Abstract
The backbone of all sphingolipids (SLs) is a sphingoid long-chain base (LCB) to which a fatty acid is N-acylated. Considerable variability exists in the chain length and degree of saturation of both of these hydrophobic chains, and recent work has implicated ceramides with different LCBs and N-acyl chains in distinct biological processes; moreover, they may play different roles in disease states and possibly even act as prognostic markers. We now demonstrate that the half-life, or turnover rate, of ceramides containing diverse N-acyl chains is different. By means of a pulse-labeling protocol using stable-isotope, deuterated free fatty acids, and following their incorporation into ceramide and downstream SLs, we show that very-long-chain (VLC) ceramides containing C24:0 or C24:1 fatty acids turn over much more rapidly than long-chain (LC) ceramides containing C16:0 or C18:0 fatty acids due to the more rapid metabolism of the former into VLC sphingomyelin and VLC hexosylceramide. In contrast, d16:1 and d18:1 ceramides show similar rates of turnover, indicating that the length of the sphingoid LCB does not influence the flux of ceramides through the biosynthetic pathway. Together, these data demonstrate that the N-acyl chain length of SLs may not only affect membrane biophysical properties but also influence the rate of metabolism of SLs so as to regulate their levels and perhaps their biological functions.
Keywords: ceramide, sphingomyelin, hexosylceramide, turnover, lipidomics, fatty acids, stable isotopes, lipid metabolism
Sphingolipids (SLs) are important membrane components that have become the focus of great interest over the past couple of decades due to their roles in membrane structure, cellular signaling, and human disease (1). In human disease, SL levels are altered either as the cause of the disease (such as in the SL storage diseases) or as a secondary response in diseases as diverse as cancer (2, 3), epilepsy (4), cystic fibrosis (5), diabetes (6), and CVDs (7, 8). Interestingly, saturated ceramides with different N-acyl chain lengths, namely long-chain (LC) compared with very-long-chain (VLC) ceramides, appear to play specific roles in diabetes (9, 10) and in CVD (11), in which LC ceramides are more deleterious than VLC ceramides. Moreover, the ratio of plasma LC ceramides versus d18:1/C24:0 ceramide can act as a prognostic biomarker for CVD and appears to be a better marker than the commonly used low-density lipoprotein cholesterol. In addition, the complexity of SL structures is becoming more apparent by advances in analytical mass spectrometry, along with significant progress in identifying most, if not all, of the enzymes related to SL metabolism or those that impinge upon SL metabolism (12).
Ceramide is the basic building block of all complex SLs and consists of a sphingoid long-chain base (LCB) to which an FA is N-acylated. Ceramide is generated by ceramide synthases (13, 14), of which six exist in mammals, each of which is defined by its use of specific fatty acyl CoAs for the generation of ceramides with defined N-acyl chain lengths. Despite significant advances in delineating SL structure and understanding how they are generated, little information is available about the metabolic fate of different SLs or their rate of turnover and flux through the biosynthetic pathway. Nonnatural substrates, or stable isotopes of palmitate (15, 16), serine (15, 17, 18), sphingoid LCBs (19, 20) (including both phytosphingosine and dihydrosphingosine), glucose (17), and water (18) have been used to examine SL metabolism. By way of example, upon labeling MCF7 cells with stable isotope d17 sphinganine, dihydroceramide generation took place within 15 min, whereas the synthesis of more complex SLs, such as SM and hexosylceramide (HexCer), occurred up to 1 h after the pulse (20).
Essentially no studies have attempted to determine whether SLs with different N-acyl chain lengths are metabolized at different rates. We now label SLs using stable-isotope, deuterated FFAs and show that the length of the N-acyl chain affects the rate of ceramide turnover, whereas the length of the LCB has no effect.
MATERIALS AND METHODS
Materials
Stable-isotope, deuterated FFAs [C16:0(d9), hexadecanoic acid (13,13,14,14,15,15,16,16,16-d9); C18:0(d9), octadecanoic acid (15,15,16,16,17,17,18,18,18-d9); C24:0(d4), tetracosanoic acid (12,12,13,13-d4); and C24:1(d7), tetracosanoic acid 15Z (22,22,23,23,24,24,24-d7)] were from Avanti Polar Lipids (Alabaster, AL). l-serine (2,3,3-d3, 98%; 15N, 98%) was from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA). Defatted BSA, FFAs, serine, and a protease inhibitor cocktail were from Sigma-Aldrich (St. Louis, MO). DMEM and FCS were from Gibco (Paisley, UK), and supplementary antibiotics and sodium pyruvate were from Biological Industries (Beit HaEmek, Israel).
Cell culture
Hek293T cells were cultured in DMEM supplemented with 10% FCS, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 110 µg/ml sodium pyruvate.
Pulse-chase labeling of SLs
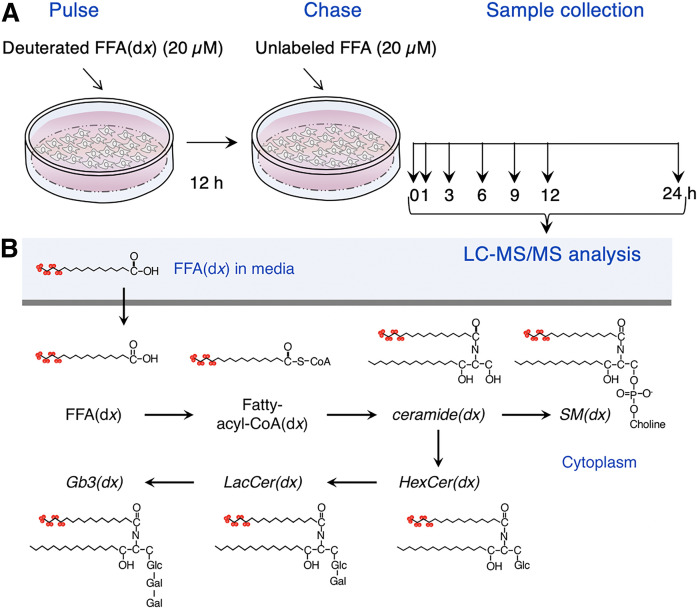
Cells cultured at 37°C in 5% CO2 were pulsed with 20 µM deuterated FFAs(dx) (Fig. 1A) complexed with 20 µM defatted BSA in DMEM supplemented with 10% FCS, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 110 µg/ml sodium pyruvate for 12 h (pulse). The medium was then replaced with unlabeled FFAs (20 µM) for appropriate times (chase) (Fig. 1A) before the cells were removed from the dishes by scraping into PBS. The medium was centrifuged twice at 800 g for 4 min. Cell homogenates were prepared in 20 mM HEPES-potassium hydroxide (pH 7.2), 25 mM potassium chloride, 250 mM sucrose, and 2 mM MgCl2 containing a protease inhibitor cocktail. Protein concentration was determined using the BCA protein kit (Thermo Fisher Scientific, Waltham, MA).
Fig. 1.
Experimental design. A: Cells were incubated with deuterated FFAs [C16:0(d9), C18:0(d9), C24:0(d4), and C24:1(d7)] for 12 h followed by a chase with unlabeled FFAs for various times prior to the analysis of the SLs shown in the scheme. B: The metabolic pathway by which C16:0(d9) [hexadecanoic acid (13,13,14,14,15,15,16,16,16-d9)] was incorporated into SLs. The SL classes analyzed in this study are in italics.
Lipidomics
SLs were extracted from cell homogenates by resuspension in PBS followed by a modified Folch extraction (21) using a Hamilton Microlab Star system (Hamilton Robotic AB, Kista, Sweden) (22). Glycerophospholipids were degraded by mild alkaline hydrolysis. An aliquot was evaporated under N2 followed by the addition of 1 ml 0.1 M sodium hydroxide-methanol for 2 h at room temperature. Sodium phosphate (100 µl; pH 7.0) and hydrogen chloride (100 µl) were added and evaporated under N2 overnight. The dried pellet was dissolved in 600 µl chloroform-methanol (2:1; v/v) followed by 300 µl 20 mM acetic acid and reextraction by the Folch procedure (22). Ceramide, HexCer, lactosylceramide (LacCer), globotriaosylceramide (Gb3), and SM were analyzed using a targeted approach by UHPLC-MS (23, 24). An Acquity BEH C18 2.1 × 50 mm column with a particle size of 1.7 μm (Waters, Milford, MA) was used with the mobile phases containing 10 mM ammonium acetate in water with 0.1% formic acid (solvent A) and 10 mM ammonium acetate in acetonitrile-isopropanol (4:3; v/v) containing 0.1% formic acid (solvent B). SLs were analyzed on a hybrid triple quadrupole/linear ion trap mass spectrometer (5500 QTRAP) equipped with a UHPLC system (Ekspert ultraLC 110-XL or Shimadzu Nexera X2) using multiple reaction monitoring in positive ion mode. Lipids were quantified using internal standards (25). Data processing was performed by MultiQuant software and SAS.
RESULTS
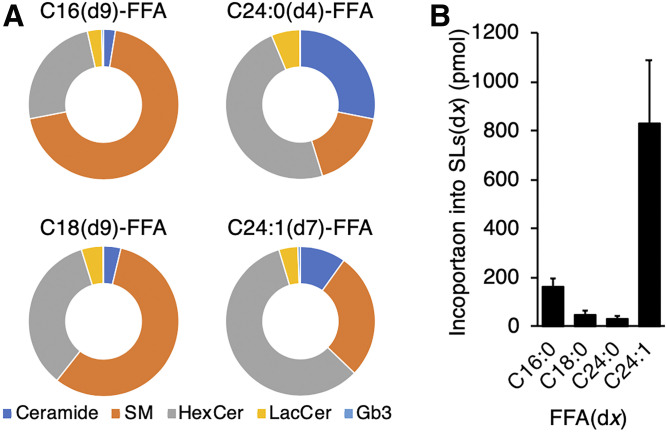
Using UHPLC-MS, ∼80 SL species were detected in Hek cells, with five major groups analyzed (Fig. 1B), namely ceramide, SM, HexCer, LacCer, and Gb3. As expected, the major SL was SM, which was found at ∼10-fold higher levels than ceramide, whereas HexCer levels were somewhat lower than SM but significantly higher than their downstream glycosylated derivatives. Levels of these SLs were similar upon incubation with each of the FFAs(dx) (Table 1), which were incorporated into each SL class at a somewhat different level. For instance, after the 12 h pulse, C16:0(d9), C18:0(d9), and C24:0(d4) FAs made up ∼0.2% to 5% of each SL class, whereas C24:1(d7) FAs made up ∼10% of SM, LacCer, and Gb3 but ∼30% of ceramide and HexCer (Table 1). However, we conclude that the incubation of Hek cells with FFAs(dx) did not differentially affect the global SL composition of the cells.
TABLE 1.
SL profile upon incubation of Hek293T cells with FFAs(dx)
| Ceramide | SM | HexCer | LacCer | Gb3 | ||
| FFA(dx) Incubation | pmol/mg protein | |||||
| C16:0(d9) | Total | 297 ± 102 | 3,106 ± 312 | 2,041 ± 647 | 484 ± 212 | 58.1 ± 9.8 |
| SLs(dx) | 6.3 ± 0. 4 | 172 ± 18.0 | 60.8 ± 6.3 | 7.6 ± 2.7 | 1.0 ± 0.2 | |
| C18:0(d9) | Total | 366 ± 128 | 3,967 ± 964 | 2,671 ± 1,330 | 602 ± 149 | 69.9 ± 24.8 |
| SLs(dx) | 2.7 ± 0.9 | 40.6 ± 13.8 | 24.7 ± 7.9 | 3.3 ± 1.0 | 0.1 ± 0.1 | |
| C24:0(d4) | Total | 413 ± 98 | 4,075 ± 1,345 | 2,783 ± 1,594 | 638 ± 165 | 82.1 ± 43.7 |
| SLs(dx) | 17.1 ± 5.0 | 10.4 ± 1.3 | 29.5 ± 6.9 | 3.8 ± 2.0 | 0.1 ± 0.1 | |
| C24:1(d7) | Total | 402 ± 135 | 3,845 ± 1,064 | 2,807 ± 1,755 | 617 ± 124 | 76.4 ± 35.6 |
| SLs(dx) | 140 ± 67 | 388 ± 122 | 827 ± 1057 | 58.3 ± 11.2 | 7.8 ± 4.6 | |
Data are means ± SDs of three individual experiments performed in triplicate. Cells were incubated with each of the FFAs(dx) for 12 h, and SLs (ceramide, SM, HexCer, LacCer, and Gb3) were measured, including both deuterated (dx) and nondeuterated SLs, the values of which are given in the top rows (Total). The second row [SLs(dx)] shows the levels of deuterated SLs in each SL class.
The incorporation of different FAs(dx) into each lipid class showed significant differences between FFA(dx) treatment. For instance, C16:0(d9) and C18:0(d9) were incorporated mainly into SM and C24:0(d4) was incorporated into ceramide, whereas both C24:0(d4) and C24:1(d7) were incorporated at high levels into HexCer (Fig. 2A). This was unrelated to the initial amount of FAs(dx) incorporated into the SLs (Fig. 2A, B) and may reflect a difference in the flux rate of SLs with different acyl chain lengths into downstream SLs. While these data suggest that different enzymes in the SL pathways have specific preferences for substrates with a particular N-acyl chain length, they do not address the rate of turnover of specific SLs with defined acyl chain lengths.
Fig. 2.
Incorporation of FFAs(dx) into SLs. A: mol% of SL(dx) classes. B: Levels of all SLs(dx) (ceramide, SM, HexCer, LacCer, and Gb3 of all chain lengths) after incubation with different FFAs(dx) at the end of the 12 h pulse. Data are means ± SDs of three individual experiments in triplicate.
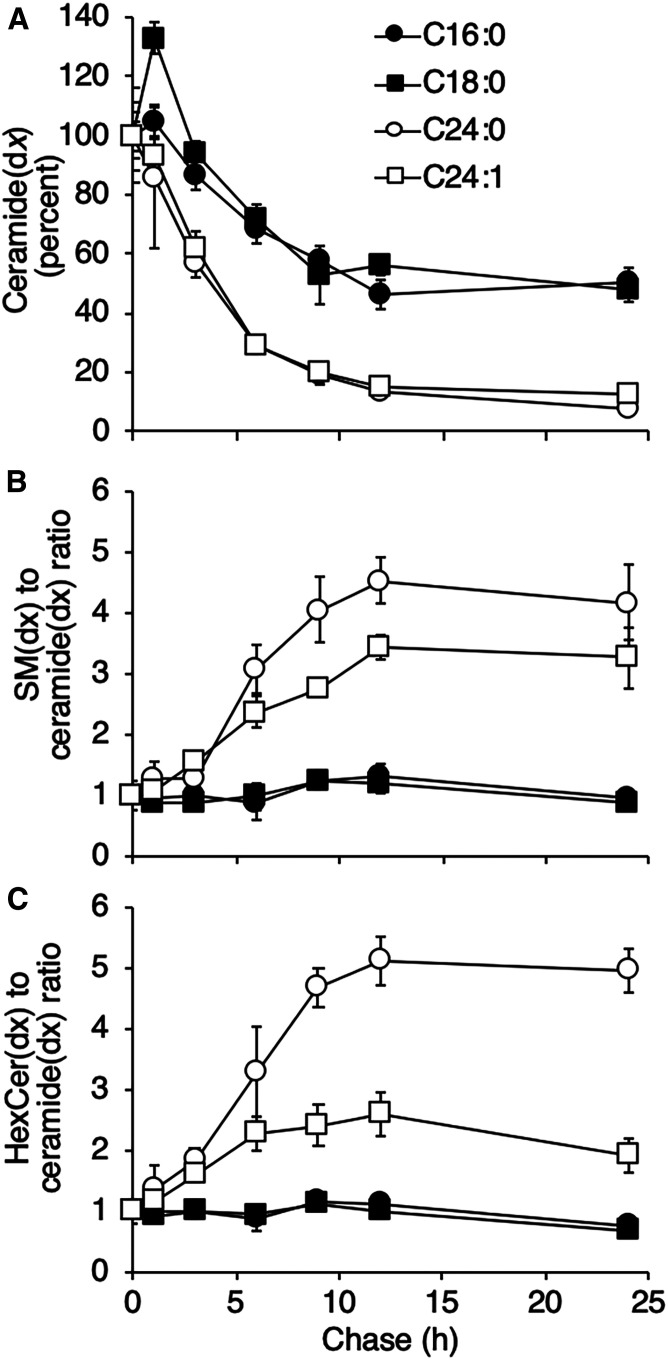
To determine the latter, we examined levels of SLs(dx) after various times of chase with unlabeled FFAs. Ceramide levels decreased over the 24 h since the beginning of the chase (Fig. 3A), but there was a noticeable difference in the rate of turnover of LC compared with VLC ceramide. Thus, ∼50% of the C16:0(d9) ceramide and C18:0(d9) ceramide persisted after the 24 h chase, whereas <∼20% of C24:0(d4) ceramide and C24:1(d7) ceramide persisted after the same time period. For the latter two VLC ceramides, it was possible to calculate half-lives of turnover, which were 4.4 ± 1.6 and 4.5 ± 0.8 h, respectively. The reason for the more rapid turnover of the VLC ceramides is likely to be their metabolism to C24:0(d4) SM and C24:1(d7) SM (Fig. 3B) and C24:0(d4) Hex Cer and C24:1(d7) HexCer (Fig. 3C), whereas the metabolism of C16:0(d9) ceramide and C18:0(d9) ceramide was much slower. Similar results were obtained when examining the turnover of FAs generated by FA elongation. Thus, when cells were incubated with the FFAs(dx), C16:0(d9) FA was elongated to both C18:0(d9) and C20:0(d9), C18:0(d9) FA was elongated to C20:0(d9) FA, C24:0(d4) FA was elongated to C26:0(d4) FA, and C24:1(d7) FA was elongated to C26:1(d7). A precise half-life for C18:0(d9) ceramide generated by elongation could not be calculated, but ∼70% of the C18:0(d9) ceramide remained after the 24 h pulse; however, a half-life could be measured for C26:0(d4) ceramide and C26:1(d7) ceramide (6.1 ± 1 and 5.0 ± 0.7 h, respectively).
Fig. 3.
VLC ceramides turn over more rapidly than LC ceramides. Hek293T cells were pulsed with FFAs(dx) for 12 h followed by a chase with FFAs for the indicated times. A: Ceramide(dx) levels during the chase with FFAs normalized (by percentage) to the beginning of the chase. B, C: Levels of SM(dx) and HexCer(dx), respectively, as a ratio to ceramide(dx). Representative data from one experiment are shown (means ± SDs; n = 3), which was repeated three times with similar results. SL levels (pmol/mg) at the beginning of the chase were as follows: C16:0(d9) ceramide, 5.43 ± 0.68; C18:0(d9) ceramide, 0.91 ± 0.07; C24:0(d4) ceramide, 16.7 ± 2.66; C24:1(d7) ceramide, 64.8 ± 7.36; C16:0(d9) SM, 139 ± 12.69; C18:0(d9) SM, 13.8 ± 0.23; C24:0(d4) SM, 10.2 ± 1.62; C24:1(d7) SM, 371 ± 17.0; C16:0(d9) HexCer, 58.0 ± 2.31; C18:0(d9) HexCer, 12.3 ± 1.62; C24:0(d4) HexCer, 25.0 ± 1.45; and C24:1(d7) HexCer, 174.5 ± 15.2.
Together, these data show that the flux of LC ceramide into downstream SLs is significantly slower than VLC ceramide. This conclusion was supported by an independent approach in which Hek293T cells were incubated with 4.2 mg/ml l-serine(d3,15N)/37.8 mg/ml l-serine for 12 h followed by a chase with 42 mg/ml l-serine. After the 24 h chase, levels of C16:0(d3,15N) and C18:0(d3,15N) ceramide were ∼50% of that measured at the beginning of the chase, whereas levels of C24:0(d3,15N) ceramide and C24:1(d3,15N) ceramide were <25% of the initial level, irrespective of the level of the different ceramide(dx) species at the beginning of the chase, confirming that VLC ceramides turn over more rapidly than LC ceramides.
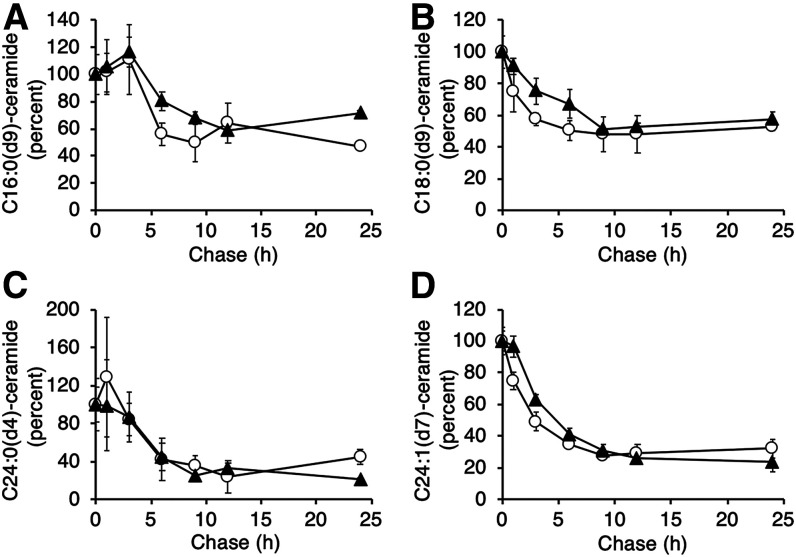
While the acyl chain length of the FA in ceramide significantly affected the rate of ceramide turnover, the length of the sphingoid LCB had no effect on ceramide turnover. Thus, during the 24 h chase, the rate of turnover of d16:1 and d18:1 ceramides, irrespective of the N-acyl chain length, was similar (Fig. 4).
Fig. 4.
LCB composition does not affect ceramide turnover. The turnover rate of d16:1 (white circles) and d18:1 (black triangles) ceramides containing the respective FAs(dx) is shown, normalized (by percentage) to the beginning of the chase. Representative data from one experiment are shown (means ± SDs; n = 3), which was repeated three times in triplicate with similar results.
DISCUSSION
While the cellular sphingolipidome is relatively well studied, far less is known about how levels of individual SL species or of different SL classes are regulated. The modes of regulation of some of the enzymes in both the biosynthetic and degradative pathways are becoming clear, but an integrated picture of SL flux is lacking (26). The current study is a step in addressing this problem, inasmuch as we have shown, by two independent pulse-labeling protocols, that VLC ceramides turn over more rapidly than LC ceramides, at least in cultured Hek cells. Preliminary data from another cell type, induced pluripotent stem cell-derived hepatocyte-like cells (HLCs), is somewhat consistent with the data in Hek cells. Thus, the turnover of C24:0(d4) ceramide and C24:1(d7) ceramide was significantly more rapid than that of C16:0(d9) ceramide due to the more rapid incorporation of the former into C24:0(d4) SM and C24:1(d7) SM and into C24:0(d4) HexCer and C24:1(d7) HexCer (data not shown). However, the rate of C18:0(d9) ceramide turnover in HLCs was more rapid than that detected in Hek cells (i.e., half-life of ∼4.7 h in HLCs), although there was some variability between different experiments. Whether this is due to different levels of CerS4 [which generates C18 ceramide (27)] in the two cell types is unclear at present.
In contrast, the sphingoid LCB, at least d16:1 versus d18:1, does not affect the rate of ceramide turnover. The enzyme that generates the LCB, serine palmitoyl transferase (SPT), is a promiscuous heteromeric enzyme that can not only use a number of amino acids rather than serine but also fatty acyl CoAs other than palmitoyl CoA. There are several subtypes of SPT subunits, with combinations of different SPT subunits conferring different specificities toward acyl CoAs (28). Thus, the complex of LCB1/LCB2/ssSPTa shows a strong preference for C16-CoA, whereas LCB1/LCB3/ssSPTa can also use C14-CoA. Irrespective of the choice, or the availability of fatty acyl CoAs used by SPT, the LCBs thereby generated do not affect the rate of turnover of the respective ceramide species into which they are incorporated.
The finding that VLC ceramides turn over more rapidly than LC ceramides may be of significance for understanding the biological functions of these SLs, not least in some of the human diseases in which they have been implicated. Noticeable among these are CVD, in which ratios of ceramides with different N-acyl chain lengths are used as predictors of death resulting from CVD. Thus, plasma CVD risk-related ceramides (d18:1/C16:0 ceramide, d18:1/C18:0 ceramide, and d18:1/C24:1 ceramide) and their ratios with d18:1/C24:0 ceramide, has emerged as potential risk stratifications for CVD patients (29). The source of these different ceramides and the route by which they are released into plasma are not fully understood, and our current data suggest that an additional factor needs to be considered when considering the role of LC versus VLC ceramides in CVD and other diseases, namely their distinct rates of cellular turnover.
Data availability
The data supporting this study are available in the article and are available from the corresponding author upon reasonable request.
Acknowledgments
The authors thank Ron Rotkopf from the Life Sciences Core Facilities at the Weizmann Institute of Science for his help with statistical analysis and Shani Blumenreich for helpful discussions.
Footnotes
Author contributions—A.H.F. project supervision; I.D.Z., G.V., L.E.V., and K. A-S. experiments; T.A. assays; I.D.Z. data analysis; D.K. and R.L. lipidomics analysis; I.D.Z. and A.H.F writing.
Author ORCIDs—Anthony H. Futerman 0000-0003-0013-0115
Funding and additional information—This work was supported in part by the Atheroflux consortium (European Union Grant PF7-602222-2). A. H. F. is the Joseph Meyerhoff Professor of Biochemistry at the Weizmann Institute of Science.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- Gb3
- globotriaosylceramide
- HexCer
- hexosylceramide
- HLC
- hepatocyte-like cell
- LacCer
- lactosylceramide
- LC
- long-chain
- LCB
- long-chain base
- SL
- sphingolipid
- SPT
- serine palmitoyl transferase
- VLC
- very-long-chain
Manuscript received June 17, 2020, and in revised form July 8, 2020. Published, JLR Papers in Press, July 10, 2020, DOI 10.1194/jlr.RA120000984.
REFERENCES
- 1.Futerman A. H. Sphingolipids In Biochemistry of Lipids, Lipoproteins and Membranes. 6th edition N. D. Ridgway and R. S. McLeod, editors. Elsevier, Boston: 297 –326. [Google Scholar]
- 2.Saddoughi S. A., and Ogretmen B.. 2013. Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 117: 37–58. [DOI] [PubMed] [Google Scholar]
- 3.Jensen S. A., Calvert A. E., Volpert G., Kouri F. M., Hurley L. A., Luciano J. P., Wu Y., Chalastanis A., Futerman A. H., and Stegh A. H.. 2014. Bcl2L13 is a ceramide synthase inhibitor in glioblastoma. Proc. Natl. Acad. Sci. USA. 111: 5682–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosbech M-B., Olsen A. S. B., Neess D., Ben-David O., Klitten L. L., Larsen J., Sabers A., Vissing J., Nielsen J. E., Hasholt L., et al. 2014. Reduced ceramide synthase 2 activity causes progressive myoclonic epilepsy. Ann. Clin. Transl. Neurol. 1: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grassmé H., Riethmüller J., and Gulbins E.. 2013. Ceramide in cystic fibrosis. Handb. Exp. Pharmacol. 216: 265–274. [DOI] [PubMed] [Google Scholar]
- 6.Adams J. M., Pratipanawatr T., Berria R., Wang E., DeFronzo R. A., Sullards M. C., and Mandarino L. J.. 2004. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes. 53: 25–31. [DOI] [PubMed] [Google Scholar]
- 7.Peterson L. R., Xanthakis V., Duncan M. S., Gross S., Friedrich N., Völzke H., Felix S. B., Jiang H., Sidhu R., Nauck M., et al. 2018. Ceramide remodeling and risk of cardiovascular events and mortality. J. Am. Heart Assoc. 7: e007931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J., Pan W., Shi R., Yang T., Li Y., Yu G., Bai Y., Schuchman E. H., He X., and Zhang G.. 2015. Ceramide is upregulated and associated with mortality in patients with chronic heart failure. Can. J. Cardiol. 31: 357–363. [DOI] [PubMed] [Google Scholar]
- 9.Turpin S. M., Nicholls H. T., Willmes D. M., Mourier A., Brodesser S., Wunderlich C. M., Mauer J., Xu E., Hammerschmidt P., Brönneke H. S., et al. 2014. Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab. 20: 678–686. [DOI] [PubMed] [Google Scholar]
- 10.Raichur S., Wang S. T., Chan P. W., Li Y., Ching J., Chaurasia B., Dogra S., Öhman M. K., Takeda K., Sugii S., et al. 2014. CerS2 haploinsufficiency inhibits β-oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab. 20: 687–695. [DOI] [PubMed] [Google Scholar]
- 11.Laaksonen R., Ekroos K., Sysi-Aho M., Hilvo M., Vihervaara T., Kauhanen D., Suoniemi M., Hurme R., März W., Scharnagl H., et al. 2016. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur. Heart J. 37: 1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannun Y. A., and Obeid L. M.. 2018. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pewzner-Jung Y., Ben-Dor S., and Futerman A. H.. 2006. When do lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281: 25001–25005. [DOI] [PubMed] [Google Scholar]
- 14.Zelnik I. D., Rozman B., Rosenfeld-Gur E., Ben-Dor S., and Futerman A. H.. 2019. A stroll down the CerS lane. Adv. Exp. Med. Biol. 1159: 49–63. [DOI] [PubMed] [Google Scholar]
- 15.Wigger D., Gulbins E., Kleuser B., and Schumacher F.. 2019. Monitoring the sphingolipid de novo synthesis by stable-isotope labeling and liquid chromatography-mass spectrometry. Front. Cell Dev. Biol. 7: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noto D., Di Gaudio F., Altieri I. G., Cefalù A. B., Indelicato S., Fayer F., Spina R., Scrimali C., Giammanco A., Mattina A., et al. 2020. Automated untargeted stable isotope assisted lipidomics of liver cells on high glucose shows alteration of sphingolipid kinetics. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1865: 158656. [DOI] [PubMed] [Google Scholar]
- 17.Skotland T., Ekroos K., Kavaliauskiene S., Bergan J., Kauhanen D., Lintonen T., and Sandvig K.. 2016. Determining the turnover of glycosphingolipid species by stable-isotope tracer lipidomics. J. Mol. Biol. 428: 4856–4866. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Berejnaia O., Liu J., Wang S-P., Daurio N. A., Yin W., Mayoral R., Petrov A., Kasumov T., Zhang G-F., et al. 2018. Quantifying ceramide kinetics in vivo using stable isotope tracers and LC-MS/MS. Am. J. Physiol. Endocrinol. Metab. 315: E416–E424. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Montañés F., and Schneiter R.. 2016. Following the flux of long-chain bases through the sphingolipid pathway in vivo using mass spectrometry. J. Lipid Res. 57: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snider J. M., Snider A. J., Obeid L. M., Luberto C., and Hannun Y. A.. 2018. Probing de novo sphingolipid metabolism in mammalian cells utilizing mass spectrometry. J. Lipid Res. 59: 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 22.Heiskanen L. A., Suoniemi M., Ta H. X., Tarasov K., and Ekroos K.. 2013. Long-term performance and stability of molecular shotgun lipidomic analysis of human plasma samples. Anal. Chem. 85: 8757–8763. [DOI] [PubMed] [Google Scholar]
- 23.Merrill A. H. Jr., Sullards M. C., Allegood J. C., Kelly S., and Wang E.. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 36: 207–224. [DOI] [PubMed] [Google Scholar]
- 24.Jung H. R., Sylvänne T., Koistinen K. M., Tarasov K., Kauhanen D., and Ekroos K.. 2011. High throughput quantitative molecular lipidomics. Biochim. Biophys. Acta. 1811: 925–934. [DOI] [PubMed] [Google Scholar]
- 25.Simons B., Kauhanen D., Sylvänne T., Tarasov K., Duchoslav E., and Ekroos K.. 2012. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites. 2: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Vasquez F., Sims K. J., Cowart L. A., Okamoto Y., Voit E. O., and Hannun Y. A.. 2005. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 433: 425–430. [DOI] [PubMed] [Google Scholar]
- 27.Riebeling C., Allegood J. C., Wang E., Merrill A. H., and Futerman A. H.. 2003. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278: 43452–43459. [DOI] [PubMed] [Google Scholar]
- 28.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Harmon J. M., and Dunn T. M.. 2009. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA. 106: 8186–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarasov K., Ekroos K., Suoniemi M., Kauhanen D., Sylvänne T., Hurme R., Gouni-Berthold I., Berthold H. K., Kleber M. E., Laaksonen R., et al. 2014. Molecular lipids identify cardiovascular risk and are efficiently lowered by simvastatin and PCSK9 deficiency. J. Clin. Endocrinol. Metab. 99: E45–E52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting this study are available in the article and are available from the corresponding author upon reasonable request.