Abstract
Early disease detection through point-of-care (POC) testing is vital for quickly treating patients and preventing the spread of harmful pathogens. Disease diagnosis is generally accomplished using quantitative polymerase chain reaction (qPCR) to amplify nucleic acids in patient samples, permitting detection even at low target concentrations. However, qPCR requires expensive equipment, trained personnel, and significant time. These resources are not available in POC settings, driving researchers to instead utilize isothermal amplification, conducted at a single temperature, as an alternative. Common isothermal amplification methods include loop-mediated isothermal amplification, recombinase polymerase amplification, rolling circle amplification, nucleic acid sequence-based amplification, and helicase-dependent amplification. There has been a growing interest in combining such amplification methods with POC detection methods to enable the development of diagnostic tests that are well suited for resource-limited settings as well as developed countries performing mass screenings. Exciting developments have been made in the integration of these two research areas due to the significant impact that such approaches can have on healthcare. This review will primarily focus on advances made by North American research groups between 2015 and June 2020, and will emphasize integrated approaches that reduce user steps, reliance on expensive equipment, and the system's time-to-result.
Keywords: Isothermal amplification, Nucleic acids, Point-of-care detection, Review, LAMP, RPA
Highlights
-
•
Summarized technologies that integrated isothermal amplification with point-of-care detection.
-
•
Primarily focused on research in North America between 2015 and June 2020.
-
•
Discussed isothermal amplification mechanisms.
-
•
Elucidated the challenges of integration and provided future perspectives.
1. Introduction
Early disease detection through point-of-care (POC) testing is vital for quickly treating patients and preventing the spread of harmful pathogens. POC testing occurs at the time and place of patient care, allowing for early-stage diagnoses and quick medical decisions. Such tests are well suited for resource-limited settings, as they are inexpensive, require minimal equipment, and eliminate the need for patient follow-up visits.
However, the diagnosis of many diseases is accomplished using quantitative polymerase chain reaction (qPCR), which requires trained laboratory personnel, numerous liquid-handling steps, and expensive equipment, such as a thermocycler. Isothermal amplification offers an alternative to qPCR, allowing for nucleic acid amplification to occur at a single, constant temperature, eliminating the need for a thermocycler. Common isothermal amplification approaches include loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), rolling circle amplification (RCA), nucleic acid sequence-based amplification (NASBA), and helicase-dependent amplification (HDA). A number of alternative detection methods have recently been developed, ranging from colorimetric detection to paper-based detection methods. This expanding toolbox enables POC detection that does not depend on expensive laboratory equipment or complicated procedures, and reduces the time-to-result. Despite significant advances in the rapidly developing field of POC testing, most recent reviews focus only on POC detection or isothermal amplification, with limited reviews interfacing the two. A paper by Lau and Botella reviewed isothermal amplification and POC detection, but focused on plant diseases (Lau and Botella, 2017), while another review by Li and Macdonald discussed isothermal amplification, but included non-POC detection methods such as gel electrophoresis (Li and Macdonald, 2015). An additional review by Giuffrida and Spoto focused on the integration of isothermal amplification methods into microfluidic devices, including many that were not POC due to their high complexity (Giuffrida and Spoto, 2017). Similarly, three reviews by Abaibou and coworkers, Bau and coworkers, and Kuhlmeier and coworkers focused on the integration of isothermal amplification and POC detection using microfluidic devices (Maffert et al., 2017; Mauk et al., 2018; Tröger et al., 2015). This review is a comprehensive survey of recent advances that merge isothermal amplification and POC detection, as such an integration can benefit low-resource areas as well as developed countries requiring mass screening of individuals.
In this review, research investigations that have combined isothermal amplification approaches and POC detection methods are discussed to provide an overview of recent developments (primarily January 1, 2015–June 30, 2020). This review is part of a special issue focusing on biosensing research in North America. In addition, we have focused on research that is related to human diseases, and therefore, have not included veterinary, environmental, biowarfare, and most foodborne pathogen applications. The mechanisms for each of the isothermal amplification methods (LAMP, RPA, RCA, NASBA, and HDA) will be overviewed, followed by a summary of advances in POC detection systems. Two additional isothermal amplification methods (strand displacement amplification and nicking enzyme amplification reaction) will also be briefly discussed.
2. Loop-mediated isothermal amplification
2.1. Introduction
Since its invention by Hase and coworkers in 2000, LAMP has been a major focus for the development of POC technologies due to its simplicity, high sensitivity, and rapidity (Notomi et al., 2000). LAMP occurs between 60 and 65 °C and can amplify DNA up to 109-1010 times in 15–60 min (Chen et al., 2016; Itou et al., 2014). In contrast to PCR, which generates amplicons of a uniform size, LAMP produces amplicons of various lengths, ranging from approximately 300 bp to 10 kb (Notomi et al., 2000).
The LAMP reaction mixture includes Bst DNA polymerase, deoxynucleoside triphosphates (dNTPs), double-stranded target DNA, and four primers specific to each target sequence. These four primers consist of two outer and two inner primers, the latter of which are each composed of two complementary sequences (Notomi et al., 2000). Amplification occurs through strand displacement activity by Bst DNA polymerase. The reaction is initiated when a forward inner primer (FIP) hybridizes to the target sequence and is extended, replacing one of the original strands. This strand displacement process repeats with the forward outer primer (F3) so that the strand containing the FIP is released. The released strand then hybridizes to itself due to the inclusion of the complementary sequence, forming a loop structure. The same process repeats on the opposite side of the released strand with the backward inner primer (BIP) and backward outer primer (B3), resulting in a dumbbell structure with a self-hybridizing loop on each end (Fig. 1-I).
Fig. 1.
Steps in the LAMP mechanism. (I) Initiation of LAMP results in the generation of a dumbbell structure. (II) Stem-loop structures are generated in the cyclic stage of LAMP. (III) Stem loop structures are elongated; amplification occurs exponentially (Notomi et al., 2000) (CC BY 4.0 license: https://creativecommons.org/licenses/by/4.0/).
This dumbbell structure has multiple sites for initiation and serves as a starting point for the second cyclic stage of LAMP. Through self-primed DNA synthesis, the dumbbell is converted to a stem-loop structure, from which more dumbbell structures are generated (Fig. 1-II). The stem-loop structures themselves are elongated and amplified exponentially in the third stage of LAMP (Fig. 1-III). The final products are duplex stem-loop DNAs of various lengths, with each single strand containing inverted repeats of the target sequence.
The longer primer length and the use of four primers derived from six regions of the target sequence make LAMP highly specific and less sensitive to non-target DNA than conventional PCR (Itou et al., 2014). LAMP is relatively simple, as it requires only one type of enzyme and fewer DNA processing steps (Tomita et al., 2008). For example, Bst DNA polymerases adapted for LAMP are relatively tolerant of blood proteins that strongly inhibit DNA polymerases used in PCR (Mohon et al., 2016). It is important to note that the concentrations of LAMP reagents such as magnesium sulfate, dNTPs, and betaine may be optimized for maximum assay sensitivity and efficiency (Njiru et al., 2011). These optimal concentrations are dependent on the specific target and primer sequences, so the development of a general LAMP kit that only requires the addition of primers may not be possible (Njiru, 2012). Another consideration is that LAMP amplicons are of a wide range of lengths, so the detection method applied must be able to identify target-specific products independent of sequence length (Chen et al., 2016). Additionally, the length of the target sequence should typically be less than 300 bp, as LAMP functions poorly for target DNA of more than 500 bp due to the rate-limiting step of strand displacement DNA synthesis (Notomi et al., 2000).
2.2. Detection methods
2.2.1. Colorimetric
Endpoint detection of LAMP products has been achieved through the use of colorimetric indicators. One such indicator is hydroxynaphthol blue (HNB), which changes color from violet to sky blue as the concentration of magnesium ions decreases as a result of amplification (Goto et al., 2009). Unlike some intercalating dyes that must be added after amplification is complete, such as SYBR Green, HNB can be present in the initial LAMP reaction solution. This feature of HNB is advantageous because opening the tube following amplification increases the risk of cross-contamination of subsequent LAMP reaction solutions via DNA aerosols. HNB-based LAMP assays provided detection of Loa loa down to 0.1 pg/mL of crude DNA lysate from blood spot and spiked microfilaria samples, and Chlamydia trachomatis down to 4.5 × 103 copies in 3 μL of crude DNA lysate from endocervical samples (Choopara et al., 2016; Drame et al., 2014). The Liu group enhanced the HNB colorimetric signal by loading preprocessed samples into a microfluidic chip inside a “smart” heating cup, where they were analyzed by a smartphone app (Yin et al., 2019). The smart cup produced heat through an exothermic reaction and maintained the temperature using a phase changing material. When the system was used for human papilloma virus (HPV)-associated cancer screening in clinical samples, the results were consistent with those obtained from qPCR and the conventional Pap smear test.
Visual detection of LAMP products has also been realized using pH-sensitive dyes, which indicate the decrease in pH caused by DNA polymerase activity (Tanner et al., 2015). LAMP assays using neutral red and phenol red to target DNA sequences of three different filarial parasites were able to detect 1/10–1/5000th of one microfilaria in 2 μL of a purified DNA sample (Poole et al., 2017).
2.2.2. Turbidity
One of the by-products of LAMP is magnesium pyrophosphate, a white salt precipitate that forms when pyrophosphate ions from dNTPs bind with magnesium ions from magnesium sulfate (Tomita et al., 2008). The resultant turbidity can serve as a measure for detection. The Li laboratory developed an inexpensive, battery-powered spectrophotometric system for analysis on a microfluidic chip (Dou et al., 2016). When the system was tested using the LAMP turbidity signal for the detection of Neisseria meningitidis, the limit of detection (LOD) was 15.3 ng/μL of a purified DNA sample.
2.2.3. On-paper
Due to its low cost and ease of production, the most common implementation of LAMP-based paper detection is the lateral-flow immunoassay (LFA). In addition to being cheap and easy to manufacture, the LFA is rapid, user friendly, and requires no equipment. However, it suffers from poor sensitivity due to having a relatively high detection limit. To address this problem, LAMP can be used to amplify a target nucleic acid sequence at an originally low concentration to enable detection by LFA, thereby increasing LFA sensitivity (Rodriguez et al., 2015). The Klapperich Group translated LAMP and LFA detection into an integrated paper chip format for the detection of HPV (Rodriguez et al., 2016). The modular, foldable apparatus was composed of tape and paper, and LAMP was conducted in a polyethersulfone membrane, reducing the amount of liquid-handling steps required. Within 45 min, the device achieved an LOD of 104 copies/mL from DNA standards that were purified with QIAquick Extraction Kits.
Linnes and coworkers developed a microfluidic rapid and autonomous analytical device (microRAAD) consisting of a microfluidic paper-based analytical device containing an electronic chip to maintain the appropriate temperature (Fig. 2 ) (Phillips et al., 2019). The paper membrane doubled as a filter, using size-based separation to extract HIV from whole blood. The device was assembled by placing the electronic and paper components into their plastic housing, after which the wash buffer and sample could be added to their respective inlets. HIV RNA extraction and detection from whole blood was completed within 90 min with an LOD of 2.3 × 107 copies/mL in whole blood.
Fig. 2.
Structure of microRAAD. A) Workflow for HIV detection: (1) The user assembles the device in the plastic housing. (2) Wash buffer and sample are deposited into inlets and sealed with tape. (3) Heating is initiated by powering with a cellular phone. (4) Temperature was maintained at 65 °C for 90 min (5) Results were analyzed on LFA. B) The assembled device connected to phone power. Reprinted with permission from Phillips et al. (2019) - Published by The Royal Society of Chemistry.
The Yoon Group used a printed paper-wax channel to filter out RNA from solutions containing lysed Zika virus (ZIKV) or other flaviviruses (Kaarj et al., 2018). Due to electrostatic and size properties, only RNA molecules were able to migrate to the end. A reverse transcription LAMP (RT-LAMP) reaction mixture containing phenol red pH indicator was then dispensed onto the filtrate, and the paper underwent a color change due to an increase in acidity from proton byproducts released by DNA polymerase during phosphodiester bond formation. While the color change was visible to the naked eye, results could be further quantified with smartphone imaging. When tested with ZIKV-spiked urine and plasma samples, this method achieved accurate results within 10 min of amplification, and an LOD of 1 copy/μL. However, the color changes were inconsistent until concentrations were increased to at least 100 copies/mL.
Another test for ZIKV used a novel device to manage its liquid-handling steps (Jiang et al., 2018). This device consisted of three sections connected linearly along a rail: buffer, mixing, and paper-based detection units. The buffer unit contained four reservoirs filled with lysis buffer, binding buffer, and two with wash buffer. When the mixing and detection units moved along the rail, a pin would trigger a ball to release liquid. After RNA lysis and enrichment, the mixture was washed into the detection unit which was then separated and allowed to undergo LAMP in a battery-powered coffee thermos. Results were visualized by the naked eye with SYBR Green and a blue LED flashlight. This device demonstrated an LOD of 0.5 plaque-forming units (PFU)/140 μL in urine or saliva samples.
Ellington and coworkers used LAMP amplicons to capture human chorionic gonadotropin (hCG), the easily detected protein indicator of pregnancy (Du et al., 2017). As a result, over-the-counter LFA pregnancy tests could be readily purchased and used as a means to test amplified products. Tests were performed on solutions composed of purified genomic samples spiked into water, human serum, and saliva. Using the store-bought devices to detect tagged products, they were able to detect a minimum of 20 copies for Zaire Ebola Virus (ZEBOV) and 2000 copies for the melanoma-associated SNP allele, BRAF V600E. Dehydrated hCG-DNA probes could be stored for 90 days with no significant loss in detection capability.
In a recent study, paper-based LAMP diagnostics were used in conjunction with CRISPR Cas-12 to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Broughton et al., 2020). This method simultaneously performed reverse transcription and amplification of RNA from nasopharyngeal or oropharyngeal swabs in universal transport medium. Following amplification, reaction products were introduced to a CRISPR Cas-12 assay. The gRNAs specific to the E and N genes of SARS-CoV-2 complexed with Cas-12, allowing it to cleave single-stranded DNA (ssDNA) probes when in the presence of the target sequence. Uncleaved probes were captured on the control line, while cleaved probes were captured on the test line, making the presence of two lines indicative of a positive result. The amplification, Cas-12 reaction, and detection process were all completed within 45 min if done manually, achieving an LOD of 104 copies/mL.
2.2.4. Fluorescent
Fluorescent dyes are commonly used for the visual detection of LAMP products. EvaGreen dye can be present during the LAMP reaction because it causes less inhibition of DNA polymerase compared to indicators such as SYBR Green 1, which must be added after amplification (Lee et al., 2017). Dye compounds that must be added after amplification are not as ideal for POC diagnostics due to the risk of contamination when opening the tube and the increase in liquid-handling steps. It should also be noted that intercalating fluorescent dyes, which indirectly detect LAMP amplicons, may potentially cause false positives by binding to non-target double-stranded DNA (dsDNA), especially after extended reaction times (Hardinge and Murray, 2019).
Multiple groups have incorporated EvaGreen into microfluidic smartphone-based LAMP systems. The Bashir laboratory developed an RT-LAMP microchip platform for the multiplexed detection of Zika, dengue, and chikungunya viruses in whole blood (Ganguli et al., 2017). Samples were processed and amplified on a microfluidic card, which was then inserted into a battery-powered heating device with a smartphone cradle for real-time fluorescence imaging. The system was able to detect ZIKV at concentrations as low as 10 copies/μL. An earlier prototype of this assay was able to detect as few as three RNA copies derived from whole blood in a 60 nL reaction droplet (Damhorst et al., 2015).
The Liu group adapted their previously discussed smart cup device for a fluorescence-based test (Liao et al., 2016). Using a smartphone camera to monitor EvaGreen fluorescence in real time, this assay detected down to 100 copies of HSV-2 DNA in a 22 μL reaction mixture. Similarly, Cunningham and coworkers developed a smartphone-based microchip device for the endpoint detection of five equine respiratory infectious diseases (Sun et al., 2020). The LOD was 5.5 × 104 DNA copies/mL of a processed, spiked horse nasal swab solution.
EvaGreen was incorporated into a one-pot RT-LAMP assay for the detection of avian influenza H5N1 in blood, achieving an LOD of 16.2 50% embryo infectious dose/mL (Tang et al., 2016). However, this system had multiple tube-opening steps, making it less optimal for use at the POC.
Another intercalating dye, SYTO 81, was part of a LAMP assay that involved real-time, smartphone-based detection and a microfluidic card that utilized an “airlock” mechanism to load samples (Stedtfeld et al., 2015). This assay was able to detect down to 20 copies of DNA extracted from Escherichia coli (E. coli) per ~1.8 μL reaction well.
Another fluorescent indicator used with LAMP is calcein, which is quenched upon initial binding with manganese ions (Tomita et al., 2008). Pyrophosphate ions produced during LAMP deprive calcein of the manganese ions, allowing calcein to emit green fluorescence. This fluorescence signal is further strengthened when calcein then complexes with magnesium ions in the solution. Calcein was integrated with an instrument-free microfluidic platform, which was heated by a battery-powered device, for the rapid detection of Bordetella pertussis (B. pertussis) (Dou et al., 2019). This approach demonstrated an LOD of 5 copies originating from 3 μL of a processed, spiked nasopharyngeal swab sample.
A smartphone-based LAMP system incorporated calcein for the real-time quantitative detection of bacterial pathogens in urinary sepsis patients (Barnes et al., 2018). This platform analyzed urine samples with results matching those of clinical diagnostics but with a much shorter turnaround time. The LOD of Salmonella typhimurium (S. typhimurium) in buffer was ≤10 colony forming units (CFU)/mL. Note that the sample preparation steps in this assay could be simplified to make the assay more POC-friendly.
Other systems utilized fluorescently labeled oligonucleotide strand displacement probes that are incorporated into LAMP amplicons (Bhadra et al., 2015, 2018; Jiang et al., 2017; Priye et al., 2017). In an RT-LAMP assay for the multiplexed detection of Zika, dengue, and chikungunya viruses in urine samples, probes for each virus were labeled with a different dye to produce a color-coded fluorescent readout (Yaren et al., 2017). The LODs were ~0.71 PFU, ~1.22 PFU, and ~38 copies of Zika, dengue, and chikungunya viral RNA, respectively, for 1 μL of purified and extracted viral RNA sample that was mixed with urine.
LAMP has demonstrated potential for POC testing for SARS-CoV-2. Using SYBR Green 1 fluorescence, an RT-LAMP assay achieved detection of 304 viral copies in a 2 μL sample volume within 30–45 min (Lamb et al., 2020). This test was found to be compatible with various human specimens, including serum, urine, saliva, oropharyngeal swabs, and nasopharyngeal swabs.
2.2.5. Electrochemical
The Li group took a unique detection approach by translating positive amplification into the production of glucose, which was then measured by a commercial glucose meter (Du et al., 2015). Each of four types of single-stranded loops in LAMP amplicons initiated a specific strand exchange reaction that released invertase, which then converted sucrose to glucose. This RT-LAMP system accomplished detection of Middle East respiratory syndrome coronavirus and ZEBOV down to 20–100 RNA copies/μL in preprocessed buffer, sputum, and saliva samples.
3. Recombinase polymerase amplification
3.1. Introduction
Developed by Armes and coworkers in 2006, RPA takes advantage of recombinant proteins to amplify DNA with specificity and sensitivity comparable to PCR (Piepenburg et al., 2006). RPA is currently commercialized by TwistDxTM, which offers kits for fluorescent and LFA detection. Alongside standard liquid reagents, TwistDxTM offers lyophilized kits with a longer shelf life intended for POC use. The effectiveness of an RPA reaction is dependent on a number of factors: temperature, reaction time, primer and probe design, and amplicon size. RPA performs optimally at temperatures between 37 and 42 °C, with the reaction run for approximately 20 min (Sun et al., 2016). RPA is advantageous in that it only needs two primers, while LAMP requires at least four primers. Compared to PCR, RPA primers are relatively long, typically between 32 and 35 nucleotides. Shorter primers between 18 and 25 nucleotides have been used, albeit with decreased reaction speed and sensitivity (Fuller et al., 2017; Mayboroda et al., 2016; Wang et al., 2017a). Most RPA literature employs amplicons with sizes between 100 and 250 nucleotides, but amplicons as small as 79 nucleotides and as large as 1941 nucleotides have been reported (Jauset-Rubio et al., 2016a, 2016b; 2017; Martorell et al., 2018; Wang et al., 2017b).
RPA can amplify both DNA and RNA with great specificity. While RPA is highly specific amongst non-closely related species (100% in most cases), its tolerance of mismatches makes multiplex detection among closely related species less successful (Mondal et al., 2016). However, this slight mismatch tolerability can be advantageous, as it allows for more flexibility in primer design for highly polymorphic targets (Boyle et al., 2013). Additionally, the sensitivity of RPA can be negatively affected by the presence of the crowding agent Carbowax20M (a standard RPA reagent) at low target copy levels since Carbowax20M can increase the viscosity of the solution, which in turn, decreases the rate of diffusion. However, a short mixing step, which can be as simple as 8–10 inversions followed by a spin down in a microcentrifuge, can resolve this issue by decreasing the length scales for diffusion (Lillis et al., 2016b). Although RPA is robust in the presence of traditional PCR inhibitors (hemoglobin, ethanol, and heparin), amplification can be inhibited by high background DNA concentrations (Kersting et al., 2014; Rohrman and Richards-Kortum, 2015).
To begin RPA, the T4 UvsX recombinase binds to the oligonucleotide primers with the assistance of T4 UvsY protein (loading factor) to form recombinase-primer complexes (Fig. 3 ). These complexes scan dsDNA for homologous sequences and initiate strand invasion at cognate sites. The displaced DNA strand is stabilized by single-stranded binding proteins in order to prevent the expulsion of the primer complex by branch migration. After recombinase disassembly, a strand displacing DNA polymerase (the large fragment of Bacillus subtilis POL I, Bsu) binds to the 3’ end of the primer and primer extension ensues in the presence of dNTPs. Exponential amplification is achieved through the cyclic repetition of this mechanism.
Fig. 3.
RPA reaction mechanism. The reaction is initiated by the binding of the T4 UvsX recombinase (yellow) to the primers with the help of the T4 UvsY loading factor (orange). The recombinase-primer complexes scan dsDNA for homologous sequences, and they invade these sequences with the strand-displacement activity of the recombinase. Single stranded binding proteins (purple) stabilize the displaced strand and the recombinase dissociates, leaving a 3′ group available for extension. Strand displacing DNA polymerase (green) binds the 3′ end of the primers and extension begins, eventually resulting in two DNA duplexes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Detection methods
3.2.1. Colorimetric
Colorimetric detection can be integrated with RPA due to its low cost, short assay time, and visual readout. Standard colorimetric detection typically takes advantage of the reaction between 3,3′,5,5′-tetramethylbenzidine (TMB) and horseradish peroxidase (HRP) in the presence of hydrogen peroxide (H2O2), which produces a blue-colored product visible to the naked eye.
A TMB-HRP system was developed for the detection of long non-coding HOX transcript antisense intergenic RNA in ovarian cancer cells with an LOD of 10 cells/mL from purified RNA samples using naked-eye detection (Islam et al., 2018). Biotinylated amplicons were produced and then bound by a streptavidin-HRP complex. The reaction between TMB and HRP produced a visible color change that served as the signal (Fig. 4 ). The multiplex colorimetric detection of 16 pathogens, including but not limited to HIV, HPV, and ZIKV, was demonstrated by Bau and coworkers using an RPA-LAMP based microfluidic device (Song et al., 2017). The chip was composed of a main chamber for the first-stage RPA reaction which branched into numerous chambers for the second-stage LAMP reactions. Following RPA, the solution diffused into the LAMP chambers where it was labeled with either an intercalating fluorescent dye or a colorimetric dye, allowing for detection by smartphone or the naked eye. While colorimetric detection is typically limited to qualitative measurements, the implementation of a smartphone allowed for more quantitative measurements by comparison to a control and the application of preprogrammed thresholds to determine positive and negative results.
Fig. 4.
Colorimetric detection using a TMB-HRP system. The target nucleic acid sequence is amplified with RPA using biotinylated deoxyuridine triphosphates (dUTPs). Streptavidin-HRP and streptavidin-magnetic beads bind to the biotinylated amplicons which are then magnetically isolated and purified. In the presence of TMB, a color change occurs allowing for the naked eye detection of positive samples. Reprinted with permission from (Islam et al., 2018). Copyright 2018 Royal Society of Chemistry. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2.2. On-paper
By far, the most common platform utilized for endpoint detection of RPA products is the LFA. Results can be achieved rapidly and with a highly intuitive visual readout using gold nanoparticles (GNs). To achieve detection with an LFA, an additional probe is required alongside the two standard primers (Fig. 5 a). This probe is typically 46–52 nucleotides long and contains a 5′ label (carboxyfluorescein, digoxin, etc.), an internal abasic site (tetrahydrofuran or dSpacer), and a 3′ block. An endonuclease IV is used to cleave the abasic site when the probe forms dsDNA, and an extendable 3′ hydroxyl group is formed, transforming the probe into a primer. To produce a dual-labeled amplicon, an opposing primer with a different 5’ label is required. This dual-labeled amplicon can then be detected by antibodies in a sandwich assay format.
Fig. 5.
Modified probes for RPA detection. A) The nfo probe is composed of a 5′ label, an internal abasic site such as tetrahydrofuran (THF), and a 3′ block. The probe is exchanged at the cognate site using recombinase proteins and the abasic site is cleaved by an endonuclease (yellow). The probe is converted to a primer and DNA polymerase (purple) begins extension. B) The fpg probe is cleaved by glycosylase/lyase E coli. fpg at the dR site (the deoxyribose of the abasic site via a C-O-C linker) to release the fluorophore and allow a fluorescent signal to develop. C) The exo probe is cleaved by the E. coli exonuclease III at the abasic site (THF) releasing the fluorophore from the quencher and creating a fluorescent signal. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Detection of RPA amplicons using an LFA was first demonstrated by Armes and coworkers in 2006 (Piepenburg et al., 2006). This LFA technology for RPA is currently commercialized by TwistDxTM in the form of the Milenia HybriDetect 1 and the Milenia HybriDetect 2 strips, by far the most popular strips used in the literature. Numerous publications detail detection of various pathogens - including Orientia tsutsugamushi, Plasmodium falciparum, Rickettsia typhi, Leishmania Viannia spp., and Entamoeba histolytica – using the aforementioned commercial LFA strips (Chao et al., 2015; Lalremruata et al., 2020; Nair et al., 2015; Saldarriaga et al., 2016). For all cases mentioned, results were obtained within 1 h for both amplification and detection with LODs as low as 10 DNA copies per reaction. An RPA-LFA detection scheme (LOD of 1 pg/μL purified DNA sample) has also been shown to improve the detection of purified Fasciola hepatica DNA from adult stool samples compared to PCR (LOD of 1.6 pg/μL purified DNA sample) (Cabada et al., 2017). The LFA has also shown successful multiplex detection, with the Macdonald group demonstrating hepta-plex detection using seven distinct 5’ labels: biotin, digoxigenin, TAMRATM, Texas Red-X, Cascade Blue C6-NH, DNP-X C6-NH, and Dansyl C6-NH (Li et al., 2019). They found that LODs for Rift Valley fever virus S, in water, using Cascade Blue (5.11 × 102 copies/μL), DNP (5.11 × 103 copies/μL), and Texas Red (5.11 × 104 copies/μL) were 10–1000 times higher than those using digoxigenin, Dansyl, and TAMRA (all three at 5.11 copies/μL) and biotin (511 copies/μL), highlighting the need for further optimization (Fig. 6 ). To further optimize lateral-flow detection for application at the POC, paper and plastic devices have been developed for the detection of Plasmodium DNA and HIV RNA with the addition of reverse transcriptase (Cordray and Richards-Kortum, 2015; Rohrman and Richards-Kortum, 2012). These devices store lyophilized enzymes and facilitate mixing of reaction components, and are small, lightweight, and easy to use, making them particularly suitable for the POC.
Fig. 6.
Septa-plex amplification and detection using RPA-LFA. A) Positioning of the antibodies in the 7 segment display. B) Number displays based on amplicon tags. C) Corresponding 5′ forward primer labels necessary for the aforementioned number displays. D) Successful detection of targets with visible numbers 0–9 and control lines (composed of rabbit anti-mouse antibody). Reprinted with permission from (Li et al., 2019). Copyright 2019 American Chemical Society.
3.2.3. Fluorescent
RPA can also be monitored in real-time as opposed to an endpoint detection assay. Real-time RPA typically uses fluorescent labels and fluorometers to measure signal output over time. While these fluorometers are not inherently portable, rechargeable versions have been created that are more POC-friendly. Although no longer sold by TwistDxTM, the Twista® Portable Real-Time Fluorometer is one example of a portable fluorometer that has been used in POC applications (Jiang et al., 2020).
Exo and fpg probes are commonly used in place of nonspecific fluorophores (Fig. 5b and c). The exo probe is composed of a blocked 3′ end and a dT-fluorophore and dT-quencher next to a dSpacer, such that a fluorescent signal is initiated by the cleaving of the dSpacer site once the probe binds a complementary target. The fpg probe, while similar to the exo, differs in the location of the fluorophore and quencher. It contains the same blocked 3′ end, but the quencher is attached to the 5’ end and the fluorophore is linked to an abasic nucleotide situated 4–5 nucleotides away. Once the probe binds homologous DNA, the E. coli exonuclease III or the glycosylase/lyase E. coli fpg cleaves at the abasic site and a fluorescent signal is generated.
Real-time fluorescent detection was demonstrated for DNA and RNA products in singleplex and multiplex applications. Reverse transcription RPA was used for the cross-subtype detection of HIV-1 in a comparison of fluorescent detection using the exo probe and LFA detection using the nfo probe (Lillis et al., 2016a). Both fluorescent and lateral-flow detection produced an LOD of 30 copies of HIV-1 RNA using samples extracted from blood using a QIAamp DNA Blood Mini Kit and were able to accurately detect up to seven HIV-1 subtypes. Additionally, the Lee laboratory demonstrated multiplex fluorescent detection of Campylobacter coli (C. coli) and Campylobacter jejuni (C. jejuni) in various food samples including chicken broth, chicken meat, and eggs (Kim and Lee, 2017). The assay exhibited an LOD of 1 CFU per reaction using purified genomic DNA, as well as 1 CFU/mL and 103 CFU/g in chicken broth and egg/meat samples, respectively. It was capable of specifically detecting C. coli and C. jejuni when tested against 33 strains, of which 12 were Campylobacter spp. and the other 21 were non-Campylobacter organisms. Although most real-time RPA reactions are carried out in microcentrifuge tubes, the detection of methicillin-resistant Staphylococcus aureus (MRSA) DNA has been shown using a microfluidic chip powered by a fully integrated vacuum battery (Yeh et al., 2017). This chip was able to amplify and detect DNA from whole blood samples in approximately 30 min between a range of 10-105 copies/μL, the most critical range for determining antibiotic treatment effectiveness and predicting mortality rate. Furthermore, a modified 3D printer capable of nucleic acid extraction, amplification, and detection has been shown to provide a limit of detection as low as 0.5 PFU/extraction for ZIKV RNA in saliva samples (Chan et al., 2018).
3.2.4. Electrochemical
While electrochemical detection of RPA products is not typically POC-friendly, the utilization of portable potentiostats and galvanostats, as well as screen-printed carbon electrodes has made it amenable to POC use. RPA-electrochemical detection is highly sensitive and is often used as a quantitative detection method (Islam et al., 2018).
The Whitesides Group developed a handheld electrochemical device capable of detecting up to 19 members of the genus Mycobacterium (Tsaloglou et al., 2018). This device takes advantage of [Ru(NH3)6]3+ as an electroactive mediator for electrochemical detection of DNA. When the ruthenium complex binds dsDNA, the number of free ruthenium complexes drops in proportion to the amount of DNA present in the initial sample, decreasing the cathodic current that can be measured voltametrically. This device is particularly useful for samples that require elaborate purification and provides rapid and sensitive detection down to approximately 0.040 ng Mycobacterium tuberculosis (M. tuberculosis)/μL of purified genomic DNA. Although electrochemical detection is not currently the best suited detection method for resource poor settings, it shows great potential for future use in POC applications.
4. Rolling circle amplification
4.1. Introduction
RCA is one of the earliest documented isothermal amplification techniques. It was developed in 1989 by Salas and coworkers, based on the principles of rolling circle replication (Blanco et al., 1989; Liu et al., 1996). These reactions are simple to design and well suited for POC applications due to a mild operating temperature of 25–37 °C and rapid amplification, occurring within 1 h (Ali et al., 2014). RCA requires a circular template and a DNA or RNA polymerase with high processivity and strand-displacing activity (most frequently ϕ29 DNA polymerase). The polymerase synthesizes long ssDNA or ssRNA that is complementary to the circular template, containing tandem repeats of the target. This allows for a 107-fold amplification with high specificity, as the system can amplify a single copy of target DNA with 100,000 copies of background DNA (Nelson et al., 2002). This robust system has been applied to the amplification of DNA, RNA and microRNA (miRNA), infectious pathogens, and single nucleotide polymorphisms. Following the 2003 SARS outbreak, sensitive RCA-based assays were designed to detect SARS-CoV down to a single target viral template (Wang et al., 2005). This demonstrates the potential for RCA to improve the sensitivity of rapid tests for SARS-CoV-2. RCA is unique in that it requires a circular template, making it compatible with amplification of circular viral genomes without need for a circularization step. Furthermore, RCA can be conducted in solution as an in-tube reaction, or solid phase, in which probes are bound to a solid support, such as a microarray. Additionally, because of repeated sequences in the product, RCA is well suited for hybridization-based assays.
There are three primary approaches to RCA: linear, ligation, and hyperbranched (HRCA) (Fig. 7 ). RCA reactions contain the following components: a DNA or RNA polymerase, a short oligonucleotide primer, a circular template, and dNTPs. The strand-displacing polymerases most commonly used are Bst, ϕ29, and vent (exo-) DNA polymerases or T7 for RNA. The polymerase binds to the primer and synthesizes the strand complementary to the circular template using dNTPs.
Fig. 7.
Schematics of RCA mechanisms. A) Ligation RCA. B) Linear RCA. C) Hyperbranched RCA.
Linear RCA is the most basic form, using a single-stranded circular template that is between 15 and 200 nucleotides (Fig. 7b). A short DNA or RNA primer anneals to the template, and the polymerase adds nucleotides complementary to the template in the 5′ to 3’ direction using dNTPs in the solution. When the polymerase circles the template back to the primer binding site, the synthesized strand is displaced and the polymerase continues synthesizing the complementary strand. This results in long ssDNA or ssRNA with repeated sequences corresponding to the entire circular template.
Ligation RCA is used to convert linear DNA or RNA into a circular template which then enters the RCA cycle by using a padlock probe (Banér et al., 1998; Jonstrup et al., 2006). The single-stranded padlock probe has ends that are complementary to the 3′ and 5’ ends of the target strand. Both ends of the padlock probe are hybridized to the target sequence with DNA ligase, circularizing the template (Fig. 7a). This process is highly specific; a single nucleotide mismatch is enough to prevent circularization (Xu et al., 2012). Once circularized, it is amplified by either linear RCA or HRCA.
HRCA uses additional primers to achieve exponential amplification (Lizardi et al., 1998). As with linear RCA, the forward primer is extended by the polymerase to create ssDNA with tandem repeats complementary to the circular template. The reverse primer binds to the newly synthesized DNA and is extended by the polymerase to create a dsDNA branch at each repeat (Fig. 7c).
4.2. Detection methods
4.2.1. Colorimetric
Several approaches have been taken to allow for colorimetric detection of RCA products. Colorimetric detection is primarily achieved using enzyme substrates, dyes, and pH indicators. The most common enzyme substrates are 2,2′-azinobis[3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) and TMB.
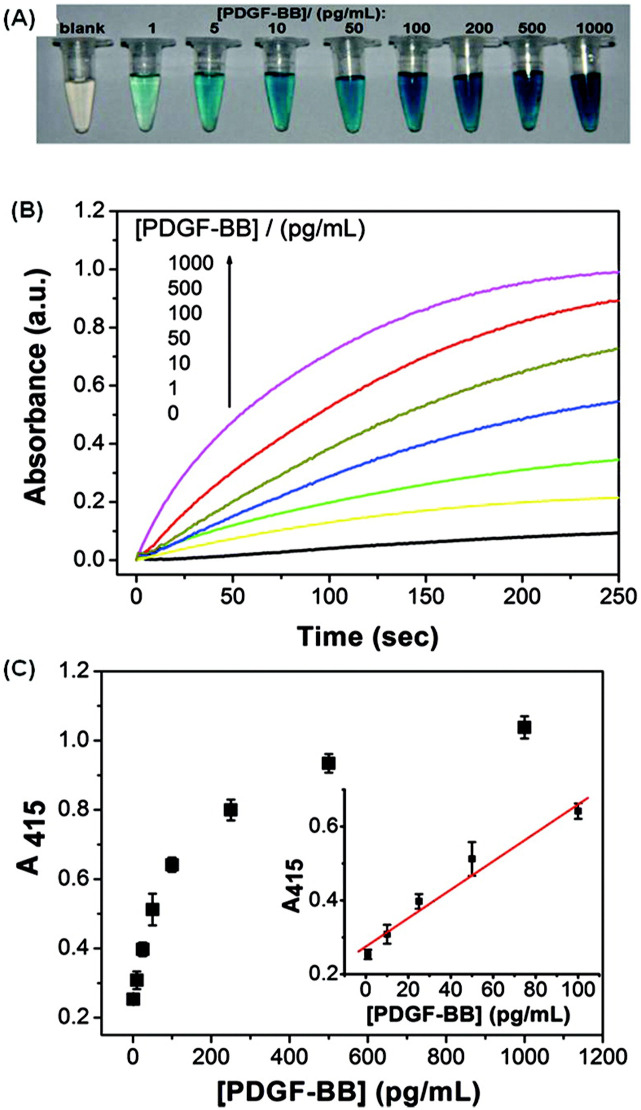
ABTS oxidation results in a colorimetric signal that can be detected with the naked eye. The RCA template can be designed to form G-quadruplex structured products that associate with hemin, creating a DNAzyme with peroxidase-like activity (Roembke et al., 2013). In the presence of H2O2, these DNAzymes catalyze the oxidation of ABTS, leading to a visible shift in color. An ABTS-based system employing peptide nucleic acids (PNAs) was developed by Gomez and coworkers, in which the PNAs allowed the remainder of the DNA to remain double stranded, reducing background signal. The PNAs complexed with the target DNA, creating the site for padlock probe ligation, which circularized the template for subsequent HRCA (Fig. 8 ) (Gomez et al., 2014). The template contained multiple nicking enzyme sites, which produced new sites for strand displacement reactions, facilitating exponential amplification. The colorimetric readout allowed for discrimination of E. coli, S. typhimurium, and Clostridium difficile (C. difficile) with single-base specificity and LODs of 10 and 100 fM purified DNA in buffer from C. difficile and Salmonella, respectively. In another ABTS-based design, Li and coworkers used aptamer-functionalized microbeads to capture, purify, and concentrate platelet-derived growth factor-BB (PDGF-BB) (Tang et al., 2012). A sandwich complex was formed as a secondary aptamer, containing the RCA primer and template, bound to the target. Upon target recognition, RCA was initiated, with rational template design allowing for the formation of a G-quadruplex-based DNAzyme to catalyze ABTS oxidation. Absorbance increased as a function of concentration from 1 to 1000 pg/mL, as the solution became deepening shades of blue-green (Fig. 9 ). As proof of concept, the system was used to test for PDGF-BB in serum with an LOD of 0.2 pg/mL.
Fig. 8.
Schematic of the PNA-based padlock probe system. A) The bis-PNAs bind to one strand of the target DNA, leaving the opposite strand open for padlock probe ligation. B) The linear padlock probe hybridizes to the displaced strand to form a PD-loop. C) The circularized template undergoes HRCA. D) Nicking enzymes cut one strand of the double-stranded product into multiple nicked pieces. Primer extension subsequently displaces the nicked strands which then form G-quadruplex structures to complex with hemin to form DNAzymes. Reprinted with permission from (Gomez et al., 2014). Copyright 2014 American Chemical Society.
Fig. 9.
DNAzyme-mediated ABTS colorimetric detection of PDGF-BB, with concentrations ranging from 0 to 1000 pg/mL. A) Photographs captured 5 min after ABTS oxidation. B) Changes in absorbance as a function of time. C) Absorbance at 415 nm as a function of PDGF-BB concentration. Reprinted with permission from (Tang et al., 2012). Copyright 2012 American Chemical Society.
Yu and colleagues took a unique approach to colorimetric output through the use of liquid crystals decorated with the cationic surfactant octadecyltrimethylammonium bromide (Qi et al., 2019). Magnetic beads were used as a probe, on which ligation RCA proceeded in the presence of the target biomarker in blood. Following amplification, the product was transferred to the liquid crystal. In a positive sample, the interaction between the amplified DNA on the magnetic beads and adsorbed surfactant induced a planar orientation of the liquid crystal, leading to a visible change in brightness (Fig. 10 ). The brightness was quantified by imaging the liquid crystal to determine the bright area coverage ratio. As a proof of concept, this method was used to detect PDGF-BB and adenosine in Tris-HCl buffer with LODs of 0.12 pM and 31 pM, respectively, and also proved effective in heparin-anticoagulated human blood. To determine specificity of the assay, the target—either PDGF-BB or adenosine—was replaced with other proteins including glucose oxidase, IgG, and thrombin. All other proteins resulted in a bright area coverage ratio approximately equal to blank, while the target resulted in a ratio of nearly 100%, indicating a high specificity. This demonstrated the ability of this method to detect both macromolecular and small molecule biomarkers with high specificity and sensitivity.
Fig. 10.
Schematic of liquid crystal colorimetric detection of PDGF-BB or adenosine. (A) Ligation-RCA amplifies the target on the magnetic bead, while (B) RCA does not proceed in the absence of the target. (C) Following RCA, the positive sample induces a planar orientation of the liquid crystal, which (D) then appears bright. (E) In the absence of the RCA product, the liquid crystal interface retains a homeotropic arrangement, (F) appearing dark. Reprinted with permission from (Qi et al., 2019). Copyright 2019 American Chemical Society.
Finally, pH-responsive RCA detection appears to be a rapidly developing approach to POC detection. During amplification, H+ ions are released as byproducts proportionately with amplification efficacy, and can result in large changes in pH. Ligation RCA was used by Hamidi and Perreault to amplify viral targets (H5N1 virus or M13 bacteriophage) in plasma with high sensitivity. Colorimetric changes as a function of viral concentration were observed using the phenol red pH indicator (Hamidi and Perreault, 2019). Unfortunately, the use of linear RCA manifested in a narrow range of colors, making it difficult to detect with the naked eye.
4.2.2. On-paper
Surprisingly, there has been less integration of RCA with detection on paper. The Li group, however, developed a paper-based device that is compatible with four different detection strategies (Liu et al., 2016). As proof of concept, this assay was used to detect a region of the hepatitis C virus genome and miRNA. A biotinylated DNA-streptavidin conjugate, which acted as the RCA primer, was printed on a nitrocellulose membrane, accompanied by pullulan-stabilized RCA reagents. Detection was enabled by either (1) radiolabeling the RCA product, (2) hybridizing the RCA product with a fluorophore-labeled probe, (3) functionalizing GNs with a complementary probe for colorimetric detection, or (4) designing the modified circular DNA template to create a peroxidase-like DNAzyme, which in the presence of hemin and H2O2 oxidizes TMB for a colorimetric output (Fig. 11 ).This work demonstrated that, due to the restriction of reaction components to a nearly two-dimensional space, the length scales for diffusion were significantly reduced. Since the components could more easily encounter each other, diffusion no longer limited the reaction, leading to higher efficiency than in-tube RCA. This robust paper-based system is compatible with four different detection strategies and can be tailored to various targets, making it well suited for POC applications.
Fig. 11.
Paper-based detection of RCA products using (A) a radioactive tracer, (B) a fluorophore-labeled oligonucleotide, (C) functionalized GNs for colorimetric detection, and (D) DNAzyme-mediated oxidation of TMB for colorimetric detection. TP1: non-fluorescently labeled primer; CDT1: circular DNA template. Reprinted with permission from (Liu et al., 2016). Copyright 2016 WILEY-VCH Verlag GmbH & Co.
4.2.3. Fluorescent
By combining fluorescence with RCA, biomarkers can be detected with high sensitivity. RCA was combined with a graphene-based biosensor to detect thrombin, ATP, and a region of the hepatitis C virus to demonstrate its versatility (Liu et al., 2014). In the presence of a reduced graphene oxide monolayer, which has a high affinity for single-stranded DNA and is a strong fluorescence quencher, a FAM-labeled aptamer was reversibly adsorbed and quenched, eliminating fluorescence in the absence of target biomolecules. When the target was present, the DNA aptamers underwent a conformational change and were released from the graphene oxide surface. Upon release, the aptamers served as the primer for RCA. The RCA products were then combined with a molecular beacon to enable fluorescence proportional to target concentration. The double stranded, label-hybridized RCA product was not adsorbed to the graphene oxide surface, thus attenuating the quenching.
In addition, the Brennan group developed both in-solution and paper-based one-pot assays with SYBR Gold and QuantiFluor dyes to detect platelet derived growth factors with LODs of 10 nM in solution and 6.8 nM on paper (Bialy et al., 2020). The same scheme was applied to thrombin, and LODs of 100 pM in solution and 240 pM on paper were achieved. In this work, without addition of a protein target, a protein-binding aptamer would be able to bind a complementary circular template, and therefore initiate RCA in the presence of ϕ29 DNA polymerase and dNTPs. Resulting products could bind to fluorescence dyes to generate a signal. However, in the presence of a target protein, an aptamer-protein complex would form, preventing aptamer binding with the template strand, inhibiting RCA and causing no signal to be produced.
Finally, another approach used gel microparticles with miRNA-specific probes for multiplexed fluorescent detection (Chapin and Doyle, 2011). A sample was added to the microparticles and the probes captured their specific miRNA target—either miRNA 141, miRNA 210, or miRNA 221, which have their dysregulation associated with cancer development. After target binding, RCA proceeded and the product was labeled with fluorescent reporters. By hybridizing each product with several labels, amplification bias, in which final target concentration does not reflect initial relative abundance, was mitigated. This system allowed for single-molecule resolution, which could be expanded to a detection range of 300 aM to 40 pM in serum. The assay was slightly modified to allow this detection in serum—which proved compatible with the fouling-resistant microparticles—for integrated RNA extraction, target amplification, and detection.
4.2.4. Electrochemical
Electrochemical detection has seen growth in recent years as a POC detection method for RCA products. Xue and colleagues developed an amperometric biosensor to detect Vibrio parahaemolyticus (V. parahaemolyticus), a foodborne pathogen (Teng et al., 2017). The V. parahaemolyticus was captured by immobilized antibodies on the surface of a gold electrode, followed by single-stranded aptamer probe binding to the captured target, forming a hetero-sandwich structure. Ligation RCA proceeded, followed by product hybridization with ssDNA-functionalized GNs. The hybridized product was labeled with methylene blue, which has a higher affinity for dsDNA, to enable electrochemical detection. The signal was characterized using differential pulse voltammetry, cyclic voltammetry, and electrochemical impedance spectroscopy. The device had an LOD of 2 CFU/mL in buffer, which is a drastic improvement compared to other recently developed nanodevices to detect V. parahaemolyticus. This assay also exhibited high specificity, and despite similarities, it did not result in a false positive for Staphylococcus aureus (S. aureus), Shigella sonnie, E. coli, Salmonella, C. jejuni, or Vibrio mimicus. This work shows the potential of an aptamer-immuno hybrid electrochemical biosensor for highly sensitive detection, with the possibility of expanding its applications to POC disease detection.
5. Nucleic acid sequence-based amplification
5.1. Introduction
Nucleic acid sequence-based amplification (NASBA) is a well-established isothermal amplification method developed by Compton in 1991 (Compton, 1991). The amplification reaction is carried out at 41 °C, and within 90 min, target nucleic acid sequences of 100–250 nucleotides can be amplified by approximately 1012-fold (Farkas and Holland, 2009).
Three enzymes are used in the NASBA reaction mixture: T7 RNA polymerase, RNase H, and avian myeloblastosis virus (AMV) reverse transcriptase. The reaction requires two specific primers. The first primer (primer 1) contains a promoter sequence at its 5′ end that is recognized by T7 RNA polymerase, and its 3′ end is complementary to the 3′ end of the target sequence. Upon introduction of a target RNA molecule in the noncyclic phase of the reaction, primer 1 initiates reverse transcription of the RNA sequence to form an RNA-DNA hybrid, followed by the addition of RNase H that degrades the original RNA of RNA:DNA hybrids, leaving the cDNA for the second primer (primer 2) to anneal. The 5′ end of primer 2 is complementary to the 3′ end of the cDNA. AMV reverse transcriptase, which also acts as DNA-dependent DNA polymerase, will extend the 3’ end of primer 2 to form dsDNA, which is then transcribed by T7 RNA polymerase to make the antisense RNA. Each newly synthesized dsDNA can create up to 100 copies of antisense RNA, which then enter the cyclic phase of NASBA, resulting in amplification of the antisense RNA strand (Compton, 1991). The NASBA mechanism is shown in Fig. 12 below.
Fig. 12.
Schematic of the NASBA mechanism. A target RNA is introduced to the noncyclic phase, resulting in an RNA (-) product that acts as a template for cyclic phase amplification, which in turn forms more RNA (-) products.
The optimal temperature for NASBA is 41 °C, allowing cheaper, easy-to-use heating methods to be utilized. NASBA can also selectively amplify RNA in the presence of background DNA, as well as DNA target sequences if they are denatured in advance (Deiman et al., 2008; Malek et al., 1994). However, NASBA may be limited by nucleic acid extraction and purification steps prior to amplification and imperfect primer design (Asghar et al., 2019; Morabito et al., 2013). Additionally, due to the low reaction temperature, primer dimerization and non-specific hybridization are prone to occur, which dramatically increase the rate of false positives.
5.2. Detection methods
5.2.1. Colorimetric
Improved and user-friendly colorimetric detection has been combined with NASBA to determine the concentration of different analytes. The Gerasimova group utilized Trizol LS (Life Technologies) to extract and purify RNA, NASBA to amplify analytes, and a color-generating deoxyribozyme (Dz) to detect a highly conserved region of ZIKV RNA (Reed et al., 2019). Following NASBA, their probes, which contained a split phosphodiesterase Dz and a peroxidase-like Dz (PDz), were introduced into the reaction mixture. The split phosphodiesterase Dz received its name from being composed of two Dz subunits, which bind to adjacent fragments of the RNA amplicons in a positive sample, forming a catalytic core. The core catalyzed the cleavage of an RNA phosphodiester bond in the inhibited form of PDz (IPDz). Upon IPDz cleavage, a PDz reporter sequence was released, which catalyzed H2O2-mediated oxidation of ABTS, resulting in color generation. Without the presence of target amplicons, no oxidation reaction would occur, and therefore no color change would be detected. Using Trizol LS and this technology, ZIKV RNA could be detected at concentrations as low as 106 copies/mL in blood, urine, and saliva (Reed et al., 2019). Furthermore, the Gerasimova group used the same scheme to detect 16S rRNA of Mycobacterium tuberculosis complex (MTC) with an LOD of 8.3 pg/mL and distinguish katG mRNA of MTC from its single-base variant using an 830 pg/mL pre-amplified sample solution (Dhar et al., 2020). Though multiple user steps are needed for this colorimetric detection method, its low-cost, portability, and rapid time-to-result demonstrate its potential for POC testing using NASBA.
5.2.2. On-paper
In order to make the detection method more cost-effective and readily available for resource-limited areas, Collins and coworkers combined their previously designed technologies—programmable RNA sensors, toehold switches, and a freeze-dried, paper-based, cell-free protein expression platform—to develop a paper-based sensor for colorimetric detection of ZIKV (Fig. 13 ) (Pardee et al., 2016). Viremic plasma diluted in water was boiled at 95 °C for 2 min for viral RNA extraction. NASBA was then used to amplify ZIKV RNA, and toehold switches that were dried on the paper were used to control regulation of the lacZ gene and expression of the reporter enzyme β-galactosidase, which mediated a visible color change of a paper disc (Pardee et al., 2016). Furthermore, they developed a portable electronic reader using an open-source code, a laser-cut acrylic housing, and a rechargeable lithium ion battery, costing less than $250, to provide a quantitative signal output. Additionally, using the Collins laboratory's NASBA-CRISPR cleavage approach, it was possible to discriminate between African and American viral strains with only a single base difference. This technology utilized a strain-specific protospacer adjacent motif (PAM), which only exists in American strains, as the cleavage sites for Cas9 to selectively cleave DNA. In the presence of an appropriate PAM sequence, Cas9 would cleave the dsDNA formed during NASBA, resulting in truncated RNA products with no color change. In the absence of the appropriate PAM sequence, full length NASBA products would be generated to activate the toehold switch, resulting in a color change of the paper disc. The use of this toehold switch technology was expanded to the detection of a panel of 10 bacteria related to inflammatory bowel disease, childhood malnutrition, and cancer immunotherapy, along with C. difficile toxin mRNA to demonstrate its potential to detect RNA (Takahashi et al., 2018).
Fig. 13.
Workflow of Zika virus detection. Online databases are used for primer design. In less than 7 h, sequence-specific toehold sensors can be assembled and validated. In 1 day, toehold sensors can be freeze-dried and embedded into a paper disc with a shelf life of more than 1 year. The paper-based sensor can then be used to detect RNA of interest with a change of color, using the NASBA technique. Reprinted from (Pardee et al., 2016), Copyright 2016, with permission from Elsevier. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The paper-based cell-free RNA toehold switch technology was also used by Green and coworkers to detect the norovirus GII.4 Sydney strain which was extracted with the QIAamp DSP Viral RNA Mini Kit; however, it was also shown that a simple extraction step of boiling the PBS-diluted sample at 95 °C for 2 min was also effective (Ma et al., 2018a). The Green laboratory also employed a synbody (a bivalent peptide that performs antibody-like functions)-based magnetic bead capture assay to increase the concentration of norovirus prior to detection to achieve a 103-fold improvement in the detection limit. According to the group, shortened assay time and reduced costs could be achieved by decreasing the quantity of magnetic beads used, optimizing toehold switches, improving the binding affinity of synbodies, and implementing new reporter proteins for faster activation.
The most frequently used paper-based endpoint detection method for NASBA is the LFA. In recent years, improvements have been made to solve the disadvantages of LFA. For example, the Ozcan group developed a cellphone-based rapid-diagnostic-test reader platform to read the signals of malaria, tuberculosis, and HIV analytes to achieve quantitative results for better interpretation of LFA results in resource-limited areas (Mudanyali et al., 2012). In order to improve the sensitivity of LFA, researchers have also looked into enzyme-based signal enhancement, thermal contrast, and fluidic control (He et al., 2018; Phillips et al., 2016; Qin et al., 2012).
NASBA with LFA detection was successfully demonstrated for Bacillus anthracis, where RNA was extracted with RNeasy mini kits and amplified by NASBA (Baeumner et al., 2004; Hartley and Baeumner, 2003). Following NASBA, the amplified RNA was mixed with running buffer and dye-entrapped liposomes tagged with reporter probes. The solution flowed across the membrane via capillary flow, where the capture probe was located. Target amplicons hybridized with both reporter probe and capture probe, giving rise to a qualitative signal. However, excess free liposomes needed to be washed off by adding a running buffer to reduce background signal.
In more recent experiments, metallic nanoparticles, especially GNs, have been used for better sensitivity, ease of operation, and more intense signal output. The Richards-Kortum group developed a GN-based LFA-NASBA system to successfully detect 50 synthetic copies of the HIV gag region of RNA generated through transcription with the MEGAscript T7 kit (Applied Biosystems), which corresponded to a plasma viral load of approximately 1000 copies/mL based on the assumptions that 100 μL of plasma sample were used and half of the viral RNA participated in the NASBA reaction (Rohrman et al., 2012). Gold enhancement solution was used to amplify the test signal to improve the LOD, and the LFA setup achieved consistent results 28 days after strip fabrication.
In addition, the Alocilja laboratory developed an LFA setup for the visual detection of dengue type 1 virus (DENV-1) RNA extracted from test samples with the QIAamp Viral RNA Mini Kit following NASBA (Yrad et al., 2019). They achieved an LOD of 0.01 μM using synthetic DENV-1 target and 1.2 × 104 PFU/mL using DENV-1 infected pooled human sera within 20 min. These results showed that the LFA-NASBA system will be quite promising for DENV-1 detection in resource-limited areas in the future.
Another approach used by the Baeumner group incorporated a micro-total analysis system (μTAS) with poly(methyl methacrylate) (PMMA) microchannels to reduce sample volume, cost, and user steps (Reinholt et al., 2014). Baeumner and coworkers developed a simple, single-channel for isolation and amplification of Cryptosporidium parvum (C. parvum) hsp70 mRNA. They applied UV/ozone treatment to carboxylate the surface of PMMA, followed by immobilization of polyamidoamine dendrimers to increase the immobilization efficiency of phosphate-modified thymidine oligonucleotide capture probes. This allowed for a greater efficiency in isolating and amplifying mRNA within the microfluidic device. In this work, several sample preparation steps were required to make 150 μL of C. parvum lysate prior to amplification. With use of an LFA, they successfully detected 30 C. parvum oocysts, showing equivalent performance to a benchtop device. Although this method required several liquid-handling steps, their μTAS has the potential to detect analytes in a sample-to-result fashion upon further improvements.
The combination of LFA with NASBA has shown potential for use in resource-limited settings. With integration of microfluidic devices, the multiple user steps may be eliminated to achieve a sample-to-result device that would be ideal for POC testing.
5.2.3. Fluorescent
Unlike previous endpoint detection methods, real-time NASBA measures the real-time fluorescent signal in order to achieve quantitative results. However, this method usually exhibits false-positive results and requires expensive equipment for signal readout. To avoid primer dimerization and nonspecific hybridization, the Morabito group manually redesigned the individual bases in the primers at 41 °C to eliminate unnecessary adenine-thymine and guanine-cytosine hydrogen bonds (Morabito et al., 2013). They also proved that shortening the T7 promoter sequence can reduce unwanted primer-dimers and hybrids. Furthermore, adjusting the salt ion concentration to create an optimal buffer solution could maximize primer specificity.
The Kolpashchikov group designed a split dapoxyl aptamer hybridization probe based on a DNA aptamer, which was divided into two strands, each of which was equipped with an analyte binding arm. These strands have low affinity to the dapoxyl dye unless specific targets bind to adjacent positions and form binding sites for the dye to generate fluorescent signals. In this work, they used these strands to successfully distinguish a fragment of the inhA gene of Mycobacterium tuberculosis from its single-base variant and detect 10 nM of Z-147 RNAs (a fragment of the ZIKV envelope gene) extracted using Trizol LS (Kikuchi et al., 2019). With portable fluorometers, this probe has the potential to achieve POC diagnosis.
In addition, Engelhart and coworkers introduced a competitor duplex sequence that is unrelated to the amplicon and does not contain T7 promoter and aptamer-encoding sequences into the Apta-NASBA system to prohibit primer dimerization to avoid false positives (Aufdembrink et al., 2020). Furthermore, they developed a Raspberry Pi-based detection system that could read fluorescent signals using a cell phone camera, eliminating the need for expensive readers. In the Apta-NASBA system, a conventional NASBA primer and a second primer that contains a fluorogenic aptamer that binds to the amplicon were introduced into the reaction mixture. As more amplicons were amplified, the amount of RNA aptamers increased, resulting in aptamer folding that generated fluorescent signals. The Engelhardt group designed a green aptamer to detect the E.coli estH gene, a yellow aptamer to detect the E.coli estP gene and a red aptamer to detect E.coli aggR gene.
Similarly, the Unrau laboratory developed Nested Mango NASBA that featured an outer primer NASBA reaction where amplification of target analytes and primer artifacts occurred, followed by a second inner Mango NASBA reaction where primers containing a Mango III tag were added to specifically bind to the products of interest to generate fluorescent signals (Abdolahzadeh et al., 2019). This system allowed them to reduce amplification of primer artifacts, resulting in greater sensitivity and specificity, and allowed them to achieve LODs of 25 pM for E. coli ClpB RNA and 2.5 aM for the Pseudomonas fluorescens ClpB target.
Moreover, the Libera group investigated solid-phase NASBA, which has potential for multiplexed microarray-based detection (Ma et al., 2018b). In this work, they first created poly(ethylene glycol) (PEG) microgels using electron beams to crosslink PEG and also cast thin films on a silicon support. Subsequently, they tethered molecular beacons and amplification primers on the microgels for target amplification and fluorescent detection.
Furthermore, it has been demonstrated that microfluidic chip devices can be integrated with real-time NASBA to lower the LOD, minimize user steps, and reduce sample-to-result time (Dimov et al., 2008; Esch et al., 2001). With further improvements to simplify the procedures and complexity of equipment associated with real-time NASBA, it will become a promising quantitative POC detection method.
6. Helicase-dependent amplification
6.1. Introduction
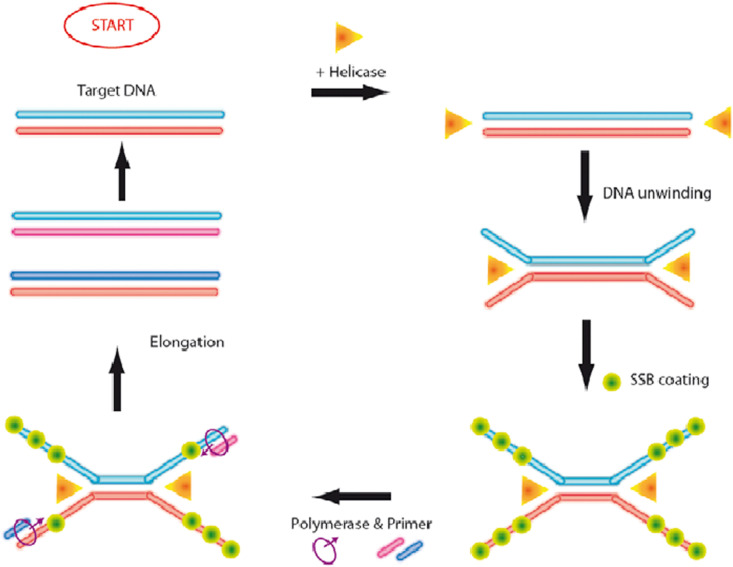
HDA is an isothermal amplification method introduced in 2004 by the Kong group (Vincent et al., 2004). HDA uses helicase, an enzyme that unwinds DNA duplexes during DNA replication, removing the need for denaturation via thermocycling (Cao et al., 2013). HDA can also be used to amplify RNA when coupled with reverse transcription (Goldmeyer et al., 2007). Thermophilic HDA (tHDA) is the most common type of HDA used by researchers, which uses thermostable Tte-UvrD helicase with Bst-DNA polymerase and operates at a single uniform temperature between 60 and 65 °C (An et al., 2005). Mesophilic HDA (mHDA) is another type of HDA that uses E. coli UvrD helicase, operates at 37 °C, and yields lower specificity than tHDA (An et al., 2005; Vincent et al., 2004). The size of an HDA amplicon is generally less than 150 nucleotides (New England BioLabs, 2020). The amplicons can be readily detected by gel electrophoresis, real-time HDA with fluorescent reporter dyes (e.g., EvaGreen), and oligonucleotide probes (Cao et al., 2013).
The mechanism of HDA is relatively simple compared to other isothermal amplification methods (Fig. 14 ). Initially, helicase unwinds the dsDNA, and ssDNA binding proteins bind to the displaced DNA strands to prevent the strands from annealing. Next, a pair of forward and reverse primers bind to target sequences and are extended by DNA polymerase at the 3’ ends. The amplified dsDNA products then enter future rounds of amplification (Cao et al., 2013; Vincent et al., 2004).
Fig. 14.
Schematic of the HDA mechanism. Reprinted with permission from (Tröger et al., 2015).
HDA requires only 2 primers, but due to the lack of a heating step, unwanted artifacts such as primer dimers, non-target specific hybrids, and non-canonical folds can result in loss of signal, high signal background, and false test results (Yang et al., 2015). Therefore, the HDA primer sequences need to be carefully designed, and many primer combinations must be screened, which adds additional time and cost (Yang et al., 2015; Deng and Gao, 2015).
6.2. Detection methods
6.2.1. Colorimetric
Colorimetric detection methods are suitable for POC diagnostics because they are simple, rapid, and cost-effective. The Jenison group developed a biosensor that combined tHDA of the mecA gene in MRSA from a positive blood culture with detection using an optically-coated chip (Jenison et al., 2014). Initially, a 10 min cell lysis step of the blood samples was achieved at room temperature. Amplification was then performed by adding tHDA reagents and crude lysates into a 96 well plate, with the bottom of each well containing a silicon chip whereupon amplicons were hybridized with complementary probes. TMB was then added, resulting in a transduced enzymatic reaction depositing a thin film onto the surface of the chip. The hybridized probes altered the path length of reflection resulting in a visible color change from gold to blue (Fig. 15 ). This biosensor only required a heat block, and could detect as few as 10 copies of input DNA in under 60 min. This input DNA came from 1 μL of crude extract that originated from 10 μL of blood culture.
Fig. 15.
The sensitivity of the MRSA detection device. (a) Colorimetric results from the detection of MRSA with 0 to 106 copies of genomic DNA. (b) Color difference as a function of target DNA copy number. (c) Real-time tHDA amplification plot with each color representing the quantity of input genomic DNA: no template control (gray), 102 copies (orange), 103 copies (blue), 104 copies (purple), 105 copies (green), 106 copies (red). Reprinted with permission from (Jenison et al., 2014). Copyright 2014 Royal Society of Chemistry. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Similar approaches utilized a modified silicon chip for the colorimetric detection of S. aureus and rpoB gene mutations in M. tuberculosis, as well as identification of Staphylococcus species following tHDA (Ao et al., 2012; Ao and Jenison, 2013; Frech et al., 2012; Pasko et al., 2012). However, some of these approaches had an arduous sample preparation procedure and also required instruments for result imaging, making them less POC-friendly.
6.2.2. On-paper
The LFA is the most common detection method for HDA products at the POC due to its portability, rapidness, and ease-of-use. Using tHDA and the LFA, detection and identification of five species of Plasmodium from unprocessed blood required minimal instrumentation (Li et al., 2013). After tHDA, the amplicons of all five species were annealed with digoxigenin-labeled probes, and Plasmodium falciparum was hybridized with an additional FAM-labeled probe. The labeled amplicons were applied to a test strip and captured by anti-digoxigenin or anti-FAM antibody lines in a BESt cassette, a portable and disposable detection device that contains a vertical LFA. Overall, this assay gave an analytical sensitivity of 200 parasites/μL of blood, and its clinical performance with 88 patient samples had 96.6% sensitivity and 100% specificity when compared to microscopy and a NASBA malaria test.
In addition, toxigenic C. difficile, HSV, HIV, Streptococcus equi subspecies equi, and S. aureus and its methicillin resistance from positive blood cultures were successfully detected utilizing handheld BESt cassettes after tHDA (Artiushin et al., 2011; Chow et al., 2008; Goldmeyer et al., 2008; Lemieux et al., 2012; Tang et al., 2010).
A paperfluidic platform that combined sample cell lysis, DNA extraction, tHDA, and detection using an LFA to test for N. gonorrhoeae had a turnaround time of 80 min (Horst et al., 2018). Initially, the sample was mixed with a lysis buffer. The mix was then pipetted into a reaction chamber and dried for 30 min in preparation for the subsequent steps of detection. The platform yielded 95% sensitivity and 100% specificity in vaginal and urethral swab samples when compared to qPCR with an LOD of 500 cells/5 μL of sample.
6.2.3. Fluorescent
Fluorescent detection of HDA amplicons is another common method. The Doseeva group described a detection assay for C. trachomatis and N. gonorrhoeae that integrated sample preparation using magnetic beads and endpoint detection (Doseeva et al., 2011). This detection approach utilized a closed-tube reaction to omit procedures after tHDA, decreasing risk of contamination. The magnetic beads were conjugated with monoclonal antibodies to capture RNA probes that hybridized specifically with target DNA, allowing the removal of extraneous DNA by washing. The tHDA reagents and dual-labeled fluorescent probes were then added to initiate amplification, and the amplicon-hybridized probes subsequently generated an endpoint fluorescence. This assay could be multiplexed using various dual-labeled probes, offer an economic alternative to qPCR, and give high sensitivity and specificity.
Endpoint fluorescent detection with tHDA was also applied in the IsoGlow typing assay for HSV-1 and HSV-2 (Tong et al., 2012). The endpoint analysis utilized cycling probe technology, which uses a chimeric probe in the form of DNA-RNA-DNA that anneals to a target sequence and gets cleaved off by RNAse H, triggering fluorescence of the probe while the target binds to new probes. A lost-cost, portable fluorescence reader was used for detection. The assay also included an internal control to detect potential amplification inhibitors, decreasing the number of false negatives. The turnaround time was approximately 1 h, with less than 5 min hands-on time, and achieved an LOD of 10 copies of HSV target per 5 μL of viral transport medium collected from oral and genital swabs.
The Klapperich group developed a microfluidic chip that integrated sample preparation using a micro solid phase extraction column, tHDA, and fluorescent detection of E. coli DNA using a UV illumination device (Mahalanabis et al., 2010, 2011). The chip contained a micro solid phase extraction column for lysing live bacteria cells and extracting DNA, and triplicate tHDA reaction chambers for amplifying and detecting target DNA. The entire procedure starting with sample preparation was under 50 min. Additionally, the chip design eliminated multiple liquid-handling steps and allowed for multiplexing.
A droplet-based microfluidic device utilized silica superparamagnetic particles (SSPs) for DNA extraction and magnetic actuation for the detection of ovarian cancer biomarkers Rsf-1 and E. coli (Zhang et al., 2011). Initially, the sample was mixed with an SSP-containing buffer droplet. By using a magnetic bar under the chip, the position of the droplet could be manipulated to mix with other reagents to achieve real-time tHDA. Fluorescent detection coupled with a melting curve analysis that helped determine specificity were conducted using an avalanche photodiode. Conducting the assay within discrete droplets allowed for high portability and reduced fabrication costs.
Lastly, the Krull group introduced a portable, paper-based test to fluorescently detect probe-target hybridization after tHDA (Noor et al., 2015). The paper surface contained immobilized quantum dot (QD)-probe oligonucleotide conjugates that emit green light when excited by a UV light. After tHDA, the amplicons hybridized with the probes, resulting in a fluorescence resonance energy transfer (FRET)-based transduction between Cy3 acceptor dye and QDs, from which an emission was then quantitatively captured using an iPad camera for ratiometric detection. This system had a turnaround time of 3 h including amplification, and offered an improved LOD in the pmol range of synthetic DNA markers for spinal muscular atrophy disorder.
6.2.4. Electrochemical
While HDA has been coupled with electrochemical detection techniques to achieve highly sensitive and robust detection of pathogens, the long detection time of such systems limits their relevance at the POC. In addition, no North American researcher has performed electrochemical detection of tHDA amplicons in the timeframe of interest for this review.
6.3. General developments of tHDA towards POC systems
As tHDA does not involve thermocycling steps, primer artifacts can be easily generated. To address this problem, the Benner group designed a “self avoiding molecular recognition system” (SAMRS) consisting of base analogues A*, T*, G*, and C* that could only pair with respective natural bases but not SAMRS bases (Yang et al., 2015). For example, A* paired only with natural T but not T*. Consequently, by incorporating SAMRS bases near the 3’ end of primers, primer-dimers would be eliminated. This system was shown to be effective in the amplification of 3 cancer genes, rpoB gene in M. tuberculosis, and HIV-1 viral RNA. Additionally, by using two different sets of SAMRS-containing primers, HIV-1 viral RNA and rpoB DNA were simultaneously detected, indicating that multiplexing capability for both DNA and RNA targets could be achieved using SAMRS.
While tHDA is a simple, rapid amplification method, many of the described detection platforms still require extensive preparation and extraction steps for unprocessed cell samples prior to amplification. The Kamei group described a one-pot isothermal platform that incorporates cell lysis, DNA extraction, and tHDA utilizing an aqueous two-phase system (ATPS) (Cheung et al., 2018). A Triton X-100 micellar ATPS was combined with tHDA reagents and a cell suspension containing intact E. coli cells. This system was able to lyse the cells and extract and concentrate DNA into the top phase as phase separation was induced at 68 °C for 15 min, followed by amplification of the target gene for another 45 min (Fig. 16 ). The top phase was then extracted after amplification. The one-pot reaction combined with gel electrophoresis yielded an LOD of 102 CFU/mL of original cell solution, considerably lower than that of the conventional tHDA performed in parallel (107 CFU/mL). Although this system has yet to be combined with POC detection methods, it eliminated several liquid-handling steps and is a promising direction for future work at the POC.
Fig. 16.
Comparison of DNA amplification by the one-pot reaction and the conventional tHDA reaction detected by gel electrophoresis. Whole cell samples were used in these experiments. Reprinted by permission from Springer Nature: Springer Nature, Analytical and Bioanalytical Chemistry (Cheung et al., 2018), Copyright 2018.
7. Strand displacement amplification
Strand displacement amplification (SDA) was developed in 1992 by Walker and coworkers; however, it required a restriction enzyme cleavage step prior to amplification (Walker et al., 1992a). To remedy this issue, the same group developed an updated SDA mechanism that eliminated this step, instead utilizing an exo-Klenow fragment to generate the necessary target copies with defined 5′ and 3′ ends (Walker et al., 1992b). This mechanism begins with a denaturation step in the presence of four primers. These primers are annealed to 5’ adjacent sequences to form a nick, and amplification begins at a fixed temperature of approximately 37 °C. As polymerase synthesizes DNA, newly synthesized strands are nicked by a restriction enzyme and polymerase starts synthesis again, kicking off the previous strand, resulting in DNA amplification.
Due to the initial denaturation step, SDA is not a truly isothermal amplification technique. In response to this problem, the Toley laboratory developed a fully isothermal SDA reaction for the amplification and detection of S. aureus using both LFA and fluorescence (Toley et al., 2015). To circumvent the initial denaturation step, amplification initiates at sites of DNA breathing and continues in a similar fashion to standard SDA reactions. LFA detection was found to detect as low as 10 copies of purified target DNA spiked in a reaction mixture and 50 copies spiked in a reaction mixture containing high concentrations of background DNA and mucins in less than 30 min. Thus, the high sensitivity, rapid time-to-response, and low operating temperature of the SDA assay make it a viable choice for POC detection of pathogens.
8. Nicking enzyme amplification reaction
Nicking enzyme amplification reaction (NEAR), also known as exponential amplification reaction (EXPAR), is an isothermal amplification technique developed by the Galas group in 2003 (Van Ness et al., 2003). The process utilizes a single-strand nicking enzyme and a DNA polymerase with strand-displacement activity. Each cycle of amplification consists of polymerization, single-strand nicking, and release, returning to the original primer template. In order to make the process exponential, the oligonucleotide forms a transient duplex that is stabilized via extension by DNA polymerase. This then acts as a stable primer for the further production of oligonucleotides.
The reaction takes place at a constant temperature of 60 °C, and it is best suited for small amplicons 10–22 nucleotides long (Menova et al., 2013). A key issue with NEAR, common with many other forms of isothermal amplification, is the relatively high rate of nonspecific amplification (Tan et al., 2008). As such, precautions such as physically separating the template and polymerase prior to reaching optimal temperatures are an important consideration for the application of NEAR in diagnostic devices.
Recent developments have led to NEAR amplification devices entering the market, warranting a brief discussion on the reaction. The most prominent is the ID NOW Abbott machine. When used to detect Influenza A/B, the ID NOW was able to amplify and detect the virus within 10 min with a very wide degree of success, with specificities between 52.2 and 93.8% for Influenza A cases and 72.2–93.3% for Influenza B cases (Kanwar et al., 2020; Chapin et al., 2015; Young et al., 2017; Nolte et al., 2016). When evaluating high viral loads, the ID NOW provides accurate results, but precipitously drops in reliability when approaching lower concentrations around Ct (threshold cycle) = 27–30. When evaluated against negative controls, the ID NOW has been shown to be resistant to false amplification, with almost all studies obtaining at or near 100% specificity.
The ID NOW test for SARS-CoV-2 relies on fluorescent reporter probes to detect the presence of the RdRp gene from nasopharyngeal swabs in viral transport medium (Smithgall et al., 2020). The Green group found that the ID NOW was able to reliably detect the disease at moderate to high concentrations with a sensitivity of 100%. At low concentrations (Ct > 30), it was unable to detect many cases and its sensitivity dropped to 34.3%. A New York University study reflected this continued trend, showing that the ID NOW was unable to detect positive cases at reduced viral loads (Basu et al., 2020). While the device displayed consistent results at high concentrations, it was only able to detect 33% of positive cases detected by a reference method. Those cases were sampled via nasopharyngeal swabs, and since Abbott indicated that the preferred technique of sampling was through the use of dry nasal swabs, the same research group examined detection efficacy using the preferred sampling method. However, they were only able to attain a sensitivity of 54.8%, where a similar pattern emerged, as all missed cases were exclusively of low viral load (Ct > 33.7). At the time of this review, the ID NOW for SARS-CoV-2 was still being extensively examined by different research groups to determine its overall performance.
9. Summary and conclusions
This systematic review focused on the mechanisms of different isothermal amplification methods, along with recent advances made by North American research groups to improve existing and potential POC detection methods that can be integrated with these methods. As researchers begin thinking about such systems to use, a wide variety of factors must be considered. For example, if the technology is to be used in regions that can vary in temperature and humidity, paper-based detection methods incorporating dehydration of reagents and desiccants may be applied. On the other hand, if small sample volume is an important parameter, on-paper, microfluidic, and in-tube detection setups are all applicable. With regard to isothermal amplification, if it is desirable to use the lowest power, RPA and NASBA may be good choices due to their low amplification temperatures. Many other factors, including sensitivity and specificity, which can vary per target, must also be considered. Accordingly, we have summarized the literature that was reviewed for this manuscript in Supplementary Table 1 in terms of important design criteria, with the idea that this compiled source of extensive information may help the researcher in the decision-making.
10. Future perspectives
POC detection methods have been trending towards colorimetric assays, often dependent upon changes in pH or oxidation, and paper-based assays, which are advantageous due to built-in reagent storage and increased reaction rate. Such systems are particularly well suited for resource-limited areas due to their low cost, short assay time, and visual readout. Other research has focused on reducing user steps, reliance on expensive equipment, and the system's time-to-result. Several systems have incorporated smartphones or iPads for a user-friendly colorimetric or fluorescent readout. Although numerous developments have been made in recent years, further improvements are still needed to better integrate isothermal amplification and POC detection for greater sensitivity, availability, user-friendliness, and cost effectiveness for resource-limited areas. Firstly, many of the manuscripts that were reviewed used extensive upstream sample processing steps to extract and purify the nucleic acids, which prevent those methods from being POC. Therefore, future research should focus on integrating sample preparation into POC diagnostic devices. Diagnostic systems may potentially be made more POC-friendly by the implementation of aqueous two-phase system-based sample preparation, on-chip microfluidic sample preparation, and/or paper-based filtration. Secondly, efforts must be made to mitigate the drawbacks of current isothermal amplification methods, such as nonspecific binding and false positives. Additionally, further improvements need to be made to make microfluidic devices better suited to POC testing. Potential improvements include simplifying the microfluidic design and fabricating using a lower-cost material. Other detection modalities should be investigated for their synergy with POC testing. Finally, POC-compatible technologies (colorimetric, on-paper, turbidity, fluorescent, electrochemical) should be further explored in the context of improving multiplexing capabilities.
CRediT authorship contribution statement
Elizabeth A. Pumford: Writing - original draft, Writing - review & editing. Jiakun Lu: Writing - original draft, Writing - review & editing. Iza Spaczai: Writing - original draft, Writing - review & editing. Matthew E. Prasetyo: Writing - original draft, Writing - review & editing. Elaine M. Zheng: Writing - original draft, Writing - review & editing. Hanxu Zhang: Writing - original draft, Writing - review & editing. Daniel T. Kamei: Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112674.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdolahzadeh A., Dolgosheina E.V., Unrau P.J. RNA detection with high specificity and sensitivity using Nested Fluorogenic Mango NASBA. RNA. 2019;25:1806–1813. doi: 10.1261/rna.072629.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.M., Li F., Zhang Z., Zhang K., Kang D., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- An L., Tang W., Ranalli T.A., Kim H., Wytiaz J., Kong H. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J. Biol. Chem. 2005;280:28952–28958. doi: 10.1074/jbc.M503096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao W., Jenison R. Detection of rpoB gene mutations using helicase-dependent amplification. In: Kolpashchikov D.M., Gerasimova Y.V., editors. Nucleic Acid Detection: Methods and Protocols. Humana Press; New Jersey: 2013. pp. 89–98. [DOI] [PubMed] [Google Scholar]
- Ao W., Aldous S., Woodruff E., Hicke B., Rea L., Kreiswirth B., Jenison R. Rapid detection of rpoB gene mutations conferring rifampin resistance in. Mycobacterium Tuberculosis. J. Clin. Microbiol. 2012;50(7):2433–2440. doi: 10.1128/JCM.00208-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin S., Tong Y., Timoney J., Lemieux B., Schlegel A., Kong H. Thermophilic helicase-dependent DNA amplification using the IsoAmpTM SE experimental kit for rapid detection of Streptococcus equi subspecies equi in clinical samples. J. Vet. Diagn. Invest. 2011;23:909–914. doi: 10.1177/1040638711416968. [DOI] [PubMed] [Google Scholar]
- Asghar W., Sher M., Khan N.S., Vyas J.M., Demirci U. Microfluidic chip for detection of fungal infections. ACS Omega. 2019;4:7474–7481. doi: 10.1021/acsomega.9b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufdembrink L.M., Khan P., Gaut N.J., Adamala K.P., Engelhart A.E. 2020. Highly Specific, Multiplexed Isothermal Pathogen Detection with Fluorescent Aptamer Readout. RNA. rna-075192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeumner A.J., Leonard Æ.B., Mcelwee Æ.J., Montagna R.A. A rapid biosensor for viable B. anthracis spores. Anal. Bioanal. Chem. 2004;380:15–23. doi: 10.1007/s00216-004-2726-7. [DOI] [PubMed] [Google Scholar]
- Banér J., Nilsson M., Mendel-Hartvig M., Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes L., Heithoff D.M., Mahan S.P., Fox G.N., Zambrano A., Choe J., Fitzgibbons L.N., Marth J.D., Fried J.C., Soh H.T., Mahan M.J. Smartphone-based pathogen diagnosis in urinary sepsis patients. EBioMedicine. 2018;36:73–82. doi: 10.1016/j.ebiom.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Zinger T., Inglima K., Woo K., Atie O., Yurasits L., See B., Aguero-Rosenfeld M.E. Performance of Abbott ID Now COVID-19 rapid nucleic acid test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01136-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) PloS One. 2015;10 doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S., Saldaña M.A., Han H.G., Hughes G.L., Ellington A.D. Simultaneous detection of different Zika Virus lineages via molecular computation in a point-of-care assay. Viruses. 2018;10:714. doi: 10.3390/v10120714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialy R.M., Ali M.M., Li Y., Brennan J.D. Protein-mediated suppression of rolling circle amplification for biosensing with an aptamer-containing DNA primer. Chem. Eur J. 2020;26:5085–5092. doi: 10.1002/chem.202000245. [DOI] [PubMed] [Google Scholar]
- BioLabs New England. Isothermal amplification application overview. 2020. https://www.neb.sg/applications/dna-amplification-pcr-and-qpcr/isothermal-amplification accessed 30 June 2020.
- Blanco L., Bernad A., Lazaro J.M., Martin G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage Φ29 DNA polymerase. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- Boyle D., Lehman D., Lillis L., Peterson D., Singhal M., Armes N., Parker M., Piepenburg O., Overbaugh J. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio. 2013;4 doi: 10.1128/mBio.00135-13. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzales A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabada M., Malaga J., Castellanos-Gonzalez A., Bagwell K., Naeger P., Rogers H., Maharsi S., Mbaka M., Clinton White A. Recombinase polymerase amplification compared to real-time polymerase chain reaction test for the detection of Fasciola hepatica in human stool. Am. J. Trop. Med. Hyg. 2017;96:341–346. doi: 10.4269/ajtmh.16-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Kim H.J., Li Y., Kong H., Lemieux B. Helicase‐dependent amplification of nucleic acids. Curr. Protoc. Mol. Biol. 2013;104 doi: 10.1002/0471142727.mb1511s104. 15.11.1-15.11.12. [DOI] [PubMed] [Google Scholar]
- Chan K., Wong P.Y., Parikh C., Wong S. Moving toward rapid and low-cost point-of-care molecular diagnostics with a repurposed 3D printer and RPA. Anal. Biochem. 2018;545:4–12. doi: 10.1016/j.ab.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C., Belinskaya T., Zhang Z., Ching W. Development of recombinase polymerase amplification assays for detection of Orientia tsutsugamushi or Rickettsia typhi. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin S.C., Doyle P.S. Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal. Chem. 2011;83:7179–7185. doi: 10.1021/ac201618k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin K.C., Flores-Cortez E.J. Performance of the molecular Alere i Influenza A&B test compared to that of the Xpert Flu A/B assay. J. Clin. Microbiol. 2015;53:706–709. doi: 10.1128/JCM.02783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cheng N., Xu Y., Huang K., Luo Y., Xu W. Point-of-care and visual detection of P. aeruginosa and its toxin genes by multiple LAMP and lateral flow nucleic acid biosensor. Biosens. Bioelectron. 2016;81:317–323. doi: 10.1016/j.bios.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Cheung S.F., Yee M.F., Le N.K., Wu B.M., Kamei D.T. A one-pot, isothermal DNA sample preparation and amplification platform utilizing aqueous two-phase systems. Anal. Bioanal. Chem. 2018;410:5255–5263. doi: 10.1007/s00216-018-1178-4. [DOI] [PubMed] [Google Scholar]
- Choopara I., Arunrut N., Kiatpathomchai W., Dean D., Somboonna N. Rapid and visual Chlamydia trachomatis detection using loop-mediated isothermal amplification and hydroxynaphthol blue. Lett. Appl. Microbiol. 2016;64:51–56. doi: 10.1111/lam.12675. [DOI] [PubMed] [Google Scholar]
- Chow W., McCloskey C., Tong Y., Hu L., You Q., Kelly C., Kong H., Tang Y., Tang W. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J. Mol. Diagn. 2008;10:452–458. doi: 10.2353/jmoldx.2008.080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Cordray M., Richards-Kortum R. A paper and plastic device for the combined isothermal amplification and lateral flow detection of Plasmodium DNA. Malar. J. 2015;14:472. doi: 10.1186/s12936-015-0995-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damhorst G.L., Duarte-guevara C., Chen W., Ghonge T., Cunningham T., Bashir R. Smartphone-imaged HIV-1 reverse-transcription loop-mediated isothermal amplification (RT-LAMP) on a chip from whole blood. Eng. Times. 2015;1:324–335. doi: 10.15302/J-ENG-2015072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiman B., Jay C., Zintilini C., Vermeer S., van Strijp D., Venema F., van de Wiel P. Efficient amplification with NASBA® of hepatitis B virus, herpes simplex virus and methicillin resistant Staphylococcus aureus DNA. J. Virol. Methods. 2008;151:283–293. doi: 10.1016/j.jviromet.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Deng H., Gao Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta. 2015;853:30–45. doi: 10.1016/j.aca.2014.09.037. [DOI] [PubMed] [Google Scholar]
- Dhar, B.C., Reed, A.J., Mitra, S., Sanchez, P.R., Nedorezova, D.D., Connelly, R.P., Rohde, K.H., Gerasimova, Y.V., Cascade of deoxyribozymes for the colorimetric analysis of drug resistance in Mycobacterium tuberculosis. Biosens. Bioelectron. 165. 112385. [DOI] [PMC free article] [PubMed]
- Dimov I.K., Garcia-Cordero J.L., O'Grady J., Poulsen C.R., Viguier C., Kent L., Daly P., Lincoln B., Maher M., O'Kennedy R., Smith T.J., Ricco A.J., Lee L.P. Integrated microfluidic tmRNA purification and real-time NASBA device for molecular diagnostics. Lab Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- Doseeva V., Forbes T., Wolff J., Khripin Y., Neil D.O., Rothmann T., Nazarenko I. Multiplex isothermal helicase-dependent amplification assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Diagn. Microbiol. Infect. Dis. 2011;71:354–365. doi: 10.1016/j.diagmicrobio.2011.08.021. [DOI] [PubMed] [Google Scholar]
- Dou M., Lopez J., Rios M., Garcia O., Xiao C., Eastman M., Li X. A fully battery-powered inexpensive spectrophotometric system for high-sensitivity point-of-care analysis on a microfluidic chip. Analyst. 2016;141:3898–3903. doi: 10.1039/c6an00370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou M., Sanchez J., Tavakoli H., Gonzalez J.E., Sun J., Dien J., Li X. A low-cost microfluidic platform for rapid and instrument-free detection of whooping cough. Anal. Chim. Acta. 2019;1065:71–78. doi: 10.1016/j.aca.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drame P.M., Fink D.L., Kamgno J., Herrick J.A., Nutman B. Loop-mediated isothermal smplification for rapid and semiquantitative detection of Loa loa infection. J. Clin. Microbiol. 2014;52:2071–2077. doi: 10.1128/JCM.00525-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Hughes R.A., Bhadra S., Jiang Y.S., Ellington A.D., Li B. A sweet spot for molecular diagnostics: coupling isothermal amplification and strand exchange circuits to glucometers. Sci. Rep. 2015;5:1–14. doi: 10.1038/srep11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Pothukuchy A., Gollihar J.D., Nourani A., Li B., Ellington A.D. Coupling sensitive nucleic acid amplification with commercial pregnancy test strips. Angew Chem. Int. Ed. Engl. 2017;56:992–996. doi: 10.1002/anie.201609108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch M., Locascio L., Tarlov M., Durst R. Detection of viable Cryptosporidium parvum using DNA-modified liposomes in a microfluidic chip. Anal. Chem. 2001;73:2952–2958. doi: 10.1021/ac001508n. [DOI] [PubMed] [Google Scholar]
- Farkas D.H., Holland C.A. Overview of molecular diagnostic techniques and instrumentation. Cell and Tissue Based Mol. Path. 2009:19–32. [Google Scholar]
- Frech G.C., Munns D., Jenison R.D., Hicke B.J. Direct detection of nasal Staphylococcus aureus carriage via helicase-dependent isothermal amplification and chip hybridization. BMC Res. Notes. 2012;5:430. doi: 10.1186/1756-0500-5-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S., Savory E., Weisberg A., Buser J., Gordon M., Putnam M., Chang J. Isothermal amplification and lateral-flow assay for detecting crown-gall-causing Agrobacterium spp. Phytopathology. 2017;107:1062–1068. doi: 10.1094/PHYTO-04-17-0144-R. [DOI] [PubMed] [Google Scholar]
- Ganguli A., Ornob A., Yu H., Damhorst G.L., Chen W., Sun F., Bhuiya A., Cunningham B.T., Bashir R. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed. Microdevices. 2017;19:73. doi: 10.1007/s10544-017-0209-9. [DOI] [PubMed] [Google Scholar]
- Giuffrida M.C., Spoto G. Integration of isothermal amplification methods in microfluidic devices: recent advances. Biosens. Bioelectron. 2017;90:174–186. doi: 10.1016/j.bios.2016.11.045. [DOI] [PubMed] [Google Scholar]
- Goldmeyer J., Kong H., Tang W. Development of a novel one-tube isothermal reverse transcription thermophilic helicase - dependent amplification platform for rapid RNA detection. J. Mol. Diagn. 2007;9:639–644. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmeyer J., Li H., McCormac M., Cook S., Stratton C., Lemieux B., Kong H., Tang W., Tang Y. Identification of Staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J. Clin. Microbiol. 2008;46:1534–1536. doi: 10.1128/JCM.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A., Miller N.S., Smolina I. Visual detection of bacterial pathogens via PNA-based padlock probe assembly and isothermal amplification of DNAzymes. Anal. Chem. 2014;86:11992–11998. doi: 10.1021/ac5018748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., Honda E., Ogura A., Nomoto A., Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- Hamidi S.V., Perreault J. Simple rolling circle amplification colorimetric assay based on pH for target DNA detection. Talanta. 2019;201:419–425. doi: 10.1016/j.talanta.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Hardinge P., Murray J.A.H. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci. Rep. 2019;9:7400. doi: 10.1038/s41598-019-43817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley H.A., Baeumner A.J. Biosensor for the specific detection of a single viable B. anthracis spore. Anal. Bioanal. Chem. 2003;376:319–327. doi: 10.1007/s00216-003-1939-5. [DOI] [PubMed] [Google Scholar]
- He Y., Zhang S., Zhang Xibao, Baloda M., Gurung A.S., Xu H., Zhang Xueji, Liu G. Ultrasensitive nucleic acid biosensor based on enzyme – gold nanoparticle dual label and lateral flow strip biosensor. Biosens. Bioelectron. 2018;26:2018–2024. doi: 10.1016/j.bios.2010.08.079. [DOI] [PubMed] [Google Scholar]
- Horst A., Rosenbohm J., Kolluri N., Hardick J., Gaydos C., Cabodi M., Klapperich C., Linnes J. A paperfluidic platform to detect Neisseria gonorrhoeae in clinical samples. Biomed. Microdevices. 2018;20:35. doi: 10.1007/s10544-018-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M., Moriam S., Umer M., Phan H., Salomon C., Kline R., Nguyen N., Shiddiky M. Naked-eye and electrochemical detection of isothermally amplified HOTAIR long non-coding RNA. Analyst. 2018;143:3021–3028. doi: 10.1039/c7an02109g. [DOI] [PubMed] [Google Scholar]
- Itou T., Markotter W., Nel L.H. Reverse transcription-loop-mediated isothermal amplification system for the detection of rabies virus. In: Rupprecht C., Nagarajan T., editors. Current Laboratory Techniques in Rabies Diagnosis, Research and Prevention. Academic Press; Massachusetts: 2014. pp. 85–95. [Google Scholar]
- Jauset-Rubio M., Svobodova M., Mairal T., McNeil C. Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep37732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauset-Rubio M., Svobodová M., Mairal T., McNeil C., Keegan N., El-Shahawi M., Bashammakh A., Alyoubi A., O'Sullivan C. Aptamer lateral flow assays for ultrasensitive detection of β-conglutin combining recombinase polymerase amplification and tailed primers. Anal. Chem. 2016;88:10701–10709. doi: 10.1021/acs.analchem.6b03256. [DOI] [PubMed] [Google Scholar]
- Jauset-Rubio M., Sabaté Del Río J., Mairal T., Svobodová M., El-Shahawi M., Bashammakh A., Alyoubi A., O'Sullivan C. Ultrasensitive and rapid detection of β-conglutin combining aptamers and isothermal recombinase polymerase amplification. Anal. Bioanal. Chem. 2017;409:143–149. doi: 10.1007/s00216-016-9973-2. [DOI] [PubMed] [Google Scholar]
- Jenison R., Jaeckel H., Klonoski J., Latorra D., Wiens J. Rapid amplification/detection of nucleic acid targets utilizing a HDA/thin film biosensor. Analyst. 2014;139:3763–3769. doi: 10.1039/c4an00418c. [DOI] [PubMed] [Google Scholar]
- Jiang Y.S., Stacy A., Whiteley M., Ellington A.D. Amplicon competition enables end-point quantitation of nucleic acids following isothermal amplification. Chembiochem. 2017;18:1692–1695. doi: 10.1002/cbic.201700317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Loeb J.C., Manzanas C., Lednicky J.A., Fan Z.H. Valve-enabled sample preparation and RNA amplification in a coffee mug for Zika virus detection. Angew. Chem. Int. Ed. 2018;57:17211–17214. doi: 10.1002/anie.201809993. [DOI] [PubMed] [Google Scholar]
- Jiang L., Ching P., Chao C.-C., Dumler J.S., Ching W.-M. Development of a sensitive and rapid recombinase polymerase amplification assay for detection of Anaplasma phagocytophilum. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.01777-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonstrup S.P., Koch J., Kjems J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA. 2006;12:1747–1752. doi: 10.1261/rna.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarj K., Akarapipad P., Yoon J. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci. Rep. 2018;8:12438. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar N., Michael J., Doran K., Montgomery E., Selvarangan R. Comparison of the ID Now Influenza A & B 2 , cobas Influenza A/B , and Xpert Xpress flu point-of-care nucleic acid amplification tests for Influenza A/B virus detection in children. J. Clin. Microbiol. 2020;58:e01611–e01619. doi: 10.1128/JCM.01611-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting S., Rausch V., Bier F., von Nickisch-Rosenegk M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar. J. 2014;13:99. doi: 10.1186/1475-2875-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi N., Reed A., Gerasimova Y.V., Kolpashchikov D.M. Split dapoxyl aptamer for sequence-selective analysis of nucleic acid sequence based amplification amplicons. Anal. Chem. 2019;91(4):2667–2671. doi: 10.1021/acs.analchem.8b03964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J. Development of a multiplex real-time recombinase polymerase amplification (RPA) assay for rapid quantitative detection of Campylobacter coli and jejuni from eggs and chicken products. Food Contr. 2017;73:1247–1255. [Google Scholar]
- Lalremruata A., Nguyen T.T., McCall M.B.B., Mombo-Ngoma G., Agnandji S.T., Adegnika A.A., Lell B., Ramharter M., Hoffman S.L., Kremsner P.G., Mordmüller B. Recombinase polymerase amplification and lateral flow assay for ultrasensitive detection of low-density Plasmodium falciparum infection from controlled human malaria infection studies and naturally acquired infections. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.01879-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PloS One. 2020;15 doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H.Y., Botella J.R. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci. 2017;8:2016. doi: 10.3389/fpls.2017.02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Baek Y., Kim Y., Choi Y., Song M., Ahn J. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front. Microbiol. 2017;7:2166. doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B., Li Y., Kong H., Tang Y. Near instrument-free, simple molecular device for rapid detection of herpes simplex viruses. Expert Rev. Mol. Diagn. 2012;12:437–443. doi: 10.1586/erm.12.34. [DOI] [PubMed] [Google Scholar]
- Li J., Macdonald J. Advances in isothermal amplification: novel strategies inspired by biological processes. Biosens. Bioelectron. 2015;64:196–211. doi: 10.1016/j.bios.2014.08.069. [DOI] [PubMed] [Google Scholar]
- Li Y., Kumar N., Gopalakrishnan A., Ginocchio C., Manji R., Bythrow M., Lemieux B., Kong H. Detection and species identification of malaria parasites by isothermal THDA amplification directly from human blood without sample preparation. J. Mol. Diagn. 2013;15:634–641. doi: 10.1016/j.jmoldx.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Pollak N., Macdonald J. Multiplex detection of nucleic acids using recombinase polymerase amplification and a molecular colorimetric 7-segment display. ACS Omega. 2019;4:11388–11396. doi: 10.1021/acsomega.9b01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Peng J., Mauk M., Awasthi S., Song J., Friedman H., Bau H., Liu C. Smart cup: a minimally-instrumented, smartphone-based point-of-care molecular diagnostic Device. Sensor. Actuator. B Chem. 2016;229:232–238. doi: 10.1016/j.snb.2016.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillis L., Lehman D., Siverson J., Weis J., Cantera J., Parker M., Piepenburg O., Overbaugh J., Boyle D. Cross-subtype detection of HIV-1 using reverse transcription and recombinase polymerase amplification. J. Virol. Methods. 2016;230:28–35. doi: 10.1016/j.jviromet.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillis L., Siverson J., Lee A., Cantera J., Parker M., Piepenburg O., Lehman D., Boyle D. Factors influencing Recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol. Cell. Probes. 2016;30:74–78. doi: 10.1016/j.mcp.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Daubendiek S.L., Zillman M.A., Ryan K., Kool E.T. Rolling circle DNA synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 1996;118:1587–1594. doi: 10.1021/ja952786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Song J., Shuang S., Dong C., Brennan J.D., Li Y. A graphene-based biosensing platform based on the release of DNA probes and rolling circle amplification. ACS Nano. 2014;8:5564–5573. doi: 10.1021/nn5007418. [DOI] [PubMed] [Google Scholar]
- Liu M., Hui C.Y., Zhang Q., Gu J., Kannan B., Jahanshahi-Anbuhi S., Filipe C.D.M., Brennan J.D., Li Y. Target-induced and equipment-free DNA amplification with a simple paper device. Angew. Chem. Int. Ed. 2016;55:2709–2713. doi: 10.1002/anie.201509389. [DOI] [PubMed] [Google Scholar]
- Lizardi P.M., Huang X., Zhu Z., Bray-Ward P., Thomas D.C., Ward D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Ma D., Shen L., Wu K., Diehnelt C.W., Green A.A. Low-cost detection of Norovirus using paper-based cell-free systems and synbody-based viral enrichment. Synth. Biol. 2018;3:ysy018. doi: 10.1093/synbio/ysy018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Teng F., Libera M. Solid-phase NASBA and length-scale effects during RNA amplification. Anal. Chem. 2018;90:6532–6539. doi: 10.1021/acs.analchem.8b00058. [DOI] [PubMed] [Google Scholar]
- Maffert P., Reverchon S., Nasser W., Rozand C., Abaibou H. New nucleic acid testing devices to diagnose infectious diseases in resource-limited settings. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1717–1731. doi: 10.1007/s10096-017-3013-9. [DOI] [PubMed] [Google Scholar]
- Mahalanabis M., Do J., ALMuayad H., Zhang J.Y., Klapperich C.M. An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed. Microdevices. 2010;12:353–359. doi: 10.1007/s10544-009-9391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalanabis M., Do J., ALMuayad H., Zhang J.Y., Klapperich C.M. Erratum to: an integrated disposable device for DNA extraction and helicase dependent amplification. Biomed. Microdevices. 2011;13:599–602. doi: 10.1007/s10544-011-9518-6. [DOI] [PubMed] [Google Scholar]
- Malek L., Sooknanan R., Compton J. Nucleic acid sequence-based amplification (NASBA™) In: Isaac P.G., editor. Protocols for Nucleic Acid Analysis by Nonradioactive Probes, Methods in Molecular Biology. Humana Press; New Jersey: 1994. pp. 253–260. [DOI] [PubMed] [Google Scholar]
- Martorell S., Palanca S., Maquieira Á., Tortajada-Genaro L. Blocked recombinase polymerase amplification for mutation analysis of PIK3CA gene. Anal. Biochem. 2018;544:49–56. doi: 10.1016/j.ab.2017.12.013. [DOI] [PubMed] [Google Scholar]
- Mauk M., Song J., Liu C., Bau H. Simple approaches to minimally-instrumented, microfluidic-based point-of-care nucleic acid amplification tests. Biosensors. 2018;8:17. doi: 10.3390/bios8010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayboroda O., Benito A.G., del Rio J.S., Svobodova M., Julich S., Tomaso H., O'Sullivan C., Katakis I. Isothermal solid-phase amplification system for detection of Yersinia pestis. Anal. Bioanal. Chem. 2016;408:671–676. doi: 10.1007/s00216-015-9177-1. [DOI] [PubMed] [Google Scholar]
- Menova P., Raindlova V., Hocek M. Scope and limitations of the nicking enzyme amplification reaction for the synthesis of base-modified oligonucleotides and primers for PCR. Bioconjugate Chem. 2013;24:1081–1093. doi: 10.1021/bc400149q. [DOI] [PubMed] [Google Scholar]
- Mohon A., Lee L., Bayih A., Folefoc A., Guelig D., Burton R., LaBarre P., Chan W., Meatherall B., Pillai D. NINA-LAMP compared to microscopy, RDT, and nested PCR for the detection of imported malaria. Diagn. Microbiol. Infect. Dis. 2016;85:149–153. doi: 10.1016/j.diagmicrobio.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal D., Ghosh P., Khan M.A.A., Hossain F., Böhlken-Fascher S., Matlashewski G., Kroeger A., Olliaro P., Abd El Wahed A. Mobile suitcase laboratory for rapid detection of Leishmania donovani using recombinase polymerase amplification assay. Parasites Vectors. 2016;9:281. doi: 10.1186/s13071-016-1572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito K., Wiske C., Tripathi A. Engineering insights for multiplexed real-time nucleic acid sequence-based amplification (NASBA): implications for design of point-of-care diagnostics. Mol. Diagn. Ther. 2013;17:185–192. doi: 10.1007/s40291-013-0029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudanyali O., Dimitrov S., Sikora U., Padmanabhan S., Navruz I., Ozcan A. Integrated rapid-diagnostic-test reader platform on a cellphone. Lab Chip. 2012;12:2678–2686. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G., Rebolledo M., Clinton White A., Crannell Z., Rebecca Richards-Kortum R., Elizabeth Pinilla A., Ramírez J., Consuelo López M., Castellanos-Gonzalez A. Detection of Entamoeba histolytica by recombinase polymerase amplification. Am. J. Trop. Med. Hyg. 2015;93:591–595. doi: 10.4269/ajtmh.15-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.R., Cai Y.C., Giesler T.L., Farchaus J.W., Sundaram S.T., Ortiz-Rivera M., Hosta L.P., Hewitt P.L., Mamone A., Palaniappan C., Fuller C.W. TempliPhi, ϕ29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques. 2002;32:S44–S47. [PubMed] [Google Scholar]
- Njiru Z.K. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiru Z.K., Traub R., Ouma J.O., Enyaru J.C., Matovu E. Detection of group 1 Trypanosoma brucei gambiense by loop-mediated isothermal amplification. J. Clin. Microbiol. 2011;49:1530–1536. doi: 10.1128/JCM.01817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte F.S., Gauld L., Barrett S.B. Direct comparison of Alere i and cobas Liat Influenza A and B tests for rapid detection of Influenza virus infection. J. Clin. Microbiol. 2016;54:2763–2766. doi: 10.1128/JCM.01586-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M., Hrovat D., Moazami-Goudarzi M., Espie G., Krull U.J. Ratiometric fluorescence transduction by hybridization after isothermal amplification for determination of zeptomole quantities of oligonucleotide biomarkers with a paper-based platform and camera-based detection. Anal. Chim. Acta. 2015;885:156–165. doi: 10.1016/j.aca.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N.M. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Pasko C., Hicke B., Dunn J., Jaeckel H., Nieuwlandt D., Weed D., Woodruff E., Zheng X., Jenison R. Staph ID/R: a rapid method for determining Staphylococcus species identity and detecting the mecA gene directly from positive blood culture. J. Clin. Microbiol. 2012;50:810–817. doi: 10.1128/JCM.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips E., Shen R., Zhao S., Linnes J. Thermally actuated wax valves for paper-fluidic diagnostics. Lab Chip. 2016;21:4230–4236. doi: 10.1039/c6lc00945j. [DOI] [PubMed] [Google Scholar]
- Phillips E., Moehling T., Ejendal K., Hoilett O., Byers K., Basing A., Jankowski L., Bennett J., Lin L., Stanciu L., Linnes J. Microfluidic rapid and autonomous analytical device (microRAAD) to detect HIV from whole blood samples. Lab Chip. 2019;19:3375–3386. doi: 10.1039/c9lc00506d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C., Stemple D., Armes N. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole C.B., Li Z., Alhassan A., Guelig D., Diesburg S., Tanner A., Zhang Y., Evans T.C., Labarre P., Wanji S., Burton A., Carlow C.K.S. Colorimetric tests for diagnosis of filarial infection and vector surveillance using non- instrumented nucleic acid loop-mediated isothermal amplification (NINA-LAMP) PloS One. 2017;12 doi: 10.1371/journal.pone.0169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priye A., Bird S.W., Light Y.K., Ball C.S., Negrete O.A., Meagher R.J. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 2017;7:44778. doi: 10.1038/srep44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Hu Q., Kang Q., Bi Y., Jiang Y., Yu L. Detection of biomarkers in blood using liquid crystals assisted with aptamer-target recognition triggered in situ rolling circle amplification on magnetic beads. Anal. Chem. 2019;91:11653–11660. doi: 10.1021/acs.analchem.9b02186. [DOI] [PubMed] [Google Scholar]
- Qin Z., Chan W.C.W., Boulware D.R., Akkin T., Butler E.K., Bischof J.C. Significantly improved analytical sensitivity of lateral flow immunoassays by using thermal contrast. Angew. Chem. Int. Ed. 2012;51:4358–4361. doi: 10.1002/anie.201200997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed A.J., Connelly R.P., Williams A., Tran M., Shim B., Choe H., Yulia V. Label-free pathogen detection by a deoxyribozyme cascade with visual signal readout. Sensor. Actuator. B Chem. 2019;282:945–951. doi: 10.1016/j.snb.2018.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholt S., Behrent A., Greene C., Kalfe A., Baeumner A.J. Isolation and amplification of mRNA within a simple microfluidic lab on a chip. Anal. Chem. 2014;86:849–856. doi: 10.1021/ac403417z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N., Linnes J., Fan A., Ellenson C., Nira R., Klapperich C. Paper-based RNA extraction, in situ isothermal amplification, and lateral flow detection for low-cost, rapid diagnosis of Influenza A (H1N1) from clinical specimens. Anal. Chem. 2015;87:7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N., Wong W., Liu L., Dewar R., Klapperich C. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip. 2016;16:753–763. doi: 10.1039/c5lc01392e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roembke B.T., Nakayama S., Sintim H.O. Nucleic acid detection using G-quadruplex amplification methodologies. Methods. 2013;64:185–198. doi: 10.1016/j.ymeth.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrman B., Richards-Kortum R. A paper and plastic device for performing recombinase polymerase amplification of HIV DNA. Lab Chip. 2012;12:3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrman B., Richards-Kortum R. Inhibition of recombinase polymerase amplification by background DNA: a lateral flow-based method for enriching target DNA. Anal. Chem. 2015;87:1963–1967. doi: 10.1021/ac504365v. [DOI] [PubMed] [Google Scholar]
- Rohrman B.A., Leautaud V., Molyneux E., Richards-Kortum R.R. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PloS One. 2012;7 doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldarriaga O., Castellanos-Gonzalez A., Porrozzi R., Baldeviano G., Lescano A., de Los Santos M., Fernandez O., Saravia N., Costa E., Melby P., Travi B.L. An innovative field-applicable molecular test to diagnose cutaneous Leishmania Viannia spp. infections. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithgall M.C., Scherberkova I., Whittier S., Green D.A. Comparison of cepheid xpert xpress and Abbott ID now to roche cobas for the rapid detection of SARS-CoV-2. J. Clin. Virol. 2020;128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Liu C., Mauk M., Rankin S., Lok J., Greenberg R., Bau H. Two-stage isothermal enzymatic amplification for concurrent multiplex molecular detection. Clin. Chem. 2017;63:714–722. doi: 10.1373/clinchem.2016.263665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld R.D., Liu Y.-C., Stedtfeld T.M., Kostic T., Kronlein M., Srivannavit O., Khalife W.T., Tiedje J.M., Gulari E., Hughes M., Etchebarne B., Hashsham S.A. Static self-directed sample dispensing into a series of reaction wells on a microfluidic card for parallel genetic detection of microbial pathogens. Biomed. Microdevices. 2015;17:89. doi: 10.1007/s10544-015-9994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Xing W., Yu X., Fu W., Wang Y., Zou M., Luo Z., Xu D. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of Schistosoma japonicum. Parasites Vectors. 2016;9:476. doi: 10.1186/s13071-016-1745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirschberg D., Wheeler M., Bashir R., Nash D., Cunningham B. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip. 2020;20:1621–1627. doi: 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Tan X., Dy A., Braff D., Akana R., Furuta Y., Ananthakrishnan A., Collins J. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 2018;9:3347. doi: 10.1038/s41467-018-05864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E., Erwin B., Dames S., Ferguson T., Buechel M., Irvine B., Voelkerding K., Niemz A. Specific versus nonspecific isothermal DNA amplification through thermophilic polymerase and nicking enzyme activities. Biochem. 2008;47:9987–9999. doi: 10.1021/bi800746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Chow W., Li Y., Kong H., Tang Y., Lemieux B. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low‐resource settings. J. Infect. Dis. 2010;201:S46–S51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Liu Y., Ali M.M., Kang D.K., Zhao W., Li J. Colorimetric and ultrasensitive bioassay based on a dual-amplification system using aptamer and DNAzyme. Anal. Chem. 2012;84:4711–4717. doi: 10.1021/ac203274k. [DOI] [PubMed] [Google Scholar]
- Tang Y., Yu X., Chen H., Diao Y. An immunoassay-based reverse-transcription loop-mediated isothermal amplification assay for the rapid detection of avian influenza H5N1 virus viremia. Biosens. Bioelectron. 2016;86:255–261. doi: 10.1016/j.bios.2016.06.063. [DOI] [PubMed] [Google Scholar]
- Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58 doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- Teng J., Ye Y., Yao L., Yan C., Cheng K., Xue F., Pan D., Li B., Chen W. Rolling circle amplification based amperometric aptamer/immuno hybrid biosensor for ultrasensitive detection of Vibrio parahaemolyticus. Microchim. Acta. 2017;184:3477–3485. [Google Scholar]
- Toley B.J., Covelli I., Belousov Y., Ramachandran S., Kline E., Scarr N., Vermeulen N., Mahoney W., Lutz B.R., Yager P. Isothermal strand displacement amplification (iSDA): a rapid and sensitive method of nucleic acid amplification for point-of-care diagnosis. Analyst. 2015;140 doi: 10.1039/c5an01632k. 7540‐7549. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Tong Y., Mccarthy K., Kong H., Lemieux B. Development and comparison of a rapid isothermal nucleic acid amplification test for typing of herpes simplex virus types 1 and 2 on a portable fluorescence detector. J. Mol. Diagn. 2012;14:569–576. doi: 10.1016/j.jmoldx.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröger V., Niemann K., Gärtig C., Kuhlmeier D. Isothermal amplification and quantification of nucleic acids and its use in microsystems. J. Nanomed. Nanotechnol. 2015;6:1. [Google Scholar]
- Tsaloglou M., Nemiroski A., Camci-Unal G., Christodouleas D., Murray L., Connelly J., Whitesides G. Handheld isothermal amplification and electrochemical detection of DNA in resource-limited settings. Anal. Biochem. 2018;543:116–121. doi: 10.1016/j.ab.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Van Ness J., Van Ness L., Galas D. Isothermal reactions for the amplification of nucleotides. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Xu Y., Kong H. Helicase‐dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G.T., Little M.C., Nadeau J.G., Shank D.D. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc. Natl. Acad. Sci. Unit. States Am. 1992;89:392‐396. doi: 10.1073/pnas.89.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G.T., Fraiser M.S., Schram J.L., Little M.C., Nadeau J.G., Malinowski D.P. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691‐1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Potter S.J., Lin Y., Cunningham A.L., Dwyer D.E., Su Y., Ma X., Hou Y., Saksena N.K. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J. Clin. Microbiol. 2005;43:2339–2344. doi: 10.1128/JCM.43.5.2339-2344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang J., Geng Y., Yuan W. A recombinase polymerase amplification-based assay for rapid detection of African swine fever virus. Can. J. Vet. Res. 2017;81:308–312. [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang F., Wang L., Qian W., Qian C., Wu J., Ying Y. Instant, visual, and instrument-free method for on-site screening of GTS 40-3-2 soybean based on body-heat triggered recombinase polymerase amplification. Anal. Chem. 2017;89:4413–4418. doi: 10.1021/acs.analchem.7b00964. [DOI] [PubMed] [Google Scholar]
- Xu W., Xie X., Li D., Yang Z., Li T., Liu X. Ultrasensitive colorimetric DNA detection using a combination of rolling circle amplification and nicking endonuclease-assisted nanoparticle amplification (NEANA) Small. 2012;8:1846–1850. doi: 10.1002/smll.201200263. [DOI] [PubMed] [Google Scholar]
- Yang Z., Mclendon C., Hutter D., Bradley K.M., Hoshika S., Frye C.B., Benner S.A. Helicase-dependent isothermal amplification of DNA and RNA by using self-avoiding molecular recognition systems. Chembiochem. 2015;16:1365–1370. doi: 10.1002/cbic.201500135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaren O., Alto B.W., Gangodkar P.V., Ranade S.R., Patil K.N., Bradley K.M., Yang Z., Phadke N., Benner S.A. Point of sampling detection of Zika virus within a multiplexed kit capable of detecting dengue and chikungunya. BMC Infect. Dis. 2017;17:293. doi: 10.1186/s12879-017-2382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Fu C., Hu L., Thakur R., Feng J., Lee L. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K., Pandian V., Kadimisetty K., Ruiz C., Cooper K., You J., Liu C. Synergistically enhanced colorimetric molecular detection using smart cup: a case for instrument-free HPV-associated cancer screening. Theranostics. 2019;9:2637–2645. doi: 10.7150/thno.32224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S., Illescas P., Nicasio J., Sickler J.J. Diagnostic of real-time PCR cobas Liat Influenza A/B assay and the Alere i Influenza A&B NEAR isothermal nucleic acid amplification assay for the detection of influenza using adult nasopharyngeal specimens. J. Clin. Virol. 2017;94:86–90. doi: 10.1016/j.jcv.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Yrad F.M., Castañares J.M., Alocilja E.C. Visual detection of Dengue-1 RNA using gold nanoparticle-based lateral flow biosensor. Diagnostics. 2019;9:74. doi: 10.3390/diagnostics9030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Park S., Liu K., Tsuan J., Yang S., Wang T. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip. 2011;11:398–406. doi: 10.1039/c0lc00296h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.