of disease, to include relatively weak stimulation of innate immune responses, adaptive antibody and T cell responses, and potentially strong activation of proinflammatory chemokines. Garcia urges increased monitoring of immunological responses to better understand the checkpoints that prevent progression of or promote disease severity, noting that much of what we understand comes from comparing immunological findings in moderate and severe cases, with few reports following asymptomatic or mild cases. Finally, Ziegler et al. (2020) recently described regulation of ACE2 cell surface expression in upper airway cells by IFN-α with significant upregulation in humans and NHPs, but not in mice. This highlights the need to further understand the risk/benefit of antiviral or IFN therapy treatments in infected humans. Further understanding of this dynamic is critical to balancing the immune response and efficacious treatments (Ziegler et al., 2020).
Descriptions of viral loads determined by PCR in clinical infections note that respiratory viral loads (sputum or nasopharyngeal swabs) were significantly higher, peaked later, and were detected longer in severe than mild infections (Liu et al., 2020b; Zheng et al., 2020). While virus could be detected for longer periods from stool samples, there was not a difference between mild and severe disease (Zheng et al., 2020). A larger study of hospitalized cases demonstrated that viral load detected by PCR from nasopharyngeal swabs was a significant predictor of mortality, with mean log10 viral loads of 5.2 copies per mL for survivors versus 6.4 copies per mL for those who died (Pujadas et al., 2020).
Gulati and colleagues published a comprehensive review of coronavirus disease, comparing SARS-CoV, MERS-CoV and SARS-CoV-2, focusing not just on pulmonary disease but encompassing other organ systems as well: cardiovascular, hepatobiliary, gastrointestinal, renal, neurologic, musculocutaneous, and hematologic (Gulati et al., 2020). The COVID-19 reports were retrospective, clinicopathologic, or case reports and generally noted increased frequency of extrapulmonary involvement with more severe disease (ICU) and comorbidities. Dyspnea is predictive of ICU admission, and many patients exhibited abnormal chest-computed tomography, predominantly ground glass opacities. Histopathology demonstrated interstitial fibrosis and inflammatory infiltrates. Severe disease requiring intensive care was associated with lymphopenia, leukocytosis, and neutrophilia and with increases in alanine aminotransferase, aspartate aminotransferase, lactate dehydrogenase, total bilirubin, creatinine, creatinine kinase, blood urea nitrogen, troponin-I, and D-dimer (Gulati et al., 2020). These extrapulmonary organ systems are not well understood in most animal models; hence, the primary endpoints of animal models have been testing of viral load and immune and inflammatory responses.
Current Status of Animal Models for Covid-19 Research
No single animal model has ever been predictive of human infection or exactly modeled it. Human infection happens by chance, and very little is known about how long a person needs to be in contact with the pathogen or how much pathogen is needed prior to symptoms presenting. For animal models to be useful, we need to identify the most reproducible methods for challenge that lead to infection and symptoms that emulate those seen in humans. Host tropism for viruses is complicated by the need for host factors to facilitate virus entry and replication, the host’s immune response, and pathways by which viruses evade immune responses (Douam et al., 2015). Models for the closely related SARS-CoV and MERS-CoV viruses have paved the way for SARS-CoV-2 infection and pathogenesis studies and have bolstered expertise in studying coronavirus vaccines and therapeutics. Angiotensin-converting enzyme 2, or ACE2, was discovered as the receptor for SARS-CoV and has proven to be the receptor for SARS-CoV-2 as well (Zhou et al., 2020b). Phylogenetic comparisons of ACE2 proteins allowed for predictions of species that are susceptible to SARS-CoV-2 infection (Liu et al., 2020a; Luan et al., 2020; Qiu et al., 2020; Wan et al., 2020), as ACE2 is a key determinant of infectivity (Liu et al., 2020a).
Mice have mismatched ACE2 receptors, and two approaches have been used to solve this problem: 1) mice that express the human ACE2 receptor; and 2) a SARS-CoV-2 strain that is adapted to recognize the murine ACE2 receptor. Although both approaches provide a way to model COVID-19 infection in mice, neither is a perfect substitute for human infection. The goal for animal models is to understand and/or provide the most accurate and complete predictor of results in a variety of human clinical settings.
At this time, there are several models for SARS-CoV-2 in development (Table 1 ). NHP models are traditionally considered the most translational models to humans. NHPs bear close similarities to human genetic, neurological, cognitive, physiological, reproductive, anatomical, and immunological systems. Their susceptibility to most human pathogens is not surprising; they are therefore models for many of the most intractable acute and persistent pathogens. Their size and longevity make them excellent models for pathogenesis, allowing repeated sampling and imaging in vivo for longitudinal studies. However, their limited supply in the face of numerous drug and vaccine candidates makes them an even more precious resource. It is therefore imperative to prioritize agents to be tested by demonstrating tolerability and efficacy in smaller mammalian models: mice, hamsters, and ferrets are currently the small animal species of choice. Small animal models are presented first as the tractable models used in early discovery and development. Mice and hamsters predominate, given that there are many choices that are becoming available. Guinea pigs do not appear to be productively infected, as virus cannot be detected despite presenting early (day 3) pulmonary histological changes; they therefore are not the best model of disease (Dick Bowen, personal communication).
Table 1.
Summary of COVID-19 Animal Models
| Species | Details | Vaccines | Antivirals | Neutralizing Antibodies | Other Therapies | Infectivity | Transmission | Disease Enhancement | Disease Manifestation & Pathology | Extent of Disease |
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | Tg(CAG-ACE2)AC70Ctkt | TBD | TBD | |||||||
| Mouse | C3B6.Cg-Tg(FOXJ1-ACE2)1Rba/Mmnc | ✓ | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; weight loss; varied death and recovery | Mild to severe | |||
| Mouse | B6.Cg-Tg(K18-ACE2)2Prlmn/J | ✓ | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; weight loss; death | Severe | |||
| Mouse | ICR-Tg(Ace2-ACE2)1Cqin/J | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild | |||||
| Mouse | C57BL/6-Ace2em1(ACE2)Yowa | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild | ||||
| Mouse | BALB/c (adapted virus) | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild | ||||
| Mouse | Adenovirus transduced hACE2 | ✓ | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; weight loss; recovery | Mild | |||
| Hamster | Syrian Golden | ✓ | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild to moderate | |||
| Hamster | Tg(K18-hACE2) | TBD | TBD | |||||||
| Guinea Pig | Wild type | – | – | – | Lung lesions | None to minimal | ||||
| Ferret | outbred | ✓ | ✓ | ✓ | ✓ | Viral titers in nasal washes; fever | Mild | |||
| AGM | Wild caught, St. Kitts | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild to moderate | ||||
| Aged AGM | Wild caught, St. Kitts | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; cytokine storm, ARDS; varied death and recovery | Severe | ||||
| Cynomolgus macaque | Cambodian | ✓ | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild | |||
| Rhesus macaque | Chinese or Indian | ✓ | ✓ | ✓ | ✓ | Lung lesions; interstitial pneumonia; recovery | Mild |
This listing of animal models indicates features of disease and efficacy testing that have been tested or are in testing, with check marks indicating positive results and dashes indicating negative results. This table is adapted from the NCATS Open Portal table (https://opendata.ncats.nih.gov/covid19/animal), which will be updated as new information becomes available. AGM, African green monkey; TBD, to be determined; ARDS, acute respiratory distress syndrome.
Small Animal Models
Mice
Mice hold a prominent position in disease research. Their small size, ease of use, rapid breeding, and ability to be inbred as well as readily genetically modified have made the mouse the go-to model in biomedical research. Mice can be used to rapidly screen vaccines, antivirals, and other therapeutics in a relatively high-throughput, pipeline approach. There are several mouse models for SARS-CoV-2 infection listed in Table 1, each with advantages and limitations. It bears emphasizing that mouse models that are not available from public repositories or commercial vendors cannot be scaled effectively to meet the high demand for animals and supply the high throughput needs of the community. Those models that remain in private labs, no matter how good they may be, will be of limited value to the large-scale efforts needed to evaluate the many vaccines, antivirals, and other therapeutics being investigated for the pandemic response, unless they are made available through commercial suppliers or repositories.
The use of the standard laboratory mouse for infection of SARS-CoV and SARS-CoV-2 has been limited due to amino acid differences in the ACE2 receptor between mouse and humans that result in reduced susceptibility to infection in this model. Older BALB/c mice are more susceptible to infection, but the infectivity is still modest, and the requirement of an aged animal can be a significant disadvantage in screening therapeutics (Roberts et al., 2005a; Vogel et al., 2007). One solution to the lack of infectivity in laboratory mice is to use an adapted virus in which multiple passages/or and selected mutations in the virus make the mice more susceptible to infection. A recent publication demonstrated a mouse adapted virus Q498T/P499Y that was not only able to readily infect mice but was used to demonstrate the effectiveness of neutralizing antibodies to reduce viral replication in vivo (Li et al., 2020). A second solution to the lack of infectivity in common laboratory mice is to express the human ACE2 gene (hACE2), either by viral transduction or genetic engineering. Two labs have used adenovirus transduction to express human ACE2 in the lungs of mice, resulting in mild disease noted by viral replication and weight loss; this could be ameliorated by neutralizing monoclonal antibodies (Hassan et al., 2020) or antivirals and convalescent sera (Sun et al., 2020a).
Several transgenic mouse models carrying the human ACE2 gene were created to study SARS-CoV infection. Five transgenic models currently exist and are rapidly being assessed for their use in the study of COVID-19. The first model is a transgenic mouse developed in China by Chuan Qin in which the human ACE2 cDNA is under the control of the mouse ACE2 promoter ICR-Tg(Ace2-ACE2)1Cqin/J. SARS-CoV-2 infection in these mice leads to weight loss and virus replication in lung with histopathology indicative of interstitial pneumonia. SARS-CoV and SARS-CoV-2 cause mild disease, and no death was reported in this model (Bao et al., 2020b). A second model, developed by Clarence Peters in 2007, employed another transgenic construct, Tg(CAG-ACE2)AC70Ctkt, with the human ACE2 gene expressed under the control of a ubiquitous CAG promoter. These mice are highly susceptible to SARS-CoV infection and demonstrate weight loss along with other clinical manifestations before reaching 100% mortality within 8 days after intranasal infection (Tseng et al., 2007; Yoshikawa et al., 2009). These mice are currently being bred in order to determine how they respond to infection with SARS-CoV-2 (Kent Tseng, personal communication). Two other transgenic mouse models also expressing the human ACE2 gene using different epithelial based promoters, Tg(FOXJ1-ACE2)1Rba and B6.Cg-Tg(K18-ACE2)2Prlmn/J, were developed by Ralph Baric and Stanley Perlman, respectively (McCray et al., 2007; Menachery et al., 2016). Both models are considered severe models of SARS-CoV, with significant weight loss, disease pathology, evidence of encephalitis, and death by 5–7 days post infection. Interestingly, they differ in severity for COVID-19; the B6C3Tg(FOXJ1-ACE2)1Rba model displayed a variable response to infection with SARS-CoV-2, whereas the B6.Cg-Tg(K18-ACE2)2Prlmn/J model develops a uniformly severe response. Half of the infected B6C3Tg(FOXJ1-ACE2)1Rba mice recovered from infection, demonstrating no significant signs of weight loss or illness; the other half of the infected B6C3Tg(FOXJ1-ACE2)1Rba mice demonstrated severe illness and body weight loss and required euthanasia (Jiang et al., 2020). The B6.Cg-Tg(K18-ACE2)2Prlmn/J mouse develops a severe infection and lung pathology in response to SARS-CoV-2. Reports from multiple independent research groups report that upon infection, mice lose significant body weight, have a hunched posture, and require euthanasia 5–7days post challenge in 100% of infected mice (Golden et al., 2020; Moreau et al., 2020; Oladunni et al., 2020; Rathnasinghe et al., 2020; Winkler et al., 2020). Importantly, these mice are not hypersensitive to infection, and lower doses of the virus result in less severe disease progression (Golden et al., 2020). Viral loads are detected in the brain of these mice, but initial reports indicate the brain infection to be less than the severity-associated encephalitis seen with SARS-CoV infection in this model, with minimal histopathological changes observed (Oladunni et al., 2020). The Tg(FOXJ1-ACE2)1Rba and B6.Cg-Tg(K18-ACE2)2Prlmn/J mice are available for distribution to research scientists by the MMRRC and The Jackson Laboratory, respectively. Another mouse model was recently developed by Sun and colleagues: C57BL/6-Ace2em1(ACE2)Yowa. This model utilizes CRISPR/Cas9 to knock the human ACE2 cDNA into the mouse ACE2 locus, utilizing the endogenous mouse Ace2 promoter. This model also results in infectivity; however, the mice recover from body weight loss post infection with SARS-CoV-2 with mild lung pathology (Sun et al., 2020b).
Given the ease of genetic engineering in mice, one can expect to see a variety of new models developed in the immediate future. These models will differ in terms of random transgenesis, targeted knockins, variations in constructs, reporter tags, and new ways to deliver the human ACE2 gene, such as through recently reported viral transgenesis (Hassan et al., 2020; Sun et al., 2020a). Humanized mice represent another potential approach, and while there are distinct advantages, there are additional challenges that impact rigorous testing of vaccine and therapeutic efficacy—namely, the required technical expertise, preparation time, immunocompromised status of mice, variability, and limited throughput (Skelton et al., 2018). Xenografts of human lung tissue into immunodeficient mice have resulted in lung-only mice or bone marrow/liver/thymus-lung (BLT-L) mice, which demonstrated infection with MERS-CoV, RSV, Zika, and cytomegalovirus, and in the case of BLT-L mice, produced a human immune response (Wahl et al., 2019). The use of NOD scid gamma (NSG) mice for engraftment of human immune cells will prove to be a powerful tool in studying the interaction of the human immune system with the virus. New models incorporating the NSG genetic background and the human ACE2 gene are underway. While the current models will be critical for high-throughput screening of therapeutics, the newer and milder models will be critical to understanding how infection responds to comorbidities such as type 1 diabetes, hypertension, obesity, etc. These comorbidities can be induced but also achieved through genetic crosses to existing mouse models engineered with mutations in key genes to produce these desired phenotypes. Furthermore, differences between mice and humans can be manipulated to more faithfully represent human disease—for example, exploiting differential regulation of ACE2 by interferons (Ziegler et al., 2020). Other applications of the mouse models include exploring the effects of genetic diversity and various genetic backgrounds, again helping to translate why some individuals are more susceptible to severe disease manifestations while others recover quickly or are asymptomatic.
Hamsters
Golden Syrian hamsters have been shown to have distinct advantages as models for diseases involving respiratory viral infections including influenza virus, adenovirus, and SARS-CoV (Miao et al., 2019; Roberts et al., 2005b). Following infection by the intranasal route, golden Syrian hamsters demonstrate clinical features, viral kinetics, histopathological changes, and immune responses that closely mimic the mild to moderate disease described in human COVID-19 patients (Chan et al., 2020; Imai et al., 2020; Sia et al., 2020). In this form of non-lethal disease, the clinical signs include rapid breathing, decreased activity, and weight loss that is most severe by day 6 post infection. Airway involvement is evident, with histopathology showing progression from the initial exudative phase of diffuse alveolar damage with extensive apoptosis to the later proliferative phase of tissue repair. Micro-CT analysis of infected hamsters revealed severe lung injury with the degree of lung abnormalities related to the infectious dose. Commonly reported imaging features of COVID-19 patients with pneumonia were present in all infected animals (Imai et al., 2020). High-dose SARS-CoV-2 infection led to severe weight loss and partial mortality (Tostanoski et al., 2020), while older hamsters appear to exhibit more pronounced and consistent weight loss (Osterrieder et al., 2020). Other findings include intestinal mucosal inflammation, myocardial degenerative changes, viral RNA in brain stem, and lymphoid necrosis. There is a marked activation of the innate immune response, with high levels of chemokines/cytokines induced by the infection (Chan et al., 2020).
Transmission of COVID-19 from infected hamsters to naive cage mates suggests utility of the model for studying transmission (Chan et al., 2020a; Chan et al., 2020b; Sia et al., 2020). In addition, passive transfer studies with either convalescent sera (Chan et al., 2020b) or neutralizing monoclonal antibodies (Rogers et al., 2020) show great promise for studies related to immunity and vaccine development. The golden Syrian hamster model of SARS-CoV-2 infection appears to be a suitable model for the evaluation of antiviral agents (Kaptein et al., 2020; Rosenke et al., 2020) and candidate vaccines (Tostanoski et al., 2020).
Hamsters carrying the hACE2 receptor under the control of the epithelial K18 promoter are also being evaluated as a model. In an initial study of SARS-CoV-2 infection of hACE2-hamsters, clinical signs were observed, including elevated body temperatures, slow or reduced mobility, weight loss, and mortality (1 out of 4 animals). Virus titers were detected in lungs, heart, and brain tissues, with the highest titers observed in lungs on days 1–3 (≥6 logs) (Bart Tarbet, personal communication). Hamsters with immune systems compromised by either cyclophosphamide treatment or RAG2-deficiency demonstrated more severe disease, longer in duration (cyclophosphamide induction) or resulting in mortality (RAG2 −/−), and could be protected by human antibody given prophylactically (Brocato et al., 2020).
Ferrets
Ferrets are considered good models for respiratory diseases, as the physiology of their lung and airways are close to humans, and they have been used extensively to model disease caused by many respiratory viruses including influenza (Thangavel and Bouvier, 2014), RSV (Stittelaar et al., 2016) and SARS-CoV (van den Brand et al., 2008). Unlike rodents, ferrets cough and possess a sneeze reflex, making them a particularly useful model in the study of disease transmission. Ferrets exhibit lethargy and appetite loss following infection with SARS-CoV-2 via the intranasal route, but the disease does not progress to acute respiratory disease, and the animals recover from the infection (Kim et al., 2020; Shi et al., 2020). Virus shedding from the upper respiratory tract (nasal washes, saliva) can persist for up to 21 days post infection; the length of shedding appears to be dependent on the initial viral challenge dose and can be intermittent after 14 days. Mild multifocal bronchopneumonia is observed early post infection (day 3 in animals receiving 4 to 6 logs of virus), with mild multifocal bronchopneumonia developing after one week. Fever has been reported in some studies, but neither coughing nor dyspnea have been observed (Kim et al., 2020; Shi et al., 2020). Ferrets re-challenged after 28 days post initial infection appear to be completely protected (Ryan et al., 2020). SARS-CoV-2 was transmitted readily to naive direct contact ferrets but less efficiently to naive indirect contact ferrets (Shi et al., 2020) and efficiently via the air, resulting in a productive infection and the detection of infectious virus in indirect recipients (Richard et al., 2020). Disease in ferrets following SARS-CoV-2 infection appears to be very mild—less severe than ferrets infected with SARS-CoV.
The use of small animals for preclinical research in the study of SARS-CoV-2 infection involves a broad spectrum of models, from infecting wild-type animals with adapted viruses to multiple methods of introducing human ACE2 receptors. Disease is typically mild, although transgenic mice present with more severe disease. Each model offers selected advantages that will be useful not only for the testing of therapies and diseases but in the understanding of disease enhancement and related comorbidities.
Primates
NHPs are indispensable models for evaluating medical countermeasures against infectious diseases and are considered the gold standard animal model for modeling human infectious diseases. The lack of suitable substitutes for NHP models for predicting response in humans serves as a bottleneck for the development of countermeasures against infectious disease like SARS-CoV-2. Summarized below is the progress to date to establish models for COVID-19 in different primate species.
Rhesus Macaques
Rhesus macaques (M. mulatta) exposed to SARS-CoV-2 become infected and display a mild, non-lethal shedding disease phenotype, which includes few to no clinical observations. If clinical observations are reported, they are typically transient and include reduced appetite, mild dehydration, tachypnea, piloerection, and dyspnea (Munster et al., 2020). When reported, fever is mild and transient beginning shortly after exposure (i.e., day 2 PE) and resolving within 2 or 3 days (Munster et al., 2020). Body weight loss findings, if reported, are mild, transient drops in weight followed by recovery (Munster et al., 2020). Clinical chemistry and hematology are generally unremarkable. However, transient leukocytosis, neutrophilia, monocytosis, and lymphopenia are reported (Munster et al., 2020; Singh et al., 2020).
Imaging (radiographs or PET/CT) confirm rhesus macaques are infected, with infiltrates and ground-glass appearances in radiographs beginning early after exposure (day 2 or 3) and resolution occurring by day 10–14 post-exposure (PE) (Munster et al., 2020; Singh et al., 2020). Anecdotal evidence suggests that older rhesus macaques develop a chronic infection in which infiltrates persists throughout the study (Yu et al., 2020b). When available, PET/CT images corroborate the radiograph findings (Singh et al., 2020).
Virus is detected in nasal, throat, rectal, and ocular swabs and in bronchoalveolar lavage (BALs) via median tissue culture infectious dose (TCID50) beginning at approximately day 2 PE, peaking around day 4/5 PE, and decreasing after day 6 PE (Chandrashekar et al., 2020; Shan et al., 2020; Deng et al., 2020; Lu et al., 2020; Munster et al., 2020; Yu et al., 2020b). Finally, exposed rhesus macaques seroconvert, as demonstrated by a SARS-CoV-2 anti-spike ELISA and neutralization assays to various endpoint titers, depending on the laboratory and assay format utilized, and are protected from reinfection (Bao et al., 2020a; Munster et al., 2020).
Cynomolgus Macaque
Cynomolgus macaques (M. fascicularis) have been used to study the pathogenesis of SARS-CoV in which aged animals were more likely to develop disease. When exposed to SARS-CoV-2, they become infected, but show no overt clinical signs of disease (Rockx et al., 2020). Weight loss is not observed, but in some studies infected animals have a fever on day 2 to 3 (Johnston et al., 2020; Lu et al., 2020). Virus shedding from the upper respiratory tract occurs, peaking early at day 1 post infection in young animals and day 4 in aged (15–20 years) animals, then decreasing rapidly but still detected intermittently up to day 10 post infection (Rockx et al., 2020). Overall, higher levels of virus shedding were measured in aged animals than young animals (Rockx et al., 2020). They develop mild to moderate lung abnormalities and macroscopic lesions in the lungs including alveolar and bronchiolar epithelial necrosis, alveolar edema, hyaline membrane formation, and accumulation of immune cells (Finch et al., 2020; Johnston et al., 2020). While self-limiting, the disease in cynomolgus macaques does recapitulate many aspects of human COVID-19 and could be utilized to test preventative and therapeutic strategies (Rockx et al., 2020).
African Green Monkeys
African green monkeys (AGMs) exposed to SARS-CoV-2 as young adults display a mild, non-lethal shedding disease phenotype that includes few to no clinical observations (Hartman et al., 2020). If clinical observations, such as fever, are reported, they are typically transient and mild with no serious manifestations (Hartman et al., 2020; Woolsey et al., 2020). Body weight findings are generally unremarkable. Clinical chemistry and hematology reveal mild and transient shifts in leukocyte populations, mild thrombocytopenia, and selected liver enzymes (Woolsey et al., 2020). A measure of acute inflammation, CRP, is elevated early in infection (Woolsey et al., 2020).
Imaging (radiographs or PET/CT) confirms that AGMs are infected with infiltrates and ground-glass appearances in radiographs beginning early after exposure (day 2 or 3) and resolving by day 10–14 PE. When available, PET/CT images corroborate the radiograph findings. Finally, plethysmography suggests respiratory disease, but there is no consistent trend (Hartman et al., 2020).
Presence of SARS-CoV-2 in BAL via RT-PCR and plaque assay is detected by approximately day 3 PE and is present at least through day 7 PE (Hartman et al., 2020; Woolsey et al., 2020). Finally, exposed AGMs do seroconvert, as demonstrated by a SARS-CoV-2 anti-spike ELISA and neutralization assays to various endpoint titers, depending on the laboratory and assay format utilized (Hartman et al., 2020; Woolsey et al., 2020). Of note, one study (Hartman et al., 2020) used a viral isolate from Munich, which had the D614G amino acid substitution in the spike protein that has been demonstrated to be more infectious to cells in culture (Zhang et al., 2020) and was reported to be more infectious in humans (Korber et al., 2020). Disease in those animals was similarly mild, as was seen in AGMs infected with the Washington isolate without the D614G substitution.
A limited dataset suggests that age is a co-morbidity for SARS-CoV-2 disease severity in AGMs. Investigators at Tulane National Primate Research Center infected 4 older AGMs, two by multiple routes of infection including intranasal, intratracheal, and conjunctival routes and two animals by aerosol route with SARS-CoV-2. One of two animals from each group exhibited acute respiratory disease syndrome, ARDS, with radiologic and histologic abnormalities observed primarily within the right caudal lung lobes (Blair et al., 2020). Of the two animals with severe disease, one met the criteria for euthanasia at day 8 (aerosol) and the other at day 22 (multi-route infection) (Blair et al., 2020). There did not appear to be significant differences in viral replication or disease pathogenesis associated with aerosol versus multi-route infection (Blair et al., 2020).
Of the surviving older AGMs, clinical disease was mild. Some transient fevers and lethargy were observed, but no serious manifestations occurred. Clinical chemistries and hematologies were generally unremarkable. Radiographs confirm that the older AGMs are infected with infiltrates and ground-glass appearances in radiographs beginning early after exposure (day 2 or 3) through at least day 14 PE. PET/CT images corroborate the radiograph findings. Finally, viral shedding (RT-PCR) in pharyngeal, nasal, buccal, bronchial brush, rectal, and vaginal swabs begins ~D2–D7/14 PE in exposed animals (Blair et al., 2020).
In view of the severe disease observed in 2 of 4 animals as well as the observation of cytokine storm in 3 of 4 animals, older AGMs may serve as faithful if impractical models of severe disease to evaluate therapeutic strategies such as immune modulators and may also provide insights into SARS-CoV-2 pathogenesis. However, additional studies are clearly needed to corroborate these initial findings.
Other Non-human Primates
Finally, limited data is available describing disease presentation following SARS-CoV-2 exposure in pigtail macaques, baboons, and marmosets. PE data shows that these animals can be infected, but they are not as widely used. Baboons had more severe pathology and shed virus longer than macaques, and they may be a good model for cardiovascular and diabetic comorbidities (Singh et al., 2020); they have also been used for immunogenicity studies (Tian et al., 2020). Marmosets did not exhibit fever, were difficult to monitor by radiograph, and did not mount an immune response even though they were positive for virus by nasal swabs (Lu et al., 2020); pathology was reduced compared to macaques (Singh et al., 2020). Thus far, pigtail macaques have only been used for immunogenicity studies (Erasmus et al., 2020).
Summary of Animal Models
The authors recognize that the development of new and refined animal models for COVID-19 disease is a rapidly evolving field and that this summary is not a complete review but rather a description of the currently available data and the resulting guidance that can be drawn from it. Some themes are emerging and warrant monitoring. Mice expressing hACE2 and hamsters are reasonable models for early testing of potential countermeasures, displaying mostly mild disease with reproducible endpoints of weight loss and viral burden and, in transgenic mice, severe disease resulting in lethality. Intranasal inoculation is generally the route of infection, and the dose of virus varies and has an impact on the course of disease. Given the availability of animals and the ability to rigorously test for statistical significance using groups of 10 or more animals, small models should be utilized to the greatest extent possible and may be sufficient for moving into clinical studies. Based on available NHP data from multiple challenge routes (intratracheal, intranasal, oral, ocular, and combinations of these along with small-particle aerosol) and multiple dose ranges (10e3–10e6), routes and challenge do not appear to have significant impact on disease presentation (i.e., mild yet reproducible). As the NHP model matures, focused efforts will be required to demonstrate any dose response and different disease presentation following a range of doses and routes. Anecdotal evidence suggests severe disease may be more common; however, focused studies are needed to confirm. Based on a Fisher’s exact test with alpha level 0.05, a sample size of n = 8 per group ensures 80% power to detect 77% vaccine efficacy in a comparison of protection between vaccinated and sham animals (measured by absence of pathology in lung, assuming that 90% of sham animals become infected). Analysis of selected NHP studies shows that n = 8 per group ensures 80% power to detect a mean difference of ~1.88 log10 viral burden between groups (as measured by genomic RT-PCR in BAL on day 1 post challenge; Allan deCamp, personal communication). Additional data are needed to allow these calculations to be applicable to across studies and laboratories. Young adult rhesus macaques have extremely mild symptoms that may be difficult to associate with symptomatic benefit for interventions; however, viral load differences may prove a useful biomarker. African green monkeys may prove to be more useful, and data is still being collected for cynomolgus macaques. While there may be a desire to select the best animal model for comparison across studies, in practice, all good models should be utilized to meet the demand for countermeasure testing. Few models faithfully recapitulate severe disease, and further exploration is needed, including assessment of comorbidities.
Discussion
To understand the landscape and to facilitate rapid data sharing, ACTIV WG members created an inventory tool to collect information on animal studies from published literature, preprints, and unpublished studies in progress. This tool started as a spreadsheet to collect the most important information about studies, such as information on the animals, the virus isolates used, the challenge process, parameters measured in the studies, and relevant observations/endpoints along with the laboratory performing the study. The group then made assessments as to whether disease was none-to-minimal, mild, moderate, or severe. A few species that are not infected are included to prevent unnecessary duplication or perhaps even to spur enhancements—for example, the development of adapted viruses. A summary of the information is available on the NCATS Open Portal under the Animal Models tab (https://opendata.ncats.nih.gov/covid19/animal). As we learn more about models, this site will be updated.
ACTIV Preclinical WG members have curated this information by combing peer reviewed publications and preprints along with information that is not publicly available but is available to us. Additional efforts are currently underway to develop a Nonhuman Primate Studies COVID-19 Coordinating Center (NHPSCCC) that will bridge the COVID-19 work at the National Primate Research Centers and that could be a repository for data from many animal models. Recognizing that ACTIV does not constitute the entire universe of activity in this area, we invite others to submit data to ACTIV, particularly as it fills gaps or further informs the models we have summarized. The information on submitting such animal model data will be found on the NCATS Open Portal Animal Model page when it becomes available.
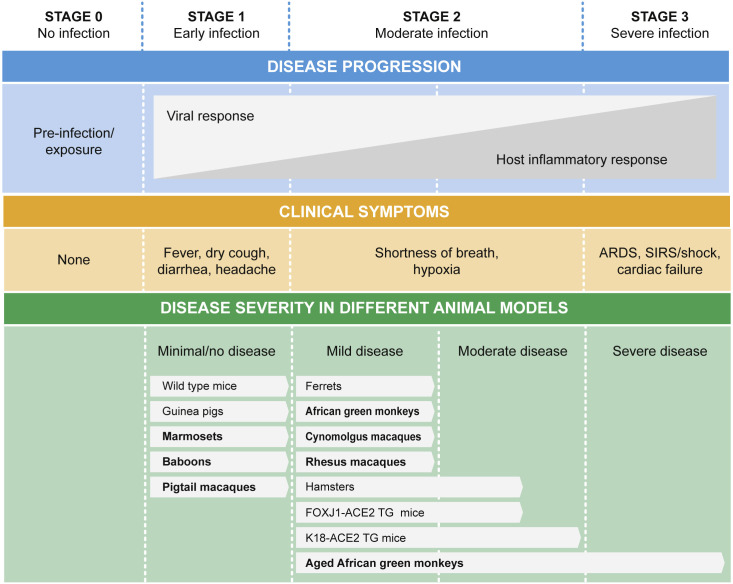
It is important to consider these animal models in the context of human disease. We adapted a framework for human disease from Siddiqi and Mehra (Siddiqi and Mehra, 2020) and placed the animal models summarized here in that framework (Figure 1 ). This graphical framework includes the full spectrum of disease but does not indicate the frequency of severity. The authors noted that 81% of cases recovered after mild disease, while 5% progressed to the most severe form, with half of those cases succumbing to disease for a 2.3% mortality rate (Siddiqi and Mehra, 2020). It is therefore not surprising and completely consistent with human disease that most animal models present with mild disease and recover. Some aged mice and aged African green monkeys present with more severe disease than younger animals; additional comorbidities have not yet been explored but would be expected to model more severe disease.
Figure 1.
Alignment of Animal Models with Course of COVID-19 Disease
The top panels represent the course of disease in humans, adapted from Siddiqi and Mehra (2020), along with clinical symptoms. The bottom panel aligns animal models with the typical course of disease seen in those models. Minimal to no disease means that there is no or limited viral replication, such that these models are not tractable for testing interventions. Bold text indicates non-human primate models. Models that are not widely available or are currently in testing are not represented here. ARDS, acute respiratory distress syndrome; SIRS, systemic inflammatory response syndrome.
Each of the current models in development is yielding valuable information about infectivity, routes of infection, viral persistence, reinfection, and relative level and types of pathogenesis per species. As researchers around the world seek to discover effective therapies and vaccines to treat or to prevent COVID-19, judicious choices will be needed to assure that the models are available for comparative studies. The complex interplay between the host and the virus means that each model is unlikely to represent every aspect of disease in humans, and thus different models may be recommended for therapeutics compared with vaccines due to animal availability and endpoints of studies. Indeed, investigators have begun testing vaccines and therapeutics as soon as species are known to get disease, more rapidly than under normal circumstances, in which additional studies might be performed to better understand the disease model prior to testing countermeasures.
Table 1 indicates which models could be used for testing vaccines, antivirals, neutralizing antibodies, or other therapies, but does not make specific recommendations. This is based on the relevant endpoints one can measure that would inform vaccine or therapeutic efficacy: viral load (swabs, BAL, etc.), body weight, body temperature, lung imaging, lung function, and cytokines. Antivirals have been tested in mice (Sheahan et al., 2020), hamsters (Kaptein et al., 2020; Rosenke et al., 2020), cynomolgus macaques (Maisonnasse et al., 2020), and rhesus macaques (Rosenke et al., 2020; Williamson et al., 2020). Antibodies have been assessed in hamsters (Brocato et al., 2020; Rogers et al., 2020) and mice (Dinnon et al., 2020). Finally, vaccines have been tested for immunogenicity in small animals, but efficacy studies have largely been done in rhesus macaques (Gao et al., 2020; van Doremalen et al., 2020; Yu et al., 2020a). The ACTIV Animal Model pages on the NCATS Open Portal will be updated with publications reflecting the utility of various models. Master protocols currently in development will define key parameters and time points to follow along with group sizes, allowing comparison across studies.
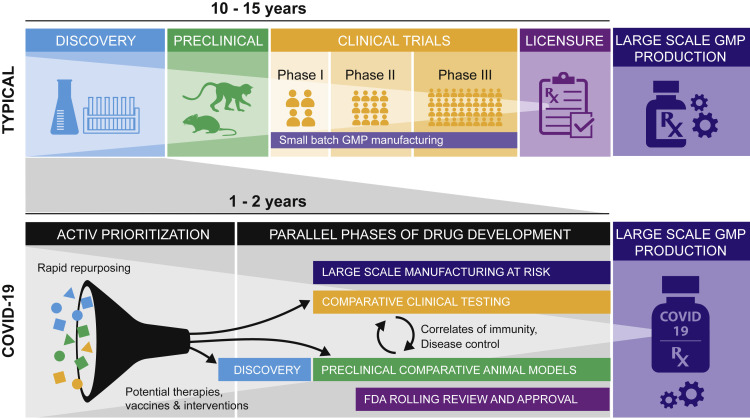
The severity of disease, rapid transmission, and global spread of this new virus make the development of vaccines and therapeutics an urgent priority. Typical development plans have been greatly accelerated by performing many activities in parallel, often at risk, shortening timelines with unprecedented speed (Figure 2 ). This includes animal model development and testing, which can contribute to the identification and confirmation of correlates and surrogate markers for use in the clinic. The vaccines that have been chosen by OWS can be compared in animal models while phase I and phase II clinical work continues (Corey et al., 2020).
Figure 2.
Animal Models in Context of Accelerated Product Development
The top panel represents the typical course of product development as sequential activities with the process taking 10–15 years. The bottom panel shows the pandemic or COVID-19 response, focusing on rapid repurposing and multiple activities occurring concurrently, in order to compress timelines into 1–2 years. The phase of compounds being repurposed are color coded with the phases of drug development, from discovery through preclinical to clinical. GMP, good manufacturing practices.
Selection of an appropriate animal model depends on the questions to be addressed. Table 2 presents our recommendations on which of the currently available animal models to select, focusing on testing classes of therapeutics and vaccines for reducing various aspects of COVID-19. For example, carboxylesterase activity is high in rodents, but not humans (Bahar et al., 2012; Li et al., 2005; Rudakova et al., 2011), impacting the pharmacokinetics of antivirals such as Remdesivir and the suitability of models for preclinical testing of specific drugs (Warren et al., 2016). It is clear that many models are available which faithfully reproduce the early phases of infection and lung disease, followed by recovery; recovery is a prevalent feature of COVID-19 in humans. Models of severe disease are largely determined by fatal outcomes when untreated but may not capture all signs and symptoms associated with human COVID-19 disease—for example, coagulopathies. One difference between some models and human disease is the observation of virus in the brain in mice and hamsters, which requires further exploration. There is a report of virus in CSF in a single clinical case (Zhou et al., 2020a), though most neurological complications are considered to be inflammatory responses (Gulati et al., 2020). Comorbidities have not been rigorously explored in animal models thus far, yet we know the strong association of age, hypertension, diabetes, lung and heart disease with poor prognoses in COVID-19. Immune responses likely play a key role in determining the severity of disease, yet are not as easily manipulated in animal species that are phylogenetically closer to humans as they are in mice.
Table 2.
Recommended Animal Models for Specific Stages of Human COVID-19 Disease
| COVID-19 Medical Need | Disease aspect | Recommended model(s) | Advantages | Limitations |
|---|---|---|---|---|
| Drugs and immunomodulators | Lung lesions, Interstitial pneumonia | Mouse models | Ease of handling; availability for statistical significance | May not be reflective of human pharmacokinetics; may not metabolize antiviral prodrugs as humans |
| Syrian Golden hamsters | Ease of handling; availability for statistical significance; historically used by coronavirus researchers due to natural ACE2 permissiveness & tissue distribution | May not be reflective of human pharmacokinetics; may not metabolize antiviral prodrugs as humans | ||
| AGMs, Aged AGMs | Immune modulators can be tested | Availability of animals | ||
| Rhesus macaque | Pharmacokinetics similar to humans | Limited supply to test drugs; narrow window for treatment | ||
| Cough, fever | Ferret | Symptoms seen in humans | Disease is mild | |
| Cytokine storm | Aged AGMs | Cytokine response well characterized; Immune modulators can be tested | Availability of aged animals | |
| K18-ACE2 mice | Cytokine response well characterized; Immune modulators can be tested | – | ||
| ARDS | Aged AGMs | Immune modulators can be tested | Availability of aged animals | |
| K18-ACE2 mice | Drugs could show promise; disease presentation resembles ARDS | Not true ARDS; Narrow treatment window | ||
| Coagulation | None to date | – | – | |
| Post-exposure prophylaxis with mAbs | Limit viremia | Rhesus or cynomolgus macaques | PK similar and valuable information about dosing; reagents available | Anti-drug antibody may develop |
| Mouse models | Potential for quick screens to choose cocktails | May not be reflective of human dosing or efficacy | ||
| Syrian Golden hamsters | Potential for quick screens to choose cocktails | May not be reflective of human dosing or efficacy | ||
| Vaccines designed to generate neutralizing antibodies | Limit viremia; Prevent infection; reduced pathology | Mouse, Syrian golden hamster | Proof of concept, required prior to NHP studies; multiple models are equivalent | – |
| Rhesus macaque, cynomolgus macaque | B cell responses similar to humans; can quantify correlates of protection; reagents available | Low level of viremia may be easier to clear than humans | ||
| Antibody-dependent enhancement | None to date | – | – | |
| Vaccines designed to generate T cell response | Limit viremia or peak of viremia; reduced pathology | Rhesus macaque, cynomolgus macaque | T cell responses similar to humans and can be studied in depth; identify correlates; reagents available | T cell ‘only’ vaccines unlikely to protect from infection but can test concept |
Within each medical need, the disease aspects are presented in order of increasing complexity, while animal models are presented in the order in which they should be approached. Advantages and limitations are also presented to help in the selection of the appropriate animal model. AGM, African green monkey; ARDS, acute respiratory distress syndrome; mAbs, monoclonal antibodies; NHP, non-human primates; PK, pharmacokinetics
In order for these preclinical studies to have the most impact, their design is critically important to assure that results are rigorous and reproducible. Studies will require statistical justification based on clinical and laboratory measures such that studies are appropriately powered to prevent getting uninterpretable results that can be misleading and wasteful of animal resources. One method to enhance reproducibility that is currently under development is the adoption of shared Master Protocols for design and sampling. Not only can experiments be compared across sites, but controls located at different sites can also be combined for increased power while retaining contemporaneous infection controls at each site. This strategy can reduce the number of controls used overall, conserving precious resources. If assays and challenge stocks can be harmonized across experimental sites, many of the variables can be further reduced.
Defining animal models and their use is a prerequisite to performing studies to compare vaccines and therapeutics so that the most promising ones advance to the next phase, whether it is testing in NHPs or the clinic. The prioritization schemes for the development and testing of treatments and vaccines are beyond the scope of this manuscript. This aim of this manuscript is defining the utility of animal models to inform the requisite prioritization of animal studies, particularly in NHPs, in which resources are limited relative to the number of candidate vaccines and therapeutics.
Conclusion
COVID-19 is a multi-faceted, multi-factorial, multi-systemic, highly infectious disease that evokes wide-ranging responses in humans, from asymptomatic to severe disease with respiratory, gastrointestinal, circulatory, neurological, and normal to hyper immune responses. No single animal model recapitulates the totality of pathogenesis or predicts interventional responses faithfully as in the human. Yet as with the response to other emerging infections, animal models will play a key role, even as candidate vaccines and therapeutics are entering clinical trials at a record pace. We have presented various animal models that are being developed and the role they may play in responding to COVID-19. Our summary table of animal models and their applications has been posted to the NCATS Open Portal (https://opendata.ncats.nih.gov/covid19/animal) and will be continuously updated as models are advanced and further interrogated. The key models appear to be mice (various), hamsters, and for NHPs, rhesus macaques and African green monkeys. Most mimic the mild form of COVID-19 disease, with the exception of transgenic mice; additional research may result in development of more severe disease models. ACTIV invites submission of information on animal models to be included in our assessment as we work toward standardized and harmonized animal models while balancing resources and availability.
Acknowledgments
This work was done on behalf of the ACTIV Preclinical Working Group (https://www.nih.gov/research-training/medical-research-initiatives/activ/preclinical-working-group). The authors would like to thank the ACTIV Preclinical Working Group for discussions and the spark for this manuscript. We would also like to thank Annaliesa Anderson for critical review, along with Michael Diamond, Prabha Fernandes, and Tomas Cihlar. We would especially like to thank Kara Carter and Jay Grobler for their review and discussions on figures and shared content with the companion manuscript. We thank Katinka Vigh-Conrad for assistance with figures.
Author Contributions
Conceptualization: N.H., J.H., C.L.; Writing – Original Draft: W.F., N.H., J.N., C.L., L.P., S.R.; Writing – Review & Editing: W.F., N.H., J.H., C.L., L.P., S.R., J.R.; Visualization: N.H., J.H.
Declaration of Interests
Srinivas Rao is an employee and shareholder of Sanofi.
References
- Bahar F.G., Ohura K., Ogihara T., Imai T. Species difference of esterase expression and hydrolase activity in plasma. J. Pharm. Sci. 2012;101:3979–3988. doi: 10.1002/jps.23258. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y. Lack of Reinfection in Rhesus Macaques Infected with SARS-CoV-2. bioRxiv. 2020 2020.2003.2013.990226. [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Blair R.V., Vaccari M., Doyle-Meyers L.A., Roy C.J., Russell-Lodrigue K., Fahlberg M., Monjure C.J., Beddingfield B., Plante K.S., Plante J.A. “ARDS and Cytokine Storm in SARS-CoV-2 Infected Caribbean Vervets”. bioRxiv. 2020 2020.2006.2018.157933. [Google Scholar]
- Brocato R.L., Principe L.M., Kim R.K., Zeng X., Williams J.A., Liu Y., Li R., Smith J.M., Golden J.W., Gangemi D. Disruption of Adaptive Immunity Enhances Disease in SARS-CoV-2 Infected Syrian Hamsters. bioRxiv. 2020 doi: 10.1128/JVI.01683-20. 2020.2006.2019.161612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Zhang A.J., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Fan Z., Li C., Liang R., Cao J. Surgical mask partition reduces the risk of non-contact transmission in a golden Syrian hamster model for Coronavirus Disease 2019 (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa644. Published online May 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa325. Published online March 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S. Comprehensive review of coronavirus disease 2019 (COVID-19) Biomed J. 2020;43:334–340. doi: 10.1016/j.bj.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S., Stoffels P. Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV): An Unprecedented Partnership for Unprecedented Times. JAMA. 2020;323:2455–2457. doi: 10.1001/jama.2020.8920. [DOI] [PubMed] [Google Scholar]
- Corey L., Mascola J.R., Fauci A.S., Collins F.S. A strategic approach to COVID-19 vaccine R&D. Science. 2020;368:948–950. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- Deng W., Bao L., Gao H., Xiang Z., Qu Y., Song Z., Gong S., Liu J., Liu J., Yu P. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in Rhesus macaques. bioRxiv. 2020 doi: 10.1038/s41467-020-18149-6. 2020.2003.2013.990036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. bioRxiv. 2020 2020.2005.2006.081497. [Google Scholar]
- Douam F., Gaska J.M., Winer B.Y., Ding Q., von Schaewen M., Ploss A. Genetic Dissection of the Host Tropism of Human-Tropic Pathogens. Annu. Rev. Genet. 2015;49:21–45. doi: 10.1146/annurev-genet-112414-054823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus J.H., Khandhar A.P., O’Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B. An <em>Alphavirus</em>-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Science Translational Medicine. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . Spotlight Series (FDA) 2020. Translating In Vitro Antiviral Activity to the In Vivo Setting: A Crucial Step in Fighting COVID-19. [Google Scholar]
- Finch C.L., Crozier I., Lee J.H., Byrum R., Cooper T.K., Liang J., Sharer K., Solomon J., Sayre P.J., Kocher G. Characteristic and quantifiable COVID-19-like abnormalities in CT- and PET/CT-imaged lungs of SARS-CoV-2-infected crab-eating macaques (<em>Macaca fascicularis</em>) bioRxiv. 2020 2020.2005.2014.096727. [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Rapid development of an inactivated vaccine for SARS-CoV-2. bioRxiv. 2020 doi: 10.1126/science.abc1932. 2020.2004.2017.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. bioRxiv. 2020 doi: 10.1172/jci.insight.142032. 2020.2007.2009.195230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler J.A., Anderson A.S., Fernandes P., Diamond M.S., Colvis C.M., Menetski J.P., Alvarez R.M., Young J.A.T., Carter K.L. Accelerated preclinical paths to support rapid development of COVID-19 therapeutics. Cell Host Microbe. 2020;28:638–645. doi: 10.1016/j.chom.2020.09.017. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati A., Pomeranz C., Qamar Z., Thomas S., Frisch D., George G., Summer R., DeSimone J., Sundaram B. A Comprehensive Review of Manifestations of Novel Coronaviruses in the Context of Deadly COVID-19 Global Pandemic. Am. J. Med. Sci. 2020;360:5–34. doi: 10.1016/j.amjms.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Nambulli S., McMillen C.M., White A.G., Tilston-Lunel N.L., Albe J.R., Cottle E., Dunn M., Frye L.J., Gilliland T.H. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and prolonged shedding of infectious virus from both respiratory and gastrointestinal tracts. bioRxiv. 2020 doi: 10.1371/journal.ppat.1008903. 2020.2006.2020.137687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HHS . U.S. Department of Health & Human Services; 2020. Fact Sheet: Explaining Operation Warp Speed. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Jay A., Raymond J.L., Rossi F., Zeng X., Scruggs J., Dyer D., Frick O., Moore J., Berrier K. Development of a Coronavirus Disease 2019 Nonhuman Primate Model Using Airborne Exposure. bioRxiv. 2020 doi: 10.1371/journal.pone.0246366. 2020.2006.2026.174128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S.J.F., Jacobs S., Langendries L., Seldeslachts L., ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Maas E. Antiviral treatment of SARS-CoV-2-infected hamsters reveals a weak effect of favipiravir and a complete lack of effect for hydroxychloroquine. bioRxiv. 2020 2020.2006.2019.159053. [Google Scholar]
- Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E., Bhattacharya T., Parker M. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 2020.2004.2029.069054. [Google Scholar]
- Li B., Sedlacek M., Manoharan I., Boopathy R., Duysen E.G., Masson P., Lockridge O. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem. Pharmacol. 2005;70:1673–1684. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Li W., Drelich A., Martinez D.R., Gralinski L., Chen C., Sun Z., Schäfer A., Leist S.R., Liu X., Zhelev D. Rapid selection of a human monoclonal antibody that potently neutralizes SARS-CoV-2 in two animal models. bioRxiv. 2020 2020.2005.2013.093088. [Google Scholar]
- Liu Y., Hu G., Wang Y., Zhao X., Ji F., Ren W., Gong M., Ju X., Li C., Hong J. Functional and Genetic Analysis of Viral Receptor ACE2 Orthologs Reveals a Broad Potential Host Range of SARS-CoV-2. bioRxiv. 2020 doi: 10.1073/pnas.2025373118. 2020.2004.2022.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. 2020 2020.2004.2008.031807. [Google Scholar]
- Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020;526:165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonnasse P., Guedj J., Contreras V., Behillil S., Solas C., Marlin R., Naninck T., Pizzorno A., Lemaitre J., Gonçalves A. Hydroxychloroquine in the treatment and prophylaxis of SARS-CoV-2 infection in non-human primates. 2020. 10.21203/rs.3.rs-27223/v1 Researchsquare. [DOI] [PubMed]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Chard L.S., Wang Z., Wang Y. Syrian Hamster as an Animal Model for the Study on Infectious Diseases. Front. Immunol. 2019;10:2329. doi: 10.3389/fimmu.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau G.B., Burgess S.L., Sturek J.M., Donlan A.N., Petri W.A., Mann B.J. Evaluation of K18-<em>hACE2</em> mice as a model of SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.4269/ajtmh.20-0762. 2020.2006.2026.171033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH . ACTIV; 2020. Accelerating COVID-19 Therapeutic Interventions and Vaccines. [DOI] [PubMed] [Google Scholar]
- Oladunni F.S., Park J.-G., Tamayo P.P., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C. Lethality of SARS-CoV-2 infection in K18 human angiotensin converting enzyme 2 transgenic mice. bioRxiv. 2020 doi: 10.1038/s41467-020-19891-7. 2020.2007.2018.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A., Nadkarni G., Glicksberg B.S., Houldsworth J., Cordon-Cardo C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zhao Y.B., Wang Q., Li J.Y., Zhou Z.J., Liao C.H., Ge X.Y. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020;22:221–225. doi: 10.1016/j.micinf.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R., Strohmeier S., Amanat F., Gillespie V.L., Krammer F., García-Sastre A., Coughlan L., Schotsaert M., Uccellini M. Comparison of Transgenic and Adenovirus hACE2 Mouse Models for SARS-CoV-2 Infection. bioRxiv. 2020 doi: 10.1080/22221751.2020.1838955. 2020.2007.2006.190066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Kok A., de Meulder D., Bestebroer T.M., Lamers M.M., Okba N.M.A., Fentener van Vlissingen M., Rockx B., Haagmans B.L., Koopmans M.P.G. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Paddock C., Vogel L., Butler E., Zaki S., Subbarao K. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 2005;79:5833–5838. doi: 10.1128/JVI.79.9.5833-5838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Vogel L., Guarner J., Hayes N., Murphy B., Zaki S., Subbarao K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005;79:503–511. doi: 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-t., Limbo O., Smith C., Song G., Woehl J. Rapid isolation of potent SARS-CoV-2 neutralizing antibodies and protection in a small animal model. bioRxiv. 2020 doi: 10.1126/science.abc7520. 2020.2005.2011.088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Jarvis M.A., Feldmann F., Schwarz B., Okumura A., Lovaglio J., Saturday G., Hanley P.W., Meade-White K., Williamson B.N. Hydroxychloroquine Proves Ineffective in Hamsters and Macaques Infected with SARS-CoV-2. bioRxiv. 2020 2020.2006.2010.145144. [Google Scholar]
- Rowley A.H. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat. Rev. Immunol. 2020;20:453–454. doi: 10.1038/s41577-020-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudakova E.V., Boltneva N.P., Makhaeva G.F. Comparative analysis of esterase activities of human, mouse, and rat blood. Bull. Exp. Biol. Med. 2011;152:73–75. doi: 10.1007/s10517-011-1457-y. [DOI] [PubMed] [Google Scholar]
- Ryan K.A., Bewley K.R., Fotheringham S.A., Brown P., Hall Y., Marriott A.C., Tree J.A., Allen L., Aram M.J., Brunt E. Dose-dependent response to infection with SARS-CoV-2 in the ferret model: evidence of protection to re-challenge. bioRxiv. 2020 2020.2005.2029.123810. [Google Scholar]
- Shan C., Yao Y.-F., Yang X.-L., Zhou Y.-W., Wu J., Gao G., Peng Y., Yang L., Hu X., Xiong J. Infection with Novel Coronavirus (SARS-CoV-2) Causes Pneumonia in the Rhesus Macaques. Cell Res. 2020;30:670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi H.K., Mehra M.R. COVID-19 illness in native and immunosuppressed states: A clinical-therapeutic staging proposal. J. Heart Lung Transplant. 2020;39:405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., Ganatra S.R., Singh B., Cole J., Alfson K.J., Clemmons E., Gazi M., Gonzalez O., Escobedo R., Lee T.-H. SARS-CoV-2 infection leads to acute infection with dynamic cellular and inflammatory flux in the lung that varies across nonhuman primate species. bioRxiv. 2020 2020.2006.2005.136481. [Google Scholar]
- Skelton J.K., Ortega-Prieto A.M., Dorner M. A Hitchhiker’s guide to humanized mice: new pathways to studying viral infections. Immunology. 2018;154:50–61. doi: 10.1111/imm.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittelaar K.J., de Waal L., van Amerongen G., Veldhuis Kroeze E.J., Fraaij P.L., van Baalen C.A., van Kampen J.J., van der Vries E., Osterhaus A.D., de Swart R.L. Ferrets as a Novel Animal Model for Studying Human Respiratory Syncytial Virus Infections in Immunocompetent and Immunocompromised Hosts. Viruses. 2016;8:168. doi: 10.3390/v8060168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., Huang J., He J., Zhu A., Zhao J. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., Liu S.S., Zhang N.N., Li X.F., Xiong R. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel R.R., Bouvier N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.-H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Lague J., Portnoff A.D., Norton J., Guebre-Xabier M. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 elicits immunogenicity in baboons and protection in mice. bioRxiv. 2020 doi: 10.1038/s41467-020-20653-8. 2020.2006.2029.178509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020 doi: 10.1038/s41591-020-1070-6. Published online September 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brand J.M., Haagmans B.L., Leijten L., van Riel D., Martina B.E., Osterhaus A.D., Kuiken T. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet. Pathol. 2008;45:551–562. doi: 10.1354/vp.45-4-551. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1038/s41586-020-2608-y. 2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel L.N., Roberts A., Paddock C.D., Genrich G.L., Lamirande E.W., Kapadia S.U., Rose J.K., Zaki S.R., Subbarao K. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV) Vaccine. 2007;25:2173–2179. doi: 10.1016/j.vaccine.2006.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A., De C., Abad Fernandez M., Lenarcic E.M., Xu Y., Cockrell A.S., Cleary R.A., Johnson C.E., Schramm N.J., Rank L.M. Precision mouse models with expanded tropism for human pathogens. Nat. Biotechnol. 2019;37:1163–1173. doi: 10.1038/s41587-019-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00120–e00127. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., Doremalen N.v., Leighton I., Yinda C.K., Pérez-Pérez L. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. bioRxiv. 2020 doi: 10.1038/s41586-020-2423-5. 2020.2004.2015.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P. SARS-CoV-2 infection in the lungs of human ACE2 transgenic mice causes severe inflammation, immune cell infiltration, and compromised respiratory function. bioRxiv. 2020 2020.2007.2009.196188. [Google Scholar]
- Woolsey C., Borisevich V., Prasad A.N., Agans K.N., Deer D.J., Dobias N.S., Heymann J.C., Foster S.L., Levine C.B., Medina L. Establishment of an African green monkey model for COVID-19. bioRxiv. 2020 doi: 10.1038/s41590-020-00835-8. 2020.2005.2017.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yoshikawa N., Yoshikawa T., Hill T., Huang C., Watts D.M., Makino S., Milligan G., Chan T., Peters C.J., Tseng C.T. Differential virological and immunological outcome of severe acute respiratory syndrome coronavirus infection in susceptible and resistant transgenic mice expressing human angiotensin-converting enzyme 2. J. Virol. 2009;83:5451–5465. doi: 10.1128/JVI.02272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 2020.06.12.148726. [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhang M., Wang J., Gao J. Sars-Cov-2: Underestimated damage to nervous system. Travel Med. Infect. Dis. 2020;36:101642. doi: 10.1016/j.tmaid.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]