Abstract
High-resolution Ca2+ imaging to study cellular Ca2+ behaviors has led to the creation of large datasets with a profound need for standardized and accurate analysis. To analyze these datasets, spatio-temporal maps (STMaps) that allow for 2D visualization of Ca2+ signals as a function of time and space are often used. Methods of STMap analysis rely on a highly arduous process of user defined segmentation and event-based data retrieval. These methods are often time consuming, lack accuracy, and extremely variable between users. We designed a novel automated machine-learning based plugin for the analysis of Ca2+ STMaps (STMapAuto). The plugin includes optimized tools for Ca2+ signal preprocessing, automated segmentation, and automated extraction of key Ca2+ event information such as: duration, spatial spread, frequency, propagation angle, and intensity in a variety of cell types including the Interstitial cells of Cajal (ICC). The plugin is fully implemented in Fiji and able to accurately detect and expeditiously quantify Ca2+ transient parameters from ICC. The plugin’s speed of analysis of large-datasets was 197-fold faster than the commonly used single pixel-line method of analysis. The automated machine-learning based plugin described dramatically reduces opportunities for user error and provides a consistent method to allow high-throughput analysis of STMap datasets.
Keywords: Ca2+ imaging analysis, Ca2+ signaling, Interstitial cell of Cajal
1. Introduction
Intracellular Ca2+ signaling mediates diverse cellular functions such as muscle contraction, metabolism, neurotransmitter release, and activation of nuclear transcription factors.[1, 2] Tight temporal and spatial control of Ca2+ signaling is required to sustain these distinct signaling mechanisms. This control is achieved by maintaining intracellular and extracellular Ca2+ pools through Ca2+ release, Ca2+ influx and extrusion mechanisms. Several cellular components including: Ca2+ pumps, Ca2+ transporters, and Ca2+ store channels are integral participants in Ca2+ signaling and homeostasis.[1, 3–6] Intracellular Ca2+ signals can vary in terms of shape, kinetics, firing frequency, origin, and spatial propagation. Ca2+ signals can be temporally and spatially localized (e.g. sparks, puffs and sparklets) that can occur on tens of millisecond timescales and spread less than 5 μm. [7–12] Ca2+ signals may also take the form of Ca2+ waves that propagate across cells or cell-to-cell and spread for distances of 100 μm or more and last for several seconds or even minutes. [10, 13–17]
Spatial and temporal parameters inherent to intracellular Ca2+ signals such as: duration, spatial spread, amplitude, frequency, and propagation angle can provide the required biological information to encode downstream cellular signaling. [18–22] Thus, Ca2+ imaging and the analysis of intracellular Ca2+ dynamics allows for effective study of a myriad of complex and physiologically essential cellular behaviors. Spatio-Temporal Maps (STMap) are a common method used to analyze and quantify Ca2+ dynamics in a range of both single cell and intact tissues by plotting intracellular Ca2+ transients as a function of space occupied over time. [10, 22–30] STMaps can thus provide a platform to effectively quantify cellular Ca2+ dynamics and its associated parameters: duration, spatial spread, frequency, event angle, and event intensity. [31–35]
Extracting quantifiable measurements of Ca2+ transients from STMaps is challenging as this information is most often manually defined in traditional forms of analysis. Opportunities for user error are prevalent, which translates to inaccuracy when quantifying STMap Ca2+ events. These fundamental issues arise from the requirement of user designation of single pixel measurement lines that are assumed to be representative of the entire Ca2+ event. This user-dependent manual process is highly variable, time consuming, and labor intensive.
To overcome this inconsistency, we developed an efficient software plugin method implemented in Fiji to achieve automated, fast and accurately characterize spatio-temporal Ca2+ activity patterns. The plugin incorporates the Waikato Environment for Knowledge Analysis (Weka) as the segmentation framework for detection and analysis of STMaps Ca2+ signals. [36] The plugin consists of modules for STMap preprocessing, automated event segmentation into regions of interest (ROIs) corresponding to single Ca2+ events, and automated data output. The machine-learning based approach in combination with automated processes is a significant step towards a robust standardization and high-throughput analysis of cellular Ca2+ dynamics.
To test our methodology, we have analyzed Ca2+ signaling in interstitial cells of Cajal (ICC). ICC act as pacemakers and neuroeffector cells in the gastrointestinal (GI) tract that display dynamic intracellular Ca2+ signaling. [37] [38–40] Ca2+ transients in ICC are fundamental in regulation of intestinal excitability and motility. The small intestine possesses two major classes of ICC that have a distinct morphology and Ca2+ dynamics: the Myenteric ICC (ICC-MY) and Deep Muscular Plexus ICC (ICC-DMP). The ICC-MY lay within the myenteric plexus, between the circular and longitudinal smooth muscle layers. [41, 42] ICC-MY function as pacemaker cells throughout the gut and generate electrical slow wave activity that paces and coordinates the regular patterns of GI smooth muscle contraction. [43–46] In contrast, ICC-DMP are found in close relation to motor neuron terminals in the deep muscular plexus.[42] ICC-DMP can modulate and transduce signals from the enteric motor neurons. [47–50] Interestingly, ICC-MY and ICC-DMP are unique from one another in their morphology and Ca2+ dynamics. ICC-MY networks exhibit entrained and temporally clustered Ca2+ signals that propagate from cell-to-cell and produce a tissue wide Ca2+ wave. [51] ICC-DMP Ca2+ signals manifest as brief and spatially localized Ca2+ release events that occur stochastically. [41]
We used intracellular Ca2+ signals recorded from ICC-MY and ICC-DMP in situ to test our Fiji plugin to analyze Ca2+ transients (STMaps) in an automated fashion. These cells provide an ideal model for this testing as they exhibit dynamic Ca2+ events that vary in intensity, spatial spread and firing behaviors, thus testing the ability of our plugin to detect a range of cellular activity (localized Ca2+ events to propagating Ca2+ waves) amongst different cell types rather than a single behavior.
2. Materials and Methods
2.1. Animals
All animals used and the protocols carried out in this study were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All procedures were approved by the Institutional Animal Use and Care Committee at the University of Nevada, Reno.
GCaMP6f-floxed mice (B6.129S-Gt(ROSA)26Sortm38(CAG-GCaMP3)Hze/J) and their wild-type siblings (C57BL/6) were acquired from Jackson Laboratories (Bar Harbor, MN, USA) and crossed with Kit-Cre mice (c-Kit+/Cre-ERT2), provided by Dr. Dieter Saur (Technical University Munich, Munich, Germany). Kit-Cre-GCaMP6f mice were injected with tamoxifen at 6–8 weeks of age (2 mg for three consecutive days), as previously described [18]. 15 days after tamoxifen injection, Kit-Cre-GCaMP3 mice were anaesthetized by isoflurane inhalation (Baxter, Deerfield, IL, USA) and killed by cervical dislocation.
2.2. Tissue preparation
jejunum tissues were removed from animals and cut into 2 cm in length segments and incubated in Krebs-Ringer bicarbonate solution (KRB) As previously described [18]. Tissues were cut along the mesenteric region and contents were removed. Sharp dissection was used to remove both The mucosa and submucosal layers, and the remaining muscle tunica muscularis was secured into a 60 mm Sylgard coated dish and pinned flat with the serosal side up.
2.3. Calcium imaging
Muscle sheets isolated from the jejunum (~5.0 × 10.0 mm) were pinned down and perfused with 37°C KRB solution. Tissues were equilibration for a period of 1–2 hours. As previously described [18],31; We used a spinning-disk confocal microscope (CSU-W1 spinning disk; Yokogawa Electric Corporation) for our Ca2+ imaging experiments. The confocal head is connected to Nikon Eclipse FN1 microscope equipped with a 40× 0.8 NA CFI Fluor lens (Nikon instruments INC, NY, USA). Laser at 488 nm wavelength was directed using a Borealis system (ANDOR Technology, Belfast, UK). EMCCD Camera (Andor iXon Ultra; ANDOR Technology, Belfast, UK) was used to capture the GCaMP6f emission. Images were acquired at 33 frames per second using MetaMorph software (Molecular Devices INC, CA, USA). Nicardipine (100 nM) was used during the imaging experiments to minimize contractile artifacts.
2.4. Calcium event analysis
Analysis of Ca2+ activity in ICC was performed as previously described. [18],31 Briefly, movies of Ca2+ activity in ICC (30 s long) were converted to a stack of TIFF (tagged image file format) images and imported into custom software (Volumetry G8c) for analysis. Whole cell ROIs were used to generate STMaps of Ca2+ activity in individual ICC. STMaps presented in the results were generated by rotating image stacks so that ICC-DMP/MY were oriented vertically. Single cells from the FOV were masked using a flood-fill routine and ST maps of Ca2+-induced fluorescence changes (averaged across the diameter of the cell within the mask) were constructed (Baker et al., 2016; Drumm et al., 2019; Drumm et al., 2017; Hennig et al., 1999). STMaps were then imported as TIFF files into Fiji (version 2.0.0-rc-69/1.52, National Institutes of Health, MD, USA, https://fiji.sc/ for pixel-line quantification analysis of Ca2+ events as previously described by. [29],31 The pixel-line method of STMap analysis is a series of measurements performed within Fiji to quantify the behavior of Ca2+ represented by STMaps. To detect the mean intensity or amplitude of a single Ca2+, a single-pixel line was drawn through each Ca2+ event, such to bisect the event. The “plot profile” function within Fiji was then selected to generate distinct intensity profiles for each Ca2+ event. Here, the peak intensity represents the amplitude of the event. The x-value(s) at the y-value halfway point to peak amplitude in the plot profile were then recorded. The difference between the two generated x-values (s) from a single plot profile was the source of Full Duration at Half Maximum (FDHM) as visualized in Figure 1. Additionally, individual single pixel lines were drawn over STMaps in Fiji such that the “measure” function could determine the x (s) and y (um) dimensions of an individual Ca2+event.
Figure 1: Generation of spatio-temporal maps (STMaps).
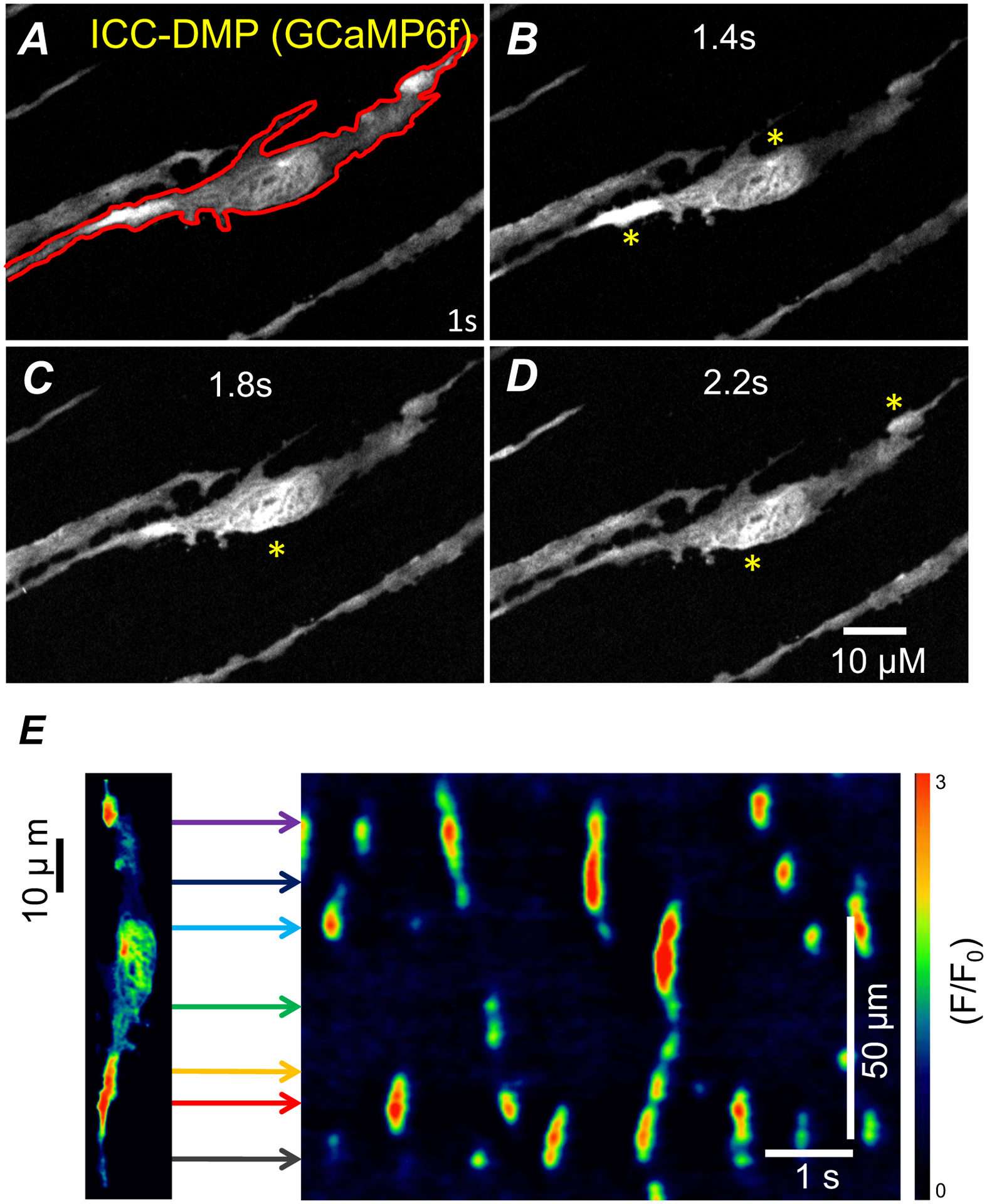
A) Representative image of ICC-DMP from the small intestine of a Kit-Cre-GCaMP6f mouse in situ. A single ICC-DMP within the FOV outlined (red) defines cellular ROI for STMap. (A-D) Representative time series of Ca2+ transients from ICC-DMP. Asterisks (yellow) indicate transient Ca2+ events. E) Representative color-coded STMap of Ca2+ activity in ICC-DMP. Horizontal lines (colored) from cell correspond to locations of Ca2+ sites within the cell plotted on STMap.
2.5. Statistics
Statistics of Ca2+ activity was performed as previously described [18],31. Briefly, Ca2+ event frequency in ICC was expressed as the number of events fired per cell per second (sec−1). Ca2+ event amplitude was expressed as F/F0, the duration of Ca2+ events were expressed as full duration (ms), and Ca2+ event spatial spread was expressed as μm of cell propagated per Ca2+ event. Unless otherwise stated, data is represented as mean ± standard error (S.E.M.). Statistical analysis was performed using either a student’s t-test or with an ANOVA with a Dunnett post hoc test where appropriate. In all statistical analyses, P<0.05 was taken as significant. P values <0.0001 are represented by four asterisks (****). When describing data throughout the text, n refers to the number of STMaps used in that dataset.
2.6. Download and installation
Fiji (Fiji) download and install is required for use of the plugin. Fiji can be downloaded from: https://Fiji.nih.gov/ij/download.html. The plugin and classifiers can be accessible via this link: https://github.com/gdelvalle99/STMapAuto or from our lab website: https://med.unr.edu/directory/sal-baker?v=bio#Biography. Once downloaded, the plugin “STMapAuto.jar” is moved to the “plugins” folder of Fiji for installation. To use the plugin, open Fiji and choose “STMapAuto” from the “Plugins” tab on the menu bar.
2.7. Plugin development
The code for The Plugin was written in Java release version: 9. The plugin incorporates Fiji Application Program Interfaces (APIs) provided by the National Institutes of Health, U.S. Department of Health and Human Services. The plugin uses features available within the Fiji version: 2.0.0-rc-69/1.52 and trainable weka segmentation for microscopy designed by [36].
2.8. Plugin testing
The classifiers used in the plugin were manually trained by two groups of 100 STMaps from ICC-MY and ICC-DMP. Final classifiers were optimized through user experimentation and as detailed by. [36] Testing of the plugin with 40 unseen STMaps was performed with a Mac Pro desktop (Mac Pro 2010; Apple inc; USA) for further assessment.
3. Results
3.1. STMaps are generated for representation of Ca2+ events from ICC.
Imaging ICC from small intestine (SI) muscles of Kit-Cre-GCaMP6f mice using spinning-disk confocal microscopy allowed monitoring of Ca2+ signals in situ with high spatial resolution (Figure 1 A). One cell type within the SI, ICC-DMP, exhibited variable and complex patterns of Ca2+ signals that occurred in a stochastic fashion (Figure 1A–D). Therefore, to effectively quantify the changes in Ca2+ transients in ICC-DMP over time, STMaps were used. STMaps allow for a more complete representation of individual cell Ca2+ signals through a 2D image that describes both time (x-axis) and space (y-axis) (Figure 1 E). Within the STMap, Ca2+ events were viewed as discrete events with different intensities (color coded; Figure 1E), and their features were quantified to describe the cellular Ca2+ dynamics and behaviors.
3.2. Current limitations of STMap analysis need resolution.
Data extraction and quantification of Ca2+ signals from STMaps is challenging due to the complex nature of the Ca2+ events, their variability and large number. Current methods of STMaps analysis focus on extracting the Ca2+ transient parameters such as duration, spatial spread, frequency, and intensity. [31–35] To retrieve these data parameters, a common research practice in STMap analysis requires user defined drawing of representative single pixel-lines through individual Ca2+ events (Figure 2B). These pixel-lines are used to produce an intensity plot profile that represents Ca2+ event amplitude as a function of time (Figure 2C). Amplitude, or peak event intensity, and event duration are both determined from the plot profile by manual measurement of profile peak and full duration at half maximum (FDHM) (Figure 2C). To characterize the spatial spread of a Ca2+ event, another pixel-line is drawn to measure the space occupied by the Ca2+ event (Figure 2D). Although these methods are useful for data generation, we recognized the opportunity for user error, inconsistency, and subjectivity in single-pixel line STMap analysis (Figure 2A–H).
Figure 2: Analysis of Ca2+ events in ICC-DMP using STMaps.
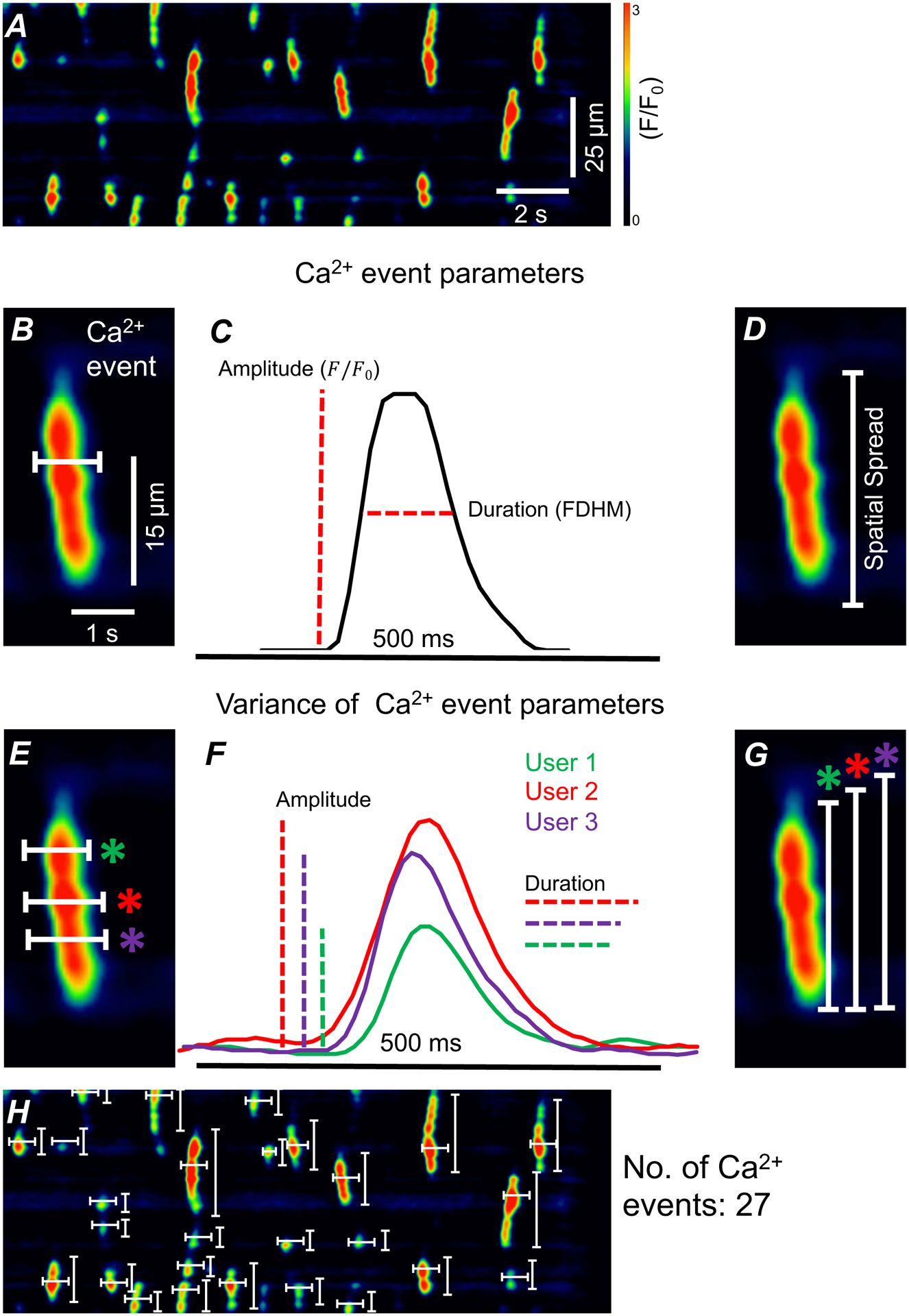
A) Representative STMap of Ca2+ transients in ICC-DMP. B) Enlarged single Ca2+ event from panel A. Representative single pixel-line drawn (white) through Ca2+ event in Fiji.. Line is drawn parallel to time (x-axis) C) Plot profile generated from single pixel-line from panel B. Amplitude and event duration (full duration of half maximum) are measured from plot profile. D) Enlarged single Ca2+ event from panel A with representative measurement of Ca2+ event spatial spread (white). E) Single pixel-lines (white) drawn by three users across Ca2+ event. Users 1,2, and 3 are indicated by green, red, and purple respectively. F) Plot profile generated by the three user defined pixel-lines from panel E. Variation is present in both amplitude and event duration. G) Variable spatial spread present in three user defined single pixel-line measurements (white) of single Ca2+ event. H) 54 single pixel-lines (white) required to analyze event duration, spatial spread, and amplitude of 27 Ca2+ events from ICC-DMP STMap. Scale bars in panel A and panel B apply to panels H and D, E, G respectively.
To highlight this problem, single pixel-lines drawn by three different users may be drawn in separate locations on an individual STMap Ca2+ events (Figure 2E). The variation in the location of line drawing can lead to variance of Ca2+ event data extraction which includes event duration (user 1 = 0.324 ms, user 2 = 0.471 ms, user 3 = 0.546 ms), event intensity (user 1 = 1.50, user 2 = 1.76 user 3 = 0.91 F/F0), and spatial spread (user 1 = 23.85 μm, user 2 = 25.35 μm user 3 = 26.85 μm) (Figure 2 E–G). Although plausible for a single Ca2+ event, quantification of many Ca2+ events within an STMap requires the user to accurately bisect each individual Ca2+ event manually. This means that for a typical STMap of ICC-DMP (30 s recording), which can contain an average of ~50 Ca2+ events, users can expect to draw ~100 single-pixel lines for both spatial and temporal axis for quantification of Ca2+ event parameters (Figure 2H). The large amount of user defined lines needed for analysis is a significant area of concern, as it increasingly exposes STMaps to high risk of user error and inconsistency. In addition, single pixel line-based analysis is an incredibly laborious task, often requiring several analysts for larger datasets. Thus, current forms of analysis are constrained by user error, data inaccuracy, and prohibitive speed of analysis.
3.3. “STMapAuto” plugin provides automation, consistency, speed, and accuracy in STMap analysis.
These constraints were ameliorated by developing an efficient plugin written using the JavaScript language that integrates the analysis tools made available within Fiji into a unified and robust pipeline. The main contribution of the proposed software is to provide a platform for STMap analysis by enhancing accuracy of detection of Ca2+ events, ensuring consistent results, automating Ca2+ event parameterization output and enabling high-throughput analysis of large datasets.
The plugin consists of three distinct automated phases: 1) STMap Preprocessing 2) Weka segmentation and Thresholding 3) ROI Generation and Data Output (Figure 3). To detect and extract Ca2+ event parameters from STMaps from a cell of interest, the plugin gives the user the option to import single or multiple STMaps to Fiji. The user can then calibrate the STMaps and determine Ca2+event size detection parameters. This step removes background noise signals from detection and allows the researcher to study events within a certain dimensional range (e.g. space and time). Following the preprocessing step, the subsequent processes leading to data extraction are entirely automated.
Figure 3: Representative workflow of automated Plugin based STMap analysis.
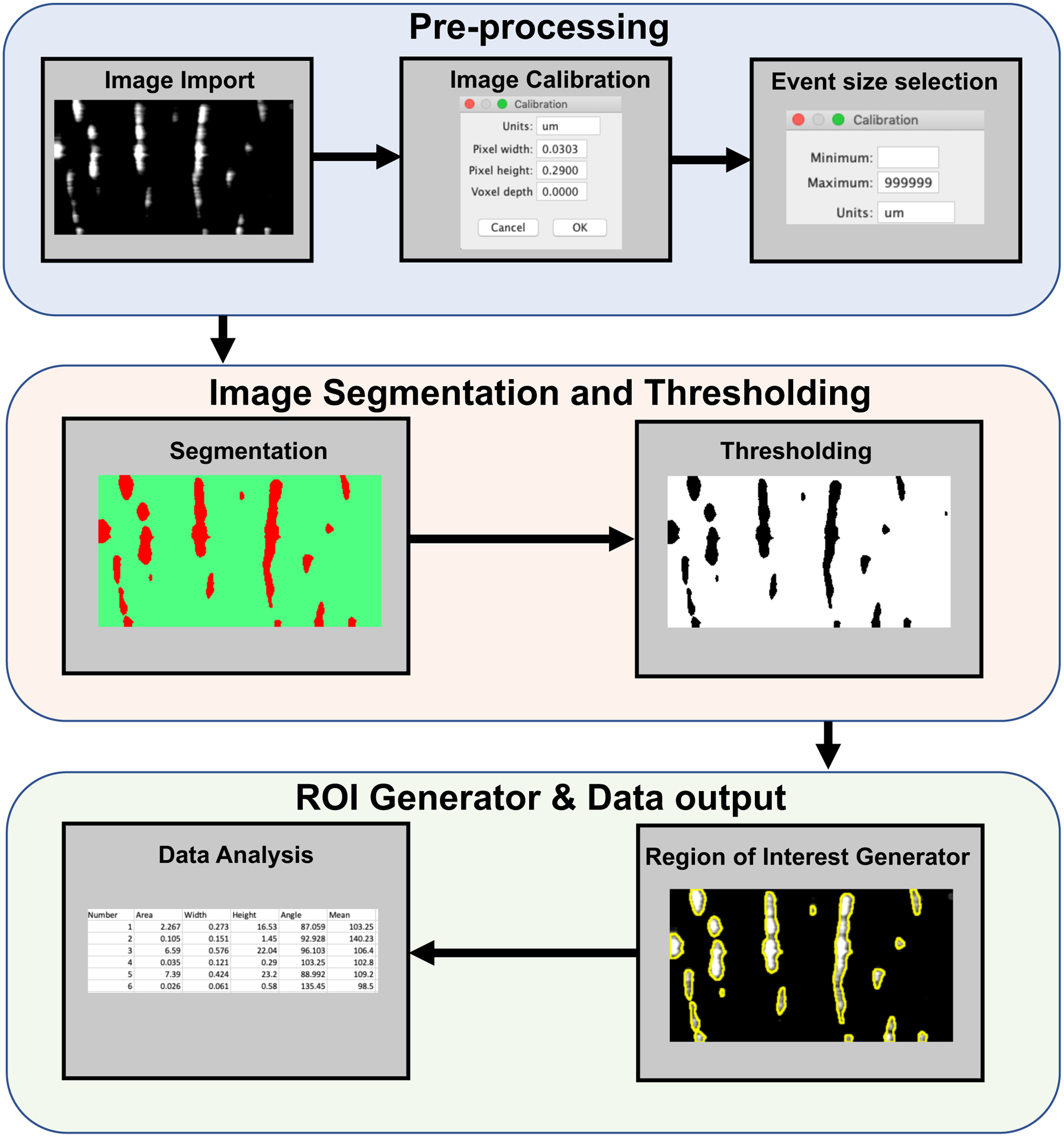
STMap is imported into Plugin for spatio-temporal calibration and Ca2+event detection selection. (Image Segmentation and Thresholding) STMap is segmented through Trainable Weka Segmentation through one of two classifiers. Intermodes threshold is automatically applied to segmented image. (Region of Interest Generator) Plugin uses image threshold from segmented image to generate distinct ROIs. ROIs are then overlaid on original image. (Data extraction) Original non-colored image is used for data extraction through Fiji API and Ca2+ event parameters are exported to excel spread sheet.
3.4. Machine-learning approach and validation
We used machine-learning protocols to enhance the efficiency of the segmentation step of the plugin. The machine-learning classifier component of the plugin utilizes a trainable weka segmentation algorithm that contains a Fast-Random Forest technique which uses different extracted features such as Sobel filter, Gaussian blur, Membrane projections etc. as inputs to detect and segment contours and patterns of Ca2+ events in STMaps. [52–54] The Fast-Random Forest Classifier functions by generating unique uncorrelated trees whose predictions are compared to generate an overall model prediction. The integration of Fast-Random Forest in trainable weka segmentation functions by classifying pixels which are learned from extracted feature vectors.[36] The plugin uses Fast-Random Forest with 200 trees and 2 random features per node from a list of features extracted from: Gaussian Blur, Hessian, Membrane projections, Anisotropic diffusion Lipschitz, Gabor, Entropy, Sobel Filter, Difference of Gaussians, Variance, Bilateral, Kuwahara, Neighbors.
When training the weka segmentation algorithm, the user inputs annotated ground truths from STMaps to train the weka classifier. During the training phase, all completed within Fiji, the classifier gradually improves as more ground truths are stored for subsequent training. It is important to note that for pixel level segmentation, the classifier uses each pixel as a data point rather than the whole image as a single data point; 100 STMaps is a significant dataset as one idle image consists of 100×1000 pixels = 100,000 data points per image. Thus, large datasets were not required for classifier training. The machine learning approach allows for segmentation through trainable machine-learning that is adaptable to different datasets.
As our datasets consist of a mosaic of Ca2+ activity that includes both rhythmic and stochastic signals, we optimized two classifiers to accurately detect a variety of Ca2+behaviors. We split our dataset into train/test splits followed by a 5-fold cross validation of the training data. 100 STMaps were manually classified and saved to train the weka classifier. [36] The classifier training steps were completed on the backbone of the machine-learning trainable weka segmentation algorithm within Fiji. [36]
The plugin functions autonomously by first segmenting the STMap through one of two classifier models we optimized for STMap analysis: 1) Stochastic Ca2+ Event Classifier 2) Rhythmic Ca2+ Event Classifier (Figure 4 A–B). For STMaps that contain localized Ca2+ transient events, the Stochastic Ca2+ Event Classifier is most effective because it includes segmentation filters that account for a number of mechanisms including: Gaussian Blur, Hessian, Membrane Projections, Sobel Filter, Difference of Gaussians (Figure 4 Ai & Bi). For STMaps that contain a rhythmic oscillation of large Ca2+ events, the Rhythmic Ca2+ Event Classifier is recommended. This classifier uses the following filters to optimize event Ca2+ detection: Gaussian Blur, Hessian, Membrane projections, Anisotropic diffusion, Lipschitz, Gabor, Entropy, Sobel Filter, Difference of Gaussians, Variance, Bilateral, Kuwahara, Neighbors. By conducting these computations, the Ca2+ events within STMap are fully segmented (Figure 4 Aii & Bii). When the classifiers are applied appropriately, Ca2+ events in both STMaps can be segmented to generate a classified image with events clearly separated from background noise (Figure 4 Aii, Bii).
Figure 4: Creation of trainable event classifiers optimized for stochastic and rhythmic Ca2+ activity.
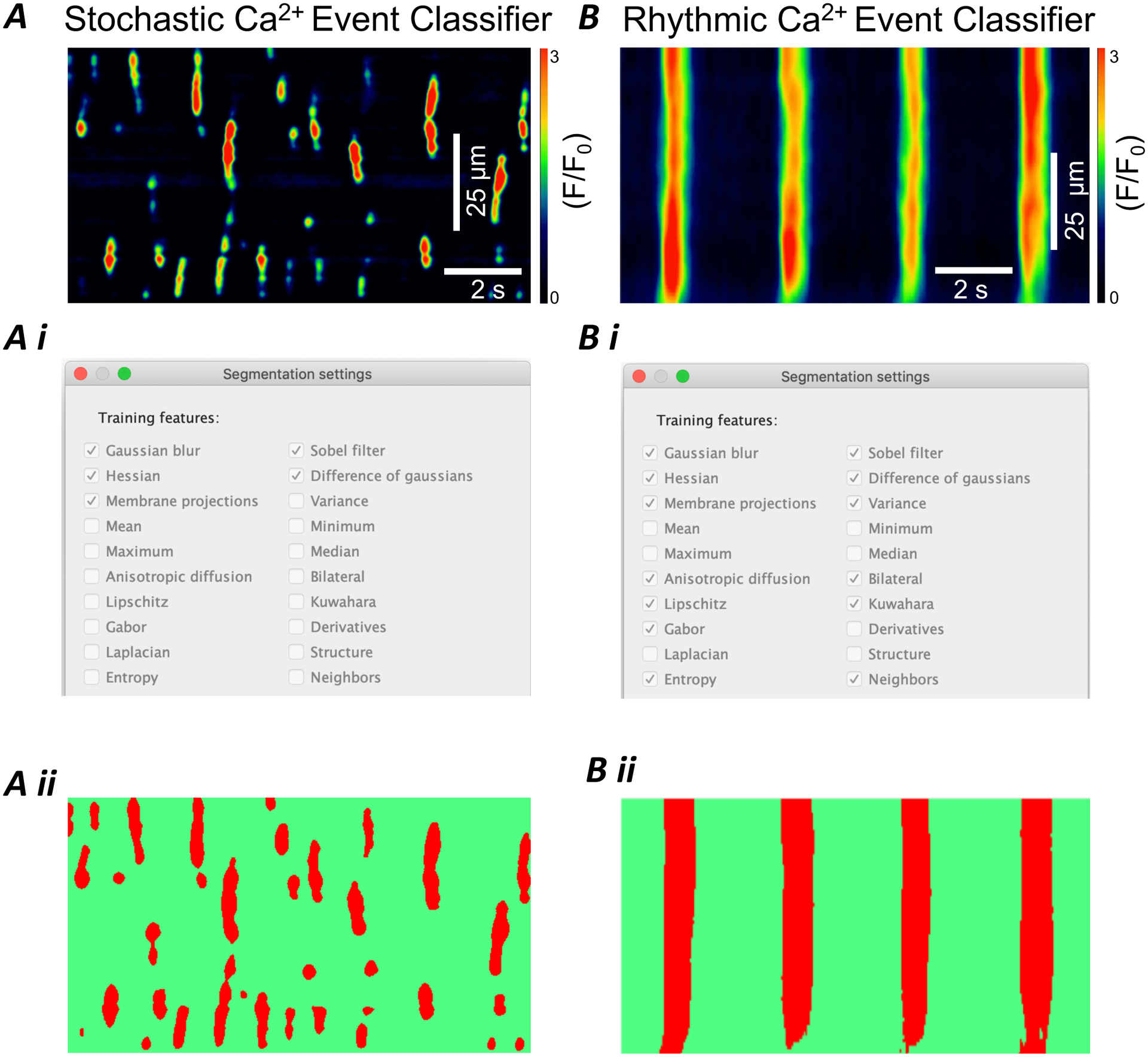
A) Representative STMap of stochastic Ca2+ transients from ICC-DMP Ai) Segmentation settings used by Ca2+ event stochastic classifier to optimize training of smaller, less defined Ca2+ events. Aii) Ca2+ event segmentation of STMap from panel A generated by plugin. Segmented events are highlighted (red) from background (green). B) Representative STMap of rhythmic Ca2+ transients from ICC-MY of the small intestine. Bi) Segmentation settings used by Ca2+ event rhythmic classifier to optimize training of large, rhythmic Ca2+ events Bii) Segmentation settings used by Ca2+ event rhythmic classifier to optimize training of larger, nosier, and rhythmic Ca2+ events. Scale bars in panel A and B also apply to panels Aii and Bii respectively.
To test our two unique classifiers, we split the data into train/test splits followed by a 5-fold cross validation of the training data. Although pixel-based accuracy could be used as a measure of validation, for any semantic segmentation task pixel-based accuracy has shown to be more error-prone. Moreover, it prioritizes background and foreground segmentation over object segmentation. For example, if the number of background pixels is around 80% and the model achieves a pixel-accuracy of 70% and largely classifies those background pixels, then our true aim to segment Ca2+ events (objects) is not achieved. That is why Mean Intersection Over Union (Mean-IOU; equation 1) is used as a measure to see how precisely ground truth masks overlap with predicted segmentation masks. The equation for Mean IOU is as follows:
| (1) |
Here, Nclass is the number of classes (class 0=background, class 1= Ca2+ events), Σi Nii the number of correctly classified pixels (true positives), ΣjNij is the number of wrongly classified pixels (false positives) and ΣjNji the number of pixels not wrongly classified (false negatives). We use a cross validation scheme for training on 100 samples of Ca2+ events: 50 samples for Rhythmic Ca2+ event detection and 50 for Stochastic Ca2+ event detection. The data is split into 5 equal portions for 5-fold cross validation (Table 1 and 2). For each fold, a classifier is trained from the beginning, trained on 40 unique samples, and validated on the remaining 10 samples. We found that a Rhythmic classifier (Fold 4) which was trained on samples 1–30 and 41–50 and then validated on samples 31–40 achieved a mean-IOU of 82.79% (Table1). For the Stochastic classifier, we used the same training procedure as followed for Rhythmic classifiers and trained on the samples 1–40 (Fold 1) and further validated on samples 41–50 to achieve a mean-IOU of 79.48% (Table 2). These classifiers were used for separate Rhythmic and Stochastic test data samples. The greatest mean-IOU scores achieved for test data of Rhythmic and Stochastic are 75.87 % and 76.69 % respectively.
Table 1:
Cross Validation Result for Rhythmic STMap Datasets
| Fold | Training Samples | Validation Samples | MeanIOU |
|---|---|---|---|
| 1 | 11–50 | 1–10 | 80.37% |
| 2 | 1–10,21–50 | 11–20 | 75.53% |
| 3 | 1–20,31–50 | 21–30 | 72.17% |
| 4 | 1–30,41–50 | 31–40 | 82.79% |
| 5 | 1–40 | 41–50 | 72.29% |
Table 2:
Cross Validation Result for Stochastic STMap Datasets
| Fold | Training Samples | Validation Samples | MeanIOU |
|---|---|---|---|
| 1 | 11–50 | 1–10 | 79.48% |
| 2 | 1–10,21–50 | 11–20 | 69.08% |
| 3 | 1–20,31–50 | 21–30 | 72.18% |
| 4 | 1–30,41–50 | 31–40 | 70.52% |
| 5 | 1–40 | 41–50 | 78.92% |
Following segmentation, the plugin automatically detects the segmented Ca2+ event areas by using the intermodes threshold function in Fiji (Figure 5 A–C). It is important to note that the proposed step functions by a histogram with two obvious relative modes, or data peaks and in our case: 2 overlapping Ca2+events.The threshold is then computed as half of the sum of 2 peaks. This step, when paired with initial detection, enables separation between two closely overlapping events and allows for proper Ca2+ event segmentation. The plugin then uses the thresholded Ca2+ events to generate distinct regions of interest (ROIs), while also incorporating the event size parameters selected upon image import. These ROIs are labeled in the sequence at which they appeared and with their cartesian coordinates on the STMap by the ROI manager (Figure 5 A–D). The plugin automatically overlays and saves the generated ROIs on the original STMap image (either grey scale or intensity color-coded) (Figure 5 E–F). The overlay steps are accomplished by automating a sequence of processes within the ROI Manager. This process is essential as it offers the user the opportunity to validate the segmentation process visually to ensure accuracy in Ca2+ event detection and segmentation steps. This method allows for accurate detection of Ca2+ events within STMaps without interference from background artifacts.
Figure 5: Generation of Ca2+ Events threshold and validation.
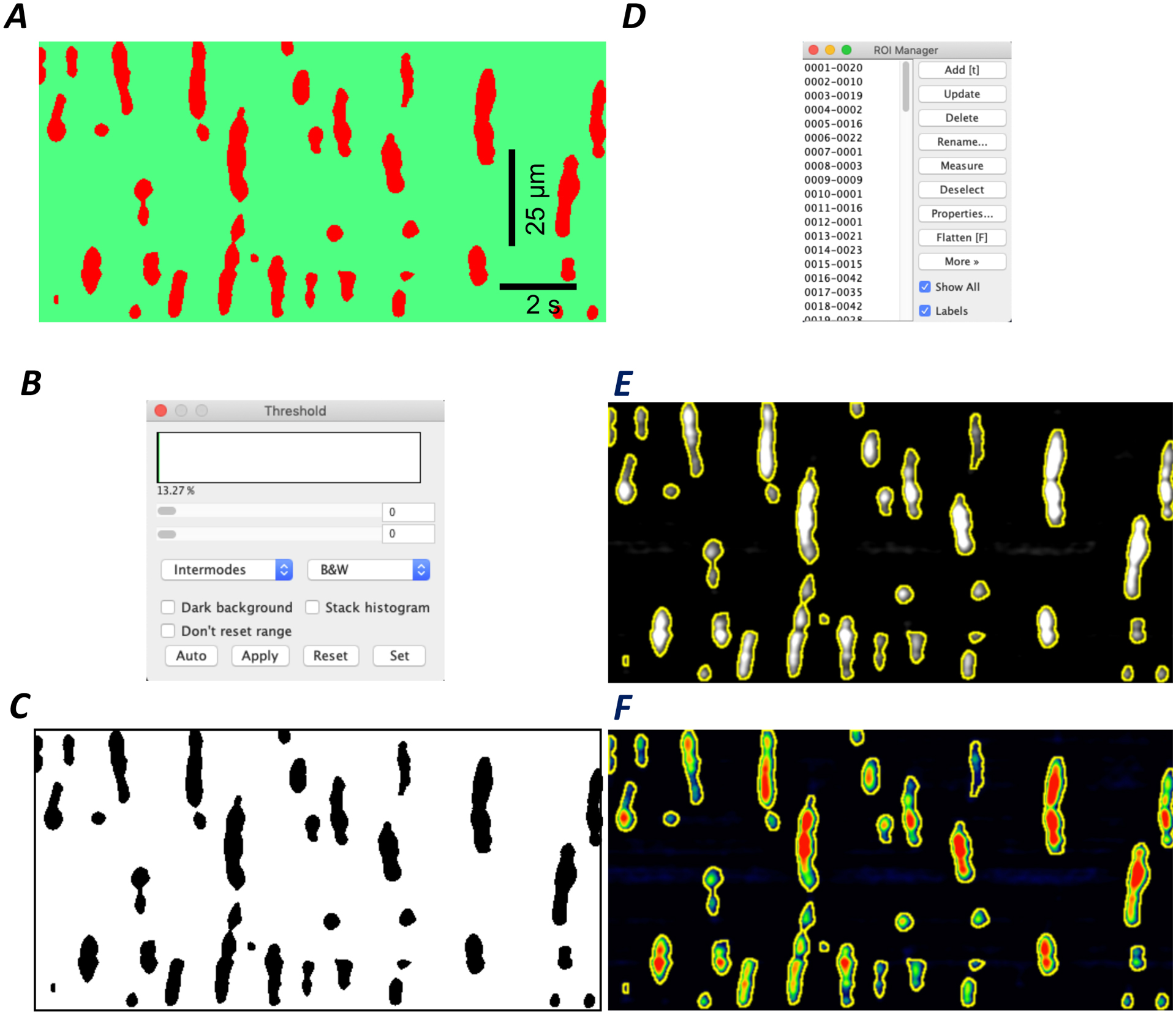
A) Representative image of a segmented STMap of an ICC-DMP from the small intestine. Scale bars in A applies to C, E and F. B) Intermodes threshold used to further segment closely linked Ca2+ events C) Resulting thresholded STMap: Ca2+ events (black) are distinct from background (white) D) The ROI Manager incorporates calibration and Ca2+ event size selection parameters while identifying Ca2+ event ROIs from thresholded STMap. E-F) Plugin identifies Ca2+ event ROIs and overlays them on original greyscale or colored STMap for user validation.
3.5. Ca2+ event parameter analysis
To allow proper quantification of Ca2+ events within STMaps, the plugin uses the generated ROI overlay of Ca2+ events on the original STMap image to extract the following Ca2+ event parameters: event duration, spatial spread, event area, frequency, angle, and average intensity (Figure 6 C – F). These parameters have been shown to be useful descriptors of Ca2+ event dynamics from STMaps.[31–35] Duration of individual Ca2+ events is calculated by the Box Rectangle function within Fiji. This function works by generating a measurement bounding box around each event whose width spans from the most left justified pixel of the event to the most right justified pixel of the event. The width represents the duration of the event (Figure 6 C). The height of the measurement box extends from the smallest y-axis value pixel of the event to the largest y-axis value pixel and represents the spatial spread (Figure 6 D). Together, the dimensions of the measurement box detail both the duration (x-axis) and spatial spread (y-axis) of the event. In addition to the Box Rectangle function, the plugin automates the retrieval of data parameters from each generated ROI overlaid on the original image such that the entire event is used for generation of event area and intensity measurements (Figure 6 D,G). Further, to achieve a measurement of Ca2+ event angle, the plugin uses the Fiji ellipse function to autofit an ellipse to each event (Figure 6 F). From each ellipse, an angle with respect to the y-axis is computed based on the deviation of the ellipse’s major axis and the horizontal axis. The angle measurement would provide a clear information on the direction of Ca2+ events; it also provides an estimation of the overall velocity of spread.
Figure 6: Analysis of STMap Ca2+ event parameters.
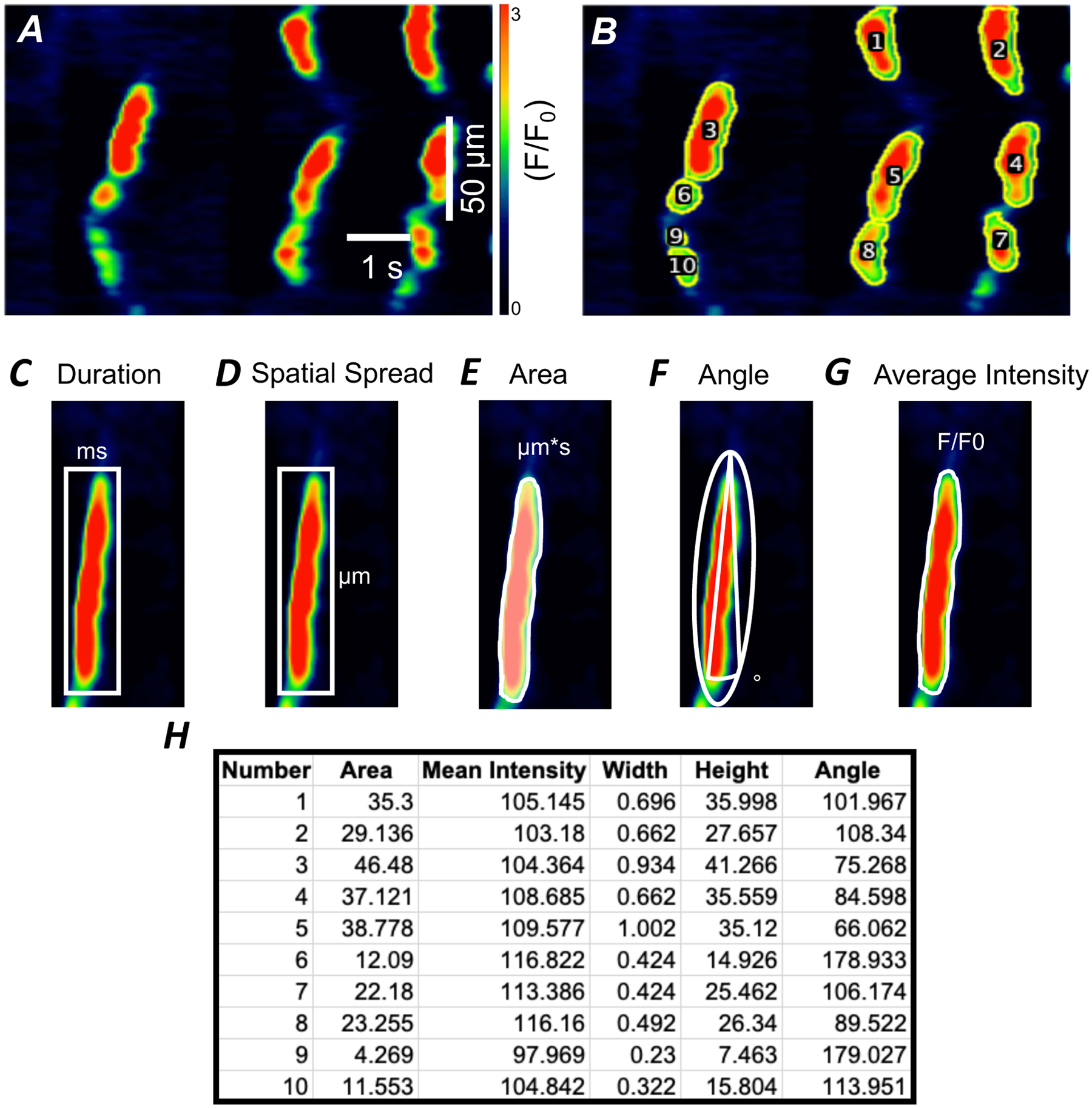
A) Representative STMap of Ca2+ transients from ICC-DMP. Scale bars in panel A also apply to panel B. B) STMap with overlaid ROI (yellow) with individual Ca2+ events labeled (1–10) after plugin segmentation process. C) Representation of Fiji mediated Box Rectangle measurement (white) of event duration and spatial spread (D). Box rectangle includes entire Ca2+ event used for data measurement. E) Representation of Fiji mediated identification of event area through plugin generated Ca2+ event ROIs (white). F) Fiji fitted ellipse (white) to determine Ca2+ event angle. G) Representation of identification of event mean intensity through plugin generated Ca2+ event ROIs (white). H) An example of Ca2+ events data generated from ICC-DMP STMap that were automatically extracted and tabulated by plugin and then exported to Excel spread sheet.
Our software plugin method of analysis uses automated detection of Ca2+ events that yield Ca2+ event data parameters from the entire Ca2+ event (Figure 6 A–G). This approach is in direct contrast with the single-pixel line method used widely to analyze STMap Ca2+ event parameters, which lack accuracy and ignores data within the Ca2+ event not present on the indicator pixel line (Figure 2 & Figure 6). Therefore, the subsequent data generation from our plugin provides a more accurate representation of STMap Ca2+ events when compared to single-pixel line analysis.
Finally, upon completion of ROI generation and Ca2+ event parameter data extraction, the plugin automatically exports all data parameters individually and as an average to an Excel sheet (.xls) for figure generation (Figure 6 H). This provides the user the opportunity for rapid interpretation and evaluation of STMaps.
3.6. Fast and high-throughput analysis achieved by the automated plugin
As outlined, the segmentation, ROI generation, and data extraction of STMaps through the plugin is done automatically. Therefore, the intensive task of drawing a single-pixel line through each individual Ca2+ event is not needed. To evaluate changes in speed of analysis, we compared the efficiency of our plugin with the pixel-line method in analysis of Ca2+ events from STMaps derived from imaging of ICC. Analysis using our plugin averaged 2.69 ± 0.5 s per event while the pixel-line method of analysis was 95.6 ± 7.5 s per Ca2+ event (n=30; Figure 7). Although significant, this difference is most apparent when comparing the speed of analysis of large STMap datasets. Plugin based analysis of 30 ICC STMaps was achieved with a speed of: 0.39 ± 0.06 hrs, while pixel-line analysis of the same STMaps was completed in 77 ± 3.4 hrs (n=4; Figure 7). This enhanced speed of plugin analysis is achieved through batch processing of STMaps for high-throughput analysis. Multiple STMaps can be analyzed and data output can be pooled together providing statistical measurements of large data sets at once. When compared to STMap single-pixel line analysis, it is clear that the plugin approach is not constrained by significant user error, low accuracy, or computational bottlenecks.
Figure 7: Speed comparison of plugin and single pixel-line methods of STMap analysis.
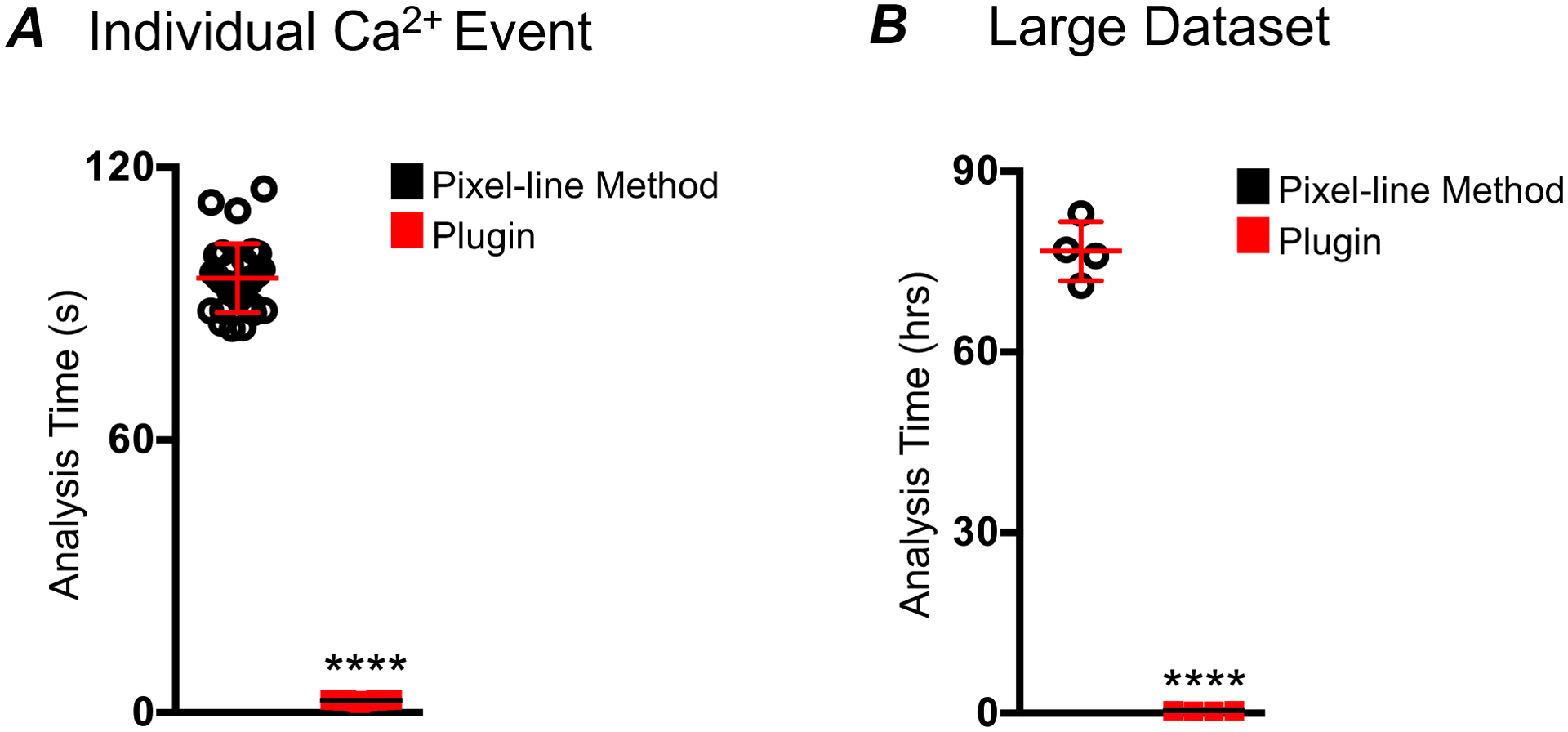
A) Comparison of time elapsed for analysis of per individual Ca2+ events from 30 ICC-DMP STMaps, using plugin vs pixel-line method. The pixel-line method required an average of 95.6 ± 7.5 s per Ca2+ event (black). The plugin spent an average of 2.69 ± 0.5 s per Ca2+ event (red; n=30). B) Comparison of time elapsed for analysis of large dataset of 30 STMaps of Ca2+ events from ICC-DMP. The pixel-line method (black) required 77 hours ± 3.4, while the plugin required 0.39 ± 0.06 hrs (30 STMap analysis; n=4, **** = P < 0.0001). All Ca2+ STMaps analyzed were 30 s recordings.
3.7. ICC Ca2+ transients analysis using the plugin.
The plugin has substantial application in STMap based analysis of variable patterns of Ca2+ transients. Because we optimized two distinct classifiers, the plugin is able to accurately analyze a wide range of Ca2+ transient patterns. Cell types such as the ICC-MY, which often generate large rhythmic firing patterns, can be quickly and accurately analyzed to yield data parameters that describe ICC-MY Ca2+ dynamics (Figure 8 A–C). These parameters are generated from automated ROI generation of STMaps as shown in (Figure 8 B–C). ICC-MY in the small intestine displays rhythmic Ca2+ transients (Figure 8 A–C). We observed that the Ca2+ transients had an average: frequency of 11 ± 1.55 per STMap (STMaps were recorded for 30 s, n=5; Figure 8 D), area of 29.4 ± 0.89 μm*s (Figure 8 E), duration of 822 ± 8.75 ms (Figure 8 F), and spatial spread of 37.43 ± 1.88 μm (Figure 8 G). Other cell types, such as the ICC-DMP often show a more transient, stochastic pattern of Ca2+ transients [31, 55] (Figure 9 A). These cells can also be readily analyzed by using the stochastic classifier without significant user modification (Figure 8 B,C). In the ICC-DMP, the plugin is able to segment STMap Ca2+ events and automatically retrieve data parameters that are useful for ICC-DMP behavior characterization in an efficient and accurate manner. We observe that, the plugin can clearly detect Ca2+ events in ICC-DMP with different Ca2 event shapes and kinetics (Figure 9 C). The plugin based STMap analysis of Ca2+ transients in ICC-DMP demonstrated that Ca2+ transients had an average: duration of 401.4 ± 6.97 ms (Figure 9 D), frequency of 29.25 ± 1.31 per STMap (STMaps were recorded for 30 s, n=5; Figure 9 E), spatial spread of 29.01 ± 1.1 μm (Figure 9 F), event angle of: 152.41 ± 5.04 (Figure 9 G), area of 20.16 ± 0.47 μm*s (Figure 9 H), and event intensity of: 99.8 ± 1.23 F/F0 (Figure 9 I).
Figure 8: High-throughput generation of Ca2+ dynamic parameters from ICC-MY STMap.
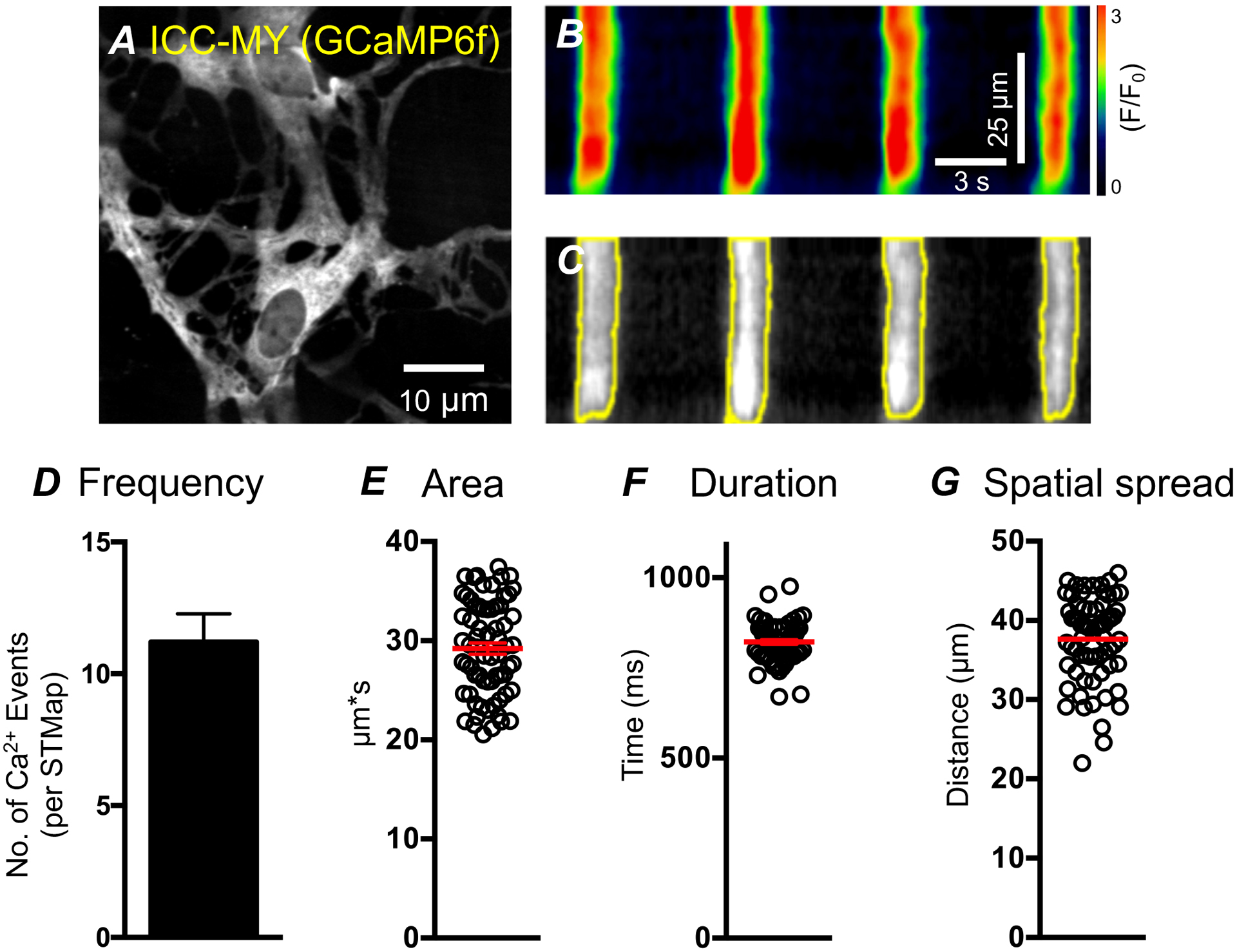
A) Representative image of ICC-MY from the small intestine Kit-Cre-GCaMP6 mouse B) STMap of rhythmic ICC-MY Ca2+ events generated from panel A. Scale bars in panel B also apply to panel C. C) Plugin generated Ca2+ event ROIs (yellow) overlaid on greyscale STMap. D) Plugin generated Ca2+ event frequency per STMap. (All Ca2+ STMaps analyzed were 30 s recordings, n=5) E-G) Plugin generated Ca2+ events parameters from ICC-MY STMaps: area, duration, and spatial spread spatial spread.
Figure 9: High-throughput extraction of Ca2+ dynamic parameters from ICC-DMP STMaps.
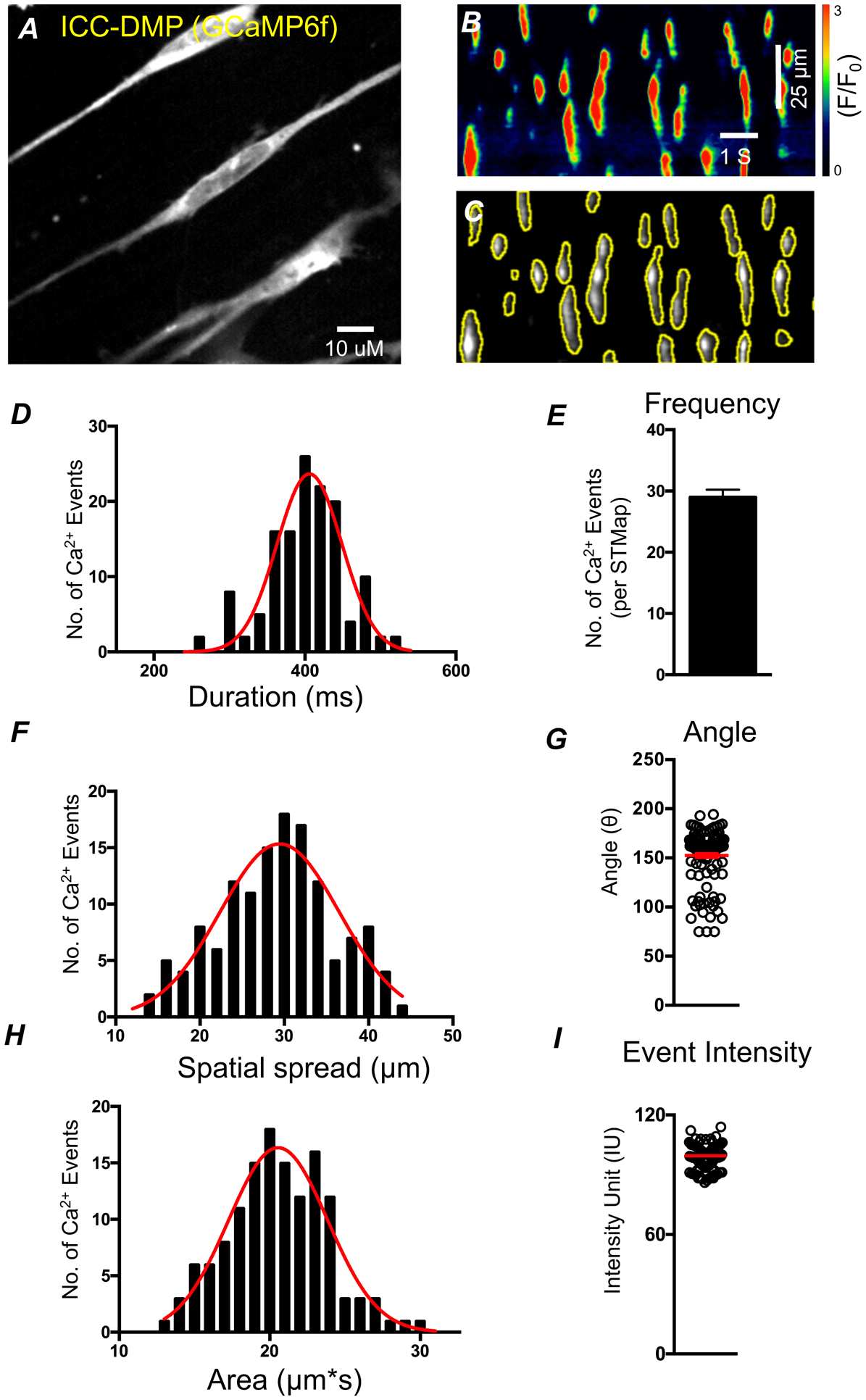
A) Representative image of ICC-DMP from the small intestine of a Kit-Cre-GCaMP6 mouse B) STMap of Ca2+ events generated from panel A. Scale bars in panel B also apply to panel C. C) Plugin generated ROIs (yellow) overlaid on greyscale STM. D,F,H) Plugin generated data of Ca2+ event parameters represented by summary histogram (black) fitted with a gaussian distribution (red) showing Ca2+ event duration, spatial spread, and area respectively. E,G,I) Plugin generated data of Ca2+ event parameters: event frequency per STMap, angle, event intensity from ICC-DMP STMaps respectively. (All Ca2+ STMaps analyzed were 30 s recordings, n=5).
4. Discussion
Understanding cellular Ca2+ dynamics requires an intensive approach of both experimental and analytical methods due to the complex patterning and kinetics of Ca2+ transients produced in different cell types. To better represent these Ca2+ signals, STMaps of Ca2+ transients are frequently used to describe and quantify Ca2+ events. [22, 26–35] STMaps are excellent for monitoring Ca2+ fluorescence changes over time because they plot cellular Ca2+ transients spatially and temporally. However, challenges exist to effectively analyze STMaps in a fast, accurate, and standardized method.
As explored in this manuscript, current methods of STMap analysis including the pixel-line method rely on manual quantification of Ca2+ transients that are heavily restricted by speed of analysis, consistency, and user input. [29, 31, 32, 56] The pixel-line method of extracting and quantifying Ca2+ transient parameters lacks accuracy by allowing data to be increasingly introduced to user influence (Figure 2). Large datasets that use the pixel-line method are extremely slow, subjective, and error prone method of analysis. This underscores the intensive need for an automated workflow of STMap analysis.
In the present study, we designed a software plugin to address limitations of pixel-line STMaps analysis. One major obstacle in STMap Ca2+ transient analysis is accurate event detection. This obstacle is often compounded as user-based detection methods are inconsistent and highly subject to bias between users. Therefore, we incorporated a machine-learning approach in the plugin to provide trainable segmentation as it has been detailed as a way to enhance image segmentation in microscopy. [36] We created two distinct classifier options for effective segmentation of STMaps with different Ca2+ transient patterns. The plugin quickly and accurately analyzed Ca2+ STMaps from two distinct intestinal ICC populations: the pacemaker ICC-MY and the neuromediator ICC-DMP that have distinct patterns of Ca2+ firing: rhythmic and stochastic, respectively (Figure 4). Because we include a trainable component in the plugin, it may be applied to a variety of cell types that exhibit local Ca2+ signals, propagating Ca2+ signals or a combination of different signaling patterns.
Machine-learning integration can provide robust advantages in image processing. Machine-learning driven approaches have been emerging heavily in the biomedical field through medical diagnosis, histopathological analysis, and CT image processing. [57–59] Researchers have found that machine-learning has the potential to dramatically aid in speed of analysis, consistency and accuracy in data extraction. [60, 61] However, a machine-learning method for STMap analysis comes with its own concerns; validation of the approach is critical to ensure accuracy of analysis. Without the opportunity to validate the output data, the user could unknowingly be improperly segmenting Ca2+ events and subsequently generating inaccurate data. Our plugin allows for validation of the output data after the process of machine-learning segmentation. This process allows the researcher to confirm the validity of the plugin machine-learning based event detection, ROI generation, and data extraction. Collectively, the automated workflow with the incorporation of a trainable event detection dramatically enhances STMap analysis.
As demonstrated in this study, analysis of STMaps is heavily constrained by the method of Ca2+ detection and measurement (Figure 2). We aimed to significantly reduce this impediment to STMap analysis. We found that the automated plugin-based approach was able to expeditiously quantify Ca2+ transients from STMaps. The difference in single event analysis speed was substantial compared to the pixel-line method of analysis. While somewhat feasible for smaller datasets, the pixel-line method is even more inadequate for larger datasets as it is increasingly time exhaustive. Indeed, these temporal distinctions emphasize the substantial value of the plugin in both small and large dataset analysis.
We also highlight that a significant problem in the accuracy of pixel-line analysis is that the method introduces user error and bias to results. Because the pixel-lines are laborious to draw and subjective, high variability in pixel-line drawing can result in misrepresentative measurements and data output. Unlike the pixel-line method, the plugin reduces this variability by limiting opportunity for user intervention; allowing different users to achieve the same results from a particular dataset. This consistency in data generation enhances accuracy of data output and is directly a result of automating user defined processes: preprocessing, event segmentation, ROI generation, data measurement. Additionally, a consequence of incorporated machine-learning to an automated workflow is that there are fundamental differences in how data parameters are measured. Trainable segmentation offers an added advancement in accuracy by permitting data generation from entire Ca2+ events. For example, the pixel-line method of analysis estimates the duration of a Ca2+ event, by measuring the Full Duration at Half Maximum (FDHM). [18, 29, 35] This measurement is intrinsically inaccurate as it is not a true measurement of event start and end; it is a measurement of median event duration. Our plugin can analyze the totality of a Ca2+ event using the machine-learning generated ROI’s as boundaries for event duration and spatial spread. Further, these plugin generated ROI’s use all data points within the ROI to generate event intensity and area. Therefore, the implementation of our plugin can provide rapid results that enhance accuracy of STMap analysis.
Although STMaps are commonly used to study Ca2+ dynamics, to date there are currently no automated machine-learning implemented plugin-based tools for STMap analysis. We provide a novel solution for STMap analysis that is comprised of an automated trainable workflow that quantifies key Ca2+ event parameters. However, automated video analysis does exist. [62–64] Though, these workflows are highly specialized, not adaptable through a machine-learning based classifier, and are not designed for STMap analysis. [65–69] Thus, the inclusion of a trainable classifier in an optimized STMap analysis workflow is a unique feature of our plugin not shared by other methods.
4.1. Conclusions
The plugin-based approach we describe functions to address the limitations of single-pixel analysis by incorporating a machine-learned based classifier to: expedite segmentation, extract STMap Ca2+ event data, and provide the investigator with consistency in analysis. Our plugin resolves the core of single pixel-line based analysis limitations by consistently automating a large portion of user input; reducing both time to completion and sources of user error while maintaining a high level of accuracy. The plugin’s performance in STMap Ca2+ event analysis demonstrate that it can be used in a high-throughput approach to quantify complex Ca2+ signaling in ICC and possibly be implemented as a solid tool for Ca2+ analysis in other cell types. Our plugin based method provides a robust new platform for efficient and accurate Ca2+ transient analysis
Highlights.
We designed a new automated machine-learning based plugin for the analysis of Ca2+ Spatio-Temporal Maps (STMaps).
The plugin is fully implemented in Fiji and able to accurately detect and quantify a variety of Ca2+ transient signals.
The plugin includes optimized tools for automated extraction of key Ca2+ events.
The plugin is extremely fast and provide an efficient method in the analysis of Ca2+ large-datasets.
The automated plugin, reduces user error and provides a consistent high-throughput analysis
Funding:
This project was supported by R01 DK-120759 from the NIDDK that supported the primary experiments.
Abbreviations
- FOV
Field of view
- GI
Gastrointestinal
- ICC
Interstitial Cells of Cajal
- ICC-DMP
Interstitial cells of Cajal at the level of the deep muscular plexus
- ICC-IM
Intramuscular interstitial cells of Cajal
- ICC-MY
Interstitial cells of Cajal at the level of the myenteric plexus
- GCaMP
Genetically encoded Ca2+ indicator composed of a single GFP
- KRB
Krebs Ringer Bicarbonate
- ROI
Region of interest
- STMap
Spatio-Temporal Map
- STMapAuto
Spatial-temporal maps automation plugin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no competing financial interests.
Declaration of Competing Interest
The authors declare no conflict of interest.
References:
- [1].Berridge MJ, Lipp P, Bootman MD, The versatility and universality of calcium signalling, Nature Reviews Molecular Cell Biology, 1 (2000) 11–21. [DOI] [PubMed] [Google Scholar]
- [2].Bootman MD, Collins TJ, Peppiatt CM, Prothero LS, MacKenzie L, De Smet P, Travers M, Tovey SC, Seo JT, Berridge MJ, Ciccolini F, Lipp P, Calcium signalling—an overview, Seminars in Cell & Developmental Biology, 12 (2001) 3–10. [DOI] [PubMed] [Google Scholar]
- [3].Konrad KR, Maierhofer T, Hedrich R, Spatio-temporal aspects of Ca2+ signalling: lessons from guard cells and pollen tubes, Journal of Experimental Botany, 69 (2018) 4195–4214. [DOI] [PubMed] [Google Scholar]
- [4].Berridge MJ, Smooth muscle cell calcium activation mechanisms, The Journal of Physiology, 586 (2008) 5047–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lee HT, Hennig GW, Fleming NW, Keef KD, Spencer NJ, Ward SM, Sanders KM, Smith TK, The Mechanism and Spread of Pacemaker Activity Through Myenteric Interstitial Cells of Cajal in Human Small Intestine, Gastroenterology, 132 (2007) 1852–1865. [DOI] [PubMed] [Google Scholar]
- [6].Egdell RM, MacLeod KT, Calcium Extrusion During Aftercontractions in Cardiac Myocytes: The Role of the Sodium-calcium Exchanger in the Generation of the Transient Inward Current, Journal of Molecular and Cellular Cardiology, 32 (2000) 85–93. [DOI] [PubMed] [Google Scholar]
- [7].Marchant JS, Parker I, Role of elementary Ca2+ puffs in generating repetitive Ca2+ oscillations, The EMBO Journal, 20 (2001) 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Santana LF, Navedo MF, Amberg GC, Nieves-Cintrón M, Votaw VS, Ufret-Vincenty CA, Calcium Sparklets in Arterial Smooth Muscle, Clinical and Experimental Pharmacology and Physiology, 35 (2008) 1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guo T, Gillespie D, Fill M, Ryanodine Receptor Current Amplitude Controls Ca2+ Sparks in Cardiac Muscle, Circulation Research, 111 (2012) 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cheng H, Lederer MR, Lederer WJ, Cannell MB, Calcium sparks and [Ca2+]i waves in cardiac myocytes, American Journal of Physiology-Cell Physiology, 270 (1996) C148–C159. [DOI] [PubMed] [Google Scholar]
- [11].Niggli E, Localized intracellular calcium signaling in muscle: Calcium Sparks and Calcium Quarks, Annual Review of Physiology, 61 (1999) 311–335. [DOI] [PubMed] [Google Scholar]
- [12].Sun X-P, Callamaras N, Marchant JS, Parker I, A continuum of InsP3-mediated elementary Ca2+ signalling events in Xenopus oocytes, The Journal of Physiology, 509 (1998) 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Drumm BT, Large RJ, Hollywood MA, Thornbury KD, Baker SA, Harvey BJ, McHale NG, Sergeant GP, The role of Ca2+ influx in spontaneous Ca2+ wave propagation in interstitial cells of Cajal from the rabbit urethra, The Journal of Physiology, 593 (2015) 3333–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Straub SV, Giovannucci DR, Yule DI, Calcium Wave Propagation in Pancreatic Acinar Cells, The Journal of General Physiology, 116 (2000) 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hennig GW, Spencer NJ, Jokela-willis S, Bayguinov PO, Lee H.t., Ritchie LA, Ward SM, Smith TK, Sanders KM, ICC-MY coordinate smooth muscle electrical and mechanical activity in the murine small intestine, Neurogastroenterology & Motility, 22 (2010) e138–e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Baker SA, Hennig GW, Salter AK, Kurahashi M, Ward SM, Sanders KM, Distribution and Ca(2+) signalling of fibroblast-like (PDGFR(+)) cells in the murine gastric fundus, The Journal of physiology, 591 (2013) 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Boittin F-X, Macrez N, Halet G, Mironneau J, Norepinephrine-induced Ca2+waves depend on InsP3 and ryanodine receptor activation in vascular myocytes, American Journal of Physiology-Cell Physiology, 277 (1999) C139–C151. [DOI] [PubMed] [Google Scholar]
- [18].Baker SA, Drumm BT, Saur D, Hennig GW, Ward SM, Sanders KM, Spontaneous Ca(2+) transients in interstitial cells of Cajal located within the deep muscular plexus of the murine small intestine, J Physiol, 594 (2016) 3317–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Berridge MJ, Elementary and global aspects of calcium signalling, The Journal of physiology, 499 (Pt 2) (1997) 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Berridge MJ, Dupont G, Spatial and temporal signalling by calcium, Current Opinion in Cell Biology, 6 (1994) 267–274. [DOI] [PubMed] [Google Scholar]
- [21].Gu X, Spitzer NC, Distinct aspects of neuronal differentiation encoded by frequency of spontaneous Ca2+ transients, Nature, 375 (1995) 784–787. [DOI] [PubMed] [Google Scholar]
- [22].Lee HT, Hennig GW, Park KJ, Bayguinov PO, Ward SM, Sanders KM, Smith TK, Heterogeneities in ICC Ca2+ Activity Within Canine Large Intestine, Gastroenterology, 136 (2009) 2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Colman MA, Pinali C, Trafford AW, Zhang H, Kitmitto A, A computational model of spatio-temporal cardiac intracellular calcium handling with realistic structure and spatial flux distribution from sarcoplasmic reticulum and t-tubule reconstructions, PLOS Computational Biology, 13 (2017) e1005714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roome CJ, Kuhn B, Simultaneous dendritic voltage and calcium imaging and somatic recording from Purkinje neurons in awake mice, Nature Communications, 9 (2018) 3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Waadt R, Krebs M, Kudla J, Schumacher K, Multiparameter imaging of calcium and abscisic acid and high-resolution quantitative calcium measurements using R-GECO1-mTurquoise in Arabidopsis, New Phytologist, 216 (2017) 303–320. [DOI] [PubMed] [Google Scholar]
- [26].Bolton Gordienko, Confocal imaging of calcium release events in single smooth muscle cells, Acta Physiologica Scandinavica, 164 (1998) 567–575. [DOI] [PubMed] [Google Scholar]
- [27].Cobine CA, Hannigan KI, McMahon M, Ni Bhraonain EP, Baker SA, Keef KD, Rhythmic calcium transients in smooth muscle cells of the mouse internal anal sphincter, Neurogastroenterology & Motility, 0 (2019) e13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dargan SL, Schwaller B, Parker I, Spatiotemporal patterning of IP3-mediated Ca2+ signals in Xenopus oocytes by Ca2+-binding proteins, The Journal of Physiology, 556 (2004) 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Drumm BT, Hennig GW, Baker SA, Sanders KM, Applications of Spatio-temporal Mapping and Particle Analysis Techniques to Quantify Intracellular Ca2+ Signaling In Situ, JoVE, (2019) e58989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Park KJ, Hennig GW, Lee H-T, Spencer NJ, Ward SM, Smith TK, Sanders KM, Spatial and temporal mapping of pacemaker activity in interstitial cells of Cajal in mouse ileum in situ, American Journal of Physiology-Cell Physiology, 290 (2006) C1411–C1427. [DOI] [PubMed] [Google Scholar]
- [31].Baker SA, Drumm BT, Cobine CA, Keef KD, Sanders KM, Inhibitory Neural Regulation of the Ca (2+) Transients in Intramuscular Interstitial Cells of Cajal in the Small Intestine, Frontiers in physiology, 9 (2018) 328–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Drumm BT, Sergeant GP, Hollywood MA, Thornbury KD, McHale NG, Harvey BJ, The role of cAMP dependent protein kinase in modulating spontaneous intracellular Ca2+ waves in interstitial cells of Cajal from the rabbit urethra, Cell Calcium, 56 (2014) 181–187. [DOI] [PubMed] [Google Scholar]
- [33].Fedigan S, Bradley E, Webb T, Large RJ, Hollywood MA, Thornbury KD, McHale NG, Sergeant GP, Effects of new-generation TMEM16A inhibitors on calcium-activated chloride currents in rabbit urethral interstitial cells of Cajal, Pflügers Archiv - European Journal of Physiology, 469 (2017) 1443–1455. [DOI] [PubMed] [Google Scholar]
- [34].Sancho M, Bradley E, Garcia-Pascual A, Triguero D, Thornbury KD, Hollywood MA, Sergeant GP, Involvement of cyclic nucleotide-gated channels in spontaneous activity generated in isolated interstitial cells of Cajal from the rabbit urethra, European Journal of Pharmacology, 814 (2017) 216–225. [DOI] [PubMed] [Google Scholar]
- [35].Sergeant GP, Johnston L, McHale NG, Thornbury KD, Hollywood MA, Activation of the cGMP/PKG pathway inhibits electrical activity in rabbit urethral interstitial cells of Cajal by reducing the spatial spread of Ca2+ waves, The Journal of Physiology, 574 (2006) 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, Sebastian Seung H, Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification, Bioinformatics, 33 (2017) 2424–2426. [DOI] [PubMed] [Google Scholar]
- [37].Sanders KM, Ward SM, Koh SD, Interstitial cells: regulators of smooth muscle function, Physiological reviews, 94 (2014) 859–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Drumm BT, Rembetski BE, Baker SA, Sanders KM, Tonic inhibition of murine proximal colon is due to nitrergic suppression of Ca2+ signaling in interstitial cells of Cajal, Scientific Reports, 9 (2019) 4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhu MH, Sung TS, O’Driscoll K, Koh SD, Sanders KM, Intracellular Ca2+ release from endoplasmic reticulum regulates slow wave currents and pacemaker activity of interstitial cells of Cajal, American Journal of Physiology-Cell Physiology, 308 (2015) C608–C620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhu MH, Kim TW, Ro S, Yan W, Ward SM, Koh SD, Sanders KM, A Ca2+-activated Cl−conductance in interstitial cells of Cajal linked to slow wave currents and pacemaker activity, The Journal of Physiology, 587 (2009) 4905–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Baker SA, Drumm BT, Saur D, Hennig GW, Ward SM, Sanders KM, Spontaneous Ca2+ transients in interstitial cells of Cajal located within the deep muscular plexus of the murine small intestine, The Journal of Physiology, 594 (2016) 3317–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Komuro T, Structure and organization of interstitial cells of Cajal in the gastrointestinal tract, The Journal of physiology, 576 (2006) 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sanders KM, Koh SD, Ward SM, INTERSTITIAL CELLS OF CAJAL AS PACEMAKERS IN THE GASTROINTESTINAL TRACT, Annual Review of Physiology, 68 (2006) 307–343. [DOI] [PubMed] [Google Scholar]
- [44].Hulzinga JD, Thuneberg L, Klüppel M, Malysz J, Mikkelsen HB, Bernstein A, W/kit gene required for interstitial cells of Cajal and for intestinal pacemaker activity, Nature, 373 (1995) 347–349. [DOI] [PubMed] [Google Scholar]
- [45].Ordög T, Ward SM, Sanders KM, Interstitial cells of cajal generate electrical slow waves in the murine stomach, The Journal of physiology, 518 (1999) 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ward SM, Burns AJ, Torihashi S, Sanders KM, Mutation of the proto-oncogene c-kit blocks development of interstitial cells and electrical rhythmicity in murine intestine, The Journal of physiology, 480 (Pt 1) (1994) 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ward SM, McLaren GJ, Sanders KM, Interstitial cells of Cajal in the deep muscular plexus mediate enteric motor neurotransmission in the mouse small intestine, J Physiol, 573 (2006) 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Iino S, Ward SM, Sanders KM, Interstitial cells of Cajal are functionally innervated by excitatory motor neurones in the murine intestine, The Journal of Physiology, 556 (2004) 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Blair PJ, Rhee P-L, Sanders KM, Ward SM, The significance of interstitial cells in neurogastroenterology, J Neurogastroenterol Motil, 20 (2014) 294–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhou DS, Komuro T, The cellular network of interstitial cells associated with the deep muscular plexus of the guinea pig small intestine, Anat Embryol (Berl), 186 (1992) 519–527. [DOI] [PubMed] [Google Scholar]
- [51].Drumm BT, Hennig GW, Battersby MJ, Cunningham EK, Sung TS, Ward SM, Sanders KM, Baker SA, Clustering of Ca(2+) transients in interstitial cells of Cajal defines slow wave duration, The Journal of general physiology, 149 (2017) 703–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH, The WEKA data mining software: an update, SIGKDD Explor. Newsl., 11 (2009) 10–18. [Google Scholar]
- [53].Belgiu M, Drăguţ L, Random forest in remote sensing: A review of applications and future directions, ISPRS Journal of Photogrammetry and Remote Sensing, 114 (2016) 24–31. [Google Scholar]
- [54].Misha D, David M, Nando De F, Narrowing the Gap: Random Forests In Theory and In Practice, PMLR, 2014, pp. 665–673. [Google Scholar]
- [55].Baker SA, Drumm BT, Skowronek KE, Rembetski BE, Peri LE, Hennig GW, Perrino BA, Sanders KM, Excitatory Neuronal Responses of Ca(2+) Transients in Interstitial Cells of Cajal in the Small Intestine, eNeuro, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sergeant GP, Hollywood MA, Thornbury KD, Spontaneous Activity in Urethral Smooth Muscle, in: Hashitani H, Lang RJ (Eds.) Smooth Muscle Spontaneous Activity: Physiological and Pathological Modulation, Springer; Singapore, Singapore, 2019, pp. 149–167. [DOI] [PubMed] [Google Scholar]
- [57].Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL, Artificial intelligence in radiology, Nat Rev Cancer, 18 (2018) 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Komura D, Ishikawa S, Machine Learning Methods for Histopathological Image Analysis, Computational and Structural Biotechnology Journal, 16 (2018) 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Samant P, Agarwal R, Machine learning techniques for medical diagnosis of diabetes using iris images, Computer Methods and Programs in Biomedicine, 157 (2018) 121–128. [DOI] [PubMed] [Google Scholar]
- [60].Hesamian MH, Jia W, He X, Kennedy P, Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges, Journal of Digital Imaging, 32 (2019) 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Maier A, Syben C, Lasser T, Riess C, A gentle introduction to deep learning in medical image processing, Zeitschrift für Medizinische Physik, 29 (2019) 86–101. [DOI] [PubMed] [Google Scholar]
- [62].Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur A, Nguyen TN, Yoo JC, Judge LM, Spencer CI, Chukka AC, Russell CR, So P-L, Conklin BR, Healy KE, Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales, Tissue Engineering Part C: Methods, 21 (2014) 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Levin-Schwartz Y, Sparta DR, Cheer JF, Adalı T, Parameter-free automated extraction of neuronal signals from calcium imaging data, 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2017, pp. 1033–1037. [Google Scholar]
- [64].Maltsev AV, Parsons SP, Kim MS, Tsutsui K, Stern MD, Lakatta EG, Maltsev VA, Monfredi O, Computer algorithms for automated detection and analysis of local Ca2+ releases in spontaneously beating cardiac pacemaker cells, PLOS ONE, 12 (2017) e0179419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pnevmatikakis EA, Kelleher K, Chen R, Saggau P, Josić K, Paninski L, Fast Spatiotemporal Smoothing of Calcium Measurements in Dendritic Trees, PLOS Computational Biology, 8 (2012) e1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Prada J, Sasi M, Martin C, Jablonka S, Dandekar T, Blum R, An open source tool for automatic spatiotemporal assessment of calcium transients and local ‘signal-close-to-noise’ activity in calcium imaging data, PLOS Computational Biology, 14 (2018) e1006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Steele Elliot M., Steele Derek S., Detection Automated and Analysis of Ca2+ Sparks in x-y Image Stacks Using a Thresholding Algorithm Implemented within the Open-Source Image Analysis Platform ImageJ, Biophysical Journal, 106 (2014) 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Francis M, Qian X, Charbel C, Ledoux J, Parker JC, Taylor MS, Automated region of interest analysis of dynamic Ca2+ signals in image sequences, American Journal of Physiology-Cell Physiology, 303 (2012) C236–C243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Janicek R, Hotka M, Zahradníková A Jr., Zahradníková A, Zahradník I, Quantitative Analysis of Calcium Spikes in Noisy Fluorescent Background, PLOS ONE, 8 (2013) e64394. [DOI] [PMC free article] [PubMed] [Google Scholar]


