Abstract
Introduction:
Germline variants in androgen metabolism genes may influence clinical response to androgen deprivation therapy (ADT) in advanced prostate cancer. We sought to investigate the prognostic significance of germline variants in androgen metabolism genes with respect to overall survival (OS) after ADT, and to associate germline variants with tumor genomic features.
Methods:
Germline and somatic whole-genome sequencing (WGS) data were evaluated in a cohort of 101 men with metastatic castration-resistant prostate cancer (mCRPC). Survival analyses were performed to identify polymorphisms associated with impaired OS after primary ADT. Germline variants found to be prognostic of OS were associated with tumor somatic DNA-sequence alterations based on WGS performed on paired metastasis biopsies from the same 101 patients. Gene set enrichment analysis was performed based on tumor RNA-sequencing data to identify genomic pathways differentially expressed in patients with germline variants.
Results:
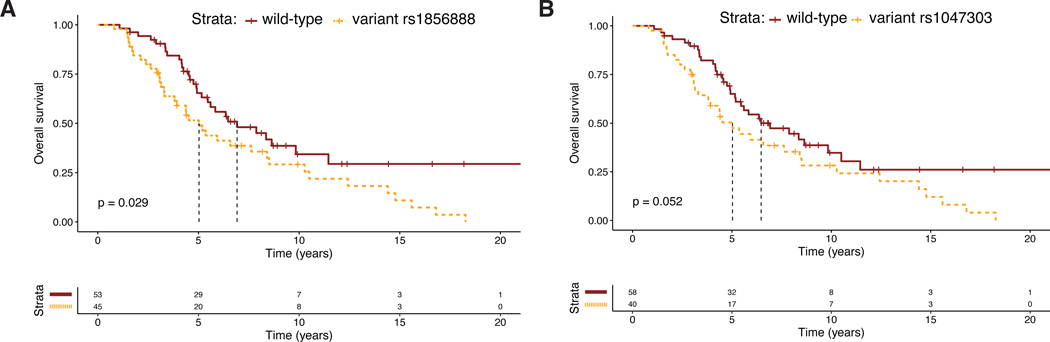
A comprehensive literature review identified 17 candidate polymorphisms in nine androgen metabolism genes that have been previously shown to have an association with response to ADT in prostate cancer. Of these, the variant rs1856888 allele located 13kb upstream of HSD3B1 was found to be significantly associated with impaired OS (P=0.029). Variant rs1856888 was commonly co-inherited with the well-characterized HSD3B1(1245A>C) polymorphism, and there was a trend towards shorter median OS in patients with HSD3B1(1245A>C) compared to homozygous wild-type patients (P=0.052). While HSD3B1 germline variants were not associated with common somatic tumor DNA alterations, they were associated with increased tumor expression of cell proliferation and cell cycle genes.
Conclusions:
This study presents a comprehensive assessment of germline variants in androgen metabolism genes and highlights HSD3B1 polymorphisms as prognostic of OS after ADT and associated with an aggressive gene expression tumor profile in mCRPC.
INTRODUCTION
Genetic variants have been implicated in both the development and progression of prostate cancer1–3. Of particular clinical interest is the relationship between germline polymorphisms in androgen metabolism genes and response to androgen deprivation therapy (ADT)4. ADT is often used as an effective therapy for metastatic castration-sensitive prostate cancer (mCSPC). However, while mCSPCs generally demonstrate a favorable initial response to ADT, most tumors eventually progress to a castration-resistant state. A major mechanism by which tumors develop ADT resistance is synthesis of androgens from extragonadal adrenal precursor steroids5. In particular, overexpression of the SRD5A1 gene product leads to increased conversion of the adrenal steroid androstenedione to 5α-androstanedione, which is then converted to dihydrotestosterone (DHT). Increased DHT synthesis can then drive the progression of metastatic castration-resistant prostate cancer (mCRPC)5.
Agents such as abiraterone acetate that inhibit the androgen biosynthesis pathway have thus been of great interest and have demonstrated clinical promise. Abiraterone inhibits the enzyme cytochrome P450 17 alpha-hydroxylase (CYP17), which drives the synthesis of androgens in both the testes and adrenal glands6. Abiraterone acetate has been shown to significantly improve survival outcomes in mCRPC and mCSPC7,8. However, despite the overall clinical benefit of abiraterone acetate, response to treatment is highly variable among patients. Motivated by the intuition that innate patient-to-patient variation in androgen biosynthesis may explain this variability in response to treatment, a recent study demonstrated that the commonly observed HSD3B1(1245A>C) germline polymorphism alters the metabolism of abiraterone acetate9. In addition, clinical outcomes studies have demonstrated significant associations between specific germline polymorphisms in androgen-related genes such as CYP19A1, HSD3B1, and HSD17B4 and survival outcomes from time of initiating primary ADT10–13. Altogether, these studies highlight the importance of germline variants in androgen biosynthesis genes as potential predictors of ADT response.
To our knowledge, no study has comprehensively assessed all of these germline variants in a cohort of mCRPC patients to see if they are predictive of ADT response or overall survival. Additionally, little is known about the association between prostate cancer-related germline variants and molecular tumor characteristics, as none of the published studies to date have coupled germline sequencing results with genomic profiling of the patients’ clinical tumor samples. We performed the first comprehensive analysis of germline polymorphisms integrated with solid tumor RNA-sequencing and DNA-sequencing results to investigate germline predictors of ADT response in metastatic castration-resistant prostate cancer (mCRPC).
MATERIALS AND METHODS
Literature review of germline polymorphisms associated with ADT in prostate cancer
SciMiner14 was used to perform a comprehensive review of genes in which polymorphisms were associated with ADT and prostate cancer. Specifically, the following Pubmed search query was input into SciMiner: “polymorphism” AND “prostate cancer” AND “androgen deprivation therapy.” The output list of genes was then manually reviewed, and all androgen synthesis genes and their corresponding polymorphisms were included in a final filtered list of candidate polymorphisms.
Data processing and variable definitions
A cohort of 101 men with mCRPC was used to investigate the prognostic significance of all candidate polymorphisms. Biopsy tissue and clinical data were obtained as part of a prospective IRB-approved trial (ClinicalTrials.gov identifier: NCT02432001), and informed consent was obtained from all study participants. Germline and somatic DNA-sequencing and RNA-sequencing data were processed as previously reported15,16. For each SNP identified from literature review, each patient was classified as “wild-type” (homozygous wild-type) or “variant” (either heterozygous or homozygous variant). Somatic DNA alterations were assessed for genes commonly altered in mCRPC as previously described15,16. For loss-of-function alterations in RB1, PTEN, TP53, CDK12, BRCA2, and CHD1, samples were considered altered if two or more DNA alterations (mutations, copy number loss, inactivating structural variants) were present, i.e., presumed bi-allelic loss. ETS fusions included fusions involving ERG, ETV1, ETV4, and ETV5 as the 3’ partner. For SPOP, MYC, and AR, the following known driver alterations were considered: SPOP mutations, MYC amplifications, and AR amplifications (including AR upstream enhancer amplifications). All DNA-sequencing and RNA-sequencing data are available in dbGaP (accession number: phs001648).
Statistical methods
Retrospective, single-cohort survival analyses were performed to examine the prognostic significance of each candidate germline variant. The primary endpoint was overall survival (OS) from time of initiating primary ADT. Kaplan-Meier curves were plotted, and patients with wild-type and variant SNPs were compared using a log-rank test. Multivariable analysis was conducted by including genomic variants significantly associated with impaired OS on univariate analysis as well as the following well-characterized clinicopathologic prognostic factors: ECOG performance status, Gleason score, PSA laboratory value, metastatic disease at time of initiating primary ADT, and treatment with enzalutamide or abiraterone acetate. Association of germline variants with common DNA alterations was performed using a Fisher’s exact test. RNA-seq differential expression analyses were performed by first performing a Wilcoxon signed-rank test comparing the wild-type and variant patients. The –log10 of the P-values (positive score if expression was higher in the variant samples, negative score if higher in the wild-type samples) was used to rank genes in order from most overexpressed to least in the tumors of patients harboring a polymorphism. Gene set enrichment analysis (GSEA) was performed using this ranked list of genes and the previously published GSEA Preranked tool17 to identify differentially expressed gene sets. Gene sets were considered significantly differentially expressed if they had FDR q-value < 0.25. All statistical comparisons were performed at a two-sided significance level of 0.05 unless otherwise stated. R version 3.5.0 was used to perform all statistical analyses, and code is available upon request.
RESULTS
Comprehensive literature review of all previously reported germline polymorphisms associated with ADT response identified 17 polymorphisms in the following nine genes: HSD17B410, HSD17B218, SRD5A219,20, SLCO2B121–23, SLCO1B323,24, CYP19A110,25,26, CYP17A118,27, AKR1C328,29, and HSD3B111–13,30,31. Assessing the prognostic significance of these polymorphisms in our cohort, we found that the only gene with a polymorphism significantly associated with OS was HSD3B1.
Two polymorphisms at the HSD3B1 locus were identified a priori from review of the literature: rs1047303, i.e., HSD3B1(1245A>C), and rs1856888, located 13kb upstream of the HSD3B1 promoter. At the rs1047303 locus, 58% (59/101) of patients were homozygous wild-type, 35% (35/101) were heterozygotes, and 7% (7/101) were homozygous variant. At the rs1856888 locus, 53% (54/101) of patients were homozygous wild-type, 36% (36/101) were heterozygotes, and 11% (11/101) were homozygous variant (Table 1). The variant rs1047303, i.e., HSD3B1(1245A>C), and variant rs1856888 alleles were commonly co-inherited, as all patients with HSD3B1(1245A>C) also demonstrated variant rs1856888 and only five patients with variant rs1856888 had homozygous wild-type rs1047303. The variant rs1856888 polymorphism was significantly associated with shorter OS (median OS [95% CI]: 5.0 [3.3–8.4] vs. 6.9 [5.2–9.8] years, P=0.029; Figure 1A). The HSD3B1(1245A>C) polymorphism was borderline significantly associated with OS differences between the wild-type and variant groups (median OS [95% CI]: 5.0 [3.1–8.4] vs. 6.5 [5.2–9.8] years, P=0.052; Figure 1B). Examining additional clinical endpoints, we found that variant rs1856888 was associated with borderline-significant shorter time to progression (TTP) from ADT initialization to mCRPC (median TTP [95% CI]: 2.0 [1.1–3.8] vs. 3.1 [1.7–7.3] years, P=0.077; Figure S1A) and shorter OS from time of mCRPC diagnosis (median OS [95% CI]: 2.5 [1.6–3.4] vs. 3.9 [3.4–4.9] years, P=0.036; Figure S1C). Variant rs1047303 was associated with borderline-significant shorter TTP from ADT initialization to mCRPC (median TTP [95% CI]: 2.3 [1.1 – 3.8] vs. 3.1 [1.7–7.3] years, P=0.084; Figure S1B) and a trend toward shorter OS from time of mCRPC diagnosis (median OS [95% CI]: 2.5 [1.6–4.1] vs. 3.5 [2.9–4.2] years, P=0.13; Figure S1D).
Table 1.
Patient demographics and clinicopathologic features
| Characteristic | Pts. sequenced (N=101) |
|---|---|
| Age – years (SD) | 71 (8.4) |
| Race | |
| Asian | 4 (4.3) |
| Black or African American | 5 (5.3) |
| White | 85 (90.4) |
| Missing | 7 (6.9) |
| Gleason score at diagnosis | |
| 8+ | 52 (57.1) |
| < 8 | 39 (42.9) |
| Missing | 10 (9.9) |
| ECOG performance status | |
| 0 | 57 (56.4) |
| 1 | 41 (40.6) |
| 2+ | 3 (3) |
| Missing | 0 (0) |
| Metastatic disease at time of ADT initiation | |
| Yes | 51 (51.5) |
| No | 48 (48.5) |
| Unknown | 2 (2.0) |
| Metastatic sites at time of biopsy | |
| Liver | 15 (14.9) |
| Visceral metastases (non-liver) | 12 (11.9) |
| Bone +/− lymph node | 64 (63.4) |
| Lymph node only | 11 (10.9) |
| Missing | 0 (0) |
| Prior treatment status (abiraterone / enzalutamide) | |
| Naïve / Naive | 38 (38) |
| Naïve / Treated | 27 (27) |
| Treated / Naive | 15 (15) |
| Treated / Treated | 20 (20) |
| Missing | 1 (1) |
| rs1856888 | |
| Homozygous wild-type | 54 (53) |
| Heterozygous | 36 (36) |
| Homozygous variant | 11 (11) |
| rs1047303, i.e., HSD3B1(1245A>C) | |
| Homozygous wild-type | 59 (58) |
| Heterozygous | 35 (35) |
| Homozygous variant | 7 (7) |
| Laboratory values – median (IQR) | |
| PSA, ng/mL | 43 (15–148) |
| Alkaline phosphatase, U/L | 91 (65–140) |
| LDH, IU/L | 203 (166–291) |
| Hemoglobin, g/dL | 13 (12–14) |
Note: all clinicopathologic variables were measured at time of biopsy and are presented as “Number (%)” unless otherwise noted.
Figure 1:
Kaplan-Meier curves demonstrating overall survival differences between patients with 1–2 variant alleles and homozygous wild-type patients in the A) rs1856888 and B) rs1047303 (1245C) HSD3B1 polymorphisms.
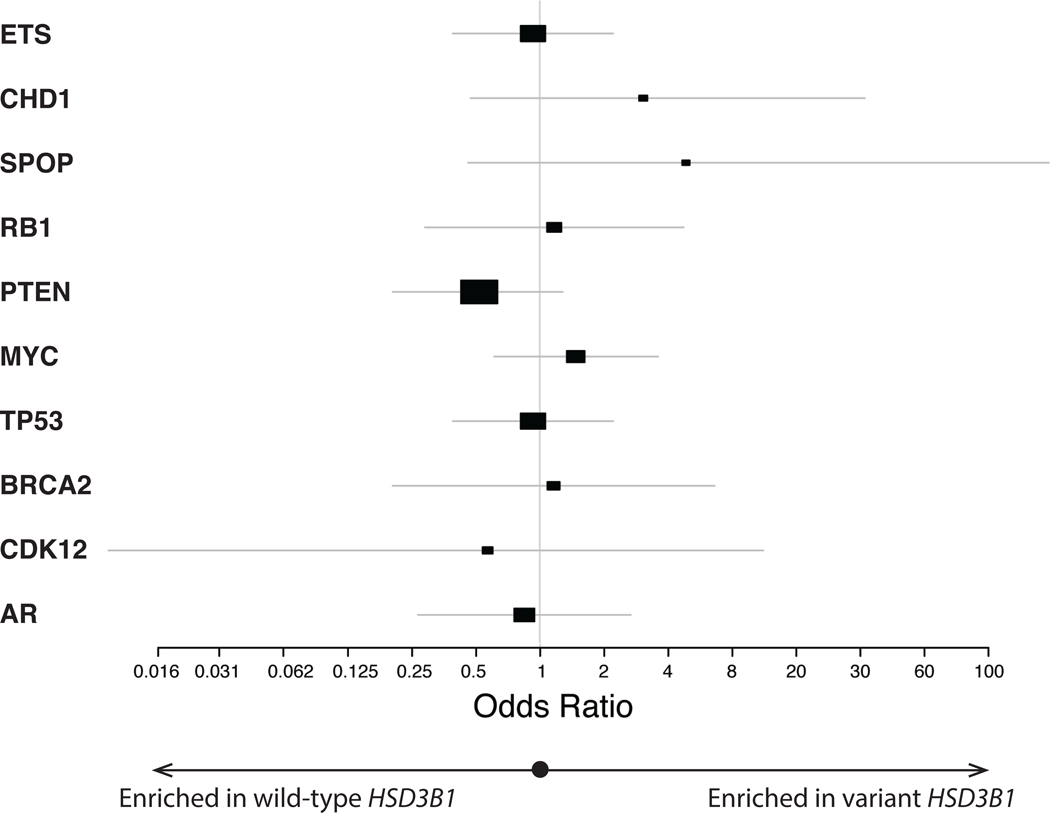
We next sought to assess the association of these HSD3B1 germline variants with molecular changes in the mCRPC tumors themselves by utilizing WGS and RNA-seq on metastatic tumor biopsies obtained from these patients. Given the frequent joint inheritance of the HSD3B1(1245A>C) and rs1856888 variant polymorphisms and prior studies suggesting both may be important in ADT response10,31, we grouped the two together for the purposes of assessing associations with somatic tumor alterations. We identified the most common somatic DNA alterations in mCRPC15 and found that the HSD3B1 germline variant was not significantly associated with any of these (Figure 2, Table S1). To assess whether the HSD3B1 germline variant was prognostic of OS independently of bi-allelic RB1 loss in the tumor, which has been recently highlighted as the key somatic DNA alteration associated with poor prognosis in mCRPC16,32, we performed a multivariable survival analysis including both variant rs1856888 and bi-allelic RB1 loss along with well-characterized, adverse clinicopathologic features. The variant HSD3B1 allele was associated with shorter OS after ADT independently of bi-allelic RB1 loss and other clinicopathologic features (HR:1.85[1.01–3.37], P=0.045; Table 2).
Figure 2:
Forest plot showing the association between variant germline HSD3B1 and common somatic DNA alterations in mCRPC tumor biopsies. A odds ratio > 1 indicates positive association between variant HSD3B1 and the somatic tumor alteration of interest. Details on the prevalence of each somatic alteration stratified by HSD3B1 status as well as exact odds ratios including 95% confidence intervals can be found in Table S1.
Table 2.
Multivariable analysis including genomic and clinicopathologic features
| HR | LHR | UHR | P-val | |
|---|---|---|---|---|
| Bi-allelic RB1 loss | 3.01 | 1.40 | 6.46 | 0.005* |
| Germline variant HSD3B1 | 1.85 | 1.01 | 3.37 | 0.045* |
| Metastases present at ADT initiation | 3.21 | 1.72 | 5.98 | 0.0002* |
| ECOG ≥ 1 | 1.25 | 0.70 | 2.21 | 0.45 |
| Gleason ≥ 8 | 0.95 | 0.55 | 1.64 | 0.85 |
| Treatment with enzalutamide / abiraterone | 1.00 | 0.54 | 1.85 | 0.995 |
| log(PSA) at biopsy | 1.54 | 1.01 | 2.35 | 0.046* |
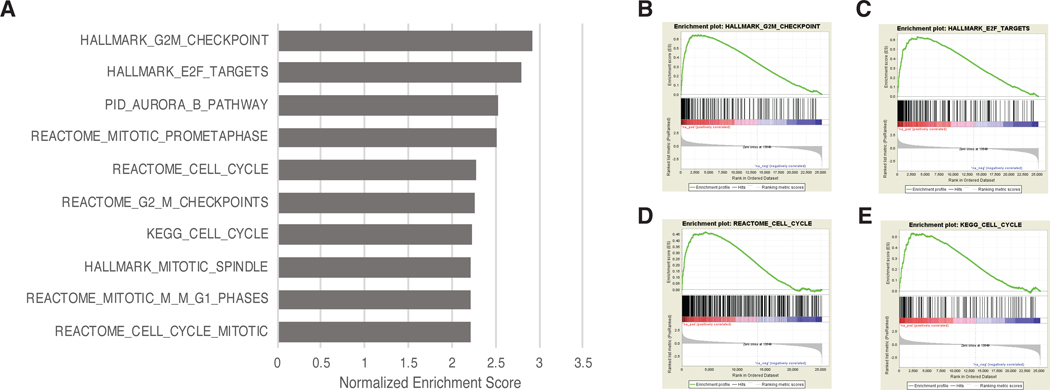
While the HSD3B1 germline variant was not associated with any of the common somatic DNA alterations in mCRPC, we wanted to examine if there were differences in the tumor gene expression patterns between the HSD3B1 variant and wild type patients. We performed differential expression analysis comparing the HSD3B1 variant and wild type groups and used GSEA to perform pathway analysis. Strikingly, 10 out of the top 10 pathways differentially overexpressed in the tumors of patients with variant HSD3B1 were related to proliferation and cell cycle regulation (Figure 3) suggesting an interplay between the germline genomics and the tumor gene expression.
Figure 3:
A) Top 10 gene sets significantly overexpressed in the tumors of patients with variant HSD3B1 based on Gene Set Enrichment Analysis (GSEA) performed on tumor RNA-seq data using the GSEA Preranked tool17. B-D) GSEA plots of four cell-cycle related pathways significantly overexpressed in tumors of patients with variant HSD3B1.
DISCUSSION
We performed a comprehensive assessment of all androgen synthesis germline variants previously suggested to be associated with ADT response and found that one germline HSD3B1 variant (rs1856888) was significantly associated with impaired survival outcomes and that the HSD3B1(1245A>C) polymorphism was borderline significantly associated. We also leveraged somatic variant data generated from whole genome sequencing and RNA-seq experiments to perform the first assessment of HSD3B1 germline variants in relation to somatic tumor DNA-seq and RNA-seq alterations.
Of all the germline variants interrogated in nine androgen metabolism genes, only variant HSD3B1 was found to be associated with impaired OS. The differences in findings between the present and prior studies may be attributable to the relatively modest sample sizes in each study. Additionally, the ethnic composition of the present cohort and those examined in many prior studies was different, with many of the previous cohorts comprised predominantly of East Asian patients19–21,27 who are known to have different prevalence of germline variants and potentially different genomic risk factors than Caucasian patients13,21. Additional studies are needed to validate the clinical relevance of these germline variants across different ethnic groups.
HSD3B1(1245A>C) is the most well-characterized polymorphism with respect to its effects on androgen synthesis9,33,34 and response to ADT12,13,26,31 in prostate cancer. Furthermore, HSD3B1(1245A>C) is associated with shorter OS in men with low volume metastatic CSPC in a phase III trial of ADT vs. ADT and docetaxel35. HSD3B1(1245A>C) was investigated in a recent Japanese study, which found that HSD3B1(1245A>C) was associated with impaired progression-free survival but no difference in OS in mCSPC, and improved response to abiraterone acetate in mCRPC26. However, given the low number of patients with HSD3B1(1245A>C) in that prior study and known low prevalence of the HSD3B1(1245A>C) polymorphism in patients of East Asian descent13, it is unclear whether these findings were generalizable to cohorts of different ethnic composition. In our study of predominantly Caucasian patients, we observed that 42% of patients demonstrated the HSD3B1(1245A>C) variant, which was higher than the 14.1% observed in the Japanese mCRPC study and comparable to the 47.9% observed in a different predominantly Caucasian mCRPC cohort13. Of note, all cases of variant HSD3B1(1245A>C) were attributable to germline alterations, as there were no somatic mutations identified at this locus in our cohort. We found in our cohort that median OS was shorter in patients with variant HSD3B1 than those homozygous wild-type, with a borderline-significant survival difference (P=0.052).
While most studies have focused on the HSD3B1(1245A>C) polymorphism, one prior study in metastatic CSPC highlighted rs1856888, a SNP 13kb upstream of HSD3B1 as a prognostic factor associated with significantly shorter time to progression on primary ADT10. Our study found variant rs1856888 to be associated with poor prognosis with respect to OS as well. Additionally, our study is the first to report a high frequency of HSD3B1(1245A>C) and variant rs1856888 co-inheritance. Given the high frequency of co-inheritance, the relationship between the two variants and their individual contributions to the clinically-aggressive mCRPC phenotype are difficult to resolve. While the mechanistic significance of the missense-encoding HSD3B1(1245A>C) polymorphism and its effects on enzyme stabilization have been elucidated, the mechanistic consequences of rs1856888 are unknown. Additional studies are needed to assess the functional and clinical relevance of the rs1856888 variant independent of HSD3B1(1245A>C) status.
We found that inheritance of a HSD3B1 germline variant was not associated with common somatic DNA-sequencing alterations in mCRPC. Somatic DNA alterations would not necessarily be expected to be associated with germline variants in androgen metabolism genes, since, unlike variant HSD3B1, none of the common somatic alterations examined (with the exception of AR amplification36,37, TP53 mutations, and RB1 loss15,38) are known to be enriched in mCRPC relative to mCSPC. As recent studies found that RB1 loss was the key somatic alteration associated with impaired outcomes in mCRPC16,39, we performed a multivariable analysis including both RB1 loss and variant HSD3B1 along with other clinicopathologic factors and found that both were independently prognostic of shorter OS after primary ADT. This further supports the independence between the germline HSD3B1 variant and somatic RB1 loss. Investigation in larger cohorts is needed to confirm these findings and to examine the association between HSD3B1 germline variants and lower-prevalence DNA-seq alterations. In our RNA expression-based gene set enrichment analysis of metastasis biopsies, we found that the HSD3B1 germline variant was associated with increased expression of cell proliferation genes. These findings demonstrate that variant HSD3B1 may be associated with more molecularly as well as clinically aggressive tumors.
There are several limitations of the present study. First, we report on a multi-institutional cohort of relatively modest sample size, and no external validation cohort was available for this analysis. With this in mind, we chose to focus on a relatively succinct set of 17 germline variants previously reported to be associated with impaired clinical outcomes. Still yet, multiple survival comparisons were made, and the nominal P-values reported should be interpreted accordingly. Future work in independent cohorts is needed to validate the prognostic significance of HSD3B1 polymorphisms. Another limitation of our study is that data on PSA dynamics (such as PSA doubling time) after ADT initiation were unavailable. Thus, we were unable to directly assess the prognostic significance of variant HSD3B1 with respect to response to primary ADT. Further investigation of the clinical implications of variant HSD3B1 is needed to potentially guide treatment decisions.
In summary, we applied whole-genome germline and somatic sequencing approaches to paired normal tissue and tumor biopsy samples to perform an integrated analysis of both inherited and acquired predictors of clinical outcomes. Future studies may shed further insights into the interplay between the germline HSD3B1 variants in advanced prostate cancer.
Supplementary Material
ACKNOWLEDGEMENTS
RA, NS, ES, and SGZ are supported by the Prostate Cancer Foundation. This work was supported in part with support from the National Cancer Institute (R01CA172382 and R01CA190289 to N.S.).
Stand Up To Cancer-Prostate Cancer Foundation Prostate Cancer Dream Team Award (SU2C-AACR-DT0812 to E.J.S.). Stand Up To Cancer (SU2C) is a division of the Entertainment Industry Foundation. This research grant was administered by the American Association for Cancer Research, the scientific partner of SU2C.
CONFLICTS OF INTEREST
SGZ has patents filed unrelated to this work with Decipher Biosciences and Celgene. A patent for 3β-hydroxysteroid dehydrogenase in steroid-dependent disease has been filed by Cleveland Clinic.
JJA has performed consulting or held an advisory role with Astellas Pharma, Bayer, Merck, and Janssen Biotech, Inc.
KNC has received grant support and honoraria from Astellas Pharma, Janssen, Sanofi, AstraZeneca, Bayer, Essa Pharma, Pfizer, and Roche.
REFERENCES
- 1.Eeles RA. Genetic predisposition to prostate cancer. Prostate Cancer Prostatic Dis. 2, 9–15 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, Graff RE, et al. The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol. Biomarkers Prev. 23, 2303–2310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dias A, Kote-Jarai Z, Mikropoulos C & Eeles R. Prostate Cancer Germline Variations and Implications for Screening and Treatment. Cold Spring Harb Perspect Med 8, a030379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimoto N, Shiota M, Tomisaki I & Minato A. Gene Polymorphism-related Individual and Interracial Differences in the Outcomes of Androgen Deprivation Therapy for Prostate Cancer. Clinical Genitourinary Cancer 15, 337–342 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Chang K-H, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 13728–13733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehman Y & Rosenberg JE Abiraterone acetate: oral androgen biosynthesis inhibitor for treatment of castration-resistant prostate cancer. Drug Des Devel Ther 6, 13–18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James ND, de Bono JS, Spears MR, Clarke NW, Mason MD, Dearnaley DP, et al. Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N Engl J Med 377, 338–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alyamani M, Emamekhoo H, Park S, Taylor J, Almassi N, Upadhyay S, et al. HSD3B1(1245A>C) variant regulates dueling abiraterone metabolite effects in prostate cancer. Journal of Clinical Investigation 128, 3333–3340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross RW, Oh WK, Xie W, Pomerantz M, Nakabayashi M, Sartor O, et al. Inherited variation in the androgen pathway is associated with the efficacy of androgen-deprivation therapy in men with prostate cancer. J. Clin. Oncol. 26, 842–847 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Shiota M, Narita S, Akamatsu S, Fujimoto N, Sumiyoshi T, Fujiwara M, et al. Association of Missense Polymorphism in HSD3B1 With Outcomes Among Men With Prostate Cancer Treated With Androgen-Deprivation Therapy or Abiraterone. JAMA Netw Open 2, e190115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almassi N, Reichard C, Li J, Russell C, Perry J, Ryan CJ, et al. HSD3B1 and Response to a Nonsteroidal CYP17A1 Inhibitor in Castration-Resistant Prostate Cancer. JAMA Oncol 4, 554–557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hearn JWD, AbuAli G, Reichard CA, Reddy CA, Magi-Galluzzi C, Chang K-H, et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 17, 1435–1444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hur J, Schuyler AD, States DJ & Feldman EL SciMiner: web-based literature mining tool for target identification and functional enrichment analysis. Bioinformatics 25, 838–840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 174, 758–769.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen WS, Aggarwal R, Zhang L, Zhao SG, Thomas GV, Beer TM, et al. Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. European Urology S0302283819302064 (2019) doi: 10.1016/j.eururo.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lévesque É, Huang S-P, Audet-Walsh É, Lacombe L, Bao B-Y, Fradet Y, et al. Molecular markers in key steroidogenic pathways, circulating steroid levels, and prostate cancer progression. Clin. Cancer Res. 19, 699–709 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Shiota M, Fujimoto N, Yokomizo A, Takeuchi A, Itsumi M, Inokuchi J, et al. SRD5A gene polymorphism in Japanese men predicts prognosis of metastatic prostate cancer with androgen-deprivation therapy. Eur. J. Cancer 51, 1962–1969 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Shiota M, Fujimoto N, Yokomizo A, Takeuchi A, Kashiwagi E, Dejima T, et al. The prognostic impact of serum testosterone during androgen-deprivation therapy in patients with metastatic prostate cancer and the SRD5A2 polymorphism. Prostate Cancer Prostatic Dis. 19, 191–196 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto N, Kubo T, Inatomi H, Bui HTT, Shiota M, Sho T, et al. Polymorphisms of the androgen transporting gene SLCO2B1 may influence the castration resistance of prostate cancer and the racial differences in response to androgen deprivation. Prostate Cancer Prostatic Dis. 16, 336–340 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Harshman LC, Xie W, Nakabayashi M, Qu F, Pomerantz MM, et al. Association of SLCO2B1 Genotypes With Time to Progression and Overall Survival in Patients Receiving Androgen-Deprivation Therapy for Prostate Cancer. J. Clin. Oncol. 34, 352–359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Xie W, Mostaghel E, Nakabayashi M, Werner L, Sun T, et al. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J. Clin. Oncol. 29, 2565–2573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharifi N, Hamada A, Sissung T, Danesi R, Venzon D, Baum C, et al. A polymorphism in a transporter of testosterone is a determinant of androgen independence in prostate cancer. BJU Int. 102, 617–621 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanda S, Tsuchiya N, Narita S, Inoue T, Huang M, Chiba S, et al. Effects of functional genetic polymorphisms in the CYP19A1 gene on prostate cancer risk and survival. Int. J. Cancer 136, 74–82 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Shiota M, Fujimoto N, Tsukahara S, Ushijima M, Takeuchi A, Kashiwagi E, et al. The impact of genetic polymorphism on CYP19A1 in androgen-deprivation therapy among Japanese men. Cancer Chemother. Pharmacol. 83, 933–938 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Yamada T, Nakayama M, Shimizu T, Nonen S, Nakai Y, Nishimura K, et al. Genetic polymorphisms of CYP17A1 in steroidogenesis pathway are associated with risk of progression to castration-resistant prostate cancer in Japanese men receiving androgen deprivation therapy. Int. J. Clin. Oncol. 18, 711–717 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Yu C-C, Huang S-P, Lee Y-C, Huang C-Y, Liu C-C, Hour T-C, et al. Molecular markers in sex hormone pathway genes associated with the efficacy of androgen-deprivation therapy for prostate cancer. PLoS ONE 8, e54627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karunasinghe N, Zhu Y, Han DY, Lange K, Zhu S, Wang A, et al. Quality of life effects of androgen deprivation therapy in a prostate cancer cohort in New Zealand: can we minimize effects using a stratification based on the aldo-keto reductase family 1, member C3 rs12529 gene polymorphism? BMC Urol 16, 48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal N, Hahn AW, Gill DM, Farnham JM, Poole AI & Cannon-Albright L. Independent Validation of Effect of HSD3B1 Genotype on Response to Androgen-Deprivation Therapy in Prostate Cancer. JAMA Oncol 3, 856–857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hearn JWD, Xie W, Nakabayashi M, Almassi N, Reichard CA, Pomerantz M, et al. Association of HSD3B1 Genotype With Response to Androgen-Deprivation Therapy for Biochemical Recurrence After Radiotherapy for Localized Prostate Cancer. JAMA Oncol 4, 558–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen WS, Alshalalfa M, Zhao SG, Liu Y, Mahal BA, Quigley DA, et al. Novel RB1-Loss Transcriptomic Signature Is Associated with Poor Clinical Outcomes across Cancer Types. Clin Cancer Res 25, 4290–4299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang K-H, Li R, Kuri B, Lotan Y, Roehrborn CG, Liu J, et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell 154, 1074–1084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabharwal N & Sharifi N. HSD3B1 Genotypes Conferring Adrenal-Restrictive and Adrenal-Permissive Phenotypes in Prostate Cancer and Beyond. Endocrinology 160, 2180–2188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hearn JWD, Sweeney C, Almassi N, Reichard CA, Reddy CA, Hobbs B, et al. HSD3B1 and overall survival (OS) in men with low-volume (LV) metastatic prostate cancer (PCa) treated with androgen deprivation therapy (ADT) or chemohormonal therapy in the CHAARTED Randomized trial. JCO 37, 5020–5020 (2019). [Google Scholar]
- 36.Chi K, Hotte SJ, Joshua AM, North S, Wyatt AW, Collins LL, et al. Treatment of mCRPC in the AR-axis-targeted therapy-resistant state. Ann Oncol 26, 2044–2056 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.