Abstract
Psoralen is a furocoumarin natural product that intercalates within DNA and forms covalent adducts when activated by ultraviolet radiation. It is well known that this property contributes to psoralen’s clinical efficacy in several disease contexts, which include vitiligo, psoriasis, graft-versus-host disease, and cutaneous T-cell lymphoma. Given the therapeutic relevance of psoralen and its derivatives, we attempted to synthesize psoralens with even greater potency. In this study, we report a library of 73 novel psoralens, the largest collection of its kind. When screened for the ability to induce cell death, we identified two derivatives even more cytotoxic than 4’-aminomethyl-4,5’,8-trimethylpsoralen (AMT), one of the most potent psoralens identified to date. Using MALDI-TOF MS, we studied the DNA adduct formation of a subset of novel psoralens and found that in most cases enhanced DNA binding correlated well with cytotoxicity. Generally, our most active derivatives contain positively charged substituents, which we believe increase DNA affinity and enhance psoralen intercalation. Thus, we provide a rational approach to guide efforts toward further optimizing psoralens to fully capitalize on this drug class’ therapeutic potential. Finally, the structure-activity insights we have gained shed light on several opportunities to study currently underappreciated aspects of psoralen’s mechanism.
Keywords: Psoralen, SAR, Cheminformatics, MALDI-TOF, Mass spectrometry
Graphical Abstract
A library of psoralen derivatives was assessed for cytotoxicity and DNA adduct formation. Using this library, we identified several highly potent compounds and established SAR insights to guide the development of next-generation psoralens.
INTRODUCTION
Psoralen and its natural derivatives have been used therapeutically since ancient times (1). Unknowingly, folk healers applied poultices of psoralen-containing plants to their patients’ skin and exposed these areas to sunlight to treat many dermatologic ailments (2). Centuries later, El-Mofty identified psoralens as the natural products responsible for these therapeutic effects (3–5). In the decades that followed, psoralen’s formal clinical uses expanded to include vitiligo, psoriasis [using psoralen + ultraviolet A (PUVA) therapy], graft-versus-host disease (GvHD), cutaneous T-cell lymphoma (CTCL), and the pathogen reduction of blood for transfusion (6–9). Progress in chemistry enabled the discovery of additional natural and synthetic psoralens for these applications [8-Methoxypsoralen (8-MOP), 4,5’,8-Trimethylpsoralen (TMP), 4’-Aminomethyl-4,5’,8-trimethylpsoralen (AMT), and Amotosalen, to name a few] (Table S1). As psoralen’s clinical applications paralleled advances in molecular biology, it was shown that many psoralens bound covalently at 5’-TpA-3’ sites in DNA following UVA activation (10–12). Firstly, the planar structure of psoralen allows it to intercalate between DNA base pairs. Absorption of a UV photon causes reaction of the 4’,5’ double bond with a closely situated thymine, forming a 4’,5’-monoadduct. Absorption of a second UV photon causes reaction at the 3,4 double bond with another thymine, forming an interstrand crosslink (ICL). 254 nm radiation can ‘photoreverse’ some ICLs back to 4’,5’- or 3,4-monoadducts (13, 14). 3,4-monoadducts can form as the initial monoadduct species but with lower efficiency (15, 16). The formation of monoadducts and ICLs throughout the genome is the most well-characterized property believed responsible for psoralen’s clinical efficacy (Figure S1) (17, 18). Thus, it follows that optimizing psoralens for increased DNA adduct formation should translate to greater potency and therapeutic effects.
The first effort to enhance psoralen potency synthetically was made by Hearst and colleagues, who identified AMT in 1977 (19). In 1993, the Cerus Corporation patented Amotosalen for its INTERCEPT™ pathogen inactivation system (20, 21). Both AMT and Amotosalen are 4’-primaryamino-substituted psoralens, suggesting that this modification is generally beneficial to psoralen’s mechanism. However, few comprehensive structure-activity relationship (SAR) studies on psoralen exist to validate such claims. The pertinent studies report only a few compounds at a time, making it difficult to establish any generalizable principles (16, 22–25). Moreover, applying modern medicinal chemistry approaches to screen a library of psoralens could identify derivatives with unprecedented potencies and minimal undesired off-target effects. Given that psoralen’s clinical uses are well-established, such derivatives would also have immediate therapeutic relevance. In this report, we return to the pursuit of psoralen optimization and seek answers to the following questions: (1) What substituents improve psoralen activity and what mechanism do such enhancing modifications leverage? (2) What are the structural and chemical considerations that define the sensitivity of psoralen to derivatization? Beyond answering such questions, we hoped to show that opportunities still exist to optimize psoralens and gain a more complete understanding of their mechanism.
We began by synthesizing one of the largest and most diverse psoralen libraries reported to date. Several novel derivatives emerged from this library that are among the most cytotoxic UV-activated psoralens ever identified. Using cheminformatics approaches, we identified substituent properties (charge, lipophilicity, substitution position, sterics) that are generally beneficial or detrimental to psoralen potency. Lastly, we develop a model to explain the enhanced potency of our derivatives in the context of their DNA adduct formation using a high-throughput mass spectrometry method. For several derivatives, we show that DNA adduct formation may not be the only pathway contributing to cytotoxicity. Taken together, our findings indicate that the pursuit of psoralen optimization is far from over and that further synthetic and screening efforts are warranted to fully realize the therapeutic potential of this drug class. We anticipate that the synthetic derivatives in this report and those emerging from future studies will serve as valuable research tools and potential therapeutic candidates in this promising area of translational medicine (26).
MATERIALS AND METHODS
Syntheses and Characterization of Psoralen Derivatives.
Full syntheses and characterization of all psoralen derivatives can be found in the chemical supporting information. 8-MOP (M3501), TMP (T6137), and AMT (A4330) were all purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. For all in vitro and cellular assays, derivatives were prepared as DMSO stock solutions and stored at −20°C. UV-visible spectra for several psoralen derivatives (dissolved at 0.5 mM in DMSO) were obtained using a NanoDrop™ ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Cytotoxicity Screen – Psoralen Treatment, UV Irradiation, Cell Proliferation and Viability Assay.
UV irradiation was carried out in 6- or 96-well plates using a Stratalinker® UV Crosslinker Model 2400 (Statagene, La Jolla, CA) at the UVA doses indicated in the figures. The B16 murine melanoma cell line (ATCC® CRL-6322™) was obtained from ATCC® and cultured according to recommended guidelines. All cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C and 5% CO2. Typically, 200 cells were plated in each well of a 96-well plate and allowed to adhere/grow overnight at 37°C and 5% CO2. Each psoralen required four independent wells, corresponding to our two experimental conditions (No UVA and +1 J UVA). All psoralen derivatives were screened in duplicate. All cells were exposed to 1.0 μM psoralen derivative and allowed to incubate for up to 60 min away from light at 37°C and 5% CO2. Appropriate wells were then irradiated with 1 J UVA (365 nm). At this light dose, approximately 3 × 104 photons are delivered per psoralen molecule. After irradiation, plates were returned to 37°C and 5% CO2 before cell proliferation quantification. Media was discarded and cells were washed once with phosphate buffered saline. Cell proliferation was assayed using the WST-1 kit and protocol without further modification (Abcam, Cambridge, United Kingdom).
Screening Psoralen DNA Adduct Formation using MALDI-TOF MS.
An oligonucleotide with sequence 5’-TATATATATATA-3’ [herein referred to as (TA)6] was synthesized by Integrated DNA Technologies (Coralville, IA) and purified by standard desalting procedures. A 1 mM stock solution of (TA)6 was prepared in ultrapure water. In a UV-transparent 96-well plate, three 100 μL reactions were prepared for each psoralen by coincubating (TA)6 and psoralen at equimolar concentration (100 μM final) in ultrapure water. We observed that conducting reactions in the presence of a Tris-EDTA buffering system interfered with downstream sample analysis by Matrix-Assisted Laser Desorption/Ionization Time‐of‐Flight Mass Spectrometry (MALDI-TOF MS), possibly due to the additional salts. The plate was then pre-incubated away from light for 30 minutes at 4°C to allow for psoralens to intercalate within (TA)6 dsDNA species. Following pre-incubation, all reactions were exposed to 10 J UVA (365 nm) on ice to limit dissociation of dsDNA species. Irradiations were performed using a Stratalinker® UV Crosslinker Model 2400.
3,4-Diaminobenzophenone (191760) was purchased from Sigma-Aldrich and used without further purification. Sample preparation was based on a previous report utilizing 3,4-Diaminobenzophenone as a MALDI-TOF matrix for oligonucleotides (27). To summarize, matrix solutions were prepared freshly each day by dissolving 3,4-Diaminobenzophenone at 15 mg/mL in acidified methanol (methanol/HCl = 80:3 v/v). 2 μL of each psoralen/DNA reaction was mixed with an equal volume of matrix solution and allowed to incubate for 3 min. 2 μL of this mixture was then applied to a MTP 384 polished steel MALDI-TOF MS target plate and allowed to dry under ambient conditions.
All mass spectrometry analysis was conducted with an autoflex speed MALDI-TOF system running flexControl (Bruker Daltonics, Billerica, MA). Prior to all analyses, the instrument was calibrated using Bruker’s mass calibration standard. All MALDI-TOF spectra presented are the sum of 10 separate laser shots (50% power) throughout the dried sample. All downstream data analysis (baseline subtraction, mass identification, intensity quantification, etc.) was performed using Bruker’s flexAnalysis software (version 3.4).
Cheminformatics and Visualization Software.
Activity Cliff analyses, Similarity analyses, and Self-organizing maps were generated using the open-source DataWarrior V5.0.0 program. 3D conformations for modeled psoralen derivatives were obtained in MarvinSketch, which uses a Dreiding force field approximation. Molecular modeling of psoralen monoadducts and ICLs was performed in UCSF Chimera using the published NMR structures for a 4’-Hydroxymethyl-4,5’,8-trimethylpsoralen (HMT) 4’,5’-monoadduct (PDB 203D) and a HMT ICL (PDB 204D) (28).
RESULTS
Psoralen Library Design and Synthesis
A library of 73 psoralens was synthesized with diverse substituents positioned around the psoralen core (Table 1 and Table S2). To our knowledge, this is the largest and most diverse psoralen library reported to date. We represented compounds with alkyl, aromatic, halide, and H-bond donors/acceptors at synthetically feasible positions (Figure S2A). Molecular similarity analyses showed that most compounds can be grouped into three major networks: (1) alkylation throughout the psoralen core, (2) alkoxylation at the 8-position, and (3) addition of charged substituents at the 5- or 8-positions (Figure S2B). In addition, several compound pairs were designed to study the effects of subtle methyl modifications (Figure S2C) (29). Lastly, some derivatives bear ring substituents fused to the 4’,5’- or 3,4-double bonds. We proposed that these compounds would either lose the ability to intercalate within DNA or be restricted to forming monoadducts only. Conversely, if these derivatives somehow maintained potency without forming DNA adducts, they could be a useful system to study alternate mechanisms of action.
Table 1. Novel Psoralen Library.
A library of 73 novel psoralen derivatives was synthesized. Structures and cytotoxicities (± UV) are provided.
| Cytotoxicity Rank | Identifier | Structure | Cell Survival (%), No UVA | Cell Survival (%), + 1J UVA |
|---|---|---|---|---|
| 19 | 1A | 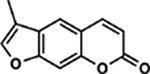 |
96.6 ± 1.9% | 27.6 ± 3.0% |
| 46 | 2A | 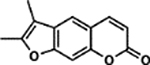 |
91.7 ± 4.1% | 91.5 ± 3.5% |
| 54 | 3A | 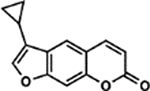 |
99.8 ± 3.3% | 95.4 ± 2.4% |
| 63 | 4A | 96.4 ± 2.3% | 98.9 ± 3.5% | |
| 69 | 5A | 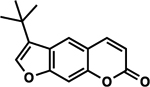 |
97.2 ± 4.0% | 102.8 ± 3.9% |
| 70 | 6A | 101.5 ± 2.9% | 105.0 ± 3.6% | |
| 55 | 7A | 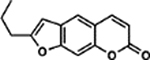 |
99.2 ± 3.6% | 95.4 ± 3.2% |
| 29 | 8A | 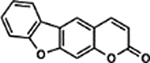 |
97.1 ± 2.5% | 71.0 ± 4.4% |
| 62 | 9A | 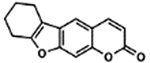 |
89.2 ± 2.9% | 97.1 ± 3.0% |
| 59 | 10A | 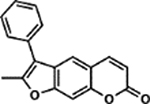 |
92.3 ± 2.8% | 96.5 ± 4.7% |
| 11 | 1B | 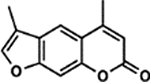 |
93.1 ± 4.6% | 2.20 ± 0.81% |
| 27 | 2B | 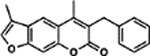 |
100.7 ± 3.0% | 70.7 ± 3.0% |
| 10 | 3B | 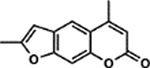 |
106.3 ± 3.4% | 1.60 ± 0.69% |
| 56 | 4B | 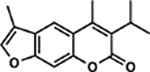 |
99.4 ± 4.6% | 95.6 ± 4.3% |
| 17 | 5B | 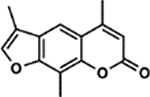 |
98.7 ± 4.8% | 21.4 ± 3.7% |
| 16 | 6B | 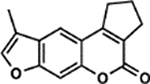 |
109.6 ± 5.2% | 8.6 ± 1.7% |
| 14 | 7B | 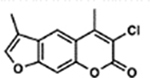 |
95.8 ± 4.0% | 3.6 ± 1.5% |
| 15 | 8B |  |
97.2 ± 6.2% | 4.1 ± 2.2% |
| 22 | 9B | 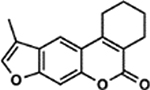 |
96.5 ± 2.8% | 39.4 ± 1.2% |
| 35 | 10B | 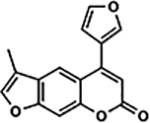 |
82.2 ± 7.4% | 83.7 ± 8.0% |
| 33 | 1C | 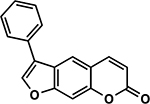 |
107.5 ± 4.8% | 83.4 ± 5.4% |
| 47 | 2C | 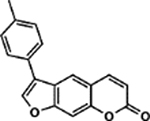 |
110.5 ± 5.9% | 91.7 ± 3.6% |
| 64 | 3C |  |
102.0 ± 3.0% | 99.8 ± 3.1% |
| 28 | 4C |  |
103.6 ± 3.3% | 70.9 ± 8.0% |
| 36 | 5C |  |
106.4 ± 4.4% | 83.9 ± 3.8% |
| 45 | 1D |  |
102.0 ± 4.2% | 90.5 ± 4.9% |
| 50 | 2D |  |
107.8 ± 5.2% | 93.6 ± 5.4% |
| 67 | 3D |  |
96.3 ± 4.9% | 102.7 ± 4.6% |
| 34 | 4D |  |
99.8 ± 4.4% | 83.5 ± 5.7% |
| 68 | 5D |  |
96.1 ± 4.8% | 102.7 ± 6.1% |
| 52 | 6D |  |
97.5 ± 5.1% | 94.4 ± 3.6% |
| 60 | 7D |  |
86.3 ± 3.9% | 96.6 ± 5.4% |
| 31 | 8D |  |
95.2 ± 4.8% | 79.2 ± 5.8% |
| 41 | 9D |  |
95.3 ± 4.6% | 89.2 ± 5.7% |
| 43 | 10D |  |
101.6 ± 5.5% | 90.1 ± 4.0% |
| 73 | 1E (Xanthotoxol) | 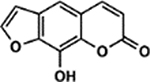 |
101.9 ± 3.6% | 111.6 ± 4.4% |
| 38 | 2E | 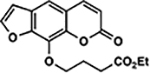 |
103.0 ± 2.9% | 85.7 ± 2.6% |
| 72 | 3E | 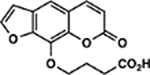 |
111.3 ± 4.2% | 108.2 ± 5.1% |
| 49 | 4E | 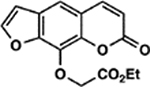 |
101.5 ± 5.8% | 93.0 ± 7.1% |
| 71 | 5E | 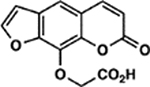 |
110.2 ± 4.3% | 107.7 ± 3.3% |
| 2 | 6E | 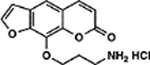 |
109.9 ± 5.4% | 0.23 ± 0.16% |
| 24 | 7E | 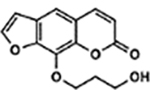 |
117.2 ± 3.5% | 54.0 ± 7.0% |
| 6 | 8E | 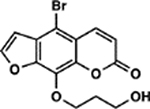 |
104.5 ± 4.0% | 0.62 ± 0.15% |
| 74 | 9E | 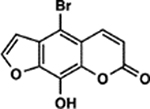 |
108.1 ± 3.8% | 112.6 ± 4.2% |
| 25 | 10E | 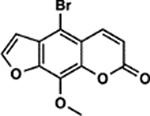 |
110.9 ± 4.7% | 60.4 ± 4.4% |
| 3 | 1F | 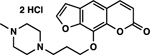 |
100.7 ± 3.9% | 0.43 ± 0.22% |
| 5 | 2F | 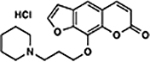 |
92.5 ± 1.9% | 0.50 ± 0.28% |
| 30 | 3F | 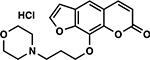 |
91.4 ± 3.7% | 71.2 ± 3.1% |
| 51 | 4F |  |
96.0 ± 2.5% | 94.0 ± 5.5% |
| 13 | 5F |  |
99.4 ± 4.2% | 2.7 ± 0.6% |
| 23 | 6F (Psoralen) |  |
97.4 ± 2.3% | 48.3 ± 3.6% |
| 26 | 7F |  |
103.8 ± 5.1% | 64.0 ± 6.0% |
| 8 | 8F |  |
97.1 ± 3.2% | 0.90 ± 0.35% |
| 57 | 9F |  |
99.2 ± 5.2% | 96.3 ± 3.1% |
| 66 | 10F | 90.9 ± 2.9% | 101.6 ± 2.4% | |
| 40 | 1G |  |
94.3 ± 3.1% | 85.9 ± 3.8% |
| 44 | 2G | 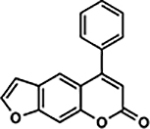 |
85.8 ± 2.7% | 90.3 ± 3.5% |
| 39 | 3G | 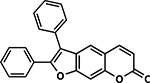 |
88.5 ± 7.1% | 85.8 ± 4.2% |
| 12 | 4G | 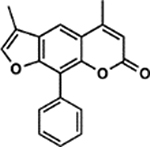 |
86.8 ± 2.5% | 2.7 ± 1.0% |
| 37 | 5G | 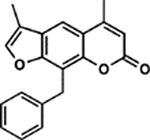 |
103.4 ± 3.7% | 85.0 ± 6.6% |
| 9 | 6G |  |
105.5 ± 5.2% | 0.94 ± 0.47% |
| 42 | 7G |  |
95.2 ± 5.8% | 89.5 ± 6.0% |
| 61 | 8G |  |
103.2 ± 5.1% | 96.9 ± 4.6% |
| 53 | 9G |  |
93.9 ± 4.7% | 95.0 ± 6.4% |
| 58 | 10G | 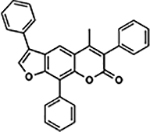 |
98.6 ± 3.7% | 96.3 ± 2.1% |
| 18 | 1H | 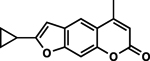 |
92.8 ± 2.4% | 22.1 ± 3.4% |
| 32 | 2H | 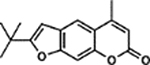 |
94.1 ± 4.1% | 81.2 ± 3.0% |
| 48 | 3H | 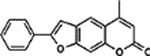 |
92.5 ± 2.3% | 92.4 ± 2.8% |
| 7 | 4H | 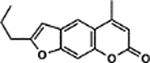 |
97.5 ± 5.1% | 0.85 ± 0.37% |
| 20 | 5H | 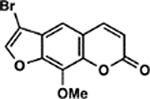 |
93.4 ± 4.8% | 31.1 ± 4.7% |
| Control Compounds | ||||
| 65 | DMSO | - | 100.0 ± 4.6% | 101.3 ± 4.6% |
| 1 | TMP | 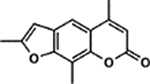 |
103.6 ± 3.6% | 0.15 ± 0.15% |
| 4 | AMT | 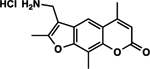 |
103.3 ± 2.8% | 0.43 ± 0.22% |
| 21 | 8-MOP |  |
88.1 ± 2.8% | 31.3 ± 3.7% |
Screening Derivatives for UV-dependent Cytotoxicity
All psoralen derivatives were screened for their UV-dependent ability to induce cell death. We screened compounds in B16 murine melanoma cells, a cell line commonly employed for the study of human melanomas and cancer metastasis. To rule out any off-target toxicity, we first screened the library in the absence of UVA (termed ‘dark toxicity’). In general, psoralen derivatives were fairly inert without UVA activation (Figure 1, inset). Among all derivatives, the greatest dark toxicity observed was ~18% cell death from a psoralen bearing a 4’-methyl and 4-furan group (10B). As expected, a 1 J dose of UVA dramatically enhanced the toxicity of many compounds and led us to identify several novel psoralens with high potencies (Figure 1).
Figure 1.
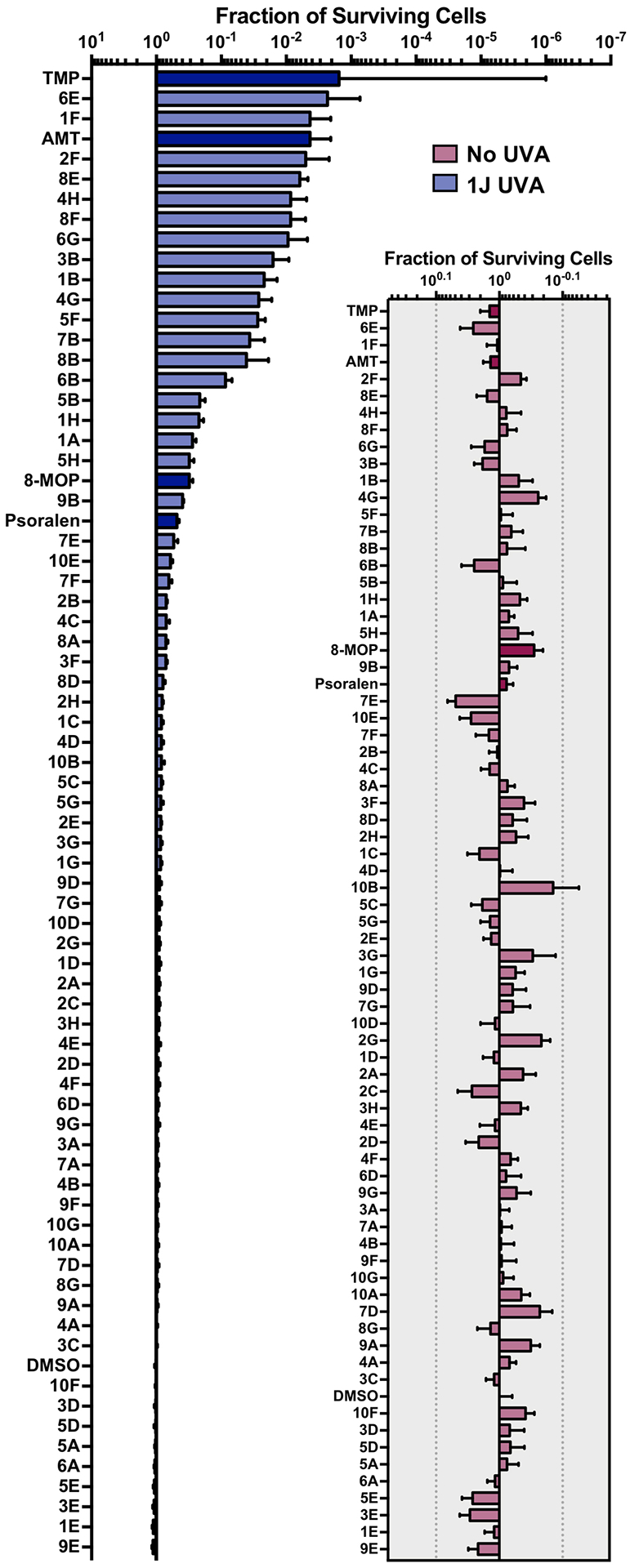
Screening Psoralens for Enhanced UV‐dependent Cytotoxicity. 73 psoralen derivatives were screened for their ability to reduce cell proliferation in the B16 murine melanoma cell line. 1.0 μm of each compound was incubated with cultured cells followed by irradiation with 1J UVA (365 nm). 4,5′,8‐Trimethylpsoralen (TMP), 4’‐Aminomethyl‐4,5’,8‐trimethylpsoralen (AMT), 8‐Methoxypsoralen (8‐MOP) and unmodified psoralen were included as benchmarks for assessing increases or decreases in potency (bars with darker shading). Psoralens are ranked according to their mean UV‐dependent toxicity. 1H and 4B correspond to the 25th and 75th percentiles for our dataset, respectively. (Inset) Without UVA activation, psoralen derivatives are generally inert. Among all derivatives, 10B was the most toxic without UVA activation, inducing ~18% cell death. All psoralen derivatives were screened in duplicate (n = 2).
Electropositive Substituents Enhance Psoralen Potency
When examining the structural motifs among our highly active psoralens, we observed that diverse charged and lipophilic substituents at several positions could enhance potency. The top three novel psoralens (6E, 1F, and 2F) all contain 8-position substituents that are positively charged under physiologic conditions (Figure S3). AMT, similarly, bears a positively charged 4’-aminomethyl group. Interestingly, an analog (3F) bearing a neutral morpholino group showed weak activity in our cell viability assay (Figure 2A). This finding argues that psoralen’s UV-dependent mechanism of action is enhanced by electropositive substituents. Given the polyanionic DNA backbone, such substituents may enhance DNA affinity and intercalation efficiency via electrostatic interactions (Figure 2B). Conversely, negatively charged substituents might be expected to exhibit ionic repulsion with the DNA backbone, resulting in reduced potency. 7G, 3E, and 5E all bear carboxylic acid groups that are negatively charged at physiologic pH and all showed negligible toxicity in our screen (Table 1).
Figure 2.
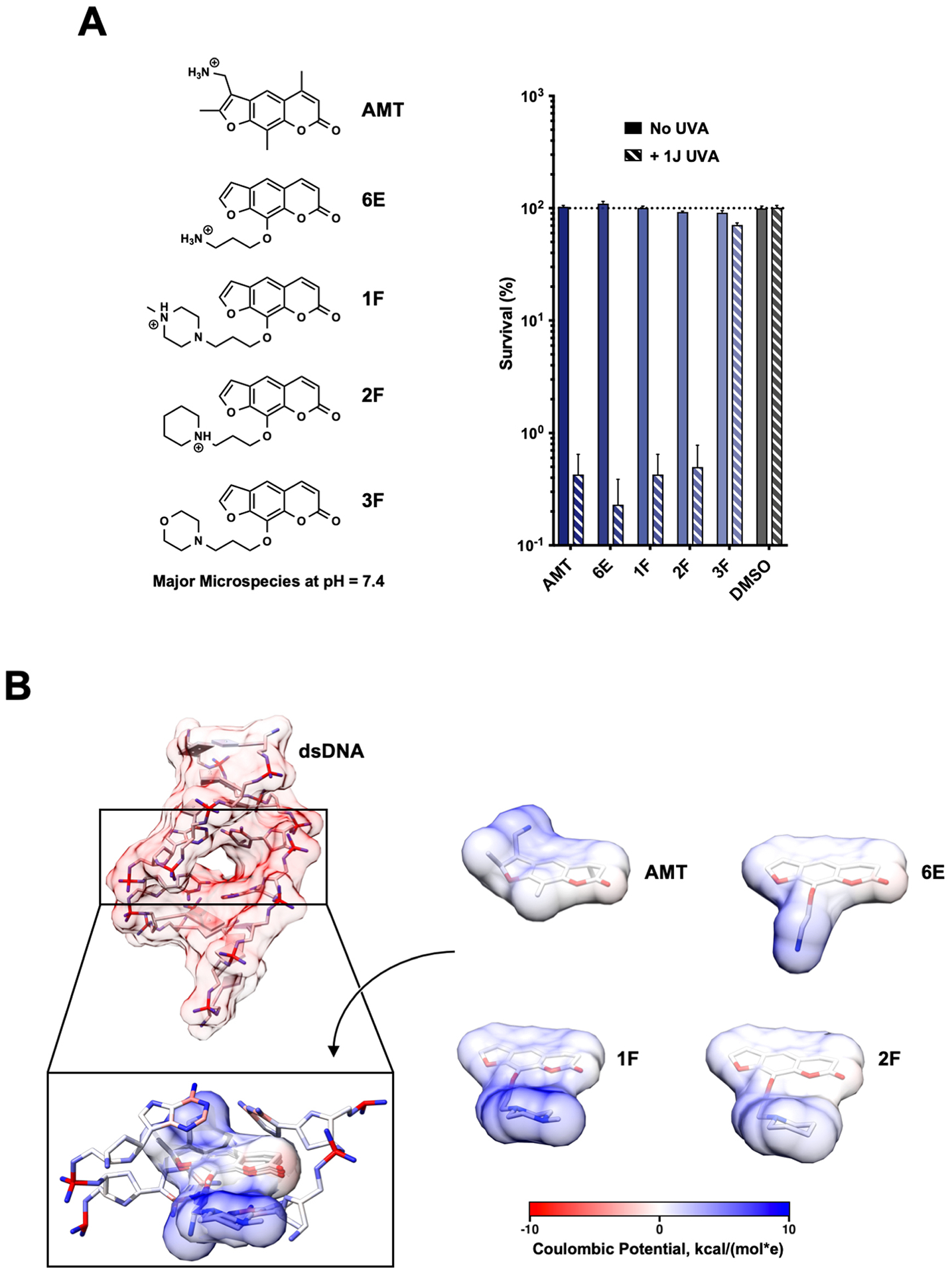
Electropositive Substituents Increase Potency. (a) The cytotoxicities for several of the most potent psoralens identified in our screen. 3F, a psoralen with a similarly sized but neutral 8‐position substituent at physiological pH, is also included. The major microspecies for each psoralen compound at pH 7.4 is illustrated. (b) Calculated Coulombic potentials mapped onto the atoms/surface of a dsDNA helix (PDB 204D) and the surfaces of several potent psoralens. 3D conformations of psoralen derivatives were optimized using the Dreiding force field (calculated in MarvinSketch). Surface coloring is based on the potential 1.4 Å away from the surface (calculating using UCSF Chimera) and assuming a distance‐dependent dielectric (ε = 4).
SAR Insights from Psoralen Library
Although several highly potent psoralens were identified in our screen, most derivatives lost activity after modification. This afforded an opportunity to study the properties of substituents that disrupt psoralen activity. Our first attempt to use cLogP, a computationally derived measure of compound hydrophilicity, to predict psoralen activity was unsuccessful, indicating that factors more complex than cell permeability contribute to potency (Figure S4). Since the importance of psoralen’s DNA interaction has been well validated, we proposed that the need for intercalation might impose steric constraints on effective psoralens. Based on the predicted binding mode, certain sites on psoralen are predicted to be more sensitive to modification. Generally, substituents on the 4’,5’ and 3,4 double bonds would need to be sufficiently compact to allow for proper intercalation geometries (Figure 3A). Additionally, it is thought that the 4’,5’-monoadduct forms most efficiently, which suggests that ICL formation may be more sensitive to 4’,5’ modifications (Figure S1) (30–33). The 5- and 8-positions of psoralen are solvent exposed, so we expected that these sites would be more tolerant to a wider range of substituent sizes. The overwhelming majority of natural psoralens are also modified at these sites, suggesting the tolerance of psoralen’s endogenous functions to such substituents (Table S1).
Figure 3.
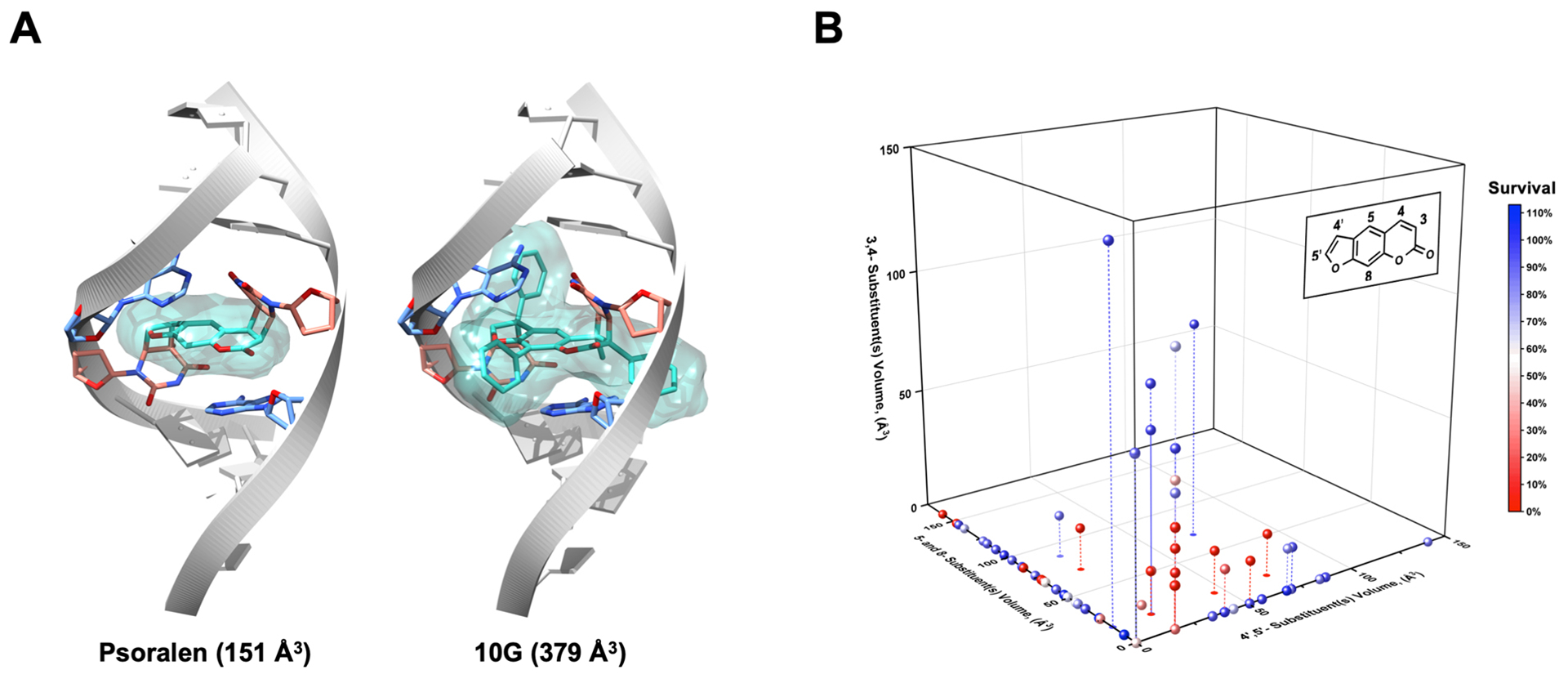
Size and Positional Constraints on Effective Psoralen Substituents. (a) Using PDB 204D, models of psoralen and 10G ICLs were built to illustrate the possibility for steric clashes with the dsDNA helix when various modifications are made to psoralen. (b) To stratify derivatives according to the sites of their substituents, the psoralen scaffold was divided in three sections: (1) 4′,5′‐ substituents, (2) 5‐ and 8‐ substituents and (3) 3,4‐ substituents. The van der Waals volumes for all substituents were computed in MarvinSketch, and all derivatives were graphed in three dimensions based on the magnitude and position of their substituents’ volumes. To provide context, the volume of a methyl substituent was computed as ~17 Å3 and that of a phenyl was ~80 Å3. Points are colored based on their overall cytotoxicity. Droplines are included when necessary to clarify the three‐dimensional position of several points.
To study the relationship between substituent size, positioning, and psoralen potency, we computed the Van der Waals volumes of all substituents in our library and stratified them according to position (Figure 3B). Our first observation was any 4’,5’ substituents beyond a methyl group (i.e. 1A) resulted in a nearly complete loss of potency when they were the only modification made to psoralen. Our library contains two derivatives with only 3,4 substituents: 1G, and 2G. These derivatives, which bear relatively large phenyl substituents at the 3- and 4-position showed minimal cytotoxicity (~86–90% survival). 5F, which bears a fused pyridine ring on the 3,4 double bond, was surprisingly potent (2.7 ± 0.6% survival) for a derivative seemingly restricted to 4’,5’-monoadduct formation. However, both 6B and 9B bear fused rings on the 3,4 double bond and these derivatives also exhibited reasonable potency (8.6 ± 1.7% and 39.4 ± 1.2% survival, respectively). The 8-position was indeed amenable to substituents of different sizes with our most potent derivatives possessing large, positively charged groups at this site (e.g. 6E, 1F, 2F) (Figure 3B). However, in a series of compounds where the length of an 8-position alkoxy chain was increased stepwise, we observed losses in potency as chain length increased (Figure S5). This may be explained by steric or cell permeability issues that arise when long alkyl chains are appended to psoralen, even at solvent exposed sites. In the cases of our most potent derivatives, the benefits of positively charged groups may outweigh the negative effect of placing long alkyl chains at the 8-position. To minimize the influence of properties besides size, we analyzed only derivatives with alkyl substituents. We found that psoralens could remain potent when alkyl substituents of <50 Å3 total volume were added at either the 3,4 or 4’,5’ double bonds (Figure S6). Despite the tolerance of both double bonds to equally sized substituents, differences arose among derivatives with rings fused to the 3,4 and 4’,5’ double bonds. Compounds with a 3,4 fused ring could still be cytotoxic, while their 4’,5’ fused ring counterparts could not. In addition, when rings are fused to the 4’,5’ double bond, DNA adduct formation is lost, supporting the model that the 4’,5’-monoadduct is favored (Figure S7).
Interestingly, we observed several instances where psoralens with previously detrimental 4’,5’ modifications could be rescued by addition of a 4-methyl group. An Activity Cliff analysis enabled us to hone in on several examples of this phenomenon (Figure 4A) (34). For example, 7A and 4H both contain a 5’-n-propyl group, but addition of a 4-methyl group in 4H results in a compound that induces 99% cell death compared to 7A, which induces only 5% cell death. 6G is an intriguing compound because addition of 4- and 8-position methyl groups results in a psoralen that induces >99% cell death. For the psoralen that only bears the 4’-phenyl group without these methyl substitutions (1C), only 17% cell death is observed. There are, however, some compounds that are not profoundly impacted by the addition of a 4-methyl group (6A vs. 2H and 10F vs. 3H), but this may result from the magnitude of preexisting bulk at the 5’-position preventing any 4’,5’-monoadduct formation (Figure 4B). A prior report showed that the 4-methyl group dramatically predisposes psoralens to 4’,5’-monoadducts and diminishes 3,4-monoadducts (16). Thus, how the 4-methyl group enhances previously detrimental 4’,5’ substitutions remains to be understood and will require further biochemical or photophysical analyses. In compounds where larger pyrone modifications were made in addition to the 4-methyl, activity was lost (e.g. 2B, 4B, 9G). This suggests that gains in potency due to pyrone modification are a balancing act and can be nullified when the added bulk is too great (35).
Figure 4.
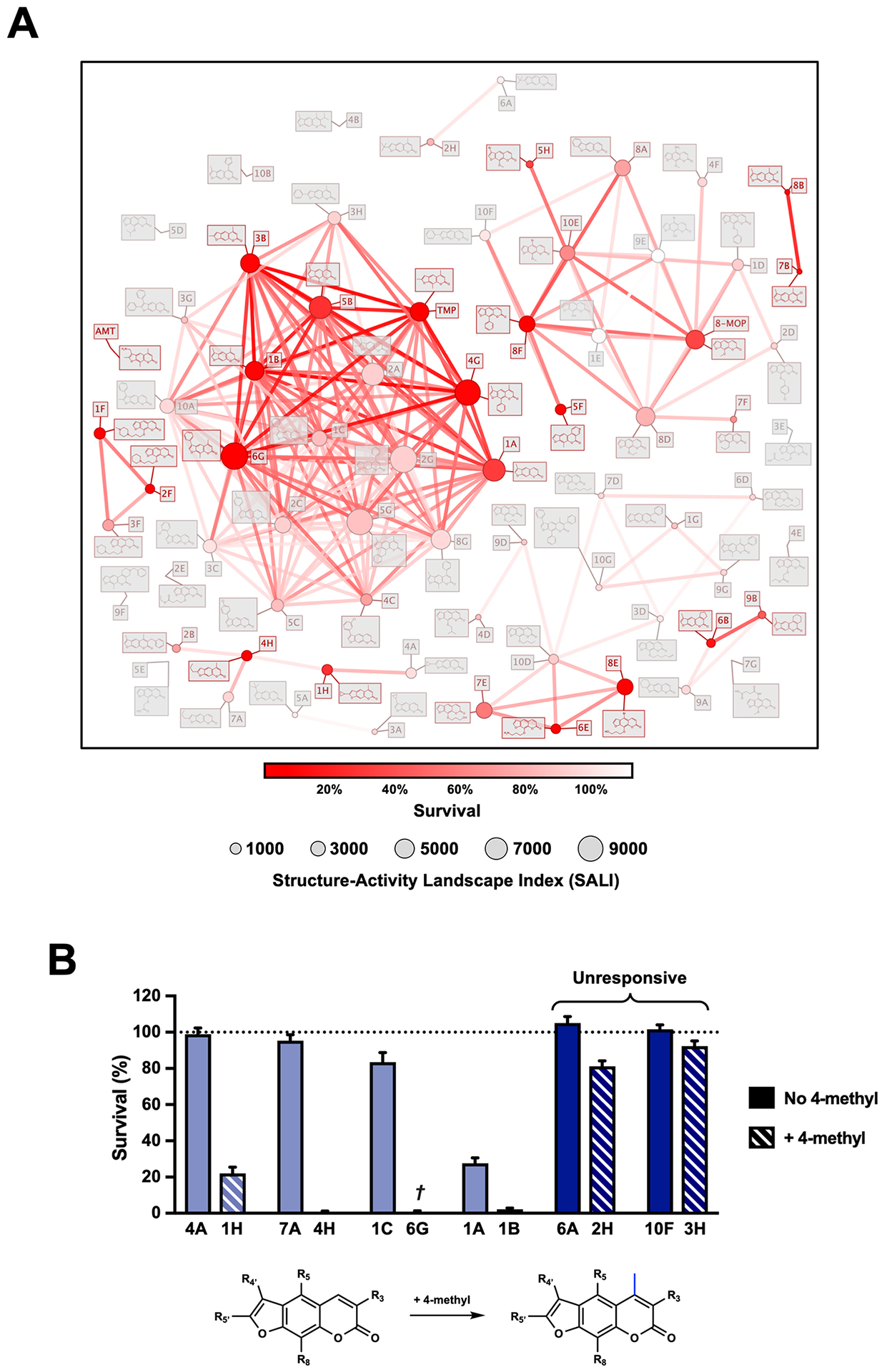
Cheminformatics Approaches for Psoralen SAR. (a) An activity cliff analysis of our library was performed using DataWarrior. This analysis enables one to visualize potency gained (or lost) when various modifications are made to psoralen. All points are graphed in two‐dimensional space based on a Rubberbanding Forcefield approach to position derivatives closest to their most similar neighbors (akin to a similarity analysis). Points and edges are colored based on the derivatives’ cytotoxicities. The structure–activity landscape index (SALI) is calculated between all pairs of similar molecules and is denoted by a point’s size. SALI is calculated based on the molecules’ activities (a1 and a2) and similarity score (s): SALI = |a1‐a2|/(1‐s). (b) A recurrent activity cliff is observed when a 4‐methyl group dramatically enhances the potency of several derivatives. †6G also bears an 8‐methyl in addition to the 4‐methyl. Darker shaded bars represent the two exceptions to this observation.
Lastly, our library afforded an opportunity to study the mechanistic basis of psoralen cellular toxicity. It is still unclear whether psoralens exert their toxicity primarily via DNA adduct formation or whether other pathways are equally contributory. To better assess such theories, we measured the in vitro DNA adduct formation of several highly potent psoralens we identified.
DNA Adduct Formation Reveals Basis for Enhanced Activity
To study the effects of substitution on DNA adduct formation, we selected a panel of novel psoralens with varied activities in our initial screen (Table 1). We first obtained UV-visible spectra for all derivatives to determine how these modifications changed spectroscopic properties (Figure S8). We found that our modifications changed psoralen’s absorbance in the UVA region moderately. Previously, we reported a high-throughput method to detect psoralen adducts using Surface‐Enhanced Laser Desorption/Ionization Time‐of‐Flight Mass Spectrometry (SELDI‐TOF MS) (36). SELDI-TOF MS offers the advantage of on-chip sample purification, which we leveraged in our previous method (37, 38). However, given the depreciation and obsolescing of SELDI-TOF MS, we developed an assay using state-of-the-art MALDI-TOF MS due to easy instrument accessibility and improved reproducibility (39). 6-Aza-2-thiothymine, the first MALDI-TOF matrix we tested for its efficacy with oligonucleotides, suffered from cation adduct species and required high laser intensities (40). Of all matrices tested, we found that 3,4-Diaminobenzophenone showed the best reproducibility, signal-to-noise ratio, and suppression of cation adducts (Figure S9).
All psoralens were incubated with a 12bp oligonucleotide composed of alternating dT:dA base pairs [5’-TATATATATATA-3’, referred to as (TA)6]. This oligonucleotide should anneal with itself and afford many possible hybrid species with sites for psoralen intercalation. Following exposure to 10 J UVA, all psoralen reactions were analyzed by MALDI-TOF MS (Figure 5A). Both 6E and 1F showed significantly higher adduct formation than AMT (p<0.0001 and p=0.0128, respectively) (Figure 5B). Our results indicate that for many derivatives, increases in DNA adduct formation lead to greater cytotoxicity (Figure 5C). This relationship is most clearly observed in the cases of 6E and 1F, which testify to the importance of DNA modification among psoralen’s cytotoxic mechanisms. Surprisingly, several highly toxic psoralens exhibited low DNA adduct levels (8E, 8B, 5H, and 8F). Interestingly, 8E, 8B, and 5H all contain halogen substituents. Indeed, 8E is significantly more toxic than its non-brominated counterpart (7E), indicating that the bromine substituent may account for this added effect (Table S2). 8F, which bears a phenyl at the 8-position, forms DNA adducts slightly less efficiently than 8-MOP yet is significantly more cytotoxic (0.90 ± 0.35% survival). This discrepancy may point towards a non-DNA mediated mechanism of action. Thus, our approach is capable of parsing out whether DNA adduct formation or alternate pathways are responsible for a given psoralen’s toxicity. In cases where DNA adduct formation cannot account for a psoralen’s toxicity, it will be interesting to assess whether additional cellular components are identified as specific targets of these psoralens.
Figure 5.
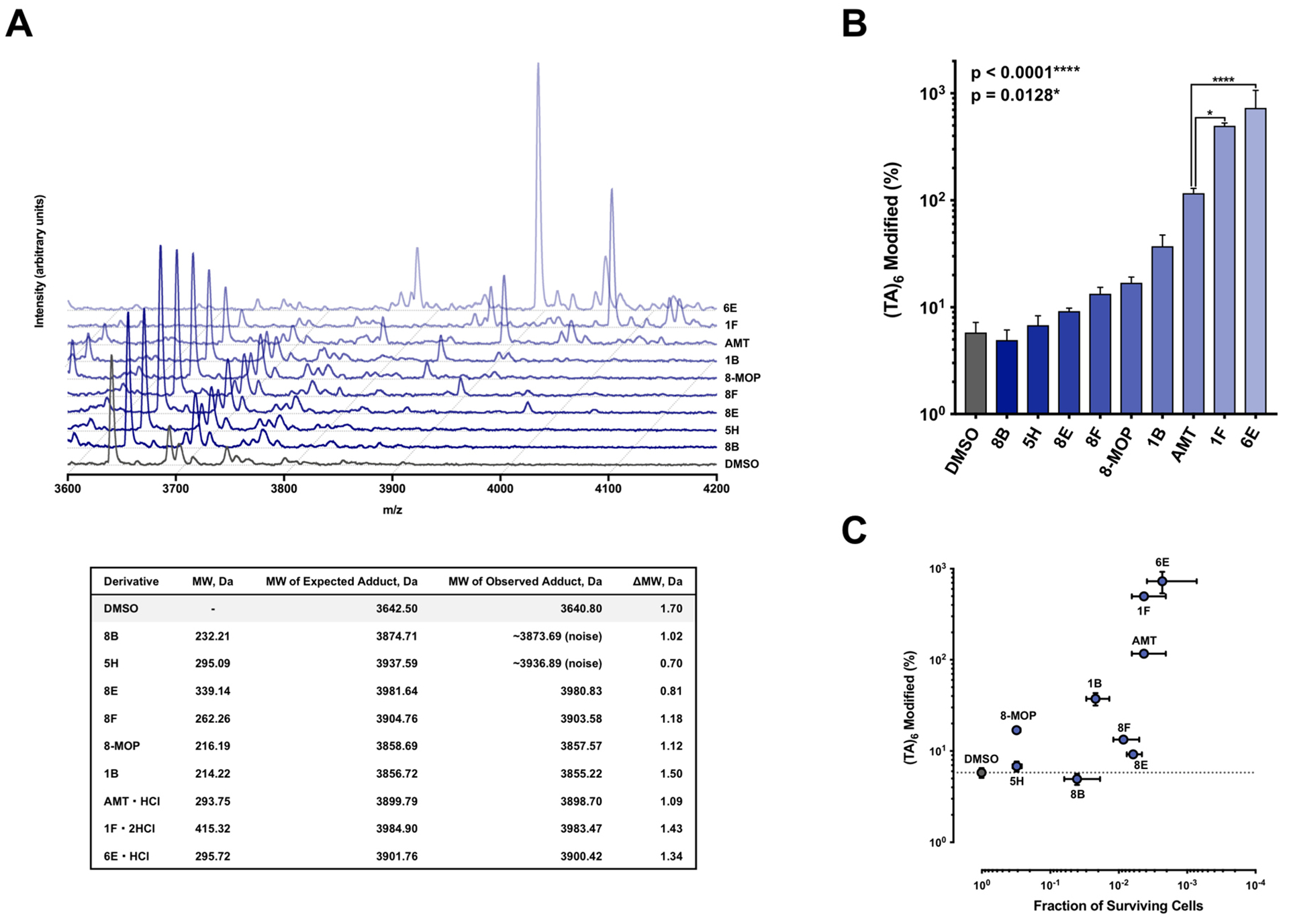
MALDI‐TOF Analysis of Psoralen‐DNA Photoadduct Formation. (a) Psoralen derivatives were incubated in the presence of a 12‐mer alternating dT:dA oligonucleotide [(TA)6, 3642.50 m/z] and irradiated with a dose of 10 J UVA (365 nm) followed by MALDI‐TOF MS analysis. Representative MS spectra for all psoralen compounds tested are overlaid in a waterfall plot. Table below provides expected and observed masses for all adduct species along with the measurement error. (b) Mean DNA adduct levels for each psoralen derivative (n = 3). Statistically significant differences were observed between AMT and 6E (Unpaired t‐test, P = 0.0128) and between AMT and 1F (Unpaired t‐test, P < 0.0001). (c) A log–log plot of DNA adduct formation versus cytotoxicity.
DISCUSSION
Given the well-established therapeutic value of psoralen and its derivatives, we sought to revisit the space of psoralen optimization and gain SAR insights into this drug class. From a library of 73 psoralens, we discovered several that are among the most cytotoxic ever identified. Next, we used cheminformatic approaches to study the substituent properties that generally increase or decrease psoralen activity. We found that substituent size and charge exerted dramatic effects that might have been underappreciated from past SAR studies. Lastly, we assessed whether the enhanced cytotoxicity could be explained by increased DNA adduct formation. For several psoralens, the direct correlation between DNA modification and cytotoxicity held true. However, there are a few psoralens that defy this expectation and exhibit extraordinary potency despite low DNA adducts. Thus, while DNA adduct formation may be a potent driver of psoralen activity, we cannot rule out alternate means by which these molecules function. Consequently, many avenues could exist for identifying potent psoralens.
Our most toxic psoralens bear positively charged substituents that enhance their ability to intercalate within the polyanionic dsDNA helix. We have not yet identified a psoralen bearing a positively charged substituent that performs poorly in our cytotoxicity or DNA adduct formation assays. Therefore, a library focused on positively charged substituents may be the most straightforward approach to identify even more cytotoxic psoralens. Interestingly, among the psoralens believed to function primarily by modifying DNA, enhances in adduct formation appear to show diminishing returns for cytotoxicity. For example, AMT shows significantly fewer DNA adducts than 1F or 6E, yet all three derivatives have >99% toxicity. It may be that all these derivatives meet a necessary threshold of DNA adducts needed to induce cell death. In addition, the screening concentration may have been too high to detect subtler differences among these highly potent psoralens. Work is underway to establish the EC50 (concentration of half-maximal cytotoxicity) for 6E and 1F relative to AMT. Nonetheless, it will be interesting to follow whether these more potent derivatives have a better therapeutic window than prior psoralens in preclinical models. AMT was recently implemented in a novel therapy for solid tumors termed X-PACT (X-ray Psoralen Activated Cancer Therapy) (41). 1F and 6E may be attractive alternatives to AMT in X-PACT if the same therapeutic effect can be induced at lower drug concentrations and X-ray doses.
Using our library, we also studied the contribution of substituent size and position to psoralen potency. First, our dataset shows that large substituents (>150 Å3) can be tolerated at the 8-position without losses in potency. However, the most potent such substituents have positively charged functional groups, which may outweigh any detrimental effects due to lipophilic bulk at the 8-position. Indeed, when the alkoxy chain of 8-MOP was systematically lengthened, significant losses in potency were observed. Early studies on psoralen photobiology showed that the 4’,5’-monoadduct forms most efficiently in the sequence of ICL formation. This led us to propose that the 4’,5’ double bond would be more sensitive to modification than the 3,4 double bond. Surprisingly, we found that the size limitations were roughly equal for both double bonds (<50–70Å3). However, only the 3,4 double bond could bear a fused ring and still retain potency. For derivatives with fused rings on the 4’,5’ double bond, a complete loss of DNA adduct formation and cytotoxicity was observed. Interestingly, 4’,5’ modifications alone were universally detrimental to psoralen activity (1A is the only exception). However, when a 4-methyl group was added, many previously disabled derivatives gained dramatic potency. The explanation for this empirical phenomenon may require further biological or photophysical characterization. We cannot exclude that our substituents alter the spectrotemporal properties of psoralen and the quantum yield of DNA photoaddition (42). Diekmann et al. have shown that photoaddition of intercalated AMT to thymine proceeds via the triplet state with quantum yield 0.12 (43). It may be valuable to measure the triplet quantum yields of psoralens identified by the current study. Though we have focused on intercalation efficiency as the basis for our molecules’ efficacy, triplet quantum yields should be investigated in future synthetic efforts. Of note, approximately 3 × 104 photons were delivered per psoralen molecule in our cellular experiments, a dose that should permit significant psoralen photoactivation despite the aforementioned low quantum yields.
In addition, we found several highly cytotoxic compounds with lipophilic substituents (4H, 8F, 6G, and 3B, to name a few). Such substituents (primarily methyl and phenyl groups) may enhance interactions within the locally hydrophobic environment between DNA base pairs. However, when the in vitro DNA adduct formation of 8F (8-position phenyl) was studied, it performed surprisingly poorly compared with comparably cytotoxic psoralens (those inducing >99% cell death). 8-MOP, which performed slightly better than 8F in our DNA adduct assay, only resulted in ~31% survival. This suggests that there may be non-DNA mediated pathways by which 8F achieves its potency. For example, activated psoralens react with oxygen to produce reactive oxygen species, which may contribute to their skin photosensitization properties (44). It was theorized long ago that psoralens could engage in specific interactions with other biological macromolecules (45, 46). Alteration of cell surface proteins may explain the drug’s remarkable immunogenic properties when used to treat CTCL (47–49). Despite these long-standing theories, surprisingly few specific protein targets of psoralen binding have been identified. Among the many possible protein targets of psoralen, the most exciting for oncology applications have been EGFR and HER2 (50–53). Given the structural conservation among kinase domains, particularly within the EGFR family, it is compelling to speculate that additional kinases may be psoralen targets. DiscoverX’s KINOMEscan® or KiNativ™ in situ kinase profiling should be appealing, straight-forward technologies to assess this theory (54–56). For a proteome-wide approach, immunoprecipitation or fluorescent labeling approaches followed by MS profiling should be capable of identifying additional protein targets of psoralens (57). Using such methods, compounds like 8F and others emerging from our screen may be promising leads to begin exploring these non-DNA mediated mechanisms of action.
Perhaps the most sophisticated approach for studying genome-wide localization of psoralen adducts was performed by Anders et al., who showed that psoralen bound preferentially at the transcriptional start sites of active genes (58). Functional annotation of these genes and the consequences of psoralen binding at these sites would be highly informative and point to a more refined picture of psoralen’s pharmacological targets. Looking forward, positively charged psoralens may be the lowest hanging fruit in the field of psoralen optimization. Efforts to derivatize psoralens with positively charged substituents will undoubtedly lead to potent molecules that bind DNA extremely well. Yet, such compounds may be optimized for a sliver of the many possible biological effects psoralens can exert. With the availability of modern genomic and proteomic tools, these previously understudied aspects of psoralen biochemistry are poised to be understood in the near future.
Supplementary Material
Acknowledgements
The authors acknowledge the generous support of Immunolight LLC throughout the preparation of this manuscript. We thank the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University for the synthesis of all oligonucleotides. ADB is supported by a Medical Scientist Training Program grant (T32 GM008152) awarded to his institution by the National Institute of General Medical Sciences.
Footnotes
Financial Disclosure(s)
The authors declare the following competing financial interest(s): HW and WFB are employees of Immunolight LLC. FPG has a consulting arrangement with Immunolight LLC, which has supported his research.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article:
Tables S1–S2, Figures S1–S9, raw data, and chemical syntheses.
REFERENCES
- 1.Ortonne JP (1989) Psoralen therapy in vitiligo. Clinics in dermatology 7, 120–135. [DOI] [PubMed] [Google Scholar]
- 2.Kitamura N, Kohtani S and Nakagaki R (2005) Molecular aspects of furocoumarin reactions: Photophysics, photochemistry, photobiology, and structural analysis. Journal of Photochemistry and Photobiology C: Photochemistry Reviews 6, 168–185. [Google Scholar]
- 3.El-Mofty AM (1952) Observations on the use of Ammi majus Linn. In vitiligo. The British journal of dermatology 64, 431–441. [DOI] [PubMed] [Google Scholar]
- 4.El-Mofty AM (1964) THE TREATMENT OF VITILIGO WITH A COMBINATION OF PSORALENS AND QUINOLINES. The British journal of dermatology 76, 56–62. [DOI] [PubMed] [Google Scholar]
- 5.El-Mofty AM (1952) Further study on treatment of leucodermia with Ammi mafus linn. The Journal of the Egyptian Medical Association 35, 1–2. [PubMed] [Google Scholar]
- 6.Melski JW, Tanenbaum L, Parrish JA, Fitzpatrick TB and Bleich HL (1977) Oral methoxsalen photochemotherapy for the treatment of psoriasis: a cooperative clinical trial. The Journal of investigative dermatology 68, 328–335. [DOI] [PubMed] [Google Scholar]
- 7.Volc-Platzer B, Honigsmann H, Hinterberger W and Wolff K (1990) Photochemotherapy improves chronic cutaneous graft-versus-host disease. Journal of the American Academy of Dermatology 23, 220–228. [DOI] [PubMed] [Google Scholar]
- 8.Edelson R, Berger C, Gasparro F, Jegasothy B, Heald P, Wintroub B, Vonderheid E, Knobler R, Wolff K, Plewig G and et al. (1987) Treatment of cutaneous T-cell lymphoma by extracorporeal photochemotherapy. Preliminary results. The New England journal of medicine 316, 297–303. [DOI] [PubMed] [Google Scholar]
- 9.van Rhenen D, Gulliksson H, Cazenave JP, Pamphilon D, Ljungman P, Kluter H, Vermeij H, Kappers-Klunne M, de Greef G, Laforet M, Lioure B, Davis K, Marblie S, Mayaudon V, Flament J, Conlan M, Lin L, Metzel P, Buchholz D and Corash L (2003) Transfusion of pooled buffy coat platelet components prepared with photochemical pathogen inactivation treatment: the euroSPRITE trial. Blood 101, 2426–2433. [DOI] [PubMed] [Google Scholar]
- 10.Dall’Acqua F, Marciani S and Rodighiero G (1970) Inter-strand cross-linkages occurring in the photoreaction between psoralen and DNA. FEBS letters 9, 121–123. [DOI] [PubMed] [Google Scholar]
- 11.Marciani S, Dall’acqua F, Vedaldi D and Rodighiero G (1976) Receptor sites of DNA for the photoreaction with psoralen. Il Farmaco; edizione scientifica 31, 140–151. [PubMed] [Google Scholar]
- 12.Dall’Acqua F, Vedaldi D, Bordin F and Rodighiero G (1979) New studies on the interaction between 8-methoxypsoralen and DNA in vitro. The Journal of investigative dermatology 73, 191–197. [DOI] [PubMed] [Google Scholar]
- 13.Cimino GD, Shi YB and Hearst JE (1986) Wavelength dependence for the photoreversal of a psoralen-DNA cross-link. Biochemistry 25, 3013–3020. [DOI] [PubMed] [Google Scholar]
- 14.Shi YB and Hearst JE (1987) Wavelength dependence for the photoreactions of DNA-psoralen monoadducts. 1. Photoreversal of monoadducts. Biochemistry 26, 3786–3792. [DOI] [PubMed] [Google Scholar]
- 15.Kanne D, Straub K, Rapoport H and Hearst JE (1982) Psoralen-deoxyribonucleic acid photoreaction. Characterization of the monoaddition products from 8-methoxypsoralen and 4,5’8-trimethylpsoralen. Biochemistry 21, 861–871. [DOI] [PubMed] [Google Scholar]
- 16.Kanne D, Rapoport H and Hearst JE (1984) 8-Methoxypsoralen-nucleic acid photoreaction. Effect of methyl substitution on pyrone vs. furan photoaddition. Journal of medicinal chemistry 27, 531–534. [DOI] [PubMed] [Google Scholar]
- 17.Holtick U, Wang XN, Marshall SR, Scheid C, von Bergwelt-Baildon M and Dickinson AM (2012) In vitro PUVA treatment preferentially induces apoptosis in alloactivated T cells. Transplantation 94, e31–34. [DOI] [PubMed] [Google Scholar]
- 18.El-Domyati M, Moftah NH, Nasif GA, Abdel-Wahab HM, Barakat MT and Abdel-Aziz RT (2013) Evaluation of apoptosis regulatory proteins in response to PUVA therapy for psoriasis. Photodermatol Photoimmunol Photomed 29, 18–26. [DOI] [PubMed] [Google Scholar]
- 19.Isaacs ST, Shen CK, Hearst JE and Rapoport H (1977) Synthesis and characterization of new psoralen derivatives with superior photoreactivity with DNA and RNA. Biochemistry 16, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 20.Wollowitz SWC, CA), Isaacs Stephen T. (Orinda, CA), Rapoport Henry (Berkeley, CA), Spielmann Hans P. (Lexington, KY), Nerio Aileen (Santa Clara, CA) (1997) Method for inactivating pathogens in blood using photoactivation of 4’-primary amino-substituted psoralens. Cerus Corporation (Concord, CA), United States. [Google Scholar]
- 21.Wollowitz SL, CA, US), Isaacs Stephen T. (Orinda, CA, US), Rapoport Henry (Berkeley, CA, US), Spielmann Hans Peter (Lexington, KY, US) (2002) Compounds for the photodecontamination of pathogens in blood. WOLLOWITZ SUSAN,ISAACS STEPHEN T.,RAPOPORT HENRY,SPIELMANN HANS PETER, United States. [Google Scholar]
- 22.Niu C, Pang GX, Li G, Dou J, Nie LF, Himit H, Kabas M and Aisa HA (2016) Synthesis and biological evaluation of furocoumarin derivatives on melanin synthesis in murine B16 cells for the treatment of vitiligo. Bioorganic & medicinal chemistry 24, 5960–5968. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Wen Y, Liang CG, Liu J, Ding YB and Zhang WH (2017) Design, Synthesis and Antifungal Activity of Psoralen Derivatives. Molecules (Basel, Switzerland) 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchman CD and Hurley TD (2017) Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives. Journal of medicinal chemistry 60, 2439–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu C, Yin L and Aisa HA (2018) Novel Furocoumarin Derivatives Stimulate Melanogenesis in B16 Melanoma Cells by Up-Regulation of MITF and TYR Family via Akt/GSK3beta/beta-Catenin Signaling Pathways. International journal of molecular sciences 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM and Levens D (2013) Transcription-dependent dynamic supercoiling is a short-range genomic force. Nature structural & molecular biology 20, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Xu S, Pan C, Ye M and Zou H (2007) 3,4-diaminobenzophenone matrix for analysis of oligonucleotides by MALDI-TOF mass spectrometry. Current protocols in nucleic acid chemistry Chapter 10, Unit 10.12. [DOI] [PubMed] [Google Scholar]
- 28.Spielmann HP, Dwyer TJ, Hearst JE and Wemmer DE (1995) Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics. Biochemistry 34, 12937–12953. [PubMed] [Google Scholar]
- 29.Peltason L and Bajorath J (2007) Molecular similarity analysis uncovers heterogeneous structure-activity relationships and variable activity landscapes. Chemistry & biology 14, 489–497. [DOI] [PubMed] [Google Scholar]
- 30.Kanne D, Straub K, Hearst JE and Rapoport H (1982) Isolation and characterization of pyrimidine-psoralen-pyrimidine photodiadducts from DNA. Journal of the American Chemical Society 104, 6754–6764. [Google Scholar]
- 31.Shi YB and Hearst JE (1987) Wavelength dependence for the photoreactions of DNA-psoralen monoadducts. 2. Photo-cross-linking of monoadducts. Biochemistry 26, 3792–3798. [DOI] [PubMed] [Google Scholar]
- 32.Gasparro FP, Saffran WA, Cantor CR and Edelson RL (1984) Wavelength dependence for AMT crosslinking of pBR322 DNA. Photochemistry and photobiology 40, 215–219. [DOI] [PubMed] [Google Scholar]
- 33.Spielmann HP, Sastry SS and Hearst JE (1992) Methods for the large-scale synthesis of psoralen furan-side monoadducts and diadducts. Proceedings of the National Academy of Sciences of the United States of America 89, 4514–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guha R and Van Drie JH (2008) Structure--activity landscape index: identifying and quantifying activity cliffs. Journal of chemical information and modeling 48, 646–658. [DOI] [PubMed] [Google Scholar]
- 35.Vedaldi D, Dall’Acqua F, Caffieri S and Rodighiero G (1979) 3-(alpha, alpha-Dimethylallyl)-psoralen: a linear furocoumarin forming mainly 4’,5’-monofunctional adducts with DNA. Photochemistry and photobiology 29, 277–281. [DOI] [PubMed] [Google Scholar]
- 36.Buhimschi AD and Gasparro FP (2014) UVA and UVB-Induced 8-Methoxypsoralen Photoadducts and a Novel Method for their Detection by Surface-Enhanced Laser Desorption Ionization Time-of-Flight Mass Spectrometry (SELDI-TOF MS). Photochemistry and photobiology 90, 241–246. [DOI] [PubMed] [Google Scholar]
- 37.Liu C (2011) The application of SELDI-TOF-MS in clinical diagnosis of cancers. Journal of biomedicine & biotechnology 2011, 245821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Bock M, de Seny D, Meuwis MA, Chapelle JP, Louis E, Malaise M, Merville MP and Fillet M (2010) Challenges for biomarker discovery in body fluids using SELDI-TOF-MS. Journal of biomedicine & biotechnology 2010, 906082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan C, Xu S, Zhou H, Fu Y, Ye M and Zou H (2007) Recent developments in methods and technology for analysis of biological samples by MALDI-TOF-MS. Analytical and bioanalytical chemistry 387, 193–204. [DOI] [PubMed] [Google Scholar]
- 40.Lecchi P, Le HM and Pannell LK (1995) 6-Aza-2-thiothymine: a matrix for MALDI spectra of oligonucleotides. Nucleic acids research 23, 1276–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldham M, Yoon P, Fathi Z, Beyer WF, Adamson J, Liu L, Alcorta D, Xia W, Osada T, Liu C, Yang XY, Dodd RD, Herndon JE 2nd, Meng B, Kirsch DG, Lyerly HK, Dewhirst MW, Fecci P, Walder H and Spector NL (2016) X-Ray Psoralen Activated Cancer Therapy (X-PACT). PloS one 11, e0162078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frobel S, Levi L, Ulamec SM and Gilch P (2016) Photoinduced Electron Transfer between Psoralens and DNA: Influence of DNA Sequence and Substitution. Chemphyschem 17, 1377–1386. [DOI] [PubMed] [Google Scholar]
- 43.Diekmann J, Gontcharov J, Frobel S, Torres Ziegenbein C, Zinth W and Gilch P (2019) The Photoaddition of a Psoralen to DNA Proceeds via the Triplet State. J Am Chem Soc 141, 13643–13653. [DOI] [PubMed] [Google Scholar]
- 44.Pathak MA and Joshi PC (1984) Production of active oxygen species (1O2 and O2-.) by psoralens and ultraviolet radiation (320–400 nm). Biochim Biophys Acta 798, 115–126. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt IM, Chimenti S and Gasparro FP (1995) Psoralen-protein photochemistry--a forgotten field. Journal of photochemistry and photobiology. B, Biology 27, 101–107. [DOI] [PubMed] [Google Scholar]
- 46.Anthony FA, Laboda HM and Costlow ME (1997) Psoralen-fatty acid adducts activate melanocyte protein kinase C: a proposed mechanism for melanogenesis induced by 8-methoxypsoralen and ultraviolet A light. Photodermatology, photoimmunology & photomedicine 13, 9–16. [DOI] [PubMed] [Google Scholar]
- 47.Gasparro FP, Malane MS, Maxwell VM and Tigelaar RE (1993) The treatment of mastocytoma cells with 8-methoxypsoralen and long-wavelength ultraviolet radiation enhances cellular immunogenicity: preliminary results. Photochemistry and photobiology 58, 682–688. [DOI] [PubMed] [Google Scholar]
- 48.Maeda A, Schwarz A, Kernebeck K, Gross N, Aragane Y, Peritt D and Schwarz T (2005) Intravenous infusion of syngeneic apoptotic cells by photopheresis induces antigen-specific regulatory T cells. Journal of immunology (Baltimore, Md.: 1950) 174, 5968–5976. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez AL, Berger CL, Remington J, Girardi M, Tigelaar RE and Edelson RL (2014) Integrin-driven monocyte to dendritic cell conversion in modified extracorporeal photochemotherapy. Clinical and experimental immunology 175, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laskin JD, Lee E, Laskin DL and Gallo MA (1986) Psoralens potentiate ultraviolet light-induced inhibition of epidermal growth factor binding. Proceedings of the National Academy of Sciences of the United States of America 83, 8211–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang XY, Ronai ZA, Santella RM and Weinstein IB (1988) Effects of 8-methoxypsoralen and ultraviolet light A on EGF receptor (HER-1) expression. Biochemical and biophysical research communications 157, 590–596. [DOI] [PubMed] [Google Scholar]
- 52.Mermelstein FH, Abidi TF and Laskin JD (1989) Inhibition of epidermal growth factor receptor tyrosine kinase activity in A431 human epidermoid cells following psoralen/ultraviolet light treatment. Molecular pharmacology 36, 848–855. [PubMed] [Google Scholar]
- 53.Xia W, Gooden D, Liu L, Zhao S, Soderblom EJ, Toone EJ, Beyer WF Jr., Walder H and Spector NL (2014) Photo-activated psoralen binds the ErbB2 catalytic kinase domain, blocking ErbB2 signaling and triggering tumor cell apoptosis. PloS one 9, e88983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK and Zarrinkar PP (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29, 1046–1051. [DOI] [PubMed] [Google Scholar]
- 55.Schurmann M, Janning P, Ziegler S and Waldmann H (2016) Small-Molecule Target Engagement in Cells. Cell chemical biology 23, 435–441. [DOI] [PubMed] [Google Scholar]
- 56.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee JD, Gray NS and Kozarich JW (2011) In situ kinase profiling reveals functionally relevant properties of native kinases. Chemistry & biology 18, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evison BJ, Actis ML and Fujii N (2016) A clickable psoralen to directly quantify DNA interstrand crosslinking and repair. Bioorganic & medicinal chemistry 24, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Loven J, Sigova AA, Smith WB, Lee TI, Bradner JE and Young RA (2014) Genome-wide localization of small molecules. Nat Biotechnol 32, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



