Abstract
Background
Mycobacterium tuberculosis (Mtb) and human immunodeficiency virus (HIV) coinfection increases mortality, accelerates progression to acquired immune deficiency syndrome, and exacerbates tuberculosis disease. However, the impact of pre-existing Mtb infection on subsequent HIV infection has not been fully explored. We hypothesized that Mtb infection creates an immunological environment that influences the course of HIV infection, and we investigated whether pre-existing Mtb infection impacts the susceptibility of CD4+ T cells to HIV-1 infection.
Methods
Plasma and blood CD4+ T cells isolated from HIV-negative individuals across the Mtb infection spectrum and non-Mtb-infected control individuals were analyzed for inflammation markers and T-cell phenotypes. CD4+ T cells were infected with HIV-1 in vitro and were monitored for viral replication.
Results
We observed differences in proinflammatory cytokines and the relative proportion of memory T-cell subsets depending on Mtb infection status. CD4+ T cells derived from individuals with latent Mtb infection supported more efficient HIV-1 transcription, release, and replication. Enhanced HIV-1 replication correlated with higher percentages of CD4+ TEM and TTD cells.
Conclusions
Pre-existing Mtb infection creates an immunological environment that reflects Mtb infection status and influences the susceptibility of CD4+ T cells to HIV-1 replication. These findings provide cellular and molecular insights into how pre-existing Mtb infection influences HIV-1 pathogenesis.
Keywords: CD4 T cells, HIV replication, latent tuberculosis infection (LTBI), TB-HIV coinfection, TB immune responses
We investigated the impact of pre-existing Mycobacterium tuberculosis infection on the susceptibility of CD4+ T cells to HIV-1 infection. We found that CD4+ T cells derived from individuals with latent Mycobacterium tuberculosis infection support more efficient HIV-1 replication.
Mycobacterium tuberculosis (Mtb) and human immunodeficiency virus (HIV) each exert a tremendous impact on global health. In 2018, the World Health Organization reported an estimated 10 million new tuberculosis (TB) cases globally, with 8.6% occurring in people with HIV (PWH) [1, 2]. Human immunodeficiency virus and Mtb coinfection is considered a syndemic with increased morbidity and mortality, including a 20–30 times increased risk of developing active TB from Mtb infection compared with those without HIV [1, 3, 4]. Furthermore, active TB in HIV-positive individuals facilitates rapid progression to acquired immune deficiency syndrome (AIDS) and increased mortality [3, 5, 6]. Worse outcomes for PWH coinfected with Mtb persist despite antiretroviral therapy, suggesting chronic pathogenesis [7]. The TB/HIV syndemic was estimated to have claimed 250 000 lives in 2018, making TB the main cause of death in PWH [1]. The 2 diseases share geography and epidemiology, with both demonstrating peaks in early adulthood and having common associations including malnutrition and poverty [8–11]. In many high TB/HIV burden settings, such as sub-Saharan Africa, initial Mtb infection is often acquired before HIV during childhood to mid-adolescence, whereas noncongenital HIV infection is typically acquired from adolescence onwards [8, 9].
Given the clinical and epidemiological relevance of Mtb-HIV coinfection, it is essential to understand how these 2 infections interact at a cellular and molecular level [3, 12]. Mycobacterium tuberculosis infection leads to persistent immunological changes in lymphocyte activation, checkpoint molecule upregulation, T-cell and macrophage function, and proinflammatory cytokine levels, all impacting the course of HIV infection [12–18]. For example, Mtb infection induces inflammatory mediators including tumor necrosis factor-alpha (TNF-α) and interferon-gamma (IFN-γ) [19, 20]. These cytokines activate T cells through transcription factors such as NF-κB, AP-1, and NFAT, which also regulate HIV transcription and replication [12, 21, 22]. Mycobacterium tuberculosis signaling through innate immune receptors such as Toll-like receptor 2 may also positively regulate HIV-1 infection and transcription through the upregulation of relevant transcription factors [23]. Although these changes have been demonstrated in coinfected individuals with active TB, the impact of pre-existing Mtb infection, either active or latent, on subsequent HIV exposure has not been fully addressed.
We hypothesize that latent Mtb infection (LTBI) and active TB create immunological environments or footprints that render individuals exposed to HIV at a higher risk of successfully acquiring the infection. We tested this hypothesis by measuring the levels of inflammatory markers in plasma from individuals with LTBI, active TB, and non-Mtb-infected controls. We also determined whether CD4+ T cells obtained from individuals with Mtb infection were more susceptible to HIV-1 infection and/or supported greater viral replication in vitro than CD4+ T cells from controls. Results indicate that LTBI influences the course of HIV-1 replication in CD4+ T cells, implicating LTBI as an important cofactor in the TB/HIV syndemic.
METHODS
Subject Recruitment and Sample Collection
Individuals were recruited between September 2018 and April 2019 from the Tuberculosis Clinic or the Immigrant and Refugee Health Program Clinic, both at Boston Medical Center (BMC). Study participants provided informed consent. We chose the Immigrant and Refugee Health Program Clinic for recruitment of control subjects with demographics similar to subjects recruited through the Tuberculosis Clinic. Inclusion criteria were as follows: HIV-uninfected (documented within 24 months before enrollment), ≥18 years of age, no history of anti-TB treatment for LTBI, and ≤6 weeks of anti-TB treatment for active TB (n = 2). We did not observe any effects of anti-TB treatment on subsequent data analyses. Exclusion criteria were immunosuppressive medications (eg, steroids, anti-TNF inhibitors, cancer chemotherapy), acute illness, pregnancy, known chronic infections (eg, hepatitis B or C), malignancy, or autoimmune disorders. Latent Mtb infection was defined as positive tuberculin skin test ([TST] n = 8) (threshold value ≥10 mm) or IFN-γ release assay ([IGRA] n = 8) (including at least 1 TB Ag-nil value ≥0.8), but no evidence of active TB by symptoms and chest radiograph. The IGRAs consisted of QuantiFERON-TB Gold or QuantiFERON-TB Gold Plus ([QIAGEN] n = 7) and T-SPOT.TB test ([Oxford Immunotec] n = 1). Subjects tested positive for LTBI within 1 year before enrollment. Five LTBI subjects (TST n = 4, IGRA n = 1) tested positive for LTBI within the last 2 years based on negative test results in preceding years, 4 of whom were born in the United States. A higher frequency of women versus men in the LTBI group did not affect subsequent data analyses. Active TB was defined by positive mycobacterial culture; in addition, 5 subjects were acid-fast bacilli smear positive and 4 were positive on GeneXpert versions 4.8 and 6.6 (Cepheid). Control subjects had no symptoms of active TB and tested negative for LTBI within 1 (n = 8) to 3 years (n = 3) before enrollment (TST n = 1, IGRA n = 10). Subjects were presumed to have received the Bacillus Calmette-Guérin vaccination based on interview response and/or a shoulder scar, except for 6 subjects born in the United States (LTBI n = 5, control n = 1). Twenty milliliters of blood was drawn by the study team or by BMC’s phlebotomy service. The study was approved by the BMC Internal Review Board (protocol # H-36860).
Peripheral Blood Mononuclear Cells Isolation and CD4+ T-Cell Purification
Peripheral blood mononuclear cells (PBMC) were isolated using the Lymphoprep density gradient (StemCell Technologies). CD4+ T cells were enriched by negative selection with the EasySep Human CD4+ T Cell Enrichment Kit (StemCell Technologies) and routinely achieved >98% purity. Purified CD4+ T cells were cultured in RPMI 1640 medium (Gibco) supplemented with penicillin/streptomycin (Invitrogen), l-glutamine (Invitrogen), and 10% of autologous human plasma collected before cell separation.
Flow Cytometry Analysis
Antibodies for phenotyping CD4+ T cells included the following: CD3-FITC (clone HIT3a; BD Biosciences), CD4-Alexa Fluor 647 (clone RPA-T4; BioLegend), CD25-BV421 (clone 2A3; BD Biosciences), CD69-BV421 (clone FN50; BioLegend), HLA-DR-BV421 (clone L243; BioLegend), CD95-PE-Dazzle594 (clone DX2; BioLegend), CD27-APC-Fire750 (clone M-T271; BioLegend), CCR7-PE-Cy7 (clone G043H7; BioLegend), and CD45RA-PerCP-Cy5.5 (clone HI100; BioLegend). Antibody-stained peripheral blood mononuclear cells were analyzed using an LSR-II SROP analyzer and FACSDiva software 6.0 (BD Biosciences). OneComp eBeads (Thermo Fisher) stained with individual colors were used for fluorescence compensation, and Fluorescence Minus One controls (prepared with cells from control subjects) were used as gating controls [24]. Data analysis was performed using FlowJo software.
In Vitro Infection of CD4+ T Cells With Human Immunodeficiency Virus-1
Stocks of HIV-1 were generated by transfection of HEK293T cells (ATCC) with plasmid encoding the infectious molecular clone HIV-1NL4-3 as previously described [25]. CD4+ T cells were infected with viral culture supernatant at a multiplicity of infection of 1 (based on virus titrated on CEM-GFP cells; NIH AIDS Reagents Program) in a single well of a 48-well plate for 2 hours at room temperature. After incubation, unbound virus was removed by washing cells with phosphate-buffered saline (Invitrogen) supplemented with 2% fetal bovine serum (Gemini Bio-Products) and resuspended in culture medium. Infected cells were divided into separate wells depending on the experimental objective. Saquinavir (1 μM; NIH AIDS Reagents Program) was added to the wells of infected cells designated for examination of HIV-1 infectivity to prevent viral spread beyond the first round. Infected cells treated with 1 μM efavirenz (NIH AIDS Reagents Program) served as a negative control for infection. On day 3 postinfection, infected cells were harvested for measuring HIV-1 infection based on HIV-1 deoxyribonucleic acid (DNA) by quantitative polymerase chain reaction (qPCR). Wells were also collected on day 4 postinfection for ribonucleic acid (RNA) isolation. To examine virus replication, HIV-infected cells were cultured in round bottom, 96-well plates in the absence of saquinavir for 9 days. Half of the culture medium was harvested on days 3 and 6 postinfection and fresh medium was added back to the cultures. Viral replication was assessed by enzyme-linked immunosorbent assay based on the levels of HIV-1 p24 antigen and normalized to p24 levels in cultures treated with efavirenz. At day 9, cells were harvested for genomic DNA and qPCR analysis of HIV-1 infection.
Measuring Human Immunodeficiency Virus-1 Replication With Enzyme-Linked Immunosorbent Assay
Culture supernatants were lysed with 0.5% Triton-X (Thermo Fisher) and subsequently loaded onto plates coated with anti-HIV-1 p24 capture antibody (catalog no. 3957; NIH AIDS Reagents Program). Standard curves were generated by serially diluting p24 standards (model no. 77620R; Perkin Elmer). Enzyme-linked immunosorbent assays were conducted as previously described [26].
Quantification of Human Immunodeficiency Virus-1 Deoxyribonucleic Acid Intermediates by Quantitative Polymerase Chain Reaction
Total cell-associated DNA was purified using the DNeasy Blood and Tissue Kit (QIAGEN). Total HIV-1 DNA per cell was quantified by qPCR as previously described [26]. The DNA from uninfected PBMCs was used as a standard for calculating cell number [26]. Serially diluted HIV-1NL4-3 plasmid was used as an HIV-1 DNA copy standard. The reactions were run in a QuantStudio 3 (Applied Biosystems) with the following program: hot-start at 95°C for 2 minutes, followed by 50 cycles of denaturation for 15 seconds at 95°C, annealing for 30 seconds at 60°C, plate read, and extension for 1 minute at 72°C.
Human immunodeficiency virus-1 integration was measured by nested Alu-PCR as described [26], except that the master mixture for the Alu-gag amplification step used in this study contained 0.48 µM Alu forward primer, 2.4 µM gag reverse primer, and a final reaction volume of 50 µL. A parallel reaction without forward primer for Alu was prepared as a control for unintegrated HIV-1 DNA [27]. The reaction was conducted in a Biometra T-Professional thermocycler with the following program: hot start at 95°C for 4 minutes and 40 cycles of 93°C for 15 seconds, 50°C for 15 seconds, and 70°C for 4 minutes. Reaction products from samples and standards (25 μL) were transferred to another plate containing 25 μL of 2× master mixture specific for the RU5 region of the proviral promoter for nested RU5 qPCR [26, 27].
Ribonucleic Acid Extractions and Generation of Complementary Deoxyribonucleic Acid
Cell pellets were resuspended in 1 mL of TRIzol (Invitrogen); 200 μL chloroform (Thermo Fisher) was added and mixed by vortexing. The samples were incubated at room temperature for 5 minutes and spun at 12 000 ×g for 15 minutes at 4°C. The top aqueous layer was transferred to a separate tube and mixed with 1.5 volume of absolute ethanol (AmericanBio). The RNA was extracted using the RNeasy kit (QIAGEN). Five hundred nanograms of RNA was typically used to generate complementary DNA (cDNA) libraries from samples as previously described [26].
Measuring Human Immunodeficiency Virus Messenger Ribonucleic Acid Transcripts
The cDNA samples (2 μL) were mixed with 10 μL GoTaq master mixture (Promega) for PCR amplification and analysis. The master mixture was prepared as follows per sample: 7.5 μL GoTaq master mixture, 1.25 μL each forward and reverse primers prediluted at 5 μM. Primers used to amplify multiply-spliced HIV-1 messenger RNA (mRNA) were as follows: forward 5’-TCCCTCAGACCCTTTTAGTCAG-3’ and reverse 5’-CATCTGTCCTCTGTCAGTTTC-3’. Primers that recognize mRNA of the human housekeeping gene RPL13a were used as internal loading controls: forward 5’-CAAGCGGATGAACACCAAC-3’ and reverse 5’-CGCTTTTTCTTGTCGTAGGGG-3’ [28]. The reaction was performed on a QuantStudio 3 with the following program: hot start for 15 minutes at 94°C, followed by 45 cycles of 15 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 72°C and plate read. Relative levels of HIV-1 mRNA were calculated using the ΔΔCt method, which normalizes the data to both the housekeeping mRNA and the negative control [29].
Multiplex Cytokine Analysis of Plasma Samples
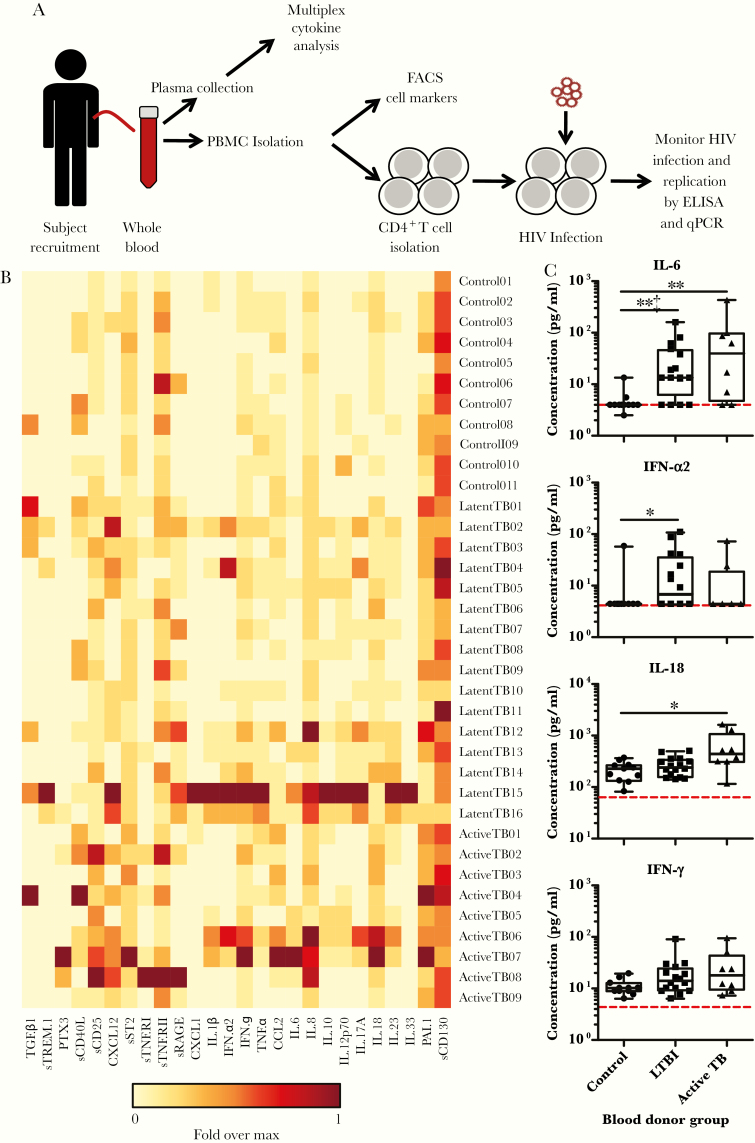
Inflammatory cytokines in plasma samples were measured using LEGENDplex Multi-Analyte Flow Assay Kits according to the manufacturer’s instructions (BioLegend nos. 740809, 740884, and 740886) using an LSRII-SROP analyzer. Plasma samples from all subjects were measured in the same plate from each kit in duplicate. Data were analyzed using LEGENDplex v8.0 software. For the preparation of the heatmap in Figure 1, each individual value was normalized by the maximum signal from each analyte, logarithmic base 2 was applied to each value to reduce the range of the data, and finally the heatmap was generated using the Los Alamos National Laboratories—HIV Database Heatmap Tool (https://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html).
Figure 1.
Mycobacterium tuberculosis infection leaves an immunological footprint detected by elevated levels of inflammation-associated analytes in plasma and that is dependent on the infection status. (A) Experimental outline. Whole blood was collected from volunteers representing the subject groups shown in Table 1. A portion of blood was kept separate for harvesting plasma to be used for multiplex cytokine analysis and for supplementing the culture media to be used for CD4+ T cells. The rest of the blood was used for isolating peripheral blood mononuclear cells (PBMC) for flow cytometry analysis and for isolating CD4+ T cells for human immunodeficiency virus (HIV)-1 infection experiments. Human immunodeficiency virus-1 infection was monitored by enzyme-linked immunosorbent assay (ELISA) of HIV-1 antigen (p24), quantitative polymerase chain reaction (qPCR) analysis of HIV-1 deoxyribonucleic acid, and qPCR analysis of HIV-1 ribonucleic acid. (B) Plasma analyte concentrations were measured by multiplex analysis and are displayed as a heatmap for each blood donor. Heatmap shows the amount of each analyte per blood donor, relative to the maximum signal for set analyte. (C) Plots of representative analytes indicating data range, median, and interquartile range. Each point represents individual donors. Dotted red line indicates the limit of detection. P values were calculated based on the Mann-Whitney U test: *, P < .05 and **, P < .01. ‡ denotes P < .0019 after Bonferroni correction. The full data set of analytes is included as Supplementary Table and Supplementary Figure 1A–C. FACS, fluorescence-activated cell sorting; IFN, interferon; IL, interleukin; LTBI, latent tuberculosis infection; TB, tuberculosis.
Statistical Analysis
Each data point represents the mean of at least 3 technical replicates for each study subject. P values were calculated with the non-parametric Mann-Whitney U test using GraphPad Prism software. We conducted one-to-one correlation analyses between the data sets of each cytokine with the HIV-1 replication data set from day 9 using the non-parametric Spearman’s rank correlation. Because we analyzed 26 cytokine measurements from plasma samples, a P value of .0019 was calculated as an adjusted cutoff for statistical significance based on the Bonferroni correction.
RESULTS
For this study, we recruited 16 subjects with LTBI, 9 with active TB, and 11 without Mtb infection (controls) (Table 1). Study participants were ethnically diverse, 56% were female, and the median age was 36 years old. The age difference between controls and LTBI subjects is not statistically significant, but subjects with active disease are statistically older (P < .05). Recruitment criteria and definitions of TB clinical categories are described in detail in the Methods. The general experimental strategy is outlined in Figure 1A.
Table 1.
Cohort Information
| Sex (n) | Race (n) | ||||||
|---|---|---|---|---|---|---|---|
| TB Status (n) | M | F | Black | Asian | Latino | White | Age (Mean ± SD) |
| Controls (11) | 5 | 6 | 10 | 1 | 38.9 ± 10.3 | ||
| LTBI (16) | 6 | 10 | 8 | 2 | 4 | 2 | 32.5 ± 12.3 |
| Active (9) | 5 | 4 | 2 | 6 | 1 | 47.4 ± 12.5a | |
Abbreviations: F, female; LTBI, latent tuberculosis infection; M, male; SD, standard deviation.
a P < .05 compared with controls.
Mycobacterium tuberculosis Infection Alters the Inflammatory Cytokine and CD4+ T-Cell Subset Profile
To understand the impact of Mtb infection on the general inflammatory state, we assessed proinflammatory cytokines present in the subjects’ plasma. Cytokines and chemokines in Mtb-infected cohorts were measured using a multianalyte flow cytometry-based assay, and results were compared with control subjects. Twenty-six analytes were assayed (Figure 1B and C, Supplementary Table, and Supplementary Figure 1). In general, proinflammatory cytokines were elevated in Mtb-positive individuals compared with controls. Analytes that were elevated in both cohorts of LTBI, and active TB subjects included interleukin (IL)-6, IL-8, and CXCL12 [30, 31]. Analytes that were elevated in the LTBI cohort compared with other groups included IFN-α2, IL-10, IL12p70, sTREM-1, and sRAGE [32]. These cytokines are indicative of macrophage proinflammatory responses to bacteria (sRAGE [33], IL-12 [34], and sTREM-1 [35]), responses to intracellular invasion by bacteria (IFN-α2 [36]), and immune downregulation (IL-10 [37]). Analytes that were elevated in the active TB cohort compared with other groups were IL-18, PAI-1, PTX3, and sCD25 [38–40]. These molecules are associated with proinflammatory signaling by epithelial cells and macrophages (IL-18 [41]), minimizing extracellular matrix degradation (PAI-1 [42]), complement activation (PTX-3 [43]), and T-cell activation (sCD25 [44]). We did not observe statistically significant changes in IFN-γ and TNF-α expression in either individuals with active TB or LTBI when compared with controls, although IFN-γ levels trended higher in both groups with Mtb infection (Figure 1C). After Bonferroni correction for multiple comparisons, the levels of IL-6 and sRAGE in LTBI subjects remain statistically higher than in control subjects (Figure 1B and C, Supplementary Table, and Supplementary Figure 1). Taken together, the changes in cytokine profiles suggest distinct inflammatory footprints for subjects depending on Mtb infection status.
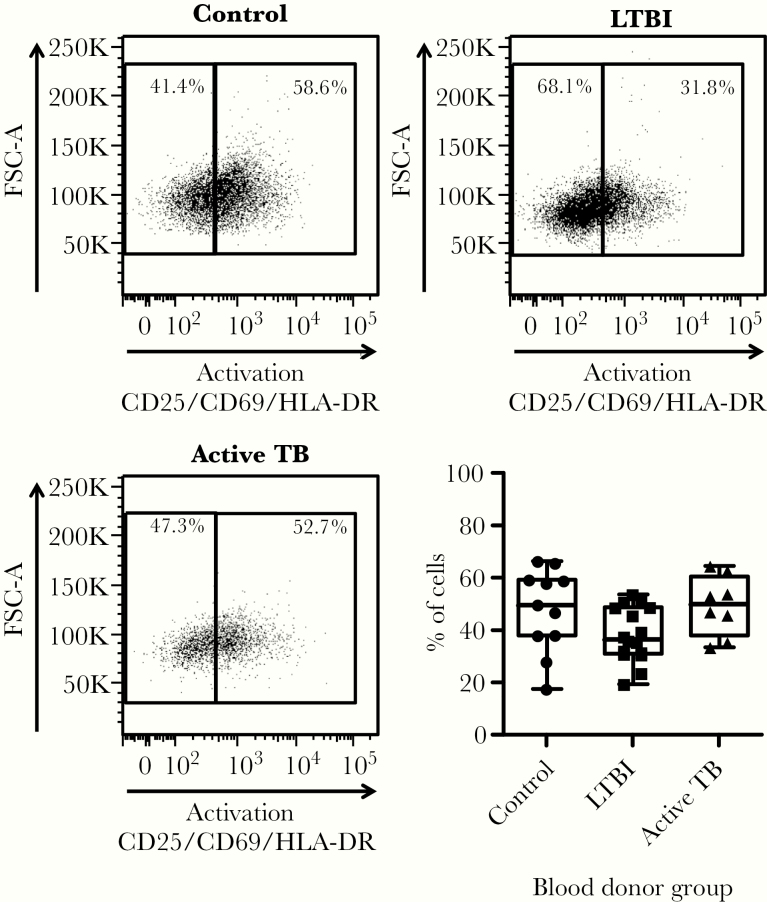
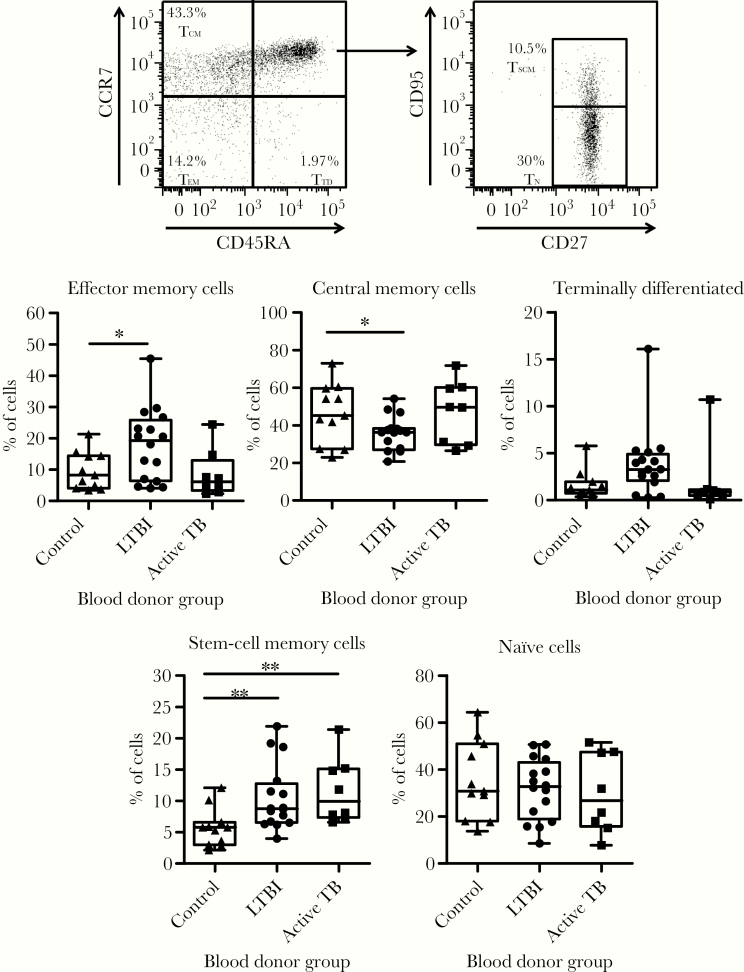
We next determined how the proportion of CD4+ T cell subsets in blood was affected by Mtb infection. Although we did not observe significant differences in the expression of activation markers across groups (Figure 2), subjects in the LTBI group had statistically higher percentages of effector memory and stem cell memory CD4+ T cells concomitantly with lower percentages of central memory CD4+ T cells (Figure 3). Individuals with active TB had higher percentages of stem cell memory CD4+ T cells. We observed similar percentages of naive T cells across all subject groups. Taken together, the differences observed in immune mediators and CD4+ T-cell subsets suggest that Mtb infection status imprints immunological signatures.
Figure 2.
Mycobacterium tuberculosis infection does not affect the overall proportion of activated CD4+ T cells in the blood. Peripheral blood mononuclear cells from study subjects were stained for flow cytometry analysis to determine the proportion of CD4+ T cells expressing any of the molecules associated with T-cell activation: CD25, CD69, and/or HLA-DR. This combination of markers captures both early and late T-cell activation states. Representative fluorescence-activated cell sorting plots for each subject group and a plot of all donors are shown. Plots indicate data range, median, and interquartile range. P values were calculated based on the Mann-Whitney U test. FSC, forward scatter; LTBI, latent tuberculosis infection; TB, tuberculosis.
Figure 3.
Mycobacterium tuberculosis infection leaves an immunological footprint detected by changes in the relative proportion of CD4+ T-cell memory subsets. Peripheral blood mononuclear cells from study subjects were stained for flow cytometry analysis to determine the proportion of CD4+ T-cell subsets based on the expression of molecules associated with memory phenotypes. A representative dot plot is shown with the gating strategy and the CD4+ T-cell subsets represented by each gate: effector memory (TEM), central memory (TCM), and terminally differentiated (TTD). CCR7+/CD45RA+ cells were further subdivided into true naive cells (TN) and stem cell memory T cells (TSCM) based on the expression of CD27 and CD95. Percentages shown for each gate indicate their proportion relative to the total CD4+ T-cell population. Plots represent the proportion of each CD4+ T-cell subset for all donors and show the data range, median, and interquartile range. P values were calculated based on the Mann-Whitney U test: *, P < .05 and **, P < .01. LTBI, latent tuberculosis infection; TB, tuberculosis.
CD4+ T Cells From Mycobacterium tuberculosis-Infected Individuals Are Similarly Susceptible to Human Immunodeficiency Virus-1 Infection
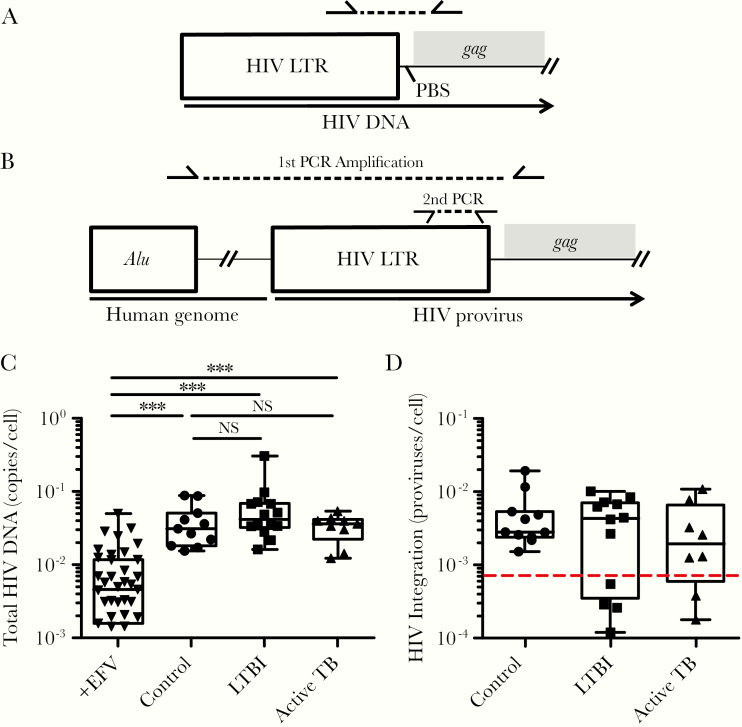
To test the susceptibility of CD4+ T cells from our TB cohorts to HIV-1 infection, CD4+ T cells were purified from PBMCs by negative selection using magnetic beads and infected with HIV-1. Purified CD4+ T cells were cultured in autologous plasma to maintain cytokine environments of individual donors. Cells were infected in vitro with HIV-1 as described in Methods in the presence of the protease inhibitor saquinavir to restrict viral infection to a single round. Negative control infections were performed in the presence of the reverse-transcriptase inhibitor efavirenz. On day 3 postinfection, DNA was isolated for qPCR analysis of total (Figure 4A and C) and integrated (Figure 4B and D) HIV-1 DNA per cell. No difference in either total HIV-1 DNA or integrated proviruses was detected between the subject groups (Figure 4C and D), suggesting that HIV-1 entry, reverse transcription (RT), and integration into CD4+ T cells are not affected by Mtb status.
Figure 4.
Pre-existing Mycobacterium tuberculosis infection does not affect the susceptibility of CD4+ T cells to human immunodeficiency virus (HIV)-1 infection. CD4+ T cells from the different subject groups were infected with HIV-1, and infection was restricted to a single round of infection with the antiretroviral drug saquinavir (1 μM). A portion of the infected cells were treated with the antiretroviral drug efavirenz (1 μM) to serve as a negative control for HIV-1 infection. Total cellular deoxyribonucleic acid (DNA) was harvested and analyzed for total and integrated HIV-1 by quantitative polymerase chain reaction (PCR) following the strategy shown in A and B. Levels of total and integrated HIV-1 DNA for each blood donor are shown in C and D respectively. Data are displayed as the number of copies per cell. The data range, median, and interquartile range are shown. The dotted red line in D indicates the limit of detection of the assay. P values were calculated based on the Mann-Whitney U test: ***, P < .001 and NS, P > .05. EFV, efavirenz; LTBI, latent tuberculosis infection; LTR, long terminal repeat; NS, not significant; PBS, primer binding site; TB, tuberculosis.
Human Immunodeficiency Virus Replicates More Efficiently in CD4+ T Cells From Latent Tuberculosis Infection Donors
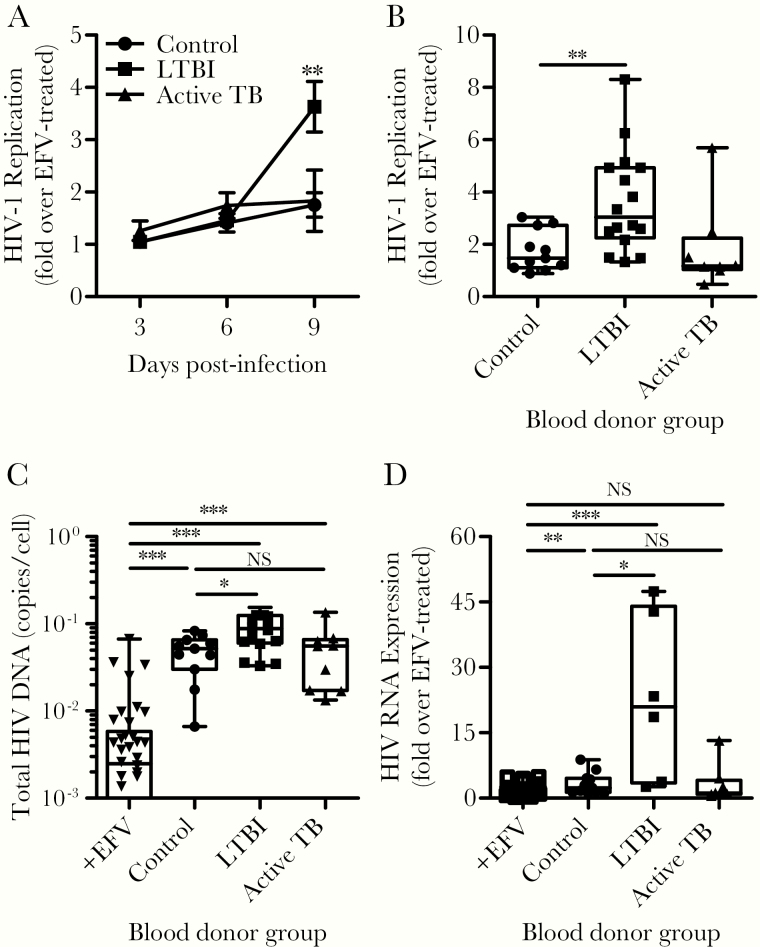
To determine whether HIV-1 replication and spread in CD4+ T cells were affected by the Mtb infection status, we infected CD4+ T cells with HIV-1 and cultured cells for 9 days. Over the course of the first 6 days, no significant differences were observed in HIV-1 p24, but a significant increase (4-fold) was observed at day 9 from CD4+ T cells derived from the LTBI group (Figure 5A and B). To confirm that CD4+ T cells derived from LTBI subjects supported viral spread more efficiently, we assessed the levels of total HIV-1 DNA at day 9 postinfection by qPCR. Although HIV-1 DNA was detected in all groups relative to efavirenz-treated cultures, infected cells from LTBI donors contained significantly more HIV-1 DNA per cell compared to the other groups (Figure 5C). These data indicate that CD4+ T cells from LTBI subjects support more robust HIV-1 replication and spread in culture.
Figure 5.
The stage of pre-existing Mycobacterium tuberculosis infection affects human immunodeficiency virus (HIV)-1 replication. CD4+ T cells from the different subject groups were infected with HIV-1, and infection was (A–C) allowed to expand or (D) restricted to a single round of infection with the antiretroviral drug saquinavir (1 μM). A portion of the infected cells were treated with the antiretroviral drug efavirenz ([EFV] 1 μM) to serve as a negative control for HIV-1 infection. (A) The HIV-1 replication was monitored by the release of HIV-1 antigen (p24) into the culture medium over the span of 9 days. Data are shown as the ratio of HIV-1 p24 signal as determined by enzyme-linked immunosorbent assay, over the signal from parallel cultures treated with EFV. Asterisks represent the statistical significance of the signal from latent tuberculosis infection (LTBI) subjects compared to the other groups on day 9. (B) Dot plot showing the distribution of HIV-1 p24 signal detected on day 9 for each blood donor group. (C) On day 9, cells were harvested and cellular deoxyribonucleic acid (DNA) was purified for quantitative polymerase chain reaction (qPCR) analysis of HIV-1 DNA. Data are displayed as copies of HIV-1 DNA per cell. (D) Cultures of cells treated with saquinavir were harvested on day 4 postinfection, and cellular ribonucleic acid (RNA) was purified for qPCR analysis of HIV-1 RNA to measure the level of proviral gene transcription. Note that RNA was available for only 6 of the donors with LTBI. Data are shown as the fold increase in signal relative to parallel cultures treated with EFV for each blood donor. Plots indicate the data range, median, and interquartile range. P values were calculated based on the Mann-Whitney U test: *, P < .05, **, P < .01, ***, P < .001, and NS P > .05. NS, not significant; TB, tuberculosis.
One explanation for more efficient viral spread in cultures of cells derived from LTBI subjects is that these cells support more robust proviral transcription. To test this, we harvested total RNA on day 4 postinfection and conducted RT-PCR analysis of HIV-1 transcripts. We restricted HIV-1 replication to a single round to measure the relative RNA transcription from proviruses integrated during initial infection. Cells from LTBI subjects supported greater proviral transcription compared with the other subject groups (Figure 5D), suggesting that, despite similar HIV-1 integration (Figure 4D), proviral gene transcription is more efficient in cells derived from LTBI individuals.
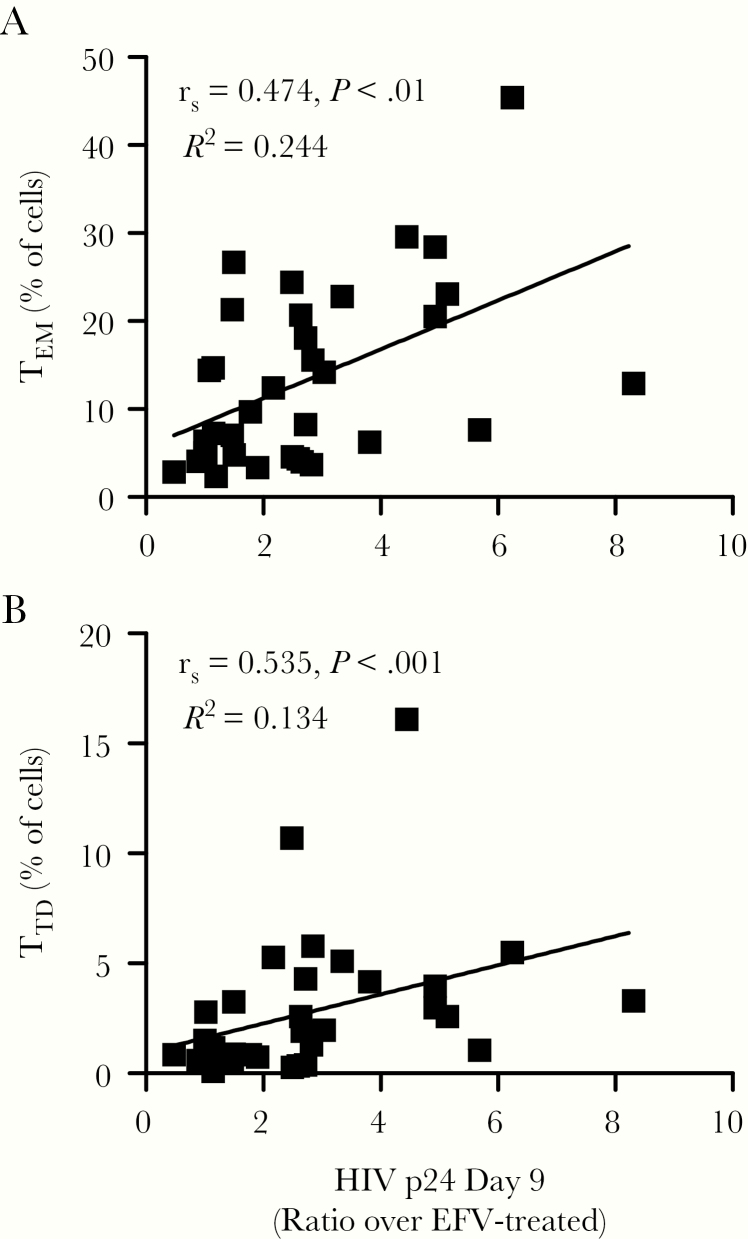
To determine whether HIV-1 replication correlated with any of the immune parameters analyzed above, we performed Spearman R correlation analyses (Figure 6 and Supplementary Figure 2). Based on one-on-one comparisons, no individual cytokine statistically correlated with higher levels of HIV-1 replication (Supplementary Figure 2). However, a significant positive correlation was observed between the percentages of TEM and TTD subsets with HIV-1 replication on day 9 (Figure 6).
Figure 6.
Higher human immunodeficiency virus (HIV)-1 replication correlates with higher percentages of TEM and TTD CD4+ T cells. We calculated one-on-one correlations between the level of HIV-1 replication on day 9 postinfection and the percentage of (A) effector memory (TEM) and (B) terminally differentiated (TTD) CD4+ T cells using Spearman’s rank correlation. Dot plots show the correlation coefficient (rs), P value from the Spearman’s correlation, and the R2 value of the best-fitting line based on linear regression. Each data point represents individual donors. Additional correlations versus plasma analytes and versus the proportion of other CD4+ T-cell subsets are included in Supplementary Figure 2A−C.
Discussion
The TB/HIV syndemic is well documented, with each infection exacerbating the prognosis of coinfected individuals. In many settings where the Mtb and HIV burdens are highest, Mtb infection occurs before HIV infection in individuals. Thus, Mtb infection may affect how the host responds to HIV exposure. We asked whether pre-existing Mtb infection, and its associated inflammatory footprints, influence the susceptibility of CD4+ T cells to HIV-1. We demonstrated that CD4+ T cells from subjects with LTBI support more active HIV-1 replication compared with those with active TB and controls.
Mycobacterium tuberculosis infection is associated with a robust immune response that effectively restricts bacterial spread. However, in many cases, bacteria resist and subvert immune responses, leading to LTBI. Despite the infection being “latent,” the stalemate between bacteria and the immune system is a dynamic process that involves constant immune effector cell turnover and proinflammatory cytokine production [45, 46], as is evident from phenotypic changes in leukocytes and the dysregulation of cytokines including TNF-α, IFN-γ, IL-6, and CCL2 (MCP-1) [32, 47, 48]. More importantly, persistent inflammation has been postulated to affect the pathogenesis and clinical outcomes of HIV coinfection [3, 5, 6]. Consistent with previous reports, we found several elevated proinflammatory cytokines, including IL-6 and sRAGE, with Mtb infection compared to controls [30, 31]. In contrast to published work, we did not observe statistically significant elevation in TNF-α, IFN-γ, or CCL2 (MCP-1) levels in either active TB or LTBI (Figure 1, Supplementary Table, and Supplementary Figure 1) [32, 48]. The discrepancy between our results and other reports may reflect sample size, assay sensitivity, or cohort variation. At the cellular level, consistent with previous reports, we observed changes in the relative proportion of CD4+ T-cell subsets that reflected Mtb infection status [49, 50]; LTBI subjects had higher percentages of CD4+ TEM cells, trends toward increased populations of CD4+ TTD cells, and diminished populations of CD4+ TCM cells (Figure 3). We did not detect major changes in overall T-cell activation in blood. We predict that higher levels of CD4+ T-cell activation will be detected at the site of Mtb infection, where antigen-specific cells would mostly be located. Our observations support that Mtb infection leaves persistent immunological footprints that may impact HIV-1 replication and pathogenesis.
Our results show that HIV-1 replicates more efficiently in CD4+ T cells derived from individuals with LTBI despite no differences in HIV-1 viral entry or integration between the different subject groups. Therefore, postintegration steps in viral replication, in particular HIV-1 transcription, are more efficient in CD4+ T cells from LTBI subjects. Mycobacterium tuberculosis infection is associated with increased inflammation and tissue damage. Combinations of cytokines associated with this Mtb-mediated inflammation influence T-cell activation, which in turn activates HIV-1 transcription [12, 21, 22]. Thus, Mtb, by initiating inflammatory mechanisms and affecting the repertoire of CD4+ T cells that persist in LTBI, may create a microenvironment that supports more robust HIV-1 transcription and HIV-associated pathogenesis.
Conclusions
In conclusion, these studies demonstrate the existence of an immunological footprint in individuals with LTBI that influences the susceptibility of CD4+ T cells to HIV-1 infection. These findings provide a cellular and molecular basis for the impact of pre-existing Mtb infection on the course of subsequent HIV-1 infection and illustrate another example of how these diseases are intertwined to contribute to the TB/HIV syndemic.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are very grateful to the study participants who made this research work possible. We thank the following: Michelle Canning and the clinical teams at the Boston Medical Center (BMC) TB; Immigration and Refugee Clinics for participant recruitment; our translators for study documents; and BMC phlebotomy services for blood draws. We also thank Manish Sagar for critical reading of our manuscript, Laura White for assistance with statistical analysis, the Boston University School of Medicine Flow Cytometry Core, and the Providence/Boston Center for AIDS Research (CFAR) Basic Science Core for support.
Financial support. This work was funded by the Providence/Boston CFAR (P30AI042853; to J. J. E. and L. M. A.), the BU Clinical HIV/AIDS Research Training Program (5T32AI052074-13; to J. J. E.), the Boston University, Clinical and Translational Science Institute, Clinical Research Training Program (UL1 TR001430; to J. J. E.), and the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R01 AI138960, R01 AI097117, R01 AI122842 [to A. J. H.] and U19 AI111276 [to K. R. J.]).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. World Health Organization. Global Tuberculosis Report 2019. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.Joint United Nations Programme on HIV and AIDS (UNAIDS). UNAIDS Data 2019. Geneva, Switzerland: UNAIDS; 2019:16–9. Available at: https://www.unaids.org/sites/default/files/media_asset/2019-UNAIDS-data_en.pdf . Accessed 07 November 2019. [Google Scholar]
- 3. Kwan CK, Ernst JD. HIV and tuberculosis: a deadly human syndemic. Clin Microbiol Rev 2011; 24:351–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization. Global Tuberculosis Control: Epidemiology, Strategy, Financing: WHO Report 2009. Geneva: World Health Organization; 2009. [Google Scholar]
- 5. Whalen C, Horsburgh CR, Hom D, Lahart C, Simberkoff M, Ellner J. Accelerated course of human immunodeficiency virus infection after tuberculosis. Am J Respir Crit Care Med 1995; 151:129–35. [DOI] [PubMed] [Google Scholar]
- 6. Pape JW, Jean SS, Ho JL, Hafner A, Johnson WD Jr. Effect of isoniazid prophylaxis on incidence of active tuberculosis and progression of HIV infection. Lancet 1993; 342:268–72. [DOI] [PubMed] [Google Scholar]
- 7. Siika AM, Yiannoutsos CT, Wools-Kaloustian KK, et al. Active tuberculosis is associated with worse clinical outcomes in HIV-infected African patients on antiretroviral therapy. PLoS One 2013; 8:e53022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdool Karim Q, Baxter C, Birx D. Prevention of HIV in adolescent girls and young women: key to an AIDS-free generation. J Acquir Immune Defic Syndr 2017; 75(Suppl 1):17–26. [DOI] [PubMed] [Google Scholar]
- 9. Bunyasi EW, Geldenhuys H, Mulenga H, et al. Temporal trends in the prevalence of Mycobacterium tuberculosis infection in South African adolescents. Int J Tuberc Lung Dis 2019; 23:571–8. [DOI] [PubMed] [Google Scholar]
- 10. Oxlade O, Murray M. Tuberculosis and poverty: why are the poor at greater risk in India? PLoS One 2012; 7:e47533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Semba RD, Darnton-Hill I, de Pee S. Addressing tuberculosis in the context of malnutrition and HIV coinfection. Food Nutr Bull 2010; 31:S345–64. [PubMed] [Google Scholar]
- 12. Bruchfeld J, Correia-Neves M, Källenius G. Tuberculosis and HIV coinfection. Cold Spring Harb Perspect Med 2015; 5:a017871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Behar SM, Carpenter SM, Booty MG, Barber DL, Jayaraman P. Orchestration of pulmonary T cell immunity during Mycobacterium tuberculosis infection: immunity interruptus. Semin Immunol 2014; 26:559–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goletti D, Weissman D, Jackson RW, et al. Effect of Mycobacterium tuberculosis on HIV replication. Role of immune activation. J Immunol 1996; 157:1271–8. [PubMed] [Google Scholar]
- 15. Toossi Z, Johnson JL, Kanost RA, et al. Increased replication of HIV-1 at sites of Mycobacterium tuberculosis infection: potential mechanisms of viral activation. J Acquir Immune Defic Syndr 2001; 28:1–8. [DOI] [PubMed] [Google Scholar]
- 16. Sullivan ZA, Wong EB, Ndung’u T, Kasprowicz VO, Bishai WR. Latent and active tuberculosis infection increase immune activation in individuals co-infected with HIV. EBioMedicine 2015; 2:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wyndham-Thomas C, Corbière V, Selis E, et al. immune activation by Mycobacterium tuberculosis in HIV-infected and -uninfected subjects. J Acquir Immune Defic Syndr 2017; 74:103–11. [DOI] [PubMed] [Google Scholar]
- 18. Olson A, Ragan EJ, Nakiyingi L, et al. brief report: pulmonary tuberculosis is associated with persistent systemic inflammation and decreased HIV-1 reservoir markers in coinfected Ugandans. J Acquir Immune Defic Syndr 2018; 79:407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol 2001; 19:93–129. [DOI] [PubMed] [Google Scholar]
- 20. Orme IM, Miller ES, Roberts AD, et al. T lymphocytes mediating protection and cellular cytolysis during the course of Mycobacterium tuberculosis infection. Evidence for different kinetics and recognition of a wide spectrum of protein antigens. J Immunol 1992; 148:189–96. [PubMed] [Google Scholar]
- 21. Garrait V, Cadranel J, Esvant H, et al. Tuberculosis generates a microenvironment enhancing the productive infection of local lymphocytes by HIV. J Immunol 1997; 159:2824–30. [PubMed] [Google Scholar]
- 22. Zhang Y, Nakata K, Weiden M, Rom WN. Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication by transcriptional activation at the long terminal repeat. J Clin Invest 1995; 95:2324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thayil SM, Ho YC, Bollinger RC, et al. Mycobacterium tuberculosis complex enhances susceptibility of CD4 T cells to HIV through a TLR2-mediated pathway. PLoS One 2012; 7:e41093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry 2001; 45:194–205. [DOI] [PubMed] [Google Scholar]
- 25. Gagne M, Michaels D, Schiralli Lester GM, Gummuluru S, Wong WW, Henderson AJ. Strength of T cell signaling regulates HIV-1 replication and establishment of latency. PLoS Pathog 2019; 15:e1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agosto LM, Hirnet JB, Michaels DH, et al. Porphyromonas gingivalis-mediated signaling through TLR4 mediates persistent HIV infection of primary macrophages. Virology 2016; 499:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liszewski MK, Yu JJ, O’Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 2009; 47:254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ledderose C, Heyn J, Limbeck E, Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res Notes 2011; 4:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8. [DOI] [PubMed] [Google Scholar]
- 30. Djoba Siawaya JF, Beyers N, van Helden P, Walzl G. Differential cytokine secretion and early treatment response in patients with pulmonary tuberculosis. Clin Exp Immunol 2009; 156:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu Y, Zhang Y, Hu S, et al. Different patterns of cytokines and chemokines combined with IFN-γ production reflect Mycobacterium tuberculosis infection and disease. PLoS One 2012; 7:e44944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. La Manna MP, Orlando V, Li Donni P, et al. Identification of plasma biomarkers for discrimination between tuberculosis infection/disease and pulmonary non tuberculosis disease. PLoS One 2018; 13:e0192664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Byun K, Yoo Y, Son M, et al. Advanced glycation end-products produced systemically and by macrophages: a common contributor to inflammation and degenerative diseases. Pharmacol Ther 2017; 177:44–55. [DOI] [PubMed] [Google Scholar]
- 34. Cooper AM, Solache A, Khader SA. Interleukin-12 and tuberculosis: an old story revisited. Curr Opin Immunol 2007; 19:441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol 2006; 7:1266–73. [DOI] [PubMed] [Google Scholar]
- 36. Donovan ML, Schultz TE, Duke TJ, Blumenthal A. Type I interferons in the pathogenesis of tuberculosis: molecular drivers and immunological consequences. Front Immunol 2017; 8:1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol 2008; 180:5771–7. [DOI] [PubMed] [Google Scholar]
- 38. Yamada G, Shijubo N, Shigehara K, Okamura H, Kurimoto M, Abe S. Increased levels of circulating interleukin-18 in patients with advanced tuberculosis. Am J Respir Crit Care Med 2000; 161:1786–9. [DOI] [PubMed] [Google Scholar]
- 39. Vankayalapati R, Wizel B, Weis SE, Samten B, Girard WM, Barnes PF. Production of interleukin-18 in human tuberculosis. J Infect Dis 2000; 182:234–9. [DOI] [PubMed] [Google Scholar]
- 40. Janssen S, Schutz C, Ward AM, et al. Hemostatic changes associated with increased mortality rates in hospitalized patients with HIV-associated tuberculosis: a prospective cohort study. J Infect Dis 2017; 215:247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaplanski G. Interleukin-18: biological properties and role in disease pathogenesis. Immunol Rev 2018; 281:138–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. J Cell Physiol 2012; 227:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mortaz E, Adcock IM, Tabarsi P, et al. Interaction of pattern recognition receptors with Mycobacterium tuberculosis. J Clin Immunol 2015; 35:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rubin LA, Kurman CC, Fritz ME, et al. Soluble interleukin 2 receptors are released from activated human lymphoid cells in vitro. J Immunol 1985; 135:3172–7. [PubMed] [Google Scholar]
- 45. Barry CE 3rd, Boshoff HI, Dartois V, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 2009; 7:845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin PL, Flynn JL. The end of the binary era: revisiting the spectrum of tuberculosis. J Immunol 2018; 201:2541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiong W, Dong H, Wang J, et al. Analysis of plasma cytokine and chemokine profiles in patients with and without tuberculosis by liquid array-based multiplexed immunoassays. PLoS One 2016; 11:e0148885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zambuzi FA, Cardoso-Silva PM, Espindola MS, et al. Identification of promising plasma immune biomarkers to differentiate active pulmonary tuberculosis. Cytokine 2016; 88:99–107. [DOI] [PubMed] [Google Scholar]
- 49. Mpande CAM, Dintwe OB, Musvosvi M, et al. Functional, antigen-specific stem cell memory (TSCM) CD4(+) T cells are induced by human Mycobacterium tuberculosis infection. Front Immunol 2018; 9:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tapaninen P, Korhonen A, Pusa L, Seppälä I, Tuuminen T. Effector memory T-cells dominate immune responses in tuberculosis treatment: antigen or bacteria persistence? Int J Tuberc Lung Dis 2010; 14:347–55. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.