Abstract
SUMMARY
Introduction and objective
Reliable urinary biomarker proteins would be invaluable in identifying children with ureteropelvic junction obstruction (UPJO) as the existing biomarker proteins are inconsistent in their predictive ability. Therefore, the aim of this study was to identify consistent and reliable urinary biomarker proteins in children with UPJO.
Methods
To identify candidate biomarker proteins, total protein from age-restricted (<2 years) and sex-matched (males) control (n=22) and UPJO (n=21) urine samples was analyzed by mass spectrometry. Proteins that were preferentially identified in UPJO samples were selected (2-step process) and ranked according to their diagnostic odds ratio value. The top ten proteins with highest odds ratio values were selected and tested individually by ELISA. The total amount of each protein was normalized to urine creatinine and the median with interquartile ranges for control and UPJO samples was determined. Additionally, fold change (UPJO/Control) of medians of the final panel of 5 proteins was also determined. Finally, we calculated the average + 3(SD) and average + 4(SD) values of each of the 5 proteins in the control samples and used it as an arbitrary cutoff to classify individual control and UPJO samples.
Results
In the first step of our selection process, we identified 171 proteins in UPJO samples that were not detected in the majority of the control samples (16/22 samples, or 72.7%). Of the 171 proteins, only 50 proteins were detected in at least 11/21 (52.4%) of the UPJO samples and hence were selected in the second step. Subsequently, these 50 proteins were ranked according to the odds ratio value and the top 10 ranked proteins were validated by ELISA. Five of the 10 proteins – prostaglandin-reductase-1, ficolin-2, nicotinate-nucleotide pyrophosphorylase [carboxylating], immunoglobulin superfamily-containing leucine-rich-repeat-protein and vascular cell adhesion molecule-1 were present at higher levels in the UPJO samples (fold-change of the median protein concentrations ranging from 2.9 – 9.4) and emerged as a panel of biomarkers to identify obstructive uropathy. Finally, the order of prevalence of the 5 proteins in UPJO samples is PTGR1>FCN2>QPRT>ISLR>VCAM1.
Conclusion
In summary, this unique screening strategy led to the identification of previously unknown biomarker proteins that when screened collectively, may reliably distinguish between obstructed vs. non-obstructed infants and may prove useful in identifying informative biomarker panels for biological samples from many diseases.
Keywords: Ureteropelvic junction obstruction, Biomarker, Prostaglandin reductase 1 (PTGR1), Ficolin-2 (FCN2), Nicotinate-nucleotide pyrophosphorylase [carboxylating] (QPRT), Immunoglobulin superfamily containing leucine-rich repeat protein (ISLR), Vascular cell adhesion molecule 1 (VCAM1)
Graphical Abstract
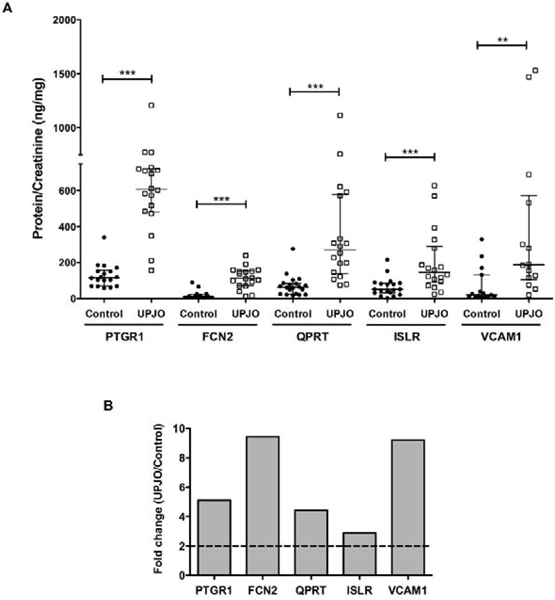
Summary Figure. PTGR1, FCN2, QPRT, ISLR and VCAM1 form a panel of UPJO-inherent urinary biomarker proteins.
(A) Normalized PTGR1, FCN2, QPRT, ISLR and VCAM1 amounts in control and UPJO samples.
(B) Fold change (UPJO/control) of medians of normalized PTGR1, FCN2, QPRT, ISLR and VCAM1 amounts.
INTRODUCTION
UPJO is one of the obstructive congenital anomalies of the kidney and urinary tract (CAKUT) and represents the most common cause of chronic kidney disease in children. Advances in maternal sonography have improved early detection of UPJO [1], however hydronephrosis does not necessarily equate to obstruction [2]. While surgical intervention is successful when nuclear medicine studies confirm obstruction, these studies are invasive. In some situations, a ‘watchful waiting’ approach is adopted for asymptomatic infants, entailing periodic assessment of function by radiotracer-extraction studies [3] that are cumbersome and often are equivocal [4]. Current therapeutic goals focus on halting progressive injury and enhancing subsequent healing. However, progress in these areas is severely hampered by the paucity of reliable biomarkers to assess the effects of obstruction.
Three proteins are currently advocated as potential urinary biomarkers of acute kidney injury, NGAL (neutrophil gelatinase-associated lipocalin), KIM-1 (kidney injury molecule-1) and cystatin C [5, 6]. However, cumulative results do not support their utility as they are inconsistent and variable in their predictive ability [5, 7–11]. We have also shown that neither NGAL nor KIM-1 were significantly different in the urine samples of control and UPJO patients [12]. Thus, further studies are necessary to identify reliable and consistent biomarkers.
In this study, we implemented quantitative label-free mass spectrometry-based proteomics analysis of UPJO patient and control urine samples to identify proteins that are preferentially present in UPJO patients (UPJO-inherent). We validated these findings with ELISA and generated a panel of 5 biomarker proteins (PTGR1, FCN2, QPRT, ISLR and VCAM1) that form a UPJO-inherent signature.
MATERIALS AND METHODS
Participants and urine samples
De-identified control (n = 22) and UPJO (n = 22) patients’ urine samples were collected with consent from parents and in accordance with an approved IRB protocol (#070–18-EP) from Children’s Hospital and Medical Center, Omaha, NE. Frozen urine samples were shipped to UConn Health according to an approved material transfer agreement. Patient cohort consists of males (<2 years) with proven UPJ obstruction (Mag-3 t½ >20 min, differential function – demonstrated <40% in the affected side) undergoing surgical repair. Urine specimens were collected upon catheterization for the Mag-3 study according to standard renogram protocol. All samples were negative for infection based on the results from urinalysis. Controls are age-matched males undergoing hypospadias repair with no associated renal anomalies.
Mass spectrometry analysis
Total protein content in the urine samples was determined and 1 mL of each sample was mixed with 8 mL of ice-cold acetone and incubated at -20°C for 1 hour. Samples were centrifuged at 11,000g for 30 minutes and supernatant was discarded. Pellets were washed with 5 mL of ice-cold acetone and centrifuged at 11,000g for 30 minutes. Protein pellets were air dried and stored at -80°C. Quantitative label-free mass spectrometry-based proteomics analysis was performed at the UConn Proteomics and Metabolomics Facility.
Urinary Proteomics Sample Preparation
Following protein precipitation and resuspension, an aliquot of each sample containing 100 ug of total protein was removed, dried to completion using a Labconco speedvac concentrator, and reconstituted in 100 μL 0.1 M ammonium bicarbonate in water (pH 8.0). Proteins were then subjected to Cys reduction and alkylation using 5 mM dithiothreitol in 0.1M ammonium bicarbonate (1.5 hours at 37°C) and 10 mM iodoacetamide in 0.1M ammonium bicarbonate (45 minutes at 37°C in the dark), respectively. Proteins were digested using sequencing grade modified trypsin (Promega, P/N V5113) at a 1:20 enzyme:protein ratio in a thermal mixer at 37°C for 16 hours. Proteolysis was quenched by the addition of formic acid to a final pH of 2.5. Tryptic peptides were desalted using Pierce C18 Desalting Spin Columns (P/N 89851) by following manufacturer’s instructions. Desalted peptides were dried in a Labconco speedvac concentrator, resuspended in 0.1% formic acid in water, and frozen at -20°C until further analysis.
Quantitative, Label-Free Mass Spectrometry-Based Proteomics Analysis
Tryptic peptides were quantified using a Nanodrop spectrophotometer (Protein A280 mode, Thermo Scientific) and diluted using 0.1% formic acid to provide uniform peptide concentrations across all samples. Peptides were injected into and separated using nanoflow ultra-high performance liquid chromatography (Dionex Ultimate 3000 RSLCnano) and immediately mass analyzed using high resolution tandem mass spectrometry (Thermo Scientific Q Exactive HF mass spectrometer). The nanoflow separation implemented a 1 hour linear gradient (Solvent A: 0.1% formic acid in water, Solvent B: 0.1% formic acid in acetonitrile) at 300 nL/min flow rates over a 2 μm, 100 Å, 75 μm x 25 cm Easy Spray PepMap C18 analytical column (Thermo Scientific) held at 35°C. Eluted peptides were directly ionized using electrospray ionization into the Q Exactive HF mass spectrometer which was operated using the following parameters: positive ESI mode, MS1 mass range 300–1800 Da with 60,000 resolution, Top 15 DDA MS/MS acquisition, MS2 resolution of 15,000, 27 NCE and charge state exclusion “on” for unassigned, +1 and >+8 charge states.
All raw files were searched against the Uniprot Homo sapiens reference proteome database (accessed 2017 Apr 22) using Andromeda and Maxquant software (v1.6.0.1) for peptide identification and label-free quantitation, respectively [13]. The following parameters were used for peptide/protein identification: 1% False Discovery Rate at the protein and peptide levels, variable modifications: oxidation of Met, N-terminal protein acetylation, and N-terminal peptide Gln to pyro Glu, fixed carbamidomethylation on Cys, trypsin cleavage specificity with 2 missed cleavages, 5 amino acids/peptide minimum, and MaxQuant LFQ “on”. All other parameters were kept at default values. Search results were uploaded into Scaffold v4.9 (Proteome Software, Inc.) for data visualization and further analysis. Average precursor intensity values of all identified proteins were determined (using Scaffold) and used for the selection of candidate biomarker proteins. Candidate biomarker proteins were identified using a 2-step process with the first step being the selection of proteins that were undetected in the majority of the control samples but were detected in UPJO samples. In the second step, proteins identified from first step were screened to select proteins that were present in at least half of the UPJO samples. Proteins (50) selected from this 2-step process were ranked according to diagnostic odds ratio values.
Diagnostic odds ratio analysis
Proteins undetected from mass spectrometric analysis had an average precursor intensity value of ‘0’, while detected proteins had a minimum value above 0.5*E+06 across all samples. Control and UPJO samples with 0 value for respective proteins were considered as ‘negative’, while samples with values >0.5*E+06 were considered as ‘positive’. Using this criteria, total number of positive and negative (control and UPJO) samples for each of the 50 proteins was determined. Subsequenlty, diagnostic odds ratio value for each protein was calculated using the following formula:
Odds ratio = [No. of positive UPJO samples × No. of negative control samples] / [No. of negative UPJO samples × No. of positive control samples]
Creatinine assay and ELISA
All the experiments were performed according to the manufacturer’s instructions. Creatinine levels were determined using creatinine assay kit (KGE005, R&D systems, MN, USA). Human FCN2 (ab213778), CFH (ab213765) and SPARCL1 (ab213826) ELISA kits were purchased from Abcam, MA, USA. Human PTGR1 (366461) and ISLR (152702) kits were purchased from US Biological, MA, USA. Human FGG (LS-F7036), BASP1 (OKCD02007), VCAM1 (KHT0601), QPRT (NBP2–60595) and SPRR3 (MBS2602661) ELISA kits were purchased from LifeSpan Biosciences, WA, USA; Aviva systems biology, CA, USA; Thermo Fisher scientific, MA, USA; Novus Biologicals, CO, USA and MyBioSource, CA, USA, respectively.
Classification of control and UPJO samples based on normalized ELISA values
To categorize individual control and UPJO samples, we used the average + 3(SD) and average + 4(SD) values of each of the 5 proteins in the control samples (Table 2) as an arbitrary cutoff to determine the number of individual control and UPJO samples that have values exceeding the cutoff value.
Table 2.
Average normalized concentrations of the 5 biomarker proteins in control samples
| Control samples | Average | SD | Average + 3(SD) | Average + 4(SD) |
|---|---|---|---|---|
| PTGR1/Creatinine (ng/mg) | 128.480 | 67.186 | 330.037 | 397.222 |
| FCN2/Creatinine (ng/mg) | 20.692 | 25.595 | 97.477 | 123.071 |
| QPRT/Creatinine (ng/mg) | 71.337 | 58.591 | 247.111 | 305.702 |
| ISLR/Creatinine (ng/mg) | 64.686 | 51.727 | 219.866 | 271.593 |
| VCAM1/Creatinine (ng/mg) | 69.849 | 100.377 | 370.981 | 471.358 |
SD – standard deviation
Statistical analyses
GraphPad Prism was used to perform statistical analyses. Values from individual experiments were tested for normal distribution by Kolmogorov-Smirnov test. Normally distributed values were compared using 2-tailed Student’s t test, while non-normally distributed values were compared using Mann-Whitney U test and significance was determined (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). All the p values from Kolmogorov-Smirnov test, Student’s t test and Mann-Whitney U test have been tabulated in the Supplementary Table 1.
RESULTS
Identification of UPJO-inherent candidate biomarker proteins
To minimize initial variability in the urine proteome [14, 15], we selected age-restricted (<2 years), infection-negative and gender-matched cohorts of UPJO patients with obvious hydronephrosis (n =21) and hypospadias repair controls (n =22) (Table 1). We analyzed bladder urine samples by mass spectrometry and proceeded with our selection strategy (Figure 1A). A two-step selection process was used to identify candidate biomarker proteins that are more likely to be present in UPJO patients and absent in controls, termed ‘UPJO-inherent’. In the first step, we identified 171 proteins in UPJO samples that were not detected in the majority of the control samples (16/22 samples, or 72.7%). Accordingly, subsequent screening demonstrated that 50 of the 171 proteins were differentially present in at least 11/21 (52.4%) UPJO samples but undetected in control samples. To further increase the stringency of the screen, we performed an odds ratio analysis to identify the top 10 ranked proteins (Figure 1B and 1C, Supplementary Table 2). NGAL/LCN2 was included in the analysis to compare the UPJO-selectivity of the top 10 ranked proteins to this acknowledged biomarker. LCN2 was present in 6/16 control samples and 5/16 UPJO samples, yielding a low odds ratio value (0.8) when compared to the odds ratios of the top 10 proteins (ranging from 7.0–52.5). Additionally, neither KIM-1 nor cystatin C were detected in our mass spectrometry analysis, potentially due to their low abundance in comparison to the 10 selected proteins, further underscoring that these top 10 proteins may be superior prognostic markers of UPJO.
Table 1.
Control and UPJO patient characteristics
| Control | UPJO | |
|---|---|---|
| Number of samples | 22 | 22 |
| Sex | M | M |
| Age - Average (SD) | 8.3 (3.0) months | 6.0 (3.8) months |
| Weight - Average (SD) | 8.5 (1.3) kg | 7.3 (1.7) kg |
| Patients | Hypospadias repair | Confirmed obstruction - Mag-3 t1/2 >20min with differential function <40% in the affected side |
| Notes | Control-12 was used for mass spectrometric analysis only. Remaining samples were used for both mass spectrometric analysis and ELISA. | UPJO-6, 7, 8 and 21 were used for mass spectrometric analysis only, while UPJO-1 was used for ELISA only. Remaining samples were used for both the analyses. |
Due to the limited availability, some of the samples were used for either mass spectrometric analysis or ELISA and have been identified in the notes.
Figure 1. Mass spectrometric identification of 10 UPJO-inherent candidate biomarker proteins.
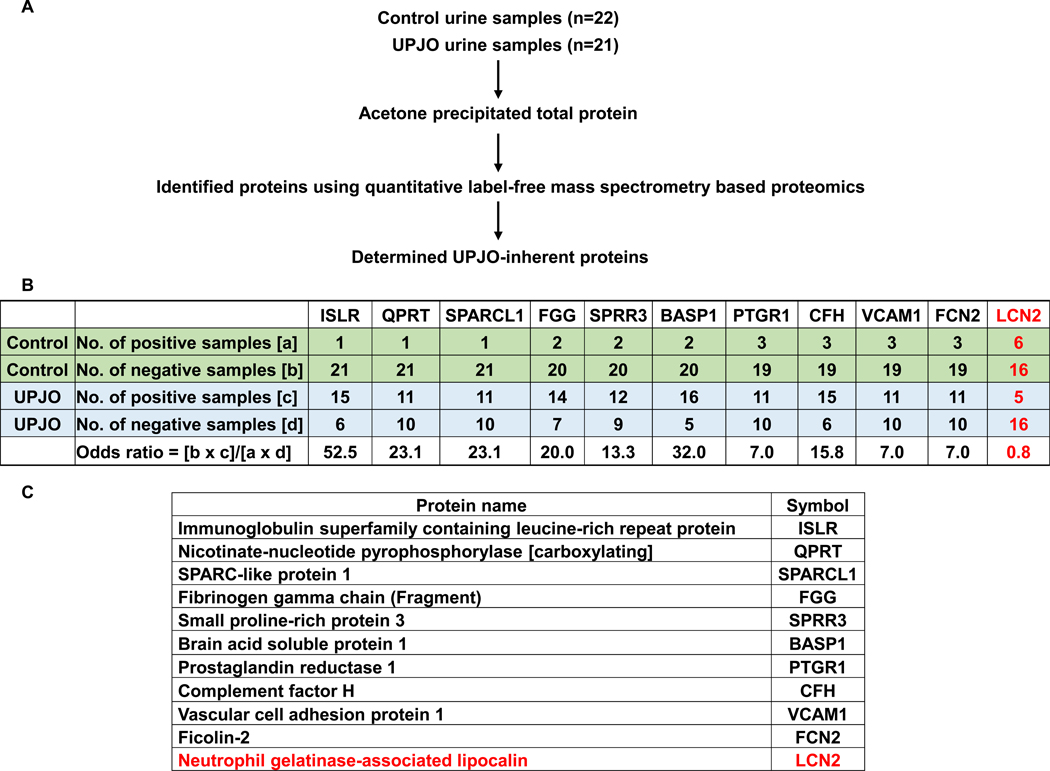
(A) Methodology employed to identify UPJO-inherent urinary proteins.
(B) Odds ratio analysis of top 10 UPJO-inherent urinary proteins identified by mass spectrometry analysis. Average precursor intensities of individual proteins was used to stratify control and UPJO samples according to the presence or absence of candidate proteins. Odds ratio was calculated for each protein as shown in the table. LCN2 was included for comparative analysis.
(C) List of the top ten ranked biomarker proteins.
PTGR1, FCN2, QPRT, ISLR and VCAM1 form a panel of UPJO-inherent biomarker proteins
To determine quantitative values and relative changes, highly sensitive ELISA assays with larger dynamic range were performed for each of the top 10 proteins and normalized to urine creatinine concentrations. As shown previously by us [12] and others [16], the average creatinine concentration in UPJO samples [Ave(SD) - 6.11(3.7)mg/dL] was consistently lower than that of the control samples [Ave(SD) – 36.42(19.5)mg/dL] (Supplementary Table 3). Concentrations of BASP1, FGG and SPRR3 proteins were below the detection limit in all the samples and subsequently excluded from the panel. Similarly, SPARCL1 and CFH were detected in only a few samples and were also removed from further analysis. However, the normalized concentrations of PTGR1, FCN2, QPRT, ISLR and VCAM1 were significantly higher in the UPJO samples (Figure 2A) with fold-change (UPJO/Control) of the median protein concentrations ranging from 2.9 – 9.4 (Figure 2B), further validating their potential as UPJO biomarkers.
Figure 2. Validation of PTGR1, FCN2, QPRT, ISLR and VCAM1 as a panel of UPJO-inherent urinary biomarker proteins.
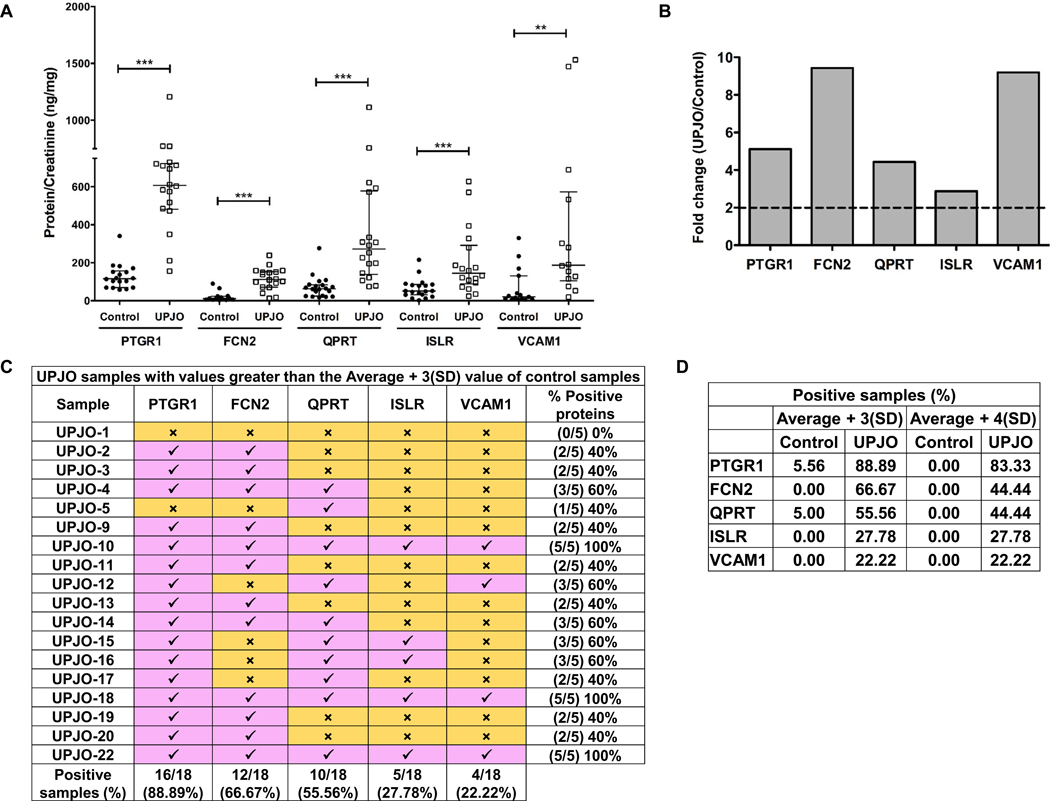
(A) Normalized PTGR1, FCN2, QPRT, ISLR and VCAM1 amounts in control and UPJO samples. Individual protein concentrations (ng/mL) were determined using commercial ELISA kits and normalized to respective urine creatinine concentrations (mg/dL) to obtain protein/creatinine in ng/mg. Data is represented as median (IQR). PTGR1 control (n = 18), median (IQR) = 118.81 (69.73 – 152.58); PTGR1 UPJO (n = 18), median (IQR) = 607.64 (493.13 – 718.67); FCN2 control (n = 18), median (IQR) = 11.86 (5.74 – 23.72); FCN2 UPJO (n = 18), median (IQR) = 111.82 (77.44 – 150.73); QPRT control (n = 20), median (IQR) = 61.30 (25.30 – 83.56); QPRT UPJO (n = 18), median (IQR) = 271.79 (157.66 – 512.80) ); ISLR control (n = 21), median (IQR) = 50.34 (31.43 – 81.37); ISLR UPJO (n = 18), median (IQR) = 144.85 (97.78 – 256.37); VCAM1 control (n = 21), median (IQR) = 20.38 (10.16 – 85.42) ) and VCAM1 UPJO (n = 18), median (IQR) = 187.30 (118.49 – 475.57). Statistical analysis on PTGR1 control and UPJO samples was performed using 2-tailed Student’s t test while the rest of the comparative analyses were performed using 2-tailed Mann Whitney test. **p ≤ 0.01, ***p ≤ 0.001, IQR – Interquartile range.
(B) Fold change (UPJO/control) of medians of normalized PTGR1, FCN2, QPRT, ISLR and VCAM1 amounts.
(C) Classification of UPJO samples based on the average + 3(SD) value of control samples. Average and SD of individual proteins in control samples as determined from ELISA was used to identify UPJO samples with values greater than the average + 3(SD) value of respective control samples. Pink boxes with a check mark indicate samples with values greater than the average + 3(SD) value of control samples, while the yellow boxes with a cross mark indicate samples with values smaller than the average + 3(SD) value of control samples. Number of positive samples for each protein as well as the number of positive proteins in each sample are enumerated in the table. SD – standard deviation.
(D) Summary of percent positive control and UPJO samples. Control and UPJO samples with values greater than the average + 3(SD) and average + 4(SD) value of respective control samples were enumerated and percent positive samples have been tabulated. SD – standard deviation.
Individual control and UPJO samples were classified using the average + 3(SD) and average + 4(SD) values of each of the 5 proteins in the control samples (Table 2) as an arbitrary cutoff to determine the number of individual control and UPJO samples that have values exceeding the cutoff value. Of the 18 UPJO samples, only one sample (UPJO-1) had a value beneath that of the control cutoff value for all 5 proteins, while the remaining 17 samples had at least 1 protein present with a value above the cutoff value (Figure 2C). Additionally, barring UPJO-5, the remaining 16 samples contain at least 2 proteins at levels above the cutoff value (Figure 2C). Alternatively, only one control sample (control-16) had PTGR1 and QPRT present at above the respective cutoff value while the remaining controls were below (Supplementary Table 4). Increasing the stringency to average + 4(SD) cutoff value eliminated the only positive control sample, while retaining a similar distribution in the UPJO samples (Supplementary Table 5A, 5B and Figure 2D). Finally, the order of prevalence of the 5 proteins in UPJO samples is PTGR1>FCN2>QPRT>ISLR>VCAM1.
DISCUSSION
In this study, we identified a panel of 5 unique proteins that form a UPJO-inherent signature. We minimized variability by selecting a specific cohort of patients who are at the greatest risk and avoided pooling of different age groups, which has been shown to confound the results [14, 15]. Additionally, only males were included since males are more frequently affected than females (2–3:1) [17]. Patient urine was also confirmed to be free of infection by urinalysis. We also established a unique methodology to identify candidate biomarker proteins by choosing proteins that are absent in most control samples rather than choosing proteins that are present in both samples at quantitatively different levels. Although these proteins were detected in some control samples by ELISA analysis, the fold difference was significantly higher with UPJO.
Urinary tract obstruction can lead to progressive cellular damage due to hypoxia, reactive oxygen species and fibrosis, ultimately resulting in tubular cell death and interstitial inflammation [18–20]. Considering the multi-dimensional effects of obstruction, it is logical that we identified a diverse biomarker protein panel. Mechanistically, these biomarker proteins are detectable in the urine due to increased release, reduced renal reabsorption or both. PTGR1 is an enzyme that mediates catabolism of eicosanoids and is highly expressed in kidney tubular cells, cancer cells and aids in cell proliferation and oxidative stress tolerance [21, 22]. Considering the increased oxidative stress on an obstructed kidney, high levels of PTGR1 could be released in the urine due to tubular cell death. Accordingly, PTGR1 was the most prevalent protein in our cohort of UPJO samples (16/18). Similarly, QPRT, an enzyme essential for the synthesis of nicotinamide adenine dinucleotide (NAD+) protects kidneys from acute injury [23]. FCN2, a plasma pattern recognition receptor inhibits epithelial to mesenchymal transition (EMT) of hepatocellular carcinoma by reducing TGF-β signaling [24]. Inhibition of EMT of tubular epithelial cells would be expected to prevent fibrosis in obstructed kidneys [25] and therefore increased FCN2 and QPRT could be early defense mechanisms seen in our cohort of young infants. Mesenchymal stromal cells (MSCs) in fetal kidney are essential for the differentiation of nephrons and the undifferentiated nature of MSCs is maintained by ISLR [26], thus the initial observation of ISLR expression in fetal but not adult kidney [27] as well as in our cohort of patients. VCAM1, an inflammation-induced endothelial cell adhesion molecule, is a urinary biomarker for kidney injury in lupus nephritis [28] and in women with preeclampsia [29].
Numerous studies have been performed to identify reliable urinary biomarkers ranging from proteins to antimicrobial peptides to polyoma virus [6, 9, 30–34]. Some of these biomarkers such as TNF-α and caspase 3 are not only increased in children with UPJO, but also show a reduction in their levels following pyeloplasty, further emphasizing their potential to monitor the recovery process [30]. Urinary NGAL and KIM-1 have long been considered as the ideal biomarkers of urinary obstruction [5, 11], but we along with others have noted that they are inconsistent across different patient groups and in the current study, NGAL showed a very weak odds ratio while KIM-1 was undetectable [5, 7–11]. Considering the diversity of the molecules that have been identified as biomarkers, it is perhaps not surprising that none of them can be hailed as the ‘gold-standard’ for detecting urinary obstruction. This may be partly attributed to the fact that these studies are primarily aimed at determining the strength of specific molecules/proteins as urinary biomarkers and not a comparative analysis of a larger pool of candidate proteins. As seen in this study and previous studies, an unbiased survey of urinary proteome could circumvent this issue and lead to the identification of a larger pool of candidate proteins that can then be further tested for their true potential [31, 35]. Our previous study with a similar objective highlights the limitation of a biased approach where we identified a panel of three proteins (CD10, CD13 and CD26) that reliably predicted UPJO [12]. However, this study specifically chose only single-pass, apically expressed proximal tubule brush border proteins to test the hypothesis that these would be shed into the urine early and rapidly and therefore could be reliable noninvasive urinary biomarkers. The current study took a more unbiased approach to identifying biomarker proteins, and CD10, CD13, and CD26 were not identified in our proteomic analysis of a more narrowly defined group. Additionally, biomarker proteins reflect the expression profile of the various kidney cells, which is constantly changing in a developing kidney in comparison to an adult kidney.
Therefore, studies that include a diverse patient pool have probably led to the misidentification of biomarker proteins or camouflaging of true biomarker proteins owing to weak statistical significance. Our prior study also included urine samples from a wide range of ages, stages and both the genders, which also may have affected our findings.
Limitations of our study include a narrow data set, consistent with the nature of pilot studies. Despite the high statistical significance observed, validation studies will clearly be needed. In addition, our biomarker panel was established by comparing the two extremes of obstruction, confirmed vs. non-obstructed in male patients. We plan further analyses comparing patients from both sexes with non-obstructive hydronephrosis vs. obstructive hydronephrosis to confirm and extend the utility of our panel. Another potential limitation is our focus on proteins in bladder urine rather than urine collected from the hydronephrotic renal pelvis during surgery. We purposely selected bladder urine because obtaining samples from the renal pelvis in patients prior to surgery is not practical. Our objective was to identify a panel of biomarkers that will perform adequately in routine specimen collection in the clinic, despite dilution from the contralateral kidney. Furthermore, it is possible that the presence of urinary tract infection could impact the reliability of our panel. In our study, we circumvented this question by choosing patients without infection, but it can be systematically addressed in the future. Similarly, potential effects of administration of prophylactic antibiotics (amoxicillin or trimethoprim/sulfamethoxazole) is unlikely since the use of these drugs at customary doses are not associated with renal injury [36], thus we do not expect alterations in the urinary proteome. This too can be tested in a controlled future study.
Despite decades of substantial effort, useful urinary biomarkers for the detection of obstruction continue to prove elusive. Utilizing a unique screening and stratification strategy we have identified a panel of previously unknown biomarker proteins that may prove useful in identifying obstruction. Evolving these biomarkers as a panel that can determine renal damage due to ongoing renal obstruction would radically change how we evaluate infants’ pre- and post-surgical intervention, reducing the use of costly and invasive studies as well as possibly reducing diagnostic delays.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ayoola Ogun and Evan Roberts from Children’s Hospital Omaha for collecting and sending samples and Claire Gerber and Miriam Harel for early contributions to the project.
FUNDING
This work was supported by the National Heart, Lung, and Blood Institute (NHLBI grant number HL127449)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Kim SY, Kim MJ, Yoon CS, Lee MS, Han KH, Lee MJ. Comparison of the reliability of two hydronephrosis grading systems: The Society for Foetal Urology grading system vs. the Onen grading system. Clin Radiol. 2013. [DOI] [PubMed] [Google Scholar]
- [2].Piepsz A Antenatal detection of pelviureteric junction stenosis: main controversies. Semin Nucl Med. 2011;41:11–9. [DOI] [PubMed] [Google Scholar]
- [3].Mesrobian HG, Mirza SP. Hydronephrosis: a view from the inside. Pediatr Clin North Am. 2012;59:839–51. [DOI] [PubMed] [Google Scholar]
- [4].Conway JJ, Maizels M. The “well tempered” diuretic renogram: a standard method to examine the asymptomatic neonate with hydronephrosis or hydroureteronephrosis. A report from combined meetings of The Society for Fetal Urology and members of The Pediatric Nuclear Medicine Council--The Society of Nuclear Medicine. J Nucl Med. 1992;33:2047–51. [PubMed] [Google Scholar]
- [5].Wasilewska A, Taranta-Janusz K, Debek W, Zoch-Zwierz W, Kuroczycka-Saniutycz E. KIM-1 and NGAL: new markers of obstructive nephropathy. Pediatr Nephrol. 2011;26:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pavlaki A, Printza N, Farmaki E, Stabouli S, Taparkou A, Sterpi M, et al. The role of urinary NGAL and serum cystatin C in assessing the severity of ureteropelvic junction obstruction in infants. Pediatr Nephrol. 2019. [DOI] [PubMed] [Google Scholar]
- [7].Liu KD, Yang W, Anderson AH, Feldman HI, Demirjian S, Hamano T, et al. Urine neutrophil gelatinase-associated lipocalin levels do not improve risk prediction of progressive chronic kidney disease. Kidney Int. 2013;83:909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nielsen SE, Andersen S, Zdunek D, Hess G, Parving HH, Rossing P. Tubular markers do not predict the decline in glomerular filtration rate in type 1 diabetic patients with overt nephropathy. Kidney Int. 2011;79:1113–8. [DOI] [PubMed] [Google Scholar]
- [9].Cost NG, Noh PH, Devarajan P, Ivancic V, Reddy PP, Minevich E, et al. Urinary NGAL levels correlate with differential renal function in patients with ureteropelvic junction obstruction undergoing pyeloplasty. J Urol. 2013;190:1462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Urbschat A, Gauer S, Paulus P, Reissig M, Weipert C, Ramos-Lopez E, et al. Serum and urinary NGAL but not KIM-1 raises in human postrenal AKI. Eur J Clin Invest. 2014;44:652–9. [DOI] [PubMed] [Google Scholar]
- [11].Holzscheiter L, Beck C, Rutz S, Manuilova E, Domke I, Guder WG, et al. NGAL, L-FABP, and KIM-1 in comparison to established markers of renal dysfunction. Clin Chem Lab Med. 2014;52:537–46. [DOI] [PubMed] [Google Scholar]
- [12].Gerber C, Harel M, Lynch ML, Herbst KW, Ferrer FA, Shapiro LH. Proximal tubule proteins are significantly elevated in bladder urine of patients with ureteropelvic junction obstruction and may represent novel biomarkers: A pilot study. J Pediatr Urol. 2016;12:120 e1–7. [DOI] [PubMed] [Google Scholar]
- [13].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- [14].Drube J, Zurbig P, Schiffer E, Lau E, Ure B, Gluer S, et al. Urinary proteome analysis identifies infants but not older children requiring pyeloplasty. Pediatr Nephrol. 2010;25:1673–8. [DOI] [PubMed] [Google Scholar]
- [15].Decramer S, Wittke S, Mischak H, Zurbig P, Walden M, Bouissou F, et al. Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nat Med. 2006;12:398–400. [DOI] [PubMed] [Google Scholar]
- [16].Alizadeh F, Taefnia AM, Haghdani S. Urinary carbohydrate antigen 19–9/creatinine ratio: A non-invasive marker for follow-up of unilateral ureteropelvic junction obstruction in children. J Pediatr Urol. 2018;14:62 e1-e4. [DOI] [PubMed] [Google Scholar]
- [17].Esmaeili M, Esmaeili M, Ghane F, Alamdaran A. Comparison Between Diuretic Urography (IVP) and Diuretic Renography for Diagnosis of Ureteropelvic Junction Obstruction in Children. Iran J Pediatr. 2016;26:e4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chevalier RL, Thornhill BA, Forbes MS, Kiley SC. Mechanisms of renal injury and progression of renal disease in congenital obstructive nephropathy. Pediatr Nephrol. 2010;25:687–97. [DOI] [PubMed] [Google Scholar]
- [19].Chevalier RL, Thornhill BA, Wolstenholme JT, Kim A. Unilateral ureteral obstruction in early development alters renal growth: dependence on the duration of obstruction. J Urol. 1999;161:309–13. [PubMed] [Google Scholar]
- [20].Forbes MS, Thornhill BA, Minor JJ, Gordon KA, Galarreta CI, Chevalier RL. Fight-or-flight: murine unilateral ureteral obstruction causes extensive proximal tubular degeneration, collecting duct dilatation, and minimal fibrosis. Am J Physiol Renal Physiol. 2012;303:F120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sanchez-Rodriguez R, Torres-Mena JE, Quintanar-Jurado V, Chagoya-Hazas V, Rojas Del Castillo E, Del Pozo Yauner L, et al. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free radical biology & medicine. 2017;102:87–99. [DOI] [PubMed] [Google Scholar]
- [22].Xue L, Zhu Z, Wang Z, Li H, Zhang P, Wang Z, et al. Knockdown of prostaglandin reductase 1 (PTGR1) suppresses prostate cancer cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Trends. 2016;10:133–9. [DOI] [PubMed] [Google Scholar]
- [23].Poyan Mehr A, Tran MT, Ralto KM, Leaf DE, Washco V, Messmer J, et al. De novo NAD(+) biosynthetic impairment in acute kidney injury in humans. Nat Med. 2018;24:1351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang G, Liang Y, Zheng T, Song R, Wang J, Shi H, et al. FCN2 inhibits epithelial-mesenchymal transition-induced metastasis of hepatocellular carcinoma via TGF-beta/Smad signaling. Cancer letters. 2016;378:80–6. [DOI] [PubMed] [Google Scholar]
- [25].Klein J, Gonzalez J, Miravete M, Caubet C, Chaaya R, Decramer S, et al. Congenital ureteropelvic junction obstruction: human disease and animal models. Int J Exp Pathol. 2011;92:168–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maeda K, Enomoto A, Hara A, Asai N, Kobayashi T, Horinouchi A, et al. Identification of Meflin as a Potential Marker for Mesenchymal Stromal Cells. Sci Rep. 2016;6:22288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nagasawa A, Kubota R, Imamura Y, Nagamine K, Wang Y, Asakawa S, et al. Cloning of the cDNA for a new member of the immunoglobulin superfamily (ISLR) containing leucine-rich repeat (LRR). Genomics. 1997;44:273–9. [DOI] [PubMed] [Google Scholar]
- [28].Singh S, Wu T, Xie C, Vanarsa K, Han J, Mahajan T, et al. Urine VCAM-1 as a marker of renal pathology activity index in lupus nephritis. Arthritis Res Ther. 2012;14:R164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Y, Gu Y, Loyd S, Jia X, Groome LJ. Increased urinary levels of podocyte glycoproteins, matrix metallopeptidases, inflammatory cytokines, and kidney injury biomarkers in women with preeclampsia. Am J Physiol Renal Physiol. 2015;309:F1009–17. [DOI] [PubMed] [Google Scholar]
- [30].Shirazi M, Eslahi A, Sharifi V, Rahimi F, Safarpour A. Evaluation of Caspase 3 Enzyme and TNF-alpha as Biomarkers in Ureteropelvic Junction Obstruction in Children- a preliminary report. Pakistan journal of medical sciences. 2017;33:315–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen H, Lin H, Xu M, Xu G, Fang X, He L, et al. Quantitative Urinary Proteome Reveals Potential Biomarkers for Ureteropelvic Junction Obstruction. Proteomics Clin Appl. 2019;13:e1800101. [DOI] [PubMed] [Google Scholar]
- [32].Papachristou F, Pavlaki A, Printza N. Urinary and serum biomarkers in ureteropelvic junction obstruction: a systematic review. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals. 2014;19:531–40. [DOI] [PubMed] [Google Scholar]
- [33].Gupta S, Jackson AR, DaJusta DG, McLeod DJ, Alpert SA, Jayanthi VR, et al. Urinary antimicrobial peptides: Potential novel biomarkers of obstructive uropathy. J Pediatr Urol. 2018;14:238 e1-e6. [DOI] [PubMed] [Google Scholar]
- [34].Assadi F, Mazaheri M. Urinary polyomavirus: novel biomarker of congenital ureteropelvic junction obstruction. J Pediatr Urol. 2019. [DOI] [PubMed] [Google Scholar]
- [35].Froehlich JW, Kostel SA, Cho PS, Briscoe AC, Steen H, Vaezzadeh AR, et al. Urinary Proteomics Yield Pathological Insights for Ureteropelvic Junction Obstruction. Mol Cell Proteomics. 2016;15:2607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rouse RL, Zhang J, Stewart SR, Rosenzweig BA, Espandiari P, Sadrieh NK. Comparative profile of commercially available urinary biomarkers in preclinical drug-induced kidney injury and recovery in rats. Kidney Int. 2011;79:1186–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


