Extended Data Figure 5. Phylogenetic analysis of hyoscyamine dehydrogenase (HDH).
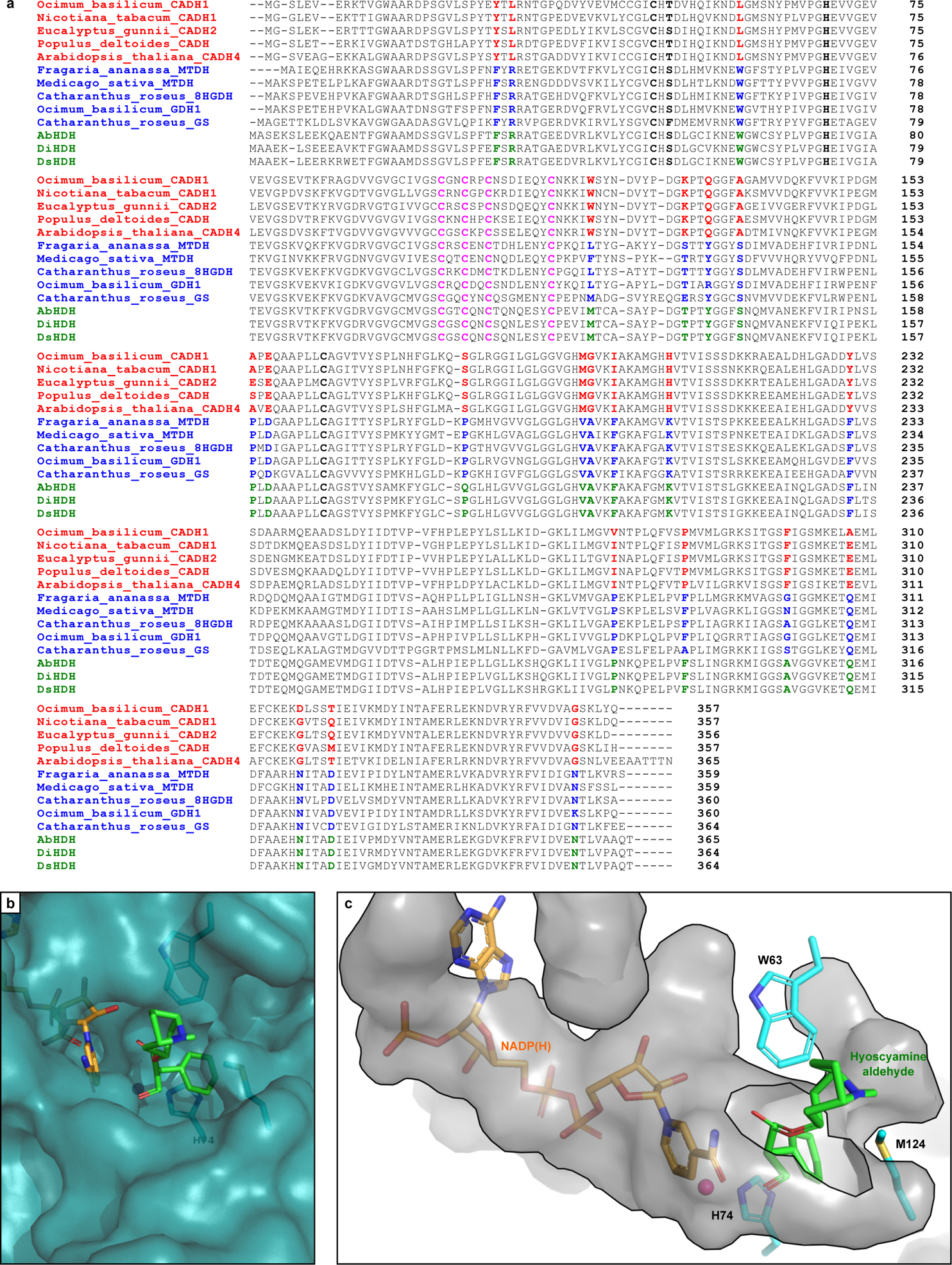
(a) Sequence alignment of AbHDH, DiHDH, and DsHDH (green) with selected BLASTp hits from the cinnamyl alcohol dehydrogenase (CAD)-like (red) and sinapyl alcohol dehydrogenase (SAD)-like (blue) families. Catalytic Zn2+ residues are highlighted in bold (black); structural Zn2+-binding residues are highlighted in pink; and key conserved residues which differentiate CAD- and SAD-like ADHs are highlighted in red/blue/green. (b) Zoomed surface view of AbHDH substrate binding pocket with bound NADPH (orange) and hyoscyamine aldehyde (green) based on docking simulations. (c) Zoomed inverse surface (cavity) view of AbHDH active site pocket with bound NADPH (orange) and hyoscyamine aldehyde (green) based on docking simulations as in b. Residues W63, H74, and M124 form a tight binding pocket for the aryl moiety of hyoscyamine aldehyde, leaving the tropine moiety to extend out of the active site.
