SUMMARY
Neural stem cells directly or indirectly generate all neurons and macroglial cells and guide migrating neurons by using a palisade-like scaffold made of their radial fibers. Here, we describe an unexpected role for the radial fiber scaffold in directing corticospinal and other axons at the junction between the striatum and globus pallidus. The maintenance of this scaffold, and consequently axon pathfinding, is dependent on the expression of an atypical RHO-GTPase, RND3/RHOE, together with its binding partner ARHGAP35/P190A, a RHO GTPase-activating protein, in the radial glia-like neural stem cells within the ventricular zone of the medial ganglionic eminence. This role is independent of RND3 and ARHGAP35 expression in corticospinal neurons, where they regulate dendritic spine formation, axon elongation, and pontine midline crossing in a FEZF2-dependent manner. The prevalence of neural stem cell scaffolds and their expression of RND3 and ARHGAP35 suggests that these observations might be broadly relevant for axon guidance and neural circuit formation.
In Brief
Kaur et al. show that the radial glial scaffold of neural stem cells from medial ganglionic eminence directs corticospinal and other axons through a previously unknown choice point at the striatopallidal junction in an RND3/ARHGAP35-dependent manner. Within corticospinal neurons, FEZF2-dependent Rnd3 expression regulates dendritic spinogenesis, axon elongation, and pontine midline crossing.
Graphical abstract
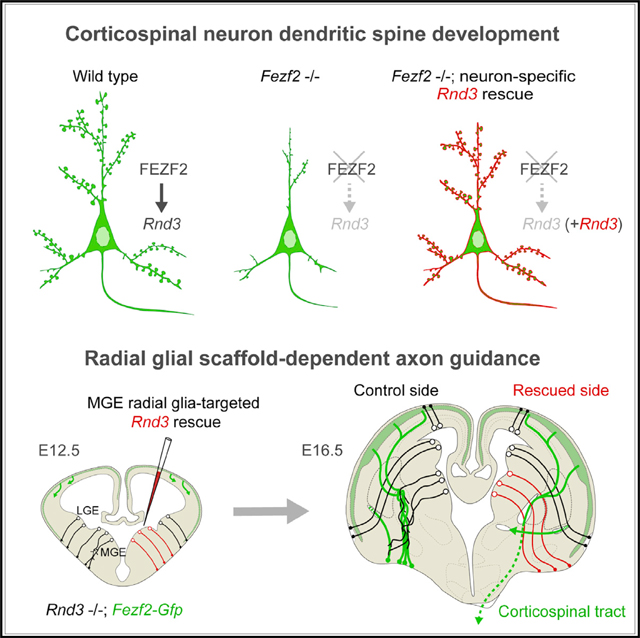
INTRODUCTION
Neural stem cells are bipolar epithelial-like cells that reside in the ventricular and subventricular zones (VZ and SVZ) and transform into highly elongated radial glial progenitors as the central nervous system (CNS) grows (Breunig et al., 2011; Kriegstein and Alvarez-Buylla, 2009; Taverna et al., 2014). These cells are responsible for two key processes of the developing CNS. First, they directly or indirectly generate all neurons and macroglial cells. Second, they guide the migration of newborn neurons to their final destinations in the developing parenchyma using their long radial (basal) fibers, which traverse the entire wall of the CNS and form a physical palisade-like scaffold (Nakagawa et al., 2019; Pilaz and Silver, 2017).
In the present study, we describe a critical role for neural stem cells in axon guidance at a distance from their cell bodies. This discovery was made in the course of investigating molecular mechanisms controlling the formation of the corticospinal tract (CST), the longest axon pathway in the CNS and one that is responsible for controlling fine voluntary movements (Lemon, 2008; Martin, 2005; Yoshida and Isa, 2018). The CST originates from a population of FEZF2-expressing glutamatergic excitatory projection (pyramidal) neurons in layer (L) 5 (L5) of the cerebral neocortex (Chen et al., 2005a, 2005b; Molyneaux et al., 2005) and extends to distant targets in the ventral forebrain, brainstem, and spinal cord (Canty and Murphy, 2008; Welniarz et al., 2017).
While investigating how CST axons navigate the long distance from the cerebral cortex to the spinal cord, we found that the radial glia-like neural stem cells within the VZ of the medial ganglionic eminence (MGE) of the ventral forebrain, through their radial process scaffold, direct these axons through a previously unknown choice point at the junction between the embryonic striatum and globus pallidus (GP; i.e., striatopallidal junction [SPJ]). To our surprise, this role was dependent on a previously unknown function of RND3 (RHOE), an atypical member of the RHO guanosine triphosphatase (GTPase) superfamily that lacks GTPase activity but regulates actin cytoskeletal organization (Azzarelli et al., 2015; Hall and Lalli, 2010; Luo, 2000), and its binding partner, ARHGAP35 (P190A), a RHO GTPase-activating protein (Wennerberg et al., 2003), specifically in radial glia-like neural stem cells, but not in the growing corticospinal (CST) axons. In addition to cell autonomous roles in axonal and dendritic growth, RND3 and ARHGAP35 regulate axon pathfinding cell non-autonomously by promoting the hitherto unexplored role of the striatopallidal radial glial scaffold as a guidepost structure.
RESULTS
FEZF2 Promotes Dendritic Spinogenesis and Axon Elongation of the Corticospinal Neurons via RND3
We sought to identify direct molecular effectors that mediate the function of FEZF2 (FEZL or ZFP312), a transcription factor that is essential for corticospinal neurons (CSNs) to acquire distinct identities, develop proper dendritic arbors and spines, and form the CST (Chen et al., 2005a, 2005b; Molyneaux et al., 2005). We therefore generated RNA sequencing (RNA-seq) data from mice in which Fezf2 was conditionally knocked out (cKO; Fezf2 fl/fl; Emx1-Cre) (Shim et al., 2012) from radial glia-like neural stem cells (hereafter referred to as radial glial cells [RGCs]) in the cerebral cortex and their progeny, the glutamatergic excitatory projection neurons (Figure S1A). These data were compared to RNA-seq data from mice in which Fezf2 was overexpressed (OE) by in utero electroporation at embryonic day 15.5 (E15.5), a time when upper-layer projection neurons, which do not typically express Fezf2, are generated (Figure S1B). This overexpression re-specifies the neurons to a CSN fate, rather than upper layers (Chen et al., 2005b). We identified 16 genes whose expression was both decreased in the neocortex of Fezf2 cKO at postnatal day 0 (P0), a time when CST axons reach the pyramidal decussation, and upregulated in Fezf2 OE upper-layer, CSN-fated neurons at P3, a time when these neurons reach their final destination (Figure 1A; Tables S1 and S2). Of these 16 candidates, Rnd3 (RhoE) was highest expressed in putative L5 CSNs, distinguished among cortical projection subtypes by the higher expression of Fezf2 (Figure 1A) in single-cell RNA-seq datasets (Li et al., 2018; Mi et al., 2018). Consistent with the potential regulation of Rnd3 by FEZF2, immunostaining of mid-fetal human frontal cortex showed colocalization of RND3 in BCL11B (CTIP2)+ L5B CSNs (Figures S1C and S1D), which at this time exhibit a characteristic mini-columnar organization (Kwan et al., 2012), as did Rnd3-Gfp BAC transgenic mice (Figures 1B and S2A). Furthermore, we observed GFP+ and, by extension, RND3 expressing CST axons in the basilar pons (Figure 1B). In addition, GFP+ cell bodies with long radial fibers were present in the VZ and SVZ of the dorsal and ventral forebrain (Figure S2A). Computational analysis of published bulk tissue and single-cell transcriptome datasets (Kang et al., 2011; Li et al., 2018; Nowakowski et al., 2017) showed higher expression of RND3 in putative L5 CSNs and RGCs within the fetal neocortex and ventral forebrain, with gradual decreases during postnatal periods (Figures S1E and S1F). Similar results were obtained at E14.5, E16.5, and P0 by in situ hybridization and in a heterozygous gene-trap (gt) mouse, Rnd3 gt, in which a LacZ-expressing gene-trap cassette was inserted in the second intron of Rnd3 (Mocholí et al., 2011), abolishing the proper expression of the trapped allele, Rnd3, but allowing for the visualization of natively expressing Rnd3 cells by LacZ staining (Figures 1C and S2B–S2H).
Figure 1. FEZF2 Transactivates Rnd3 in Developing Corticospinal Neurons (CSNs).
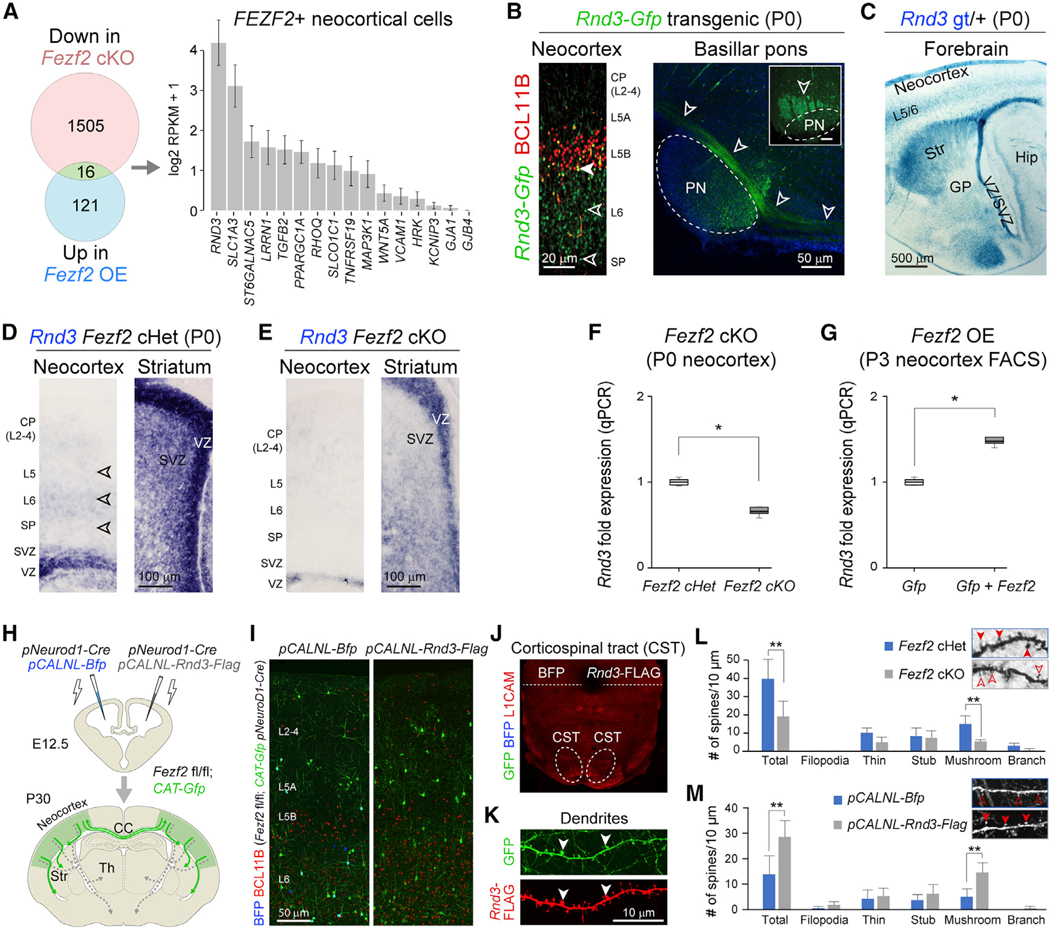
(A) Union of genes (green) downregulated in Fezf2 fl/fl; Emx1-Cre (cKO; red) and upregulated in Fezf2 overexpression (OE; blue) conditions, and their enrichment in mid-fetal human Fezf2+ neocortical excitatory projection neurons.
(B) Left panel: GFP (green) expression in deep-layer (L5 and L6) cortical neurons, including BCL11B-expressing (red) L5b pyramidal neurons in the P0 Rnd3-Gfp BAC transgenic brain. Scale bar: 20 μm. Right panel: sagittal and coronal (inset) views of GFP+ corticospinal tract (CST) in the ventral pons (PN) (arrowheads). Scale bar: 50 μm.
(C) Staining of B-galactosidase (X-gal) (blue) in Rnd3 gene-trap mice (Rnd3 gt) at P0 showing expression in neocortical deep layers (L5 and L6 [L5/6]) and VZ/SVZ and Str. GP, globus pallidus; Str, striatum. VZ/SVZ, ventricular zone/subventricular zone. Scale bar: 500 μm.
(D and E) Rnd3 expression at P0 in conditional Fezf2 fl/+; Emx1-Cre (Fezf2 Het) (D) and Fezf2 cKO (E) neocortex and subpallium (Str) detected by in situ hybridization (n = 3).
(F and G) Rnd3 expression change in Fezf2 cKO (F) and Fezf2 OE (G), as validated by qPCR. p < 0.05 (unpaired two-tailed t test, n = 3).
(H) Schematic illustration of the conditional deletion via bihemispheric in utero electroporation of Fezf2 fl/fl; CAT-Gfp with pNeuroD1-Cre and either pCALNL-Bfp (control) or pCALNL-Rnd3-Flag, at E12.5.
(I) BCL11B (red), GFP (green), and BFP (blue) immunolabeling of the pCALNL-Bfp and the pCALNL-Rnd3-Flag electroporated hemispheres. Scale bar: 50 μm.
(J) L1CAM+ CST axons lacking GFP+ or BFP+ projections at the pontine level of electroporated brains.
(K) RND3 localization, marked by FLAG tag immunolabeling (red) at the membrane, including the dendrites, and synapses of the pCALNL-Rnd3-Flag electroporated projection neurons (green). The arrowheads point at mature spines. Scale bar: 10 μm.
(L and M) Bar graph depicts number of spines in 10 μm of dendritic length (insets) in Fezf2 cKO (gray), compared to Fezf2 Het (blue) mice (means ± SEs; unpaired two-tailed t test applied. *p < 0.05, **p < 0.01, n = 11 neurons/3 brains/condition) (L) and in the pCALNL-Rnd3-Flag neurons (gray) as compared to the pCALNL-Bfp electroporated neurons (blue) (means ± SEs; unpaired two-tailed t test applied. **p < 0.01, n = 10 neurons/4 brains/condition) (M).
See also Figures S1 and S2.
Furthermore, we detected the reduced expression of Rnd3 in L5 and the VZ and SVZ of Fezf2 cKO brains at P0 by in situ hybridization (Figures 1D and 1E). qPCR confirmed both a reduction in Rnd3 expression following conditional Fezf2 deletion (Figure 1F) and an increase following Fezf2 overexpression (Figure 1G). To assess whether Rnd3 is directly or indirectly regulated by FEZF2, we performed chromatin immunoprecipitation (ChIP) sequencing using, in the absence of ChIP-grade antibodies against FEZF2, an antibody against a V5-tagged FEZF2 protein transiently expressed in N2a neuroblastoma cells. We identified 19,213 FEZF2-bound genomic regions (Figure S1G; Table S3). Three of these regions were at or adjacent to the Rnd3 locus and were found by the luciferase reporter assay to be transactivated by FEZF2 (Figure S1H). Thus, developing CSNs express Rnd3, where it is directly transactivated by FEZF2.
Because the loss of FEZF2 prevents nascent L5 neurons from acquiring a CSN molecular identity and establishing appropriate dendritic arborization, spines, and CST (Chen et al., 2005a, 2005b; Molyneaux et al., 2005), we conducted in vivo and in vitro assays to assess the contribution of RND3 to these phenotypes. We crossed Rnd3 gt/gt and Fezf2-Gfp mice, to label CSNs and CST with Fezf2-Gfp, in addition to generating Rnd3 KO mice. We found no significant differences in the laminar distribution of putative L6, TBR1+ projection neurons, L5B, BCL11B+ CSNs, L2–L4, SATB2+ intracortical projection neurons, or L1, RELN+ neurons following iododeoxyuridine/chlorodeoxyuridine (IdU/CldU) birth dating of neurons at early (E11.5/E12.5; CldU) and late (E15.5/E16.5; IdU) gestational time points (Figures S3A–S3F). However, we did identify minor differences in CldU labeling following injection at E15.5 (Figure S3F) that were consistent with a previously described migration deficit (Azzarelli et al., 2015). In addition, bihemispheric in utero electroporation of a plasmid expressing Cre under the post-mitotically active NeuroD1 promoter concurrently with either pCALNL-Rnd3-Flag (ipsilateral) or pCALNL-Bfp control (contralateral) into Fezf2 fl/fl; CAT-Gfp neocortices at E12.5 (Figure 1H) revealed that Rnd3 expression did not rescue the decreased expression of BCL11B in L5 or L6 or CST formation in a Fezf2-deficient context (Chen et al., 2005a, 2005b; Molyneaux et al., 2005) (Figures 1I and 1J).
In contrast, in utero electroporation of pCALNL-Rnd3-Flag, but not control pCALNL-Bfp, into Fezf2 fl/fl; CAT-Gfp neocortices reversed the loss of dendritic complexity and dendritic spines, particularly mature, mushroom-shaped spines, associated with the conditional deletion of Fezf2 (Figures 1L, 1M, and S3I). Similarly, after 7 days in vitro, cultured cortical neurons isolated from Rnd3 gt/gt; Fezf2-Gfp neocortices displayed reduced dendritic complexity and shorter axons compared to Rnd3 gt/+; Fezf2-Gfp neurons (Figures S3G and S3H). These neurons also exhibited reduced staining for synaptic markers, including synaptophysin (SYP) (Figure S3H). Consistent with a possible role for RND3 in neurite outgrowth and dendritic spine formation, FLAG-tagged RND3 protein localized to the soma, dendrites, and axons of electroporated neurons (Figure 1K). These results demonstrate that RND3 mediates the cell autonomous and FEZF2-dependent regulation of neurite outgrowth and dendritic spine formation, but not the molecular specification of CSNs or CST formation.
CST Axons Misroute at the SPJ in Rnd3 Mutant Mice
To further explore RND3 functions in brain development, including the CST, we analyzed the homozygous Rnd3 gt/gt gene-trap mouse (Figures S2B and S3J). In addition to hindlimb deformities, premature death within the fourth postnatal week, and reduced size of both the neocortex and body (Mocholí et al., 2011), Rnd3 gt/gt mice also lacked CST at the level of the cerebral peduncles and pons (Figure S3K). Crossing Rnd3 gt/gt mice with Fezf2-Gfp mice revealed that descending cortical GFP+ axons did not enter or form the internal capsule after passing through the striatum but were stuck at the SPJ and misrouted through the GP before aberrantly terminating near the surface of the ventral pallidum. Moreover, the GP occupied a more anterior location in the Rnd3 gt/gt; Fezf2-Gfp brain (Figures 2A, S3A, and S3B), and Fezf2-Gfp+ axon bundles exhibited slightly abnormal trajectories located more ventrolaterally within the striatum (Figures 2A1–A3 and 2B1–B3). In addition, Fezf2-Gfp+ CST axons also appeared less robust in the Rnd3 gt/gt brain, perhaps because of the role of RND3 in axon growth (Figures S3G, S8E, and S8F).
Figure 2. CST Axons Misroute at the SPJ in the Absence of RND3.
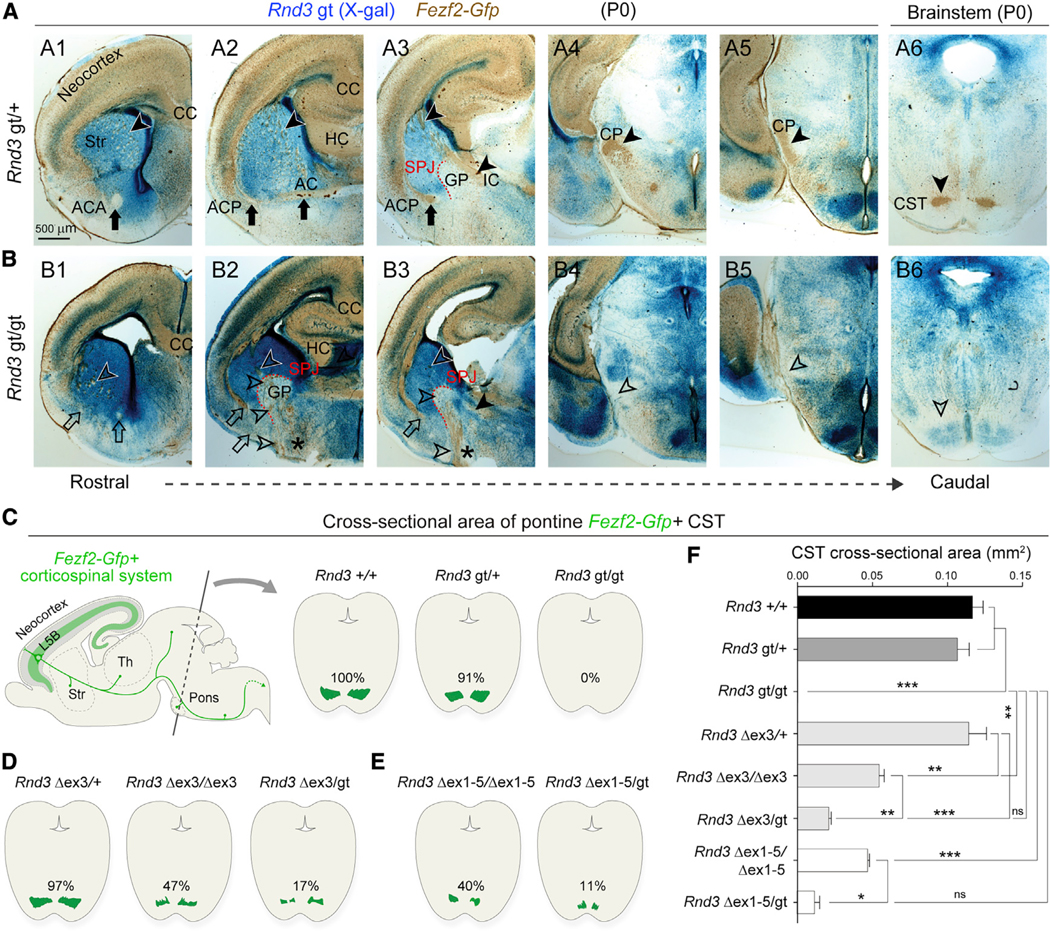
(A and B) X-gal staining (blue) and GFP (DAB; brown) on serial coronal sections of P0 Rnd3 gt/+; Fezf2-Gfp (A1–A6) and Rnd3 gt/gt; Fezf2-Gfp (B1–B6) brains depict defects in the Rnd3 gt/gt brain, including reduced cortical thickness (B1–B3); a thin ACA (B1; open arrow); the misrouted ACP to the ventral surface (B2 and B3; open arrows); and misrouted Fezf2-Gfp+ subcerebral projections within the striatum and at the SPJ through the GP (B2 and B3; asterisks and open arrowheads), at the cerebral peduncle (CP) and ventral pons (open arrowheads, B4–B6). Scale bar: 500 μm. CC, corpus callosum; HC, hippocampal commissure; AC, anterior commissure; IC, internal capsule; ACA, anterior branch of AC; ACP, posterior branch of AC.
(C–E) Schematic representations of CST area labeled by GFP (Fezf2-Gfp) in the coronal cross-sections of pontine nuclei from Rnd3 +/+, Rnd3 gt/+, and Rnd3 gt/gt mice (C); Rnd3 Δex3/+; Fezf2-Gfp, Rnd3 Δex3/Δex3; Fezf2-Gfp, and Rnd3 Δex3/gt; Fezf2-Gfp mice (D); and Rnd3 Dex1–5/Dex1–5; Fezf2-Gfp, and Rnd3 Dex1–5/gt; Fezf2-Gfp mice (E) at P0.
(F) Quantitative analysis of CST between mice bearing various Rnd3 alleles. y axis: groups. x axis: CST area in mm2. ANOVA Tukey’s test was applied (means ± SEs,*p < 0.01, **p < 0.001, and ***p < 0.0001. n = 2 for each group).
See also Figures S3–S5.
To verify these findings, we generated multiple lines of Rnd3-floxed mice by deleting alternative exons using CRISPR-Cas9 genome editing and different Cre transgenic lines (Figure S4; Table S4). In these mice (Rnd3 Δex3/Δex3; Fezf2-Gfp and Rnd3 Dex1–5/Dex1–5; Fezf2-Gfp), we observed cortical axonal defects similar to those seen in the Rnd3 gt/gt brain (Figures 2C–2F and S5A–S5E; Table S5), including a misrouted CST at the SPJ (Figures S5B and S5E). The CST was also reduced at the pons in Rnd3 Δex3/Δex3; Fezf2-Gfp and Rnd3 Dex1–5/Dex1–5; Fezf2-Gfp mice (to 47% and 40%, respectively), and in compound heterozygous mice in which the gt allele was introduced alongside these floxed alleles (Rnd3 Δex3/gt; Fezf2-Gfp, to 17% and Rnd3 Dex1–5/gt; Fezf2-Gfp, to 11%) (Figures 2C–2F, S5A–S5E). Commissural projections were also affected in Rnd3 gt/gt mice, including a dramatic reduction in the anterior branch of the anterior commissure (ACA) (Figures 2A, 2B, 3A, 3B, and S5B–S5E) and the frequent failure of the posterior branch (ACP) to develop or cross the midline, with diffuse axons exiting the external capsule and, like the CST, extending inappropriately toward the ventral surface of the forebrain (Figures 2A, 3A, 3B, and S5B–S5E). In addition, cytochrome oxidase staining on tangential sections of Rnd3 gt/+ and Rnd3 gt/gt brains at P14 revealed defective thalamocortical projections in the cortex, which failed to form the barrel fields in Rnd3 gt/gt brains (Figure 3C). Furthermore, whole-mount immunofluorescence staining for the axonal cell adhesion molecule CNTN2 (TAG1) using the iDISCO tissue clearing method (Belle et al., 2014) found that axons forming the ACA in the Rnd3 gt/gt brain were present at their origin (i.e., the anterior olfactory nucleus and anterior piriform cortex), but then defasciculated in the ventral striatum rather than projecting contralaterally (Figures 3D–3G).
Figure 3. Cortical Projections Defects in the Absence of RND3.
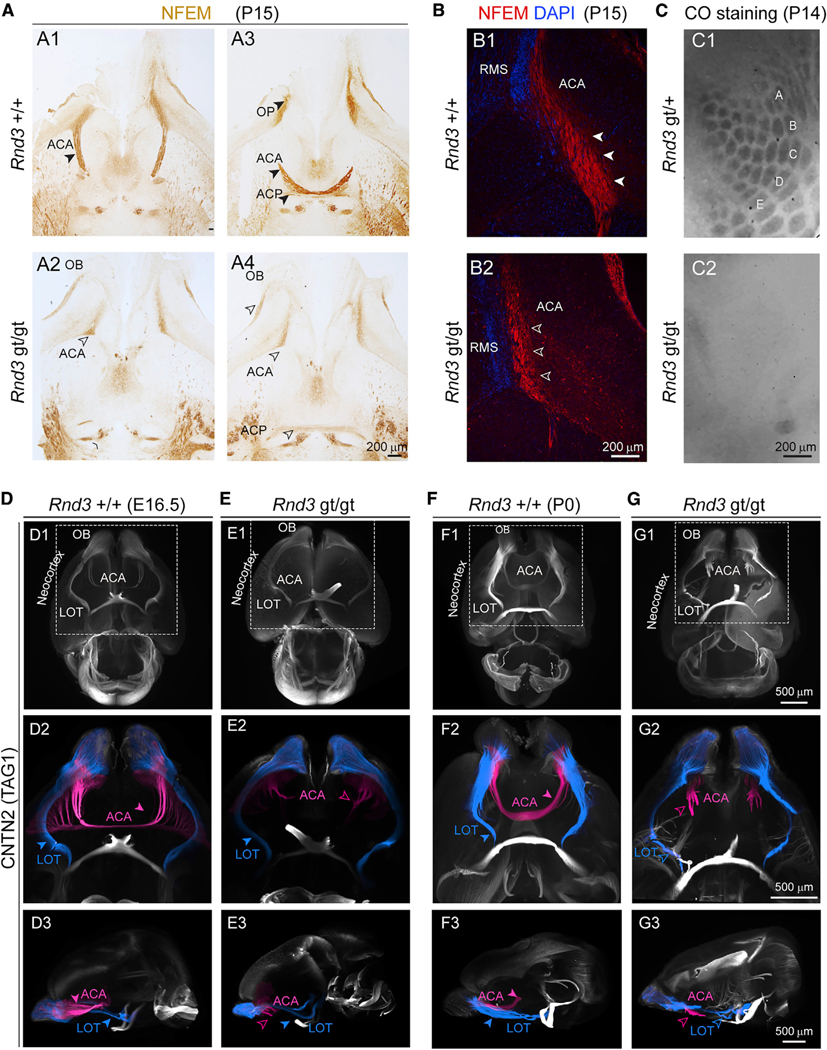
(A) Horizontal brain sections of Rnd3 +/+ (A1 and A3) and Rnd3 gt/gt (A2 and A4) from P15 mice stained for NEFM shows reduced ACP and absent ACA in Rnd3 gt/gt mice. Solid arrowheads indicate normal and open arrowheads indicate reduced ACA and OB axons. Scale bar: 200 μm. OB, olfactory bulb.
(B) The ACA (NEFM, red) runs parallel to the rostral migratory stream (RMS; DAPI; blue) at its origin from the OB, both in P15 Rnd3 +/+ (B1) and Rnd3 gt/gt (B2) mice. Solid arrowheads indicate normal and open arrowheads indicate reduced ACA. Scale bar: 200 μm.
(C) Cytochrome oxidase staining of tangential sections of Rnd3 gt/+ and Rnd3 gt/gt cortices from P14 mice depicting barrel cortex (A–E). Scale bar: 200 μm.
(D–G) Whole-mount anti-CNTN2 immunostaining of Rnd3 +/+ (D and F) and Rnd3 gt/gt (E and G) from E16.5 (D and E) and P0 (F and G) mice brains followed by 3DiSCO, show similar LOT (blue) trajectory in both (D2–G2), whereas the ACA (pink) never crossed the midline in Rnd3 gt/gt mice (E2 and G2). Solid arrowheads indicate normal and open arrowheads indicate reduced or absent ACA. Scale bar: 500 μm. LOT, lateral olfactory tract.
See also Figure S3.
In Rnd3 deletion mice, as in the Rnd3 gt/gt mouse, we found a reduction in size in the ACA and a failure by the ACP to cross the midline (Figures S5B–S5E; Table S5). Branching of the cortical subcerebrally projecting axons at the level of the internal capsule, cerebral peduncle, and pons was also less prominent compared to control mice but not absent as it was in the Rnd3 gt/gt brain (Figures S5B, S5D, and S5E; Table S5). Although the penetrance and severity of defects in rostral projections were different among Rnd3 gt/gt and Rnd3 deletion mice, these results suggest that RND3 is necessary for the pathfinding of Fezf2-Gfp+ CST and other axons through the ventral forebrain, most prominently at the SPJ.
The Striatopallidal Radial Glial Scaffold Is Disrupted in Rnd3 Mutant Mice
The misrouting and tangled bundling of GFP+ axons at and ventral to the SPJ, coupled with defects in the patterning of NKX2–1+ pallidal structures (Figures 4A–4D), may indicate that RND3 regulates ventral forebrain patterning and the cytoarchitecture of basal ganglia. Because NKX2–1 expressing neurons of the SPJ and the GP arise and migrate primarily from the MGE, we assessed the RGCs in the MGE for defects in Rnd3-deficient mice. To do so, we immunolabeled radial fibers with the molecular marker vimentin (VIM) and CST and other descending projections (i.e., CST, corticothalamic, and other projection systems) with either neurofilament medium polypeptide (NEFM) or GFP at E16.5 in control Rnd3 gt/+; Fezf2-Gfp and Rnd3 gt/gt; Fezf2-Gfp brains (Figures 4E and 4F). In Rnd3 gt/gt; Fezf2-Gfp brains, we observed severe misrouting of NEFM+ corticofugal projections (Figures 4E1, 4F1, S8A, and S8B) coincident with the disruption of a palisade-like structure of VIM+ radial glial fibers at the SPJ (Figures 4E2, 4E3, 4F2, and 4F3). To analyze the relationship between the VIM+ radial glial scaffold and Fezf2-Gfp+ axons, we performed super resolution microscopy using stimulated emission depletion (STED) at E16.5 (Figures 4G and 4H). We observed close associations and contact points between Fezf2-Gfp+ CST axons and radial glial fibers in both Rnd3 gt/+ and Rnd3 gt/gt brains (Figures 4G2, 4G3, 4H2, and 4H3). Moreover, radial glial fibers in Rnd3 gt/gt brains exhibited a disorganized and highly branched morphology (Figures 4H2 and 4H3), including the formation of loop-like structures. The bundles of Fezf2-Gfp labeled axons appeared to follow and, in some cases, intertwine with radial glial fiber loops, potentially leading to the entanglement and misrouting of CST and other axons at the SPJ (Figure 4H2). Analysis of slightly oblique tangential sections of Rnd3 gt/+; Fezf2-Gfp and Rnd3 gt/gt; Fezf2-Gfp brains at E16.5 confirmed that radial glial fibers were generally oriented in a uniform direction and maintained a palisade-like organization in controls (Figures S6A–S6D), but not Rnd3 gt/gt; Fezf2-Gfp sections (Figures S6E–S6H). These observations indicate that RND3 is essential for maintaining the organization and branching of radial glial fibers that are required for proper pathfinding of the CST and other axon projection systems through the striatopallidal region.
Figure 4. Concurrent Axonal Misrouting and Altered Radial Glial Scaffold at the Embryonic SPJ in the Absence of RND3.
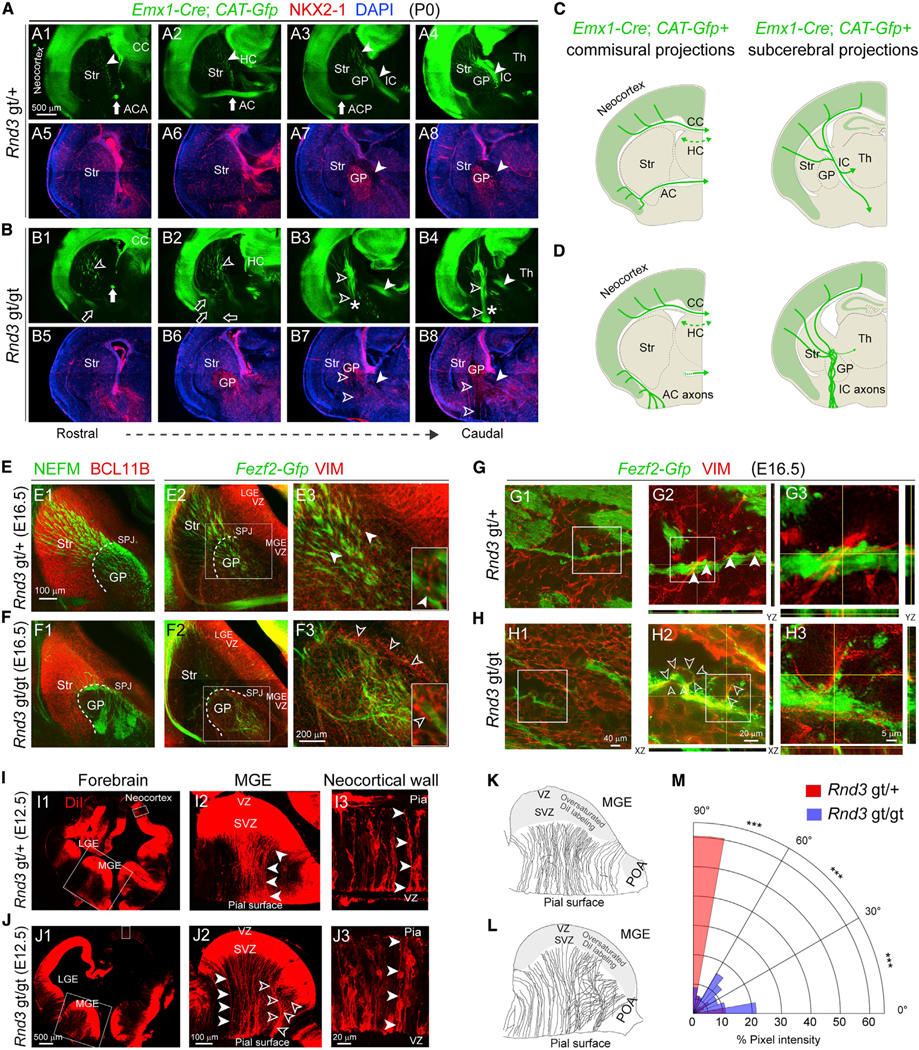
(A and B) Coronal sections of Rnd3 gt/+; Emx1-Cre; CAT-Gfp (A) or Rnd3 gt/gt; Emx1-Cre; CAT-Gfp (B) brain immunolabeled for GFP (green), NKX2–1 (red), and DAPI (blue). The solid arrows (AC/ACA/ACP) and the solid arrowheads (IC/GP) in A1–A8 mark the typical localization of these features, while the open arrows, open arrowheads, and asterisk (CST) in B1–B8 denote disruption or dislocation. The CST axons appear to project ventrolaterally in the Rnd3 gt/gt brains, as compared to their normal trajectory in Rnd3 gt/+, in addition to their misrouting through the GP toward ventral forebrain surface. Scale bar: 500 μm.
(C and D) Schematic representation of commissural and subcerebral projections in Rnd3 gt/+; Fezf2-Gfp (C) and Rnd3 gt/gt; Fezf2-Gfp (D) in (A) and (B), respectively. Th, thalamus.
(E and F) Coronal sections of E16.5 Rnd3 gt/+; Fezf2-Gfp (E) and Rnd3 gt/gt; Fezf2-Gfp (F) brains co-immunolabeled for NEFM (E1 and F1; green), BCL11B (E1 and F1; red), GFP (E2 and E3, F2 and F3; green), and VIM (E2 and E3, F2 and F3; red). Scale bars: 100 μm (E1 and E2, F1 and F2) and 200 μm (E3 and F3). The solid arrowheads indicate parallel, fasciculated VIM+ radial glial fibers and the open arrowheads indicate disorganized, non-fasciculated VIM+ radial glial fibers. LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence.
(G and H) Super resolution images acquired using STED of Rnd3 gt/+; Fezf2-Gfp (G) and Rnd3 gt/gt; Fezf2-Gfp (H) sections depicting VIM+ radial glial projections at the SPJ. The normal ascent of the radial glial fibers is shown by solid arrowheads (G) and the defective projections are shown by open arrowheads (H). Scale bars: 40 μm (G1 and H1), 20 μm (G2 and H2), and 5 μm (G3 and H3).
(I and J) DiI labeled E12.5 Rnd3 gt/+ (I) and Rnd3 gt/gt (J) brains showing radial glial fiber orientation in the MGE of Rnd3 gt/+ brains (solid arrowheads, I2) and Rnd3 gt/gt brain (open arrowheads, J2), and in the neocortex (solid arrowheads, I3 and J3). Pia: Pial surface. Scale bars: 500 μm (I1 and J1), 100 μm (I2 and J2), and 20 μm (I3 and J3).
(K andL) Tracings of the DiI+ radial glial fibers from E12.5 Rnd3 gt/+ (K) and Rnd3 gt/gt (L). POA, preoptic area.
(M) Polar histogram showing the distribution of orientation of DiI-labeled radial glial fibers in Rnd3 gt/gt; Fezf2-Gfp (blue), or Rnd3 gt/+; Fezf2-Gfp (red) brains that are orthogonal to the VZ of the MGE in the coronal sections. Pairwise comparison using two-tailed Chi-Square tests with Yates correction applied (means, ***p < 0.0001. n = 4/condition).
See also Figures S6–S8.
To determine whether the radial fibers of RGCs originating in the VZ of the MGE form a substrate mediating the directionality of CST axons in the ventral forebrain, we labeled the VZ/SVZ of the lateral ganglionic eminence (LGE), MGE, and the cortical plate with a lipophilic tracer, DiI, beginning at E12.5, a period overlapping with the generation and migration of the NKX2–1+ interneurons populating the GP (Nóbrega-Pereira et al., 2010), but before the period when subcerebral or thalamocortical projections reach the subpallium (Canty and Murphy, 2008). In Rnd3 gt/+ mice, radial fibers of RGCs from the MGE ran parallel to one another, extended to the pial surface of the brain, and formed a palisade-like scaffold, with a majority of these radial fibers (~52%) deviating by <30° from an orthogonal approach (Figures 4I, 4K, and 4M). In contrast, the radial projections of RGCs originating from the MGE in Rnd3 gt/gt brains do not maintain a consistent ascent to the pial surface, intersect with one another, exhibit increased branching, and approach the pial surface from a variety of angles (Figure 4J), with >81% of axons deviating by >30° from orthogonal (Figures 4I–4M). However, the requirement for RND3 in radial fiber orientation appears to be specific to the MGE, as we observed no similar defect in the projection of radial fibers in the dorsal forebrain and the LGE at E12.5 (Figures 4I and 4J), even though RGCs in the VZ of these 2 regions express Rnd3 (Figure 1C).
As an alternate possibility, and because Fezf2-Gfp+ axon bundles exhibited slightly abnormal trajectories within the striatum of Rnd3 gt/gt brains (Figures 2A1–2A3, 2B1-2B3, S6C, S6D, S7E, and S7F), we assessed medium spiny neurons and their striatopallidal and striatonigral projections. We found that neurons expressing DARPP32, a molecular marker for medium spiny neurons within the striatum (Anderson and Reiner, 1991) and their growing axons (Morello et al., 2015), were present in Rnd3 gt/+ and Rnd3 gt/gt striatum at both E14.5 (Figures S7C and S7D) and P0 (Figures S7E and S7F). In P0 Rnd3 gt/+ mice, DARPP32+ striatopallidal axons projected caudally in the striatum toward the SPJ and entered the GP ventrolateral to Fezf2-Gfp+ axons in the internal capsule (Figure S7E). In contrast, fewer DARPP32+ axons appeared to enter the GP from the striatum in Rnd3 gt/gt brains or to associate with the ectopic Fezf2-Gfp+ axons in the GP (Figures S7E4 and S7F4). Because the defects seen in the radial glial scaffold (Figure 4H) generally precede the entry of striatopallidal projections into the GP, which remain underdeveloped until E15.5 (Morello et al., 2015), neither medium spiny neurons nor their projections are likely to be the primary drivers of the radial glial and CST axonal phenotypes that we observed in Rnd3 mutants. This is in line with previous findings that striatal projections do not depend on axon-axon interactions with thalamocortical axons for navigation through the GP (Morello et al., 2015).
To determine whether defects in the RGCs in MGE also contribute to defects in the tangential migratory streams of interneurons toward the neocortex, we analyzed serial sections of Rnd3 gt/+; Fezf2-Gfp; Nkx2–1-Cre; Ai9 and Rnd3 gt/gt; Fezf2-Gfp; Nkx2–1-Cre; Ai9 at E14.5. In Rnd3 gt/+; Fezf2-Gfp; Nkx2–1-Cre; Ai9 brains, the tangential migratory streams of interneurons were normal (Figures S7G and S7I). However, in the Rnd3 gt/gt; Fezf2-Gfp; Nkx2–1-Cre; Ai9 brains, the interneurons in the caudal regions misrouted along the same defective CST axonal trajectory and showed reduced migration into the neocortex (Figures S7H and S7J). Although RGCs in the LGE may contribute to CST guidance in an RND3-dependent manner, our results indicate that the misrouting observed in Rnd3 gt/gt brains is primarily due to deficits in RGCs from the MGE.
We also evaluated other possible causes of the defects that we observed in the CST and other axon pathways. For example, we found that the expression of genes encoding axon guidance proteins, including SLIT1, SLIT2, ROBO1, and ROBO2 (Figures S8C and S8D), known to aid in the navigation of the CST, corticothalamic, thalamocortical, and other axons through the striatopallidal region and the formation of the internal capsule (Andrews et al., 2006; Bagri et al., 2002; Bielle et al., 2011; López-Bendito et al., 2006), appeared normal. In addition, although we found a rostral shift in the interneuronal “corridor” guidepost cells, which establish a passage for axonal navigation between proliferation zones of the MGE and GP (López-Bendito et al., 2006), we did not observe discontinuity in this path (Figures S7A, 7B, S8A, and S8B). Consequently, the phenotypes we observed at the SPJ were distinct from those reported in earlier studies in which the subcerebral projections misroute at the pallial-subpallial or at the telencephalon-diencephalon boundary (Jones et al., 2002; López-Bendito et al., 2006; Marín et al., 2002; Tuttle et al., 1999). This suggests that these corridor cells are not responsible for the axonal phenotypes seen in Rnd3 gt/gt mice.
Restoring Rnd3 in MGE Radial Glial Cells Rescues Radial Scaffold and CST Pathfinding
To rescue the orientation of radial glial projections and subsequent pathfinding defects, we re-introduced Rnd3 expression into Rnd3 gt/gt; Fezf2-Gfp RGCs of the MGE using in utero electroporation of pBLBP-Rnd3 or pBLBP-mScarlet-I at E12.5 (Figure 5A). Confirming the success of in utero electroporation, we detected faint mScarlet-I expression at E14.5 in interneurons originating from the MGE (Figures S8G and S8H). However, due to the low transcriptional activation of the mouse Blbp promoter (Schmid et al., 2006), we were unable to observe the expression of electroporated plasmids in the rescued or control hemispheres at E16.5. Nonetheless, several phenotypes were rescued in the pBLBP-Rnd3 electroporated hemispheres at E16.5, a time point chosen for the analysis of RGCs and the CST because CST axons invade the striatum and cerebral peduncle at this time (Canty and Murphy, 2008) and most of the RGCs in the MGE still maintain their orthogonal ascent from the VZ (Tan et al., 2016). For example, in the pBLBP-Rnd3 electroporated Rnd3 gt/gt; Fezf2-Gfp hemispheres, we observe rescued Fezf2-Gfp+ fibers passing through the internal capsule that were tightly fasciculated (Figures 5B2, 5D1, 5D2, S8K5, and S8L5), compared to controls that showed disorganized Fezf2-Gfp fibers at the SPJ (Figures 5B1, 5C1, 5C2, S8K2–S8K4, and S8L2–S8L4). Similarly, radial fibers originating from each GE were also arranged in a well-organized palisade-like scaffold that traversed the SPJ (Figures 5C, 5D, S8I, S8J, and S8L5). We observed that nearly 59% of radial fibers deviated <30° from orthogonal to the VZ of the LGE and MGE in pBLBP-Rnd3 rescued brains, as compared to just 35% in the Rnd3 gt/gt brain electroporated with pBLBP-mScarlet-l (Figure 5E). Fezf2-Gfp+ CST projections reached the cerebral peduncles in pBLBP-Rnd3 electroporated hemispheres and exhibited a 2.5-fold increase in GFP intensity as compared to control or pBLBP-mScarlet-I electroporated Rnd3 gt/gt; Fezf-Gfp hemispheres (Figures 5F, 5G, and S8L6). The expression of RND3 in Rnd3 gt/gt; Fezf2-Gfp brains also corrected defects in the commissural projections (Figures 5F, S8I, and S8L).
Figure 5. Striatopallidal Pathfinding of CST Axons Depends on RND3 in MGE Radial Glial Cells.
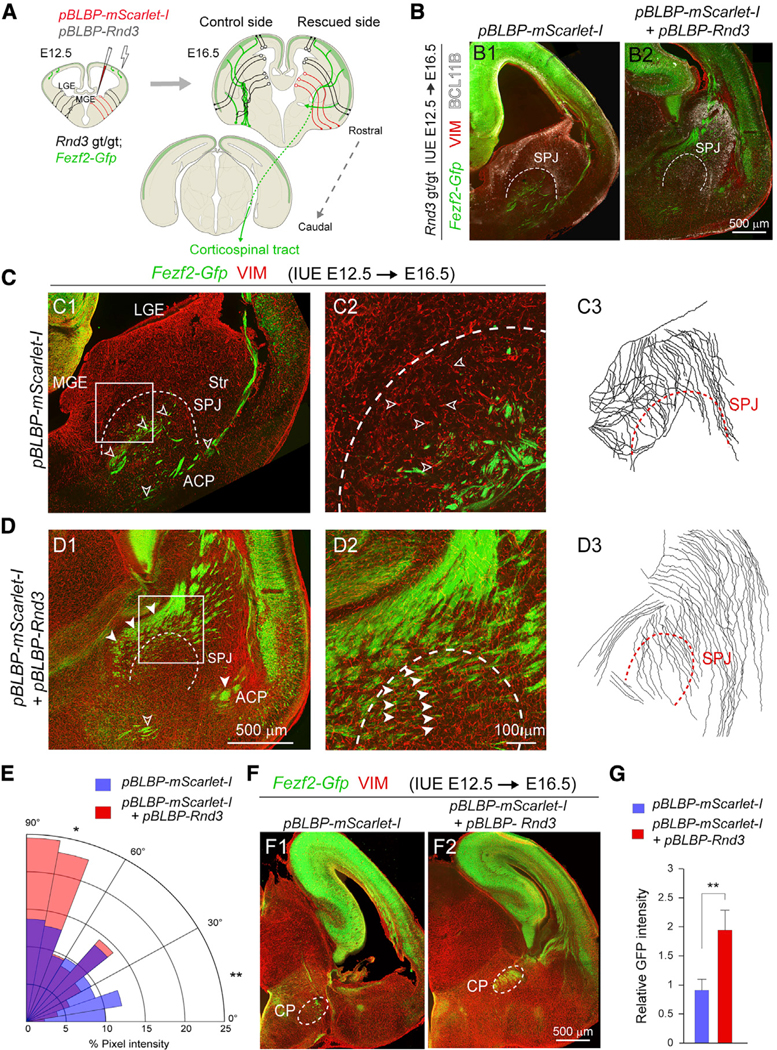
(A) Schematic showing in utero electroporation of pBLBP-Rnd3 and pBLBP-mScarlet-I to the MGE of Rnd3 gt/gt; Fezf2-Gfp brains.
(B) Coronal sections of E16.5 Rnd3 gt/gt; Fezf2-Gfp brains with pBLBP-mScarlet-I (B1) or pBLBP-mScarlet-I+pBLBP-Rnd3 electroporations (B2) at E12.5, immunolabeled for BCL11B (white). The dotted line marks the SPJ boundary. Scale bar: 500 μm.
(C and D) Z-stacked images from E16.5 Rnd3 gt/gt; Fezf2-Gfp brain electroporated with pBLBP-mScarlet-I (C1 and C2) or pBLBP-mScarlet-I-+pBLBP-Rnd3 (D1 and D2), stained for VIM and Fezf2-Gfp, and tracings of the radial glial fibers of the ganglionic eminences (C3 and D3). Scale bars: 500 mm (C1 and D1) and 100 μm (C2 and D2). The open arrowheads show misoriented Fezf2-Gfp+ projections and VIM+ radial fibers (C1 and C2). The solid arrowheads show the rescued Fezf2-Gfp+ and VIM+ radial fibers (D1 and D2).
(E) Polar histogram showing the distribution of orientation of VIM+ radial glial fibers in pBLBP-mScarlet-I (red) or pBLBP-mScarlet-I+pBLBP-Rnd3 (blue) brains of Rnd3 gt/gt; Fezf2-Gfp orthogonal to the VZ of the LGE and MGE in the coronal sections. Pairwise comparison was performed using two-tailed Chi-square test with Yates correction applied (means, *p < 0.05 and **p < 0.01. n = 3/group).
(F) Coronal sections of Rnd3 gt/gt; Fezf2-Gfp brain showing the CST axons at the CP following electroporation with pBLBP-Rnd3+pBLBP-mScarlet-I (F2), or pBLBP-mScarlet-I (F1). Scale bar: 500 μm.
(G) Quantitative analysis of the mean integrated intensity of GFP detected at the CP following pBLBP-Rnd3 in utero electroporation as compared to controls (means ± SEs, n = 3; **p < 0.01; unpaired two-tailed t test).
See also Figure S8.
In addition, we observed no visible change in the progenitor pool of the MGE in the pBLBP-Rnd3 electroporated brain as compared to the controls (Figure S8L2). Also, L1CAM+ thalamocortical axons showed normal trajectories in both pBLBP-Rnd3 and control electroporated hemispheres of Rnd3 gt/gt; Fezf2-Gfp brain sections at the telencephalon-diencephalon boundary, but were affected within the subpallium at the SPJ in E16.5 Rnd3 gt/gt brains (Figures S8B and S8L6). Thus, we conclude that RND3 expression in the RGCs of the MGE mediates the patterning of the SPJ and pallidal structures, including the GP, by providing a scaffold for the appropriate guidance of CST and anterior commissure projections.
RND3 Regulates CST Axon Guidance in an L5- and FEZF2-Independent Manner
To confirm that these phenotypes arise independently of L5- and FEZF2-dependent cell intrinsic mechanisms, we generated chimeric mice in which Rnd3 gt/gt; Fezf2-Gfp neurons reside in a wild-type context. In contrast to both the Rnd3 gt/gt and Rnd3 −/− brains, axons from the Rnd3 gt/gt; Fezf2-Gfp cortical neurons in the chimera did not misroute toward the ventral surface of the forebrain but instead descended to the basal pons at P0 (Figure 6A). Conditional deletion mouse models in which we induced the deletion of Rnd3 using several regional- and cell-type-specific CRE-recombinase transgenic lines, provided further evidence of a non-cell autonomous role for RND3 (Figures 6B–6G). In most of these mice, we found no consistent reduction in or misrouting of the CST, although we did observe misrouting of commissural projections toward the ventral pallidum in Rnd3 fl/fl; Dlx5/6-Cre brains, suggesting that SVZ progenitors and/or interneurons play a role in commissural projection pathfinding (Figures 6C–6E). Reasoning that Rnd3 might be essential in both the dorsal and ventral forebrains for CST pathfinding, we analyzed Rnd3 gt/fl; Dlx5/6-Cre; Emx1-Cre brain sections immunolabeled for L1CAM. We detected no defects in the CST of these mice (Figures 6F and 6G). Recombination driven by Gsx2-iCre, which is largely specific for RGCs in the LGE but not MGE (Kessaris et al., 2006), also failed to phenocopy either the striatal and ventral misrouting or the reduction of the CST (Figures S8M and S8N). However, the possible contribution of RGCs in LGE to CST defects in striatum cannot be excluded due to the mosaic nature of Gsx2-iCre mice (Qin et al., 2016). Similarly, direct confirmation of the involvement of RGCs from the MGE was hindered by our observations and those previous (Xu et al., 2008) of absent or incomplete recombination events in the MGE RGCs of Nkx2–1-Cre; CAT-Gfp mice. Our results suggest that VZ RGCs in the MGE are responsible for the major CST defects that we observed in Rnd3-deficient mice.
Figure 6. Rnd3 Axon Guidance Phenotypes in the Ventral Forebrain Are Independent of Cortical Fezf2 Expression and Rnd3 Expression in Intermediate Progenitors and Interneurons.
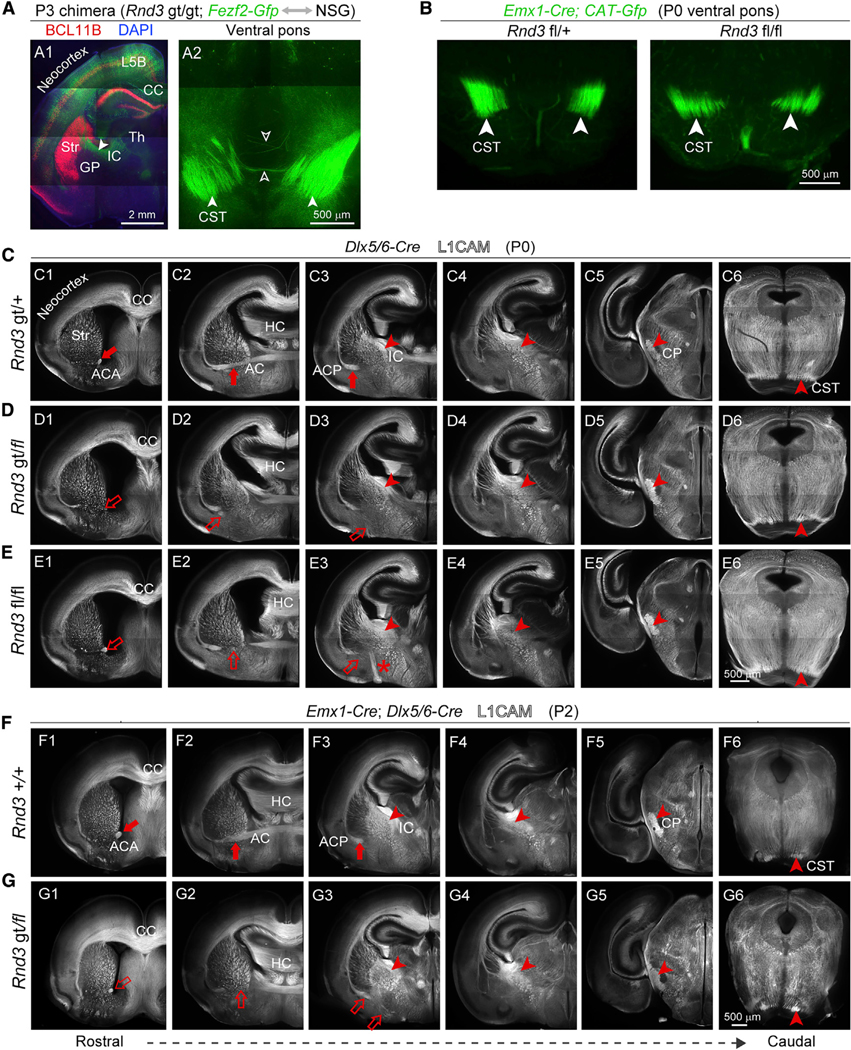
(A) Rnd3 gt/gt; Fezf2-Gfp cells in a wild-type environment, prepared by a blastocyst injection chimera and immunolabeled with antibody against BCL11B (red). A branch of CST showing the decussation defect (open arrow). Scale bars: 2 μm (A1) and 500 μm (A2).
(B) Coronal section of Rnd3 fl/fl; Emx1-Cre mice and Rnd3 fl/+; Emx1-Cre at pons exhibit the size of the CST. Scale bar: 500 μm.
(C–E) L1CAM immunolabeling of Rnd3 fl/fl; Dlx5/6-Cre and Rnd3 gt/fl; Dlx5/6-Cre showing reduced ACA (C1, D1, and E1), reduced and misrouted ACP (D2 and D3, E2 and E3), and stereotypically normal internal capsule (IC), CP, and CST, with deficits highlighted by open arrows and arrowheads and normal features highlighted by solid arrows and arrowheads. Scale bar: 500 μm.
(F and G) Rnd3 gt/fl; Emx1-Cre; Dlx5/6-Cre mice showed a reduction of the ACA and failure to cross the midline of the ACP, and no reduction in CST projections at CP and pons. Scale bar: 500 μm.
See also Figures S4 and S8.
Genetic Interaction of RND3 and ARHGAP35 in Regulating CST Formation
Because RND3 binds and increases the GAP activity of ARHGAP35 (Brouns et al., 2000; Wennerberg et al., 2003), we next assessed the contribution of ARHGAP35 to RND3-associated phenotypes. Analysis of the transcriptome from six human brain regions (Kang et al., 2011; Li et al., 2018) and the E14.5 and P0 mouse brain found that CSNs express both Arghap35 and Rnd3 in the mouse neocortex and in the VZ/SVZ of the LGE and MGE (Figures S10A–S10D). Analysis of the P0 Arghap35 −/− brain revealed that, as in the Rnd3 gt/gt brain, a branch of the CST ran through the GP and misrouted toward the ventral surface of the forebrain (Figures 7A, 7B, S9C, and S9D). Also, like the Rnd3-deficient brain, the size of the cerebral peduncle and the CST at the brainstem were both reduced, the ACA was reduced, and the ACP was absent (Figures 7A and 7B). However, some phenotypes in the Arhgap35 −/− brain were absent in the Rnd3 gt/gt brain, including the corpus callosum misrouting along the midline and the diffusion of axons from the external capsule toward the ventral surface of the forebrain (Figure 7B). Several defects present in the Rnd3 gt/gt brain were also not as severe at the SPJ in the Arhgap35 −/− brain. For example, the GP did not shift rostrally but instead slightly ventrally in Arhgap35 −/−brains (Figures S9A and S9B). In addition, there were fewer disoriented radial glial fibers at the SPJ in the Arhgap35 −/− brain (Figures S9E–S9I). Crucially, we detected defects in the CST at the SPJ (Figures S9C–S9F). These results suggest that guidance of CST axons through the ventral subpallium requires the proper orientation of the radial fibers and that the organization of this substrate requires both Rnd3 and Arhgap35 expression in the MGE.
Figure 7. Genetic Interaction between Rnd3 and Arhgap35 in Regulating CST Pathfinding.
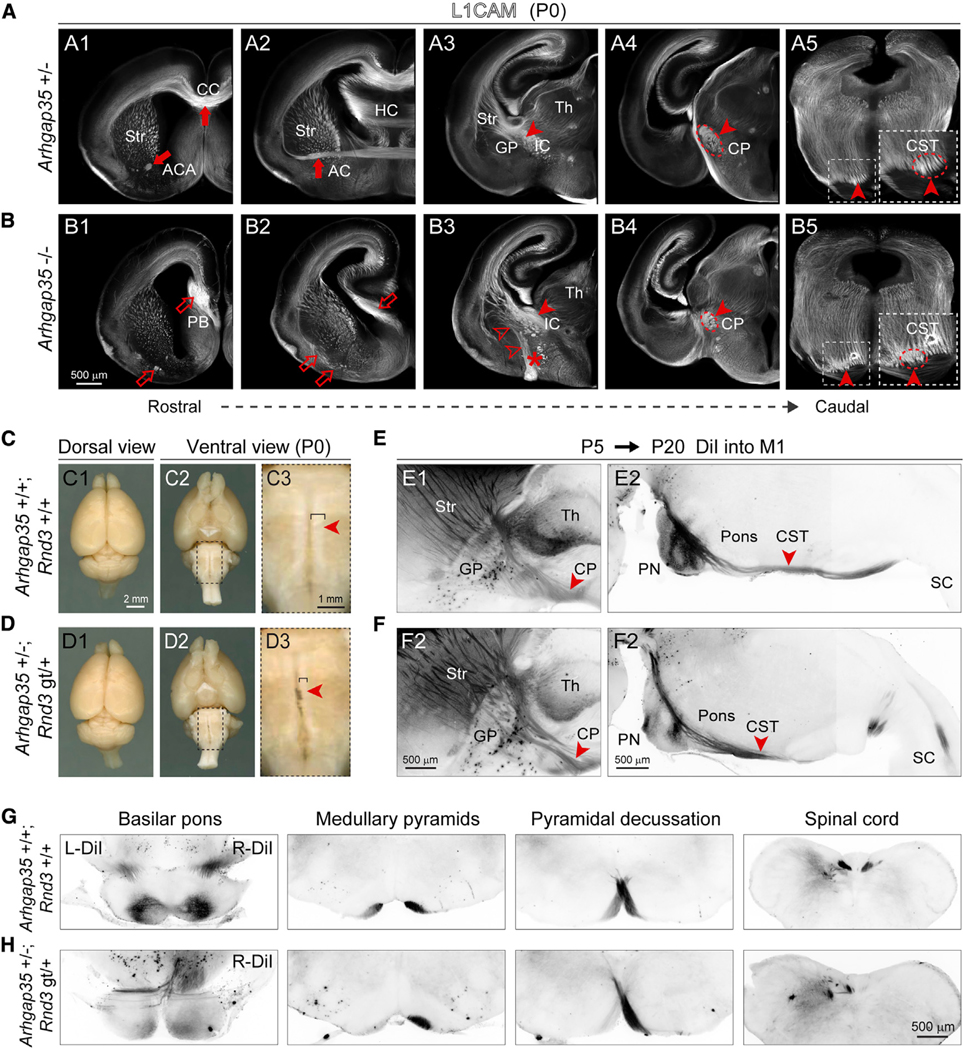
(A and B) Coronal sections from P0 Arhgap35+/− (A1–A5) and Arhgap35 −/− (B1-B5) brain stained for L1CAM reveal phenotypes, similar to Rnd3 gt/gt. The deficits are labeled with open arrows, arrowheads, and asterisks, whereas the normal trajectory is labeled with solid arrows and arrowheads. Scale bar: 500 μm.
(C and D) Gross morphology is maintained in Arhgap35 +/−; Rnd3 gt/+ brains, but the ventral brainstem shows thinner medullary pyramids (i.e., CST) as compared to wild-type controls. The closed arrowheads indicate the presence of the CST. Scale bars: 2 mm (C1 and C2, D1 and D2) and 1 mm (C3 and D3).
(E–H) DiI injection at P5 into primary motor cortex (M1) labeled the CST in CP and pons at P20 in Arhgap35 +/−; Rnd3 gt/+ brain depicting decussation defects in the sagittal (E1–F2) and in coronal sections (G and H) at medullary pyramids. The solid arrowheads indicate the CST decussation. SC, spinal cord. Scale bar: 500 μm.
See also Figures S9 and S10.
Deletion of one copy of each gene (Arhgap35 +/−; Rnd3 gt/+) resulted in thinner cerebral peduncles and medullary pyramids (i.e., the CST) (Figures 7C, 7D, S10E, and S10F), which is indicative of a cumulative gene dosage effect. Bilateral or unilateral injections of DiI into the early postnatal motor cortex (Figures 7G and 7H) and immunostaining for PRKCG (Figures S10E and S10F), a marker of CST axons (Mori et al., 1990), confirmed the reduction of the CST at multiple levels and revealed the premature and aberrant midline crossing of the CST at the basilar pons in Arhgap35 +/−; Rnd3 gt/+ mice (Figures 7E, 7F, S10G, and S10H). The unilateral injections of DiI in Arhgap35 +/−; Rnd3 gt/+ further emphasized the aberrant midline crossing and ipsilateral defasciculation of CST bundles at the basilar pons and in the spinal cord (Figures 7G, 7H, and S10H). These results show that the haploinsufficiency of Rnd3 and Arhgap35 results in early and incomplete midline crossing, suggesting that RND3 acts through ARHGAP35, likely regulating their target RHOA.
DISCUSSION
We initiated this study with the aim of identifying genetic targets of FEZF2, a transcription factor necessary for the proper specification of CSNs and CST formation. FEZF2 activates and represses gene expression in the cerebral cortex (Clare et al., 2017; Lodato et al., 2014), including in CSNs in L5. Here, we found that FEZF2 activates the expression of Rnd3, which is cell autonomously required for the proper regulation of axon elongation, dendritic complexity, and spine formation in CSNs. Commensurate with these observations, RND proteins function as RHOA antagonists by activating ARHGAP35, and this signaling has been shown to be critical for cytoskeletal rearrangements (Wennerberg et al., 2003) and responsiveness to guidance cues (Bonanomi et al., 2019) in growing axons. In addition, we identified an unexpected, cell-non-autonomous role for RND3 and ARHGAP35 in MGE RGCs in their regulation of CST axon guidance through a critical and previously unknown choice point at the SPJ. The expression of RND3 in MGE is also FEZF2 independent, as FEZF2 is not expressed by RGCs in MGE, but it may be driven by ASCL1 (MASH1), which is highly expressed by these cells. Crucially, ACSL1 regulates the expression of Rnd3 in cortical apical progenitors and is essential for properly orienting RGCs during their divisions (Azzarelli et al., 2014).
The principal conceptual finding of this study is that the RGCs use their radial fiber scaffold to facilitate and direct axon guidance, which occurs at a considerable distance from their cell bodies, in a region- and time-specific manner. This finding is consistent with several previous observations. First, the radial fiber is a key cellular and functional compartment of RGCs (Pilaz and Silver, 2017), with known roles in cell division, neuronal migration, and the maintenance of the structural integrity of the CNS (Higginbotham et al., 2011; Rasin et al., 2007). Second, electron microscopy of the developing hamster cerebral cortex revealed contacts between the radial glial fibers and growing callosal projections (Norris and Kalil, 1991). Third, and similar to our observations, previous works reported that RGCs and similar embryonic glial cells in different regions and species appear to form transient spatial compartments for orthogonally oriented axon tracts to pass through (Barry et al., 2013; Silver, 1984; Singer et al., 1979). Fourth, the radial scaffold transports RNA transcripts and proteins involved in axon elongation and guidance throughout the thickness of the wall of the growing CNS in a regionally specified and topographically preserved manner (Dominici et al., 2017; Okada et al., 2017; Varadarajan et al., 2017). The use of temporal, regional, and subcellular-specific mediation of axon guidance by RGCs therefore ensures that the structural palisade-like organization and molecular cues for neuronal migration and axon guidance provided by the cell bodies of RGCs stay regionally restricted during dynamic structural changes in the developing CNS.
In the present study, we also show that the junction between the developing striatum and the GP is a critical point in the unification of the CST and other subcerebral projecting axons into forming an internal capsule. This unification depends on the genetic interaction of Rnd3 and Arhgap35 in the MGE RGCs. We also show that these cells form regularly organized radial fibers that pass between the ventral extent of the striatum and the dorsal edge of the GP in what will be the embryonic external medullary lamina. Previous studies have described other specific features of the SPJ that suggest that it plays a distinct role during the process of axonogenesis. For example, the radial glial-like cells in the VZ of the avian equivalent of the SPJ (border) exhibit a compartmentalized expression of several important developmental genes (Bardet et al., 2006), indicating that these cells are molecularly distinct. Similarly, during early human fetal development, compact bundles of intensely CD15 immunopositive radial glial fibers are present in the external medullary lamina (SPJ) (Mai et al., 1999), indicating that these cells may also have distinct molecular features. Consistent with these findings, we observed radial glial fibers traversing the embryonic striatum and GP and coursing through the external laminar medulla.
The defects in the organization of the radial scaffold observed in Rnd3 mutant mice were specific to the developing external medullary lamina and the GP, but not the striatum. Crucially, this phenotype was rescued by the re-introduction of Rnd3 expression into the RGCs of the MGE, demonstrating that the palisade-like organization of their radial fibers is dependent specifically on RND3 and is essential for the guidance of subcerebrally projecting cortical and likely thalamocortical axons and for correct patterning of the GP. These defects were also associated with the abnormal rostral positioning of the GP in Rnd3 mutant mice, which also likely results from the altered migration of neurons in the SPJ and the GP along the disorganized radial fibers we observed from the MGE RGCs. Complementary phenotypes were also observed following the deletion of Arhgap35, a gene coding for a binding partner of RND3. Moreover, these defects could not be ascribed to any of the previously known cell populations regulating the guidance of the CST, thalamocortical, striatal, or other axons in the ventral forebrain. Given the near-global presence of radial scaffolds during brain development, the conceptual underpinnings of this work may have broad application throughout the CNS.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nenad Sestan (nenad.sestan@yale.edu).
Materials Availability
All unique reagents and resource generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The data reported in this paper is deposited to Gene Expression Omnibus (GEO) with accession number: GSE152611.
METHOD DETAILS
Experimental Animals
All experiments were carried out with the protocol approved by the Committee on Animal Research at Yale University. Rnd3 gt/gt mice were a kind gift from Ignacio Perez Roger’ and the generation and genotyping were previously described (Figure S2B; Mocholí et al., 2011). These mice were generated by inserting a LacZ cassette that disrupts the intron 2 in the Rnd3 gene trap (Rnd3 gt) mutant mouse (Mocholí et al., 2011), abolishing the proper expression of the trapped allele, Rnd3, but allowing for the visualization of natively expressing Rnd3 cells. The Arhgap35 −/− mutant mice were provided by Anthony Koleske and the details for generation and genotyping was described (Brouns et al., 2000). The generation of Emx1-Cre and CAT-Gfp mice were described previously (Iwasato et al., 2004; Kawamoto et al., 2000). Actin-Cre, Dlx6-Cre animals were kind gifts from Angeliki Louvi Lab and Jess Cardin Lab at Yale, respectively. Gsx2-iCre and Nkx2–1-Cre mouse were obtained from Jackson lab. Rnd3-Gfp mice were obtained from the GENSAT Project (http://www.gensat.org/index; (Gong et al., 2003), where green fluorescent protein (Gfp) reporter gene expression was driven by regulatory elements surrounding the endogenous Rnd3 locus. Please refer to Table S6 for genotyping details.
Recombinant DNA
For the pCALNL-Rnd3-Flag, the 3XFlag was added to pCALNL-Gfp using MfeI and NotI sites (Addgene, 13770). The mouse Rnd3 cDNA (GenBank: BC009002) and subcloned into the vector. pCAG-Bfp was a kind gift from Rakic lab, which was used to amplify Bfp sequence to clone it into pCALNL-Gfp vector after excising Gfp. pNeuroD1-Cre plasmid was made by removing the ires-Gfp cassette from pNeuroD1-ires-Gfp (Addgene, 61403) and inserting PCR amplified Cre sequence into EcoRI and BsrGI sites. To express RND3 in RGCs predominantly, pBLBP-Rnd3 and pBLBP-mScarlet-I were generated by removing the mCherry from pBLBP-mCherry (Addgene, 63721) and inserting PCR amplified sequences into AgeI and KpnI sites. For the luciferase assay plasmids, FEZF2 bound sequences was PCR amplified from the mouse genomic DNA, isolated from P0 cortices and inserted into the upstream of the Firefly luciferase in the pGL3-basic vector (Promega, E1751). Primers used for the amplification of all the regions are listed in Table S6.
Human tissue
Human post-mortem brain tissues analyzed in this study were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland. All tissue was collected after obtaining parental or next of kin consent and with approval by the institutional review boards at the Yale School of Medicine and at each institution from which tissue specimens were obtained. Tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the NIH (https://oir.nih.gov/sites/default/files/uploads/sourcebook/documents/ethical_conduct/guidelines-biospecimen.pdf) and the WMA Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medicalresearch-involving-human-subjects/).
Generation of floxed Rnd3 mice
Guide RNA sequences (gRNAs) were designed using an online program (http://zlab.bio/guide-design-resources) as previously described (Hsu et al., 2013). Guide RNAs with the minimum off-targets were selected, and sequences are provided in Table S6. DNA oligos carrying guide RNA sequences were cloned into pX330 vector (Cong et al., 2013), and used as template for in vitro transcription by MEGAscript® T7 Transcription Kit to produce ready to inject gRNAs. Cas9 mRNA were in vitro transcribed from vector px330 linearized by restriction enzyme NotI-HF (New England Biolabs, #R3189S) and purified by phenol/chloroform extraction. We made exon 3 floxed, exon 1–5 deletion, of Rnd3 by this method (Figure S4). gRNAs microinjections were performed as per established protocols. Please see the microinjection details in Table S4. Founders were genotyped and bred at least 3 generations to exclude chimeras. Genotyping of mice carrying deletions was performed using primers shown in Table S6.
In utero electroporations
For RNA seq experiments, pCAG-V5-Fezf2 and pCAG-Gfp were used, in the ratio 3:1, to perform in utero electroporations into E15.5 cortices (n = 3) for overexpression studies. pCAG-Gfp was electroporated into littermates as control. Briefly, the animals were deeply anesthesized and uterine horns were exposed. 1μl of 1μg total DNA, mixed with 0.05% fast green dye, was injected into lateral ventricle of an E15.5 CD1 mouse embryo through the uterine wall and electroporated using BTX apparatus at 35V, 5 pulses, 950 ms each pulse. At P0, pups are screened for GFP expression under a fluorescent microscope. At P3, brains were harvested for tissue preparation and GFP positive cells were FAC sorted
For the rescue experiments to determine spine formation, we performed bihemispherical injections. In one hemisphere, we injected pCALNL-Rnd3-Flag and pNeuroD1-Cre into the ventricles of Fezf2 fl/fl; CAT-Gfp mice at E12.5 to delete Fezf2 from the subcerebral neurons and at the same time, rescue with Rnd3; and into the other hemisphere with pCALNL-Bfp and pNeuroD1-Cre was injected to cause Fezf2 deletion. The electroporated mice were dissected at P30 and 70 μm thick coronal sections were obtained. After staining the sections with GFP, and FLAG antibodies, for spine analysis, the Z stack images spanning 8 μm thickness and of optical thickness 0.43 μm (~18 images/section), were acquired using LSM 800 microscope at 63X magnification. We acquired 10 images per brain from each hemisphere (n = 4 brains). Images were analyzed using reconstruct software. Please refer Golgi staining section for details of analysis (Risher et al., 2014).
For rescuing RND3 in the RGCs of MGE of Rnd3 gt/gt; Fezf2-Gfp brains, we used pBLBP-Rnd3 plasmid to restrict the expression to the RGCs (Shariati et al., 2013). We performed in utero electroporations with pBLBP-Rnd3 + pBLBP-mScarlet-I or with pBLBP-mScarlet-I in the MGE of the embryos obtained from Rnd3 gt/+; Fezf2-Gfp cross at E12.5. The expression of genes under the BLBP promoter diminished in 48 hr. Further the brains were isolated at E16.5 and fixed in 3.7%PFA O/N. Tails from the same embryos were used for genotyping and the brains were sectioned coronally at 70 μm thickness on vibratome. The sections were stained for GFP, NKX2–1, BCL11B, VIM, BLBP and L1CAM (antibodies details in Key Resources Table). Z stack images (10 stacks/image, 0.5 μm thickness, n = 3 brains/condition) from VIM stained sections were used to create a montage of the radial processes of the RGCs originating from ganglionic eminences with NeuronJ plugin of ImageJ. The orientation of the fibers was measured using Orientation J plugin as described below in section 16 of DiI injections and orientation analyses (Rezakhaniha et al., 2012). For the single hemisphere rescue, we injected pBLBP-Rnd3 into the right ventricle of the pups and directed it to MGE using the electrode paddles at E12.5. At E16.5, after embedding dissected tissue in agarose blocks, we marked the right side of the block and sectioned the brains on a vibrotome at a 70 μm thickness. The serial sections collected into 6 wells were stained with respective antibodies.
KEY RESOURCES TABLE
| Experimental Models: Animals | SOURCE | IDENTIFIER |
|---|---|---|
| Gsx2-iCre | The Jackson Laboratory | Cat#25806 |
| Nkx2–1-Cre | The Jackson Laboratory | Cat#8661 |
| Rnd3-Gfp | The GENSAT Project | ID#010612-UCD |
| Fezf2-Gfp | The GENSAT Project | (Gong et al., 2003) |
| Fezf2 fl/fl | Sestan Lab, Yale University | (Han et al., 2011) |
| Emx1-Cre | Takuji Iwasato Lab | (Iwasato et al., 2004) |
| CAG-Cat-Gfp | Melissa Colbert Lab | (Kawamoto et al., 2000) |
| Rnd3 gt/gt | Pérez-Roger Lab, Spain | (Mocholí et al., 2011) |
| Arhgap35 −/− | Koleske Lab, Yale University | (Brouns et al., 2000) |
| Actin-Cre | Louvi Lab, Yale University | N/A |
| Dlx5/6-Cre | Cardin Lab, Yale University | N/A |
| Rnd3 fl/fl | Sestan Lab, Yale University | N/A |
| Rnd3 Δex3 | Sestan Lab, Yale University | N/A |
| Rnd3 Δex1–5 | Sestan Lab, Yale University | N/A |
| Antibodies | ||
| Rat anti - BCL11B (CTIP2) | Abcam | Cat# ab18465; RRID: AB_2064130 |
| Mouse anti - V5 | Thermo Fisher Scientific | Cat# R960–25; RRID: AB_2556564 |
| Mouse anti - SATB2 | Genway | Cat# 20–372-60065; RRID: AB_1029576 |
| Mouse anti - IDU | BD PharMingen | Cat# 555627; RRID: AB_395993 |
| Rat anti - CldU | Accurate Chemical | Cat# MA3–071; RRID: AB_10986341 |
| Chicken anti - GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| Mouse anti - RND3 | Millipore | Cat# 05–723; RRID: AB_309937 |
| Mouse anti - RND3 | Abcam | Cat# ab50316; RRID: AB_2181825 |
| Rat anti - L1CAM | Millipore | Cat# MAB5272; RRID: AB_2133200 |
| Rabbit anti - NKX2–1 | Santa Cruz Biotechnology | Cat# sc-13040; RRID: AB_10015210 |
| Mouse anti - neurofilament (NEFM) | DSHB | Cat# 2H3; RRID: AB_531793 |
| Mouse anti - ISLET1 | DSHB | Cat# 39.4D5; RRID: AB_2314683 |
| Rabbit anti - PRKCG | Santa Cruz Biotechnology | Cat# sc-208; RRID: AB_2168668 |
| Goat anti - Vimentin | Santa Cruz Biotechnology | Cat# sc-7557; RRID: AB_793998 |
| Rabbit anti - Vimentin | Abcam | Cat# ab92547; RRID: AB_10562134 |
| Rabbit anti - CUX1 | Santa Cruz Biotechnology | Cat# sc-13024; RRID: AB_2261231 |
| Rabbit anti - TBR1 | Santa Cruz Biotechnology | Cat# sc-48816; RRID: AB_2287060 |
| Rabbit anti - FABP7 (BLBP) | Abcam | Cat# ab32423; RRID: AB_880078) |
| Rabbit anti - FABP7 (BLBP) | Millipore | Cat# ABN14; RRID: AB_10000325 |
| Mouse anti - FLAG M2 | Sigma Aldrich | Cat# F1804; RRID: AB_262044 |
| Mouse anti - RELN | Millipore Sigma | Cat# 553731–50UL; RRID: AB_565117 |
| Mouse anti - SYP | Sigma Aldrich | Cat# S5768; RRID: AB_477523 |
| Goat anti - CNTN2 (TAG1) | R&D System | Cat# AF4439; RRID: AB_2044647 |
| Mouse anti - MEIS2 | Santa Cruz Biotechnology | Cat# sc81986; RRID: AB_2143037 |
| Rabbit anti - RFP | Rockland Antibodies and Assays | Cat# 600–401-379; RRID: AB_2209751 |
| Rabbit anti - DARPP32 | Santa Cruz Biotechnology | Cat# sc11365; RRID: AB_639000 |
| Alexa Flour 488 donkey Anti-Chicken IgG | Jackson ImmunoResearch | Cat#703–545-155 |
| Biotin conjugate Anti- Chicken IgG | Jackson ImmunoResearch | Cat#703–065-155 |
| Cy3 donkey anti-Rabbit IgG | Jackson ImmunoResearch | Cat#711–165-152 |
| Experimental Models: Animals | SOURCE | IDENTIFIER |
| Cy3 donkey anti-Mouse IgG | Jackson ImmunoResearch | Cat#715–165-150 |
| Alexa Fluor 488 donkey Anti-Mouse IgG | Jackson ImmunoResearch | Cat#715–545-150 |
| Alexa Fluor 488 donkey Anti-Goat IgG | Jackson ImmunoResearch | Cat#714–545-147 |
| Alexa Fluor AMCA donkey Anti-Goat IgG | Jackson ImmunoResearch | Cat#705–155-147 |
| Alexa Fluor 647 donkey Anti- Rat IgG | Jackson ImmunoResearch | Cat#712–605-153 |
| Alexa Fluor 647 donkey Anti- Rabbit IgG | Jackson ImmunoResearch | Cat#711–605-152 |
| Alexa Fluor 647 donkey Anti- Mouse IgG | Jackson ImmunoResearch | Cat#715–605-150 |
| CHEMICALS | ||
| Lipofectamine 2000 | Invitrogen | Cat#11668027 |
| MEGAscript® T7 Transcription Kit | Thermofisher Scientific | Cat#AM1334 |
| Dnase | Sigma Aldrich | Cat#4716728001 |
| Papain | Worthington Biochemical Co | Cat#LS003127 |
| Dispase | Sigma Aldrich | Cat#4942078001 |
| Vectarshield | Vector Laboratories | Cat#H-1000 |
| RNase A | Sigma Aldrich | Cat#10109134001 |
| Proteinase K | Sigma Aldrich | Cat#3115801001 |
| QIAquick PCR Purification Kit | QIAGEN | Cat#28104 |
| Poly D-Lysine | Thermofisher Scientific | Cat#A3890401 |
| Illumina TruSeq mRNA kit | Illumina | Cat# RS-122–2101 |
| NuGEN Ovation V2 system | NuGEN | Cat#7102–32 |
| QIAGEN RNAeasy kit | QIAGEN | Cat#74104 |
| QIAGEN minElute RNAeasy kit | QIAGEN | Cat#74204 |
| Illumina TruSeq CHIP kit | Illumina | Cat#IP-202–1012 |
| Dual Luciferase reporter kit | Promega | Cat#E1910 |
| FD Rapid Golgi Stain kit | FD NeuroTechnologies, Inc | Cat#PK401 |
| Smarter Ultra Low Input RNA kit | Takara | Cat#634891 |
| Kapa genotyping kit | Kapabiosystems | Cat#7961804001 |
| DIG RNA labeling mix | Sigma Aldrich | Cat#11277073910 |
| BCIP/NBT substrate | Sigma Aldrich | Cat#B1911 |
| VECTASTAIN® Elite® ABC HRP Kit (Peroxidase, Universal) | Vector Laboratories | Cat#PK-6200 |
| Tissue-Tek® O.C.T. Compound, Sakura® Finetek | VWR | Cat#4583 |
| Phosphate-buffered saline | ThermoFisher | Cat#14190–144 |
| 1M Tris (pH 7.40) | American Bioanalytical | Cat#AB14044 |
| L-glutamine | ThermoFisher | Cat#25030–081 |
| Penicillin and streptomycin | ThermoFisher | Cat#15140–122 |
| Lipofectamine 2000 | ThermoFisher | Cat#11668–019 |
| NHDF Nucleofector Kit | Lonza | Cat#VPD-1001 |
| Pyruvate | Sigma Aldrich | Cat#P-2256 |
| Paraformaldehyde | J.T. Baker | Cat#S898–07 |
| Bovine serum albumin | Sigma | Cat#A9647 |
| Normal donkey serum | Jackson ImmunoResearch | Cat#017–000-121 |
| DAB-nickel | Vector Laboratories | Cat#SK-4100 |
| Cytochrome Oxidase | Sigma | Cat# C5499 |
| DAPI | ThermoFisher | Cat#D3571 |
| X-gal | American Bioanalytical | Cat#AB02400 |
| Neurobasal medium | ThermoFisher | Cat#21103–049 |
| B-27 supplement | ThermoFisher | Cat#17504–044 |
| Insulin solution, human | Sigma Aldrich | Cat#I9278 |
| MEM non-essential amino acids | ThermoFisher | Cat#11140–050 |
| Mouse Natural Laminin | ThermoFisher | Cat#23017–015 |
| Gelatin subbed slides | SouthernBiotech | Cat#SLD01 |
| Sodium azide | Alfa Aesar | Cat#14314 |
| Dil Stain | Invitrogen | Cat#D282 |
| Cresyl violet acetate | MP Biomedicals | Cat#150727 |
| RECOMBINANT DNA | ||
| pX330 | Addgene | Cat#42230 |
| pCALNL-Gfp | Addgene | Cat#13770 |
| pCAGEN | Addgene | Cat#11160 |
| pCAG-Gfp | Addgene | Cat#11150 |
| pNeuroD1-iresGfp | Addgene | Cat#45025 |
| pBLBP- mCherry | Addgene | Cat#63721 |
| pmScarlet-i_C1 | Addgene | Cat#85044 |
| pGL3 basic | Promega | Cat#E1751 |
| pRL-SV40 | Promega | Cat#E2231 |
| pCAG-Bfp | Rakic Lab, Yale University | N/A |
| pCAG-V5-Fezf2 | Sestan Lab, Yale University | N/A |
| pNeuroD1-Cre | Sestan Lab, Yale University | N/A |
| pCALNL- Rnd3-Flag | Sestan Lab, Yale University | N/A |
| pBLBP- Rnd3 | Sestan Lab, Yale University | N/A |
| pBLBP-mScarlet-I | Sestan Lab, Yale University | N/A |
| pCALNL-Bfp | Sestan Lab, Yale University | N/A |
| INSTRUMENTS | ||
| Tapestation | Agilent | N/A |
| Illumina HiSeq 2000 platform | Illumina | N/A |
| Fluidigm C1 | Fluidigm | N/A |
| BTX Harvard apparatus | Harvard Appartus | N/A |
| cryostat (Leica Biosystem) | Leica Biosystem | N/A |
| BioRad iQ5 system | Biorad | N/A |
| Illumina HiSeq 2000 platform | Illumina | N/A |
| Vibrotome | Leica | N/A |
| SDC microscope | Nikon | N/A |
| Aperio CS2 | Leica Microsystems | N/A |
| Leica TCS SP 8 3X | Leica Microsystems | N/A |
| Zeiss LSM 800 confocal | Zeiss Microscopy | N/A |
| G:Box Chemi XRQ | Syngene | N/A |
| 96/384 microplate reader | Ascent | N/A |
| Ultramicroscope | La Vision Biotec | N/A |
| SOFTWARES | ||
| Gene Tools | Syngene | N/A |
| Imaris X 64 (version 8.0.1) | Bitplane | RRID:SCR_007370 |
| ZEN | Zeiss Microscopy | N/A |
| Aperio ImageScope v12 | Leica | N/A |
| Human Brain Transcriptome exonarray data | (Kang et al., 2011) | GEO: GSE25219 |
| BrainSpan and PsychEncode RNA-seq data | (Mi et al., 2018) | GEO: GSE109796 |
| Experimental Models: Animals | SOURCE | IDENTIFIER |
| BrainSpan and PsychEncode RNA-seq data | (Li et al., 2018) | N/A |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Reconstruct | NA | https://synapseweb.clm.utexas.edu/software-0 |
| Fiji | NIH | https://imagej.net/Fiji |
| Neuron J | NIH | https://imagescience.org/meijering/software/neuronj/ |
| Orientation J | NIH | (Rezakhaniha et al., 2012) |
| MATLAB | MathWorks | https://www.mathworks.com |
| TopHat (v.1.0.13) | N/A | RRID:SCR_013035 |
| Bowtie 2 | (Langmead and Salzberg, 2012) | RRID:SCR_016368 |
| MACS | (Zhang et al., 2008) | RRID:SCR_013291 |
| R | The R Project for Statistical Computing | RRID:SCR_001905 |
| TRANSFAC | GeneXplain GmbH | RRID:SCR_005620 |
| PROMO | alggen.lsi.upc.es | RRID:SCR_016926 |
| DEPOSITED DATA | ||
| Fezf2 RNA/ChIP Seq data | This paper | GSE152611 |
| BIOLOGICAL SAMPLES | ||
| PCW22 Brain | Sestan Lab, Yale University | N/A |
Generation of Rnd3 gt/gt; Fezf2-Gfp chimeric mouse
ES cells were harvested from Rnd3 gt/gt; Fezf2-Gfp embryos and frozen in −80° C and stored in liquid N2. 2–3 days before injection, ES cells were thawed, and media was removed, cells were washed 2–4 times with 1X PBS to get rid of the dead cells and debris. 0.5 mL of 0.5% trypsin was added per 6 cm dish and incubate for 3–5 min at 37° C. Trypsin digestion was stopped by adding 1.5 mL ES cell culture medium with FBS. ES cells were dispersed into single-cell suspension by gentle pipetting, then transferred to 15 mL falcon tube with 3 mL ES medium. Cell suspension was centrifuged at 1000 rpm for 5 min and supernatant discarded. The cell pellets were re-suspended thoroughly in ES cell medium and plate on the gelatinized 6 cm dish and incubated for 20–30 min at 37° C to let the feeder cell attach to the surface, then the medium containing ES cells was collected and centrifuged 1000 rpm for 5 min. Pellets were thoroughly re-suspended in 100–200 μl ES cell injection medium and keep on ice for injection. 3.5 dpc (days post conception) C57BL/6J embryos were collect on the injection day and culture in KSOM at 37° C. For each blastocyst, 6–10 ES cells injected, and 15–20 injected blastocysts were transferred to the uterine of 2.5 dpc CD1 pseudopregnant female recipient.
Bulk RNA-seq
Cortices from P0 Fezf2 fl/fl; Emx1-Cre and littermate Fezf2 fl/+; Emx1-Cre pups were collected (n = 3). Total mRNA was extracted using QIAGEN RNAeasy Kit. For overexpression experiments, P3 cortices from in utero electroporated mice at E15.5 were collected and dissociated (n = 3). pCAG-Gfp injected into littermates were used as controls. GFP+ neurons were sorted and collected. Total mRNA was extracted from the collected neurons using QIAGEN minElute RNAeasy Kit as per manufacturer’s instructions. Libraries were prepared with Illumina TruSeq mRNA preparation kit (Illumina RS-122–2101) for whole cortices, and NuGEN Ovation V2 system (NuGEN, 0510–32) for Fezf2 overexpressing, FAC sorted cells, as per the manufacturer’s instructions. Libraries were quality-controlled by Tapestation/Bioanalyzer analysis and sequenced on Illumina HiSeq 2000 platform at Yale Center for Genome Analysis (YCGA) to generate 75 bp single reads. Sequencing data were quality controlled by FastQC and aligned to the mouse genome (NCBI37/mm9) using STAR (v2.4.0e) (https://doi.org/10.1093/bioinformatics/bts635). To improve the mapping quality of splice junction reads, mouse gene annotation retrieved from the GENCODE project (version M1) was additionally provided (https://doi.org/10.1101/gr.135350.111). Command line “–sjdbOverhang 74” was used to construct splice junction library. At least 10 million uniquely mapped reads were obtained for each sample.
Differential gene expression (DEX) analysis was performed by R package DESeq2 (https://doi.org/10.1186/gb-2010–11-10-r106) and PCA analysis was performed by R package prcomp. A false discovery rate (FDR) < 0.01 was used to detect statistically significant DEX transcripts. Integrated analysis was performed with DEX transcripts from Fezf2 fl/fl; Emx1-Cre neocortex and Fezf2-over-expressing upper layer neurons, with the genes that showed significant expression fold change toward opposite direction (down in Fezf2 fl/fl; Emx1-Cre, up in Fezf2-overexpression) selected as candidate downstream targets of Fezf2, as shown in Tables S1 and S2.
Single-cell RNA-seq analysis
Procedure of single cell capture, and data analysis was described in previous study (Li et al., 2018; Onorati et al., 2016). Briefly, cells were captured with medium-sized (10–17 μm) RNA-seq IFCs using the Fluidigm C1 system according to the manufacturer’s instruction. After capture, C1 chips were examined visually and the number of cells at each capture site was recorded manually. Cells captured by C1 were subsequently processed through lysis, reverse transcription and PCR amplification to generate single-cell cDNA libraries using the Smarter Ultra Low Input RNA kit for Fluidigm (Clontech). Libraries of single cells were sequenced for 100 cycles using a single-read recipe on Illumina’s HiSeq 2000. We employed STAR (v2.4.0) (Dobin et al., 2013) to uniquely align the sequencing reads to human reference genome with default parameters. For human single cells, GRCh38/hg38 was used. The gtf formatted annotations for human (GENCODE, released version v21) were retrieved from GENCODE project. We used RSEQTools and SAMtools to calculate the gene expression (Habegger et al., 2011; Li et al., 2009). We used HTSeq (v0.6.1) to calculate the gene reads count values for each annotation entry. The gene RPKM values were calculated using in-house R script. In addition to FastQC, we implemented a series of quality control measures as described by (Li et al., 2018; Onorati et al., 2016).
Quantitative RT-PCR
Same batch of total RNA that was used for RNA-Seq, was also used for quantitative RT-PCR validation. cDNA was synthesized using superscript III as per instructions, and quantitative RT-PCR was performed in triplicates for each sample on a BioRad iQ5 system with pre-designed primer and probe sets that targeting Rnd3 coding region. The graph was plotted and unpaired two-tailed t test was employed to determine the pvalue (p < 0.05, *; n = 3 brains each condition). Sequences for primers and probes used are provided in Table S6.
ChIP-Seq
Due to absence of antibody against the native FEZF2 protein, a V5 tagged- FEZF2 protein was overexpressed in Neuro2a (N2a) cells to perform ChIP seq. Briefly, Neuro2a (N2a) cells were transfected with pCAG-V5-Fezf2 plasmid using Lipofectamine 2000 (Invitrogen, 11668027). 1× 108 Neuro2a cells were transfected by 25 μg plasmid using Lipofectamine 2000. with a DNA: Lipid ratio = 1: 2. 24 hours (hr) post transfection, cells were dissociated with 0.25% trypsin, washed with 1X PBS and cross-linked with formaldehyde solution at a final concentration of 1% at room temperature for 10 min with constant rotation. Glycine at a final concentration of 125 mM was added to quench cross-linking for 5 min at room temperature with gentle rotation. Cells were washed by 1X PBS, disrupted by lysis buffer I (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100 in Nuclease free H2O) with freshly added protease inhibitors for 20 min at 4° C, lysis buffer II (200 mM NaCl, 1 Mm EDTA, pH8, 0.5 mM EGTA pH 8, 10 mM Tris-HCl pH 8 in Nuclease free H2O) with freshly added protease inhibitors for 10 min at room temperature. Cells are centrifuged at 3000 g, 15 min at 4° C and pellets were dissolved in 400–600μl lysis buffer III (1 mM EDTA, pH 8, 0.5 Mm EGTA, pH 8, 10 mM Tris-HCl, pH 8, 0.5% Sarkosyl in Nuclease free H2O) with freshly added protease inhibitors and sheared into 200–500 bp fragments with a Bioruptor. Dyna beads protein G (Novex, 10003D) was pre-blocked by 5 mg/ml ice-cold BSA and incubated with 5 μg anti-V5 antibody, and IgG, separately at 4° C with constant rotation for 12 hr. 25 μg chromatin was added to beads-antibody complex per reaction and incubated at 4° C for 16 hr with constant rotation. Beads were washed with ice-cold RIPA buffer for 8 times and rinsed by 1X TE, and eluted by adding 200 ml ChIP elution buffer (1% SDS in 1X TE) and incubated in a shaker for 20 min at 65° C. ChIP DNA was incubated for 12 hr at 65° C for reverse-crosslinking, and subjected to RNase A treatment at 37° C for 1 hr, and Proteinase K treatment at 55° C for 2 hr, and then purified on PCR purification columns (QIAGEN, 28104). For input control, 5 μg chromatin from each sample whole-cell-extract DNA was also treated by reverse crosslinking, RNase A and Proteinase K together with IP samples and column purified. DNA amount were quantified by PicoGreen assay (Invitrogen, P11496). 5 ng IP DNA and input from each sample were used to prepare ChIP libraries with Illumina Truseq ChIP library preparation kit (Illumina, IP-202–1012). Libraries were size selected to enrich size 300–400 bp fragments and quality controlled and sequenced on Illumina Hiseq 2000 platform. Roughly 20 million reads were obtained from each sample. FASTA files were quality-controlled and mapped to mouse genome (NCBI37/mm9) using Bowtie (Langmead and Salzberg, 2012). Peaks were called by MACS (Zhang et al., 2008). The experiments were performed in triplicates and p values were calculated using unpaired two-tailed t test. P values greater than 0.05 were considered significant.
Luciferase assays
N2a cells were plated into 24 well plates at a density of 0.05 3 106 per well and transfected as described with Firefly luciferase (pGL3-Basic) plasmids containing each of the Fezf2-bound regions in the mouse Rnd3 locus, together with pRL-SV40 (Promega). Transfected cells were lysed and assayed 24 hr later using the dual-luciferase reporter kit (Promega). Relative luciferase activity was calculated by normalizing the Firefly luciferase luminescence to the Renilla luciferase luminescence value. Experiment was performed in triplicates and p value was calculated using unpaired two-tailed t test. Primers used for amplifying the candidate enhancer and promoter regions are shown in Table S6.
Immunohistochemistry
The brains isolated from prenatal and early postnatal animals were fixed overnight in paraformaldehyde (PFA) at 4° C. Immunostaining were performed on the sections of 60–70 μm thickness cut on vibratome. Sections were blocked with blocking buffer (10% Donkey serum + 1% BSA + 0.3% Triton X-100 in 1X PBS) for 1 hr at RT. After blocking, sections were incubated overnight at 4° C with primary antibodies, Key Resources Table. Sections were washed with washing buffer (0.3% Triton X-100 in 1X PBS) thrice, 10 min each and were incubated with secondary antibodies (Jackson Immunoassay) for 1 hr at RT, followed by three times washing with washing buffer, 10 min each. Sections were mounted onto glass slides with vector shield (Vector labs, H-1000) and sealed with nail-polish. All the slides were stored at −20° C for further analyses. Antigen retrieval was performed on the brain sections prior to NKX2–1, and RND3 immunostaining (Dako Antigen Retrieval Solution). For acquiring the images, Z stack and tiling was performed using, LSM 800 (Zeiss Microsystems) was used. Analyses of the images was done using ZEN software or Fiji using BioFormats plugin.
STED (STimulated Emission Depletion) microscopy
The GFP+ and VIM+ sections were mounted using #1.5 coverslips. A super resolution optical CW STED microscope (Leica TCS SP 8 3X), equipped with white light excitation laser, 590 and 715 nm depletion lasers, 100X/1.4 oil STED objective, and gated HyD detectors, was used to obtain the detailed structures of the radial glial projections and CST axons interaction. For GFP an excitation laser wavelength of 470 nm and for the VIM a wavelength of 647 nm were used. For GFP and VIM, the detection ranges were 478 nm to 504 nm and 656 nm to 708 nm respectively, with a depletion laser at 592 and 715 nm. Stacks of fibers (3–5 μm thickness) with a slice distance of 130 nm were acquired with a 100x oil objective (Leica HCX PL APO 100x/1.4 Oil). The raw data images were processed using spectral unmixing and deconvoluted using Huygens professional (Scientific Volume Imaging).
X-gal staining
P0 brains were fixed with 4% PFA for 10 hr and then rinsed with 1X PBS and sectioned on vibrotome after embedding into agarose. X-gal solution was freshly prepared by diluting 100 μl of X-gal stock solution in a buffer containing 1 mL of 5 mM K ferri, 1 mL of 5 mM K ferro, 20 μl of 1M MgCl2, 40 μl of 25% Triton X-100 and 1X PBS to final volume of 10 ml. Stock solution of X-gal was prepared by diluting 100 mg of X-gal in 2.7 mL of DMF. The sections were stained in this freshly prepared solution for 2 hr to overnight at 30° C in dark. Sectioned were then washed and mounted onto glass slides.
Cytochrome oxidase staining
The cortices from Rnd3 gt/+ and Rnd3 gt/gt animals were isolated at P14 and were flattened between two glass slides. The flattened cortices were fixed overnight in 4% PFA at 4° C and were sectioned on vibrotome following day at thickness of 70 μm. The tangential sections were incubated at 37° C in a buffer containing 4 g sucrose, 50 mg DAB and 30 mg cytochrome C per 100 mL for 2 to 3 hr until desired staining is achieved. Sections from Rnd3 gt/+ and Rnd3 gt/gt cortices were always treated for the same amount of time. Sectioned were washed with 1X PBS and mounted into slides. Images were acquired on a brightfield microscope at 10X magnification.
Thymidine analog labeling
Timed-pregnant mice were injected intraperitoneally with 50 mg per kg of body weight IdU at E11.5 and E12.5, and with 40 mg per kg of body weight CldU at E15.5 and E16 (n = 3). For antigen retrieval, sections cut on vibrotome, were treated with 2N HCl for 30 min at 30° C and were washed thrice with 1X PBS, each wash was for 5 min. Following antigen retrieval, same procedure for immunostaining was followed as describe in section 7. Images were acquired on confocal (LSM 800) at 20X magnification. For quantification of the CldU or IdU positive cells was performed by counting total number of cells in one field (n = 5). Six layers of cortex were divided into 10 bins and number of cells per bin were manually counted for Rnd3 gt/gt and Rnd3 gt/+ animals. Percentage of the cells per bin of total number cells in all the 10 bins were plotted. Unpaired two-tailed t test per bin was employed to calculate p values for each bin.
Golgi-staining and quantifications
The Golgi staining were performed using FD Rapid Golgi Stain™ Kit (PK401) as per manufacturer’s protocol on Fezf2 fl/fl; Emx1-Cre and Fezf2 fl/+; Emx1-Cre brains and the analyses of Golgi stained sections was performed. Briefly, P30 brains were isolated after euthanizing the mice and immersed in solution A+ B for 10 days, followed by immersion in Solution C for 3 days. The brains were then cut on vibrotome in solution C at 150 μm thickness and were serially mounted onto gelatin coated slides. The sections were dried overnight, washed and developed by immersing the slides in solution D and solution E mixed in equal amounts for 10 min. The slides were washed twice with distilled water and dehydrated in grades of ethanol and xylene. Drying of slides was always pre-vented to avoid sections from cracking. After mounting the Z stacked images (~60 images/section), spanning 22 μm thickness, optical thickness 0.36 μm were acquired using Nikon microscope at 63X magnification. The Z stack images were opened in the Reconstruct software (http://synapseweb.clm.utexas.edu/software-0) and a new series was created which enabled us to move across different stacks across Z planes in same image. Dendrites with a minimum length of 10 μm were used for analysis. Whole dendrite was subdivided into segments of 10 μm and number of spines across whole thickness were traced for length and breadth of each spine. All the spines along 10 μm length of dendrites was traced. After tracing the length and breadth of the spines their ratio was used to determine the spine subtype as described earlier (Risher et al., 2014). After the analysis for each class of spine, standard error and p values are calculated using unpaired two-tailed t test.
Fluorescence-activated cell sorting (FACS)
In utero electroporated brains of either overexpression group (pCAG-V5-Fezf2 and pCAG-Gfp) or control group (pCAG-Gfp only) were quickly harvested at P3 and neocortical regions marked by GFP were precisely dissected under a fluorescent microscope and dissociated into single cell suspension using a papain-based solution, which contains DNase, Papain and Dispase (Sestan et al., 1999). Cell were washed and re-suspended in 1X PBS and filtered through a 40 μm strainer and sorted by FACS (BD Aria) with a 488 nm laser. Cell were maintained at 4° C during the whole FACS process. FAC sorted cells were centrifuged down at 1000 rpm at 4° C, flash-frozen by liquid N2 and stored at −80° C for subsequent RNA extraction.
In situ hybridization
RNA probes for were generated mouse cortical tissue cDNA as template (Rnd3, ENSMUSG00000017144; Arhgap35, ENSMUST00000075845) followed by TA cloning and in vitro transcription as per manufacturer’s instructions using similar probes as reported earlier (Visel et al., 2004). Templates were purified by phenol/chloroform extraction, and Digoxigenin-labeled probes were synthesized using T7 and RNA labeling mix (Roche) according to manufacturer’s instructions. Probes were purified by phenol/chloroform extraction, quantified and quality controlled and stored at −80° C till hybridization. Primers used for generating probes are shown in Table S6. Slide-mounted cryo-sections at 30 μm thickness were processed using a previously described protocol (Sousa et al., 2017). Briefly, brains were fixed overnight at 4° C in 4% PFA diluted in 1X PBS, equilibrated for 12 hr at 4° C in 10% sucrose, and another 12 hr at 4° C in 30% sucrose in 1X PBS. Fixed brains were then either embedded in OCT, frozen on dry ice and sliced on a cryostat (Leica Biosystem). Slides were stored at −80° C until processed for in situ hybridization. Sections were first post-fixed in 4% PFA in 1X PBS for 15 min at RT, washed with 1X PBS, treated with proteinase K and submerged in hybridization buffer (5X SSC, 50% formamide, 1% SDS, 200 μg/ml of aBSA, 500 μg/ml of yeast tRNA and 50 μg/ml of heparin) supplemented with 1000 ng/ml appropriate digoxigenin-labeled probe at 70° C overnight. Sections were washed two times 45 min at 70° C in 2X SSC, 50% formamide, 1% SDS, followed by washing in 100 mM Tris HCl pH 7.5, 150 mM NaCl, 0.1% Tween, blocked with 10% sheep inactivated serum (Sigma-Aldrich) and incubated overnight at 4° C with an anti-digoxigenin antibody conjugated to alkaline phosphatase (1:5000, Roche). Sections were then rinsed in 100 mM Tris-Cl pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween before being overlaid with BCIP/NBT substrate (Sigma Aldrich). Revelation was done at RT in the dark until the desired signal is reached. Finally, sections were rinsed in 1X PBS, post-fixed with 4% PFA in 1X PBS, washed in water and mounted with paramount medium. The slides were scanned on Aperio CS2 (Leica Biosystems).
Primary cell culture and neurite length analysis
The cortices from Rnd3 gt/gt; Fezf2-Gfp and Rnd3 gt/+; Fezf2-Gfp were harvested and dissociated for primary culture at P0 mice, as previously described (Chen et al., 2005b). Briefly, P0 mice were sacrificed and forebrains were isolated quickly. Using fine forceps, meninges were peeled off; the neocortices were dissected precisely under a dissecting microscope. The cortices were chopped into fine pieces using a sterile scalpel blade and were collected into 15 mL tube containing using a papain-based solution (Sestan et al., 1999). Tissue was incubated at 37° C for 15 min and triturated by a fire polished glass pipette for 5–10 times to dissociate the cells. Cells were centrifuged at 300 x g for 5 min at room temperature. After washing with 1X PBS twice, cells were plated onto laminin and poly-D-lysine coated glass coverslips in Neurobasal medium supplemented with B27, glutamine and antibiotic. The neurons are stained for SYP and GFP and images are acquired using LSM 800 (Leica). The neurites were traced using ImageJ plugin, Neuron J. The neurites were categorized into primary, secondary and tertiary. The total length of each category is normalized to Rnd3 gt/+; Fezf2-Gfp. Unpaired two-tailed t test is calculated for each category of neurites.
3DISCO tissue clearing
Complete 3DISCO technique was performed with the protocol described by Belle et al., 2014. Samples (Rnd3 +/+ and Rnd3 gt/gt E16.5 and P0 brains) were dehydrated using different solutions of methanol diluted in 1X PBS (50%, 80%, 100%) for 1 hr each step at RT on a rotating shaker, incubated in methanol incubated in 6% H2O2 in methanol 100% O/N at 4° C in the dark, and finally rehydrated with 100%, 80%, 50% methanol in PBS for 1.5 hr each step at RT on a rotating shaker. For whole-mount immunostaining, samples were incubated in a PBSGT solution (0.2% gelatin, 0.5% Triton X-100 and 0.01% thimerosal in PBS) for 24 hr at RT on a rotating shaker and then incubated with the primary antibody (Goat anti-Tag1 (Cntn2); 1: 500; R&D System, AF4439) diluted in 0.1% saponin (Sigma-S4521) PBSGT solution for 10 days at 37° C (with rotation at 100 rpm). Afterward, samples were washed six times (30 min each) in PBSGT at RT and incubated O/N at 37° C with conjugated secondary antibodies (anti-Goat Alexa Fluor 647, Jackson ImmunoResearch). Finally, samples were washed six times in PBSGT for 30 min at RT. For tissue clearing, samples were dehydrated in increasing concentrations (50%, 80%, 100%) of tetrahydroflurane (THF; anhydrous, Sigma-Aldrich; 186562), 1 hr in each step. Lipids were removed with dichloromethane (DCM; Sigma-Aldrich; 270997) during 20 min. Tissue was cleared and stored in dibenzylether (DBE; Sigma-Aldrich; 108014) in the dark and at RT. 3D imaging was performed with an ultramicroscope (LaVision BioTec) using InspectorPro software (LaVision BioTec). Images, 3D volume, and movies were generated using Imaris x64 software (version 8.0.1, Bitplane). Stack images were first converted to Imaris files (.ims) using ImarisFileConverter. 3D reconstruction of samples was performed using the “volume rendering” (Imaris). The aAC and LOT were segmented using the “surface” tool by creating a mask around each volume. 3D pictures were generated using the “snapshot” and tools.
DiI injections and orientation analyses
For lipophilic tracer, DiI, injections embryonic brains were isolated at E12.5. Tails of each embryo were dissected for genotyping and the brains were fixed for 4 hr in 1 % PFA in 1X PBS at 4° C. After that, DiI (1mg/ml) was pressure injected using Picospritzer III into the ventricles at 20 psi, 3 pulse/brain. Brains were then incubated at 37° C for 2 days in 4% PFA and sectioned on vibrotome at 70 μm thickness. Sections are washed in 1X PBS and mounted onto the slides. The images were acquired on LSM 800 microscope at 10X and 20X magnification. Z stack images (10 images/section, 0.5 μm thickness, n = 4 brains/condition) of the brains were acquired. The images were imported into Fiji and analyzed by Orientation J plugin (Rezakhaniha et al., 2012). The images from MGE areas from Rnd3 gt/gt and Rnd3 gt/+ brains were cropped to exclude the tracer saturated areas and DiI labeled fibers were in field of view. This cropped image was used as input for analyses by the Orientation J program that computes the structure tensor for each pixel by sliding the Gaussian analysis window over the entire image. The output is a color map in HSB (hue, saturation and brightness) mode, where hue is orientation, saturation is coherency and brightness were the source image. For quantitative measure of the orientations, the pixel that have a coherency > 1% and energy > 1% were considered to reduce the background. The software provided an output as a text file of orientation as number of pixels per degree ranging from 0° to 180°. Data was further processed using MATLAB (The MathWorks, Inc., Natick, Massachusetts, United States). Angles above 90° were renormalized to range from 0° to 90°. Pixel orientation was further binned in 10° increments for graphical analysis, and in 30° increments for statistical analysis. Pairwise comparison was performed using two-tailed Chi-square test with Yates correction to compare ratios of pixels between two experimental groups for each 30° bin.
Supplementary Material
Highlights.
Rnd3 is expressed by corticospinal neurons (CSNs) and radial glia/neural stem cells
FEZF2-driven Rnd3 expression helps CSNs to form dendritic spines and extend axons
RND3 and ARHGAP35 maintain the radial glial scaffold at the striatopallidal junction
The striatopallidal radial glial scaffold facilitates corticospinal axon pathfinding
ACKNOWLEDGMENTS
We thank Joshua Corbin, Kenneth Campbell, and Nicoletta Kessaris for providing mice. This work was supported by Labex LifeSenses grants ANR-10-LABX-65 and ANR-11-IDEX-0004-02 to A.C.; MINECO SAF2013-49176-C2-1-R and Programa Santander-FUSP to I.P.-R. and J.T.; NIH grants R01 MH115939, NS105640, and NS089662 to A.J.K.; and NIH grants MH103339, MH106934, MH110926, and MH109904 to N.S. Additional support was provided by the Kavli Foundation and the Simons Foundation. We thank the Yale Macaque Brain Resource (grant to Alvaro Duque and Lynn D. Selemon, National Institute of Mental Health [NIMH] 5R01MH113257-02) for the Aperio CS2 scanner.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.neuron.2020.06.035.
REFERENCES
- Anderson KD, and Reiner A. (1991). Immunohistochemical localization of DARPP-32 in striatal projection neurons and striatal interneurons: implications for the localization of D1-like dopamine receptors on different types of striatal neurons. Brain Res. 568, 235–243. [DOI] [PubMed] [Google Scholar]
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, and Richards LJ (2006). Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development 133, 2243–2252. [DOI] [PubMed] [Google Scholar]
- Azzarelli R, Pacary E, Garg R, Garcez P, van den Berg D, Riou P, Ridley AJ, Friedel RH, Parsons M, and Guillemot F. (2014). An antagonistic interaction between PlexinB2 and Rnd3 controls RhoA activity and cortical neuron migration. Nat. Commun 5, 3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzarelli R, Guillemot F, and Pacary E. (2015). Function and regulation of Rnd proteins in cortical projection neuron migration. Front. Neurosci 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagri A, Marín O, Plump AS, Mak J, Pleasure SJ, Rubenstein JL, and Tessier-Lavigne M. (2002). Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 33, 233–248. [DOI] [PubMed] [Google Scholar]
- Bardet SM, Cobos I, Puelles E, Martínez-De-La-Torre M, and Puelles L. (2006). Chicken lateral septal organ and other circumventricular organs form in a striatal subdomain abutting the molecular striatopallidal border. J. Comp. Neurol 499, 745–767. [DOI] [PubMed] [Google Scholar]
- Barry DS, Pakan JM, O’Keeffe GW, and McDermott KW (2013). The spatial and temporal arrangement of the radial glial scaffold suggests a role in axon tract formation in the developing spinal cord. J. Anat 222, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle M, Godefroy D, Dominici C, Heitz-Marchaland C, Zelina P, Hellal F, Bradke F, and Chédotal A. (2014). A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep. 9, 1191–1201. [DOI] [PubMed] [Google Scholar]
- Bielle F, Marcos-Mondejar P, Keita M, Mailhes C, Verney C, Nguyen Ba-Charvet K, Tessier-Lavigne M, Lopez-Bendito G, and Garel S. (2011). Slit2 activity in the migration of guidepost neurons shapes thalamic projections during development and evolution. Neuron 69, 1085–1098. [DOI] [PubMed] [Google Scholar]
- Bonanomi D, Valenza F, Chivatakarn O, Sternfeld MJ, Driscoll SP, Aslanian A, Lettieri K, Gullo M, Badaloni A, Lewcock JW, et al. (2019). p190RhoGAP Filters Competing Signals to Resolve Axon Guidance Conflicts. Neuron 102, 602–620.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, Haydar TF, and Rakic P. (2011). Neural stem cells: historical perspective and future prospects. Neuron 70, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, Silver J, Bronson RT, and Settleman J. (2000). The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development 127, 4891–4903. [DOI] [PubMed] [Google Scholar]
- Canty AJ, and Murphy M. (2008). Molecular mechanisms of axon guidance in the developing corticospinal tract. Prog. Neurobiol 85, 214–235. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, and McConnell SK (2005a). Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. USA 102, 17184–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, and Sestan N. (2005b). Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc. Natl. Acad. Sci. USA 102, 17792–17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare AJ, Wicky HE, Empson RM, and Hughes SM (2017). RNA-Sequencing Analysis Reveals a Regulatory Role for Transcription Factor Fezf2 in the Mature Motor Cortex. Front. Mol. Neurosci 10, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici C, Moreno-Bravo JA, Puiggros SR, Rappeneau Q, Rama N, Vieugue P, Bernet A, Mehlen P, and Chédotal A. (2017). Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature 545, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, and Heintz N. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, and Gerstein M. (2011). RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics 27, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, and Lalli G. (2010). Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol 2, a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MMS, Shin Y, Xu X, Zhu Y, Li M, and Sestan N. (2011). TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc. Natl. Acad. Sci. USA 108, 3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Yokota Y, and Anton ES (2011). Strategies for analyzing neuronal progenitor development and neuronal migration in the developing cerebral cortex. Cereb. Cortex 21, 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T, Nomura R, Ando R, Ikeda T, Tanaka M, and Itohara S. (2004). Dorsal telencephalon-specific expression of Cre recombinase in PAC transgenic mice. Genesis 38, 130–138. [DOI] [PubMed] [Google Scholar]
- Jones L, López-Bendito G, Gruss P, Stoykova A, and Molnár Z. (2002). Pax6 is required for the normal development of the forebrain axonal connections. Development 129, 5041–5052. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. (2011). Spatiotemporal transcriptome of the human brain. Nature 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, and Miyazaki J. (2000). A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 470, 263–268. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, and Richardson WD (2006). Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci 9, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, and Alvarez-Buylla A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci 32, 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rasin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, et al. (2012). Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon RN (2008). Descending pathways in motor control. Annu. Rev. Neurosci 31, 195–218. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. ; BrainSpan Consortium; PsychENCODE Consortium; PsychENCODE Developmental Subgroup (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Molyneaux BJ, Zuccaro E, Goff LA, Chen HH, Yuan W, Meleski A, Takahashi E, Mahony S, Rinn JL, et al. (2014). Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat. Neurosci 17, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G, Cautinat A, Sánchez JA, Bielle F, Flames N, Garratt AN, Talmage DA, Role LW, Charnay P, Marín O, and Garel S. (2006). Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell 125, 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. (2000). Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci 1, 173–180. [DOI] [PubMed] [Google Scholar]
- Mai JK, Krajewski S, Reifenberger G, Genderski B, Lensing-Höhn S, and Ashwell KW (1999). Spatiotemporal expression gradients of the carbohydrate antigen (CD15) (Lewis X) during development of the human basal ganglia. Neuroscience 88, 847–858. [DOI] [PubMed] [Google Scholar]
- Marín O, Baker J, Puelles L, and Rubenstein JL (2002). Patterning of the basal telencephalon and hypothalamus is essential for guidance of cortical projections. Development 129, 761–773. [DOI] [PubMed] [Google Scholar]
- Martin JH (2005). The corticospinal system: from development to motor control. Neuroscientist 11, 161–173. [DOI] [PubMed] [Google Scholar]
- Mi D, Li Z, Lim L, Li M, Moissidis M, Yang Y, Gao T, Hu TX, Pratt T, Price DJ, et al. (2018). Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocholí E, Ballester-Lurbe B, Arqué G, Poch E, Peris B, Guerri C, Dierssen M, Guasch RM, Terrado J, and Pérez-Roger I. (2011). RhoE deficiency produces postnatal lethality, profound motor deficits and neurodevelopmental delay in mice. PLOS ONE 6, e19236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, and Macklis JD (2005). Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47, 817–831. [DOI] [PubMed] [Google Scholar]
- Morello F, Prasad AA, Rehberg K, Vieira de Sá R, Antón-Bolaños N, Leyva-Diaz E, Adolfs Y, Tissir F, López-Bendito G, and Pasterkamp RJ (2015). Frizzled3 Controls Axonal Polarity and Intermediate Target Entry during Striatal Pathway Development. J. Neurosci 35, 14205–14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Kose A, Tsujino T, and Tanaka C. (1990). Immunocytochemical localization of protein kinase C subspecies in the rat spinal cord: light and electron microscopic study. J. Comp. Neurol 299, 167–177. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Plestant C, Yabuno-Nakagawa K, Li J, Lee J, Huang CW, Lee A, Krupa O, Adhikari A, Thompson S, et al. (2019). Memo1-Mediated Tiling of Radial Glial Cells Facilitates Cerebral Cortical Development. Neuron 103, 836–852.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega-Pereira S, Gelman D, Bartolini G, Pla R, Pierani A, and Marín O. (2010). Origin and molecular specification of globus pallidus neurons. J. Neurosci 30, 2824–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris CR, and Kalil K. (1991). Guidance of callosal axons by radial glia in the developing cerebral cortex. J. Neurosci 11, 3481–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Keino-Masu K, Nagamine S, Kametani F, Ohto T, Hasegawa M, van Kuppevelt TH, Kunita S, Takahashi S, and Masu M. (2017). Desulfation of Heparan Sulfate by Sulf1 and Sulf2 Is Required for Corticospinal Tract Formation. Sci. Rep 7, 13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, et al. (2016). Zika Virus Disrupts Phospho-TBK1 Localization and Mitosis in Human Neuroepithelial Stem Cells and Radial Glia. Cell Rep. 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, and Silver DL (2017). Moving messages in the developing brainemerging roles for mRNA transport and local translation in neural stem cells. FEBS Lett. 591, 1526–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Madhavan M, Waclaw RR, Nakafuku M, and Campbell K. (2016). Characterization of a new Gsx2-cre line in the developing mouse telencephalon. Genesis 54, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, and Sestan N. (2007). Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat. Neurosci 10, 819–827. [DOI] [PubMed] [Google Scholar]
- Rezakhaniha R, Agianniotis A, Schrauwen JT, Griffa A, Sage D, Bouten CV, van de Vosse FN, Unser M, and Stergiopulos N. (2012). Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech. Model. Mechanobiol 11, 461–473. [DOI] [PubMed] [Google Scholar]
- Risher WC, Ustunkaya T, Singh Alvarado J, and Eroglu C. (2014). Rapid Golgi analysis method for efficient and unbiased classification of dendritic spines. PLOS ONE 9, e107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid RS, Yokota Y, and Anton ES (2006). Generation and characterization of brain lipid-binding protein promoter-based transgenic mouse models for the study of radial glia. Glia 53, 345–351. [DOI] [PubMed] [Google Scholar]
- Sestan N, Artavanis-Tsakonas S, and Rakic P. (1999). Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 286, 741–746. [DOI] [PubMed] [Google Scholar]
- Shariati SA, Lau P, Hassan BA, Müller U, Dotti CG, De Strooper B, and Gärtner A. (2013). APLP2 regulates neuronal stem cell differentiation during cortical development. J. Cell Sci 126, 1268–1277. [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, and Sestan N. (2012). Cis-regulatory control of corticospinal system development and evolution. Nature 486, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. (1984). Studies on the factors that govern directionality of axonal growth in the embryonic optic nerve and at the chiasm of mice. J. Comp. Neurol 223, 238–251. [DOI] [PubMed] [Google Scholar]
- Singer M, Nordlander RH, and Egar M. (1979). Axonal guidance during embryogenesis and regeneration in the spinal cord of the newt: the blueprint hypothesis of neuronal pathway patterning. J. Comp. Neurol 185, 1–21. [DOI] [PubMed] [Google Scholar]
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. (2017). Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Liu WA, Zhang XJ, Shi W, Ren SQ, Li Z, Brown KN, and Shi SH (2016). Vascular Influence on Ventral Telencephalic Progenitors and Neocortical Interneuron Production. Dev. Cell 36, 624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Götz M, and Huttner WB (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol 30, 465–502. [DOI] [PubMed] [Google Scholar]
- Tuttle R, Nakagawa Y, Johnson JE, and O’Leary DD (1999). Defects in thalamocortical axon pathfinding correlate with altered cell domains in Mash-1-deficient mice. Development 126, 1903–1916. [DOI] [PubMed] [Google Scholar]
- Varadarajan SG, Kong JH, Phan KD, Kao TJ, Panaitof SC, Cardin J, Eltzschig H, Kania A, Novitch BG, and Butler SJ (2017). Netrin1 Produced by Neural Progenitors, Not Floor Plate Cells, Is Required for Axon Guidance in the Spinal Cord. Neuron 94, 790–799.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, and Eichele G. (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welniarz Q, Dusart I, and Roze E. (2017). The corticospinal tract: evolution, development, and human disorders. Dev. Neurobiol 77, 810–829. [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, Settleman J, Der CJ, and Hansen SH (2003). Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol 13, 1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, and Anderson SA (2008). Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol 506, 16–29. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, and Isa T. (2018). Neural and genetic basis of dexterous hand movements. Curr. Opin. Neurobiol 52, 25–32. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper is deposited to Gene Expression Omnibus (GEO) with accession number: GSE152611.


