Abstract
Neanderthal ancestry remains across modern Eurasian genomes, and introgressed sequences influence diverse phenotypes. Here we demonstrate that introgressed sequences reintroduced thousands of ancestral alleles that were lost in Eurasian populations prior to introgression. Our simulations and variant effect predictions argue that these reintroduced alleles (RAs) are more likely to be tolerated by modern humans than introgressed Neanderthal-derived alleles (NDAs) due to their distinct evolutionary histories. Consistent with this, we show enrichment for RAs and depletion for NDAs on introgressed haplotypes with expression quantitative trait loci (eQTL) and phenotype associations. Analysis of available cross-population eQTLs and massively parallel reporter assay (MPRA) data show that RAs commonly influence gene expression independent of linked NDAs. We further validate these independent effects for one RA in vitro. Finally, we demonstrate that NDAs are depleted for regulatory activity compared to RAs, while RAs have activity levels similar to non-introgressed variants. In summary, our study reveals that Neanderthal introgression reintroduced thousands of lost ancestral variants with gene regulatory activity and that these RAs were more tolerated than NDAs. Thus, RAs and their distinct evolutionary histories must be considered when evaluating the effects of introgression.
ONE SENTENCE SUMMARY
Neanderthal interbreeding with anatomically modern humans restored thousands of ancient alleles that were previously lost in Eurasian populations.
Modern Eurasian populations have lower genetic diversity than modern African populations, despite having larger census population sizes1,2. This disparity reflects the genetic bottlenecks experienced by the ancestors of Eurasian anatomically modern humans (AMH) as they moved out of Africa ~50,000 years ago2,3. The effective population size of this ancestral Eurasian population is estimated to have been less than 20% of the size of contemporaneous African populations1,4. As a result of this out of Africa (OOA) bottleneck and subsequent population dynamics, millions of ancient alleles were lost in the ancestors of modern Eurasian populations.
More than 500,000 years prior to the AMH OOA bottleneck, members of other hominin groups in Africa, including the ancestors of Neanderthals and Denisovans, moved into Eurasia5. The sequencing of ancient DNA from Neanderthal and Denisovan individuals has enabled reconstruction of their genomes5–7. Comparing Neanderthal genomes to genomes of modern humans from around the world revealed that Eurasian AMHs interbred with Neanderthals approximately 50,000 years ago5,8. The legacy of this archaic introgression is reflected in the genomes of modern Eurasians, where ~1–3% of individuals’ DNA sequences are of Neanderthal ancestry9–11.
Neanderthal introgression introduced new alleles into Eurasian populations that were derived in the Neanderthal lineage. Some of these alleles were adapted to non-African environments and thus were beneficial to Eurasian AMHs11–16. However, Neanderthal interbreeding also came with a genetic cost due to accumulation of weakly deleterious alleles in their lineage, because of their lower effective population size compared to AMHs17,18. The distribution of archaic ancestry across modern Eurasian genomes is non-random, with significant deserts of Neanderthal ancestry as well as many genomic regions in which Neanderthal ancestry is common. These patterns reflect the long term actions of selection and drift on introgressed alleles10,11,19, with negative selection acting most strongly immediately after admixture20.
Introgressed alleles on Neanderthal haplotypes that remain in modern Eurasian populations are associated with diverse traits, including risk for skin, immune, and neuropsychiatric diseases12,13,21–24. For example, an introgressed Neanderthal haplotype at the OAS1 locus is associated with innate immune response; however, this introgressed haplotype also carries an ancestral allele that may influence function25. Thus, while most studies have focused on identifying and testing the effects of Neanderthal derived alleles in AMHs, archaic admixture may also have served as a route by which more ancient, functional alleles were reintroduced into Eurasian genomes25,26.
Here, we evaluate the hypothesis that Neanderthal introgression reintroduced previously lost functional ancestral alleles into Eurasian populations by analyzing archaic, modern, and simulated genomes. Our results demonstrate that Neanderthal populations served as reservoirs of hundreds of thousands of ancestral alleles that were lost to the ancestors of Eurasians (and in some cases all modern humans), and that many of these alleles have functional effects in Eurasians after being reintroduced by Neanderthal admixture.
RESULTS
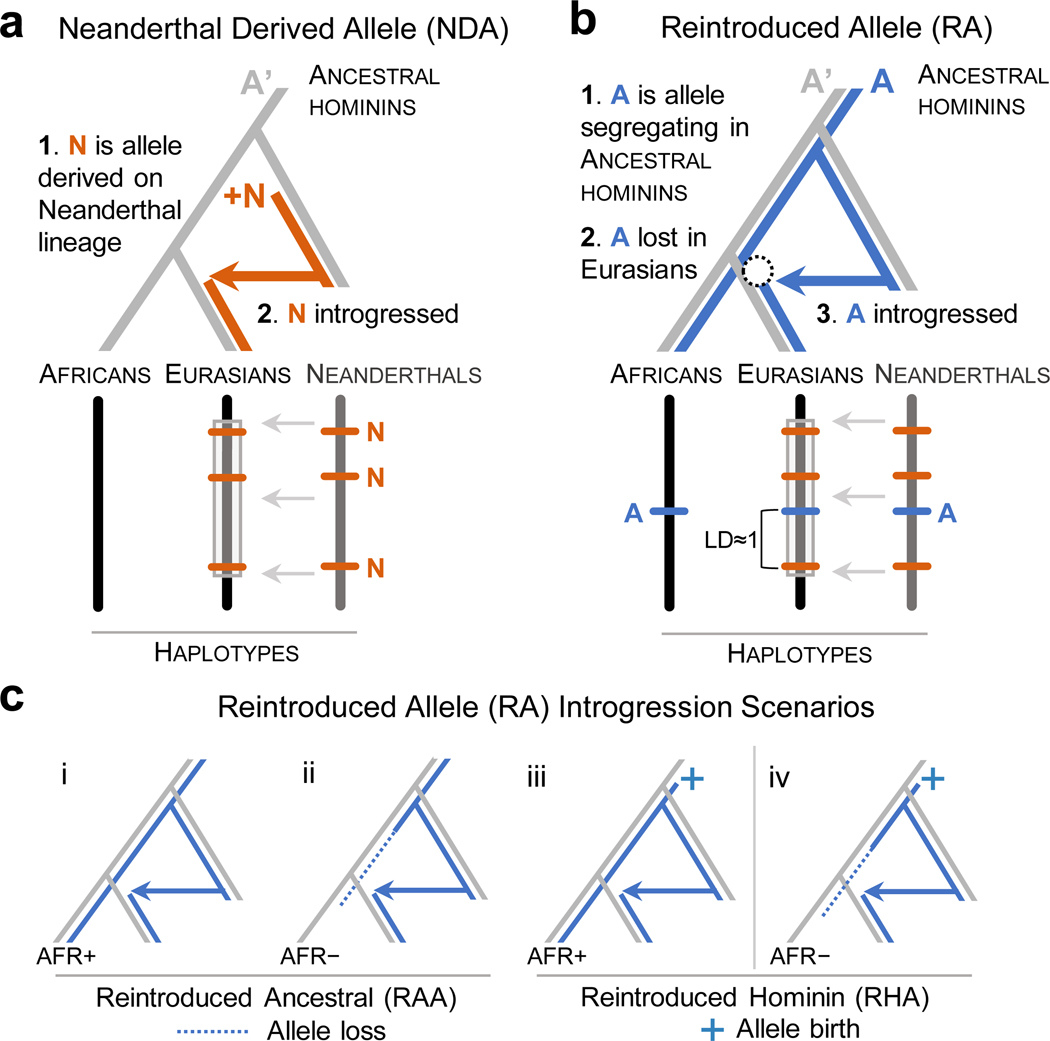
Many alleles segregating in ancestral hominins were lost in the AMH lineage after the divergence of the ancestors of AMHs and Neanderthals. Some were lost in all AMHs, while others were lost only in Eurasian populations, e.g., during the OOA bottleneck. (Figure 1). Any lost allele that persisted in archaic hominins had the potential to be reintroduced into Eurasian populations via archaic admixture. Within Eurasian populations, such reintroduced alleles would initially be exclusive to introgressed haplotypes, and many would retain high linkage disequilibrium (LD) with archaic-derived alleles over time (Figure 1b).
Figure 1. Schematic of the reintroduction of lost ancestral alleles by Neanderthal introgression.
(a) Evolutionary trajectory and resulting genomic signature of a Neanderthal derived allele N (orange) within an introgressed haplotype in modern Eurasians. A’ is the ancestral allele present in the population ancestral to anatomically modern humans (AMHs). (b) Evolutionary trajectory and resulting genomic signature of a reintroduced allele A (blue). To be an RA, A had to have been: (1) segregating (with A’) in the common ancestor of AMH and Neanderthals, (2) lost to the ancestors of modern Eurasians (dotted circle), and (3) reintroduced to modern Eurasians through Neanderthal admixture. RAs are characterized by their high linkage disequilibrium (LD≈1) with NDAs on introgressed haplotypes in modern Eurasians. (c) RAs can have different evolutionary histories. We distinguish several scenarios based on the status of the RA in the human-chimpanzee ancestor and in modern African populations. Reintroduced ancestral alleles (RAA, i and ii) are ancestral (i.e. they are the inferred ancestral allele of the human-chimpanzee ancestor) while reintroduced hominin alleles (RHA, iii and iv) appeared before the split AMH and Neanderthals, but may not have been present in the human-chimpanzee ancestor. Among RAAs, the ancestral allele may or may not be present in modern Africans (AFR+ vs. AFR–). However, the identification of RHAs requires that they are present in modern sub-Saharan African populations at reasonable frequencies (>1%). As a result, scenario iv cannot be inferred from modern genomic data alone and is not analyzed here.
In the following analysis, we will refer to alleles that were present in the most recent common ancestor of AMHs and Neanderthals as “ancestral hominin alleles.” We will refer to ancestral hominin alleles that are only observed in Eurasians on introgressed Neanderthal haplotypes as “reintroduced alleles” (RAs, Figure 1b), and introgressed alleles that first appeared on the Neanderthal lineage as “Neanderthal-derived alleles” (NDAs, Figure 1a). We also distinguish several distinct evolutionary scenarios among RAs based on their age and presence in African populations (Figure 1c). Our goal is to evaluate the presence and function of RAs in modern Eurasians and contrast them with NDAs.
Hundreds of thousands of RAs exist in modern Eurasian populations
To identify candidate RAs, we sought ancestral variants in 1000 Genomes Phase 3 Eurasian populations that are present only on introgressed haplotypes identified by S*27. For each population, we identified variants that are in perfect LD (r2=1) with a Neanderthal tag variant, but are ancestral rather than Neanderthal-derived (Extended Data Fig. 1, Methods). Our approach is robust to small fractions of apparent Neanderthal ancestry present in some sub-Saharan African populations (Methods), and forward-time evolutionary simulations suggest that false positives due to recombination artifacts or convergent mutations are rare, even at highly mutable CpG dinucleotides (Extended Data Fig. 2; Tables S1 and S2). Finally, our approach is likely conservative given that many RAs are not expected to retain perfect LD with any NDA.
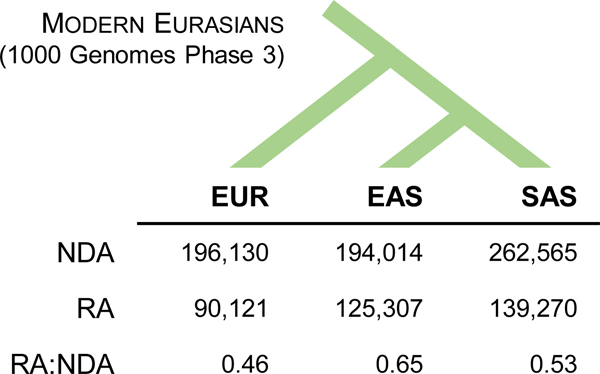
RAs are pervasive. Over 80% of introgressed haplotypes contain RAs, averaging ~17 RAs per haplotype. Altogether, we identify 209,176 RAs. The South and East Asian populations each have more RAs than the European population (Figure 2, Extended Data Figs. 1 and 3), likely reflecting differences in demographic histories28 and the greater levels of Neanderthal ancestry previously observed in East Asians29,30. The observed ratio of RA to NDA in each population (0.46–0.65) is consistent with predictions from our simulations (Figure 2, Extended Data Fig. 2, Supplementary Table 3). RAs are more clustered and less correlated with haplotype length than NDAs (Extended Data Fig. 4).
Figure 2. Neanderthal introgression reintroduced thousands of lost ancestral alleles to Eurasian populations.
The number of RAs and NDAs in each Eurasian 1000 Genomes population (EAS = East Asian; EUR = European ancestry; SAS = South Asian) identified by our pipeline (Extended Data Fig. 1; Methods). Overall, Neanderthal admixture is responsible for the presence of over 200,000 ancestral alleles lost in the human OOA bottleneck or later migrations into the ancestors of Eurasian populations.
Functional effect predictions differ for RAs and NDAs
RAs and NDAs were introduced into Eurasian populations on Neanderthal haplotypes, but they have different origins and evolutionary histories. Since NDAs first appeared outside the AMH context in Neanderthal populations with low effective population sizes, we expect that NDAs are more likely to have deleterious effects and been subject to negative selection in introgressed Eurasians. In contrast, RAs originated in larger, ancestral hominin populations suggesting they are more likely to be tolerated. Variant effect predictions support this hypothesis; both Combined Annotation Dependent Depletion (CADD) and PolyPhen2 predict RAs to be more benign than NDAs (Extended Data Fig. 5 and 6). These results are consistent with our evolutionary modeling that predicts NDAs to have significantly more negative selection coefficients than RAs (Extended Data Fig. 2). NDAs are also depleted compared to RAs for overlap of regulatory elements from RegulomeDB (Extended Data Fig. 7).
Among RAs, there are several distinct possible evolutionary histories, each suggestive of different functional expectations (Figure 1c). As expected, given their older ages, reintroduced ancestral alleles (RAAs) are predicted to be less deleterious than reintroduced hominin alleles (RHAs). For ~70% of RAAs, the corresponding allele is still segregating in African populations (RAAAFR+, Figure 1c); however, a fraction (EAS: 22%, EUR: 30%, and SAS: 28%) are absent in modern Africans (RAAAFR−). These RAAAFR− variants represent instances where the derived allele was likely at high frequency in pre-introgression AMH populations, potentially due to AMH-specific positive selection. These RAAAFR− variants are now present in AMH populations only due to introgression. Consistent with the loss of these variants in African populations, RAAAFR− variants are segregating at lower frequencies in Eurasian populations than RAAAFR+ variants (Extended Data Fig. 8). However, CADD predictions of variant effects are similar between RAAAFR+ and RAAAFR− variants (data not shown).
RA-containing introgressed haplotypes are associated with anthropometric human traits and disease risk
Previous studies have successfully used GWAS to associate variants on introgressed haplotypes with human phenotypes5,6,10. While some RAs were considered in these previous analyses (though not recognized as RAs), alleles that are present in Africans were not considered due to these studies’ focus on NDAs. Eurasian RAs tag 2,197 unique, significant associations (P < 10–8; Methods) in the GWAS Catalog31, while NDAs tag 2,547 (File S2). Patterns were similar when limiting the analysis to European introgressed haplotypes and associations. Overall, >70% of the phenotypes associated with NDAs have an equally strong association with at least one RA.
Many of the phenotypes tagged by NDAs and RAs are morphometric (e.g., cranial base width, BMI, and height), and several others influence external traits (e.g., chin dimples, male-pattern baldness, and skin pigmentation). RAs are also associated with many pathologies, including cancers (breast, esophageal, lung, prostate), Alzheimer’s disease, and neurological conditions like neuroticism and bipolar disorder (File S2).
RAAAFR− are particularly interesting because derived alleles became fixed in modern human populations at these loci after the split from ancestors of Neanderthals and these ancestral alleles are only present in AMHs due to introgression. For example, an RAAAFR− (rs11564258) near MUC19, a gel-forming mucin expressed in epithelial tissues with a potential role in interaction with microbial communities, is strongly associated with both Crohn’s disease and inflammatory bowel disease32,33. This locus has been identified in scans for potential adaptive introgression15. We also find associations between RAAAFR− and facial morphology, body mass index, sleep phenotypes, and metabolite levels in smokers34–38. Overall, these results expand the number of traits associated with introgressed haplotypes and provide evolutionary context for interpreting candidate causal introgressed variants on these haplotypes.
Introgressed haplotypes with phenotype associations are enriched for RAs and depleted for NDAs
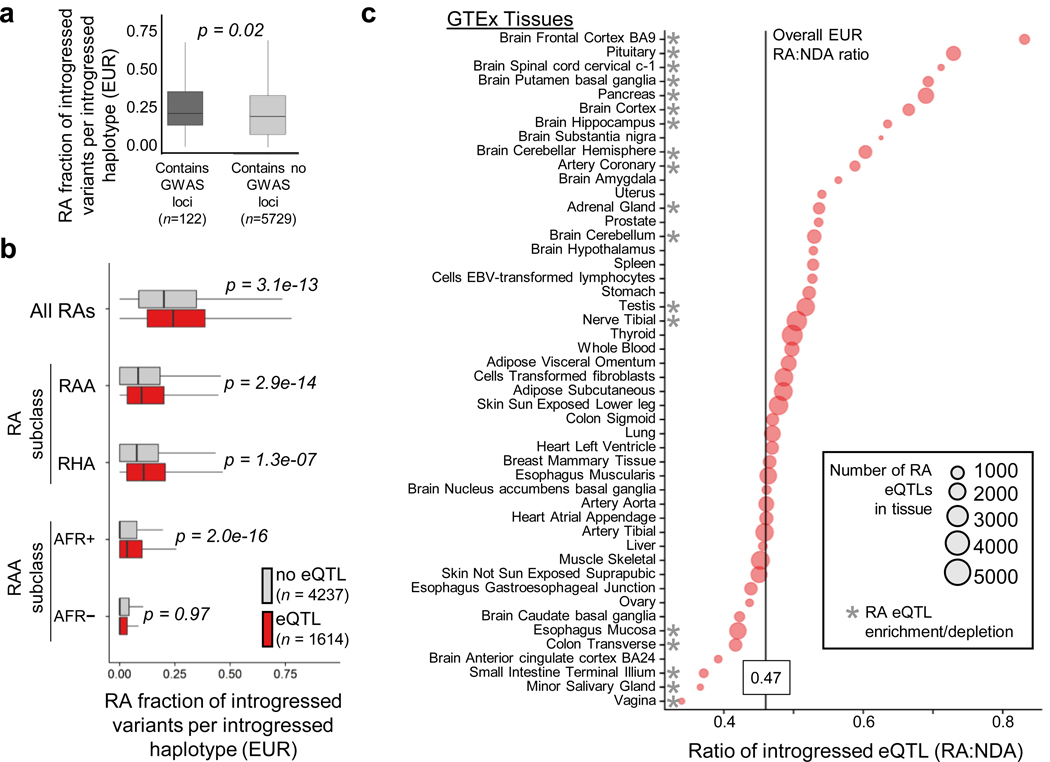
The high LD between RAs and NDAs prevents the direct identification of either introgressed variant class as causal for any association. Nevertheless, introgressed haplotypes with a significant GWAS trait association in Europeans contain a higher fraction of RAs compared to introgressed haplotypes with no associations (Figure 3a; 0.23 vs. 0.21, P = 0.02, Mann-Whitney U test). Enrichment for RAs relative to NDAs could reflect selection against functional NDAs (i.e., a depletion of NDAs) and/or a greater tolerance of functional haplotypes containing RAs. The enrichment holds when considering some subclasses of RAs (RAAs and RAAAFR+), but not others (RHAs and RAAAFR–; Extended Data Fig. 9). The lack of difference for RAAAFR– could be due to lower power as a result of the small number of these sites and their lower allele frequencies in Europeans (Extended Data Fig. 8).
Figure 3. Reintroduced alleles are more prevalent than NDAs among introgressed haplotypes with GWAS hits and eQTL.
(a) The median fraction of RAs among all classified introgressed alleles (RAs plus NDAs) on introgressed haplotypes in Europeans (EUR) with at least one genome-wide significant (P < 1.0e−8) trait association reported in the GWAS Catalog versus introgressed haplotypes without trait associations (0.23 vs. 0.21 respectively, P = 0.02, Mann-Whitney U test). Introgressed haplotypes with GWAS hits have a higher RA fraction (and, symmetrically, lower NDA fraction) than those with no associations (median RA fraction of 0.23 vs. 0.21, P = 0.02). This enrichment varies across RA subclasses (Extended Data Fig. 7). (b) The fraction of RAs among introgressed alleles on introgressed haplotypes in EUR that contain GTEx eQTL (n=1,585) versus introgressed haplotypes without eQTL (n=4,237). Introgressed haplotypes with eQTL have a higher RA fraction than those with no eQTL (median RA fraction of 0.24 vs. 0.20, P = 3.0e−13, Mann-Whitney U test). This holds for all RA subclasses except RAAAFR–. (c) The RA:NDA ratio among introgressed eQTLs in each of 48 GTEx v7 tissues. Circles are scaled by the number of RA eQTLs in each tissue. Compared to the genome wide average RA:NDA ratio (0.47; vertical black line), 13 tissues have significantly more than the expected ratio of RA:NDA among eQTL and five tissues have fewer than the expected number (P < 0.01, hypergeometric test after Bonferroni correction). Brain tissues comprise eight of the 13 tissues with significant RA enrichment among eQTL.
Introgressed haplotypes with eQTL are enriched for RAs and depleted for NDAs
Previous analyses of associations between Neanderthal haplotypes and phenotypes have highlighted a role for effects on gene expression22,39. Accordingly, we evaluated the prevalence of RAs and NDAs among eQTL in 48 tissues profiled in the Genotype-Tissue Expression (GTEx) project40. Introgressed eQTL are found in all GTEx tissues; 18% of EUR RAs (16,318) and 16% of EUR NDAs (31,822) are eQTLs in at least one tissue. While each RA is associated with at least one NDA, the numbers of RAs and NDAs on an introgressed haplotype are only somewhat correlated (Pearson r2 = 0.46; Extended Data Fig. 4).
As in the analysis of GWAS loci, there is a significantly higher fraction of RAs (and lower fraction of NDAs) in introgressed EUR haplotypes with at least one introgressed eQTL compared to introgressed haplotypes with no eQTL (Figure 3b; 0.24 vs. 0.20, P = 3×10−13). All RA subclasses, except RAAAFR– were significantly enriched as well. This result also holds when stratifying eQTL containing introgressed haplotypes by their tissue of activity (Extended Data Fig. 10).
Among introgressed eQTL, the ratios of RAs to NDAs varied across tissues; thirteen tissues have a significantly higher RA:NDA ratio than the genome as a whole (Figure 3c, P < 0.01 for each, hypergeometric test with Bonferroni correction). For example, among introgressed frontal cortex eQTL, the RA:NDA ratio is 0.83, while the overall ratio observed across all introgressed regions is 0.47. Introgressed haplotypes have been previously shown to modulate gene regulation, especially in the brain22,39. Brain tissues comprise eight of the 13 tissues enriched for RA eQTLs, likely due to shared regulatory architectures across the brain. RA eQTLs are also more abundant in the pancreas, adrenal gland, testes, and tibial nerve. RA eQTL are less abundant than expected in the introgressed eQTL from mucosal tissues and salivary gland. In summary, introgressed haplotypes containing eQTL contain a higher fraction of RAs (and symmetrically a lower fraction of NDAs), and RA eQTL are not evenly distributed among tissues.
Some RAs have conserved gene regulatory associations in European and African populations
As shown in the previous sections, the majority of introgressed haplotypes associated with traits and gene expression contain RAs. However, high LD makes it challenging to determine if a particular RA or NDA is causal. Thus, we sought to evaluate RA regulatory activity independent of associated NDAs.
First, we analyzed cross-population eQTL data from lymphoblastoid cell lines (LCLs) from European (EUR) and sub-Saharan African Yoruba (YRI) individuals41. We identified eQTL alleles that are RAs in EUR and are present in YRI in non-Neanderthal introgressed haplotypes (Figure 4a; Methods). If an allele that was reintroduced into Eurasians has similar effects on gene expression in both populations, it suggests that that the RA (rather than linked NDAs, which are not present in Yoruba) influences expression, and that introgression reintroduced an ancestral allele with gene regulatory effects.
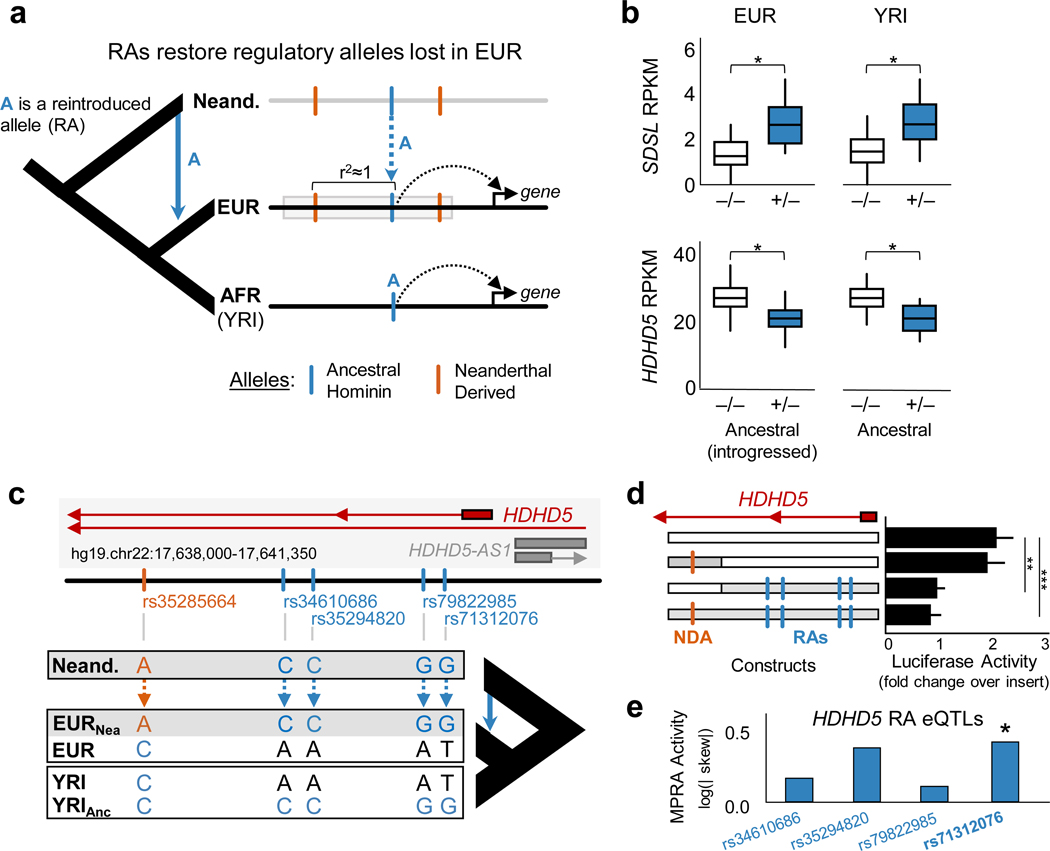
Figure 4. Reintroduced alleles restore regulatory functions lost in Eurasians.
(a) Conceptual model of restored regulatory function from Neanderthal admixture. Here, A is a cis-acting regulatory allele that is exclusively found on introgressed haplotypes (gray box) in modern Europeans (EUR). Allele A is also present in sub-Saharan Yoruba individuals (YRI) where it is not in LD with Neanderthal-derived alleles (NDAs). If this allele has similar cis-regulatory activity in both populations, it suggests that the reintroduced allele influences gene regulation independent of the associated NDAs. (b) Two examples of genes (SDSL and HDHD5) with consistent expression differences (in RPKM) associated with RA eQTLs in EUR and the corresponding allele in YRI LCLs. The RAs are present only on introgressed haplotypes in EUR, and the NDAs associated with the RAs are not present in YRI. This suggests that these RAs restore lost gene regulatory functions in EUR. (c) Schematic of the HDHD5 locus highlighting the locations of one NDA (orange) and four RA eQTLs (blue) in the introgressed haplotype and the different combinations of these alleles present in EUR, YRI, and Neanderthals. (d) Luciferase activity driven by constructs carrying different combinations of alleles present in the HDHD5 locus. A reporter construct with the EUR version of this sequence without introgression (EUR-EUR) drove significant expression above baseline (~2.0x vector with no insert, P < 0.01, t-test). We compared this activity to constructs synthesized to carry the RAs with the associated NDA (NDA-RA), the RAs without the NDA (EUR-RA), and the NDA without the RAs (NDA-EUR). This last combination never segregated in human populations, but is included here for context. As expected from the eQTL data, constructs lacking RAs drive significantly stronger expression (~2x baseline) than constructs containing RAs (~1x baseline; two-tailed t-test, P < 0.01 (**) and P < 0.001 (***)). The regulatory effect of the RAs are independent of the NDA in introgressed EUR haplotypes. (e) Regulatory activity in a massively parallel reporter assay (MPRA) for the four HDHD5 RA eQTLs reveals that rs71312076 has significant regulatory effects (RA:EUR allelic skew=2.122, P=6.6e−3, FDR=0.034) in the non-introgressed EUR background sequence.
In the LCL eQTL data, 2,564 RAs were significant eQTL in EUR, 180 were eQTL in YRI, and 42 displayed significant eQTL effects in both populations. These RA eQTLs influence the expression of nine genes (Supplementary Table 4). The RAs in EUR have the same direction of effect and similar magnitude as those observed for the corresponding allele in YRI. For example, two genes, SDSL and HDHD5, each have four cross-population RA eQTLs that have similar effects on gene expression in both EUR and YRI (Figure 4b). Thus, despite the limited cross-population eQTL data available, these results suggest that some RAs influence gene regulation in Eurasian individuals independent of NDAs.
RAs can influence expression independent of NDAs
To determine whether RAs directly influence expression in EUR individuals independently of linked NDAs, we functionally dissected the regulatory activity of combinations of introgressed alleles in one of the cross-population eQTL. The HDHD5 locus is an attractive target for in vitro examination because it contains a 2 kb region with four cross-population RA eQTLs and one introgressed NDA in EUR that is absent in YRI (Figure 4c). HDHD5 is a hydrolase domain containing protein located in chromosome 22q11 and associated with chromosomal abnormalities related to Cat Eye Syndrome (CES)42.
We performed luciferase reporter assays in LCLs using four different combinations of the NDA and RAs (Figure 4d, Supplementary Table 5 and S6). A reporter construct with the non-introgressed European sequence (EUR-EUR) drove significant luciferase expression above baseline (~2.0x, P < 0.01, t-test). We compared this activity to constructs synthesized to carry the RAs with and without the associated NDA (NDA-RA and EUR-RA respectively), and the NDA without the RAs (NDA-EUR). Both RA-containing sequences had significantly lower luciferase activity, and there was no significant difference in the activity of the NDA-RA and the EUR-RA sequences (Figure 4d). These results are consistent with the cross-population eQTL data and show that changes in activity are independent of NDAs. MPRA data from EUR and YRI LCLs (Supplementary Table 7)43 implicate one specific cross-population RA eQTL (rs71312076, Figure 4e).
Together, these cross-population eQTL, luciferase reporter, and MPRA results provide three orthogonal lines of evidence implicating RAs in the reintroduction of regulatory effects in the HDHD5 locus. Both our luciferase assays and the MPRA data show that the functional contribution of these RAs within a European genomic context is not dependent on the NDA present on the introgressed haplotype in which it occurs.
RAs are more likely than NDAs to have gene regulatory activity
A recent MPRA quantified the gene regulatory effects of 5.9 million variants in the context of ~400 bp of flanking sequence in K562 and HepG2 cells44. We identified all Eurasian introgressed alleles tested for regulatory activity. Excluding any within 400 bp of one another enabled us to characterize the independent regulatory effects of 42,016 RAs and 26,063 NDAs.
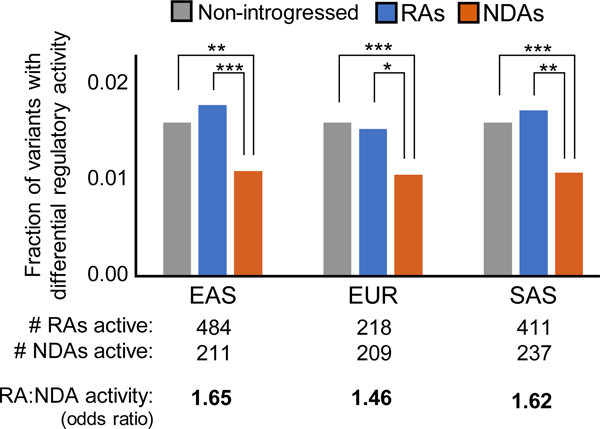
In total, 527 of the tested RAs have differential regulatory activity independent of NDAs, and 252 NDAs have differential activity independent of RAs. In each Eurasian population, the RAs are significantly more likely to have independent regulatory activity than NDAs (Figure 5; 1.5–1.7% vs. 1.0–1.1%; P<1E–3). The enrichment for activity among RAs compared to NDAs holds across a range of minimum activity thresholds, backgrounds, and RA subclasses (Supplementary Table 8, S9).
Figure 5. Hundreds of RAs have regulatory activity independent of NDAs.
The number and fraction of RAs and NDAs by population with significant differential gene regulatory activity in MPRAs. Among introgressed variants identified in each Eurasian population, RAs are significantly more likely than NDAs to have independent regulatory activity (odds ratios: 1.46–1.65, P < 1e−3 for all populations). RAs have similar levels of activity compared to non-introgressed variants (odds ratios: ~1.0, P > 0.01), while NDAs are significantly depleted for activity compared to non-introgressed variants (odds ratios: 0.65–0.68; P < 1e−6 for each population). Introgressed variants were required to have significantly different activity from the non-introgressed allele (controlling the FDR at 5%) in at least one of the two cell lines assayed (K562, HepG2; Methods). The odds ratio is the ratio of differentially active RA:NDA over the ratio of all RA:NDA tested (Supplementary Table 8). The enrichment for activity among RAs compared to NDAs held across a range of activity thresholds and when comparing only to sites with activity (Supplementary Table 9). Significance of the differences between variant sets was evaluated with Fisher’s exact test; * indicates P < 1e–3, ** indicates P < 1e–6, *** indicates P < 1e−9.
The fraction of RAs with activity is similar to the fraction for non-introgressed variants (Figure 5; 1.5–1.8% vs. 1.6%; P > 0.01), while NDAs are consistently less likely to be active than non-introgressed variants (1.0–1.1% vs. 1.6%; P < 1E–6). Matching the minor allele frequency and LD distributions of non-introgressed variants to introgressed variants did not change this result (average: 1.6%, standard deviation: 0.3%, over 1000 replicates). Thus, NDAs have significantly lower independent regulatory activity levels compared to RAs, while RAs have similar levels of regulatory activity to non-introgressed variants.
DISCUSSION
Here we demonstrate that hundreds of thousands of ancient alleles are present in modern Eurasians due exclusively to reintroduction via archaic admixture between Neanderthals and AMHs (Figures 1 and 2). We find that the majority of NDAs are in high LD with RAs, and our simulation and computational results suggest that RAs should be more tolerated than NDAs in AMH. Indeed, we show that RAs are more frequent than NDAs on introgressed haplotypes with GWAS associations and eQTL. We present multiple lines of evolutionary and experimental evidence to demonstrate that RAs on one introgressed haplotype have gene regulatory effects that are not dependent upon linked NDAs. These in vitro analyses of different combinations of introgressed alleles provide an in-depth experimental approach that disentangles LD. Finally, we use eQTL and MPRA data to show that at least 500 RAs have regulatory activity independent of NDAs. These analyses further demonstrate that NDAs are depleted of regulatory activity compared to RAs and that RAs have similar levels of activity to non-introgressed variants. This is consistent with selection against NDAs with regulatory effects. In light of these results, we conclude that the distinct evolutionary histories of introgressed alleles must be considered in analyses of archaic admixture.
RAs are enriched (and NDAs are depleted) among introgressed eQTL in some tissues—the brain in particular (Figure 3b)—but not others. These patterns are qualitatively consistent with the allele-specific downregulation of Neanderthal alleles in the brain and testes39 and the enrichment for Neanderthal eQTL in brain tissues22. We hypothesize that the regulatory effects of RAs and NDAs differ among tissues based on the genetic diversity and strength of constraint on their regulatory landscapes. Supporting this, nervous system tissues and the testes have extreme levels of selection on gene expression (high and low, respectively)45 and show significant RA:NDA differences (Figure 3b). Given the range of RA eQTL enrichments across additional tissues, including tissues without evidence of selection against Neanderthal alleles, we propose that they are the result of a mixture of selective pressures acting within the regulatory constraints of each tissue.
In particular, two non-exclusive evolutionary scenarios may explain the differences we observe between RAs and NDAs. First, the depletion of NDAs relative to RAs on introgressed haplotypes with gene regulatory functions could reflect previously demonstrated selection against NDAs in general20, in some tissues39, and in regulatory regions overall46,47. This selection would deplete regulatory regions of NDA-rich haplotypes. Indeed, two tissues with known allele-specific down-regulation of Neanderthal alleles, brain and testes, are among those significantly depleted for NDAs among introgressed eQTL. This scenario is further supported by the depletion of NDAs for regulatory activity compared to both RAs and other human variants in the MPRA analysis (Figure 5).
Second, introgressed RAs may mitigate negative selection against linked NDAs or even be selected for themselves. Under this scenario, archaic admixture restored alleles with beneficial (or at least non-deleterious) regulatory functions, and these RAs contributed to the maintenance of some introgressed haplotypes, as suggested at the OAS1 locus25. To evaluate evidence for this scenario, we analyzed introgression in two sets of regions with evidence for recent positive selection. Introgressed haplotypes that likely experienced strong, recent positive selection19 had significantly lower RA levels (Supplementary Table 10), suggesting that RAs did not drive most instances of positive selection after introgression. We also analyzed RA content in regions of the human genome predicted to be under positive selection in the human lineage after the split of AMH and archaic hominins48. Introgressed haplotypes in these regions are significantly enriched for RAs in each Eurasian population (Supplementary Table 11). This suggests that introgressed haplotypes with high RA:NDA ratios were more likely to be retained in regions important for AMH-specific biology. Nevertheless, our results point to NDAs being less tolerated overall, thus driving their depletion compared to RAs in functional contexts.
Furthermore, different evolutionary histories among RAs may influence RAs’ effects in AMH (Figure 1c). Our analyses of RAs stratified into subclasses based on presence in modern Africans (RAAAFR+ vs. RAAAFR–) and in the human-chimpanzee ancestor (RAA vs. RHA) reveal some differences between RA subclasses. For example, the RAAs present in Africans are at higher frequency in Eurasians (Extended Data Fig. 8) and are enriched on haplotypes with GWAS hits or eQTL compared to RAAs absent in Africans (Figures 3 and Extended Data Fig. 9). This could imply that, upon reintroduction, RAAAFR+ were more tolerated than RAAAFR–; however, the small number of RAAAFR– in Eurasians reduces power to detect differences. Similarly, variant effect prediction algorithms suggest that the older RAAs are slightly more benign than the younger RHAs. RAAs are also enriched for GWAS trait associations while RHAs are not.
Regardless of the differences between RA subclasses seen in some analyses, all RAs are distinct from NDAs in that they arose in a genomic background ancestral to AMHs, and were maintained in a relatively larger ancestral hominin population in which selection could act more efficiently. Previous work has implicated the small effective population size of Neanderthal populations as a key factor in their transmission of weakly deleterious NDAs into AMHs via introgression17,18,49. In contrast, RAs’ longer exposure to more efficient selection in ancestral hominin populations would explain why all RA subclasses are distinct from NDAs in most analyses. Our results also raise the question of whether we should have different expectations for the functional effects of RAs compared to non-introgressed variants of similar age. Among RAs, different evolutionary histories show distinct functional properties in some analyses. And, as noted above, RAs segregated in the Neanderthal lineage for hundreds of thousands of years. As a result, we might expect RAs to have more deleterious effects in the AMH context compared to non-introgressed alleles. However, in our analyses the RAs generally behave similarly to non-introgressed variants, especially when compared to NDAs (Figure 5). Thus, future work must consider the distinct evolutionary histories among introgressed variants when interpreting the effects of Neanderthal admixture.
Further analysis of RAs will also be relevant to studies of the genetics of ancient hominin populations. For example, tens of thousands of RAs that are present in Eurasians are not present in African populations. These ancient variants could both inform ongoing debates over differences in the efficiency of natural selection between Africa and Eurasia 50–53, as well as provide a window into ancient genetic variation that was present in Africa over a half million years ago. Finally, our focus has been on Neanderthals, but Denisovan introgression also likely reintroduced lost alleles, particularly in Asian populations with high levels of Denisovan ancestry.
In conclusion, we show that Neanderthal introgression reintroduced alleles lost in the ancestors of Eurasian populations and that hundreds of these RAs are functional. This demonstrates the importance of accounting for shared ancestral variation among hominin populations and illustrates a way that hybridization events have the potential to modulate the effects of bottlenecks on allelic diversity. RAs and their distinct evolutionary histories must be considered in analyses of Neanderthal introgression, at both the haplotype and genome scale.
METHODS
Sequence data
Genomic variants were taken from 1000 Genomes Phase 3v5a data1. Introgressed Neanderthal tag variants were downloaded from: http://akeylab.princeton.edu/downloads.html27. All analyses were conducted using GRCh37/hg19 genomic coordinates.
RA candidate identification and classification from 1000 Genomes data
To generate a set of candidate RAs, we gathered Neanderthal tag variants identified in each of the three 1000 Genomes Eurasian super-populations (EUR, EAS, SAS). These tag SNPs were identified by S*27, which compared Neanderthal genomes to those of European (EUR), East Asian (EAS), and South Asian (SAS) populations and represent variants that are rarely or never observed in African populations, yet that are present in Neanderthal introgressed haplotypes in Eurasians. We then calculated LD using vcftools54 for all variants in +/–500 kb windows around each variant across individuals from these super-populations in Phase 3 of the 1000 Genomes project. We extracted all variants that were in perfect LD (r2=1) with any Neanderthal tag SNP in any of EUR, EAS, or SAS populations.
For each candidate RA (i.e., a variant in perfect LD with a Neanderthal tag SNP that was not itself a Neanderthal tag SNP) we: 1) extracted the ancestral allele call from 1000 Genomes, 2) ascertained whether the designated REF or the ALT allele was the introgressed variant (i.e., in LD with the Neanderthal tag SNP), 3) calculated the introgressed allele frequency, 4) calculated the allele frequency for the same allele in sub-Saharan African 1000 Genomes populations, and 5) extracted the Altai Neanderthal genotype. We then assigned RA status based on this information by following the steps laid out in Extended Data Fig. 1. Specifically, for each RA candidate, if the introgressed variant matched the high-confidence, ancestral state, it was classified as a reintroduced ancestral allele (RAA). Candidate RAs that did not match the ancestral allele (or that did not have a high confidence ancestral allele call) were evaluated for presence in both the Altai Neanderthal and in sub-Saharan Africans (average allele frequency > 1% in ESN, GWD, LWK, MSL, and YRI). If the candidate variant was only present in sub-Saharan African at a frequency > 1%, it was classified as a reintroduced hominin alleles (RHA) since its origin likely predated the Neanderthal split, but its ancestral status is not assigned. Importantly, the criteria that an RHA also be present at a minimum allele frequency of 1% frequency over all 5 sub-Saharan populations insulates our results from the low levels of apparent Neanderthal ancestry that are detectible in modern Africans (0.18–0.5%) due to backflow and possible gene flow from an earlier human population to Neanderthals55,56. If the candidate variant was only present in the Altai Neanderthal and introgressed Eurasian haplotypes, it was classified as an NDA. For nearly all analyses presented here, RAAs and RHAs are combined into a single RA class. The results of this classification are summarized in Figure 2 and supplied in full in File S1.
We considered all Neanderthal haplotypes and tag SNPs identified by Vernot et al. 2016 27. Only 10% of identified RAs were found in haplotypes shorter than 10 kb or with fewer than 10 NDAs. Removal of these NDAs and RAs from our analyses did not substantially change any of our results, thus we present results with all RAs for completeness.
Approximately 90% of RAs are within the boundaries of previously characterized introgressed haplotypes; however, over half of the haplotypes in each population have at least one associated RA beyond their previous bounds. In total, extending all introgressed haplotypes to accommodate all associated RAs increases introgression estimates by 40.0, 42.6, and 51.9 megabases (Mb) in the EUR, EAS, and SAS populations, respectively. This represents an increase of ~1.5% in the amount of introgressed sequence present in each Eurasian population.
As described above, when no confident ancestral allele call was available, we used the presence of an allele in modern Africans to infer that it was present in the ancient hominin population. In such cases, if the allele was present in modern Eurasians on a Neanderthal haplotype, it was inferred to be an RHA. However, without a confident ancestral state for these alleles, RHAs may be susceptible to false positives due to independent, convergent mutations on the AMH African and Neanderthal lineages. This is of particular concern at CpG sites due to their significantly higher mutation rates compared to other dinucleotides. Therefore, to estimate how many inferred RHAs could be the result of independent C→T transitions along the Neanderthal and AMH lineages, we identified all RHAs that were either “A” or “T” with a 5’ “C” or 3’ “G” respectively. We focused on RHAs, because RAAs match ancestral allele calls supported by cross-species alignments making convergent mutation unlikely. Only 7% of RHAs were potentially subject to this bias (Supplementary Table 2). Furthermore, we carried out simulations with an estimate of the CpG mutation rate (7.0 × 10−7 mutations per site per generation57) and estimated that 3% of all CpGs would be expected to display convergent mutations between Neanderthals and AMH Eurasians. Thus, confounding due to convergent mutations, even at CpGs, is likely to be rare (Supplementary Table 2). (See “Estimating confounding with simulations” section for more details on the simulations.)
Spatial characterization of RAs and NDAs along introgressed haplotypes
The locations and distributions of RAs within introgressed haplotypes are less correlated with haplotype length and more clustered than the distribution of NDAs. The number of NDAs per haplotype is strongly positively correlated with the length of the haplotype (r2 = 0.85; Extended Data Fig. 4), but the RA number per haplotype is more variable (r2 = 0.56). Therefore, while the overall RA:NDA ratio is ~1:2 over all haplotypes (Figure 2), this ratio varies across introgressed haplotypes.
To evaluate whether RAs are more clustered on introgressed haplotypes than NDAs, we summarized the distribution of both NDAs and RAs across all RA-containing haplotypes. We first divided each RA-containing haplotype into 100 equal-size bins and counted the number of RAs in each bin. For each haplotype, the bins were then ranked from high to low in terms of RA count, and the RA contents of each corresponding percentile bin were summed over all the haplotypes. This percentile sum was then divided by the total number of all RAs present over all the haplotypes to obtain per-bin densities. By calculating per-bin densities only at the end, we mitigate the potentially confounding effect of some haplotypes containing fewer variants than others. The result is a summary of the total fraction of RAs found within increasing density percentiles across all haplotypes. We then did the same for NDAs (Extended Data Fig. 4). Overall, a larger fraction of RAs is found in the densest bins compared to NDAs. For example, in EUR, 55% of RAs are in the four densest bins, while only 26% of NDAs are in the four densest bins. These results held across each population and were maintained when down sampling to a set of haplotypes with matched NDA and RA counts. Thus, when RAs are present, they often occur in more discrete clusters along introgressed haplotypes than do NDAs. However, we note that the incomplete ascertainment of RAs and the LD thresholds used to link NDAs may contribute to these patterns.
Computational variant effect estimation
To assess the potential functional impact of RAs, we analyzed precomputed Combined Annotation-Dependent Depletion (CADD) v1.3 scores (https://cadd.gs.washington.edu/download) for all RA and NDA variants. CADD scores are available in two forms: raw and scaled. Raw CADD scores for variants are the output of a support vector machine trained to distinguish variants observed in 1000 Genomes (likely benign) from non-observed variants (likely deleterious) based on diverse annotations. To enable comparisons across sites, the raw scores for all possible mutations to the hg19 genome were ranked and PHRED-scaled (–10 * log10(Rank))58. Therefore, the scaled scores communicate how deleterious the effect of a given variant is with respect to the effects seen in all other possible variants (e.g., a scaled CADD of 20 means that that a variant is within the top 1% of variants as ranked by their predicted deleteriousness). Thus, we focused on the PHRED-scaled scores. In particular, we highlighted in Extended Data Fig. 5, scaled CADD scores at the upper range (e.g., above 10 or 15) that are most suggestive of deleteriousness. We also compared functional annotation classes downloaded for RAs and NDAs from RegulomeDB v1.1 (Extended Data Fig. 7; http://www.regulomedb.org/) and PolyPhen2 (Extended Data Fig. 6; http://genetics.bwh.harvard.edu/pph2/). For purposes of establishing background expectations (Extended Data Fig. 7) we generated 1000 sets of allele frequency- and LD density-matched EUR variants (SNPsnap 59); sets were based upon an input set consisting of the highest frequency Neanderthal tag SNP from each EUR introgressed haplotype.
Evaluation of introgressed variants in GWAS Catalog
To evaluate the prevalence of RAs among significant GWAS associations, we intersected all RAs and NDAs with variants reported in GWAS Catalog (as of January 24, 2019). To account for other variants tagged by the variant reported in the GWAS Catalog, we expanded the target set of GWAS variants to include variants in perfect LD (1000 Genomes r2=1) in each of the three Eurasian populations. We then intersected sets of introgressed variants from each population with LD-expanded GWAS hits for that population. The results of these expansions and intersections are presented in File S2.
GTEx eQTL enrichment analysis.
Expression quantitative trait loci (eQTL) data from GTEx v7 were downloaded from the GTEX portal (https://www.gtexportal.org/home/datasets) and all significant gene-eQTL pairs were extracted for each tissue. We then identified all RAs and NDAs with eQTL status. Because the GTEx cohort is >85% European ancestry, we only considered European RAs and NDAs in this analysis.
First, we evaluated whether introgressed haplotypes with eQTL activity have a different RA fraction than introgressed haplotypes without eQTL (Figure 3a). This observation held when eQTL were stratified by tissue (Extended Data Fig. 10), suggesting that at the haplotype level, RAs appear at greater frequency within regions associated with gene regulation.
Next, within each tissue, we considered only those significant introgressed (i.e. RA and NDA) eQTL and the RA:NDA ratio for each tissue in Figure 3b. To test whether there was an overrepresentation of RAs among introgressed eQTLs, we performed a hypergeometric test on each tissue’s set of introgressed eQTLs with respect to the background of all variants evaluated for eQTL status within GTEx. We applied a Bonferroni correction to account for the testing of the 48 tissues (0.01/48=0.0002).
Shared RA eQTLs between Europeans and Africans
To identify RAs with similar regulatory associations between populations with and without Neanderthal ancestry, we analyzed data from a previous study that identified eQTL across LCLs derived from 495 individuals41. The LCLs were of either European (EUR; 373 lines) or African (YRI; 89 lines) ancestry; given the smaller YRI sample size, there was much lower power to detect eQTL in the African samples. We downloaded all significant exon-level expression eQTLs from the study (https://www.ebi.ac.uk/arrayexpress/files/E-GEUV-1/analysis_results/). They found 704,157 unique eQTL in EUR and 75,742 in YRI, and of these, 52,869 are shared. Of the shared loci, 42 are RAs, and these RAs associate with expression levels for nine genes (Supplementary Table 4). For each of the 42 variants, we also confirmed that they were not in LD in YRI with any previously characterized NDA.
MPRA analysis of RAs
We analyzed data from two recent MPRA studies. While estimating the magnitudes or directions of effect in vivo from MPRA can be challenging43, they do demonstrate regulatory effects for individual variants. The first study evaluated the regulatory impact of 32,373 variants in 3,642 known eQTL and regions identified via GWAS 43. For each variant, the MPRA quantified the expression of a reporter driven by both the reference and alternate alleles (plus 150 bp of reference genomic context) in LCLs. Expression modulating variants were identified by quantifying the “allelic skew” between the expression driven by the reference and alternate allele. This enabled the identification of hundreds of variants likely to cause the observed associations between these loci and expression levels/phenotypes. We intersected European NDAs and RAs in introgressed haplotypes with the variants with significant combined skew (FDR < 0.1). In total, 11 introgressed variants were tested (6 NDAs and 5 RAs; Supplementary Table 7). This included all cross-population RA eQTLs in the introgressed haplotype that is associated with HDHD5 expression (Figure 3b).
The second MPRA study used the high-throughput survey of regulatory elements (SuRE) reporter approach to survey the effect of both alleles at 5.9 million genetic variants in K562 and HepG2 cell types44. In short, ~400 bp regions were sampled from the fragmented genomes of four individuals, barcoded, and prepared as SuRE libraries. These libraries were transfected into K562 and HepG2 cell lines and their expression was measured using Illumina paired-end sequencing. We analyzed the reported mean SuRE scores for each allele of each variant tested in each cell line. For our analysis, we excluded introgressed variants for which a variant from the other introgressed variant class (NDA or RA) was within 400 bp. This step ensured that the regulatory effects observed for a variant were independent of variants from the other class. Following the authors’ approach, we called significant differential activity between the alleles at sites based on Wilcoxon rank-sum test p-values (controlling the permutation-based FDR at 5%). We confirmed our results across several “maxSuRE” score activity thresholds (i.e. requiring that at least one of the two alleles at a locus meet a minimum SuRE score requirement), and by comparing to both the background of all variants tested or only those that met the activity thresholds (Supplementary Table 8). These results are reported in Figure 5 and Supplementary Table 8.
Experimental validation of RA regulatory function via luciferase assays
To further demonstrate that the cross-population RA eQTLs associated with HDHD5 expression function independently of the NDA in perfect LD, we evaluated the effects of four different sequences on luciferase expression in LCLs (Figure 4d).
Modified pGL4 luciferase constructs were generated via Gibson cloning (New England Biolabs) to contain an 1826 bp oligo corresponding to the region of interest in CECR5/HDHD5 with variants corresponding to a European reference (EUR-EUR), the introgressed NDA sequence (NDA-EUR), the RA sequences (EUR-RA), or both sets of introgressed variants (NDA-RA) (Supplementary Table 5). Inserts were cloned into the pGL4.27 reporter vector (Promega) as two separate blocks, as b1-EUR or b1-NDA (first 576 bp at the 3’ end of blocks containing either NDA or EUR specific sequence) and b2-EUR or b2-RA (1273 bp at the 5’ end of blocks containing either RA or EUR specific sequence) (Supplementary Table 5). b1-EUR, b1-NDA, and b2-RA sequences were generated by oligonucleotide synthesis (IDT). b2-EUR variants were generated via site-directed mutagenesis using primers with EUR specific alleles (Supplementary Table 6) and amplified directly from the b2-RA oligo as five separate sub-regions. B2-EUR sub-regions were assembled into the pGL4.27 vector and sub-cloned into EUR-EUR and NDA-EUR pGL4 constructs. Inserts were amplified to include NheI and XhoI overhangs to allow for cloning into the pGL4 reporter plasmid. The sequences of full-length inserts were confirmed by Sanger sequencing (Genewiz).
GM11831 B-cells were cultured in RPMI with penicillin/streptomycin and 15% fetal bovine serum. 1×106 GM11831 cells were transfected with 5 ug HDHD5-EUR-EUR-pGL4.27, HDHD5-NDA-EUR-pGL4.27, HDHD5-EUR-RA-pGL4.27, or HDHD-NDA-RA-pGL4.27 along with 500 ng pRL-CMV (Renilla reporter plasmid) via electroporation (Neon Transfection System, Invitrogen). Firefly and Renilla luciferase activity were analyzed using the Dual-Glo Luciferase Assay System (Promega) and Synergy HTX MicroPlate Reader (BioTek) 19 hours post electroporation. Firefly reporter expression was normalized to Renilla luciferase activity. Statistical significance was determined through a two tailed t-test comparing fold change of the normalized luciferase activity over an unmodified (no insert) pGL4.27 reporter control.
Evolutionary simulation framework
We carried out evolutionary simulations to explore RA dynamics and estimate false positive rates under a range of scenarios. SLiM (v2.6) was used for all evolutionary simulations60. We based our simulations on genomic and demographic models used in previous simulation studies of Neanderthal introgression and mutation load18. In brief, the human genome was represented by a syntenic, locus-based model that reflects the gene structures in the hg19 reference genome. Nucleotide positions of exons were modeled individually while intergenic regions and chromosomal boundaries were modeled as single sites. We used a recombination rate of 1.0 × 10−8 crossovers per site per generation with probabilities in intergenic regions scaled by their respective sizes. Chromosome boundaries had a recombination rate of 0.5. To estimate false positives, we also considered each of three mutations rates: 7.0 × 10−9, 7.0 × 10−8, and 7.0 × 10−7 mutations per site per generation. The highest rate was included to simulate the high mutability at CpG dinucleotides, while the lowest is in keeping with genome-wide estimates for non-synonymous sites in humans. We simulated fitness effects (FE) of mutations based either on neutrality (FE=0) or purifying selection (FE drawn from gamma distribution with shape parameter 0.23 and mean selection coefficient −0.043)61. We also considered Eurasian–Neanderthal admixture fractions of 0.02 and 0.04.
The general demographic model used is illustrated in Extended Data Fig. 2. Genetic diversity within the ancient human population (10,000 diploid individuals) was first established by allowing mutations to arise and evolve through a “burn in” period of 44,000 generations in the ancestral hominin population prior to subsequent migrations. To track allelic loss and reintroduction, we focused on segregating sites that were present in this simulated ancestral population immediately before the split between the human and Neanderthal lineages; we tracked all of these ancestral hominin alleles over the 18,000 subsequent generations that encompassed both the Neanderthal and Eurasian OOA bottlenecks. The ancestral Neanderthal population was subsampled to 1,000 individuals and both human and Neanderthal populations evolved separately for 16,000 generations (400,000–464,000 years assuming a generation time of 25–29 years).
The Eurasian OOA migration and Neanderthal admixture were then modeled as a simultaneous, discrete event that resulted in an admixed Eurasian population size of 1861 individuals 18,62. The admixed Eurasian population was then allowed to evolve for 2000 generations before undergoing exponential growth leading to 20,310 modern Eurasians. One hundred replicates for both neutral or purifying selection models were run to evaluate properties of RAs and estimate rates of confounding mutations (Extended Data Fig. 2). To enable further analyses, snapshots of alleles present in each population were collected at four relevant timepoints for each simulation: t1) Neanderthal OOA, t2) immediately prior to the Eurasian migration, t3) immediately following admixture, and t4) modern human populations. Mutation origin was used to establish when (generation) and where (genome location and population) a variant arose and to trace its presence/absence through successive timepoints.
To quantify the frequency of RAs in simulated modern Eurasian populations we defined “ancestral hominin variants” as those alleles segregating in the simulated population immediately prior to the Neanderthal split ~500,000 years ago (t2). We tracked segregating ancestral variants through the Neanderthal lineage and into the modern Eurasian population. We used SLiM’s mutation identifiers to track these ancestral variants through Neanderthals and into modern Eurasians over each replicate. We identified all the ancestral variants that passed into AMH exclusively through either 1) the Eurasian OOA migration or 2) archaic admixture with Neanderthals (RAs). We extracted allele counts and selection coefficients (in admixture models run with purifying selection) for these RA variants from the SLiM output. We then did the same for the simulated NDAs, the only other class of variants that entered the modern Eurasian populations exclusively through Neanderthal introgression. These data are summarized in Supplementary Table 1 and Extended Data Fig. 2b and contrasted in Extended Data Fig. 2e.
Estimating confounding factors with simulations
We explored several sources of possible confounding through simulation. First, we estimated the rate at which variants could be mis-assigned RA status as the result of independent, convergent origins in African and Neanderthal populations. To infer the frequency of such confounding events, all variants in simulated human and Neanderthal populations were compared immediately prior to admixture (t2) in each of the 100 replicates for each model. We chose 100 replicates due to the computational cost of each simulation and the fact that the variance in the output statistics stabilized well before reaching 100 replicates. Confounding variants were identified based upon a shared genomic location between existing variants in Africans and variants that arose within the Neanderthal lineage. These counts (false positives) were then contrasted with the number of non-Neanderthal derived mutations (true negatives) and found to be very rare (Extended Data Fig. 2). Moreover, because SLiM does not consider nucleotide state and allows for “stacked” mutations (i.e., mutations at the same locus), our estimates of false assignment of RA status in this model are conservative because we also considered nucleotide state in the real data. We also considered a mutation rate an order of magnitude higher than the genome-wide average (7.0 × 10−7 mutations per site per generation) to reflect the hypermutatbility of CpG dinucleotides (Extended Data Fig. 2).
Second, it is also possible that non-Neanderthal alleles could have recombined on to introgressed haplotypes and subsequently been lost outside of the introgressed context. We reasoned that this scenario would be very unlikely, especially given our requirement of perfect LD between RAs and NDAs in modern Eurasian populations in the inference of RA status. To test this, we examined each of the simulated Eurasians (t4) and extracted all variants in perfect LD with an NDA in modern Eurasians. We then queried the simulation data from t2 to count how many of these candidate RA variants were not present on a Neanderthal haplotype. These variants in perfect LD with an NDA in modern Eurasians that were not present in Neanderthals (and that had not independently appeared within Eurasians) would be incorrectly inferred to be RAs by our approach. As expected, these events were very rare (1% of RAs or fewer) for each admixture fraction (Supplementary Table 3). Furthermore, these are likely overestimates since in the real data, RAs most frequently appear within introgressed haplotypes, with linked NDAs present on both sides. This would suggest two recombination events, with all the confounding alleles then being subsequently lost on all non-introgressed haplotypes to maintain perfect LD. In the future, we anticipate that these simulations can be refined to confidently identify more RAs that retain lower LD with NDAs.
Data analysis and visualization
Evolutionary simulations and primary data analysis were conducted on Vanderbilt’s computing cluster (ACCRE). Results were parsed and analyzed with custom python and bash scripts. Statistical tests were performed with R. Plots were generated in R using ggplot2.
DATA AVAILABILITY
All results reported in this paper are available in supplementary material and/or on the project’s github repository (https://github.com/DaRinker/Neanderthal.reintroduction).
CODE AVAILABILITY
Full code used in this analysis are available on the project’s github (https://github.com/DaRinker/Neanderthal.reintroduction).
Extended Data
Extended Data Fig. 1. Introgressed allele class assignment decision tree and allele count summary.
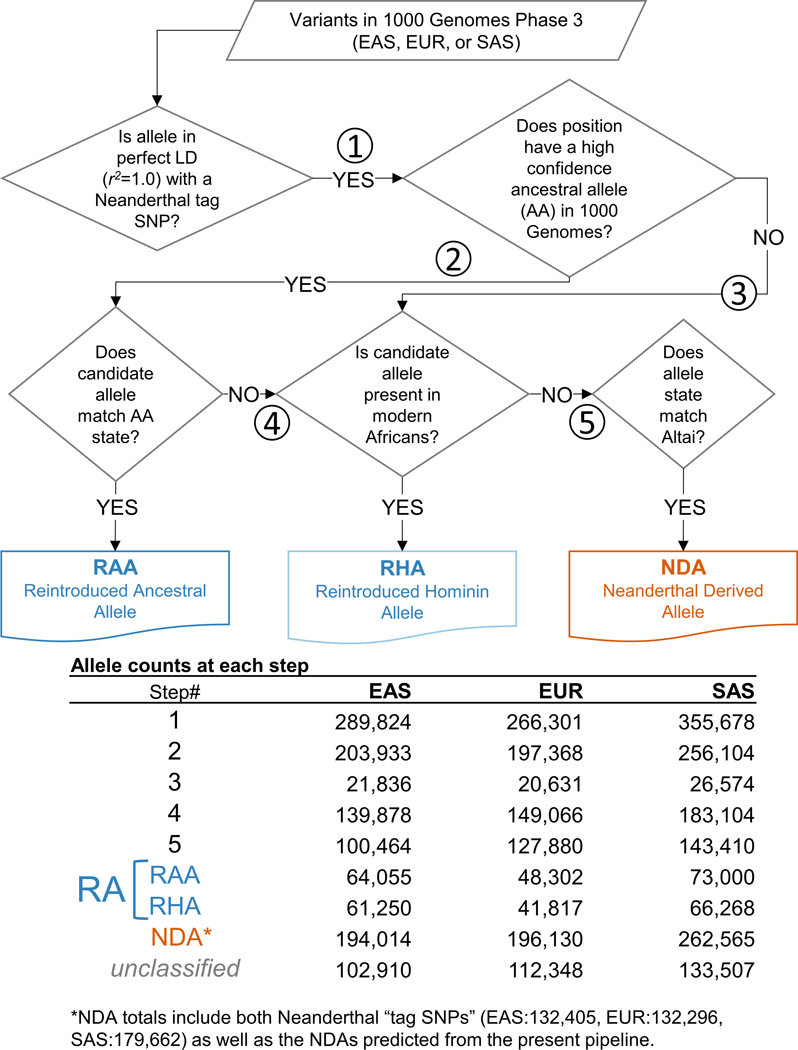
Decision tree by which 1000 Genomes variants in perfect LD with Neanderthal tag SNPs were classified as RAs and NDAs. The counts of variants making it to each of the numbered steps (1–5) is summarized in the lower table.
Extended Data Fig. 2. Evolutionary simulations suggest that RAs are common and more tolerated than NDAs.
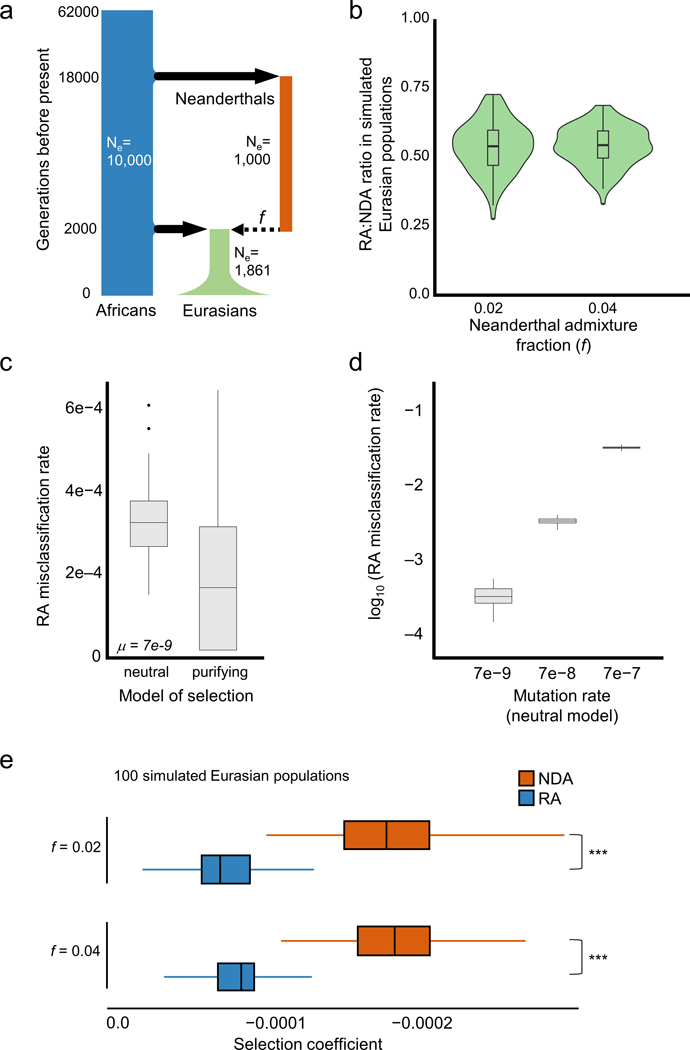
a) The demographic model used to simulate human–Neanderthal admixture and quantify the reintroduction of lost alleles. The model and effective population sizes (Ne) were based on previous simulations of Neanderthal admixture. We considered models in which mutations incurred a fitness cost (mildly purifying selection) or no fitness cost (strict neutrality). Two different admixture fractions (f=0.02 and f=0.04) and three mutation rates were used in the simulations (Methods). b) The ratios of RAs to NDAs over 100 simulated Eurasian populations. The simulations predict approximately one RA for every two NDAs, and these estimates are robust to changes in the simulated Neanderthal admixture fraction. Misclassification of non-RAs as RAs due to independent, convergent mutations is extremely rare and the overall false discovery rate for LD-based RA identification is below ~1% (Table S3). While these forward time simulations only approximate the demographic histories of these populations, the observed RA-to-NDAs ratio are qualitatively consistent with the simulations (Figure 2). (c) Boxplots summarize the frequencies of these potentially confounding NDAs among all sites that would be called as RAs at the time of admixture (c.f. Figure 1). The incidence of these confounding mutations is slightly higher under a purely neutral model (left) than under a model in which new mutations can be deleterious (right). (d) Comparison of the effect of elevated mutation rates on the incidence of potentially confounding variants. Under a neutral model, the false positive rate scales with the mutation rate. The highest rate (μ= 7e-7) provides an estimate for CpG sites and results in a 3% false positive rate. Each boxplot represents 100 simulated populations. e) Selection coefficients in Eurasians from SLiM simulations with high (0.04) and low (0.02) admixture fractions. Each boxplot summarizes the average selection coefficient of all alleles in each introgressed class in each of 100 simulated modern Eurasian populations. The differences between the selection coefficients between RAs and NDAs is large and not dependent upon admixture fraction (~2.5x, P ≈ 0, Mann Whitney U test test).
Extended Data Fig. 3. Introgressed allele sharing across three Eurasian populations.
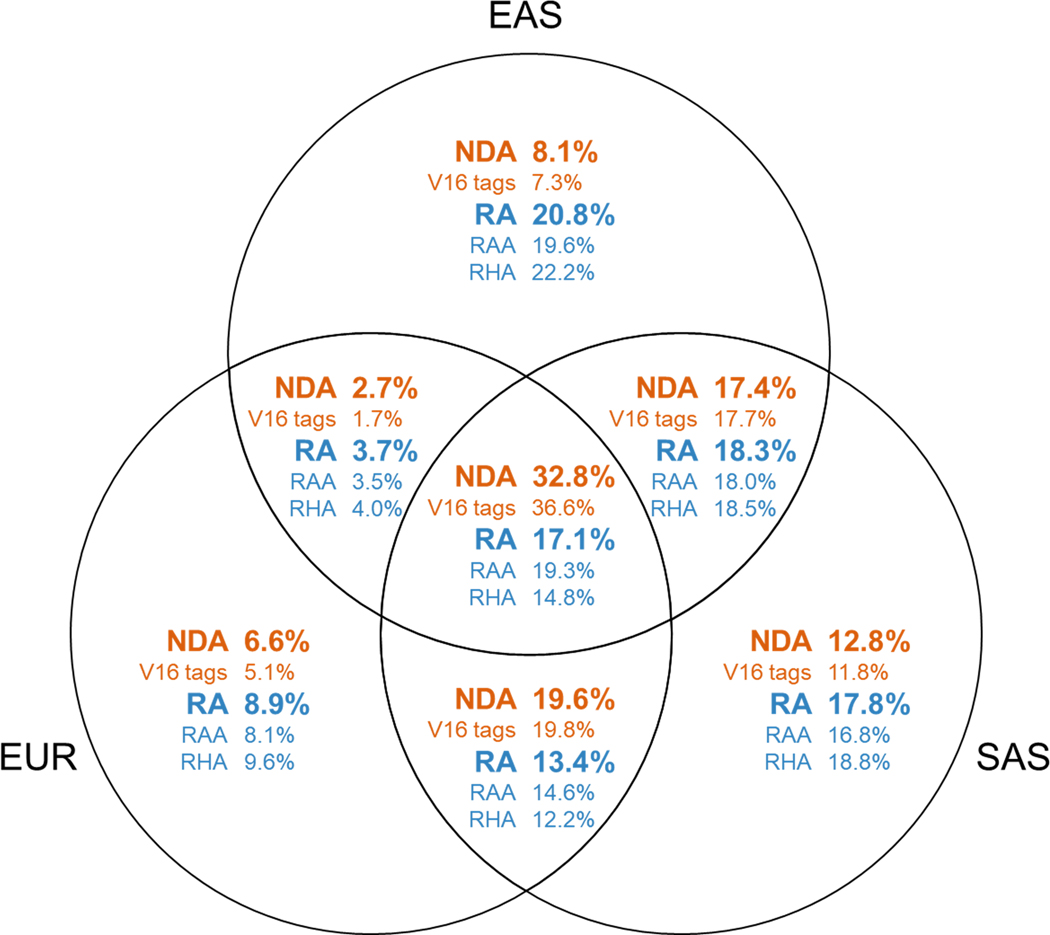
Venn diagram showing the fractions of each introgressed variant class that are shared between populations.
Extended Data Fig. 4. Reintroduced alleles cluster within introgressed Neanderthal haplotypes.
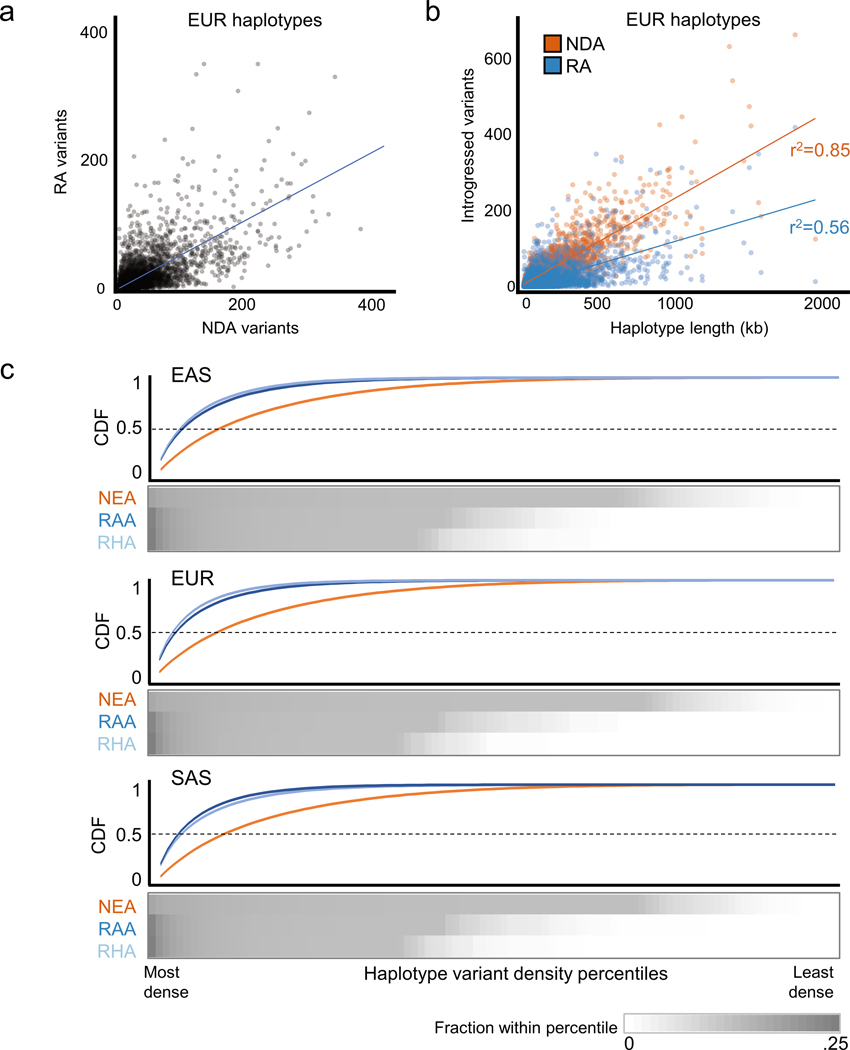
(a) Scatter plot of the numbers of RAs and NDAs contained on all introgressed haplotypes in EUR. The correlation between the NDA and RA content is moderate (Pearson’s r2=0.46), with 18% of the haplotypes containing no RAs and 10% having more RAs than NDAs. (b) Scatter plot of the number of introgressed variants on each haplotype vs. haplotype length. The NDA content of a haplotype is proportional to its length (r2 = 0.85), but the number of RAs in each haplotype is less strongly correlated with length (r2 = 0.56). (c) Heatmap of the fraction of NDAs and RAs in density percentiles (high to low, left to right) averaged over all introgressed Eurasian haplotypes. This information is summarized in a cumulative density function (CDF) above the heatmaps. A higher fraction of all RAs are found in the most dense percentiles; this reflects the fact that RAs are often present in more dense clusters than are NDAs.
Extended Data Fig. 5. Reintroduced alleles have different predicted fitness effects than Neanderthal-derived alleles.
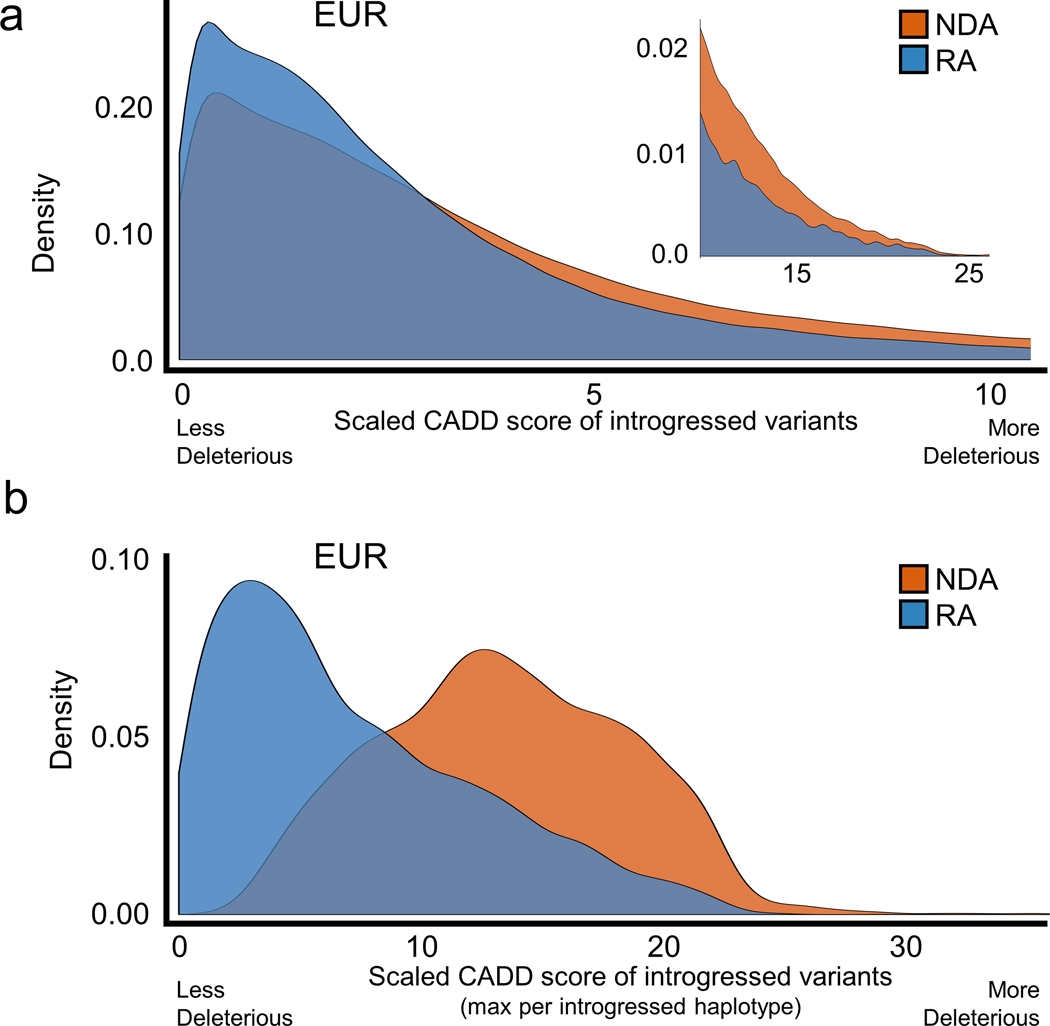
(a) In modern European (EUR) populations, RAs are predicted to be significantly less deleterious than NDAs by CADD (median scaled CADD: NDA=2.67; RA=2.1; P ≈ 0). The upper tail of highly deleterious mutations (CADD score >10) is highlighted in the inset. Results are similar for unscaled scores. (b) At the haplotype level, the maximum RA CADD score per introgressed haplotype is significantly lower than for NDAs (median scaled max CADD: NDA=13.3; RA=5.8; P ≈ 0). This is in part due to the overall difference demonstrated in (a) and to the greater number of NDAs per haplotype. RAs are rarely the most deleterious variant per haplotype. Results in East Asian and South Asian populations are similar (data not shown).
Extended Data Fig. 6. PolyPhen2 predicts RAs to be less damaging than NDAs.
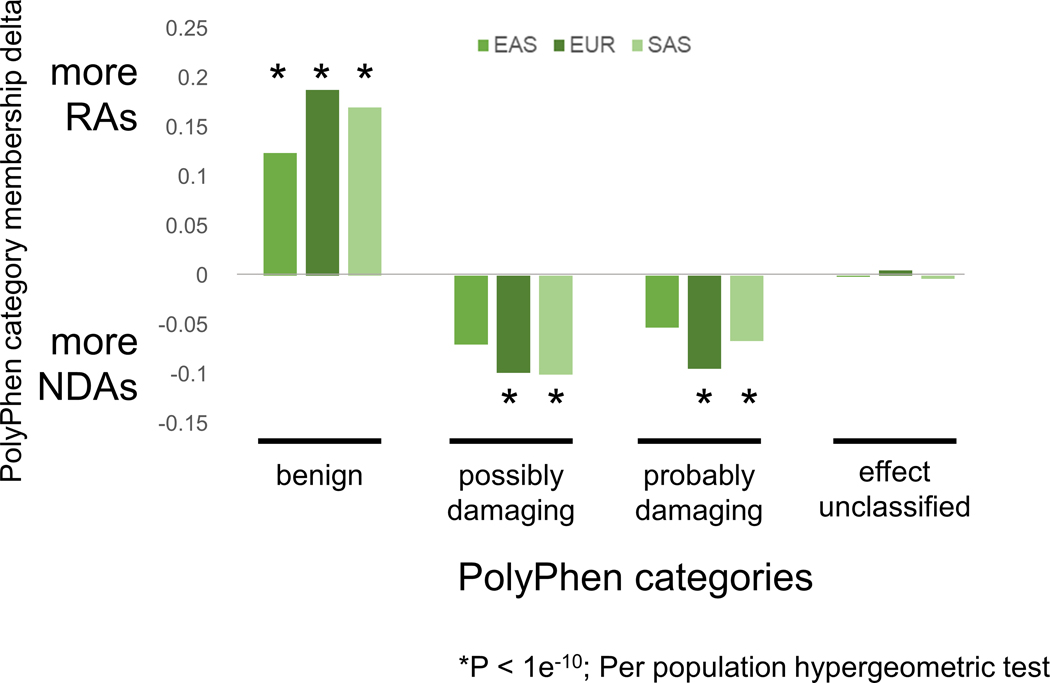
PolyPhen2 is more likely to classify RAs as “benign” in all three Eurasian populations. Conversely, NDAs are significantly more likely to be classified as “damaging” in both EUR and SAS populations. The y-axis reports the difference in PolyPhen category membership for each population (i.e., the fraction all RAs in the population in the PolyPhen category minus the fraction of all NDAs in population in the PolyPhen category). Per population hypergeometric test is calculated on the enrichment (positive delta) or depletion (negative delta) for RA content within each PolyPhen category.
Extended Data Fig. 7. Distribution of RegulomeDB scores among all introgressed Eurasian SNPs.
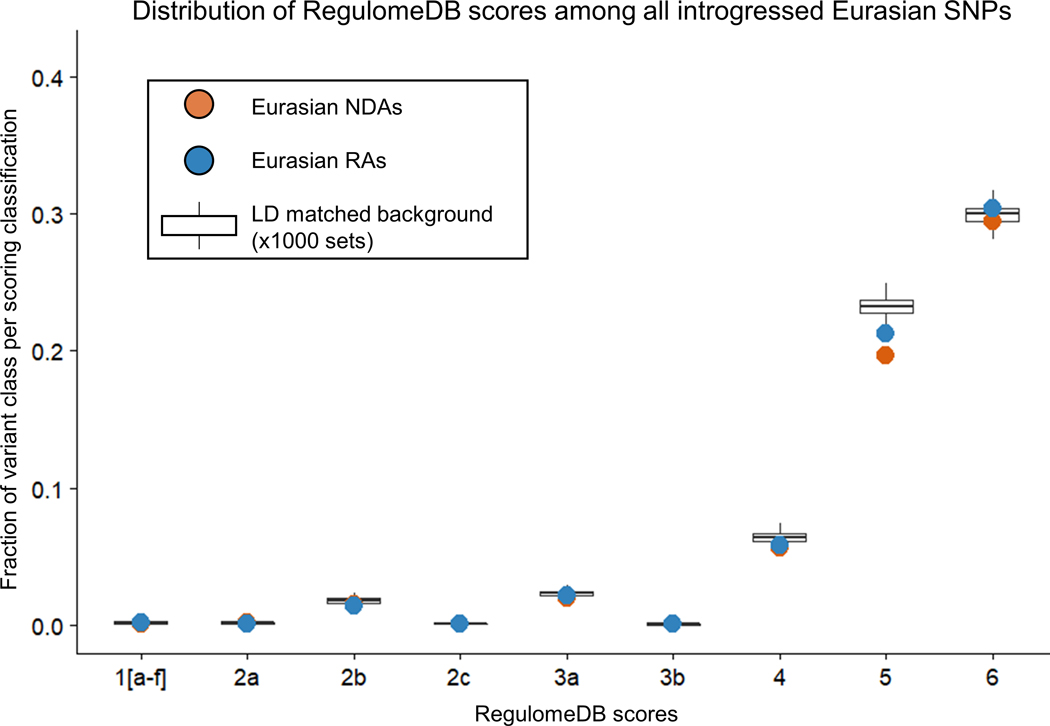
Comparison of the composition of NDAs and RAs within in each functional category of RegulomeDB. Boxplots refer to 1000 independently samples sets of background variiants matched on the allele frequency and local LD structure of the highest frequency Neanderthal tag SNP per introgressed EUR haplotype (Vernot 2016).
Extended Data Fig. 8. Comparison of allele frequencies across Eurasian populations.
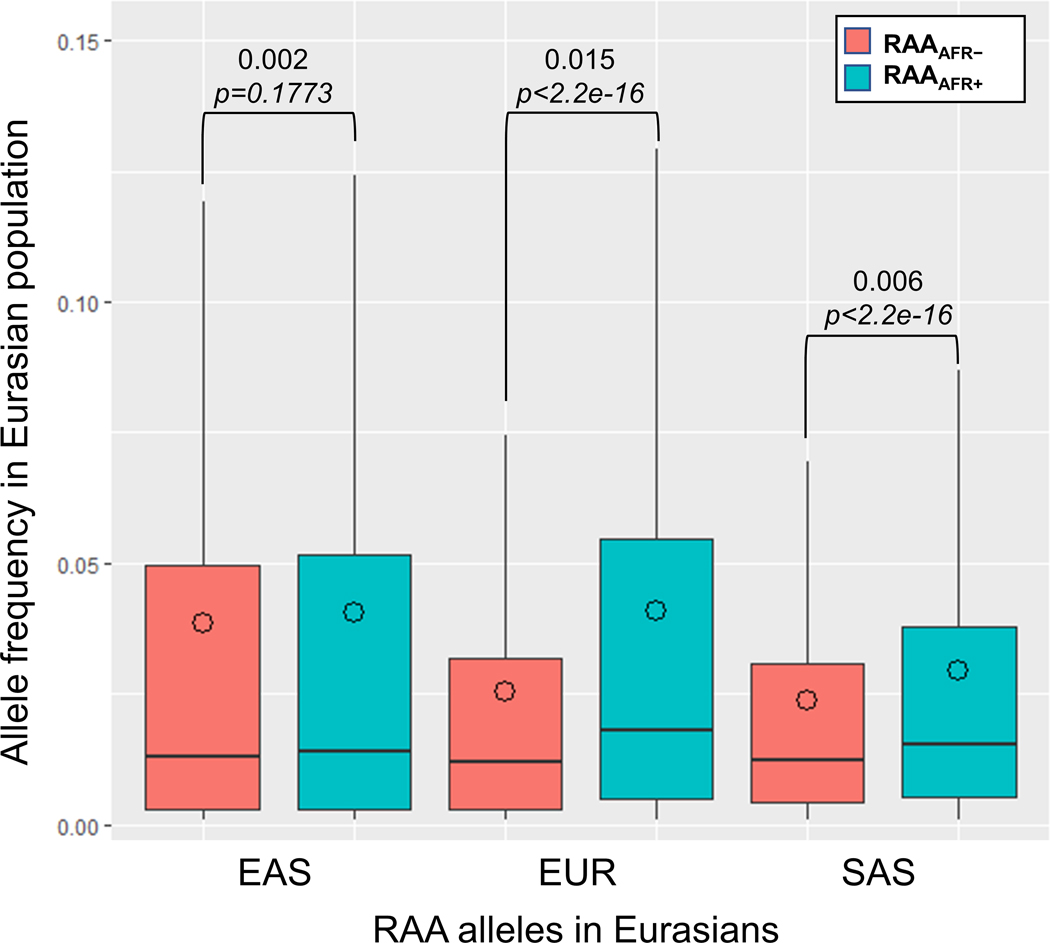
Stratified by presence/absence of allele in modern sub-Saharan African populations. Reintroduced Ancestral Alleles (RAAs) that are also present in modern African (AFR) populations segregate at higher allele frequencies (AF) in all Eurasian populations than RAAs for which the allele is absent in AFR. Intra-population median differences in AF are displayed along with P-values (Mann Whitney U test). Outliers are not shown. Circles indicate mean AF.
Extended Data Fig. 9. RA sublass composition of introgressed haplotypes containing GWAS hits.
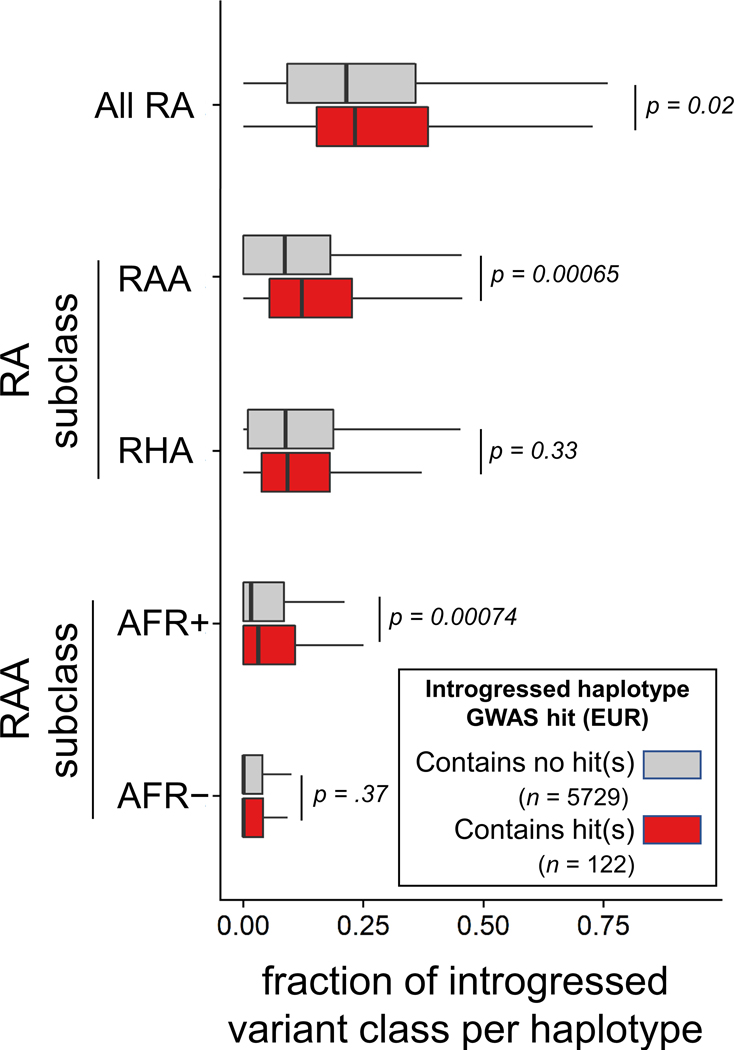
The fraction of RAs among introgressed alleles on introgressed haplotypes in EUR that contain GWAS Catalog associations in Europeans versus introgressed haplotypes having no reported GWAS associations.
Extended Data Fig. 10. RA fraction in introgressed haplotypes containing eQTL in GTEx tissues.
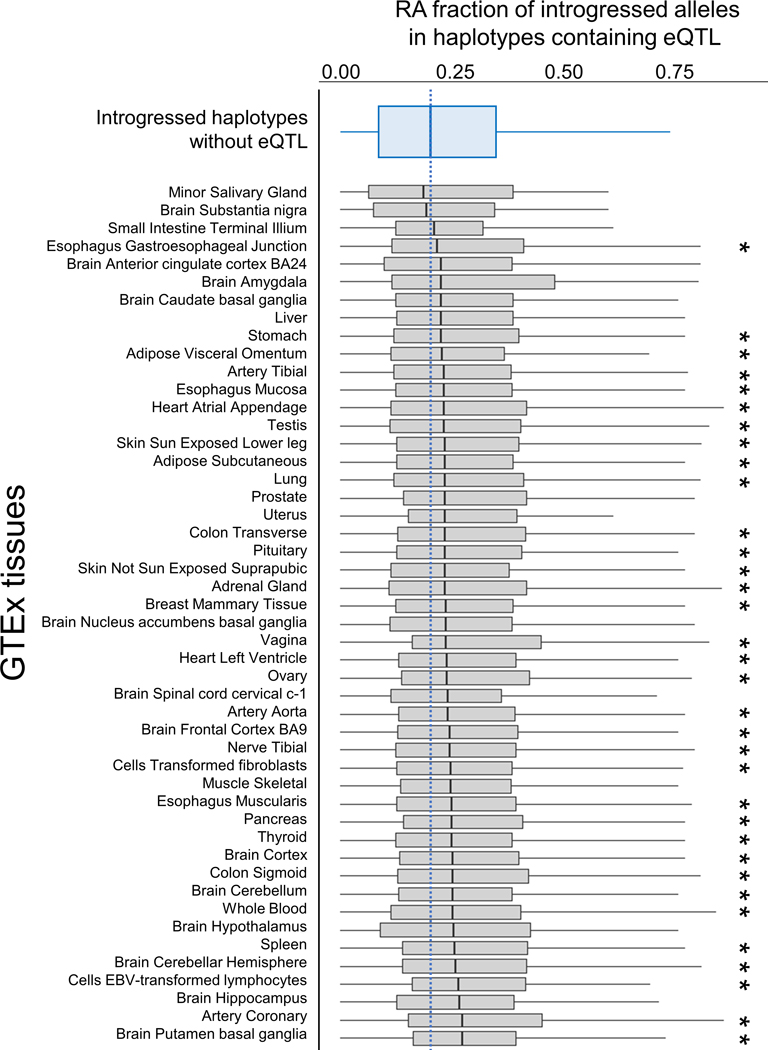
Summary of the RA fraction among introgressed variants in Neanderthal haplotypes in Europeans (EUR). Boxplots show the distributions RA fractions of all haplotypes containing at least one introgressed eQTL (RA or NDA) in the given GTEx tissue (gray box plots). These distributions are then compared pairwise with distribution for introgressed haplotypes that contain no introgressed GTEx eQTL (top, blue boxplot; n=4237). Haplotypes containing GTEx eQTL have RA contents higher than non-eQTL containing haplotypes in 46 tissues, with 34 of the tissues (*) having a significantly higher the RA fraction (P<0.05, Mann Whitney U Test).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ben Haller, Phillip Messer, and Kelley Harris for advice on evolutionary simulations. We thank Ryan Tewhey for discussions of MPRA results. We thank Laura Colbran and other members of the Capra Lab for helpful comments on the figures and manuscript. This work was supported by the National Institutes of Health: T32EY021453 to CNS; T32GM080178 to DS; K22CA184308 to EH; and R01GM115836 and R35GM127087 to JAC. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallick S. et al. The Simons genome diversity project: 300 genomes from 142 diverse populations. Nature 538, 201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pagani L. et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature 538, 238–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henn BM, Botigué LR, Bustamante CD, Clark AG & Gravel S Estimating Mutation Load in Human Genomes. Nat. Rev. Genet 16, 333–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prüfer K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prüfer K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science (80-. ). 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science (80-. ). 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green RE et al. A draft sequence of the neandertal genome. Science (80-. ). 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankararaman S, Mallick S, Patterson N & Reich D. The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans. Curr. Biol 26, 1241–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sankararaman S. et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernot B & Akey JM Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science (80-. ). 343, 1017–1021 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Abi-Rached L. et al. The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans. Science (80-. ). 334, 89–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendez FL, Watkins JC & Hammer MF A Haplotype at STAT2 Introgressed from Neanderthals and Serves as a Candidate of Positive Selection in Papua New Guinea. Am. J. Hum. Genet 91, 265–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dannemann M, Prüfer K & Kelso J. Functional implications of Neandertal introgression in modern humans. Genome Biol. 18, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Racimo F, Marnetto D & Huerta-Sánchez E. Signatures of archaic adaptive introgression in present-day human populations. Mol. Biol. Evol (2017). doi: 10.1093/molbev/msw216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Racimo F, Sankararaman S, Nielsen R & Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet 16, 359–371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juric I, Aeschbacher S & Coop G. The Strength of Selection against Neanderthal Introgression. PLOS Genet. 12, e1006340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris K & Nielsen R. The Genetic Cost of Neanderthal Introgression. Genetics (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gittelman RM et al. Archaic Hominin Admixture Facilitated Adaptation to Out-of-Africa Environments. Curr. Biol 26, 3375–3382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petr M, Pääbo S, Kelso J & Vernot B. Limits of long-term selection against Neandertal introgression. Proc. Natl. Acad. Sci. U. S. A. 201814338 (2019). doi: 10.1073/pnas.1814338116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dannemann M & Kelso J. The Contribution of Neanderthals to Phenotypic Variation in Modern Humans. Am. J. Hum. Genet 101, 578–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonti CN et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science (80-. ). 351, 737–741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nédélec Y. et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell (2016). doi: 10.1016/j.cell.2016.09.025 [DOI] [PubMed] [Google Scholar]
- 24.Quach H. et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell (2016). doi: 10.1016/j.cell.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sams AJ et al. Adaptively introgressed Neandertal haplotype at the OAS locus functionally impacts innate immune responses in humans. Genome Biol. 17, 246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Ding Q, He Y, Xu S & Jin L. Reintroduction of a Homocysteine Level-Associated Allele into East Asians by Neanderthal Introgression. Mol. Biol. Evol 32, msv176 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Vernot B. et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science (80-. ). 352, 235–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BY & Lohmueller KE Selection and Reduced Population Size Cannot Explain Higher Amounts of Neandertal Ancestry in East Asian than in European Human Populations. Am. J. Hum. Genet 96, 454–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villanea FA & Schraiber JG Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol 3, 39–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wall JD et al. Higher Levels of Neanderthal Ancestry in East Asians than in Europeans. Genetics 194, 199–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacArthur J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franke A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet 42, 1118–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jostins L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee MK et al. Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. PLoS One 12, e0176566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SL et al. Mercapturic Acids Derived from the Toxicants Acrolein and Crotonaldehyde in the Urine of Cigarette Smokers from Five Ethnic Groups with Differing Risks for Lung Cancer. PLoS One 10, e0124841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spada J. et al. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. J. Sleep Res 25, 690–701 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Kulminski AM et al. Strong impact of natural-selection-free heterogeneity in genetics of age-related phenotypes. Aging (Albany. NY). 10, 492–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutz SM et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet. 16, 138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy RC, Wakefield J & Akey JM Impacts of Neanderthal-Introgressed Sequences on the Landscape of Human Gene Expression. Cell 168, 916–927.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium GTEx. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lappalainen T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.OMIM. CAT EYE SYNDROME; CES. Available at: https://www.omim.org/entry/115470. (Accessed: 1st November 2018) [Google Scholar]
- 43.Tewhey R. et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell 165, 1519–1529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Arensbergen J. et al. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet 51, 1160–1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brawand D. et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Colbran LL et al. Inferred divergent gene regulation in archaic hominins reveals potential phenotypic differences. Nat. Ecol. Evol 3, 1598–1606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Telis N, Aguilar R & Harris K. Selection against archaic DNA in human regulatory regions. bioRxiv 708230 (2019). doi: 10.1101/708230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyregne S, Boyle MJ, Dannemann M & Prufer K. Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 27, 1563–1572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castellano S. et al. Patterns of coding variation in the complete exomes of three Neandertals. Proc. Natl. Acad. Sci 111, 6666–6671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henn BM et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. U. S. A. 113, E440–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohmueller KE The distribution of deleterious genetic variation in human populations. Curr. Opin. Genet. Dev 29, 139–146 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Lohmueller KE et al. Proportionally more deleterious genetic variation in European than in African populations. Nature 451, 994–997 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simons YB, Turchin MC, Pritchard JK & Sella G. The deleterious mutation load is insensitive to recent population history. Nat. Genet 46, 220–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen L, Wolf AB, Fu W, Li L & Akey JM Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals. Cell 180, 677–687.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Bergström A. et al. Insights into human genetic variation and population history from 929 diverse genomes. Science (80-. ). 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodgkinson A & Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet 12, 756–766 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Kircher M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pers TH, Timshel P & Hirschhorn JN SNPsnap: a Web-based tool for identification and annotation of matched SNPs. Bioinformatics 31, 418–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haller BC & Messer PW SLiM 2: Flexible, interactive forward genetic simulations. Mol. Biol. Evol (2017). doi: 10.1093/molbev/msw211 [DOI] [PubMed] [Google Scholar]
- 61.Eyre-Walker A, Woolfit M & Phelps T. The Distribution of Fitness Effects of New Deleterious Amino Acid Mutations in Humans. Genetics 173, 891–900 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gravel S. et al. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci 108, 11983–11988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All results reported in this paper are available in supplementary material and/or on the project’s github repository (https://github.com/DaRinker/Neanderthal.reintroduction).