Figure 1. Engineered biosynthetic pathway for de novo production of scopolamine in yeast and optimization of PLA-glucoside biosynthesis.
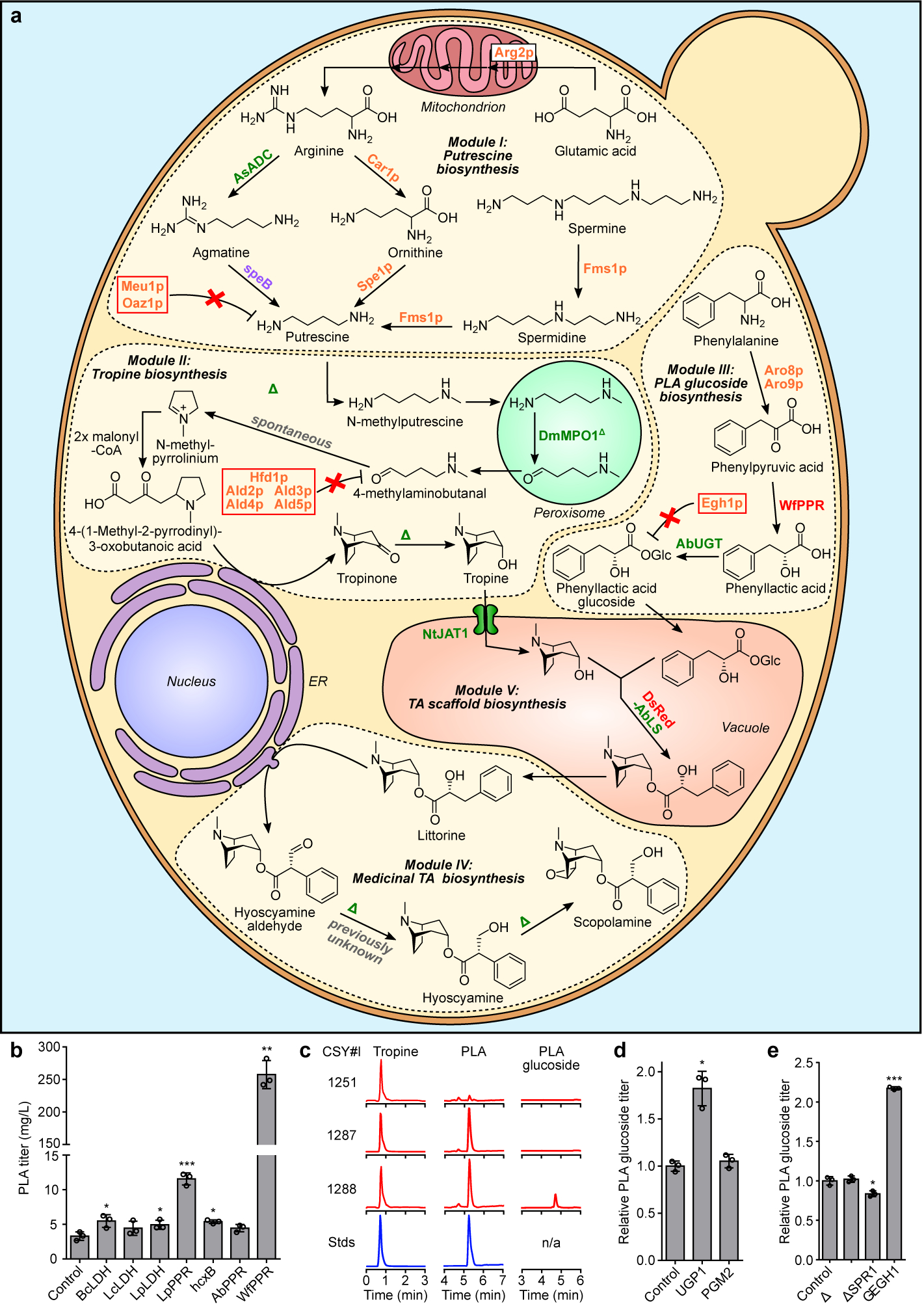
(a) Modular pathway construction for scopolamine biosynthesis in yeast. Enzyme/protein color scheme: orange, yeast (overexpressed); green, plant; purple, bacteria; red, other eukaryote; grey, spontaneous/non-enzymatic. Red boxes indicate disrupted yeast proteins; dotted lines/solid line of vacuole membrane delineate functional biosynthetic modules. Enzymes: Arg2p, glutamate N-acetyltransferase; Car1p, arginase; AsADC, Avena sativa arginine decarboxylase; speB, agmatine ureohydrolase; Spe1p, ornithine decarboxylase; Fms1p, polyamine oxidase; Meu1p, methylthioadenosine phosphorylase; Oaz1p, ornithine decarboxylase antizyme-1; AbPMT1/DsPMT1, Atropa belladonna/Datura stramonium putrescine N-methyltransferase 1; DmMPO1ΔC-PTS1, Datura metel N-methylputrescine oxidase 1 with peroxisome targeting sequence 1 and truncated C-terminus; AbPYKS, A. belladonna pyrrolidine ketide synthase; AbCYP82M3, A. belladonna tropinone synthase; AtATR1, Arabidopsis thaliana NADP+-cytochrome P450 reductase; DsTR1, D. stramonium tropinone reductase I; Hfd1p/Ald2p-Ald5p, aldehyde dehydrogenases; Aro8p/Aro9p, aromatic amino acid aminotransferases; WfPPR, Wickerhamia fluorescens 3-phenylpyruvate reductase; AbUGT, A. belladonna UDP-glucosyltransferase 84A27; Egh1p, steryl-β-glucosidase; DsRed-AbLS, Discosoma sp. red fluorescent protein fused to the N-terminus of A. belladonna littorine synthase; NtJAT1, Nicotiana tabacum jasmonate-inducible alkaloid transporter 1; AbCYP80F1, A. belladonna littorine mutase; DsHDH, D. stramonium hyoscyamine dehydrogenase; DsH6H, D. stramonium hyoscyamine 6β-hydroxylase/dioxygenase. (b) PLA production in yeast engineered for expression of phenylpyruvate reductases (PPRs) or lactate dehydrogenases (LDHs). Heterologous enzymes or negative control (BFP) were expressed from low-copy plasmids in strain CSY1251. (c) Multiple reaction monitoring (MRM) and extracted ion chromatogram (EIC) traces from culture media of yeast engineered for step-wise reconstitution of PLA glucoside biosynthesis via Module III. Chromatogram traces are representative of three biological replicates. (d) Relative PLA glucoside titers in yeast engineered for overexpression of UDP-glucose biosynthetic enzymes. Enzymes or negative control (BFP) were expressed from low-copy plasmids in strain CSY1288. (e) Relative PLA glucoside titers in CSY1288 with disruptions to endogenous glucosidases. For d and e, PLA glucoside accumulation was compared using relative titers due to lack of an authentic chemical standard. Strains were cultured for 72 h before LC-MS/MS analysis of metabolites in culture supernatant. Data in b, d, and e represent the mean of n = 3 biologically independent samples (open circles), error bars show standard deviation. Student’s two-tailed t-test: *P < 0.05, **P < 0.01, ***P < 0.001. Statistical significance is shown relative to controls. Exact P-values are in Supplementary Table 5.
