Abstract
Epidermal growth factor (EGF) and its receptor (EGFR) play a paramount role in lung carcinogenesis. The polymorphism in the EGF promoter region EGF+61A>G (rs4444903) has been associated with cancer susceptibility, but its role in lung cancer patients treated with tyrosine kinase inhibitors (TKIs) remains unknown. Here, we aimed to evaluate the predictive and prognostic role of EGF+61A>G SNP in lung cancer from Brazilian EGFR‐mutated TKI‐treated patients. Herein, patients carrying EGFR‐sensitizing mutations submitted to TKI treatment (gefitinib/erlotinib) were analyzed (n = 111) for EGF+61A>G genotype by TaqMan genotyping assay. TKI treatment was classified as partial response (PR), stable disease (SD), and disease progression (DP), according to RECIST1.1. Association analysis was assessed by chi‐square and Fisher's test (univariate) and multinomial model (multivariate) and survival analysis by Kaplan‐Meier method and log‐rank test. The EGF+61A>G genotype frequencies observed were: AA = 31.5% (n = 35), AG = 49.6% (n = 55) and GG = 18.9% (n = 21). The allelic frequencies were 56.3% for A, and 43.7% for G and the population was in Hardy‐Weinberg equilibrium (P = 0.94). EGF+61A>G codominant model (AA vs. AG vs. GG) was associated with a response to TKIs (P = 0.046), as well as a recessive model (AA vs. AG + GG; P = 0.023). The multinomial regression showed an association between the codominant model (AG) and recessive model (AG + GG) with SD compared with DP (P = 0.01;OR = 0.08; 95% CI = 0.01–0.60 and P = 0.02;OR = 0.12; 95% CI = 0.20–0.72, respectively). No association between genotypes and progression‐free or overall survival was observed. In conclusion, the EGF+61 polymorphism (AG and AG + GG) was independently associated with stable disease in lung cancer patients although it was not associated with the overall response rate to first‐generation TKIs or patient outcome.
Keywords: EGF+61 A>G polymorphism, EGFR, first‐generation TKI SNP, lung cancer, TKI
• EGF+61 SNP was assessed in TKI‐treated NSCLC patients harboring EGFR‐sensitizing mutations.
• EGF+61 SNP (AG + GG) was independently associated stable in EGFR‐mutated TKI‐treated NSCLC patients.
• EGF+61 SNP (AG + GG) cannot predict progression‐free and overall survival or overall response rate to first generation TKIs.
Introduction
Epidermal growth factor (EGF) and its receptor (EGFR) play a key role in the pathogenesis of several tumors, including non‐small cell lung cancer (NSCLC). 1 , 2 EGFR is a critical therapeutic target, and the presence of EGFR mutations has previously successfully guided clinical management in NSCLC. 3 , 4 EGFR mutations located at the tyrosine kinase domain, sensitizes NSCLC to treatment with anti‐EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib. 3 , 4 , 5 , 6 However, most TKI‐treated patients will experience disease progression due to resistance mechanisms, such as the presence of EGFR resistance mutations. 7
A single nucleotide polymorphism (SNP) in the 5′ untranslated region of the EGF gene (EGF+61 A>G — rs4444903) has been associated with increased levels of EGF and consequently is a risk for cancer development of distinct tumors. 2 In lung cancer, conflicting results have been reported in different populations. 8 , 9 , 10 , 11 Importantly, in advanced rectal cancer, EGF+61 A>G SNP predicted a complete pathologic response to cetuximab, an anti‐EGFR monoclonal antibody. 12
Although cetuximab and TKIs are two different forms of therapies directed at EGFR, they both result in blocking signal transduction. 13 Thus, we hypothesized that EGF+61 A>G SNP might be associated with a response to first‐generation TKIs in lung cancer. Thereafter, we investigated the association between EGF+61 A>G genotype and TKI response in a Brazilian series comprised of 111 TKI‐treated lung adenocarcinoma patients harboring EGFR‐sensitizing mutations.
Methods
Study population
This was a retrospective and observational study, which included patients with NSCLC, diagnosed between 2001 and 2017 at Barretos Cancer Hospital. The present study was approved by the Barretos Cancer Hospital IRB (Project No. 630/2012), and all methods were performed following the Helsinki declaration.
From a series of lung adenocarcinoma cases (n = 870), 183 patients presented with EGFR mutations and 127 out of 183 patients were TKI‐treated (Fig 1). The response to TKI‐treatment was determined using RECIST 1.1, and the clinical response was categorized as partial response (PR) for overall response rate (ORR), stable disease (SD) for disease control rate (DCR), or disease progression (DP). NSCLC patients harboring EGFR resistance mutations (eg, T790M) are not currently eligible for erlotinib, and gefitinib treatment; thus, these patients (n = 7) were not evaluated (Fig 1). Nine patients were excluded due to inconclusive genotyping results (Fig 1 ). The demographic and clinicopathological data from eligible patients (n = 111) were obtained by reviewing the medical records (Table 1).
Figure 1.
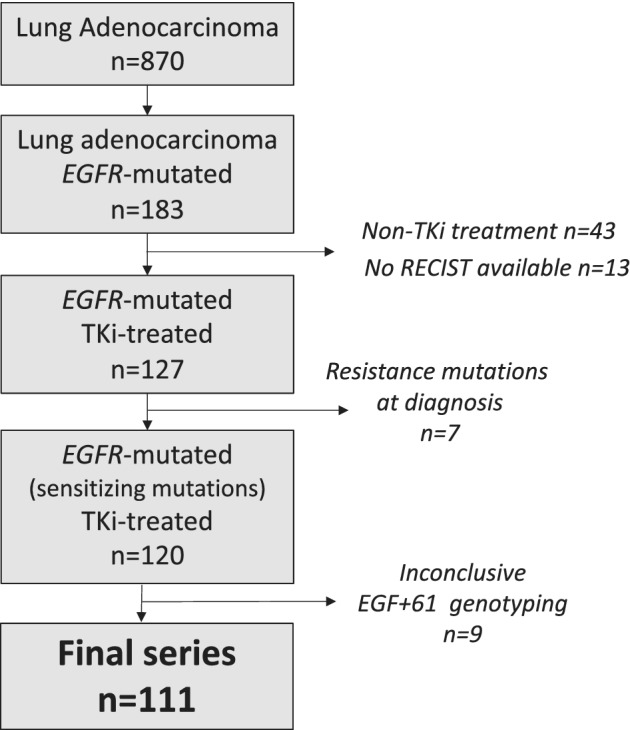
Workflow of sample procurement according to eligibility criteria.
Table 1.
Demographic and clinicopathological features of EGFR‐mutated TKI‐treated NSCLC patients enrolled in the study (n = 111)
| Characteristics | n = 111 | Frequencies | |
|---|---|---|---|
| Sex | Male | 47 | 42.3% |
| Female | 64 | 57.7% | |
| Age | Median (SD) | 61.75 (11.48) | |
| ≤63 years | 58 | 52.3% | |
| >63 years | 53 | 47.7% | |
| Self‐reported skin color | White | 96 | 86.5% |
| Other | 15 | 13.5% | |
| Smoking status | Never | 65 | 58.6% |
| Current | 17 | 15.3% | |
| Former | 28 | 25.2% | |
| NA | 1 | 0.9% | |
| ECOG PS | 0–1 | 78 | 70.3% |
| 2 | 20 | 18.0% | |
| 3–4 | 8 | 7.2% | |
| NA | 5 | 4.5% | |
| Weight loss in the last six months prior to diagnosis | >10% | 18 | 16.2% |
| <10% | 46 | 41.4% | |
| None | 39 | 35.1% | |
| NA | 8 | 7.3% | |
| TNM stage | I/II/III | 12 | 10.8% |
| IV | 99 | 89.2% | |
| Histological subtype | Acinar | 42 | 37.8% |
| Mucinous | 1 | 0.9% | |
| Lepidic | 8 | 7.3% | |
| Papillary | 10 | 9.0% | |
| Solid | 10 | 9.0% | |
| Others | 7 | 6.3% | |
| Not defined | 33 | 29.7% | |
| Central nervous system metastasis | Yes | 32 | 28.8% |
| No | 79 | 71.2% | |
| Current status | Alive | 24 | 21.6% |
| Dead due to cancer | 80 | 72.1% | |
| Dead due to comorbidity | 1 | 0.9% | |
| NA | 6 | 5.4% | |
| Response to TKI treatment | Partial response | 78 | 70.3% |
| Stable disease | 18 | 16.2% | |
| Disease progression | 15 | 13.5% | |
| EGFR mutation according to exon location | Exon 19 | 69 | 62.2% |
| Exon 20 | 3 | 2.7% | |
| Exon 21 | 25 | 22.5% | |
| Exon 18 and exon 21 | 2 | 1.8% | |
| Exon 19 and exon 20 | 9 | 8.1% | |
| Exon 20 and exon 21 | 3 | 2.7% |
NA, data unavailable; SD, standard deviation.
DNA isolation and EGFR mutational status
EGFR mutational status was assessed by examining formalin‐fixed paraffin‐embedded (FFPE) tumor tissue from NSCLC patients. DNA was isolated using QIAamp DNA Micro Kit (Qiagen) according to the manufacturer's instructions, and EGFR mutations were evaluated by Sanger sequencing as previously described. 14 A subset (n = 444) of this series comprised of EGFR‐mutated patients has been previously reported. 14
EGF+61 A>G genotyping
EGF+61 A>G polymorphism was analyzed in the same DNA samples used to access EGFR mutation. Genotyping was performed using quantitative real‐time PCR, with a commercially available TaqMan genotyping assay (ThermoFisher, USA) (C_27031637_10 [EGF+61 A>G]) in the QuantStudio 6 Flex Real‐Time PCR System (ThermoFisher, USA) and under standard cycling, as previously reported. 8
Statistical analysis
Categorical variables are presented as percentages. Hardy‐Weinberg equilibrium was verified. EGF+61 A>G genotype was analyzed using three different models: codominant model – AA versus AG versus GG; recessive model – AA versus AG + GG; dominant model – AA+AG versus GG. χ2 test was applied to verify the association between TKI treatment response and EGF+61 A>G genotype and demographic and clinicopathological characteristics. The Kaplan‐Meier method and log‐rank test were employed for survival analysis, considering for progression‐free survival (PFS), disease progression as the event of interest and, for overall survival (OS), death as the event of interest. Patients who had not experienced disease progression were considered as censored for PFS and alive patients and those lost to follow up were considered as censored for OS.
Multinomial regression adjusted by age, TNM and PS‐ECOG were used to access the association between EGF+61 A>G genotype and TKI treatment response by computing the odds ratio (ORs) with 95% confidence interval (CI) considering DP as the reference category. All statistical analyses were performed using IBM SPSS Statistics, version 21.0 (IBM Corp, Armonk, NY, USA). A P‐value lower than 0.05 was considered statistically significant.
Results
The demographic and clinicopathological characteristics of the 111 eligible patients (Fig 1) are detailed in Table 1. The median age was 61.75 years (SD ± 11.48), 47 were males and 64 females, most patients were never smokers (n = 65), and the majority were diagnosed at disease stage IV (n = 99) (Table 1). Notably, most patients harboring EGFR‐sensitizing mutations presented a partial response as the best TKI response (n = 78, 70.3%; Table 1), and none of the patients presented with a complete response (CR) (Table 1). The association between response to TKI treatment and demographic and clinicopathological characteristics from TKI‐treated NSCLC patients harboring EGFR‐sensitizing mutations (n = 111) was also assessed, and no association was revealed (Table S1).
EGF+61 A>G genotypic frequencies were 31.5% (n = 35) for AA, 49.6% (n = 55) for AG and 18.9% (n = 21) for GG. The allelic distribution was 56.3% for the A allele and 47.7% for the G allele (Table S2). The present series (n = 111) was in Hardy‐Weinberg equilibrium, according to EGF+61 A>G polymorphism (P = 0.941).
The patients were grouped into three categories according to response to treatment (PR vs. SD vs. DP) and three genotypic models to assess the association between EGF+61 A>G polymorphism and response to TKI treatment. There was an association between response to TKI treatment and EGF+61 A>G polymorphism for a codominant and recessive model (P = 0.046 and P = 0.023, respectively; Table 2). No association was observed between response to TKI treatment and the dominant model (P = 0.601, Table 2). In addition, no association was observed between response to TKI treatment and allele frequencies (P = 0.163) (Table 2).
Table 2.
Association between EGF+61 genotype and alleles from TKI‐treated NSCLC patients harboring EGFR‐sensitizing mutations (n = 111) and TKI treatment response
| PR | SD | DP | |||||
|---|---|---|---|---|---|---|---|
| Response to TKI treatment | n | % | n | % | n | % | P‐value |
| Genotype (codominant model) | |||||||
| AA | 23 | 29.5% | 10 | 55.6% | 2 | 13.3% | 0.046 |
| AG | 42 | 53.8% | 4 | 22.2% | 9 | 60.0% | |
| GG | 13 | 16.7% | 4 | 22.2% | 4 | 26.7% | |
| Genotype (recessive model) | |||||||
| AA | 23 | 29.5% | 10 | 55.6% | 2 | 13.3% | 0.023 |
| AG + GG | 55 | 70.5% | 8 | 44.4% | 13 | 86.7% | |
| Genotype (dominant model) | |||||||
| AA + AG | 65 | 83.3% | 14 | 77.8% | 11 | 73.3% | 0.601 |
| GG | 13 | 16.7% | 4 | 22.2% | 4 | 26.7% | |
| Allele frequency | |||||||
| A | 56.4% | 66.7% | 43.3% | 0.163 | |||
| G | 43.6% | 33.3% | 56.7% | ||||
Bold P‐values indicate P ≤ 0.05 from univariate analysis (χ2 test).
DP, disease progression; PR, partial response; SD, stable disease.
Since EGF+61 A>G genotypes (codominant and recessive model) were associated with response to TKI treatment, we assessed whether any clinical parameter could be influenced by this result as a confounder variable and a multivariate analysis (multinomial analysis) was conducted adjusted for age, TNM, and PS‐ECOG. We observed that in the codominant model, AG genotype was independently associated with SD (P = 0.01; OR = 0.08; 95% CI: 0.01–0.60). Likewise, we observed that in the recessive model, AG + GG genotypes were independently associated with SD (P = 0.02; OR = 0.12; 95% CI: 0.20–0.72) (Table 3).
Table 3.
Multivariate analysis for association between EGF+61 genotype and alleles from TKI‐treated NSCLC patients harboring EGFR‐sensitizing mutations (n = 111) and TKI treatment response
| Response to TkI treatment* | n | OR | 95% CI | P‐value | ||
|---|---|---|---|---|---|---|
| Codominant model | PR | AA | 23 | 1 | — | Ref. |
| AG | 42 | 0.50 | 0.09–2.80 | 0.43 | ||
| GG | 13 | 0.39 | 0.05–2.62 | 0.33 | ||
| SD | AA | 10 | 1 | — | Ref. | |
| AG | 4 | 0.08 | 0.01–0.60 | 0.01 | ||
| GG | 4 | 0.19 | 0.02–1.58 | 0.12 | ||
| Recessive model | PR | AA | 23 | 1 | — | Ref. |
| AG + GG | 55 | 0.47 | 0.91–2.44 | 0.37 | ||
| SD | AA | 10 | 1 | — | Ref. | |
| AG + GG | 8 | 0.12 | 0.20–0.72 | 0.02 | ||
Bold P‐values indicate P ≤ 0.05 from multivariate analysis by multinomial adjusted by age, TNM and PS‐ECOG. Reference category: DP, disease progression.
PR, partial response; SD, stable disease; Ref., reference category.
*According to RECIST criteria.
In addition, we evaluated the association between EGF+61 A>G genotypes and post‐TKI PFS and OS. No association was observed between EGF+61 A>G codominant model (AA vs. AG vs. GG; P = 0.562), recessive model (AA vs. AG + GG; P = 0.709) and dominant model (AA +AG vs. GG; P = 0.401) and post‐TKI PFS (Fig S1A–C, respectively). Likewise, no association was observed between EGF+61 A>G codominant model (AA vs. AG vs. GG; P = 0.493), recessive model (AA vs. AG
+GG; P = 0.318) and dominant model (AA + AG vs. GG; P = 0.342) and OS (Fig S1–F, respectively). Thus, the results show that the EGF+61 A>G polymorphism does not influence disease outcomes, both PFS and OS from NSCLC TKI‐treated patients harboring EGFR‐sensitizing mutations.
Discussion
Molecular therapies have revolutionized clinical management in NSCLC patients, with particular emphasis on EGFR‐TKIs. 3 , 4 , 5 , 15 , 16 , 17 Herein, we report the impact of EGF+61 A>G genotype over ORR and DCR to first‐generation TKIs (erlotinib and gefitinib) on 111 EGFR‐mutated Brazilian lung adenocarcinoma patients.
The EGF+61 polymorphism (AG and AG + GG) was associated with stable disease in both univariate and multivariate in NSCLC patients treated with first‐generation TKIs harboring EGFR‐sensitizing mutations. Nevertheless, the SNP was not associated with ORR or disease outcome (progression‐free and overall survival). It is well known that EGFR‐sensitizing mutations can predict a response to first‐generation TKIs, and TK‐treated EGFR‐mutated patients can benefit from a substantial improvement in progression‐free and overall survival. 6 , 17 However, most patients will experience disease progression due to acquired resistance mechanisms. 7 Currently, there is an effective third‐generation TKI, osimertinib, for patients harboring EGFR‐resistance mutations. 18 Nonetheless, TKIs are considered high‐cost drugs for low‐middle income countries, and only first‐generation TKIs (erlotinib/gefitinib) are supported by the Brazilian public health system for EGFR‐mutated NSCLC patients. Thus, a biomarker for predicting both ORR and DCR in patients treated with first‐generation TKIs harboring EGFR‐sensitizing mutations would be valuable when financial resources are limited.
In conclusion, the present study revealed that the EGF+61 polymorphism (AG and AG + GG) was independently associated with stable disease, although the identification of this SNP cannot predict ORR, DCR or disease outcome in patients treated with first‐generation TKIs harboring EGFR‐sensitizing mutations.
Disclosure
The authors declare that they have nothing to disclose.
Supporting information
Figure S1. Survival analysis, according to EGF+61 genotypes. Progression‐free survival for codominant (a); recessive (b); and dominant (c) models. Overall survival for codominant (c); recessive (d); and dominant (e) models.
Table S1. Demographic and clinicopathological features of TKI‐treated NSCLC patients harboring EGFR‐sensitizing mutations (n = 111) and association with treatment response.
Table S2. Genotypic and allelic frequencies of TKI‐treated NSCLC patients harboring EGFR sensitizing mutations (n = 111).
Acknowledgments
This study was partially supported by the Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer), FINEP ‐ CT‐INFRA (02/2010), Barretos Cancer Hospital Research Fund (PAIP) and National Council for Scientific and Technological Development (CNPq, Brazil). Funding sources have no contribution to filling out authorship for the present study.
We thank all members of the GTOP group (Translational Group of Pulmonary Oncology, Barretos Cancer Hospital, Brazil) for scientific discussion and suggestions. We also thank Aline Larissa Virginio da Silva and Lázaro Antônio Campanha Novaes for all their support with data collection from medical records.
References
- 1. da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011; 6 (1): 49–69. [DOI] [PubMed] [Google Scholar]
- 2. Li T‐F, Ren K‐W, Liu P‐F. Meta‐analysis of epidermal growth factor polymorphisms and cancer risk: Involving 9,779 cases and 15,932 controls. DNA Cell Biol 2012; 31 (4): 568–74. [DOI] [PubMed] [Google Scholar]
- 3. Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non–small‐cell lung cancer. J Clin Oncol 2007; 25 (5): 587–95. [DOI] [PubMed] [Google Scholar]
- 4. Kris MG, Natale RB, Herbst RS et al Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non–small cell lung cancer. JAMA 2003; 290 (16): 2149–58. [DOI] [PubMed] [Google Scholar]
- 5. Lynch TJ, Bell DW, Sordella R et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350 (21): 2129–39. [DOI] [PubMed] [Google Scholar]
- 6. Wu Y‐L, Zhou C, Liam C‐K et al First‐line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer: Analyses from the phase III, randomized, open‐label, ENSURE study. Ann Oncol 2015; 26 (9): 1883–9. [DOI] [PubMed] [Google Scholar]
- 7. Ma C, Wei S, Song Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. J Thorac Dis 2011; 3 (1): 10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laus AC, de Paula FE, de Lima MA et al EGF+61 A>G polymorphism is not associated with lung cancer risk in the Brazilian population. Mol Biol Rep 2019; 46 (2): 2417–25. [DOI] [PubMed] [Google Scholar]
- 9. Lim YJ, Kim J‐W, Song JY et al Epidermal growth factor gene polymorphism is different between schizophrenia and lung cancer patients in Korean population. Neurosci Lett 2005; 374 (3): 157–60. [DOI] [PubMed] [Google Scholar]
- 10. de Mello RA, Ferreira M, Costa S et al Association between EGF +61 genetic polymorphisms and non‐small cell lung cancer increased risk in a Portuguese population: A case–control study. Tumour Biol 2012; 33 (5): 1341–8. [DOI] [PubMed] [Google Scholar]
- 11. Chen Q, Zheng Y, Wu B, Chen X, Ge P, Wang P. Association between polymorphisms of epidermal growth factor 61 and susceptibility of lung cancer: A meta‐analysis. Medicine (Baltimore) 2020; 99 (17): e19456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu‐lieskovan S, Vallbohmer D, Zhang W et al EGF61 polymorphism predicts complete pathologic response to cetuximab‐based chemoradiation independent of KRAS status in locally advanced rectal cancer patients. Clin Cancer Res 2011; 17 (15): 5161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mukohara T, Engelman JA, Hanna NH et al Differential effects of gefitinib and cetuximab on non–small‐cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst 2005; 97 (16): 1185–94. [DOI] [PubMed] [Google Scholar]
- 14. Leal LF, de Paula FE, De Marchi P et al Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci Rep 2019; 9 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Remon J, Steuer CE, Ramalingam SS, Felip E. Osimertinib and other third‐generation EGFR TKI in EGFR‐mutant NSCLC patients. Ann Oncol 2018; 29: i20–7. [DOI] [PubMed] [Google Scholar]
- 16. Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene 2000; 19 (56): 6550–65. [DOI] [PubMed] [Google Scholar]
- 17. Maemondo M, Inoue A, Kobayashi K et al Gefitinib or chemotherapy for non–small‐cell lung cancer with mutated EGFR. N Engl J Med 2010; 362 (25): 2380–8. [DOI] [PubMed] [Google Scholar]
- 18. Jänne PA, Yang JC‐H, Kim D‐W et al AZD9291 in EGFR inhibitor–resistant non–small‐cell lung cancer. N Engl J Med 2015; 372 (18): 1689–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Survival analysis, according to EGF+61 genotypes. Progression‐free survival for codominant (a); recessive (b); and dominant (c) models. Overall survival for codominant (c); recessive (d); and dominant (e) models.
Table S1. Demographic and clinicopathological features of TKI‐treated NSCLC patients harboring EGFR‐sensitizing mutations (n = 111) and association with treatment response.
Table S2. Genotypic and allelic frequencies of TKI‐treated NSCLC patients harboring EGFR sensitizing mutations (n = 111).


