Abstract
The transitions between developmental stages are critical points in the Plasmodium life cycle. The development of Plasmodium in the livers of their mammalian hosts bridges malaria transmission and the onset of clinical symptoms elicited by red blood cell infection. The egress of Plasmodium parasites from the liver must be a carefully orchestrated process to ensure a successful switch to the blood stage of infection. Cysteine protease activity is known to be required for liver-stage Plasmodium egress, but the crucial cysteine protease(s) remained unidentified. Here, we characterize a member of the papain-like cysteine protease family, Plasmodium berghei serine repeat antigen 4 (PbSERA4), that is required for efficient initiation of blood-stage infection. Through the generation PbSERA4-specific antisera and the creation of transgenic parasites expressing fluorescently tagged protein, we show that PbSERA4 is expressed and proteolytically processed in the liver and blood stages of infection. Targeted disruption of PbSERA4 results in viable and virulent blood-stage parasites. However, upon transmission from mosquitoes to mice, Pbsera4(-) parasites displayed a reduced capacity to initiate a new round of asexual blood-stage replication. Our results from cultured cells indicate that this defect results from an inability of the PbSERA4-deficient parasites to egress efficiently from infected cells at the culmination of liver-stage development. Protection against infection with wildtype P. berghei could be generated in animals in which Pbsera4(-) parasites failed to establish infection. Our findings confirm that liver-stage merozoite release is an active process and demonstrate that this parasite-encoded cysteine protease contributes to parasite escape from the liver.
Author summary
Plasmodium parasites cause over 200 million cases of malaria every year. When parasites are transmitted by mosquito bite, they initially colonize the liver before they move into the blood and cause disease. During successful transition from the liver into the blood, Plasmodium cloak themselves in host plasma membrane as they egress from the liver cells. Although some aspects of how Plasmodium exit their host hepatocytes appear unique, certain attributes are shared across diverse pathogens. For example, protease activity is required not only for multiple stages of Plasmodium exit, but is also involved in the egress of some bacteria and other protozoan. Here we characterize a protease in Plasmodium berghei that is expressed in the liver and conserved across Plasmodium species. Through gene targeting, we found PbSERA4 is required for efficient egress of Plasmodium from the liver. In the absence of this protease the transition between the liver and blood stages of growth is prolonged due to inefficient parasite release from liver cells. These findings provide new insights into the function of a conserved Plasmodium protease and into the process of Plasmodium escape from the liver.
Introduction
For a parasitic disease to be maintained in a population, each step of an often complex life cycle must be sustained, and successful transitions between these steps are essential. Plasmodium parasites, which are responsible for malaria, alternate between a vertebrate and an insect host. Inside the vertebrate hosts, Plasmodium parasites enter suitable target cells and proliferate within an intracellular compartment termed the parasitophorous vacuole [1, 2]. As a consequence, parasites have to actively enter and exit their host cell in order to progress in the life cycle. Pathogen exit from host cells is a complex series of events, typically involving both pathogen-encoded factors as well as host-derived proteins [3, 4].
A hallmark of the Plasmodium life cycle in the mammalian host is the initial obligatory parasite population expansion phase inside the mammalian liver. Prior to the pathogenic asexual parasite propagation in red blood cells, Plasmodium parasites infect hepatocytes, where they transform from the motile sporozoite transmission form into spherical trophozoites that initiate multiple rounds of closed mitosis and form thousands of first-generation liver-stage merozoites [5, 6]. In order to egress from the liver, these merozoites escape the parasitophorous vacuole to the host hepatocyte cytoplasm, which requires a Plasmodium phospholipase [7, 8]. Upon parasitophorous vacuole membrane breakdown, the host cell architecture is altered to allow the budding of host-cell-derived extracellular vesicles containing infectious merozoites [7, 9]. These so-called merosomes emerge directly into the liver sinusoids and arrest in the lung capillaries where merozoites are discharged and invade neighboring erythrocytes to commence blood infection [6], the exclusive cause of clinical malaria symptoms.
Inhibitor studies revealed a crucial role for cysteine protease activity in egress of either liver-stage or blood-stage merozoites [7, 10–13], but the identity of the essential liver-stage cysteine proteases remains undetermined. A family of papain-like proteases termed the serine-rich antigen (SERA) family is encoded by all Plasmodium species and includes members that are expressed in the liver stage of infection [10, 14]. Several of the SERA proteins are important for exit of Plasmodium at different stages of development. For example, the Plasmodium berghei PbSERA5/ECP1 is required for egress of sporozoites from oocysts in the mosquito midgut [15], whereas the Plasmodium falciparum PfSERA5 and PfSERA6 are important for exit of merozoites from blood-stage schizonts [16, 17]
SERA family members have been shown to be processed by the Plasmodium subtilisin-like protease 1, SUB1, in the parasitophorous vacuole prior to parasite egress [18]. In infected erythrocytes, cleavage and activation of SERAs is triggered by secretion of SUB1 into the parasitophorous vacuole. This proteolytic cascade is followed by breakdown of the parasitophorous vacuole and eventual release of blood-stage merozoites [11, 12]. SUB1 is not only required for egress of blood-stage Plasmodium; P. berghei SUB1 was also shown to be essential for liver-stage parasite egress [19, 20]. One of the SERA genes that shows high transcript levels during late liver-stage development is PbSERA4 [10, 14]. PbSERA4 is a cysteine-type SERA that is well conserved across the known rodent- and primate-infecting Plasmodium species [21]. Here, targeted PbSERA4 gene disruption reveals a role for this putative protease in the process of Plasmodium escape from the liver.
Results
The Plasmodium berghei SERA4 cysteine protease is expressed at multiple stages in the parasite life cycle
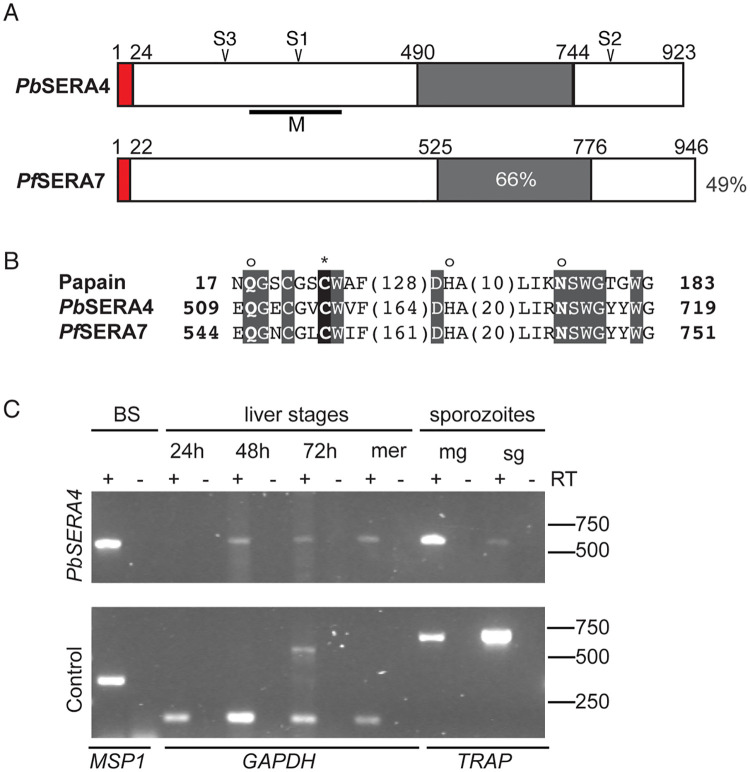
The Plasmodium berghei PbSERA4 (PBANKA_0304800) gene encodes a cysteine-type SERA and is conserved and syntenic across the primate and rodent Plasmodium species [21]. The predicted orthologue of PbSERA4 in Plasmodium falciparum is PfSERA7 (PF3D7_0207400), and the proteins encoded by these genes are 44% identical and 57% similar (Fig 1A). Within the signature papain family cysteine protease domain (PFAM: PF00112), PbSERA4 and PfSERA7 share 66% amino acid identity (Fig 1A) and include the strictly conserved catalytic residues, i.e. an amino-terminal glutamine, the catalytic cysteine residue, a central histidine, and a carboxy-terminal asparagine (Fig 1B).
Fig 1. Plasmodium berghei SERA4 is a papain-like cysteine protease expressed across the Plasmodium life cycle.
(A) Schematic structure of P. berghei PbSERA4 (PBANKA_0304800) and its ortholog in P. falciparum PfSERA7 (PF3D7_0207400). The putative signal sequences and the central papain-like cysteine protease domains are boxed in red and grey, respectively. Putative cleavage sites S1, S2 and S3 are as indicated at amino acids 295 (IKGEDDLD), 806 (ISGQTDNQ) and 173 (LTAKIEED), respectively. Antibodies were generated against the portions of PbSERA4 indicated by the black line labeled M. Amino acid sequence identities between PbSERA4 and PfSERA7 are indicated as percent for both the overall sequence and the catalytic domain. (B) The catalytic residues of the papain family of cysteine proteases are conserved in PbSERA4 and PfSERA7. Strictly conserved amino acid residues are boxed in grey, and the putative active-site cysteine is boxed in black and marked with an asterisk. The catalytic histidine and flanking glutamine and asparagine residues, which form the oxyanion hole, are marked with an ‘o’. Papain is from Carica papaya. Bold numbers represent the amino acid positions within the protein before and after the catalytic domain; numbers in parenthesis represent the number of intervening amino acids at the indicated positions. (C) Expression profiling of PbSERA4 in P. berghei ANKA. RT-PCR analysis from mixed blood stages (BS), liver stages (at 24, 48, and 72 hours after infection), liver stage merozoites (mer), and midgut (mg) and salivary gland (sg) sporozoites. Control transcripts were merozoite surface protein 1 (MSP1), glyceraldehyde dehydrogenase (GAPDH) or thrombospondin-related anonymous protein (TRAP) for blood, liver, and mosquito stages, respectively. Products run at the expected sizes in comparison to these products amplified from P. berghei genomic DNA (S1A Fig).
To profile PbSERA4 expression throughout the Plasmodium life cycle, we employed standard RT-PCR analysis (Fig 1C and S1A Fig). PbSERA4 transcripts are detectable in blood-stage P. berghei and can be detected in mid and late liver-stage parasites but are below the detection threshold in early liver stages. In the insect vector, PbSERA4 transcripts can be detected in sporozoites isolated from the midguts or salivary glands of infected mosquitoes. Together, these data support our previous quantification of PbSERA transcripts in the mammalian host [10, 14] and confirm global transcript profiling data of both P. berghei [22] and P. falciparum [23–25]. Expression of PbSERA4 in several developmental stages indicates functions during multiple points of the Plasmodium life cycle.
Expression and localization of PbSERA4 in late liver-stage P. berghei
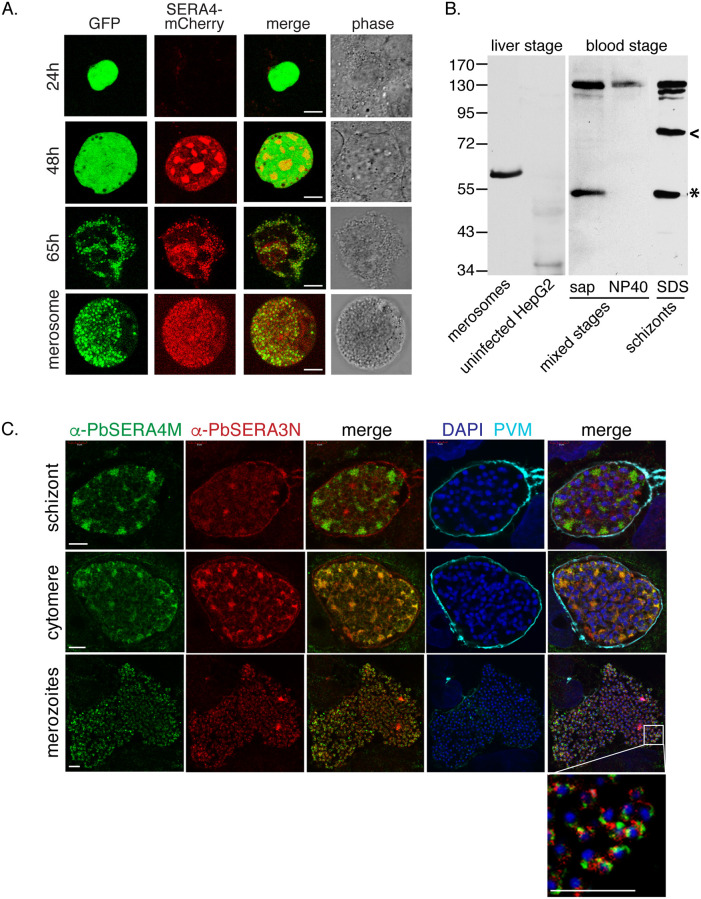
To examine protein expression and assess the localization of PbSERA4, we generated a transgenic P. berghei ANKA line that expresses fluorescently tagged PbSERA4 from the native PbSERA4 promoter (S1B Fig). Diagnostic PCR confirmed the integration of the targeting vector, which results in C-terminal tagging of PbSERA4 with mCherry (S1C Fig). We then followed the life cycle progression of the resulting transgenic PbSERA4-mCherry parasite line, which expresses both constitutive cytoplasmic green fluorescence [26], and the red fluorescently tagged PbSERA4 protein. PbSERA4-mCherry parasites progressed normally through the Plasmodium life cycle, including transmission to Anopheles mosquitoes and back to mice, indicating that expression of endogenously tagged PbSERA4 is compatible with parasite viability in all stages. Consistent with reported transcriptome data [27, 28], we detected PbSERA4-mCherry signal in late asexual blood stages and gametocytes (S1D Fig), and we could observe red fluorescence throughout exflagellation of male gametes. We failed to detect a red fluorescent signal in PbSERA4-mCherry sporozoites isolated from mosquito midguts or salivary glands despite the presence of PbSERA4 transcripts (Fig 1C)[25]. We could confirm the absence of protein expression in sporozoites using a parasite line we generated in which the PbSERA4 promoter drives GFP expression (S2 Fig). In these parasites, low levels of GFP are detected in oocysts but not in sporozoites isolated from salivary glands. In contrast, GFP expressed under control of the promoter of PbSERA5, which is known to function in the egress of sporozoites from oocysts, is clearly detectable in sporozoites. Neither GFP expressed from the PbSERA4 promoter (S2 Fig), nor mCherry signal in PbSERA4-mCherry parasites (Fig 2A) could be detected in early liver-stage P. berghei. However fluorescent protein expression in both lines is evident later in the liver stage. PbSERA4-mCherry could be detected 48 hours after infection of cultured cells with PbSERA4-mCherry sporozoites (Fig 2A). The PbSERA4-mCherry signal progressively increased, and late liver schizonts and liver merozoites exhibited bright fluorescence.
Fig 2. Proteolytic processing and localization of PbSERA4 in late liver-stage P. berghei.
(A) PbSERA4-mCherry is detected in late liver-stage P. berghei. Huh7 hepatoma cells were infected with the PbSERA4-mCherry P. berghei ANKA line, fixed at the indicated time points and imaged by confocal microscopy. The parasites were detected with constitutive, cytoplasmic GFP expression (green). Scale bars, 8 μm. (B) The α-PbSERA4M antibody detects full length PbSERA4 and PbSERA4 cleavage products. Liver-stage lysates were prepared from merosomes and detached cells (lane 1) collected from infected HepG2 cells and compared to uninfected HepG2 cells (lane 2). Proteins from mixed purified blood-stage parasites were initially solubilized with saponin and subsequently NP40 and compared to whole-cell SDS lysates of purified mature blood-stage schizonts. The predicted catalytic domain released by proteolytic processing is marked by the asterisk and appears to run slower in liver-stage compared to blood-stage samples. All displayed lanes are from one gel. (C) P. berghei ANKA-infected HepG2 cells were fixed at different time points (45-58h) post infection and stained with α-PbSERA4M (green) in comparison to α-PbSERA3N (red), α-PbEXP1 (cyan), which labels the parasitophorous vacuole membrane (PVM), and the DNA dye DAPI (blue). For easier interpretation, images obtained with α-PbSERA3N and α-PbSERA4M were merged individually. Additional merges with images obtained with DAPI and α-PbEXP1 are shown for orientation. Scale bar: 5μm.
In parallel to this tagging approach, we generated an antibody against the central domain of PbSERA4 (α-PbSERA4M) (Fig 1A). Because the central region of the SERA proteins is relatively conserved, we tested the specificity of the α-PbSERA4M antiserum against recombinant purified central domains of the P. berghei proteins PbSERA1-4 and found α-PbSERA4M to exclusively recognize PbSERA4 (S3 Fig). When lysates of liver-stage and blood-stage P. berghei were probed by western blot with α-PbSERA4M, the detection of multiple fragments indicates that, like other SERA proteins, PbSERA4 is proteolytically processed during infection. We predicted putative SUB1 cleavage sites within PbSERA4 based on the cleavage pattern of other SERA proteins, and on the previously reported consensus recognition motif of SUB1 [18] (Fig 1A). In addition to the full-length protein, which is predicted to be 107kDa, the α-PbSERA4M antibody recognizes a protein in the lysates of blood-stage schizonts that may represent the central catalytic domain, which is predicted to be 59kDa, as well as an intermediate cleavage product. If the order of PbSERA4 processing occurs as described for PfSERA5 [29, 30], we would expect this intermediate cleavage product to represent the product C-terminal to the predicted S1 cleavage site and have the predicted size of ~73 kDa. The intermediate cleavage product we detected corresponds to a slightly larger size (Fig 2B and S4C Fig, arrowhead) and could represent the N-terminal product that would be generated by cleavage only at the S2 cleavage site.
Proteolytic processing of other Plasmodium SERAs has been shown to occur in the parasitophorous vacuole prior to schizont rupture [18, 31]. To biochemically distinguish the parasitophorous vacuole from the parasite, purified mixed blood stages, which comprises mostly trophozoites, were treated with saponin to lyse the red blood cell and parasitophorous vacuole (Fig 2B). The putative catalytic domain-containing cleavage product appears in the saponin-soluble fraction, whereas the lysate of the remaining parasite-containing fraction contained only the full-length PbSERA4 protein. This data suggests that in blood-stage P. berghei, the proteolytic processing of PbSERA4 occurs in the parasitophorous vacuole. We also probed lysates from merosomes generated from sporozoite-infected hepatoma cells (Fig 2B). Here, only one band of approximately 60kDa is detected. In contrast to the sample containing blood-stage schizonts, we did not detect full length PbSERA4 protein. This result may be due to greater enrichment of late-stage parasites in the merosome sample in comparison to the blood-stage schizont sample, which contains late trophozoites and early schizonts. The band detected in the merosomes appears to run higher on the gel than the putative PbSERA4 proteolytic domain from the blood-stage parasites, which may be due to different properties of the samples or may indicate modifications or alternate processing. We were unable to detect protein with the α-PbSERA4M antibody in late liver stage-infected cells, likely due to the low infection rate relative to the sensitivity of the antibody. Still, these results suggest that PbSERA4 likely undergoes proteolytic processing by the end of the liver infection stage.
Using this antibody for immunofluorescence allowed detection of PbSERA4 in late liver-stage P. berghei and direct comparison to the localization of PbSERA3, which is also abundantly expressed during late liver infection [10] (Fig 2C). In P. berghei-infected hepatoma cells the α-PbSERA4M antibody recognized distinct regions in liver-stage schizonts, and in cytomeres the two signals overlapped in the parasite. Upon release of parasites from the parasitophorous vacuole, both PbSERA4 and PbSERA3 are found inside the first-generation merozoites with distinct localizations (Fig 2C). While PbSERA3 is also detected surrounding the parasite in the parasitophorous vacuole, the α-PbSERA4M signal appears restricted to the parasite. Together our findings show that PbSERA4 is expressed and proteolytically processed in the liver and blood stages of infection and that protein expression is limited to the intermediate vertebrate host.
Absence of PbSERA4 delays the onset of blood-stage infection
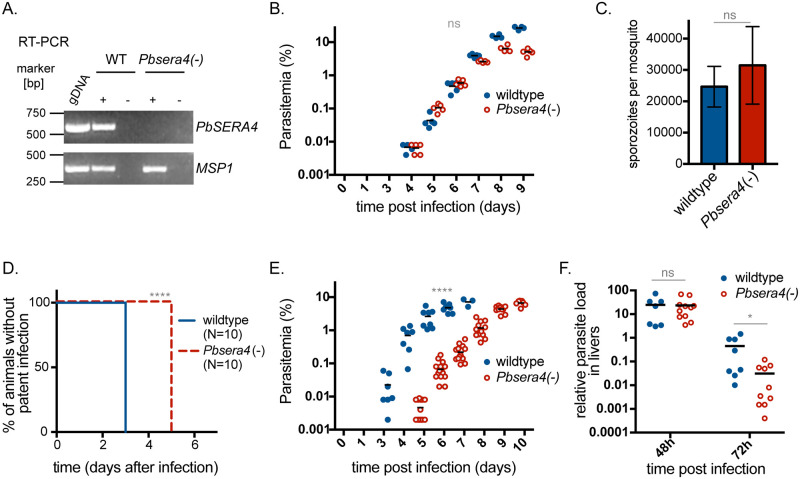
To analyze the contribution of PbSERA4 to parasite development and life cycle progression, we generated a Pbsera4 knockout line. In a P. berghei ANKA line constitutively expressing GFP [26] the PbSERA4 gene was replaced by the Tgdhfr/ts positive selection cassette through double homologous recombination (S4A Fig). After pyrimethamine-driven selection and cloning by limited dilution, the gene deletion in the Pbsera4(-) line was confirmed by diagnostic PCR, which showed the expected alteration of the genomic DNA (S4B Fig), RT-PCR, which confirmed absence of PbSERA4 transcripts (Fig 3A), and western blot, which verified absence of the PbSERA4 protein (S4C Fig).
Fig 3. Absence of PbSERA4 delays the time to appearance of Plasmodium-infected red blood cells.
(A) Depletion of PbSERA4 transcripts in Pbsera4(-)-ANKA parasites. cDNA from WT and Pbsera4(-) late-stage schizonts were used to verify the SERA4 specific PCR reaction. MSP1 transcript was used as a positive control. RT-PCR was performed in the presence (+) or absence (-) of reverse transcriptase (RT). Parasite genomic DNA (gDNA) was included as an amplification control. (B) Blood stage growth of Pbsera4(-)-ANKA P. berghei parasites. Naïve NMRI mice (n = 5) were injected intravenously with 1,000 wildtype or 1,000 Pbsera4(-) P. berghei-infected erythrocytes. Infection was then monitored by daily examination of Giemsa-stained blood smears to determine the parasitemia. ns, not significant (unpaired t-test, day 6 values). (C) Development of Pbsera4(-)-ANKA P. berghei in mosquitoes. Sporozoites extracted from salivary glands of infected mosquitoes were quantified 17–25 days after infection. Data is presented as the mean +/- SD. ns, not significant (paired t-test, n = 4 independent experiments). (D and E) Delayed patency following Pbsera4(-)-ANKA sporozoite infection. C57BL/6 mice were infected intravenously with 10,000 sporozoites, and Giemsa-stained blood smears were monitored daily to determine presence of parasites (D) ****, p<0.0001, Log-rank (Mantel-Cox) test, n = 10) and parasitemia (E) ****, p <0.0001 (unpaired t test, day 6 values). (F) Pbsera4(-)-ANKA parasites exhibit no growth defects in infected livers. RNA was extracted from livers of C57BL/6 mice infected with 50,000 to 100,000 sporozoites at the indicated time points. Levels of Pb18s rRNA were normalized to levels of mouse HPRT RNA by qPCR. ns, not significant; *, p < 0.05 (unpaired t-test).
Successful gene deletion indicated that although PbSERA4 transcripts were detected at high levels during late blood stage development, PbSERA4 is not essential for asexual blood-stage development. The effect of PbSERA4 deletion on blood-stage growth was specifically tested by initiating infection of NMRI mice with intravenous delivery of infected red blood cells (Fig 3B). The time until appearance of infected cells in the blood and the growth rate of the Pbsera4(-) line were indistinguishable from wildtype P. berghei. Gametocytes were readily detected in the blood from mice infected with the Pbsera4(-) line, and we observed no defect in transmission of this line to mosquitoes, as evidenced by high numbers of sporozoites in the salivary glands of Pbsera4(-)-infected mosquitoes (Fig 3C). These findings indicate that PbSERA4 is dispensable for Plasmodium blood-stage development as well as transmission to and sporogony in the mosquito vector.
To assess the impact that absence of PbSERA4 has on early stages of P. berghei development in mammalian hosts, C57BL/6 mice were injected intravenously with 10,000 Pbsera4(-) sporozoites. While wildtype-infected red blood cells appear in the blood three days after infection with sporozoites, Pbsera4(-) parasites did not appear in blood smears until five days after infection (Fig 3D; S2 Table). After appearance in the blood, Pbsera4(-) parasites grew at rates comparable to wildtype P. berghei (Fig 3E). Infection using 10x more sporozoites or infection of SD rats with Pbsera4(-) sporozoites resulted in similar, approximately two-day, delays in appearance of blood-stage infection in comparison to infections with wildtype sporozoites (S2 Table).
To determine whether this delay in patency observed upon infection with Pbsera4(-) sporozoites is due to inefficient infection of hepatocytes or reduced development of parasites in the liver, we assessed the parasite burden in the livers of sporozoite-infected C57BL/6 mice using quantitative PCR (Fig 3F). Pbsera4(-) parasites reached levels equivalent to wildtype parasites in livers of infected animals 48 hours after infection. The parasite burden in livers infected by wildtype parasites drops by 72 hours after infection due to egress and merozoite release. Although the appearance of Pbsera4(-) parasites in the blood of sporozoite-infected animals is delayed, the Pbsera4(-) parasites do not appear to be retained in the liver at 72 hours after infection. The higher levels of detectable parasite RNA in livers of wildtype-infected animals at 72 hours post-infection in comparison to Pbsera4(-)-infected animals is likely due to contamination with blood-stage parasites that have grown in the blood of wildtype-infected individuals by 72 hours post-infection. The comparable parasite burden near the onset of egress, however, indicates that the delay in time to patency occurring during Pbsera4(-) infection is not due to reduced growth of the parasites in the liver.
PbSERA4 is required for efficient egress of liver-stage P. berghei
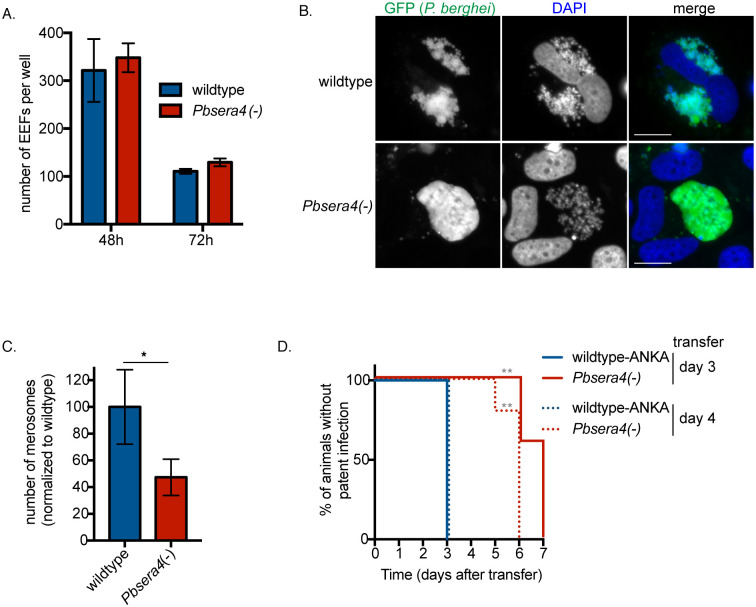
To further investigate the stage-conversion phenotype we see in Pbsera4(-)-infected mice, we confirmed the ability of Pbsera4(-)-ANKA sporozoites to efficiently infect and grow in cells in in vitro infection experiments (Fig 4). When Huh7 cells were infected, Pbsera4(-) sporozoites developed into exo-erythrocytic forms (EEF) in numbers similar to those formed by wildtype sporozoites (Fig 4A). Imaging of infected cells late in liver-stage development revealed that while several wildtype parasites existed as merozoites that were detaching from one another and were within the host cell cytoplasm, the Pbsera4(-) parasites that were imaged appeared to remain as cytomeres (Fig 4B). When merosomes and parasite-containing detached cells were collected from the supernatants of infected cultured cells following liver-stage infection, notably fewer Pbsera4(-) merosomes were detected (Fig 4C, S3 Table). Pbsera4(-)-infected cultures generated approximately half the numbers of merosomes produced by wildtype-infected cultures. When equal numbers of wildtype or Pbsera4(-) merosomes produced by cultured cells were injected into mice, all mice developed a blood-stage infection (S3 Table), indicating that the Pbsera4(-) liver-stage merozoites that are generated are as capable of infecting red blood cells and establishing a blood-stage infection as wildtype merozoites. Together, these data indicate that liver-stage Pbsera4(-) parasites are released inefficiently from infected cells, but that they are infectious to red blood cells.
Fig 4. PbSERA4 is not required for liver-stage growth, but is needed for efficient egress from cultured cells.
(A) Huh7 cells were infected with sporozoites of the indicated P. berghei ANKA lines and fixed at the indicated time points. P. berghei exoerythrocytic forms (EEF) were visualized and quantified by fluorescence microscopy. Data is presented as the mean +/- SD. (B) Infected Huh7 cells were fixed and imaged at 65 hours post-infection. At least 50 cells infected with each strain were visualized. More than a quarter of wildtype EEFs were at the advanced stage shown in the upper panel, whereas in the Pbsera4(-)-infected cells, no individual merozoites could be detected in any cell, and none of the Pbsera4(-) parasites had progressed further than the cytomere stage. (C) Merosomes and parasite-containing detached cells were collected from the supernatants of infected Huh7 cells between 65 and 72 hours of liver-stage infection and quantified. Data compiled from five experiments are shown and presented as means +/- SEM. * P<0.05 (ratio paired two-tailed t-test, n = 5 independent experiments). (D) Sub-inoculation from pre-patent Pbsera4(-)-ANKA infected animals. C57BL/6 mice were infected intravenously with 10,000 wildtype-ANKA or Pbsera4(-)-ANKA sporozoites. Blood was transferred from these infected mice to naïve NMRI mice at either 3 or 4 days after infection, and Giemsa-stained blood smears were monitored daily to determine presence of parasites. **, p<0.01, (Log-rank (Mantel-Cox) test, n = 5).
If liver-stage merozoite egress is impeded in vivo, this could manifest either as a reduction in the numbers of merozoites released from the liver or a delay in their release, and either could cause the delay in the onset of blood-stage infection we observed in infected mice. To distinguish between these two possibilities, we transferred blood from sporozoite-infected animals into naïve animals three days or four days after infection. At these time points, although patent infection is not yet visible in blood smears of Pbsera4(-)-infected animals, all animals that received the blood sub-inoculation developed a blood-stage infection (Fig 4D). This indicates that merozoites have been released from the livers by this time, but that blood-stage infection is below the level of detection by Giemsa-stained blood smears. Blood-stage parasites did not appear in the Pbsera4(-)-sub-inoculated animals until three days after the appearance of blood-stage parasites in animals receiving the wildtype sub-inoculation. These results illustrate that the numbers of blood-stage parasites present at this time in the Pbsera4(-)-sporozoite infected animals is remarkably fewer in comparison to wildtype-sporozoite-infected animals. These data together with the observed decrease in detectable parasite RNA in the livers of Pbsera4(-)-infected animals at three days post-infection (Fig 3F) suggest that the timing of liver-stage egress is not markedly delayed in a Pbsera4(-) infection, but rather that only few liver-stage merozoites reach the blood.
Absence of PbSERA4 prolongs the prepatent period of the NK65 P. berghei strain and leads to the upregulation of PbSERA3 during liver stage development
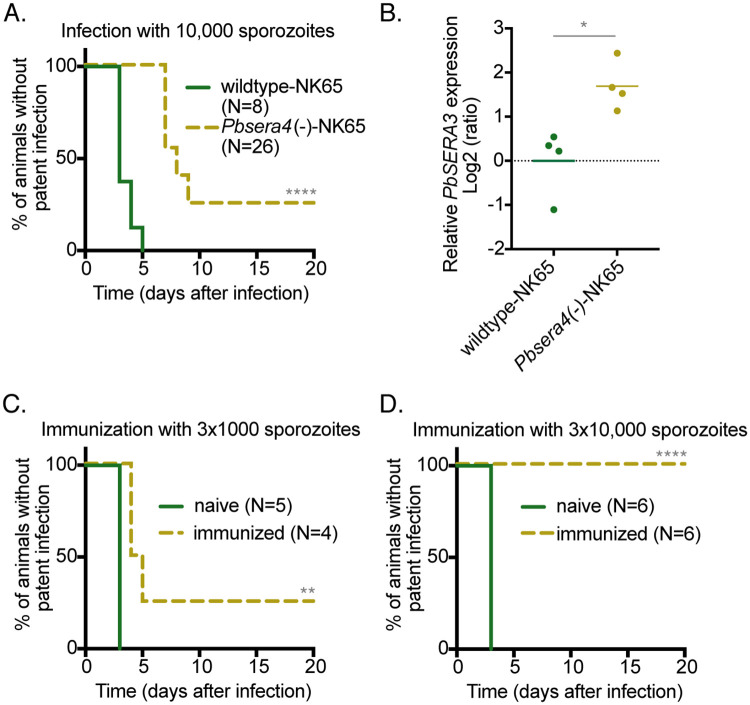
To confirm that PbSERA4 is required for efficient P. berghei stage conversion from liver- to blood-stage growth, we generated a second knockout line in the NK65 strain of P. berghei using the same approach we used for the knockout line generated in the ANKA strain (S4D Fig). When sporozoites of the resulting Pbsera4(-)-NK65 line were used to infect mice, the Pbsera4(-)-NK65 parasites not only took longer to appear in the blood of infected animals in comparison to wildtype NK65 parasites, but some animals infected with the knockout line never developed a blood-stage infection (Fig 5A). The four-day delay in time to patent blood-stage infection in the absence of PbSERA4 was likewise seen in P. berghei NK65-infected SD rats and also following infection by mosquito bite (S2 Table).
Fig 5. Pbsera4(-)-NK65 parasites inefficiently initiate blood-stage infection and can confer protection against sporozoite challenge infection.
(A) Delayed patency following Pbsera4(-)-NK65 sporozoite infection. C57BL/6 mice were infected intravenously with 10,000 sporozoites, and Giemsa-stained blood smears were monitored daily to determine the presence of parasites. ****, p<0.0001, Log-rank (Mantel-Cox) test. (B) PbSERA3 is upregulated in the liver in the absence of PbSERA4. PbSERA3 gene expression was analyzed in livers of C57BL/6 mice two days after infection with wildtype and Pbsera4(-)-NK65 sporozoites. Relative gene expression was normalized to Pb18s and the average of the wildtype-NK65 samples using the Pfaffl method. *, p<0.05 (unpaired t test, n = 4). (C and D) C57BL/6 mice were immunized three times with 1,000 or 10,000 Pbsera4(-)-NK65 sporozoites and were challenged two months after the last immunization with wildtype NK65 sporozoites. Giemsa-stained blood smears were monitored daily to determine the presence of parasites. **, p<0.01; ****, p<0.0001, Log-rank (Mantel-Cox) test.
Targeted deletion of SERA genes has been shown to influence the expression of other SERA genes [14, 32]. Absence of the serine-type PfSERA4 leads to upregulation of the essential serine-type PfSERA5 [32], and deletion of the serine-type PbSERA1 and PbSERA2 results in upregulation of cysteine-type PbSERA3 [14]. We, therefore, examined the expression of the other liver stage-expressed cysteine-type SERA, PbSERA3, in the absence of PbSERA4 at the end of liver-stage development (Fig 5B). PbSERA3 is upregulated in Pbsera4(-)-NK65 in comparison to the wildtype line in the livers of sporozoite-infected animals. Altering the expression of PbSERA3 may represent a compensatory mechanism by the parasite and may contribute to the emergence of parasites in the blood in the absence of PbSERA4.
Infection with PbSERA4-deficient NK65 P. berghei sporozoites can confer protection against reinfection
Following our observation that some mice stayed malaria-free once infected with either 1,000 or 10,000 Pbsera4(-)-NK65 sporozoites, we used these mice to test whether immunization with Pbsera4(-)-NK65 sporozoites can confer sterile protection against challenge with wildtype NK65 sporozoites. These protection assays could not be performed with the Pbsera4(-)-ANKA line because all animals infected with this line developed a blood-stage infection, albeit delayed. The two groups of mice (N = 10) were immunized with another two doses of either 1,000 or 10,000 Pbsera4(-)-NK65 sporozoites, respectively (S4 Table, Fig 5C and 5D). Half of the immunized mice were challenged by infection with wildtype sporozoites two months after the last immunization. All mice immunized with 10,000 Pbsera4(-)-NK65 sporozoites were protected and did not develop detectable blood-stage infections (Fig 5D). The mice immunized with low doses of 1,000 Pbsera4(-)-NK65 sporozoites were modestly protected; three mice of four developed a delayed blood-stage infection when challenged two months after the last immunization (Fig 5C).
To assess long-term protection, the remaining immunized mice were challenged by injection of wildtype sporozoites 20 months after the last immunization (S4 Table). The six mice that had shown protection after challenge at 2 months were also re-challenged at this time. None of the mice that had received only three immunizations were protected at 20 months; blood-stage parasites were detected in all of these animals after challenge. In contrast, all five mice that received immunization doses of 10,000 Pbsera4(-)-NK65 sporozoites and were protected from the challenge at two months, were also protected against the re-challenge infection at 20 months. This result indicates that the fourth exposure to pre-erythrocytic P. berghei conferred long-lasting protection in these mice.
Our results with the NK65 knockout line confirm that the absence of PbSERA4 interferes with the parasites’ ability to establish blood-stage infection. We further demonstrate that although protection wanes over time, immunization with Pbsera4(-)-NK65 sporozoites can confer sterile protection.
Discussion
Our studies have identified a role for PbSERA4 in the escape of Plasmodium parasites from the liver stage of infection. All functional data on the Plasmodium SERA family thus far indicate roles for these proteases in parasite egress [15, 17, 33]. Until now, of the Plasmodium SERA family, only the contributions of PbSERA1 and PbSERA2 to liver-stage egress have been tested [14]. Neither of these serine-type SERAs are necessary for Plasmodium exit from the liver [14], and the specific cysteine proteases known to be needed for liver-stage egress [7] have remained unidentified. The central finding of this study is that loss of PbSERA4 function reduces the efficiency of liver-stage egress and weakens the transition from liver-stage growth to blood-stage infection.
PbSERA4 is one of the cysteine-type SERAs, meaning that unlike the serine-type SERAs, the papain-like catalytic site is intact and includes all key catalytic amino acid residues [21, 34]. The cysteine-type SERA genes have clear orthologs across Plasmodium species and the cysteine-type SERA we describe here appears to have emerged in a common ancestor to all primate- and rodent-infecting Plasmodium [21]. The predicted ortholog of PbSERA4 in P. falciparum is PfSERA7, which is syntenic with PbSERA4 and shares the highest sequence identity with PbSERA4 of the P. falciparum SERA genes. Furthermore, PfSERA7 has been shown to be dispensable for blood-stage P. falciparum growth [16], which is consistent with our data showing the PbSERA4(-) P. berghei lines grow proficiently in the blood.
Our expression and localization studies of PbSERA4 using a specific antibody and a parasite line in which the endogenously encoded protein is fluorescently tagged show that PbSERA4 is expressed in blood-stage and late-liver-stage Plasmodium. Previous studies demonstrated that SERA family members in P. falciparum and P. berghei are proteolytically processed to their active form [10, 18, 31, 33], and our data demonstrate that PbSERA4 appears also to be proteolytically processed during infection. Processing of at least some SERA family members occurs in the parasitophorous vacuole upon secretion of SUB1 [18]. In contrast to other liver-stage-expressed SERAs [10, 14], PbSERA4 was only detected inside the parasite, but the localization in liver-stage schizonts detected by both antibody staining and fluorescent tagging highly resembles parasite endoplasmic reticulum [35]. This localization and the predicted N-terminal signal peptide suggest that PbSERA4 is engaged by the parasites’ secretory pathway. The region of PbSERA4 used to generate the α-PbSERA4M antibody contains a putative SUB1 cleavage site; therefore, there is a possibility that this antibody may only recognize uncleaved protein when the protein is in its native state after PFA fixation. Thus, if PbSERA4 cleavage products are in the parasitophorous vacuole during liver infection, the antibody may have failed to detect them.
Targeted deletion of PbSERA4 hindered the ability of P. berghei to transition from the liver into the blood of infected animals. We observed that time until the appearance of blood-stage parasites in animals following liver-stage infection increased in the absence of PbSERA4. The length of this time delay depended on the P. berghei strain. The Pbsera4(-)-NK65 line was more severely impaired than the Pbsera4(-)-ANKA line; patent infection was delayed by approximately four days with the NK65 knockout line and two days with the ANKA knockout line. In some animals, the Pbsera4(-)-NK65 line even failed to establish blood-stage infection. Pbsera4(-) liver-stage growth was unobstructed either in vivo or in cultured cells. Furthermore, blood-stage growth of Pbsera4(-) parasites proceeded with normal growth rates regardless of the route of infection. These results indicate that the restriction of Pbsera4(-) parasites occurs between these two growth stages. Pbsera4(-)-infected cultured cells produce fewer merosomes, which suggests a defect in egress of liver-stage merozoites from infected cells. This conclusion would be consistent with the known egress functions of other SERA family members. The levels of Pbsera4(-) parasite RNA in infected livers drop by 72 hours post-infection, indicating that parasite-containing cells do not remain detectable in the liver in spite of few merozoites reaching the bloodstream. An alternative possibility is that PbSERA4 is required for egress of liver-stage merozoites from merosomes after they have entered the blood stream. PbSERA4 is also expressed in blood-stage parasites, which is reported as well for PfSERA7. Our data confirm that PbSERA4 is not essential for asexual blood-stage development, as demonstrated for PfSERA7 [16], and reveal that PbSERA4 is also expendable for gametocyte egress, which was demonstrated by efficient infection of mosquitoes by the Pbsera4(-) lines. Whether PbSERA4 contributes to egress with functions for which other factors can compensate in its absence remains to be determined.
The cysteine-type SERA genes form three phylogenetically distinct groups [36]. Group one, represented by PbSERA5 and PfSERA8, in P. berghei and P. falciparum, respectively, is required for egress of sporozoites from oocysts [15]. Group three, represented by PbSERA3 and PfSERA6, is essential for egress of merozoites from red blood cells [17]. PbSERA4 is within group two, and our data suggests that this group contributes to egress of liver-stage merozoites from the liver. In contrast to the studied representatives of the other two groups, PbSERA4 is not vital to the process of parasite egress. In the absence of PbSERA4, a few parasites escape the liver and successfully reach the blood. Another enzymatic protein that contributes to liver-stage Plasmodium egress, P. berghei phospholipase, is also not strictly required for parasite transition into the blood [8]. Because of the complexity and importance of this process for establishing an infection in the mammalian host, Plasmodium may have multiple mechanisms by which to promote its transition from the liver into the blood. The upregulation of PbSERA3 in liver-stage Pbsera4(-)-NK65 parasites suggests that this other cysteine-type SERA may be able to, in part, compensate for the loss of PbSERA4 in these parasites. While the localizations of PbSERA3 and PbSERA4 do not always overlap, they do colocalize in the parasite in the cytomere stage. While the function of PbSERA3 in the liver remains undefined, together these data indicate that PbSERA3 could play a role in the ability of PbSERA4-deficient parasites to transition to the blood and may therefore contribute to liver-stage egress in wildtype parasites as well.
Because multiple SERA proteins are expressed at the same time in both the blood and liver, it has been proposed that they may influence the function of one another. For example, one SERA could alter the function of another by affecting the recognition of its substrate or by influencing its activation by proteolytic processing [34]. While this has been proposed in particular for the serine-type SERA proteins, we cannot formally exclude this as a possibility for the cysteine-type SERAs. However, because PbSERA3 is essential for blood-stage development, and processing of the corresponding P. falciparum ortholog, PfSERA6 is essential for its activity [17], we suspect that the processing and function of PbSERA3 is unaltered in parasites lacking PbSERA4, at least in the blood stage of infection.
The failure of the Pbsera4(-)-NK65 line to establish blood-stage infection in some animals provided an opportunity to assess the ability of this parasite line to elicit an immune response that can protect animals against reinfection. Late-liver-stage-arresting P. berghei often confer strong protection against re-infection [37], but often present the risk of breakthrough infections. The ability of these parasites to develop into late liver stages and elicit protective responses indicates that perhaps this SERA gene could be deleted in combination with other genes to achieve iterative attenuation at multiple checkpoints in the formation of first-generation merozoites, and this approach could lead to a safe and efficacious whole sporozoite vaccine strategy.
Taken together, these studies have revealed a role for PbSERA4 in exit of Plasmodium from the liver. While not strictly essential for stage-conversion, PbSERA4 contributes to efficient transition between liver- and blood-stage growth. As in egress of blood-stage merozoites, a proteolytic cascade involving a distinct member, PbSERA4/PfSERA7, of the conserved family of papain-like cysteine proteases contributes to exit of merozoites from the liver.
Materials and methods
Ethics statement
All animal work was conducted in accordance with the German ‘Tierschutzgesetz in der Fassung vom 18. Mai 2006 (BGBl. I S. 1207)’, which implements directive 86/609/EEC from the European Union and the European Convention for the protection of vertebrate animals used for experimental and other scientific purposes. All appropriate measures were taken to reduce the pain or discomfort of the animals, and the protocol was approved by the ethics committee of the University of Heidelberg and Max Planck Institute for Infection Biology and by the state authorities (Regierungspräsidium Karlsruhe G154/05 and Landesamt für Gesundheit und Soziales G0469/09).
Experimental animals, cells and parasite lines
Naval Medical Research Institute (NMRI) mice, C57BL/6 mice, and Sprague Dawley (SD) rats were purchased from Charles River Laboratories (Sulzfeld, Germany). Anopheles stephensi mosquitoes were raised under a 14 h light/ 10 h dark cycle, 75% humidity and at 28 °C (non-infected) or 20 °C (infected). The Huh7 hepatoma cell line was cultured in RPMI supplemented with 10% FCS, 100 U penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 10 mM HEPES and 1× non-essential amino acids (NEAA) (Gibco). The HepG2 hepatoma cells were cultivated in EMEM (Gibco) containing 10% fetal calf serum, 1% L-Glutamine, 1% Penicillin/Streptomycin and 1% MEM Non-essential amino acids (PAA Laboratories GmbH, Pasching, Austria). Wild-type P. berghei were either the P. berghei ANKA strain, ANKA clone 507 [26], which expresses GFP, or P. berghei NK65 [38].
Transcript detection and quantitation
Transcript detection in the various stages was performed as described [14]. RNA purification was followed by cDNA synthesis and products were amplified using specific primers described in S1 Table. Standard curves were generated for all primers using WT cDNA serial dilutions and gave amplification efficiencies of 90–100%. Relative PbSERA3 gene expression was normalized to Pb18s, and calculated with the Pfaffl method to account for differing primer efficiencies [39]. The average Ct values from the wildtype-NK65-infected livers were used as calibrators in this analysis.
Generation of sera4(-) and mCherry-tagged SERA4 parasites
Transgenic P. berghei lines were generated as described previously [14, 26]. For mCherry tagging, the PbSERA4 locus was amplified with primers mCherry-SERA4for and mCherry-SERA4rev and integrated into B3D+mCherry [40] in frame with the mCherry coding sequence. For targeted disruption of PbSERA4, a replacement vector was generated through amplification of genomic regions flanking PbSERA4 using primers SERA4rep1for and SERA4rep2rev or SERA4rep3for and SERA4rep4rev and cloned into b3D.DT^H.^D (provided by Dr. Andrew Waters, Glasgow University) flanking the Tgdhfr/TS expression cassette. P. berghei parasites were transfected with linearized plasmids as described [14, 26], injected into naïve NMRI mice and selected by oral pyrimethamine. Integration of the constructs was confirmed by PCR with specific primer pairs (see supplementary information for details), and clonal lines were isolated via limiting dilution. All oligonucleotide sequences are listed in S1 Table.
Generation of anti-PbSERA4 antibodies
The central (M) (Phe216-His365) region of PbSERA4 was amplified from P. berghei ANKA cDNA. The resulting PCR product was ligated into pGEX6P-1 (Amersham, Bucking-hampshire, England) and expressed in Escherichia coli BL21 cells (Stratagene) as glutathione S-transferase (GST) fusion proteins. Bacteria were lysed by sonication in PBS containing 10 mM EDTA and CompleteTM protease inhibitor (Roche Molecular Biochemicals). GST-fusion proteins were purified from the supernatant using glutathione-agarose as described by the manufacturer (Amersham Biosciences). 20μg of purified PbSERA4-N-GST or PbSERA4-M-GST fusion protein was used to immunize six week old female NMRI mice by intraperitoneal injection along with complete Freund adjuvant, followed by multiple boosting immunizations. For anti-PbSERA3N, blood samples were collected following immunization, to test serum reactivity against the SERA4-N protein. From a positive mouse spleen, cells were isolated and fused to the mouse myeloma cell line X63Ag8.653. Supernatants were screened by indirect ELISA and single-cell clones were isolated by limited dilution.
Immunofluorescence analysis
For analysis of PbSERA4 localization in late liver stages, infected HepG2 cells were fixed with 4% formaldehyde, permeabilized with ice-cold methanol and incubated with mouse anti-PbSERA4M serum, rat anti-PbSERA3N serum [10] (1:200 in 10% FCS diluted in PBS) or a chicken anti-Exp1 antibody. Bound antibodies were detected using fluorescently-conjugated secondary antibodies (Molecular Probes, Leiden, The Netherlands). Labeled cells were examined by confocal microscopy using the Olympus FV1000 (SIM- scanner and spectral detection).
SDS-PAGE and western blot analysis
Blood-stage-schizont-infected red blood cells were enriched on a 55% Nycodenz gradient and lysed in 0.15% saponin in PBS. Pelleted parasites were subsequently lysed in a TRIS-buffered solution containing 150mM NaCl and 1.0% NP-40. For the merosome sample, loaded lysates contained merosomes and detached cells collected from at least two wells of a 24-well dish containing infected HepG2 cells. Merosomes and detached cells were lysed directly in Laemmli sample buffer. Lysates were separated on 10% SDS-PAGE reducing gels and transferred to nitrocellulose membranes. Membranes were probed with mouse α-PbSERA4-M antisera (1: 4000). Horseradish peroxidase-conjugated goat anti-mouse (1: 20.000, Pierce) were used for detection, and bands were visualized by enhanced chemiluminescence Pico or Femto Detection Kit (Pierce).
Parasite phenotyping during Plasmodium life cycle progression
To quantify patency or blood stage development, daily Giemsa-stained blood smears of infected NMRI and C57BL/6 mice were counted. Exflagellation of microgametes were examined prior to mosquito feeding. For analysis of liver stage development, sporozoites were added to cultured hepatoma cells, either Huh7 or HepG2 as indicated, incubated for two hours at 37°C, and washed. After fixation, exoerythrocytic forms (EEFs) were visualized by their GFP expression or by using an anti-PbHSP70 antibody [41]. Merosomes and parasite-containing detached cells were collected 65–70 hours post infection when merosomes were visible in the culture by light microscopy. To collect and quantify merosomes, the culture was gently agitated by gently tapping the plate, and the culture medium was collected and centrifuged for 3 minutes at 2000 rpm. The pellet was resuspended in a low volume and merosomes were quantified using a Neubauer chamber and fluorescence microscopy, which allowed identification based on parasite GFP fluorescence. The parasite burden of Pbsera4(-) and WT infected livers was determined as described [42, 43]. Briefly, C57Bl/6 mice were injected with 104 sporozoites intravenously, and livers of infected animals were harvested at the indicated time points. Total RNA was extracted and relative parasite levels were determined with quantitative–PCR by comparing the mean Ct value of the P. berghei 18s ribosomal subunit to the mean Ct value of the Mus musculus hypoxanthine phosphoribosyl transferase (HPRT) in the generated cDNA. Subinoculation was performed through transfer of blood taken from mice at day three or four after sporozoite inoculation. Blood from infected mice was transferred into naïve mice by intravenous injection. About 100μl PBS with one drop Heparin was mixed with two drops of infected blood. The blood-PBS mixture then was injected intravenously into the tail of naïve mice. The development of a patent parasitemia was analyzed by daily examination of Giemsa-stained smears.
Immunization and parasite challenge experiments
Age-matched female C57BL/6 mice were immunized with 1,000 or 10,000 Pbsera4(-)-NK65 sporozoites, injected intravenously. Animals that remained malaria-free after the first immunization were given a second dose. Only animals that remained blood stage parasite-negative after the first immunization and subsequent boost were used for the challenge experiments 2 and 20 months after the last immunization. Mice were challenged by intravenous injection of 10,000 WT sporozoites. At least three age-matched naïve animals were included to verify infectivity of sporozoites during all challenge experiments. Parasitemia was monitored by daily Giemsa-stained thin blood smears, starting from day 3 after immunization or challenge infection until at least day 14.
Statistical analyses
For assays quantifying parasites, RNA or merosomes, transgenic and wildtype P. berghei were compared using two-tailed t-tests, and details are defined in the figure legends. Assays evaluating time to patency were analyzed using the Mantel-Cox test. All statistical analyses were performed in GraphPad Prism version 7.
Supporting information
(PDF)
(PDF)
Huh7 cells in 6-well dishes were infected with P. berghei sporozoites of the indicated lines, and merosomes and parasite-infected detached cells (collectively referred to as merosomes in table headings) were collected, quantified and the numbers indicated were injected intravenously into naïve NMRI mice.
(PDF)
Animals were infected intravenously with Pbsera4(-)-NK65 sporozoites. Those that remained negative for blood-stage infection were boosted as indicated.
(PDF)
(A) Control PCRs for detection of SERA4 and control transcripts. P. berghei ANKA genomic DNA was used as a template in PCRs run in parallel to those using cDNA as the template shown in Fig 1C. The product sizes are as expected and match those amplified from cDNA. (B) Generation of an endogenously encoded SERA4-mCherry fusion protein using a single cross-over insertion strategy. An integration plasmid containing the region encoding the C-terminus of SERA4 (dark grey box) fused to mCherry (red box) is targeted into the PbSERA4 genomic locus. The targeting plasmid contains the 3’ UTR of PbDHFR/TS (light grey box) and the Tgdhfr/TS selectable marker (white box). Integration results in a PbSERA4 gene that is expressed from its endogenous promoter and tagged with mCherry followed by non-expressed truncated duplication of the 3’ end of the gene. (C) Genotyping of the transgenic PbSERA4-mCherry parasite line with the primers indicated in panel B resulted in expected products. (D) Confocal fluorescence microscopy reveals expression of the SERA4-mCherry fusion protein (red) in blood stage parasites, including trophozoites (arrowheads) and gametocytes (asterisk). Scale bars, 8 μm.
(TIF)
The indicated 5’ regions upstream of the PbSERA4 or PbSERA5 coding sequences were cloned in front of GFP into the P. berghei transfection plasmid pL0031 (right). GFP expression in P. berghei ANKA parasites transfected with these constructs was monitored across the life cycle stages: midgut-oocysts (oo), salivary gland sporozoites (sp), HepG2 cells infected with transgenic exo-erythrocytic parasites at indicated timepoints (EE) and erythrocytic stage (ES) were stained with Hoechst 33342 and visualized by live microscopy.
(TIF)
Recombinant PbSERA proteins (1μg) spanning the central papain-like protease domain (M) were separated on a polyacrylamide gel and stained with Coomassie (top panel). Tags of the MBP-fusion proteins (lane1,2) were cleaved off. The MBP-tag (grey arrow) and resulting proteins (black arrow) are indicated. GST fusion proteins were insoluble and thus were purified by Prep-Cell purification (lane 3, 4) GST-tags could therefore not be cleaved off. Thirty nanograms of these recombinant proteins were run and transferred to a membrane that was subsequently probed with α-PbSERA4M polyclonal antisera. α-PbSERA4M specifically recognized PbSERA4 and not other SERA proteins.
(TIF)
(A) Experimental strategy for targeted disruption of PbSERA4. The wildtype (WT) PbSERA4 genomic locus is targeted with KpnI/SacII-linearized replacement plasmid containing 5’ and 3’ untranslated regions adjacent to the SERA4 open reading frames and the dhfr/ts-positive selectable marker. Upon a double cross-over homologous recombination, the PbSERA4 open reading frame is replaced by the selectable marker, resulting in PbSERA4 knockout parasites termed Pbsera4(-). For genotyping, replacement-specific test and WT test primer combination are indicated by arrows and expected fragments as lines. (B) Genotyping of the Pbsera4(-) disruption in P. berghei ANKA using PCR analysis. The primer combinations that amplify signals in the recombinant locus (test1 and 2) of purified genomic DNA verified the successful replacement event. The absence of a WT specific signal from Pbsera4(-) parasites confirms the purity of the clonal populations. (C) PbSERA4 protein is not detected in the Pbsera4(-) line. Lysates of mixed-stage infected erythrocytes were analysed by western blot using the anti-PbSERA4M antibody (top). Anti-HSP70 antibodies were used as a loading control (bottom). Full length PbSERA4 and intermediate cleavage products are detected exclusively in the wildtype samples. (D) Genotyping of the Pbsera4(-) disruption in P. berghei NK65 using PCR analysis. The primer combinations that amplify signals in the recombinant locus (test 1 for and test 2 rev) of purified genomic DNA verified the successful replacement event. The absence of a WT specific signal from Pbsera4(-)-NK65 parasites confirms the purity of the clonal populations.
(TIF)
(XLSX)
Acknowledgments
We thank Carolin Rauch and Manuel Rauch for technical support.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the European Commission (EC):Kai Matuschewski EviMalaR,#34; Deutsche Forschungsgemeinschaft (DFG):Volker Heussler HE 4497/1-2; Deutscher Akademischer Austauschdienst (DAAD):Elyzana Dewi Putrianti. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Matz JM, Beck JR, Blackman MJ. The parasitophorous vacuole of the blood-stage malaria parasite. Nat Rev Microbiol. 2020; 18(7): 379–91. 10.1038/s41579-019-0321-3 . [DOI] [PubMed] [Google Scholar]
- 2.Nyboer B, Heiss K, Mueller AK, Ingmundson A. The Plasmodium liver-stage parasitophorous vacuole: A front-line of communication between parasite and host. Int J Med Microbiol. 2018; 308(1):107–17. 10.1016/j.ijmm.2017.09.008 . [DOI] [PubMed] [Google Scholar]
- 3.Hybiske K, Stephens RS. Exit strategies of intracellular pathogens. Nat Rev Microbiol. 2008; 6(2):99–110. 10.1038/nrmicro1821 . [DOI] [PubMed] [Google Scholar]
- 4.Friedrich N, Hagedorn M, Soldati-Favre D, Soldati T. Prison break: pathogens' strategies to egress from host cells. Microbiol Mol Biol Rev. 2012; 76(4):707–20. 10.1128/MMBR.00024-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prudencio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: the Plasmodium liver stage. Nat Rev Microbiol. 2006; 4(11):849–56. 10.1038/nrmicro1529 . [DOI] [PubMed] [Google Scholar]
- 6.Baer K, Klotz C, Kappe SH, Schnieder T, Frevert U. Release of hepatic Plasmodium yoelii merozoites into the pulmonary microvasculature. PLoS Pathog. 2007; 3(11):e171 10.1371/journal.ppat.0030171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, Rennenberg A, et al. Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science. 2006; 313(5791):1287–90. 10.1126/science.1129720 . [DOI] [PubMed] [Google Scholar]
- 8.Burda PC, Roelli MA, Schaffner M, Khan SM, Janse CJ, Heussler VT. A Plasmodium phospholipase is involved in disruption of the liver stage parasitophorous vacuole membrane. PLoS Pathog. 2015; 11(3):e1004760 10.1371/journal.ppat.1004760 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burda PC, Caldelari R, Heussler VT. Manipulation of the host cell membrane during Plasmodium liver stage egress. mBio. 2017; 8(2):e00139–17. 10.1128/mBio.00139-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt-Christensen A, Sturm A, Horstmann S, Heussler VT. Expression and processing of Plasmodium berghei SERA3 during liver stages. Cell Microbiol. 2008; 10(8):1723–34. 10.1111/j.1462-5822.2008.01162.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmon BL, Oksman A, Goldberg DE. Malaria parasite exit from the host erythrocyte: a two-step process requiring extraerythrocytic proteolysis. Proc Natl Acad Sci U S A. 2001; 98(1):271–6. 10.1073/pnas.011413198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickham ME, Culvenor JG, Cowman AF. Selective inhibition of a two-step egress of malaria parasites from the host erythrocyte. J Biol Chem. 2003; 278(39):37658–63. 10.1074/jbc.M305252200 . [DOI] [PubMed] [Google Scholar]
- 13.Glushakova S, Mazar J, Hohmann-Marriott MF, Hama E, Zimmerberg J. Irreversible effect of cysteine protease inhibitors on the release of malaria parasites from infected erythrocytes. Cell Microbiol. 2009; 11(1):95–105. 10.1111/j.1462-5822.2008.01242.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putrianti ED, Schmidt-Christensen A, Arnold I, Heussler VT, Matuschewski K, Silvie O. The Plasmodium serine-type SERA proteases display distinct expression patterns and non-essential in vivo roles during life cycle progression of the malaria parasite. Cell Microbiol. 2010; 12(6):725–39. 10.1111/j.1462-5822.2009.01419.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aly AS, Matuschewski K. A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J Exp Med. 2005; 202(2):225–30. 10.1084/jem.20050545 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller SK, Good RT, Drew DR, Delorenzi M, Sanders PR, Hodder AN, et al. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J Biol Chem. 2002; 277(49):47524–32. 10.1074/jbc.M206974200 . [DOI] [PubMed] [Google Scholar]
- 17.Thomas JA, Tan MSY, Bisson C, Borg A, Umrekar TR, Hackett F, et al. A protease cascade regulates release of the human malaria parasite Plasmodium falciparum from host red blood cells. Nat Microbiol. 2018; 3(4):447–55. 10.1038/s41564-018-0111-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeoh S, O'Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, et al. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007; 131(6):1072–83. 10.1016/j.cell.2007.10.049 . [DOI] [PubMed] [Google Scholar]
- 19.Suarez C, Volkmann K, Gomes AR, Billker O, Blackman MJ. The malarial serine protease SUB1 plays an essential role in parasite liver stage development. PLoS Pathog. 2013; 9(12):e1003811 10.1371/journal.ppat.1003811 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tawk L, Lacroix C, Gueirard P, Kent R, Gorgette O, Thiberge S, et al. A key role for Plasmodium subtilisin-like SUB1 protease in egress of malaria parasites from host hepatocytes. J Biol Chem. 2013; 288(46):33336–46. 10.1074/jbc.M113.513234 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arisue N, Kawai S, Hirai M, Palacpac NM, Jia M, Kaneko A, et al. Clues to evolution of the SERA multigene family in 18 Plasmodium species. PLoS One. 2011; 6(3):e17775 10.1371/journal.pone.0017775 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldelari R, Dogga S, Schmid MW, Franke-Fayard B, Janse CJ, Soldati-Favre D, et al. Transcriptome analysis of Plasmodium berghei during exo-erythrocytic development. Malar J. 2019; 18(1):330 10.1186/s12936-019-2968-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003; 1(1):e5 10.1371/journal.pbio.0000005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003; 301(5639):1503–8. 10.1126/science.1087025 . [DOI] [PubMed] [Google Scholar]
- 25.Lindner SE, Swearingen KE, Shears MJ, Walker MP, Vrana EN, Hart KJ, et al. Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat Commun. 2019; 10(1):4964 10.1038/s41467-019-12936-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janse CJ, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006; 145(1):60–70. 10.1016/j.molbiopara.2005.09.007 . [DOI] [PubMed] [Google Scholar]
- 27.Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010; 76(1):12–24. 10.1111/j.1365-2958.2009.07026.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasonder E, Rijpma SR, van Schaijk BC, Hoeijmakers WA, Kensche PR, Gresnigt MS, et al. Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 2016; 44(13):6087–101. 10.1093/nar/gkw536 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debrabant A, Maes P, Delplace P, Dubremetz JF, Tartar A, Camus D. Intramolecular mapping of Plasmodium falciparum P126 proteolytic fragments by N-terminal amino acid sequencing. Mol Biochem Parasitol. 1992; 53(1–2):89–95. 10.1016/0166-6851(92)90010-h . [DOI] [PubMed] [Google Scholar]
- 30.Delplace P, Fortier B, Tronchin G, Dubremetz JF, Vernes A. Localization, biosynthesis, processing and isolation of a major 126 kDa antigen of the parasitophorous vacuole of Plasmodium falciparum. Mol Biochem Parasitol. 1987; 23(3):193–201. 10.1016/0166-6851(87)90026-0 . [DOI] [PubMed] [Google Scholar]
- 31.Li J, Mitamura T, Fox BA, Bzik DJ, Horii T. Differential localization of processed fragments of Plasmodium falciparum serine repeat antigen and further processing of its N-terminal 47 kDa fragment. Parasitol Int. 2002; 51(4):343–52. 10.1016/s1383-5769(02)00042-9 . [DOI] [PubMed] [Google Scholar]
- 32.McCoubrie JE, Miller SK, Sargeant T, Good RT, Hodder AN, Speed TP, et al. Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design. Infect Immun. 2007; 75(12):5565–74. 10.1128/IAI.00405-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins CR, Hackett F, Atid J, Tan MSY, Blackman MJ. The Plasmodium falciparum pseudoprotease SERA5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes. PLoS Pathog. 2017; 13(7):e1006453 10.1371/journal.ppat.1006453 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodder AN, Drew DR, Epa VC, Delorenzi M, Bourgon R, Miller SK, et al. Enzymic, phylogenetic, and structural characterization of the unusual papain-like protease domain of Plasmodium falciparum SERA5. J Biol Chem. 2003; 278(48):48169–77. 10.1074/jbc.M306755200 . [DOI] [PubMed] [Google Scholar]
- 35.Kaiser G, De Niz M, Zuber B, Burda PC, Kornmann B, Heussler VT, et al. High resolution microscopy reveals an unusual architecture of the Plasmodium berghei endoplasmic reticulum. Mol Microbiol. 2016; 102(5):775–91. 10.1111/mmi.13490 . [DOI] [PubMed] [Google Scholar]
- 36.Arisue N, Palacpac NMQ, Tougan T, Horii T. Characteristic features of the SERA multigene family in the malaria parasite. Parasit Vectors. 2020; 13(1):170 10.1186/s13071-020-04044-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreutzfeld O, Muller K, Matuschewski K. Engineering of genetically arrested parasites (GAPs) for a precision malaria vaccine. Front Cell Infect Microbiol. 2017; 7:198 10.3389/fcimb.2017.00198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoeli M, Most H, Bone G. Plasmodium berghei: Cyclical transmissions by experimentally infected Anopheles quadrimaculatus. Science. 1964; 144(3626):1580–1. 10.1126/science.144.3626.1580 . [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29(9):e45 10.1093/nar/29.9.e45 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silvie O, Goetz K, Matuschewski K. A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PLoS Pathog. 2008; 4(6):e1000086 10.1371/journal.ppat.1000086 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potocnjak P, Yoshida N, Nussenzweig RS, Nussenzweig V. Monovalent fragments (Fab) of monoclonal antibodies to a sporozoite surface antigen (Pb44) protect mice against malarial infection. J Exp Med. 1980; 151(6):1504–13. 10.1084/jem.151.6.1504 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruna-Romero O, Hafalla JC, Gonzalez-Aseguinolaza G, Sano G, Tsuji M, Zavala F. Detection of malaria liver-stages in mice infected through the bite of a single Anopheles mosquito using a highly sensitive real-time PCR. Int J Parasitol. 2001; 31(13):1499–502. 10.1016/s0020-7519(01)00265-x . [DOI] [PubMed] [Google Scholar]
- 43.Friesen J, Silvie O, Putrianti ED, Hafalla JC, Matuschewski K, Borrmann S. Natural immunization against malaria: causal prophylaxis with antibiotics. Sci Transl Med. 2010; 2(40):40ra9 10.1126/scitranslmed.3001058 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Huh7 cells in 6-well dishes were infected with P. berghei sporozoites of the indicated lines, and merosomes and parasite-infected detached cells (collectively referred to as merosomes in table headings) were collected, quantified and the numbers indicated were injected intravenously into naïve NMRI mice.
(PDF)
Animals were infected intravenously with Pbsera4(-)-NK65 sporozoites. Those that remained negative for blood-stage infection were boosted as indicated.
(PDF)
(A) Control PCRs for detection of SERA4 and control transcripts. P. berghei ANKA genomic DNA was used as a template in PCRs run in parallel to those using cDNA as the template shown in Fig 1C. The product sizes are as expected and match those amplified from cDNA. (B) Generation of an endogenously encoded SERA4-mCherry fusion protein using a single cross-over insertion strategy. An integration plasmid containing the region encoding the C-terminus of SERA4 (dark grey box) fused to mCherry (red box) is targeted into the PbSERA4 genomic locus. The targeting plasmid contains the 3’ UTR of PbDHFR/TS (light grey box) and the Tgdhfr/TS selectable marker (white box). Integration results in a PbSERA4 gene that is expressed from its endogenous promoter and tagged with mCherry followed by non-expressed truncated duplication of the 3’ end of the gene. (C) Genotyping of the transgenic PbSERA4-mCherry parasite line with the primers indicated in panel B resulted in expected products. (D) Confocal fluorescence microscopy reveals expression of the SERA4-mCherry fusion protein (red) in blood stage parasites, including trophozoites (arrowheads) and gametocytes (asterisk). Scale bars, 8 μm.
(TIF)
The indicated 5’ regions upstream of the PbSERA4 or PbSERA5 coding sequences were cloned in front of GFP into the P. berghei transfection plasmid pL0031 (right). GFP expression in P. berghei ANKA parasites transfected with these constructs was monitored across the life cycle stages: midgut-oocysts (oo), salivary gland sporozoites (sp), HepG2 cells infected with transgenic exo-erythrocytic parasites at indicated timepoints (EE) and erythrocytic stage (ES) were stained with Hoechst 33342 and visualized by live microscopy.
(TIF)
Recombinant PbSERA proteins (1μg) spanning the central papain-like protease domain (M) were separated on a polyacrylamide gel and stained with Coomassie (top panel). Tags of the MBP-fusion proteins (lane1,2) were cleaved off. The MBP-tag (grey arrow) and resulting proteins (black arrow) are indicated. GST fusion proteins were insoluble and thus were purified by Prep-Cell purification (lane 3, 4) GST-tags could therefore not be cleaved off. Thirty nanograms of these recombinant proteins were run and transferred to a membrane that was subsequently probed with α-PbSERA4M polyclonal antisera. α-PbSERA4M specifically recognized PbSERA4 and not other SERA proteins.
(TIF)
(A) Experimental strategy for targeted disruption of PbSERA4. The wildtype (WT) PbSERA4 genomic locus is targeted with KpnI/SacII-linearized replacement plasmid containing 5’ and 3’ untranslated regions adjacent to the SERA4 open reading frames and the dhfr/ts-positive selectable marker. Upon a double cross-over homologous recombination, the PbSERA4 open reading frame is replaced by the selectable marker, resulting in PbSERA4 knockout parasites termed Pbsera4(-). For genotyping, replacement-specific test and WT test primer combination are indicated by arrows and expected fragments as lines. (B) Genotyping of the Pbsera4(-) disruption in P. berghei ANKA using PCR analysis. The primer combinations that amplify signals in the recombinant locus (test1 and 2) of purified genomic DNA verified the successful replacement event. The absence of a WT specific signal from Pbsera4(-) parasites confirms the purity of the clonal populations. (C) PbSERA4 protein is not detected in the Pbsera4(-) line. Lysates of mixed-stage infected erythrocytes were analysed by western blot using the anti-PbSERA4M antibody (top). Anti-HSP70 antibodies were used as a loading control (bottom). Full length PbSERA4 and intermediate cleavage products are detected exclusively in the wildtype samples. (D) Genotyping of the Pbsera4(-) disruption in P. berghei NK65 using PCR analysis. The primer combinations that amplify signals in the recombinant locus (test 1 for and test 2 rev) of purified genomic DNA verified the successful replacement event. The absence of a WT specific signal from Pbsera4(-)-NK65 parasites confirms the purity of the clonal populations.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.