Abstract
Microsatellite instability-high (MSI-H) and tumor mutational burden (TMB) are predictive biomarkers for immune-checkpoint inhibitors (ICIs). Still, the relationship between the underlying cause(s) of MSI and TMB in tumors remains poorly defined. We investigated associations of TMB to mismatch repair (MMR) protein expression patterns by immunohistochemistry (IHC) and MMR mutations in a diverse sample of tumors. Hypothesized differences were identified by the protein/gene affected/mutated and the tumor histology/primary site. Overall, 1057 MSI-H tumors were identified from the 32 932 tested. MSI was examined by NGS using 7000+ target microsatellite loci. TMB was calculated using only nonsynonymous missense mutations sequenced with a 592-gene panel; a subset of MSI-H tumors also had MMR IHC performed. Analyses examined TMB by MMR protein heterodimer impacted (loss of MLH1/PMS2 vs. MSH2/MSH6 expression) and gene-specific mutations. The sample was 54.6% female; mean age was 63.5 years. Among IHC tested tumors, loss of co-expression of MLH1/PMS2 was more common (n = 544/705, 77.2%) than loss of MSH2/MSH6 (n = 81/705, 11.5%; P < .0001), and was associated with lower mean TMB (MLH1/PMS2: 25.03 mut/Mb vs MSH2/MSH6 46.83 mut/Mb; P < .0001). TMB also varied by tumor histology: colorectal cancers demonstrating MLH1/PMS2 loss had higher TMBs (33.14 mut/Mb) than endometrial cancers (20.60 mut/Mb) and other tumors (25.59 mut/Mb; P < .0001). MMR gene mutations were detected in 42.0% of tumors; among these, MSH6 mutations were most common (25.7%). MSH6 mutation patterns showed variability by tumor histology and TMB. TMB varies by underlying cause(s) of MSI and tumor histology; this heterogeneity may contribute to differences in response to ICI.
Keywords: microsatellite instability, mismatch repair, tumor mutational burden, MSI, TMB, immune checkpoint inhibitors
1 |. INTRODUCTION
DNA mismatch repair (MMR) is crucial to ensure the integrity of the genome. The primary function of the MMR pathway is to identify and correct base-base mismatches and insertions/deletions (indels) generated during DNA replication and recombination.1 A direct consequence of MMR deficiency is instability (ie, gains or losses) in microsatellite regions of the genome, consisting of mono- and di-nucleotide base pair repeats. The microsatellite instability (MSI) phenotype is widely used as a diagnostic marker for MMR deficiency in tumor cells and generally equates to reduced or deficient activity of the mutSα protein complex (responsible for initial recognition of DNA mismatches) due to mutation or inactivation of the heterodimer partners MSH2 or MSH6, or the mutLα protein complex (which subsequently nicks DNA at sites of mismatch to begin the repair) due to mutation or inactivation of the heterodimer partners MLH1 or PMS2. Clinically, the integrity of MMR is frequently assessed using immunohistochemical (IHC) staining for expression of the four MMR proteins, with loss of MLH1/PMS2 and/or MSH2/MSH6 expression being the most common patterns seen in tumors with MMR deficiency. The integrity of MMR may also be assessed by fragment analysis for MSI at five predefined genomic regions (ie, Bethesda panel), or, more recently, by next-generation sequencing (NGS).2,3 While MSI-H along with tumor mutational burden (TMB), measured on NGS tumor profiles has emerged as a relevant biomarker for predicting tumor response to immune checkpoint inhibitors (ICIs),4–6 studies have shown TMB varies significantly among MSI-H tumors,7,8 and their independent and combined impact on responsiveness to ICI remains under investigation.8–10
To date, studies of MSI as a predictive therapeutic marker have not examined patients according to the MMR protein/heterodimer pair impacted (MLH1/PMS2 vs MSH2/MSH6) or the individual MMR gene(s) mutated.11–13 Analyses have also not characterized how underlying cause of MMR deficiency may impact potentially relevant molecular and clinical factors such as the rate of TMB-H and TMB as measured by total mutations per megabase (mut/Mb).
The aim of our study is to characterize a large commercial clinical sample of MSI-H tumors, and specifically to characterize how MMR protein expression patterns (by IHC) and MMR gene mutations (by NGS) are associated with TMB. Our goal is to better elucidate how relevant clinical predictors of response to ICI, such as MSI-H and TMB, are related across a broad spectrum of clinically tested tumor histologies. We hypothesized TMB would show significant differences depending on whether the MSI-H phenotype was associated with loss of co-expression of MLH1/PMS2 or MSH2/MSH6. Further, we also hypothesized that overall TMB magnitude would show significant variability by MMR gene(s) mutated.
2 |. METHODS
2.1 |. Patients
A retrospective analysis was conducted on consecutive MSI-H tumors from an unselected international sample of patients undergoing commercial tumor genomic profiling using a single platform between January 30, 2015, and January 18, 2018. IHC protein expression of the four MMR proteins was also conducted on 66.7% of tumors [primarily colorectal cancer (CRC) and endometrial cancer (EC) due to clinical relevance].
2.2 |. Next-generation sequencing
NGS was performed on genomic DNA isolated from formalin-fixed paraffin-embedded (FFPE) tumor samples using the NextSeq platform (Illumina, Inc., San Diego, California). Tumor enrichment was achieved by harvesting targeted tissue using manual microdissection techniques. Only tumor was sequenced (no matched normal). A custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies, Santa Clara, California). All variants were detected with >99% confidence based on allele frequency and amplicon coverage, with an average sequencing depth of coverage of >500 and an analytic sensitivity of 5%. Only pathogenic/presumed pathogenic variants were included. No amplification of MMR genes was identified. Analyses of copy number variation and gene fusions were not available on this dataset.
2.3 |. MMR protein expression by immunohistochemistry
MMR protein expression was tested by immunohistochemistry (IHC) using antibody clones (MLH1, M1 antibody; MSH2, G2191129 antibody; MSH6, 44 antibody3; and PMS2, EPR3947 antibody, Ventana Medical Systems, Inc., Tucson, Arizona). The complete absence of protein expression of any of the four proteins tested (0+ in 100% of cells) was considered deficient MMR. Because the MMR proteins function as heterodimers, we conducted analyses of MSH2/MSH6 deficient tumors vs MLH1/PMS2.
2.4 |. MSI testing
MSI was examined using over 7000 target microsatellite loci and compared to the reference genome hg19 from the University of California, Santa Cruz (UCSC) Genome Browser database. The number of microsatellite loci that were altered by insertion or deletions (indels) was counted for each sample. Only indels that increased or decreased the number of repeats were considered. Genomic variants in the microsatellite loci were detected using the same depth and frequency criteria as we used for mutation detection. MSI-NGS results were compared to results from over 2000 matching clinical cases analyzed with traditional PCR-based methods. The threshold to determine MSI-High by NGS was determined to be 46 or more loci with indels to generate a sensitivity of >95% and specificity of >99%.3
2.5 |. Tumor mutational burden
TMB was measured by counting all nonsynonymous missense mutations found per tumor that had not been previously described as germline alterations in gnomAD, dbSNP and in house Caris databases (592 genes and 1.4 megabases [MB] sequenced per tumor). TMB was considered as both a binary variable (TMB-L vs TMB-H defined as ≥17 mut/Mb) and a continuous variable measuring TMB magnitude. The threshold to define TMB-H was established by comparing TMB with MSI by fragment analysis in CRC cases.8
2.6 |. Statistical analysis
Comparisons of TMB among molecular groups were tested with Analysis of Variance (ANOVA) and confirmed with nonparametric Wilcox test using JMP (13.2.1); biomarker alteration prevalence comparisons were done by Fisher-Exact test. A nonparametric test of association between microsatellite loci and TMB was performed using Spearman’s rank-based measure of association using R statistical software version 3.4.3. P < .05 was considered statistically significant for all tests performed.
2.7 |. Ethics statement
All human subjects’ data were de-identified. Thus, this research was determined to be exempt from the requirement for informed consent per the Western Institutional Review Board (WIRB).
3 |. RESULTS
3.1 |. Sample characteristics
In total, of 32 932 tumors tested, 1057 (3.2%) were found to be MSI-H by NGS testing. This sample included 283 CRCs, 449 ECs, and 325 other tumors (OTs) from 29 other cancer types (Table 1). The sample was 54.6% female, and median patient age of the time of tumor testing was 63.5 years.
TABLE 1.
Characteristics of MSI-H tumors
| Tumor histology | n | n (%) with complete IHC | n (%) with complete NGS | n (%) with both complete IHC and NGS | Average of TMB (mt/MB) |
|---|---|---|---|---|---|
| Endometrial cancer | 449 | 333 (74) | 272 (61) | 210 (47) | 22.8 |
| Colorectal adenocarcinoma | 283 | 246 (87) | 170 (60) | 148 (52) | 38.8 |
| Other cancers | |||||
| Ovarian surface epithelial carcinomas | 47 | 27 (57) | 24 (51) | 16 (34) | 42.6 |
| Gastric adenocarcinoma | 45 | 28 (62) | 24 (53) | 14 (31) | 30.2 |
| Lung nonsmall cell lung cancer (NSCLC) | 42 | 1 (2) | 25 (60) | 0 | 31.6 |
| Cancer of unknown primary | 26 | 8 (31) | 13 (50) | 3 (12) | 45.5 |
| Small intestinal malignancies | 23 | 13 (57) | 14 (61) | 8 (35) | 26.7 |
| Pancreatic adenocarcinoma | 19 | 13 (68) | 8 (42) | 5 (26) | 23.2 |
| Breast carcinoma | 16 | 6 (38) | 5 (31) | 2 (13) | 24.8 |
| Prostatic adenocarcinoma | 16 | 6 (38) | 3 (19) | 0 | 27.0 |
| Esophageal and esophagogastric junction carcinoma | 12 | 6 (50) | 6 (50) | 3 (25) | 25.4 |
| Cholangiocarcinoma | 11 | 5 (45) | 7 (64) | 3 (27) | 28.1 |
| Neuroendocrine tumors | 10 | 2 (20) | 4 (40) | 1 (10) | 23.8 |
| Cervical cancer | 9 | 1 (11) | 7 (78) | 1 (11) | 21.4 |
| Glioma | 7 | 3 (43) | 4 (57) | 1 (14) | 48.1 |
| Soft tissue tumors | 6 | 0 | 2 (33) | 0 | 16.0 |
| Uterine sarcoma | 6 | 3 (50) | 2 (33) | 1 (17) | 16.3 |
| Bladder cancer | 5 | 2 (40) | 3 (60) | 0 | 35.6 |
| Nonmelanoma skin cancers—squamous cell skin cancer | 4 | 0 | 3 (75) | 0 | 38.3 |
| Female genital tract malignancy-other | 3 | 1 (33) | 1 (33) | 1 (33) | 18.0 |
| Kidney cancer | 3 | 0 | 2 (67) | 0 | 17.0 |
| Other | 3 | 0 | 1 (33) | 0 | 18.0 |
| Head and neck squamous carcinoma | 2 | 0 | 2 (100) | 0 | 16.0 |
| Liver hepatocellular carcinoma | 2 | 0 | 1 (50) | 0 | 15.5 |
| Thymic carcinoma | 2 | 0 | 2 (100) | 0 | 15.0 |
| Bone cancer | 1 | 0 | 0 | 0 | 4.0 |
| Lung small cell cancer (SCLC) | 1 | 0 | 1 (100) | 0 | 11.0 |
| Lymphoma | 1 | 0 | 1 (100) | 0 | 89.0 |
| Non epithelial ovarian cancer (non-EOC) | 1 | 0 | 1 (100) | 0 | 14.0 |
| Thyroid carcinoma | 1 | 0 | 1 (100) | 0 | 20.0 |
| Uveal melanoma | 1 | 1 (100) | 1 (100) | 1 (100) | 17.0 |
| Total | 1057 | 705 (67) | 610 (58) | 418 (40) | 29.6 |
Paired IHC of the four MMR proteins was conducted on 66.7% (705/1057) of the sample [MLH1 67.9%; PMS2 67.6%, MSH2 68.5%, MSH6 68.1%]. NGS DNA sequencing of the four MMR proteins was conducted in nearly 100% of the sample [MLH1, 99.6%; MSH2, 96.5%; MSH6, 98.2%; PMS2, 59.1%]. PMS2, notably, had more indeterminate results due to poor sequencing of large germline deletions and result ambiguity due to structural complications of the gene including PMS2CL pseudogene.
3.2 |. Baseline variability in MMR IHC patterns
Sex, age, and tumor histology variability in IHC patterns was seen. For example, in CRC, where a female:male ratio of 50:50 would be anticipated, females had higher rates of loss of MLH1/PMS2 co-expression by IHC (57.6%) than males (42.4%), and lower rates of loss of MSH2/MSH6 co-expression (45.0%) than males (55.0%). Patients with loss of MSH2/MSH6 co-expression were also substantially younger than those with loss of MLH1/PMS2 co-expression among CRCs (48.9 vs 64.8 years; P < .0001), with differences less striking among ECs (59.1 vs 64.5 years; P = .14) and other cancers (59.2 vs 65.9 years; P = .33).
3.3 |. Association of MMR IHC patterns with TMB
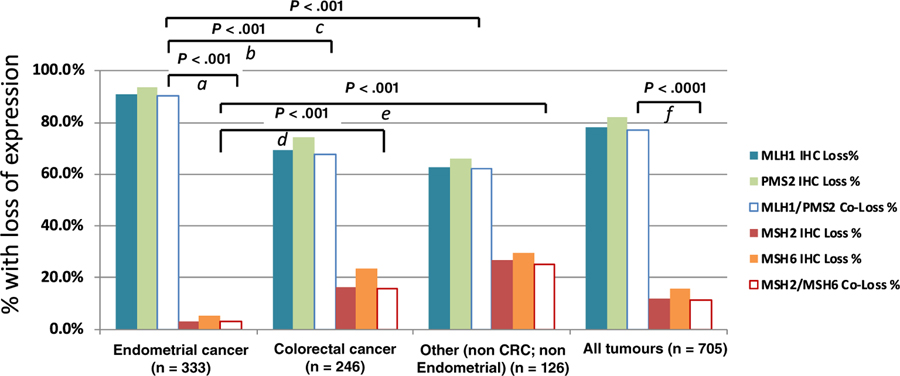
We examined patterns of MMR protein losses in the subset of MSI-H tumor undergoing IHC (705/1057 tumors), and the relationship of those patterns to TMB. Loss of MLH1 expression (78.2%) and/or PMS2 expression (82.0%) occurred at higher frequency than loss of MSH2 (12.1%) or MSH6 (15.9%; P < .0001), and loss of MLH1/PMS2 co-expression (n = 544/705, 77.2%) was more common than loss of MSH2/MSH6 co-expression (n = 81/705, 11.5%; P < .0001; Figure 1). When examined by tumor histology, ECs were more likely to have loss of co-expression of MLH1/PMS2 (90.1%) than CRCs (67.5%; P < .001) and OTs (61.9%; P < .001), and less likely to have loss of co-expression of MSH2/MSH6 (3.0%) than CRCs (15.9%; P < .001) and OTs (25.4%; P < .001). Overall, abnormalities in expression of the MLH1/PMS2 proteins were >fivefold more common than abnormalities in expression of the MSH2/MSH6 proteins in our sample.
FIGURE 1.
Rates of MMR protein expression by IHC stratified by tumor histology. (a) Difference in rate of MLH1/PMS2 protein expression loss vs MSH2/MSH6 in EC. (b) Difference in rate of MLH1/PMS2 protein expression loss in EC vs MLH1/PMS2 protein expression loss in CRC. (c) Difference in rate of MLH1/PMS2 protein expression loss vs MSH2/MSH6 in OT. (d) Difference in rate of MSH2/MSH6 protein expression loss in EC vs CRC. (e) Difference in rate of MSH2/MSH6 protein expression loss in EC vs OT. (f) Difference in rate of MLH1/PMS2 protein expression loss vs MSH2/MSH6 in all tumors
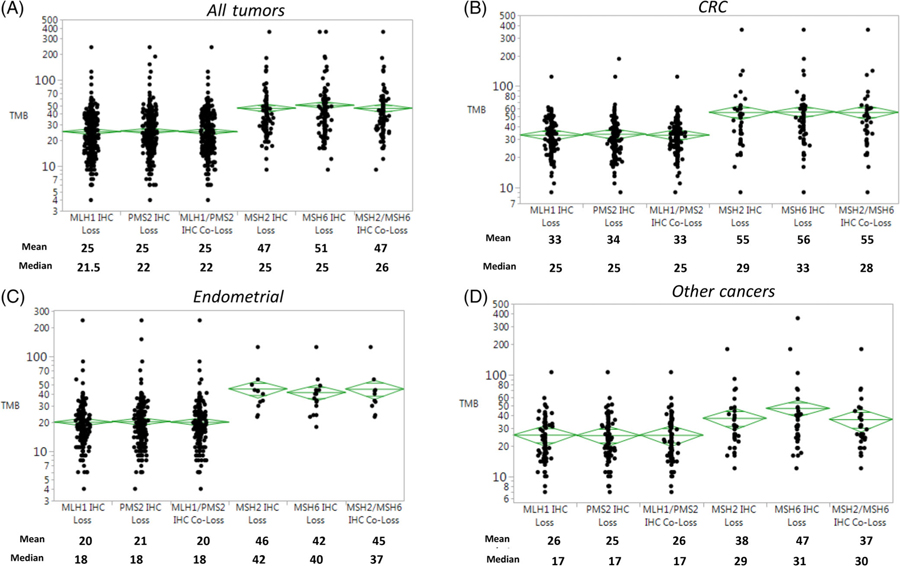
Associations of patterns of MMR protein losses and TMB were then investigated (Figure 2). TMB ranged from 4 to 361 mut/Mb over the entire sample (9–360 mut/Mb in CRCs, 4–239 mut/Mb in EC and 7–361 mut/Mb in OTs). Among all tumors, loss of co-expression of MSH2/MSH6 was associated with higher mean TMB (46.83 mut/Mb) than loss of MLH1/PMS2 (25.03 mut/Mb; P < .0001). Similarly, CRCs that exhibited loss of MSH2/MSH6 (n = 40, 55.45 mut/Mb) tended to have higher TMBs than ECs (n = 10, 45.30 mut/Mb) and OTs (n = 32, 36.53 mut/Mb; P = .09). As noted above, TMB associated with loss of MLH1/PMS2 was generally lower than that associated with loss of MSH2/MSH6. Among tumors with loss of MLH1/PMS2, CRCs had higher TMBs (n = 170, 33.14 mut/Mb) than ECs (n = 303, 20.60 mut/Mb) and OTs (n = 80, 25.59 mut/Mb; P < .0001).
FIGURE 2.
Tumor mutational burden (TMB) examined by mismatch repair (MMR) protein expression and disease site. CRC, Colorectal cancer; IHC, immunohistochemistry; mt/Mb, mutations per megabase
3.4 |. Association of MMR gene mutations with TMB
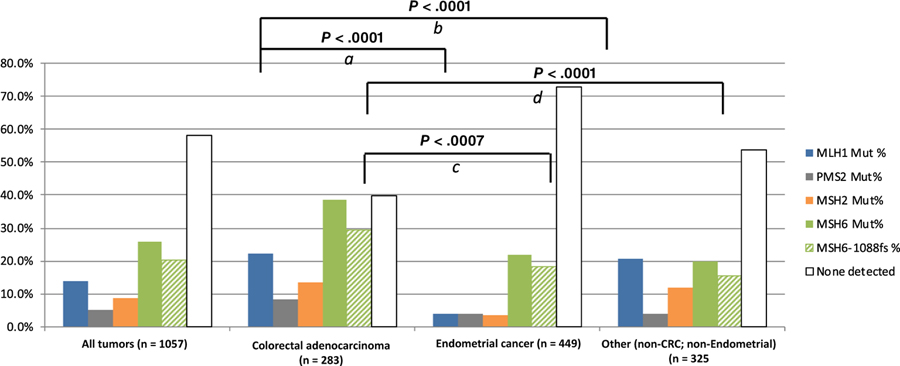
Mutations were heterogeneously distributed among the MMR genes and by tumor histology. Overall, 58% (614/1057) tumors had no detectable MMR mutation. A substantial minority (41.9%, 443/1057) of tumors had at least one MMR mutation, and many tumors bore multiple mutations (Figure S1). ECs (27.3%; 123/449) had lower rates of MMR mutations compared to CRC (59.7%; 169/283) and OTs (46.1%; 150/325; P < .0001).
MSH6 mutations were identified in 25.7% (267/1038) tumors, and were the most common MMR mutations detected [60.3% (267/443) of total mutations]. MSH6 mutations were more common in CRCs (106/275, 38.5%) than ECs (98/449, 21.9%, P < .0001) or OTs (63/325, 19.9%, P < .0001) (Figure 3). A previously described14 recurrent passenger mutation in an MSH6 exon 5 coding microsatellite, MSH6 F1088fs, was found in 20.0% (212/1057) of all tumors, comprising a substantial minority 39.6% (212/535) of all MMR mutations detected overall and nearly 64.6% of all MSH6 mutations observed (Figure S2). The MSH6 F1088fs mutation was more prevalent in CRC than OTs (18.3% of all EC, 29.5% of all CRCs and 15.5% of OTs; P = .0007 for CRC vs ECs; P < .0001 for CRC vs OTs; Figure 3)
FIGURE 3.
Rate of mismatch repair gene mutations by specific gene and tumor histology. (a) Rate of mutation in MLH1 in CRC vs EC. (b) Rate of mutation in MLH1 in CRC vs OT. (c) Rate of mutation in MSH6 in CRC vs EC. (d) Rate of mutation in MSH6 in CRC vs OT
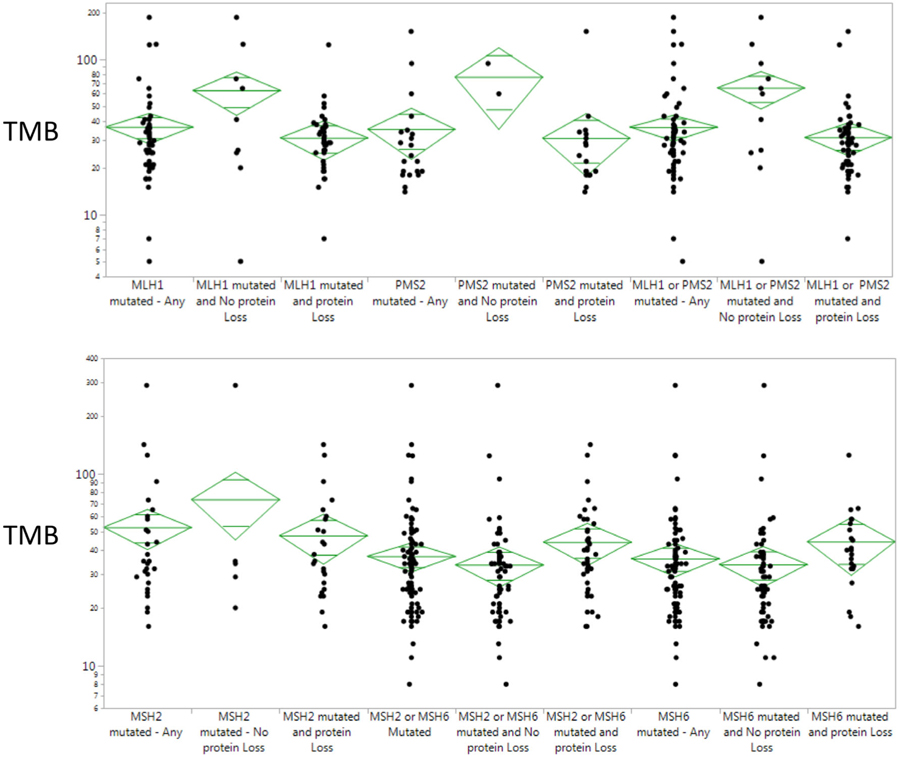
A sizable subset of tumors (418/1057 or 39.5%) underwent complete 4-gene MMR NGS testing and 4-gene MMR IHC staining (Table 1 and Figure S3). Gene-specific IHC protein expression and NGS mutations for MLH1 and PMS2 demonstrated high levels of discordance. The majority of tumors that showed loss of MLH1 (67%) and PMS2 expression (78%) had no detectable mutations in the MLH1 or PMS2 genes—only 11% of tumors showed a concordance between MLH1 protein loss and an MLH1 mutation, and only 5% of PMS2 tumors showed concordance. MSH6 in comparison frequently showed normal expression even when an underlying MSH6 mutation was detected—among 599 tumors with intact MSH6 expression on IHC, 21.4% (n = 128) had underlying MSH6 mutations.
Finally, we examined associations between MMR mutations and TMB, and stratified results by the presence or absence of concurrent protein losses by IHC. Overall, substantial variability was seen depending on whether gene-specific mutations were concurrent with IHC protein losses (Figure 4). When examined by histology, ECs bearing MMR mutations generally had lower TMBs than CRCs and OTs. For example, TMB associated with MSH6 mutations in CRCs (44.5 mut/Mb) and OTs (52.0 mut/Mb) was significantly higher than in ECs with MSH6 mutations (26.7 mut/Mb; P = .0001). It was interesting to note that TMB among all tumors that bore the passenger MSH6 F1088fs mutation (n = 212, 36.34 mut/Mb) was lower than the TMB of tumors bearing other MSH6 mutations (n = 55, 52.96 mut/Mb; P = .0063). This difference varied by disease site: TMB associated with MSH6 F1088fs in ECs (25.8 mut/Mb) were significantly lower than those in CRCs (43.2 mut/Mb) (P = .0008) and OTs (42.7 mut/Mb; P = .0008; Table S1).
FIGURE 4.
Tumor mutational burden stratified by MMR mutation and MMR protein expression. MMR, mismatch repair; TMB, tumor mutational burden
4 |. DISCUSSION
In the current study, we demonstrate MMR gene and protein expression associated variation in TMB; our data show that tumors with loss of MLH1/PMS2 co-expression by IHC are associated with a lower TMB compared to tumors with loss of MSH2/MSH6 co-expression, a relationship that is consistent across tumor histologies. We hypothesize this variability may be related to (a) gene-specific and tissue-specific differences in the molecular events that precede loss of MLH1/PMS2 vs MSH2/MSH6 co-expression (eg, MLH1 promoter methylation as the dominant cause of loss of MLH1 expression) or (b) gene-specific and tissue-specific events that occur in tumors subsequent to the loss of MMR function when the MLH1/PMS2 or the MSH2/MSH6 heterodimers are rendered nonfunctional. Whether the underlying cause of MSI is, in parallel to TMB, important in determining differential responses to ICI therapy, response durability or development of resistance remains uncertain, but further research is warranted due to the potential relevance of this finding to clinical practice. While early studies in CRC have not detected differential responses to ICI in the Lynch syndrome population vs MSI-H patients with MLH1 promoter methylation, differences in outcomes or toxicities may still be observed in future studies and with longer follow-up.11–13
Our results additionally demonstrate histology-specific patterns of MMR IHC protein co-expression and associated TMB. Compared to the OTs examined, ECs exhibited more frequent loss of MLH1/PMS2 by IHC, lower histology-specific TMBs, and lower TMBs associated with loss of MLH1/PMS2 co-expression and loss of MSH2/MSH6 co-expression. In comparison, CRCs demonstrated the highest rates of loss of MSH2/MSH6 co-expression and overall highest mean TMBs relative to EC and OTs. The contribution of inactivation of the MLH1/PMS2 heterodimer in ECs (vs CRCs) due largely to methylation of the MLH1 promoter is well-documented, but the consistent association of MLH1/PMS2 inactivation with lower TMBs than seen in comparison to inactivation of the MSH2/MSH6 heterodimer is new. Our findings suggest that the role of the mutSα protein complex (MSH2/MSH6) due to poor recognition of sites of nucleotide mismatches may be more critical to ensuring intact MMR than the mutLα complex (MLH1/PMS2). Malfunction of mutSα may lead then to a higher burden of mutations in the tumor overall that may be mitigated in some tumor histology types by secondary DNA repair pathways better than in others (leading to histology-specific differences in TMBs across similar IHC protein loss patterns). Taken as a whole, these data reveal the diversity of gene- and histology-specific heterogeneity within MSI-H tumors.
Sequencing of the MMR genes demonstrated several interesting findings. Our results showed that MMR mutations occur in less than half of MSI-H tumors—nearly 60% of the tumors examined here did not carry an MMR mutation. Recently published research examining unselected tumors via paired tumor/normal sequencing identified germline MMR mutations in only 16.3% MSI-H tumors overall, with CRC and EC representing over half of this group, and Lynch syndrome spectrum tumors over 90%.15 Unfortunately, our results lack paired germline data to conduct similar analyses. However, among those tumors in our sample without an MMR mutation, we found the most common abnormality by IHC was loss of MLH1/PMS2 across all tumor types. This suggests that loss of function of the MLH1/PMS2 protein heterodimer, most often caused by methylation of the MLH1 gene promoter inhibiting MLH1 gene transcription and MLH1 protein expression, is likely the major mechanism that drives MSI in diverse adult tumors, not just CRC and EC. Moreover, it further suggests the contribution of functional MLH1 to MMR may be particularly vulnerable to inactivation across a wide variety of tumors, although this does not exclude the possibility that other mechanisms also exist. Research demonstrating that hypomethylating agents can reactivate an MLH1 gene silenced by promoter methylation may support future novel approaches to cancer treatment and prevention.16
An additional finding was the significant histology-specific variability in TMB associated with the MSH6 F1088fs passenger mutation. This mutation of a C8 coding microsatellite in exon 5 of MSH6 has been previously described in tumors and germline.16,17 In our study, this was the only MMR mutation seen in 35% of tumors. Further, it was differentially distributed by histology and differentially associated with TMB by histology—that is it represented a greater portion of the total mutations seen in CRC (compared to EC or OT; Figure 3) but in EC was associated with a lower TMB than in CRC and OTs (Table S1). We found other MSH6 mutations identified in this cohort (data not shown) were more often associated with POLE mutations, high TMB (>100 mut/mb), and a high frequency of premature stop codons. Taken together, these findings suggest that MSI and TMB could be modulated by tissue and cause-specific factors that ultimately impact a tumor’s individual mutational spectrum. Individual variability in causes of MSI, TMB, and the resultant downstream passenger mutational patterns may then also impact each tumor’s neo-antigen profile and, perhaps, ICI responsiveness, although further research is needed to demonstrate the latter.
Our findings may have direct impact on our clinical practice. In fact, recent literature data showed that overall response rate (ORR) to ICI (eg, pembrolizumab) are quite different among MSI-H tumors based on tumor types. For example, ORR was 57% in MSI-H EC,18 33% in CRC19 and 34% in non-CRC.20 In addition, among non-CRC tumor types, a huge variability in ORR was noticed, such as 18.2% for pancreatic cancer and 0% for CNS cancers.20,21 Our data shown here may explain the variability in ICI benefit among MSI-H tumors, based on primary sites.
It is worth mention that not all MSI-H tumors are associated with TMB-H, as we already showed elsewhere.7,8 This phenomenon may have direct impact of response to ICI in clinical practice and deserve further evaluation in prospective studies.
Our study has several limitations. As we analyzed a convenience sample of commercial tumor profiling data, the only available clinical data were age, sex, and tumor histology gathered from a nonclinical test requisition. Because a matched blood sample was not collected, our ability to distinguish germline mutations from tumor-only mutations was diminished, increasing the chances of inaccurate calls. This is particularly a problem among individuals from racial/ethnic subpopulations that have undergone limited germline and tumor genetic sequencing and may be underrepresented in public sequencing databases that are used to determine whether observed variants are benign vs damaging. Also, MLH1 promoter methylation testing was not available at the time these data were collected, limiting our ability to more fully characterize MLH1 expression loss by IHC. Finally, small sample sizes necessitated analyses of non-CRC non-EC as one group of OTs.
5 |. CONCLUSION
In summary, we identified substantial MMR gene- and histology-specific heterogeneity among MSI-H tumors and among variables that are considered predictors of response to ICIs. TMB varies significantly according to the MMR protein heterodimer affected and the MMR gene affected, and inactivation of MLH1/PMS2 appears to be the dominant mechanism driving MSI in this diverse tumor sample. These findings offer insight into what underlying molecular events are most prevalent in MSI-H tumors and how these events impact variability in TMB.
Supplementary Material
What’s new?
Immunotherapy based on checkpoint inhibitors shows promising results in a variety of cancer types, but still benefits a minority of patients. High microsatellite instability (MSI) and tumor mutational burden (TMB) have both been identified as biomarkers predictive of response to checkpoint inhibitors. Here, the authors investigated how the underlying causes of MSI influence TMB. Tumors lacking the mismatch repair protein duo MLH1/PMS2 had lower TMB than those lacking a different protein heterodimer, MLH2/MSH6. Even among tumors lacking the same mismatch repair proteins, the tissue of origin influenced mutational burden.
ACKNOWLEDGEMENT
This work was supported in part by the National Institutes of Health through the Core Grant to Fox Chase Cancer Center (P30CA006927 for support of M. J. H and J. N. B).
Funding information
NIH Clinical Center, Grant/Award Number: P30CA006927
Abbreviations:
- CRC
colorectal cancer
- EC
endometrial cancer
- FFPE
formalin-fixed paraffin-embedded
- ICIs
immune-checkpoint inhibitors
- IHC
immunohistochemistry
- Indels
insertions/deletions
- MMR
DNA mismatch repair
- MSI-H
microsatellite instability-high
- Mut/Mb
mutations per megabase
- NGS
next-generation sequencing
- ORR
overall response rate
- OTs
other tumors
- TMB
tumor mutational burden
- TMB-L
tumor mutational burden-low
- TMB-H
tumor mutational burden-high
Footnotes
CONFLICT OF INTEREST
Drs J. N. B., A. P., E. S. K., A. F. S., A. G. and W. M. W. declare no potential conflicts of interest. Dr J. X. is employed by Caris Life Sciences. Dr W. M. K. is employed by Caris Life Sciences and stock option holder with Caris Life Sciences, consultant for Merck and Sharp & Dohme. Dr J. L. M. was an interim CMO in 2018 for Caris Life Sciences, is consultant and speaker for Merck, Genentech, Bayer, Taiho, Amgen, Celgene. Dr R. M. G. is consultant for Merck, Taiho, Genentech, Novartis. Dr M. E. S. received travel support from Caris Life Sciences, and is consultant and speaker for Taiho, BMS, Merck. Dr H.-F. L. is advisory board for Caris Life Sciences and BMS. Dr M. J. H. received travel support from Caris Life Sciences, and performed collaborative research with Myriad Genetics, Invitae, Gene Dx, Ambry Genetics Corporation and Caris Life Sciences. His institution received payments for the Keynote-177 trial (Merck) and for the POLO trial (AstraZeneca) based on patients accrual as PI site.
DATA ACCESSIBILITY
The data that supports the findings of this study are available from the corresponding author upon reasonable request.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. [DOI] [PubMed] [Google Scholar]
- 2.Nowak JA, Yurgelun MB, Bruce JL, et al. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn. 2017;19(1): 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7(3):746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salem ME, Xiu J, Lenz H-J, et al. Characterization of tumor mutation load (TML) in solid tumors. J Clin Oncol. 2017;35(15 suppl):11517–11517. [Google Scholar]
- 8.Salem ME, Puccini A, Grothey A, et al. Landscape of tumor mutation load, mismatch repair deficiency, and PD-L1 expression in a large patient cohort of gastrointestinal cancers. Mol Cancer Res. 2018;16(5): 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129(7–8):391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah SN, Hile SE, Eckert KA. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 2010;70(2):431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT, Uram JN, Wang H, et al. PD-1 blockade in Tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357 (6349):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy AJ, Capo-Chichi JM, Spence T, et al. Heterogenous loss of mismatch repair (MMR) protein expression: a challenge for immunohistochemical interpretation and microsatellite instability (MSI) evaluation. J Pathol Clin Res. 2019;5(2):115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hesson LB, Patil V, Sloane MA, et al. Reassembly of nucleosomes at the MLH1 promoter initiates Resilencing following Decitabine exposure. PLoS Genet. 2013. July;9(7):e1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodfellow PJ, Buttin BM, Herzog TJ, et al. Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc Natl Acad Sci USA. 2003;100(10):5908–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omalley D, Marabelle A, De Jesus-Acosta A, et al. Pembrolizumab in patients with MSI-H advanced endometrial cancer from the keynote-158 study. Ann Oncol. 2019;30(suppl 5):v403–v434. [Google Scholar]
- 19.Le DT, Kim TW, Van Cutsem E. Phase II open-label study of Pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J Clin Oncol. 2020;38(1):11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair–deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidaway P MSI-H: a truly agnostic biomarker? Nat Rev Clin Oncol. 2020;17:68 10.1038/s41571-019-0310-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.