Abstract
Intracellular pathogens have evolved multiple ways to manipulate their host cells in order to survive and replicate in a hostile environment. They often exploit membrane trafficking pathways at multiple steps of their infection cycle: entry, establishment of a replicative niche, avoid degradation and immune response, acquire nutrients and lastly, egress. Recent studies on membrane trafficking exploitation by intracellular pathogens have led to the discovery of novel and fascinating cell biology, including a non-canonical mechanism of ubiquitination and a novel mitophagy receptor. Thus, studying how pathogens target host cell membrane trafficking pathways is not only important for the development of new therapeutics, but also helps understanding fundamental mechanisms of cell biology.
Introduction
Cellular membrane trafficking pathways comprise of a series of highly dynamic endocytic, secretory and autophagic pathways. These processes require the combined actions of different SNAREs and GTPases as well as the actin cytoskeleton. They are crucial to maintain organelle identity, homeostasis and localization and to ensure intracellular transport. Intracellular pathogens, such as bacteria and viruses, have to cross the host cell membrane to enter the cell, avoid degradation by the lysosome, hide from innate immune sensors, establish a safe niche for their replication and survival and finally have to cross the cell membrane again to leave the host cell. It is therefore not surprising that these pathogens have evolved diverse ways to manipulate and exploit host cell membrane trafficking pathways. In fact, a recent screen with ~200 secreted effectors from gram-negative bacteria revealed that more than 30 % of these effectors localized to host cell membranes [1]. Different pathogens use different strategies during their journey throughout the cell, depending on their requirements for survival and replication and the way they co-evolved with their host. For instance, certain bacteria reside in and hijack the endocytic pathway for their replication, while other bacteria avoid the endocytic pathway and establish their intracellular niche elsewhere [2]. In addition to using host cell membranes or membrane compartments, viruses exploit host cell machineries, such as the translation machinery, for their replication [3]. However, the most crucial points of contact for the manipulation of host cell membranes are quite similar between different pathogens: the cell membrane, the actin cytoskeleton, the endocytic pathway, the secretory pathway and autophagy [2–4]. New technologies and high-throughput methods such as whole genome CRISPR/Cas9 screens and Cryo-EM have strongly advanced our understanding of the interactions between pathogens and host cell membranes. A detailed overview of how pathogens hijack host membrane trafficking pathways can be found elsewhere [2–4]. In this review, we will discuss the most recent advances in the exploitation of the secretory and autophagy pathways by intracellular bacteria and viruses.
Manipulation of the secretory pathway by pathogens - getting the ER into the right shape
The secretory pathway has crucial functions for protein and lipid metabolism and homeostasis in the host cell and is therefore an attractive target for intracellular pathogens. The regulation of membrane trafficking in the secretory pathway is mediated by specialized small GTPases of the ARF and Rab families, fusion factors such as soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) and membrane-shaping proteins. Many of these proteins are targeted and/or exploited by different intracellular pathogens, including bacteria, viruses, parasites and fungi. We will highlight select examples of recently discovered microbial strategies to exploit the host cell secretory pathway.
Legionella pneumophila
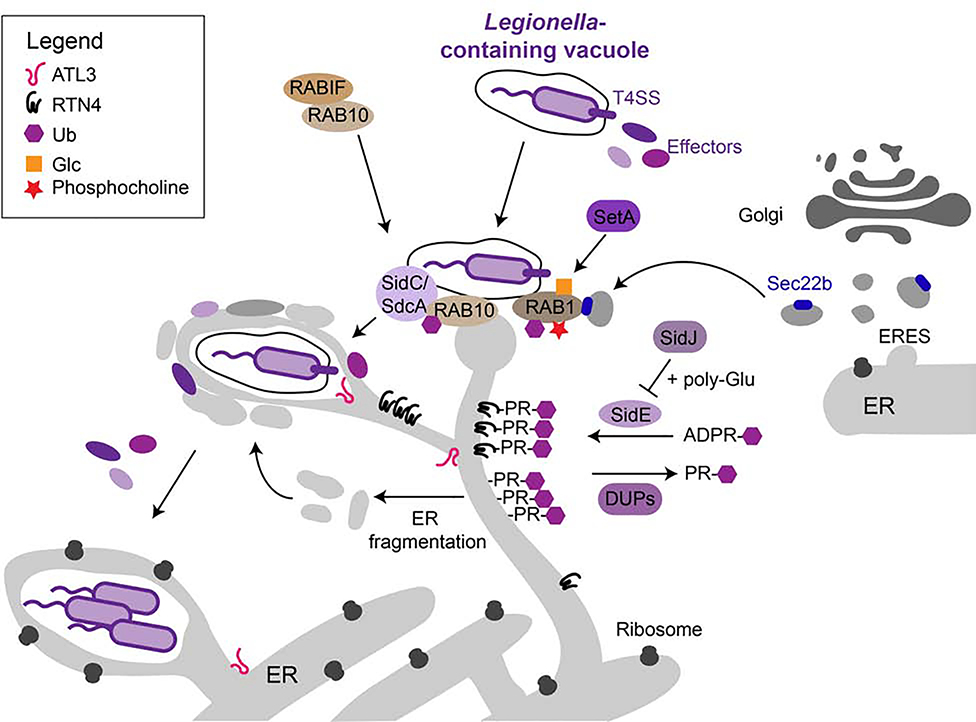
One master manipulator of the secretory pathway is the gram-negative, facultative intracellular bacterium Legionella pneumophila (L.p.), the causative agent of Legionnaires’ disease. Upon entry into the host cell, L.p. secretes an unprecedented number (> 330) of bacterial effector proteins via its Dot/Icm Type IV secretion system (T4SS) [5]. Many of these effectors regulate the function of several host proteins via post-translational modifications (PTMs) such as phosphorylation, ubiquitylation, and methylation and have led to the discovery of novel PTMs such as phosphocholination or biochemical mechanisms such as non-canonical protein ubiquitination [6–9]. Immediately after infection, a subset of effectors prevents fusion of the phagosome with the endosomal/lysosomal system, while other effectors help building and remodeling the L.p.’s intracellular niche, the plasma membrane-derived Legionella-containing vacuole (LCV). These effectors establish contact with ER-derived membranes and convert the LCV into an ER-like organelle required for L.p. replication. The LCV seems to first interact with the tubular ER, ER fragments and ER-derived vesicles and later acquire rough ER markers and ribosomes on its surface, but the precise sequence of events is not fully understood [10,11] (Figure 1). The most well studied host cell factor in this context is Rabí, a small GTPase involved in ER-to-Golgi transport, which is targeted and modified by multiple L.p. effectors at different stages of infection in order to recruit ER-derived vesicles to the LCV [6,12]. Previous work has shown that Rab1 activity is regulated by effector-mediated AMPylation (and de- AMPylation) and phosphocholination (and de-phosphocholination) of its switch region [6,12]. In addition to AMPylating Rab1, the effector DrrA has recently been shown to recruit the exocyst components Sec5, Sec6, and Sec15 to the LCV to mediate its fusion with ER-derived vesicles [13]. Adding another level of complexity in the regulation of Rab1, the effector SetA has been found to glucosylate Rab1’s switch domain to inhibit its function [14]. Rab1 also seems to be a target of the SidE effector family (SidEs), further highlighting the crucial role of Rab1 for LCV formation [7,15]. SidEs have been the major focus of current research as they revealed a novel, non-canonical mechanism of protein ubiquitination. In contrast to canonical ubiquitination, which requires the concerted actions of E1-, E2- and E3-ligases and ATP, SidEs are “all-in-one” ubiquitin ligases which first modify ubiquitin with NAD-derived ADP-ribose and subsequently transfer phosphoribosyl (PR)-ubiquitin to serine residues on the target protein [7,8,15–17]. The activity of SidEs is counterbalanced by the effector SidJ, which inhibits SidEs through polyglutamylation of their mono-ADP ribosyl transferase domain in a calmodulin-dependent manner [18–21]. Initially it was suggested that SidEs mainly target small GTPases such as RAB1, RAB6 (intra-Golgi transport), RAB33b (intra-Golgi transport) and RAB30 (maintenance of Golgi infrastructure, biogenesis of autophagosomes) [7]. A follow-up study then identified the ER-tubule forming protein reticulon 4 (RTN4) as an additional substrate for SidE-mediated PR- ubiquitination [10]. Modification of RTN4 with PR-Ubs seems to induce its oligomerization and the formation of pseudovesicles with protrusions around the LCV (Figure 1). Two recent independent studies finally revealed that PR-Ub can be removed by the effectors DupA and DupB and, using complementary approaches, identified hundreds of potential SidEs targets [11,22]. Interestingly, among the targets were proteins implicated in ER remodeling and ER- phagy, such as LNP1, RTN1, RTN3, FAM134A-C, SEC62 and TEX264, suggesting an important role of these processes for LCV biogenesis. Indeed, the activity of SidEs led to fragmentation of the ER and accumulation of the modified ER proteins around the LCV, indicating that ER fragmentation might be an additional mechanism of L.p. to recruit ER membranes to the LCV [11,22] (Figure 1). In line with these findings, the large, ER-resident GTPase atlastin 3 (ATL3), which has functions in ER shaping and ERphagy [23], has been shown to promote recruitment of the tubular ER to LCV and to mediate homotypic fusion of ER- derived membranes around the LCV [24]. A whole genome host cell CRISPR screen from our laboratory identified RAB10 and RABIF as additional host cell factors required for recruitment of the tubular ER to the LCV [25]. We showed that Rab10, which is stabilized by RabIF, is recruited to the LCV by the L.p. effectors SidC/SdcA and ubiquitinated by effectors yet to be identified. Knockout of both RAB10 and RABIF inhibited co-localization of RTN4 with the LCV. Interestingly, RAB10 has been shown to induce growth of ER tubules in vesicle-like, dynamic ER domains [26] (Figure 1). Whether these structures are identical to the RTN4-positive pseudovesicles previously observed around the LCV [10] remains to be established. Thus, the LCV seems to acquire ER membranes through several independent mechanisms including the recruitment of ER-derived vesicles from ERES by modification of small GTPases and of ER fragments induced by SidE-mediated PR-ubiquitination of ER shaping proteins.
Figure 1: Subversion of the secretory pathway by L. pneumophila.
Upon entry into the host cell, L. p. secretes more than 300 bacterial effectors via its T4SS. Some of these effectors manipulate host cell proteins to recruit ER membranes to the LCV. The small GTPase RAB1 is modified by multiple effectors, such as SetA, to recruit Sec22b-positive ER-derived vesicles to the LCV. These modifications include phosphocholination, (PR-)ubiquitination, and glucosylation. The effectors SidC and SdcA recruit the GTPase RAB10, normally stabilized by RABIF, to the LCV which is then ubiquitinated by unknown effectors. This promotes the recruitment of RTN4-positive ER to the LCV. The fusion of ER derived vesicles around the LCV requires the large GTPase ATL3. The SidE family of effectors mediates PR-ubiquitination of proteins implicated in ER shaping and ERphagy, resulting in the formation of ER fragments that might contribute to LCV maturation. SidE activity is controlled through glutamylation by the effector SidJ and counterbalanced by the deubiquitinases DupA and DupB (DUPs). The mature LCV resembles the rough ER and allows for bacterial replication.
Flaviviruses
Members of the genus Flavivirus, including Dengue virus (DENV), Zika virus (ZIKV), West Nile virus (WNV) and yellow fever virus, also have an intimate connection with the secretory pathway, particularly with the ER [27]. The genome of these viruses consists of positive-strand RNA ((+) RNA), largely mimicking host cell mRNA. The RNA is translated in a cap-dependent manner at host cell ribosomes [28] and a single polypeptide is synthesized at the rough ER membrane (Figure 2) where it is then cleaved into three structural (SPs) and seven non-structural proteins (NSPs), most of them being transmembrane multipass proteins, by viral and host proteases [27]. Recent high-through put screens revealed a central role fom9.
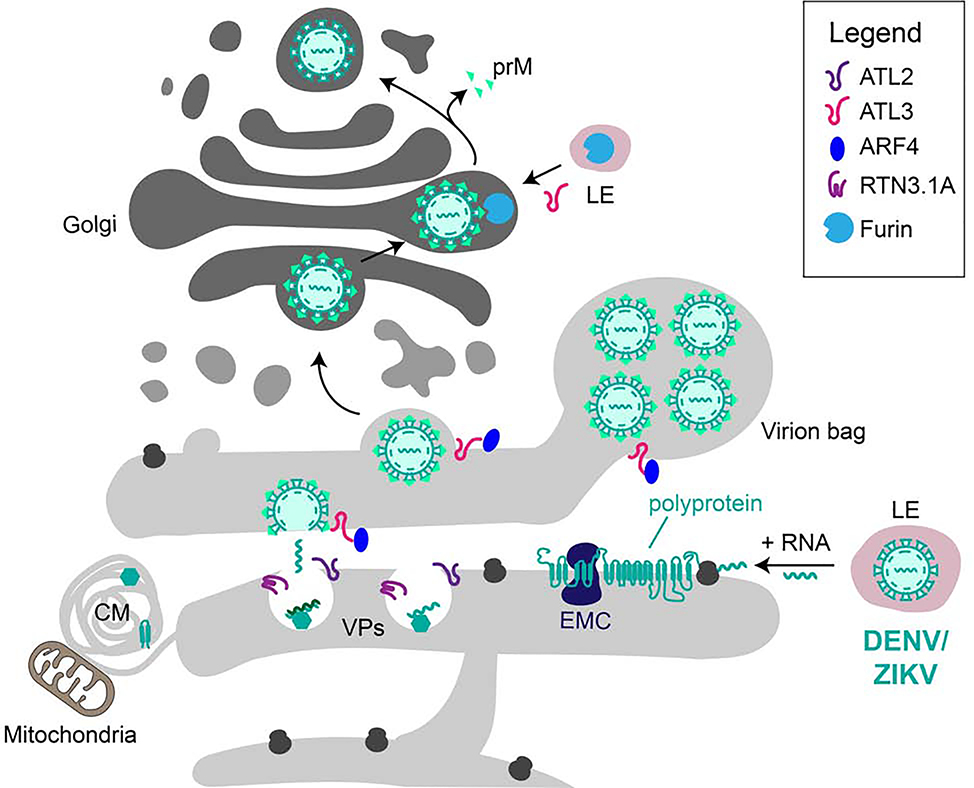
Figure 2: Exploitation of the secretory pathway by DENV/ZIKV.
DENV/ZIKV reside in the endocytic pathway and, upon maturation to a late endosome (LE) and the concomitant drop in pH, viral (+) RNA is released into the cytosol. The RNA is translated into a single polyprotein at host cell ribosomes attached to the ER. The EMC is required for the biogenesis of viral proteins. NSPs and SPs, which are generated by cleavage of the polyprotein by viral and host cell proteases, mediate viral replication or virion assembly, respectively. Upon viral infection, the ER is remodeled to form distinct structures such as CMs, VPs and virion bags. The replication complex is found in VPs whose formation requires the host cell ER shaping proteins RTN3.1A and ATL2. In contrast, ATL3 and its interaction partner ARF4 are required for virion maturation and trafficking. The virion matures on its way through the Golgi where it is processed by the host cell protease furin. Recycling of furin from late endosomes to the Golgi and subsequent cleavage of the viral protein prM was found to be dependent on ATL3.
[poiu3r ER-resident host cell proteins or protein complexes for viral replication or maturation, such as the translocon component SEC61, the oligosaccharyltransferase (OST) complex or protein complexes involved in ER-associated degradation [29–31]. Components of the ER membrane complex (EMC) have been identified as essential host cell factors for DENV and ZIKV replication and are required for efficient biogenesis of the essential viral non-structural proteins NS4A and NS4B by promoting their folding and stability [32–34]. In order to protect the newly forming virus particles and the replication complex from the host cell cytosol and innate immune detection, the NSPs dramatically remodel the ER, creating an interconnected network of invaginated vesicles packets (VPs) in the rough ER and convoluted membranes (CMs) in the smooth ER [28]. The function of CMs, which are located close to VPs and host cell mitochondria is unclear and their formation seems to be cell-type specific, but it has been proposed that these membrane structures play a role for the maturation of viral proteins or are a site of lipid storage [28]. In contrast, VPs appear to serve as the replication organelles (ROs) in which the viral RNA is replicated by the viral replication complex and the newly synthesized (+) RNA is then transferred to a budding virion on opposing ER cisternae (Figure 2). However, how these membrane structures form and which host cell factors are required for their biogenesis is poorly understood. Recent studies revealed that, similar to L.p., DENV and ZIKV exploit ER membrane shaping proteins for their replication, intracellular trafficking and maturation [35–37]. The atlastins ATL2 and ATL3, two ER-resident large GTPases that mediate the fusion of ER tubules, seem to play a central role in the formation of VPs and the maturation of the virion [35,36].While ATL2 is involved in VP biogenesis, ATL3 plays a role in virion trafficking and maturation. Interestingly, the interactome of both proteins dramatically changed upon infection [35]. For instance, the binding of the ADP-ribosylation factor 4 (ARF4) to ATL3 was enhanced during infection and ARF4 was required for assembly and release of the virus. Furthermore, ATL3 was required for recycling of the host cell protease furin to the Golgi where it cleaves the viral envelope protein prM, an important step in virus maturation. In addition, the ER tubulation protein and ERphagy receptor RTN3, specifically its isoform RTN3.1A, is recruited to replication organelles and promotes viral replication [37]. Knockdown of RTN3.1A results either in a reduction of vesicles in VPs (ZIKV and WNV) or their elongation (DENV).
Manipulation of autophagy pathways by pathogens
The autophagic pathway possibly resembles the most threatening membrane trafficking pathway for intracellular pathogens as it can be used by the host cell to engulf and degrade the invaders in a process termed xenophagy. Therefore, most pathogens avoid fusion of the phagosome with any components of the autophagic pathway. However, certain pathogens have developed strategies to exploit this antimicrobial pathway. Here, we will highlight recent insights into mechanisms of either autophagy inhibition or exploitation by pathogens.
Pathogens inhibiting autophagic pathways
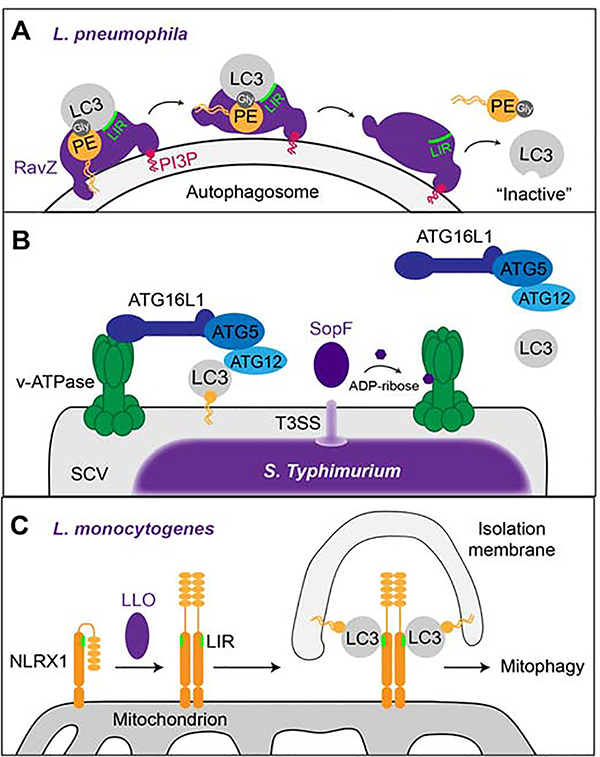
As one of the pathogens avoiding autophagy, L.p. uses several bacterial effectors to shut down this pathway. The effector RavZ has a cysteine protease activity that irreversibly cleaves the amide bond between phosphatidylethanolamine (PE) and LC3 during which LC3 loses its terminal glycine [38]. Thus, the cellular pool of RavZ-processed LC3 cannot be re-lipidated, resulting in a strong inhibition of autophagy. Until recently, it was unclear how RavZ recognizes LC3 and how it can extract it from autophagosomal membranes. The current model is that RavZ possesses i) a C-terminal PI3P-binding domain which allows its recruitment to the autophagosomal membrane [39] and ii) an N-terminal LC3-interacting region (LIR) motif that binds to phosphatidylethanolamine (PE)-anchored LC3 on the autophagosomal surface [40] (Figure 3A). It has been proposed that an N-terminal a-Helix of RavZ inserts into the autophagosomal membrane and is essential for extracting the PE-anchor from the membrane and for shifting it towards its catalytic site, where cleavage of PE from LC3 can take place [40]. Another mechanism of L.p. to inhibit autophagy is cleavage of the host SNARE protein Stx17 by the effector Lpg1137 [41]. This leads to the loss of PtIns3P at mitochondria-associated membranes, thereby causing the abrogation of autophagy. In order to protect itself from autophagic degradation in immune cells, the intracellular bacterium Rickettsia parkeri (R. parkeri) shields itself from recognition by the autophagy machinery with its outer membrane protein OmpB [42]. OmpB both forms a protective layer around the bacterium and prevents ubiquitination of other bacterial outer membrane proteins, thereby enabling the bacterium to evade recognition by autophagy receptors. A recent study on xenophagy evasion by Salmonella enterica serovar Typhimurium (S. Typhimurium) has identified the vacuolar type H+- translocating (v)- ATPase as a novel player in this antimicrobial host defense pathway [43,44]. The study revealed that, upon bacterial infection, v-ATPase recruits ATG16L1 to the bacteria- containing vacuole via its WD40 domain. This interaction is crucial to bring the LC3 lipidation machinery (e.g. ATG5, ATG12) in close contact to the vacuole and initiate xenophagy (Figure 3B). The authors further showed that the S. Typhimurium effector SopF, a phosphoinositol- binding protein required for Salmonella-containing vacuole (SCV) maintenance [45], ADP- ribosylates a subunit of the v-ATPase, thereby blocking the recruitment ATG16L1 onto the and autophagic clearance of S. Typhimurium [43].
Figure 3: Manipulation of autophagic pathways by intracellular pathogens.
(A) The L.p. effector RavZ binds to PI3P on membranes (e.g. autophagosome) and binds to PE-anchored LC3 via its LIR motif. The N-terminal α-helix of RavZ then moves the PE moiety towards its catalytic site where the bond between LC3 and PE is irreversibly cleaved at the terminal glycine residue, thereby releasing LC3 proteins that cannot be re-lipidated. (B) Upon bacterial infection, the v-ATPase on the bacterial vacuole recruits ATG16L1 to induce xenophagy via ATG5- ATG12-mediated lipidation and membrane anchoring of LC3. The T3SS-secreted S. Typhimurium effector SopF ADP-ribosylates the v-ATPase on the SCV which prevents recruitment of the autophagic machinery and, hence, xenophagy. (C) The L. monocytogenes toxin LLO induces the oligomerization of NLRX1 on host cell mitochondria which leads to a conformational shift that exposes the LIR motif on NLRX1. This enables the binding of LC3 to NLRX1 and mitophagy.
Pathogens subverting autophagic pathways
Listeria monocytogenes (L.monocytogenes) produces the pore-forming toxin listeriolysin O (LLO) which is a crucial virulence factor at different steps during the infection cycle. Studying the intracellular functions of LLO has recently led to the discovery of a novel receptor for the autophagic degradation of mitochondria (mitophagy) [46]. LLO induces the oligomerization of Nod-like receptor X1 (NLRX1) which exposes a LIR motif on NLRX1, thereby promoting its binding to LC3 and induction of mitophagy (Figure 3). This has a protective effect on L.monocytogenes by reducing mitochondrial production of reactive oxygen species. Also flaviruses have been shown to hijack selective autophagy pathways for their replication. The viral protease NS3 cleaves the ERphagy receptor FAM134B, which results in the inhibition of ERphagy and promotes virus production [47]. Furthermore, the DENV protein NS4A binds to the host protein AUP1 on lipid droplets. This interaction activates the acetyltransferase activity of AUP1, thereby promoting lipophagy to support production of virus particles [48]. Enterovirus D68 (EV-D68), has recently been shown to actively induce autophagy for its own benefit. The autophagosomal SNARE SNAP29 and the orphan SNARE SNAP47 are required for viral replication. However, later during infection the EV-D68 protease C3 inhibits autophagosomal- lysosomal fusion by cleaving SNAP29 [49]. Autophagy also has proviral functions on human cytomegalovirus (hCMV) and seems to be important for efficient assembly of viral particles [50,51]. A recent report suggested the incorporation of LC3 homologs into viral particles, indicating that hCMV uses autophagic membranes for its maturation [51].
Conclusion and perspective
In order to survive in the host cell, intracellular pathogens have developed diverse strategies to avoid or subvert host cell membrane trafficking pathways. The exploitation of membrane trafficking pathways is often achieved through modifications (i.e. PTMs) or cleavage of key host cell proteins. Novel high-throughput technologies such as CRISPR/Cas9 screens have enabled the identification of previously unexplored host cell targets. For instance, recent advances have highlighted the emerging role of proteins involved in ER remodeling and ERphagy for pathogenesis. As the detailed mechanism of ERphagy is still poorly understood, studying intracellular pathogens targeting ERphagy might shed light into this pathway and help identifying its key components. It will be highly informative to study how PTMs of proteins involved in ERphagy, such as PR-ubiquitination, affect their function in this pathway. As presented in this review, other forms of selective autophagy, such as mitophagy, lipophagy and xenophagy are targeted and manipulated by pathogens which has resulted, e.g., in the discovery of a novel mitophagy receptor. Thus, pathogens are invaluable cell biology tools and will continue to help our understanding of crucial cellular membrane trafficking pathways.
Acknowledgements
S.M. is supported by the National Institutes of Health RO1 grant AI118974 and an award from the Pew Charitable Trust (A129837). We apologize that we could not include all relevant studies in this review due to space constraints.
Footnotes
Conflict of interest statement
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Weigele BA, Orchard RC, Jimenez A, Cox GW, Alto NM: A systematic exploration of the interactions between bacterial effector proteins and host cell membranes. Nat Commun 2017, 8:532.• Study showing that about 30% of effectors from six different gram-negative bacterial species localize to host cell membranes.
- 2.Asrat S, de Jesús DA, Hempstead AD, Ramabhadran V, Isberg RR: Bacterial pathogen manipulation of host membrane trafficking. Annu Rev Cell Dev Biol 2014, 30:79–109. [DOI] [PubMed] [Google Scholar]
- 3.Robinson M, Schor S, Barouch-Bentov R, Einav S: Viral journeys on the intracellular highways. Cell Mol Life Sci 2018, 75:3693–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber MM, Faris R: Subversion of the Endocytic and Secretory Pathways by Bacterial Effector Proteins. Frontiers in cell and developmental biology 2018, 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu J, Luo Z-Q: Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 2017, 15:591–605. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE, Roy CR: Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 2011,477:103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo Z-Q: Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 2016, 533:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalayil S, Bhogaraju S, Bonn F, Shin D, Liu Y, Gan N, Basquin J, Grumati P, Luo Z-Q, Dikic I: Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 2018, 557:734–738. ··(Together with 17): This study uncovers the catalytic mechanism of the non-canonical ubiquitination mediated by the L. pneumophila SidE family of effectors.
- 9.Michard C, Doublet P: Post-translational modifications are key players of the Legionella pneumophila infection strategy. Front Microbiol 2015, 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotewicz KM, Ramabhadran V, Sjoblom N, Vogel JP, Haenssler E, Zhang M, Behringer J, Scheck RA, Isberg RR: A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host Microbe 2017, 21:169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin D, Mukherjee R, Liu Y, Gonzalez A, Bonn F, Liu Y, Rogov VV, Heinz M, Stolz A, Hummer G, et al. : Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB. Mol Cell 2019, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilbi H, Nagai H, Kubori T, Roy CR: Subversion of Host Membrane Dynamics by the Legionella Dot/Icm Type IV Secretion System. Curr Top Microbiol Immunol 2017, 413:221–242. [DOI] [PubMed] [Google Scholar]
- 13.Arasaki K, Kimura H, Tagaya M, Roy CR: Legionella remodels the plasma membrane- derived vacuole by utilizing exocyst components as tethers. J Cell Biol 2018, 217:3863–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, McCloskey A, Cheng S, Wu M, Xue C, Yu Z, Fu J, Liu Y, Luo Z-Q, Liu X: Regulation of the small GTPase Rab1 function by a bacterial glucosyltransferase. Cell Discov 2018, 4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhogaraju S, Kalayil S, Liu Y, Bonn F, Colby T, Matic I, Dikic I: Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 2016, 167:1636–1649.e13. [DOI] [PubMed] [Google Scholar]
- 16.Qiu J, Luo Z-Q: Hijacking of the Host Ubiquitin Network by Legionella pneumophila. Front Cell Infect Microbiol 2017, 7:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Shi M, Feng H, Zhu Y, Liu S, Gao A, Gao P: Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE. Cell 2018, 173:1231–1243.e16. [DOI] [PubMed] [Google Scholar]
- 18.Black MH, Osinski A, Gradowski M, Servage KA, Pawtowski K, Tomchick DR, Tagliabracci VS: Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE- family ubiquitin ligases. Science 2019, 364:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhogaraju S, Bonn F, Mukherjee R, Adams M, Pfleiderer MM, Galej WP, Matkovic V, Lopez-Mosqueda J, Kalayil S, Shin D, et al. : Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 2019, 572:382–386.• (Together with 18,20–21): Study revealing the mechanism by which the L. pneumophila SidJ opposes SidE-mediated toxicity.
- 20.Sulpizio A, Minelli ME, Wan M, Burrowes PD, Wu X, Sanford EJ, Shin J-H, Williams BC, Goldberg ML, Smolka MB, et al. : Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan N, Zhen X, Liu Y, Xu X, He C, Qiu J, Liu Y, Fujimoto GM, Nakayasu ES, Zhou B, et al. : Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 2019, 572:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan M, Sulpizio AG, Akturk A, Beck WHJ, Lanz M, Faça VM, Smolka MB, Vogel JP, Mao Y: Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase- domain-containing Legionella effectors. Proc Natl Acad Sci U S A 2019, doi: 10.1073/pnas.1916287116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q, Xiao Y, Chai P, Zheng P, Teng J, Chen J: ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr Biol 2019, 29:846–855.e6. [DOI] [PubMed] [Google Scholar]
- 24.Steiner B, Swart AL, Welin A, Weber S, Personnic N, Kaech A, Freyre C, Ziegler U, Klemm RW, Hilbi H: ER remodeling by the large GTPase atlastin promotes vacuolar growth of Legionella pneumophila. EmBo Rep 2017, 18:1817–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeng EE, Bhadkamkar V, Ibe NU, Gause H, Jiang L, Chan J, Jian R, Jimenez-Morales D, Stevenson E, Krogan NJ, et al. : Systematic Identification of Host Cell Regulators of Legionella pneumophila Pathogenesis Using a Genome-wide CRISPR Screen. Cell Host Microbe 2019, doi: 10.1016/j.chom.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.English AR, Voeltz GK: Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol 2013, 15:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothan HA, Kumar M: Role of Endoplasmic Reticulum-Associated Proteins in Flavivirus Replication and Assembly Complexes. Pathogens 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R: Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 2018, 16:125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanini F, Pu S-Y, Bekerman E, Einav S, Quake SR: Single-cell transcriptional dynamics of flavivirus infection. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ooi YS, Majzoub K, Flynn RA, Mata MA, Diep J, Li JK, van Buuren N, Rumachik N, Johnson aG, Puschnik AS, et al. : An RNA-centric dissection of host complexes controlling flavivirus infection. Nat Microbiol 2019, doi: 10.1038/s41564-019-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, et al. : Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535:159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, et al. : Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep 2016, 16:232–246. [DOI] [PubMed] [Google Scholar]
- 33.Lin DL, Inoue T, Chen Y-J, Chang A, Tsai B, Tai AW: The ER Membrane Protein Complex Promotes Biogenesis of Dengue and Zika Virus Non-structural Multi-pass Transmembrane Proteins to Support Infection. Cell Rep 2019, 27:1666–1674.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngo AM, Shurtleff MJ, Popova KD, Kulsuptrakul J, Weissman JS, Puschnik AS: The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neufeldt CJ, Cortese M, Scaturro P, Cerikan B, Wideman JG, Tabata K, Moraes T, Oleksiuk O, Pichlmair A, Bartenschlager R: ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat Microbiol 2019, doi: 10.1038/s41564-019-0586-3.• First study showing the requirement of host cell atlastins for efficient flavivirus infection.
- 36.Monel B, Rajah MM, Hafirassou M-L, Sid-Ahmed S, Burlaud-Gaillard J, Zhu P-P, Nevers Q, Buchrieser J, Porrot F, Meunier C, et al. : The Atlastin ER-shaping proteins facilitate Zika virus replication. J Virol 2019, doi: 10.1128/JVI.01047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM: The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep 2017, 21:1639–1654. [DOI] [PubMed] [Google Scholar]
- 38.Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR: The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 2012, 338:1072–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horenkamp FA, Kauffman KJ, Kohler LJ, Sherwood RK, Krueger KP, Shteyn V, Roy CR, Melia TJ, Reinisch KM: The Legionella Anti-autophagy Effector RavZ Targets the Autophagosome via PI3P- and Curvature-Sensing Motifs. Dev Cell 2015, 34:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang A, Pantoom S, Wu Y-W: Elucidation of the anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arasaki K, Mikami Y, Shames SR, Inoue H, Wakana Y, Tagaya M: Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat Commun 2017, 8:15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engström P, Burke TP, Mitchell G, Ingabire N, Mark KG, Golovkine G, lavarone AT, Rape M, Cox JS, Welch MD: Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nat Microbiol 2019, doi: 10.1038/s41564-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp A-K, Li L, Shi X, Zhou Z, Gao W, et al. : A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell 2019, 178:552–566.e20. ··This study reveals a crucial step of xenophagy that is inhibited by the S. Typhimurium effector SopF.
- 44.Wen X, Klionsky DJ: How bacteria can block xenophagy: an insight from Salmonella. Autophagy 2019, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau N, Haeberle AL, O’Keeffe BJ, Latomanski EA, Celli J, Newton HJ, Knodler LA: SopF, a phosphoinositide binding effector, promotes the stability of the nascent Salmonella-containing vacuole. PLoS Pathog 2019, 15:e1007959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Yao Y, Qiu X, Wang G, Hu Z, Chen S, Wu Z, Yuan N, Gao H, Wang J, et al. : Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat Immunol 2019, 20:433–446. ··By studying the L. monocytogenes toxin LLO, the authors discovered a novel mitophagy receptor that is exploited by the bacterium.
- 47.Lennemann NJ, Coyne CB: Dengue and Zika viruses subvert reticulophagy by NS2B3- mediated cleavage of FAM134B. Autophagy 2017, 13:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Lan Y, Li MY, Lamers MM, Fusade-Boyer M, Klemm E, Thiele C, Ashour J, Sanyal S: Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host Microbe 2018, 23:819–831.e5. [DOI] [PubMed] [Google Scholar]
- 49.Corona AK, Saulsbery HM, Corona Velazquez AF, Jackson WT: Enteroviruses Remodel Autophagic Trafficking through Regulation of Host SNARE Proteins to Promote Virus Replication and Cell Exit. Cell Rep 2018, 22:3304–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouna L, Hernandez E, Bonte D, Brost R, Amazit L, Delgui LR, Brune W, Geballe AP, Beau I, Esclatine A: Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1-binding proteins. Autophagy 2016, 12:327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taisne C, Lussignol M, Hernandez E, Moris A, Mouna L, Esclatine A: Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci Rep 2019, 9:4560. [DOI] [PMC free article] [PubMed] [Google Scholar]