Abstract
Protein arginine methyltransferase (PRMT) enzymes play a crucial role in RNA splicing, DNA damage repair, cell signaling, and differentiation. Arginine methylation is a prominent post-transitional modification of histones and various non-histone proteins that can either activate or repress gene expression. The aberrant expression of PRMTs has been linked to multiple abnormalities, notably cancer. Herein, we review a number of non-histone protein substrates for all nine members of human PRMTs and how PRMT-mediated non-histone arginine methylation modulates various diseases. Additionally, we highlight the most recent clinical studies for several PRMT inhibitors.
Keywords: PRMT, arginine methylation, non-histone protein, PRMT inhibitor, cancer, epigenetic modifications
1. INTRODUCTION
Epigenetic modifications, including methylation, ubiquitination, phosphorylation, and acetylation, are crucial for regulating gene expression, cellular differentiation and function in a heritable fashion without changing the DNA code [1]. Targeting epigenetic abnormalities represents a feasible approach for treating various diseases, notably cancer [2]. Four histone deacetylase enzyme (HDAC) inhibitors are currently utilized as anticancer drugs [3], such as vorinostat (SAHA) which is used for the treatment of the cutaneous T-cell lymphoma (CTCL) [4], romidepsin (FK228) for CTCL [5], belinostat (PXD101) for peripheral T-cell lymphoma (PTCL) [6], and panobinostat for multiple myeloma [7]. Recently, several PRMT1 and PRMT5 inhibitors have entered clinical trials for hematological and solid tumors.
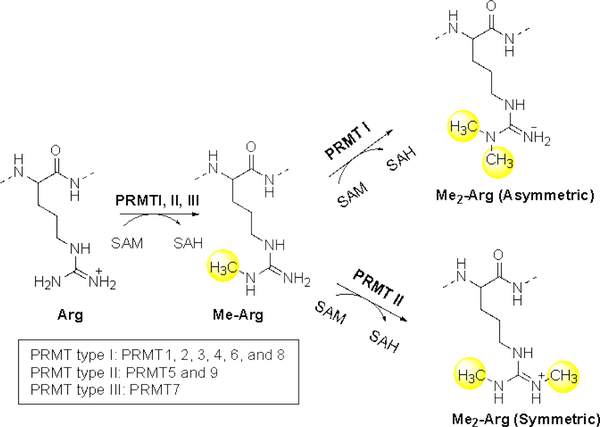
Arginine methylation is a prevalent post-translational modification that plays crucial roles in transcriptional regulation, RNA splicing, DNA damage repair, cell differentiation, and apoptosis [8]. Protein arginine methyltransferases (PRMTs) are responsible for arginine methylation by transferring the methyl group from S-adenosylmethionine (AdoMet or SAM) to the guanidinium nitrogen atoms of the arginine residue. Based on their product specificity, PRMTs are divided into three types: type I includes PRMT1, 2, 3, 4, 6 and 8; type II includes PRMT5 and 9; and type III includes only PRMT7 (Figure 1). All PRMTs are able to catalyze the monomethylation of arginine, but type I and II can further proceed to introduce a second methyl group asymmetrically and symmetrically on the guanidino group of the arginine, respectively. Enhanced levels of PRMTs are detected in cancer, cardiovascular diseases, inflammatory diseases, metabolic disorders, and diabetes [9–15]. Consequently, emerging efforts have been pursued to modulate PRMTs as new approaches to interrogate numerous abnormalities. A myriad of arginine methylation sites on histones have been characterized. Depending on the site of methylation and the effector protein, the methylated histone can either activate or repress transcription. Besides histones, PRMTs also methylate various functionally important non-histone proteins. This review will focus on the non-histone protein substrates of PRMTs and the function of arginine methylation on non-histone proteins.
Figure 1.
Arginine methylation
2. PRMT ENZYMES AND NON-HISTONE SUBSTRATES
PRMTs share a conserved seven-stranded Rossmann-fold domain that interacts with SAM and a ß-barrel domain that supports substrate binding [16]. The formation of homodimers is essential for the catalytic activities of most PRMTs except for PRMT7. Although PRMT7 acts as a monomer, it does contain two SET domains that are capable of forming a pseudo dimer [17]. Recently, more evidence has pointed to the formation of PRMT oligomers between different PRMT members. Type II PRMT5 and most type I PRMT enzymes except PRMT4 methylate substrates containing glycine and arginine (GAR) [18,19]. The residues distal to the GAR motif can modulate the methylation efficiency, which has been shown in both PRMT1 and 5 [20,21]. The other PRMTs recognize their own unique substrate recognition motifs. For example, PRMT4 specifically methylates arginine residues in proline, glycine, and methionine (PGM) rich motifs [22]. In addition, PRMT7 shows specificity for substrates that are enriched in RXR sequences (X is any amino acid) [23], while PRMT9 binds specifically to the FKRKY sequence of Splicing Factor 3b Subunit 2 (SF3B2) [24]. With the exception of PRMT7 as a monomethylase, all other PRMTs involve a sequential two-step mechanism to introduce two methyl groups on the arginine side chain. A multiple step methylation reaction undergoes either a processive or distributive mechanism. The dimethylation of arginine catalyzed by most PRMT enzymes proceeds in a distributive manner, where the mono-methylated intermediate is released after the first turnover [25–27]. Then, monomethylated arginine can subsequently rebind to the enzyme to liberate the dimethylated product. Arginine methylation does not change the charge state of the arginine residue, nor does it affect the ability to form electrostatic interactions. However, methylation does increase the size and hydrophobicity of arginine, and decreases the capacity of arginine as a hydrogen bond donor. Thus, arginine methylation has profound impacts on protein-DNA/RNA and protein-protein interactions, consequently modulating innumerable biological pathways. Applications of bioorthogonal profiling of protein methylation and global proteomic profiling have greatly advanced the identification of the physiological substrates of individual PRMT isoforms [18,28–30]. However, the functional study of specific arginine methylation remains underexplored. Below we will discuss the non-histone protein substrates for each PRMT enzyme (Table 1).
Table 1.
PRMT Enzymes and Their Substrates
| PRMT enzyme | Type | Arginine methylation | Cellular location | Histone substrate | Non-histone substrates |
|---|---|---|---|---|---|
| PRMT1 | I | MMA and ADMA | Cytoplasm and nucleus | H4R3, H3R3, H2AR11 | TWIST1 [35], TAF15 [36], RUNX1 [37], FOXO1 [38], E2F1 [39], C/EBP [40], SMAD6 [41], SMAD7 [42], eIF4A1 [43], PABPN1 [44], hnRNPR [45], ICP27 [46], 53BP1 [47], MRE11 [48], BRCA1 [49], TOP3B [50], ERα [51], EGFR [52], HMGA1 [53], SMURF2 [54], DP [55], FLT3 [56]. |
| PRMT2 | I | MMA and ADMA | Nucleus | H3R8 | RB [65]. |
| PRMT3 | I | MMA and ADMA | Cytoplasm | H4 | RPS2 [68], HMGA1 [53], PABPN1 [44], hnRNPA1 [69], TCOF1, MYO18B, KRT6A, TUBA1C, TUBB4A, TPI1 [28], CSTF-64 [45], TOP3B [50]. |
| PRMT4 | I | MMA and ADMA | Nucleus | H3R2, R17, R26, R42 | p300/CREB-binding protein [70], BAF155 [71], SRC-3 [72], CA150, SAP49, SmB, U1C, HuD [22], MED12 [73], PABPN1 [74], HuR [22,75], CBP KIX [70], GAPDH [76]. |
| PRMT5 | II | MMA and SDMA | Cytoplasm and nucleus | H2AR3, H3R2, H3R8, H4R3 | p53 [93], NF-kB [94], KLF4 [95], E2F1 [96], CFLARL [97], DDX5 [98]. |
| PRMT6 | I | MMA and ADMA | Nucleus | H3R2, H2AR11, H2AR29 | TAT [80], FOXO3 [81], HMGA1 [53], PABPN1 [44]. |
| PRMT7 | III | MMA | Cytoplasm | H3R2, H4R3, H4R17 | eIF2α [105], p38MAPK [106]. |
| PRMT8 | I | MMA and ADMA | Plasma membrane | - | NIFK [83], EWS [85], SOX2 [86], γ-H2AX [87]. |
| PRMT9 | II | MMA and SDMA | Cytoplasm and nucleus | - | SAP145 [101]. |
2.1. Type I PRMTs
2.1.1. PRMT1
PRMT1 is the most extensively studied PRMT enzyme, as it was the first member identified within this family. PRMT1 mediates more than 85% of the reported arginine methylation events [30]. The active form of PRMT1 is either a homodimer or oligomer [31]. Residues M48, E100, E144, E153, M155, and H293, located in the PRMT1 active site, are essential for substrate and cofactor binding [32,33]. Mutation of these residues can decrease the catalytic activity or binding of PRMT1 to its substrates. For example, the M48A and M155A mutants reduce the catalytic efficiency about 350-fold and 10-fold compared to the wild type PRMT1, respectively [32,33]. Similarly, the mutation of E144 and E153 to Ala decreased the catalytic efficiency for the AcH4–21 peptide substrate by 14- and 190-fold, respectively. In addition, the single mutation M48F can alter the product specificity of PRMT1 to generate both SDMA and ADMA, while the M155A mutant decreased enzyme processivity, with a slower formation of ADMA relative to wild type [34].
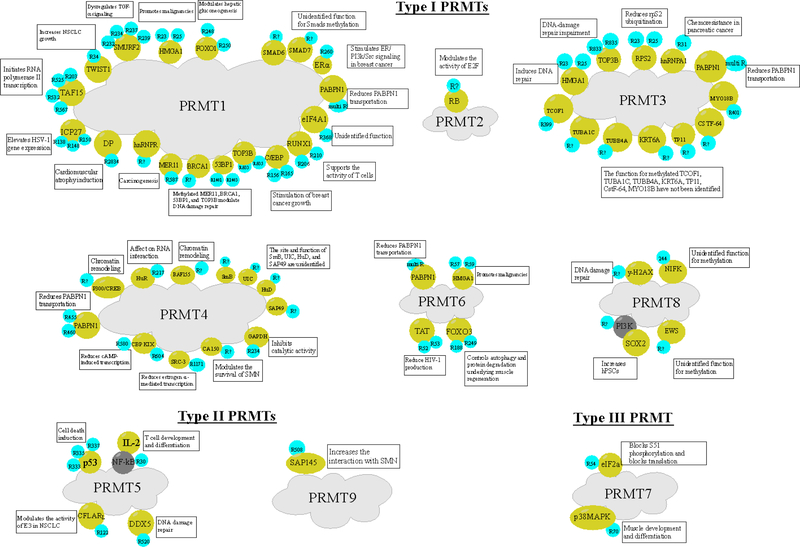
PRMT1 methylates a myriad of non-histone proteins. Based on their molecular functions, these substrates are categorized into four classes. Class 1 are transcriptional factors including TWIST 1 [35], TAF15 [36], Runt-related transcription factor 1 (RUNX1) [37], forkhead box other (FOXO) proteins [38], E2F1 [39], CCAAT/enhancer binding proteins (C/EBP) [40], SMAD6 [41] and SMAD7 [42]. Class 2 are RNA-binding proteins including eukaryotic initiation factor 4A1 (eIF4A1) [43], poly(A)-binding nuclear protein 1 (PABPN1) [44], heterogeneous nuclear ribonucleoprotein R (hnRNPR) [45], and the viral replication modulator ICP27 [46]. Class 3 are involved in DNA damage repair, like the p53-binding protein 1 (53BP1) [47], the double-strand break repair protein MRE11 [48], breast cancer type 1 susceptibility protein (BRCA1) [49], and DNA topoisomerase 3 beta (TOP3B) [50]. Class 4 are proteins that play roles in signal transduction and other functions, including estrogen receptor α (ERα) [51], epidermal growth factor receptor (EGFR) [52], the chromatin-associated proteins high-mobility group AT-hook 1 (HMGA1) [53], SMAD ubiquitination regulatory factor 2 (SMURf2) [54], the structural maintaining protein desmoplakin (DP) [55], and FMS-like tyrosine kinase 3 (FLT3) [56] (Figure 2). Among the aforementioned substrates, the site of arginine methylation is currently unknown for SMADs and hnRNPR [41,42]. Although PRMT1-mediated arginine methylation sites of eIF4A1 (R368), HMGA1 (R23 and R25) and SMURF2 (R232, R234, R237, and R239) have been identified [43,53,54], the function of this methylation remains unclear.
Figure 2.
The Interaction Between PRMT Enzymes with Various Non-Histone Proteins.
2.1.1.1. Transcription Factors TWIST 1, TAF15, C/EBPα, RUNX1, and FOXO as PRMT1 Non-histone Protein Substrates
The transcription factor TWIST1 plays an important role in cancer metastasis by repressing E-cadherin and upregulating N-cadherin, which are characteristic of epithelial-mesenchymal transition (EMT) [57]. PRMT1-mediated methylation of TWIST1 at R34 led to the decrease in E-cadherin expression and increased in N-cadherin expression seen in the epithelial-mesenchymal transition (EMT), implying the regulating role of PRMT1 in cancer progression and metastasis [35]. Knockdown of PRMT1 was able to reduce cell migration and invasion in lung cancer cells [35], indirectly confirming the downstream effects of targeting PRMT1-mediated TWIST1 methylation as an attractive approach for NSCLC treatment.
TAF15, a nuclear RNA-binding protein, is also subject to PRMT1-mediated methylation. TAF15 also binds to RNA targets, notably RNA polymerase II, to initiate transcription. This protein contains both an RNA recognition motif and multiple Arg-Gly-Gly (RGG) repeats at its C-terminal end. PRMT1 selectively methylated the R203, R525, R532, and R567 residues of TAF15 on the C-terminal RGG motifs [36]. The methylation of TAF15 affects its cellular localization between the nucleus and cytoplasm, and positively regulates the expression of TAF15-targeted genes [36]. RUNX1, also known as acute myeloid leukemia 1 (AML1), is essential for hematopoiesis and lymphocyte growth [37]. Methylation of the R206 and R210 residues in RUNX1 enhanced the transcription activity for several gene promotors that are important for the maintenance of a proper peripheral T cell count in mice [37].
C/EBPα belongs to a family of basic leucine zipper DNA-binding proteins and acts as a tumor suppressor by inhibiting cell proliferation [40]. Three arginine residues (R35, R156, and R165) are methylated by PRMT1, which exerts a negative impact on the interaction between C/EBPα and its corepressor HDAC3 to promote cyclin D1 expression [40]. The FOXO proteins are pioneering factors that can directly bind to condensed chromatin, and play important roles in regulating cellular differentiation, growth, survival, and metabolism. The FOXO forkhead box subunit, which contains around 100 amino acids, is essential for DNA binding. Arginine methylation of both FOXO and DP proteins demonstrated a negative impact on the phosphorylation of their respective serine residues [38,55]. PRMT1 methylates the highly conserved R248 and R250 of FOXO1 [38]. The methylation on R248 and R250 blocked the Akt-dependent phosphorylation of the neighboring residue S253, which consequently enhanced cell apoptosis and gluconeogenesis in mouse models [38,58]. The crosstalk between R248/R250 methylation and S253 phosphorylation is possibly caused by the steric hindrance imposed by arginine methylation, which interferes with the recognition of S253 by Akt kinase.
2.1.1.2. RNA-binding Proteins (eIF4A1, ICP27, and PABPN1) as PRMT1 Non-histone Protein Substrates
A substrate profiling study indicated that the R368 residue of the eIF4A1 protein is selectively methylated by PRMT1, while the other eIF4A isoforms (eIF4A2 and eIF4A3) are not methylated by PRMT1 despite over 80% similarity of their sequences to that of eIF4A1 [43]. However, the biochemical significance of eIF4A1 methylation remains ambiguous. The ICP27 protein plays a vital role in the gene expression and replication of herpes simplex virus type-1 (HSV-1) [46]. The PRMT1-catalyzed arginine methylation of ICP27 on R138, R148, and R150 residues is responsible for the nuclear foci-like structure formation, RNA-binding affinity, and SRPK interactions of ICP27. Hypomethylation of ICP27 significantly inhibited the replication of HSV-1 [46], which implies the potential of PRMT inhibitors in HSV-1 treatment. The R289 residue of PABPN1 is preferably methylated by PRMT1 [59]. RNA and transportin are known to compete for binding to PABPN1. Methylation on R289 of PABPN1 reduces the binding between PABPN1 and transportin by about 10-fold and consequently promotes the PABPN1-RNA interaction [59].
2.1.1.3. DNA Damage Repair
Arginine methylation has demonstrated a central role in regulating early response to the DNA damage repair pathway. MRE11, 53BP1, and BRCA1 are three identified protein substrates that exemplify the role of PRMT1-mediated methylation in DNA damage repair. MRE11 is the component of one of the early DNA repair complexes that is recruited at DNA double-strand breaks [48]. The R587 residue of its C-terminal GAR domain is subject to PRMT1 methylation, which is important for its exonuclease activity and association with nuclear structures [48]. Knockdown of PRMT1 resulted in MER11 hypomethylation and consequently induced the intra-S-phase checkpoint defect. 53BP1 is another important DNA damage response factor, which contains a canonical GAR motif in its kinetochore-binding domain that is methylated by PRMT1 [47]. The arginine methylation of 53BP1 is required for its interaction with single and double-stranded DNA, although it did not affect the relocalization to the DNA damage sites [47]. The 504–802 region of tumor suppressor protein BRCA1 is also subject to PRMT1 methylation. Arginine methylation in this region influences the interaction of BRCA1 to specific promoters or proteins [49].
2.1.1.4. DP, ERα, EGFR, and FLT3 as PRMT1 Non-histone Protein Substrates
The aforementioned negative impact of arginine methylation on serine phosphorylation in FOXO was also observed in DP. The most abundant component of desmosomes, DP functions to provide strong adhesion for intercellular links and mechanical strength to tissues like skin and heart [60]. There are four arginine residues (R2826, R2834, R2838, and R2846) on the C-terminus of DP that undergo methylation. Among them, PRMT1-mediated R2834 methylation impedes the adjacent S2849 phosphorylation that is dependent on the glycogen synthase kinase 3, and results in cardiomuscular atrophy in mice [55]. Additionally, cooperative effects between arginine methylation and phosphorylation have been observed in signaling events mediated by ERα, EGFR, and FLT3. PRMT1 specifically methylates R260 in the DNA-binding domain of ERα upon estrogen stimulation [51]. This arginine methylation of ERα activates the cytoplasmic signaling events by forming the Src-ERα-p85 or Src-ERα-PI3K complex in breast cancer cells [51,61]. Although the molecular mechanism of how ERα methylation recruits Src and assemble the complex remains elusive, arginine methylation of ERα plays an important role in triggering its non-genomic signaling cascades in breast cancers. EGFR, a receptor tyrosine kinase, is activated by forming a dimer to mediate numerous signal transduction pathways. Methylation of the R198 and R200 located at the extracellular domain of EGFR promotes its binding to EGF and subsequent dimerization, thus activating the signal pathway in both colorectal and triple-negative breast cancers [52,62]. On the contrary, the arginine methylation at R972 and R973 in FLT3 is dependent on PRMT1 [56,63], which has a positive effect to facilitate the phosphorylation of Y969. In addition, this methylation also induces progression of mixed-lineage leukemia-rearranged Acute Lymphoblastic Leukemia (MLL-r ALL) through an alternative mechanism [56].
2.1.2. PRMT2
PRMT2, also called human arginine methyltransferase (HRMT1L1), primarily localizes in the nucleus. The SH3 domain at the N-terminal of PRMT2 is essential for the interaction with its substrate E1B-AP5, which is a heterogeneous nuclear ribonucleoprotein and interacts with adenovirus early protein E1B [64]. PRMT2 is an underexplored member of the PRMT family and its physiological substrates need to be systematically investigated. PRMT2 is reported to interact with few regulatory proteins, as well as to form an oligomer with PRMT1. PRMT2 directly interacted with retinoblastoma (RB) via its cofactor binding subunit (Figure 2). The interaction between PRMT2 and RB downregulated the activity of E2F through several mechanisms, including histone methylation, transcription factor methylation, and RNA splicing [65].
2.1.3. PRMT3
PRMT3 is abundantly located in the cytoplasm [66]. Its structural features include a catalytic core domain in its central region, and a single zinc-finger domain at its N-terminus that binds to Zn2+ at a 1:1 ratio [67]. The zinc-finger domain contributes not only to the recognition of RNA-associated substrates, but also serves to modulate the catalytic activity of PRMT3 through the recruitment of a different interacting partner, such as the tumor suppressor DAL-1/4.1B [28]. Besides 40S RPS2 [68], the validated non-histone protein substrates of PRMT3 also include HMGA1 proteins [53], PABPN1 [44], hnRNPA1 [69], treacle ribosome biogenesis factor 1 (TCOF1), myosin-XVIIIb (MYO18B), keratin 6A (KRT6A) [28], tubulin alpha 1C (TUBA1C), tubulin beta 4A (TUBB4A), triosephosphate isomerase 1 (TPI1) [28], cleavage stimulation factor 64 kDa subunit (CSTF-64) [45], and DNA topoisomerase 3B (TOP3B) [50] (Figure 2). Among them, arginine methylation sites have been identified, such as the R23 and R25 residues of HMGA1 [53]. PRMT3-mediated arginine methylation at the R31 residue of hnRNPA1 reduces its RNA binding activity to ATP-binding cassette subfamily G member 2 (ABCG2) mRNA, which consequently results in the upregulation of ABCG2 and chemoresistance in pancreatic cancer [69]. However, a deep understanding of the function of arginine methylation on PRMT3 substrates remains lacking, as the elucidation of its biological significance and identification of methylation sites are still under-explored, which delays a deep understanding of the function of PRMT3-mediated arginine methylation.
2.1.4. PRMT4 (CARM1)
PRMT4, also known as CARM1 (coactivator-associated arginine methyltransferase 1), is predominantly located in the nucleus and normally has a positive impact on transcription. PRMT4 consists of a unique N-terminal EVH1 domain (residues 28–140) that can bind to proline-rich sequence [18], which is necessary to substrate recognition and catalytic activity of PRMT4. This also explains why PRMT4 preferentially recognizes arginine residues in PGM-enriched sequences. Based on their known functions, PRMT4-mediated non-histone proteins can be categorized into two classes: class 1 substrates are involved in chromatin remodeling, such as histone acetyltransferase p300/CREB-binding protein [70], critical component of SWI/SNF chromatin remodeling component BAF155 [71], steroid receptor coactivator-3 (SRC-3) [72], and PRMT4 itself at the C-terminal R551 residue; class 2 substrates are involved in RNA processing, such as splicing factors CA150, SAP49, SmB, and U1C [22], R1899 of mediator complex subunit 12 (MED12) [73], the RNA-binding proteins PABP1 [74], HuR, and HuD [22,75] (Figure 2).
PRMT4-mediated methylation has demonstrated diverse functions in gene expression and glucose metabolism. PRMT4-mediated methylation on R580 and R604 residues of the CBP KIX domain [70] selectively diminished cAMP-induced transcription. Methylation of CA150 and MED12 facilitates their interaction with the tudor domain of the survival of motor neurons protein (SMN) to promotes exon skipping, and the tudor domain-containing protein 3 to activate long noncoding RNAs [22], respectively. PRMT4 methylates the R234 residue located in the catalytic pocket of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Methylation of R234 may affect the interaction of coenzyme to GAPDH, which consequently inhibits its catalytic activity [76]. Both R455 and R460 residues of PABP1 were efficiently methylated by PRMT4, but the molecular function of these two arginine methylation on PABP1 remains obscure [74]. The methylation of the R217 residue of HuR is solely dependent on PRMT4 [75]. Since R217 locates in the hinge region of HuR between tandem RNA recognition motif domains, methylation is likely to affect its interaction with RNA or cellular localization through its shuttling mechanism.
2.1.5. PRMT6
PRMT6 is able to automethylate its R35 residue, which increases the stability of PRMT6 and inhibits human immunodeficiency virus type 1 (HIV-1) replication [77]. PRMT6 is found predominantly in the cell nucleus [78], and is known as a transcriptional repressor. Besides its ability to methylate H2A protein on its R11 and R29 residues [79], TAT [80], FOXO3 [81], HMGA1 [53], and PABPN1 [44] have been reported to be non-histone protein substrates for PRMT6 (Figure 2). For example, the PRMT6-dependent methylation at R52 and R53 residues of the TAT protein impairs the interaction of TAT with the Tat transactivation region of human immunodeficiency virus type 1 (HIV-1), as well as cyclin T1-dependent TAT transcriptional activation [80]. Hence, overexpression of wild-type PRMT6 reduced TAT transactivation of HIV-1, and the knockdown of PRMT6 elevated viral replication [80]. In this context, PRMT6 activators would be beneficial to combat HIV replication.
Additionally, PRMT6 methylates the R57 and R59 residues of chromatin protein HMGA1 in the second AT-hook [53], which may perturb the HMGA1-involved protein-DNA and protein-protein interactions. PRMT6-mediated FOXO3 methylation at R188 and R249 residues can activate the activity of FOXO3 to induce protein degradation and result in muscle atrophy [81]. However, PRMT6 expression can be negatively regulated by PRMT1, which consequently serves an important role in skeletal muscle metabolism regeneration [81]. Therefore, the side effects of PRMT1 inhibitor on skeletal muscle need to be monitored.
2.1.6. PRMT8
PRMT8 shows a high tissue-specific expression pattern in brain and neuron tissues [82]. Although PRMT1 and PRMT8 share over 80% homology similarity [82], the recent tetrameric structure of PRMT8 in solution implies the regulatory role on substrate selectivity by an allosteric beta strand [83]. The PRMT8 N-terminal region supports the association of PRMT8 to the plasma membrane [82]. Additionally, the N-terminus contains the substrate binding site and is essential for PRMT8 methylation activity. Besides histone proteins, PRMT8 can automethylate its N-terminal R108 and R182 residues [84], as well as the R244 residue of Nucleolar protein interacting with an RNA-binding protein, the forkhead-associated domain of Ki-67 (NIFK) [83], and the C-terminal RGG region of pro-oncoprotein Ewing sarcoma (EWS) [85] (Figure 2). However, the impact of those methylation events is still obscure. As a less characterized member, PRMT8 has been suggested to involve in PI3K/AKT-dependent Sox2 induction through its interaction with the p85 subunit of PI3K in human embryonic stem cells [86]. In addition, knockout of PRMT8 in mice resulted in progressive muscle atrophy, axonal transport impairment, γ-H2AX accumulation, and reduced level of cAMP response-element-binding protein 1 (CREB1) [87].
2.2. Type II PRMTs
2.2.1. PRMT5
PRMT5 is a founding member of type II PRMT, which interacts with various proteins. PRMT5 was initially named Janus kinase-binding protein 1 (JBP1) because of its binding to the Janus Kinase [88]. Recent studies manifest the role of PRMT5 as a master epigenetic regulator in various cellular processes. PRMT5 consists of four major domains: a N-terminal TIM-barrel domain, the Rossmann-fold in the middle, a ß-barrel subunit at the C-terminal end, and a dimerization arm that contains around 60 residue that is close to the ß-barrel domain [89]. The conserved F379 residue in the PRMT5 active site governs the product specificity of PRMT5 to produce the symmetric di-methylated arginine. The F379M mutation not only increased methylation activity of PRMT5, but also altered the product specificity by producing both symmetric and asymmetric di-methylation of H4R3. Conversely, both F379G and F379A mutation reduced the PRMT5 activity significantly [89]. Specific methylation on H4R3 instead of R17 or R19 on a 21-mer H4 peptide (SGRGKGGKGLGKGGAKRHRKV) corroborated that PRMT5 preferably methylates substrates containing GRG motifs. In addition, the reduction of H4R3 methylation by 40–50 fold was caused by the H41–21 (R17A) or (H18Q) mutation, further exemplifying the contribution of the distal residues to the substrate recognition through the electronic and spatial changes in the peptide structure [21]. PRMT5 interacts with various partners, including cooperator of PRMT5 (COPR5) [90], methylosome protein 50 (MEP50/WDR77) [91], transcriptional factor Sp1, and ATP-dependent chromatin remodeler BRG1, to regulate the substrate recognition and methylation activity [92].
PRMT5 has been implicated in several cellular processes such as RNA processing, signaling, and gene regulation. PRMT5 modulates gene transcription through the methylation of H2AR3, H3R8, and H4R3. Besides histones and well-known Sm protein substrates, PRMT5 also methylates several non-histone proteins, including p53 [93], nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) [94], Krüppel-like factor (KLF4) [95], E2F1 [96], CFLARL [97], and DEAD-box helicase 5 (DDX5) [98] (Figure 2).
PRMT5 is implicated in DNA damage response through installation of methylation on p53, DDX5, and KLF4. PRMT5 negatively regulates DNA damage-induced apoptosis through the modulation of the tumor suppressor protein, p53 [93]. The PRMT5-dependent methylation of p53 at residues R333, R335 and R337 represses the p53-DNA interaction. Hence the p53 methylation alters the promoter specificity and leads to functional outcome impairment. Knockdown of PRMT5 induced p53-dependent apoptosis in U2OS cells [93], which infers the application of PRMT5 inhibitors in activation of the p53 pathway. The DDX5 is an ATP-dependent RNA helicase that is involved in the suppression of the formation of a DNA:RNA hybrid to minimize the DNA damage. Recently, PRMT5 has been shown to upregulate DDX5 through methylation at the R520 residue, within the C-terminal GRG-rich region of DDX5 [98]. Therefore, repression of PRMT5 led to the accumulation of DNA:RNA hybrid and subsequent DNA double-strand damage. The transcription factor KLF4 regulates DNA damage response, inflammation, apoptosis, stem cell renewal, and genome integrity maintenance. Three arginine residues (R374, R376, and R377) of KLF4 can be methylated by PRMT5 [95]. KLF4 has a short half-life at about 4 hours [95]. The arginine methylation leads to the conformation change of KLF4, which subsequently affects the recognition by von Hippel-Lindau (VHL)/VBC E3 ligase and counteracts the ubiquitination to increase the stability of KLF4 [95]. The accumulated and defective KLF4 results in attenuation of DNA damage response, inhibition of apoptosis, and breast tumor progression [95]. The KLF4 arginine methylation demonstrates an interesting interplay between arginine methylation and ubiquitylation, where arginine methylation plays a negative role in ubiquitination.
In addition to KLF4, NF-kB transcription factors involved in inflammation, immune responses, and malignancies also undergo arginine methylation by PRMT5 [94]. Aberrant expression of PRMT5 promotes the arginine methylation on the R30 residue of p65 subunit of NF-kB and increases the NF-kB activity in immune responses and tumorigenesis. PRMT5 knockdown or the p65 subunit R30A mutation results in NF-kB repression [94], which further highlights the importance of PRMT5-mediated methylation in NF-kB activity.
PRMT5-mediated SDMA and PRMT1-mediated ADMA can exert disparate effects on the same protein. One example is the transcription factor E2F, which plays a master role in controlling the cell cycle progression. The disparate effects resulted from two different methylation states on E2F1 illustrate the importance of specific methylation events. The R111 and R113 residues of E2F1 are subject to symmetric dimethylation by PRMT5, while its R109 is modified by PRMT1 to introduce asymmetric dimethylation on the R109 site [39,96]. PRMT5-dependent methylation of E2F promotes cell growth and regulates alternative splicing of E2F target genes [39,96]. On the other hand, PRMT1-mediated arginine methylation has distinct effects on stimulating apoptosis in DNA damaged cells. Another example is CFLARL, an essential anti-apoptotic protein that represses the activity of caspase 8 [99]. In this case, PRMT1 and PRMT5 methylate the same R122 residue of CFLARL, but produce opposite impacts on the CFLARL protein level in NSCLC cells. Specifically, PRMT5-mediated symmetric dimethylation of CFLARL elevates the expression level of CFLARL by reducing its ubiquitination [97]. Such crosstalk between symmetric arginine methylation and ubiquitination has also been observed in KLF4 as discussed above. In contrast, asymmetric dimethylation of R122 by PRMT1 downregulated CFLARL levels through the elevation of CFLARL ubiquitination and degradation [97]. The distinct downstream effects that are caused by different methylation states on the same arginine residue is another example to clearly demonstrate the need to develop selective modulators for each PRMT isoform.
2.2.2. PRMT9
The latest identified member of the PRMT enzymes, PRMT9, was initially known as FLJ12673 or FBXO11 [100]. PRMT9 methylates fewer substrates than the other type II PRMT, PRMT5 [24]. Like PRMT7, PRMT9 also contains two MTase domains and forms a pseudodimer for substrate binding. The FKRKY motif is essential for PRMT9 substrate recognition at the N-terminal domain [101]. PRMT9 barely exhibited any activity on histones, but methylates the R508 residue of intact spliceosome-associated protein SAP145 (SF3b2) to produce MMA and SDMA [101] (Figure 2). This arginine methylation is important for the interaction of SAP145 to the survival motor neuron protein [24].
2.3. Type III PRMT7
PRMT7 was originally identified as a type II PRMT to methylate both MMA and SDMA [102], but established its uniqueness as the only type III PRMT as PRMT7 strictly monomethylates its substrates, including H4R17 [103]. This specific monomethylation may be caused by a more constricted substrate-binding site pocket in PRMT7 compared to other types of PRMT [17]. PRMT7 plays important roles in transcriptional regulation, DNA damage response, RNA splicing, cell differentiation, and metastasis [104]. PRMT7 has been reported to methylate eukaryotic translation initiation factor 2 alpha (eIF2α) and p38 mitogen-activated protein kinase (p38MAPK) [105,106] (Figure 2).
The eIF2α arginine methylation at R54 has been implied as the methylation site for PRMT7, which can affect stress-induced phosphorylation on serine 51 (Ser51) and block translation initiation [105]. The p38MAPK is a class of mitogen-activated protein kinases that is essential for myogenic differentiation, immune modulation, and inflammatory reactions [107]. PRMT7-mediated methylation of p38MAPK on its R70 residue promotes the activation of p38MAPK and MyoD-mediated myoblast differentiation. The p38MAPK mutation A70R or PRMT7 deficiency resulted in a decrease in myogenic differentiation and a retardation of muscle regeneration in mice [106].
3. THE ROLE OF PRMTS IN DISEASES
The dysregulation of various PRMT subtypes is implicated in various diseases, including cancer, metabolic dysfunction, inflammation, and cardiovascular diseases. Compared with the pharmacological roles of PRMTs in cancer, the information is sparse in other diseases. Here, this review mainly focuses on cancers involved the arginine methylation on non-histone proteins.
The overexpression of type I PRMTs leads to the enhancement of asymmetric dimethylarginine levels and is associated with several malignancies. For example, PRMT1 overexpression has been reported in melanoma, leukemia, lymphoma, breast and lung cancers. Additionally, PRMT4 is upregulated in lung cancer and osteosarcoma (OS) [108]. The overexpression of PRMT8 in breast, cervical, and ovarian malignancies is well-known [109]. Either depletion or inhibition of those PRMTs has exhibited anticancer activities. In non-transformed lung epithelial cells, PRMT1 has been shown to trigger the epithelial-mesenchymal transition (EMT), a process integral to cancer progression, through the upregulation of N-cadherin and the repression of the tumor suppressor E-cadherin [35]. The knockdown of PRMT1 is correlated with the upregulation of E-cadherin levels and results in the reversal of EMT. PRMT1 overexpression is also common in human melanoma, where it regulates growth and metastasis by interaction with the activated leukocyte cell adhesion molecule (ALCAM). The reduction of ALCAM levels upon PRMT1 knockdown in A375 and Hs294T melanoma cell lines resulted in the suppression of tumor cell growth suppression and reduced metastatic ability [110]. These results suggest that PRMT inhibition may have antiproliferative effects that can be exploited in the development of future cancer treatments.
PRMT5 plays an important role in governing cancer cell proliferation and tumorigenesis through activating some proliferative signaling including WNT/β-CATENIN, AKT-involved pathway. Aforementioned PRMT5-mediated KLF4 methylation has been proposed as a targeted therapy for triple negative breast cancers. Indeed, specific disrupting the PRMT5 and KLF4 interaction suppresses tumor progression and stimulates tumor cell death in both cell-based and animal studies in triple negative breast cancer [111]. Elevated levels of PRMT5 are observed in hepatocellular carcinoma (HCC) tissues. PRMT5 knockdown inhibits the growth of HCC cells and selectively abrogates their proliferation, with no effect on the growth of normal liver cells. Additionally, PRMT5 silencing significantly downregulated the HCC-associated signaling protein β-catenin, which suggests that PRMT5 is a regulator of HCC tumor growth and proliferation [112]. PRMT9 suppression by miR-543 promotes the proliferation of osteosarcoma cells [113]. Conversely, the overexpression of the type I PRMT4 has been shown to promote osteosarcoma cell proliferation by increasing the phosphorylation of GSK3β to activate β-catenin/cyclin D1 signaling [114]. PRMT7 expression is markedly upregulated in breast cancer cells and elevates the expression of the breast cancer metastasis mediator matrix metalloproteinase 9 (MMP9). The inhibition of PRMT7 with siRNA is correlated with the reduction of breast cancer cell invasion, metastasis, and progression. Targeting PRMT7 may support the development of selective therapies for breast cancer and prevent the possibility for metastasis and recurrence [115].
Both type I and II PRMT mainly methylate substrates containing GAR motifs, like E2F1. Thus, it is not surprising about the crosstalk existing between these two types of PRMTs. Recent study identified PRMT1 as a significant hit to function synergistically with PRMT5 inhibitor through a combination screening with a CRISPR/Cas9 library [116]. Combination of both PRMT1 and 5 inhibitors display a strong anti-tumor activity in a synergistic manner. The knockdown of PRMT1 and PRMT6 resulted in the suppression of bladder and lung cancer proliferation [117]. Meanwhile, the interplay between arginine methylation and other modifications have been observed, including phosphorylation and ubiquitination. Thus, dual inhibition of both PRMT5 and AKT by their respective inhibitors exhibited lethal effects on diffuse large B cell lymphoma cells [118]. Dual inhibition of both PRMT1 and FLT3 inhibitors enhanced abolishment of MLL-r ALL cells in patient-derived mouse xenografts [64]. Such synergistic effect has also been observed in PRMT1-FLT3 involved pathway in acute myeloid leukemia [119].
4. PRMT INHIBITORS IN CLINICAL TRIALS
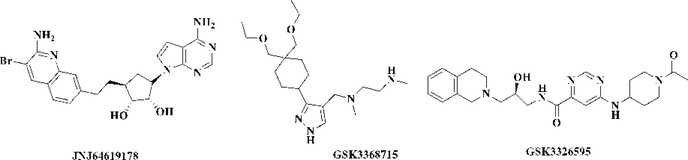
The physiological and pharmacological functions of PRMTs imply their potentials as novel therapeutic targets. As such, there exists a pressing need for the development of selective, potent, and cell-active PRMT inhibitors. Hence, a plethora of PRMT inhibitors have been developed in order to target different PRMT-involved abnormalities. As discussed earlier, different arginine methylation states (ADMA or SDMA) on the same residue may have dichotomic downstream effects, accentuating the importance of the discovery of isoform-specific inhibitors to target each PRMT. Recent progress on the elucidation of the substrate recognition and molecular pathways has greatly advanced the development of specific inhibitors for PRMTs except PRMT2, PRMT8, and PRMT9. Currently, multiple clinical trials are ongoing for the PRMT1 and 5 inhibitors JNJ46419178, GSK3368715 [120], GSK3326595 [121], and PF-06939999 (Figure 3, Table 2). Moreover, several PRMT3, PRMT4 and PRMT6 selective inhibitors [122–125] have shown superior activity in preclinical studies. Phase I trials are underway for a potent and selective type I PRMT inhibitor, GSK3368715, for the targeting of several tumors including the refractory pancreatic, NSCLC, and bladder solid tumors, and the hematologic tumor of diffuse large B-cell lymphoma (DLBCL). The goal of these trials is to assess the therapeutic activity and the maximally tolerated oral dose for GSK3368715 in the treated patients [120,126]. Currently, the competition in the development of PRMT5 inhibitors is intensive, with three different inhibitors under clinical investigation. The SAM analog PRMT5 inhibitor, JNJ46419178, was developed by Janssen Research & Development, LLC. Phase I studies are currently being conducted to evaluate the safety, pharmacokinetics, and pharmacodynamics of JNJ46419178 in patients with refractory B cell non-Hodgkin lymphoma (NHL) or advanced solid tumors [127]. Meanwhile, two clinical studies are currently ongoing for a selective substrate competitive PRMT5 inhibitor, GSK3326595. Solo safety, pharmacokinetics, and pharmacodynamics phase I trials are being conducted on subjects with recurrent solid tumors and NHL to investigate the maximally tolerated oral dose for further clinical studies [128]. Another phase II study for GSK3326595 is being carried out in combination with 5-azacitidine to treat AML and myelodysplastic syndrome (MDS) [129]. In addition, phase I trials for the PRMT5 inhibitor PF-06939999 (structure not disclosed) are underway to evaluate the safety, pharmacokinetics, and pharmacodynamics of PF-06939999 in patients with advanced or metastatic solid tumors [130].
Figure 3.
The Chemical Structure for PRMT Inhibitors in Clinical Trials
Table 2.
PRMT Inhibitors in Clinical Trials.
| Name | Target | Company | Starting date | Phase | Treated cancer | Clinical trial identifier |
|---|---|---|---|---|---|---|
| JNJ46419178 | PRMT5 | Janssen | Jul-18 | Phase I | Solid tumor, adult NHL, and myelodysplastic syndromes | NCT03573310 [127] |
| GSK3368715 | Type I PRMTs | GlaxoSmithKline | Oct-18 | Phase I | Solid tumors and DLBCL | NCT03666988 [126] |
| GSK3326595 | PRMT5 | GlaxoSmithKline | Aug-16 | Phase I | Solid tumors and NHL | NCT02783300 [128] |
| GSK3326595 | PRMT5 | GlaxoSmithKline | Oct-18 | Phase II | Myelodysplastic syndrome and acute myeloid leukemia | NCT03614728 [129] |
| PF-06939999 | PRMT5 | Pfizer | Mar-19 | Phase I | Advanced solid tumors | NCT038544227 [130] |
5. OPPORTUNITIES
PRMT enzymes regulate the activity of histones and a wide spectrum of non-histone proteins that are essential for cellular regulation, including RNA binding proteins, signal transduction modulators, and transcription factors. Depending on the site of modification, PRMTs either act as coactivators or repressors for the effector proteins. Protein arginine methylation modulates protein-protein, protein-DNA, and protein-RNA interactions. PRMT enzymes interact with a huge number of macromolecules including themselves in the form of dimers and oligomers to profoundly regulate the substrate recognition and enzymatic activity. Recently, a new array of non-histone protein substrates and interacting partners has been unveiled, expanding the scope and complexity of arginine methylation to a different dimension. For example, the substrate specificity of PRMT5 varies in presence of different cofactors. Additionally, oligomers display different activity and sensitivity to recognize histones versus non-histone proteins. These additional complexities must therefore be taken into consideration throughout any new investigations. The elucidation of each PRMT involved-network is imperative to pave the road to develop selective and cell-potent inhibitors as novel therapeutic agents.
Many reports on the aberrant levels of various PRMT enzymes have been implicated in several pathological conditions such as cancer, diabetes, cardiovascular and inflammatory diseases. However, the mechanisms and roles of the dysregulation of PRMT enzymes in specific diseases are still poorly understood. Highly subtype-selective inhibitors may have a high impact on deciphering the underlying functions and network control of PRMT-related diseases. On the other hand, emerging links to cellular dysfunction strengthen that PRMTs are attractive targets for a number of diseases. Significant efforts have been applied with multiple clinical trials ongoing for PRMT1 and 5 inhibitors. Future directions of inhibitor discovery should include but not be limited to specifically targeting newly identified and unique pathways.
ACKNOWLEDGEMENTS
This work was funded by the National Institutes of Health R01GM117275 (R.H.), U01CA214649 (R.H.), and Purdue University.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- [1].Dawson MA; Kouzarides T Cancer Epigenetics: From Mechanism to Therapy. Cell, 2012, 150, 12–27. [DOI] [PubMed] [Google Scholar]
- [2].Johann PD; Erkek S; Zapatka M; Kerl K; Buchhalter I; Hovestadt V; Jones DTW; Sturm D; Hermann C; Segura Wang M; Korshunov A; Rhyzova M; Gröbner S; Brabetz S; Chavez L; Bens S; Gröschel S; Kratochwil F; Wittmann A; Sieber L; Geörg C; Wolf S; Beck K; Oyen F; Capper D; van Sluis P; Volckmann R; Koster J; Versteeg R; von Deimling A; Milde T; Witt O; Kulozik AE; Ebinger M; Shalaby T; Grotzer M; Sumerauer D; Zamecnik J; Mora J; Jabado N; Taylor MD; Huang A; Aronica E; Bertoni A; Radlwimmer B; Pietsch T; Schüller U; Schneppenheim R; Northcott PA; Korbel JO; Siebert R; Frühwald MC; Lichter P; Eils R; Gajjar A; Hasselblatt M; Pfister SM; Kool M Atypical Teratoid/Rhabdoid Tumors are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell, 2016, 29, 379–393. [DOI] [PubMed] [Google Scholar]
- [3].Falkenberg KJ; Johnstone RW Histone Deacetylases and Their Inhibitors in Cancer, Neurological Diseases and Immune Disorders. Nat. Rev. Drug Discov, 2014, 2, 673–691. [DOI] [PubMed] [Google Scholar]
- [4].Kelly WK; O’connor OA; Krug LM; Chiao JH; Heaney M; Curley T; Gregore-Cortelli B; Tong W; Secrist JP; Schwartz L; Richardson S; Chu E; Olgac S; Marks PA; Scher HI; Richon VM Phase I Study of an Oral Histone Deacetylase Inhibitor, Suberoylanilide Hydroxamic Acid, in Patients with Advanced Cancer. J. Clin. Oncol, 2005, 23, 3923–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].VanderMolen KM; McCulloch W; Pearce CJ; Oberlies NH Romidepsin (Istodax, NSC 630176, FR901228, FK228, Depsipeptide): A Natural Product Recently Approved for Cutaneous T-Cell Lymphoma. J. Antibiot. (Tokyo), 2011, 64, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Campbell P; Thomas CM Belinostat for the Treatment of Relapsed or Refractory Peripheral T-Cell Lymphoma. J. Oncol. Pharm. Pract, 2016, 7, 209–218. [DOI] [PubMed] [Google Scholar]
- [7].Garnock-Jones KP Panobinostat: First Global Approval. Drugs, 2015, 75, 695–704. [DOI] [PubMed] [Google Scholar]
- [8].Jahan S; Davie JR Advances in Biological Regulation Protein Arginine Methyltransferases ( PRMTs ): Role in Chromatin Organization. Adv. Biol. Regul, 2015, 57, 173–184. [DOI] [PubMed] [Google Scholar]
- [9].Jarrold J; Davies CC PRMTs and Arginine Methylation : Cancer’s Best-Kept Secret? Trends Mol. Med, 2019, just accep, 1–16. [DOI] [PubMed] [Google Scholar]
- [10].Yang Y; Bedford MT Protein Arginine Methyltransferases and Cancer. Nat. Rev. Cancer, 2012, 13. [DOI] [PubMed] [Google Scholar]
- [11].Matsuguma K; Ueda S; Yamagishi S; Matsumoto Y; Kaneyuki U; Shibata R; Fujimura T; Matsuoka H; Kimoto M; Kato S; Imaizumi T; Okuda S Molecular Mechanism for Elevation of Asymmetric Dimethylarginine and Its Role for Hypertension in Chronic Kidney Disease. J. Am. Soc. Nephrol, 2006, 17, 2176–2183. [DOI] [PubMed] [Google Scholar]
- [12].Parry RV; Ward SG Protein Arginine Methylation : A New Handle on T Lymphocytes? Trends Immunol, 2010, 31, 164–169. [DOI] [PubMed] [Google Scholar]
- [13].Han H; Choi D; Choi S; Koo S Roles of Protein Arginine Methyltransferases in the Control of Glucose Metabolism. Endocrinolgy Metab, 2014, 29, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang L; Liu J; Zhang X; Sibley K; Najjar SM; Lee MM; Wu JQ Inhibition of Protein Arginine Methyltransferase 5 Enhances Hepatic Mitochondrial Biogenesis. J. Biol. Chem, 2018, 293, 10884–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Iwasaki H Impaired PRMT1 Activity in the Liver and Pancreas of Type 2 Diabetic Goto–Kakizaki Rats. Life Sci, 2009, 85, 161–166. [DOI] [PubMed] [Google Scholar]
- [16].Schapira M; de Freitasa RF Structural Biology and Chemistry of Protein Arginine Methyltransferases. Med. Chem. Commun, 2014, 5, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hasegawa M; Toma-Fukai S; Kim JD; Fukamizu A; Shimizu T Protein Arginine Methyltransferase 7 Has a Novel Homodimer-like Structure Formed by Tandem Repeats. FEBS Lett, 2014, 588, 1942–1948. [DOI] [PubMed] [Google Scholar]
- [18].Shishkova E; Zeng H; Liu F; Kwiecien NW; Hebert AS; Coon JJ; Xu W Global Mapping of CARM1 Substrates Defines Enzyme Specificity and Substrate Recognition. Nat. Commun, 2017, 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Morales Y; Cáceres T; May K; Hevel JM Biochemistry and Regulation of the Protein Arginine Methyltransferases (PRMTs). Arch. Biochem. Biophys, 2016, 590, 138–152. [DOI] [PubMed] [Google Scholar]
- [20].Osborne TC; Obianyo O; Zhang X; Cheng X; Thompson PR Protein Arginine Methyltransferase 1: Positively Charged Residues in Substrate Peptides Distal to the Site of Methylation are Important for Substrate Binding and Catalysis. Biochemistry, 2007, 46, 13370–13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang M; Xu R-M; Thompson PR Substrate Specificity, Processivity, and Kinetic Mechanism of Protein Arginine Methyltransferase 5. Biochemistry, 2013. 52,5430–5440. [DOI] [PubMed] [Google Scholar]
- [22].Cheng D; Côté J; Shaaban S; Bedford MT The Arginine Methyltransferase CARM1 Regulates the Coupling of Transcription and MRNA Processing. Mol. Cell, 2007, 25, 71–83. [DOI] [PubMed] [Google Scholar]
- [23].Feng Y; Maity R; Whitelegge JP; Hadjikyriacou A; Li Z; Zurita-Lopez C; Al-Hadid Q; Clark AT; Bedford MT; Masson JY; Clarke SG Mammalian Protein Arginine Methyltransferase 7 (PRMT7) Specifically Targets RXR Sites in Lysine- and Arginine-Rich Regions. J. Biol. Chem, 2013, 288, 37010–37025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang Y; Hadjikyriacou A; Xia Z; Gayatri S; Kim D; Zurita-Lopez C; Kelly R; Guo A; Li W; Clarke SG; Bedford MT PRMT9 is a Type II Methyltransferase that Methylates the Splicing Factor SAP145. Nat. Commun, 2015, 6, 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fulton MD; Brown T; Zheng YG Mechanisms and Inhibitors of Histone Arginine Methylation. Chem. Rec, 2018, 18, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kölbel K; Ihling C; Bellmann-Sickert K; Neundorf I; Beck-Sickinger AG; Sinz A; Kühn U; Wahle E Type I Arginine Methyltransferases PRMT1 and PRMT-3 Act Distributively. J. Biol. Chem, 2009, 284, 8274–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang M; Fuhrmann J; Thompson PR Protein Arginine Methyltransferase 5 Catalyzes Substrate Dimethylation in a Distributive Fashion. Biochemistry, 2014, 53, 7884–7892. [DOI] [PubMed] [Google Scholar]
- [28].Guo H; Wang R; Zheng W; Chen Y; Blum G; Deng H; Luo M Profiling Substrates of Protein Arginine N-Methyltransferase 3 with S-Adenosyl-L-Methionine Analogues. ACS Chem. Biol, 2014, 9, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Musiani D; Bok J; Massignani E; Wu L; Tabaglio T; Ippolito MR; Cuomo A; Ozbek U; Zorgati H; Ghoshdastider U; Robinson RC; Guccione E; Bonaldi T Proteomics Profiling of Arginine Methylation Defines PRMT5 Substrate Specificity. Sci. Signal, 2019, 12,eaat8388. [DOI] [PubMed] [Google Scholar]
- [30].Obianyo O; Causey CP; Jones JE; Thompson PR Activity-Based Protein Profiling of Protein Arginine Methyltransferase 1. ACS Chem. Biol, 2011, 6, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lim Y; Kwon YH; Nam HW; Min BH; Park IS; Woon KP; Kim S Multimerization of Expressed Protein-Arginine Methyltransferases During the Growth and Differentiation of Rat Liver. Biochim. Biophys. Acta - Gen. Subj, 2005, 172, 240–247. [DOI] [PubMed] [Google Scholar]
- [32].Gui S; Wooderchak WL; Daly MP; Porter PJ; Johnson SJ; Hevel JM Investigation of the Molecular Origins of Protein-Arginine Methyltransferase I (PRMT1) Product Specificity Reveals a Role for Two Conserved Methionine Residues. J. Biol. Chem, 2011, 286, 29118–29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rust HL; Zurita-Lopez C; Clarke S; Thompson PR Mechanistic Studies on Transcriptional Coactivator Protein Arginine Methyltransferase 1. Biochemistry, 2011, 50, 3332–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gui S; Gathiaka S; Li J; Acevedo O; Hevel JM A Remodeled Protein Arginine Methyltransferase 1 (PRMT1) Generates Symmetric Dimethylarginine. J. Biol. Chem, 2014, 289, 9320–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Avasarala S; Van Scoyk M; Rathinam MKK; Zerayesus S; Zhao X; Zhang W; Pergande MR; Borgia JA; DeGregori J; Port JD; Winn RA; Bikkavilli RK PRMT1 is a Novel Regulator of Epithelial-Mesenchymal-Transition in Non-Small Cell Lung Cancer. J. Biol. Chem, 2015, 290, 13479–13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jobert L; Argentini M; Tora L PRMT1 Mediated Methylation of TAF15 is Required for Its Positive Gene Regulatory Function. Exp. Cell Res, 2009, 315, 1273–1286. [DOI] [PubMed] [Google Scholar]
- [37].Mizutani S; Yoshida T; Zhao X; Nimer SD; Taniwaki M; Okuda T Loss of RUNX1/AML1 Arginine-Methylation Impairs Peripheral T Cell Homeostasis. Br. J. Haematol, 2015, 170, 859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yamagata K; Daitoku H; Takahashi Y; Namiki K; Hisatake K; Kako K; Mukai H; Kasuya Y; Fukamizu A Arginine Methylation of FOXO Transcription Factors Inhibits Their Phosphorylation by Akt. Mol. Cell, 2008, 32, 221–231. [DOI] [PubMed] [Google Scholar]
- [39].Roworth AP; Carr SM; Liu G; Barczak W; Miller RL; Munro S; Kanapin A; Samsonova A; La Thangue NB Arginine Methylation Expands the Regulatory Mechanisms and Extends the Genomic Landscape under E2F Control. Sci. Adv, 2019, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu L-M; Sun W-Z; Fan X-Z; Xu Y-L; Cheng M-B; Zhang Y Methylation of C/EBPα by PRMT1 Inhibits Its Tumor-Suppressive Function in Breast Cancer. Cancer Res, 2019, 79, 2865 LP–2877. [DOI] [PubMed] [Google Scholar]
- [41].Inamitsu M; Itoh S; Hellman U; Kato M Methylation of Smad6 by Protein Arginine N-Methyltransferase 1. FEBS Lett, 2006, 580, 6603–6611. [DOI] [PubMed] [Google Scholar]
- [42].Katsuno Y; Qin J; Oses-Prieto J; Wang H; Jackson-Weaver O; Zhang T; Lamouille S; Wu J; Burlingame A; Xu J; Derynck R Arginine Methylation of SMAD7 by PRMT1 in TGF-ß–Induced Epithelial–Mesenchymal Transition and Epithelial Stem-Cell Generation. J. Biol. Chem, 2018, 293, 13059–137072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wooderchak WL; Zang T; Zhou ZS; Acuña M; Tahara SM; Hevel JM Substrate Profiling of PRMT1 Reveals Amino Acid Sequences That Extend beyond the “RGG” Paradigm. Biochemistry, 2008, 47, 9456–9466. [DOI] [PubMed] [Google Scholar]
- [44].Fronz K; Otto S; Kölbel K; Kühn U; Friedrich H; Schierhorn A; Beck-Sickinger AG; Ostareck-Lederer A; Wahle E Promiscuous Modification of the Nuclear Poly(A)-Binding Protein by Multiple Protein-Arginine Methyltransferases Does Not Affect the Aggregation Behavior. J. Biol. Chem, 2008, 283, 20408–20420. [DOI] [PubMed] [Google Scholar]
- [45].Kim C; Lim Y; Yoo BC; Won NH; Kim S; Kim G Regulation of Post-Translational Protein Arginine Methylation during HeLa Cell Cycle. Biochim. Biophys. Acta-Gen. Subj, 2010, 1800, 977–985. [DOI] [PubMed] [Google Scholar]
- [46].Yu J; Shin B; Park ES; Yang S; Choi S; Kang M; Rho J Protein Arginine Methyltransferase 1 Regulates Herpes Simplex Virus Replication through ICP27 RGG-Box Methylation. Biochem. Biophys. Res. Commun, 2010, 391, 322–328. [DOI] [PubMed] [Google Scholar]
- [47].Boisvert FM; Rhie A; Richard S; Doherty AJ The GAR Motif of 53BP1 is Arginine Methylated by PRMT1 and is Necessary for 53BP1 DNA Binding Activity. Cell Cycle, 2005, 4, 1834–1841. [DOI] [PubMed] [Google Scholar]
- [48].Boisvert FM; Déry U; Masson JY; Richard S Arginine Methylation of MRE11 by PRMT1 is Required for DNA Damage Checkpoint Control. Genes Dev, 2005, 19, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Guendel I; Carpio L; Pedati C; Schwartz A; Teal C; Kashanchi F; Kehn-Hall K Methylation of the Tumor Suppressor Protein, BRCA1, Influences Its Transcriptional Cofactor Function. PLoS One, 2010, 5, e11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huang L; Wang Z; Narayanan N; Yang Y Arginine Methylation of the C-Terminus RGG Motif Promotes TOP3B Topoisomerase Activity and Stress Granule Localization. Nucleic Acids Res, 2018, 46, 3061–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Romancer ML; Treilleus I; Leconte N; Robin-Lespinasse Y; Sentis S; Bouchekioua-Bouzaghou K; Goddard S; Gobert-Gosse S; Corbo L Regulation of Estrogen Rapid Signaling through Arginine Methylation by PRMT1. Mol. Cell, 2008, 31, 212–221. [DOI] [PubMed] [Google Scholar]
- [52].Nakai K; Xia W; Liao HW; Saito M; Mien-chie Hung MC; Yamaguchi H The Role of PRMT1 in EGFR Methylation and Signaling in MDA-MB-468 Triple-Negative Breast Cancer Cells. Breast Cancer, 2017, 7, 2587–2599. [DOI] [PubMed] [Google Scholar]
- [53].Zou Y; Perna AD; Zhang Q; Wang Y; Webb K; Clarke S A Mass Spectrometric Study on the in Vitro Methylation of HMGA1a and HMGA1b Proteins by PRMTs: Methylation Specificity, the Effect of Binding to AT-Rich Duplex DNA, and the Effect of C-Terminal Phosphorylation. Biochemistry, 2007, 46, 7896–7906. [DOI] [PubMed] [Google Scholar]
- [54].Cha B; Park Y; Hwang BN; Kim S; Jho E Protein Arginine Methyltransferase 1 Methylates Smurf2. Mol. Cells, 2015, 38, 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Albrecht LV; Zhang L; Shabanowitz J; Purevjav E; Towbin JA; Hunt DF; Green KJ GSK3- and PRMT-1–Dependent Modifications of Desmoplakin Control Desmoplakin–Cytoskeleton Dynamics. J. Cell Biol. Biol, 2015, 208, 597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhu Y; He X; Lin Y-C; Dong H; Zhang L; Chen X; Wang Z; Shen Y; Li M; Wang H; Sun J; Nguyen LX; Zhang H; Jiang W; Yang Y; Chen J; Müschen M; Chen CW; Konopleva MY; Sun W; Jin J; Carlesso N; Marcucci G; Luo Y; Li L Targeting PRMT1-Mediated FLT3 Methylation Disrupts Maintenance of MLL- Rearranged Acute Lymphoblastic Leukemia. Blood, 2019, blood.2019002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhao Z; Rahman MA; Chen ZG; Shin DM Multiple Biological Functions of Twist1 in Various Cancers. Oncotarget, 2017, 8, 20380–20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Choi D; Oh KJ; Han HS; Yoon YS; Jung CY; Kim ST; Koo SH Protein Arginine Methyltransferase 1 Regulates Hepatic Glucose Production in a FoxO1-Dependent Manner. Hepatology, 2012, 56, 1546–1556. [DOI] [PubMed] [Google Scholar]
- [59].Fronz K; Güttinger S; Burkert K; Kühn U; Stöhr N; Schierhorn A; Wahle E Arginine Methylation of the Nuclear Poly(A) Binding Protein Weakens the Interaction with Its Nuclear Import Receptor, Transportin. J. Biol. Chem, 2011, 286, 32986–32994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Choi HJ; Weis WI Purification and Structural Analysis of Desmoplakin. Methods Enzymol, 2016, 569, 197–213. [DOI] [PubMed] [Google Scholar]
- [61].Poulard C; Treilleux I; Lavergne E; Bouchekioua-Bouzaghou K; Goddard-Léon S; Chabaud S; Trédan O; Corbo L; Le Romancer M Activation of Rapid Oestrogen Signalling in Aggressive Human Breast Cancers. EMBO Mol. Med, 2012, 4, 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Liao HW; Hsu JM; Xia W; Wang HL; Wang YN; Chang WC; Arold ST; Chou CK; Tsou PH; Yamaguchi H; Fang YF; Lee HJ; Lee HH; Tai SK; Yang MH; Morelli MP; Malabika S; Ladbury JE; Chen CH; Grandis JR; Kopetz S; Huang MC PRMT1-Mediated Methylation of the EGF Receptor Regulates Signaling and Cetuximab Response. J. Clin. Invest, 2015, 125, 4529–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhu Y; He X; Dong H; Sun J; Wang H; Zhang L; Miao Y; Jin J; Shen Y; Chen J; Muschen M; Chen C-W; Konopleva MY; Sun W; Zhang B; Kuo Y-H; Carlesso N; Marcucci G; Li L Inhibition of PRMT1 Mediated FLT3 Arginine Methylation as a Potent Therapeutic Strategy for MLL-r ALL. Blood, 2018, 132, 892 LP–892.29997221 [Google Scholar]
- [64].Kzhyshkowska J; Schütt H; Liss M; Kremmer E; Stauber R; Wolf H; Dobner T Heterogeneous Nuclear Ribonucleoprotein E1B-AP5 is Methylated in Its Arg-Gly-Gly (RGG) Box and Interacts with Human Arginine Methyltransferase HRMT1L1. Biochem. J, 2001, 358, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yoshimoto T; Boehm M; Olive M; Crook MF; San H; Langenickel T; Nabel EG The Arginine Methyltransferase PRMT2 Binds RB and Regulates E2F Function. Exp. Cell Res, 2006, 312, 2040–2053. [DOI] [PubMed] [Google Scholar]
- [66].Tang J; Gary JD; Clarke S; Herschman HR PRMT 3, a Type I Protein Arginine N-Methyltransferase That Differs from PRMT1 in Its Oligomerization, Subcellular Localization, Substrate Specificity, and Regulation. J. Biol. Chem, 1998, 273, 16935–16945. [DOI] [PubMed] [Google Scholar]
- [67].Papers JBC; Doi M; Frankel A; Clarke S PRMT3 is a Distinct Member of the Protein Arginine N-Methyltransferase Family. J. Biol. Chem, 2000, 275, 32974–32982. [DOI] [PubMed] [Google Scholar]
- [68].Swiercz R; Person MD; Bedford MT Ribosomal Protein S2 is a Substrate for Mammalian PRMT3 (Protein Arginine Methyltransferase 3). Biochem. J, 2005, 386, 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hsu MC; Pan MR; Chu PY; Tsai YL; Tsai CH; Shan YS; Chen LT; Hung WC Protein Arginine Methyltransferase 3 Enhances Chemoresistance in Pancreatic Cancer by Methylating HnRNPA1 to Increase ABCG2 Expression. Cancers (Basel)., 2019, 11, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xu W; Chen H; Du K; Asahara H; Tini M; Emerson BM; Montminy M; Evans RM A Transcriptional Switch Mediated by Cofactor Methylation. Science, 2001, 294, 2507–2511. [DOI] [PubMed] [Google Scholar]
- [71].Wang L; Zhao Z; Meyer MB; Saha S; Yu M; Guo A; Wisinski KB; Huang W; Cai W; Pike JW; Yuan M; Ahlquist P; Xu W CARM1 Methylates Chromatin Remodeling Factor BAF155 to Enhance Tumor Progression and Metastasis. Cancer Cell, 2014, 25, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Feng Q; Yi P; Wong J; O’Malley BW Signaling within a Coactivator Complex: Methylation of SRC-3/AIB1 is a Molecular Switch for Complex Disassembly. Mol. Cell. Biol, 2006, 26, 7846–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cheng D; Vemulapalli V; Lu Y; Shen J; Aoyagi S; Fry CJ; Yang Y; Foulds CE; Stossi F; Treviño LS; Mancini MA; O’Malley BW; Walker CL; Boyer TG; Bedford MT CARM1 Methylates MED12 to Regulate Its RNA-Binding Ability. Life Sci. Alliance, 2018, 1, e201800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lee J; Bedford MT PABP1 Identified as an Arginine Methyltransferase Substrate Using High-Density Protein Arrays. EMBO Rep, 2002, 3, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Li H; Park S; Kilburn B; Jelinek MA; Henschen-Edman A; Aswad DW; Stallcup MR; Laird-Offringa IA Lipopolysaccharide-Induced Methylation of HuR, an MRNA-Stabilizing Protein, by CARM1. J. Biol. Chem, 2002, 277, 44623–44630. [DOI] [PubMed] [Google Scholar]
- [76].Zhong XY; Yuan XM; Xu YY; Yin M; Yan WW; Zou SW; Wei LM; Lu HJ; Wang YP; Lei QY CARM1 Methylates GAPDH to Regulate Glucose Metabolism and is Suppressed in Liver Cancer. Cell Rep, 2018, 24, 3207–3223. [DOI] [PubMed] [Google Scholar]
- [77].Singhroy DN; Mesplède T; Sabbah A; Quashie PK; Falgueyret JP; Wainberg MA Automethylation of Protein Arginine Methyltransferase 6 (PRMT6) Regulates Its Stability and Its Anti-HIV-1 Activity. Retrovirology, 2013, 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Herrmann F; Pably P; Eckerich C; Bedford MT; Fackelmayer FO Human Protein Arginine Methyltransferases in Vivo - Distinct Properties of Eight Canonical Members of the PRMT Family. J. Cell Sci, 2009, 122, 667–677. [DOI] [PubMed] [Google Scholar]
- [79].Waldmann T; Izzo A; Kamieniarz K; Richter F; Vogler C; Sarg B; Lindner H; Young NL; Mittler G; Garcia BA; Schneider R Methylation of H2AR29 is a Novel Repressive PRMT6 Target. Epigenetics and Chromatin, 2011, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Xie B; Invernizzi CF; Richard S; Wainberg MA Arginine Methylation of the Human Immunodeficiency Virus Type 1 Tat Protein by PRMT6 Negatively Affects Tat Interactions with Both Cyclin T1 and the Tat Transactivation Region. J. Virol, 2007, 81, 4226–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Choi S; Jeong HJ; Kim H; Choi D; Cho SC; Seong JK; Koo SH; Kang JS Skeletal Muscle-Specific Prmt1 Deletion Causes Muscle Atrophy via Deregulation of the PRMT6-FOXO3 Axis. Autophagy, 2019, 15, 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lee J; Sayegh J; Daniel J; Clarke S; Bedford MT PRMT8, a New Membrane-Bound Tissue-Specific Member of the Protein Arginine Methyltransferase Family. J. Biol. Chem, 2005, 280, 32890–32896. [DOI] [PubMed] [Google Scholar]
- [83].Lee WC; Lin WL; Matsui T; Chen ESW; Wei TYW; Lin WH; Hu H; Zheng YG; Tsai MD; Ho MC Protein Arginine Methyltransferase 8: Tetrameric Structure and Protein Substrate Specificity. Biochemistry, 2015, 54, 7514–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sayegh J; Webb K; Cheng D; Bedford MT; Clarke SG Regulation of Protein Arginine Methyltransferase 8 (PRMT8) Activity by Its N-Terminal Domain. J. Biol. Chem, 2007, 282, 36444–36453. [DOI] [PubMed] [Google Scholar]
- [85].Kim U-D; Kako K; Kakiuchi M; Park GG; Fukamizu A EWS is a Substrate of Type I Protein Arginine Methyltransferase, PRMT8. Int. J. Mol. Med, 2008, 22, 309–315. [PubMed] [Google Scholar]
- [86].Jeong HC, Park SJ, Choi JJ, Go YH, Hong SK, Kwon OS, Shin JG, Kim RK, Lee MO, Lee SJ, Shin HD, Moon SH, C.H. PRMT8 Controls the Pluripotency and Mesodermal Fate of Human Embryonic Stem Cells By Enhancing the PI3K/AKT/SOX2 Axis. Stem Cells, 2017, 35, 2037–2049. [DOI] [PubMed] [Google Scholar]
- [87].Simandi Z; Pajer K; Karolyi K; Sieler T; Jiang L-L; Kolostyak Z; Sari Z; Fekecs Z; Pap A; Patsalos A; Contreras GA; Reho B; Papp Z; Guo X; Horvath A; Kiss G; Keresztessy Z; Vámosi G; Hickman J; Xu H; Dormann D; Hortobagyi T; Antal M; Nógrádi A; Nagy L Arginine Methyltransferase PRMT8 Provides Cellular Stress Tolerance in Aging Motoneurons. J. Neurosci, 2018, 38, 7683–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Branscombe TL; Frankel A; Lee JH; Cook JR; Yang Z; Pestka S; Clarke S PRMT5 (Janus Kinase-Binding Protein 1) Catalyzes the Formation of Symmetric Dimethylarginine Residues in Proteins. J. Biol. Chem, 2001, 276, 32971–32976. [DOI] [PubMed] [Google Scholar]
- [89].Sun L; Wang M; Lv Z; Yang N; Liu Y; Bao S; Gong W; Xu R Structural Insights into Protein Arginine Symmetric Dimethylation by PRMT5. Proc. Natl. Acad. Sci, 2011, 108, 20538–20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lacroix M; El Messaoudi S; Rodier G; Le Cam A; Sardet C; Fabbrizio E The Histone-Binding Protein COPR5 is Required for Nuclear Functions of the Protein Arginine Methyltransferase PRMT5. EMBO Rep, 2008, 9, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Antonysamy S; Bonday Z; Campbell RM; Doyle B; Druzina Z; Gheyi T Crystal Structure of the Human PRMT5 : MEP50 Complex. PNAS, 2012, 109, 17960–17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Deng X; Shao G; Zhang HT; Li C; Zhang D; Cheng L; Elzey BD; Pili R; Ratliff TL; Huang J; Hu CD Protein Arginine Methyltransferase 5 Functions as an Epigenetic Activator of the Androgen Receptor to Promote Prostate Cancer Cell Growth. Oncogene, 2017, 36, 1223–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Jansson M; Durant ST; Cho E; Sheahan S; Edelmann M; Kessler B; La Thangue NB Arginine Methylation Regulates the P53 Response. Nat. Cell Biol, 2008, 10, 1431–1439. [DOI] [PubMed] [Google Scholar]
- [94].Wei H; Wang B; Miyagi M; She Y; Gopalan B; Huang D; Ghosh G PRMT5 Dimethylates R30 of the P65 Subunit to Activate NF-ΚB. PNAS, 2013, 110, 13516–13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hu D; Gur M; Zhou Z; Gamper A; Hung MC; Fujita N; Lan L; Bahar I; Wan Y Interplay between Arginine Methylation and Ubiquitylation Regulates KLF4-Mediated Genome Stability and Carcinogenesis. Nat. Commun, 2015, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zheng S; Moehlenbrink J; Lu YC; Zalmas LP; Sagum CA; Carr S; McGouran JF; Alexander L; Fedorov O; Munro S; Kessler B; Bedford MT; Yu Q; La Thangue NB Arginine Methylation-Dependent Reader-Writer Interplay Governs Growth Control by E2F-1. Mol. Cell, 2013, 52, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Li M; An W; Xu L; Lin Y; Su L; Liu X The Arginine Methyltransferase PRMT5 and PRMT1 Distinctly Regulate the Degradation of Anti-Apoptotic Protein CFLARL in Human Lung Cancer Cells. J. Exp. Clin. Cancer Res, 2019, 38, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mersaoui SY; Yu Z; Coulombe Y; Karam M; Busatto FF; Masson J-Y; Richard S Arginine Methylation of the DDX5 Helicase RGG/RG Motif by PRMT5 Regulates Resolution of RNA:DNA Hybrids. EMBO J, 2019, 38, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].An W; Yao S; Sun X; Hou Z; Lin Y; Su L; Liu X Glucocorticoid Modulatory Element-Binding Protein 1 (GMEB1) Interacts with the de-Ubiquitinase USP40 to Stabilize CFLAR L and Inhibit Apoptosis in Human Non-Small Cell Lung Cancer Cells. J. Exp. Clin. Cancer Res, 2019, 38, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lee J; Yang Z; Krause CD; Herth N; Ho R; Pestka S FBXO11/PRMT9, a New Protein Arginine Methyltransferase, Symmetrically Dimethylates Arginine Residues. 2006, 342, 472–481. [DOI] [PubMed] [Google Scholar]
- [101].Hadjikyriacou A; Yang Y; Espejo A; Bedford MT; Clarke SG Unique Features of Human Protein Arginine Methyltransferase 9 (PRMT9) and Its Substrate RNA Splicing Factor SF3B2. J. Biol. Chem, 2015, 290, 16723–16743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lee J-H; Cook JR; Yang Z-H; Mirochnitchenko O; Gunderson SI; Felix AM; Herth N; Hoffmann R; Pestka S PRMT7, a New Protein Arginine Methyltransferase That Synthesizes Symmetric Dimethylarginine. J. Biol. Chem, 2005, 280, 3656–3664. [DOI] [PubMed] [Google Scholar]
- [103].Jain K; Jin CY; Clarke SG Epigenetic Control via Allosteric Regulation of Mammalian Protein Arginine Methyltransferases. Proc. Natl. Acad. Sci, 2017, 114, 201706978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Cura V; Troffer-Charlier N; Wurtz JM; Bonnefond L; Cavarelli J Structural Insight into Arginine Methylation by the Mouse Protein Arginine Methyltransferase 7: A Zinc Finger Freezes the Mimic of the Dimeric State into a Single Active Site. Acta Crystallogr. Sect. D Biol. Crystallogr, 2014, 70, 2401–2412. [DOI] [PubMed] [Google Scholar]
- [105].Haghandish N; Baldwin RM; Morettin A; Dawit HT; Adhikary H; Masson J-Y; Mazroui R; Trinkle-Mulcahy L; Côté J PRMT7 Methylates Eukaryotic Translation Initiation Factor 2α and Regulates Its Role in Stress Granule Formation. Mol. Biol. Cell, 2019, 30, 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jeong H-J; Lee S-J; Lee H-J; Kim H-B; Anh Vuong T; Cho H; Bae G-U; Kang J-S Prmt7 Promotes Myoblast Differentiation via Methylation of P38MAPK on Arginine Residue 70. Cell Death Differ, 2019, Just accep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Cuenda A; Rousseau S P38MAP-Kinases Pathway Regulation, Function and Role in Human Diseases. Biochimica et Biophysica Acta-Molecular Cell Research, 2007, 1773, 1358–1375. [DOI] [PubMed] [Google Scholar]
- [108].Elakoum R; Gauchotte G; Oussalah A; Wissler M; Clément-duchêne C; Vignaud J; Guéant J; Namour F Biochimie CARM1 and PRMT1 are Dysregulated in Lung Cancer without Hierarchical Features. 2014, 97, 210–218. [DOI] [PubMed] [Google Scholar]
- [109].Hernandez SJ; Dolivo DM; Dominko T PRMT8 Demonstrates Variant‑Specific Expression in Cancer Cells and Correlates with Patient Survival in Breast, Ovarian and Gastric Cancer. Oncilogy Lett., 2017, 13, 1983–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Li L; Zhang Z; Ma T; Huo R PRMT1 Regulates Tumor Growth and Metastasis of Human Melanoma via Targeting ALCAM. Mol. Med. Rep, 2016, 14, 521–528. [DOI] [PubMed] [Google Scholar]
- [111].Zhou Z; Feng Z; Hu D; Yang P; Gur M; Bahar I; Cristofanilli M; Gradishar WJ; Xie X.qun; Wan Y A Novel Small-Molecule Antagonizes PRMT5-Mediated KLF4 Methylation for Targeted Therapy. EBioMedicine, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhang B; Dong S; Li Z; Lu L; Zhang S; Chen X; Cen X; Wu Y Targeting Protein Arginine Methyltransferase 5 Inhibits Human Hepatocellular Carcinoma Growth via the Downregulation of Beta-Catenin. J. Transl. Med, 2015, 13, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zhang H; Guo X; Feng X; Wang T; Hu Z MiRNA-543 Promotes Osteosarcoma Cell Proliferation and Glycolysis by Partially Suppressing PRMT9 and Stabilizing HIF-1α Protein. Oncotarget, 2017, 8, 2342–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Li S; Cheng D; Zhu B; Yang Q The Overexpression of CARM1 Promotes Human Osteosarcoma Cell Proliferation through the Pgsk3β/β-Catenin/CyclinD1 Signaling Pathway. Int. J. Biol. Sci, 2017, 13, 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Baldwin RM; Haghandish N; Daneshmand M; Paris G; Falls TJ; Bell JC; Islam S; Côté J Protein Arginine Methyltransferase 7 Promotes Breast Cancer Cell Invasion through the Induction of MMP9 Expression. Oncotarget, 2014, 6, 3013–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Fedoriw A; Rajapurkar SR; O’Brien S; Gerhart SV; Mitchell LH; Adams ND; Rioux N; Lingaraj T; Ribich SA; Pappalardi MB; Shah N; Laraio J; Liu Y; Butticello M; Carpenter CL; Creasy C; Korenchuk S; McCabe MT; McHugh CF; Nagarajan R; Wagner C; Zappacosta F; Annan R; Concha NO; Thomas RA; Hart TK; Smith JJ; Copeland RA; Moyer MP; Campbell J; Stickland K; Mills J; Jacques-O’Hagan S; Allain C; Johnston D; Raimondi A; Porter Scott M; Waters N; Swinger K; Boriack-Sjodin A; Riera T; Shapiro G; Chesworth R; Prinjha RK; Kruger RG; Barbash O; Mohammad HP Anti-Tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss. Cancer Cell, 2019, 36, 100–114. [DOI] [PubMed] [Google Scholar]
- [117].Yoshimatsu M; Toyokawa G; Hayami S; Unoki M; Tsunoda T; Field HI Dysregulation of PRMT1 and PRMT6, Type I Arginine Methyltransferases, is Involved in Various Types of Human Cancers. 2011, 1, 562–573. [DOI] [PubMed] [Google Scholar]
- [118].Zhu F; Guo H; Bates PD; Zhang S; Zhang H; Nomie KJ; Li Y; Lu L; Seibold KR; Wang F; Rumball I; Cameron H; Hoang NM; Yang DT; Xu W; Zhang L; Wang M; Capitini CM; Rui L PRMT5 is Upregulated by B-Cell Receptor Signaling and Forms a Positive-Feedback Loop with PI3K/AKT in Lymphoma Cells. Leukemia, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].He X; Zhu Y; Lin Y-C; Li M; Du J; Dong H; Sun J; Zhu L; Wang H; Ding Z; Zhang L; Zhang L; Zhao D; Wang Z; Wu H; Zhang H; Jiang W; Xu Y; Jin J; Shen Y; Perry J; Zhao X; Zhang B; Liu S; Xue S-L; Shen B; Chen C-W; Chen J; Khaled S; Kuo Y-H; Marcucci G; Luo Y; Li L PRMT1-Mediated FLT3 Arginine Methylation Promotes Maintenance of FLT3-ITD+ Acute Myeloid Leukemia. Blood, 2019, 134, 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Fedoriw A; Rajapurkar SR; Brien SO; Kruger RG; Barbash O; Mohammad HP Anti-Tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss. Cancer Cell, 2019, 36, 100–114. [DOI] [PubMed] [Google Scholar]
- [121].Gerhart SV; Kellner WA; Thompson C; Pappalardi MB; Zhang X; Monte R; Oca D; Penebre E; Duncan K; Boriack-sjodin A; Le B; Majer C; Mccab MT; Carpenter C; Johnson N; Kruger RG; Barbash O Activation of the P53-MDM4 Regulatory Axis Defines the Anti- Tumour Response to PRMT5 Inhibition through Its Role in Regulating Cellular Splicing. Sci. Rep, 2018, 8, 9711–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kaniskan HÜ; Szewczyk MM; Yu Z; Eram MS; Yang X; Schmidt K; Luo X; Dai M; He F; Zang I; Lin Y; Kennedy S; Li F; Dobrovetsky E; Dong A; Smil D; Min SJ; Landon M; Lin-Jones J; Huang XP; Roth BL; Schapira M; Atadja P; Barsyte-Lovejoy D; Arrowsmith CH; Brown PJ; Zhao K; Jin J; Vedadi M A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3). Angew. Chemie - Int. Ed, 2015, 54, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kaniskan HÜ; Eram MS; Zhao K; Szewczyk MM; Yang X; Schmidt K; Luo X; Xiao S; Dai M; He F; Zang I; Lin Y; Li F; Dobrovetsky E; Smil D; Min S-J; Lin-Jones J; Schapira M; Atadja P; Li E; Barsyte-Lovejoy D; Arrowsmith CH; Brown PJ; Liu F; Yu Z; Vedadi M; Jin J Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3). J. Med. Chem, 2018, 61, 1204–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Mitchell LH; Drew AE; Ribich S. a.; Rioux N; Swinger KK; Jacques SL; Lingaraj T; Boriack-Sjodin PA; Waters NJ; Wigle TJ; Moradei O; Jin L; Riera T; Porter-Scott M; Moyer MP; Smith JJ; Chesworth R; Copeland R. a. Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound. ACS Med. Chem. Lett, 2015, 6, 655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Nakayama K; Szewczyk MM; Sena C. dela; Wu H; Dong A; Zeng H; Li F; de Freitas RF; Eram MS; Schapira M; Baba Y; Kunitomo M; Cary DR; Tawada M; Ohashi A; Imaeda Y; Saikatendu KS; Grimshaw CE; Vedadi M; Arrowsmith CH; Barsyte-Lovejoy D; Kiba A; Tomita D; Brown PJ TP-064, a Potent and Selective Small Molecule Inhibitor of PRMT4 for Multiple Myeloma. Oncotarget, 2018, 9, 18480–18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].National Institutes of Health, United States National Library of Medicine, Clinical Trials. https://clinicaltrials.gov/ct2/show/NCT03666988 (Accessed August 5, 2019).
- [127].National Institutes of Health, United States National Library of Medicine, Clinical Trials. https://clinicaltrials.gov/ct2/show/NCT03573310 (Accessed August 5, 2019).
- [128].National Institutes of Health, United States National Library of Medicine, Clinical Trials. https://clinicaltrials.gov/ct2/show/NCT02783300 (Accessed August 5, 2019).
- [129].National Institutes of Health, United States National Library of Medicine, Clinical Trials. https://clinicaltrials.gov/ct2/show/NCT03614728 (Accessed August 5, 2019).
- [130].National Institutes of Health, United States National Library of Medicine, Clinical Trials. https://clinicaltrials.gov/ct2/show/NCT03854227 (Accessed August 5, 2019).