Abstract
The hypothalamic-pituitary-adrenal (HPA) axis has been of interest in attempts to identify genetic vulnerability for posttraumatic stress disorder (PTSD). Although numerous HPA-axis genes have been implicated in candidate gene studies, the findings are mixed and interpretation is limited by study design and methodological inconsistencies. To address these inconsistencies in the PTSD candidate gene literature, we conducted meta-analyses of HPA-related genes from both a traditional single nucleotide polymorphism (SNP)–level analysis and a gene-level analysis, using novel methods aggregating markers in the same gene. Database searches (PubMed and PsycINFO) identified 24 unique articles examining six HPA-axis genes in PTSD; analyses were conducted on four genes (ADCYAP1R1, CRHR1, FKBP5, NR3C1) that met study eligibility criteria (original research, human subjects, main effect association study of selected genes, PTSD as an outcome, trauma-exposed control group) and had sufficient data and number of studies for use in meta-analysis, within 20 unique articles. Findings from SNP-level analyses indicated that two variants (rs9296158 in FKBP5 and rs258747 in NR3C1) were nominally associated with PTSD, ps = .001 and .001, respectively, following multiple testing correction. At the gene level, significant relations between PTSD and both NR3C1 and FKBP5 were detected and robust to sensitivity analyses. Although study limitations exist (e.g., varied outcomes, inability to test moderators), taken together, these results provide support for FKBP5 and NR3C1 in risk for PTSD. Overall, this work highlights the utility of meta-analyses in resolving discrepancies in the literature and the value of adopting gene-level approaches to investigate the etiology of PTSD.
Exposure to traumatic events is common, with approximately 70% of individuals reporting exposure to at least one traumatic event in their lifetime (Benjet et al., 2016) and 5%–31% of exposed individuals meeting criteria for posttraumatic stress disorder (PTSD; Kessler, Sonnega, Bromet, Hughes, & Nelson, 1995). As not all trauma-exposed individuals develop PTSD, researchers have sought to examine the factors that may distinguish between those who do and do not develop PTSD, including underlying potential biological vulnerability. Characterization of these biological underpinnings is expected to facilitate identification of individuals who are most at risk, with an aim to effectively intervene and reduce or prevent clinically significant posttraumatic symptomatology.
In terms of biological vulnerability, the role of the hypothalamic-pituitary-adrenal (HPA) axis has been a source of interest. Specifically, exposure to an acute stressor initiates a “fight-or-flight” response, prompting the hypothalamus to secrete corticotrophin-releasing hormone (CRH), which stimulates release of adrenocorticotrophic hormone (ACTH) from the pituitary gland, which, in turn, stimulates the subsequent release of cortisol from the adrenal glands. These hormones, particularly cortisol, then exert effects via a negative feedback loop within the HPA axis to regulate hormone release in the presence of future stressors (Munck & Guyre, 1986). Following exposure to a traumatic event, however, some individuals demonstrate more pronounced alterations in HPA-axis functioning, with these alterations consequently associated with risk for the development of PTSD (Delahanty & Nugent, 2006). Researchers have suggested that these individual differences in modulators of response to stress, in part impacted by unique genetic background, might help explain PTSD risk (Yehuda, Koenen, Galea, & Flory, 2011). Initial stress system reactivity and HPA axis dysfunction leading to “turning off” of the stress response have been proposed as key mechanisms that influence the effect of trauma exposure on the development of PTSD (Carvalho, Coimbra, Ota, Mello, & Belangero, 2017).
Twin studies have demonstrated that components of the basic stress response are moderately heritable (Holsboer, Lauer, Schreiber, & Krieg, 1995), as is PTSD (e.g., True et al., 1993); these findings were supported by a recent molecular genetic investigation (Duncan et al., 2018). The role of the HPA axis in the stress response and its relation to PTSD risk has led genetic researchers to focus on genes involved in this system. Evidence for a link between PTSD and variants within this system, such as a variant in the PAC1 receptor of pituitary adenylate cyclase-activating polypeptide (ADCYAP1R1) and variants in the steroid receptor chaperone FK506 binding protein 5 (FKBP5) has been compelling (for reviews, see Banerjee, Morrison, & Ressler, 2017; Smoller, 2016). However, as with studies of other candidate genes, some investigations have found no significant results, and the literature is mixed (Skelton, Ressler, Norrholm, Jovanovic, & Bradley-Davino, 2012).
Although candidate gene studies still represent the most commonly used approach in the identification of genetic variants that contribute to PTSD risk, this approach has several limitations. Specifically, findings often are not replicated (Smoller, 2016), which leads to conflicting results (Cornelius et al., 2010) and likely high rates of false-positive findings (Koenen, 2007). These limitations stem from small sample sizes and low power (Banerjee et al., 2017), differences in study design and methodology (Cornelius et al., 2010), flaws in interpretation, and publication practices (Sullivan, 2007). Thus, researchers are advised to use caution in interpreting candidate gene findings, as they may overestimate the true genetic effect size (for a review, see Sullivan, 2007). Despite these concerns, candidate gene studies warrant continued investigation given their dense coverage of targeted genes and ability to test mechanistic hypotheses (Koenen, 2007).
One approach to address the inconsistencies is to meta-analyze these studies (Koenen, 2007). Meta-analyses of PTSD-implicated genes have increased in recent years with the investigation of other frequently studied PTSD candidate genes: serotonin (Gressier et al., 2013), brain-derived neurotrophic factor (BDNF; Bountress et al., 2017), pituitary adenylate cyclase-activating polypeptides (PACAP) receptor (ADCYAP1R1; Lind et al., 2017), dopamine (Li et al., 2016), and gene-environment interactions of FKBP5 (Hawn et al., 2019). One challenge in meta-analyzing a broad candidate gene system is the analysis of different markers within genes, as relatively few studies examine the same variants, making interpretation of findings across studies difficult (Koenen, 2007). In his commentary, Sullivan (2007) posed the question of whether it is necessary to require precise replication of the same genetic marker, genotype, and direction of association, or if less precise definitions of replication suffice (e.g., any significant marker in the same gene). The approach of meta-analyzing numerous markers within the same gene is a useful way to summarize existing evidence and attempt to make broader conclusions with regard to systems of interest. Thus, developing an approach to examining numerous markers within the same gene may serve as a useful method in which to aggregate extant, and mixed, candidate gene information with regard to the HPA axis.
The present study aimed to perform meta-analyses at both the level of single-nucleotide polymorphisms (SNP) and of the genes they comprise. Specifically, genetic studies of PTSD that focused on any markers within the HPA axis were identified, and meta-analyses that examined variation within several genes involved in the HPA axis—ADCYAP1R1, CRHR1, FKBP5, and NR3C1—were conducted. We first performed SNP-level meta-analyses, utilizing existing methods. Next, because traditional meta-analytic approaches do not translate to a gene-level analytic approach, as different SNPs are measured in different studies for a given gene, novel methods were developed to aggregate markers in the same gene. Given decades of support for the role of the HPA axis in PTSD, we expected both SNP- and gene-level significance for at least some genes.
Method
Search Strategy
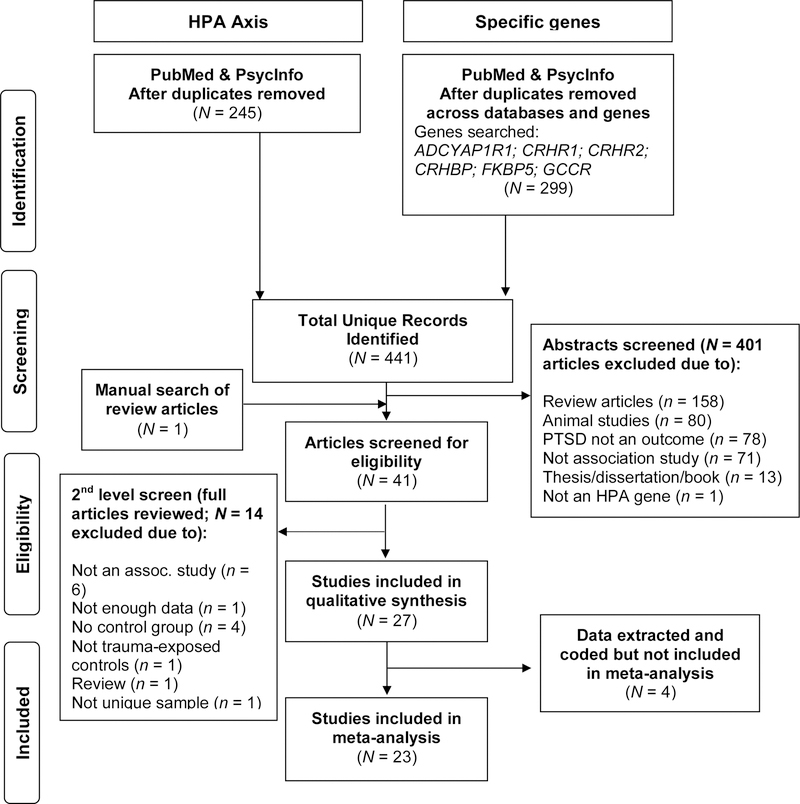
We identified existing candidate gene studies that examined the main effects of HPA axis–related genes associated with PTSD. A total of six HPA axis genes (ADCYAP1R1, CRHBP, CRHR1, CRHR2, FKBP5, and NR3C1 [the glucocorticoid receptor gene; also known as GCCR or GR]) were selected based on current reviews of genes linked to this system and associated with PTSD (Almli, Fani, Smith, & Ressler, 2014; Sheerin, Lind, Bountress, Nugent, & Amstadter, 2017; Voisey, Young, Lawford, & Morris, 2014). Following a two-step search strategy of association studies for (a) HPA axis genes broadly and (b) targeted genes more specifically, potential studies were identified through the databases PubMed, which is the primary database for biomedical and genetic studies in psychiatry, and PsycINFO, given its focus on traumatic stress and PTSD. Search terms were as follows: [posttraumatic stress disorder OR PTSD OR traumatic stress] AND [gene OR genetic] AND [HPA axis OR hypothalamic pituitary adrenal axis]. The targeted gene search included the following search terms (replacing the HPA axis terms): [ADCYAP1R1 OR PAC1 OR pituitary adenylate cyclase-activating polypeptide type 1 receptor]; [CRHBP OR corticotropin-releasing hormone binding protein OR corticotrophin-releasing hormone binding protein]; [CRHR1 OR corticotropin releasing hormone receptor 1 OR corticotrophin releasing hormone receptor 1]; [CRHR2 OR corticotropin releasing hormone receptor 2]; [FKBP5 OR FK506 binding protein 5]; [GCCR OR glucocorticoid receptor OR NR3C1 OR nuclear receptor subfamily 3 group C member 1]. An initial search that included articles published up to June 2016 was conducted; additional date-restricted searches were conducted to identify any new studies that were published between this initial search and January 11, 2017, and again on July 1, 2019. We identified 276 unique articles in the initial search and 62 and 103 unique articles, respectively, in the two date-restricted follow-up searches (see Figure 1 for details on search and screening steps). We also examined the reference sections of PTSD genetics review articles.
Figure 1.
PRISMA-style flowchart presenting the search and screening process for studies included in the meta-analysis of hypothalamic-pituitary-adrenal (HPA) axis genes and posttraumatic stress disorder (PTSD). Although some studies met multiple exclusion criteria, the number shown reflects the higher-level exclusion criterion for each excluded study. Studies for which data were extracted and coded but ultimately not included in the final analysis were omitted for various reasons (e.g., data determined to be insufficient for analyses and unable to be obtained from study authors, k too small to meta-analyze for CRHR2).
Inclusion and Exclusion Criteria
Two review authors (ML and CS) independently screened results to select studies for possible inclusion. The following inclusion criteria were applied to titles and abstracts: 1) Original research; 2) Use of human subjects; 3) Association study including one of six genes (ADCYAP1R1, CRHBP, CRHR1, CRHR2, FKBP5, NR3C1); 4) PTSD as an outcome; and 5) Trauma-exposed control group (with the exception of studies of the Grady Trauma Project which were retained given high level of trauma exposure overall). The primary outcome was effect size resulting from main effects of an HPA axis gene on PTSD (interactions were excluded). When the criteria were unclear, articles were more thoroughly examined and a consensus determination was made. Supplemental material was also reviewed for relevant data. See Figure 1 for details on screening, eligibility, and final N.
Data Extraction and Coding
The two review authors extracted the following information from each study, based on a predetermined coding manual developed and agreed upon by all authors: citation, sample size, gene and SNP examined, alleles, ancestry, gender, outcome assessed (diagnostic status or severity count), timing (current or lifetime), statistics (type of analysis, estimate, p value), recruitment type, and trauma type. The quality of the included studies was evaluated in four key areas: methodological, clinical, genetic, and statistical. Study authors were contacted to request further information when data necessary for effect-size computation were missing. Following data extraction and entry, the reviewers separately checked the information for agreement across coders. There was 94% agreement between the two reviewers for articles from the initial search. Discrepancies were resolved through discussion.
Data Harmonization Across Papers
Individual studies used different analytic approaches (e.g., regression, ANCOVA) and presented varying types of data (e.g., odds ratios, regression coefficients, raw frequency or mean data). To put these analyses on a common scale in order to meta-analyze the effect sizes, all summary results (e.g., p values) were converted to Pearson correlation coefficients as measures of effect size, which, after a multiplication by , is equivalent to a Gaussian z score. In addition, data were standardized to have the same risk alleles whenever markers were identified by authors or literature to be putative “risk markers.” In some included studies, SNPs were only reported as “nonsignificant.” As such, their sign and sometimes magnitude were unknown. In these studies, we conservatively set the z score for such SNPs to 0.
Many of the included studies also reported multiple findings, such as effect sizes for both PTSD severity and diagnosis, PTSD outcomes across the lifetime and isolated to specific time periods, or both additive and dominant models. To adhere to the assumption of independence, which refers to the assumption that each measure of effect is representative of independent studies, we prioritized studies with sufficient data to calculate effect size and implemented a protocol to handle studies with multiple effect sizes (see Supplementary Table S1). The resulting studies included in the meta-analyses nonetheless still varied with regard to outcome (i.e., diagnosis or symptom severity; current or lifetime) and modes of inheritance (i.e., some used an additive model, examining the effect of having 0, 1, or 2 risk alleles, whereas others compared a high-risk genotype status to a low-risk genotype status). The same allele was chosen as the reference allele in both additive and dominant models. Due to variations in both trait and mode of inheritance, our meta-analytic approach used random-effects modeling, an approach that is useful when traits and modes of inheritance vary, to combine summary SNP statistics among different studies.
Data Analysis
SNP-level meta-analyses.
All analyses were conducted in R (Version 3.6.1). As variables describing differences between studies were numerous (e.g., genetic model tested, diagnosis or severity outcomes, gender and ethnicity distributions), meta-analyses by SNP were performed using a random effect in the call to omni function in R, an integrated meta-analysis package for conducted correlational research synthesis. This function outputs the Gaussian (under the null hypothesis) z scores for each SNP. Due to the limited number of studies for most SNPs, moderator analyses are unlikely to be very informative and, thus, were not carried out. To address the likelihood that negative findings were never reported, the sensitivity of our findings was tested for a range of such unreported studies (i.e., from 0 to 0.6, or 60%, of the meta-analysis presented herein).
Gene-level meta-analyses.
Custom R scripts, based on formulas developed specifically to conduct these analyses, were developed for the study aims. In addition to the considerations mentioned earlier, additional steps were taken for the gene-level analyses. To conduct gene-level analyses, z scores for all SNPs from a gene were combined in a Mahalanobis-type statistic. However, to accurately estimate the covariance (i.e., correlation) matrix of the z scores used in the Mahalanobis test, two major issues needed to be overcome. First, not all SNPs were measured in all studies, resulting in decreased correlations between SNP meta-analysis statistics as compared to the scenario in which all SNPs are measured across all studies. Second, even when measured in all studies, their linkage disequilibrium (LD) structure (i.e., correlations) was different among different ethnicities.
Addressing LD between SNPs that were not measured in all studies.
In order to conduct gene-level testing, a z score for meta-analysis of each SNP was computed as a weighted sum of statistics from individual studies. Based on the assumption that z scores between different studies were independent or uncorrelated, the correlation between SNP meta-analysis statistics could be estimated from a reference panel. However, an increased false positive rate would be likely when the missing pattern of SNPs within studies was not accounted for. To prevent against such an increase in Type I errors, we computed the exact correlation between SNP meta-analysis statistics in the presence of missingness, as briefly described later and in the Supplementary Materials.
Let , , be the trait vector for the kth cohort of subjects, and , , be the matrix of genotypes for the subjects at m SNPs, and the corresponding vector of z scores of testing the association of SNPs with the trait. Let be the correlation matrix of , which, near the null hypothesis of no association between trait and genotype , is also the covariance matrix of . However, as some SNPs were not measured in all studies, let be the indicator of SNP j being measured in study k (i.e., if SNP j is measured in study k and 0 otherwise). The meta-analysis z score is of the form , where are the weights used for combining statistics from individual studies. Then, under , given that z scores between different studies are independent or uncorrelated, the correlation between and is
This formula is further described in the Supplementary Materials (Supplemental Methods, Formula 1).
LD estimation in mixed-ancestry cohorts.
The correlations for the largest observed ancestral groups (Europeans and African Americans) were computed using 1000 genome Phase I release (Version 3) as the reference panel (Abecasis et al., 2010). Subsequently, the overall covariance of the statistics in mixed-ancestry studies were computed as detailed in the Supplementary Materials (Supplemental Methods, Formula 2; Lee et al., 2015).
Results
As shown in Figure 1, 415 article abstracts were screened. The full text of 41 articles were then reviewed, and 27 met initial eligibility criteria. Following further assessment, 23 unique articles across four genes were included in the present analyses. Two genes, CRHBP and CRHR2, were excluded from meta-analyses as only one published study for each gene was found. Studies of the four remaining genes included sufficient data from unique samples for SNP and/or gene-based meta-analysis, resulting in a total of 10 samples (nine manuscripts) included in the meta-analysis for ADCYAP1R1, six samples in the meta-analysis of CRHR1, 10 samples (nine manuscripts) in the meta-analysis of FKBP5, and four samples in the meta-analysis of NR3C1 (presented in Table 1).
Table 1.
Descriptive Information on Studies Included in the Meta-Analysis
| Author (year) | Gene(s) Examined | Sample | Outcomeb | Outcome Measure | Trauma Measure | Total N | % AAd | % Female |
|---|---|---|---|---|---|---|---|---|
| Almli (2013) | ADCYAP1R1 | Outpatient Clinic | Severity | PSS | TEIc,e/CTQ | 1,163 | 100.0 | 74.0 |
| Chang (2012) | ADCYAP1R1 | Population-based | Diagnosis | Otherc | Otherc,e/CTQ | 2,528 | 0.0 | 100.0 |
| Lowe (2015) | ADCYAP1R1 | Outpatient Clinic | Severity | PSS | TEI | 1,361 | 94.1 | 100.0 |
| Ressler (2011a) | ADCYAP1R1 | Outpatient Clinic | Diagnosis | Otherc | TEI/CTQ | 798 | 91.9 | 63.0 |
| Ressler (2011) | ADCYAP1R1 | Outpatient Clinic | Diagnosis | Otherc | TEI/CTQ | 439 | 92.0 | 59.2 |
| Rothbaum (2014) | ADCYAP1R1 | Outpatient Clinic | Severity | PDS | TEI/CTQ | 34 | 83.1 | 61.5 |
| Solovieff (2014) |
ADCYAP1R1 CRHR1 GR (NR3C1) FKBP5 |
Population-based | Lifetime | Otherc | Otherc | 2,538 | 0.0 | 100.0 |
| Stevens (2014) | ADCYAP1R1 | Hospital Admission | Severity | PSS | TEI/CTQ | 49 | 100.0 | 100.0 |
| Uddin (2013) | ADCYAP1R1 | Population-based | Diagnosis | Otherc | Otherc,e | 401 | 83.1 | 100.0 |
| Wang (2013) | ADCYAP1R1 | Epidemiologic Exposure | Severity | PCL | NR | 319 | 0.0 | 70.5 |
| Amstadter (2011) | CRHR1 | Hospital Admission | Severity | UCLA PTSD | Injury Severity Scoree | 103 | 45.6 | 26.2 |
| Davydow (2014) | CRHR1 | Hospital Admission | Severity | PCL | THS | 76 | 0.0 | 43.0 |
| Dunn (2014) |
CRHR1 FKBP5 |
Epidemiologic Exposure | Severity | IES | Hurricane Exposure Severity | 205 | 100.0 | 96.1 |
| Jaksic (2019) |
CRHR1 FKBP5 |
Population-based, war-exposed areas | Severity | CAPS | CAPS | 719 | 100.0 | 32.3 |
| White (2013) | CRHR1 | Outpatient Clinics | Severity | NWS PTSD module | Hurricane Exposure Severity | 564 | 0.0 | 64.0 |
| Comasco (2015) | FKBP5 | Population-based | Severity | TSSC | JVQ | 394 | 0.0 | 45.7 |
| Fani (2014) | FKBP5 | Outpatient Clinics | Severity | PSS | TEI/CTQ | 82 | 100.0 | 100.0 |
| Kang (2019) | FKBP5 | Outpatient Clinics (Military) | Diagnosis | CAPS | CAPS | 239 | Korean | 0.0 |
| Kohrt (2015) | FKBP5 | Population-based | Severity | PCL | TEI/CTQ/SLERS | 682 | Southeast Asian | 49.1 |
| Watkins (2016a) | FKBP5 | Population-based | Lifetime Severity | PCL | THS | 1,585 | 0.0 | 7.5 |
| Watkins (2016b) | FKBP5 | Population-based | Lifetime Severity | PCL | THS | 577 | 0.0 | 8.6 |
| Young (2018) | FKBP5 | Outpatient Clinics (Military) | Diagnosis | CAPS | CAPS, combat | 266 | 13.9 | 13.9 |
| Bachman (2005) | GR (NR3C1) | Outpatient Clinics (Military) | Diagnosis | CAPS | CAPS, combat | 160 | 0.0 | 0.0 |
| Hauer (2011) | GR (NR3C1) | Hospital Admission | Severity | PTSS | NR | 126 | 0.0 | 23.8 |
| Lian (2014) | GR (NR3C1) | Outpatient Clinics | Diagnosis | Otherc | TLEQ | 1,618 | Han Chinese | 43.4 |
Note. PTSD = posttraumatic stress disorder; AA = African American; CAPS = Clinician Administered PTSD Scale; CTQ = Childhood Trauma Questionnaire; IES = Impact of Events Scale–Revised; JVQ = Juvenile Violence Questionnaire; NR = not reported; NWS = National Women’s Study PTSD Module; PCL = PTSD Checklist; PDS = Posttraumatic Stress Diagnostic Scale; PSS = Posttraumatic Symptom Scale; PTSS = Posttraumatic Stress Scale; SLERS = Stressful Life Event Rating Scale for Cross Cultural Study; TEI = Traumatic Events Inventory; THS = Trauma History Screen; TLEQ = Traumatic Life Events Questionnaire; TSCC = Trauma Symptom Checklist for Children.
Outpatient clinics include medical or nonmedical clinics and Veteran Affairs clinics. Hospital admission includes intensive care units, emergency departments, and inpatient units.
All diagnoses are current unless specified.
“Other” in the Outcome Measure column refers to unspecified or study-specific interviews as well as other diagnostic measures, all DSM-IV criteria (exception of Lian 2014, which used DSM-5); “Other” in the Trauma Measure column refers to unspecified of study-specific determinations of trauma exposure.
Remainder is European ancestry unless otherwise specified.
Denotes studies that only presented trauma- or covariate-adjusted results; in the event that a study presented both adjusted and unadjusted results, the unadjusted results were analyzed.
Quality Assessment
Information regarding quality evaluation of the studies is available in Supplementary Table S2. Briefly, all included studies clearly described recruitment processes but only half clearly described inclusion/exclusion criteria. All studies except for two identified a psychometrically sound self-report instrument or clinical interview; the two that did not reported the use of diagnostic criteria to measure PTSD. All but one study described sample ethnicity, all but one study assessed for deviations from Hardy–Weinberg, and the majority of studies assessed for comorbidities and applied corrections for multiple comparisons. We do not believe that differences in study quality affected our results.
SNP-Level Results
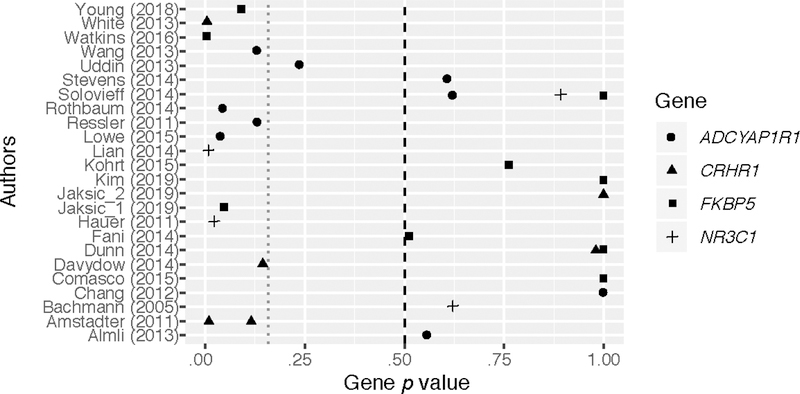
The SNP meta-analyses indicated that some variants within all four genes attained nominal significance: FKBP5 (rs9296158), p = .001; CRHR1 (rs4074461), p = .020; NR3C1 (rs258747) p = .001; and ADCYAP1R1 (rs2267735), p = .003 (see Supplementary Table S3 for further detailed results). However, only two of these variants (FKBP5 rs9296158 and NR3C1 rs258747) remained significant after Bonferroni adjustment for multiple testing: .05/total number of SNPs, p = .001. The SNPs did not retain significance in a sensitivity analysis when assuming a nontrivial rate of unreported studies. Furthermore, homogeneity tests using Cochran’s Q were conducted and did not show evidence for significant heterogeneity across SNPs that were analyzed in more than one study (Supplementary Table S3). The one exception was for rs12938931 in CRHR1, which was not significant in the SNP-level meta-analysis. Thus, no additional analyses were conducted. A forest plot of p values for each study organized by gene is presented in Figure 2.
Figure 2.
Forest plot of p values by gene and study. This plot presents the p values from the single-nucleotide polymorphism (SNP)–based meta-analysis, organized by study and gene of interest. The magenta line is set at a threshold of p = .160, which is considered suggestive as corresponding to the Akaike information criterion (AIC). The points at the p value of 1.0 are those that were set conservatively at 1.0 in studies that did not provide a p value (i.e., reported only that results were “not significant”). It is noted that due to the novelty of combining various numbers of measured SNPs across genes and studies, this forest plot is presents p values and not effect sizes, as is standard practice.
Gene-Level Results
Gene-level meta-analyses showed that NR3C1, CRHR1 and FKBP5 yielded significant signals following Bonferroni correction: at .05/4 genes, p = .0125 (Table 2). Sensitivity analyses (i.e., examination of different thresholds of percentage of unreported null findings) suggested that the signal in CRHR1 was rather marginal as it did not retain significance if there were unreported null studies of a sample size larger than 15% of the sample size in this meta-analysis. More robust signals were found for NR3C1 and FKBP5, which were found to retain significance in the context of unreported null studies of a sample size 40% of the meta-analysis sample size. Given that SNP-level tests did not show significant heterogeneity (with one exception noted previously), it is not likely to be of concern for the gene-level analyses, as they represent a combination of SNPs
Table 2.
Gene-Level Meta-Analytic Overall Results
| Gene | Number of Unique SNPs Analyzed | p | Sensitivitya |
|---|---|---|---|
| ADCYAP1R1 | 17 | .860 | NA |
| CRHR1 | 22 | .017 | 14.0 |
| FKBP5 | 4 | .011 | 29.0 |
| NR3C1 | 5 | .003 | 41.0 |
Note. SNP = single-nucleotide polymorphism.
Represents the percentage of the meta-analysis sample size at which unreported null study sample sizes would make the signal nonsignificant.
Discussion
The goal of the present work was to conduct meta-analyses of HPA-related genes, including both traditional SNP-level analysis and new gene-level analysis methods developed for the present research. The findings from the current investigation suggest a significant relation between PTSD and NR3C1 and FKBP5 at the gene level. A significant association with PTSD was also observed in the gene-level meta-analysis of CRHR1. However, sensitivity analyses suggested this finding is not robust to consideration of unreported null studies even when assuming a relatively small number of unreported samples. Although the gene-level analyses did not support an overall effect of ADCYAP1R1 on PTSD risk, we recently published a more extensive analysis of ADCYAP1R1 in which we focused on the SNP rs2267735 and expected sex differences in this gene (Lind et al., 2017). Two additional genes (CRHBP and CRHR2) were excluded from analyses due to inadequate numbers of published studies at the time of this research.
A generally similar pattern of findings was observed in the SNP-based analyses, which is not surprising given that gene-level analyses aggregate the statistical information from the individual SNPs. Specifically, our findings supported significant associations between PTSD and FKBP5 (rs9296158) and NR3C1 (rs258747), although these effects were near borderline in significance after adjustments for multiple testing and would not withstand sensitivity analyses aimed at estimating whether findings are robust to unpublished null findings. Further, ADCYAP1R1 (rs2267735) and CRHR1 (rs4074461) showed suggestive associations with PTSD but did not survive multiple-testing correction. However, if these genes would have been analyzed alone such that the SNP p value was adjusted only for the number of variants in these genes, both would have been deemed significant, replicating the findings of existing studies (e.g., Lind et al., 2017).
Perhaps the most important finding observed herein is the valuable contribution of adopting gene-level approaches to examining the associations of markers with PTSD. It is perhaps not surprising to observe more robust effects using a gene-based approach, as many markers examined within genes are explicitly intended to “tag” an area of influence, and this approach benefits from enhanced power of these multiple markers.
Importantly, there are a number of limitations in the gene-based analyses conducted here, such as challenges in considering directionality of each SNP within a gene and a nontraditional forest plot as well as a host of common considerations in molecular genetic studies related to accounting for ancestry, incorporating linkage disequilibrium, and challenges in modeling the potential presence of interactions of markers. Further, the included studies varied with regard to outcome and mode of inheritance. We also recognize that type of trauma exposure may influence results, but because most studies consisted of samples with varied trauma histories, we could not examine this in the present study. Due to the limited number of studies for most SNPs, moderator analyses could not be examined here. The examination of moderators, such as potential differences as a function of trauma type or outcome (i.e., symptom count compared to diagnosis), as well as sensitivity analyses examining the impact of PTSD outcome (e.g., diagnosis, symptom severity) would be important future work if larger samples become available. It is worth note that the strongest findings were observed for the two genes with the fewest supporting studies; although efforts were made to counteract the file-drawer problem, it is still possible this meta-analytic approach was nonetheless impacted by Type 1 error. Finally, this study examined main effects of HPA genes. Gene-by-environment interaction studies (GxE), although of interest, have methodological considerations and limitations (e.g., coding of outcome and risk allele, failure to check and correct for statistical interactions; Eaves, 2006), which can lead to difficulties with interpretation and are outside the scope of this paper. Research that can address these and other intricacies is called for; our group recently applied a gene-level approach to GxE effects of FKBP5 interaction with trauma exposure in a meta-analysis of PTSD (Hawn et al., 2019).
The findings from the present investigation provide support at the gene level for HPA markers, particularly FKBP5 and NR3C1, as candidate genes associated with PTSD and help to resolve apparent inconsistencies in the candidate-gene literature. Importantly, other markers included in the original intent for analysis either did not have sufficient numbers of studies (CRHBP, CRHR2), were observed to approach significant (though not robust) effects (CRHR1), or were not significant with tests of main effects but have been previously been shown by our group to demonstrate effects (ADCYAP1R1) in sex-stratified approaches. Accordingly, it is expected that as new publications permit better power for incorporation of moderation, future meta-analytic studies that adopt gene-based, and even likely SNP-based, approaches will continue to inform our understanding of the potential for variation within the HPA axis to be associated with PTSD. The longstanding critique of candidate approaches is not addressed using this approach. However, meta-analyses, as used here, may begin to address some critiques through enhancing power to detect small individual gene effects and explicitly testing the degree to which findings may be robust to the “file drawer problem” of unpublished null results.
Although outside the scope of this study, the methods used herein could be applied to a range of candidate genes and systems. In future studies, these methods may be adapted to incorporate markers from other molecular approaches, such as genome-wide association studies (GWAS). We note that to date, the majority of significant variants identified in GWAS of PTSD are found in genes not traditionally included in previous candidate-gene approaches, and, in fact, none of the candidate genes have been directly supported in the GWAS literature.
Supplementary Material
Acknowledgments
Dr. Sheerin’s time is funded the National Institute on Alcohol Abuse and Alcoholism (NIAAA; K01 AA025692). Ms. Lind was supported by the National Institute of Mental Health (NIMH; T32 MH020030). Dr. Amstadter’s time was partially funded by the NIAAA (K02 AA023239). Dr. Nugent’s time was funded by the NIMH (R01MH105379, R01MH108641). Dr. Bacanu’s effort was also partially funded by the NIMH (R01MH113665). The authors have no conflicts of interest to disclose. We thank all authors of included studies who contributed their data for use in analysis and responded to our e-mail requests for additional information concerning their samples. We also thank the undergraduate research assistants in the Amstadter lab, particularly Katie Hummel and Laurel Kovalchick, for their help with reliability coding.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, … McVean GA (2010). A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Fani N, Smith AK, & Ressler KJ (2014). Genetic approaches to understanding post-traumatic stress disorder. International Journal of Neuropsychopharmacology, 17, 355–370. 10.1017/s1461145713001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Mercer KB, Kerley K, Feng H, Bradley B, Conneely KN, & Ressler KJ (2013). ADCYAP1R1 genotype associates with post-traumatic stress symptoms in highly traumatized African-American females. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 162b, 262–272. 10.1002/ajmg.b.32145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstadter AB, Nugent NR, Yang BZ, Miller A, Siburian R, Moorjani P, … Koenen KC (2011). Corticotrophin-releasing hormone type 1 receptor gene (CRHR1) variants predict posttraumatic stress disorder onset and course in pediatric injury patients. Disease Markers, 30, 89–99. 10.3233/dma-2011-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann AW, Sedgley TL, Jackson RV, Gibson JN, Young RM, & Torpy DJ (2005). Glucocorticoid receptor polymorphisms and post-traumatic stress disorder. Psychoneuroendocrinology, 30, 297–306. 10.1016/j.psyneuen.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Banerjee SB, Morrison FG, & Ressler KJ (2017). Genetic approaches for the study of PTSD: Advances and challenges. Neuroscience Letters, 649, 139–146. 10.1016/j.neulet.2017.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjet C, Bromet E, Karam EG, Kessler RC, McLaughlin KA, Ruscio AM, … Koenen KC (2016). The epidemiology of traumatic event exposure worldwide: Results from the World Mental Health Survey Consortium. Psychological Medicine, 46, 327–343. 10.1017/s0033291715001981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bountress KE, Bacanu SA, Tomko RL, Korte KJ, Hicks T, Sheerin C, … Amstadter AB (2017). The effects of a BDNF Val66Met polymorphism on posttraumatic stress disorder: A meta-analysis. Neuropsychobiology, 76, 136–142. 10.1159/000489407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Coimbra BM, Ota VK, Mello MF, & Belangero SI (2017). Single-nucleotide polymorphisms in genes related to the hypothalamic-pituitary-adrenal axis as risk factors for posttraumatic stress disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 174, 671–682. 10.1002/ajmg.b.32564 [DOI] [PubMed] [Google Scholar]
- Chang SC, Xie P, Anton RF, De Vivo I, Farrer LA, Kranzler HR, … Koenen KC (2012). No association between ADCYAP1R1 and posttraumatic stress disorder in two independent samples. Molecular Psychiatry, 17, 239–241. 10.1038/mp.2011.118 [DOI] [PubMed] [Google Scholar]
- Comasco E, Gustafsson PA, Sydsjo G, Agnafors S, Aho N, & Svedin CG (2015). Psychiatric symptoms in adolescents: FKBP5 genotype–early life adversity interaction effects. European Child & Adolescent Psychiatry, 24, 1473–1483. 10.1007/s00787-015-0768-3 [DOI] [PubMed] [Google Scholar]
- Cornelius JR, Kirisci L, Reynolds M, Clark DB, Hayes J, & Tarter R (2010). PTSD contributes to teen and young adult cannabis use disorders. Addiction Behavior, 35, 91–94. 10.1016/j.addbeh.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydow DS, Kohen R, Hough CL, Tracy JH, Zatzick D, & Katon WJ (2014). A pilot investigation of the association of genetic polymorphisms regulating corticotrophin-releasing hormone with posttraumatic stress and depressive symptoms in medical-surgical intensive care unit survivors. Journal of Critical Care, 29, 101–106. 10.1016/j.jcrc.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahanty DL, & Nugent NR (2006). Predicting PTSD prospectively based on prior trauma history and immediate biological responses. Annals of the New York Academy of Sciences, 1071, 27–40. 10.1196/annals.1364.003 [DOI] [PubMed] [Google Scholar]
- Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, … Koenen KC (2018). Largest GWAS of PTSD (N = 20, 070) yields genetic overlap with schizophrenia and sex differences in heritability. Molecular Psychiatry, 23, 666–673. 10.1038/mp.2017.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EC, Solovieff N, Lowe SR, Gallagher PJ, Chaponis J, Rosand J, … Smoller JW (2014). Interaction between genetic variants and exposure to Hurricane Katrina on post-traumatic stress and post-traumatic growth: A prospective analysis of low income adults. Journa of Affective Disorders, 152–154, 243–249. 10.1016/j.jad.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ (2006). Genotype x Environment interaction in psychopathology: Fact or artifact? Twin Research and Human Genetics, 9, 1–8. 10.1375/183242706776403073 [DOI] [PubMed] [Google Scholar]
- Fani N, King TZ, Reiser E, Binder EB, Jovanovic T, Bradley B, & Ressler KJ (2014). FKBP5 genotype and structural integrity of the posterior cingulum. Neuropsychopharmacology, 39, 1206–1213. 10.1038/npp.2013.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressier F, Calati R, Balestri M, Marsano A, Alberti S, Antypa N, & Serretti A (2013). The 5-HTTLPR polymorphism and posttraumatic stress disorder: A meta-analysis. Journal of Traumatic Stress, 26, 645–653. 10.1002/jts.21855 [DOI] [PubMed] [Google Scholar]
- Hauer D, Weis F, Papassotiropoulos A, Schmoeckel M, Beiras-Fernandez A, Lieke J, … Schelling G (2011). Relationship of a common polymorphism of the glucocorticoid receptor gene to traumatic memories and posttraumatic stress disorder in patients after intensive care therapy. Critical Care Medicine, 39, 643–650. 10.1097/CCM.0b013e318206bae6 [DOI] [PubMed] [Google Scholar]
- Hawn SE, Sheerin CM, Lind MJ, Hicks TA, Marraccini ME, Bountress K, … Amstadter AB (2019). G x E effects of FKBP5 and traumatic life events on PTSD: A meta-analysis. Journl of Affective Disorders, 243, 455–462. 10.1016/j.jad.2018.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsboer F, Lauer CJ, Schreiber W, & Krieg JC (1995). Altered hypothalamic-pituitary-adrenocortical regulation in healthy subjects at high familial risk for affective disorders. Neuroendocrinology, 62, 340–347. 10.1159/000127023 [DOI] [PubMed] [Google Scholar]
- Jaksic N, Sabic Dzananovic E, Aukst Margetic B, Rudan D, Cima Franc A, Bozina N, … Jakovljevic M (2019). A candidate gene association study of FKBP5 and CRHR1 polymorphisms in relation to war-related posttraumatic stress disorder. Psychiatria Danubia, 31, 269–275. 10.24869/psyd.2019.269 [DOI] [PubMed] [Google Scholar]
- Kang JI, Kim TY, Choi JH, So HS, & Kim SJ (2019). Allele-specific DNA methylation level of FKBP5 is associated with posttraumatic stress disorder. Psychoneuroendocrinology, 103, 1–7. 10.1016/j.psyneuen.2018.12.226 [DOI] [PubMed] [Google Scholar]
- Kessler RC, Sonnega A, Bromet E, Hughes M, & Nelson CB (1995). Posttraumatic stress disorder in the National Comorbidity Survey. Archives of General Psychiatry, 52, 1048–1060. 10.1001/archpsyc.1995.03950240066012 [DOI] [PubMed] [Google Scholar]
- Koenen KC (2007). Genetics of posttraumatic stress disorder: Review and recommendations for future studies. Journal of Traumatic Stress, 20, 737–750. 10.1002/jts.20205 [DOI] [PubMed] [Google Scholar]
- Kohrt BA, Worthman CM, Ressler KJ, Mercer KB, Upadhaya N, Koirala S, … Binder EB (2015). Cross-cultural gene-environment interactions in depression, post-traumatic stress disorder, and the cortisol awakening response: FKBP5 polymorphisms and childhood trauma in South Asia. International Rev Psychiatry, 27, 180–196. 10.3109/09540261.2015.1020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Bigdeli TB, Williamson VS, Vladimirov VI, Riley BP, Fanous AH, & Bacanu SA (2015). DISTMIX: Direct imputation of summary statistics for unmeasured SNPs from mixed ethnicity cohorts. Bioinformatics, 31, 3099–3104. 10.1093/bioinformatics/btv348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bao Y, He S, Wang G, Guan Y, Ma D, … Yang J (2016). The association between genetic variants in the dopaminergic system and posttraumatic stress disorder: A meta-analysis. Medicine, 95, e3074 10.1097/md.0000000000003074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Y, Xiao J, Wang Q, Ning L, Guan S, Ge H, … Liu J (2014). The relationship between glucocorticoid receptor polymorphisms, stressful life events, social support, and post-traumatic stress disorder. BMC Psychiatry, 14, 232 10.1186/s12888-014-0232-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind MJ, Marraccini ME, Sheerin CM, Bountress K, Bacanu SA, Amstadter AB, & Nugent NR (2017). Association of posttraumaic stress disorder with rs2267735 in the ADCYAP1R1 gene: A meta-analysis. Journal of Traumatic Stress, 30, 389–398. 10.1002/jts.22211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SR, Pothen J, Quinn JW, Rundle A, Bradley B, Galea S, … Koenen KC (2015). Gene-by-social-environment interaction (GxSE) between ADCYAP1R1 genotype and neighborhood crime predicts major depression symptoms in trauma-exposed women. Journal of Affective Disorders, 187, 147–150. 10.1016/j.jad.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck A, & Guyre PM (1986). Glucocorticoid physiology, pharmacology and stress. Advances in Experimental Medicine and Biology, 196, 81–96. 10.1007/978-1-4684-5101-6_6 [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, … May V (2011). Posttraumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature, 470, 492–497. 10.1038/nature09856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbaum BO, Kearns MC, Reiser E, Davis JS, Kerley KA, Rothbaum AO, … Ressler KJ (2014). Early intervention following trauma may mitigate genetic risk for PTSD in civilians: A pilot prospective emergency department study. Journal of Clinical Psychiatry, 75, 1380–1387. 10.4088/JCP.13m08715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin CM, Lind MJ, Bountress K, Nugent NR, & Amstadter AB (2017). The genetics and eEpigenetics of PTSD: Overview, recent advances, and future directions. Current Opinions in Psychology, 14, 5–11. 10.1016/j.copsyc.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton K, Ressler KJ, Norrholm SD, Jovanovic T, & Bradley-Davino B (2012). PTSD and gene variants: New pathways and new thinking. Neuropharmacology, 62, 628–637. 10.1016/j.neuropharm.2011.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW (2016). The genetics of stress-related disorders: PTSD, depression, and anxiety disorders. Neuropsychopharmacology, 41, 297–319. 10.1038/npp.2015.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Roberts AL, Ratanatharathorn A, Haloosim M, De Vivo I, King AP, … Koenen KC (2014). Genetic association analysis of 300 genes identifies a risk haplotype in SLC18A2 for post-traumatic stress disorder in two independent samples. Neuropsychopharmacology, 39, 1872–1879. 10.1038/npp.2014.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JS, Almli LM, Fani N, Gutman DA, Bradley B, Norrholm SD, … Ressler KJ (2014). PACAP receptor gene polymorphism impacts fear responses in the amygdala and hippocampus. Proceedings of the National Academy of Sciences, 111, 3158–3163. 10.1073/pnas.1318954111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF (2007). Spurious genetic associations. Biological Psychiatry, 61, 1121–1126. 10.1016/j.biopsych.2006.11.010 [DOI] [PubMed] [Google Scholar]
- True WR, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, & Nowak J (1993). A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Archives of General Psychiatry, 50, 257–264. 10.1001/archpsyc.1993.01820160019002 [DOI] [PubMed] [Google Scholar]
- Uddin M, Chang SC, Zhang C, Ressler K, Mercer KB, Galea S, … Koenen KC (2013). ADCYAP1R1 genotype, posttraumatic stress disorder, and depression among women exposed to childhood maltreatment. Depression & Anxiety, 30, 251–258. 10.1002/da.22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey J, Young RM, Lawford BR, & Morris CP (2014). Progress toward understanding the genetics of posttraumatic stress disorder. Jounal of Anxiety Disorders, 28, 873–883. 10.1016/j.janxdis.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Wang L, Cao C, Wang R, Qing Y, Zhang J, & Zhang XY (2013). PAC1 receptor (ADCYAP1R1) genotype is associated with PTSD’s emotional numbing symptoms in Chinese earthquake survivors. Journal of Affective Disorders, 150, 156–159. 10.1016/j.jad.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Watkins LE, Han S, Harpaz-Rotem I, Mota NP, Southwick SM, Krystal JH, … Pietrzak RH (2016). FKBP5 polymorphisms, childhood abuse, and PTSD symptoms: Results from the National Health and Resilience in Veterans Study. Psychoneuroendocrinology, 69, 98–105. 10.1016/j.psyneuen.2016.04.001 [DOI] [PubMed] [Google Scholar]
- White S, Acierno R, Ruggiero KJ, Koenen KC, Kilpatrick DG, Galea S, … Amstadter AB (2013). Association of CRHR1 variants and posttraumatic stress symptoms in hurricane-exposed adults. Journal of Anxiety Disorders, 27, 678–683. 10.1016/j.janxdis.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Koenen KC, Galea S, & Flory JD (2011). The role of genes in defining a molecular biology of PTSD. Disease Markers, 30(2–3), 67–76. 10.3233/dma-2011-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DA, Inslicht SS, Metzler TJ, Neylan TC, & Ross JA (2018). The effects of early trauma and the FKBP5 gene on PTSD and the HPA axis in a clinical sample of Gulf War veterans. Psychiatry Research, 270, 961–966. 10.1016/j.psychres.2018.03.037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.