Abstract
The PF74 binding site in HIV-1 capsid protein (CA) is a compelling antiviral drug target. Although PF74 confers mechanistically distinct antiviral phenotypes by competing against host factors for CA binding, it suffers from prohibitively low metabolic stability. Therefore, there has been increasing interest in designing novel sub-chemotypes of PF74 with similar binding mode and improved metabolic stability. We report herein our efforts to explore the inter-domain interacting indole moiety for designing novel CA-targeting small molecules. Our design includes simple substitution on the indole ring, and more importantly, novel sub-chemotypes with the indole moiety replaced with a few less electron-rich rings. All 56 novel analogs were synthesized and evaluated for antiviral activity, cytotoxicity, and impact on CA hexamer stability. Selected analogs were tested for metabolic stability in liver microsomes. Molecular modeling was performed to verify compound binding to the PF74 site. In the end, 5-hydroxyindole analogs (8,9 and 12) showed improved potency (up to 20-fold) over PF74. Of the novel sub-chemotypes, α- and β-naphthyl analogs (33 and 27) exhibited sub micromolar antiviral potencies comparable to that of PF74. Interestingly, although only moderately inhibiting HIV-1 (single-digit micromolar EC50s), analogs of the 2-indolone sub-chemotype consistently lowered the melting point (Tm) of CA hexamers, some with improved metabolic stability over PF74.
Keywords: HIV-1, capsid-targeting antivirals, PF74, metabolic stability
Graphical Abstract
1. Introduction
Although dozens of antivirals and fixed dose combinations [1] have been approved to treat the infection of human immunodeficiency virus type 1 (HIV-1), these drugs are not curative and HIV-1 remains a global healthcare challenge. Managing HIV-1 infection requires lifelong treatment, and hence, the virus will eventually develop resistance to current drug classes. Combating resistant viruses entails antivirals with novel molecular targets. The multifunctional HIV-1 capsid protein (CA) [2–4], which is a key component of the HIV-1 gag polyprotein [5], represents an emerging and highly attractive target in HIV-1 drug discovery [6–8]. CA is the building block of the mature HIV-1 capsid core [9, 10], and core stability critically depends on CA-CA interactions. Disrupting these interactions in the early stages of viral replication can perturb core stability to result in premature uncoating, impaired reverse transcription, and loss of infection [11, 12]. In addition, CA also interacts with multiple cellular factors [13] including TRIM5α [14, 15], cleavage and polyadenylation specific factor 6 (CPSF6) [16, 17], nucleoporins 153 [18–20] and 358 [21, 22] (NUP153, NUP358), MxB [23, 24], and Cyclophilin A (CypA) [25–27]. These CA-host interactions regulate multiple post entry events, such as uncoating, cytoplasmic trafficking, reverse transcription, nuclear transport, integration site distribution, and the evasion of innate immunity [28]. During the late stage of viral replication, CA-CA interactions also drive the assembly and maturation of new infectious viral particles [29]. Therefore, CA-targeting small molecules could confer both early and late stage antiviral phenotypes.
Efforts targeting HIV-1 CA have identified a few chemotypes with distinct binding sites (Figure 1) [6, 7, 33]. CA is highly helical consisting of an N terminal domain (CANTD) and a C terminal domain (CACTD) with a flexible linker in-between [30, 34, 35]. Although CACTD-targeting compounds, such as polypeptide CAI [32], arylquinazoline (AQ) [36], and the covalently binding ebselen [37], have been reported, considerably more efforts have been directed toward targeting CANTD. At the base of the CANTD is the binding site for three chemical classes represented by BM-4 [31], BD-3 [31] and CAP-1 [38]. Interestingly, the backbone of BM-4 is a benzoimidazole core flanked by a pyrazole ring and a phenyl ring (Figure 1B). However, when the flanking moieties are reversed, the resulting compound C-4 [39] binds to a completely different site near the top of the CANTD and around the base of the cyclophilin A binding loop [40] (Figure 1A). In general, compounds binding to these two sites perturbed CA assembly in vitro and moderately inhibited HIV-1 in cell culture [6]. There was no evidence that they act in the early steps of viral replication, and no direct evidence linking the antiviral activity to the targeting of CA. By contrast, by far the most interesting CA-targeting compounds are the three chemical classes that bind to the PF74 binding site, which also include the BI compounds [41] and the GS-CA compounds [42] (Figure 1B). These compounds all inhibited HIV-1 at both the early and the late stages of viral replication. Not surprisingly, there appears to be a positive correlation between the antiviral potency and the structural complexity: the simplest BI compounds exhibited moderate (low micromolar) antiviral activity and the structurally highly complex GS-CA compounds demonstrated sub-nanomolar antiviral activity, whereas the structurally elaborate, yet highly synthetically accessible peptidomimetic PF74 inhibited HIV-1 in the sub-micromolar range. The binding mode [30] and the activity profile [43, 44] of PF74 have been particularly well-characterized, revealing a dual antiviral mechanism of action: at low concentrations PF74 competes against host factors for capsid binding; at high concentrations it induces premature uncoating, and consequently, impairs reverse transcription [11].
Figure 1.
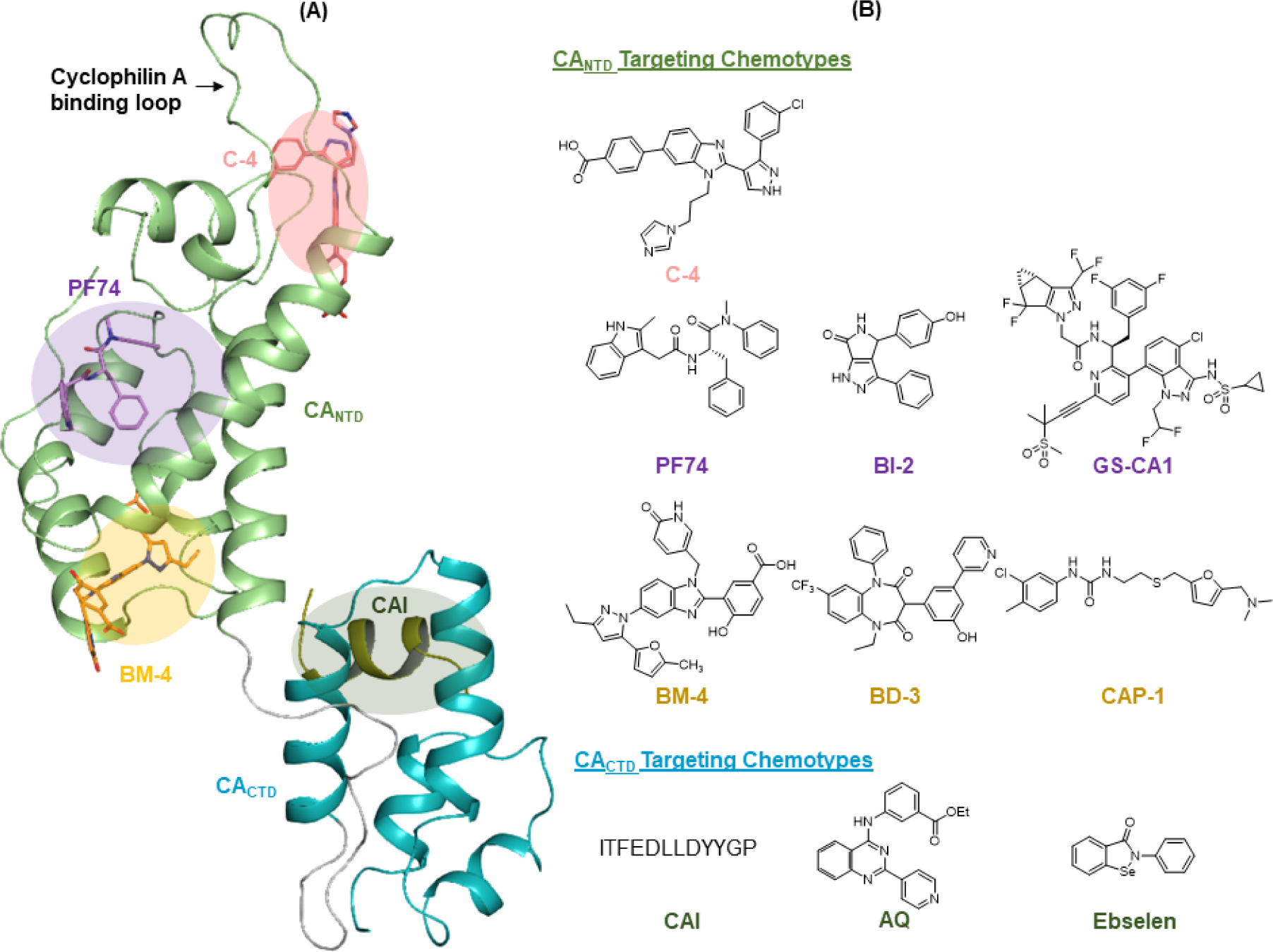
Compound binding sites and chemotypes targeting HIV-1 CA. (A) Ligand binding sites. Three distinct small molecule binding sites at CANTD are known for C-4, PF74 and BM-4, respectively. Host factor cyclophilin A binds to the loop on top. Polypeptide CAI binds to the CACTD. Binding modes were reproduced in Maestro based on PDB 4XFZ [30] with ligands C-4 & BM-4 (PDB: 4E92 [31]) and CAI (PDB: 2BUO [32]) aligned. The picture was created in PyMOL while shadows of binding sites were rendered in PowerPoint; (B) major chemotypes binding to each site: BI-2 and GS-CA1 bind to the PF74 site; BD-3 and CAP-1 bind to the BM-4 site; arylquinazoline (AQ) and ebselen bind to CACTD.
The PF74 binding pocket is lined with residues in H3 and H4 of the CANTD (cyan) and H8 and H9 of the adjacent CACTD (blue), where an extensive network of molecular interactions defines the PF74 binding mode (Figure 2A) [30]. Structurally, PF74 features a phenylalanine core, connected by an aniline moiety at the carboxylate end and an indole-3-acetic acid at the amino end (Figure 2B). Although all three components provide key molecular interactions for CA binding, the indole ring (boxed, Figure 2A) uniquely interacts with both the CANTD (via H-bond with Q63 and π-cation interaction with K70) and the adjacent CACTD (via Y169, R173 and K182) [30]. These interactions are lacking with the BI compounds, which do not have a structural equivalent to the indole moiety of PF74, likely accounting for their weaker antiviral potency. The core and the aniline moiety of PF74 engage extensively with the CANTD [30] (Figure 2A): 1) the phenylalanine core forms two H-bonds with N57, and an additional one with K70; 2) the aniline moiety makes contact with N53 (via the N-methyl group) and A105, T107, and Y130 (via the phenyl ring); and 3) the phenyl ring of the phenylalanine core forms hydrophobic interactions with residues M66 and L69. Since the indole moiety provides the unique inter-domain binding interactions, there have been reported efforts replacing the indole moiety with a simpler and synthetically more accessible 1,2,3-triazole ring, though the replacement resulted in substantially decreased potency (by >10-fold) [45, 46]. However, a recent report showed that replacing the indole ring with a piperazinone moiety yielded compounds with better antiviral activity (up to 6-fold) than PF-74 [47]. We have previously conducted a comprehensive SAR on PF74 with the synthesis of a large number of analogs [48], and have identified a structurally novel and metabolically stable CA-targeting small molecule [49]. In the current report, we describe our own efforts targeting the two-domain interactions (Figure 2B). We first synthesized a series of PF74 analogs with a substituent on the indole ring, and then designed and synthesized analogs of a few novel sub-chemotypes, each featuring a distinct ring system to replace the indole. In the end, 56 analogs were synthesized and tested, many of which demonstrated significant anti-HIV-1 activity and interesting effects on CA hexamer stability, and some exhibited improved metabolic stability over PF74.
Figure 2.
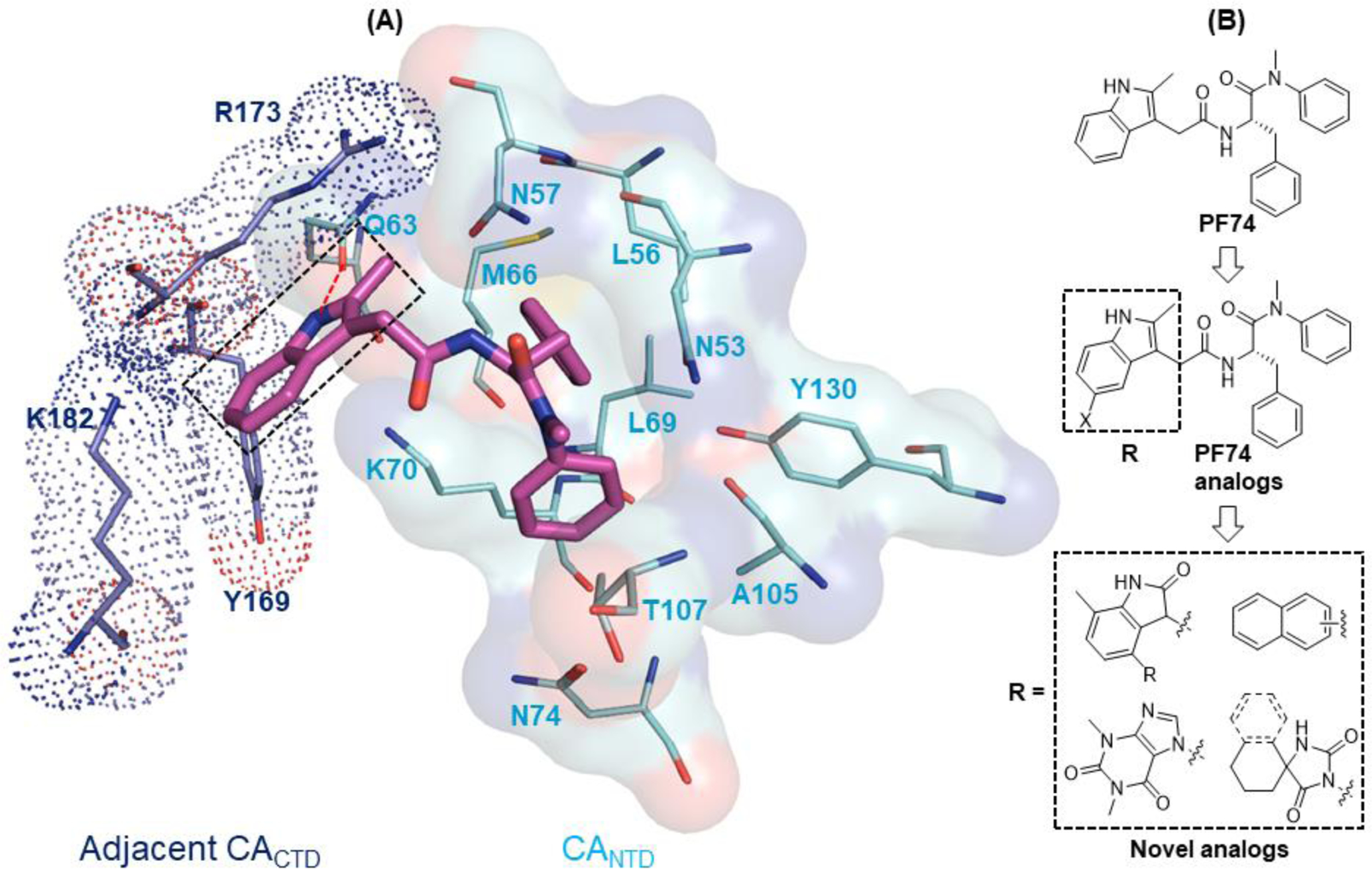
Binding mode of PF74 based on PDB 4XFZ [30] and the design of novel analogs. (A) Detailed molecular interaction network of PF74 (magenta sticks) with residues (lines) in both the CANTD (transparent cyan surface) and the adjacent CACTD (blue dot). The indole moiety of PF74 (boxed) interacts with Q63, K70 of CANTD and Y169, R173, and K182 of the adjacent CACTD; (B) Novel analogs designed to explore the two-domain interactions of the indole moiety.
2. Results and discussion
2.1. Chemistry
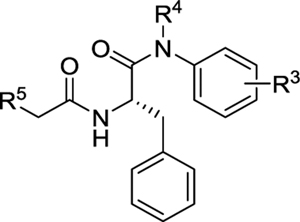
Briefly, commercially available (tert-butoxycarbonyl)-L-phenylalanine (58) was treated with various amines under a well-established method using T3P or HATU as the coupling agent in the presence of DIPEA as base to afford 59. After removal of Boc protecting group using TFA, amine salts 60 were obtained which were further reacted with commercially available acids 61 to produce analogs 1–57. Details about the synthesis of 59 and 60 were described in the Supporting Information.
2.2. Biological assays and SAR
All analogs were first evaluated in a thermal shift assay, where the effect of a compound on protein stability was measured by the change in protein melting point compared to DMSO control (ΔTm). A positive value in ΔTm indicates a stabilizing effect and a negative value a destabilizing effect on the protein. Of note, the CA protein used in the thermal shift assay is in a covalently crosslinked hexameric state. Thus, the ΔTm values likely reflect local changes that may affect stabilization and exclude inter-hexamer effects, which are important correlates of overall capsid core stability. To simplify presentation of the data, we refer to the effects of these compounds as stabilization or destabilization of “CA hexamer.” All compounds were also screened at 20 μM in a cell-based antiviral assay against HIV-1. Compounds demonstrating significant inhibition were then tested at 2 μM. Promising compounds were further assessed in dose response fashion for antiviral EC50 values. All compounds were also tested for cytotoxicity either by screening at 100 or 50 μM, or determination of CC50 values. For some compounds, % inhibition at two concentrations (2 μM and 20 μM) was reported instead of the EC50. PF74 was resynthesized and tested in these assays (1, ΔTm = 7.4 °C, EC50 = 0.61 μM, CC50 = 76 μM).
2.2.1. Substitutions at the indole (R1 and R2) and aniline ring (R3)
This series features PF74 analogs with the indole ring substituted (R1) with an OMe (3-7) or an OH (8-15). It is noteworthy that all these analogs lack the methyl group at R2 (R2 = H). This is because when compared to PF74 (R2 = Me, ΔTm = 7.4 °C, EC50 = 0.61 μM) analog 2 (R2 = H, ΔTm = 6.1 °C, EC50 = 0.46 μM) showed a largely comparable activity profile. Hence, the SAR within this series concerns mainly the effect of the H-bond enabling and electron donating groups at R1, in combination with the substitution effects on the aniline ring (R3). Notably, an OMe substitution at R1 by itself did not improve the activity profile (3 vs 2), though slight improvement was observed when R3 is a para-Cl. This chlorine effect was significant as revealed by the direct comparison between analogs 7 (R3 = 4-Cl, ΔTm = 8.0 °C, EC50 = 0.31 μM) and 3 (R3 = H, ΔTm = 4.9 °C, EC50 = 0.56 μM), possibly due to a halogen bond (see molecular modeling for similar analogs). Among the compounds, 8-15, substituted at R1 with OH, were found the most potent ones of the series. Particularly, analogs 8 (EC50 = 0.053 μM), 9 (EC50 = 0.035 μM) and 12 (EC50 = 0.032 μM) conferred up to ~20-fold higher potency than PF74 (EC50 = 0.61 μM) and analog 2 (EC50 = 0.46 μM). Furthermore, 9 (CC50 > 100 μM) and 12 (CC50 > 100 μM) showed lower toxicity than PF74 (CC50 = 76 μM). These results suggest that an R1 substituent capable of both H-bond donating and H-bond accepting (e.g. OH) confers better antiviral potency than does an H-bond accepting group (e.g. OMe). However, the significantly improved antiviral potency with 8, 9 and 12 over PF74 is not correlated with results from the thermal shift assay, where very similar ΔTm (6.3–6.8 °C for 8, 9, 12 vs 7.4 °C for PF74) was observed. It is possible that the afore-mentioned OH group could affect capsid assembly via unknown post-binding molecular mechanisms. In addition, PF74 is known to inhibit both the early and the late stages of HIV-1 replication cycle. Mechanistic studies are currently underway to determine if the most potent compounds (e.g. 8, 9, 12) from this series display similar antiviral profile.
2.2.2. Novel sub-chemotypes with indole bioisosteres (R5) and modifications at the aniline ring (R3 and R4)
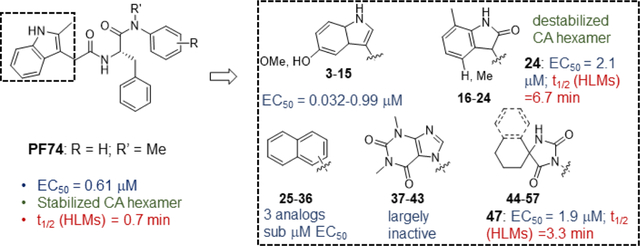
Table 2 summarizes our SAR studies on a few novel sub-chemotypes featuring different ring structures as the indole replacement. The first new sub-chemotype includes nine analogs (16-24) in which the indole moiety is replaced by an indolin-2-one ring. The first four analogs (16-19) bear a 7-methyl group and the next five (20-24) are 4,7-dimethyl substituted. Two prominent SAR trends were observed: the additional 4-methyl group (20-24) led to improved antiviral potency (20 vs 16, 22 vs 19, 23 vs 17); and both 3-Cl and 4-Cl on the aniline moiety (R3) also enhanced potency (18 vs 16, 24 / 23 vs 20). A beneficial chlorine effect conferred by both a 3-Cl and a 4-Cl of the aniline moiety (R3) was observed with this sub-chemotype only (see molecular modeling). With PF74 and all other sub-chemotypes, only the 4-Cl is properly positioned for halogen bonding. Overall, analogs of this series exhibited low micromolar potencies, and more intriguingly, seem to destabilize the CA hexamer. The next series consists of twelve analogs (25-36) with the indole moiety replaced by a naphthyl ring, either via a β substitution (25–30) or an α substitution (31–36). The substitution site (β vs α) did not appear to impact antiviral activity (32 vs 26, 33 vs 27) as prominently as the R3 group, where a 4-Me (26) or 4-Cl (27) conferred submicromolar activities, though in the α series (31–36) two additional compounds (31 and 36, R3 = H) also demonstrated low micromolar antiviral activity. Next we synthesized seven analogs (37-43) where R5 is a 1,3-dimethyl-purine-2,6-dione ring system. Unfortunately, none of them inhibited HIV-1 significantly at 2 μM. Discernible impact on CA hexamer stability was not observed either. The last two sub-chemotypes (44-57) synthesized both feature a [6,5] spiro ring system. Overall, analogs of these two sub-chemotypes did not significantly impact CA hexamer stability. As for antiviral potency, while many of the bicyclic analogs (44-53) inhibited HIV-1, such as compounds 44, 46, 47, 50, and 53, the four tricylic analogs (54-57) did not show any antiviral activity.
Table 2.
Anti-HIV-1 activity, cytotoxicity, and CA hexamer stability profiles of novel analogs (R3, R4, R5).
 |
|||||||
|---|---|---|---|---|---|---|---|
| Compd | R5 | R4 | R3 | Inhibition% at 2 μM / 20 μM | EC50a (μM) | CC50b (μM) | TSAc ΔTm (°C) |
| PF74 (1) | 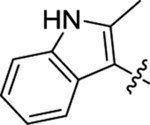 |
Me | H | 91/98 | 0.61 ± 0.2 | 76 ± 9 | 7.4 |
| 16 | 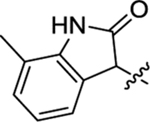 |
Me | H | 0/91 | ND | > 50 | −1.2 |
| 17 | 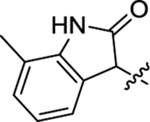 |
Me | 4-Cl | 17/96 | ND | ~ 50 | −0.4 |
| 18 | 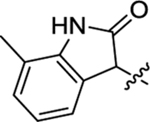 |
Me | 3-Cl | 14/85d | 3.3 ± 0.3 | > 100 | −1.7 |
| 19 |  |
Me | 3-F | 0/93 | ND | > 50 | −0.9 |
| 20 |  |
Me | H | 0/72d | 4.9 ± 0.3 | > 100 | −1.8 |
| 21 |  |
Me | 4-F | 0/70d | 5.4 ± 0.5 | > 100 | −1.7 |
| 22 |  |
Me | 3-F | 0/70d | 5.2 ± 0.4 | 60 ± 6 | −1.8 |
| 23 | 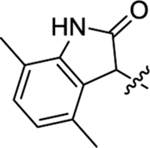 |
Me | 4-Cl | 10/98d | 2.8 ± 0.1 | 63 ± 13 | −1.3 |
| 24 | 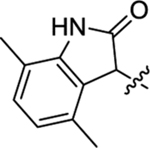 |
Me | 3-Cl | 27/99d | 2.1 ± 0.2 | 43 ± 2 | −2.4 |
| 25 | 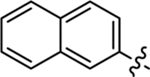 |
Me | H | 40/98 | ND | < 50 | 4.8 |
| 26 | 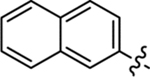 |
Me | 4-Me | 80/99 | 0.99 ± 0.005 | < 50 | 6.4 |
| 27 | 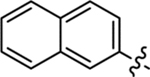 |
Me | 4-Cl | 95/100 | 0.63 ± 0.02 | < 50 | 7.0 |
| 28 | 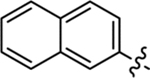 |
Me | 3-Cl | 0/94 | ND | < 50 | 4.3 |
| 29 | 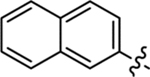 |
Me | 3-F | 0/97 | ND | < 50 | 5.2 |
| 30 | 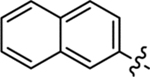 |
Et | H | 46/97 | ND | < 50 | 4.3 |
| 31 | 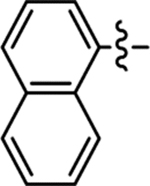 |
Me | H | 66/100 | 1.1 ± 0.04 | < 50 | 6.0 |
| 32 | 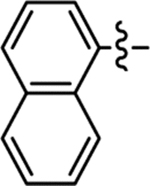 |
Me | 4-Me | 96/100 | 1.0 ± 0.2 | < 50 | 5.7 |
| 33 | 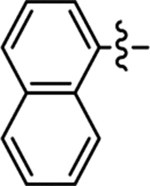 |
Me | 4-Cl | 98/100 | 0.83 ± 0.03 | < 50 | 7.2 |
| 34 | 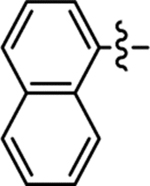 |
Me | 3-Cl | 53/96 | ND | < 50 | 4.4 |
| 35 | 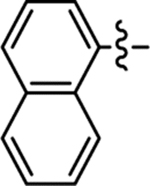 |
Me | 3-F | 44/97 | ND | < 50 | 5.4 |
| 36 | 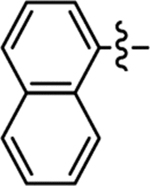 |
Et | H | 53/98 | 1.8 ± 0.09 | < 50 | 4.9 |
| 37 | 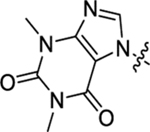 |
Me | H | 10/94 | ND | > 50 | 0.9 |
| 38 | 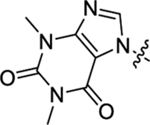 |
Me | 4-Me | 18/98 | ND | > 50 | 1.6 |
| 39 | 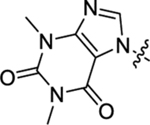 |
Me | 4-F | 14/85 | ND | > 50 | 0 |
| 40 | 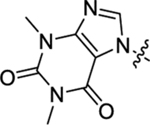 |
Me | 3-F | 1/67 | ND | > 50 | 0.5 |
| 41 | 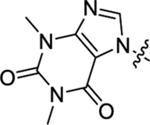 |
Me | 4-Cl | 6/96 | ND | > 50 | 1.5 |
| 42 | 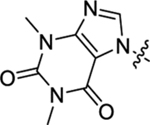 |
Me | 3-Cl | 0/71 | ND | > 50 | 0 |
| 43 | 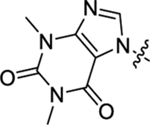 |
Et | H | 0/94 | ND | > 50 | 0.8 |
| 44 | 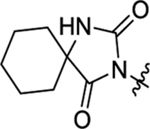 |
Me | H | −/89 | 8.9 ± 1.5 | >100 | 1.3 |
| 45 | 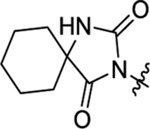 |
H | H | −/0 | >20 | >100 | 0 |
| 46 | 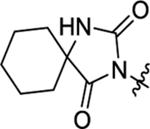 |
Me | 4-OMe | −/93 | 5.1 ± 2 | >100 | 1.8 |
| 47 | 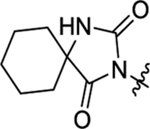 |
Me | 4-Me | −/92 | 1.9 ± 0.5 | >100 | 2.7 |
| 48 | 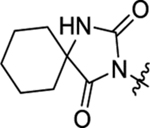 |
Me | 4-F | 9/99 | ND | > 50 | 3.1 |
| 49 | 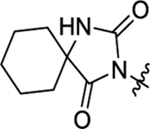 |
Me | 3-F | 24/98 | ND | > 50 | 1.3 |
| 50 | 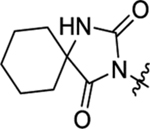 |
Me | 4-Cl | −/96 | 2.5 ± 0.5 | >100 | 2.2 |
| 51 | 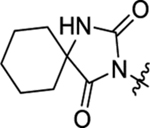 |
Me | 3-Cl | 9/98 | ND | < 50 | 3.3 |
| 52 | 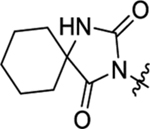 |
Me | 3-Br | −/36 | > 20 | > 50 | 1.5 |
| 53 | 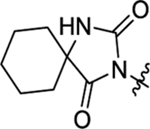 |
Et | H | −/92 | 7.6 ± 0.9 | >100 | 2.0 |
| 54 | 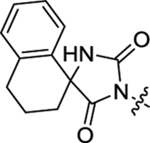 |
Me | H | −/32 | > 20 | > 100 | 0 |
| 55 |  |
H | H | −/22 | > 20 | > 100 | 0.6 |
| 56 |  |
Me | 4-OMe | −/20 | > 20 | > 100 | 0.8 |
| 57 |  |
Me | 4-Me | −/41 | > 20 | > 100 | 0.8 |
Concentration of compound inhibiting HIV-1 replication by 50%, expressed as the mean ± standard deviation from at least two independent experiments.
Concentration of compound causing 50% cell death, expressed as the mean ± standard deviation from at least two independent experiments.
TSA: thermal shift assay. ΔTm: change of CA hexamer melting point in presence of compound compared to DMSO control.
For these compounds, % inhibition is reported at 1 μM / 10 μM.
2.3. Metabolic stability in liver microsomes
Despite its potent antiviral activity, well-characterized mode of binding and easy synthetic accessibility, PF74 is seriously flawed as an antiviral lead due to its prohibitively poor metabolic stability. In human liver microsomes (HLMs), the half-life (t1/2) of PF74 was less than 1 min [46, 50]. We have previously tested around 20 PF74 analogs in HLMs and they all showed very poor metabolic stability (t1/2 = 0.6–2.1 min) [48]. However, an analog featuring a quinazoline-2,4-dione as the indole replacement was found to be metabolically stable in HLMs (t1/2 = 31 min) [49]. To gauge the metabolic stability of the sub-chemotypes described herein, we tested selected compounds in HLMs and mouse liver microsomes (MLMs) and the results are summarized in Table 3. Consistent with literature reports, the t1/2 of PF74 in our metabolic assays was 0.7 min in HLMs and 0.6 min in MLMs. Peptidomimetics, a compound class to which PF74 belongs, are particularly liable toward phase I metabolism, presumably because they are good substrates for liver metabolizing enzyme subfamily cytochrome P450 3A (CYP3A) [51]. This extensive liver metabolism constitutes a major pharmacokinetic (PK) barrier for many FDA-approved HIV-1 protease inhibitors [52]. For example, darunavir was reported to have a half-life (t1/2) of 3.6 min in a human liver microsomal incubation system with less CYP protein content compared to ours (0.1 mg/mL vs 0.5 mg/mL) [53]. The main mitigating strategy has been to co-administer the antiviral drugs with a CYP3A inhibitor, such as ritonavir [54] or Cobicistat (Cobi) [55], as a PK enhancer [52]. Therefore, our metabolic stability assays were performed under two distinct sets of conditions: with PK enhancer Cobi or without Cobi (Table 3). Without Cobi, PF74 analogs (3 and 7) exhibited very short half-life (1.2 min and 1.8 min, respectively). The metabolic stability remained poor for analogs of novel sub-chemotypes, though a few of them, such as 16, 24, 37 and 46, did show substantially (7 to 10-fold) improved half-life over PF74 in HLMs. For the five selected compounds tested in the presence of PK enhancer Cobi, drastically improved metabolic stability was observed with all (Table 3). Collectively, these results demonstrated that replacement of the indole moiety with a less electron-rich ring could lead to metabolically more stable compounds, and that inhibiting CYP3A could mitigate the phase I metabolic liability of this type of compounds.
Table 3.
Phase I metabolic stability in liver microsomes t1/2 (min).
| Compd | HLMa | HLMa (+Cobic) | MLMb | MLMb (+Cobic) |
|---|---|---|---|---|
| PF74c | 0.7 | 91 | 0.6 | 34 |
| 3 | 1.2 | -- | 0.5 | -- |
| 7 | 1.8 | -- | 0.6 | -- |
| 16 | 7.0 | -- | 1 | -- |
| 20 | 1.5 | -- | 0.6 | -- |
| 24 | 6.7 | 107 | 1.4 | 34 |
| 25 | 1.8 | -- | 0.5 | -- |
| 27 | 2.7 | >120 | 1.4 | 18 |
| 31 | 0.6 | -- | 0.5 | -- |
| 33 | 1.1 | 102 | 0.6 | 7.4 |
| 37 | 4.7 | -- | 1.2 | -- |
| 46 | 5.2 | -- | 0.7 | -- |
| 47 | 3.3 | -- | 1.2 | -- |
| 48 | 1.1 | -- | 0.5 | -- |
| 50 | 2.5 | >120 | 1.1 | 20 |
HLM: human liver microsome
MLM: mouse liver microsome
Microsomal stability measured in the presence of CYP3A inhibitor Cobi.
2.4. Molecular modeling
To evaluate the binding mode of the new analogs, we performed molecular modeling of key compounds based on the co-crystal structure of native HIV-1 capsid protein bound to PF74 (PDB code: 4XFZ [30]). Analog 12 is found to orient in a similar fashion as the parent compound PF74 (Figure 3A), and share common key interactions on CANTD domain including (i) hydrogen bonding with N57 via the carbonyl and NH of phenylalanine core, with Q63 via indole N-H, and with K70 via the other carbonyl moiety; (ii) Cation-π interaction with K70 via the indole moiety (Figure 3A). However, unlike PF74, the two additional substituents of 12, 4-Cl on the aniline moiety and 5-OH of the indole ring, confer a halogen bond (pink dashed line) and another hydrogen bond with K182 on the adjacent CACTD domain (black dotted line), respectively (Figure 3A). These additional interactions are consistent with improved target binding affinity (Glide score: −7.6 kcal/mol for 12 vs −5.8 kcal/mol for PF74) and largely increased potency (EC50: 0.032 μM for 12 vs 0.61 μM for PF74). Importantly, while the halogen bond conferred by the 4-Cl of the aniline moiety appeared to be less important for potency than the hydrogen bond by the 5-OH (2 vs 8 vs 12), moving the Cl from 4 position to 3 (compound 13) led to a 10-fold potency drop, likely due to the potential steric clash posed by the 3-Cl. Similar effects were observed with 3-Br (14) and 3-CF3 (15). The potency difference between 3-Cl and 4-Cl aniline analog was prominent throughout SAR studies (27 vs 28, 33 vs 34, and 50 vs 51). The only exception is with the 4,7-dimethylindolin-2-one sub-chemotype where a beneficial halogen effect was observed for both the 4-Cl analog 23 (Figure 3B) and the 3-Cl analog 24 (Figure 3C). In both cases, the loss of the key H-bonding with Q63 and the cation-π interactions with K70, was compensated by a halogen-bond (pink dashed line) with N74 (4-Cl) and N53 (3-Cl), respectively. Similarly, compound 27 having the 2-naphthyl ring in place of indole ring exhibited an identical potency and protein binding affinity to PF74, despite the loss of a key hydrogen-bonding with Q63 (Figure 3D). Possible orientation of the aniline ring of the compound 27 as shown in Figure 3D could improve hydrophobic contacts with N53, G106, and Y130, as a result of halogen bond between G106 and 4-Cl aniline, and could explain the observed potency for this and another similar compound 33.
Figure 3.
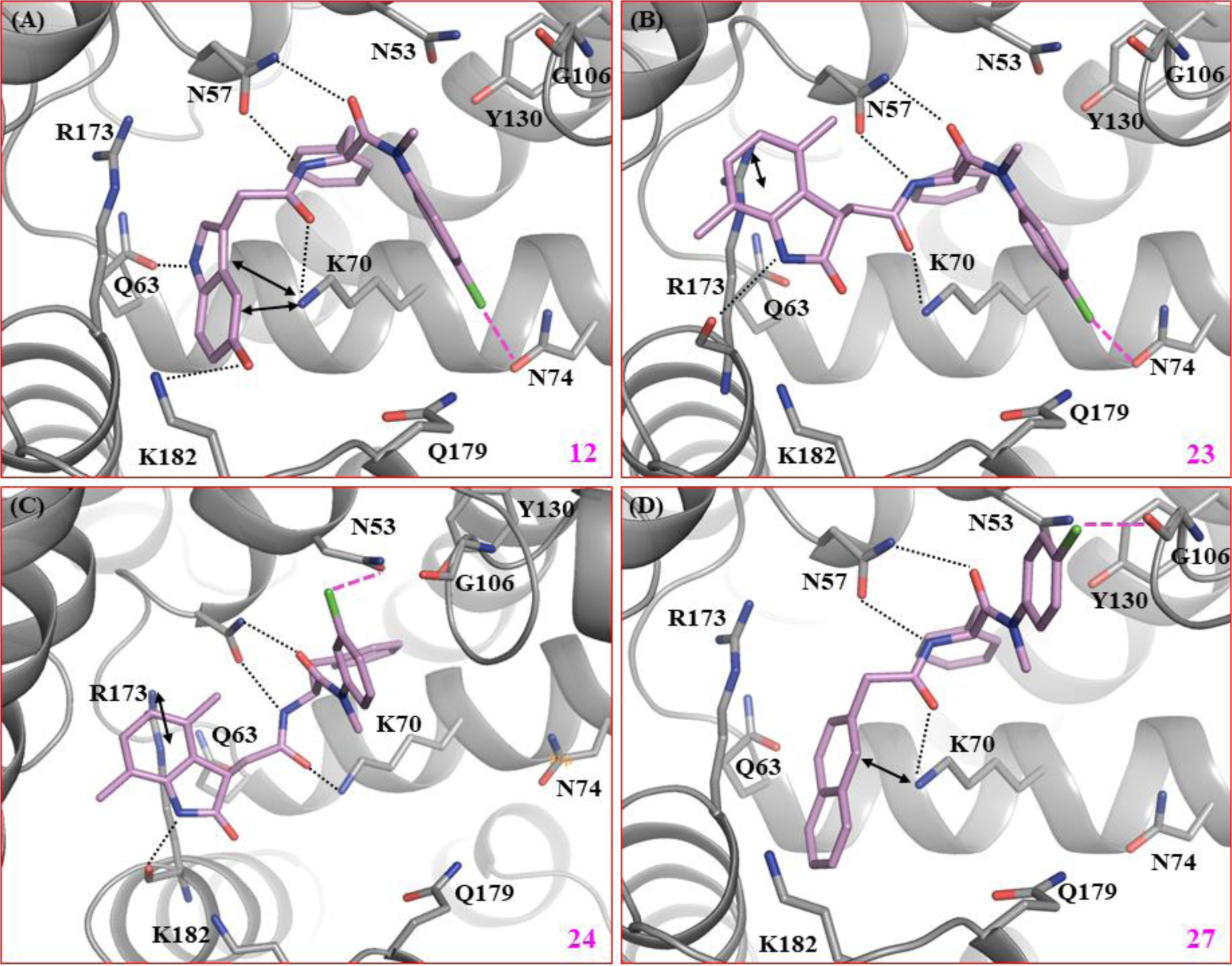
Docking poses of key compounds based on native HIV-1 capsid protein bound to PF74 (Glide score: −5.8) (PDB code: 4XFZ [30]). (A) Predicted binding mode of compound 12 (Glide score: −7.6 kcal/mol). (B) Predicted binding mode of 23 (Glide score: −6.4 kcal/mol). (C) Predicted binding mode of 24 (Glide score: −6.8 kcal/mol). (D) Predicted binding mode of 27 (Glide score: −6.1 kcal/mol). Hydrogen-bonding, halogen-bonding and cation-π interactions are depicted as black dotted lines, pink dashed lines, and double headed arrows, respectively. CA with key residues around the binding site, and ligands 12, 23, 24, and 27 are colored grey and violet, respectively. The nitrogen, oxygen, and chlorine atoms are colored blue, red, and green, respectively.
3. Conclusions
In this work, we have designed, synthesized and evaluated 56 compounds consisting of PF74 analogs and a few novel PF74-like chemotypes featuring rings less electron-rich than the indole ring. In the end, 5-hydroxyindole analogs (8, 9, 12) were up to 20-fold more potent than the prototype PF74. Analogs with low to sub micromolar antiviral activities were also identified from all sub-chemotypes, except for the 1,3-dimethyl-purine-2,6-dione sub-chemotype. Of all sub-chemotypes studied herein, the naphthalene ring conferred the best potency as both the α- and β-naphthyl analogs (33 and 27) exhibited antiviral potencies comparable to that of PF74. Interestingly, the 2-indolone sub-chemotype lowered the Tm of CA hexamers, indicating a unique HIV-1 CA-destabilizing mechanism. The best 2-indolone analog, 24, also showed 10-fold longer half-life in HLMs than PF74. These findings necessitate future medicinal chemistry to further optimize these novel sub-chemotypes, particularly the CA-destabilizing 2-indolone chemotype.
4. Experimental section
4.1. Chemistry
General Procedures.
All commercial chemicals were used as supplied unless otherwise indicated. Flash chromatography was performed on a Teledyne Combiflash RF-200 with RediSep columns (silica) and indicated mobile phase. 1H and 13C NMR spectra were recorded on a Varian 600 MHz or Bruker 400 spectrometer. Diastereomeric ratio (dr) was determined by 1H NMR analysis. Mass data were acquired using an Agilent 6230 TOF LC/MS spectrometer. All NMR and mass spectrometers are located in the shared instrument rooms at the Center for Drug Design, University of Minnesota.
4.1.1. Synthesis of 59 and 60
Synthesis of intermediates 59 and 60 are described in Supporting Information.
4.1.2. Synthesis of (S)-N-Methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (1)
To a solution of commercially available 2-(2-methyl-1H-indol-3-yl)acetic acid (100 mg, 0.53 mmol, 1 equiv.) in DMF (3 mL), HATU (402 mg, 1.06 mmol, 2 equiv.) and DIPEA (205 mg, 1.59 mmol, 3 equiv.) were added and the mixture was stirred at room temperature for 20 min before (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 235 mg, 0.64 mmol, 1.2 equiv.) was added. The mixture was further stirred overnight at room temperature. Upon completion, H2O was added and the reaction mixture was extracted with EtOAc (3×30 mL). The organic phases were combined and washed with brine, dried over anhydrous MgSO4, filtered and concentrated. The product was purified by Combiflash on silica gel using EtOAc/hexane (1:2 to 6:1) as eluent. Yield 70%. 1H NMR (600 MHz, DMSO-d6) δ 10.68 (s, 1H), 8.27 (d, J = 7.0 Hz, 1H), 7.38 (dd, J = 17.5, 6.5 Hz, 3H), 7.27 (dd, J = 18.7, 6.7 Hz, 3H), 7.18 (d, J = 7.6 Hz, 1H), 7.12 (s, 3H), 6.98 – 6.89 (m, 1H), 6.84 (d, J = 6.9 Hz, 1H), 6.79 (s, 2H), 4.41 (s, 1H), 3.51 – 3.31 (m, 2H), 3.15 (s, 3H), 2.82 (d, J = 11.5 Hz, 1H), 2.67 (d, J = 11.5 Hz, 1H), 2.23 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 171.5, 171.0, 143.3, 138.0, 135.4, 133.3, 130.0, 129.2, 128.8, 128.4, 128.2, 128.0, 126.7, 120.3, 118.4, 118.4, 110.5, 105.3, 52.2, 37.6, 37.4, 31.3, 11.7; HRMS (ESI) m/z calcd for C27H26N3O2 [M − H]− 424.2031, found 424.2029.
(S)-2-(2-(1H-Indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (2).
The synthetic method was similar to that of compound 1 except that 2-(1H-indol-3-yl)acetic acid (100 mg, 0.57 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 252 mg, 0.68 mmol, 1.2 equiv.) were used as starting materials. Yield 65%. 1H NMR (600 MHz, CD3OD) δ 7.42 (d, J = 7.9 Hz, 1H), 7.37 – 7.31 (m, 4H), 7.18 – 7.05 (m, 5H), 7.03 – 6.94 (m, 3H), 6.75 (d, J = 7.3 Hz, 2H), 4.66 (t, J = 7.2 Hz, 1H), 3.61 (dd, J = 22.4, 15.8 Hz, 2H), 3.16 (s, 3H), 2.87 (dd, J = 13.3, 6.6 Hz, 1H), 2.64 (dd, J = 13.3, 6.6 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 172.7, 171.7, 142.5, 136.7, 136.5, 129.4, 128.7, 128.0, 127.9, 127.2, 127.0, 126.4, 123.5, 121.1, 118.6, 118.0, 110.9, 107.7, 51.9, 37.7, 36.6, 32.2; HRMS (ESI) (−) m/z calcd for C26H24N3O2 [M − H]− 410.1874, found 410.1878.
(S)-2-(2-(5-Methoxy-1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (3).
The synthetic method was similar to that of compound 1 except that 2-(5-methoxy-1H-indol-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 217 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. Yield 42%. 1H NMR (600 MHz, CDCl3) δ 8.34 (s, 1H), 7.36 − 7.31 (m, 3H), 7.26 − 7.24 (m, 1H), 7.13 (t, J = 7.4 Hz, 1H), 7.07 − 7.04 (m, 2H), 6.97 − 6.88 (m, 5H), 6.66 (d, J = 7.4 Hz, 2H), 6.30 (d, J = 8.0 Hz, 1H), 4.81 – 4.77 (m, 1H), 3.79 (s, 3H), 3.67 – 3.60 (m, 2H), 3.18 (s, 3H), 2.76 (dd, J = 13.3, 7.0 Hz, 1H), 2.56 (dd, J = 13.3, 7.1 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ 171.3, 171.0, 154.4, 142.4, 136.0, 131.4, 129.7, 129.1, 128.3, 128.2, 127.4, 127.3, 126.7, 124.4, 113.0, 112.1, 108.4, 100.1, 55.8, 51.3, 38.8, 37.6, 33.3; HRMS (ESI) m/z calcd for C27H26N3O3 [M − H]− 440.1980, found 440.1983.
(S)-2-(2-(5-Methoxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (4).
The synthetic method was similar to that of compound 1 except that 2-(5-methoxy-1H-indol-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 225 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. Yield 63%. 1H NMR (600 MHz, CDCl3) δ 8.60 (s, 1H), 7.22 (d, J = 8.7 Hz, 1H), 7.13 – 7.12 (m, 3H), 7.08 − 7.05 (m, 2H), 6.88 − 6.69 (m, 7H), 6.33 (d, J = 8.2 Hz, 1H), 4.82 − 4.79 (m, 1H), 3.79 (s, 3H), 3.66 − 3.59 (m, 2H), 3.15 (s, 3H), 2.77 (dd, J = 13.3, 7.0 Hz, 1H), 2.58 (dd, J = 13.3, 6.9 Hz, 1H), 2.35 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.5, 170.9, 154.3, 139.9, 138.1, 136.1, 131.5, 130.3, 129.2, 128.3, 127.4, 127.0, 126.7, 124.5, 112.8, 112.2, 108.2, 100.1, 55.8, 51.2, 38.8, 37.7, 33.4, 21.1; HRMS (ESI) m/z calcd for C28H28N3O3 [M − H]− 454.2136, found 454.2139.
(S)-N-(4-Fluorophenyl)-2-(2-(5-methoxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (5).
The synthetic method was similar to that of compound 1 except that 2-(5-methoxy-1H-indol-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 228 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. Yield 62%. 1H NMR (600 MHz, CD3OD) δ 7.24 (d, J = 8.8 Hz, 1H), 7.17 − 7.12 (m, 3H), 7.06 (s, 1H), 7.01 − 6.98 (m, 3H), 6.86 − 6.76 (m, 5H), 4.57 (t, J = 7.4 Hz, 1H), 3.79 (s, 3H), 3.57 (s, 2H), 3.11 (s, 3H), 2.89 (dd, J = 13.3, 7.5 Hz, 1H), 2.67 (dd, J = 13.3, 7.3 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.15, 173.20, 163.35 (d, JCF = 246.9 Hz), 155.20, 139.96, 137.82, 133.26, 130.67 (d, JCF = 7.8 Hz), 130.19, 129.50, 128.75, 127.92, 125.67, 117.37 (d, JCF = 23.0 Hz), 113.01 (d, JCF = 9.3 Hz), 108.97, 101.28, 56.27, 53.23, 39.26, 38.07, 33.66; HRMS (ESI) m/z calcd for C27H25FN3O3 [M − H]− 458.1885, found 458.1886.
(S)-N-(3-Fluorophenyl)-2-(2-(5-methoxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (6).
The synthetic method was similar to that of compound 1 except that 2-(5-methoxy-1H-indol-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 228 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. Yield 59%. 1H NMR (600 MHz, CDCl3) δ 8.36 (s, 1H), 7.31 − 7.26 (m, 2H), 7.18 (t, J = 7.3 Hz, 1H), 7.12 − 7.10 (m, 2H), 7.03 − 7.00 (m, 2H), 6.90 − 6.87 (m, 2H), 6.77 − 6.73 (m, 3H), 6.28 (d, J = 7.9 Hz, 1H), 4.79 – 4.75 (m, 1H), 3.81 (s, 3H), 3.69 − 3.62 (m, 2H), 3.14 (s, 3H), 2.75 (dd, J = 13.1, 8.0 Hz, 1H), 2.61 (dd, J = 13.1, 6.5 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ 171.3, 170.9, 162.8 (d, JCF = 249.0 Hz), 154.4, 143.8, 135.8, 131.5, 130.7 (d, JCF = 9.0 Hz), 129.2, 128.4, 127.4, 126.9, 124.4, 123.3, 115.2 (d, JCF = 21.0 Hz), 114.7 (d, JCF = 22.8 Hz), 113.0, 112.2, 108.3, 100.1, 55.8, 51.4, 39.2, 37.5, 33.4; HRMS (ESI) m/z calcd for C27H25FN3O3 [M − H]− 458.1885, found 458.1887.
(S)-N-(4-Chlorophenyl)-2-(2-(5-methoxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (7).
The synthetic method was similar to that of compound 1 except that 2-(5-methoxy-1H-indol-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 237 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. Yield 80%. 1H NMR (600 MHz, CD3OD) δ 7.25 − 7.22 (m, 3H), 7.17 − 7.12 (m, 3H), 7.06 (s, 1H), 6.99 (s, 1H), 6.82 − 6.75 (m, 5H), 4.57 (t, J = 7.4 Hz, 1H), 3.78 (s, 3H), 3.57 (s, 2H), 3.09 (s, 3H), 2.88 (dd, J = 13.2, 7.7 Hz, 1H), 2.67 (dd, J = 13.2, 7.1 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 174.1, 173.0, 155.3, 142.5, 137.8, 135.0, 133.3, 130.8, 130.2, 129.5, 128.8, 127.9, 125.7, 113.0, 113.0, 109.0, 101.4, 56.3, 53.3, 39.4, 37.9, 33.7; HRMS (ESI) m/z calcd for C27H25ClN3O3 [M − H]− 474.1590, found 474.1595.
(S)-2-(2-(5-Hydroxy-1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (8).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 232 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 49%. 1H NMR (600 MHz, CDCl3) δ 8.42 (s, 1H), 7.30 − 7.27 (m, 3H), 7.13 − 7.11 (m, 2H), 7.08 − 7.06 (m, 2H), 6.90 (s, 1H), 6.86 − 6.77 (m, 4H), 6.68 (d, J = 7.3 Hz, 2H), 6.60 (d, J = 8.1 Hz, 1H), 4.79 (q, J = 7.3 Hz, 1H), 3.55 (s, 2H), 3.17 (s, 3H), 2.79 (dd, J = 13.4, 7.0 Hz, 1H), 2.59 (dd, J = 13.4, 7.3 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 171.7, 171.6, 150.4, 142.3, 136.0, 131.4, 129.8, 129.2, 128.3, 128.2, 127.8, 127.3, 126.7, 124.7, 112.5, 112.0, 107.6, 103.0, 51.4, 38.7, 37.8, 33; HRMS (ESI) m/z calcd for C26H24N3O3 [M − H]− 426.1823, found 426.1826.
(S)-2-(2-(5-Hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (9).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 241 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 80%. 1H NMR (600 MHz, CDCl3) δ 8.27 (s, 1H), 7.26 (s, 1H), 7.16 − 7.08 (m, 6H), 6.91 (s, 1H), 6.79 − 6.78 (m, 3H), 6.72 (d, J = 7.1 Hz, 2H), 6.53 (d, J = 8.2 Hz, 1H), 6.27 (s, 1H), 4.83 − 4.79 (m, 1H), 3.57 (s, 2H), 3.15 (s, 3H), 2.80 (dd, J = 13.4, 6.9 Hz, 1H), 2.60 (dd, J = 13.4, 7.2 Hz, 1H), 2.33 (s, 3H); 13C NMR (150 MHz, CDCl3) δ 171.7, 171.3, 150.2, 139.7, 138.2, 136.1, 131.4, 130.3, 129.2, 128.3, 127.8, 127.0, 126.7, 124.6, 112.5, 112.0, 107.8, 103.1, 51.2, 38.7, 37.8, 33.2, 21.1; HRMS (ESI) m/z calcd for C27H26N3O3 [M − H]− 440.1980, found 440.1984.
(S)-N-(4-Fluorophenyl)-2-(2-(5-hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (10).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 241 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 49%. 1H NMR (600 MHz, CD3OD) δ 7.19 − 7.14 (m, 5H), 7.03 − 7.00 (m, 3H), 6.86 − 6.82 (m, 4H), 6.69 (d, J = 8.6 Hz, 1H), 4.56 (t, J = 7.4 Hz, 1H), 3.54 (s, 2H), 3.11 (s, 3H), 2.88 (dd, J = 13.2, 7.7 Hz, 1H), 2.68 (dd, J = 13.2, 7.1 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.2, 163.4 (d, JCF = 246.9 Hz), 151.5, 140.96, 139.94, 137.8, 133.0, 130.7 (d, JCF = 8.8 Hz), 130.3, 129.5, 129.2, 127.9, 125.8, 117.4 (d, JCF = 23.0 Hz), 112.8 (d, JCF = 9.7 Hz), 108.3, 103.6, 53.2, 39.4, 38.1, 33.7; HRMS (ESI) m/z calcd for C26H23FN3O3 [M − H]− 444.1729, found 444.1732.
(S)-N-(3-Fluorophenyl)-2-(2-(5-hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (11).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 241 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 39%. 1H NMR (600 MHz, CDCl3) δ 8.30 (s, 1H), 7.27 − 7.11 (m, 5H), 6.99 − 6.94 (m, 2H), 6.85 − 6.75 (m, 5H), 6.53 (d, J = 8.0 Hz, 1H), 6.47 (s, 1H), 6.32 (s, 1H), 4.78 – 4.74 (m, 1H), 3.59 (s, 2H), 3.12 (s, 3H), 2.78 (dd, J = 13.2, 8.1 Hz, 1H), 2.63 (dd, J = 13.2, 6.6 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ 171.6, 171.5, 162.7 (d, JCF = 249.4 Hz), 150.4, 143.6, 135.8, 131.4, 130.8 (d, JCF = 9.5 Hz), 129.2, 128.4, 127.7, 126.9, 124.6, 123.3, 115.3 (d, JCF = 21.2 Hz), 114.6 (d, JCF = 20.9 Hz), 112.6, 112.1, 107.7, 103.0, 51.5, 39.0, 37.6, 33.2; HRMS (ESI) m/z calcd for C26H23FN3O3 [M − H]− 444.1729, found 444.1730.
(S)-N-(4-Chlorophenyl)-2-(2-(5-hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (12).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 254 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 60%. 1H NMR (600 MHz, CDCl3) δ 8.16 (s, 1H), 7.25 − 7.12 (m, 6H), 7.00 (s, 1H), 6.82 − 6.71 (m, 5H), 6.50 (d, J = 8.0 Hz, 1H), 6.05 (s, 1H), 4.74 − 4.70 (m, 1H), 3.60 (s, 2H), 3.12 (s, 3H), 2.79 (dd, J = 13.2, 8.0 Hz, 1H), 2.62 (dd, J = 13.2, 6.7 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ 171.5, 171.4, 150.3, 140.8, 135.9, 134.0, 131.3, 129.8, 129.3, 128.7, 128.4, 127.7, 126.9, 124.6, 112.6, 112.0, 107.8, 103.0, 51.3, 38.9, 37.7, 33.2; HRMS (ESI) m/z calcd for C26H23ClN3O3 [M − H]− 460.1433, found 460.1436.
(S)-N-(3-Chlorophenyl)-2-(2-(5-hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (13).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 254 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 40%. 1H NMR (600 MHz, CD3OD) δ 7.23 (d, J = 8.1 Hz, 1H), 7.18 (t, J = 7.9 Hz, 1H), 7.12 − 7.06 (m, 5H), 6.96 (s, 1H), 6.77 − 6.74 (m, 4H), 6.60 (d, J = 8.6 Hz, 1H), 4.47 (t, J = 7.4 Hz, 1H), 3.49 − 3.43 (m, 2H), 3.02 (s, 3H), 2.78 (dd, J = 13.1, 8.1 Hz, 1H), 2.60 (dd, J = 13.1, 6.7 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.0, 151.5, 145.1, 137.6, 135.9, 133.1, 131.9, 130.2, 129.6, 129.5, 129.2, 128.7, 128.1, 127.2, 125.8, 112.8, 112.8, 108.3, 103.6, 53.4, 39.5, 37.9, 33.7; HRMS (ESI) m/z calcd for C26H23ClN3O3 [M − H]− 460.1433, found 460.1430.
(S)-N-(3-Bromophenyl)-2-(2-(5-hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (14).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-(3-bromophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 282 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 65%. 1H NMR (600 MHz, CD3OD) δ 7.47 (d, J = 8.0 Hz, 1H), 7.22 − 7.17 (m, 6H), 7.06 (s, 1H), 6.87 − 6.83 (m, 4H), 6.69 (d, J = 8.6 Hz, 1H), 4.57 (t, J = 7.4 Hz, 1H), 3.59 − 3.53 (m, 2H), 3.10 (s, 3H), 2.87 (dd, J = 13.0, 8.2 Hz, 1H), 2.70 (dd, J = 13.1, 6.7 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.0, 151.5, 145.2, 137.6, 133.0, 132.4, 132.1, 131.5, 130.2, 129.6, 129.2, 128.1, 127.7, 125.8, 123.6, 112.8, 112.8, 108.3, 103.6, 53.4, 39.5, 38.0, 33.7; HRMS (ESI) m/z calcd for C26H23BrN3O3 [M − H]− 504.0928, found 504.0930.
(S)-2-(2-(5-Hydroxy-1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(3-(trifluoromethyl)phenyl)propanamide (15).
The synthetic method was similar to that of compound 1 except that 2-(5-hydroxy-1H-indol-3-yl)acetic acid (100 mg, 0.52 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(3-(trifluoromethyl)phenyl)propanamide (TFA salt, 275 mg, 0.63 mmol, 1.2 equiv.) were used as starting materials. Yield 49%. 1H NMR (600 MHz, CD3OD) δ 7.60 (d, J = 7.7 Hz, 1H), 7.47 (t, J = 7.8 Hz, 1H), 7.18 − 7.12 (m, 6H), 7.04 (s, 1H), 6.86 (s, 1H), 6.81 (d, J = 7.2 Hz, 2H), 6.68 (d, J = 8.6 Hz, 1H), 4.52 (t, J = 7.4 Hz, 1H), 3.58 − 3.52 (m, 2H), 3.13 (s, 3H), 2.87 (dd, J = 12.9, 8.3 Hz, 1H), 2.68 (dd, J = 13.0, 6.7 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.1, 151.4, 144.6, 137.6, 133.0, 132.7, 131.8, 130.1, 129.6, 129.10, 129.09, 128.0, 126.0, 125.7, 125.4, 124.9 (q, JCF = 271.2 Hz), 112.8, 112.8, 112.7, 108.3, 53.4, 39.5, 38.0, 33.7; HRMS (ESI) m/z calcd for C27H23F3N3O3 [M − H]− 494.1697, found 494.1693.
(2S)-N-Methyl-2-(2-(7-methyl-2-oxoindolin-3-yl)acetamido)-N,3-diphenylpropanamide (16).
The synthetic method was similar to that of compound 1 except that 2-(7-methyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 215 mg, 0.58 mmol, 1.2 equiv.) were used as starting materials. (Yield 70%, dr 1.1:1). 1H NMR (600 MHz, CD3OD) δ 7.36 − 7.31 (m, 3H), 7.22 − 7.17 (m, 3H), 6.99 − 6.95 (m, 2H), 6.88 − 6.83 (m, 3H), 6.75 (t, J = 7.6 Hz, 1H), 6.65 (d, J = 7.4 Hz, 1H), 4.73 − 4.63 (m, 1H), 3.72 − 3.69 (m, 1H), 3.18 − 3.15 (m, 1H), 2.96 − 2.82 (m, 2H), 2.71 − 2.65 (m, 1H), 2.54 − 2.47 (m, 1H), 2.22 (s, 3H); 13C NMR (100 MHz, CD3OD) δ 181.8, 181.7, 173.1, 172.3, 172.0, 144.0, 143.9, 142.0, 141.9, 138.2, 138.1, 130.8, 130.5, 130.4, 130.3, 130.2, 130.0, 129.5, 129.3, 128.6, 127.9, 127.8, 123.3, 123.3, 122.9, 122.6, 120.5, 120.4, 53.5, 53.1, 39.5, 39.4, 38.1, 38.0, 37.1, 37.0, 16.7; HRMS (ESI) m/z calcd for C27H26N3O3 [M − H]− 440.1980, found 440.1985.
(2S)-N-(4-Chlorophenyl)-N-methyl-2-(2-(7-methyl-2-oxoindolin-3-yl)acetamido)-3-phenylpropanamide (17).
The synthetic method was similar to that of compound 1 except that 2-(7-methyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 238 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. (Yield 70%, dr 1.1:1). 1H NMR (600 MHz, CD3OD) δ 7.30 − 7.21 (m, 5H), 6.99 − 6.86 (m, 4H), 6.81 − 6.75 (m, 2H), 6.62 (s, 1H), 4.63 − 4.55 (m, 1H), 3.73 − 3.68 (m, 1H), 3.13 − 3.08 (m, 3H), 2.96 − 2.82 (m, 2H), 2.71 − 2.53 (m, 2H), 2.23 − 2.22 (m, 3H); 13C NMR (100 MHz, CD3OD) δ 181.8, 181.7, 173.1, 173.0, 172.2, 171.9, 142.6, 142.5, 142.2, 142.0, 138.02, 134.92, 130.72, 130.50, 130.4, 130.3, 130.28, 130.19, 130.1, 130.0, 129.60, 128.0, 123.32, 122.8, 122.6, 120.5, 53.4, 52.9, 44.3, 44.2, 39.71, 39.5, 38.0, 37.8, 37.1, 36.9, 16.7; HRMS (ESI) m/z calcd for C27H25ClN3O3 [M − H]− 474.1590, found 474.1595.
(2S)-N-(3-Chlorophenyl)-N-methyl-2-(2-(7-methyl-2-oxoindolin-3-yl)acetamido)-3-phenylpropanamide (18).
The synthetic method was similar to that of compound 1 except that 2-(7-methyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 238 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. (Yield 72%, dr 1:1). 1H NMR (600 MHz, CD3OD) δ 7.31 − 7.24 (m, 6H), 7.01 − 6.88 (m, 5H), 6.81 − 6.77 (m, 1H), 6.67 (s, 1H), 4.62 − 4.55 (m, 1H), 3.76 − 3.71 (m, 1H), 3.13 − 3.09 (m, 3H), 2.96 − 2.84 (m, 2H), 2.73 − 2.69 (m, 1H), 2.55 − 2.51 (m, 1H), 2.23 (s, 3H); 13C NMR (100 MHz, CD3OD) δ 181.75, 173.0, 172.9, 172.3, 172.0, 145.2, 145.0, 142.1, 142.0, 137.9, 135.9, 131.8, 130.5, 130.3, 130.1, 130.0, 129.7, 129.6, 129.4, 128.8, 128.1, 127.24, 123.4, 123.3, 122.9, 122.7, 122.6, 120.5, 53.6, 53.1, 44.3, 39.8, 39.7, 38.0, 37.8, 37.1, 36.9, 16.7; HRMS (ESI) m/z calcd for C27H25ClN3O3 [M − H]− 474.1590, found 474.1594.
(2S)-N-(3-Fluorophenyl)-N-methyl-2-(2-(7-methyl-2-oxoindolin-3-yl)acetamido)-3-phenylpropanamide (19).
The synthetic method was similar to that of compound 1 except that 2-(7-methyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.49 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 227 mg, 0.59 mmol, 1.2 equiv.) were used as starting materials. (Yield 69%, dr 1:1). 1H NMR (600 MHz, CD3OD) δ 7.34 − 7.22 (m, 4H), 7.08 − 7.05 (m, 1H), 7.00 − 6.87 (m, 5H), 6.79 − 6.73 (m, 2H), 6.57 (s, 1H), 4.67 − 4.59 (m, 1H), 3.75 − 3.70 (m, 1H), 3.15 − 3.11 (m, 3H), 2.97 − 2.82 (m, 2H), 2.73 − 2.68 (m, 1H), 2.55 − 2.50 (m, 1H), 2.22 (s, 3H); HRMS (ESI) m/z calcd for C27H25FN3O3 [M − H]− 458.1885, found 458.1890.
(2S)-2-(2-(4,7-Dimethyl-2-oxoindolin-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (20).
The synthetic method was similar to that of compound 1 except that 2-(4,7-dimethyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.46 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 203 mg, 0.55 mmol, 1.2 equiv.) were used as starting materials. (Yield 58%, dr 1.2:1). 1H NMR (600 MHz, CD3OD) δ 7.30 − 7.24 (m, 2H), 7.20 − 7.14 (m, 4H), 6.92 − 6.82 (m, 3H), 6.69 − 6.62 (m, 1H), 6.45 (s, 1H), 4.58 − 4.52 (m, 1H), 3.68 − 3.67 (m, 1H), 3.14 − 3.06 (m, 3H), 3.01 − 2.81 (m, 3H), 2.65 − 2.59 (m, 1H), 2.23 − 2.16 (m, 6H); 13C NMR (100 MHz, CD3OD) δ 182.1, 182.0, 173.1, 172.9, 171.8, 171.3, 143.9, 143.6, 142.5, 142.2, 138.1, 133.1, 132.8, 130.8, 130.6, 130.5, 130.4, 130.3, 130.2, 129.4, 129.2, 129.1, 128.5, 127.8, 125.0, 124.9, 117.9, 117.8, 53.1, 52.6, 39.8, 39.5, 38.0, 37.8, 36.1, 36.0, 18.6, 16.4; HRMS (ESI) m/z calcd for C28H28N3O3 [M − H]− 454.2136, found 454.2145.
(2S)-2-(2-(4,7-Dimethyl-2-oxoindolin-3-yl)acetamido)-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (21).
The synthetic method was similar to that of compound 1 except that 2-(4,7-dimethyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.46 mmol, 1 equiv.) and (S)-2-amino-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 212 mg, 0.55 mmol, 1.2 equiv.) were used as starting materials. (Yield 65%, dr 1.1:1). 1H NMR (600 MHz, CD3OD) δ 8.27 − 8.14 (m, 1H), 7.31 − 7.18 (m, 3H), 6.99 − 6.82 (m, 6H), 6.70 − 6.64 (m, 1H), 6.31 (s, 1H), 4.53 – 4.46 (m, 1H), 3.69 − 3.65 (m, 1H), 3.10 − 3.01 (m, 3H), 2.99 − 2.85 (m, 3H), 2.65 − 2.61 (m, 1H), 2.23 − 2.17 (m, 6H); HRMS (ESI) m/z calcd for C28H27FN3O3 [M − H]− 472.2042, found 472.2047.
(2S)-2-(2-(4,7-Dimethyl-2-oxoindolin-3-yl)acetamido)-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (22).
The synthetic method was similar to that of compound 1 except that 2-(4,7-dimethyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.46 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 212 mg, 0.55 mmol, 1.2 equiv.) were used as starting materials. (Yield 68%, dr 1:1). 1H NMR (600 MHz, CD3OD) δ 7.28 − 7.11 (m, 4H), 7.05 − 6.86 (m, 4H), 6.69 − 6.41 (m, 2H), 6.19 − 6.04 (m, 1H), 4.56 − 4.50 (m, 1H), 3.70 − 3.65 (m, 1H), 3.11 − 3.02 (m, 3H), 3.00 − 2.85 (m, 3H), 2.67 − 2.64 (m, 1H), 2.21 − 2.17 (m, 6H); HRMS (ESI) m/z calcd for C28H27FN3O3 [M − H]− 472.2042, found 472.2046.
(2S)-N-(4-Chlorophenyl)-2-(2-(4,7-dimethyl-2-oxoindolin-3-yl)acetamido)-N-methyl-3-phenylpropanamide (23).
The synthetic method was similar to that of compound 1 except that 2-(4,7-dimethyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.46 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 222 mg, 0.55 mmol, 1.2 equiv.) were used as starting materials. (Yield 72%, dr 1:1). 1H NMR (600 MHz, CD3OD) δ 7.24 − 7.18 (m, 4H), 7.08 (d, J = 8.6 Hz, 1H), 6.92 – 6.85 (m, 3H), 6.70 − 6.63 (m, 2H), 6.23 (s, 1H), 4.51 − 4.45 (m, 1H), 3.68 − 3.63 (m, 1H), 3.09 − 2.99 (m, 3H), 2.91 − 2.84 (m, 3H), 2.65 − 2.60 (m, 1H), 2.23 − 2.16 (m, 6H); 13C NMR (100 MHz, CD3OD) δ 182.1, 181.9, 173.0, 172.7, 171.8, 171.1, 142.7, 142.5, 142.2, 138.0, 134.9, 134.7, 133.0, 132.8, 130.7, 130.6, 130.5, 130.4, 130.3, 130.1, 129.5, 127.9, 127.8, 127.7, 125.0, 124.9, 117.9, 117.8, 53.0, 52.4, 44.2, 44.1, 39.9, 39.6, 37.9, 37.6, 36.0, 18.6, 16.4; HRMS (ESI) m/z calcd for C28H27ClN3O3 [M − H]− 488.1746, found 488.1750.
(2S)-N-(3-Chlorophenyl)-2-(2-(4,7-dimethyl-2-oxoindolin-3-yl)acetamido)-N-methyl-3-phenylpropanamide (24).
The synthetic method was similar to that of compound 1 except that 2-(4,7-dimethyl-2-oxoindolin-3-yl)acetic acid (100 mg, 0.46 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 222 mg, 0.55 mmol, 1.2 equiv.) were used as starting materials. (Yield 79%, dr 1:1). 1H NMR (600 MHz, CD3OD) δ 7.27 − 7.07 (m, 6H), 6.91 − 6.85 (m, 3H), 6.68 − 6.63 (m, 1H), 6.29 (s,1H), 4.51 − 4.46 (m, 1H), 3.70 − 3.65 (m, 1H), 3.07 − 2.99 (m, 3H), 2.96 − 2.84 (m, 3H), 2.66 − 2.63 (m, 1H), 2.21 − 2.16 (m, 6H); 13C NMR (100 MHz, CD3OD) δ 182.0, 181.9, 172.9, 172.7, 171.8, 171.3, 145.0, 144.7, 142.5, 142.1, 137.9, 137.8, 135.8, 135.6, 133.0, 132.8, 131.8, 131.6, 130.5, 130.3, 129.6, 129.3, 128.6, 128.0, 127.8, 127.2, 127.1, 125.0, 124.9, 117.9, 117.8, 53.1, 52.6, 44.2, 39.8, 37.9, 37.7, 36.1, 18.6, 16.5; HRMS (ESI) m/z calcd for C28H27ClN3O3 [M − H]− 488.1746, found 488.1751.
(S)-N-Methyl-2-(2-(naphthalen-2-yl)acetamido)-N,3-diphenylpropanamide (25).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-2-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 239 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 79%. 1H NMR (600 MHz, CD3OD) δ 8.26 (d, J = 7.5 Hz, 1H), 7.81 − 7.74 (m, 3H), 7.64 (s, 1H), 7.46 − 7.42 (m, 2H), 7.35 − 7.34 (m, 3H), 7.28 − 7.27 (m, 1H), 7.15 − 7.03 (m, 4H), 6.81 (d, J = 7.2 Hz, 2H), 4.69 – 4.65 (m, 1H), 3.66 − 3.61 (m, 2H), 3.19 (s, 3H), 2.95 (dd, J = 13.4, 6.4 Hz, 1H), 2.72 (dd, J = 13.4, 8.4 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 173.4, 173.3, 144.0, 138.1, 135.0, 134.2, 133.9, 130.9, 130.1, 129.4, 129.4, 129.1, 128.8, 128.7, 128.6, 128.6, 128.3, 127.8, 127.1, 126.7, 53.6, 43.5, 39.0, 38.1; HRMS (ESI) m/z calcd for C28H25N2O2 [M − H]− 421.1922, found 421.1927.
(S)-N-Methyl-2-(2-(naphthalen-2-yl)acetamido)-3-phenyl-N-(p-tolyl)propanamide (26).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-2-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 249 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 58%. 1H NMR (600 MHz, CDCl3) δ 7.83 − 7.77 (m, 3H), 7.63 (s, 1H), 7.49 − 7.45 (m, 2H), 7.28 − 7.26 (m, 1H), 7.14 − 7.05 (m, 5H), 6.79 − 6.75 (m, 4H), 6.17 (d, J = 8.2 Hz, 1H), 4.84 − 4.80 (m, 1H), 3.67 − 3.62 (m, 2H), 3.17 (s, 3H), 2.81 (dd, J = 13.4, 6.9 Hz, 1H), 2.62 (dd, J = 13.4, 7.1 Hz, 1H), 2.35 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.4, 170.0, 139.9, 138.1, 136.1, 133.6, 132.5, 132.2, 130.4, 129.2, 128.6, 128.3, 128.1, 127.7, 127.7, 127.3, 127.0, 126.7, 126.2, 125.9, 51.0, 43.8, 38.8, 37.7, 21.1; HRMS (ESI) m/z calcd for C29H27N2O2 [M − H]− 435.2079, found 435.2083.
(S)-N-(4-Chlorophenyl)-N-methyl-2-(2-(naphthalen-2-yl)acetamido)-3-phenylpropanamide (27).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-2-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 262 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 70%. 1H NMR (600 MHz, CD3OD) δ 7.80 − 7.74 (m, 3H), 7.65 (s, 1H), 7.45 − 7.41 (m, 2H), 7.30 − 7.26 (m, 3H), 7.18 − 7.13 (m, 3H), 6.88 − 6.87 (m, 4H), 4.59 (t, J = 7.5 Hz, 1H), 3.65 − 3.61 (m, 2H), 3.12 (s, 3H), 2.95 (dd, J = 13.3, 7.5 Hz, 1H), 2.74 (dd, J = 13.3, 7.5 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 173.3, 173.2, 142.6, 137.9, 135.0, 134.2, 133.9, 130.8, 130.3, 129.5, 129.1, 128.8, 128.7, 128.6, 128.3, 128.0, 127.1, 126.7, 53.5, 43.4, 39.2, 38.0; HRMS (ESI) m/z calcd for C28H24ClN2O2 [M − H]− 455.1532, found 455.1531.
(S)-N-(3-Chlorophenyl)-N-methyl-2-(2-(naphthalen-2-yl)acetamido)-3-phenylpropanamide (28).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-2-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 262 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 90%. 1H NMR (600 MHz, CD3OD) δ 7.79 − 7.74 (m, 3H), 7.67 (s, 1H), 7.44 − 7.40 (m, 2H), 7.32 − 7.24 (m, 3H), 7.19 − 7.13 (m, 3H), 6.93 − 6.76 (m, 4H), 4.58 (t, J = 7.4 Hz, 1H), 3.64 (s, 2H), 3.11 (s, 3H), 2.95 (dd, J = 13.1, 7.9 Hz, 1H), 2.75 (dd, J = 13.1, 7.3 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 173.4, 173.1, 145.1, 137.8, 135.9, 135.0, 134.2, 133.9, 131.9, 130.2, 129.6, 129.4, 129.1, 128.8, 128.7, 128.7, 128.6, 128.3, 128.1, 127.2, 127.1, 126.7, 53.6, 43.4, 39.4, 38.0; HRMS (ESI) m/z calcd for C28H24ClN2O2 [M − H]− 455.1532, found 455.1533.
(S)-N-(3-Fluorophenyl)-N-methyl-2-(2-(naphthalen-2-yl)acetamido)-3-phenylpropanamide (29).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-2-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 251 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 79%. 1H NMR (600 MHz, CD3OD) δ 7.79 − 7.73 (m, 3H), 7.66 (s, 1H), 7.44 − 7.40 (m, 2H), 7.31 − 7.27 (m, 2H), 7.17 − 7.11 (m, 3H), 7.06 (t, J = 7.9 Hz, 1H), 6.87 − 6.81 (m, 3H), 6.62 (s, 1H), 4.63 (t, J = 7.4 Hz, 1H), 3.66 − 3.61 (m, 2H), 3.13 (s, 3H), 2.95 (dd, J = 13.2, 7.5 Hz, 1H), 2.74 (dd, J = 13.2, 7.6 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 173.4, 173.2, 164.2 (d, JCF = 247.4 Hz), 145.4 (d, JCF = 9.7 Hz), 137.9, 135.0, 134.2, 133.9, 132.1 (d, JCF = 9.1 Hz), 130.2, 129.5, 129.1, 128.8, 128.7, 128.6, 128.3, 128.0, 127.1, 126.7, 124.7, 116.2 (d, JCF = 21.3 Hz), 115.9 (d, JCF = 23.1 Hz), 53.6, 43.4, 39.3, 38.0; HRMS (ESI) m/z calcd for C28H24FN2O2 [M − H]− 439.1827, found 439.1830.
(S)-N-Ethyl-2-(2-(naphthalen-2-yl)acetamido)-N,3-diphenylpropanamide (30).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-2-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-ethyl-N,3-diphenylpropanamide (TFA salt, 249 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 80%. 1H NMR (600 MHz, CDCl3) δ 7.83 − 7.78 (m, 3H), 7.64 (s, 1H), 7.50 − 7.45 (m, 2H), 7.36 − 7.34 (m, 3H), 7.27 − 7.26 (m, 1H), 7.14 (t, J = 7.4 Hz, 1H), 7.06 (t, J = 7.6 Hz, 2H), 6.91 − 6.86 (m, 1H), 6.73 (d, J = 7.3 Hz, 2H), 6.08 (d, J = 8.1 Hz, 1H), 4.72 − 4.68 (m, 1H), 3.81 − 3.75 (m, 1H), 3.67 − 3.62 (m, 2H), 3.59 − 3.53 (m, 1H), 2.82 (dd, J = 13.4, 6.9 Hz, 1H), 2.60 (dd, J = 13.4, 7.1 Hz, 1H), 1.06 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 170.7, 169.9, 140.8, 136.1, 133.6, 132.5, 132.2, 129.7, 129.3, 128.6, 128.4, 128.3, 128.1, 127.7, 127.7, 127.3, 126.8, 126.2, 125.9, 51.4, 44.6, 43.8, 38.8, 12.8; HRMS (ESI) m/z calcd for C29H27N2O2 [M − H]− 435.2079, found 435.2081.
(S)-N-Methyl-2-(2-(naphthalen-1-yl)acetamido)-N,3-diphenylpropanamide (31).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-1-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 239 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 70%. 1H NMR (600 MHz, CD3OD) δ 7.86 − 7.83 (m, 2H), 7.76 (d, J = 8.2 Hz, 1H), 7.46 − 7.42 (m, 2H), 7.39 – 7.36 (m, 1H), 7.33 − 7.29 (m, 4H), 7.17 − 7.10 (m, 3H), 7.00 (m, 2H), 6.77 (d, J = 7.3 Hz, 2H), 4.68 − 4.66 (m, 1H), 3.94 (s, 2H), 3.17 (s, 3H), 2.92 (dd, J = 13.4, 6.3 Hz, 1H), 2.70 (dd, J = 13.4, 8.4 Hz, 2H); 13C NMR (150 MHz, CD3OD) δ 173.3, 173.2, 143.9, 138.0, 135.3, 133.5, 132.8, 130.8, 130.1, 129.6, 129.4, 129.3, 129.0, 128.8, 128.6, 127.8, 127.3, 126.7, 126.5, 124.9, 53.5, 40.9, 39.0, 38.1; HRMS (ESI) m/z calcd for C28H25N2O2 [M − H]− 421.1922, found 421.1928.
(S)-N-Methyl-2-(2-(naphthalen-1-yl)acetamido)-3-phenyl-N-(p-tolyl)propanamide (32).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-1-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 249 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials.
Yield 69%. 1H NMR (600 MHz, CDCl3) δ 7.87 − 7.85 (m, 2H), 7.80 (d, J = 8.2 Hz, 1H), 7.50 − 7.41 (m, 3H), 7.35 (d, J = 6.9 Hz, 1H), 7.13 − 7.08 (m, 3H), 7.01 (t, J = 7.6 Hz, 2H), 6.75 (s, 1H), 6.60 (d, J = 7.4 Hz, 2H), 6.08 (d, J = 8.3 Hz, 1H), 4.82 − 4.78 (m, 1H), 3.99 − 3.90 (m, 2H), 3.12 (s, 3H), 2.68 (dd, J = 13.3, 6.9 Hz, 1H), 2.51 (dd, J = 13.4, 7.0 Hz, 1H), 2.35 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.2, 169.9, 139.8, 138.1, 136.0, 133.9, 132.1, 131.0, 130.4, 129.1, 128.7, 128.3, 128.2, 128.2, 127.0, 126.6, 126.5, 126.0, 125.7, 123.8, 51.0, 41.6, 38.7, 37.6, 21.1; HRMS (ESI) m/z calcd for C29H27N2O2 [M − H]− 435.2079, found 435.2085.
(S)-N-(4-Chlorophenyl)-N-methyl-2-(2-(naphthalen-1-yl)acetamido)-3-phenylpropanamide (33).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-1-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 262 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 80%. 1H NMR (600 MHz, CD3OD) δ 7.90 − 7.84 (m, 2H), 7.77 (d, J = 8.2 Hz, 1H), 7.46 − 7.45 (m, 2H), 7.40 – 7.37 (m, 1H), 7.32 (d, J = 6.9 Hz, 1H), 7.25 − 7.15 (m, 5H), 6.86 − 6.85 (m, 4H), 4.59 (t, J = 7.5 Hz, 1H), 3.98 − 3.93 (m, 2H), 3.12 (s, 3H), 2.93 (dd, J = 13.3, 7.4 Hz, 1H), 2.74 (dd, J = 13.3, 7.6 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 173.3, 173.2, 142.6, 138.0, 135.4, 135.0, 133.6, 132.8, 130.8, 130.3, 130.2, 129.7, 129.6, 129.1, 128.9, 128.0, 127.3, 126.8, 126.6, 124.9, 53.5, 40.8, 39.2, 38.0; HRMS (ESI) m/z calcd for C28H24ClN2O2 [M − H]− 455.1532, found 455.1534.
(S)-N-(3-Chlorophenyl)-N-methyl-2-(2-(naphthalen-1-yl)acetamido)-3-phenylpropanamide (34).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-1-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 262 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 82%. 1H NMR (600 MHz, CD3OD) δ 7.91 − 7.90 (m, 1H), 7.85 − 7.83 (m, 1H), 7.76 (d, J = 8.2 Hz, 1H), 7.47 − 7.44 (m, 2H), 7.40 − 7.16 (m, 8H), 6.91 − 6.85 (m, 3H), 4.60 − 4.57 (m, 1H), 3.99 − 3.93 (m, 2H), 3.11 (s, 3H), 2.93 (dd, J = 13.1, 7.8 Hz, 1H), 2.74 (dd, J = 13.1, 7.3 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 173.3, 173.1, 145.1, 137.8, 135.9, 135.3, 133.6, 132.8, 131.9, 130.2, 129.7, 129.6, 129.4, 129.0, 128.9, 128.7, 128.1, 127.3, 127.2, 126.8, 126.6, 124.9, 53.6, 40.8, 39.3, 38.0; HRMS (ESI) m/z calcd for C28H24ClN2O2 [M − H]− 455.1532, found 455.1535.
(S)-N-(3-Fluorophenyl)-N-methyl-2-(2-(naphthalen-1-yl)acetamido)-3-phenylpropanamide (35).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-1-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 251 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 65%. 1H NMR (600 MHz, CD3OD) δ 8.23 (d, J = 6.7 Hz, 1H), 7.90 − 7.76 (m, 3H), 7.46 − 7.26 (m, 6H), 7.20 − 7.14 (m, 3H), 7.05 (t, J = 7.8 Hz, 1H), 6.85 − 6.80 (m, 3H), 6.61 (s, 1H), 4.66 − 4.63 (m, 1H), 3.96 (s, 2H), 3.13 (s, 3H), 2.94 (dd, J = 13.2, 7.3 Hz, 1H), 2.74 (dd, J = 13.1, 7.7 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 173.4, 173.1, 164.2 (d, J = 247.5 Hz), 145.4 (d, J = 9.5 Hz), 137.9, 135.3, 133.6, 132.7, 132.1 (d, J = 9.2 Hz), 130.2, 129.7, 129.5, 129.0, 128.8, 128.0, 127.3, 126.8, 126.5, 124.9, 124.7, 116.2 (d, J = 21.1 Hz), 115.9 (d, J = 22.7 Hz), 53.5, 40.8, 39.2, 37.9; HRMS (ESI) m/z calcd for C28H24FN2O2 [M − H]− 439.1827, found 439.1831.
(S)-N-Ethyl-2-(2-(naphthalen-1-yl)acetamido)-N,3-diphenylpropanamide (36).
The synthetic method was similar to that of compound 1 except that 2-(naphthalen-1-yl)acetic acid (100 mg, 0.54 mmol, 1 equiv.) and (S)-2-amino-N-ethyl-N,3-diphenylpropanamide (TFA salt, 249 mg, 0.65 mmol, 1.2 equiv.) were used as starting materials. Yield 69%. 1H NMR (600 MHz, CDCl3) δ 7.87 − 7.85 (m, 2H), 7.80 (d, J = 8.2 Hz, 1H), 7.50 − 7.41 (m, 3H), 7.36 − 7.32 (m, 4H), 7.09 (t, J = 7.4 Hz, 1H), 7.02 − 6.99 (m, 2H), 6.83 (s, 1H), 6.60 (d, J = 7.4 Hz, 2H), 6.07 (d, J = 8.3 Hz, 1H), 4.70 − 4.66 (m, 1H), 3.99 − 3.90 (m, 2H), 3.74 − 3.71 (m, 1H), 3.53 − 3.47 (m, 1H), 2.69 (dd, J = 13.3, 6.9 Hz, 1H), 2.50 (dd, J = 13.3, 7.0 Hz, 1H), 1.01 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 170.5, 169.9, 140.7, 136.0, 133.9, 132.1, 131.0, 129.7, 129.2, 128.7, 128.4, 128.3, 128.2, 128.2, 126.6, 126.5, 126.0, 125.7, 123.8, 51.3, 44.6, 41.6, 38.8, 12.7; HRMS (ESI) m/z calcd for C29H27N2O2 [M − H]− 435.2079, found 435.2083.
(S)-2-(2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (37).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 184 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 77%. 1H NMR (600 MHz, CD3OD) δ 7.85 (s, 1H), 7.31 − 7.29 (m, 4H), 7.19 − 7.15 (m, 3H), 6.91 − 6.87 (m, 3H), 5.06 − 5.00 (m, 2H), 4.66 (t, J = 7.4 Hz, 1H), 3.53 (s, 3H), 3.30 (s, 3H), 3.17 (s, 3H), 2.98 (dd, J = 13.4, 7.4 Hz, 1H), 2.75 (dd, J = 13.4, 7.5 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 172.9, 168.1, 156.5, 153.2, 149.7, 144.6, 143.8, 137.9, 130.8, 130.2, 129.5, 129.3, 128.6, 127.9, 108.4, 53.4, 49.5, 39.3, 38.1, 30.2, 28.2; HRMS (ESI) m/z calcd for C25H25N6O4 [M − H]− 473.1943, found 473.1945.
(S)-2-(2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (38).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 191 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 72%. 1H NMR (600 MHz, CD3OD) δ 7.82 (s, 1H), 7.14 − 7.07 (m, 5H), 6.87 − 6.77 (m, 4H), 5.00 (s, 2H), 4.66 (t, J = 7.4 Hz, 1H), 3.47 (s, 3H), 3.24 (s, 3H), 3.13 (s, 3H), 2.94 (dd, J = 13.4, 7.3 Hz, 1H), 2.72 (dd, J = 13.4, 7.5 Hz, 1H), 2.27 (s, 3H); 13C NMR (100 MHz, CD3OD) δ 172.9, 168.0, 156.4, 153.2, 149.7, 144.6, 141.2, 139.5, 137.9, 131.3, 130.2, 129.4, 128.2, 127.9, 108.4, 53.3, 39.4, 38.1, 30.2, 28.2, 21.1; HRMS (ESI) m/z calcd for C26H27N6O4 [M − H]− 487.2100, found 487.2104.
(S)-2-(2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (39).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 193 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 67%. 1H NMR (600 MHz, CD3OD) δ 8.53 (d, J = 7.5 Hz, 1H), 7.85 (s, 1H), 7.21 − 7.20 (m, 3H), 7.01 − 6.93 (m, 5H), 5.06 − 5.00 (m, 2H), 4.61 (q, J = 7.4 Hz, 1H), 3.52 (s, 3H), 3.29 (s, 3H), 3.13 (s, 3H), 2.99 (dd, J = 13.2, 8.1 Hz, 1H), 2.78 (dd, J = 13.3, 7.0 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 172.9, 168.0, 163.3 (d, J = 247.0 Hz), 156.4, 153.2, 149.7, 144.6, 139.9 (d, J = 3.2 Hz), 137.8, 130.6, 130.3, 129.6, 128.0, 117.3 (d, J = 23.0 Hz), 108.4, 53.3, 49.4, 39.4, 38.1, 30.2, 28.2; HRMS (ESI) m/z calcd for C25H24FN6O4 [M − H]− 491.1849, found 491.1845.
(S)-2-(2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (40).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 193 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 75%. 1H NMR (600 MHz, CD3OD) δ 8.57 (d, J = 7.3 Hz, 1H), 7.86 (s, 1H), 7.29 − 7.20 (m, 4H), 7.03 (t, J = 8.3 Hz, 1H), 6.94 − 6.93 (m, 2H), 6.71 − 6.54 (m, 1H), 5.04 (s, 2H), 4.67 − 4.63 (m, 1H), 3.52 (s, 3H), 3.30 (s, 3H), 3.14 (s, 3H), 2.99 (dd, J = 13.1, 8.4 Hz, 1H), 2.79 (dd, J = 13.2, 6.7 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 172.7, 168.0, 164.1 (d, J = 247.4 Hz), 156.5, 153.2, 149.7, 145.2 (d, J = 9.6 Hz), 144.6, 137.7, 132.0 (d, J = 9.2 Hz), 130.3, 129.6, 128.0, 124.6, 116.1 (d, J = 21.2 Hz), 115.9 (d, J = 22.6 Hz), 108.4, 53.4, 49.4, 39.6, 37.9, 30.2, 28.2; HRMS (ESI) m/z calcd for C25H24FN6O4 [M − H]− 491.1849, found 491.1843.
(S)-N-(4-Chlorophenyl)-2-(2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-methyl-3-phenylpropanamide (41).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 201 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 90%. 1H NMR (600 MHz, CDCl3) δ 7.67 (d, J = 8.1 Hz, 1H), 7.63 (s, 1H), 7.27 − 7.25 (m, 2H), 7.20 − 7.15 (m, 3H), 6.90 − 6.89 (m, 2H), 6.75 (s, 1H), 4.98 − 4.87 (m, 2H), 4.78 (q, J = 7.7 Hz, 1H), 3.59 (s, 3H), 3.37 (s, 3H), 3.18 (s, 3H), 2.96 (dd, J = 13.4, 7.9 Hz, 1H), 2.80 (dd, J = 13.4, 7.1 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 171.2, 165.2, 155.5, 151.5, 148.7, 142.3, 140.8, 135.8, 134.1, 129.9, 129.2, 128.7, 128.4, 127.0, 106.8, 51.4, 49.2, 39.0, 37.7, 29.8, 29.7, 28.0; HRMS (ESI) m/z calcd for C25H24ClN6O4 [M − H]− 507.1553, found 507.1548.
(S)-N-(3-Chlorophenyl)-2-(2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-methyl-3-phenylpropanamide (42).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 201 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 82%. 1H NMR (600 MHz, CDCl3) δ 7.63 (s, 1H), 7.50 (d, J = 8.0 Hz, 1H), 7.28 − 7.17 (m, 5H), 6.90 − 6.89 (m, 3H), 6.58 (s, 1H), 4.96 − 4.84 (m, 2H), 4.79 − 4.75 (m, 1H), 3.59 (s, 3H), 3.38 (s, 3H), 3.17 (s, 3H), 2.95 (dd, J = 13.3, 8.3 Hz, 1H), 2.80 (dd, J = 13.3, 6.8 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 170.9, 164.9, 155.5, 151.5, 148.8, 143.4, 142.2, 135.6, 135.0, 130.6, 129.1, 128.5, 127.5, 127.1, 125.7, 106.8, 51.4, 49.4, 39.1, 37.6, 29.8, 28.0; HRMS (ESI) m/z calcd for C25H24ClN6O4 [M − H]− 507.1553, found 507.1559.
(S)-2-(2-(1,3-Dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetamido)-N-ethyl-N,3-diphenylpropanamide (43).
The synthetic method was similar to that of compound 1 except that 2-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)acetic acid (100 mg, 0.42 mmol, 1 equiv.) and (S)-2-amino-N-ethyl-N,3-diphenylpropanamide (TFA salt, 191 mg, 0.50 mmol, 1.2 equiv.) were used as starting materials. Yield 75%. 1H NMR (600 MHz, CDCl3) δ 7.57 (s, 1H), 7.37 − 7.33 (m, 4H), 7.15 − 7.09 (m, 3H), 6.90 − 6.81 (m, 3H), 4.95 – 4.76 (m, 2H), 4.74 − 4.70 (m, 1H), 3.86 − 3.80 (m, 1H), 3.62 −3.58 (m, 1H), 3.58 (s, 3H), 3.38 (s, 3H), 2.94 (dd, J = 13.7, 6.7 Hz, 1H), 2.73 (dd, J = 13.7, 7.8 Hz, 1H), 1.09 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 170.3, 164.8, 155.5, 151.6, 148.7, 142.0, 140.6, 136.0, 129.7, 129.1, 128.4, 128.4, 128.2, 126.7, 106.7, 51.4, 49.5, 44.7, 38.7, 29.8, 28.0, 12.8; HRMS (ESI) m/z calcd for C26H27N6O4 [M − H]− 487.2100, found 487.2105.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (44).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 195 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 65%. 1H NMR (600 MHz, DMSO-d6) δ 8.65 (s, 1H), 8.53 (d, J = 7.8 Hz, 1H), 7.45 – 7.32 (m, 3H), 7.20 – 7.12 (m, 4H), 6.82 – 6.77 (m, 2H), 4.42 – 4.38 (m, 1H), 3.95 – 3.82 (m, 2H), 3.13 (s, 3H), 2.81 (dd, J = 13.2, 6.2 Hz, 1H), 2.61 (dd, J = 13.5, 9.2 Hz, 1H), 1.64 – 1.59 (m, 4H), 1.57 – 1.45 (m, 5H), 1.29 – 1.24 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 176.9, 171.0, 166.1, 155.8, 143.1, 137.7, 130.0, 129.2, 129.2, 128.6, 128.3, 127.9, 126.9, 61.5, 52.1, 37.8, 37.6, 33.7, 24.8, 21.2; HRMS (ESI) m/z calcd for C26H31N4O4 [M + H]+ 463.2340, found 463.2337.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N,3-diphenylpropanamide. (45).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N,3-diphenylpropanamide (TFA salt, 188 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 70%. 1H NMR (600 MHz, DMSO-d6) δ 10.04 (s, 1H), 8.67 (s, 1H), 8.55 (d, J = 7.9 Hz, 1H), 7.54 (d, J = 7.8 Hz, 2H), 7.30 – 7.24 (m, 4H), 7.20 – 7.15 (m, 1H), 7.04 (t, J = 7.4 Hz, 1H), 4.64 – 4.59 (m, 1H), 4.00 – 3.89 (m, 2H), 3.02 (dd, J = 13.7, 5.8 Hz, 1H), 2.88 (dd, J = 13.7, 8.5 Hz, 1H), 1.65 – 1.55 (m, 4H), 1.54 – 1.46 (m, 5H), 1.29 – 1.25 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 177.0, 170.1, 166.4, 155.9, 139.1, 137.7, 129.6, 129.1, 128.6, 126.9, 123.9, 119.9, 61.5, 55.4, 38.3, 33.7, 33.7, 24.8, 21.2; HRMS (ESI) m/z calcd for C25H29N4O4 [M + H]+ 449.2183, found 449.2186.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (46).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (TFA salt, 211 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 62%. 1H NMR (600 MHz, DMSO-d6) δ 8.65 (s, 1H), 8.50 (d, J = 7.9 Hz, 1H), 7.20 – 7.14 (m, 3H), 7.07 – 6.99 (m, 2H), 6.93 (d, J = 8.3 Hz, 2H), 6.84 (d, J = 7.0 Hz, 2H), 4.41 – 4.35 (m, 1H), 3.95 – 3.79 (m, 2H), 3.76 (s, 3H), 3.08 (s, 3H), 2.83 (dd, J = 13.5, 5.3 Hz, 1H), 2.61 (dd, J = 13.4, 8.8 Hz, 1H), 1.64 – 1.58 (m, 4H), 1.54 – 1.42 (m, 5H), 1.30 – 1.23 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 176.9, 171.2, 166.0, 158.9, 155.8, 137.7, 135.8, 129.5, 129.1, 128.8, 128.4, 127.1, 115.3, 114.9, 61.5, 55.9, 55.7, 52.0, 51.8, 37.9, 33.7, 24.9, 21.2; HRMS (ESI) m/z calcd for C27H33N4O5 [M + H]+ 493.2445, found 493.2449.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (47).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 203 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 61%. 1H NMR (600 MHz, DMSO-d6) δ 8.65 (s, 1H), 8.50 (d, J = 7.9 Hz, 1H), 7.20 – 7.15 (m, 4H), 6.98 (d, J = 7.1 Hz, 2H), 6.85 – 6.80 (m, 2H), 4.44 – 4.39 (m, 1H), 3.96 – 3.82 (m, 2H), 3.09 (s, 3H), 2.82 (dd, J = 12.5, 7.4 Hz, 1H), 2.61 (dd, J = 13.2, 8.9 Hz, 1H), 2.31 (s, 3H), 1.66 – 1.59 (m, 4H), 1.54 – 1.44 (m, 5H), 1.31 – 1.24 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 176.9, 171.0, 166.0, 155.8, 140.6, 137.7, 137.7, 130.5, 129.7, 129.3, 128.6, 127.6, 126.9, 61.5, 51.9, 38.0, 37.6, 33.7, 24.8, 21.2, 21.1; HRMS (ESI) m/z calcd for C27H33N4O4 [M + H]+ 477.2496, found 477.2494.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (48).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 205 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 69%. 1H NMR (600 MHz, CD3OD) δ 7.32 – 7.23 (m, 5H), 7.03 – 7.00 (m, 2H), 6.98 – 6.96 (m, 2H), 4.60 − 4.58 (m, 1H), 4.16 − 4.04 (m, 2H), 3.13 (s, 3H), 2.97 (dd, J = 13.1, 8.4 Hz, 1H), 2.75 (dd, J = 13.1, 6.6 Hz, 1H), 1.83 − 1.77 (m, 4H), 1.66 − 1.63 (m, 3H), 1.58 − 1.53 (m, 2H), 1.43 − 1.37 (m, 1H); 13C NMR (150 MHz, CD3OD) δ 172.9, 168.0, 163.3 (d, J = 246.8 Hz), 157.7, 139.9, 139.9, 137.9, 130.6 (d, J = 7.6 Hz), 130.4, 129.6, 128.0, 117.4 (d, J = 23.0 Hz), 63.3, 53.2, 40.9, 39.6, 38.1, 34.6, 25.8, 22.5; HRMS (ESI) m/z calcd for C26H28FN4O4 [M − H]− 479.2100, found 479.2106.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (49).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 205 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 70%. 1H NMR (600 MHz, CD3OD) δ 7.33 − 7.23 (m, 5H), 7.06 (t, J = 8.3 Hz, 1H), 6.97 − 6.96 (m, 2H), 6.73 (s, 1H), 6.47 (s, 1H), 4.64 − 4.61 (m, 1H), 4.17 − 4.05 (m, 2H), 3.14 (s, 3H), 2.97 (dd, J = 13.0, 8.6 Hz, 1H), 2.77 (dd, J = 13.1, 6.4 Hz, 1H), 1.84 − 1.79 (m, 4H), 1.67 − 1.64 (m, 3H), 1.59 − 1.52 (m, 2H), 1.43 − 1.37 (m, 1H); 13C NMR (150 MHz, CD3OD) δ 178.9, 172.7, 168.1, 164.2 (d, J = 247.4 Hz), 157.8, 145.3 (d, J = 9.6 Hz), 137.8, 132.1 (d, J = 9.1 Hz), 130.4, 129.6, 128.1, 124.6, 116.1 (d, J = 21.1 Hz), 115.8 (d, J = 22.9 Hz), 63.3, 53.4, 40.9, 39.8, 37.9, 34.6, 25.8, 22.5; HRMS (ESI) m/z calcd for C26H28FN4O4 [M − H]− 479.2100, found 479.2100.
N-(4-Chlorophenyl)-2-(2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-methyl-3-phenylpropanamide (50).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 213 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 56%. 1H NMR (400 MHz, DMSO-d6) δ 8.67 (s, 1H), 8.60 (d, J = 7.7 Hz, 1H), 7.45 (d, J = 8.5 Hz, 2H), 7.22 – 7.18 (m, 3H), 7.10 (d, J = 8.1 Hz, 2H), 6.91 – 6.87 (m, 2H), 4.40 – 4.32 (m, 1H), 3.98 – 3.82 (m, 2H), 2.90 – 2.83 (m, 1H), 2.66 (dd, J = 13.2, 8.3 Hz, 1H), 1.70 – 1.60 (m, 4H), 1.56 – 1.41 (m, 5H), 1.31 – 1.28 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 177.0, 170.9, 166.2, 155.8, 142.0, 137.5, 132.8, 130.0, 129.8, 129.3, 128.7, 127.1, 61.6, 52.0, 38.0, 37.5, 33.7, 24.8, 21.3; HRMS (ESI) m/z calcd for C26H30ClN4O4 [M + H]+ 497.1950, found 497.1952.
N-(3-Chlorophenyl)-2-(2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-methyl-3-phenylpropanamide (51).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 213 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 72%. 1H NMR (600 MHz, CD3OD) δ 7.32 – 7.25 (m, 7H), 6.98 – 6.96 (m, 2H), 4.60 − 4.58 (m, 1H), 4.17 − 4.05 (m, 2H), 3.12 (s, 3H), 2.97 (dd, J = 13.0, 9.0 Hz, 1H), 2.77 (dd, J = 13.0, 6.2 Hz, 1H), 1.84 − 1.78 (m, 4H), 1.68 − 1.64 (m, 3H), 1.59 − 1.53 (m, 2H), 1.44 − 1.37 (m, 1H); HRMS-ESI (−) m/z calcd for C26H28ClN4O4 [M − H]− 495.1805, found 495.1810.
N-(3-Bromophenyl)-2-(2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-methyl-3-phenylpropanamide (52).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-(3-bromophenyl)-N-methyl-3-phenylpropanamide (TFA salt, 237 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 68%. 1H NMR (600 MHz, DMSO-d6) δ 8.65 (s, 1H), 8.60 (d, J = 7.3 Hz, 1H), 7.54 (d, J = 7.9 Hz, 1H), 7.33 (t, J = 8.0 Hz, 1H), 7.20 – 7.13 (m, 5H), 6.88 – 6.87 (m, 2H), 4.34 − 4.31 (m, 1H), 3.95 − 3.84 (m, 2H), 3.08 (s, 3H), 2.86 (dd, J = 13.4, 5.9 Hz, 1H), 2.66 (dd, J = 14.4, 7.2 Hz, 1H), 1.63 – 1.48 (m, 9H), 1.29 − 1.23 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 177.0, 171.0, 166.2, 155.8, 144.6, 137.5, 131.7, 131.3, 130.7, 129.3, 128.8, 127.2, 127.1, 122.3, 61.6, 52.2, 38.1, 37.5, 33.7, 24.9, 21.3; HRMS (ESI) m/z calcd for C26H28BrN4O4 [M − H]− 539.1299, found 539.1297.
2-(2-(2,4-Dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetamido)-N-ethyl-N,3-diphenylpropanamide (53).
The synthetic method was similar to that of compound 1 except that 2-(2,4-dioxo-1,3-diazaspiro[4.5]decan-3-yl)acetic acid (100 mg, 0.44 mmol, 1 equiv.) and (S)-2-amino-N-ethyl-N,3-diphenylpropanamide (TFA salt, 203 mg, 0.53 mmol, 1.2 equiv.) were used as starting materials. Yield 58%. 1H NMR (400 MHz, DMSO-d6) δ 8.65 (s, 1H), 8.49 (d, J = 7.9 Hz, 1H), 7.46 – 7.39 (m, 3H), 7.20 – 7.16 (m, 3H), 7.08 – 7.04 (m, 2H), 6.86 – 6.81 (m, 2H), 4.35 – 4.28 (m, 1H), 3.98 – 3.83 (m, 2H), 3.69 – 3.55 (m, 2H), 2.86 (dd, J = 13.4, 5.5 Hz, 1H), 2.62 (dd, J = 13.4, 8.5 Hz, 1H), 1.65 – 1.53 (m, 4H), 1.56 – 1.45 (m, 5H), 1.34 – 1.25 (m, 1H), 0.97 (t, J = 7.1 Hz, 3H); 13C NMR (100 MHz, DMSO-d6) δ 170.0, 170.4, 166.0, 155.8, 141.3, 137.7, 130.0, 129.4, 128.9, 128.6, 128.5, 127.0, 61.5, 52.3, 44.2, 38.0, 33.7, 24.9, 21.3, 13.1; HRMS (ESI) m/z calcd for C27H33N4O4 [M + H]+ 477.2496, found 477.2500.
2-(2-(2,5-Dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (54).
The synthetic method was similar to that of compound 1 except that 2-(2,5-dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetic acid (100 mg, 0.36 mmol, 1 equiv.) and (S)-2-amino-N-methyl-N,3-diphenylpropanamide (TFA salt, 158 mg, 0.43 mmol, 1.2 equiv.) were used as starting materials. Yield 52%. 1H NMR (600 MHz, DMSO-d6) δ 8.83 (s, 1H), 8.63 (d, J = 8.0 Hz, 1H), 7.46 – 7.32 (m, 3H), 7.31 – 7.04 (m, 9H), 6.84 (d, J = 6.4 Hz, 2H), 4.46 – 4.41 (m, 1H), 4.09 – 3.94 (m, 2H), 3.12 (s, 3H), 2.86 (dd, J = 13.3, 5.3 Hz, 1H), 2.75 – 2.60 (m, 2H), 2.63 – 2.60 (m, 1H), 2.09 – 1.82 (m, 4H); HRMS (ESI) m/z calcd for C30H31N4O4 [M + H]+ 511.2340, found 511.2340.
2-(2-(2,5-Dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetamido)-N,3-diphenylpropanamide (55).
The synthetic method was similar to that of compound 1 except that 2-(2,5-dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetic acid (100 mg, 0.36 mmol, 1 equiv.) and (S)-2-amino-N,3-diphenylpropanamide (TFA salt, 152 mg, 0.43 mmol, 1.2 equiv.) were used as starting materials. Yield 72%. 1H NMR (600 MHz, DMSO-d6) δ 10.09 (s, 1H), 8.87 (s, 1H), 8.68 (s, 1H), 7.54 (s, 2H), 7.26 – 6.94 (m, 10H), 4.72 (s, 1H), 4.15 – 4.05 (m, 2H), 3.05 (s, 2H), 2.91 (s, 1H), 2.76 (s, 2H), 2.08 – 1.81 (m, 4H); 13C NMR (150 MHz, DMSO-d6) δ 176.6, 170.0, 166.4, 155.7, 139.0, 138.1, 137.7, 134.6, 129.6, 129.6, 129.5, 129.1, 128.6, 128.5, 128.0, 119.9, 126.9, 123.9, 62.8, 55.4, 38.5, 34.1, 28.9, 18.9; HRMS (ESI) m/z calcd for C29H29N4O4 [M + H]+ 497.2183, found 497.2182.
2-(2-(2,5-Dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (56).
The synthetic method was similar to that of compound 1 except that 2-(2,5-dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetic acid (100 mg, 0.36 mmol, 1 equiv.) and (S)-2-amino-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (TFA salt, 171 mg, 0.43 mmol, 1.2 equiv.) were used as starting materials. Yield 67%. 1H NMR (600 MHz, DMSO-d6) δ 8.83 (s, 1H), 8.61 – 8.57 (m, 1H), 7.25 – 7.11 (m, 7H), 7.06 – 6.96 (m, 2H), 6.95 – 6.92 (m, 2H), 6.91 – 6.86 (m, 2H), 4.44 – 4.34 (m, 1H), 4.10 – 3.98 (m, 2H), 3.77 (s, 3H), 3.08 (s, 3H), 2.87 (dd, J = 13.1, 5.5 Hz, 1H), 2.77 – 2.75 (m, 2H), 2.66 – 2.61 (m, 1H), 2.15 – 2.02 (m, 2H), 1.92 – 1.80 (m, 2H); HRMS (ESI) m/z calcd for C31H33N4O5 [M + H]+ 541.2445, found 541.2446.
2-(2-(2,5-Dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (57).
The synthetic method was similar to that of compound 1 except that 2-(2,5-dioxo-3’,4’-dihydro-2’H-spiro[imidazolidine-4,1’-naphthalen]-1-yl)acetic acid (100 mg, 0.36 mmol, 1 equiv.) and (S)-2-amino-N-methyl-3-phenyl-N-(p-tolyl)propanamide (TFA salt, 164 mg, 0.43 mmol, 1.2 equiv.) were used as starting materials. Yield 55%. 1H NMR (600 MHz, DMSO-d6) δ 8.83 (s, 1H), 8.60 (d, J = 7.9 Hz, 1H), 7.29 (d, J = 7.8 Hz, 1H), 7.29 – 7.12 (m, 8H), 6.95 (d, J = 7.1 Hz, 2H), 6.87 (d, J = 7.4 Hz, 2H), 4.46 – 4.40 (m, 1H), 4.13 – 3.92 (m, 2H), 3.08 (s, 3H), 2.87 (dd, J = 13.5, 5.8 Hz, 1H), 2.77 – 2.72 (m, 2H), 2.64 (dd, J = 13.3, 8.4 Hz, 1H), 2.32 (s, 3H), 2.08 – 1.80 (m, 4H); 13C NMR (150 MHz, DMSO-d6) δ 176.6, 171.0, 166.0, 155.7, 138.1, 137.7, 137.6, 134.6, 130.4, 129.5, 129.4, 128.6, 128.5, 127.6, 126.9, 62.7, 51.8, 38.1, 37.6, 34.0, 28.9, 21.1, 18.9; HRMS (ESI) m/z calcd for C31H33N4O4 [M + H]+ 525.2496, found 525.2498.
4.2. Thermal Shift Assays (TSAs) to test compounds for HIV-1 CA hexamer stability
Compounds were screened for their effect on CA stability using purified covalently-crosslinked hexameric CAA14C/E45C/W184A/M185A (CA121). CA121 cloned in a pET11a expression plasmid was kindly provided by Dr. Owen Pornillos (University of Virginia, Charlottesville, VA). Protein was expressed in E. coli BL21(DE3)RIL and purified as reported previously [34]. The TSA has been previously described [56–58]. Briefly, the TSA was conducted on the PikoReal Real-Time PCR System (Thermo Fisher Scientific) or the QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific). Each reaction contained 7.5 μM CA121, 1x Sypro Orange Protein Gel Stain (Life Technologies), 50 mM sodium phosphate buffer (pH 8.0) and 1% DMSO (control) or 20 μM compound. The plate was heated from 25 to 95 °C with a heating rate of 0.2 °C every 10 sec. The fluorescence intensity was measured with an Ex range of 475–500 nm and Em range of 520–590 nm. The differences in the melting temperature (ΔTm) of CA hexamer in DMSO (T0) verses in the presence of compound (Tm) were calculated using the following formula: ΔTm=Tm−T0.
4.3. Virus Production
The wild-type laboratory HIV-1 strain, HIV-1NL4–3 [59], was produced using a pNL4–3 vector that was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. HIV-1NL4–3 was generated by transfecting HEK 293FT cells in a T75 flask with 10 μg of the pNL4–3 vector and FuGENE®HD Transfection Reagent (Promega). Supernatant was harvested 48–72 hours post-transfection and transferred to MT2 cells for viral propagation. Virus was harvested when syncytia formation was observed, which took 3–5 days. The viral supernatant was then concentrated using 8% w/v PEG 8,000 overnight at 4 °C, followed by centrifugation for 40 min at 3,500 rpm. The resulting viral-containing pellet was concentrated 10-fold by resuspension in DMEM without FBS and stored at −80 °C.
4.4. Anti-HIV-1 and Cytotoxicity assays
Anti-HIV-1 activity of PF74 and PF74-related analogs were examined in TZM-GFP cells. The potency of HIV-1 inhibition by a compound was based on its inhibitory effect on viral LTR-activated GFP expression compared with that of compound-free (DMSO) controls. Briefly, TZM-GFP cells were plated at density of 1 × 104 cells per well in a 96-well plate. 24 h later, media was replaced with increasing concentrations of compound. 24 h post treatment, cells were exposed to an HIV-1 strain (MOI = 1). After incubation for 48 h, anti-HIV-1 activity was assessed by counting the number of GFP positive cells on a Cytation™ 5 Imaging Reader (BioTek) and 50% effective concentration (EC50) values were determined.
Cytotoxicity of each compound was also determined in TZM-GFP cells. Cells were plated at a density of 1× 104 cells per well in a 96-well plate and were continuously exposed to increasing concentrations of a compound for 72 hours. The number of viable cells in each well was determined using a Cell Proliferation Kit II (XTT), and 50% cytotoxicity concentration (CC50) values were determined. All the cell-based assays were conducted in duplicate and in at least two independent experiments.
For the EC50 and CC50 dose responses, values were plotted in GraphPad Prism 5 and analyzed with the log (inhibitor) vs. normalized response – variable slope equation. Final values were calculated in each independent assay and the average values were determined. Statistical analysis (calculation of standard deviation) was performed by using Microsoft Excel.
4.5. Microsomal stability assay.
The in vitro microsomal stability assay was conducted in duplicate in mouse and human liver microsomal systems, which were supplemented with nicotinamide adenine dinucleotide phosphate (NADPH) as a cofactor. Briefly, a compound (1 μM final concentration) was pre-incubated, in the absence or presence of 0.5 μM Cobicistat (CYP 3A inhibitor, purchased from medchemexpress.com and verified with LCMS), with the reaction mixture containing liver microsomal protein (0.5 mg/mL final concentration) and MgCl2 (1 mM final concentration) in 0.1 M potassium phosphate buffer (pH 7.4) at 37 °C for 15 minutes. The reaction was initiated by addition of 1 mM NADPH, followed by incubation at 37 °C. A negative control was performed in parallel in the absence of NADPH to measure any chemical instability or non-NADPH dependent enzymatic degradation for each compound. At various time points (0, 5, 15, 30, and 60 min), 1 volume of reaction aliquot was taken and quenched with 3 volumes of acetonitrile containing an appropriate internal standard and 0.1% formic acid. The samples were then vortexed and centrifuged at 15,000 rpm for 5 min at 4 °C. The supernatants were collected and analyzed by LC-MS/MS to determine the in vitro metabolic half-life (t1/2).
4.6. Molecular modeling
Molecular modeling was performed using the Schrödinger small molecule drug discovery suite 2019–1 [60]. The crystal structure of native full length HIV-1 capsid protein in complex with PF74 was retrieved from the protein data bank (PDB code: 4XFZ) [30]. The above structure was analyzed using Maestro [61] (Schrödinger Inc.) and subjected to a docking protocol that involves several steps including preparing the protein of interest, grid generation, ligand preparation, and docking. The crystal structure was refined using the protein preparation wizard [62] (Schrödinger Inc.) in which missing hydrogen atoms, side chains, and loops were added using prime and minimized using the OPLS 3e force field [63] to optimize the hydrogen bonding network and converge the heavy atoms to an rmsd of 0.3 Å. The receptor grid generation tool in Maestro (Schrödinger Inc.) was used to define an active site around the native ligand PF74 to cover all the residues within 12 Å. All the compounds were drawn using Maestro and subjected to LigPrep [64] to generate conformers, possible protonation at pH of 7±2 that serves as an input for docking process. All the dockings were performed using Glide XP [65](Glide, version 8.2) with the van der Waals radii of nonpolar atoms for each of the ligands scaled by a factor of 0.8. The solutions were further refined by post docking and minimization under implicit solvent to account for protein flexibility. The residue numbers of HIV-1 capsid protein used in the discussion and the figures were based on the native full length HIV-1 capsid protein.
Supplementary Material
Scheme 1. Synthesis of novel PF74 analogs.
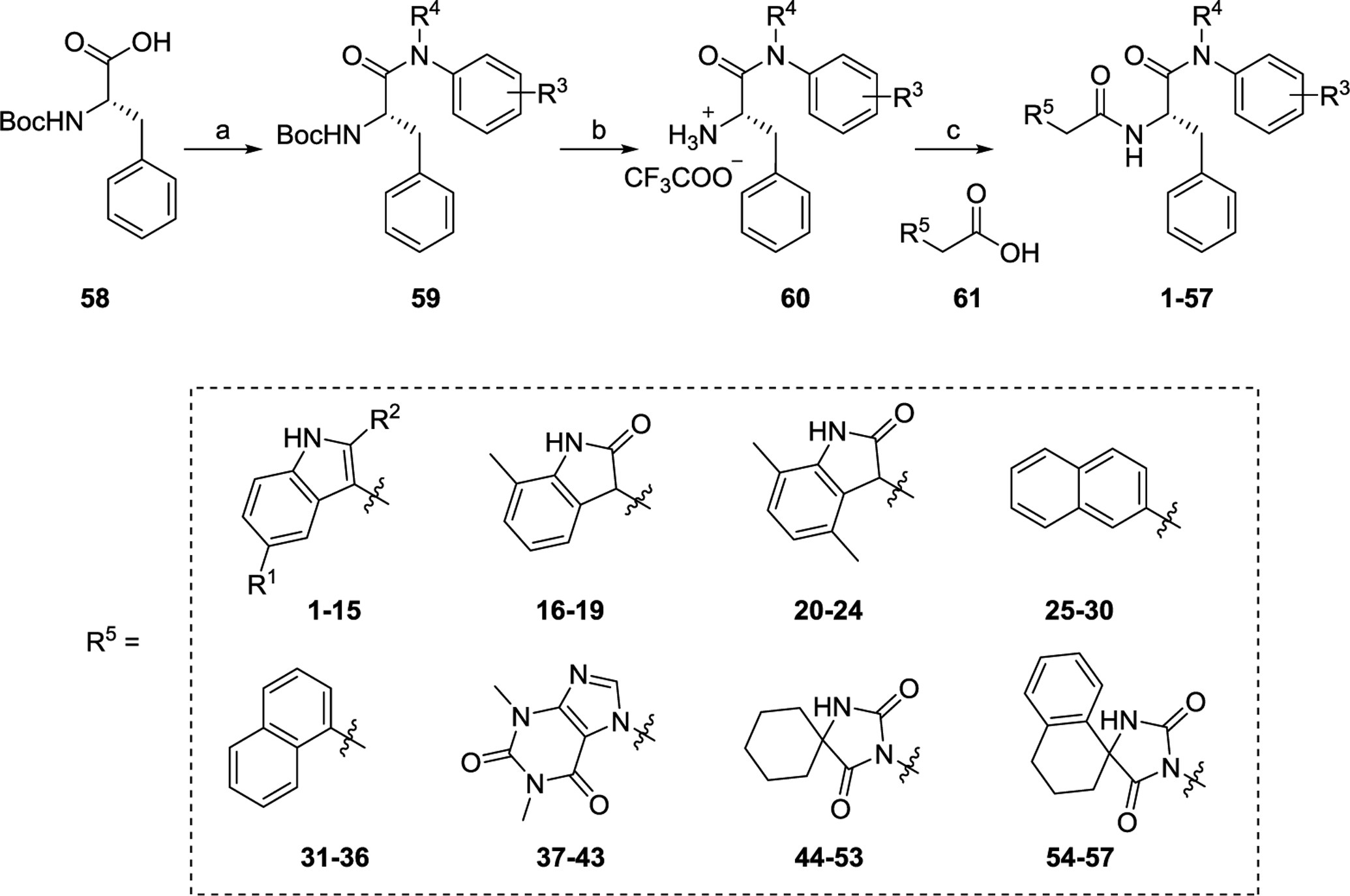
Reagents and conditions: (a) amine, HATU (or T3P), DIPEA, DMF, rt, 12 h; (b) TFA; DCM, rt, 4–6 h; (c) HATU, DIPEA, DMF, rt, 12 h.
Table 1.
Anti-HIV-1 activity, cytotoxicity, and CA hexamer stability profiles of PF74 analogs (modifications at R1, R2, R3).
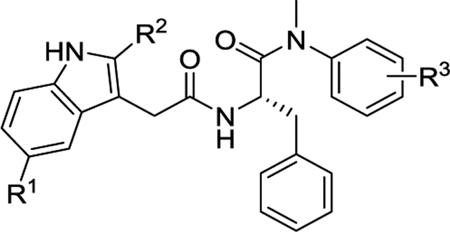 |
||||||
|---|---|---|---|---|---|---|
| Compd | R1 | R2 | R3 | EC50a(μM) | CC50b(μM) | TSAc ΔTm (°C) |
| PF74 (1) | H | Me | H | 0.61 ± 0.2 | 76 ± 9 | 7.4 |
| 2 | H | H | H | 0.46 ± 0.1 | > 100 | 6.1 |
| 3 | OMe | H | H | 0.56 ± 0.3 | 48 ± 5 | 4.9 |
| 4 | OMe | H | 4-Me | 0.28 ± 0.02 | > 50 | 5.4 |
| 5 | OMe | H | 4-F | 0.57 ± 0.02 | > 50 | 6.6 |
| 6 | OMe | H | 3-F | 0.99 ± 0.002 | > 50 | 4.2 |
| 7 | OMe | H | 4-Cl | 0.31 ± 0.02 | > 50 | 8.0 |
| 8 | OH | H | H | 0.053 ± 0.02 | 55 ± 2 | 6.8 |
| 9 | OH | H | 4-Me | 0.035 ± 0.004 | > 100 | 6.3 |
| 10 | OH | H | 4-F | 0.46 ± 0.03 | > 50 | 3.1 |
| 11 | OH | H | 3-F | 0.32 ± 0.02 | 47 ± 6 | 4.4 |
| 12 | OH | H | 4-Cl | 0.032 ± 0.01 | > 100 | 6.6 |
| 13 | OH | H | 3-Cl | 0.31 ± 0.01 | > 100 | 6.9 |
| 14 | OH | H | 3-Br | 0.38 ± 0.1 | > 100 | 5.8 |
| 15 | OH | H | 3-CF3 | 0.48 ± 0.02 | 65 ± 18 | 6.8 |
Concentration of compound inhibiting HIV-1 replication by 50%, expressed as the mean ± standard deviation from at least two independent experiments.
Concentration of compound causing 50% cell death, expressed as the mean ± standard deviation from at least two independent experiments.
TSA: thermal shift assay. ΔTm: change of CA hexamer melting point in presence of compound compared to DMSO control.
Highlights.
Design of novel analogs and sub-chemotypes of HIV-1 CA-targeting antiviral PF74.
5-Hydroxyindole analogs (8,9 and 12) showed much improved potency over PF74
2-Indolone analogs (16-24) decreased the Tm of CA hexamers
The potencies of α- and β-naphthyl analogs (33 and 27) were comparable to PF74
Acknowledgments
This research was supported by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, grant number R01AI120860 (to SGS and ZW). We thank the Minnesota Supercomputing Institute for molecular modeling resources.
Abbreviations
- HIV
human immunodeficiency virus
- CA
capsid protein
- CPSF6
cleavage and polyadenylation specific factor 6
- CypA
Cyclophilin A
- CANTD
CA N-terminal domain
- CACTD
CA C-terminal domain
- SAR
structure-activity-relationship
- HATU
1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- T3P
2,4,6-Tripropyl-1,3,5,2,4,6-trioxatriphosphorinane-2,4,6-trioxide
- DIPEA
N,N-Diisopropylethylamine
- TFA
Trifluoroacetic acid
- TSA
thermal shift assay
- HLM
human liver microsome
- MLM
mouse liver microsome
- EC50
50% effective concentration
- CC50
50% cytotoxicity concentration
References
- 1.AIDSinfo, ‘FDA-Approved HIV Medicines’ <https://aidsinfo.nih.gov/understanding-hiv-aids/fact-sheets/21/58/fda-approved-hiv-medicines> [Accessed March 2020.
- 2.Campbell EM, and Hope TJ, HIV-1 capsid: the multifaceted key player in HIV-1 infection, Nat Rev Microbiol, 13 (2015), 471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Sage V, Mouland AJ, and Valiente-Echeverria F, Roles of HIV-1 capsid in viral replication and immune evasion, Virus Res, 193 (2014), 116–29. [DOI] [PubMed] [Google Scholar]
- 4.Fassati A, Multiple roles of the capsid protein in the early steps of HIV-1 infection, Virus Res, 170 (2012), 15–24. [DOI] [PubMed] [Google Scholar]
- 5.Bell NM, and Lever AM, HIV Gag polyprotein: processing and early viral particle assembly, Trends Microbiol, 21 (2013), 136–44. [DOI] [PubMed] [Google Scholar]
- 6.Carnes SK, Sheehan JH, and Aiken C, Inhibitors of the HIV-1 capsid, a target of opportunity, Curr Opin HIV AIDS, 13 (2018), 359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thenin-Houssier S, and Valente ST, HIV-1 Capsid Inhibitors as Antiretroviral Agents, Curr Hiv Res, 14 (2016), 270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JY, Liu XY, and De Clercq E, Capsid (CA) Protein as a Novel Drug Target: Recent Progress in the Research of HIV-1 CA Inhibitors, Mini-Rev Med Chem, 9 (2009), 510–18. [DOI] [PubMed] [Google Scholar]
- 9.Ganser BK, Li S, Klishko VY, Finch JT, and Sundquist WI, Assembly and analysis of conical models for the HIV-1 core, Science, 283 (1999), 80–3. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Hill CP, Sundquist WI, and Finch JT, Image reconstructions of helical assemblies of the HIV-1 CA protein, Nature, 407 (2000), 409–13. [DOI] [PubMed] [Google Scholar]
- 11.Rankovic S, Ramalho R, Aiken C, and Rousso I, PF74 Reinforces the HIV-1 Capsid To Impair Reverse Transcription-Induced Uncoating, J Virol, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambrose Z, and Aiken C, HIV-1 uncoating: connection to nuclear entry and regulation by host proteins, Virology, 454–455 (2014), 371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita M, and Engelman AN, Capsid-Dependent Host Factors in HIV-1 Infection, Trends Microbiol, 25 (2017), 741–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayah DM, Sokolskaja E, Berthoux L, and Luban J, Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1, Nature, 430 (2004), 569–73. [DOI] [PubMed] [Google Scholar]
- 15.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, and Sodroski J, The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys, Nature, 427 (2004), 848–53. [DOI] [PubMed] [Google Scholar]
- 16.Achuthan V, Perreira JM, Sowd GA, Puray-Chavez M, McDougall WM, Paulucci-Holthauzen A, Wu X, Fadel HJ, Poeschla EM, Multani AS, Hughes SH, Sarafianos SG, Brass AL, and Engelman AN, Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration, Cell Host Microbe, 24 (2018), 392–404 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejarano DA, Peng K, Laketa V, Borner K, Jost KL, Lucic B, Glass B, Lusic M, Mueller B, and Krausslich HG, HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex, Elife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodward CL, Prakobwanakit S, Mosessian S, and Chow SA, Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1, J Virol, 83 (2009), 6522–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matreyek KA, Yucel SS, Li X, and Engelman A, Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity, PLoS Pathog, 9 (2013), e1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buffone C, Martinez-Lopez A, Fricke T, Opp S, Severgnini M, Cifola I, Petiti L, Frabetti S, Skorupka K, Zadrozny KK, Ganser-Pornillos BK, Pornillos O, Di Nunzio F, and Diaz-Griffero F, Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells, J Virol, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dharan A, Talley S, Tripathi A, Mamede JI, Majetschak M, Hope TJ, and Campbell EM, KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection, PLoS Pathog, 12 (2016), e1005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meehan AM, Saenz DT, Guevera R, Morrison JH, Peretz M, Fadel HJ, Hamada M, van Deursen J, and Poeschla EM, A cyclophilin homology domain-independent role for Nup358 in HIV-1 infection, PLoS Pathog, 10 (2014), e1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, and Diaz-Griffero F, MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1, Retrovirology, 11 (2014), 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu B, Pan Q, and Liang C, Role of MxB in Alpha Interferon-Mediated Inhibition of HIV-1 Infection, J Virol, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke EK, Yuan HE, and Luban J, Specific incorporation of cyclophilin A into HIV-1 virions, Nature, 372 (1994), 359–62. [DOI] [PubMed] [Google Scholar]
- 26.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, and Gottlinger HG, Functional association of cyclophilin A with HIV-1 virions, Nature, 372 (1994), 363–5. [DOI] [PubMed] [Google Scholar]
- 27.Kim K, Dauphin A, Komurlu S, McCauley SM, Yurkovetskiy L, Carbone C, Diehl WE, Strambio-De-Castillia C, Campbell EM, and Luban J, Cyclophilin A protects HIV-1 from restriction by human TRIM5alpha, Nat Microbiol, 4 (2019), 2044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novikova M, Zhang Y, Freed EO, and Peng K, Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle, Virol Sin, 34 (2019), 119–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed EO, HIV-1 assembly, release and maturation, Nat Rev Microbiol, 13 (2015), 484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, and Sarafianos SG, STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability, Science, 349 (2015), 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemke CT, Titolo S, von Schwedler U, Goudreau N, Mercier JF, Wardrop E, Faucher AM, Coulombe R, Banik SS, Fader L, Gagnon A, Kawai SH, Rancourt J, Tremblay M, Yoakim C, Simoneau B, Archambault J, Sundquist WI, and Mason SW, Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein, J Virol, 86 (2012), 6643–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ternois F, Sticht J, Duquerroy S, Krausslich HG, and Rey FA, The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor, Nat Struct Mol Biol, 12 (2005), 678–82. [DOI] [PubMed] [Google Scholar]
- 33.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao JA, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, and Butler SL, HIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention, Plos Pathogens, 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, and Yeager M, X-ray structures of the hexameric building block of the HIV capsid, Cell, 137 (2009), 1282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, and Zhang P, Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics, Nature, 497 (2013), 643–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machara A, Lux V, Kozisek M, Grantz Saskova K, Stepanek O, Kotora M, Parkan K, Pavova M, Glass B, Sehr P, Lewis J, Muller B, Krausslich HG, and Konvalinka J, Specific Inhibitors of HIV Capsid Assembly Binding to the C-Terminal Domain of the Capsid Protein: Evaluation of 2-Arylquinazolines as Potential Antiviral Compounds, J Med Chem, 59 (2016), 545–58. [DOI] [PubMed] [Google Scholar]
- 37.Thenin-Houssier S, de Vera IM, Pedro-Rosa L, Brady A, Richard A, Konnick B, Opp S, Buffone C, Fuhrmann J, Kota S, Billack B, Pietka-Ottlik M, Tellinghuisen T, Choe H, Spicer T, Scampavia L, Diaz-Griffero F, Kojetin DJ, and Valente ST, Ebselen, a Small-Molecule Capsid Inhibitor of HIV-1 Replication, Antimicrob Agents Chemother, 60 (2016), 2195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, and Hill CP, Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein, J Mol Biol, 373 (2007), 355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goudreau N, Lemke CT, Faucher AM, Grand-Maitre C, Goulet S, Lacoste JE, Rancourt J, Malenfant E, Mercier JF, Titolo S, and Mason SW, Novel inhibitor binding site discovery on HIV-1 capsid N-terminal domain by NMR and X-ray crystallography, ACS Chem Biol, 8 (2013), 1074–82. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z, Pan Q, Liang Z, Qiao W, Cen S, and Liang C, The highly polymorphic cyclophilin A-binding loop in HIV-1 capsid modulates viral resistance to MxB, Retrovirology, 12 (2015), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamorte L, Titolo S, Lemke CT, Goudreau N, Mercier JF, Wardrop E, Shah VB, von Schwedler UK, Langelier C, Banik SSR, Aiken C, Sundquist WI, and Mason SW, Discovery of Novel Small-Molecule HIV-1 Replication Inhibitors That Stabilize Capsid Complexes, Antimicrob Agents Ch, 57 (2013), 4622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yant SR, Mulato A, Hansen D, Tse WC, Niedziela-Majka A, Zhang JR, Stepan GJ, Jin D, Wong MH, Perreira JM, Singer E, Papalia GA, Hu EY, Zheng J, Lu B, Schroeder SD, Chou K, Ahmadyar S, Liclican A, Yu H, Novikov N, Paoli E, Gonik D, Ram RR, Hung M, McDougall WM, Brass AL, Sundquist WI, Cihlar T, and Link JO, A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model, Nat Med, 25 (2019), 1377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito A, Ferhadian D, Sowd GA, Serrao E, Shi J, Halambage UD, Teng S, Soto J, Siddiqui MA, Engelman AN, Aiken C, and Yamashita M, Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74, J. Virol, 90 (2016), 5808–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Zhou J, Shah VB, Aiken C, and Whitby K, Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization, J Virol, 85 (2011), 542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L, Huang T, Dick A, Meuser ME, Zalloum WA, Chen CH, Ding X, Gao P, Cocklin S, Lee KH, Zhan P, and Liu X, Design, synthesis and structure-activity relationships of 4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as novel HIV-1 capsid inhibitors with promising antiviral activities, Eur J Med Chem, 190 (2020), 112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu G, Zalloum WA, Meuser ME, Jing L, Kang D, Chen CH, Tian Y, Zhang F, Cocklin S, Lee KH, Liu X, and Zhan P, Discovery of phenylalanine derivatives as potent HIV-1 capsid inhibitors from click chemistry-based compound library, Eur J Med Chem, 158 (2018), 478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun L, Dick A, Meuser ME, Huang T, Zalloum WA, Chen C-H, Cherukupalli S, Xu S, Ding X, Gao P, Kang D, Clercq ED, Pannecouque C, Cocklin S, Lee K-H, Liu X, and Zhan P, Design, Synthesis, and Mechanism Study of Benzenesulfonamide-Containing Phenylalanine Derivatives as Novel HIV-1 Capsid Inhibitors with Improved Antiviral Activities, J. Med. Chem, 63 (2020), 4790–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Casey MC, Vernekar SKV, Do HT, Sahani RL, Kirby KA, Du H, Hachiya A, Zhang H, Tedbury PR, Xie J, Sarafianos SG, and Wang Z, Chemical profiling of HIV-1 capsid-targeting antiviral PF74, Eur. J. Med. Chem, 200 (2020), 112427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernekar SKV, Sahani RL, Casey MC, Kankanala J, Wang L, Kirby KA, Du H, Zhang H, Tedbury PR, Xie J, Sarafianos SG, and Wang Z, Toward Structurally Novel and Metabolically Stable HIV-1 Capsid-Targeting Small Molecules, Viruses, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu JP, Francis AC, Meuser ME, Mankowski M, Ptak RG, Rashad AA, Melikyan GB, and Cocklin S, Exploring Modifications of an HIV-1 Capsid Inhibitor: Design, Synthesis, and Mechanism of Action, J Drug Des Res, 5 (2018). [PMC free article] [PubMed] [Google Scholar]
- 51.Wacher VJ, Silverman JA, Zhang Y, and Benet LZ, Role of P-glycoprotein and cytochrome P450 3A in limiting oral absorption of peptides and peptidomimetics, J Pharm Sci, 87 (1998), 1322–30. [DOI] [PubMed] [Google Scholar]
- 52.Xu L, and Desai MC, Pharmacokinetic enhancers for HIV drugs, Curr Opin Investig Drugs, 10 (2009), 775–86. [PubMed] [Google Scholar]
- 53.Midde NM, Gong YQ, Cory TJ, Li JH, Meibohm B, Li WH, and Kumar S, Influence of Ethanol on Darunavir Hepatic Clearance and Intracellular PK/PD in HIV-Infected Monocytes, and CYP3A4-Darunavir Interactions Using Inhibition and in Silico Binding Studies, Pharm Res-Dordr, 34 (2017), 1925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buss N, Snell P, Bock J, Hsu A, and Jorga K, Saquinavir and ritonavir pharmacokinetics following combined ritonavir and saquinavir (soft gelatin capsules) administration, Br J Clin Pharmacol, 52 (2001), 255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu LH, Liu HT, Murray BP, Callebaut C, Lee MS, Hong A, Strickley RG, Tsai LK, Stray KM, Wang YJ, Rhodes GR, and Desai MC, Cobicistat (GS-9350): A Potent and Selective Inhibitor of Human CYP3A as a Novel Pharmacoenhancer, Acs Med Chem Lett, 1 (2010), 209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, and Ellestad G, Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery, Anal Biochem, 332 (2004), 153–9. [DOI] [PubMed] [Google Scholar]
- 57.Miyazaki Y, Doi N, Koma T, Adachi A, and Nomaguchi M, Novel In Vitro Screening System Based on Differential Scanning Fluorimetry to Search for Small Molecules against the Disassembly or Assembly of HIV-1 Capsid Protein, Front Microbiol, 8 (2017), 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, and Salemme FR, High-density miniaturized thermal shift assays as a general strategy for drug discovery, J Biomol Screen, 6 (2001), 429–40. [DOI] [PubMed] [Google Scholar]
- 59.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, and Martin MA, Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone, J Virol, 59 (1986), 284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrödinger Small-Molecule Drug Discovery Suite 2019–1, Schrödinger, LLC, New York, NY, 2019. [Google Scholar]
- 61.Schrödinger Release 2019–1: Maestro, Schrödinger, LLC, New York, NY, 2019. [Google Scholar]
- 62.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, and Sherman W, Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments, J Comput Aid Mol Des, 27 (2013), 221–34. [DOI] [PubMed] [Google Scholar]
- 63.Jorgensen WL, Maxwell DS, and TiradoRives J, Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids, J Am Chem Soc, 118 (1996), 11225–36. [Google Scholar]
- 64.Schrödinger Release 2019–1: LigPrep, Schrödinger, LLC, New York, NY, 2019. [Google Scholar]
- 65.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, and Shenkin PS, Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J Med Chem, 47 (2004), 1739–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


