SUMMARY
Transposable elements (TEs) comprise nearly half of the human genome and are often transcribed or exhibit cis-regulatory properties with unknown function in specific processes such as heart development. In the case of endogenous retroviruses (ERVs), a TE subclass, experimental interrogation is constrained as many are primate-specific or human-specific. Here, we use primate pluripotent stem-cell-derived cardiomyocytes that mimic fetal cardiomyocytes in vitro to discover hundreds of ERV transcripts from the primate-specific MER41 family, some of which are regulated by the cardiogenic transcription factor TBX5. The most significant of these are located within BANCR, a long non-coding RNA exclusively expressed in primate fetal cardiomyocytes. Functional studies reveal that BANCR promotes cardiomyocyte migration in vitro and ventricular enlargement in vivo. We conclude that recently evolved TE loci such as BANCR may represent potent de novo developmental regulatory elements that can be interrogated with species-matching pluripotent stem cell models.
In Brief
Endogenous retroviruses (ERVs) have unknown function in heart development. Wilson et al. studied ERV-derived BANCR, a long non-coding RNA expressed in primate fetal cardiomyocytes. Using pluripotent stem cell and rodent models, BANCR was found to promote cardiomyocyte migration and cardiac enlargement, arguing for the importance of ERVs in heart development.
Graphical Abstract
INTRODUCTION
Cardiovascular disease is the most common cause of death worldwide (Mozaffarian et al., 2015) and includes a spectrum of adult-onset conditions as well as congenital phenotypes that are the most common category of severe birth defects (Yang et al., 2006). Causes of heart disease include environmental risk factors, common genomic variants, and rare and de novo mutations (Ashley et al., 2012; Ma et al., 2019). Candidate gene sequencing has proven useful (Rehm, 2013), though current approaches are often limited to the approximately 2% of the genome that is protein coding and ignore the 98% of the genome that is non-coding. This is important because the non-coding genome accounts for the majority of trait-associated single-nucleotide polymorphisms (SNPs) in the human genome (Leslie et al., 2014). Whole-genome sequencing (WGS) can detect non-coding variation; however, without robust annotation linking these loci to phenotype, these findings are mostly uninterpretable. Because of these pitfalls, the non-coding genome of the heart has been largely ignored, particularly in non-conserved regions that may be primate specific or even human specific. Complicating matters is the fact that non-coding elements can be active only in certain cell types or at particular developmental stages and are therefore missed by large-scale sequencing studies of adult tissues. Studies are needed that identify critical non-coding elements, followed by validation of their function through robust and repeatable experimentation, in species- and state-specific model systems.
One source of non-coding variation in the human genome is a subclass of non-coding RNAs, long non-coding RNAs (lncRNAs), that function as regulatory molecules via multiple mechanisms and have thus become an active area of study for human disease (Batista and Chang, 2013). Among other functions, lncRNAs direct protein complexes, genes, and chromosomes to appropriate cellular locations, where they are then activated or deactivated to influence fundamental processes, such as pluripotency, differentiation, and development (Batista and Chang, 2013). Given their critical functions, it is surprising that lncRNAs are only modestly conserved across species, with primates estimated to carry thousands of lncRNAs not found in other clades (Necsulea et al., 2014). This lack of conservation is due, in part, to transposable elements (TEs), mobile DNA sequences that are ubiquitous throughout the genome and found in two-thirds of lncRNA transcripts (Kapusta et al., 2013). For example, the endogenous retrovirus (ERV) subclass of TEs comprises 8%–10% of the human genome, several of which are active in evolutionary genetics (Hughes and Coffin, 2001), cancer (Gibb et al., 2015), embryo implantation (Grow et al., 2015), and immunity (Chuong et al., 2016). However, the vast majority of their functions, if any, remain largely unknown. A critical challenge to understanding ERV function is that many human ERVs only recently invaded the primate genome and cannot be studied in murine knockout models.
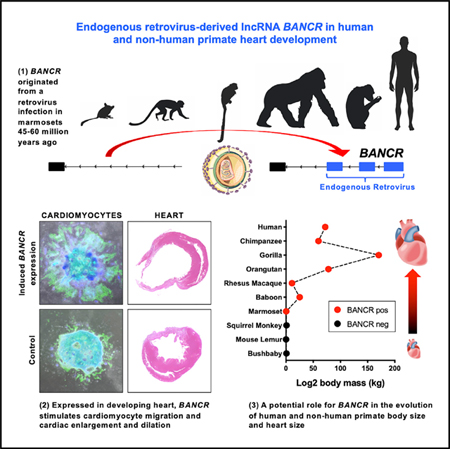
Here, we report that MER41, a family of primate-specific ERVs, directly impacted human and non-human primate heart development via a MER41-derived lncRNA that induces migration in fetal cardiomyocytes. This pro-migratory phenotype is outside the more typical contractility phenotypes associated with cardiomyocytes and highlights the singular ability of TEs, such as ERVs to integrate into genomes and create novel functional elements with potentially profound phenotypic effects. In our functional experiments, we use human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) that mimic fetal cardiomyocytes in vitro and discover hundreds of ERV transcripts from the MER41 family. The most significant of these are located within the lncRNA BANCR, which is exclusively expressed in fetal cardiomyocytes. We then derive induced pluripotent stem cells (iPSCs) from a male silverback gorilla, a chimpanzee, and a rhesus macaque, and confirm expression of orthologous MER41-derived BANCR in the corresponding species-specific differentiated cardiomyocytes. This demonstrates conservation of BANCR transcription at both the human and non-human primate fetal heart stage, which suggests a role for BANCR in primate heart development. Employing custom bioengineered micropatterning experiments, we find that BANCR knockdown, knockout, and over-expression in human embryonic stem cell-derived cardiomyocytes (hESC-CMs) markedly affects cardiomyocyte migration. Importantly, chromatin immunoprecipitation sequencing (ChIP-seq) and assay for transposase-accessible chromatin (ATAC-seq) reveal that a key cardiogenic transcription factor, TBX5, binds a regulatory enhancer upstream of BANCR and induces its expression specifically in fetal heart. Interestingly, several key factors (TEAD4/YAP1) in Hippo signaling and mechanotransduction (Dupont et al., 2011) also bind to the BANCR enhancer and have been shown previously to control organ size and regeneration (Heallen et al., 2011; Totaro et al., 2018). In line with these known TEAD4/YAP1 signaling effects, BANCR knockin murine embryos exhibit increased heart size compared with littermate controls. Our results therefore position BANCR as a potential member of TEAD/YAP signaling, and its co-regulation by TBX5 induces its cardiac-specific expression. We conclude that recently evolved TE loci, such as BANCR may represent potent de novo developmental regulatory elements that can be interrogated with species-matching pluripotent stem cell models to understand their roles in organogenesis.
RESULTS
BANCR Is an ERV-Derived lncRNA Exclusively Expressed in Fetal Heart and Pluripotent Cell-Derived Cardiomyocytes
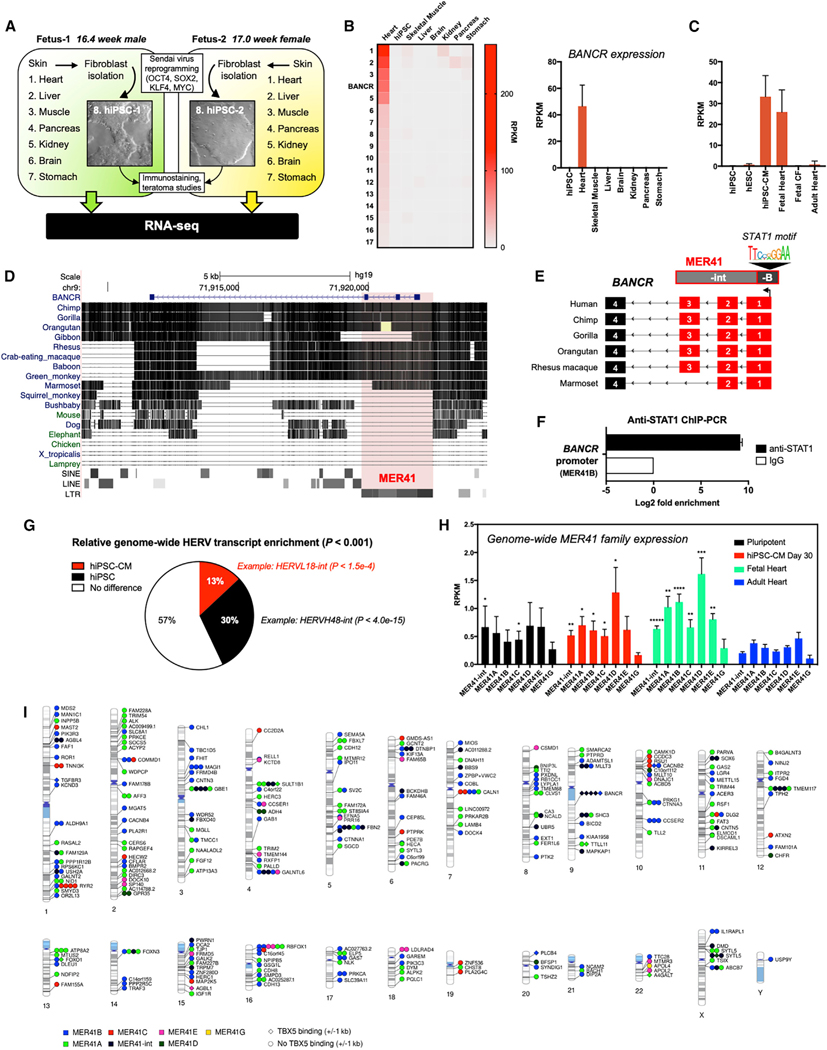
As both lncRNAs and ERVs are frequently active in pluripotency and development, we performed RNA sequencing (RNA-seq) of organs and tissues obtained from two electively aborted fetuses, as well as hiPSCs reprogrammed from each fetus’s skin fibroblasts, to determine heart-specific expression (Figures 1A, S1A, and S1B). Whole exome sequencing did not detect known non-synonymous exonic mutations in genes related to congenital heart disease, cardiomyopathy, or channelopathy (Wilson et al., 2015) that could bias our results (Figure S1C). We focused on lncRNA expression (Figure S1D) and found 17 transcripts with a predominant expression in the fetal heart relative to other tissues and hiPSCs (Figures 1B [left panel] and S1E). One of these, BANCR, was also expressed in hiPSC-CMs but was absent or minimally expressed in pluripotent cells, fetal cardiac fibroblasts, and adult tissues including adult heart (Figures 1B [right panel], 1C, and S1F), suggesting a role in fetal cardiomyocyte development rather than normal adult heart physiology.
Figure 1. BANCR Is Expressed Exclusively in Fetal Heart and Pluripotent Stem-Cell-Derived Cardiomyocytes.
(A) Overview of fetal tissue and hiPSC RNA-seq.
(B) Heatmap (left panel) showing average RPKM expression of 17 lincRNAs with predominant expression in fetal heart (see Figure S1E for table). A graph of RPKM expression values for BANCR specifically is also shown (right panel). n = 4 biological replicates (fetal hearts) and n = 2 biological fetal tissues (all other samples). Error bars ± 1 standard deviation (SD).
(C) Quantitative BANCR expression across hiPSC-CMs, undifferentiated hESCs and hiPSCs, fetal and adult heart tissues, and fetal cardiac fibroblasts (CFs). There is high BANCR expression in fetal hearts (n = 4 biological replicates) and hiPSC-CMs (n = 3 biological replicates) but not in hiPSCs (n = 2 biological replicates), hESCs (n = 6 biological replicates), fetal CFs (n = 4 biological replicates), or adult heart (n = 4 biological replicates). Error bars ± 1 SD.
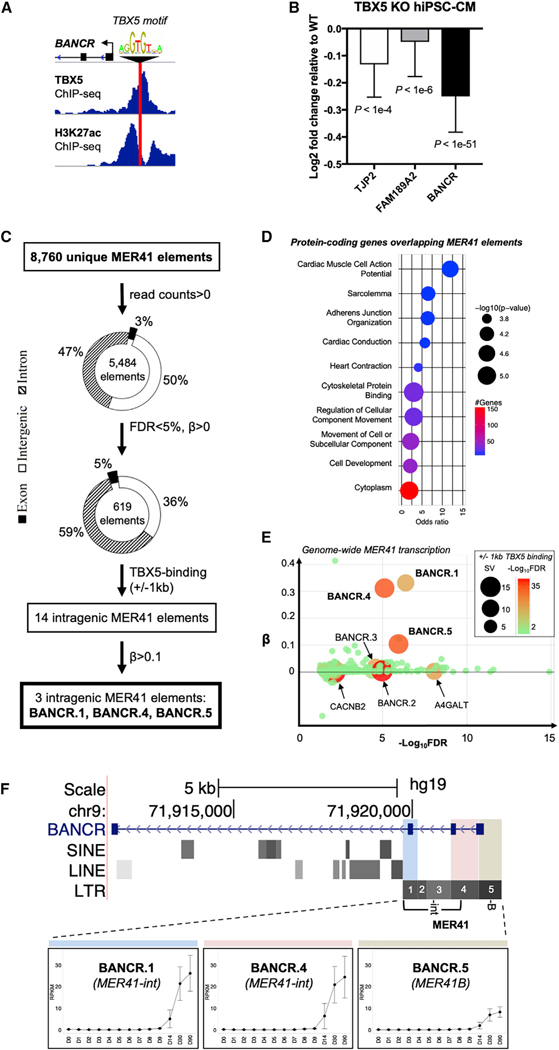
(D) BANCR conservation in primates with MER41 locus highlighted.
(E) Schematic of MER41B (-B and -int subfamilies) in primate BANCR homologous regions. A STATI-binding motif in the MER41B-derived promoter is shown.
(F) STAT1 ChIP-PCR shows STAT1 binding in BANCR promoter. Error bars ± 1 SD.
(G) Global HERV transcript enrichment in hiPSCs and hiPSC-CMs. n = 3 biological replicates.
(H) Increased genome-wide expression of MER41 subfamilies in human fetal heart and hiPSC-CMs compared with corresponding subfamily in adult heart. *p value < 0.01, **p < 0.001, ***p < 0.0001, ****p < 0.00001, two-sided t test. n = 4 biological replicates, error bars are ± 1 SD.
(I) Virtual karyogram showing intragenic MER41 element expression across all chromosomes with indicated overlapping RefSeq genes. Elements are categorized by subfamily (color) and based on whether TBX5 binds within 1 kb (diamond) or if there is no nearby TBX5 binding (circle). Note that multiple MER41 elements can bind within the same gene.
BANCR arose in a common ancestor with marmosets from the primate-specific ERV medium reiteration frequency inter-spersed repeats 41 (MER41) (Figure 1D). Importantly, because BANCR does not exist in mouse (Figure S1G), it would not be possible to generate a transgenic BANCR knockout model to assess its impact on murine phenotype. In contrast, greater than 99% of nucleotides in homologous BANCR exons across Homininae genomes are conserved (Table S1). MER41 is a remnant of an endogenized gammaretroviral infection some 45–60 million years ago (Bao et al., 2015; Chuong et al., 2016; Kapusta et al., 2013; Marmoset Genome Sequencing and Analysis Consortium, 2014) and has rapidly expanded to cover approximately 0.1% of the human genome (Schmid and Bucher, 2010). Although there is evidence for its role in innate immunity (Chuong et al., 2016), MER41 influence on primate cardiac biology, if any, is unknown. Interestingly, MER41-derived BANCR is active in melanoma (Flockhart et al., 2012), suggesting that co-option of this primate fetal heart-specific gene may have pro-oncogenic potential. The MER41 family consists of 8,760 unique elements throughout the human genome that comprise six subfamilies (MER41A, B, C, D, E, and G). The BANCR promoter is derived from MER41B and binds the transcription factor STAT1 at the binding motif (Figures 1E and 1F), similar to other MER41B elements (Chuong et al., 2016). Human ERV (HERV)-derived transcripts, as a whole, are approximately twice as likely to be enriched in hiPSCs relative to hiPSC-CMs (Figure 1G); however, MER41 transcripts are mildly enriched in hiPSC-CMs and fetal heart (Figure 1H) and are expressed from loci on all human chromosomes (Figure 1I).
Homologous BANCR Is Expressed in Non-human Primate (NHP) iPSC-CMs
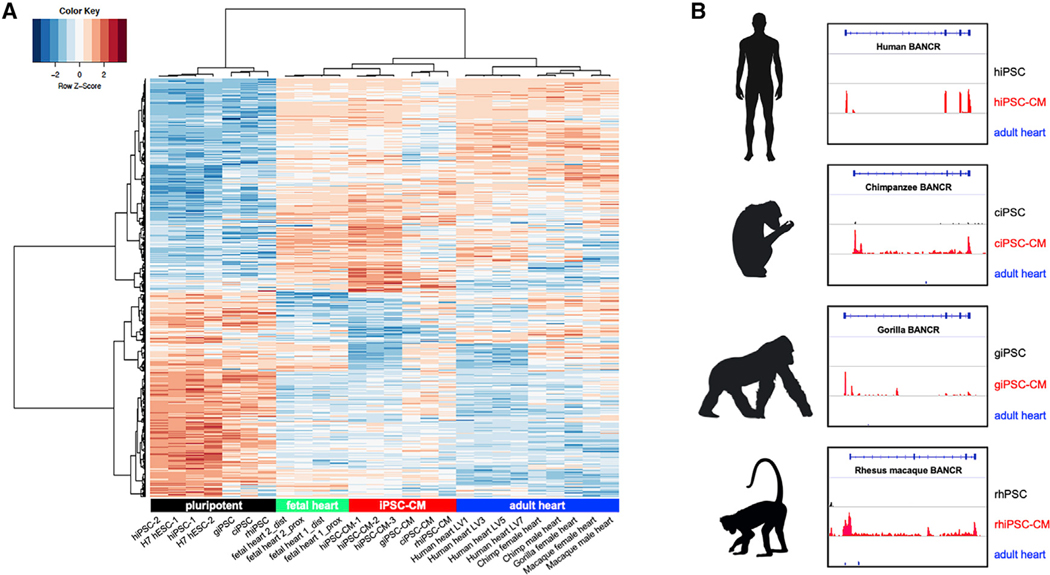
Because NHP fetal tissues were not available, we generated iPSCs from chimpanzee, gorilla, and rhesus macaque primary cells, followed by cardiac differentiation (Burridge et al., 2014) (Videos S1 and S2). Pearson correlation hierarchical clustering of RNA-seq data from undifferentiated primate iPSCs, derived cardiomyocytes, and adult primate heart tissues revealed clustering of iPSC-CMs from all species with human fetal heart (Figure 2A), as expected. We then confirmed BANCR transcription within NHP homologous regions: as in humans, NHP iPSC-CMs expressed BANCR, whereas the corresponding species-specific adult heart tissues and iPSCs do not (Figure 2B). These findings suggest that when MER41 elements inserted at a unique position on chromosome 1 of the ancestral marmoset genome, a de novo non-coding gene arose that is active specifically during heart development in primates. Importantly, MER41 was not epigenetically silenced or mutated, as is more typical after ERV invasion (Stoye, 2012).
Figure 2. BANCR Is Expressed in Human, Chimpanzee, Gorilla, and Rhesus Macaque iPSC-Derived Cardiomyocytes.
(A) Heatmap showing Pearson correlation hierarchical clustering of RNA-seq data for human (hiPSC), chimpanzee (ciPSCs), gorilla (giPSCs), rhesus macaque (rhiPSCs), and corresponding cardiac derivatives and adult hearts. iPSC-CMs from all species cluster with (human) fetal heart, as expected, and are distinct from adult heart tissues and pluripotent cells. Bonferroni corrected p value < 0.05.
(B) Homologous BANCR regions show iPSC-CM-specific transcription in human, chimpanzee, gorilla, and rhesus macaque.
BANCR Knockdown and Over-Expression in hESCs Elucidates Its Pro-Migratory Function in Fetal Cardiomyocytes
To understand why BANCR remained active in primate evolution, it was necessary to determine its functional role, if any, in cardiomyocytes. We knocked down BANCR (“BKD”) using lentiviral shRNA and over-expressed BANCR (“BOE”) using a custom lentiviral vector in hESCs (Figure S2A). After cardiac differentiation, we observed no significant differences in protein markers of cardiac differentiation (TNNT2) or proliferation (Aurora B), nor differences in sarcomeric immunostaining (TNNT2, Actinin) or electrophysiology by whole cell patch-clamp (Figures 3A and S2B-S2F). However, RNA-seq revealed enrichment of extracellular matrix organization, collagen formation, cell movement and migration, organization of cytoskeleton, and integrin interaction pathways in BOE hESC-CMs relative to BKD hESC-CMs (Figures 3B [left panel], S3A, and S3B). Integrin, interleukin-8 (IL-8), ephrin, and Rho family GTPase signaling genes were some of the canonical signaling pathways enriched in BOE hESC-CMs (Figure 3B, right panel). Proteins in the Rho/Rac subfamily of small GTP-binding proteins regulate cell migration and cytoskeletal reorganization (Raftopoulou and Hall, 2004), and we confirmed the activation of Rac1, a core member of this pathway, in BOE hESC-CMs and concomitant inactivation in BKD hESC-CMs by ELISA (Figure S3C). Because BANCR transcript is present in the nuclear fraction (though at lower levels than cytoplasm) of hESC-CMs (Figure S3D), we performed ATAC-seq to assess nuclear genome-wide chromatin states at promoter regions and found 10-fold more accessible promoters in BOE hESC-CMs relative to BKD hESC-CMs (Figure S3E). The most significant of these were pro-migratory ephrin receptor signaling genes (Kania and Klein, 2016) (Figure S3F), similar to our RNA-seq findings (Figure S3G).
Figure 3. BANCR Promotes Cardiomyocyte Migration.

(A) Confocal images of α-actinin and TNNT2 immunostaining of cardiac sarcomeres in BANCR knockdown (BKD), scramble control plasmid (SCR), and BANCR over-expressing (BOE) hESC-CMs. No differences in TNNT2 immunostaining were observed. Scale bar, 20 μm.
(B) Bubble plots of ingenuity pathway analysis (IPA) biofunctions (left, Benjamini-Hochberg-adjusted –log10p value > 22, Z score > 3) and canonical pathways (right, Benjamini-Hochberg-adjusted –log10p value >2, Z score > 3) generated from corresponding RNA-seq data. Relative to BKD hESC-CMs, cell movement biofunction pathways are significantly enriched in BOE hESC-CMs as well as integrin and Rho GTPase signaling canonical pathways.
(C) Wound healing assay demonstrates diminished gap closure after 1 week in BKD hESC-CMs compared with SCR control hESC-CMs. Bar graph shows quantitative data. Two-sided t test. n = 4 biological replicates, error bars are ± 1 SD. Scale bar, 0.5 mm.
(D) Circular hESC-CM micropattern arrays are an effective alternative method to the wound healing assay for assessing migration in differentiated cardiomyocytes. hESC-CMs are seeded on micropatterned stencils in tissue culture plates. Stencils are removed the next day, and the resulting circular hESC-CM clusters are cultured for 14 days followed by immunostaining and imaging by fluoroscopy to assess overall micropattern size. The right panel shows TNNT2/DAPI fluorescence (top) and photo (bottom) of a single well of 24-well plate with 48 beating micropatterns.
(E) BOE hESC-CM micropatterns have high migratory potential and thus are significantly larger than SCR and BKD hESC-CM micropatterns after 14 days of culture due to heightened outward radial cardiomyocyte migration. In contrast, BKD micropatterns are retracted due to diminished hESC-CM migratory potential. Bar graph shows quantitative data. Two-sided t test. n = 4 biological replicates, error bars are ± 1 SD. Dashed circles overlying DAPI images depict 2-mm-starting diameters.
(F) Circular micropattern of undifferentiated hESCs followed by 16 days of cardiac differentiation after removal of stencils, resulting in beating “cardioids” that mimic heart development in vitro. BOE cardioids showed dramatic outward radial movement of TNNT+ cardiomyocytes whereas SCR cardioids exhibit minimal radial migration. Scale bar, 1 mm.
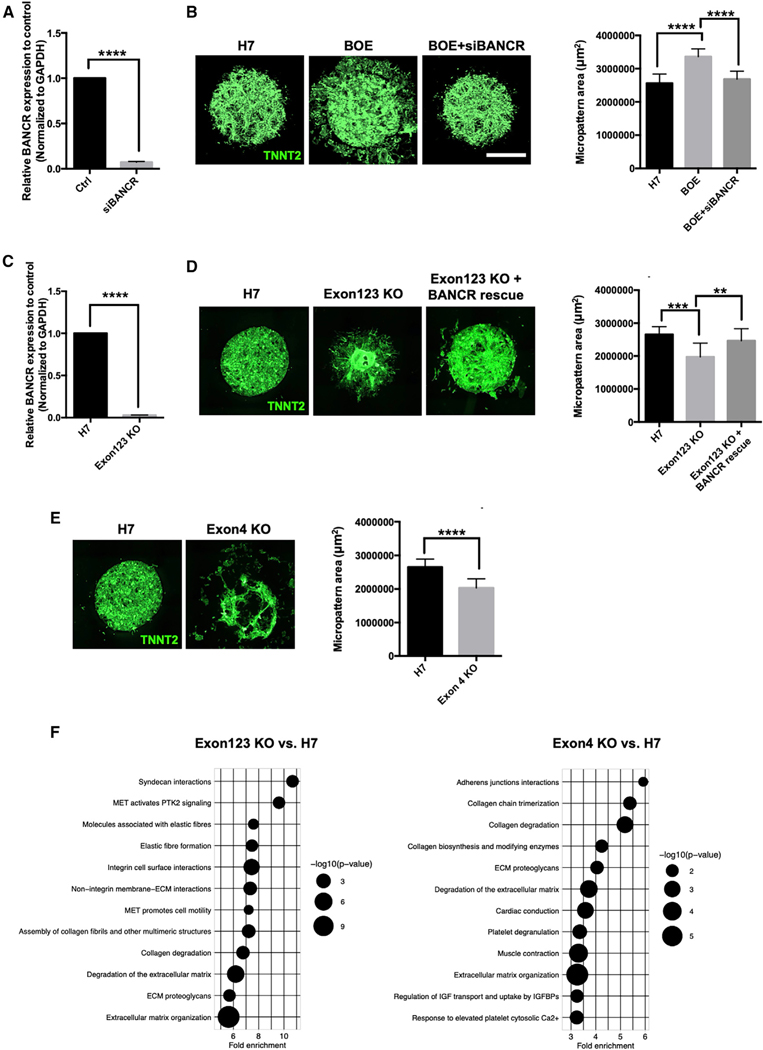
Based on these initial results, we suspected that BANCR may be regulating fetal cardiomyocyte migration, and we, therefore, performed a standard wound healing assay that revealed a subtle reduction in BKD hESC-CM migration after 1 week of culture (Figure 3C). hESC-CM migration is a known (Moyes et al., 2013) but poorly understood phenomenon in cardiomyocytes. To better elicit this phenotype, we modeled heart development in vitro using custom circular micropattern arrays (Figure 3D). Circular micropatterning approaches have been shown to recapitulate embryonic germ layer patterning (Warmflash et al., 2014) and cardiogenesis (Myers et al., 2013). Compared with scramble control (SCR), BOE micropatterns exhibited marked radial migration of TNNT2+ cardiomyocytes, whereas BKD micropatterns substantially retracted (Figure 3E). We also created circular micropatterns of undifferentiated hESCs followed by cardiac differentiation into two-dimensional, mixed cardiovascular lineage beating “cardioids” that more closely mimic heart development. BOE cardioids formed dramatic starburst patterns with “rafts” of outwardly moving TNNT2+ cardiomyocytes (Figure 3F; Videos S3 and S4), whereas BKD cardioids retracted into highly disorganized configurations (Figure S4). We also performed a rescue experiment utilizing siRNA knockdown of BANCR in BOE micropatterns that reduced the hyper-migratory potential of BOE cardiomyocytes to normal control levels (Figures 4A and 4B). Overall, based on these in vitro observations, as well as our gene expression and chromatin studies, we hypothesized that BANCR is a key regulator of fetal cardiomyocyte migration.
Figure 4. Reduced Migratory Potential of hESC-CMs after Targeted Deletion of BANCR Exons 1–3 and 4.
(A) qPCR showing significant knockdown of BANCR expression in H7 hESC-CMs 48 h after transfection with siRNA targeting BANCR (“siBANCR”). n = 3 biological replicates, error bars are ± 1 SD.
(B) TNNT2 immunostaining of beating micropatterns after 2 weeks of culture shows marked radial migration of cardiomyocytes in BANCR over-expressing hESC-CMs (BOE) that is reduced with knockdown of BANCR expression via siRNA. n = 2 biological replicates (32 micropatterns per group), error bars are ± 1 SD. Scale bar, 1 mm.
(C) qPCR shows significant loss of BANCR expression in Exon123 KO hESC-CMs relative to isotype WT control cells. Two-sided t test. n = 3 biological replicates, error bars are ± 1 SD.
(D) TNNT2 immunostaining of beating micropatterns after 2 weeks of culture shows marked loss of migratory potential in Exon123 KO hESC-CMs relative to H7 hESC-CM controls that is restored after ectopic BANCR transfection (“BANCR rescue”). Two-sided t test. n = 2 biological replicates (32 micropatterns per group), error bars are ± 1 SD.
(E) TNNT2 immunostaining of beating micropatterns after 2 weeks of culture shows reduced migration of Exon4 KO hESC-CMs relative to H7 hESC-CM controls. n = 2 biological replicates (32 micropatterns per group), error bars are ± 1 SD (see Figure S5D for RNA-seq data showing knockout of exon 4 expression).
(F) Biological pathway analysis of RNA-seq data from comparisons of Exon123 KO versus H7 hESC-CMs (left panel) and Exon4 KO versus H7 (right panel). Bubble plots show the top 12 significant pathways (FDR < 0.05, [Fold enrichment] > 1.0) for each comparison. Similar to BKD/BOE comparisons (Figures 3B and S3B), these pathways reveal significant enrichment of ECM organization, collagen degradation and integrin signaling and are consistent with a migratory phenotype. ****p < 0.00001. ***p < 0.0001, **p < 0.001.
All Four Exons within BANCR, Including Those that Are MER41-Derived, Have Biological Function
To determine whether MER41-derived exons of BANCR have higher, lower, or the same pro-migratory activity as the more well-conserved exon 4 in primates (see Figures 1D and 1E), we created two hESC lines using CRISPR-Cas9 (Figures S5A-S5C): (1) specific deletion of the BANCR MER41-derived exonic region (which includes exons 1, 2, and 3, hereafter referred to as “Exon123 KO”) and (2) specific deletion of the non-MER41-derived exonic region (which includes exon 4, hereafter referred to as “Exon4 KO”). After cardiac differentiation, Exon123 KO hESC-CMs have negligible expression of BANCR exons 1–3, as confirmed by qPCR primers targeting exons 1–2 (Figure 4C) but do maintain residual exon 4 expression by RNA-seq (Figure S5D). Similar to the BANCR shRNA (BKD) micropatterns shown previously, Exon123 KO hESC-CMs exhibit significantly reduced radial migration in circular micropatterns (Figure 4D). We performed a rescue experiment of Exon123 KO micropatterns via transfection of (wild type) BANCR plasmid and observed restoration of hESC-CM migratory potential. Likewise, targeted deletion of the more well-conserved exon 4 in hESCs resulted in residual expression of exons 1–3 but not exon 4 (Figure S5D) and reduced migration in hESC-CM circular micropatterns (Figure 4E). Biological pathway analysis of Exon123 KO and Exon4 KO hESC-CM transcriptomes relative to H7 wild-type hESC-CMs revealed enrichment of ECM degradation, collagen degradation, and integrin pathways (Figure 4F), highly similar to those seen with BANCR over-expression and knockdown (see Figures 3B and S3B). These findings suggest that all four exons, regardless of ERV (MER41) origin, contribute to the potency of BANCR in larger primates. Furthermore, based on the significant loss of migratory phenotype seen with specific knockout of BANCR MER41-derived exons in cardiomyocytes, we hypothesize that MER41 invasion of the ancestral genome played a role in larger primate heart development, particularly cardiomyocyte migration.
BANCR Is Regulated by an Upstream Enhancer in Cardiomyocytes
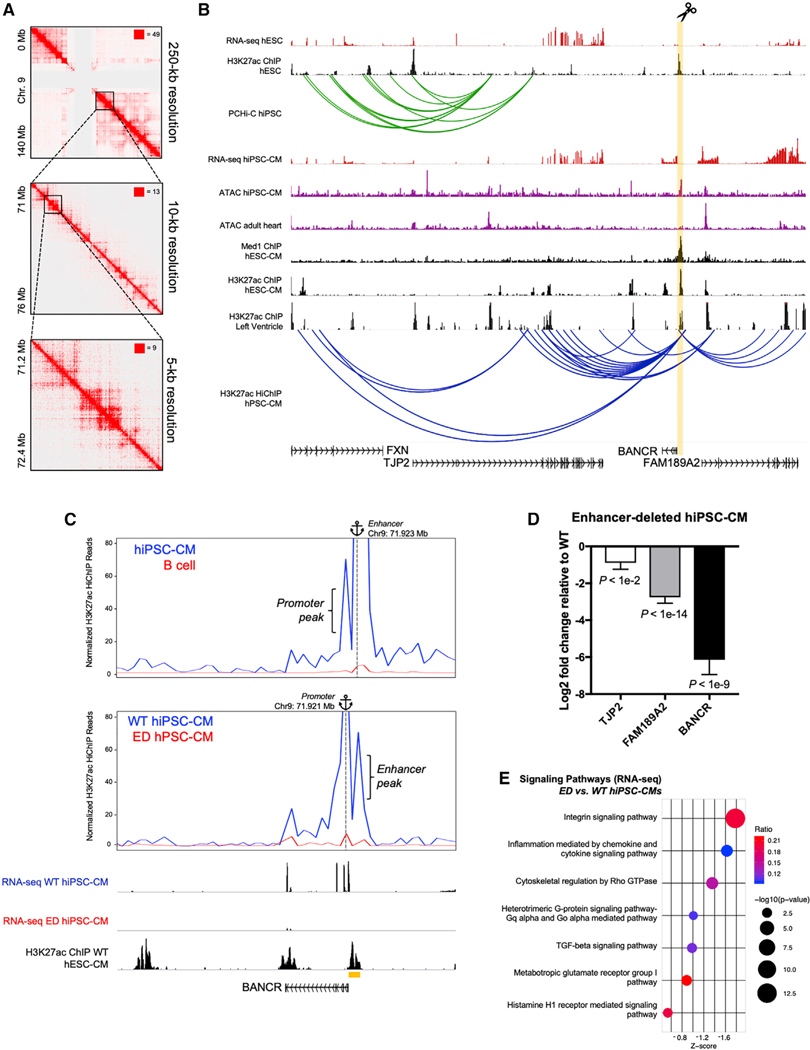
We next sought to understand how BANCR expression is regulated in primate cardiomyocytes so that it reaches maximal potency during development. Employing H3K27ac HiChIP (Mumbach et al., 2016), H3K27ac, and Med1 ChIP-, ATAC-, and RNA-seq, we found a highly active enhancer in pluripotent stem cell-derived CMs (Figure 5A, highlighted in orange) less than 1 kb upstream of BANCR that has significant physical interactions (“looping”) with distal loci including BANCR (Figures 5A and 5B) (note that pluripotent stem cells [PSCs] includes both hESCs and iPSCs in our experiments). Note that this enhancer is upstream of BANCR MER41 elements and, thus, does not contain MER41 sequence. It also exhibits minimal activity in PSCs and adult heart. This enhancer loops with BANCR and two nearby genes, tight junction protein 2 (TJP2) and FAM189A2, that represent a putative topologically associated domain (TAD). TJP2 en-codes a known structural protein within cardiomyocyte intercalated discs (Soni et al., 2016), whereas FAM189A2 has unknown function. The interaction between this enhancer and BANCR promoter is best visualized by virtual 4C (v4C), in which a specific genomic position is set (“anchored”), and all interactions occurring at that anchor are then visualized in two dimensions (Figure 5C, top panel). CRISPR-Cas9 deletion of this enhancer (Figures S5E-S5H) resulted in both loss of BANCR expression and interactivity at its promoter in enhancer-deleted (ED) hiPSC-CMs (Figure 5C, middle and bottom panels). The TAD genes TJP2 and FAM189A2 are likewise downregulated in ED hiPSC-CMs (Figure 5D). Finally, similar to our RNA-seq studies of BOE and BKD hESC-CMs, we found that ED hiPSC-CMs have altered integrin and Rho GTPase signaling (Figure 5E), underscoring the correlation of these pathways with BANCR expression. Of note, the BKD and ED cardiomyocyte transcriptomes did not significantly correlate overall, nor did the latter exhibit reduced migratory potential (Figure S5I). The additional genes regulated by this enhancer (TJP2 and FAM189A2) likely contribute to a different transcriptome and phenotype than those cardiomyocytes with specific knockdown of BANCR.
Figure 5. BANCR Is Regulated by an Upstream Enhancer in Cardiomyocytes.
(A) H3K27ac HiChIP identifies high-resolution chromosome conformation in human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) at the BANCR enhancer. Knight-Ruiz (KR) matrix-balanced interaction maps are shown at 250-, 10-, and 5-kb resolution. n = 2 biological replicates.
(B) H3K27ac HiChIP, promoter capture Hi-C (PCHi-C), Med1, and H3K27ac ChIP-seq, ATAC-seq, and RNA-seq data for the indicated cell and tissue types. Orange highlight demarcates BANCR putative enhancer that loops with nearby genes TJP2 and FAM189A2 in hPSC-CMs, as seen by H3K27ac HiChIP (blue loops), but is not present in PCHi-C of hESCs (green loops), suggesting stage-specific activity. Note that this enhancer was deleted in hiPSCs using CRISPR-Cas9 in the subsequent figure panels. This enhancer exhibits strong enhancer marker signal (H3K27ac and Med1 ChIP-seq) and open chromatin in hiPSC-CMs but not adult heart by ATAC-seq, indicating fetal cardiomyocyte-specific activity.
(C) Top panel: HiChIP interaction profile anchored on enhancer (chr9: 71.923 Mb) in hiPSC-CMs and human B cell lymphocytes. Middle panel: HiChIP interaction profile anchored on BANCR promoter (chr9:71.921 Mb) in wild-type hiPSC-CMs and ED hiPSC-CMs. Interactivity between promoter and enhancer is lost in ED hiPSC-CMs. Bottom panel: RNA-seq and H3K27ac ChIP-seq demonstrates diminished BANCR expression in ED hiPSC-CMs.
(D) BANCR, FAM189A2, and TJP2 are significantly downregulated in ED hiPSC-CMs relative to wild-type isotype control hiPSC-CMs, suggesting a TAD at this locus. Two-sided t test. n = 2 biological replicates, error bars are ± 1 SD.
(E) Bubble plot of significant signaling pathways (ED versus wild-type hiPSC-CMs) reveals enrichment of integrin and Rho GTPase signaling, similar to results for BOE versus BKD hESC-CMs (see Figure 2B). Benjamini-Hochberg-adjusted –log10 p value > 1.3.
BANCR Is Regulated by TBX5
With the discovery of the regulatory region of BANCR, we next sought to understand how BANCR is specifically expressed in a fetal heart. To address this question, we looked for key cardiogenic transcription factor binding at the enhancer using ChIP-seq and found TBX5 binding at a predicted motif (Figure 6A; Table S2). BANCR and its TAD genes TJP2 and FAM189A2 are significantly downregulated in TBX5-knockout hiPSC-CMs, further corroborating BANCR regulation by TBX5 (Figure 6B). To understand whether TBX5 regulation is a general phenomenon of MER41 transcripts genome-wide or is specific to BANCR, we quantified genome-wide MER41 longitudinal transcription (0 to 90 days of cardiac differentiation) and calculated the reads per kilobase million (RPKM) slope (β) at each genomic position. We found 5,484 MER41 genomic elements with at least one mapped read at any time point during differentiation, of which approximately 50% were intergenic and 50% intragenic (3% exonic and 47% intronic) (Figure 6C). Of these, 619 MER41 elements exhibited significant longitudinal up-regulation (false discovery rate [FDR] < 5%, β > 0), and protein-coding genes that overlapped significant intragenic elements were enriched for cardiac muscle cell action potential, conduction, contraction, and other biological processes (Figure 6D). We then overlaid hESC-CM TBX5 ChIP-seq data with these 619 MER41 elements and found 14 intragenic elements that were within 1 kb from putative TBX5 binding sites, suggesting coordinated regulation by this essential cardiac development transcription factor. Within this group of 14, three MER41 elements that originate from the first three exons of BANCR (BANCR.1, BANCR.4, and BANCR.5) had the highest longitudinal up-regulation during cardiac differentiation (Figure 6E). Thus, the three most salient MER41 elements revealed by this analysis all originate from BANCR (Figure 6F). We therefore conclude that, among MER41 transcripts, TBX5-regulated BANCR exhibits the strongest cardiac up-regulation.
Figure 6. BANCR Is Uniquely Regulated by TBX5.
(A) TBX5 ChIP-seq data for hPSC-CMs show TBX5 binding at BANCR’s upstream H3K27ac+ enhancer. A TBX5-binding motif is also present at this locus.
(B) TAD genes BANCR, FAM189A2, and TJP2 are significantly downregulated in TBX5 KO hiPSC-CMs relative to wild-type isotype control. n = 3 biological replicates, error bars are ± 1 SD.
(C) Flowchart showing the results at each step of RNA-seq and TBX5 ChIP-seq analysis of MER41 element expression in differentiating hiPSCs. hiPSCs were differentiated to cardiomyocytes and RNA-seq performed on days 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 14, 30, and 90 of differentiation. n = 3 biological replicates at each time point. RPKM slope (β) calculation denotes rates of up-regulation over time. Overlaid TBX5 ChIP-seq data with MER41 elements revealed three TBX5-bound MER41 elements (BANCR.1, BANCR.4, and BANCR.5) that originate from the first three exons of BANCR and exhibit significant up-regulation in cardiomyocytes.
(D) Bubble plot of Gene Ontology (GO) terms associated with protein-coding genes that overlap MER41 elements with FDR < 5% and β >0. The top ten pathways with lowest p values are shown and include cardiac muscle cell action potential, conduction, contraction, and other biological processes.
(E) β (rate of change in gene expression over 90 days of differentiation) versus FDR plot of 5,484 MER41 elements with overlaid TBX5 ChIP-seq data. SV, signal value. n = 3 biological replicates.
(F) Integrative genomics viewer (IGV) screenshot of BANCR and RepeatMasker elements that underlie three of its four exons. The MER41 element is composed of four -int elements and one -B LTR. Note that MER41 elements 2 and 3 are located within an intron and are transcriptionally silent. Time course expression of three MER41 elements within BANCR is shown in bottom panels. Error bars are ± 1 standard error.
BANCR Is Regulated by TEAD/YAP Signaling
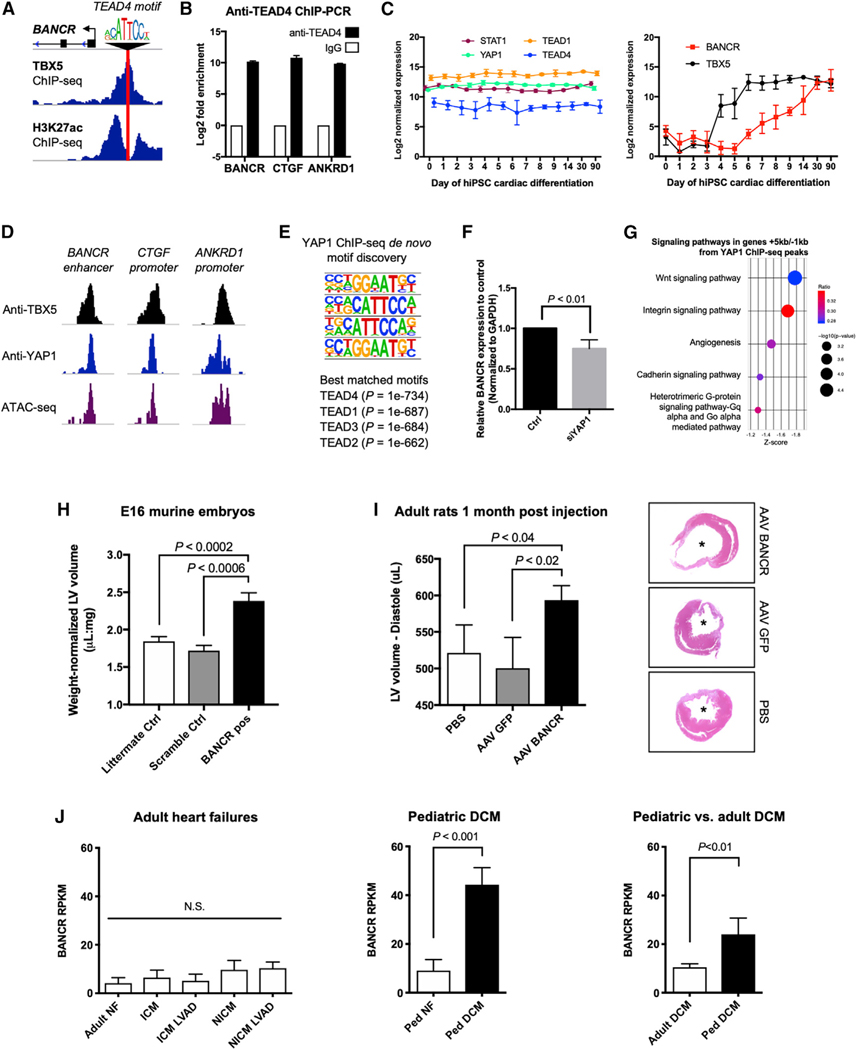
Interestingly, we also observed a TEAD4 binding motif at BANCR’s regulatory enhancer that overlaps with TBX5 ChIP-seq (Figure 7A). TEAD4 and its co-factor YAP1 are central downstream effectors of the evolutionarily conserved Hippo signaling pathway in addition to being relays of mechanical signals exerted by ECM rigidity and cell shape and play myriad roles in development, regeneration, mechanotransduction, cancer, and even organ size (Dupont et al., 2011; Totaro et al., 2018). TEAD4 ChIP-PCR confirmed the binding of TEAD4 protein at BANCR’s enhancer as well as at promoters for the canonical downstream effector genes ANKRD1 and CTGF (Figure 7B), suggesting that BANCR is regulated by TEAD4 in addition to TBX5. TEAD and YAP gene expression is unchanged during cardiac differentiation of hiPSCs, suggesting that TBX5 imparts fetal heart-specific expression to BANCR (Figure 7C). We also performed YAP1 ChIP-seq on hiPSC-CMs and found overlapping peaks with those of TBX5 and open chromatin regions (ATAC-seq) in BANCR, CTGF, and ANKRD1 (Figure 7D). As expected, the top four motifs enriched in YAP1 ChIP-seq peaks all belong to the TEA domain family (Figure 7E). Knockdown of YAP1 results in significantly reduced BANCR expression in hESC-CMs (Figure 7F), confirming its direct regulation by TEAD/YAP signaling. Pathway analysis of genes near YAP1 peaks (Figure 7G) revealed Wnt and integrin signaling, the latter of which was observed throughout our BOE/BKD/Exon123/Exon4/ED RNA-seq analysis for cardiomyocytes (see Figures 3B, 4F, and 5E). This indicates a shared mechanistic effect between BANCR and YAP1/TEAD4. Importantly, the interplay between Wnt and Hippo signaling has previously been shown to regulate heart size in mice (Heallen et al., 2011), and YAP1, TBX5, and β-catenin (Wnt signaling member) are known to form a tumorigenic transcriptional complex (Rosenbluh et al., 2012). Taken together, we propose that BANCR is a novel downstream effector gene of TEAD/YAP signaling.
Figure 7. BANCR Is Regulated by TEAD/YAP Signaling and Induces Ventricular Enlargement In Vivo.
(A) ChIP-seq data for hPSC-CMs shows a TEAD4-binding motif at BANCR TBX5-binding H3K27ac+ enhancer.
(B) TEAD4 ChIP-PCR revealsTEAD4 binding at BANCR’s enhancer as well as the promoters for CTGF and ANKRD1 (canonical downstream targets of TEAD/YAP signaling). n = 3 biological replicates, error bars ± 1 SD.
(C) Longitudinal log2 normalized expression for STAT1, YAP1, TEAD1, and TEAD4 (left panel), and BANCR and TBX5 (right panel) during cardiac differentiation of hiPSCs. n = 3 biological replicates, error bars are ± 1 SD.
(D) YAP1 and TBX5 ChIP-seq demonstrating co-occupancy at BANCR’s enhancer and the promoters for CTGF and ANKRD1 in hiPSC-CMs.
(E) Top four significant motifs in YAP1 ChIP-seq peaks are TEA domain family members.
(F) siRNA-mediated knockdown ofYAP1 in hESC-CMs causes significant reduction in BANCR expression after 24h.n = 3 biological replicates, error bars are ± 1 SD.
(G) Signaling pathways in genes +5 or −1 kb from YAP1 ChIP-seq peaks. Wnt and integrin signaling pathways are the most significantly enriched.
(H) Larger weight-normalized LV volumes calculated by magnetic resonance imaging (MRI) in BANCR-positive E16 embryos (n = 9 biological replicates) relative to scramble (n = 6 biological replicates) and PCR-negative (n = 13 biological replicates) littermate controls. One-way ANOVA, p value < 0.05, error bars are ± 1 SEM.
(I) Adult rats show increased LV diastolic dilation by echocardiography 1 month after infarction and AAV delivery of BANCR plasmids (n = 14 biological replicates) relative to GFP plasmids (n = 9 biological replicates) and PBS (n = 9 biological replicates). One-way ANOVA, p value < 0.05, error bars are ± 1 SEM. Representative H&E staining of explanted rat hearts are shown in the right panel (asterisks denote left ventricle).
(J) BANCR is highly expressed in pediatric dilated cardiomyopathy (DCM) but not adult heart failures. Left panel: BANCR expression in adult non-failing (NF) hearts (n = 8 individuals), ischemic cardiomyopathy (ICM) before and after left ventricular assist device (LVAD) (n = 8 patients), and non-ischemic cardiomyopathy (NICM) before and after LVAD (NICM LVAD) (n = 8 patients). p value < 0.05, one-way ANOVA, error bars are ± 1 SEM. Middle panel: BANCR expression in pediatric DCM (n = 7 patients) and age-matched healthy NF controls (n = 7 patients). Right panel: BANCR expression in pediatric DCM (n = 18 patients) and adult DCM (n = 33 patients). BANCR expression was significantly higher in pediatric versus adult DCM, suggesting high BANCR expression uniquely affects children. One sided t test, error bars are ± 1 SEM.
Given the role of TEAD/YAP signaling in development, organ size, and regeneration, we wondered whether BANCR could induce cardiac changes in living organisms. Transgenic E16 murine embryos carrying a murine β-myosin heavy chain (bMHC) promoter (primarily active in fetal mouse heart [Weiss and Leinwand, 1996]) driving expression of human BANCR have larger weight-normalized left ventricles than controls have (Figures 7H and S6). This finding is unlikely to be a result of cardiomyocyte hypertrophy as cellular nuclear:cytoplasm ratios are unchanged across all three study groups (BANCR-positive, Scramble- and Littermate-control embryos) (Figure S6E, left panel). Ki67 staining is also unchanged across the groups (Figure S6E, right panel), though precise quantification of Ki67+ cells can be inaccurate in tissue sections if the overall differences are small. Alternatively, increased cardiomyocyte proliferation in BANCR-positive embryos may have occurred before E16 when the embryos were harvested, which may be due to the murine bMHC promoter (that drives BANCR expression in the plasmid) having maximum activity during mesodermal or very early embryonic stages. We found additional evidence for an in vivo functional role of BANCR after left anterior descending (LAD) coronary artery ligation followed by direct cardiac injection of Adeno-associated virus (AAV) BANCR plasmids in rats. Compared with controls (AAV GFP and PBS injections), significant end-diastolic left ventricular (LV) dilation in the AAV BANCR cohort was observed 1 month after LAD ligation and peri-infarct injection (Figures 7I and S7).
Finally, in humans, BANCR is significantly upregulated in pediatric dilated cardiomyopathy (DCM) patients but not in adult patients with ischemic or non-ischemic cardiomyopathies (Figure 7J). Future studies are needed to determine the link between in vitro cardiomyocyte migration and in vivo cardiac dilation/enlargement and whether BANCR’s relationship to DCM in children is causative or associative. However, our results presented here suggest that BANCR may be a unique pathogenic factor in pediatric heart disease.
DISCUSSION
Our findings highlight the largely unknown activities of TEs that constitute nearly 50% of the human genome (Chuong et al., 2017; Göke and Ng, 2016; Kapusta et al., 2013). Within TE-derived genomic elements, ERVs such as MER41, which represent an historic record of germline retroviral infections, comprise a substantial portion. Many of these loci are unique to primates, thereby reflecting more recent evolution that cannot be captured in rodent models, nor can they be functionally studied in classical transgenic knockout models. More recently, human PSCs and primate iPSCs that model heart development in vitro, in combination with next generation sequencing (NGS) methods to assess expression, chromatin accessibility, and conformation, can interrogate primate-specific TE loci critical to cardiac development and disease. Here, we focus on one primate-specific ERV family, MER41, and use RNA-seq to identify hundreds of MER41 elements that are actively transcribed during cardiac differentiation. The profile of MER41 expression in hiPSC-CMs and fetal hearts is remarkably similar and reflects the general immaturity of hiPSC-CMs. In contrast, adult hearts express significantly fewer MER41 transcripts. Some of these MER41 transcripts are expressed from loci that are near TBX5 ChIP-seq bindings sites, suggesting coordinated activation during heart development. The fetal heart-specific lncRNA BANCR contains three of the most significantly up-regulated MER41 elements, and TBX5 binds an enhancer less than 1 kb from its transcription start site. 3D chromatin analysis using H3K27ac HiChIP experiments reveal that this same enhancer loops with two neighboring genes (TJP2 and FXN) as part of a putative TAD, which are entirely lost after CRISPR-Cas9 knockout of this same enhancer and results in significantly reduced BANCR expression.
BANCR originated in ancestral marmosets and has subsequently been conserved in all related primate lineages, including humans. Similar to human cells, NHP iPSC-CMs actively express BANCR, suggesting that not only is this genomic element conserved but so is its regulation and transcription. Surprisingly, BANCR knockdown and over-expression affect cardiomyocyte migration, an important phenotype in cardiac cells particularly during development. The predominant molecular pathway appears to be TEAD/YAP signaling, which has gained significant interest of late, in particular in the heart where its activity has been shown to control organ size and regeneration (Heallen et al., 2011; Totaro et al., 2018). Although it is currently unknown whether Hippo signaling or mechanotransduction specifically induces TEAD/YAP binding at BANCR’s enhancer, our results do suggest that BANCR is a downstream effector of TEAD/YAP signaling in primates, and its co-regulation by TBX5 induces its cardiac-specific expression.
In line with known TEAD/YAP signaling downstream effects, BANCR knockin murine embryos exhibit increased heart size compared with littermate controls. These findings raise an intriguing question regarding the potential for a relationship between fetal cardiomyocyte migration and cardiac size, particularly since the latter requires increased cellular proliferation that was not detected in our murine embryo experiments. As discussed in the Results, this apparent discrepancy may be due to technical issues in accurately quantifying proliferating cells in tissue sections and may also be due to cellular proliferation occurring before the time of embryo harvest (E16). However, we also did not observe increased proliferation in in vitro engineered BANCR cell lines (BKD and BOE) (Figure S2B), which argues against a proliferative mechanism mediated directly by BANCR. In terms of heart development, it is possible that the increased migratory potential of BANCR-expressing fetal cardiomyocytes concomitantly stimulates cardiomyocyte division in vivo through secondary mechanisms not seen in an artificial in vitro environment in which cells are removed from their native context. We do believe our findings raise the possibility of a connection between cardiomyocyte migration and heart size that warrants follow-up investigation.
We also performed AAV BANCR gene therapy of infarcted rat hearts and found that BANCR causes ventricular dilation. These rat data underscore our findings that BANCR is highly expressed in pediatric (but not adult) dilated cardiomyopathies, a devastating disease that results in enlargement of the heart and is the most common cause of heart transplants in children. BANCR may, therefore, present a therapeutic molecular target in young patients for whom heart transplant is the only curative therapy. Given our findings of an in vitro cardiomyocyte migratory phenotype that leads to an in vivo dilatory/enlargement cardiac phenotype with BANCR expression, this study presents a general argument for the utility of PSCs that should be emphasized. With access to species- and disease-specific PSCs, it is now possible to interrogate relatively pure differentiated cell populations in vitro to yield insights into development and disease that may not make immediate sense in the context of our historical understanding of these processes. This is particularly true given that nearly all knowledge of human heart development, particularly its genetic underpinnings, has been inferred from rodent and zebrafish models. BANCR is one example of a primate-specific genomic locus that, when studied in a context-appropriate primate cellular model system, has yielded phenotypic results (migration) that would likely have been missed if only rodent models were employed.
With regard to delineating a specific molecular mechanism, lncRNAs do present significant technical challenges, particularly highly repetitive ones such as BANCR that have endogenous retroviral origins and share sequence homology with elements throughout the genome. These repeat sequences, which constitute more than 70% of the BANCR transcript, prevented us from specifically and sensitively targeting BANCR with oligonucleotides for pull-down assays that could identify protein or DNA-binding moieties. Due to these technical challenges, we are primarily left with indirect measures of BANCR effects on cardiomyocytes, such as RNA-seq and ATAC-seq after knockdown/knockout/over-expression. These indirect measures consistently show perturbation of pro-migratory biological pathways, such as integrin signaling, extracellular matrix dynamics, collagen degradation, Rho GTPase signaling, etc. However, because it is impossible to conclusively confirm a direct link between BANCR and these pathways, we are reticent to make claims about the importance of any one in particular. Importantly, we are confident in the TEAD/YAP signaling pathway as a regulatory mechanism of BANCR expression, as there is clear interaction between these factors and BANCR’s enhancer. The involvement of TEAD/YAP regulation is robust and makes sense in light of the cardiomyocyte and heart phenotypes seen with BANCR expression and knockdown. Interestingly, integrin signaling is highly significant in the YAP1 ChIP-seq analysis of genes near to YAP1 peaks (that include the BANCR enhancer), which we interpret to suggest that BANCR and YAP1 overlap in regulating this important cytoskeleton/ECM pathway in cardiomyocytes.
In broad terms, BANCR emerged after an ancient primate viral infection led to the creation of this non-coding gene with unique, and presumably necessary, pro-migratory function in the developing primate heart. This potent pro-migratory effect on fetal cardiomyocytes, under regulation by TBX5 and TEAD/YAP signaling, emphasizes the ability of ERVs to rapidly invade host genomes and create functional elements that can alter development, disease, and possibly primate evolution (see Graphical Abstract). It is remarkable to consider the series of molecular and evolutionary events necessary to accomplish this feat, especially in light of the fact that a de novo gene (BANCR) was created that took advantage of existing cis-regulatory machinery to specifically affect cardiomyocyte development. Starting from a single germline insertion, the primate MER41 LTR copy-and-pasted itself to cover approximately 0.1% of the human genome over 40–65 million years of primate evolution. Several of these LTR copies, either stochastic or carefully orchestrated, inserted into a favorable locus that ultimately became useful for primate heart development. In mammalian genomes, ERVs have previously been shown to regulate the interferon (IFN) response, a branch of innate immunity, via generation of distributed IFN-inducible enhancers (Chuong et al., 2016). Overall, however, functional studies of TEs in primate-specific organismal development and evolution are lacking.
In conclusion, functional studies of primate-specific TEs such as BANCR/MER41 have been lacking due to ineffective organismal models. Human and NHP PSCs that model development and physiology in vitro, when combined with methods for assessing gene expression, chromatin accessibility, and conformation, have the potential to address many of the challenges in interrogating primate- and human-specific genomes. We expect studies such as ours will continue to elucidate how primate retroviral elements have fundamentally shaped our origins and our health.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Joseph C. Wu (joewu@stanford.edu).
Materials Availability
Plasmids and cell lines generated in this study are available upon request from the lab of Dr. Joseph C. Wu (Stanford University). Requests for the chimpanzee and gorilla iPSC lines used in this study should be sent to Dr. Sebastian Diecke (Berlin Institute of Health, sebastian.diecke@mdc-berlin.de).
Data and Code Availability
Sequencing data sets generated for this study are deposited in the Gene Expression Omnibus under accession number GSE111930. This study did not generate code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
NHP Tissue and Skin Fibroblast Isolation
Skin biopsies were taken on the medial side of the upper leg of a 42-year-old male Western lowland gorilla (G. gorilla) and 37-year-old male chimpanzee (P. troglodytes) under general anesthesia at the Zoological Garden Berlin, Germany. The male rhesus macaque (M. mulatta) (unknown age) skin biopsy was obtained from University of California, Davis (Zhao et al., 2018). Fibroblast isolation was performed with the Whole Skin Dissociation Kit, Human (130–101-540, Miltenyi Biotech) and an overnight incubation step using the gentleMACS Octo Dissociator (Miltenyi Biotech).
Human Fetal Tissue and Skin Fibroblast Isolation
All protocols were approved by the Stanford Institutional Review Board (IRB). Fetal tissues with estimated gestational ages of 16.4 (male) and 17.0 weeks (female) were purchased from StemExpress Foundation (Placerville, CA). Tissues were delivered to Stanford in RPMI and were immediately dissected, washed with PBS, then snap frozen in liquid nitrogen. Skin tissue was also acquired from each fetus, rinsed with PBS, and transferred to a 35 mm dish. Tissue was minced in 1 ml collagenase I (1 mg/ml in Dulbecco’s modified Eagle medium (DMEM), Invitrogen, Carlsbad, CA) and allowed to digest for 2 hr at 37°C. Dissociated dermal fibroblasts were pelleted and re-plated with DMEM containing 10% FBS (Invitrogen), Glutamax (Invitrogen), 4.5 g/L glucose (Invitrogen), 110 mg/L sodium pyruvate (Invitrogen), 100 U/mL penicillin (Invitrogen), and 100 ng/mL streptomycin (Invitrogen) at 37°C, 95% air, and 5% CO2 in a humidified incubator.
Human and NHP iPSC Reprogramming and Culture
For human and NHP reprogramming, passage 3–5 fibroblasts were infected with the CytoTune-iPS 2.0 Sendai Reprogramming Kit (A16517, Thermo Fisher) according to the manufacturer’s instructions. Single embryonic-like colonies were identified and manually picked on 15–25 days post infection. Human and NHP pluripotent stem cell lines were maintained as described (Burridge et al., 2014). ln brief, pluripotent stem cells were grown on Matrigel-coated plates (1:100 Matrigel in E8 medium) using chemically defined E8 medium. The medium was changed daily and cells were passaged every 3–4 days using EDTA or Accutase.
Transgenic E16 Murine Embryos
Plasmid vector construction of pRP[Exp]-Beta MHC>hBANCR and pRP[ncRNA]-Beta MHC>Stuffer_300bp and purification, pronuclear plasmid injection of FVB embryos, harvesting and genotyping of E16 embryos were performed by Cyagen Biosciences (Santa Clara, CA, USA). Fixed embryos were then sent to a histology service provider (Histo-Tec Laboratory, Hayward, CA) for H&E, Ki67, and Trichrome staining. Image analysis performed with ImageJ and Keyence BZ-X700 fluorescence microscope system.
Sprague Dawley Rats Used for BANCR AAV Studies
Ten-week old male Sprague Dawley rats (Charles River) were used for left anterior descending (LAD) coronary artery ligation to induce myocardial infarction, followed by BANCR AAV intra-cardiac injections. All animal procedures were approved by the Stanford University Institutional Animal Care and Use Committee.
METHOD DETAILS
Teratoma Formation
5×106 undifferentiated hiPSCs were harvested, pelleted, and re-suspended in 50 μL Matrigel (BD Biosciences). Cells in Matrigel were injected into the gastrocnemius muscles of 8–10 week-old male SCID Beige mice (n = 2) using a 28.5-gauge insulin syringe. Eight weeks after cell delivery, tumors were explanted for hematoxylin and eosin (H&E) staining.
Lentivirus Production and hESC Transduction
shRNA BANCR knockdown (BKD) and scramble control (SCR) plasmids were acquired from Paul Khavari lab at Stanford University (Flockhart et al., 2012). BANCR over-expression (BOE) plasmid pCDH-MSCV-BANCR-EF1-copGFP-T2A-Puro (.dna plasmid map available upon request, see also Extended Data 5a) was designed by the Neuroscience Gene Vector and Virus Core at Stanford. To package plasmid in lentivirus, HEK293T cells were plated in 10 cm dishes and transfected with target plasmid and packaging plasmids (pMD2G and pPAX2) using Lipofectamine 2000 (11668019, Life Technologies). Transfected HEK293T cells were incubated for 3 days and media collected daily. Media was then centrifuged at 3,000 g for 15 min to remove cells and cell debris. The supernatant was concentrated using PEG-it Virus Precipitation Solution according to the manufacturer’s protocol (LV810A, System Biosciences). H7 hESCs were seeded on 6-well plates and infected with the different lentivirus (BKD, SCR, and BOE). Cells were selected by puromycin (2 μg/ml) treatment for 2–3 days.
siRNA Transfection
80 μl of BANCR siRNA (Dharmacon) was added into a master mix of 3.2 μl DharmaFECT1 (Dharmacon) transfection reagent and 236.8 μl OptiMEM (Thermo Fisher Scientific) and incubated for 20 min before addition to a 6-well plate of hESC-CMs. The cell media was then changed after 24 hr and cells were harvested at 48 hr.
Adeno-Associated Virus (AAV) Production
The BANCR over-expression (BOE) plasmid used for lentivirus was modified for AAV by the Neuroscience Gene Vector and Virus Core at Stanford, then packaged into AAV serotype 9 (AAV9) vector particles. For control, green fluorescent protein expressing plasmid packaged into AAV9 was prepared by the virus core facility. Prior to injection into rat hearts, each AAV stock was diluted with PBS and aliquoted to 6.8e11 vg in 60 μL volume.
Human and NHP Cardiomyocyte Differentiation
Cardiomyocytes were differentiated from human and NHP pluripotent stem cells using our previously published methods (Burridge et al., 2014). Briefly, a differentiation medium consisted of RPMI-1640 media (11875–085, Life Technologies) supplemented with B27® minus insulin (A1895601, Life Technologies) (RPMI + B27 minus). To this medium, various small molecules were added over a week-long timetable as previously described (Burridge et al., 2014). On the first day (D0) of differentiation, 6 μM CHIR 99021 (C-6556, LC Laboratories) was added. On D2, the medium was aspirated and replaced with RPMI + B27 minus. On D3, the medium was aspirated and replaced with 5 μM of IWR-1 (S7086, Selleck Chemicals) in RPMI + B27 minus. The medium was replaced with RPMI + B27 minus on D5 and RPMI plus B27 supplemented with insulin (17504–044, Life Technologies) (RPMI + B27) on D7. Cardiomyocytes were maintained in RPMI + B27 with media change every other day. Cardiomyocytes generally began spontaneously beating between D7-D10. A glucose starvation step further purified cardiomyocyte culture. For NHP cardiac differentation, after 10–12 days of differentiation, cardiomyocytes were purified using a metabolic selection step based on lactate treatment. TBX5 knockout hiPSCs have been previously described (Karakikes et al., 2017).
Immunostaining
Pluripotent stem cells or differentiated cardiomyocytes were grown on coverslips and fixed in 4% paraformaldehyde at room temperature for 10 min, then blocked with 5% goat serum in PBS and 0.1% Tween 20 for one hr at room temperature. Fixed cells were stained with SSEA4 antibody (clone MC-813–70, R&D systems), OCT3/4 antibody (sc-9081, Santa Cruz Biotechnology), NANOG antibody (sc-33759, Santa Cruz Biotechnology), TRA-1–60 antibody (MAB4360, Millipore), Hoechst 33342 (Life Technologies), cardiac troponin T antibody (ab45932, Abcam), cardiac myosin antibody (MA1–83347, Thermo Fisher), or alpha-actinin antibody (ab18061, Abcam) at 4°C overnight. The sections were incubated with Alexa Fluor 488 or 594 secondary antibodies (Life Technologies) to visualize the specific stains. Image acquisition was performed on an Eclipse 80i fluorescent microscope (Nikon instruments, New York, USA), Keyence BZ-X700 (Keyence Corporation, Osaka, Japan), or confocal microscope (Zeiss LSM 710).
Fluorescent Activated Cell Sorting (FACS)
hiPSC-CMs were dissociated and stained with anti-Aurora B antibody (ab2254, Abcam), cardiac troponin T antibody (ab45932, Abcam), PE (P-2771MP, Thermo Fisher), and Pacific Blue (P-10993, Thermo Fisher) secondary antibodies using Fixation/Permeabilization Kit (554714, BD Biosciences). The stained cells were filtered through a 35 μm cell strainer snap cap and collected in a 5 ml FACS tube (Corning). The cells were analyzed on a BD Biosciences FACS Aria II instrument using FACSDiva software. The cells were gated on the basis of forward scatter and side scatter. Flow cytometric gates were set using parental cells.
ELISA
Protein lysates from day 20–30 hESC-CMs were assessed for Rho GTPase activation using G-LISA kits (Cytoskeleton, Inc): RhoA (BK124-S), Rac1 (BK128-S), and Cdc42 (BK127-S). Fluorescent signals were measured at 490 nm absorbance using a BioTek Synergy HTX plate reader.
Electrophysiology
Whole cell action potentials were recorded with the use of standard patch-clamp technique, as previously described (Burridge et al., 2014). Cultured hESC-CMs were dissociated using TrypLE for 10 min at 37°C, centrifuged at 200 g, suspended in the RPMI media supplemented with B27 (RPMI+B27) and filtered through a 100 μM cell strainer (BD Biosciences), and plated as single cells (1×105 cells per well of a 24-well plate) on No. 1 8 mm glass cover slips (Warner Instruments) coated with Matrigel (1:50 ratio) in RPMI+B27 media supplemented with 2 μM thiazovivin and allowed to attach for 48–72 hr, changing the media every other day. Cells were then placed in a RC-26C recording chamber (Warner) and mounted onto the stage of an inverted microscope (Nikon). The chamber was continuously perfused with the perfused with warm (35–37°C) extracellular solution of following composition: (mM) 150 NaC1, 5.4 KCl, 1.8 CaCl2, 1.0 MgCl2, 1.0 Na pyruvate, 15 HEPES, and 15 glucose; pH was adjusted to 7.4 with NaOH. Glass micropipettes (2–3 MΩ tip resistance) were fabricated from standard wall borosilicate glass capillary tubes (Sutter BF 100–50–10, Sutter Instruments) using a programmable puller (P-97; Sutter Instruments) and filled with the following intracellular solution: (mM) 120 KCl, 1.0 MgCl2, 10 HEPES, 10 EGTA, and 3 Mg-ATP. pH was adjusted to 7.2 with KOH. Single beating cardiomyocytes were selected and action potentials (APs) were recorded in whole cell current clamp mode using an EPC-10 patch-clamp amplifier (HEKA). Data were acquired using PatchMaster software (HEKA) and digitized at 1.0 kHz. The following are the criteria used for classifying observed APs into ventricular-, atrial-, and nodal-like hESC-CMs. For ventricular-like, the criteria were a negative maximum diastolic membrane (MDP) potential (< −50 mV), a rapid AP upstroke, a long plateau phase, AP amplitude > 90 mV, and AP duration at 90% repolarization/AP duration at 50% repolarization (APD90/APD50) < 1.4. For atrial-like, the criteria were an absence of a prominent plateau phase, a negative diastolic membrane potential (< −50 mV), and APD90/APD50 > 1.7. For nodal-like, the criteria were a more positive MDP, a slower AP upstroke, a prominent phase 4 depolarization, and APD90/APD50 between 1.4 and 1.7 (Burridge et al., 2014). The ventricular-like cardiomyocytes were selected for assessing the effects of norepinephrine (NE; 1 μM).
Wound Healing Assay
hESC-CMs were glucose-starved (before and after re-plating) prior to using the CytoSelect 24-Well Wound Healing Assay (Cell Bio-Labs) according to the manufacturer’s instructions. On days 1 and 7, 5–8 fields of view were image-captured per scratch of BKD and SCR hESC-CM cultures. On day 7, cultures were fixed and stained for alpha-actinin and DAPI. Image J was used for quantitative image analysis.
Micropatterns
Single cell suspensions of hESC lines were prepared by harvesting cells that had reached approximately 75–80% confluence to ensure cells were in the optimal growth phase upon re-plating. Optimal cell seeding density was determined for all tested lines and was chosen based on the density that would allow the cells to grow over the course of 48 hr into the geometry of stencils (circular stencils with 2 mm holes for patterning single or arrayed colonies in each well of a 24-well plate) prior to cardiac differentiation, as described (Myers et al., 2013). On day −2, 5–10×103 cells were incubated on stencils in 37°C for a minimum of 1 hr to allow cells to settle onto a previously deposited Matrigel matrix within stencil holes. After this time, 500 uL of E8 medium + 10 uM ROCK Inhibitor (Y-27632, Sigma Aldrich) were added per well and stencils then carefully removed with forceps, leaving a 2 mm single colony or 48 arrayed colonies in each well. Media was changed the following day and cells were allowed to fill in each stencil over the following two days. The confluence of the cells was carefully tracked to ensure that cells reached 95–100% confluence at the start of differentiation. The cardiac differentiation protocol used for micropatterns was identical to that for normal cell cultures (Burridge et al., 2014).
Gene Editing of hiPSCs (Enhancer Region)
CRISPR/Cas9 gene editing was performed using two single-guide RNA (sgRNAs) flanking the enhancer region upstream of BANCR (Chr9: 71.293 Mb). The guide DNA oligos were designed using a web-based tool (crispr.mit.edu/) and chosen based on a high score for on-target binding and the lowest off-target score. The gRNAs were cloned into the pSpCas9(BB)-2A-GFP vector (PX458; a gift from Feng Zhang; Addgene plasmid #48138) using annealed reverse complementary guide DNA oligos. The sequences of the sgRNAs were gRNA_3’: AGAAGGTCGGCACAAGATAT and gRNA_3’: TTAGCTCTGGAATTTTCCCC. Two CRISPR/Cas9 vectors (1 ug each) were transfected in hiPSCs (SCVI015) using the Lipofectamine Stem Transfection Reagent (Thermo Fisher Scientific). The cells were dissociated using TrypLE express 1× (Thermo Fisher Scientific) and GFP+ cells were sorted by flow cytometry 24 hr post-transfection. GFP+ cells were seeded at a density of 1,000 cells per well in a 6-well plate to generate clonal isolates. Ten to fourteen days after seeding, individual clones were picked for genotypic screening by PCR. (FW: CTGTGTCTTCTCTTCTGTCTCTTG, RV: CTAAACCCACCAACACCAAGA) Knock-out clones were confirmed by Sanger sequencing.
Enhancer Knockout Region (1,026 bp)
AGATATTCCCTTCCTCCACTCTGTCATGTTAATAGCCCACCTGGGGAAAATTCCAGAGCTAAAAAAGTAGCCAGGTGGCTCTTTTTTTGTTGTTGTTTTAAACAAAAGATCTCAAAGTGGGCCATATCGCCCATCTCATTGTTTGCTCCTTCCACATAAGTGCTAATGTGTTTGATTATATTCTGCTCCAAATTCCACTACAACCACTAATATAAGGCACATACAGAGAATTAACTGTGTCCAAACAGTTCTTTCCTCATGCCCTCATTCTTCTTGAATTTTAAATGCCTGTTGAACAGAAGCATTAGAGTGTTTATTTTCTGAAAACCTTCTGGAGAGCAATCACCTTGTCTTTTCAAAGAAGCCATACTGTGTCTGGTTTTTCCATTCCAAAAATAACATAGAAATTCTTAGAGACAGTCATGTGGCAGAGAACCAAGAGCTGGAATGCAAAGCTTTTGTGTTTCTTGTGGAAGGCCTCTTGAGTACTTAAGTAGTTACATTCCAGCTTCACAGGGAAGCTGAAGAGGGGCAAATGCAGTAAGGAGAGTACTTGAGGCAGGCCTGGTATTTCTCTTCCATGAAATGCCAAGTTTGGTCATGTCCTGGAGAAGTTAGGGTGCCCCACAAGAGGTGTTGGTACCGCTGTAGTCTGTGAGGCTACCTGAAGGGAGACCACGTCTTCCGAGAAATCTAATCA CCAGCTGCCTACAACTTCCCCAAACACCCACCTGAACACTGTTTTACTCCTGACCACCCTCACTCTGCAGAATCACATGACAGCCCAACATTGTTACTTCACCTAGTGGCAAGTATGCACATCAATTTTTTCCATTCCCTTTCCTACCGTAGTCTTTCtgtcagagatgtttgaaccagagcaactccatctcgaataagaactgggtaaaataaggctgagacctactgggctgcatttccaggaggttaggcattctaagtcacaggatgagatagaaggtcggcacaagatataggtcacaaaggccttgctgataaaatagc
Gene Editing of hESCs (BANCR Exons)
CRISPR/Cas9 gene editing was performed using two pairs (5’ and 3’) of gRNAs flanking (1) exons 1–3 (“BANCRexon123”) and (2) exon 4 (“BANCRexon4”). The guide DNA oligos were designed using a web-based tool (crispr.mit.edu/) and chosen based on a high score for on-target binding and the lowest off-target score. The gRNAs were cloned into the pSpCas9(BB)-2A-GFP vector (PX458; a gift from Feng Zhang; Addgene plasmid #48138) using annealed reverse complementary guide DNA oligos. CRISPR/Cas9 vectors (1 ug each) were transfected in H7 hESCs using the Lipofectamine Stem Transfection Reagent (Thermo Fisher Scientific). The cells were dissociated using Gentle Cell Dissociation Reagent (STEMCELL Technologies) and GFP+ cells were sorted by flow cytometry 24 hr post-transfection. GFP+ cells were seeded at a density of 1,000 cells per well in a 6-well plate to generate clonal isolates. Ten to fourteen days after seeding, individual clones were picked for genotypic screening by PCR. Knock-out clones were confirmed by Sanger sequencing.
| Primer name | Sequence | Note |
|---|---|---|
| BANCRexon123_5’F_CRISPR | ATAACCATAAGTATCAACAG | BANCR exon 123 KO 5’ CRISPR |
| BANCRexon123_5’R_CRISPR | CTGTTGATACTTATGGTTAT | BANCR exon 123 KO 5’ CRISPR |
| BANCRexon123_3’F_CRISPR | AAGAAGACTTACCATGAACT | BANCR exon 123 KO 3’ CRISPR |
| BANCRexon123_3’R_CRISPR | AGTTCATGGTAAGTCTTCTT | BANCR exon 123 KO 3’ CRISPR |
| BANCRexon4_5’F_CRISPR | AGAAATGTGCACAGTAAAAC | BANCR exon 4 KO 5’ CRISPR |
| BANCRexon4_5’R_CRISPR | GTTTTACTGTGCACATTTCT | BANCR exon 4 KO 5’ CRISPR |
| BANCRexon4_3’F_CRISPR | GCTGAAACCAAAGTAGACAA | BANCR exon 4 KO 3’ CRISPR |
| BANCRexon4_3’R_CRISPR | TTGTCTACTTTGGTTTCAGC | BANCR exon 4 KO 3’ CRISPR |
| BANCR_EcoRI_exon4_F | GGGGGAATTCCTGCTGAGAAGTTCAGAGTCAA | exon4 overexpression plasmids F primer |
| BANCR_NotI_R | GGGGGCGGCCGCGGATCCGATTTAAATT | exon4 overexpression plasmids R primer |
| CRISPR_31F | AAGAAGCCAGCCTAAACCC | Verify CRISPR 31 clones |
| CRISPR_31R | TCATCTTTATGCCCATGAGTACC | Verify CRISPR 31 clones |
| CRISPR_4F | CTTTGAGCAATGTCTGGTTGTC | Verify CRISPR 4 clones |
| CRISPR_4R | GCTCTTGTAGAACT CAGCAGAA | Verify CRISPR 4 clones |
| BANCR_qPCR_F | TTCCTTAGGGTCAGGGGTCT | BANCR qPCR primers |
| BANCR_qPCR_R | GATTGGGACCCTTTTCTGGT | BANCR qPCR primers |
7-Tesla Magnetic Resonance Imaging (MRI)
After formalin fixation, E16 mouse embryos were immersed in 8 mM gadolinium solutions (0.8 mmol/kg, Gadavist Bayer, Germany) for two weeks. They were then placed in a Varian millipede coil and scanned by with a 7T Bruker horizontal bore scanner (Bruker, Billerica, MA) with a shielded gradient system (600 mT/m). Embryonic hearts (n = 28) were imaged with a 3D spoiled gradient echo sequence: echo time (TE) 10 ms, repetition time (TR) 30 ms, flip angle 60°, number of signal average 7, field of view (FOV: 26×26×26 mm3, matrix size of 512 mm3, isotropic resolution of 51×51×51 mm3 per voxel, and no slice gap). Post-image acquisitions, both endocardial and epicardial borders of the embryonic hearts were manually segmented on MR images using Segment (http://segment.heiberg.se) to estimate myocardial volumes.
LAD Ligation and AAV Virus Intra-Cardiac Injection
Ten-week old male Sprague Dawley rats (n = 32) were anesthetized with isoflurane and then subjected to LAD ligation. Briefly, rats were intubated and their chests opened at the left intercostal space to expose the heart and LAD artery. 5–0 sutures were used to permanently ligate the LAD artery. With the chest still open, 60 uL of AAV BANCR or GFP plasmids (6.8×1011 vector genomes (vg)) or PBS was directly injected into the left anterior wall of each rat followed by closure of the chest with 4–0 sutures. Rats were then extubated and monitored until fully awake and active. Echocardiography was performed 30 days later, followed by heart extraction and H&E and Trichrome histology (Animal Histology Services, Stanford University). Anti-GFP antibodies (ab6673, Abcam) were used for GFP immunostaining of unstained slides.
Rat Echocardiography
Cardiac function of rats at 30 days after infarction and AAV or PBS cardiac injection was evaluated by transthoracic echocardiography (Vevo 2100 Imaging System, VisualSonics). Rats were anesthetized with 1% to 2% (v/v) isoflurane in oxygen and left ventricle internal dimensions were measured in systole and diastole from short axis M-mode images, following the guidelines of the American Society of Echocardiography (Sahn et al., 1978). Left ventricle systolic function was quantitatively measured using Vevo Lab software.
Real-time Quantitative PCR (qPCR)
RNA was isolated using miRNeasy kit (Qiagen) following the manufacturer’s protocol. RNA from each sample was then reversed transcribed with iScript cDNA Synthesis Kit (Bio-Rad) or High Capacity RNA-to-cDNA Kit (Life Technologies). qPCR was performed on a StepOnePlus System (Life Technologies) using iQ™ SYBR® Green Supermix (Thermo Fisher) and GAPDH endogenous control (Thermo Fisher). BANCR PCR primers: TTCCTTAGGGTCAGGGGTCT (Forward), GATTGGGACCCTTTTCTGGT (Reverse).
Nuclear/Cytosol Fractionation
Nuclear and cytoplasmic RNA fractions were prepared from H7 wildtype hESC-CMs using Cytoplasmic and Nuclear RNA Purification Kit (21000, Norgen Biotek) according to the manufacturer’s instructions.
RNA-seq Analysis (All Other Samples)
In addition to in-house generated RNA-seq data sets, the following published data sets were downloaded from the Sequence Read Archive database (NCBI): H9_1 (SRX027480); H9_2 (SRX243742); H1_1(SRX007165); H1_2 (SRX171595); human fetal cardiac fibroblasts (Jonsson et al., 2016) (SRP107817); female and male chimpanzee hearts (SRR306819, SRR306820); female gorilla heart (SRR306804); and female and male rhesus macaque hearts (SRR306782, SRR306783) (Brawand et al., 2011). Additionally, we included RNA-seq data from (GSE81585) for hiPSC-CMs (Days 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 14, 30, and 90 of differentiation, n = 3 biological replicates) (Churko et al., 2018), from human adult left ventricles (LV1, LV3, LV5, LV7), and from TBX5 knockout hiPSC-CMs (Karakikes et al., 2017). Human cardiomyopathy data sets were downloaded from GSE99321 (pediatric DCM (Tatman et al., 2017)), SRP108128 (pediatric vs. adult DCM (Patel et al., 2017)), and GSE46224 (adult heart failures (Yang et al., 2014)). RPKM values for supplementary published adult and fetal tissue RNA-seq datasets (Cabili et al., 2011; Duff et al., 2015; Fagerberg et al., 2014; Szabo et al., 2015) (Figure S1F) were obtained from NCBI Gene. For all in-house and public datasets, raw RNA-seq reads were trimmed for quality control by TrimGalore version 0.4.2. The trimmed RNA-seq reads were mapped to hg19, gorGor3, panTro4 or rheMac3 using STAR software. Uniquely mapped reads were counted with featureCounts based on Gencode v19. Each gene’s reads per kilobase million (RPKM) value was calculated using the following the formula RPKM=(10^9*C)/(T* L), where C is the count of reads mapped to the gene, T is the total mapped reads count, and L is the gene length. Unique mapped reads are kept for generating tiled data file (TDF) files.
LncRNA Analysis
Human Body Map large intergenic non-coding RNAs (lincRNAs) and transcripts of uncertain coding potential (TUCP) were downloaded from the UCSC Genome Browser website. The Human Body Map catalog was generated by integrating previously existing annotation sources with transcripts that were de novo assembled from RNA-seq data across 24 tissues and cell types (Cabili et al., 2011). We then constructed a unique transcript for each gene by merging the overlapped exons into a unique exon. LncRNA read counts, RPKM, and significant differential expression were determined as described (He et al., 2016). 1,343 lncRNAs showing differentially expressed among 9 samples (one-way ANOVA, P value < 0.05, total lncRNAs: 2,894) were used for the lncRNA heatmap.
ERV Expression Analysis
For genome-wide ERV analysis, uniquely mapped RNA-seq reads were assigned to ERV regions using hg19 RepeatMasker annotation. To calculate global ERV sub-family expression, assigned read counts and genomic lengths of all loci for each sub-family were added to calculate RPKM. Each local ERV repeat element was also calculated individually and linear regression was performed in R software.
Comparison of Human and NHP Transcriptomes
Differentially expressed genes between hiPSCs and hiPSC-CMs were determined using DESeq2. Afterwards, 2,022 genes with Bonferroni-corrected P values < 0.05 were then selected for transcriptome profile comparison across primate samples. Of these genes, 1,975 were successfully converted to gorGor3 (gorilla reference genome), panTro4 (chimpanzee reference genome), and rheMac3 (rhesus macaque reference genome) by LiftOver using parameters with default values. These 1,975 genes were used for hierarchical clustering in the heatmap.
CHROMATIN IMMUNOPRECIPITATION (ChIP) ANTIBODIES
The anti-YAP1 (Novus; NB110), anti-TEAD4 (Abcam; ab58310), anti-STAT1 (Cell Signaling; 9172), normal mouse IgG (Santa Cruz; sc-2025), and normal rabbit IgG (Santa Cruz; sc-2027) antibodies were used in this study.
ChIP qRT-PCR
qRT-PCR was performed by using iQ® SYBR™ Green Supermix (Bio-Rad; 170–8880) and CFX96™ real-time PCR detector (Bio-Rad). 1/50th of the ChIP sample or 0.01% of input chromatin per PCR were used to run qRT-PCR. Amplifications were performed in triplicate and standard deviation was calculated from the triplicates, and error bars are indicated accordingly. The following primers were used:
RNA-seq Analysis (BOE, BKD, and SCR hESC-CMs)
Total RNA was submitted to a core facility (Stanford Functional Genomics) for library preparation and Illumina sequencing. Each condition had three biological replicates. Each individual sample had 26–35 million 75 bp long single-end reads. FastQC (version 0.11.4) was used for sequencing quality assessment. Reads were then aligned to the human (hg19) transcriptome using Bowtie version 2.2.7 with splice junctions being defined in GTF file (obtained from UCSC). An average of 58% of reads was aligned to the reference transcriptome. Expression at gene level was determined by calculating reads per kilo base per million aligned reads (FPKM) as well as raw count using RSEM (version 1.2.30). Differently expressed genes with fold-change were further detected by DEseq2 version 1.10.1 for two comparable conditions. Gene set enrichment analysis (GSEA, version 2.2.0) was used and the gene ontology and pathway database were downloaded from the Bader Lab (http://download.baderlab.org release March_24_2016). RSEM was further used to quantify relative abundance of isoforms for each gene. Ingenuity Pathway Analysis (IPA Inc, Redwood city, CA) and PANTHER classification system were employed to determine statistically significant functional categories and pathways within lists of significant genes.
BANCR promoter: Forward primer 5’ TTAGCGTATAATGAGCAGTGAGG 3’ and reverse primer 5’ GAGATAGAAGGTCGGCACAAG 3’
BANCR enhancer: Forward primer 5’ CTGCCTCAAGTACTCTCCTTAC 3’ and reverse primer 5’ CCAAGAGCTGGAATGCAAAG 3’
ANKRD1: 5’ GAGGGGAGGACAAGCTAACC 3’ and reverse primer, 5’ CGATGTGATCACCACCAAAG 3’
CTGF: 5’ GCCAATGAGCTGAATGGAGT 3’ and reverse primer 5’ CAATCCGGTGTGAGTTGATG 3’
ChIP-seq
Anti-YAP1 antibody was incubated with Dynabeads (Life Technology; 10003D) for 12 hr at 4°C. A small portion of the crosslinked, sheared chromatin was saved as the Input, and the remainder was employed for the immunoprecipitation using antibody conjugated Dynabeads. After overnight incubation at 4°C, the incubated beads ware rinsed with sonication buffer (50 mM HEPES, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 0.5 mM PMSF), high salt buffer (50 mM HEPES, pH 7.9, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 0.5 mM PMSF), and LiCl buffer (20 mM Tris, pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 0.5 mM PMSF). The washed beads were incubated with an elution buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 1% SDS, 50 mM NaHCO3) for 1 hr at 65°Cand then de-crosslinked with 5 M NaCl for overnight at 65°C. The immunoprecipitated DNA was treated with Rnase A and Proteinase K, and purified by ChIP DNA clean and concentrator (Zymo Research; D5205). The sequencing library was generated using NEBNext Ultra DNA library prep kit (NEB; E7645S). Samples were then submitted to Stanford Functional Genomics Facility for Illumina sequencing. FASTQ files were mapped to Hg19 by Bowtie. Additional ChIP-seq data sets were included from GSE85628 (Ang et al., 2016) Med1 hESC-CMs Day 32 (SRR4032212); GSE81585 (Churko et al., 2018) TBX5 hiPSC-CMs; GSE107785 (Lee et al., 2018) H3K27ac hESC-CMs Day 9; and H3K27ac Left Ventricle (GSM908951) (NIH Roadmap Epigenomics Mapping Consortium).
ChIP-seq Data Processing
All sequencing reads were mapped to hg19 version of human reference genome using Bowtie1.2.2 with parameters –best, –strata and –m 1 to allow only unique alignment. PCR duplicates are removed using samtools 1.8 “rmdup” command (http://samtools.sourceforge.net/). Peaks were called using MACS2 version 2.1.1 with “–nomodel –extsize 147 –q 0.01”. Motif analysis on the peak regions was performed using HOMER (http://homer.ucsd.edu/homer/motif/) “findMotifsGenome.pl” script with –size given option. We used “–log10 (p-value)” to rank the enrichment level of TF motifs.
HiChIP
The HiChIP protocol was performed as previously described (Mumbach et al., 2016, 2017) for H7 hESC-CMs (n = 1), hiPSC-CM (n = 1), and enhancer-deletion hiPSC-CM (n = 1). In brief, up to 15 million crosslinked hESC-CMs or hiPSC-CMs were resuspended in 500 μL of ice-cold Hi-C lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.2% NP-40, 1X Roche protease inhibitors) and rotated at 4°C for 30 min. The nuclei were pelleted and washed once with 500 μL of ice-cold Hi-C lysis buffer. The pellet was resuspended in 100 μLof 0.5% SDS and incubated at 62°Cfor 10 min. 285 μL of water and 50 μLof 10% Triton X-100 were added and samples were rotated at 37°C for 15 min. 50 μL of NEB Buffer 2 and 15 μL of 25 U/μL MboI restriction enzyme (NEB, R0147) were then added and sample was rotated at 37°C for 2 hr. The MboI was then heat inactivated at 62°C for 20 min. 52 μL of incorporation master mix was then added: 37.5 μL of 0.4 mM biotin-dATP, 4.5 μL of a dCTP, dGTP, and dTTP mix at 10 mM each, and 10 μL of 5 U/μL DNA Polymerase I, Large (Klenow) Fragment. The reactions were rotated at 37°C for 1 hr. Wethen added 948 μL of ligation master mix: 150 μL of 10X NEB T4 DNA ligase buffer with 10 mM ATP, 125 μL of 10% Triton X-100, 3 μL of 50 mg/mL BSA, 10 μL of 400 U/μL T4 DNA Ligase, and 660 μL of water. The reactions were rotated at room temperature for 4 hr. After proximity ligation, the nuclei were pelleted and the supernatant removed. 880 uL of Nuclear Lysis Buffer (50 mM Tris-HCl pH 7.5, 10 mM EDTA, 1% SDS, 1X Roche protease inhibitors) was added and the samples were sheared using a Covaris E220 using the following parameters: Fill Level = 10, Duty Cycle = 5, PIP = 140, Cycles/Burst = 200, Time = 4 min. Clarified samples were transferred to Eppendorf tubes and 2X volume of ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 7.5, 167 mM NaCl) was added. Antibody was then added (4 μg anti-H3K27ac, Abcam, ab4729) and incubated overnight at 4°C. Chromatin–antibody complexes were captured with 34 μl of Protein A beads (Thermo Fisher). The post-ChIP DNA Qubit quantification ranged from 50–100 ng. The estimated amount of Tn5 (Illumina) needed to generate libraries was based on post-ChIP quantifications, as previously described (Mumbach et al., 2016). Libraries were size selected by PAGE purification (300–700 bp) and sequenced on an Illumina HiSeq 4000 instrument on PE75 mode to an average depth of 600 million total reads. HiChIP paired-end reads were aligned to hg19 genome using the HiC-Pro pipeline (Servant et al., 2015). Default settings were used to remove duplicate reads, assign reads to MboI restriction fragments, filter for valid interactions, and generate binned interaction matrices. HiC-Pro filtered reads were then processed into a .hic file using the hicpro2juicebox function. HiChIP interaction maps were visualized using Juicebox software at 250 kb, 10 kb, and 5 kb resolutions as indicated in each analysis (Durand et al., 2016; Rao et al., 2014). Virtual 4C plots were generated from dumped matrices generated with Juicebox, and published H3K27ac HiChIP data for B cells (Mumbach et al., 2017) were included as negative control. The Juicebox tools dump command was used to extract the chromosome of interest from the .hic file. The interaction profile of a specific 5 kb or 10 kb bin containing the anchor was then plotted in R. Fit Hi-C loop calls were loaded into the WashU Epigenome Browser along with corresponding ATAC-seq and H3K27ac ChIP-seq datasets (Ay et al., 2014). We also included a track for promoter capture Hi-C of hiPSCs (Montefiori et al., 2018) in the WashU browser shot shown in Figure 5B.
ATAC-seq
Samples were treated and processed as described previously for Assay for Transposase-Accessible Chromatin (ATAC)-seq (Buenrostro et al., 2013). Briefly, 100,000 cells were centrifuged 500 g for 5 min at room temperature. The cell pellet was resuspended in 50 ml lysis buffer (10 mM Tris–Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.01% Igepal CA-630) and centrifuged immediately 500 g for 10 min at 4°C. The cell pellet was resuspended in 50 ml transposase mixture (25 ul 2×TD buffer, 22.5 μl dH2O, and 2.5 μl Illumina Tn5 transposase or with final concentration of 100 nM Atto-590-labeled in-house Tn5) and incubated at 37°C for 30 min. After transposition, the mixture was purified with the Qiagen Mini-purification kit and eluted in 10 ul Qiagen EB elution buffer. Sequencing libraries were prepared following the original ATAC-seq protocol (Buenrostro et al., 2013). The sequencing was performed on Illumina Next-Seq at the Stanford Functional Genomics Facility. ATAC-seq reads were trimmed of adapters then mapped to hg19 genome assembly using Bowtie 2. Following QC steps to remove duplicate reads, ATAC-seq read counts were tallied with the aid of RSubRead (v.3.5). Relevant chromatin regions for each gene were within 1.5 kb (either direction) of hg19 gene transcription start sites (TSS). Differential ATAC read counts within each defined region were analyzed with the aid of DESeq2 (v.1.16.1). Heatmaps of average read intensities of ATAC-seq alignments were generated with the aid of deepTools and R/Bioconductor (v.3.2.1). Enrichment of sequence motifs among regions with differential read counts was analyzed with the aid of Homer (v.4.9). Additionally, we included ENCODE ATAC-seq data for human female adult left ventricle (ENCLB332HEY).
Whole Exome Sequencing (WES)
Genomic DNA isolated from skeletal muscle of each fetus (Fetus-1 and Fetus-2) was sent to Axeq Technologies (now Macrogen, Inc., South Korea) for whole exome sequencing and interpretation. After receipt of small in-del and single nucleotide variant profiles for each fetus, 135 genes related to cardiomyopathy, channelopathy, congenital heart disease, and sudden cardiac death were analyzed for nonsynonymous exonic variants with < 3% allele population frequency as reported by ExAC (Broad Institute, Cambridge, MA). ClinVar (Landrum et al., 2016) was used to annotate genomic variants as benign, likely benign, likely pathogenic, pathogenic, or unknown significance.
Comparison of Primate Heart and Body Masses
ln the Graphical Abstract, published male primate body masses (Smith and Jungers, 1997) were used to calculate body masses, an indirect measure of heart mass (Stahl, 1965).
QUANTIFICATION AND STATISTICAL ANALYSIS
Specific statistical details for all experiments in this study can be found in Method Details and in the Figures and Figure Legends, including statistical tests used, number (n), and SD/SEM measures. Significance was generally considered to be P < 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-YAP1 | Novus | Cat # NB110 |
| Mouse anti-TEAD4 | Abcam | Cat # ab58310 |
| Rabbit anti-STAT1 | Cell Signaling | Cat # 9172 |
| Normal mouse IgG | Santa Cruz | Cat # sc-2025 |
| Normal rabbit IgG | Santa Cruz | Cat # sc-2027 |
| Mouse anti-SSEA4 | R&D Systems | Cat # MC-813-70 |
| Rabbit anti-OCT3/4 | Santa Cruz | Cat # sc-9081 |
| Rabbit anti-NANOG | Santa Cruz | Cat # sc-33759 |
| Mouse anti-TRA-160 | Millipore | Cat # MAB-4360 |
| Rabbit anti-TNNT2 | Abcam | Cat # ab45932 |
| Mouse anti-MYH7 | Thermo Fisher | Cat # MA1-83347 |
| Mouse anti-ACTN1 | Abcam | Cat # ab18061 |
| Rabbit anti-Aurora B | Abcam | Cat # ab2254 |
| Anti-Rabbit Alexa Fluor 488 secondary antibody | Life Technologies | Cat # A32731 |
| Anti-mouse Alexa Fluor 488 secondary antibody | Life Technologies | Cat # A28175 |
| Anti-Rabbit Alexa Fluor 594 secondary antibody | Life Technologies | Cat # A32754 |
| Anti-mouse Alexa Fluor 594 secondary antibody | Life Technologies | Cat # A32740 |
| Anti-Histone H3 (acetyl K27) | Abcam | Cat # ab4729 |
| Bacterial and Virus Strains | ||
| AAV9 (for BANCR expression in rat heart infarcts) | Neuroscience Gene Vector and Virus Core (Stanford) | Custom service |
| pSpCas9(BB)-2A-GFP (PX458) | Addgene #48138 | |
| Lentivirus (pMD2G, pPAX2 target/packaging plasmids) | Neuroscience Gene Vector and Virus Core (Stanford) | Custom service |
| Biological Samples | ||
| Western lowland gorilla skin biopsy | Zoological Garden Berlin, Germany | N/A |
| Chimpanzee skin biopsy | Zoological Garden Berlin, Germany | N/A |
| Rhesus macaque skin biopsy | University of California, Davis | N/A |
| Human fetal tissues | StemExpress Foundation (Placerville, CA) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| hESC-qualified Matrigel | BD Biosciences | Cat # 354277 |
| Lipofectamine | Life Technologies | Cat # 11668019 |
| CHIR99021 | LC Laboratories | Cat # C6556 |
| IWR | Selleck Chemicals | Cat # S7086 |
| Y-27632 | Sigma Aldrich | Cat # HY-10583 |
| Critical Commercial Assays | ||
| CytoTune-iPS 2.0 Sendai Reprogramming Kit | Thermo Fisher | Cat # A16517 |
| Lincode Human BANCR siRNA | Dharmacon | 100885775 |
| siGENOME YAP1 siRNA | Dharmacon | M-012200-00-0005 |
| RhoA ELISA Kit | Cytoskeleton | Cat # BK124-S |
| Rac ELISA Kit | Cytoskeleton | Cat # BK128-S |
| Cdc42 ELISA Kit | Cytoskeleton | Cat # BK127-S |
| Zymo-Spin ChIP Kit | Zymo Research | Cat # D5210 |
| Deposited Data | ||
| All sequencing datasets deposited in GEO | This paper | GSE111930 |
| Experimental Models: Cell Lines | ||
| Human H7 ESC line | WiCell | Cat # WA07 |
| E. coli DH5 alpha | Zymo Research | Cat # T3007 |
| Gorilla iPSCs | Diecke lab (Berlin Institute of Health) | N/A |
| Chimpanzee iPSCs | Diecke lab (Berlin Institute of Health) | N/A |
| Rhesus macaque iPSCs | Wu lab (Stanford) | N/A |
| TBX5 knockout hiPSCs | Wu lab (Stanford) | 114-TBX5 KO Clone16 |
| BANCR knockdown (BKD) H7 hESCs | Wu lab (Stanford) | N/A |
| BANCR over-expression (BOE) H7 hESCs | Wu lab (Stanford) | N/A |
| BANCR enhancer deletion (ED) hiPSCs | Wu lab (Stanford) | N/A |
| BANCR Exon123 KO H7 hESCs | Wu lab (Stanford) | N/A |
| BANCR Exon4 KO H7 hESCs | Wu lab (Stanford) | N/A |
| Experimental Models: Organisms/Strains | ||
| Transgenic E16 murine FVB embryos | Cyagen Biosciences | Custom service |
| 10-week old Sprague Dawley rats | Charles River | N/A |
| Oligonucleotides | ||
| See Table S3 for list of all custom oligonucleotides and corresponding sequences | This paper | N/A |
| Recombinant DNA | ||
| pRP[Exp]-Beta MHC>hBANCR (for transgenic mouse embryo studies) | Cyagen Biosciences | Custom service |
| pCDH-MSCV-BANCR-EF1-copGFP-T2A-Puro (BANCR over-expression plasmid) | Neuroscience Gene Vector and Virus Core (Stanford) | Custom service |
| BANCR shRNA plasmid | Paul Khavari lab (Stanford) | N/A |
| Software and Algorithms | ||
| FastQC | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| DESeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| igvtools | Broad Institute | https://software.broadinstitute.org/software/igv/igvtools |
| TrimGalore | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| STAR | GitHub | https://github.com/alexdobin/STAR |
| R | The R Foundation | https://www.r-project.org/ |
| LiftOver | UC Santa Cruz | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
| Bowtie | Johns Hopkins | http://bowtie-bio.sourceforge.net/index.shtml |
| MACS2 | GitHub | https://hbctraining.github.io/Intro-to-ChIPseq/lessons/05_peak_calling_macs.html |
| HOMER | UC San Diego | http://homer.ucsd.edu/homer/ |
| featureCounts | Waltzer + Eliza Hall Institute of Medical Research | http://bioinf.wehi.edu.au/featureCounts/ |
Highlights.
Primate-specific, ERV-derived BANCR promotes fetal cardiomyocyte migration
BANCR induces cardiac enlargement in rodent models
BANCR expression is positively regulated by cardiogenic TBX5 and TEAD/YAP signaling
ACKNOWLEDGMENTS
We thank Ning-Yi Shao, Chunjiang He, and Yue Zhang for bioinformatics assistance; Praveen Shukla for patch clamping; Elina Tzatzalos for assistance with micropatterns; and John Coller, Xuhuai Ji, and Vanita Natu (Stanford Functional Genomics) for sequencing services. Sequencing datasets generated for this study are deposited in the Gene Expression Omnibus under accession number GSE111930. This work was supported by American Heart Association (AHA) 20POST35210924 (L.T.); National Institutes of Health (NIH) K08 HL119251 (K.D.W.); K01 HL130608 (O.J.A.); K99 HL135258 (M.G.); S10 OD018220 (Stanford Functional Genomics); P50 HG007735 and R35 CA209919 (H.Y.C.); and R01 HL113006, R01 HL130020, R01 HL126527, R01 HL133272, R01 HL141371, and P01 HL141084 (J.C.W.). H.Y.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
H.Y.C. is a co-founder of Accent Therapeutics and Boundless Bio and an advisor to 10x Genomics, Arsenal Biosciences, and Spring Discovery. J.C.W. is a co-founder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent. The other authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2020.07.006.
REFERENCES
- Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, et al. (2016). Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 167, 1734–1749.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Hershberger RE, Caleshu C, Ellinor PT, Garcia JG, Herrington DM, Ho CY, Johnson JA, Kittner SJ, MacRae CA, et al. (2012). Genetics and cardiovascular disease: a policy statement from the American Heart Association. Circulation 126, 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert J-P, Noble WS, and Le Roch KG (2014). Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res 24, 974–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, and Kohany O (2015). Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, and Chang HY (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. (2011).The evolution of gene expression levels in mammalian organs. Nature 478, 343–348. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, and Feschotte C (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, and Feschotte C (2017). Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Garg P, Treutlein B, Venkatasubramanian M, Wu H, Lee J, Wessells QN, Chen S-Y, Chen W-Y, Chetal K, et al. (2018). Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat. Commun. 9, 4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff MO, Olson S, Wei X, Garrett SC, Osman A, Bolisetty M, Plocik A, Celniker SE, and Graveley BR (2015). Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 521, 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183. [DOI] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, and Aiden EL (2016). Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, and Khavari PA (2012). BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 22, 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb EA, Warren RL, Wilson GW, Brown SD, Robertson GA, Morin GB, and Holt RA (2015). Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, and Ng HH (2016). CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 17, 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. (2015). Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Hu H, Wilson KD, Wu H, Feng J, Xia S, Churko J, Qu K, Chang HY, and Wu JC (2016). Systematic characterization of long noncoding RNAs reveals the contrasting coordination of cis- and trans-molecular regulation in human fetal and adult hearts. Circ. Cardiovasc. Genet. 9, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, and Martin JF (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, and Coffin JM (2001). Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29, 487–489. [DOI] [PubMed] [Google Scholar]
- Jonsson MKB, Hartman RJG, Ackers-Johnson M, Tan WLW, Lim B, van Veen TAB, and Foo RS (2016). A transcriptomic and epigenomic comparison of fetal and adult human cardiac fibroblasts reveals novel key transcription factors in adult cardiac fibroblasts. JACC: Basic Transl. Sci. 1, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, and Klein R (2016). Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat. Rev. Mol. Cell Biol. 17, 240–256. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, and Feschotte C (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long non-coding RNAs. PLoS Genet. 9, e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I, Termglinchan V, Cepeda DA, Lee J, Diecke S, Hendel A, Itzhaki I, Ameen M, Shrestha R, Wu H, et al. (2017). A comprehensive TALEN-based knockout library for generating human-induced pluripotent stem cell-based models for cardiovascular diseases. Circ. Res. 120, 1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. (2016). ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Shao NY, Paik DT, Wu H, Guo H, Termglinchan V, Churko JM, Kim Y, Kitani T, Zhao M-T, et al. (2018). SETD7 drives cardiac lineage commitment through stage-specific transcriptional activation. Cell Stem Cell 22, 428–444.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie R, O’Donnell CJ, and Johnson AD (2014). GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics 30, i185–i194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Zhang SL, and Wu JC (2019). Complex heritability in cardiomyopathy. Nat. Biomed. Eng. 3, 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmoset Genome Sequencing and Analysis Consortium (2014). The common marmoset genome provides insight into primate biology and evolution. Nat. Genet. 46, 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori LE, Sobreira DR, Sakabe NJ, Aneas I, Joslin AC, Hansen GT, Bozek G, Moskowitz IP, McNally EM, and Nóbrega MA (2018). A promoter interaction map for cardiovascular disease genetics. eLife 7, e35788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes KW, Sip CG, Obenza W, Yang E, Horst C, Welikson RE, Hauschka SD, Folch A, and Laflamme MA (2013). Human embryonic stem cell-derived cardiomyocytes migrate in response to gradients of fibronectin and Wnt5a. Stem Cells Dev. 22, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, et al. (2015). Heart disease and stroke statistics-2015 update: a report from the American Heart Association. Circulation 131, e29–e322. [DOI] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, et al. (2017). Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers FB, Silver JS, Zhuge Y, Beygui RE, Zarins CK, Lee LP, and Abilez OJ (2013). Robust pluripotent stem cell expansion and cardiomyocyte differentiation via geometric patterning. Integr. Biol. (Camb.) 5, 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, and Kaessmann H (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. [DOI] [PubMed] [Google Scholar]
- Patel MD, Mohan J, Schneider C, Bajpai G, Purevjav E, Canter CE, Towbin J, Bredemeyer A, and Lavine KJ (2017). Pediatric and adult dilated cardiomyopathy represent distinct pathological entities. JCI Insight 2, e94382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M, and Hall A (2004). Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23–32. [DOI] [PubMed] [Google Scholar]
- Rao SS., Huntley MH., Durand NC., Stamenova EK., Bochkov ID., Robinson JT., Sanborn AL., Machol I., Omer AD., Lander ES., and Aiden EL. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL (2013). Disease-targeted sequencing: a cornerstone in the clinic. Nat. Rev. Genet. 14, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. (2012). β-catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahn DJ, DeMaria A, Kisslo J, and Weyman A (1978). Recommendations regarding quantitation in M-mode echocardiography: results of a survey of echocardiographic measurements. Circulation 58, 1072–1083. [DOI] [PubMed] [Google Scholar]
- Schmid CD, and Bucher P (2010). MER41 repeat sequences contain inducible STAT1 binding sites. PLoS One 5, e11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, and Barillot E (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, and Jungers WL (1997). Body mass in comparative primatology. J. Hum. Evol. 32, 523–559. [DOI] [PubMed] [Google Scholar]
- Soni S, Raaijmakers AJ, Raaijmakers LM, Damen JM, van Stuijvenberg L, Vos MA, Heck AJ, van Veen TA, and Scholten A (2016). A proteomics approach to identify new putative cardiac intercalated disk proteins. PLoS One 11, e0152231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl WR (1965). Organ weights in primates and other mammals. Science 150, 1039–1042. [DOI] [PubMed] [Google Scholar]
- Stoye JP (2012). Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 10, 395–406. [DOI] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, and Salzman J (2015). Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatman PD, Woulfe KC, Karimpour-Fard A, Jeffrey DA, Jaggers J, Cleveland JC, Nunley K, Taylor MR, Miyamoto SD, Stauffer BL, and Sucharov CC (2017). Pediatric dilated cardiomyopathy hearts display a unique gene expression profile. JCI Insight 2, e94249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Panciera T, and Piccolo S (2018). YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20, 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A, Sorre B, Etoc F, Siggia ED, and Brivanlou AH (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, and Leinwand LA (1996). The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 12, 417–439. [DOI] [PubMed] [Google Scholar]
- Wilson KD, Shen P, Fung E, Karakikes I, Zhang A, InanlooRahatloo K, Odegaard J, Sallam K, Davis RW, Lui GK, et al. (2015). A rapid, high-quality, cost-effective, comprehensive and expandable targeted next-generation sequencing assay for inherited heart diseases. Circ. Res. 117, 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, and Nerbonne JM (2014). Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129, 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Chen H, Correa A, Devine O, Mathews TJ, and Honein MA (2006). Racial differences in infant mortality attributable to birth defects in the United States, 1989–2002. Birth Defects Res. Part A Clin. Mol. Teratol. 76, 706–713. [DOI] [PubMed] [Google Scholar]
- Zhao X, Chen H, Xiao D, Yang H, Itzhaki I, Qin X, Chour T, Aguirre A, Lehmann K, Kim Y, et al. (2018). Comparison of non-human primate versus human induced pluripotent stem cell-derived cardiomyocytes for treatment of myocardial infarction. Stem Cell Rep. 10, 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data sets generated for this study are deposited in the Gene Expression Omnibus under accession number GSE111930. This study did not generate code.