Abstract
Introduction:
Mesenchymal stem cells (MSCs) have endogenous reparative properties, and may constitute an exogenous therapeutic intervention in patients with chronic kidney disease. The microenvironment of metabolic syndrome (MetS) induces fat inflammation, with abundant expression of tumor necrosis factor (TNF)-α. MetS may also alter the content of adipose tissue-derived MSCs, and we hypothesized that the inflammatory profile of MetS manifests via upregulating MSC mRNAs and proteins of the TNF-α pathway.
Methods:
Domestic pigs were fed a 16-week Lean or MetS diet (n=4 each). MSCs were harvested from abdominal subcutaneous fat, and their extracellular vesicles (EVs) isolated. Expression profiles of mRNAs and proteins in MSCs and EVs were obtained by high-throughput sequencing and proteomics. Nuclear translocation of the pro-inflammatory transcription factor (NF)-kB was evaluated in MSC and in pig renal tubular cells (TEC) co-incubated with EVs.
Results:
We found 13 mRNAs and 4 proteins in the TNF-α pathway upregulated in MetS- vs. Lean-MSCs (fold-change>1.4, p<0.05), mostly via TNF-α receptor-1 (TNF-R1) signaling. Three mRNAs were upregulated in MetS-EVs. MetS-MSCs, as well as TECs co-incubated with MetS-EVs, showed increased nuclear translocation of NF-kB. Using qPCR, JUNB, MAP2K7 and TRAF2 genes followed the same direction of RNA-sequencing findings.
Conclusions:
MetS upregulates the TNF-α transcriptome and proteome in swine adipose tissue-derived MSCs, which are partly transmitted to their EV progeny, and are associated with activation of NF-kB in target cells. Hence, the MetS milieu may affect the profile of endogenous MSCs and their paracrine vectors and limit their use as an exogenous regenerative therapy. Anti-inflammatory strategies targeting the TNF-α pathway might be a novel strategy to restore MSC phenotype, and in turn function.
Keywords: Metabolic syndrome, mesenchymal stem cells, tumor necrosis factor, mRNA, proteins
Introduction
The prevalence of obesity has doubled in the past few decades, and it is estimated that nearly one third of the world population is either overweight or obese (1). Obesity is associated with multiple comorbidities, such as cardiovascular disease and chronic kidney disease (CKD), and can involve multiple organs. Adipose tissue is a major endocrine organ secreting various cytokines, which induces a state of chronic low-grade inflammation. In particular, adipose tissue in obesity has upregulated expression of the inflammatory cytokine tumor necrosis factor (TNF)-α, and its levels correlate with the degree of obesity and insulin resistance (2).
With the concomitant rise in the prevalence of CKD and its associated comorbidities, novel therapeutic approaches are needed. For example, delivery of mesenchymal stem cells (MSCs) has shown promise in clinical trials in patients with renovascular disease (3). These multipotent stromal cells have the ability for self-renewal, and are characterized by expression of several surface marker, such as CD44, CD73, CD90, CD105, CD166, CD271 and absence of CD14, CD34, CD45 and HLA DR (4). Additionally, they show plasticity and are able to differentiate into mesodermal cell types.
However, in patients with obesity, MSCs are exposed to an aberrant microenvironment that may impact their phenotype and function. Adipose tissue-derived MSCs harvested from animal models of obesity show altered mRNA, micro-RNA, and protein cargo compared with Lean-MSCs, with upregulation of genes involved in inflammation (5). Moreover, this inflammatory microenvironment favors MSC differentiation into adipocytes and increases senescence, which is prominently mediated by TNF-α (6). Furthermore, an inflammatory milieu can abolish the MSC immunomodulatory capacity (7). A range of MSC tissue sources for MSC collection, as well as diverse microenvironments at both the harvest and implantation sites, may therefore partly account for inconsistent effects of autologous MSCs delivered exogenously.
MSC regenerative potential relies primarily on their paracrine function, which is executed by release of soluble mediators and extracellular vesicles (EVs). EVs carry cargo from their parent cells consisting of proteins, mRNAs, and microRNAs, and activate several repair mechanisms to ameliorate injury in target organs or tissues. However, in addition to its influence on MSCs (5, 8), metabolic syndrome (MetS) may also alter the cargo of their EV progeny. Therefore, changes in the cargo of EVs derived from MetS-MSCs may impact MSC paracrine function and reparative potential (9).
TNF-α is a pivotal mediator of inflammation, and its systemic level is of the earliest to rise during development of obesity (10). Therefore, it might be involved in alterations in MSCs in experimental obesity and MetS. TNF-α belongs to the TNF superfamily (TNFSF), and signals through its receptors, TNF-R1 and TNF-R2. TNFSF is involved in many processes, and constitutes a potential target area for therapies against disease processes like atherosclerosis and ischemia (11). TNF-R1 is expressed on most cells, whereas the expression of TNF-R2 is more limited, yet includes MSC(12). TNF-R1 is associated with inflammation and apoptosis via activation of the adaptor proteins TNF-R1 associated death-domain and Fas-associated death-domain (FADD), and TNF-R2 signaling is dependent on TNF receptor-associated factor-2 (TRAF2) activation and associated with pro-survival (13) (14).
Notably, EVs from MetS-MSCs can upregulate TNF-α in target cells and target insulin signaling and metabolic complications (15-17). However, whether MetS alters TNF-α signaling pathways in MSCs and their EVs remains unclear. We therefore hypothesized that MetS upregulates TNF-α signaling in adipose tissue-derived MSCs, and this alteration would be reflected in their EVs.
Materials and Methods
Mayo Clinic Animal Care and Use Committee approved this study. Three-month-old female domestic pigs were randomized into Lean and MetS groups (n=4 each) for 16 weeks. MetS pigs were fed a high-cholesterol/carbohydrate diet (ether extract fat 43.0%, carbohydrates 40.8%, and protein 16.1%, 5B4L, Purina Test Diet, Richmond, IN) (18) and Lean pigs were fed a standard chow (13% protein, 2% fat, 6% fiber, Purina Animal Nutrition LLC, MN).
Systemic Measurements
Body weight and intra-arterial blood pressure of the pigs was recorded after 16 weeks. Total cholesterol, triglyceride and low-density lipoprotein levels were measured by enzymatic methods, and insulin resistance calculated by homeostasis model-assessment of insulin resistance (HOMA-IR). Animals were then euthanized with a lethal intravenous dose of sodium pentobarbital (100 mg/kg IV, Fatal Plus®, Vortech Pharmaceuticals, Dearborn, MI), and subcutaneous abdominal adipose tissue collected for MSC isolation.
MSC and EV isolation, characterization, and culture
Primary MSCs were isolated from abdominal fat (5-10g) using collagenase-H. As previously shown, cells were cultured for 3 weeks in advanced MEM medium (Gibco/Invitrogen, Carlsbad CA) supplemented with platelet lysate (PLTmax, Mill Creek Life Sciences, Rochester, MN), and used at the third passage (19-21). We have previously described the detailed characteristics of these MSCs, which express specific surface markers, differentiate into osteoblasts, chondroblasts, and adipocytes, and are plastic adherent (4). Characterization of MSCs was further confirmed by positive immunofluorescent staining in vitro and flow cytometry for CD44, CD73, CD90, and CD105 (22).
After a series of centrifugations and ultra-centrifugations, Lean- and MetS-EVs were harvested from supernatants of the parent MSCs (23). A total of 10x10Λ6 MSCs (a dose used for delivery) were cultured for 48 hours in advanced MEM without any supplements, and then centrifuged. This was followed by second ultracentrifugation of cell-free supernatants at 100,000g for 1 hour at 4°C, washed in serum-free media containing HEPES 25nM, and submitted to a third step of ultra-centrifugation. The nanoparticle tracking analysis size distribution exhibited by Lean- and MetS-EVs was typical, and expressed common MSC and EV markers by western blotting and fluorescence, as previously shown (23).
mRNA sequencing analysis
mRNA sequencing and analysis were performed at the Mayo Clinic Genomics and Bioinformatics Cores and followed previously described methodology (21). The libraries were prepared (TruSeq RNA Sample Prep Kit v2, Illumina, San Diego, CA) and loaded onto flow cells at cluster densities of 700,000/mm2 (Illumina cBot and cBot Paired-end cluster kit version-3). Cells were then sequenced (Illumina HiSeq 2000; TruSeq SBS kit 3.0, HCS v2.0.12), and data analyzed (MAPRSeq v.1.2.1 system and the Bioinformatics Core standard tool). R 2.6.2 was used for normalization and differential expression analysis. We normalized to one million reads (reads per kilo base pair per million mapped reads, RPKM) in order to correct for gene length. mRNAs with fold-change >1.4 and p<0.05 were considered to be upregulated in MetScompared to Lean-MSCs or EVs. To predict association among TNF-α genes, Search Tool for the Retrieval of Interacting Genes (STRING) version 9.1 (http://string-db.org/) was used. To validate the mRNA sequencing results, we measured the expression of JUNB, MAP2K7 and TRAF2 genes in Lean- and MetS-MSCs by quantitative polymerase chain reaction (qPCR) (JUNB: APPRNY2, Forward: GTGTCCCTGGGTGCCA, Reverse: AATAGCTGCTGAGATTGGTGTAGAC, Probe: CTGGACCCGCGTAGAC. MAP2K7: APNKVE4, Forward: CGACTCCAAGGCCAAAACG, Reverse: GAATGTCGTAGTCAGGCTTGGTA, Probe: ATGGCACCCGAGCGCA. TRAF2: ss06886963, GAPDH housekeeping gene: ss3375629. All from ThermoFisher, Waltham, MA, USA)(24).
Proteomic analysis
MSCs were solubilized and lysed, and protein samples denatured by incubation at 85°C for 10min for LC- MS proteomic analysis (25-27). Aliquots were re-solubilized in reducing sample buffer and samples electrophoresed in 4-20% TGX Ready gels at 200V for 30min. Gel sections were digested with trypsin and peptides extracted and transferred onto a 35cmx100μm PicoFrit column 9 (NewObjective), self-packed with Agilent Poroshell 120S 2.7μm EC-C18 stationary phase, using a Dionex UltiMate 3000 RSLC LC system (Thermo-Fisher Scientific, Waltham, MA). Peptides were separated and eluting peptides analyzed using a QExactive mass spectrometer (Thermo-Fisher Scientific, Waltham, MA). We used label-free peptide MS1 intensity-based methods to identify differentially expressed proteins between Lean- and MetS-MSCs. MaxQuant 1.5.1 software was used to assess data quality, and reversed protein sequences were appended to the database for estimating protein identification false discovery rates (FDRs). Protein group intensities of each sample were log2 transformed, normalized, and modeled using a Gaussian-linked generalized linear model. Data were normalized by protein loading, and differential p-values FDR-corrected. Proteins with fold change>1.4 and p<0.05 were considered upregulated in MetS- vs. Lean-MSCs.
MSC inflammatory potential
To explore the functional implications of these findings, we compared nuclear translocation of the pro-inflammatory transcription factor NF-kB by immunofluorescent staining (Anti NF-kB p65 antibody, ab16502, Abcam, 1:200, Cambridge, MA, USA) between Lean- and MetS-MSCs. Nuclear DNA was stained with 4’,6’-diamino-2-phenylindole (DAPI, Thermo-Fisher Scientific). Double positive (NF-kB/DAPI) areas were quantified using a computer-aided image analysis program (ZEN® 2012 blue edition; Carl Zeiss SMT, Oberkochen, Germany), and the results from all fields were averaged. Furthermore, because EVs are non-nucleated, we compared the ability of Lean- and MetS-EVs to induce NF-kB translocation in pig proximal kidney tubular epithelial cells (LLC-PK1, ATCC, Manassas). Cells were cultured in Medium-199 (Gibco BRL, USA) containing 3% FBS [19] alone or co-cultured with Lean- or MetS-EVs (5μg of EV protein) for 24 hours. NF-kB/DAPI areas were quantified using ZEN®, and results from all fields were averaged. To further delineate mechanism, we evaluated expression of the TNF-α receptors (ab1939, Abcam, 1:100, Cambridge, MA), TNF-R2 (MA5-32618, Life Technologies, 1:100), the CCL20 receptor CCR6 (abPA5-29015, 1:100, Thermo-Fisher), and VEGFR1 (absc-504,1:100, Santa Cruz, USA), by immunofluorescent staining with specific antibodies.
Results
Systemic characteristics
At sixteen weeks after commencing the diet, body weight and blood pressure were both elevated in MetS pigs in comparison to Lean pigs (Table 1). Levels of insulin, HOMA-IR, and lipid fractions were greater in MetS pigs versus Lean, whereas fasting glucose levels were comparable between the groups. These findings were consistent with development of MetS.
Table 1.
Systemic characteristic of Lean and metabolic syndrome (MetS) pigs (n=4 each).
| Parameter | Lean | MetS |
|---|---|---|
| Body Weight (Kg) | 73.1±10.5 | 91.6±1.9* |
| Mean blood pressure (mmHg) | 98.6±10.7 | 125.0±7.1* |
| Total cholesterol (mg/dl) | 81.3±6.7 | 438.5±82.4* |
| LDL cholesterol (mg/dl) | 31.6±6.6 | 342.7±145.6* |
| Triglycerides (mg/dl) | 8.0±1.2 | 18.3±4.8* |
| Fasting glucose (mg/dl) | 127.5±13.6 | 115.0±19.0 |
| Fasting insulin (μU/ml) | 0.4±0.1 | 0.7±0.1* |
| HOMA-IR score | 0.6±0.1 | 1.8±0.3* |
p≤0.05 vs. Lean. LDL: Low-density lipoprotein, HOMA-IR: Homeostasis model assessment of insulin resistance.
mRNAs and proteins upregulated in MetS-MSCs
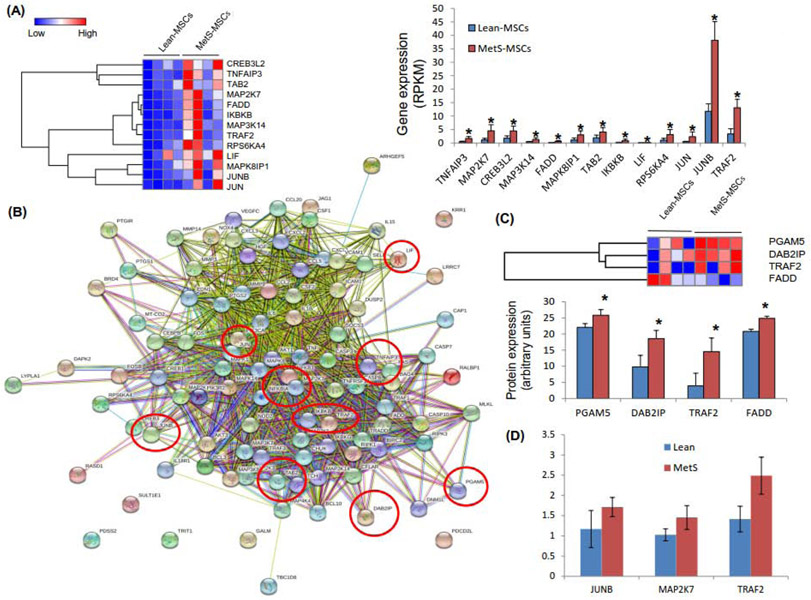
Annotated genes (n=10,413) were filtered for mRNAs involved in TNF-α pathways, including TNF-R1 and TNF-R2. Of these, we found 13 mRNAs upregulated in MetS-MSCs compared to Lean-MSCs (Figure 1A). These genes were considerably inter-linked, manifested in a significant number of known or predicted interactions with other TNF-α-related genes (Figure 1B). qPCR analysis onfirmed that MetS-MSCs expression of the TNF-α-related genes JUNB, MAP2K7, and TRAF2 followed the same direction of the mRNA sequencing findings (Figure 1D). Of the 4,933 proteins identified in MSCs, 4 TNF-α-related proteins were upregulated in MetS-MSCs compared to Lean-MSCs (Figure 2C). No TNF-α-related mRNAs or proteins were downregulated in MetS-MSCs. Most mRNAs and proteins were upregulated in MetS-MSCs were involved in TNF-R1 signaling.
Figure 1.
A: Comparison of TNF-α mRNAs in MetS-MSCs and Lean-MSCs (heat maps and bar graphs) showed 13 mRNAs upregulated in MetS-MSCs. B: Interactions among TNF-α genes derived from STRING. Nodes represent TNF-α genes, and color lines their interactions according to the functional association networks from the STRING database. Genes upregulated in MetS-MSCs are indicated with red circles. C: Comparison of TNF-α proteins (heat maps and bar graphs) in MetS-MSCs and Lean-MSCs identified 4 proteins upregulated in MetS-MSCs. *p<0.05 vs. Lean-MSCs. D: Expression of the genes JUNB, MAP2K7 and TRAF2 was concordant with the RNAseq findings.
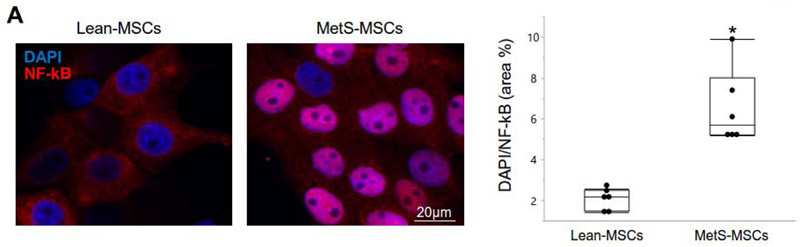
Figure 2.
Immunofluorescence staining showing subcellular distribution of nuclear factor (NF)-kB in Lean-MSCs. MetS induced a marked translocation of NFkB from the MSC cytoplasm to the cellular nucleus. *p<0.05 vs. Lean-MSCs.
As we have found before (5), in Lean MSCs NF-kB was localized in the cytoplasm, but translocated to the nucleus of MetS-MSCs (Figure 2A), suggesting a functionally significant activation of inflammatory signaling.
mRNAs upregulated in MetS-EVs
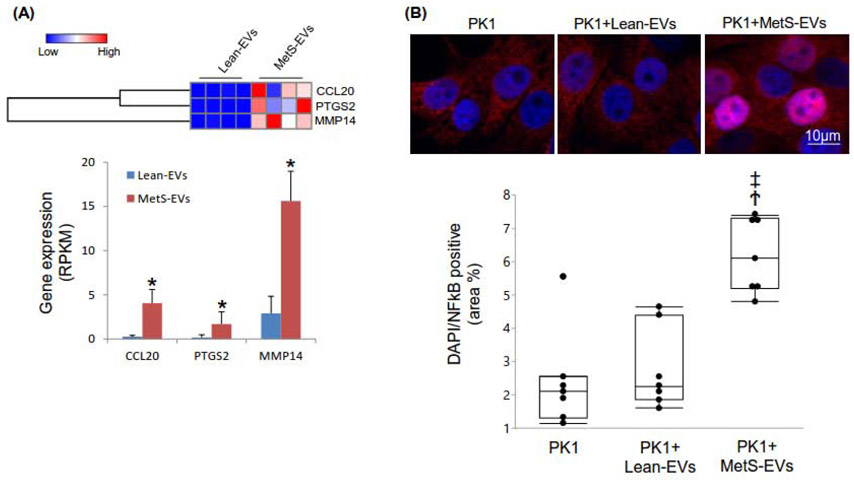
Of the genes annotated in EVs, 3 were significantly upregulated in MetS-EVs compared to Lean-EVs after filtering for TNF-α pathway (Figure 3A), but no proteins were found upregulated. Similar to findings in MSCs, no mRNAs in this pathway were downregulated in MetS-EVs.
Figure 3.
A: Three TNF-α-related mRNAs were upregulated in MetS-EVs compared to Lean-EVs (heat maps and bar graphs). B: Treatment with MetS-EVs induced translocation of NFkB to the nucleus of PK1 cells. *p<0.05 vs. Lean-MSCs, Ϯp<0.05 vs. PK1, ‡p<0.05 vs. PK1+Lean-EVs.
The effects of EVs on PK-1 cells
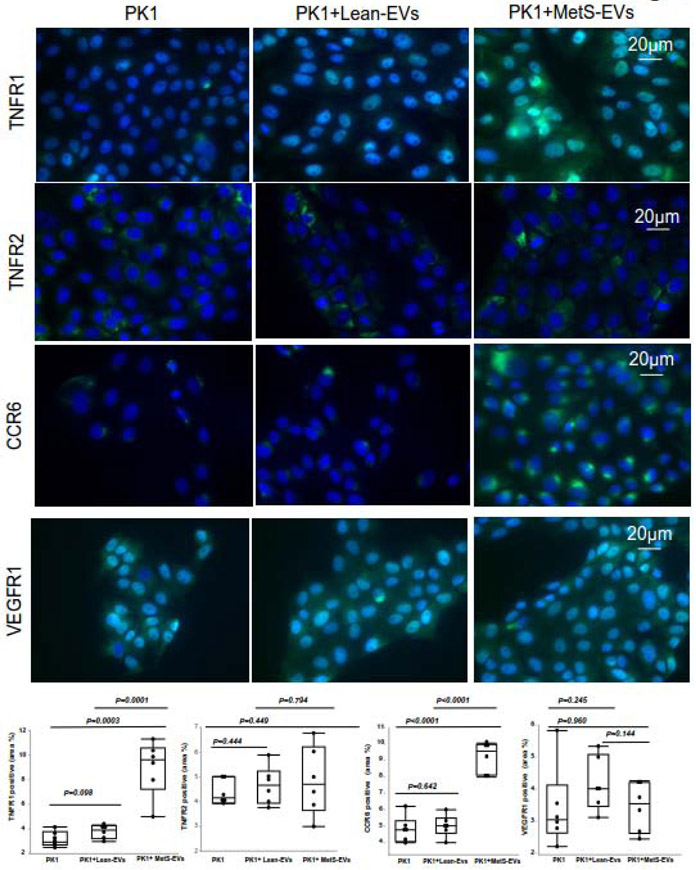
As we have shown before (28), co-culture only MetS-EVs, but not with Lean-EVs led to induced nuclear translocation of NF-kB (Figure 3B). Additionally, cells co-cultured with MetS EVs showed increased expression of TNF-R1 and the CCL20 receptor CCR6 compared to cells co-cultured with Lean EVs (Figure 4). There was no change in the expression of VEGFR1 or TNFR2 in cells co-cultured with MetS or Lean EVs.
Figure 4.
The effects of extracellular vesicles (EVs), obtained from Lean or metabolic syndrome (MetS) pigs, on PK-1 cells. A. Expression of TNF-R1 was similar in untreated PK1 and PK1+Lean-EVs, but higher in PK1+MetS-EVs compared with PK1 or PK1+Lean-EVs. TNF-R2 expression was similar among the groups. Expression of CCR6 was elevated in PK1+MetS-EVs compared with PK1 or PK1+Lean-EVs. VEGF1 expression was unchanged.
Discussion
The current study shows that MetS upregulates TNF-α signaling-related transcriptome and proteome in swine adipose tissue-derived MSCs, and may thereby activate pro-inflammatory signaling via the NF-kB pathway. Similarly, modest alterations were propagated to paracrine function of MSCs reflected by upregulated TNF-α genes in MetS-EVs. These findings suggest that the MetS microenvironment may modulate the impact of endogenous MSCs on target cells, and increase the inflammatory profile not only in MSCs, but in their EV progeny as well. This may partly account for the increased inflammation in subjects with MetS, and might limit the use of autologous MSCs as an exogenous regenerative therapy.
The prevalence of obesity continues to grow around the world, accompanied by growth in incidence and prevalence of associated comorbidities (1). MSCs may have a potential therapeutic role in decreasing target organ injury in cardiovascular disease, but the milieu of MetS may modulate their intercellular communication and blunt their reparative potential. Our prior work has shown that MetS alters the content of genes and protein in MSCs, and upregulates overall genes involved in inflammation (5). The current study extends and focuses our previous findings showing the inflammatory environment in MetS is established partly via upregulated MSC mRNAs and proteins in the TNF-α pathway. Furthermore, our study proposes a pathway by which TNF-α modulates inflammation in MetS, including upregulation of 13 mRNAs and 4 proteins mostly involved in the TNF-R1 pathway, except for TRAF2 protein, which is involved in TNF-R2 receptor signaling and in TNF-α induced NF-kB activation (29). The downstream ramification of MetS-induced changes in MSC TNF-α signaling was confirmed by increased nuclear translocation of NF-kB, as previously observed (5), underscoring increased inflammatory activation in MSCs.
The transcription factor NF-kB plays a crucial role in regulation of numerous genes involved in the inflammatory response and control of cell death. Activation of NF-kB is mediated through the phosphorylation of its inhibitory subunit I-kappa-B (IKB), and its subsequent degradation achieved at the proteasome (27). TNF-α and members of its signaling pathway, including mitogen-activated protein kinases (MAPK), TRAF2, and CCL20(30), can all regulate NF-kB translocation or transcriptional activity. Indeed, several elements in the TNF-α signaling cascade were upregulated in MetS compared to Lean EVs, as well as in PK-1 cells co-cultured with MetS EVs.
The genes found to be upregulated in MetS-MSCs suggested increased activation of NF-kB, whereas some upregulated genes may act as its suppressors, such as TNF-α-induced protein (TNFA1P)-3, which activates the ubiquitin-editing enzyme A20 (31). Many of the upregulated genes reflect the pro-inflammatory action of the MAPK pathway in regulation of NF-kB transcriptional activity (27). Therefore, MAP2K7 encodes for a protein that mediates cell responses to pro-inflammatory cytokine and environmental stresses (32). Another member of this family, MAP3K14, stimulates its activity and participates in NF-kB signaling cascade. It is essential for non-canonical NF-kB pathway as it phosphorylates IkB Kinase-alpha (IKKα). MAPK8IP1 functions as a regulator of vesicle transport, suggesting a potential impact of MetS on the paracrine activity of MetS-MSCs that includes EV release. The Fas-associated protein with death domain (FADD) gene encodes for a protein that interacts with various cell receptors and mediates apoptotic signals (33, 34), which might decrease survival of MSCs in a MetS milieu. The protein encoded by TAB2 gene activates MAPK (especially MAP3K7) and NF-KB and MAPK8 (35).
MetS-MSCs also showed upregulation of IkBKB gene, which acts through TNF-R1 in signaling and transcription of NF-kB by phosphorylating IKB inhibitors (36-38). IKBKB phosphorylates IkB molecules that inhibit NFKB transcription factors. Multiple signaling pathways that activate NF-kB, like TNF-α, converge at the level of IKBKB. The protein encoded by the LIF gene is involved in generation of regulatory lymphocytes (39), suggesting enhanced immunomodulatory function in MSCs. LIF is generally involved in phenotype differentiation, survival and response to injury, and maintains the stemness and bio-potency of MSCs (40, 41). The JUNB gene may be involved in cell growth, differentiation, and neoplastic transformation (42) and is a component of activator protein-1 that selectively regulates transcription (43). This may suggest altered cell survival of MetS-MSCs via TNF-α pathway.
Upregulation of TRAF2 gene is also consistent with inflammation. This key intracellular signaling mediator acts downstream of various members of the TNFSF(44), directly interacts with both TNF-receptors, and is required for TNF-α-mediated activation of MAPK8/JNK and NF-kB (45, 46). This fundamental requirement may account for its upregulation in multiple levels. Additional upregulated TNF-α pathway proteins in Mets-MSCs included PGAM5, DAB2IP, TRAF2 and FADD. Recruitment of FADD and signaling molecules form the death-inducing signaling complex activates downstream caspases (such as caspase-3) and leads to apoptosis. Downregulation of DAB2IP promotes neuronal differentiation of human MSCs (47), while the mitochondrial phosphatase PGAM5 promotes inflammasome activation in macrophages (48). Given upregulation of these canonical pathways that involve TNF-α, altered TNF-α signaling in MSCs might possibly play a role in apoptosis, cell differentiation, and inflammation, yet future studies will need to determine any causal relationship.
MetS-EVs exhibited upregulated CCL20, which is involved in MSCs transmigration (49, 50), as well as PTGS2 and MMP14 that belong to TNF-α pathway. On the other hand, no proteins in the TNF-α pathway were upregulated in MetS- versus Lean-MSCs, indicating that MetS primarily modulates the genetic cargo of EVs. Packaging EV is a selective process, and their cargo may therefore differ from their cells of origin. Nevertheless, MetS-induced changes in the expression of genes packed in their EVs can alter protein expression in recipient cells. Indeed, our previous (28) in-vitro study showed that co-culture with MetS-EVs induces NF-kB nuclear translocation in renal tubular epithelial cells, reflecting NF-kB activation. The present study demonstrates that this is associated with upregulated expression of TNF-R1 and the CCL20 receptor CCR6 in target cells, linking the effects of MetS-EV to upregulated CCL20 and TNF-α pathway in their parent MetS-MSC. Nevertheless, their TNF-α-related cargo was modest relative to their parent MSCs, suggesting that TNF-α may participate in MSC functions not necessarily related to EVs, such as those involving cell-cell contact (51) or release of soluble mediators. Further studies are needed to elucidate the primary mechanisms implicated in EVmediated NFkB activation.
We acknowledge several limitations to this study, including a modest number of samples. However, similar sample sizes are often employed in sequencing and proteomic studies (21). Evidently, this sample size has been sufficient to detect clear differences in TNF-α related genes and proteins in MetS-MSCs and their daughter EVs compared to their lean counterparts. Furthermore, we have validated trends observed in RNA sequencing using PCR and western blots to show MSCs at the 3rd passage possess a phenotype comparable to their parent cells (8, 52-55). The functional limitations that MetS might impose on MSCs, and the mechanisms by which it modifies genetic and protein cellular information, such as posttranslational/epigenetic changes needs to be defined by future studies. Despite limitations, our cutting-edge techniques revealed and highlighted the crosstalk between TNF-α and MSCs, as well as its impact on mRNA and proteomic cargo of adipose tissue-derived MSCs, and their EVs.
In summary, we found that MetS modifies not only the cargo of porcine MSCs, but also their EVs and upregulates proteins and genes, including those in the TNF-α signaling pathway, which might in turn modulate several important cellular functions and cell fate. These inflammatory pathways activated in MetS-MSCs may in turn be upregulated via TNF-α in adipose tissue and systemic circulation. Therefore, MetS might increase MSC inflammatory profile by upregulating the transcriptome and proteome of the TNF-α signaling pathway. Further studies are needed to determine whether these changes adversely impact MSC reparative function.
Supplementary Material
Acknowledgements
This study was partly supported by NIH grant numbers DK120292, DK102325, DK122137, and DK106427.
Footnotes
Conflict of interests
Dr. Lerman receives grant funding from Novo Nordisk, and is an advisor to Weijian Technologies and AstraZeneca. The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chooi YC, Ding C, Magkos F. The epidemiology of obesity. Metabolism. 2019;92:6–10. [DOI] [PubMed] [Google Scholar]
- 2.Tzanavari T, Giannogonas P, Karalis KP. TNF-alpha and obesity. Curr Dir Autoimmun. 2010;11:145–56. [DOI] [PubMed] [Google Scholar]
- 3.Saad A, Dietz AB, Herrmann SMS, Hickson LJ, Glockner JF, McKusick MA, et al. Autologous Mesenchymal Stem Cells Increase Cortical Perfusion in Renovascular Disease. Journal of the American Society of Nephrology : JASN. 2017;28(9):2777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. [DOI] [PubMed] [Google Scholar]
- 5.Pawar AS, Eirin A, Krier JD, Woollard JR, Zhu XY, Lerman A, et al. Alterations in genetic and protein content of swine adipose tissue-derived mesenchymal stem cells in the metabolic syndrome. Stem Cell Res. 2019;37:101423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu XY, Ma S, Eirin A, Woollard JR, Hickson LJ, Sun D, et al. Functional Plasticity of Adipose-Derived Stromal Cells During Development of Obesity. Stem Cells Transl Med. 2016;5(7):893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papadopoulou A, Yiangou M, Athanasiou E, Zogas N, Kaloyannidis P, Batsis I, et al. Mesenchymal stem cells are conditionally therapeutic in preclinical models of rheumatoid arthritis. Ann Rheum Dis. 2012;71(10):1733–40. [DOI] [PubMed] [Google Scholar]
- 8.Conley SM, Zhu XY, Eirin A, Tang H, Lerman A, van Wijnen AJ, et al. Metabolic syndrome alters expression of insulin signaling-related genes in swine mesenchymal stem cells. Gene. 2018;644:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conley SM, Shook JE, Zhu XY, Eirin A, Jordan KL, Woollard JR, et al. Metabolic Syndrome Induces Release of Smaller Extracellular Vesicles from Porcine Mesenchymal Stem Cells. Cell Transplant. 2019;28(9-10):1271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouyang S, Li W, Liu Z, Li Y, Li S, Wu J. Association between serum soluble tumor necrosis factor-alpha receptors and early childhood obesity. Endocr J. 2016;63(6):581–7. [DOI] [PubMed] [Google Scholar]
- 11.Nash M, McGrath JP, Cartland SP, Patel S, Kavurma MM. Tumour necrosis factor superfamily members in ischaemic vascular diseases. Cardiovasc Res. 2019;115(4):713–20. [DOI] [PubMed] [Google Scholar]
- 12.Kelly ML, Wang M, Crisostomo PR, Abarbanell AM, Herrmann JL, Weil BR, et al. TNF receptor 2, not TNF receptor 1, enhances mesenchymal stem cell-mediated cardiac protection following acute ischemia. Shock. 2010;33(6):602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan L, Zheng D, Xu R-H. Critical Role of Tumor Necrosis Factor Signaling in Mesenchymal Stem Cell-Based Therapy for Autoimmune and Inflammatory Diseases. Frontiers in immunology. 2018;9:1658-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, Wang J, Brand DD, Zheng SG. Role of TNF-TNF Receptor 2 Signal in Regulatory T Cells and Its Therapeutic Implications. Front Immunol. 2018;9:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng Y, Eirin A, Zhu XY, O'Brien DR, Lerman A, van Wijnen AJ, et al. The metabolic syndrome modifies the mRNA expression profile of extracellular vesicles derived from porcine mesenchymal stem cells. Diabetol Metab Syndr. 2018;10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eirin A, Zhu XY, Woollard JR, Tang H, Dasari S, Lerman A, et al. Metabolic Syndrome Interferes with Packaging of Proteins within Porcine Mesenchymal Stem Cell-Derived Extracellular Vesicles. Stem Cells Transl Med. 2019. [DOI] [PMC free article] [PubMed]
- 17.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. The metabolic syndrome alters the miRNA signature of porcine adipose tissue-derived mesenchymal stem cells. Cytometry A. 2017. [DOI] [PMC free article] [PubMed]
- 18.Pawar AS, Zhu XY, Eirin A, Tang H, Jordan KL, Woollard JR, et al. Adipose tissue remodeling in a novel domestic porcine model of diet-induced obesity. Obesity (Silver Spring). 2015;23(2):399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebrahimi B, Eirin A, Li Z, Zhu X-Y, Zhang X, Lerman A, et al. Mesenchymal Stem Cells Improve Medullary Inflammation and Fibrosis after Revascularization of Swine Atherosclerotic Renal Artery Stenosis. PLoS ONE. 2013;8(7):e67474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eirin A, Zhu X-Y, Ebrahimi B, Krier JD, Riester SM, van Wijnen AJ, et al. Intra-renal delivery of mesenchymal stem cells and endothelial progenitor cells attenuates hypertensive cardiomyopathy in experimental renovascular hypertension. Cell transplantation. 2015;24(10):2041–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O'Brien D, et al. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;551(1):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu XY, Urbieta-Caceres V, Krier JD, Textor SC, Lerman A, Lerman LO. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem Cells. 2013;31(1):117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eirin A, Zhu XY, Puranik AS, Woollard JR, Tang H, Dasari S, et al. Integrated transcriptomic and proteomic analysis of the molecular cargo of extracellular vesicles derived from porcine adipose tissue-derived mesenchymal stem cells. PLoS One. 2017;12(3):e0174303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eirin A, Herrmann SM, Saad A, Abumoawad A, Tang H, Lerman A, et al. Urinary mitochondrial DNA copy number identifies renal mitochondrial injury in renovascular hypertensive patients undergoing renal revascularization: A Pilot Study. Acta Physiol (Oxf). 2019;226(3):e13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan MC, Johnson KL, Zenka RM, Charlesworth MC, Madden BJ, Mahoney DW, et al. Subfractionation, characterization, and in-depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int. 2014;85(5):1225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogan MC, Bakeberg JL, Gainullin VG, Irazabal MV, Harmon AJ, Lieske JC, et al. Identification of Biomarkers for PKD1 Using Urinary Exosomes. J Am Soc Nephrol. 2015;26(7):1661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eirin A, Zhu XY, Puranik AS, Woollard JR, Tang H, Dasari S, et al. Comparative proteomic analysis of extracellular vesicles isolated from porcine adipose tissue-derived mesenchymal stem/stromal cells. Sci Rep. 2016;6:36120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eirin A, Zhu XY, Woollard JR, Tang H, Dasari S, Lerman A, et al. Metabolic Syndrome Interferes with Packaging of Proteins within Porcine Mesenchymal Stem Cell-Derived Extracellular Vesicles. Stem Cells Transl Med. 2019;8(5):430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JY, Morgan M, Kim DG, Lee JY, Bai L, Lin Y, et al. TNFalpha induced noncanonical NF-kappaB activation is attenuated by RIP1 through stabilization of TRAF2. J Cell Sci. 2011;124(Pt 4):647–56. [DOI] [PubMed] [Google Scholar]
- 30.Marsigliante S, Vetrugno C, Muscella A. CCL20 induces migration and proliferation on breast epithelial cells. J Cell Physiol. 2013;228(9):1873–83. [DOI] [PubMed] [Google Scholar]
- 31.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–9. [DOI] [PubMed] [Google Scholar]
- 32.Wolter S, Mushinski JF, Saboori AM, Resch K, Kracht M. Inducible expression of a constitutively active mutant of mitogen-activated protein kinase kinase 7 specifically activates c-JUN NH2-terminal protein kinase, alters expression of at least nine genes, and inhibits cell proliferation. J Biol Chem. 2002;277(5):3576–84. [DOI] [PubMed] [Google Scholar]
- 33.Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2(4):241–3. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, Gao L, Wang P, Xie Z, Cen S, Li Y, et al. TNF-α Induced the Enhanced Apoptosis of Mesenchymal Stem Cells in Ankylosing Spondylitis by Overexpressing TRAIL-R2. Stem Cells International. 2017;2017:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanayama A, Seth RB, Sun L, Ea C-K, Hong M, Shaito A, et al. TAB2 and TAB3 Activate the NF-κB Pathway through Binding to Polyubiquitin Chains. Molecular Cell. 2004; 15(4) :535–48. [DOI] [PubMed] [Google Scholar]
- 36.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. [DOI] [PubMed] [Google Scholar]
- 37.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. [DOI] [PubMed] [Google Scholar]
- 38.Scheidereit C IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25(51):6685–705. [DOI] [PubMed] [Google Scholar]
- 39.Nasef A, Mazurier C, Bouchet S, François S, Chapel A, Thierry D, et al. Leukemia inhibitory factor: Role in human mesenchymal stem cells mediated immunosuppression. Cellular Immunology. 2008;253(1):16–22. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Vaessen B, Lenvik T, Blackstad M, Reyes M, Verfaillie CM. Multipotent progenitor cells can be isolated from postnatal murine bone marrow, muscle, and brain. Exp Hematol. 2002;30(8):896–904. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz J, Van de Pavert S, Clarke I, Rao A, Ray D, Vrana K. Paracrine interactions within the pituitary gland. Ann N Y Acad Sci. 1998;839:239–43. [DOI] [PubMed] [Google Scholar]
- 42.Yang M-Y, Liu T-C, Chang J-G, Lin P-M, Lin S-F. JunB gene expression is inactivated by methylation in chronic myeloid leukemia. Blood. 2003;101(8):3205–11. [DOI] [PubMed] [Google Scholar]
- 43.Hsu J-C, Cressman DE, Taub R. Promoter-specific trans-Activation and Inhibition Mediated by JunB. Cancer Research. 1993;53(16):3789–94. [PubMed] [Google Scholar]
- 44.Lin WJ, Su YW, Lu YC, Hao Z, Chio II, Chen NJ, et al. Crucial role for TNF receptor-associated factor 2 (TRAF2) in regulating NFkappaB2 signaling that contributes to autoimmunity. Proc Natl Acad Sci U S A. 2011;108(45):18354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12(4):419–29. [DOI] [PubMed] [Google Scholar]
- 46.Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7(5):715–25. [DOI] [PubMed] [Google Scholar]
- 47.Chang SL-Y, Chou R-H, Zeng H-J, Lin Y-H, Chiu T-Y, Yang D-M, et al. Downregulation of DAB2IP Promotes Mesenchymal-To-Neuroepithelial Transition and Neuronal Differentiation of Human Mesenchymal Stem Cells. PLOS ONE. 2013;8(9):e75884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriwaki K, Farias Luz N, Balaji S, De Rosa MJ, O'Donnell CL, Gough PJ, et al. The Mitochondrial Phosphatase PGAM5 Is Dispensable for Necroptosis but Promotes Inflammasome Activation in Macrophages. Journal of immunology (Baltimore, Md. : 1950). 2016;196(1):407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith H, Whittall C, Weksler B, Middleton J. Chemokines stimulate bidirectional migration of human mesenchymal stem cells across bone marrow endothelial cells. Stem Cells Dev. 2012;21(3):476–86. [DOI] [PubMed] [Google Scholar]
- 50.Chamberlain G, Smith H, Rainger GE, Middleton J. Mesenchymal stem cells exhibit firm adhesion, crawling, spreading and transmigration across aortic endothelial cells: effects of chemokines and shear. PLoS One. 2011;6(9):e25663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horiuchi T, Mitoma H, Harashima S, Tsukamoto H, Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology (Oxford). 2010;49(7):1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. Obesity-induced mitochondrial dysfunction in porcine adipose tissue-derived mesenchymal stem cells. Journal of Cellular Physiology. 2018;233(8):5926–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SR, Eirin A, Herrmann SMS, Saad A, Juncos LA, Lerman A, et al. Preserved endothelial progenitor cell angiogenic activity in African American essential hypertensive patients. Nephrol Dial Transplant. 2018;33(3):392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng Y, Eirin A, Zhu XY, O'Brien DR, Lerman A, van Wijnen AJ, et al. The metabolic syndrome modifies the mRNA expression profile of extracellular vesicles derived from porcine mesenchymal stem cells. Diabetology & Metabolic Syndrome. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng Y, Eirin A, Zhu XY, Tang H, Chanana P, Lerman A, et al. The metabolic syndrome alters the miRNA signature of porcine adipose tissue-derived mesenchymal stem cells. Cytometry Part A. 2018;93a(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.