Abstract
Pollinator declines can leave communities less diverse and potentially at increased risk to infectious diseases. Species-rich plant and bee communities have high species turnover, making the study of disease dynamics challenging. To address how temporal dynamics shape parasite prevalence in plant and bee communities, we screened >5,000 bees and flowers through an entire growing season for five common bee microparasites (Nosema ceranae, N. bombi, Crithidia bombi, C. expoeki and neogregarines). Over 110 bee species and 89 flower species were screened, revealing 42% of bee species (12.2% individual bees) and 70% of flower species (8.7% individual flowers) had at least one parasite in or on them, respectively. Some common flowers (e.g., Lychnis flos-cuculi) harboured multiple parasite species, whilst others (e.g., Lythrum salicaria) had few. Significant temporal variation of parasite prevalence in bees was linked to bee diversity, bee and flower abundance, and community composition. Specifically, we found that bee communities had the highest prevalence late in the season, when social bees (Bombus spp. and Apis mellifera) were dominant and bee diversity was lowest. Conversely, prevalence on flowers was lowest late in the season when floral abundance was the highest. Thus, turnover in the bee community impacted community-wide prevalence, and turnover in the plant community impacted when parasite transmission was likely to occur at flowers. These results imply that efforts to improve bee health will benefit from promoting high floral numbers to reduce transmission risk, maintaining bee diversity to dilute parasites, and monitoring the abundance of dominant competent hosts.
Introduction
As the world experiences its sixth mass extinction event, communities are becoming less diverse, increasingly fragmented, and the dynamics of disease spread within these communities is being transformed 1–4. Despite a growing urgency to understand these dynamics, effective management of disease spread in wildlife is hampered by a poor understanding of parasite dynamics in species-rich communities 5–7. This is especially important for the conservation of complex pollinator communities, which in addition to suffering from disease-linked declines and extinctions, have outstanding value to the environment and economy 8–11.
Temporal changes in multi-species communities present significant challenges in our understanding of disease ecology due to the turnover of hosts, transmission sites, and parasite species over time 7,12, and ignoring these dynamics can lead to incorrect conclusions about disease risk. Three key parameters that can shape disease prevalence and spread in multi-host communities are host contact rates, density of disease spreaders, and frequency of transmission sites. The contact rate between competent hosts will change over time due to species turnover influencing host diversity and abundance, driving so-called dilution and amplification effects 13–19. To our knowledge, few studies have addressed temporal dynamics of amplification/dilution in species-rich communities. Heterogeneity in species’ ability to harbour and transmit parasites is common 12, so community changes over time can alter the relative abundance of disease spreaders. Indeed, the identification and targeting of such superspreaders often determines the success of disease control programs 20. Finally, the presence and location of transmission hotspots such as shared food resources can change over time, influencing risk of disease spread 12.
Plant and bee communities are complex multi-host, multi-parasite systems, where solitary and social bees experience differing levels of host-host contact rates, and where flowers can act as microbial transmission and dispersal hubs 21–24. Plant and bee communities also exhibit a high degree of species turnover throughout the year with overlapping periods of floral blooms and bee activities 25. With so many factors changing over time, it is difficult to understand parasite dynamics without temporal sampling. Yet to our knowledge, no studies have quantified parasite temporal dynamics in natural, species-rich pollinator communities, or on flowers acting as parasite transmission/dispersal hubs. This dearth of data prevents the development of targeted management strategies which could reduce parasite spread and increase host and ecosystem health 6,26.
Here, we screened over 5,000 samples of flowers and bees over a 26-week period at 3 old-field meadow sites. All samples were screened for five common multi-host parasite species or groups using PCR assays. Using these data of parasite prevalence on flowers and in bees, we address two main questions: 1) How does parasite prevalence change over time in species-rich plant-bee communities, and 2) Are changes in plant and bee species abundance, diversity and/or composition associated with changes in parasite prevalence? Overall, our goal was to view in high resolution the temporal dynamics of multiple parasites across entire plant and bee communities. With these data, we gain further insight into the epidemiology of species-rich communities.
Results
Parasite prevalence on flowers and in bees
In total, we screened 2,624 flowers from 89 species and 2,672 bees from at least 110 species for parasites – to our knowledge the most comprehensive screening of such plant and bee samples to date. Specifically, this study quantifies microparasites across the wildflower community, and many of the bee species have never been screened for parasites before.
Overall, we found an unexpectedly high proportion (8.7%) of individual flowers and 53.8% of flower genera to be positive for at least one of the five parasite species (see Fig. 1b and Supplementary Table 2), and 12.5% and 72.3% prevalence, respectively, for at least one of the two broad parasite groups (trypanosomes or microsporidians). Broken down by parasite species, 2.0% of flowers were positive for N. bombi, 0.7% were positive for N. ceranae, 3.2% were positive for C. bombi, 1.2% were positive for C. expoeki, and 2.9% were positive for neogregarines. From 89 flower species, 16 were sampled over 50 times, representing >60% of all flower samples. Of these, Lychnis flos-cuculi (ragged robin) had the highest prevalence of N. bombi (8%), N. ceranae (2%), C. bombi (10%) and neogregarines (6%), whilst Leucanthemum vulgare (oxeye daisy) had the highest prevalence of C. expoeki (5%). Whilst it is known that parasites can be horizontally transmitted between bees at flowers, these data confirm that bee parasites are present on a wide variety of wildflowers in natural communities.
Fig. 1 |. Parasite prevalence in bee (a) and on flower (b) genera across three old-field communities.
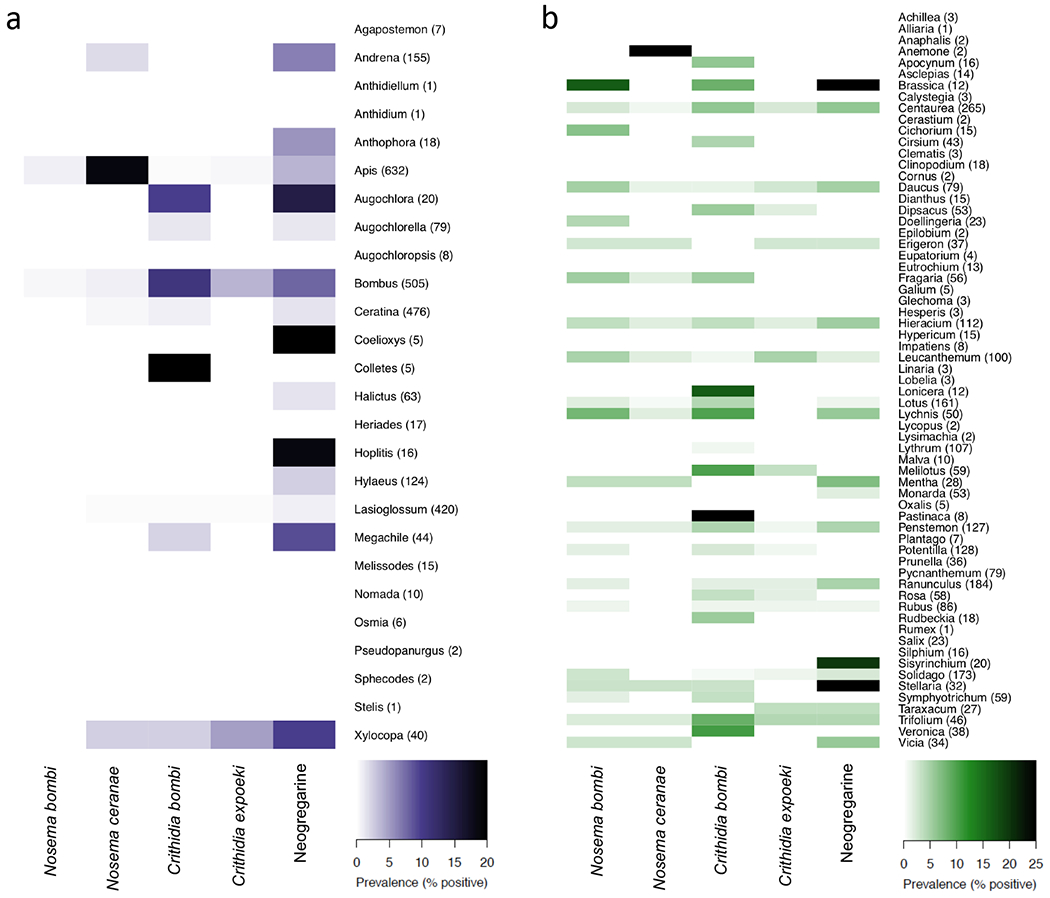
Screenings of 2,672 bees representing 26 genera (at least 110 species), and 2,624 flowers representing 65 genera (89 species). Rows represent bee (a) or plant (b) genus with sample number in parenthesis. Details of PCR protocols for parasite (Nosema bombi, N. ceranae, Crithidia bombi, C. expoeki, and neogregarines) screenings are outlined in the Methods. Full parasite information listed by species is shown in Supplementary Tables 2 and 3.
For bees, we found that 12.2% of individual bees and 57.7% of bee genera were positive for at least one parasite (see Fig. 1a and Supplementary Table 3), and 12.5% and 65.4% prevalence, respectively, for the broad parasite groups (trypanosomes, neogregarines, or microsporidians). Broken down by parasite species, 0.3% of bees were positive for N. bombi, 4.9% were positive for N. ceranae, 2.5% were positive for C. bombi, 1.0% were positive for C. expoeki, and 4.0% were positive for neogregarines. From 110 bee species, 10 were sampled over 50 times, representing 70% of all bee samples. Of these, Bombus bimaculatus had the highest prevalence of N. bombi (1.8%) and C. expoeki (12.5%), Apis mellifera the highest N. ceranae (18.8%), and Bombus impatiens had the highest prevalence of C. bombi (11%) and neogregarines (9.6%). Our understanding of these parasites is largely restricted to their presence in Apis or Bombus hosts, however these data confirm their presence across a much broader range of bee species (30 non-Apis and non-Bombus species; Fig 1a, Supplementary Table 3).
Temporal trends of parasite prevalence in bee and floral communities
Temporal trends of parasite prevalence were evaluated using generalized linear mixed models (GLMM), with parasite status as a binomial response, week number within the field season as the explanatory variable, and site as a random factor. Analyses were conducted separately for each broad parasite group, each parasite species, neogregarines, and a combined group comprising all four species together with neogregarines. We used a Bonferroni-corrected significance level of α = 0.05/Np, where Np is the number of parasite species/groups considered in each analysis. We did not use autoregressive time-series models since Durbin-Watson tests for temporal auto-correlation using scaled residuals were not significant. N. bombi was excluded from the bee analysis and N. ceranae from the flower analysis due to the small number of positives (< 20).
Prevalence of all parasite species/groups showed positive temporal trends in the bee community, with all except the trend in C. expoeki being statistically significant (Fig. 2a; χ21 = 2.9, p = 0.087 for C. expoeki, χ21 = 7.6, p = 0.006 for neogregarines, and χ21 ≥ 14, p < 0.001 for the rest, likelihood ratio test, n = 2,672; see Supplementary Table 4). In contrast, prevalence of all parasite species/groups showed negative temporal trends in the floral community, although only the trend in the combined parasite group was significant (Fig. 2b; χ21 = 8.3, p = 0.004 for the combined group, likelihood ratio test, n = 2,624; see Supplementary Table 5 for the rest). We also noted that whilst prevalence tended to be of comparable levels between bee and flower communities for most parasite species/groups, N. ceranae was much less prevalent on flowers than in bees, whereas the reverse was true for N. bombi.
Fig. 2 |. Parasite prevalence increased throughout the season in the bee community while it decreased or remained constant in the floral community.
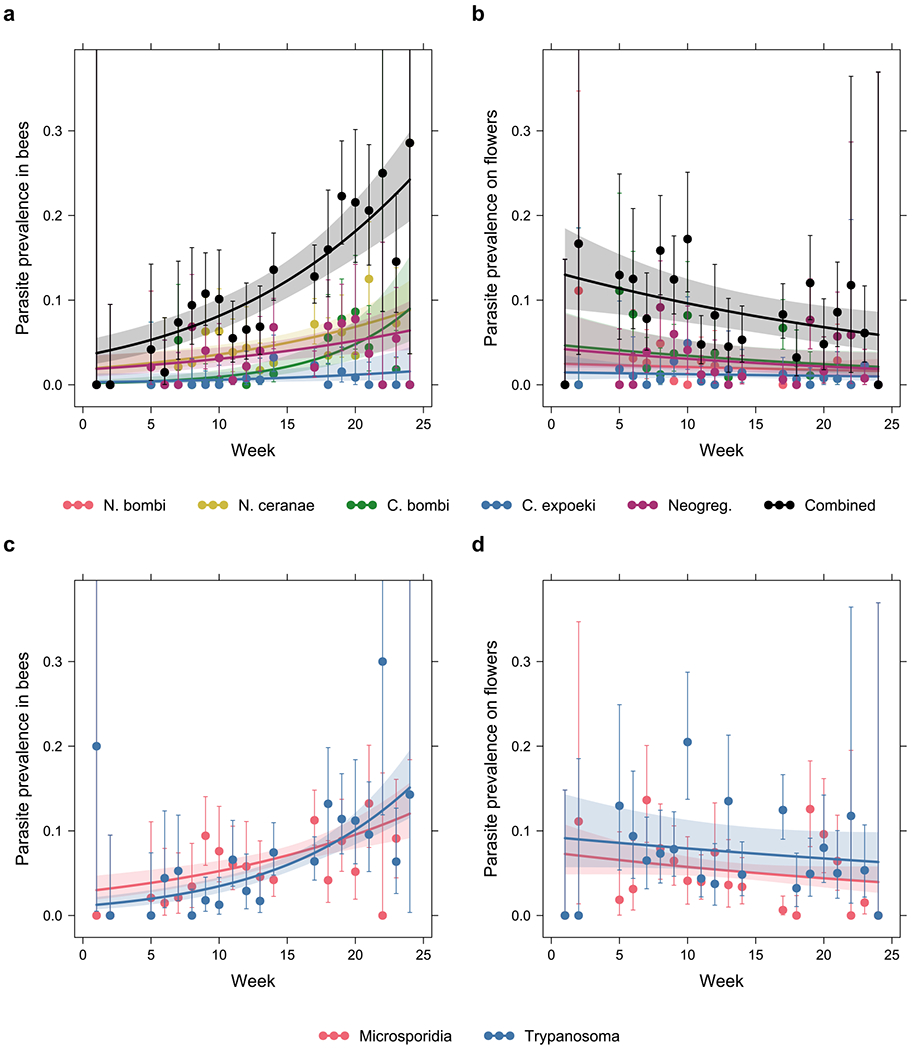
Prevalence of the specific parasite species or groups (Nosema bombi, N. ceranae, Crithidia bombi, C. expoeki, and neogregarines) in a, the bee community, and b, the floral community. Prevalence of the broad groups (microsporidians, trypanosomatids) in c, the bee community, and d, the floral community. The x-axis corresponds to week number in the field season (week 1 starting April 18, week 24 ending September 22). Error bars are 95% Clopper-Pearson confidence intervals. To reduce plot clutter, points and error bars are based on pooled data from all three sites each week; however, statistical tests and the prediction curves and confidence bands are based on GLMMs with site as a random factor.
Associations between bee community composition and parasite prevalence
The four most common bee genera - Apis, Bombus, Ceratina and Lasioglossum - comprised 76% of all surveyed and screened bees. We found a gradual turnover in the bee community across the season for all sites combined (Fig. 3a), comprised mostly of Ceratina, Lasioglossum and other genera at the start, while Apis and Bombus became dominant later in the season. Results of exact multinomial (p < 0.001 at every site, see Supplementary Fig. 1 legend for n at each site and window) and post-hoc binomial tests (Supplementary Fig. 1), evaluated at three different temporal windows, confirmed this turnover in species.
Fig. 3 |. Associations between bee community composition, diversity and parasite prevalence through time.
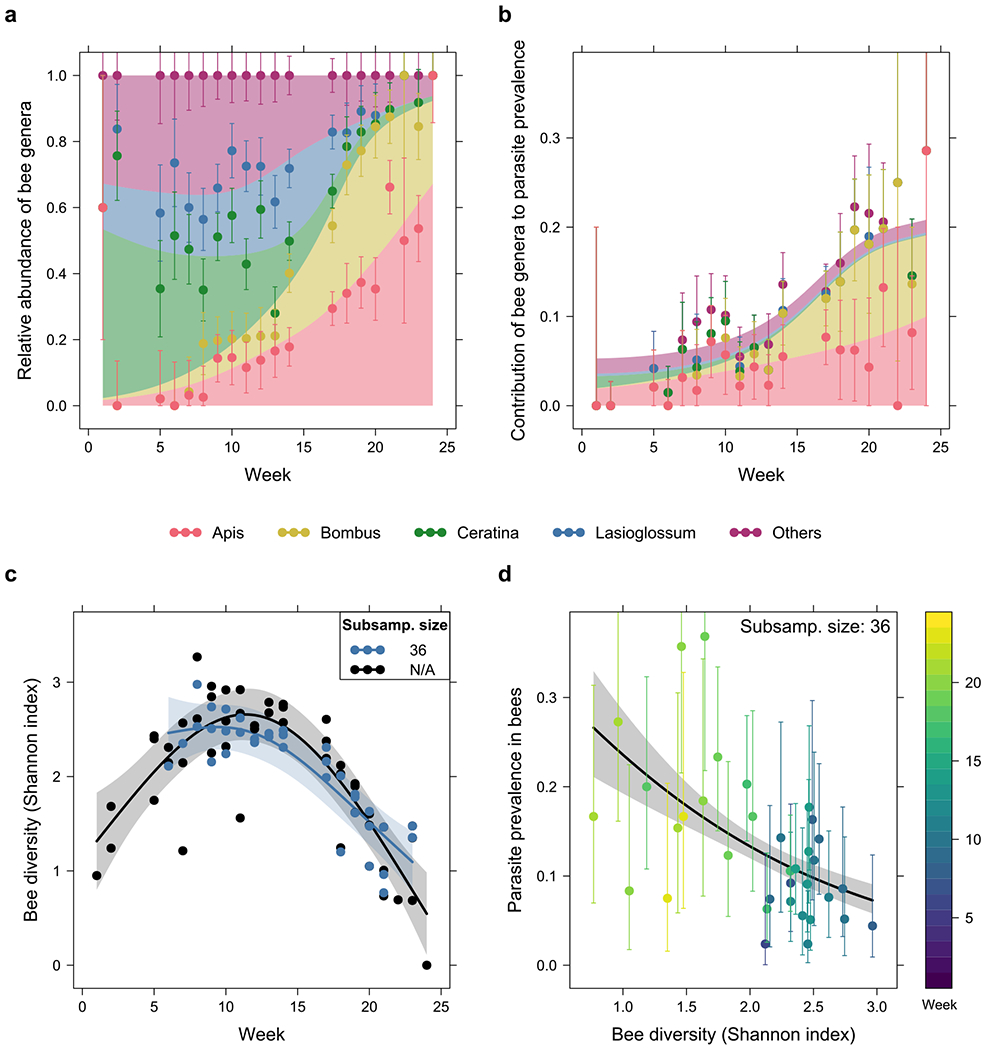
a, Relative abundance of each bee genus, and b, contributions from each genus to overall parasite prevalence in the community throughout the season. A gradual turnover in the community was observed from Ceratina, Lasioglossum and other unspecified genera early in the season, to Apis and Bombus later on. As Apis and Bombus exhibited higher parasite prevalence than other genera, this drove the observed increase in overall prevalence over time. c, Temporal trends in bee diversity (Shannon index), with rarefaction (subsample size = 36, see Methods) and without rarefaction (N/A), and d, association between parasite prevalence and rarefied Shannon index. The rarefied Shannon index decreased from weeks 6 to 23, while parasite prevalence also showed a negative association with Shannon index. Both observations are consistent with the increase in parasite prevalence over time. For b and d, the prevalence shown is that of all four parasite species and neogregarines combined as a single parasite group. Error bars are 95% Sison-Glaz multinomial confidence intervals in a, and 95% Clopper-Pearson binomial confidence intervals in b and d.
We explored parasite heterogeneity across species and sites using binomial GLMMs fitted for each parasite, with genus and its interaction with week number as additional explanatory variables. Bee genus was found to be a significant predictor of prevalence for all parasite species/groups (χ21 ≥ 28, p < 0.001, likelihood ratio test, n = 2,672, see Supplementary Table 6 and Supplementary Fig. 2). Post-hoc pairwise contrasts (Supplementary Table 7) indicated significantly higher prevalence in Apis and Bombus than other genera, with prevalence in Apis mostly associated with microsporidians and N. ceranae, and prevalence in Bombus associated with trypanosomatids, C. bombi and neogregarines. Most temporal trends of parasite prevalence in individual bee genera were not significant (Supplementary Table 8), with a notable exception being that of N. ceranae showing a negative trend in Apis (z = −4.1, p < 0.001, z-test).
In the early period of the season, solitary bee genera, particularly Ceratina and Lasioglossum, were the dominant bees yet contributed minimally to community-wide parasite prevalence (Fig. 3b). By the late period of the field season, Apis and Bombus were the dominant bees and contributed substantially to overall parasite prevalence (Fig. 3b; see Supplementary Fig. 3 for individual parasite species/groups). These results support findings that increased abundance of bumblebees and honeybees in an area is linked to increased parasite prevalence 27–30.
Associations between bee diversity and parasite prevalence
To assess how bee diversity varied across the season, we calculated the Shannon index for each week at each site, based on the collected bee samples, and assessed the temporal trend using a linear mixed model. Effects of rarefaction on Shannon index of the bee community are shown in Supplementary Figure 4. Decreasing the subsample size increased the range of weeks covered by the data (we excluded samples smaller than the subsample size), but at the cost of reducing the strength of any temporal trends. In subsequent analyses, we chose a subsample size of 36, since this affected Shannon indices by no more than 13%, yet allowed more than 70% of the site/week samples to be retained. No significant differences between sites were observed (F2,21 = 1.2, p = 0.32, one-way ANOVA, n = 35; Supplementary Fig. 5). Shannon index showed a significant decrease from weeks 6 to 23 (χ21 = 42, p < 0.001, likelihood ratio test, n = 35; Fig. 3c); this decrease is consistent with Fig. 3a, suggesting a shift in the community from one with comparable proportions among all four major genera mid-season to one mostly of Apis and Bombus late season. Shannon indices without rarefaction also suggested an initial increase in bee diversity early in the season (black line in Fig. 3c), but this trend was not robust as it could have been influenced by the much smaller bee samples in the first few weeks.
Negative associations were detected between parasite prevalence and the Shannon diversity index of bees for all parasite species/groups, with all except the associations in C. expoeki and neogregarines being statistically significant (Fig. 3d; χ21 = 7.4, p = 0.007 for microsporidians, χ21 < 0.001, p = 0.99 for C. expoeki, χ21 = 3.6, p = 0.060 for neogregarines, and χ21 ≥ 13, p < 0.001 for the rest, likelihood ratio test, n = 2,446; see Supplementary Table 9). Together with the negative trend of Shannon diversity index in Fig. 3c, these results are consistent with the increase in parasite prevalence through time in the bee community, as shown in Fig. 2a.
To explore whether these negative associations were driven entirely by the high-prevalence genera Apis and Bombus becoming more abundant later in the season, we repeated the same analysis but this time without Apis and Bombus samples when calculating parasite prevalence. We found that among the four parasite groups with sufficient number of positives for analysis, trypanosomatids retained a significant negative association but the remaining parasites did not (χ21 = 0.54, p = 0.46 for microsporidians, χ21 = 22, p < 0.001 for trypanosomatids, χ21 = 0.001, p = 0.97 for neogregarines, and χ21 = 2.6, p = 0.11 for the combined group, likelihood ratio test, n = 1,390; see Supplementary Table 9 and Supplementary Fig. 7). These results indicate that Apis and Bombus were responsible for some, but not all, associations between parasite prevalence and Shannon diversity. Focusing on the prevalence of two parasites in their known narrow host ranges, we found that N. ceranae in Apis hosts decreased over the year (Supplementary Table 8) and was positively correlated with Shannon diversity (χ21 = 8.2, p = 0.0041, likelihood ratio test, n = 586), whereas C. bombi in Bombus hosts increased over the year (Supplementary Table 8) and was negatively correlated with Shannon diversity (χ21 = 4.32, p = 0.038, likelihood ratio test, n = 470).
Associations between floral abundance and parasite prevalence
Floral abundance was measured at each site in randomly placed 10 m2 quadrats and found to increase across the season (χ21 = 132, p < 0.001, likelihood ratio test, n = 147; Fig. 4a). Prevalence of all parasite species/groups showed negative associations with floral abundance, although only the association in the combined parasite group was significant (Fig. 4b; χ21 = 8.0, p = 0.005 for the combined group, likelihood ratio test, n = 2,624; see Supplementary Table 11 and Supplementary Fig. 10 for the rest). These results hint at the possibility that parasite dispersal amongst flowers was diluted by increasing floral abundance as the year progressed. Although we do not have absolute abundance data for bees throughout the season, we postulate bee numbers were stable or decreased over the later period of the season, reducing overall bee visitation per flower, whilst at the same time parasite prevalence reduced on flowers. This positive relationship between floral visitation and the prevalence of parasites on flowers is partially supported by recent work showing that flowers in high visitation areas (apiaries) have a higher prevalence of bee viruses 31.
Fig. 4 |. Increase in floral abundance over time may dilute parasite prevalence on flowers.
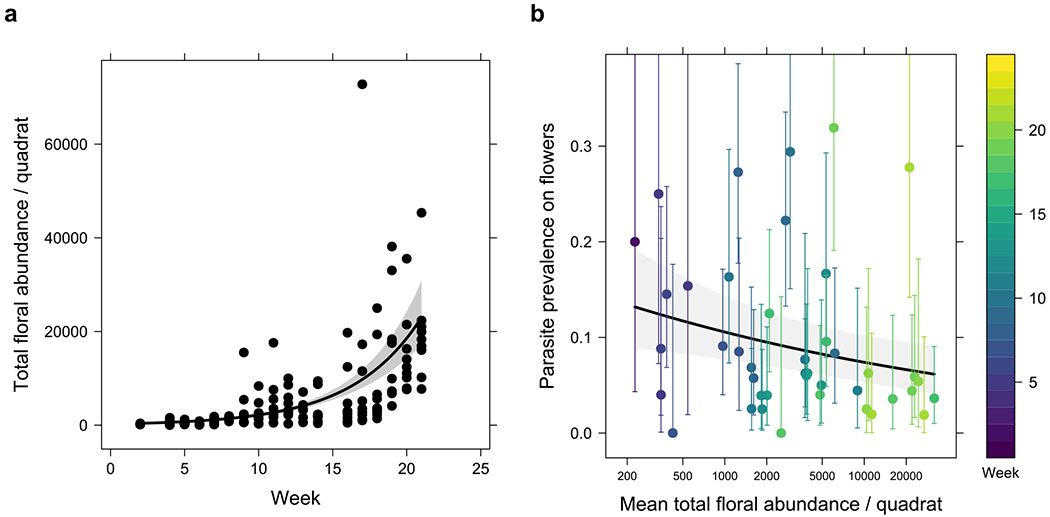
a, Total floral abundance within each quadrat showed a significant positive temporal trend. b, association between parasite prevalence, floral abundance per quadrat and week number of samples. The prevalence shown in b is that of all four parasite species and neogregarines combined. Error bars are 95% Clopper-Pearson confidence intervals.
The diversity of flowers surveyed within the quadrats did not differ between the field sites (F2,31 = 1.1, p = 0.36, one-way ANOVA, n = 49; Supplementary Fig. 5), nor was there any significant temporal trend (χ21 = 0.035, p = 0.85, likelihood ratio test, n = 49). Low Shannon indices were consistent with observations that the floral abundance was dominated at any point of time by a small number of species (Supplementary Fig. 8). No significant relationships were found between parasite prevalence on flowers and floral diversity for all parasite species/groups (Supplementary Table 10 and Supplementary Fig. 9).
Discussion
In this study, we found that parasite prevalence in plant and bee communities varies over the season and is linked to bee diversity, bee and flower abundance, and community composition. The three most common bee parasites were most prevalent late in the year, when bee communities were least diverse and social bees (Bombus spp. and Apis) were dominant. This pattern is consistent with both an overall dilution effect and/or increased importance of within-colony transmission and dominance of social bee hosts. We also found that bee parasites are prevalent on numerous wildflower species and the overall prevalence on flowers decreased with increasing floral abundance later in the season. These data suggest that the risk of parasite transmission among bee species may be reduced when the abundance of Apis and Bombus bees are low (as in early season), and when floral abundance is high (as in late season). Thus, species turnover and abundance across the bee-flower community is important to consider when identifying super-spreaders, disease hotspots, and key periods when transmission risk is likely to be high.
Over time, parasite prevalence in the community and within particular species changed, sometimes with contradictory trends, highlighting the importance of repeated temporal sampling to reveal an accurate determination of disease risk. Temporally, diversity of the bee community exhibited a unimodal distribution and as diversity declined from its peak, parasite prevalence increased. This coincided with increasing dominance of two social host groups, Apis and Bombus. During this period, parasites may benefit from increased contact rates of competent hosts (driven by increased density and within-colony transmission) and reduced overall host diversity, both increasing transmission rates and driving an increase in parasite prevalence. The combined increases in parasite prevalence and relative abundance of dominant, competent hosts suggests the health of these two dominant social groups may play an influential role in driving parasite prevalence across the community. For example, N. ceranae prevalence over the year decreased in its primary host, Apis, although the increasing relative abundance of Apis in the community drove an overall increase of the parasite at the community level. Similarly, prevalence of C. bombi and neogregarines increased in Bombus throughout the year, coinciding with an increase in prevalence of these parasites at the community level. It is also notable that the diversity of parasites in wild solitary bees are understudied, and there may be additional parasite species in these bees that have dynamics less influenced by social bees.
Whilst the ubiquity of the dilution/amplification effect in nature is still debated 32, here we find evidence these effects may be present in species-rich communities of bees. Various mechanisms, including changes in host density, contact rates, or parasite contagiousness have been proposed to cause dilution/amplification in several other systems 14,16,33–35. At a community level, our data show a negative association between bee diversity and parasite prevalence – supportive of a dilution effect. Conversely, despite rarely being found on flowers, the prevalence of N. ceranae in its primary host, A. mellifera, was positively correlated with local bee diversity - supportive of an amplification effect. However reductions in host diversity were intrinsically linked to increased densities of competent hosts, and the social nature of these hosts facilitates within-colony transmission as the season progresses 36,37. These confounding factors prevent a conclusive determination of amplification/dilution effects. To see if these forces could be teased apart, we tested for associations between host diversity and parasite prevalence excluding Apis and Bombus – although this result could be influenced by spillover, the absence of any associations in the remaining community would indicate dilution is not a driver. We found that trypanosomes remained negatively associated with diversity (supportive of a dilution effect) although other parasites did not. Thus, we find support for both dominant species and dilution influencing parasite prevalence in the community. Given these patterns, the foundations are laid for manipulative studies to elucidate the mechanisms involved.
We found a range of bee parasites on a high number and diversity of flowers. Though this has not been shown before in wildflower communities, it is common in disease ecology to find that shared feeding areas can be sites of disease spread (e.g., watering holes) 12,38,39. We found that parasite prevalence differed by flower species, supporting earlier manipulative studies that have identified transmission variation between flower species/traits 21,23,40,41. This supports the prediction that risk of parasite transmission to bees will vary by flower availability and foraging choice. Furthermore, we found that parasite prevalence on transmission hubs (i.e., flowers that can potentially transmit and disperse microbes) declined over time, though unlike in bee hosts, the diversity of transmission hub species remained constant whilst their abundance increased. This increase in transmission hub abundance may have led to dilution via density, where an increase in the ratio of parasite-free to contaminated hubs resulted from increases in floral abundance compared to bee abundance over time. Theory indicates that increasing flower numbers will reduce parasite transmission in plant-pollinator networks 42. Whilst our data cannot directly support this finding, we did see that increasing flower numbers reduced the prevalence of parasites on flowers. Therefore, floral composition and abundance, in addition to the foraging patterns and abundance of bees, likely play an important role in parasite transmission. More work is required to understand the underlying mechanics behind these complex interactions and, in particular, whether the introduction of managed honeybees or bumblebees to a landscape and the resulting change in bee/flower ratios influences parasite spread across wider geographic areas.
Species-rich bee and plant communities are complex, and our findings of multiple factors influencing parasite prevalence and transmission risk pose challenges for conservation and management. In various systems the health of animals at risk from disease have been shown to improve following targeted disturbance of transmission pathways and direct removal/killing of parasites 5,6,26,43–46. By performing this temporal study of multi-parasite prevalence across plant-bee communities, we identified patterns and species that may be targeted. An important conservation target is the maintenance of bee biodiversity, with reductions likely to exacerbate parasite threats to the community. Furthermore, the identification of honeybees and dominant bumblebee species as key drivers of parasite prevalence and transmission risk indicates their health likely has knock-on effects to others. During times of pandemic threat, it is the control of such high dispersers that can be the key to reducing disease spread 12,43,47–52. Parasite management strategies that target these host groups could greatly increase the efficiency of disease control efforts within local bee communities, allowing control efforts to be less intrusive and more cost effective than a system-wide approach 43. Finally, to foster healthy bee communities, our data support the promotion of high floral numbers to reduce parasite prevalence and risk of transmission at flowers. Further work is now required to understand the role that foraging patterns and floral traits have on parasite spread, and to determine the full host range and virulence of parasites across various bee species.
Methods
Bee and flower collections
To determine parasite prevalence within bees and upon the surfaces of flowers, we collected samples weekly across three old-field sites in upstate New York, between 18th April 2017 and 22nd September 2017. The sites were named Lansing (Lat: 42° 32’ 24.4932’’ N, Long: 76° 29’ 47.9076’’ W), McDaniels (Lat: 42° 32’ 11.5872’’ N, Long: 76° 25’ 3.7668’’ W), and Whipple (Lat: 42° 29’ 23.6328’’ N, Long: 76° 25’ 49.818’’ W). The McDaniels and Whipple sites are managed by Cornell University and required no permits for their use. The Lansing site is privately owned and we obtained permission to use the site. The distance between sites ranges from 5.3km to 7.5 km. Weather depending, we dedicated a day of collecting (~7 hours) per site, every week. On a typical collection day 3 members would be collecting samples from the same site (21 person-hours per site per week). In total we spent 58 days collecting samples over the five-month period (equivalent of approx. 1,200 person-hours). We collected bees in sterile tubes either directly or with the use of a net. Variety of floral form meant that ‘flowers’ were either single flowers or inflorescence depending on the plant species. The number of florets used per inflorescence was the same number a bee would forage upon in a single interaction with that species. We placed flowers that the bees were foraging upon directly into separate sterile tubes using sterilized forceps. Bee and flower samples were placed on dry ice immediately after collection, then transported to the lab and stored at −80C until we commenced DNA extraction.
Floral abundance and diversity
Each week throughout the growing season, we randomly selected three quadrats (each 10 m × 10 m) at each site, within which we identified the floral species in bloom and counted the number of stems for each (two out of 26 weeks were not surveyed due to weather). Per species, we estimated the number of floral units per stem and averaged this across the sites and weeks the species was in bloom (minimum five measurements per species). We define a floral unit as being the typical unit (single flower or inflorescence) that a bee typically foraged from 53. This definition of a floral unit was also consistent with the amount of plant material we used for each sample during parasite screening. We therefore define floral abundance of each plant species within the quadrat as the product of stem counts with the estimated number of floral units per stem. The raw floral survey data are held on Dryad [https://doi.org/10.6086/D1X09V].
Plant and bee identification
We identified plants either in the field or via specimens and/or photos brought back to the lab and keyed out 54–57. We confirmed the identity of each individual plant screened for parasites before DNA extraction occurred. Overall, we screened 2,624 flowers from 89 species for parasites (Supplementary Table 2). We identified bees after the gut dissections occurred for each specimen using reference materials located in the Cornell University Insect Collection (CUIC: http://cuic.entomology.cornell.edu/) and published keys 58–61. All identifications were conducted by P. Muñiz, taxa verifications were conducted by M. Arduser, and all voucher specimens are housed in the McArt lab or the CUIC. Overall, we screened 2,672 bees from at least 110 species for parasites (Supplementary Table 3).
Bee parasites
We selected to screen samples for parasites with known effects on bee health and links to population declines 62,63. Historically, research has focused on parasites of honeybees and a small number of bumblebee species. This research bias has contributed to a fundamental lack in our understanding of the full host ranges of bee parasites. Because we were interested in screening across bee taxa, we specifically sought to screen for parasites that have been identified in multiple bee species. We considered common microparasites of bees as three groups; microsporidians, trypansomatids, and neogregarines. The microsporidians are comprised mostly of Nosema species whose spores are transmitted via the faecal-oral route 64. The effects of microsporidia infections in honeybees and bumblebees includes wing deformity, reduced foraging efficiency, reduced colony fitness and increased mortality, with N. bombi being found in multiple species of bumblebees and N. ceranae found in bumblebees and honeybees 65–71 In addition N. ceranae may be able to infect the mason bee, Osmia bicornis72. While this study did not find impacts on survival, a similar study that assessed impacts of N. ceranae on larval O. bicornis did find negative impacts on survival73. The trypanosomatids are mostly Crithidia species whose cells are transmitted via the fecal-oral route. The effects of trypanosomatid infection in bumblebees include reduced foraging efficiency, reduced queen fitness, and increased mortality of infected bees. Crithidia spp. can infect bumblebees, honeybees and several solitary bee species 74–82. The neogregarines are an understudied group, with one described bee parasite, Apicystis bombi. This neogregarine has been detected in honeybees, a range of bumblebee species and solitary bees. Infected bumblebees have reduced fat bodies, increased mortality, and queen bumblebees are less likely to survive hibernation 30,83–88.
Parasite DNA extraction from individual bees and inflorescences
Parasite detection does not confirm an active infection, and although we endeavoured to reduce the likelihood of reporting uninfected bees by eliminating parasites on the outer cuticle via surface sterilization, and only report parasites in tissues known to harbour the selected parasites, some of the detections may be due to transient parasite material and not an active infection 89. We performed dissections in bleached, UV sterilized hoods, and sterilised instruments between samples using a dry bead steriliser set to 250°C. We carefully removed the alimentary canal from the mid-gut to the rectum using standard techniques 90. If the gut broke apart inside the bee, in addition to dissecting out the torn gut, we washed 10μl of Phosphate-buffered saline in and out of the bee cavity with a pipette to recapture any spilled gut contents before adding it to the gut tissue for DNA extraction. We performed dissections with minimal destruction to the cuticle to allow accurate species identification.
Similarly with parasite detections in bees, we do not know if parasites molecularly detected on flowers are viable, or pathogenic to all foraging bees. We term flowers as potential ‘transmission hubs’ because of their role in providing a physical platform for microbes to be deposited by bees and then acquired by subsequent foraging bees21,24,40. Using the 96-well plate Qiagen DNeasy blood and tissue Extraction Kit protocol, we washed single flowers/inflorescences in 600μl ATL buffer by pulse vortexing for 30 seconds. We then transferred 450μl of the wash to a 2ml screwcap tube with ~100 µL of 0.10mm zirconia beads and one 5mm steel bead. The wash was then lysed for 30 seconds at 6.5M/s on a Omni Bead Ruptor 24 homogenizer. For bee guts, we added 180 µl buffer ATL, ~ 100 µL 0.1 mm zirconia beads, and one 5 mm steel bead to each sample. We then homogenized the bee guts at 30 hz for 3 minutes on a Qiagen Tissue Lyser II. For both flower washes and bee gut homogenate, we next added 50 µL of Proteinase K before allowing the samples to incubate at 56°C overnight. Following incubation, we followed the standard “Quick Start” protocol provided by Qiagen, until the DNA was eluted in 100 µL of AE buffer.
Parasite screening
We took a 2-step approach to determine the parasite prevalence on flowers and in bee guts; (1) we screened for the presence of two broad taxonomic groups (trypanosomatids and microsporidians) known to contain multiple parasite genera, before (2) screening these positively identified groups for likely species of bee parasites within those groups. In addition, the multiplex PCR also identified samples containing neogregarine parasites. The broad multiplex panel was designed to efficiently screen flowers and insect guts for the most common bee-infecting parasites within a single reaction 91. Samples positive for Crithidia or Nosema were then diagnosed with species-specific multiplex panels (C. bombi, C. expoeki, and N. bombi, N. ceranae, respectively). Primers were either newly designed (see supplementary methods) or chosen from existing literature 91–93. The concentrations of each reagent and the thermocycling conditions are given in Supplementary Table 1. PCR products were run alongside a size standard on a 2% agarose gel, stained with GelRed® to visualise and confirm amplicon size. Each assay included a negative and a positive control. The raw data for broad range and species-specific parasite screens plus Sanger sequences of a subset can be found on Dryad [https://doi.org/10.6086/D1X09V] and via NCBI database with accession numbers MT212154-MT212159, MT296581-MT296586, MT302779-MT302784, MT359894-MT359896, MT366919, MT387450 and MT387451).
Statistical analysis
1. Parasite prevalence of the bee community through time
For each of the broad taxonomic groups and parasite species that we screened for, temporal trends in the overall parasite prevalence of the bee communities were evaluated using generalized linear mixed models (GLMM), with parasite status of the samples as binomial response, week number within the field season as the explanatory variable, and site as a random factor. All subsequent GLMMs also used site as the random factor. Many issues can potentially arise from the low parasite prevalence in the data (e.g., prevalence less than 1% for some parasite species); for instance, maximum likelihood estimates (MLE) are only asymptotically unbiased, meaning that they can be significantly biased if the number of positives is small 94. Therefore, we excluded any parasite groups or species with less than 20 positives in the analysis, which in this case meant leaving out N. bombi. The same criterion was used in all subsequent analysis involving parasite status. In addition, we repeated the analysis for the four species and neogregarines combined as a single group of bee parasites. The broad groups microsporidia and trypanosomatids were not included in this combination since they also contain species that may not infect bees. To account for multiple testing given the number of parasite species/groups being considered, throughout this manuscript we use a Bonferroni-corrected significance level of α = 0.05/Np, where Np is the number of parasite species/groups considered in each analysis. To justify the use of GLMMs instead of autoregressive time-series models, we checked for temporal auto-correlation using Durbin-Watson tests on scaled residuals.
Next we investigated bee community composition and diversity as possible drivers of any observed temporal trends. These are discussed in details below.
a. Bee community composition
Should there be significant heterogeneity in host competence among taxa, community turnover may drive changes in parasite prevalence. As the four most common bee genera Apis, Bombus, Ceratina and Lasioglossum comprised as much as 76% of the bee samples, we explored how the individual contributions of these genera (as well as all other genera as a single group) to the overall parasite prevalence of the community varied over time. First, temporal trends in the relative abundance of these genera were visualized with smoothing splines fitted using multinomial generalized additive models (GAM) with site included as a random factor. Relative abundances were further quantified by partitioning the season into three equal windows of 8 weeks each, and performing for each site and window an exact multinomial goodness-of-fit test (against the hypothesis of equal multinomial proportions), followed by post-hoc tests (with Bonferroni-corrected α given the number of genera) to identify the genera that fall above or below the expected proportions.
Second, to evaluate heterogeneity in parasite prevalence across genera, binomial GLMMs were again fitted for each parasite, but this time with genus and its interaction with week number as additional explanatory variables. Main effects of genus were evaluated regardless of whether the interactions were significant. Testing for main effects in the presence of significant interactions is known to violate the principle of marginality 95,96. More specifically, the main effects of genera are given by the differences in fitted logit links at week 0 between genera (i.e. differences in y-intercepts); since significant interactions imply different temporal trends between genera (different slopes), the main effects of genera become ambiguous since they now depend on what date we choose to assign as week 0. To address this, we defined week 0 of each genus to be the median week number among all samples of that genus; week numbers of the samples were then shifted accordingly (the shifts were not shown in Supplementary Fig. 2 to avoid confusion). By doing so, the main effects could be unambiguously interpreted as the difference in parasite prevalence between genera, evaluated for each genus at a characteristic date of that genus. Post-hoc tests of pairwise contrasts (with multiple testing corrections for the number of contrasts) were performed whenever the main effects of genus turned out to be significant. Significance of temporal trends in the parasite prevalence of each genus were also assessed (with multiple testing corrections for the number of genera).
Finally, temporal trends in the individual contributions of each genus to the overall parasite prevalence of the community (relative abundance × parasite prevalence of genus) were visualized using smoothing splines fitted using GAMs with site as a random factor.
b. Bee diversity
Should bee diversity affect parasite prevalence, temporal trends in the former may drive that of the latter. To assess how the bee diversity varied across the season, we calculated the weekly Shannon index at each site, based on the collected bee samples, and assessed the temporal trend using a linear mixed model. Before fitting the model, rarefaction was implemented for the bee Shannon indices to address non-uniformity in bee collection efforts. Note that rarefaction tended to reduce larger indices more than smaller ones, hence potentially affecting the strength of any temporal trends. Therefore, we evaluated a range of subsample sizes, rather than simply have the size be determined by the smallest site/week sample. Each time, samples smaller than the size being considered were discarded. This allowed us to identify an optimal size large enough for most indices to remain close to their non-rarefied values, but yet small enough to minimize the number of discarded samples. All analyses were conducted using rarefied indices at this optimal size; robustness of any results to the choice of subsample size were also assessed. We also assessed whether there were differences in bee diversity between sites using one-way ANOVA.
Next, we evaluated the relation between parasite prevalence and diversity using a GLMM with parasite status of the weekly bee samples as binomial response, and rarefied Shannon indices calculated using the samples as the explanatory variable. To explore whether any observed associations were entirely driven by temporal trends in the abundance of Apis and Bombus, we also repeated the analysis, but this time without Apis and Bombus samples when calculating parasite prevalence.
2. Parasite prevalence of the floral community through time
As was done for bees, temporal trends in the overall parasite prevalence of the floral community were evaluated using GLMMs. Flowers were not obtained at random, instead their collections were directed by the co-collection of foraging bees upon them. Hence the floral community being screened should be thought of as being biased to some extent by bee foraging preferences – unlike the floral surveys used for abundance/diversity analyses. N. ceranae was excluded from the analysis since less than 20 flower samples tested positive.
Temporal trends in the total floral abundance within quadrats were also evaluated using GLMMs, with counts as negative binomial response. Negative binomial was chosen to allow for overdispersion due to clustering. To investigate whether floral abundance can dilute parasite prevalence on flowers, we evaluated the relation between parasite prevalence and floral abundance using a binomial GLMM, with parasite status on flower samples as binomial response and log10(mean total floral abundance per quadrat) as explanatory variables. We chose to take the logarithm of floral abundance, first because of model convergence issues, and second because it was the more appropriate linear predictor for the logit link if we assumed prevalence to be inversely proportional to floral abundance.
Finally, temporal trends in the floral diversity (Shannon index based on the quadrat floral abundance surveys) were also evaluated using linear models, and the relation between parasite prevalence on flowers and floral diversity evaluated using a binomial GLMM.
Software and packages used
All analyses were performed in R v3.5.1 97. Packages used were lme4 98 and glmmTMB 99 for fitting GLMMs, DHARMa 100 for performing Durbin-Watson tests using scaled residuals, DescTools 101 for generating binomial and multinomial confidence intervals in the plots, mgcv 102 for fitting smoothing splines using GAM, XNomial 103 for exact multinomial tests, multcomp 104 for pairwise contrasts, and Vegan 2.54 for calculating Shannon indices 105.
Data availability
All raw data including site surveys and screening results are data found on Dryad in addition to all analysis code used [https://doi.org/10.6086/D1X09V]. Sequence data is also deposited in the NCBI database with accession numbers MT212154-MT212159, MT296581-MT296586, MT302779-MT302784, MT359894-MT359896, MT366919, MT387450 and MT387451).
Supplementary Material
Acknowledgements
Timothy Salazar and David Lewis assisted with field work, Jeffrey Teague helped with bee dissections, Mike Arduser confirmed bee identifications, and James Strange (USDA-ARS-PIRU) provided support in the development of diagnostic primers. The research group of Dr. Richard Gill provided comments on the manuscript. We would also like to thank four anonymous reviewers whose comments have greatly improved this manuscript
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH; award R01GM122062). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests
We declare that none of the authors have competing financial or non-financial interests as defined by Nature Research. Dr. Tripodi contributed to this article in her personal capacity. The views expressed are her own and do not necessarily represent the views of the Agricultural Research Service or the United States Government.
References
- 1.Pongsiri MJ et al. Biodiversity loss affects global disease ecology. Bioscience 59, 945–954 (2009). [Google Scholar]
- 2.Dirzo R et al. Defaunation in the Anthropocene. Science 345, 401–406 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Barnosky AD et al. Has the Earth’s sixth mass extinction already arrived? Nature 471, 51–57 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Sala OE et al. Accelerated modern human – induced species losses: entering the sixth mass extinction. Sci. Adv 1, 1–5 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson PK et al. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol 19, 535–44 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Daszak P, Cunningham AA & Hyatt AD Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science 287, 443–9 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Johnson PTJJ, de Roode JC & Fenton A Why infectious disease research needs community ecology. Science 349, 1259504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulson D, Lye GC & Darvill B Decline and conservation of bumble bees. Annu. Rev. Entomol 53, 191–208 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Williams PH & Osborne JL Bumblebee vulnerability and conservation world-wide. Apidologie 40, 367–387 (2009). [Google Scholar]
- 10.Goulson D, Nicholls E, Botias C & Rotheray EL Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science (2015). doi: 10.1126/science.1255957 [DOI] [PubMed] [Google Scholar]
- 11.Gallai N, Salles JM, Settele J & Vaissiere BE Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ 68, 810–821 (2009). [Google Scholar]
- 12.Paull SH et al. From superspreaders to disease hotspots: Linking transmission across hosts and space. Front. Ecol. Environ 10, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood CL et al. Does biodiversity protect humans against infectious disease? Ecology 95, 817–832 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Salkeld DJ, Padgett KA & Jones JH A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol. Lett 16, 679–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood CL & Lafferty KD Biodiversity and disease: A synthesis of ecological perspectives on Lyme disease transmission. Trends Ecol. Evol 28, 239–247 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Luis AD, Kuenzi AJ & Mills JN Species diversity concurrently dilutes and amplifies transmission in a zoonotic host–pathogen system through competing mechanisms. Proc. Natl. Acad. Sci 115, 7979–7984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keesing F, Holt RD & Ostfeld RS Effects of species diversity on disease risk. Ecol. Lett 9, 485–98 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Ostfeld RS & Keesing F Biodiversity and disease risk: the case of Lyme disease. Conserv. Biol 14, 722–728 (2000). [Google Scholar]
- 19.Schmidt KA & Ostfeld RS Biodiversity and the dilution effect in disease ecology. Ecology 82, 609–619 (2001). [Google Scholar]
- 20.Woolhouse MEJ, Dye C & Etard J . Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc. Natl. Acad. Sci 94, 338–342 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graystock P, Goulson D & Hughes WOH Parasites in bloom: flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. R. Soc. B Biol. Sci 282, 20151371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigaud T, Perrot-Minnot M-J & Brown MJF Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc. R. Soc. B Biol. Sci 277, 3693–702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler LS et al. Disease where you dine: plant species and floral traits associated with pathogen transmission in bumble bees. Ecology 99, 2535–2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McFrederick QS et al. Flowers and Wild Megachilid Bees Share Microbes. Microb. Ecol 73, 188–200 (2017). [DOI] [PubMed] [Google Scholar]
- 25.CaraDonna PJ et al. Interaction rewiring and the rapid turnover of plant–pollinator networks. Ecol. Lett 20, 385–394 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Jones KE et al. Global trends in emerging infectious diseases. Nature 451, 990–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piot N et al. Establishment of wildflower fields in poor quality landscapes enhances micro-parasite prevalence in wild bumble bees. Oecologia 189, 149–158 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Theodorou P et al. Pollination services enhanced with urbanization despite increasing pollinator parasitism. Proc. R. Soc. B Biol. Sci 283, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graystock P, Goulson D & Hughes WOH The relationship between managed bees and the prevalence of parasites in bumblebees. PeerJ 2, e522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graystock P, Blane EJ, McFrederick QS, Goulson D & Hughes WOH Do managed bees drive parasite spread and emergence in wild bees? Int. J. Parasitol. Parasites Wildl 5, 64–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alger SA, Burnham PA, Boncristiani HF & Brody AK RNA virus spillover from managed honeybees (Apis mellifera) to wild bumblebees (Bombus spp.). PLoS One 14, e0217822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randolph SE & Dobson ADM Pangloss revisited: A critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology 139, 847–863 (2012). [DOI] [PubMed] [Google Scholar]
- 33.LoGiudice K et al. Impact of host community on Lyme disease risk. Ecology 89, 2841–2849 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Keesing F et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson PTJ, Lund PJ, Hartson RB & Yoshino TP Community diversity reduces Schistosoma mansoni transmission, host pathology and human infection risk. Proc. R. Soc. B Biol. Sci 276, 1657–1663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell CE, Tilman D & Groth JV Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology 83, 1713–1726 (2013). [Google Scholar]
- 37.Johnson PTJ & Thieltges DW Diversity, decoys and the dilution effect: how ecological communities affect disease risk. J. Exp. Biol 213, 961–970 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Becker DJ, Streicker DG & Altizer S Linking anthropogenic resources to wildlife-pathogen dynamics: a review and meta-analysis. Ecol. Lett 18, 483–495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunn CL, Thrall PH & Kappeler PM Shared resources and disease dynamics in spatially structured populations. Ecol. Modell 272, 198–207 (2014). [Google Scholar]
- 40.Durrer S & Schmid-Hempel P Shared use of flowers leads to horizontal pathogen transmission. Proc. R. Soc. B Biol. Sci 258, 299–302 (1994). [Google Scholar]
- 41.Figueroa LL et al. Landscape simplification shapes pathogen prevalence in plant-pollinator networks. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truitt LL, McArt SH, Vaughn AH & Ellner SP Trait-based modeling of multihost pathogen transmission: Plant-pollinator networks. Am. Nat 193, E149–E167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lloyd-Smith JO, Schreiber SJ, Kopp PE & Getz WM Superspreading and the effect of individual variation on disease emergence. Nature 438, 355–359 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daszak P et al. Interdisciplinary approaches to understanding disease emergence: the past, present, and future drivers of Nipah virus emergence. Proc. Natl. Acad. Sci 110 Suppl, 3681–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lafferty KD & Gerber LR Good medicine for conservation biology: the intersection of epidemiology and conservation theory. Conserv. Biol 16, 593–604 (2002). [Google Scholar]
- 46.Cottam EM et al. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc. R. Soc. B Biol. Sci 275, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhatt S et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyšek P & Richardson DM Invasive species, environmental change and management, and health. Annu. Rev. Environ. Resour 35, 25–55 (2010). [Google Scholar]
- 49.Malone JD et al. U.S. airport entry screening in response to pandemic influenza: modeling and analysis. Travel Med. Infect. Dis 7, 181–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatem AJ, Rogers DJ & Hay SI Global transport networks and infectious disease spread. Adv. Parasitol 62, 293–343 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicolaides C, Cueto-Felgueroso L, González MC & Juanes R A metric of influential spreading during contagion dynamics through the air transportation network. PLoS One 7, e40961 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardner L & Sarkar S A global airport-based risk model for the spread of dengue infection via the air transport network. PLoS One 8, e72129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urbanowicz CM, Muñiz PA & McArt SH Honey bees and wild bees differ in their preference for and use of introduced floral resources. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiegand KM & Eames AJ The flora of the Cayuga Lake Basin, New York: (The University, 1926). doi: 10.5962/bhl.title.59518 [DOI] [Google Scholar]
- 55.Medina BF & Medina V Central appalachian wildflowers. (2002). [Google Scholar]
- 56.House HD The Wild Flowers of New York. (University of New York Albany, 1918). [Google Scholar]
- 57.Niering WA, Olmstead NC, Rayfield S & Nehring C National Audubon Society Field Guide to North American Wildflowers (Eastern Region). (1979). [Google Scholar]
- 58.Ascher JS & Pickering J DiscoverLife Bee Species Guide and World Checklist. Available at: http://www.discoverlife.org/mp/20q?guide=Bee_genera. [Google Scholar]
- 59.Gibbs J Revision of the metallic Lasioglossum (Dialictus) of eastern North America (Hymenoptera: Halictidae: Halictini). Zootaxa 216, 1–216 (2011). [Google Scholar]
- 60.Grixti JC, Wong LT, Cameron SA & Favret C Decline of bumble bees (Bombus) in the North American Midwest. Biol. Conserv 142, 75–84 (2009). [Google Scholar]
- 61.Sheffield CS, Ratti C, Packer L & Griswold T Leafcutter and Mason Bees of the Genus Megachile Latreille(Hymenoptera: Megachilidae) in Canada and Alaska. Can. J. Arthropod Identif 18, 1–107 (2011). [Google Scholar]
- 62.Schwarz RS & Evans JD Single and mixed-species trypanosome and microsporidia infections elicit distinct, ephemeral cellular and humoral immune responses in honey bees. Dev. Comp. Immunol 40, 300–10 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Meeus I, Brown MJF, de Graaf DC & Smagghe G Effects of invasive parasites on bumble bee declines. Conserv. Biol 25, 662–71 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Solter LF Epizootiology of microsporidiosis in invertebrate hosts. in Microsporidia: Pathogens of Opportunity: First Edition 165–194 (2014). doi: 10.1002/9781118395264.ch4 [DOI] [Google Scholar]
- 65.Otti O & Schmid-Hempel P Nosema bombi: A pollinator parasite with detrimental fitness effects. J. Invertebr. Pathol 96, 118–124 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Graystock P, Yates K, Darvill B, Goulson D & Hughes WOH Emerging dangers: deadly effects of an emergent parasite in a new pollinator host. J. Invertebr. Pathol 114, 114–119 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Fürst MA, McMahon DP, Osborne JL, Paxton RJ & Brown MJF Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506, 364–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Otti O & Schmid-Hempel P A field experiment on the effect of Nosema bombi in colonies of the bumblebee Bombus terrestris. Ecol. Entomol 33, 577–582 (2008). [Google Scholar]
- 69.Higes M, Martín-Hernández R & Meana A Nosema ceranae in Europe: an emergent type C nosemosis. Apidologie 41, 375–392 (2010). [Google Scholar]
- 70.Li J et al. Diversity of Nosema associated with bumblebees (Bombus spp.) from China. Int. J. Parasitol. Parasites Wildl 42, 49–61 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Sinpoo C, Disayathanoowat T, Williams PH & Chantawannakul P Prevalence of infection by the microsporidian Nosema spp. In native bumblebees (Bombus spp.) in northern Thailand. PLoS One 14, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Müller U, McMahon DP & Rolff J Exposure of the wild bee Osmia bicornis to the honey bee pathogen Nosema ceranae. Agric. For. Entomol 21, 363–371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bramke, Müller, McMahon & Rolff. Exposure of Larvae of the Solitary Bee Osmia bicornis to the Honey Bee Pathogen Nosema ceranae Affects Life History. Insects 10, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown MJF, Schmid-Hempel R & Schmid-Hempel P Strong context-dependent virulence in a host-parasite system: reconciling genetic evidence with theory. J. Anim. Ecol 72, 994–1002 (2003). [Google Scholar]
- 75.Yourth CP, Brown MJF & Schmid-Hempel P Effects of natal and novel Crithidia bombi (Trypanosomatidae) infections on Bombus terrestris hosts. Insectes Soc. 55, 86–90 (2008). [Google Scholar]
- 76.Brown MJF, Loosli R & Schmid-Hempel P Condition-dependent expression of virulence in a trypanosome infecting bumblebees. Oikos 91, 421–427 (2000). [Google Scholar]
- 77.Gegear RJ, Otterstatter MC & Thomson JD Bumble-bee foragers infected by a gut parasite have an impaired ability to utilize floral information. Proc. R. Soc. B Biol. Sci 273, 1073–1078 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imhoof B & Schmid-Hempel P Patterns of local adaptation of a protozoan parasite to its bumblebee host. Oikos 82, 59–65 (1998). [Google Scholar]
- 79.Dill LM Costs of energy shortfall for bumble bee colonies: Predation, social parasitism, and brood development. Can. Entomol 123, 283–293 (1991). [Google Scholar]
- 80.Strobl V, Yañez O, Straub L, Albrecht M & Neumann P Trypanosomatid parasites infecting managed honeybees and wild solitary bees. Int. J. Parasitol 49, 605–613 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Ravoet J et al. Differential diagnosis of the honey bee trypanosomatids Crithidia mellificae and Lotmaria passim. J. Invertebr. Pathol 130, 21–27 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Ngor L et al. Cross-Infectivity of Honey and Bumble Bee-Associated Parasites Across Three Bee Families. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lipa JJ & Triggiani O Apicystis gen nov and Apicystis bombi (Liu, Macfarlane & Pengelly) comb nov (Protozoa: Neogregarinida), a cosmopolitan parasite of Bombus and Apis (Hymenoptera: Apidae). Apidologie 27, 29–34 (1996). [Google Scholar]
- 84.Graystock P, Meeus I, Smagghe G, Goulson D & Hughes WOH The effects of single and mixed infections of Apicystis bombi and deformed wing virus in Bombus terrestris. Parasitology 143, 358–365 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Maharramov J et al. Genetic variability of the neogregarine Apicystis bombi, an etiological agent of an emergent bumblebee disease. PLoS One 8, e81475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rutrecht ST & Brown MJF The life-history impact and implications of multiple parasites for bumble bee queens. Int. J. Parasitol 38, 799–808 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Plischuk S, Meeus I, Smagghe G & Lange CE Apicystis bombi (Apicomplexa: Neogregarinorida) parasitizing Apis mellifera and Bombus terrestris (Hymenoptera: Apidae) in Argentina. Environ. Microbiol. Rep 3, 565–568 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Tian T, Piot N, Meeus I & Smagghe G Infection with the multi-host micro-parasite Apicystis bombi (Apicomplexa: Neogregarinorida) decreases survival of the solitary bee Osmia bicornis. J. Invertebr. Pathol 158, 43–45 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Lacey LA Manual of Techniques in Insect Pathology Manual of Techniques in Insect Pathology (Academic Press, 1997). doi: 10.1016/b978-0-12-432555-5.x5000-3 [DOI] [Google Scholar]
- 90.Fries I et al. Standard methods for Nosema research. J. Apic. Res 52, 1–28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mullins JL, Strange JP & Tripodi AD Why Are Queens Broodless? Failed Nest Initiation Not Linked to Parasites, Mating Status, or Ovary Development in Two Bumble Bee Species of Pyrobombus (Hymenoptera: Apidae: Bombus). J. Econ. Entomol (2019). doi: 10.1093/jee/toz330 [DOI] [PubMed] [Google Scholar]
- 92.Schmid-Hempel R & Tognazzo M Molecular divergence defines two distinct lineages of Crithidia bombi (Trypanosomatidae), parasites of bumblebees. J. Eukaryot. Microbiol 57, 337–45 (2010). [DOI] [PubMed] [Google Scholar]
- 93.Tripodi AD, Szalanski AL & Strange JP Novel multiplex PCR reveals multiple trypanosomatid species infecting North American bumble bees (Hymenoptera: Apidae: Bombus). J. Invertebr. Pathol 153, 147–155 (2018). [DOI] [PubMed] [Google Scholar]
- 94.King G & Zeng L Logistic Regression in Rare Events Data. Polit. Anal 9, 137–163 (2001). [Google Scholar]
- 95.Nelder JA A Reformulation of Linear Models. J. R. Stat. Soc. Ser. A 140, 48 (1977). [Google Scholar]
- 96.Venables WN Exegeses on Linear Models. in Paper presented to the SPlus User’s Conference, Washington DC, 8-9 October 1998 (2000). [Google Scholar]
- 97.R Core Team. R: A language and environment for statistical computing R Found. Stat. Comput Vienna, Austria: URL http//www.R-project.org/ (2018). [Google Scholar]
- 98.Bates D, Mächler M, Bolker B & Walker S Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw 67, (2015). [Google Scholar]
- 99.Brooks Mollie E., et al. glmmTMB Balances Speed and Flexibility Among Packages for Zero-inflated Generalized Linear Mixed Modeling. R J 9, 378 (2017). [Google Scholar]
- 100.Hartig F DHARMa: Residual Diagnostics for Hierarchical (Multi-Level / Mixed) Regression Models. R Packag. version 0.2.0 https://CRAN.R-project.org/package=DHARMa (2018). doi: 10.1016/j.diagmicrobio.2015.06.021 [DOI] [Google Scholar]
- 101.Signorell A DescTools: Tools for Descriptive Statistics. (2019). [Google Scholar]
- 102.Wood SN, Pya N & Säfken B Smoothing Parameter and Model Selection for General Smooth Models. J. Am. Stat. Assoc 111, 1548–1563 (2016). [Google Scholar]
- 103.Engels B XNomial: Exact Goodness-of-Fit Test for Multinomial Data with Fixed Probabilities. (2015). [Google Scholar]
- 104.Hothorn T, Bretz F & Westfall P Simultaneous inference in general parametric models. Biometrical J. 50, 346–363 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Oksanen J et al. vegan: Community Ecology Package R package version 2.4-3. (2017). Available at: https://cran.r-project.org/package=vegan. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data including site surveys and screening results are data found on Dryad in addition to all analysis code used [https://doi.org/10.6086/D1X09V]. Sequence data is also deposited in the NCBI database with accession numbers MT212154-MT212159, MT296581-MT296586, MT302779-MT302784, MT359894-MT359896, MT366919, MT387450 and MT387451).


