Abstract
Rationale & Objective:
Biomarkers that provide reliable evidence of future diabetic kidney disease (DKD) are needed to improve disease management. In a cross-sectional study, we previously identified thirteen urine metabolites that were reduced in DKD compared with healthy controls. Here, we evaluated associations of these thirteen metabolites with future DKD progression.
Study Design:
Prospective cohort.
Setting & Participants:
1001 Chronic Renal Insufficiency Cohort (CRIC) participants with diabetes with estimated glomerular filtration rate (eGFR) between 20 and 70 ml/min/1.73m2 were followed prospectively for a median of 8 (range: 2–10) years.
Predictors:
Thirteen urine metabolites, age, race, sex, smoked >100 cigarettes in lifetime, body mass index, hemoglobin A1c, blood pressure, urine albumin, and eGFR.
Outcomes:
Annual eGFR slope and time to incident kidney failure with replacement therapy (KFRT; ie, initiation of dialysis or receipt of transplant).
Analytical Approach:
Several clinical-metabolite models were developed for eGFR slope as the outcome via stepwise selection and penalized regression, and further tested on the time-to-KFRT outcome. A best cross-validated (final) prognostic model was selected based on high prediction accuracy for eGFR slope and high concordance statistic for incident KFRT.
Results:
During follow-up, mean eGFR slope was −1.83 +/−1.92 (SD) ml/min/1.73m2 per year; 359 (36%) subjects experienced KFRT. Median time-to-KFRT was 7.45 years from the time of entry to the CRIC Study. In our final model, after adjusting for clinical variables, metabolites 3-hydroxyisobutyrate (3-HIBA) and 3-methylcrotonyglycine had a significant negative association with eGFR slope, whereas citric and aconitic acid were positively associated. Further, 3-HIBA and aconitic acid were associated with higher and lower risk of KFRT, respectively (HRs of 2.34 [95% CI, 1.51–3.62] and 0.70 [95% CI, 0.51–0.95]).
Limitations:
Subgroups for whom metabolite signatures may not be optimal, non-targeted metabolomics by flow-injection analysis and two-stage modeling approaches.
Conclusions:
Urine metabolites may offer insights into DKD progression. If replicated in future studies, aconitic acid and 3-HIBA could identify individuals with diabetes at high risk of GFR decline, potentially leading to improved clinical care and targeted therapies.
INDEX WORDS: Biomarker, chronic kidney disease (CKD), Chronic Renal Insufficiency Cohort (CRIC), diabetes, end-stage renal disease (ESRD), estimated glomerular filtration rate (eGFR), incident kidney failure, kidney disease progression, kidney function decline, longitudinal study, metabolomics, multivariate model, prediction, prognosis, risk factor
Introduction
Diabetes accounts for nearly 40% of chronic kidney disease (CKD) cases in the United States general adult population for the past two decades, with reportedly > 90% of diabetic cases being type 2 diabetes1–3. Patients with diabetic kidney disease (DKD) are at high risk for hospitalization, morbidity, and mortality4. While clinical markers such as albuminuria level are prognostic for kidney disease progression, these markers exhibit high intraindividual variability5 and patients may exhibit nonalbuminuric kidney disease with progressive glomerular filtration rate (GFR) decline even preceding the onset of albuminuria6. Hence, there is an urgent need for identifying new biomarkers that are prognostic for future DKD and/or provide biological insights that could inform clinical care.
The metabolome provides a direct and sensitive measure of the phenotype at the molecular level, which makes it a powerful tool for informing physiological and pathological effects of chronic diseases7. Urine metabolomics offers direct insights into biochemical pathways potentially associated with kidney dysfunction since the kidney is responsible for concentrating and excreting a variety of metabolites from the human body. Recent systematic reviews noted several candidate urine metabolites that discriminated DKD from controls8–12, but few studies have prospectively examined urine metabolomics for DKD progression. In one prospective study of 90 patients with type 2 diabetes13, tyrosine, hexose, and glutamine were associated with progression from moderate to severe albuminuria, but none were associated with progression from normal to moderate albuminuria. We previously identified thirteen metabolites that were consistently and reliably reduced in patients with DKD compared with healthy controls in cross-sectional analysis14. Given the cross-sectional design, we could not infer whether metabolomic differences were precursors to or a consequence of DKD. Hence, in the current study, we evaluated associations of these previously identified thirteen metabolites with future kidney function decline. Our objective was to develop multivariate models of DKD progression, to identify which among the thirteen metabolites were associated with GFR decline, after adjusting for known clinical variables, with the ultimate goal of improving risk prediction and offering insights into the pathophysiology of DKD.
Methods
Study Cohort
Our study cohort is a subsample of the Chronic Renal Insufficiency Cohort (CRIC) Study. Details on the rationale and design of the CRIC Study have been previously published15,16,17. Briefly, the CRIC Study recruited a racially and ethnically diverse group of adults aged 21 to 74 years with a broad spectrum of kidney disease severity, half of whom had diagnosed diabetes. Participants underwent extensive clinical evaluation at baseline and at annual intervals. Sociodemographic information, medical and family history, medications used in the previous 30 days, anthropometric parameters (weight, height), resting blood pressure, and heart rate were collected from CRIC participants. In addition, blood specimens and 24-h urine samples were also obtained. Our analytic cohort comprised 1001 enrolled CRIC participants with diabetes, sampled across albuminuria and eGFR categories essentially equivalent to CKD A1–A3 and CKD G2–G4. Specifically, these categories were urinary albumin levels of <30, 30-<300, and >=300 mg/d, and eGFR categories of 60-<70, 45-<60, 30-<45, and 20-<30 mL/min/1.73 m2. Participants were followed prospectively for a median of 8 (range: 2–10) years.
Individual-level informed consent was obtained on all CRIC participants. The protocol/methods were approved by each local center’s Institutional Review Board (IRB) as well as the Scientific and Data Coordinating Center (SDCC; IRB approval number, 807882).
Non-targeted Metabolomics
Non-targeted metabolome profiling in urine was performed for 1001 samples collected at study entry. Aliquots of urine samples stored at −80 °C and limited to less than 3 freeze-thaw cycles were used in this study. Samples were thawed at room temperature, centrifuged for 5 min at 5000g and precipitate-free supernatants were diluted 1:50 in double-distilled H2O and stored at −80°C in 96-well polypropylene storage microplates (Abgene) sealed with easy-peel heat sealing foil (Abgene) until measurement. Quantification of relative ion abundance was carried out with an MPS3xt autosampler (Gerstel) coupled to an Agilent 6550 Q-TOF mass spectrometer (Agilent Technologies) by non-targeted flow injection analysis as described previously18. Briefly, the flow rate was 150 μL/min of mobile phase consisting of isopropanol and water (with a volume ratio of 60:40) buffered with 5 mM ammonium fluoride and for online mass axis correction, homo-taurine and hexakis (1H, 1H, 3H-tetrafluoropropoxy) phosphazine (Agilent Technologies) were added to the mobile phase. Profile mass spectra were recorded in 4Ghz acquisition mode from 50 to 1000 m/z in negative ionization mode with the following source settings: temperature 225° C, drying gas 11 l/min, nebulizer 20 psig, sheath gas temperature 350° C, sheath gas flow 10 l/min, Vcap 3500 V, nozzle 2000 V, fragmentor 350 V and Oct 1 RF Vpp 750V. All steps of data processing and analysis were performed with Matlab R2017b (The Mathworks) using functions embedded in the bioinformatics, statistics, database and parallel computing toolboxes. After sample alignment, correction for ion intensity drift over time and between plates was performed and the common mass axis was recalibrated using known frequently occurring ions. Raw mass spectrometry data was normalized based on creatinine ion abundances. Final annotation of approximately 15k ions common to all datasets were done based on accurate mass comparison using 1 mDa mass tolerance against Human Metabolome Database HMDBv4.0 assuming single deprotonation.
The relative, creatinine normalized ion abundances for the thirteen metabolites of interest were extracted and used for the current analysis. The metabolites (and related pathways) were: aconitic acid, citric acid (nucleotide metabolism: tricarboxylic acid [TCA] cycle); uracil (nucleotide metabolism: purine); 3-hydroxyisobutyrate, 2-methylacetoacetate, 3-hydroxyisovalerate, 2-ethyl-3-hydroxypropionate, 3-methylcrotonyglycine, tiglyglycine (amino acid metabolism: valine, leucine, and isoleucine); homovanillic acid (amino acid metabolism: phenylalanine and tyrosine); glycolic acid, 3-methyladipic acid, and 3-hydroxypropionate. A single ion could annotate multiple metabolites, resulting in ambiguities in the assignments. In our analysis, the following four pairs were indistinguishable: 3-hydroxypropionate or lactic acid, 3-hydroxyisobutyrate or 2-hydroxybutyrate, 2-ethyl-3-hydroxypropionate or 3-hydroxyisovalerate and 3-methylcrotonyglycine or tiglyglycine. Lactic acid and 2-hydroxybutyrate were not part of the thirteen urine metabolite set14, and, to simplify notation in subsequent sections, we will generally only refer to a single feature in each pair according to its membership in the 13-metabolite set.
Disease Outcomes: eGFR Slope and Time to Incident Kidney Failure With Replacement Therapy
We evaluated two outcomes: the annual rate of eGFR change (eGFR slope) ml/min/1.73m2 per year, and time to incident kidney failure with replacement therapy (KFRT) from entry into the CRIC Study, with drop-out or death prior to KFRT as censoring events. We used the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) creatinine equation19 to calculate eGFR. For sensitivity analyses, we replicated our analysis using the CRIC eGFR equation20, which was developed specifically for the CRIC Study, and compared results to those obtained using the CKD-EPI equation.
In order to calculate eGFR slope for our cohort, we fit a linear mixed-effects model21 using repeated GFR estimates over time with a median of 7 (range, 1–11; IQR, 4–9) repeats per participant. The model consisted of a fixed effect for time (i.e. year of follow-up) and random intercept and slope terms. Based on previous findings,22,23 we included race as a covariate in the linear mixed-effects model. Model-based estimates of individual-level slopes, namely the best linear unbiased predictor slopes, were derived and constituted the outcome for developing prognostic models for annual rate of eGFR decline.
We evaluated Pearson correlations between the thirteen (log-transformed) metabolites and eGFR slope and examined hazard ratios (HR), unadjusted and adjusted for clinical variables, of each of the thirteen metabolites for KFRT. The clinical variables are baseline data of age, race, sex, smoked >100 cigarettes in lifetime, BMI, HbA1c, mean arterial pressure, urine albumin, and eGFR. We used a 2-sided 5% significance level for hypothesis testing.
Multivariate Model Development and Validation Strategy
Linear Models
Several types of prognostic models for DKD progression, using the eGFR slope outcome, were developed: clinical variables only (C), clinical variables + metabolites (CM), and metabolites only (M). The first set of models (C or CM) included all the clinical variables, and then performed variable selection on the thirteen metabolites. Next, to evaluate the prognostic value of metabolites alone (M), we fit a second set of models that excluded clinical variables, and only selected from among the thirteen metabolites. Stepwise selection under Akaike Information Criteria (AIC,-S) and penalized regression under the L1-penalty, i.e. LASSO (-L)24, were used for metabolite selection. Models that included all metabolites (-A) was also fitted for comparison. Accurate model(s) to predict eGFR slope were identified as having low 5-fold cross-validated mean square error (MSE).
Cox Models
The set of clinical variables and selected metabolites from the most accurate (i.e., lowest MSE) eGFR-slope model(s) were identified and used as predictors (without further model selection) of KFRT as the right-censored outcome in Cox regression models. A best cross-validated (final) prognostic model was selected based on high prediction accuracy reflected by high concordance (Harrell’s C-statistic) for incident KFRT. We conducted F- and likelihood ratio tests for assessing the goodness-of-fit of the final model compared to the clinical variables-only model. These statistical tests are the most powerful when comparing nested model25. In the incident KFRT analysis, no further tuning or variable selection was conducted. Thus the overfitting problem associated with variable selection would be mitigated and coefficient estimates for the incident KFRT models should have minimal bias.
Validation Strategy
We conducted one-hundred repeats of 5-fold cross-validation for MSE and the C-statistic to correct for potential overfitting, and provide optimism-corrected estimates of model performance on future samples.
To aid in interpretation of the final multivariate model(s), we calculated Pearson, point biserial and polyserial correlations between metabolites and clinical variables.
Statistical Software
All statistical analysis was conducted using the R (version 3.6.1) programming environment26.
Results
Baseline Characteristics and Disease Outcomes
Baseline characteristics (Table 1) indicate that at study entry, subjects (N=1001) had a mean age of 59.89 +/− 9.44 (SD) years, 44% were white, 44% were female and 57% smoked > 100 cigarettes in lifetime. Furthermore, subjects had mean BMI 34.19 +/−7.93 kg/m2, HbA1c 7.57% +/− 1.54% and eGFR 40.59 +/− 11.19 ml/min/1.73m2. A large proportion had moderate or severe albuminuria (69%) and hypertension (93%). The eGFR slope outcome, derived from the mixed model analysis, showed notable variation [Figure 1] with mean of −1.83 +/1− .92 ml/min/1.73m2 per year. During the 10 years of follow-up, with median 7.45 (IQR, 4.46–9.13) years to KFRT event, there were 359 cases of incident KFRT observed (36%); 159 deaths prior to KFRT (16%), 78 of which were attributed to cardiovascular disease (CVD); and 28 (3%) drop-outs/lost-to-follow-up.
Table 1:
Baseline clinical and metabolomica characteristics of 1001 CRIC Study participants with diabetes.
| Characteristic | Value |
|---|---|
| Age (years) | 59.89 ± 9.44 |
| Other | 136 (14) |
| Male Sex | 564 (56) |
| Smoked >100 cigarettes | 568 (57) |
| BMI (kg/m2) | 34.19 ± 7.93 |
| HbAlc (%) | 7.57 ± 1.54 |
| Diastolic BP (mmHg) | 68.78 ± 12.27 |
| Systolic BP (mmHg) | 132.23 ± 21.42 |
| Urine Creatinine (mg/dL) | 66.21 ± 32.13 |
| ≥300 mg/d | 421 (42) |
| Baseline eGFR (ml/min/1.732) | 40.59 ± 11.19 |
| Hypertension | 926 (93) |
| ACE Inhibitor or ARB use** | 804 (80) |
| Aconitic acid | 13.16 ± 0.44 |
| Homovanillic acid | 12.29 ± 0.6 |
| Citric acid | 14.8 ± 0.79 |
| 3-hydroxyisobutyrate* | 10.51 ± 0.31 |
| Uracil | 10.93 ± 0.56 |
| 2-methylacetoacetate | 9.87 ± 0.31 |
| 2-ethyl-3-hydroxypropionate* | 10.19 ± 0.38 |
| Glycolic acid | 9.72 ± 0.3 |
| 3-methylcrotonyglycine* | 10.54 ± 0.37 |
| 3-methyladipic acid | 11.28 ± 0.36 |
Values are expressed as mean ± SD or count (%). BMI, body mass index; HbA1c, hemoglobin A1c; BP, blood pressure; eGFR, estimated glomerular filtration rate; ACE, angiotensin-converting enzyme; ARB, angiotensin-receptor blocker; CRIC, Chronic Renal Insufficiency Cohort.
Metabolite ion abundances were creatinine normalized and natural log transformed.
Single ion can correspond to multiple metabolites, which resulted in some ambiguities for identifying metabolites.
information missing for 3 participants
Figure 1:
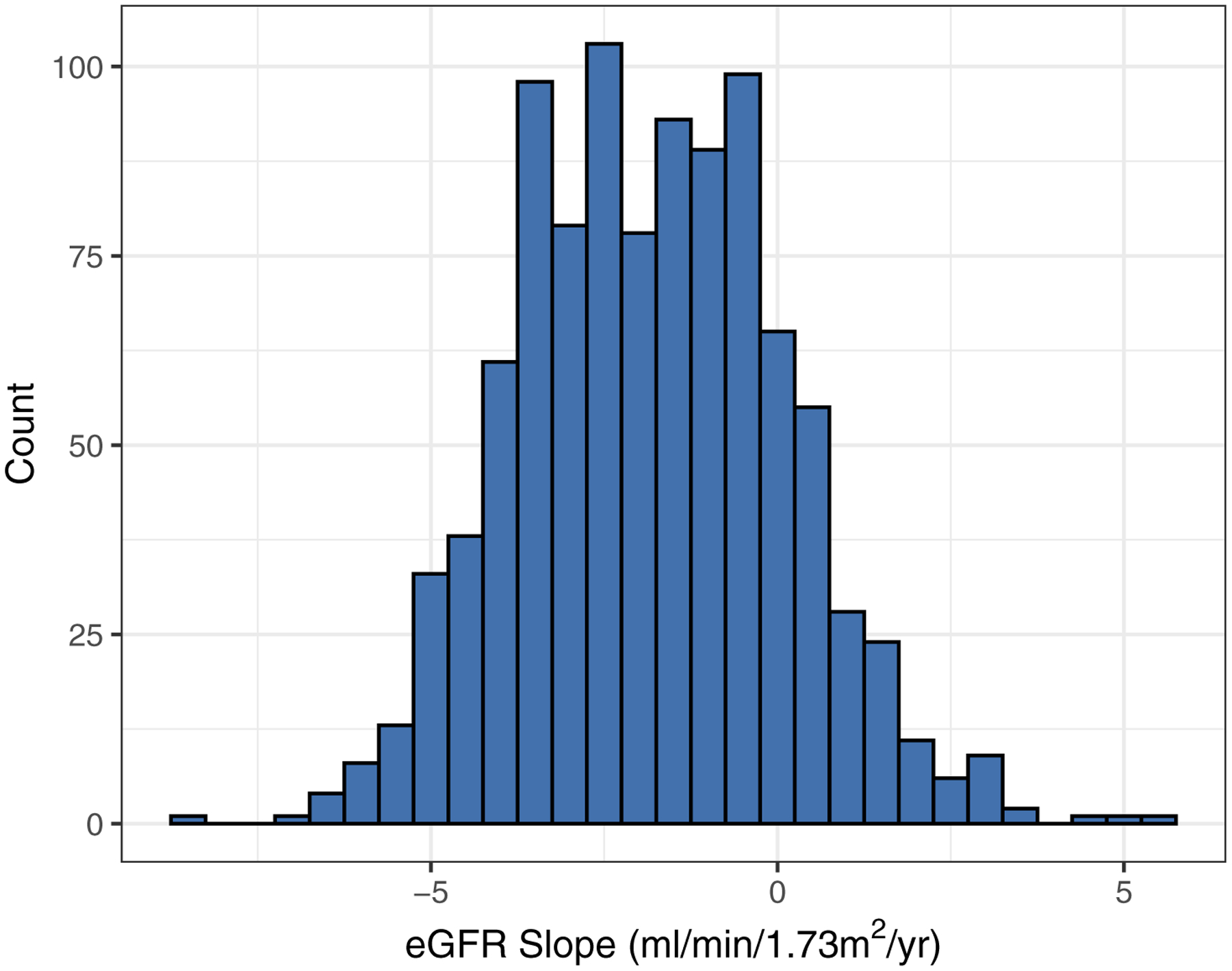
Histogram of predicted eGFR slopes (N=1001). The eGFR slopes were estimated via linear mixed effects model with a mean of −1.83 +/− 1.92 (SD) ml/min/1.73m2 per year.
Metabolite Univariate Association with Kidney Disease Progression
Seven metabolites were significantly correlated with eGFR slope, five with risk of incident KFRT unadjusted for clinical variables and six when adjusted for clinical variables (p < 0.05) in the single-metabolite univariate analysis [Table 2]. Of these, 3-hydroxypropionate and 3-hydroxyisobutyrate were significant risk factors for eGFR decline in all single-metabolite analyses. Aconitic acid, citric acid, and uracil had significant positive correlations with eGFR slope and unadjusted HRs for incident KFRT < 1, indicating that these features associate with renal protection when not controlling for clinical variables. Associations of these three metabolites with incident KFRT were attenuated after adjusting for clinical variables; however, in contrast, 2-methylacetoacetate, glycolic acid, 3-methylcrotonylglycine, and 3-methyladipic acid had significant adjusted associations with incident KFRT with HRs > 1. Clinical variable-metabolite (Figure S1) correlations shed some light on these discrepancies: e.g., aconitic acid, citric acid, and uracil were positively associated with baseline eGFR (Pearson correlation > 0.2), so that inclusion of eGFR in the models attenuated the associations of these metabolites with outcomes. Similarly, 2-methylacetoacetate, glycolic acid, 3-methylcrotonylglycine, and 3-methyladipic acid are all associated with race (point biserial correlation > 0.15), suggesting confounding by race in the unadjusted models.
Table 2:
Associations of baseline urinary metabolite excretion with eGFR slope and time to incident KFRT, and multivariate model selection.
| Metabolite Iona | Pearson Corr. w/ eGFR slope (95% CI); p | Unadj HR for incident KFRT (95% CI); p | Adj HR for incident KFRTb (95% CI); p | Multivariate Model Selectionc | |||
|---|---|---|---|---|---|---|---|
| CM-L | CM-S | M-L | M-S | ||||
| 3-hydroxypropionate* | −0.13 (−0.19, −0.06); <0.001+ |
1.34 (1.14, 1.58); <0.001+ |
1.35 (1.12, 1.62); 0.001+ |
✘ | ✘ | ✓ | ✓ |
| Aconitic acid | 0.13 (0.07, 0.19); <0.001+ |
0.52 (0.41, 0.65); <0.001+ |
0.85 (0.64, 1.13); 0.3 |
✓ | ✓ | ✓ | ✓ |
| Homovanillic acid | 0.00 (−0.06, 0.06); 0.9 |
1.12 (0.95, 1.33); 0.2 |
1.14 (0.96, 1.36); 0.1 |
✘ | ✘ | ✓ | ✘ |
| Citric acid | 0.12 (0.06, 0.18); <0.001+ |
0.69 (0.61, 0.78); <0.001+ |
0.96 (0.84, 1.11); 0.6 |
✓ | ✓ | ✓ | ✓ |
| 3-hydroxyisobutyrate* | −0.12 (−0.18, −0.06); <0.001+ |
1.4 (1.01, 1.95); 0.04 |
2.11 (1.43, 3.11); <0.001+ |
✓ | ✓ | ✓ | ✓ |
| Uracil | 0.08 (0.01, 0.14); 0.02 |
0.79 (0.64, 0.97); 0.02 |
1.11 (0.91, 1.34); 0.3 |
✓ | ✘ | ✓ | ✓ |
| 2-methylacetoacetate | −0.04 (−0.1, 0.02); 0.2 |
1.24 (0.88, 1.73); 0.2 |
1.6 (1.12, 2.29); 0.01 |
✘ | ✘ | ✓ | ✘ |
| 2-ethyl-3-hydroxypropionate* | −0.1 (−0.16, −0.04); 0.001+ |
1.03 (0.78, 1.36); 0.8 |
1.28 (0.97, 1.7); 0.08 |
✘ | ✘ | ✓ | ✘ |
| Glycolic acid | −0.03 (−0.09, 0.03); 0.4 |
1.21 (0.85, 1.7); 0.3 |
2.14 (1.49, 3.07); <0.001+ |
✓ | ✘ | ✓ | ✓ |
| 3-methylcrotonyglycine* | −0.06 (−0.13, 0.00); 0.04 |
1.18 (0.9, 1.55); 0.2 |
1.37 (1.03, 1.83); 0.03 |
✓ | ✓ | ✓ | ✘ |
| 3-methyladipic acid | −0.01 (−0.07, 0.05); 0.7 |
1.05 (0.8, 1.39); 0.7 |
1.51 (1.13, 2.01); 0.005 |
✓ | ✘ | ✘ | ✘ |
Metabolite ion abundances were creatinine normalized and natural log transformed.
Values adjusted for age, race, sex, smoked >100 cigarettes, BMI, HbA1c, mean arterial pressure, urine albumin, and baseline eGFR.
LASSO, least absolute shrinkage and selection operator; AIC, Akaike information criterion.
Model type:
CM-L: LASSO – Forced clinical-variables + Selection from 13 metabolites
CM-S: Stepwise AIC – Forced clinical-variables + Selection from 13 metabolites
M-L: LASSO – Selection from 13 metabolites
M-S: Stepwise AIC - Selection from 13 metabolites
KFRT: kidney failure with replacement therapy; unadj, unadjusted; adj, adjusted
Selected in model,
Not selected in model
Single ion can correspond to multiple metabolites, which resulted in some ambiguities for identifying metabolites.
Significant p-value after Bonferroni correction (p < 0.05/11).
Multivariate Models and Metabolite Selection
Metabolites selected in the multivariate analysis for the eGFR slope outcome using stepwise selection under AIC (model CM-S) and penalized regression under the L1-penalty, i.e. LASSO (CM-L) for metabolite selection are shown in Table 2. In the CM-S model, selected metabolites were aconitic acid, citric acid, 3-hydroxyisobutyrate, and 3-methylocrotonyglycine. In addition to these, uracil, glycolic acid and 3-methyladipic acid were also selected in the CM-L model. The metabolite-only models (which did not adjust for clinical variables) selected additional metabolite features as shown in Table 2. Of note, aconitic acid, citric acid, and 3-hydroxyisobutyrate were consistently selected in the stepwise and penalized approaches regardless of adjusting for clinical variables.
Model Prediction Performance
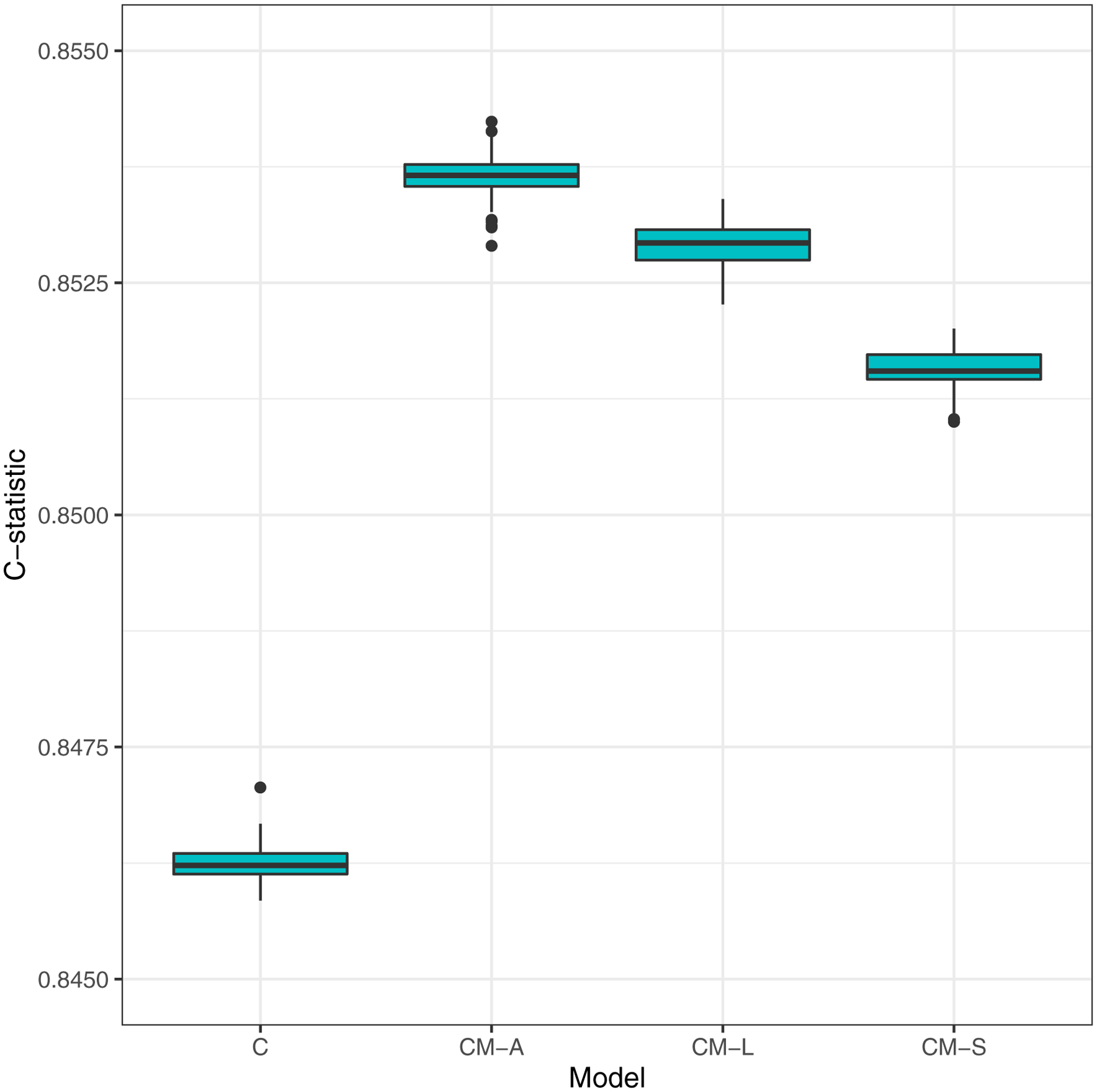
One-hundred repeats of 5-fold cross-validated MSEs for eGFR slope noticeably stratifies the prediction performance into two similarly performing groups: (1) Models with clinical variables and (2) models with only metabolite predictors [Figure 2A]. The models with clinical variables displayed greater cross-validated test accuracy, MSE range 2.15–2.26 across the four models (C, CM-A, CM-L and CM-S), than those with only metabolite predictors, MSE range 3.38–3.51 across the three models (M-A, M-L and M-S). Since all four models with clinical variables had equally competitive prediction accuracies for eGFR slope, we evaluated these in the survival setting with incident KFRT as the right-censored outcome. The comparison of one-hundred repeats of 5-fold cross-validated C-statistics for time-to-incident-KFRT revealed that all four models (C, CM-A, CM-L and CM-S) also had comparable C-statistics with value range 0.846–0.854 [Figure 2b].
Figure 2:
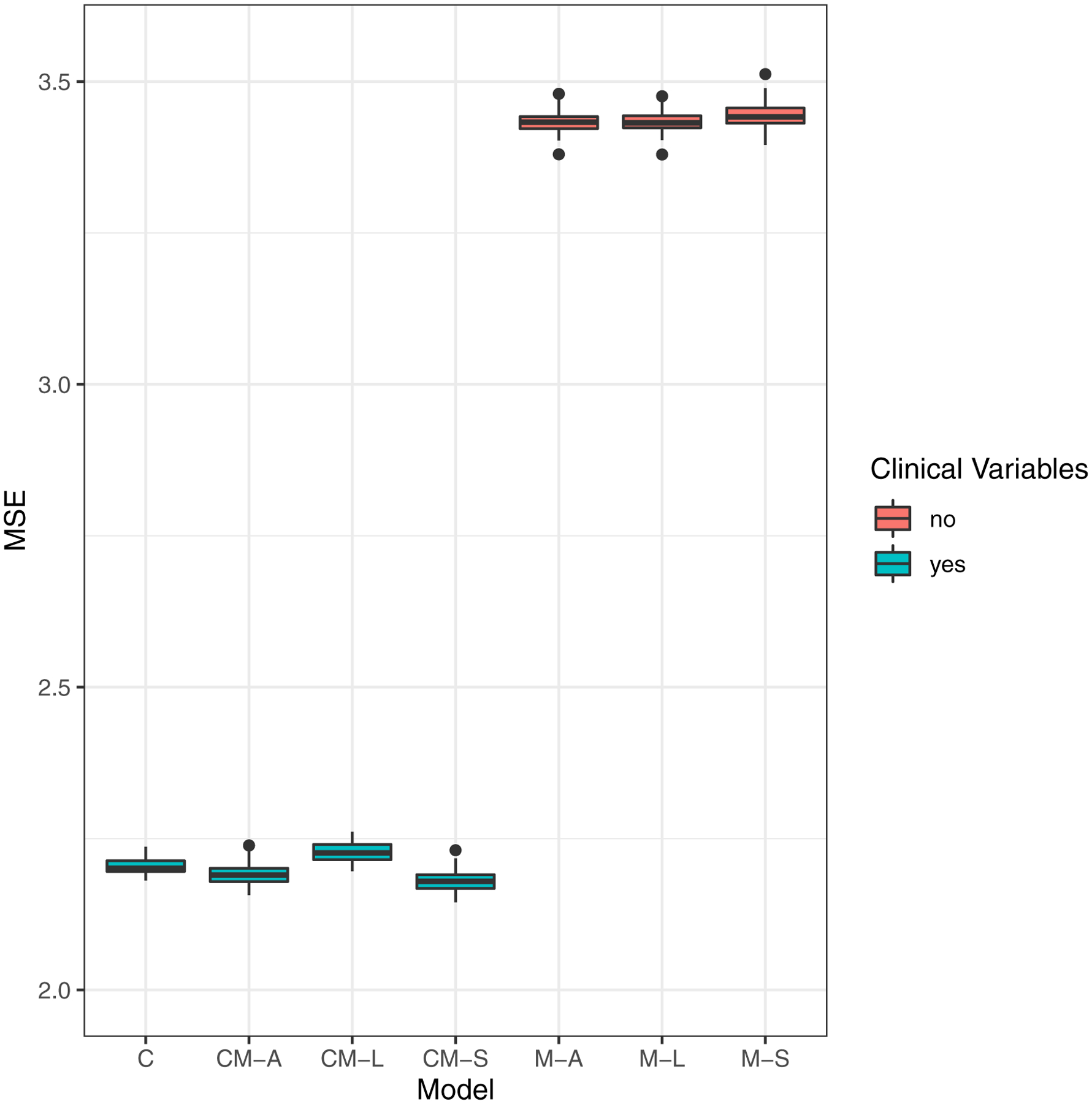
Boxplots of model prediction performance: 100 repeats of 5-fold cross-validated (a) Mean-squared error (MSE) for eGFR slopes and (b) C-statistics for time-to-incident KFRT.
Model type:
C: Clinical-variables only
CM-A: Clinical-variables + All 13 metabolites
CM-L: LASSO – Forced clinical-variables + Selection from 13 metabolites
CM-S: Stepwise AIC – Forced clinical-variables + Selection from 13 metabolites
M-A: All 13 Metabolites
M-L: LASSO – Selection from 13 metabolites
M-S: Stepwise AIC - Selection from 13 metabolites
Multivariate Metabolite Models and Association with Kidney Disease Progression
The CM-A, CM-L, and the CM-S models all displayed similar model prediction performances for the eGFR slope and incident KFRT outcomes, with CM-S having the fewest predictors, i.e., the CM-S was the most parsimonious model. Hence we selected the CM-S model as our final best prognostic model [Table 3]. The clinical variables race, mean arterial pressure, and urine albumin were significantly associated with eGFR decline and incident KFRT, controlling for all other variables. As noted previously, Blacks had worse prognosis compared to Whites, and as expected high blood pressure and moderate or severe albuminuria (compared to normal albuminuria) were associated with greater eGFR decline and higher hazard of incident KFRT.
Table 3:
Final model based on stepwise selection with annual eGFR slope outcome and time-to-incident KFRT outcome (right).
| Variable | Linear Regression Coef. for annual eGFR slope (95% CI); p | HR (95% CI) for time to incident KFRT; p |
|---|---|---|
| Age, per 1-y older | 0.00 (−0.01, 0.01); 0.5 | 0.98 (0.97, 0.99); <0.001 |
| Other | −0.46 (−0.76, −0.16); 0.003 | 1.28 (0.91, 1.81); 0.2 |
| Female sex | −0.21 (−0.43, 0.00); 0.05 | 1.02 (0.8, 1.3); 0.9 |
| Smoked >100 cigarettes | −0.02 (−0.22, 0.17); 0.8 | 0.98 (0.79, 1.23); 0.9 |
| BMI per 1-kg/m2 greater | 0.01 (0.00, 0.02); 0.2 | 0.98 (0.97, 1.00); 0.05 |
| HbAlc, per 1 percentage point greater | −0.08 (−0.14, −0.01); 0.02 | 0.99 (0.92, 1.06); 0.8 |
| MAP, per 1 mmHg greater | −0.02 (−0.02, −0.01); <0.001 | 1.02 (1.01, 1.03); <0.001 |
| ≥300 mg/d | −2.14 (−2.4, −1.87); <0.001 | 11.79 (6.85, 20.31); <0.001 |
| Baseline eGFR, per 1 ml/min/1.732 | 0.00 (−0.01, 0.01); 0.6 | 0.92 (0.91, 0.93); <0.001 |
| 3-methylcrotonyglycine* | −0.35 (−0.62, −0.08); 0.01 | 1.16 (0.84, 1.6); 0.4 |
| Citric acid | 0.16 (0.03, 0.3); 0.02 | 0.97 (0.84, 1.12); 0.6 |
| 3-hydroxyisobutyrate* | −0.57 (−0.94, −0.21); 0.002 | 2.33 (1.51, 3.62); <0.001 |
| Aconitic acid | 0.32 (0.06, 0.58); 0.01 | 0.7 (0.51, 0.95); 0.02 |
Linear regression coefficient and hazard ratio are per unit increase in (log)-metabolite abundance. MAP, Mean Arterial Pressure
Single ion can correspond to multiple metabolites, which resulted in some ambiguities for identifying metabolites.
Associations of the four selected metabolites with eGFR slope were consistent in direction between the adjusted [Table 3] and unadjusted [Table 2] models, whereby higher levels of aconitic and citric acids were associated with slower decline, whereas higher levels of 3-hydroxyisobutyrate and 3-methylocrotonyglycine acid were associated with faster decline. When evaluating incident KFRT, two of these four metabolites retained significant associations after adjusting for clinical variables: higher aconitic acid levels were associated with lower hazard and higher levels of 3-hydroxyisobutyrate with higher hazard of incident KFRT. Of note, both the F- and likelihood ratio tests revealed that the CM-S model was a better fit for the data than the C (F = 6.79, p < 0.001; χ2 = 21.16, p < 0.001), CM-L (F = 0.54, p = 0.7; χ2 = 7.37, p = 0.06), and CM-A (F = 0.42, p = 0.9; χ2 = 12.17, p = 0.1) models for both eGFR slope and time-to-incident KFRT outcomes. Thus adding these metabolites to the clinical model improved prognostication25.
Sensitivity Analysis
Using our final CM-S model, results did not change substantially when a competing risks analysis was undertaken, with (i) any death and (ii) death-due-to-CVD each defining the competing risk. The metabolite hazard ratios were similar in the competing risks and crude analyses. Also, given the similar C-statistics for the CM-S and CM-L models, we also examined the CM-L model on the incident KFRT outcome. Of the seven features selected in the CM-L model, glycolic acid (HR, 1.84 [95% CI, 1.09–3.09]; p = 0.02) and aconitic acid (HR, 0.65 [95% CI, 0.47–0.90]; p = 0.009) were significantly associated with incident KFRT, whereas in contrast, the 3-hydroxyisobutyrate association with incident KFRT was attenuated (HR, 1.57 [95% CI, 0.91–2.70]; p = 0.1) compared to that of the CM-S model (HR, 2.33 [95% CI, 1.51–3.62]; p < 0.001).
Using the CRIC equation to estimate GFR did not change our results. Therefore, we report the results with the CKD-EPI equation since it is more widely used in clinical and research settings.
Discussion
Our study extended earlier work by evaluating associations between thirteen (previously identified14) urine metabolites and future kidney disease. Multivariate analysis identified several metabolites associated with eGFR decline, after adjusting for clinical variables. Survival analysis provided further validation of these models for a clinically important outcome, time-to-incident KFRT. Cross-validated accuracy of models were comparable, suggesting that clinical variables alone can achieve high model discrimination. Nevertheless, inclusion of selected metabolites improved overall fit of the models, as evidenced by likelihood ratio and F statistics, indicating that these markers improved prognostication and might provide potentially relevant biological information over and above clinical variables.
Two of the selected metabolites, citric acid and aconitic acid, are part of the TCA pathway. Reduction of these two metabolites in the twenty-four hour urine may reflect a reduction of TCA cycle activity due to reduced mitochondrial function or content14,27. Additional studies to evaluate genes involved in citrate production and conversion to aconitate will shed light on our current findings. The other two metabolite features are annotated as (i) 3-hydroxyisobutyrate (3-HIBA) or 2-hydroxybutyrate and (ii) 3-methylcrotonylglycine or tiglylglycine. In a previous study with a small sample-size14, 3-HIBA was reduced in patients with established DKD, whereas in the present study, higher 3-HIBA was associated with progressive DKD and time-to-incident KFRT. 3-HIBA is a catabolic intermediate of branched chain amino acid (BCAA) valine and previous studies have shown that it drives vascular fatty acid transport and uptake into cardiac endothelial cells28,29. Furthermore, in endothelial cells 3-HIBA levels increase in response to pathways induced by the transcriptional coactivator PGC1a, and thus 3-HIBA may be a biomarker for PGC1a activity28. Elevated plasma levels of BCAA30 and 3-HIBA29 have been shown to be associated with obesity and incident type 2 diabetes in humans29 and rodents31. 3-HIBA has also been associated with excess angiogenesis which could be a contributing factor to progressive DKD. Notably, 3-HIBA is indistinguishable from 2-hydroxybutyrate in our assay, and the latter, a hydroxy acid involved in propanoate metabolism, has a well-known positive association with insulin resistance and ketoacidosis32–34. Finally, the two metabolite features annotated as 3-methylcrotonylglycine or tiglylglycine are metabolites of BCAA degradation and were reduced in our prior study in established DKD14. Elevation of these metabolites indicates that shunting of BCAA catabolism is a risk factor for kidney disease progression. Detailed pathway information on these selected metabolites has been previously published by our group (see the second and third figures in Sharma et al14). Additional research using independent cohorts and targeted assays are needed to clarify these associations and shed further light on biological mechanisms underlying metabolite levels and DKD progression.
Our study addresses several gaps in current DKD research. While a few prospective studies7,12,13,35,36 have evaluated urine or plasma metabolites for future kidney disease, small sample-sizes have hampered the ability to validate results. Furthermore, statistical methods for building cross-validated multivariate models of multiple urine metabolites to classify and predict DKD, have not been fully exploited37,38. Our CRIC sample of diabetic patients is one of the largest in the US, with comprehensive data on clinical factors, metabolite profiles, and extended follow-up with multiple longitudinal assessments of kidney function. We implemented rigorous statistical methods to select metabolites for eGFR decline, and these selected features were then tested for associations with incident KFRT. Notably, models were not trained on the incident KFRT outcome, so that overfitting is less likely for this outcome, and thus our hazard ratio estimates should be unbiased. We used likelihood ratio tests, the most powerful statistical approach for evaluating the prognostic value of metabolite features when added to a model with clinical variables alone. We also calculated a cross-validated C-statistic, a popular measure of model discrimination; use of cross-validation reduces overfit and provides a more accurate assessment of how well the model will perform on independent test samples. For future work, we will use independent cohorts with varying clinical risk profiles to further evaluate our findings.
We acknowledge some limitations. First, given the substantial heterogeneity of DKD progression, there may be subgroups for whom our signatures may not be optimal. A future aim is to evaluate our models on clinically distinct patient populations (e.g., by albuminuria group), and develop new signatures using an expanded set of metabolites. Second, non-targeted metabolomics by flow-injection analysis cannot distinguish between metabolites with identical sum formula and only reports relative abundance change. In future work, we will use targeted gas chromatography-mass spectrometry or liquid chromatography–tandem mass spectrometry methods to replicate these findings. Third, our “two-stage” approach to modeling kidney disease progression via eGFR slope has the advantage of being easy to interpret and analyze since standard statistical methods can be employed. However, this approach could entail an efficiency loss when there are missing data or irregular spaced repeated measures. A future aim is to develop appropriate weighting methods for unbalanced data. Finally, eGFR decline may not always be linear. However, despite the linearity assumption, eGFR slope offers ease of biological interpretation, and permits application of standard statistical modeling methods. Importantly, the CRIC Study has produced notable findings with eGFR slope as a marker for kidney disease progression39,40. Nevertheless, we aim to examine nonlinear trajectories in future work.
While our study demonstrated that the clinical-variables only model had competitively good discrimination compared to other metabolite models, we identified several metabolites that offer biological insights in DKD. We identified key pathways including TCA cycle features and amino acid metabolism, further implicating mitochondrial function as a key parameter to monitor chronic organ dysfunction, as well as features linked with angiogenesis and/or insulin resistance and ketoacidosis. If replicated in other cohorts, these metabolites and related pathways could inform therapeutic targets for DKD, and improve clinical management of DKD.
Supplementary Material
Figure S1: Heatmap correlation matrix of clinical variables vs metabolite ions.
Acknowledgements:
We thank Lisa E. Wesby, MS for her assistance as CRIC project manager in managing and facilitating the manuscript among all the authors. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650112.
Support: Funding for the CRIC Study was obtained under a cooperative agreement from National Institute of Diabetes and Digestive and Kidney Diseases (U01DK060990, U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK060980, U01DK060963, and U01DK060902). In addition, this work was supported in part by: the Perelman School of Medicine at the University of Pennsylvania Clinical and Translational Science Award NIH/NCATS UL1TR000003, Johns Hopkins University UL1 TR-000424, University of Maryland GCRC M01 RR-16500, Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, Michigan Institute for Clinical and Health Research (MICHR) UL1TR000433, University of Illinois at Chicago CTSA UL1RR029879, Tulane COBRE for Clinical and Translational Research in Cardiometabolic Diseases P20 GM109036, Kaiser Permanente NIH/NCRR UCSF-CTSI UL1 RR-024131. BK, TF, JZ, MDa, DM, KS, LN were partially supported by NIDDK 1R01DK110541-01A1. BK was also partially supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650112. JZ, MDa, DM, LN, KS were also partially supported by DP3DK094352. MDo was supported by 5R01HL141846-02. CH was supported by U01DK60902. TNK was partially supported by NIDDK R01DK101505-01A1. DSR was supported by National Institute of Health grants R01 DK073665-01A1, 1U01DK099924-01 and 1U01DK099914-01. PSR was supported by U01DK061028. SLS was supported in part by the National Heart, Lung and Blood Institute (K23HL125984 and R03HL146788). JS was supported in part by a pilot and feasibility award from the Diabetes Complications Consortium (U24DK115255). SSW was supported by U01DK085660, U01DK104308, R01DK103784, UG3DK114915. The funders of this study had no role in study design; collection, analysis, and interpretation of data; writing the report; and the decision to submit the report for publication.
Financial Disclosure: Dr de Boer is consulting for Boehringer-Ingelheim, Ironwood, George Clinical and Goldfinch Bio and has research equipment and supplies from Medtronic and Abbott. Dr Dobre had received consulting fees from Relypsa and Tricida. SLS is a consultant for Novartis and a speaker for Global Blood Therapeutics. JS has modest research support for clinical event committee activities Sanofi and Glaxo Smith Kline, and is part of the advisory board for Tricida. SSW reports personal fees from Harvard Clinical Research Institute, Cerus, Strataca, Venbio, Takeda, CVS, Janssen, Mass Medical International, GSK, grants and personal fees from Allena, personal fees from Wolters Kluwer, outside the submitted work; and Expert witness consultation for litigation related to Granuflo, Omniscan, statins, cisplatin nephrotoxicity and mercury exposure. KS was on advisory board for Janssen and served on DSMB for Sanofi in past year. The other authors declare that they have no relevant financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer: Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
References
- 1.Bailey RA, Wang Y, Zhu V, Rupnow MF. Chronic kidney disease in US adults with type 2 diabetes: An updated national estimate of prevalence based on Kidney Disease: Improving Global Outcomes (KDIGO) staging. BMC Res Notes. 2014;7(1):1–7. doi: 10.1186/1756-0500-7-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koro CE, Lee BH, Bowlin SJ. Antidiabetic medication use and prevalence of chronic kidney disease among patients with type 2 diabetes mellitus in the United States. Clin Ther. 2009;31(11):2608–2617. doi: 10.1016/j.clinthera.2009.10.020 [DOI] [PubMed] [Google Scholar]
- 3.Saran R, Robinson B, Abbott KC, et al. US Renal Data System 2018 Annual Data Report: epidemiology of kidney disease in the United States. Am J Kidney Dis. 2019;73(3) (suppl 1): A7–A8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grams ME, Yang W, Rebholz CM, et al. Risks of Adverse Events in Advanced CKD: The Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis. 2017;70(3):337–346. doi: 10.1053/j.ajkd.2017.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waikar SS, Rebholz CM, Zheng Z, et al. Biological Variability of Estimated GFR and Albuminuria in CKD. Am J Kidney Dis. 2018;72(4):538–546. doi: 10.1053/j.ajkd.2018.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krolewski AS, Niewczas MA, Skupien J, et al. Early progressive renal decline precedes the onset of microalbuminuria and its progression to macroalbuminuria. Diabetes Care. 2014;37(1):226–234. doi: 10.2337/dc13-0985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena MJ, De Zeeuw D, Mischak H, et al. Prognostic clinical and molecular biomarkers of renal disease in type 2 diabetes. Nephrol Dial Transplant. 2015;30: iv86–iv95. doi: 10.1093/ndt/gfv252 [DOI] [PubMed] [Google Scholar]
- 8.Abbiss H, Maker G, Trengove R. Metabolomics Approaches for the Diagnosis and Understanding of Kidney Diseases. Metabolites. 2019;9(2):34. doi: 10.3390/metabo9020034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colhoun HM, Marcovecchio ML. Biomarkers of diabetic kidney disease. Diabetologia. 2018;61(5):996–1011. doi: 10.1007/s00125-018-4567-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirayama A, Nakashima E, Sugimoto M, et al. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Anal Bioanal Chem. 2012;404(10):3101–3109. doi: 10.1007/s00216-012-6412-x [DOI] [PubMed] [Google Scholar]
- 11.Kalim S, Rhee EP. An overview of renal metabolomics. Kidney Int. 2017;91(1):61–69. doi: 10.1016/j.kint.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang S, Wang G. Metabolomic biomarkers in diabetic kidney diseases - A systematic review. J Diabetes Complications. 2015;29(8):1345–1351. doi: 10.1016/j.jdiacomp.2015.06.016 [DOI] [PubMed] [Google Scholar]
- 13.Pena MJ, Lambers Heerspink HJ, Hellemons ME, et al. Urine and plasma metabolites predict the development of diabetic nephropathy in individuals with Type 2 diabetes mellitus. Diabet Med. 2014;31(9):1138–1147. doi: 10.1111/dme.12447 [DOI] [PubMed] [Google Scholar]
- 14.Sharma K, Karl B, Mathew AV, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24(11):1901–1912. doi: 10.1681/ASN.2013020126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lash JP, Go AS, Appel LJ, et al. Chronic Renal Insufficiency Cohort (CRIC) Study: Baseline Characteristics and Associations with Kidney Function. Clin J Am Soc Nephrol. 2009;4(8):1302–1311. doi: 10.2215/cjn.00070109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denker M, Boyle S, Anderson AH, et al. Chronic Renal Insufficiency Cohort Study (CRIC): Overview and Summary of Selected Findings. Clin J Am Soc Nephrol. 2015;10(11):2073–2083. doi: 10.2215/cjn.04260415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman HI, Appel LJ, Chertow GM, et al. The Chronic Renal Insufficiency Cohort (CRIC) Study: Design and Methods. J Am Soc Nephrol. 2003;14(90002):148S–153. doi: 10.1097/01.asn.0000070149.78399.ce [DOI] [PubMed] [Google Scholar]
- 18.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal Chem. 2011;83(18):7074–7080. doi: 10.1021/ac201267k [DOI] [PubMed] [Google Scholar]
- 19.Levey AS, Stevens LA, Schmid CH, et al. A New Equation to Estimate Glomerular Filtration Rate. Artic Ann Intern Med. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson AH, Yang W, Hsu CY, et al. Estimating GFR among participants in the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis. 2012;60(2):250–261. doi: 10.1053/j.ajkd.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2011. doi: 10.1002/9781119513469 [DOI] [Google Scholar]
- 22.Fischer MJ, Lora CM, Ricardo AC, et al. CKD progression and mortality among hispanics and non-hispanics. J Am Soc Nephrol. 2016;27(11):3488–3497. doi: 10.1681/ASN.2015050570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu CY, Lin F, Vittinghoff E, Shlipak MG. Racial Differences in the Progression from Chronic Renal Insufficiency to End-Stage Renal Disease in the United States. J Am Soc Nephrol. 2003;14(11):2902–2907. doi: 10.1097/01.ASN.0000091586.46532.B4 [DOI] [PubMed] [Google Scholar]
- 24.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York, NY, USA: Springer New York; 2009. doi: 10.1007/978-0-387-84858-7 [DOI] [Google Scholar]
- 25.Pepe MS, Kerr KF, Longton G, Wang Z. Testing for improvement in prediction model performance. Stat Med. 2013;32(9):1467–1482. doi: 10.1002/sim.5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team. R: A Language and Environment for Statistical Computing. 2019. https://www.r-project.org/.
- 27.Hallan S, Afkarian M, Zelnick LR, et al. Metabolomics and Gene Expression Analysis Reveal Down-regulation of the Citric Acid (TCA) Cycle in Non-diabetic CKD Patients. EBioMedicine. 2017;26:68–77. doi: 10.1016/j.ebiom.2017.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang C, Oh SF, Wada S, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22(4):421–426. doi: 10.1038/nm.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mardinoglu A, Gogg S, Lotta LA, et al. Elevated Plasma Levels of 3-Hydroxyisobutyric Acid Are Associated With Incident Type 2 Diabetes. EBioMedicine. 2018; 27:151–155. doi: 10.1016/j.ebiom.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newgard CB, An J, Bain JR, et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neinast MD, Jang C, Hui S, et al. Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell Metab. 2019;29(2):417–429.e4. doi: 10.1016/j.cmet.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gall WE, Beebe K, Lawton KA, et al. A-Hydroxybutyrate Is an Early Biomarker of Insulin Resistance and Glucose Intolerance in a Nondiabetic Population. PLoS One. 2010;5(5):e10883. doi: 10.1371/journal.pone.0010883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamerling JP, Gerwig GJ, Duran M, Ketting D, Wadman SK. The absolute configuration of urinary 2-hydroxybutyric acid in patients with ketosis and lactic acidosis. Clin Chim Acta. 1978;88(1):183–188. doi: 10.1016/0009-8981(78)90168-7 [DOI] [PubMed] [Google Scholar]
- 34.Landaas S, Pettersenm. Clinical conditions associated with urinary excretion of 2-hydroxybutyric acid. Scand J Clin Lab Invest. 1975;35(3):259–266. doi: 10.1080/00365517509095738 [DOI] [PubMed] [Google Scholar]
- 35.Breit M, Weinberger KM. Metabolic biomarkers for chronic kidney disease. Arch Biochem Biophys. 2016;589:62–80. doi: 10.1016/j.abb.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 36.Niewczas MA, Sirich TL, Mathew AV., et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: Metabolomic study. Kidney Int. 2014;85(5):1214–1224. doi: 10.1038/ki.2013.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pencina MJ, Parikh CR, Kimmel PL, et al. Statistical methods for building better biomarkers of chronic kidney disease. Stat Med. 2019;38(11):1903–1917. doi: 10.1002/sim.8091 [DOI] [PubMed] [Google Scholar]
- 38.Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and diabetes: Analytical and computational approaches. Diabetes. 2015;64(3):718–732. doi: 10.2337/db14-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harhay MN, Xie D, Zhang X, et al. Cognitive Impairment in Non–Dialysis-Dependent CKD and the Transition to Dialysis: Findings From the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis. 2018;72(4):499–508. doi: 10.1053/j.ajkd.2018.02.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koye DN, Magliano DJ, Reid CM, et al. Risk of Progression of Nonalbuminuric CKD to End-Stage Kidney Disease in People With Diabetes: The CRIC (Chronic Renal Insufficiency Cohort) Study. Am J Kidney Dis. 2018;72(5):653–661. doi: 10.1053/j.ajkd.2018.02.364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Heatmap correlation matrix of clinical variables vs metabolite ions.


