Abstract
PURPOSE
High-grade neuroendocrine cervical cancer (HGNECC) is an uncommon malignancy with limited therapeutic options; treatment is patterned after the histologically similar small-cell lung cancer (SCLC). To better understand HGNECC biology, we report its genomic landscape.
PATIENTS AND METHODS
Ninety-seven patients with HGNECC underwent comprehensive genomic profiling (182-315 genes). These results were subsequently compared with a cohort of 1,800 SCLCs.
RESULTS
The median age of patients with HGNECC was 40.5 years; 83 patients (85.6%) harbored high-risk human papillomavirus (HPV). Overall, 294 genomic alterations (GAs) were identified (median, 2 GAs/sample; average, 3.0 GAs/sample, range, 0-25 GAs/sample) in 109 distinct genes. The most frequently altered genes were PIK3CA (19.6% of cohort), MYC (15.5%), TP53 (15.5%), and PTEN (14.4%). RB1 GAs occurred in 4% versus 32% of HPV-positive versus HPV-negative tumors (P < .0001). GAs in HGNECC involved the following pathways: PI3K/AKT/mTOR (41.2%); RAS/MEK (11.3%); homologous recombination (9.3%); and ERBB (7.2%). Two tumors (2.1%) had high tumor mutational burden (TMB; both with MSH2 alterations); 16 (16.5%) had intermediate TMB. Seventy-one patients (73%) had ≥ 1 alteration that was theoretically druggable. Comparing HGNECC with SCLC, significant differences in TMB, microsatellite instability, HPV-positive status, and in PIK3CA, MYC, PTEN, TP53, ARID1A, and RB1 alteration rates were found.
CONCLUSION
This large cohort of patients with HGNECC demonstrated a genomic landscape distinct from SCLC, calling into question the biologic and therapeutic relevance of the histologic similarities between the entities. Furthermore, 73% of HGNECC tumors had potentially actionable alterations, suggesting novel treatment strategies for this aggressive malignancy.
INTRODUCTION
The treatment of solid malignancies has evolved and is perhaps best exemplified by the approach to non–small-cell lung cancer, for which molecular characterization and use of targeted agents have emerged as standard therapeutic paradigms. Recently, The Cancer Genome Atlas (TCGA) completed and published the integrated genomic and molecular characterization of cervical cancer.1 In addition to data previously released for both ovarian (high-grade serous) and endometrial (endometrioid and serous) cancers, this publication completed the molecular and genomic evaluation of the most common gynecologic malignancies.2,3
Context
Key Objective
To define the molecular landscape of high-grade neuroendocrine cervical cancer in a large cohort of patients.
Knowledge Generated
High-grade neuroendocrine cervical cancer appears molecularly distinct from the histologically similar small-cell lung cancer. Up to 73% of patients’ samples harbored potentially actionable alterations, informing novel treatment strategies.
Relevance
Continued understanding of the molecular underpinnings of high-grade neuroendocrine cervical carcinoma will be critical to driving drug discovery for this disease.
Traditionally, cervical cancer clinical trials have excluded less common histologies such as high-grade neuroendocrine cervical carcinoma (HGNECC). Despite the low incidence of HGNECC (< 2% of all cervical cancers) the oncologic impact is significant because these tumors exhibit more aggressive clinical characteristics.4,5 Unfortunately, the 5-year overall survival rate for patients with early-stage disease is only a 36%, and those with metastatic spread face an even more dismal prognosis. Given these poor outcomes, patients with HGNECC represent an area of unmet clinical need.
Developing therapeutic options for patients with rare tumors is challenging, relying on international collaboration, as well as small case series or retrospective reports rather than prospective clinical trials. The current therapeutic paradigm for the treatment of HGNECC was adopted from the more common, morphologically similar, small-cell lung cancer (SCLC) and includes surgical resection if feasible, followed by platinum plus etoposide-based combination chemotherapy, and possibly radiation.6,7 There are few studies informing treatment of recurrent disease, and there are no drugs approved by the Food and Drug Administration (FDA) specifically for HGNECC.8
Recently, 2 reports of exceptional responses to immune checkpoint inhibition in patients with recurrent HGNECC were published.9,10 To better understand these responses and to identify molecular aberrations underlying this uncommon malignancy, we examined the genomic landscape of HGNECC. Here, we report the identified molecular alterations in a cohort of HGNECC specimens, several of which may serve as actionable therapeutic targets, and compare the genomic landscape of this disease to the histologically similar SCLC, which has been historically used to model treatment of HGNECC.
PATIENTS AND METHODS
We evaluated a fully informative genomic profile of patients diagnosed with poorly differentiated (G3) neuroendocrine cervical carcinomas inclusive of both small- or large-cell subtypes (HGNECCs) whose cancers were submitted for hybrid capture–based next-generation sequencing (NGS) testing from March 2013 to December 2017 (N = 97). A cohort of 1,800 similarly tested cases of SCLC from the same period were subsequently evaluated to allow for comparison of genomic alterations (GAs). The submitting physicians provided specification of a poorly differentiated, neuroendocrine tumor type of cervical origin, which was then independently reviewed by a gynecologic pathologist (J.E.) to confirm high-grade neuroendocrine pathologic features in the pathology report and/or the representative sample of tumor submitted for sequencing (grade 3 cytomorphologic features, some component of small-cell or large-cell carcinoma histology, and/or positivity for neuroendocrine markers). The database was de-identified with only the diagnosis available. NGS data were generated by FoundationOne (Foundation Medicine; Cambridge, MA). The study was performed in accordance with University of California, San Diego, Institutional Review Board guidelines for a de-identified database. Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, were also obtained from the Western Institutional Review Board (Protocol No. 20152817).
Tissue Samples and Mutational Analysis
Available tissue from diagnostic or therapeutic procedures was used to determine oncogenic molecular alterations. Sequencing information was collected on 97 patients with HGNECC and 1,800 with SCLC, whose formalin-fixed, paraffin-embedded tumor samples were submitted to Foundation Medicine for genomic profiling. The test sequences the entire coding region of 182 or, more recently, 236 or 315 cancer-related genes plus up to 47 introns of up to 19 genes often rearranged or altered in cancer to an average depth of coverage of > 500×.11 The pathologic diagnosis of each case was confirmed on routine hematoxylin- and eosin-stained slides and all samples forwarded for DNA extraction contained a minimum of 20% tumor nuclear area. Microsatellite instability (MSI) status was evaluable in 75 HGNECC and 1,573 SCLC cases.
The sequencing methods used for comprehensive genomic profiling have been validated and reported previously (Appendix).12,13 The optimized loci used to evaluate MSI status were selected from a total set of 1,897 that have adequate coverage on all versions of the assay. Each locus is intronic and has a reference repeat length of 10-20 bp, which allows for analysis with the read length used by FoundationOne testing. Principal components analysis is used to produce an NGS-based MSI score. There was no need to extend beyond the first principal component, because it explained approximately 50% of the total data variance, whereas none of the other principal components explained > 4% each. Ranges of the MSI score were assigned MSI-High (MSI-H), MSI ambiguous, or microsatellite stable (MSS). MSI-Low calls are not made because there was no gold-standard test set, but we presume such samples would significantly overlap with the MSI-ambiguous category reported here. For samples in which MSI-specific quality control criteria were not met (n = 22 HGNECC; n = 227 SCLC), a status of MSI unknown was assigned,14 and these cases were excluded from additional MSI analysis.
Tumor Mutational Burden
The number of somatic mutations detected on NGS (interrogating up to 1.2 Mb of the genome) were quantified and that value extrapolated to the whole exome, using a validated algorithm.15 Alterations likely or known to be germline polymorphisms or bona fide oncogenic drivers were excluded. Tumor mutational burden (TMB) was measured in mutations per megabase. TMB levels were grouped into 3 bins: TMB-low (TMB-L; 1-5 mutations/Mb), intermediate (TMB-I; 6-19 mutations/Mb), and high (TMB-H; ≥ 20 mutations/Mb). The cutoff of 20 coding mutations/Mb is approximately equal to 400 nonsynonymous mutations per exome.15
Human Papillomavirus Detection
In addition, the presence of high-risk human papillomavirus (HPV) was examined in submitted specimens, as previously reported.16 Hybrid-capture reagents included baits designed to capture unique regions of select viral genomes including HPV-16 and -18. Sequence read pairs were aligned to the reference genome of the respective viral genomes, and the number of pairs mapping to each viral genome was counted. A total HPV-16/18 aligned read count of ≥ 5 reads per million was considered a positive HPV status, and < 5 reads per million was considered HPV not detected.16
End Points and Statistical Methods
Descriptive statistics were used to summarize the baseline patient characteristics. Fisher exact test was used to determine the association between categorical variables in univariate analysis and the Z-test was used to assess population differences, where appropriate. All tests were 2 sided. All statistical tests were carried out using GraphPad Prism, version 6.0 (GraphPad Software, San Diego, CA).
RESULTS
Characterization of GAs in HGNECC
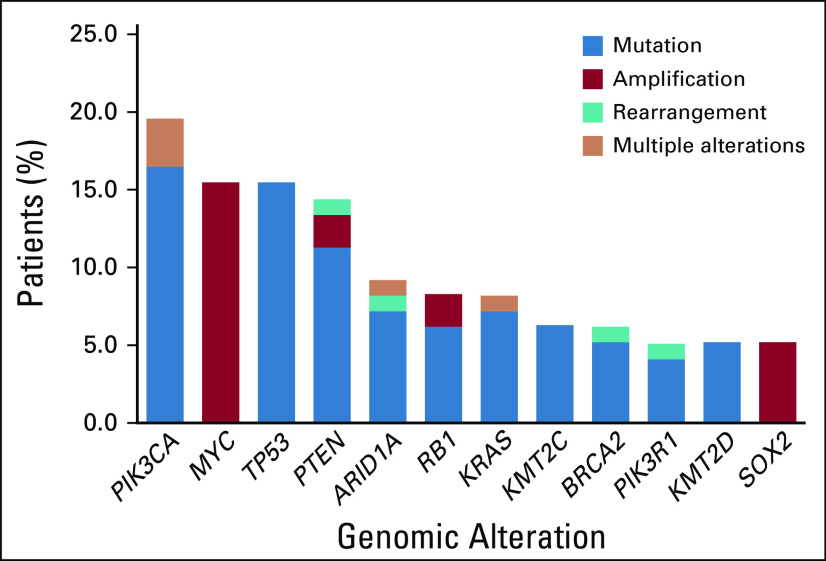
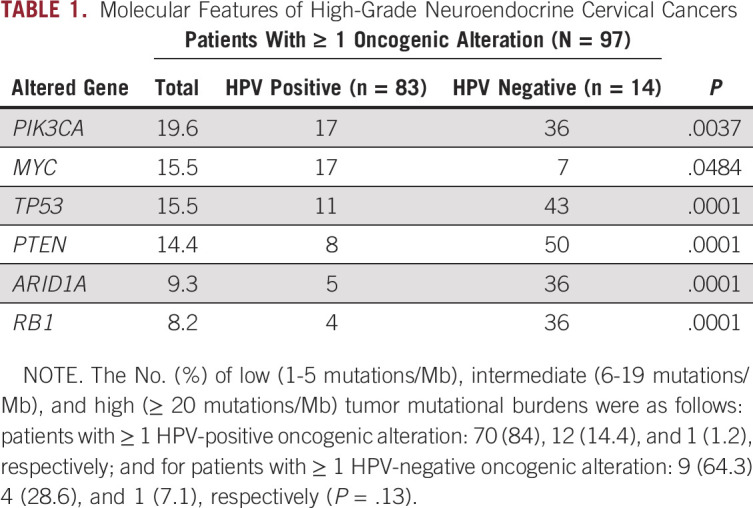
The median age of the cohort was 40.5 years (range, 25-77 years). Of the 97 patients, 83 were high-risk HPV positive (85.6%) and 14 were negative (14.4%). All samples were reflective of HGNECC, including both small-cell and large-cell HGNECC cases. Among the HGNECC cohort (N = 97), the most frequently identified GAs (discerned in > 10% of the cohort) involved PIK3CA (19.6% of patients), MYC (15.5%), TP53 (15.5%), and PTEN (14.4%) (Fig 1; a detailed list of all GAs can be found in Appendix Table A1).
FIG 1.
Most common genomic alterations in patients with high-grade neuroendocrine cervical cancer (HGNECC), reflected as percentage of patients (N = 97). Only genes altered in > 5% of samples are depicted. Variants of unknown significance excluded.
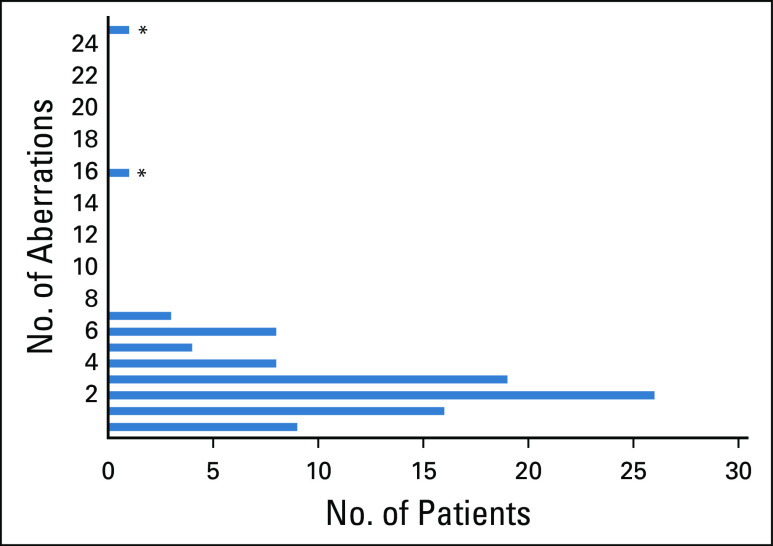
A total of 109 different genes were mutated in the 97 patient samples evaluated (variants of unknown significance were excluded from all analyses). The most frequently reported number of GAs per sample was 2, with a range of 0-25 (average, 3.0 GAs/sample; Fig 2). When evaluating TMB, 2 cases (2.1%) were TMB-H and 16 cases (16.5%) were TMB-I. Most patients’ tumors (n = 79; 81.4%) were TMB-L (Table 1). Nine patient samples had no known or likely GAs on comprehensive genomic profiling. Of the 88 patients who had an alteration, 72 had at least 1 alteration for which there currently existed an agent potentially targeting that alteration.
FIG 2.
Number of reported genomic alterations per patient. Variants of unknown significance are excluded. (*) Tumor mutational burden high (MSH2 mutation).
TABLE 1.
Molecular Features of High-Grade Neuroendocrine Cervical Cancers
Genomics in HPV-Positive Versus -Negative Patients
When examining distribution of GAs on the basis of HPV detection status, a significant difference was identified in the frequency of several GAs, including PIK3CA, TP53, PTEN, ARID1A, and RB1, all of which were more frequent in the HPV-negative subgroup (Table 1). There was no difference in the frequency of TMB-H between HPV-positive and HPV-negative or unknown samples (P = .2691).
Mismatch Repair Gene Aberrations Associated With MSI-High
Three cases (3%) had pathogenic MSH2 alterations. Two of the 3 cases were both MSI-H and TMB-H and harbored the highest numbers of identifiable GAs in the cohort.17,18 The third case, with a nonsense mutation near the 3′ end of the coding sequence (MSH2 R929*), was MSS and TMB-L. The 2 MSH-2–mutant MSI-H cases were both of small-cell histology and accounted for all MSI-H cases out of the 75 cases of HGNECC with evaluable microsatellite status (2.7%).
Less Frequent GAs
Additional genomic characterization was performed in which we specifically explored the homologous recombination deficiency (HRD), RAS, PI3K/AKT/mTOR, and ERBB pathways. Nine cases (9.3%) were had GAs in HRD-related genes, with the most frequent alterations noted in BRCA2 (n = 6 of 9; 66.7%).17,19 Three additional patient samples had BRCA1, ATM, and PALB2 mutations (n = 1 in each case) case (n = 3 of 9; 33.3%).
Eleven patient samples (11.3%) had alterations in the RAS pathway, with KRAS and BRAF mutations being the most frequent (72.7% [n = 8 of 11] and 27.3% [n = 3 of 11], respectively). Of the identified BRAF mutations, only 1 was a V600E alteration. Furthermore, a total of 40 patient samples (41.2%) harbored mutations in the PI3K/AKT/mTOR pathway; mutations in PIK3CA were identified in 47.5% of these samples (n = 19), and PTEN mutations were reported in 35% (n = 14). Last, 7 patients (7.2%) had mutations in the ERBB pathway, with ERBB2 mutations occurring in tumors of 4 individuals (57.1%).
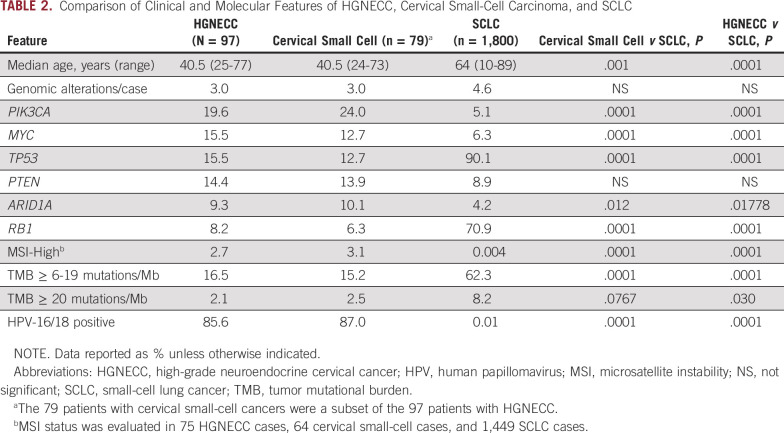
Comparison of HGNECC and SCLC
Given the histologic similarity between SCLC and HGNECC, tumor samples from a cohort of 1,800 patients with SCLC were compared with the HGNECC samples (Table 2). The SCLC samples featured significantly lower frequencies of GAs in PIK3CA, MYC, and ARID1A. In contrast, the HGNECC samples featured significantly lower frequencies of GAs in TP53 and RB1. High-risk HPV was identified in much less than 1% of SCLC tumor samples compared with 85.6% of HGNECC tumor samples. There was a single MSI-H SCLC case (n = 1 of 1,449; < 0.001%), whereas MSI-H status was found in 2 HGNECC cases (2.7%). Last, TMB was significantly higher in the SCLC samples compared with the HGNECC samples with respect to both intermediate and high TMB levels. The small-cell subset of HGNECC samples showed analogous gene mutation differences from SCLC.
TABLE 2.
Comparison of Clinical and Molecular Features of HGNECC, Cervical Small-Cell Carcinoma, and SCLC
DISCUSSION
Neuroendocrine carcinoma is an uncommon but aggressive variant accounting for approximately 1.5% of all newly diagnosed cervical cancers.20 The great majority of these lesions are high-grade large- or small-cell subtypes, with only rare reports of well-differentiated cervical carcinoid tumors.20 The treatment of patients with HGNECC remains clinically challenging, with limited response rates to chemotherapy; however, anecdotal reports of exceptional responders have been described.9,10
The paradigm for management of HGNECC has been informed by the treatment of the more commonly diagnosed (and histologically similar) SCLC, which accounts for approximately 15% of all lung cancer cases. In prior studies, whole-genome sequencing of 110 SCLC specimens identified essentially ubiquitous TP53 and RB1 inactivating mutations, with biallelic losses of each gene respectively in 100% and 93% of cases without chromothripsis.21
In an effort to better define the molecular landscape of HGNECC, we evaluated the comprehensive genomic profiling of 97 patient samples. The most frequently identified GA was PIK3CA mutation, occurring in 19.6% of submitted samples (n = 19). At least 1 characterized alteration was identified in 88 patient samples (90.7%) and of these, 72 had a potentially pharmacologically tractable alteration.
Interestingly, the frequency and distribution of GAs identified in this cohort of patients are similar and distinct from mutational patterns described in the more common HPV-related cervical cancer histologies.1 As detailed in TCGA’s integrated genomic characterization of cervical cancer (ie, squamous, adenocarcinoma, and adenosquamous histologies), mutations in the PIK3CA gene were the most frequently identified aberration, occurring in 26% of samples, approximating the nearly 20% rate in our cohort. In addition, significantly mutated genes reported by the TCGA, identified in similar proportions in this patient cohort, included ARID1A (7% in TCGA and 9.3% in our cohort) and KRAS (6% in TCGA and 8.2% in our cohort). Conversely, the examined neuroendocrine cohort had a greater frequency of PTEN mutations (8% in TCGA v 14.4% in our cohort). These molecular differences may be reflective of the varying histologies or, potentially, the differential high-risk HPV detection rates (85.6% in our cohort v 95% in the TCGA).1 Importantly, the high-risk HPV rate in our cohort should be interpreted with caution because the assay used has not undergone formal concordance study with gold standard tests such as hybrid capture and can detect only HPV 16/18.
Our own, much larger cohort of SCLC samples (n = 1,800) recapitulates prior studies and had a strikingly different molecular portfolio when compared with HGNECC samples. The frequency of TP53 and RB1 alterations in the SCLC cohort significantly exceeded that seen in our HGNECC cohort (15.5% and 8.2%, respectively), the HPV16/18 positive subset (11% and 4%, respectively), and the subset where HPV16/18 was not detected (43% and 36%, respectively; Table 2). Furthermore, mutations affecting the NOTCH pathway were identified in 25% of the examined SCLC samples; the NOTCH pathway is hypothesized to function as a regulator of neuroendocrine differentiation. In our examined HGNECC cohort, only 7 patients (7.2%) had NOTCH alterations. Alterations in PIK3CA, MYC, and PTEN were significantly more common in HGNECC when compared with SCLC (Table 2). MSI-H status was also more common in the HGNECC cohort whereas TMB-I/TMB-H was more common in SCLC (despite the lack of MSI-H status). Finally, HPV positivity was discerned in 85.6% of our HGNECC samples, but in only 0.01% of our SCLC samples (P < .0001). No parallels in molecular alterations were identified when comparing our findings for HGNECC with those of prior SCLC studies, supporting our premise that the similarity between these entities is largely morphologic and the treatment approaches for HGNECC can likely be improved through improved molecular granularity.
Despite the infrequency of HGNECC, the identification of potentially actionable GAs may inform treatment of a subset of patients with historically limited therapeutic options.18,22-25 In this cohort of patients, aberrations in the PI3K/AKT/mTOR pathway were commonly seen (PIK3CA [19.6%]; PTEN [14.4%]). The use of everolimus, or an alternate mTOR or PIK3CA inhibitor, may be considered in such circumstances, although the utility of a PIK3CA mutation in predicting response to single-agent everolimus in the presence of multiple GAs remains limited.26,27
Although less frequently identified, alterations in the HRD pathway were detected in 9.3% of patient samples, potentially supporting use of a poly-ADP ribose polymerase inhibitor. The identification of both TMB-H (n = 2) and GAs in mismatch repair genes (n = 3) may also inform the use of immune checkpoint inhibition.28 In May 2017, the FDA approved pembrolizumab for the treatment of mismatch repair–deficient or MSI-H solid tumors that progressed after prior therapy. This disease site–agnostic approval allows for a promising therapeutic option for patients with a previously unmet clinical need. More recently, the FDA accepted and granted priority review to a supplemental Biologics License Application for pembrolizumab for the treatment of adult and pediatric patients with unresectable or metastatic solid tumors with tissue TMB-H whose disease has progressed after prior treatment and who have no satisfactory alternative treatment options, supported by data from the phase II Keynote-158 trial. Notably, there are 2 published case reports of patients with recurrent, treatment-refractory HGNECC with exceptional and durable responses to checkpoint inhibition; 1 of these tumors was from our current HGNECC cohort and had a mismatch repair defect and the other lacked correlative genomic testing.9,10
Last, the identification of ARID1A (9.3%) and SMARCA4 (4.1%) mutations may predict sensitivity to an alternate therapeutic strategy.29 Homeostasis requires balanced ARID1A and EZH2 activity, facilitated via chromatin-mediated gene expression. Loss of ARID1A expression results in imbalanced EZH2 activity, and use of an EZH2 inhibitor such as tazemetostat may capitalize on this oncogene addiction. Importantly, 2 of the 4 SMARCA4 aberrations were identified in patients with MSI-H lesions, possibly reflecting that the SMARCA4 may be a passenger mutation resulting from the underlying MSI. Furthermore, of the 4 cases with SMARCA4 alterations, 1 was HPV-18 positive and another was p16 positive by immunohistochemical assessment.
Despite the large sample size and robust genomic data, this study has limitations. The retrospective design and use of archival tumor tissues from various time points during therapy may make interpretation of GAs difficult. In addition, the lack of demographic and clinical data, as well as treatment history, precludes exploratory assessments of response to a selected targeted agent. Last, HPV status was determined using molecular surrogates that differ from the assays used in clinical practice. It remains unclear if HPV infection is a prerequisite for neuroendocrine cervical carcinoma, although recent publications suggest > 85% of neuroendocrine cervical carcinomas are HPV positive, with HPV-16 and HPV-18 accounting for > 95% of the identified high-risk HPV strains.30
This report highlights the potential therapeutic utility of genomic testing in patients with this uncommon disease.27 Of interest, despite the histologic similarity between HGNECC and SCLC, which has led to the latter being used as a model for treating the former, the molecular portfolio of these 2 entities is strikingly different. Therefore, it is plausible that patients with HGNECC may benefit from alternative therapeutic strategies.
It is not anticipated that traditional prospective trials will accrue sufficient patient numbers in this disease setting, and novel study designs, including umbrella, basket, and platform trials, should be considered given the presence of actionable targets. Interestingly, the first reported cohort of the DART trial (ClinicalTrials.gov identifier: NCT02834013)31 was the neuroendocrine cohort, with a 44% overall response rate in those with high-grade disease. Ultimately, comprehensive genomic characterization may catalyze the investigation and identification of effective therapies, allowing us to improve oncologic outcomes in this aggressive disease.
Appendix
Sequencing Methods Used for Comprehensive Genomic Profiling
Sample processing and sequencing were performed in a Clinical Laboratory Improvement Amendments– and College of American Pathologists–accredited laboratory. Briefly, after pathologic review to confirm sufficient tumor nuclei (minimum, 20%) and mitigate pathologic inconsistencies, at least 50 ng of DNA was extracted from 40 microns of tumor samples provided as formalin-fixed, paraffin-embedded tissue blocks. The samples were assayed using adaptor-ligation and hybrid-capture next-generation sequencing (FoundationOne) for all coding exons from 182 (version 1), 287 (version 2), or 315 (version 3) cancer-related genes plus selected introns from 14 (version 1), 19 (version 2), or 28 (version 3) genes frequently rearranged in cancer.
Sequencing of captured libraries was performed using HiSEquation 2500/4000 (Illumina, San Diego, CA) to a mean exon coverage depth of > 500×, and resultant sequences were analyzed using both an algorithmic pipeline and manual curation for base substitutions, small insertions or deletions (indels), copy number alterations (focal amplifications and homozygous deletions), and selected gene fusions, as previously described.13 Clinically relevant genomic alterations were defined as alterations targetable by anticancer drugs currently available on the market or in registered clinical trials. Germline variants documented in the dbSNP database (dbSNP142; http://www.ncbi.nlm.nih.gov/SNP/), with ≥ 2 counts in the ExAC database (http://exac.broadinstitute.org/), or recurrent variants of unknown significance that were predicted by an internally developed algorithm to be germline were removed, with the exception of known driver germline events.12 Known, confirmed somatic alterations deposited in the Catalog of Somatic Mutations in Cancer (version 62) were highlighted as biologically significant, as were inactivating events in tumor suppressor genes. To maximize mutation-detection accuracy (sensitivity and specificity) in impure clinical specimens, the test was previously optimized and validated to detect base substitutions at a ≥ 5% mutant allele frequency (MAF), indels with a ≥ 10% MAF with ≥ 99% accuracy, and fusions occurring within baited introns/exons with > 99% sensitivity.12 Each tumor sample was analyzed alongside an internally validated mixture of 10 heterozygous diploid HapMap control samples, which custom algorithms used to normalize the sequence coverage distribution across baited targets.
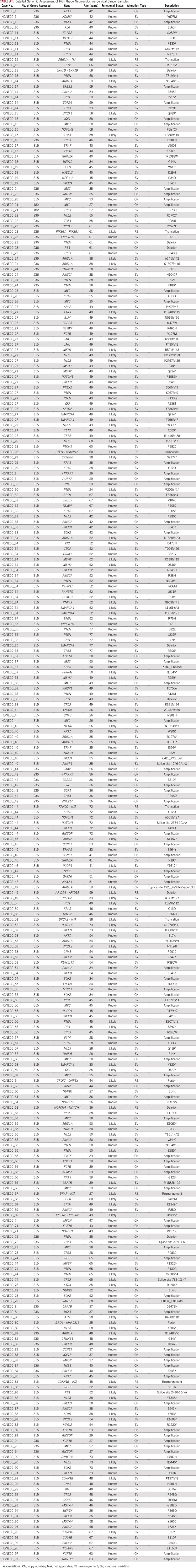
TABLE A1.
Detailed Genomic Assessment of High Grade Neuroendocrine Cervical Cancer Samples
R.N.E. and J.E. contributed equally to this work.
SUPPORT
Supported in part by National Cancer Institute (Grant No. P30 CA023100 [R.N.E.]) and the Joan and Irwin Jacobs Fund philanthropic fund.
AUTHOR CONTRIBUTIONS
Conception and design: Ramez N. Eskander, Julia Elvin, Jeffrey S. Ross, Vincent A. Miller
Collection and assembly of data: Ramez N. Eskander, Julia Elvin, Laurie Gay, Jeffrey S. Ross
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Ramez N. Eskander
Consulting or Advisory Role: Pfizer, Clovis Oncology, AstraZeneca/MedImmune, Tesaro, Merck
Speakers' Bureau: Clovis Oncology, AstraZeneca/MedImmune, Roche
Travel, Accommodations, Expenses: Clovis Oncology, AstraZeneca/MedImmune, Roche, Merck, Pfizer
Julia Elvin
Employment: Foundation Medicine
Stock and Other Ownership Interests: Hoffman-LaRoche
Laurie Gay
Employment: Foundation Medicine, Invitae
Stock and Other Ownership Interests: Foundation Medicine, Naveris, Invitae
Consulting or Advisory Role: Invitae, Naveris
Jeffrey S. Ross
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Consulting or Advisory Role: Celsius Therapeutics
Research Funding: Foundation Medicine
Vincent A. Miller
Employment: Foundation Medicine
Leadership: Foundation Medicine, Revolution Medicines
Stock and Other Ownership Interests: Foundation Medicine, Mirati Therapeutics, Revolution Medicines
Patents, Royalties, Other Intellectual Property: Receive periodic royalties related to T790M patent awarded to Memorial Sloan Kettering Cancer Center
Razelle Kurzrock
Leadership: CureMatch, CureMetrix
Stock and Other Ownership Interests: CureMatch, IDbyDNA, Soluventis
Honoraria: Roche, EUSA Pharma, NeoGenomics Laboratories, Biocom, NeoMed Therapeutics, Advanced Therapeutics, LEK, AACR, Chugai Pharma USA, Wiley
Consulting or Advisory Role: Actuate Therapeutics, Loxo, XBiotech, Neo-Med, Roche, Gaido, Soluventis, Pfizer, Merck
Speakers' Bureau: Roche, Guardant Health (Inst)
Research Funding: Sequenom (Inst), Merck Serono (Inst), Genentech (Inst), Pfizer (Inst), Foundation Medicine (Inst), Incyte (Inst), Konica Minolta (Inst), Grifols (Inst), OmniSeq (Inst), Debiopharm Group (Inst), Boerhinger Ingelheim (Inst)
Travel, Accommodations, Expenses: Roche, EUSA Pharma, NeoGenomics Laboratories, Biocom, NeoMed Therapeutics, Advanced Therapeutics, LEK, AACR, Chugai Pharma USA, Wiley
No other potential conflicts of interest were reported.
REFERENCES
- 1. doi: 10.1038/nature21386. Cancer Genome Atlas Research Network: Integrated genomic and molecular characterization of cervical cancer. Nature 543:378-384, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. doi: 10.1038/nature12113. Kandoth C, Schultz N, Cherniack AD, et al: Integrated genomic characterization of endometrial carcinoma. Nature 497:67-73, 2013 [Erratum: Nature 500:242, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. doi: 10.1038/nature10166. Cancer Genome Atlas Research Network: Integrated genomic analyses of ovarian carcinoma. Nature 474:609-615, 2011 [Erratum: Nature 490:298, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner GJ, Reidy-Lagunes D, Gehrig PA. Neuroendocrine tumors of the gynecologic tract: A Society of Gynecologic Oncology (SGO) clinical document. Gynecol Oncol. 2011;122:190–198. doi: 10.1016/j.ygyno.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 5. Satoh T, Takei Y, et al: Gynecologic Cancer InterGroup (GCIG) consensus review for small cell carcinoma of the cervix. Int J Gynecol Cancer 24:S102-S108, 2014 (9 Suppl 3) [DOI] [PubMed]
- 6.Fukuoka M, Masuda N, Furuse K, et al. A randomized trial in inoperable non-small-cell lung cancer: Vindesine and cisplatin versus mitomycin, vindesine, and cisplatin versus etoposide and cisplatin alternating with vindesine and mitomycin. J Clin Oncol. 1991;9:606–613. doi: 10.1200/JCO.1991.9.4.606. [DOI] [PubMed] [Google Scholar]
- 7.Kubota K, Hida T, Ishikura S, et al. Etoposide and cisplatin versus irinotecan and cisplatin in patients with limited-stage small-cell lung cancer treated with etoposide and cisplatin plus concurrent accelerated hyperfractionated thoracic radiotherapy (JCOG0202): A randomised phase 3 study. Lancet Oncol. 2014;15:106–113. doi: 10.1016/S1470-2045(13)70511-4. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa M, Kasamatsu T, Tsuda H, et al. Prognostic factors and optimal therapy for stages I-II neuroendocrine carcinomas of the uterine cervix: A multi-center retrospective study. Gynecol Oncol. 2018;148:139–146. doi: 10.1016/j.ygyno.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 9.Paraghamian SE, Longoria TC, Eskander RN. Metastatic small cell neuroendocrine carcinoma of the cervix treated with the PD-1 inhibitor, nivolumab: A case report. Gynecol Oncol Res Pract. 2017;4:3. doi: 10.1186/s40661-017-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharabi A, Kim SS, Kato S, et al. Exceptional response to nivolumab and stereotactic body radiation therapy (SBRT) in neuroendocrine cervical carcinoma with high tumor mutational burden: Management considerations from the Center for Personalized Cancer Therapy at UC San Diego Moores Cancer Center. Oncologist. 2017;22:631–637. doi: 10.1634/theoncologist.2016-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foundation Medicine: FoundationOne CDx. http://foundationone.com/docs/FoundationOne_tech-info-and-overview.pdf.
- 12.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross JS, Fakih M, Ali SM, et al. Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer. 2018;124:1358–1373. doi: 10.1002/cncr.31125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4:959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechner M, Frampton GM, Fenton T, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh CS. Two decades beyond BRCA1/2: Homologous recombination, hereditary cancer risk and a target for ovarian cancer therapy. Gynecol Oncol. 2015;137:343–350. doi: 10.1016/j.ygyno.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Chae YK, Pan AP, Davis AA, et al. Path toward precision oncology: Review of targeted therapy studies and tools to aid in defining “actionability” of a molecular lesion and patient management support. Mol Cancer Ther. 2017;16:2645–2655. doi: 10.1158/1535-7163.MCT-17-0597. [DOI] [PubMed] [Google Scholar]
- 19.Carey JPW, Keyomarsi K. Leveraging MYC as a therapeutic treatment option for TNBC. Oncoscience. 2018;5:137–139. doi: 10.18632/oncoscience.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tempfer CB, Tischoff I, Dogan A, et al. Neuroendocrine carcinoma of the cervix: A systematic review of the literature. BMC Cancer. 2018;18:530. doi: 10.1186/s12885-018-4447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel M, Kato SM, Kurzrock R. Molecular tumor boards: Realizing precision oncology therapy. Clin Pharmacol Ther. 2018;103:206–209. doi: 10.1002/cpt.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kato S, Ross JS, Gay L, et al:Analysis of MDM2 amplification: Next-generation sequencing of patients with diverse malignancies. JCO Precis Oncol 10.1200/PO.17.00235. [DOI] [PMC free article] [PubMed]
- 24.Kato S, Kurasaki K, Ikeda S, et al. Rare Tumor Clinic: The University of California San Diego Moores Cancer Center experience with a precision therapy approach. Oncologist. 2018;23:171–178. doi: 10.1634/theoncologist.2017-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hainsworth JD, Meric-Bernstam F, Swanton C, et al. Targeted therapy for advanced solid tumors on the basis of molecularp: Results from MyPathway, an open-label, phase IIa multiple basket study. J Clin Oncol. 2018;36:536–542. doi: 10.1200/JCO.2017.75.3780. [DOI] [PubMed] [Google Scholar]
- 26.Janku F, Hong DS, Fu S, et al. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep. 2014;6:377–387. doi: 10.1016/j.celrep.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsimberidou AM, Hong DS, Ye Y, et al: Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): An MD Anderson precision medicine study. JCO Precis Oncol 10.1200/PO.17.00002. [DOI] [PMC free article] [PubMed]
- 28.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alldredge JK, Eskander RN. EZH2 inhibition in ARID1A mutated clear cell and endometrioid ovarian and endometrioid endometrial cancers. Gynecol Oncol Res Pract. 2017;4:17. doi: 10.1186/s40661-017-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alejo M, Alemany L, Clavero O, et al. Contribution of human papillomavirus in neuroendocrine tumors from a series of 10,575 invasive cervical cancer cases. Papillomavirus Res. 2018;5:134–142. doi: 10.1016/j.pvr.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.