Abstract
Background
The combination of daptomycin (DAP) plus ampicillin (AMP), ertapenem (ERT), or ceftaroline has been demonstrated to be efficacious against a DAP-tolerant Enterococcus faecium strain (HOU503). However, the mechanism for the efficacy of these combinations against DAP-resistant (DAP-R) E. faecium strains is unknown.
Methods
We investigated the efficacy of DAP in combination with AMP, ERT, ceftaroline, ceftriaxone, or amoxicillin against DAP-R E. faecium R497 using established in vitro and in vivo models. We evaluated pbp expression, levels of penicillin-binding protein (PBP) 5 (PBP5) and β-lactam binding affinity in HOU503 versus R497.
Results
DAP plus AMP was the only efficacious regimen against DAP-R R497 and prevented emergence of resistance. DAP at 8, 6, and 4 mg/kg in combination with AMP was efficacious but showed delayed killing compared with 10 mg/kg. PBP5 of HOU503 exhibited amino acid substitutions in the penicillin-binding domain relative to R497. No difference in pbp mRNA or PBP5 levels was detected between HOU503 and R497. labeling of PBPs with Bocillin FL, a fluorescent penicillin derivative, showed increased β-lactam binding affinity of PBP5 of HOU503 compared with that of R497.
Conclusions
Only DAP (10 mg/kg) plus AMP or amoxicillin was efficacious against a DAP-R E. faecium strain, and pbp5 alleles may be important contributors to efficacy of DAP plus β-lactam therapy.
Keywords: Vancomycin-resistant E. faecium, E. faecium, Daptomycin, β-lactam, PBP
We present evidence that the success of daptomycin–β-lactam combination therapy against multidrug-resistant Enterococcus faecium with LiaFSR substitutions is likely dependent on β-lactam interactions with penicillin-binding proteins. Our findings open a possible new approach to severe enterococcal infections.
Enterococci are major nosocomial pathogens with an important ability to acquire resistance determinants [1–4]. Infections caused by multidrug-resistant Enterococcus faecium are a challenge owing to the paucity of efficacious antibiotics [5]. Daptomycin (DAP) is a cell membrane acting lipopeptide antibiotic that has become a key drug in the treatment of multidrug-resistant E. faecium infections. DAP displays concentration-dependent bactericidal activity against most E. faecium, and the area under the concentration-time curve (AUC) from 0 to 24 hours (AUC0–24h) divided by the minimum inhibitory concentration (MIC) is the pharmacokinetic-pharmacodynamic (PK/PD) parameter that best predicts the in vivo efficacy of the antibiotic against these organisms [6].
Emergence of in vivo of DAP resistance in E. faecium seems to occur commonly, mainly mediated by changes in the liaFSR system [2, 7–9], a 3-component regulatory system that controls cell membrane adaptation. Resistance development can be prevented by combining DAP with β-lactams resulting in higher relative exposures (AUC0–24h/MIC) because the DAP MICs are lower with the combination [10–13]. We previously demonstrated [14] that DAP monotherapy (doses from 6 to 10 mg/kg) against a DAP-tolerant strain of E. faecium (HOU503; DAP MIC of 2 µg/mL, designated as DAP-susceptible (DAP-S) dose dependent by the current break points) was marginally effective [15, 16]) This strain harbors liaFSR substitutions, resulting in the emergence of resistance with all DAP monotherapy regimens. In contrast, addition of ampicillin (AMP), ceftaroline (CPT) or ertapenem (ERT) led to increased killing and abolished emergence of resistance over the 14-day in vitro model experiments [14].
Although these combinations seem promising, there are few data to suggest which β-lactam works best with DAP and whether these combinations would be effective against DAP-resistant (DAP-R) strains (including those with mutations in the liaFSR system). In the current study, we evaluated the effect of DAP monotherapy (10 mg/kg) and DAP in combination with AMP, CPT, or ERT (and others) using the simulated endocardial vegetation (SEV) PK/PD model, validating the results in a rat model of infective endocarditis [14]. We demonstrated that the combination of DAP plus AMP was the only regimen that achieved therapeutic efficacy and prevented development of resistance against DAP-R R497.
To gain insights into the mechanistic basis of the so-called see-saw effect (resensitization to β-lactams in DAP-R strains), we characterized the possible role of penicillin-binding proteins (PBPs) in the phenotype. We found that the synergistic activity of the DAP plus AMP combination was correlated with the pbp5 allele sequence and the binding affinity of Bocillin-FL (BOC-FL), a fluorescent penicillin derivative, to PBP 5 (PBP5). No alterations in transcript levels of the pbp genes or protein levels of PBP5 were observed. Our results suggest that β-lactam interactions with PBP5 isotypes may be important mediators of the see-saw phenomenon in enterococci.
MATERIALS AND METHODS
Bacterial Strains
E. faecium R497 is a DAP-R (DAP MIC, 16 µg/mL) clinical isolate [17–19] that harbors the T120S and W73C substitutions in LiaS and LiaR, respectively, and was used in all SEV and in vivo experiments. The other strains investigated in this research include HOU503, a vancomycin-resistant E. faecium and DAP-tolerant clinical isolate (MIC, 2 µg/mL; DAP-S dose dependent), S447 a vancomycin-resistant E. faecium DAP-S (dose dependent) strain, lacking substitutions in the LiaFSR system, and R496, a DAP-R with LiaFSR substitutions (Supplementary Table 1).
Antimicrobial Agents and Media
DAP and ERT were obtained from Merck; AMP, ceftriaxone (CRO), and amoxicillin (AMX) powder were purchased from Sigma-Aldrich; and CPT was obtained from Allergan Pharmaceuticals. Mueller-Hinton broth II (MHB; Difco) with 50 µg/mL of calcium and 12.5 µg/mL magnesium was used for susceptibility testing. Because of the dependency of DAP on calcium for antimicrobial activity and calcium loss from the media due to calcium binding to albumin, MHB supplemental to a concentration of 75 µg/mL was used in the in vitro SEV model experiments, as described elsewhere [20]. Colony counts were determined using brain-heart infusion (BHI) agar supplemented with 50 µg/mL of calcium.
Susceptibility Testing
All MICs were determined in duplicate using microbroth dilution method [21] at approximately 5 × 105 colony-forming units (CFUs)/mL, following the Clinical and Laboratory Standards Institute guidelines [15]. Combination MIC values for DAP in the presence of β-lactams were determined by supplementing the broth with concentrations of β-lactam at their respective free peak concentration biological (Cmax) and included AMP (70 µg/mL), CPT (17 µg/mL), and ERT (15.5 µg/mL). The combination MICs were performed at one-half, one-quarter, and one-eighth times the peaks, respectively, to investigate the impact of varying concentrations. The DAP MIC fold reduction from baseline was calculated by dividing DAP MICs by the MIC obtained in the presence of listed β-lactams.
Time-kill experiments were performed in the broth containing approximately 3 to 3.5 g/dL of albumin and equivalent of 50 µg/mL calcium in 24 hours to reproduce the protein binding characteristic of the aforementioned antibiotics. Time-kill plots were generated by plotting mean colony counts versus time to compare 24-hour time killing effects of the different antibiotic combinations. Bactericidal activity was defined as a decrease of ≥3 log10 CFUs/mL, and synergy between 2 agents was defined as ≥2 log10 CFUs/mL reduction at the end of 24 hours in comparison with the most potent single agent alone [15]. Antibiotic carryover was addressed by serial dilutions of the samples, and the lower limit of detection was 100 CFUs/g.
In Vitro PK/PD Model
A SEV PK/PD model [22] was used for all antibiotic experiments and has been described elsewhere [23, 24]. DAP was administered once daily via an injection port. The simulated DAP regimens with a targeted half-life (t½) of 8 hours were 6, 8, 10, and 14 mg/kg/d, with peaks of 93.9, 123.3 141.1, and 197.54 µg/mL, respectively [14]. The DAP regimens were tested alone and in combination with AMP (2 g) continuous infusion (70 µg/mL) and AMX (2 g) continuous infusion (16.1 µg/mL), using AMP and AMX supplied daily, to ensure drug stability, CRO (2 g) (Cmax, 257 µg/mL; t½, 8 hours) every 24 hours, ERT (1 g) (Cmax ,155 µg/mL; t½, 4 hours) every 24 hours, or CPT (600 mg) (Cmax, 20.448 µg/mL; t½, 2.66 hours) every 12 hours.
SEV samples were removed in duplicate from each model over the sampling period of 0–336 hours. All samples were plated for CFU counts (CFUs per gram), and emergence of resistance was tested by plating on DAP-containing agar at 3 times the baseline DAP MIC. Any colonies detected on drug plates were tested for changes in MIC, using microbroth dilution MIC testing according to Clinical and Laboratory Standards Institute guidelines [15].
Humanized Model of Rat Infective Endocarditis
Aortic valve endocarditis induction, infective dose (ID90–100) determination, bacterial inoculation in anesthetized male Sprague-Dawley, jugular vein catheterized rats, and the use of a programmable pump to infuse the test drugs intravenously via jugular vein were carried out, following methods published elsewhere [14, 25]. Animals were inoculated with ≥107 CFUs per rat, inoculum representing ≥10 times the ID90 and antibiotic infusion therapy starting 24 hours after bacterial inoculation. At the time of therapy initiation, 2–3 animals per each experiment were euthanized as baseline untreated controls to determine the CFU counts of bacteria in vegetations, as described elsewhere [14, 25, 26].
We used a humanized DAP dose of 6 mg/kg/d (45.3 mg/kg/d for rats) for 3 days, as published elsewhere [14, 25]. Experimental efficacious doses used in published rat endocarditis or murine models were used for CPT (40 mg/kg; intravenous infusion via jugular vein for 30 minutes, every 8 hours for 3 days) [26] AMP (333.33 mg/kg; intravenous infusion for 30 minutes, every 8 hours for 3 days) [27] and ERT (20 mg/kg; infusion for 30 minutes, every 8 hours for 3 days) [28]. Animals were euthanized ≥16 hours after the last antibiotic dose and CFU counts of vegetation (CFUs per gram) were determined and compared with each other and with controls. The unpaired t test was used for statistical analysis with Prism 4 software for Windows (GraphPad). The minimum detection limit was 101 CFUs/g of tissue. The ID90 values were determined using the method outlined by Reed and Muench [29, 30].
PBP5 Sequence Alignment, Protein Quantification, and Gene Expression Analyses
Whole-genome sequences for S447, HOU503, and R497 are available on the National Center for Biotechnology Information Web site and were used to retrieve the pbp5 sequence. DNA and amino acid sequence multiple alignments were performed using the MUSCLE tool from the European Bioinformatics Institute Web site (http://www.ebi.ac.uk/Tools/msa/muscle/). The results of the bioinformatics analysis were confirmed by means of polymerase chain reaction (PCR) amplification and Sanger sequencing. For expression analyses, all strains were grown in BHI broth at 37°C to the exponential phase (optical density at 600 nm, approximately 0.8), and RNA was extracted using a PureLink RNA Mini Kit (Ambion) in 3 technical and 3 biological replicates.
Subsequently, treatment with Turbo DNAse kit (Ambion) was performed to remove genomic DNA. The complementary DNA was generated from approximately 1 µg of purified RNA, using SuperScript II Reverse Transcriptase (Invitrogen). Gene expression was evaluated with 5 ng of complementary DNA using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Relative expression ratios were calculated by normalizing to the housekeeping genes gyrB and ddl.
Because the efficiency was different with the primers investigated, fold changes were calculated using an efficiency-corrected calculation model described by Pfaffl [31]. Primer efficiency was determined by the LinRegPCR software, version 11.0, program in each reaction. Differences in gene expression between pairs of strains were calculated using the normalized expression for each gene and considered significant at P < .05 (based on 2-tailed unpaired Student t test). Results are an average of 3 independent experiments with 3 biological replicates each. S447 gene expression was considered baseline for relative comparison with HOU503 and R497 levels.
BOC-FL Labeling of PBPs
Cells grown overnight were inoculated at a 1:50 dilution into fresh 50 mL of BHI and grown until the midexponential phase (optical density at 600 nm, approximately 0.8–1). Cells were spun down and washed in 1× phosphate-buffered saline before being disrupted with glass beads in the FastPrep24 instrument. Cell debris were spun down, and supernatants centrifuged at 100 000g for 1 hour. Membrane pellets were resuspended in 150 µL of phosphate-buffered saline with protease inhibitor cocktails (Roche) and mechanically disrupted for optimal homogenization without chemical agents.
Whole-protein content was quantified using a Pierce bicinchoninic assay (Thermo Fisher Scientific). Next, 100 µg of whole-membrane extracts was incubated for 30 minutes at 37°C with 100 µmol/L of BOC-FL in the dark. The reaction was stopped with 0.1% Triton-X 100 for 20 minutes at 4°C before spinning down at 15 000 rpm for 10 minutes and collection of the supernatants for Western blotting. Then 25 µL of the samples were loaded and run on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and visualized with the Bio-Rad ChemiDoc Alpha Imager under the Alexa 488 filter. Individual PBP proteins were identified by bands corresponding to their known molecular weights, based on previous findings [32, 33].
RESULTS
DAP MIC Reduction in Presence of β-Lactams and Synergy with DAP Plus AMP Against R497
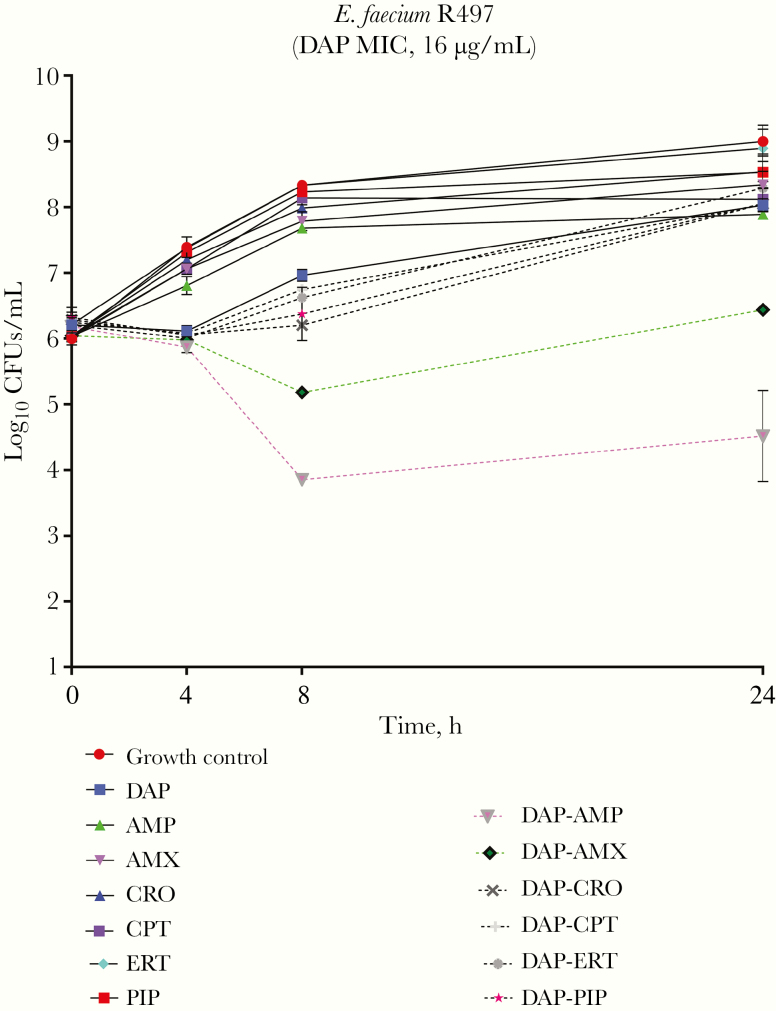
The presence of β-lactams (AMP, CPT, ERT, AMX, piperacillin, or CRO) reduced the DAP MIC between 16- and 32-fold (Supplementary Table 2). The MICs against E. faecium R497 were as follows: DAP, 16 μg/mL; CPT, >64 μg/mL; AMP, >64 μg/mL; ERT, >64 μg/mL; DAP plus CPT, 4 μg/mL; DAP plus AMP, 2 μg/mL; and DAP plus ERT, 4 μg/mL. The concentration of the β-lactam antibiotic in the medium did not seem to affect the DAP MIC change, because concentrations below the Cmax (one-half, one-quarter, and one-eighth times the Cmax) produced similar reductions in the DAP MIC, compared with the Cmax. Nonetheless, despite the important effect of all the β-lactam antibiotics on the DAP MIC, only the combination of AMP plus DAP was synergistic in time-kill experiments (Figure 1), although the effect was not bactericidal.
Figure 1.
Results of 24-hour time-kill experiments against Enterococcus faecium R497 using various daptomycin (DAP) and β-lactam combinations. Abbreviations: AMP, ampicillin; AMX, amoxicillin; CPT, ceftaroline; CRO, ceftriaxone; ERT, ertapenem; PIP, piperacillin.
β-Lactam-Dependence of DAP Combinations Against E. faecium R497
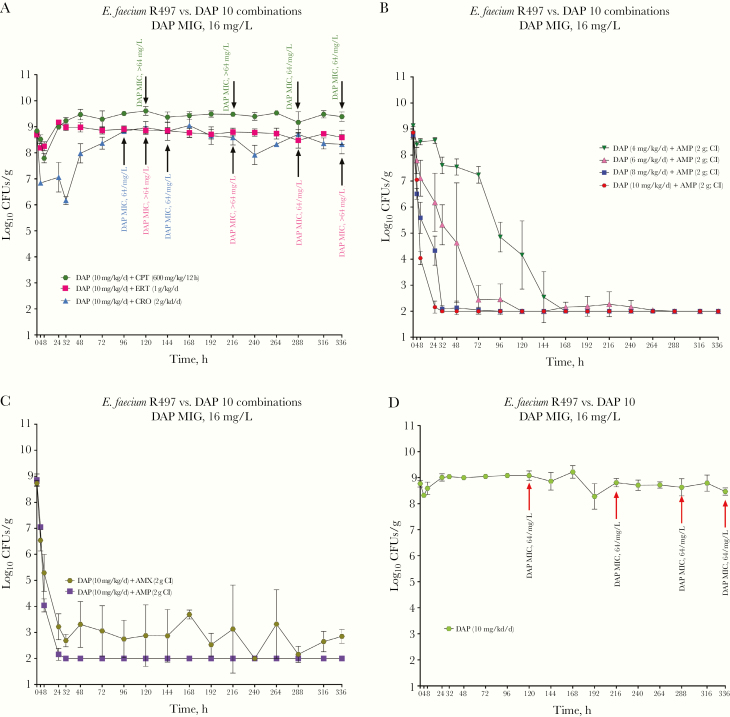
DAP monotherapy (at 10 mg/kg/d) and DAP in combination with CPT, ERT, or CRO showed no activity against R497 in the SEV PK/PD models, with selection of resistant isolates with elevated MICs as high as >64 µg/mL (Figure 2 and Supplementary Table 3). Even regimens of DAP at 14 mg/kg with ERT or CPT exhibited no efficacy. In contrast, the combination of DAP (10 mg/kg/d) plus AMP demonstrated enhanced killing activity compared with DAP alone. This regimen reached CFU counts below detection limits at 24 hours and maintained that level for the duration of the in vitro model. Of note, DAP doses <10 mg/kg combined with AMP had similar killing activity and, compared with 10 mg/kg/d, only a delay in reaching CFU counts below the detection limit was observed (Figure 2B). DAP in combination with AMX caused a reduction in CFU count, to at or slightly above the level of detection during the 14-day experiments (Figure 2C). Emergence of DAP resistance was prevented in all experiments using DAP in combination with AMP or AMX. The achieved pharmacokinetics parameters for DAP, CPT, AMP, AMX, CRO and ERT are shown in Supplementary Table 4 and coefficients of variation between all standards for each assay are listed in Supplementary Table 5.
Figure 2.
Results of simulated endocardial vegetation pharmacokinetic-pharmacodynamic model A, Comparison of daptomycin (DAP) activity against Enterococcus faecium R497 with ceftaroline (CPT), ceftriaxone (CRO), or ertapenem (ERT); arrows show development of resistance at various time points. B, DAP–ampicillin (AMP) combinations using dose deescalation of DAP. C, Comparison between combinations of DAP-AMP and DAP–amoxicillin (AMX) D, DAP monotherapy (10 mg/kg/d); arrows show development of resistance at various time points. Abbreviation: CI, continuous infusion.
Agreement Between Humanized Rat Endocarditis Model and SEV PK/PD Model Results
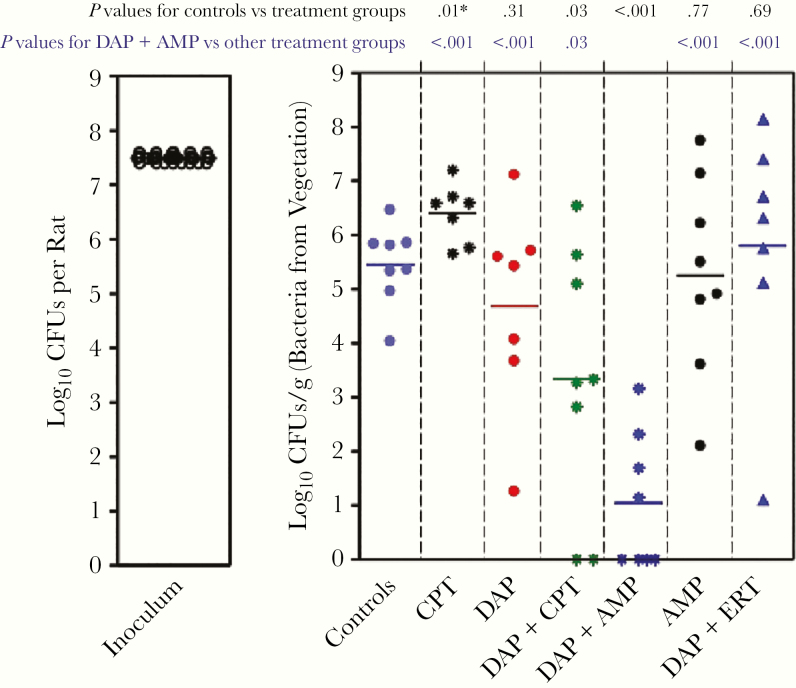
To validate the in vitro findings in the SEV PK/PD model, we tested the in vivo efficacy of DAP combinations with AMP, ERT, or CPT in the rat infective endocarditis model. DAP was used at a humanized dose of 6 mg/kg/d for 3 days. Results for controls (infected; no treatment for 24 hours) and all infected animals plus treatment bacterial counts (log10 CFUs per gram) after 3 days of monotherapy or combination therapy are shown in Figure 3. DAP plus AMP was the most efficacious combination, showing sterile vegetation in 50% (4 of 8 rats) and a statistically significant (P < .001) reduction in geometric mean (standard deviation) CFU count (1 [1] log10 CFUs) compared with controls (5.4 [0.7] log10 CFUs) and compared with all the other regimens (Table 1).
Figure 3.
The results of in vivo efficacy of daptomycin (DAP) in combination with ampicillin (AMP), ertapenem (ERT), or ceftaroline (CPT) in the rat infective endocarditis model. CPT (40 mg/kg) and DAP (45.3 mg/kg; both by 30-minute intravenous infusion via the jugular vein, every 8 hours for 3 days) were used against Enterococcus faecium strain R497. Horizontal lines represents geometric mean colony-forming unit (CFU) counts. Previously published experimental efficacious doses of AMP (333.33 mg/kg/8 h), ERT (20 mg/kg/8 h), and CPT (40 mg/kg/8 h) for 3 days were used. Controls represent no antibiotic treatment.
Table 1.
Animal Model Results
| Treatmenta | Vegetation, Mean (SD), Log10 CFUs/g | Comparison | Difference Between Means (CFU/g)b | P Valuec |
|---|---|---|---|---|
| DAP + AMP | 1 (1) | Versus baseline untreated controls (5.4 [0.7]) | −4.4 (0.5) | <.001 |
| DAP + CPT | 3.3 (1) | −2.1 (0.8) | .03 | |
| DAP | 4.6 (1) | −0.7 (0.7) | .31 | |
| CPT | 6.4 (0.1) | +0.9 (0.3) | .01 | |
| DAP + AMP | 1 (1) | Versus DAP + CPT | −2.3 (0.9) | .03 |
| Versus DAP | −3.6 (0.8) | <.001 | ||
| Versus CPT | −5.3 (0.5) | <.001 | ||
| Versus AMP | −4.2 (0.7) | <.001 | ||
| Versus DAP + ERTd | −4.7 (0.9) | <.001 | ||
| DAP | 4.6 (1) | Versus CPT | −1.7 (0.7) | .04 |
| AMP | 5.2 (1) | Versus CPT | −1.1 (0.7) | .13 |
Abbreviation: AMP, ampicillin; CFUs, colony-forming units; CPT, ceftaroline; DAP, daptomycin; ERT, ertapenem.
a All antibiotics were administered daily day for 3 days against Enterococcus faecium strain R497. The dosages were as follows: DAP, 45.3 mg/kg (7-hour intravenousinfusion every 24 hours); AMP, 333 mg/kg (30-minute infusion every 8 hours); and CPT, 40 mg/kg (30-minute infusion every 8 hours).
b Negative values in this column indicate that CFU counts were lower than in the comparison group (control or other treatment group); positive values, that CFU counts were higher than in controls.
c Data were log-transformed, and unpaired t tests were performed to obtain P values.
d ERT dosage: 20 mg/kg (infusion for 30 minutes, every 8 hours for 3 days).
NOTE: The first 4 rows are compared with controls.
Effect of PBP Sequence on Affinity of β-Lactams for PBP5
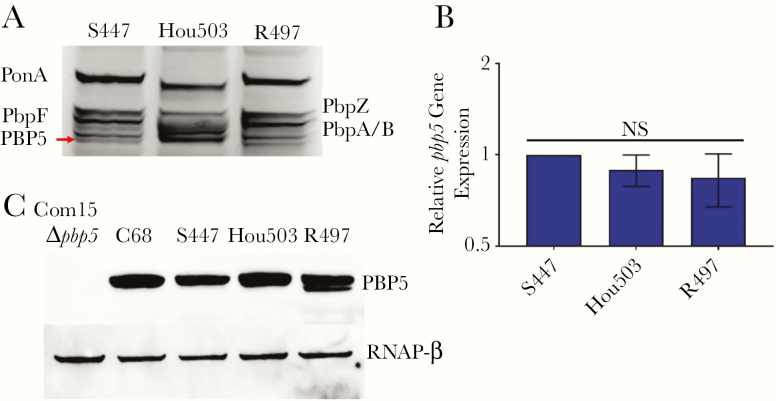
We determined the PBP5 sequence to evaluate the mechanistic basis for the varying efficacies of DAP and β-lactam combinations in killing and preventing the emergence of DAP resistance in HOU503 versus R497. We hypothesized that R497 responds only to the DAP plus AMP combination owing to decreased PBP5 β-lactam binding affinity relative to that of HOU503. BOC-FL labeling of PBPs embedded in the membrane revealed that the low-molecular-weight PBP5 of HOU503 had increased β-lactam binding affinity relative to PBP5 from a DAP-S strain S447, or the DAP-R, R497 (Figure 4A). Relative gene expression levels of all pbp genes were evaluated and compared between S447, HOU503, and R497, by means of quantitative reverse-transcription PCR with normalization to the housekeeping genes, gyrB and ddl. There were no significant changes in pbp transcript levels in the strains (Figure 4B). Immunoblotting also indicated no significant changes in PBP5 levels between the strains (Figure 4C).
Figure 4.
Penicillin-binding protein (PBP) 5 (PBP5) of HOU503 has increased β-lactam binding affinity, independent of gene expression and protein levels. A, PBP β-lactam affinity determined by Bocillin FL labeling of PBPs embedded in enterococcal membrane fractions, followed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and visualization of fluorescence intensities. PBP5 (red arrow) has increased band intensity in HOU503 relative to S447 and R497. B, There are no significant differences in pbp5 relative gene expression, as determined with quantitative reverse-transcription. Statistics were calculated with data from 3 biological replicates and 3 internal technical replicates. Data represent means with standard deviations (error bars). C, PBP5 expression levels determined by Western blotting cell lysates. Com15, a clade B strain lacking pbp5, was used as a negative control. C68, a hospital-associated clade A1 strain used to purify PBP5 for antibody production, served as a positive control. An antibody against the RNA polymerase β subunit (RNAP-β) was used as the loading control.
These results suggested that pbp5 allele sequence variation is likely contributing to alterations in β-lactam binding affinity. Thus, we compared the pbp5 sequence of the strains. All 3 clinical strains (S447, HOU503, and R497) had classic mutations widely associated with high-level AMP-resistance (M485A and 466’S) [17, 34–36]. Interestingly, the pbp5-predicted amino acid sequences of the DAP-S S447 were identical to those of the DAP-R R497. In contrast, PBP5 of HOU503 had substitutions in the transpeptidase domain (H408Q, A462V, T546N, T558A, S582G, and V586L) and the penicillin-binding domain (Q632K and L642P), compared with that of HOU503 (Supplementary Figure 1). Thus, HOU503 harbors, to our knowledge, a previously unreported pbp5 allele that includes both classic AMP-susceptible and AMP resistance–related mutations in the E. faecium population. We speculate that these substitutions produce a hybrid pbp5 allele and contribute to increased β-lactam binding affinity to PBP5. Thus, pbp5 alleles are likely to correlate with the activity of specific β-lactams in the see-saw effect associated with nonsusceptibility to DAP.
DISCUSSION
Treatment of infections caused multidrug-resistant E. faecium is challenging, and previous in vitro and limited clinical data suggest that the combination of DAP plus β-lactams is a promising strategy. We have previously shown that DAP combined with AMP, CPT, or ERT not only was efficacious in vitro and in vivo against a DAP-tolerant strain of E. faecium (HOU503) harboring known substitutions in liaFSR but also prevented emergence of resistance during therapy [16]. Moreover, the therapeutic efficacy of DAP was similar even at doses as low as 6 mg/kg when any of the β-lactams tested (AMP, CPT, or ERT) was added to the regimen. Although the mechanistic basis of the phenomenon is unknown, these encouraging results with a DAP-tolerant strain (exhibiting MICs within the susceptible range) raised the possibility of using the same strategy for fully DAP-R E. faecium strains exhibiting high MICs.
In the current study, using E. faecium (R497) (a DAP-R strain with MIC of 16 µg/mL) and harboring liaFSR changes (among others previously associated with DAP-R), we show that the only efficacious regimen against this strain, both in vitro and in vivo, was the combination of DAP plus AMP. DAP plus AMX (a derivative of AMP) had therapeutic efficacy similar to that of DAP plus AMP, albeit with a less prominent reduction in bacterial counts. Furthermore, the success of this combination was dependent on DAP exposure; the time required for killing to the limits of detection was reliant on the dose (AUC/MIC) exposure to DAP. Interestingly, CPT and ERT, which were previously shown to improve DAP therapeutic efficacy (E. faecium HOU503), failed against E. faecium R497. Thus, our current results implicate the importance of β-lactam specific structure differences and DAP concentration exposure dependence in the final efficacy of DAP–β-lactam combination.
These results were highly reproducible in both our SEV PK/PD model and a humanized model of rat endocarditis. Moreover, in vitro MIC determinations did not predict the SEV PK/PD model results, because MIC reduction was shown in all DAP–β β-lactam combinations. Of interest, cation-adjusted MHB with 50 mg/L of calcium ion was used for MIC experiments and albumin broth in time-kill experiments and SEV models. Thus, the discrepancy between the results may be due to additional protein binding of β-lactams in the medium containing albumin. In time-kill experiments, only the DAP plus AMP combination displayed synergy, while the DAP plus AMX combination stayed at the initial inoculum level at the end of 24-hour exposure. These results confirm one of the major limitations of DAP MIC determination and the lack of translatability of this test to in vivo situations. Furthermore, the bacterial density used in our models is significantly higher than that for MIC determination, and both DAP and β-lactams are affected by the inoculum [16].
The discrepancy in the effect of the DAP plus β-lactam combinations between E. faecium HOU503 and R497 prompted us to provide insights into the actual mechanism of the DAP plus β-lactam synergism. Both HOU503 and R497 harbor the classic substitutions in the response regulator LiaR (W73C) and the LiaS sensor histidine kinase (T120S) previously associated with activation of the LiaFSR system [35].
In E. faecalis, we recently showed [9] that the major effector of the LiaFSR response is through LiaX, a novel sentinel protein that detects the presence of DAP and antimicrobial peptides in the extracellular milieu leading to activation of cell membrane adaptation and redistribution of anionic phospholipid microdomains. LiaX-mediated resistance to DAP and innate immune antimicrobial peptides also leads to increased virulence in vivo [9]. Interestingly, LiaX has a homolog in E. faecium (designated PBP5-binding protein, and also regulated by LiaR) [35, 36], which seems to be involved in β-lactam resistance, connecting the LiaFSR system not only to membrane homeostasis but also to peptidoglycan synthesis. Thus, LiaX-mediated changes in the cell membrane can hypothetically affect the functions or affinity of PBPs and, ultimately, lead to the see-saw effect or β-lactam resensitization. Indeed, the differential affinity of β-lactams to PBPs has been shown before in Staphylococcus aureus [11, 37].
Using the available sequences of PBP5 of R497 and HOU503, we show that there are 7 changes in the PBP5 sequence of R497 compared with HOU503 (Supplementary Figure 1). All the amino acid substitutions are located in the transpeptidase and penicillin-binding domains. The changes did not seem to affect the expression or amount of PBP5 available. However, using BOC-FL labeling of PBPs in the membrane, we showed that PBP5 of HOU503 had increased β-lactam binding affinity compared with R497 and a DAP-S strain (S447). Of note, R497 and S447 have identical pbp5 alleles. Thus, our results suggest the possibility that the PBP5 allele sequence is a major determinant of the see-saw effect in terms of β-lactam selectivity. Indeed, because AMP seems to have the highest affinity for PBP5 [38, 39], it seems reasonable to select this compound as the best partner for DAP when treating deep-seated infections due to E. faecium. Moreover, our data suggest that a sequence-based approach (pbp5 sequence) could be used to determine the ability of β-lactams to synergize with DAP. Our findings also support the notion that the initial activation of liaFSR is the critical genetic event for the evolution of resistance both in vivo and in vitro, and, after such occurrence, multiple pathways are plausible.
In summary, we present evidence that the success of DAP–β-lactam combination therapy against multidrug-resistant E. faecium with LiaFSR substitutions is likely dependent on β-lactam interactions with PBPs. The combination of DAP (even at 4 mg/kg) with AMP, is effective in infections of high bacterial density, preventing developing of resistance. Our findings suggest a possible new approach to severe enterococcal infections using a combination of DAP plus AMP antibiotics initially, as well as the potential to use lower DAP doses.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R01 AI121400 to M. J. R. and grants K24-AI121296, R01AI134637, and R21AI143229 to C. A. A.)
Potential conflicts of interest. B. E. M. and C. A. A. have received grant support from Merck, MeMed Diagnostics, and Entasis Pharmaceuticals. MJR has received support from, consulted for, or provided lectures on behalf of Allergan and Merck. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Mishra NN, Bayer AS, Tran TT, et al. . Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 2012; 7:e43958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Diaz L, Tran TT, Munita JM, et al. . Whole genome analyses of Enterococcus faecium with diverse daptomycin minimal inhibitory concentrations. Antimicrob Agents Chemother 2014; 58:4527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Humphries RM, Kelesidis T, Tewhey R, et al. . Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother 2012; 56:6051–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004; 39:309–17. [DOI] [PubMed] [Google Scholar]
- 5. Arias CA, Contreras GA, Murray BE. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect 2010; 16:555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Safdar N, Andes D, Craig WA. In vivo pharmacodynamic activity of daptomycin. Antimicrob Agents Chemother 2004; 48:63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arias CA, Panesso D, McGrath DM, et al. . Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 2011; 365:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prater AG, Mehta HH, Kosgei AJ, et al. . Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob Agents Chemother 2019; 63: e00790-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khan A, Davlieva M, Panesso D, et al. . Antimicrobial sensing coupled with cell membrane remodeling mediates antibiotic resistance and virulence in Enterococcus faecalis. Proc Natl Acad Sci USA 2019; 116:26925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith JR, Barber KE, Raut A, Aboutaleb M, Sakoulas G, Rybak MJ. β-Lactam combinations with daptomycin provide synergy against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J Antimicrob Chemother 2015; 70:1738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob Agents Chemother 2008; 52:2223–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berti AD, Sakoulas G, Nizet V, Tewhey R, Rose WE. β-Lactam antibiotics targeting PBP1 selectively enhance daptomycin activity against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2013; 57:5005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barber KE, Werth BJ, Rybak MJ. The combination of ceftaroline plus daptomycin allows for therapeutic de-escalation and daptomycin sparing against MRSA. J Antimicrob Chemother 2015; 70:505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kebriaei R, Rice SA, Singh KV, et al. . Influence of inoculum effect on the efficacy of daptomycin monotherapy and in combination with β-lactams against daptomycin-susceptible Enterococcus faecium harboring LiaSR substitutions. Antimicrob Agents Chemother 2018; 62:e00315-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 26th ed. M100 2016. Clinical Laboratory Standards Institute,2018. [Google Scholar]
- 16. Humphries RM. The new, new daptomycin breakpoint for Enterococcus spp. J Clin Microbiol 2019; 57:e00600-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Panesso D, Reyes J, Gaston EP, et al. . Deletion of liaR reverses daptomycin resistance in Enterococcus faecium independent of the genetic background. Antimicrob Agents Chemother 2015; 59:7327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reyes J, Panesso D, Tran TT, et al. . A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis 2015; 211:1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munita JM, Tran TT, Diaz L, et al. . A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 2013; 57:2831–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steed ME, Vidaillac C, Rybak MJ. Novel daptomycin combinations against daptomycin-nonsusceptible methicillin-resistant Staphylococcus aureus in an in vitro model of simulated endocardial vegetations. Antimicrob Agents Chemother 2010; 54:5187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008; 3:163–75. [DOI] [PubMed] [Google Scholar]
- 22. Marron A, Carratalà J, Alcaide F, Fernández-Sevilla A, Gudiol F. High rates of resistance to cephalosporins among viridans-group streptococci causing bacteraemia in neutropenic cancer patients. J Antimicrob Chemother 2001; 47:87–91. [DOI] [PubMed] [Google Scholar]
- 23. Hall AD, Steed ME, Arias CA, Murray BE, Rybak MJ. Evaluation of standard- and high-dose daptomycin versus linezolid against vancomycin-resistant Enterococcus isolates in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations. Antimicrob Agents Chemother 2012; 56:3174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hershberger E, Coyle EA, Kaatz GW, Zervos MJ, Rybak MJ. Comparison of a rabbit model of bacterial endocarditis and an in vitro infection model with simulated endocardial vegetations. Antimicrob Agents Chemother 2000; 44:1921–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith D, Singh K, Tran T, Arias C, Murray B. Daptomycin plus ceftaroline against daptomycin-resistant Enterococcus faecium in a rat model of experimental endocarditis. Open Forum Infect Dis 2016; 3. doi: 10.1093/ofid/ofw172.1528 [DOI] [Google Scholar]
- 26. Singh KV, Tran TT, Nannini EC, Tam VH, Arias CA, Murray BE. Efficacy of ceftaroline against methicillin-susceptible Staphylococcus aureus exhibiting the cefazolin high-inoculum effect in a rat model of endocarditis. Antimicrob Agents Chemother 2017; 61:e00324-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pefanis A, Thauvin-Eliopoulos C, Eliopoulos GM, Moellering RC Jr. Activity of ampicillin-sulbactam and oxacillin in experimental endocarditis caused by beta-lactamase-hyperproducing Staphylococcus aureus. Antimicrob Agents Chemother 1993; 37:507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Catena F, Ansaloni L, Gazzotti F, et al. . Effect of early antibiotic prophylaxis with ertapenem and meropenem in experimental acute pancreatitis in rats. J Hepatobiliary Pancreat Surg 2009; 16:328–32. [DOI] [PubMed] [Google Scholar]
- 29. Ramakrishnan MA. Determination of 50% endpoint titer using a simple formula. World J Virol 2016; 5:85–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 1938; 27:493–7. [Google Scholar]
- 31. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Henry X, Amoroso A, Coyette J, Joris B. Interaction of ceftobiprole with the low-affinity PBP 5 of Enterococcus faecium. Antimicrob Agents Chemother 2010; 54:953–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arbeloa A, Segal H, Hugonnet JE, et al. . Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J Bacteriol 2004; 186:1221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pietta E, Montealegre MC, Roh JH, Cocconcelli PS, Murray BE. Enterococcus faecium PBP5-S/R, the missing link between PBP5-S and PBP5-R. Antimicrob Agents Chemother 2014; 58:6978–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Montealegre MC, Roh JH, Rae M, et al. . Differential penicillin-binding protein 5 (PBP5) levels in the Enterococcus faecium clades with different levels of ampicillin resistance. Antimicrob Agents Chemother 2017; 61:e02034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rice LB, Bellais S, Carias LL, et al. . Impact of specific pbp5 mutations on expression of beta-lactam resistance in Enterococcus faecium. Antimicrob Agents Chemother 2004; 48:3028–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cha R, Rybak MJ. Daptomycin against multiple drug-resistant Staphylococcus and Enterococcus isolates in an in vitro pharmacodynamic model with simulated endocardial vegetations. Diagn Microbiol Infect Dis 2003; 47:539–46. [DOI] [PubMed] [Google Scholar]
- 38. Sifaoui F, Arthur M, Rice L, Gutmann L. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob Agents Chemother 2001; 45:2594–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rice LB, Carias LL, Hutton-Thomas R, Sifaoui F, Gutmann L, Rudin SD. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother 2001; 45:1480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.