Abstract
NMDA receptors are ligand-gated ion channels that mediate excitatory neurotransmission. Most native NMDA receptors are tetrameric assemblies of two glycine-binding GluN1 and two glutamate-binding GluN2 subunits. Co-assembly of the glycine-binding GluN1 with glycine-binding GluN3 subunits (GluN3A-B) creates glycine-activated receptors that possess strikingly different functional and pharmacological properties compared to GluN1/GluN2 NMDA receptors. The role of GluN1/GluN3 receptors in neuronal function remains unknown, in part due to lack of pharmacological tools with which to explore their physiological roles. We have identified the negative allosteric modulator EU1180–438, which is selective for GluN1/GluN3 receptors over GluN1/GluN2 NMDA receptors, AMPA, and kainate receptors. EU1180–438 is also inactive at GABA, glycine, and P2X receptors, but displays inhibition of some nicotinic acetylcholine receptors. Furthermore, we demonstrate that EU1180–438 produces robust inhibition of glycine-activated current responses mediated by native GluN1/GluN3A receptors in hippocampal CA1 pyramidal neurons. EU1180–438 is a non-competitive antagonist with activity that is independent of membrane potential (i.e. voltage-independent), glycine concentration, and extracellular pH. Non-stationary fluctuation analysis of neuronal current responses provided an estimated weighted mean unitary conductance of 6.1 pS for GluN1/GluN3A channels, and showed that EU1180–438 has no effect on conductance. Site-directed mutagenesis suggests that structural determinants of EU1180–438 activity reside near a short pre-M1 helix that lies parallel to the plane of the membrane below the agonist binding domain. These findings demonstrate that structural differences between GluN3 and other glutamate receptor subunits can be exploited to generate subunit-selective ligands with utility in exploring the roles GluN3 in neuronal function.
Keywords: N-methyl-D-aspartate (NMDA) receptors, GluN3A subunit, Negative allosteric modulator, Non-competitive antagonist, Site of action
1. Introduction
N-methyl-D-aspartate (NMDA) receptors are ligand-gated cation-selective channels that belong to the family of ionotropic glutamate receptors (Stephen F. Traynelis et al., 2010). NMDA receptors that contain GluN1 and GluN2 subunits have been extensively studied over the past several decades, and many of the structural and functional properties are now well understood (K. B. Hansen et al., 2018; Paoletti et al., 2013; Stephen F. Traynelis et al., 2010). In stark contrast to GluN1/GluN2 NMDA receptors, many key properties of GluN3-containing NMDA receptors remain elusive (reviewed in (N. A. Cavara & Hollmann, 2008a; Eriksson et al., 2007; M. A. Henson et al., 2010a; X. Huang et al., 2017; Y. H. Huang et al., 2013; Kehoe et al., 2013; Low & Wee, 2010; Pachernegg et al., 2012; Perez-Otano et al., 2016; Wesseling & Perez-Otano, 2015; T. Yuan & Bellone, 2013)). NMDA receptors composed of two GluN1 and two GluN3 subunits (GluN1/GluN3) have been consistently demonstrated in heterologous expression systems (e.g. Xenopus oocytes and HEK cells) (Awobuluyi et al., 2007; Balasuriya et al., 2014; N. A. Cavara et al., 2009; Chatterton et al., 2002; Cummings et al., 2017; Cummings & Popescu, 2016; Grand et al., 2018; Kaniakova et al., 2018; Kvist et al., 2013a; Kvist et al., 2013b; Madry et al., 2008, 2010; Madry et al., 2007; McClymont et al., 2012; Schuler et al., 2008; Smothers & Woodward, 2003, 2007, 2009; Ulbrich & Isacoff, 2007, 2008; Wada et al., 2006; Wee et al., 2010). NMDA receptors composed of GluN1, GluN2, and GluN3 subunits (GluN1/GluN2/GluN3) have also been suggested to exist in some expression systems (reviewed in (Perez-Otano et al., 2016)), but the function and subunit stoichiometry of GluN1/GluN2/GluN3 receptors have not been resolved (e.g. see (Ulbrich & Isacoff, 2008)). GluN1 and GluN3A both bind glycine (and D-serine), and GluN1/GluN3 receptors only require glycine for activation (Awobuluyi et al., 2007; Chatterton et al., 2002; Kvist et al., 2013a; Madry et al., 2007), but strongly desensitize following glycine exposure (Awobuluyi et al., 2007; Cummings & Popescu, 2016; Grand et al., 2018; Kvist et al., 2013a; Madry et al., 2007). GluN1/GluN2 receptors have high Ca2+-permeability and are blocked by extracellular Mg2+ at resting membrane potentials, while recombinant GluN1/GluN3 receptors are less permeable to Ca2+ and insensitive to Mg2+ block (Chatterton et al., 2002; Pina-Crespo et al., 2010) (see also (F. Yi et al., 2018)). Thus, GluN3 subunits endow NMDA receptors with unique functional properties. In addition, the GluN3A subunit has been suggested to influence synaptic development (Das et al., 1998; Maile A Henson et al., 2012; Pérez-Otaño et al., 2006; Roberts et al., 2009), plasticity (Larsen et al., 2011), and place aversion conditioning (Otsu et al., 2019). Some reports provide evidence that GluN3 subunits can be neuroprotective (Káradóttir et al., 2005; Martínez-Turrillas et al., 2012; Micu et al., 2006; Nakanishi et al., 2009; Salter & Fern, 2005). These properties raise the possibility that GluN3 subunits could be new targets for therapeutic strategies to treat neurological disease (Nora A. Cavara & Hollmann, 2008b; Maile A. Henson et al., 2010b; Stys & Lipton, 2007). In this regard, pharmacological agents that are selective for GluN3 subunits are needed to establish proof-of-concept for therapeutic utility, and potentially serve as a starting point for drug development.
GluN1/GluN3 receptors have seemingly cryptic activation properties, since agonist binding to GluN1 triggers strong desensitization of GluN1/GluN3 receptors, whereas agonist binding to GluN3 mediates activation (Awobuluyi et al., 2007; Grand et al., 2018; Kvist et al., 2013a; Madry et al., 2007). Furthermore, glycine appears to bind GluN1 with high potency (EC50 ~0.1–1 μM) and GluN3A with low potency (EC50 ~10–60 μM) (Grand et al., 2018; Kvist et al., 2013a). Consistent with this idea, mutations within the GluN1 agonist binding pocket that block glycine binding can prevent the desensitization of GluN1/GluN3 receptors that is mediated by glycine binding to GluN1(Awobuluyi et al., 2007; Kvist et al., 2013a; Madry et al., 2007). Recent data with glycine site antagonists further support this conclusion, and show that selective block of GluN1 can enhance the glycine response from GluN1/GluN3 receptors. For example, CGP-78608, which is a glycine-site antagonist that is highly selective for GluN1 over GluN3 subunits (Yao & Mayer, 2006), can dramatically enhance activation of GluN1/GluN3 receptors (Grand et al., 2018). However, CGP-78608 blocks GluN1/GluN2 NMDARs through its action at the GluN1 glycine site, which limits its utility as a tool to evaluate the contribution of GluN3 to neurological functions. Indeed, there are currently no reliable pharmacological means to selectively inhibit or potentiate GluN3-containing receptors, apart from TK13, TK30, and TK80 antagonists, which show relatively modest preference (~5- to 10-fold) for GluN1/GluN3 over GluN1/GluN2 receptors (Kvist et al., 2013a). In this study, we describe a novel negative allosteric modulator that selectively inhibits the response of both recombinant and native GluN1/GluN3 receptors, and provide mechanistic data describing its actions.
2. Materials and Methods
2.1. Molecular Biology
cDNA encoding human wild-type (WT) GluN1–1a (GenBank accession numbers NP_015566, hereafter GluN1) and rat GluN1–1a (NP_058706.1) were synthesized and subcloned into pGEM-HE plasmid; some experiments used the rat splice variant GluN1–4a (NP_001257531.1). Human GluN3A (NP_597702.2) and rat GluN3B (NP_579842.2) were synthesized and subcloned into SP6 plasmids. For most experiments, human GluN1 was co-expressed with human GluN3A, except for mutagenesis experiments where rat GluN1 was co-expressed with human GluN3A. Rat GluN1–4a was always expressed with rat GluN3B. EU1180–438 was also tested at 20 μM for actions at AMPA (rat GluA1flip, rat GluA2flip-R607Q, human GluA3flip-L531Y, human GluA4flip-L505Y (Stern-Bach et al., 1998), human kainate (GluK1, GluK2, GluK2/GluK5), rat GABAA (α1β2γ2S), rat GABAC (ρ1), rat glycine (α1), rat nicotinic acetylcholine (nAChR, α1β1δγ, α4β2, α7), and purinergic (human P2X2) receptors expressed in Xenopus oocytes. The cDNA was linearized by restriction enzymes (Thermo Scientific, Waltham, MA), and cRNA encoding the receptor subunits was synthesized from the linear template cDNA using the mMESSAGE mMACHINE kit (Invitrogen/Thermo Fisher Scientific). The α1β1δγ nAChR subunits were injected at a 1:1:1:1 ratio while α4β2 nAChR subunits were injected at a 1:1 ratio. The cDNAs encoding GABA and glycine subunits were provided by Dr. D. Weiss (University of Texas Health Science Center at San Antonio). cDNAs encoding nAChR subunits were provided by Drs. R. Papke (University of Florida) and S. Heinemann (Salk Institute). cDNA encoding the purinergic receptor was provided by Dr. R. Hume (University of Michigan). Responses in presence of EU1180–438 were expressed as a percent of control calculated as the average of the responses before and after EU1180–438 exposure. Site-directed mutagenesis was performed using standard molecular biology protocols. All DNA constructs were verified by sequencing (Eurofins MWG Operon, Huntsville, AL).
2.2. Two-electrode voltage-clamp (TEVC) recordings
Defolliculated Xenopus laevis oocytes (stage V-VI) were obtained from Ecocyte BioScience (Austin, Texas), or prepared from commercially available Xenopus ovaries (Xenopus 1, Dexter, MI) as previously described (Kasper B. Hansen et al., 2013). A Drummond Nanoject II (Broomall, PA) was used to inject each oocyte with cRNAs encoding GluN1 and GluN3 at a 1:3 ratio in RNase-free water. The cRNA was diluted as needed to give responses with amplitudes ranging between 200–2000 nA (0.2–10 ng total cRNA). Following cRNA injection, the oocytes were stored at 15–19°C in Barth’s solution that contained (in mM) 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, 5 Tris-HCl, pH 7.4 with NaOH, supplemented with 1 μg/ml streptomycin (Invitrogen, Carlsbad, CA) and 100 μg/ml gentamicin (Fisher Scientific, Pittsburg, PA). Recordings were performed 2–5 days following cRNA microinjection at room temperature (23°C) using a two-electrode voltage-clamp amplifier (OC725, Warner Instrument, Hamilton, CT). The signal was low-pass filtered at 10–20 Hz (4-pole, −3 dB Bessel) and digitized at the Nyquist rate using PCI-6025E or USB-6212 BNC data acquisition boards (National Instruments, Austin, TX) using EasyOocyte (https://emoryott.technologypublisher.com). Oocytes were placed in a custom-made chamber and continuously perfused (2.5 ml/min) with oocyte recording solution containing (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5–1.0 BaCl2, 0.01 EDTA (pH 7.4 with NaOH). Solutions were applied by gravity, and solution exchange was controlled through a rotary valve (Hamilton, Reno, NV). Recording electrodes were filled with 0.3–3.0 M KCl, and current responses were recorded at a holding potential of −40 mV. Agonist concentration-effect curves were expressed as a percentage of the response to 1 mM glycine and fitted by
| (1) |
where nH is the Hill slope, EC50 if the concentration of agonist that produces a half-maximal response, and maximum is the maximal response predicted for saturating agonist. The IC50 values for the negative allosteric modulators were expressed as a percentage of the response to maximally effective concentration of agonist (1 mM glycine) in the absence of test ligand and fitted by
| (2) |
where nH is the Hill slope, IC50 is the concentration that produces a half-maximal effect, and minimum is the degree of residual current at a saturating concentration of the negative allosteric modulator.
2.3. Hippocampal slice preparation
All procedures were approved by the University of Montana Institutional Animal Care and Use Committee. Wild type C57Bl/6J mice and 3A-KO mice (Das et al., 1998) (B6; 129X1-Grin3atm1Nnk, Jackson lab, Stock No: 029974) were given access to food and water at libitum and housed together in one breeding pair per cage. Mice (aged P8–15) of both sexes were used in studies of glycine and NMDA responses in acute hippocampal slices. Transverse hippocampal slices were cut as previously described (Feng Yi et al., 2014). Briefly, immediately after euthanasia by deep anesthesia using isoflurane, the mouse was cardiac perfused with ice-old oxygenated high sucrose solution containing (in mM) 3 KCl, 24 NaHCO3, 1.25 NaH2PO4, 10 glucose, 230 sucrose, 0.5 CaCl2, and 10 MgSO4, saturated with 95% O2 / 5% CO2. When removed, the mouse brain was immediately submerged in ice-cold oxygenated cutting solution containing (in mM) 130 NaCl, 3 KCl, 24 NaHCO3, 1.25 NaH2PO4, 10 glucose, 1 CaCl2, and 3 MgSO4. Transverse hippocampal slices (300 μm) were cut by a vibrating microtome (VT1200S, Leica Microsystems, Buffalo Grove, IL) and then incubated in the cutting solution at room temperature for at least one hour.
2.4. Whole-cell patch-clamp recordings from hippocampal neurons
A hippocampal slice was transferred to a recording chamber (RC-26GLP, Warner Instruments, Hamden, CT) mounted on a SliceScope Pro 2000 (Scientifica, Clarksburg, NJ). The slice was continuously perfused at a rate of 2–3 ml/min with oxygenated ACSF solution containing (in mM) 130 NaCl, 3 KCl, 24 NaHCO3, 1.25 NaH2PO4, 10 glucose, 2 CaCl2, and 1 MgSO4, maintained at 32°C using dual in-line and heated platform temperature control (TC-344A, Warner Instruments). Recording electrodes with a tip resistance of 2–4 MΩ were fabricated using a micropipette puller (P-1000, Sutter Instruments, Novato, CA) and filled with internal solution containing (in mM): 120 Cs-methanesulfonate, 4.6 MgCl2, 10 HEPES, 15 BAPTA, 4 Na2-ATP, 0.4 Na-GTP, 1 QX-314, and 10 K2-creatine phosphate, pH 7.25, 280–290 mOsm. Whole-cell recordings were made using a Multiclamp 700B amplifier (Molecular Devices) with filtering at 4 kHz (Bessel) and digitized at 10 kHz using Digidata 1440A with pClamp 10 software (Molecular Devices). For recordings of glycine-induced or NMDA-induced current responses, glycine (10 mM, 100 ms duration, 5 psi.) or NMDA (0.5 mM, 15–30 ms duration, 5 psi.) was delivered using a custom-made picospritzer (openspritzer; (Forman et al., 2017)) every 30–60 s to the soma of patched cell through a borosilicate capillary tube (~10 μM in tip diameter). For recordings of glycine responses, the external solution was supplemented with 10 μM gabazine, 2 μM NBQX, 100 μM DL-APV, 50 μM strychnine to block GABAA, AMPA, GluN1/GluN2 NMDA, and glycine receptors, respectively. After at least 5 min of stable baseline recordings, 1 μM CGP-78608 was bath-applied to the slice to prevent glycine-induced desensitization of GluN1/GluN3A receptors. Subsequently, test compound EU1180–438 at 30 μM or vehicle (0.15% DMSO) was added to the recording solution and bath-applied to the recorded cells from WT mice. For the recordings of NMDA responses, the external solution was supplemented with 10 μM gabazine, 2 μM NBQX, and 3 μM glycine. After at least 5 min stable baseline recordings, test compound EU1180–438 at 30 μM or vehicle (0.15% DMSO) was added to the recording solution and bath-applied to the recorded cells from WT mice for 10 min. In the end, 400 μM DL-APV was applied for 5 min for full inhibition of GluN1/GluN2 NMDA receptors.
For all the recorded cells, series resistance (typically <20 MΩ) was monitored throughout the experiment with a 200 ms long, 5 mV hyperpolarizing voltage jump. The recording was excluded from analysis if series resistance changes of >20% were observed. For glycine and NMDA experiments in hippocampal slices, analyses of peak amplitudes were performed with Axograph (axograph.com). Data are shown as mean ± SEM. Paired/unpaired t test were used for statistical comparisons as indicated, and P<0.05 was considered significant.
Nonstationary variance analysis (R. E. Perszyk et al., 2020; S. F. Traynelis & Jaramillo, 1998) was conducted on the deactivation phase of the neuronal glycine-evoked current responses (see Fig. 2) bathed in vehicle (control) or 30 μM EU1180–438 (VHOLD: −60 mV). Ten individual responses to pressure-applied glycine were identified both during control and after inhibition by EU1180–438 had reached steady-state. The current was divided into 50 equally spaced segments, and the mean current amplitude of each segment was determined. The current was high pass filtered with a cutoff of 1 Hz, and the current variance was determined for 50 segments. The plot of variance vs. current for each response was fitted by the equation
| (3) |
where I is the current amplitude of macroscopic response, i is the weighted mean unitary current, N is the channel numbers. Virtually identical results were found if the current-variance relationship for all 10 responses for each cell were averaged and then fitted by equation 3. Chord conductance γ was determined assuming 0 mV for the reversal potential.
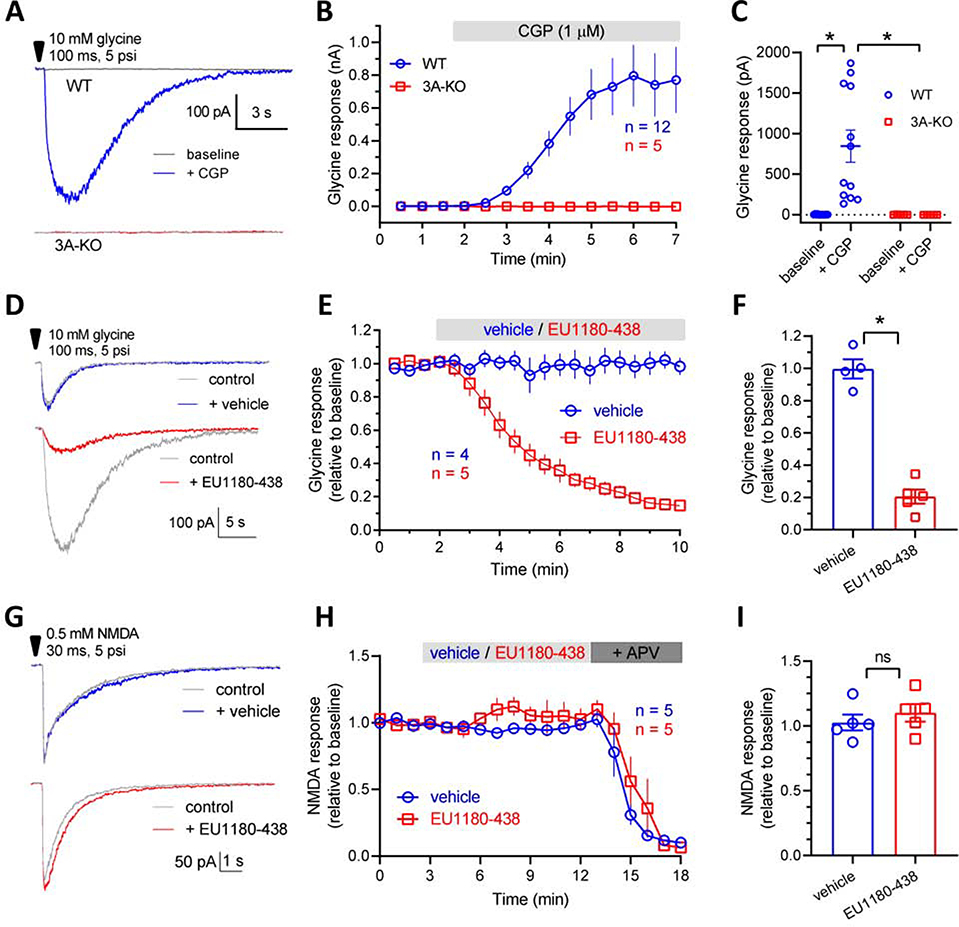
Fig. 2. Modulation of native GluN3A-containing receptors.
A, Representative recordings of current responses from GluN3A-containing receptors in hippocampal CA1 pyramidal cells from acute mouse brain slices (P8–15). The responses are activated by pressure-induced application of 10 mM glycine to the neuronal cell soma from wild type (WT) and GluN3A-deficient (3A-KO) mice in the absence or presence of bath-applied 1 μM CGP-78608 (CGP). B, Averaged development of glycine responses from WT and 3A-KO mice following addition of CGP-78608 to the recording solution as indicated by the grey bar. C, Bar graph summarizing the glycine-activated responses from GluN3A-containing NMDA receptors revealed by CGP-78608 in WT, but not 3A-KO mice. D-F, In the presence of 1 μM CGP, GluN3A-containing receptor-mediated responses were inhibited significantly by 30 μM EU1180–438. G-I, EU1180–438 (30 μM) does not affect current responses from GluN1/GluN2-containing NMDA receptors in hippocampal CA1 pyramidal cells from acute mouse brain slices (P8–15). The responses are activated by pressure-induced application of 0.5 mM NMDA to the neuronal cell soma from wild type (WT) mice. * indicates significantly different (P< 0.05; unpaired t-test). ns; not significant.
2.5. Homology modeling
Amino acids are numbered with the initiating methionine set to 1. A protein alignment was created for GluN1, GluN2, and GluN3A subunit sequences using the program Muscle (Edgar, 2004). Five GluN1/GluN3A receptor homology models made up of 3313 amino acids were generated using modeler 9v21 (ŠAli & Overington, 1994) from three different templates (molpdf scores ranged from 113309–115258). The templates consisted of two diheteromeric GluN1/GluN2B receptor structures (PDB entries; 4PE5 and 5FXH resolution of 3.96 and 5Å) and a monomer that represents the ABD domain of GluN3A (PDB entry 2RC7, resolution of 1.58Å). Regions of GluN3A for which there were no template structure coverage were removed during model building. The sequence identity and similarity are provided in Supplemental Figure S1 and full-length alignments are provided in Supplemental Figure S2. Quality analysis was performed using the PDBsum generator (http://www.ebi.ac.uk/pdbsum, (Laskowski, 2009), Schrödinger Release 2019–1 (Protein Preparation Wizard, Epik, Impact, Prime, Schrödinger, LLC, New York, NY, 2019) and Modeler (ŠAli & Overington, 1994). The model was prepared for analysis using the protein preparation wizard in which protonation states were assigned followed by an energy minimization cycle to relieve unfavorable constraints (Schrödinger Release 2019–1). The minimization cycle consisted of first minimizing only hydrogens, followed by a restrained minimization using imperf with a convergence of the RMSD of heavy atoms to 0.3Å (Schrödinger Release 2019–1).
3. Chemicals and reagents
General chemistry experimental
The compound [(1 S)-1-[[(7-bromo-1,2,3,4-tetrahydro-2,3-dioxo-5-quinoxalinyl)methyl]amino]ethyl]phosph onic acid hydrochloride (CGP-78608, Tocris) was prepared as a stock solution of 10 mM in 2.2 eq NaOH solution. DL-APV, QX-314, NBQX and gabazine were purchased from Hello Bio. All the other regents were purchased from Sigma and used directly without further purification. EU1180–438 was synthesized as described below and prepared in DMSO at stock concentrations of 20 mM. Purification by flash column chromatography was done using a Teledyne ISCO Combiflash Companion instrument using Teledyne Redisep normal phase columns. 1H and 13C NMR spectra were recorded on an INOVA-500 spectrometer. Chemical shifts were reported in ppm and referenced to the residual deuterated solvent. Reactions were monitored by thin layer chromatography on precoated aluminum plates (silica gel 60 F254, 0.25 mm) or LCMS on an Agilent Technologies 1200 series instrument. Yields are reported as percentages in both Scheme 1 and experimental descriptions. High resolution mass spectra were recorded on a VG 70-S Nier Johnson or JEOL instrument by the Emory University Mass Spectroscopy Center. Purity of EU1180–438 was established via LCMS (Varian) in two solvent systems (MeOH:water/MeOH:water) and was found to be >95% pure.
Scheme 1.
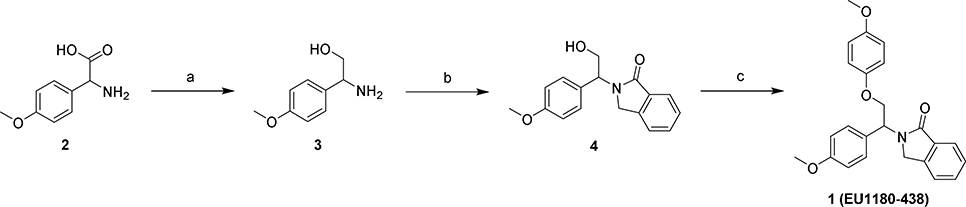
Synthesis of EU-1180–438. (a) LiAlH4, THF, 0 °C to reflux, o/n, 81%; (b) methyl (2-bromomethyl)benzoate, Et3N, EtOH, rt, o/n, 62%; (c) 4-iodoanisole, CuI, Me4Phen, Cs2CO3, MePh, reflux, o/n, 73%.
3.1. Synthesis of EU1180–438
2-amino-2-(4-methoxyphenyl)ethan-1-ol (3)
Lithium aluminum hydride (13.8 mL, 27.6 mmol, 2 M in THF, 5 eq) was added to THF (31 mL, ~0.18 M), cooled to 0 °C, and 2-amino-2-(4-methoxyphenyl)acetic acid (2) (1.00 g, 5.52 mmol, 1 eq) was added in small portions. The solution was stirred at 0 °C for 1 hour and then brought to reflux overnight. Upon completion, the reaction was brought to 0 °C and 1.05 mL of water, 1.05 mL of 15% NaOH, and 3.15 mL of water were added dropwise in succession. The mixture was stirred until a white suspension was observed. The precipitate was filtered and the combined THF extracts were concentrated, brought up in DCM, washed with brine, dried over MgSO4, and concentrated in vacuo. The crude product was purified via flash column chromatography (ISCO, Redisep 24 g column, solid load, 0–20% DCM/[MeOH w/ 6% v/v 7 M NH3 in MeOH] gradient) to afford the title compound as a white solid (0.75 g, 81%). Rf (8:2 DCM/[MeOH w/ 6% v/v 7 M NH3 in MeOH]): 0.73; 1H NMR (500 MHz, Chloroform-d) δ 7.26 – 7.20 (m, 2H), 6.90 – 6.83 (m, 2H), 3.98 (dd, J = 8.4, 4.3 Hz, 1H), 3.78 (s, 3H), 3.67 (dd, J = 10.8, 4.3 Hz, 1H), 3.52 (dd, J = 10.8, 8.4 Hz, 1H). 13C NMR (126 MHz, cdcl3) δ 158.95, 134.62, 127.59, 113.99, 67.99, 56.72, 55.28. HRMS calcd. for C9H14O2N, 168.10191 [M + H]+; found 168.10175 [M + H]+.
2-(2-hydroxy-1-(4-methoxyphenyl)ethyl)isoindolin-1-one (4)
Compound 3 (0.20 g, 1.2 mmol, 1 eq) was added to methyl 2-(bromomethyl)benzoate (0.33 g, 1.4 mmol, 1.2 eq) and triethylamine (0.33 mL, 2.4 mmol, 2 eq) in EtOH (6.0 mL, ~0.20 M) and stirred at room temperature overnight. The reaction was quenched with water, extracted with DCM (3x), washed with brine, dried over MgSO4, and concentrated in vacuo. The crude product was purified via flash column chromatography (ISCO, Redisep 12 g column, 0–100% EtOAc/hexanes gradient) to afford the title compound as a white foam (0.21 g, 62%). Rf (1:1 EtOAC:Hex): 0.54; 1H NMR (500 MHz, Chloroform-d) δ 7.77 (d, J = 7.6 Hz, 1H), 7.45 (td, J = 7.5, 1.2 Hz, 1H), 7.38 (t, J = 7.5 Hz, 1H), 7.33 (d, J = 7.5 Hz, 1H), 7.27 – 7.21 (m, 2H), 6.89 – 6.82 (m, 2H), 5.33 (dd, J = 8.6, 4.5 Hz, 1H), 4.43 (d, J = 17.1 Hz, 1H), 4.26 (dd, J = 11.8, 8.8 Hz, 1H), 4.19 – 4.11 (m, 2H), 3.77 (s, 3H). 13C NMR (126 MHz, cdcl3) δ 169.61, 159.29, 141.51, 132.65, 131.42, 129.51, 128.78, 127.93, 123.69, 122.60, 114.23, 63.34, 58.89, 55.27, 48.62. HRMS calcd. for C17H18O3N, 284.12812 [M + H]+; found 284.12799 [M + H]+.
2-(2-(4-methoxyphenoxy)-1-(4-methoxyphenyl)ethyl)isoindolin-1-one (1, EU1180–438)
An oven-dried microwave vial was charged with an oven-dried stir bar, compound 4 (100 mg, 0.350 mmol, 1 eq), copper(I) iodide (13 mg, 0.071 mmol, 0.2 eq), 3,4,7,8-tetramethyl-1,10-phenanthroline (17 mg, 0.071 mmol, 0.2 eq), 1-iodo-4-methoxybenzene (124 mg, 0.529 mmol, 1.5 eq), and cesium carbonate (173 mg, 0.529 mmol, 1.5 eq). The vessel was then capped and evacuated and back-filled with dry argon 3 times. Dry toluene (0.71 mL, ~0.5 M) was then added, the vial evacuated and back-filled with dry argon once more and the reaction stirred at 110 °C for 48 hours. The reaction mixture was then cooled to room temperature, diluted with EtOAc, and filtered through a plug of celite, washing with additional EtOAc. The crude material was then concentrated and purified via flash column chromatography (ISCO, Redisep 4 g column, 0–100% EtOAc/hexanes gradient) to afford the title compound as a yellow foam. (0.10 g, 73%). Rf (1:1 EtOAc:Hex): 0.48; 1H NMR (500 MHz, Chloroform-d) δ 7.89 (d, J = 7.4 Hz, 1H), 7.50 (td, J = 7.4, 1.3 Hz, 1H), 7.45 (td, J = 7.4, 0.9 Hz, 1H), 7.40 – 7.34 (m, 3H), 6.91 – 6.85 (m, 4H), 6.84 – 6.80 (m, 2H), 5.87 (t, J = 5.8 Hz, 1H), 4.58 (dd, J = 10.0, 6.6 Hz, 1H), 4.54 (d, J = 16.9 Hz, 1H), 4.48 (dd, J = 10.0, 5.2 Hz, 1H), 4.24 (d, J = 17.0 Hz, 1H), 3.79 (s, 3H), 3.75 (s, 3H). 13C NMR (126 MHz, cdcl3) δ 168.66, 159.26, 154.29, 152.46, 141.72, 132.61, 131.33, 129.38, 129.13, 127.92, 123.80, 122.74, 115.83, 114.72, 114.17, 68.75, 55.71, 55.27, 53.35, 47.79. HRMS calcd. for C24H24O4N, 390.16998 [M + H]+; found 390.17026 [M + H]+. Purity was established using an Agilent pump on a Zorbax XBD-C18 column (4.6 mm × 50 mm, 3.5 μm). Method 1: 85–95% MeOH in water over 5 min at 1 mL/min (retention time = 0.73 min). Method 2: 75–95% MeOH in water over 3 min at 1 mL/min (retention time 1.05 min).
3.2. Separation of (1, 1180–438) Enantiomers
Semipreparative separation of 1, 1180–438 enantiomers from racemic 1, 1180–438 (0.020 g) was done using a Kromasil 5-AmyCoat (30 mm × 250 mm) with the following conditions: 20 mL/min flow rate, 10 mL injection volume (2 mg/1 mL), 90% hexanes/10% IPA over 150 min to afford (−)-1, 1180–438, tR 72.8 min; (+)-1, 1180–438, tR 92.0 min. The enantiomeric excess (ee) was determined using an Agilent pump on a Kromasil 5-AmyCoat column (4.6 mm × 150 mm, 5 μm) with the following conditions: 1 mL/min flow rate, 10 μL injection volume, 90% hexanes/10% IPA. (−)-1, 1180–438: tR 38.1 min; >99% ee; [α]D20 = −36 (c 0.10, dry CHCl3). (+)-1, 1180–438: tR 33.0 min, >99% ee; [α]D20 = +37 (c 0.10, dry CHCl3).
4. Results
4.1. EU1180–438 inhibits GluN1/GluN3 receptors
Introduction of two mutations (F484A,T518L) in the orthosteric glycine binding pocket of GluN1 (hereafter GluN1FA,TL), alters the glycine concentration-response profile for GluN1/GluN3 receptors by completely abolishing the inhibitory component caused by glycine binding to GluN1 (Kvist et al., 2013a). Thus, GluN1FA,TL/GluN3 receptors, which contain a GluN1 subunit that is virtually incapable of binding glycine, exhibit a conventional concentration-response relationship that reaches a maximal steady-state response, allowing determination of the glycine EC50 values and thereby potential assessment of GluN3-selective ligands (Kvist et al., 2013a). We have previously synthesized more than 1000 potential allosteric NMDA receptor modulators within compound classes that have structural determinants of action within the linker connecting the agonist binding domain to the transmembrane region of GluN1/GluN2 NMDA receptors. We reasoned that the similar architecture of the pore and linker regions of GluN1/GluN3 to the GluN1/GluN2 receptors might allow some of these compounds to act as modulators of GluN1/GluN3 receptors. We therefore screened a subset of this focused library (404 compounds) on GluN1FA,TL/GluN3A receptors to determine if any compounds showed selective allosteric modulation of GluN1/GluN3A (Santangelo et al., 2012).
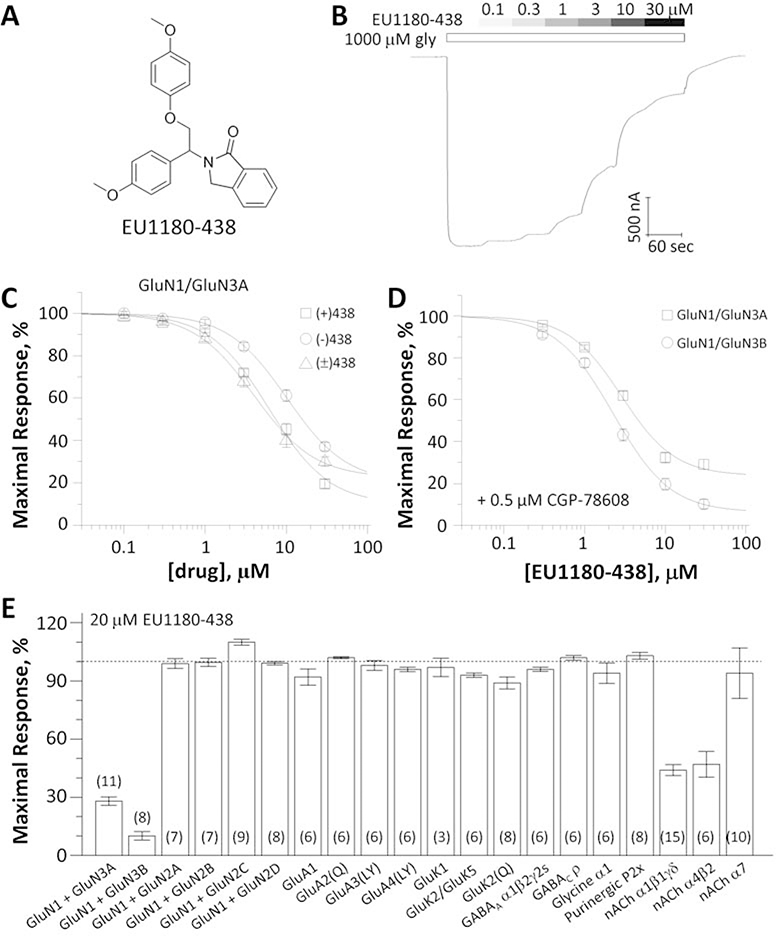
From this screen we identified EU1180–438 as an inhibitor of recombinant GluN1FA,TL/GluN3A receptors with no detectable activity at recombinant GluN1/GluN2 NMDA receptors. To evaluate inhibition of GluN1/GluN3 receptors by EU1180–438 (Fig. 1A), we established the concentration-effect relationship using a racemic mixture of EU1180–438, which inhibited GluN1FA,TL/GluN3A receptors by 82% at a fitted saturating concentration with an IC50 value of 3.5 μM (Fig. 1B,C). We subsequently isolated the (+) and (−) enantiomers of EU1180–438, which showed similar potency and effects at GluN1FA,TL/GluN3A receptors (Table 1).
Fig. 1. Evaluation of EU1180–438 as a selective GluN1/GluN3 receptor antagonist.
A, Chemical structure of EU1180–438. B, Representative two-electrode voltage-clamp recording of EU1180–438 inhibition of recombinant GluN1FA,TL/GluN3A receptor responses activated by 1000 μM glycine. C, Concentration-response curves for inhibition of GluN1FA,TL/GluN3A receptor responses activated by 1000 μM glycine by enantiomers and racemate of EU1180–438. (−) EU1180–438 IC50 13 μM, Slope 1.4, minimum 16%; (+) EU1180–438 IC50 7.1 μM, Slope 1.2, minimum 4%; (±) EU1180–438 IC50 4.3 μM, Slope 1.3, minimum 28%). Data are mean ± SEM of 18–26 oocytes. D, Concentration-response relationship for EU1180–438 in 500 nM CGP-78608 plus 100 μM glycine at human GluN1/GluN3A and rat GluN1/GluN3B receptors (IC50 1.8 μM, Slope 1.5, minimum 15% and IC50 2.2 μM, Slope 1.5, minimum 7%). Data are mean ± SEM of 8–9 oocytes. E, Receptor responses to agonists in the presence of 20 μM EU1180–438 normalized to agonist responses in vehicle. GluN1 was co-expressed with GluN2A-D and activated by 100 μM glutamate/100 μM glycine, GluA1–4, GluK1, and GluK2 were activated by 100 μM glutamate, GluK2/K5 was activated by 100 μM AMPA, GABAA and GABAC were activated by 100 μM GABA, Glycine α1 was activated by 100 μM glycine, P2X purinergic receptors were activated by 9 μM ATP, α1β1γδ nicotinic acetylcholine receptors were activated by 1 μM acetylcholine, α4β2 nicotinic acetylcholine receptors were activated by 10 μM acetylcholine, and α7-nicotinic acetylcholine receptors were activated by 300 μM acetylcholine. The numbers of oocytes recorded are in parentheses; Numerical data are given in Supplemental Table 1.
Table 1.
Pharmacological properties of EU1180–438 at human GluN1/GluN3 receptors
| GluN1FA,TL/GluN3A IC50, (95% CI) % Inhibitiona, (n) |
GluN1/GluN3A+CGP b IC50, (95% CI) % Inhibitiona, (n) |
GluN1/GluN3B+CGP b IC50, (95% CI) % Inhibitiona, (n) |
|
|---|---|---|---|
| (±) EU1180–438, pH 7.4 | 3.5 μM, (2.8–3.8) 82%, (n=18) |
1.8 μM, (1.5–2.1) 85%, (n=9) |
2.2 μM, (1.9–2.6) 93%, (n=8) |
| (+) EU1180–438, pH 7.4 | 7.1 μM, (5.7,7.7) 96%a, (n=22) |
ND | ND |
| (−) EU1180–438, pH 7.4 | 13 μM, (9.0–14) 84%a, (n=20) |
ND | ND |
| (±) EU1180–438, pH 6.8 | 4.2 μM, (3.0–5.2) 72%a, (n=7) |
ND | ND |
Data are shown as mean with 95% confidence interval given in parentheses, determined from mean of log(IC50). The fitted (i.e. predicted from curve fit) maximal inhibition in saturating EU1180–438 is given as percent of control, and (n) is the number of oocytes. Holding potential was −40 mV. Receptors contained human GluN1FA,TL/GluN3A or rat GluN1–4a/GluN3B.
Percentage inhibition determined from fitted maximum at saturating test compound, calculated as 100 × (1-IDRUG/ICONTROL)
Experiments were performed in 0.5 μM GCP-78608
ND is not determined.
4.2. EU1180–438 is selective for GluN1/GluN3 receptors
The binding of glycine to the GluN1 subunits of the GluN1/GluN3 receptors appears to cause auto-inhibition by promoting entry into a desensitized state (Awobuluyi et al., 2007; Kvist et al., 2013a; Madry et al., 2007; Smothers & Woodward, 2009). Paoletti and colleagues (Grand et al., 2018) therefore tested glycine site antagonists for their ability to enhance activation of GluN1/GluN3 by glycine by selectively preventing glycine binding to the GluN1 subunit. CGP-78608 was found to strongly enhance GluN1/GluN3A current responses, converting small and rapidly desensitizing currents from wild type GluN1/GluN3A into large and stable current responses (Grand et al., 2018). We thus evaluated the effects of EU1180–438 on human GluN1/GluN3A and rat GluN1/GluN3B receptors activated in the presence of 500 nM CGP-78608, and found 85% fitted maximal inhibition with an IC50 of 1.8 μM at GluN1/GluN3A and 93% fitted maximal inhibition with an IC50 of 2.2 μM at GluN1/GluN3B (Fig. 1D, Table 1). In addition, rat GluN1–4aFA,TL/GluN3B receptors, which contain a GluN1 splice variant that has been suggested to increase GluN3-mediated current amplitudes (N. A. Cavara et al., 2009; Cummings et al., 2017), were also inhibited by 82% at saturating concentrations determined from the fitted curve with an IC50 value of 5.2 μM (95% CI 4.4–5.9, n=6) (data not shown). EU1180–438 (20 μM) showed virtually no activity at GluN1/GluN2 NMDA receptors, GluA1–4 AMPA receptors, GluK1–2 and GluK2/GluK5 kainate receptors, GABA receptors, glycine receptors, or alpha7 nicotinic receptors; EU1180–438 modestly inhibited muscle and α2β4 nicotinic receptors (Fig. 1E; Supplemental Table S1). These data suggest EU1180–438 can be used as a potential tool for exploration of GluN1/GluN3 activity in neuronal preparations.
4.3. EU1180–438 inhibits neuronal responses mediated by GluN3A-containing receptors
To evaluate the effects of EU1180–438 on responses from neuronal GluN3A-containing NMDA receptors, we first reproduced the recent evidence for native GluN1/GluN3A receptors expressed in hippocampal CA1 pyramidal cells (Grand et al., 2018). Whole-cell recordings from hippocampal CA1 pyramidal cells in acute mouse brain slices (P8–15) were performed in the presence of strychnine, gabazine, NBQX, and APV to block inhibitory glycine receptors, GABAA receptors, AMPA/kainate receptors, and GluN1/GluN2 NMDA receptors, respectively (Fig. 2A–C). Thus, the presence of APV and CGP-78608 inhibits all “conventional” GluN1/GluN2 NMDA receptors. Responses were activated by brief pressure-induced application (hereafter puffs) of 10 mM glycine to the neuronal cell body in the absence and presence of bath-applied CGP-78608 (Fig. 2A–C). The glycine responses are abolished in slices from GluN3A-deficient mice (3A-KO) (Fig. 2A–C). These results demonstrate expression of native GluN1/GluN3A receptors in hippocampal CA1 pyramidal cells and show that these receptors are incapable of mediating glycine-activated current responses in the absence of a GluN1-selective antagonist.
Next, we evaluated allosteric modulation by EU1180–438 of neuronal current responses from native GluN1/GluN3A receptors in the presence of 1 μM CGP-78608 (Fig. 2D–F). EU1180–438 (30 μM) produced a strong inhibition of the glycine-activated responses compared to control responses in the absence of modulator (p < 0.05; paired t-test) (Fig. 2D–F). By contrast, addition of vehicle (0.15% DMSO) did not affect glycine-activated responses from native GluN1/GluN3A receptors (p > 0.05; paired t-test) (Fig. 2D–F). To further establish the selectivity of EU1180–438 for GluN3-containing NMDA receptors, we investigated potential effects on responses activated by pressure application of 0.5 mM NMDA onto the hippocampal CA1 pyramidal cells (Fig. 2G–I). These responses, which are mediated by GluN1/GluN2 NMDA receptors, were not modulated by EU1180–438 compared to control in the absence of modulator (p > 0.05; paired t-test) or compared to vehicle (0.15% DMSO; p > 0.05; unpaired t-test) (Fig. 2G–I). Thus, EU1180–438 is capable of producing strong inhibition of native GluN1/GluN3A receptors in acute hippocampal slices with high selectivity for GluN1/GluN3 over GluN1/GluN2 NMDA receptors.
4.4. EU1180–438 is a GluN1/GluN3 negative allosteric modulator
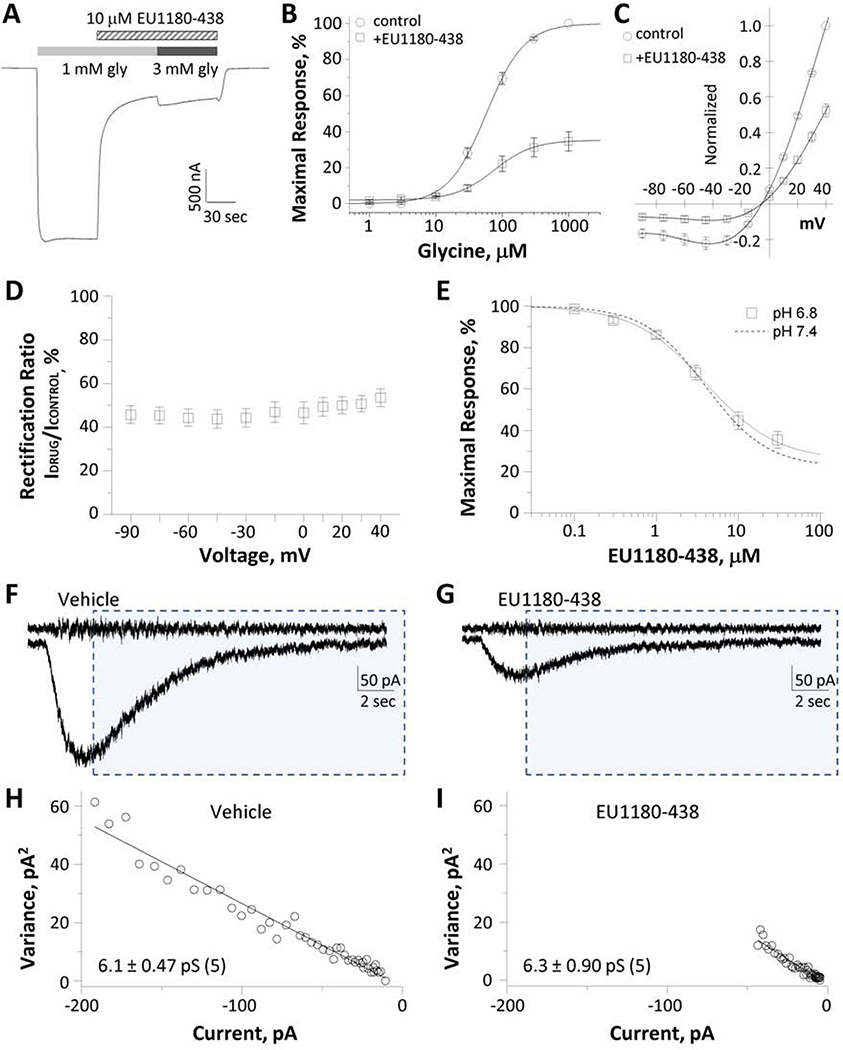
To determine the mechanism of action for EU1180–438 inhibition, we evaluated whether inhibition could be surmounted by increasing the glycine concentration. The inhibition of GluN1FA,TL/GluN3A by EU1180–438 was not altered by increasing glycine concentration, consistent with a non-competitive mechanism of action (Fig. 3A). The response in the presence of 10 μM EU1180–438 was 29 ± 4.2% of control in 1 mM glycine, and 29 ± 4.2% of control in the presence 3 mM glycine (n = 10). We subsequently recorded glycine concentration-response curves in the absence and presence of 30 μM EU1180–438 at GluN1FA,TL/GluN3A receptors (Fig. 3B). The glycine EC50 in the presence of EU1180–438 was 71 μM at GluN1FA,TL/GluN3A and the 95% CI determined from logEC50 was 50–84 μM (n = 10), compared to an EC50 value of 61 μM (95% CI 46–71 μM, n = 10) observed in the absence of inhibitor. We also recorded the current-voltage (I-V) relationship for glycine-activated currents in the absence and presence of 4 μM EU1180–438 (Fig. 3C). Analysis of the I-V relationship revealed a similar degree of inhibition of the current response by EU1180–438 at all voltages, which we interpret as evidence that the inhibition does not reflect voltage-dependent channel block (Fig. 3D). These data support the idea that EU1180–438 is a negative allosteric modulator of GluN1/GluN3 receptors.
Fig. 3. EU1180–438 inhibition of GluN1/GluN3A current responses.
A, The inhibition of GluN1FA,TL/GluN3 responses by 10 μM EU1180–438 was not altered by increasing glycine concentration (representative response; n = 10 oocytes). B,C, Effects of 4 μM EU1180–438 on glycine concentration-response curve and current-voltage relationship, normalized by current at +40 mV (mean ± SEM, n = 12 oocytes). Glycine EC50 values: 71 ± 8.8 μM (n = 10) for control, and 61 ± 6.0 μM (n = 10) for EU1180–438. D, There was no effect of holding voltage on EU1180–438 inhibition, suggesting that the inhibition does not reflect voltage-dependent channel block (n = 12). E, Composite concentration-response curve for inhibition of GluN1FA,TL/GluN3A at pH 6.8 (n = 8 oocytes), shown together with the corresponding inhibition data at pH 7.4 from Figure 1 (dashed line). F,G, Representative neuronal current responses from GluN1/GluN3A receptors activated by pressure-induced application of 10 mM glycine in vehicle (F) or EU1180–438 (G) (as shown in Figure 2). The top trace was generated from the high-pass filtered current and shaded boxes indicate portions for the response subjected to variance analysis. H,I, Current-variance plot of control (vehicle, H) or EU1180–438 (I).
Reduced extracellular pH has been shown to increase GluN1/GluN3A-mediated current amplitudes (Cummings & Popescu, 2016). We tested whether EU1180–438 can inhibit currents elicited by glycine at pH 6.8. We found that EU1180–438 inhibited GluN1FA,TL/GluN3A responses with an IC50 value of 4.2 μM, with maximal inhibition of 72% at saturating concentrations of EU1180–438 (n = 7, Fig. 3E, Table 1), compared to an IC50 value of 3.5 μM at pH 7.4. These data suggest that EU1180–438 can inhibit GluN1/GluN3A responses during activity-dependent or ischemia-dependent changes in extracellular pH.
Some allosteric modulators with molecular determinants in the pre-M1 can reduce single channel conductance for glutamate receptors (R. E. Perszyk et al., 2020; C. L. Yuan et al., 2019). To determine if EU1180–438 also changes unitary conductance of GluN3A-containing receptors, we performed nonstationary noise analysis (see Methods) on neuronal current responses (Fig. 3F,G) to estimate unitary current of GluN1/GluN3A channels from the relationship between the macroscopic current amplitude and variance. The average chord conductance of GluN1/GluN3A channels that give rise to the macroscopic response in the presence of EU1180–438 showed no significant difference compared to the vehicle (vehicle: 6.1 ± 0.47 pS, EU1180–437: 6.3 ± 0.90 pS; n = 5, p = 0.78, paired t-test; Fig. 3H,I). These data suggest that inhibition of GluN3A-containing receptor current by EU1180–438 does not alter involve changes in channel conductance.
4.5. Potential site of action for EU1180–438
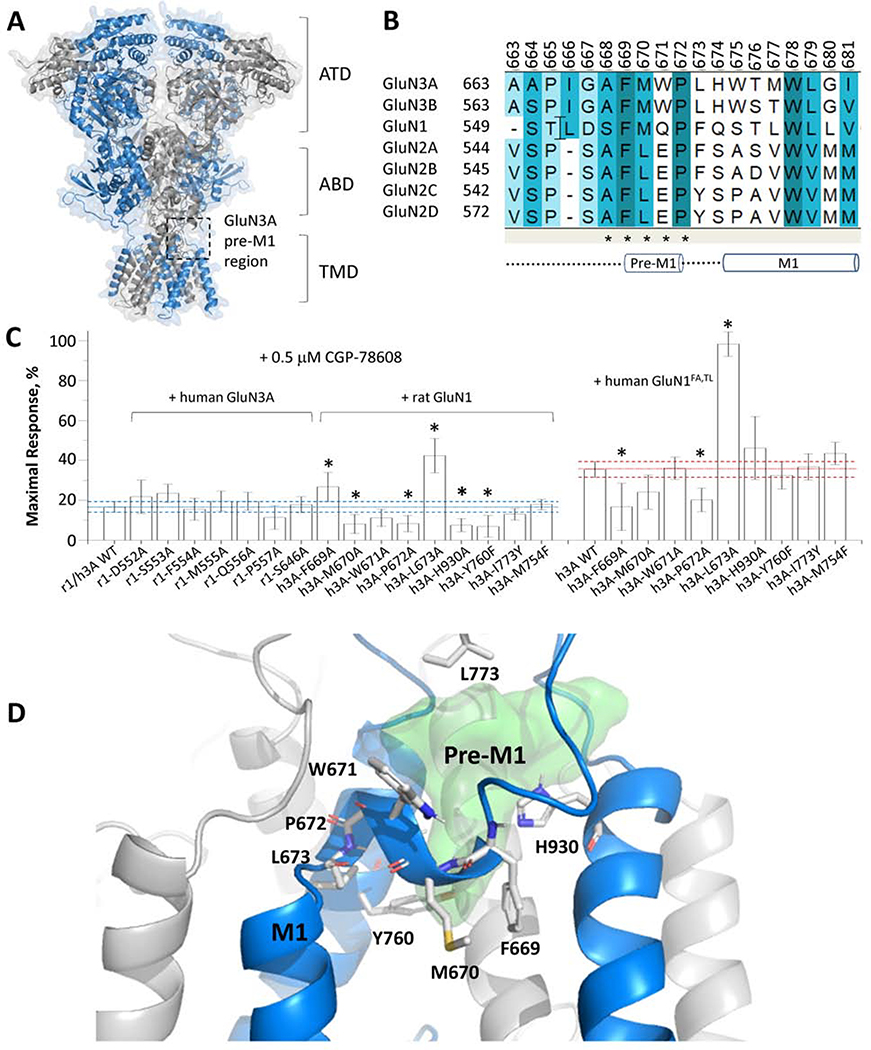
Ionotropic glutamate receptors harbor a binding site for modulators at the interface of the transmembrane region and the agonist binding domain (K. K. Ogden & Traynelis, 2013; R. Perszyk et al., 2018; R. E. Perszyk et al., 2020; Wang et al., 2017; Yelshanskaya et al., 2016). This site has been shown by crystallography of AMPA receptors to bind distinct scaffolds within the same binding site (Yelshanskaya et al., 2016). The latter pockets are located between the pre-M1 helix, the extracellular end of the M3 transmembrane helix, and parts of the M4 linker ((K. K. Ogden & Traynelis, 2013; Wang et al., 2017; Yelshanskaya et al., 2016); Fig. 4A–B). At present, NMDA receptor structures have not been resolved with modulators bound to this region. We have identified mutations affecting the binding of modulators within this region for a similar series to which EU1180–438 belong (K. K. Ogden & Traynelis, 2013). We therefore performed mutagenesis studies to narrow down the potential binding pocket for EU1180–438. Site-directed mutagenesis was performed in proximity to the pre-M1 region of GluN1, GluN1FA,TL, and GluN3A, and responses recorded from GluN1FA,TL/GluN3A receptors or GluN1/GluN3A receptor in the presence of CGP-78608to prevent desensitization. We initially determined the effect of all mutations in the presence of CGP-78608 (Fig. 4C). There were no detectable effects of any GluN1 mutations on EU1180–438 inhibition, but 6 out of 9 GluN3A mutations perturbed the effects of EU1180–438 (Fig. 4C; Supplemental Table S2). We repeated analysis of these mutations in the absence of CGP-78608, but using the GluN1FA,TL subunit to prevent desensitization, and found similar results for adjacent residues GluN3-P672A and GluN3-L673, which line a cavity that resides above the pre-M1 helix (Fig. 4D). These data are compatible with structural determinants of action for EU1180–438 in the pre-M1 region of GluN3A rather than GluN1 (Fig. 4D). The cavity in GluN3A lined by residues affecting EU1180–438 inhibition is shown in Fig. 4D. The shape and size of this cavity is permissive for EU1180–438 occupancy, suggesting that it could harbor the binding pocket for EU1180–438 in GluN3A.
Fig. 4. Structural determinants of EU-1180–438.
A, Representation of a homology model of the GluN1/GluN3A structure. A black box indicates one of two hypothetical GluN3A binding pockets. GluN3A chain is colored in blue and GluN1 in grey. B, Sequence alignment of the pre-M1 region of GluN3A-B, GluN1, and GluN2A-D. C, Inhibition by 10 μM EU1180–438 of rat GluN1/human GluN3 (r1/h3A) receptors harboring pre-M1 mutations activated by 1 mM glycine in the presence of 0.5 μM CGP-78608 are shown as mean ± 95% CI (error bars); the dashed line shows 95% CI of wild type. Experiments with GluN3A mutations were repeated at human GluN1FA,TL/GluN3A (h1-FATL/h3A) receptors in the absence of CGP-78608. * indicates non-overlapping confidence intervals. Mean data are given in Supplemental Table 2. D, A cartoon representation of the pre-M1 GluN3A region with mutated residues shown in stick representation (white sticks). The green surface represents a potential binding cavity that EU1180–438 could occupy.
5. Discussion
In this study, we have identified a novel modulator for GluN1/GluN3 receptors that shows remarkable selectivity for GluN1/GluN3 receptors over all other glutamate receptors tested. Furthermore, EU1180–438 is inactive at GABA, glycine, and P2X receptors, but displays modest inhibition of some nicotinic acetylcholine receptors. This degree of selectivity should allow use of EU1180–438 as a tool to determine whether specific neuronal functions or behaviors involve GluN3-containing receptors, with the caveat that some effects could reflect reduction in nicotinic receptor function, which can be easily controlled for by available pharmacological tools. The inhibitor EU1180–438 acts in a voltage-independent and non-competitive fashion, and has some similarities in the scaffold with the tetrahydroisoquinolines that can act on GluN1/GluN2 NMDA receptors (Santangelo Freel et al., 2013; Strong et al., 2017) with structural determinants in the GluN2 pre-M1 helix (Kevin K. Ogden & Traynelis, 2011). Given the specificity for GluN1/GluN3, we anticipate that EU1180–438 interacts primarily with GluN3 residues rather than GluN1 residues, and speculate that the pre-M1 region is a likely target of action. However, additional efforts will be required to fully identify the structural determinants for EU1180–438.
GluN1/GluN3 receptors are the least understood receptor subtype among the glutamate receptor family. This is presumably due to difficulties related to expression in heterologous systems, in part because of auto-inhibition mediated by glycine binding to the GluN1 subunit (Awobuluyi et al., 2007; Grand et al., 2018; Kvist et al., 2013a; Madry et al., 2007). This has led to multiple strategies to reveal glycine-activated currents on GluN1/GluN3 receptors, including mutation of the GluN1 glycine binding site as well as recording in the presence of inhibitors of glycine binding to GluN1 (Awobuluyi et al., 2007; Grand et al., 2018; Kvist et al., 2013a; Madry et al., 2007). Evaluation of potential extracellular modulators has revealed strong regulation by both protons and Zn2+ (Cummings & Popescu, 2016; Madry et al., 2008). Moreover, the recent study using CGP-78608 to prevent GluN1/GluN3 desensitization has revealed a remarkable sensitivity to redox reagents and Zn2+, raising the possibility that glycine auto-inhibition may occur in vivo in order to allow the GluN1/GluN3 receptor to respond to other extracellular conditions (Grand et al., 2018). In addition to uncertainty that has surrounded GluN1/GluN3 receptors, there is also a lack of data with which to understand potential triheteromeric GluN1/GluN2/GluN3 receptors. While early studies have suggested this stoichiometry to reflect a major role of GluN3, there have been few successful attempts to confirm early findings, and thus there remains a question about the potential role of GluN3 in NMDA receptors. The uncertainty surrounding GluN3-containing receptors in heterologous systems has carried over to native systems, with relatively few studies being published arguing for a role of GluN3 subunits. In this regard, the identification of a selective inhibitor of GluN3 activity could provide a valuable tool with which to evaluate the potential role of this receptor class in various neuronal functions.
Supplementary Material
Highlights.
EU1180–438 is negative allosteric modulator selective for GluN1/GluN3 receptors.
EU1180–438 inhibits current responses mediated by neuronal GluN1/GluN3A receptors.
EU1180–438 activity is independent of membrane potential and agonist concentration.
Structural determinants of EU1180–438 activity reside near the GluN3A pre-M1 helix.
EU1180–438 is a tool to investigate the physiology of native GluN1/GluN3 receptors.
Acknowledgments
We thank Phuong Le, Jing Zhang, and Gina Bullard for excellent technical assistance.
Funding and disclosure
SFT is a PI on research grants from Allergan, Biogen, and Janssen to Emory University, is a paid consultant for Janssen, is a member of the SAB for Sage Therapeutics, is co-founder of NeurOp Inc, and receives licensing fees and royalties for software. KBH is a PI on a research grant from Janssen to University of Montana. DCL is a member of the Board of Directors for NeurOp Inc. DCL, DSM, SLS, MPE, SFT are co-inventors on Emory University-owned Intellectual Property that includes allosteric modulators of glutamate receptor function.
This work was supported by the NIH (R35NS111619 to SFT; P20GM103546 to KBH; R01NS097536 to KBH; R01HD08237 to HY). The opinions expressed in this publication are those of the author(s) and do not necessarily reflect the views of the NIH.
Abbreviations
- NMDA
N-methyl-D-aspartate
- ATD
amino-terminal domain
- ABD
agonist binding domain
- CTD
carboxyl-terminal domain
- EPSC
excitatory postsynaptic current
- MD
molecular dynamics
- NAM
negative allosteric modulator
- TEVC
two-electrode voltage-clamp
- TMD
transmembrane domain
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Uncategorized References
- Awobuluyi M, Yang J, Ye Y, Chatterton JE, Godzik A, Lipton SA, & Zhang D (2007). Subunit-specific roles of glycine-binding domains in activation of NR1/NR3 N-methyl-D-aspartate receptors. Molecular pharmacology, 71(1), 112–122. 10.1124/mol.106.030700 [DOI] [PubMed] [Google Scholar]
- Balasuriya D, Takahashi H, Srivats S, & Edwardson JM (2014). Activation-induced structural change in the GluN1/GluN3A excitatory glycine receptor. Biochem Biophys Res Commun, 450(4), 1452–1457. 10.1016/j.bbrc.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Cavara NA, & Hollmann M (2008a). Shuffling the deck anew: how NR3 tweaks NMDA receptor function. Mol Neurobiol, 38(1), 16–26. 10.1007/s12035-008-8029-9 [DOI] [PubMed] [Google Scholar]
- Cavara NA, & Hollmann MJMN (2008b). Shuffling the Deck Anew: How NR3 Tweaks NMDA Receptor Function. 38(1), 16–26. 10.1007/s12035-008-8029-9 [DOI] [PubMed] [Google Scholar]
- Cavara NA, Orth A, & Hollmann M (2009). Effects of NR1 splicing on NR1/NR3B-type excitatory glycine receptors. BMC Neurosci, 10, 32 10.1186/1471-2202-10-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, … Zhang D (2002). Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature, 415(6873), 793–798. 10.1038/nature715 [DOI] [PubMed] [Google Scholar]
- Cummings KA, Belin S, & Popescu GK (2017). Residues in the GluN1 C-terminal domain control kinetics and pharmacology of GluN1/GluN3A N-methyl-d-aspartate receptors. Neuropharmacology, 119, 40–47. 10.1016/j.neuropharm.2017.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KA, & Popescu GK (2016). Protons Potentiate GluN1/GluN3A Currents by Attenuating Their Desensitisation. Sci Rep, 6, 23344 10.1038/srep23344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, … Nakanishi N (1998). Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature, 393(6683), 377–381. 10.1038/30748 [DOI] [PubMed] [Google Scholar]
- Edgar RCJBB (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. 5(1), 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Nilsson A, Samuelsson H, Samuelsson EB, Mo L, Akesson E, … Sundstrom E (2007). On the role of NR3A in human NMDA receptors. Physiol Behav, 92(1–2), 54–59. 10.1016/j.physbeh.2007.05.026 [DOI] [PubMed] [Google Scholar]
- Forman CJ, Tomes H, Mbobo B, Burman RJ, Jacobs M, Baden T, & Raimondo JV (2017). Openspritzer: an open hardware pressure ejection system for reliably delivering picolitre volumes. Scientific Reports, 7(1), 2188 10.1038/s41598-017-02301-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand T, Abi Gerges S, David M, Diana MA, & Paoletti P (2018). Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun, 9(1), 4769 10.1038/s41467-018-07236-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, … Traynelis SF (2013). Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Molecular pharmacology, 84(1), 114–127. 10.1124/mol.113.085803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, & Traynelis SF (2018). Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol, 150(8), 1081–1105. 10.1085/jgp.201812032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson MA, Larsen RS, Lawson SN, Pérez-Otaño I, Nakanishi N, Lipton SA, & Philpot B. D. J. P. o. (2012). Genetic deletion of NR3A accelerates glutamatergic synapse maturation. 7(8), e42327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson MA, Roberts AC, Perez-Otano I, & Philpot BD (2010a). Influence of the NR3A subunit on NMDA receptor functions. Prog Neurobiol, 91(1), 23–37. 10.1016/j.pneurobio.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson MA, Roberts AC, Pérez-Otaño I, & Philpot BD (2010b). Influence of the NR3A subunit on NMDA receptor functions. Prog Neurobiol, 91(1), 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ni W, & Zhang C (2017). Calcium-Impermeable NMDA Receptor: A Novel Target for Addiction. Neurosci Bull, 33(3), 357–358. 10.1007/s12264-017-0121-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Schluter OM, & Dong Y (2013). An unusual suspect in cocaine addiction. Neuron, 80(4), 835–836. 10.1016/j.neuron.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniakova M, Kleteckova L, Lichnerova K, Holubova K, Skrenkova K, Korinek M, … Horak M (2018). 7-Methoxyderivative of tacrine is a ‘foot-in-the-door’ open-channel blocker of GluN1/GluN2 and GluN1/GluN3 NMDA receptors with neuroprotective activity in vivo. Neuropharmacology, 140, 217–232. 10.1016/j.neuropharm.2018.08.010 [DOI] [PubMed] [Google Scholar]
- Káradóttir R, Cavelier P, Bergersen LH, & Attwell D (2005). NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature, 438(7071), 1162–1166. 10.1038/nature04302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe LA, Bernardinelli Y, & Muller D (2013). GluN3A: an NMDA receptor subunit with exquisite properties and functions. Neural Plast, 2013, 145387 10.1155/2013/145387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist T, Greenwood JR, Hansen KB, Traynelis SF, & Brauner-Osborne H (2013a). Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors. Neuropharmacology, 75, 324–336. 10.1016/j.neuropharm.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist T, Steffensen TB, Greenwood JR, Mehrzad Tabrizi F, Hansen KB, Gajhede M, … Brauner-Osborne H (2013b). Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site. J Biol Chem, 288(46), 33124–33135. 10.1074/jbc.M113.480210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RS, Corlew RJ, Henson MA, Roberts AC, Mishina M, Watanabe M, … Philpot BD (2011). NR3A-containing NMDARs promote neurotransmitter release and spike timing–dependent plasticity. Nature Neuroscience, 14, 338 10.1038/nn.2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA (2009). PDBsum new things. Nucleic acids research, 37(Database issue), D355–D359. 10.1093/nar/gkn860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low CM, & Wee KS (2010). New insights into the not-so-new NR3 subunits of N-methyl-D-aspartate receptor: localization, structure, and function. Molecular pharmacology, 78(1), 1–11. 10.1124/mol.110.064006 [DOI] [PubMed] [Google Scholar]
- Madry C, Betz H, Geiger JR, & Laube B (2008). Supralinear potentiation of NR1/NR3A excitatory glycine receptors by Zn2+ and NR1 antagonist. Proc Natl Acad Sci U S A, 105(34), 12563–12568. 10.1073/pnas.0805624105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Betz H, Geiger JR, & Laube B (2010). Potentiation of Glycine-Gated NR1/NR3A NMDA Receptors Relieves Ca-Dependent Outward Rectification. Front Mol Neurosci, 3, 6 10.3389/fnmol.2010.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Mesic I, Bartholomaus I, Nicke A, Betz H, & Laube B (2007). Principal role of NR3 subunits in NR1/NR3 excitatory glycine receptor function. Biochem Biophys Res Commun, 354(1), 102–108. 10.1016/j.bbrc.2006.12.153 [DOI] [PubMed] [Google Scholar]
- Martínez-Turrillas R, Puerta E, Chowdhury D, Marco S, Watanabe M, Aguirre N, & Pérez-Otaño I (2012). The NMDA receptor subunit GluN3A protects against 3-nitroproprionic-induced striatal lesions via inhibition of calpain activation. Neurobiology of Disease, 48(3), 290–298. [DOI] [PubMed] [Google Scholar]
- McClymont DW, Harris J, & Mellor IR (2012). Open-channel blockade is less effective on GluN3B than GluN3A subunit-containing NMDA receptors. Eur J Pharmacol, 686(1–3), 22–31. 10.1016/j.ejphar.2012.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micu I, Jiang Q, Coderre E, Ridsdale A, Zhang L, Woulfe J, … Stys PK (2006). NMDA receptors mediate calcium accumulation in myelin during chemical ischaemia. Nature, 439(7079), 988–992. 10.1038/nature04474 [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Tu S, Shin Y, Cui J, Kurokawa T, Zhang D, … Lipton SA (2009). Neuroprotection by the NR3A Subunit of the NMDA Receptor. 29(16), 5260–5265. 10.1523/JNEUROSCI.1067-09.2009 %J The Journal of Neuroscience [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, & Traynelis SF (2011). New advances in NMDA receptor pharmacology. Trends in Pharmacological Sciences, 32(12), 726–733. 10.1016/j.tips.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, & Traynelis SF (2013). Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Molecular pharmacology, 83(5), 1045–1056. 10.1124/mol.113.085209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu Y, Darcq E, Pietrajtis K, Matyas F, Schwartz E, Bessaih T, … Diana MA (2019). Control of aversion by glycine-gated GluN1/GluN3A NMDA receptors in the adult medial habenula. Science, 366(6462), 250–254. 10.1126/science.aax1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachernegg S, Strutz-Seebohm N, & Hollmann M (2012). GluN3 subunit-containing NMDA receptors: not just one-trick ponies. Trends Neurosci, 35(4), 240–249. 10.1016/j.tins.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, & Zhou Q (2013). NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci, 14(6), 383–400. 10.1038/nrn3504 [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Larsen RS, & Wesseling JF (2016). Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci, 17(10), 623–635. 10.1038/nrn.2016.92 [DOI] [PubMed] [Google Scholar]
- Pérez-Otaño I, Luján R, Tavalin SJ, Plomann M, Modregger J, Liu X-B, … Ehlers MD (2006). Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nature Neuroscience, 9(5), 611–621. 10.1038/nn1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk R, Katzman BM, Kusumoto H, Kell SA, Epplin MP, Tahirovic YA, … Traynelis SF (2018). An NMDAR positive and negative allosteric modulator series share a binding site and are interconverted by methyl groups. Elife, 7 10.7554/eLife.34711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RE, Swanger SA, Shelley C, Khatri A, Fernandez-Cuervo G, Epplin MP, … Traynelis SF (2020). Biased modulators of NMDA receptors control channel opening and ion selectivity. Nat Chem Biol, 16(2), 188–196. 10.1038/s41589-019-0449-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina-Crespo JC, Talantova M, Micu I, States B, Chen HS, Tu S, … Lipton SA (2010). Excitatory glycine responses of CNS myelin mediated by NR1/NR3 “NMDA” receptor subunits. J Neurosci, 30(34), 11501–11505. 10.1523/JNEUROSCI.1593-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AC, Díez-García J, Rodriguiz RM, López IP, Luján R, Martínez-Turrillas R, … Pérez-Otaño I (2009). Downregulation of NR3A-Containing NMDARs Is Required for Synapse Maturation and Memory Consolidation. Neuron, 63(3), 342–356. 10.1016/j.neuron.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ŠAli A, & Overington JP (1994). Derivation of rules for comparative protein modeling from a database of protein structure alignments. 3(9), 1582–1596. 10.1002/pro.5560030923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, & Fern R (2005). NMDA receptors are expressed in developing oligodendrocyte processes and mediate injury. Nature, 438(7071), 1167–1171. 10.1038/nature04301 [DOI] [PubMed] [Google Scholar]
- Santangelo Freel RM, Ogden KK, Strong KL, Khatri A, Chepiga KM, Jensen HS, … Liotta DC (2013). Synthesis and Structure Activity Relationship of Tetrahydroisoquinoline-Based Potentiators of GluN2C and GluN2D Containing N-Methyl-d-aspartate Receptors. Journal of Medicinal Chemistry, 56(13), 5351–5381. 10.1021/jm400177t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo RM, Acker TM, Zimmerman SS, Katzman BM, Strong KL, Traynelis SF, & Liotta DC (2012). Novel NMDA receptor modulators: an update. Expert opinion on therapeutic patents, 22(11), 1337–1352. 10.1517/13543776.2012.728587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler T, Mesic I, Madry C, Bartholomaus I, & Laube B (2008). Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-D-aspartate receptor assembly. J Biol Chem, 283(1), 37–46. 10.1074/jbc.M703539200 [DOI] [PubMed] [Google Scholar]
- Smothers CT, & Woodward JJ (2003). Effect of the NR3 subunit on ethanol inhibition of recombinant NMDA receptors. Brain Res, 987(1), 117–121. [DOI] [PubMed] [Google Scholar]
- Smothers CT, & Woodward JJ (2007). Pharmacological characterization of glycine-activated currents in HEK 293 cells expressing N-methyl-D-aspartate NR1 and NR3 subunits. J Pharmacol Exp Ther, 322(2), 739–748. 10.1124/jpet.107.123836 [DOI] [PubMed] [Google Scholar]
- Smothers CT, & Woodward JJ (2009). Expression of glycine-activated diheteromeric NR1/NR3 receptors in human embryonic kidney 293 cells Is NR1 splice variant-dependent. J Pharmacol Exp Ther, 331(3), 975–984. 10.1124/jpet.109.158493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, & Rosenmund C (1998). A Point Mutation in the Glutamate Binding Site Blocks Desensitization of AMPA Receptors. Neuron, 21(4), 907–918. 10.1016/S0896-6273(00)80605-4 [DOI] [PubMed] [Google Scholar]
- Strong KL, Epplin MP, Bacsa J, Butch CJ, Burger PB, Menaldino DS, … Liotta DC (2017). The Structure–Activity Relationship of a Tetrahydroisoquinoline Class of N-Methyl-d-Aspartate Receptor Modulators that Potentiates GluN2B-Containing N-Methyl-d-Aspartate Receptors. Journal of Medicinal Chemistry, 60(13), 5556–5585. 10.1021/acs.jmedchem.7b00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stys PK, & Lipton SA (2007). White matter NMDA receptors: an unexpected new therapeutic target? Trends in Pharmacological Sciences, 28(11), 561–566. 10.1016/j.tips.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, & Jaramillo F (1998). Getting the most out of noise in the central nervous system. Trends Neurosci, 21(4), 137–145. 10.1016/s0166-2236(98)01238-7 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, … Dingledine R (2010). Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol Rev, 62(3), 405–496. 10.1124/pr.109.002451 %J Pharmacological Reviews [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, & Isacoff EY (2007). Subunit counting in membrane-bound proteins. Nature Methods, 4(4), 319–321. 10.1038/nmeth1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, & Isacoff EY (2008). Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci U S A, 105(37), 14163–14168. 10.1073/pnas.0802075105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A, Takahashi H, Lipton SA, & Chen HS (2006). NR3A modulates the outer vestibule of the “NMDA” receptor channel. J Neurosci, 26(51), 13156–13166. 10.1523/JNEUROSCI.2552-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TM, Brown BM, Deng L, Sellers BD, Lupardus PJ, Wallweber HJA, … Hanson JE (2017). A novel NMDA receptor positive allosteric modulator that acts via the transmembrane domain. Neuropharmacology, 121, 204–218. 10.1016/j.neuropharm.2017.04.041 [DOI] [PubMed] [Google Scholar]
- Wee KS, Wee ZN, Chow NB, & Low CM (2010). The distal carboxyl terminal of rat NR3B subunit regulates NR1–1a/NR3B and NR1–2a/NR3B surface trafficking. Neurochem Int, 57(2), 97–101. 10.1016/j.neuint.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Wesseling JF, & Perez-Otano I (2015). Modulation of GluN3A expression in Huntington disease: a new n-methyl-D-aspartate receptor-based therapeutic approach? JAMA Neurol, 72(4), 468–473. 10.1001/jamaneurol.2014.3953 [DOI] [PubMed] [Google Scholar]
- Yao Y, & Mayer ML (2006). Characterization of a soluble ligand binding domain of the NMDA receptor regulatory subunit NR3A. J Neurosci, 26(17), 4559–4566. 10.1523/jneurosci.0560-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Singh AK, Sampson JM, Narangoda C, Kurnikova M, & Sobolevsky AI (2016). Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs. Neuron, 91(6), 1305–1315. 10.1016/j.neuron.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Ball J, Stoll KE, Satpute VC, Mitchell SM, Pauli JL, … Lawrence JJ (2014). Direct excitation of parvalbumin-positive interneurons by M1 muscarinic acetylcholine receptors: roles in cellular excitability, inhibitory transmission and cognition. 592(16), 3463–3494. 10.1113/jphysiol.2014.275453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Zachariassen LG, Dorsett KN, & Hansen KB (2018). Properties of Triheteromeric N-Methyl-d-Aspartate Receptors Containing Two Distinct GluN1 Isoforms. Molecular pharmacology, 93(5), 453–467. 10.1124/mol.117.111427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CL, Shi EY, Srinivasan J, Ptak CP, Oswald RE, & Nowak LM (2019). Modulation of AMPA Receptor Gating by the Anticonvulsant Drug, Perampanel. ACS Med Chem Lett, 10(3), 237–242. 10.1021/acsmedchemlett.8b00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, & Bellone C (2013). Glutamatergic receptors at developing synapses: the role of GluN3A-containing NMDA receptors and GluA2-lacking AMPA receptors. Eur J Pharmacol, 719(1–3), 107–111. 10.1016/j.ejphar.2013.04.056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.