Abstract
Quorum sensing (QS) is a mechanism by which bacteria regulate cell density-dependent group behaviors. Gram-positive bacteria generally rely on auto-inducing peptide (AIP)-based QS signaling to regulate their group behaviors. To develop synthetic modulators of these behaviors, the natural peptide needs to be identified and its structure-activity relationships (SARs) with its cognate receptor (either membrane-bound or cytosolic) need to be understood. SAR information allows for the rational design of peptides or peptide mimics with enhanced characteristics, which in turn can be utilized in studies to understand species-specific QS mechanisms and as lead scaffolds for the development of therapeutic candidates that target QS. In this review, we discuss recent work associated with the approaches used towards forwarding each of these steps in Gram-positive bacteria, with a focus on species that have received less attention.
Graphical Abstract
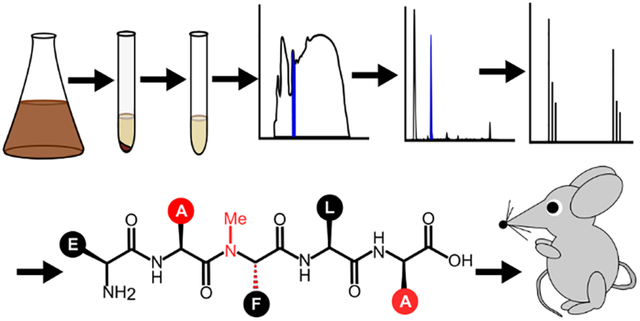
This review discusses the development of peptide-based quorum sensing modulators and their use as potential therapeutics.
Introduction
Bacterial communication pathways play a role in coordinating an assortment of bacterial group behaviors that can aid bacteria in infectivity (virulence factor production, biofilm formation), antibiotic resistance (competence, biofilm formation, swarming), and symbiotic interactions (luminescence, root nodulation).1–10 Because the phenotypes involved require bacteria to be present in sufficient numbers to be effective, these communication pathways are dependent on bacterial cell density and the process of activating these population size-dependent pathways has been termed quorum sensing (QS).1,11
In order to assess the cell density and trigger these group behaviors, bacteria rely on the production and release of signaling molecules into their surroundings. Their QS circuits include receptors that can be activated when the signaling molecules reach a certain threshold concentration indicating that there are sufficient bacteria present to begin coordinating the group behavior(s).1–3,12,5,7,13 In some cases, the receptor acts directly as a response regulator, while in others, activation of the receptor results in activation of a separate response regulator that is responsible for upregulating the genes involved in the QS circuit itself, such as genes responsible for the production and export of the signaling molecule out of the cell, to synchronize the transition from working as individual cells to working as a group. Importantly, the response regulator also activates genes involved in the group behavior(s), thus controlling the initiation of QS-dependent phenotypes.2,3,6,14,12,15,7,9
There are three major categories of QS circuits that can be divided by the type of signaling molecule used. Gram-negative bacteria generally rely on small molecules known as acyl-homoserine lactones. Gram-positive bacteria instead generally rely on peptide-based molecules, often referred to as auto-inducing peptides (AIPs). These two broad classes tend to have limited crosstalk between most species, with each species producing a version of the signal that they will respond strongly to, although there is growing evidence that these signaling molecules can be involved in interspecies communication with certain partners. The third category of signaling molecules has been implicated as being primarily used for interspecies communication and relies on a “universal” molecule known as auto-inducer 2. This review will focus on the advances that have been made in Gram-positive bacteria and their AIPs.
Gram-positive QS circuits typically include a membrane-bound histidine-kinase receptor that, when dimerized and bound by the AIP, activates the response regulator through phosphorylation. Some QS circuits instead involve a membrane bound AIP importer and an intracellular receptor that may also act as the response regulator. The signaling peptide is expressed as an immature propeptide that requires processing to become active. This processing can be coupled with the export of the peptide out of the cell, but post-export processing is sometimes involved as well.16,17 The mature peptides can be linear or macrocyclic. When macrocyclic, the macrocycle is typically formed through the formation of a thioester or ester bond between a cysteine or serine, respectively and the peptide’s C-terminus. Due to this type of cyclization, these macrocycles typically have a short “tail” sequence of exocyclic amino acids in addition to the macrocycle itself as can be seen by reviewing the Quorumpeps database.13,18
The involvement of many QS circuits in bacterial pathogenicity, coupled with the rising prevalence of resistance to conventional antibiotics, has led to a growing interest in targeting QS as an alternative therapeutic approach. Targeting QS should reduce the selective pressure on bacteria to develop resistance since disrupting the QS circuit will not directly result in cell death but will still attenuate the infection. In addition, understanding how bacteria communicate within their species and with others can expand our understanding of microbiomes. This understanding has important implications given the growing body of evidence that indicates microbiomes affect human health.19–21
The AIP signal makes a promising starting point for the development of tools to study Gram-positive QS as well as for the development of lead therapeutic compounds. Since the circuits rely on interactions between a membrane-bound protein (such as receptors or peptide importers) and the AIP, compounds based on the AIP do not necessarily need to enter cells in order to have their effect. In addition, the AIP already possesses bioactivity in its interactions with the receptor, so the development of modulators becomes more of an issue of adjusting existing interactions rather than trying to create de novo interactions, which can potentially save time when trying to develop a therapeutic agent.
In this review, we will discuss the strategies (with a focus on chemical strategies) for and recent advances in identifying new AIPs, determining SARs between AIPs and their receptors, rationally designing analogs with desired characteristics or properties, and finally how additional information can be gained using peptides that modulate QS. The discussed body of work will demonstrate workflows for the development of potential lead compounds, and/or for developing tools for gaining greater understanding of the role of QS. To reflect this, sections have been named after major steps in those workflows, with recent developments in those steps organized as subsections. In general, the workflow begins with the identification and isolation of the native peptide and associated cellular machinery. This is followed by evaluation of the SARs between the cellular machinery and the peptide signal, which then informs the design of potential lead compounds to modulate the QS response. This is often accompanied and/or followed by an assessment of the regulation response pathways and the evaluation of the therapeutic potential of lead peptides. We will focus on work that has not previously seen much review but also supply references for more heavily reviewed species (namely Staphylococcus aureus) and QS circuits.22–27
1. Identification of the Native Peptide Signal
The most common approach currently used to identify potential peptide signals is to utilize genomic data to identify species or strains that are likely to possess the QS circuit of interest. This can be accomplished through the use of sequence alignments to find gene clusters that resemble the genes already associated with functional QS circuits. The presence of such sequences can be confirmed using polymerase chain reaction (PCR) to amplify DNA regions containing complementary sequences that match with carefully designed flanking primer sequences. In addition to serving as a strong indicator of the presence of sequences that code for the AIP or its associated QS circuitry, these amplified sequences can also be used in the construction of reporter strains that can respond selectively to the AIP being studied. Predictions are confirmed by isolating and identifying the signal from bacterial cultures. Because the signals are usually processed in the cell prior to being exported, this stage can be challenging, especially for cyclic peptides since it is possible for several possible cyclization sites to exist and the sequence alone does not always indicate which site is selected during propeptide processing. However, HPLC and tandem mass spectrometry (MS/MS) methods have contributed to streamlining these identifications.15 Once a potential signal has been isolated and identified, it must then be verified through biological assays with the bacteria. This step can involve the use of a reporter strain (e.g. a strain that contains a plasmid engineered to produce a signal under the control of a promoter that should only be activated due to a response initiated by the signal peptide) or the use of phenotypic assays (e.g. detection of a signal-induced phenotype such as biofilm formation or the production of a virulence factor). Table 1 contains examples of peptide structure and modification nomenclature. Table 2 summarizes recently identified AIPs, the species they were identified from and the methods used for their identification.
Table 1.
Summary of Peptide Modification/Structural Nomenclature
| Modification | Symbol | Example Structure and Sequence |
|---|---|---|
| Nonea |
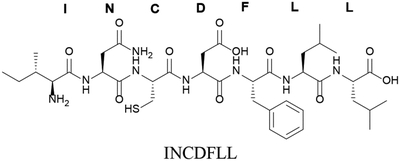 |
|
| Alanine replacement of glutamic acid at position 5 | F5A |
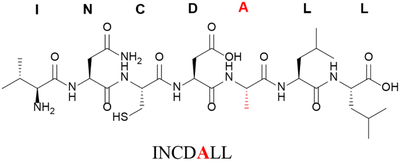 |
| Cyclization between C-terminus and cysteine in position 3b | ( ) |
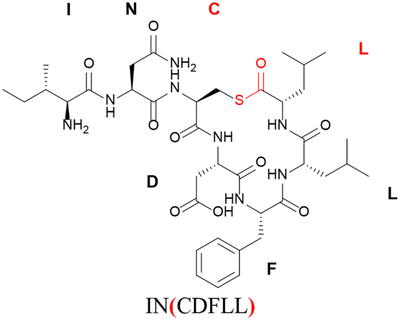 |
| D-amino acid of phenylalanine at position 5 | f5 |
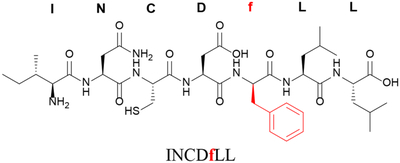 |
| Removal (truncation) of a single residue (C-terminal in this example) | Δ7L or des-L7 |
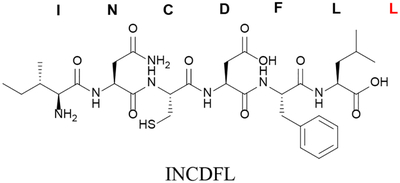 |
| Removal of multiple residues (N-terminal in this example) | des-I1N2 |
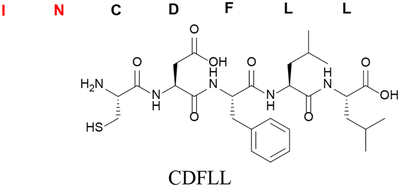 |
The linearized mature sequence for AIP-III from S. aureus is used to illustrate common peptide modification nomenclature.
In this example, the nomenclature indicates a thiolactone with the thioester linkage between Cys3 and the C-terminus (Leu7); IN is the exocyclic tail.
Table 2.
Recently Identified or Proposed AIPs
| Peptide Sequence | Type | Species | Identification Strategy | Ref |
|---|---|---|---|---|
| SKPDIVG | Mature |
Bacillus cereus Bacillus sp. strain S51107 |
Genetic analysis, standard comparison, HPLC, MS | 34,35 |
| FSLIEGFKRIa | Mature | Bacillus Licheniformis | Genetic analysis, HPLC, MS | 36 |
| SKPDI | Predicted Mature (Verified activity) |
Bacillus anthracis Bacillus thuringiensis (serovar thuringiensis, israelensis, and pondicheriensis) |
Genetic analysis, standard comparison | 37 |
| WKPDN | Predicted Mature |
Bacillus mycoides Bacillus pseudomycoides |
||
| SDIYG | Predicted Mature | Bacillus thuringiensis (serovar mycoides, kurstaki, huazhongen-sis, andsotto) | ||
| MRKLNNKVLMAVAAFATVFASVVATSACVWCSYQPEEPKCLRDKb | Propeptide (agrD) | Clostridium chauvoei | Genetic Analysis, qPCR, RNA expression | 38 |
| (CVLVTL)b | Mature | Clostridium acetobutylicum | Genetic Analysis, Knockout complementation. | 39 |
| AEPTWGW | Putative Mature (Verified Activity) | Clostridium acetobutylicum | Genetic Analysis, Knockout complementation, standard comparison | 40 |
| TSA(CLWFI) | Predicted Mature | Clostridium perfringens | Genetic Analysis, standard comparison | 41 |
| (CFMFV)b | Mature | Listeria monocytogenes | Genetic analysis, synthetic standards, co-culture complementation | 42 |
| DM(CNGYF) | Mature Slu-AIP-II | Staphylococcus lugdunensis | Genetic Analysis, Native Chemical Ligation (NCL) Trapping, HPLC, MS, standard comparison | 43 |
| KYPF(CIGYF) | Mature Ssc-AIP | Staphylococcus. schleiferi | ||
| KYNP(CLGFL) | Mature Ssi-AIP | Staphylococcus simulans | ||
| KINP(CTVFF) | Mature Shy-AIP | Staphylococcus hyicus | ||
| SINP(CTGFF) | Mature Sch-AIP | Staphylococcus chromogenes | ||
| YST(CDFIM) | Mature AIP-I | Staphylococcus argenteus | ||
| YST(CYFIM) | Mature AIP-IV | Staphylococcus schweitzeri | ||
| YSP(CTNFF) | Mature Swa-AIP | Staphylococcus warneri | ||
| VIRG(CTAFL) | Mature Svi-AIP | Staphylococcus vitulinus | ||
| TYST(CYGYF) | Mature Sho-AIP | Staphylococcus hominis | ||
| SFTP(CTTYF) | Mature Sha-AIP | Staphylococcus haemolyticus | ||
| DVGKADb | Minimal Mature | Streptococcus pneumoniae | Genetic analysis, knockout complementation, synthetic standards | 44 |
| IMDILIIVGG (10-mer) MDILIIVGG (9-mer) DILIIVGG (8-mer, most active) |
Mature SHP2 | Streptococcus pyogenes | Genetic analysis, HPLC, MS, standard comparison | 45 |
| AMDIIIIVGG (10-mer) MDIIIIVGG (9-mer) DIIIIVGG (8-mer, most active) IIIIVGG (7-mer) |
Mature SHP3 | Streptococcus pyogenes | ||
| DFLIVGPFDWLKKNHKPTK | Mature CSP | Streptococcus gallolyticus | Genetic analysis, HPLC, MS, standard comparison | 46 |
| IAILPYFAGCL | Mature ComS | Streptococcus thermophilus | Genetic analysis, HPLC, MS, standard comparison, overexpression | 16 |
Peptide has been isolated/identified from/in cell cultures but has unknown activity.
Peptide is proposed and has not been verified through isolation/identification from/in cell cultures. The actual native peptide may differ depending on how the propeptide is processed into the mature form.
1A. Overview of Two Common QS Circuits Classes
There are several forms of QS circuits (Figure 1) that have been observed in Gram positive bacteria and it is not uncommon for some species to utilize more than one. Before discussing some of the advances made in identifying the signaling peptides, it would be helpful to briefly review the circuit classes that are referenced.
Figure 1: Generalized Common Quorum Sensing Circuits.
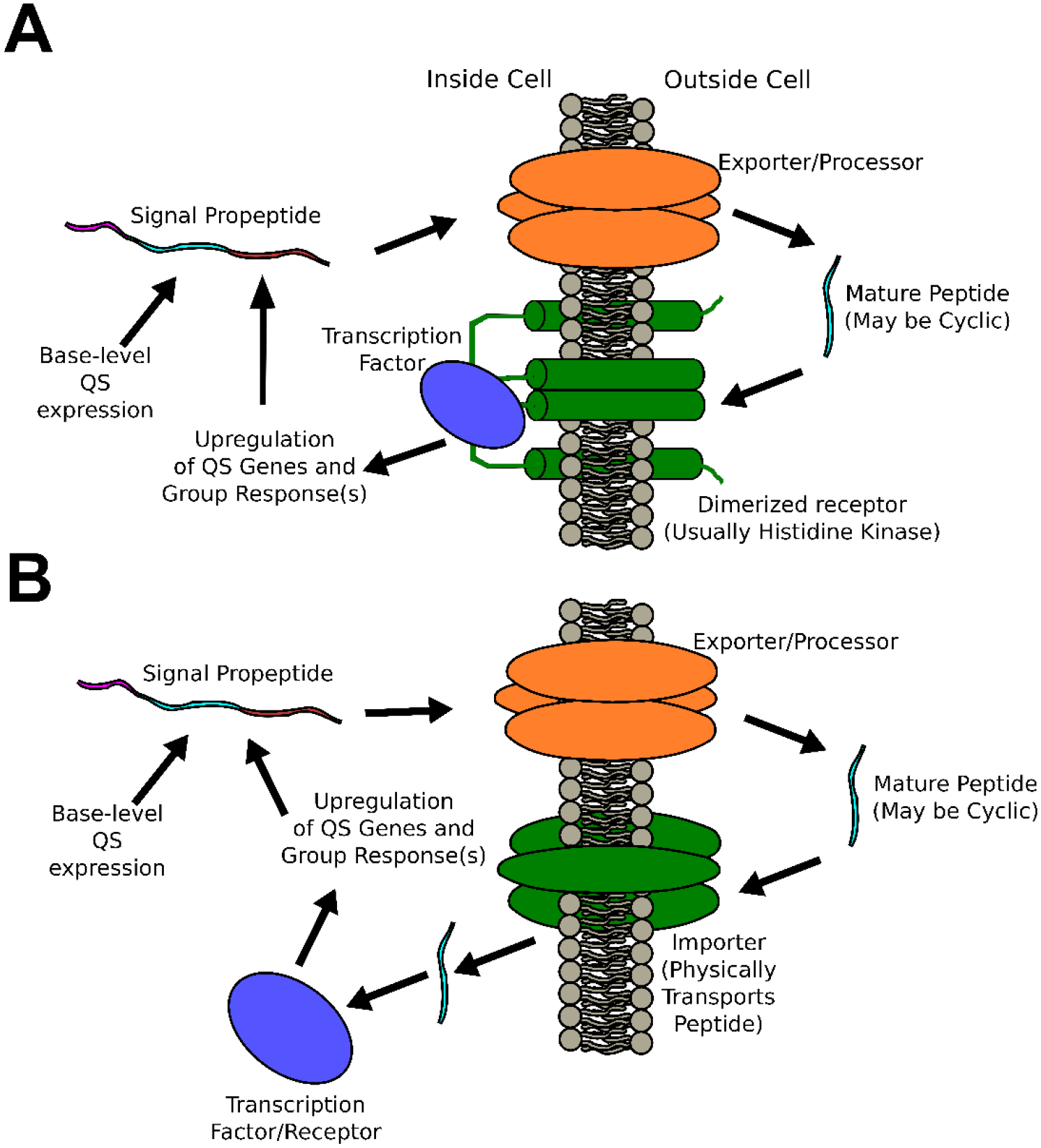
A) Agr-like circuits involve the transduction of peptide signal across the membrane without physical transport of the peptide signal, B) RNPP-like circuits involve the physical import of the peptide signal before it binds to and activates an internal transcription factor/receptor.
One of the most widely studied peptide QS circuit types are the two-component systems. These circuits typically involve a set of genes labeled A-D.3,28–30 The D-type gene product is a propeptide that is eventually processed into a mature AIP and exported from the cell. The B-type gene product is a membrane-bound protein associated with the processing and export of the propeptide in its mature form out of the cell (in some cases an additional protease is required for final processing to the mature peptide form). The C-type gene product is typically a membrane-bound histidine-kinase receptor that recognizes the mature peptide and is responsible for transducing the peptide’s signal across the membrane. Finally, the A-type gene product is a protein response regulator that triggers upregulation of both the A-D genes (resulting in auto-induction) as well as genes associated with the downstream phenotypes associated with the specific species’ QS behavior(s). A few example species where this circuit class has been studied include S. aureus (Agr system), Streptococcus pneumoniae (Com system), and Enterococcus faecalis (Fsr system).
Another common circuit class is the RRNPP circuit. This circuit class is named for the response-regulators identified from among the founding species for this circuit class: Rap (Bacillus subtilis), Rgg (Streptococcus), NprR (Bacillus cereus), PlcR (B. cereus), and PrgX (Enterococcus faecalis).31–33 As with the two-component system, a propeptide is produced that needs to be processed into the mature AIP and exported from the cell. However, in this system class, the AIPs do not bind to a membrane-bound receptor. Instead, they are reimported into signal-receiving cells where they can then form a complex with an intracellular receptor/response regulator resulting in regulation of the associated genes.
1B. Adaptation of a Common Peptide Synthesis Technique for Use in Peptide Identification
Recently, the native chemical ligation (NCL) strategy was applied in the development of a methodology for the rapid isolation and identification of macrocyclic AIPs containing a thiolactone.43,47 NCL takes advantage of the interconversion of a series of thiols and thioesters, eventually leading to a peptide bond at an N-terminal Cysteine.48 In the identification approach, an N-terminal cysteine is supplied using a L-cysteine-functionalized resin (Figure 2). When cell supernatants from a species that produces macrocyclic thioester AIPs are combined with the functionalized resin under NCL conditions, the macrocyclic ring is opened at the thioester and a peptide bond is formed between the AIP’s released C-terminus and the cysteine on the resin. Because the resin is insoluble, the peptide can be rapidly isolated simply by removing the cell supernatant with washing and then releasing the peptide from the resin. This predictably modified, linearized peptide can be analyzed via MS and compared with sequences from known or suspected propeptides in order to determine/confirm the signal’s origin.
Figure 2: Native Chemical Ligation (NCL)-Assisted Trapping of Thiolactone Peptides.
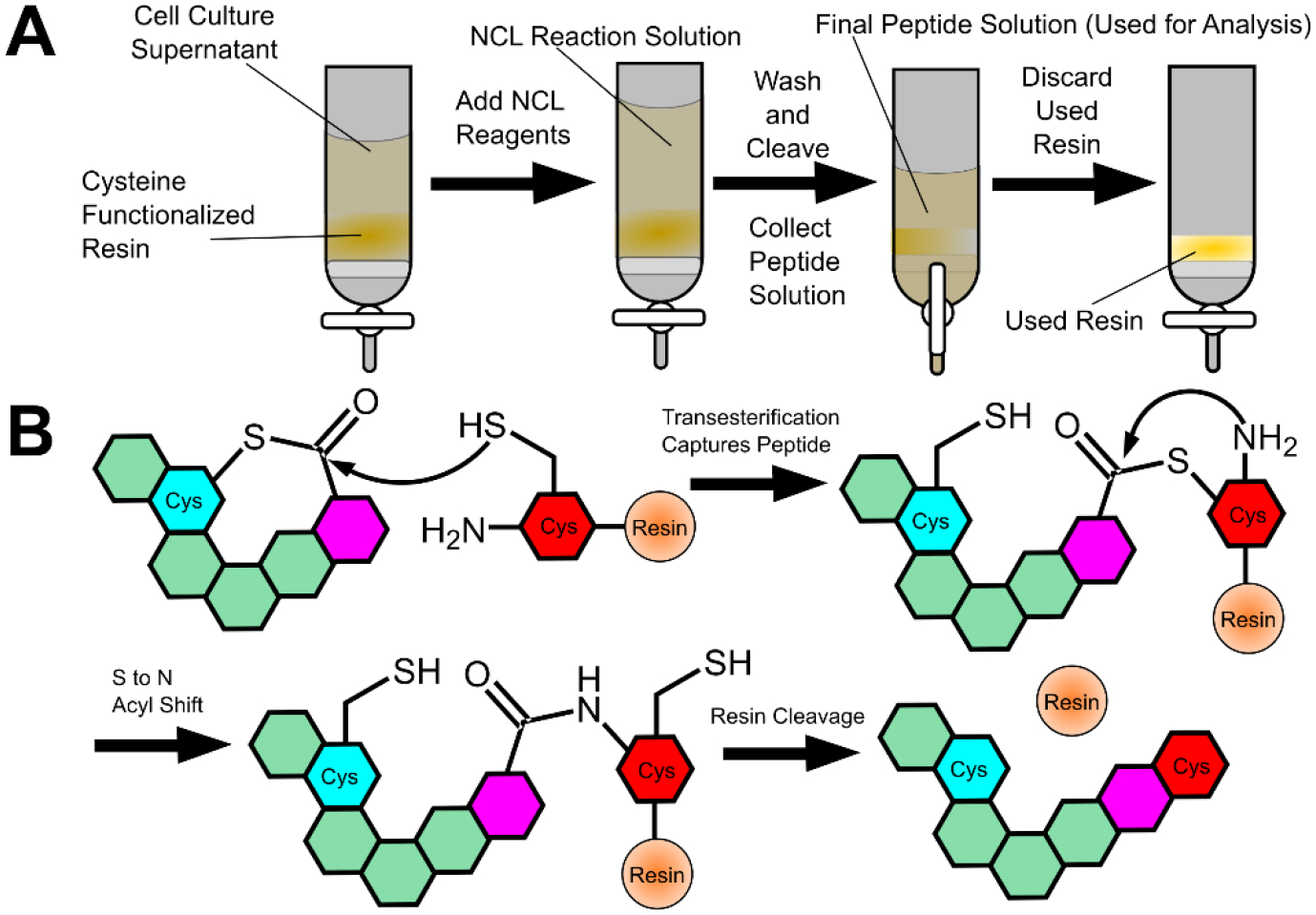
A) The general procedure involves treating cell supernatant with functionalized resin and NCL reagents in order to trap the peptide on the insoluble resin. B) A thiol-thioester exchange reaction traps the peptide on the resin. This reaction will be reversible if it occurs with other cysteines but is irreversible when reacting with the cysteine on the resin due to an S to N acyl shift.
This method was used to confirm 5 AIPs known to act on Staphylococcus aureus, and to identify 11 AIPs from non-aureus Staphylococci species. It was also applied toward attempting to confirm the identity of the AIP used in Listeria monocytogenes (see below). It is important to note that characterization of naturally occurring mature AIPs is difficult because the AIPs must be isolated from cell supernatants that are full of contaminants and that the AIP may be present in low concentrations relative to those contaminants. Furthermore, direct analysis of crude cell lysates can give misleading results due to the presence of many competing peptide fragments.
Thus, although this method only applies to thiolactone AIPs, this is a significant contribution to the QS field that allows for the rapid identification of an important subclass of AIPs.
Genetic analysis of the agr system in L. monocytogenes led to the proposal of a head-to-tail-cyclized thiolactone peptide, (CFMFV), as the mature signal (Table 1).42 Several synthetic analogs with different exocyclic tails were tested and the analog lacking a tail was shown to be the most active – leading to the hypothesis that it is the native peptide. As with Clostridium acetobutylicum (Table 1),40 the proposed peptide is potentially capable of undergoing an S to N acyl shift. Regardless, the peptide (and several additional analogs) were demonstrated to be active in reporter strains.42 The synthetic analogs could also rescue the QS phenotype in AIP deletion mutants. When the deletion reporter strains were co-cultured with wildtype L. monocytogenes, they demonstrated QS activity, implying the presence of an active, naturally produced AIP. However, efforts to isolate the proposed AIP from L. monocytogenes cultures using the new methodology (see above) were unsuccessful,43 even though it was demonstrated that the methodologies employed could detect the chemically synthesized AIP when it was doped into cell cultures at low concentration. Since the AIP-detection methodology developed is generic for any thiolactone, it is somewhat surprising that no signal was isolated (either the one proposed or a related analog). Although the potential S to N acyl shift did not appear to disrupt the ability to detect the peptide when it was doped in, if the naturally produced peptide was almost exclusively present as the lactam form instead of the thiolactone, it would not be detectable using this technique.43 At this point, it is still not clear what the exact sequence and structure of the native mature AIP in L. monocytogenes is, although the available evidence does support that the QS circuitry is active, including the gene responsible for producing the propeptide signal.
1C. Development of an AIP Prediction Model
Efforts have been made to develop tools that can aid the identification of AIPs. To this end, a computer algorithm was developed that evaluates how likely a given sequence is to be an AIP.49 The program QSPpred was trained using 220 unique and experimentally validated AIP sequences and a negative data set assembled from the combination of 5 experimentally validated Non-AIPs and 215 putative non-AIPs from UniProt. This training was used to develop predictive patterns based on a variety of characteristics including sequence features and physicochemical properties. The program was also used to make predictions for AIPs among Gram positive/negative bacterial species and even among some archaea. As these predictions are tested and the body of knowledge available to programs like this broadens, these in silico methods show the potential for being increasingly valuable in supporting QS research efforts.
2. Establishing the Structure-Activity Relationships between the Signal and Its Receptor and Peptide Lead Development
After identification of the QS signal, it is important to understand the effects of the AIP’s structural features on its interaction with the receptor. This understanding, termed structure-activity relationship (SAR), can be determined through many methods including systematic modifications (scans) of the peptide through substitution (alanine, epimer, etc.), functionalization (methylation, capping, etc.), or truncation (see Figure 3). Additional methods include modeling the binding of the signal to the receptor. From this information, other scans or analogs can be designed. Additional SAR details and context can be obtained through conformational analysis using circular dichroism (CD) or through the use of X-ray or NMR structures.
Figure 3: Common SAR Scans.
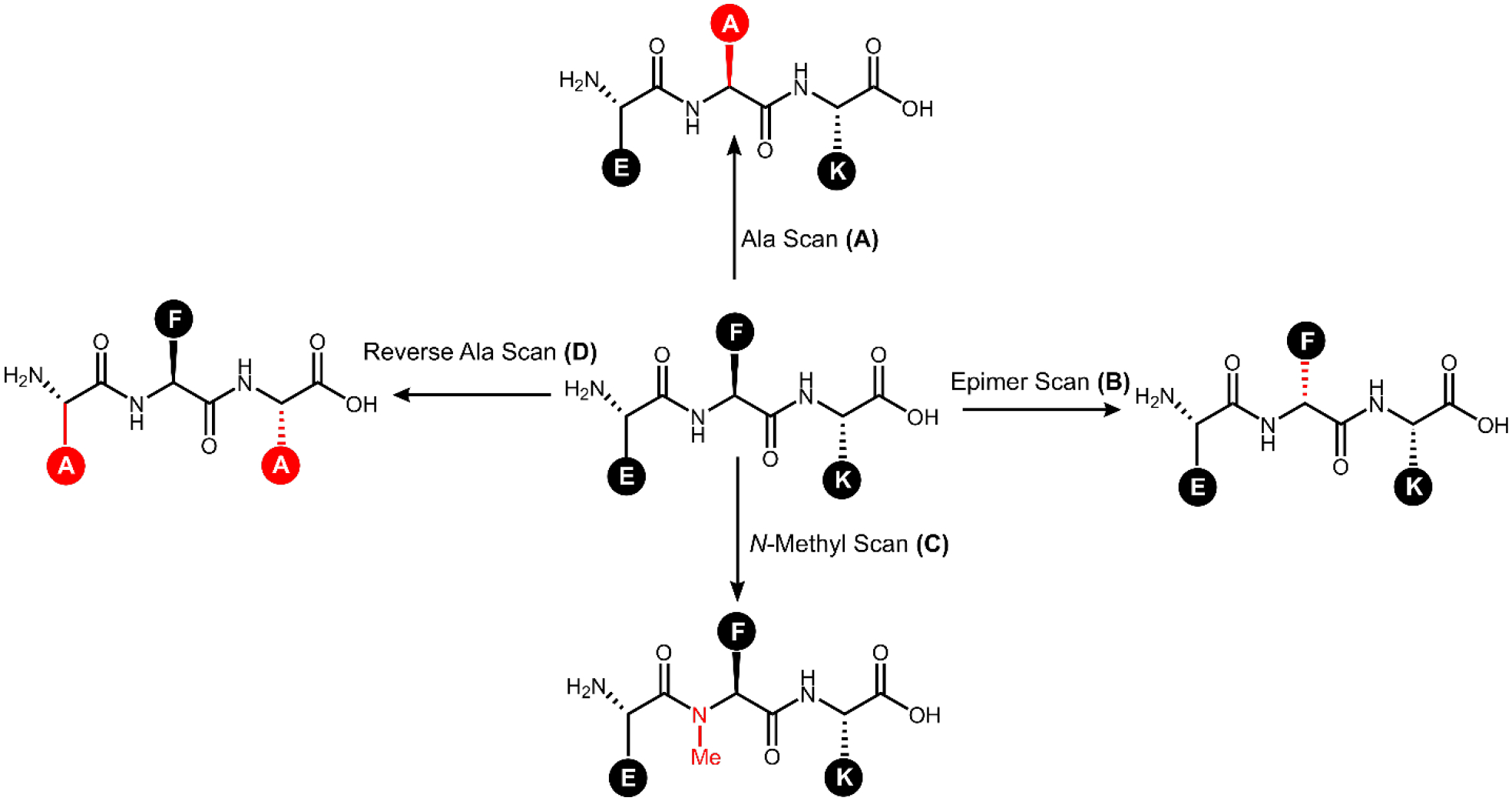
The peptide EFK is used to demonstrate several common structural scans with the associated changes colored in red. A) Alanine scans replace each non-alanine amino acid with alanine (F2A substitution shown) and provide insights into the importance of the side chain functional groups – in this example the Ala scan library would contain three peptides. B) An epimer scan replaces each amino acid with its epimer (converting L- to D-, f2 substitution shown) and provides insights into the importance of side chain orientation. C) N-methyl scans replace each amino acid with its N-methylated version (NMe-F2 substitution shown) and provide insights into the importance of backbone hydrogen bonding and steric effects. D) Reverse-alanine scans retain previously identified crucial residues (F2 in this example) and substitute alanine at all other locations; systematically back-substituting each original amino acid allows for minimal active scaffolds to be identified.
2A. Progress in Identifying Bacillus QS SARs and Lead Development
The co-evolutionary relationship of a RRNPP system receptor that resembles PlcR, NprR, and its AIP, NprRB, was investigated in the Bacillus cereus group and synthetic peptides based on the associated AIPs were used to evaluate the response of NprR in Bacillus thuringiensis.37 When SKPDI (see Table 1) and two analogs; SKPDT and SAPDT were tested for their ability to induce the expression of cry1Aa in B. thuringiensis, it was found that all three could elicit a response in a reporter assay with varying potencies, but only SKPDT gave a response above background at 100 nM.37 Experiments with hepta, octa, and nonapeptides (based on SKPDIVG, Table 1) found that the heptapeptides YSSKPDI, SSKPDIV, and SKPDIVG, all had mild inhibitory effects. The octapeptide, SSKPDIVG (Figure 4), had the most pronounced inhibitory effect (observed as a 35% drop in signal relative to the untreated control) at 20 nM while the nonapeptide YSSKPDIVG had no effect. It was found that SKPDIVG and SSKPDIVG could upregulate sporulation in the bacteria (increasing efficiency by 2.1 and 1.6-fold, respectively).
Figure 4: Inhibitor of Bacillus thuringiensis Quorum Sensing Circuitry.
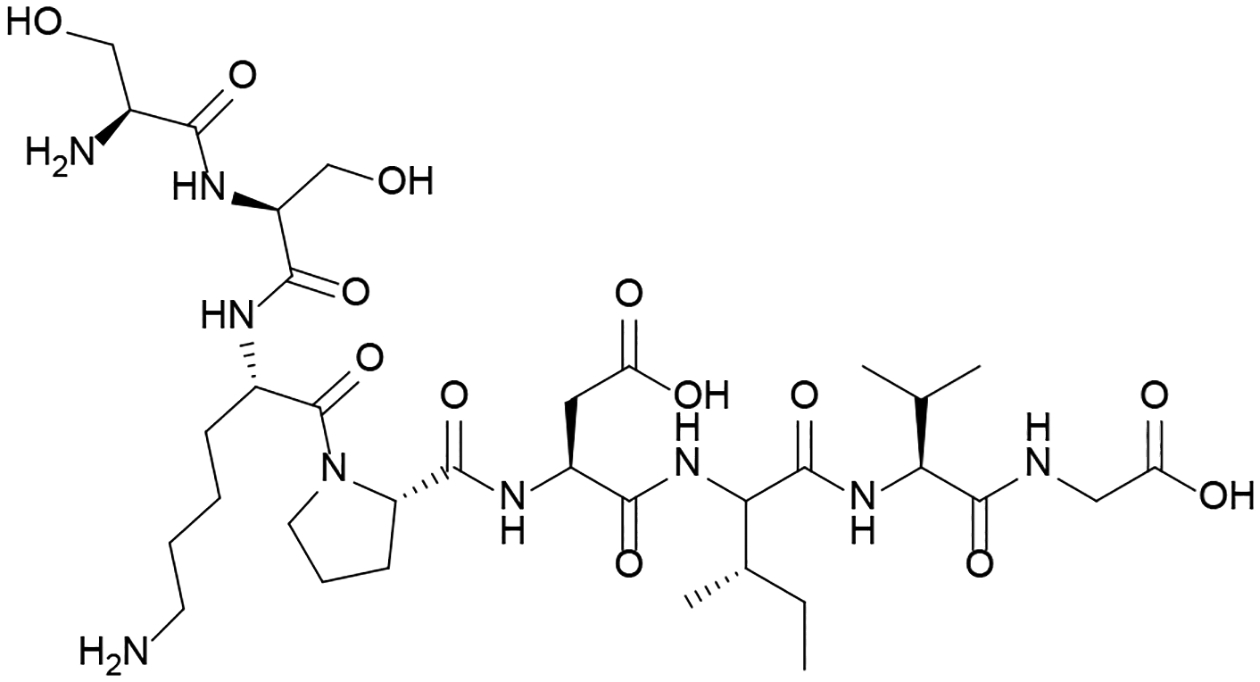
The octapeptide SSKPDIVG was found to exhibit the most potent inhibition of cry1Aa upregulation.
In order to establish an SAR between the PlcR receptor and its signal, PapR, alanine and epimer scans were performed, resulting in the discovery of one activator and five inhibitors of this RRNPP QS circuit based on the PapR7 peptide.50 PapR is processed to the mature C-terminal heptapeptide, PapR7, that is responsible for binding the PlcR receptor. The results of scans with PapR7 suggested that the side chains in positions 3–7 and their stereochemistry are critical for activation of the receptor. In addition, it was confirmed that another truncated version of PapR, the C-terminal pentapeptide, PapR5, is equally active as the PapR7 analog. Analog activity was evaluated using a reporter mutant strain and a hemolysis phenotypic assay. X-ray crystallographic analysis of a truncated version of the receptor NprR in B. cereus (with the DNA binding domain removed) bound to the AIP (SKPDIVG) has given insights into the signaling peptide’s SARs.51 As is common with such receptors, the NprΔHTH receptor was purified as a dimer. A 3.5 Å resolution crystal structure was obtained and two forms of dimerization were observed. Receptor mutations coupled with isothermal calorimetry (ITC) and transcription analysis experiments indicated that the AIP was involved in facilitating dimerization solely at one dimerization site and likely involve interaction with residues N407 and Y410 and that an active tetramer is formed through a second dimerization at another site involving residues Y223 and F225. The AIP binds in a deep cleft in the receptor with its Asp4 residue forming a distinct hydrogen bond with Arg126 on the receptor. Mutation and ITC experiments revealed that this interaction is important for maintaining high binding affinity and is crucial for activating the receptor response. ITC experiments involving the AIPs from other pherotypes (closely related strains that use AIPs with alternative sequences) indicated that positions 5 and 6 tolerate changes in amino acid content and are not crucial for binding to/activating the receptor. AIPs containing a tryptophan at position 1 did not bind as well, with steric hinderance being the most likely reason. Results also suggest that Gly7 in the native AIP is important for allowing the peptide to adopt the correct orientation when binding to the receptor and that Lys2 may also play a role in activating the receptor.
2B. Progress in Identifying Clostridium QS SARs and Lead Development
In C. perfringens, the VirSR QS circuitry controls the expression of several genes that encode toxins and is regulated by the five-membered thiolactone peptide AIPCp, (CLWFT). An alanine scan of AIPCp revealed that W3A and F4A substitutions resulted in a complete loss of activity.52 Further testing of these two modifications and their ability to inhibit QS showed that the F4A analog was able to inhibit the QS pathway, suggesting that Phe4 is associated with activation while Trp3 is required for binding.
Data obtained for AIPCp in C. perfringens was used in the rational design of two QS inhibitors (a partial agonist and a partial antagonist).52 L2A and T5A substitutions were combined to generate a partial agonist. The comparatively low activity of this partial agonist allowed it to act as a competitive inhibitor of QS. The second peptide design (Figure 5) combined modifications to residues that SAR had suggested were important for receptor activation (F4A and T5S substitutions). The resulting peptide functioned as a competitive inhibitor. However, inhibition reached a maximum threshold beyond which increasing concentration no longer had an effect, making it a partial antagonist.
Figure 5: Inhibitor of Clostridium perfringens Quorum Sensing Circuitry.
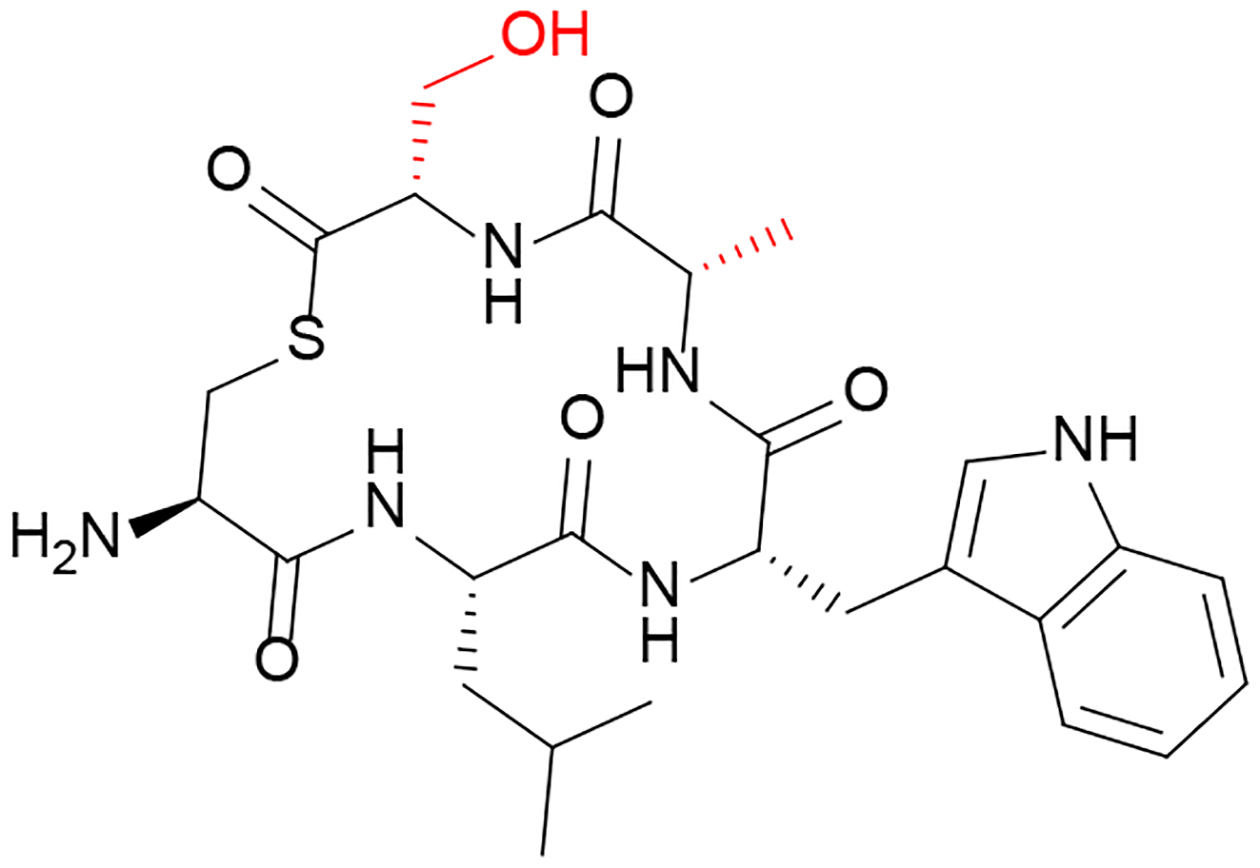
The designed analog AIPCP-F4AT5S was found to act as a partial antagonist of the VirSR QS circuitry.
2C. Progress in Identifying Enterococcus QS SARs and Lead Development
The fsr QS circuitry in E. faecalis is under the control of an AIP known as Gelatinase Biosynthesis Activating Pheromone (GBAP), QN(SPNIFGQWM). SAR experiments involving an alanine scan and an epimer scan of GBAP were conducted.53 It was found that the two N-terminal exocyclic tail residues do not contribute to bioactivity and are therefore dispensable. All residues in the macrocycle, except for Pro4, were determined to be important for activity as modifications of these residues to alanine resulted in a reduction in potency. Interestingly, the P4A analog had increased potency. An epimer scan revealed that D-substitutions in the tail residues increased potency, while chirality modifications to amino acids in the macrocycle, with the exception of Phe7, were not tolerated. In the case of Phe7, inverting the chirality did not impact the potency significantly, suggesting that the identity of the residue in the seventh position is more important than its orientation. The SARs between GBAP and its receptor were further investigated using an N-methyl scan.54 Experiments with a reporter system showed that only the modification of Phe7 in this manner was tolerated. Protease stability experiments with chymotrypsin and blood plasma indicated that this modification can be used to defend peptide analogs against digestion without sacrificing activity, which can be used when designing analogs of this peptide that may be used as drugs.
The information obtained from studying GBAP in E. faecalis was used to develop “super” agonists and antagonists.65 Substitutions shown in the systematic SAR studies to enhance binding/agonism were combined. It was found that only some of the modifications could be combined in an additive fashion, while other combinations conflicted and reduced the potency of the analog (for example the q1 modification was incompatible with the P4A substitution, resulting in reduced potency when combined). This was found to be particularly true for the antagonist analogs. Combining an n2 with a P4A substitution resulted in an agonist peptide with a 24-fold improvement in potency as determined by reporter assays. The antagonists included an unusual benzyl-tyrosine substitution at Asn5 that had previously been identified.66 It was shown that this substitution alone was sufficient to convert the peptide from a full agonist to a partial antagonist.65 This benzyl-tyrosine substitution, when combined with M11A resulted in the most potent inhibitor (Figure 6). Crystal violet biofilm phenotypic assays revealed that this inhibitor is capable of attenuating the ability of E. faecalis to form biofilms, with biofilm production reduced to the levels observed in QS-inactive mutants.
Figure 6: Inhibitor of Enterococcus faecalis Quorum Sensing Circuitry.
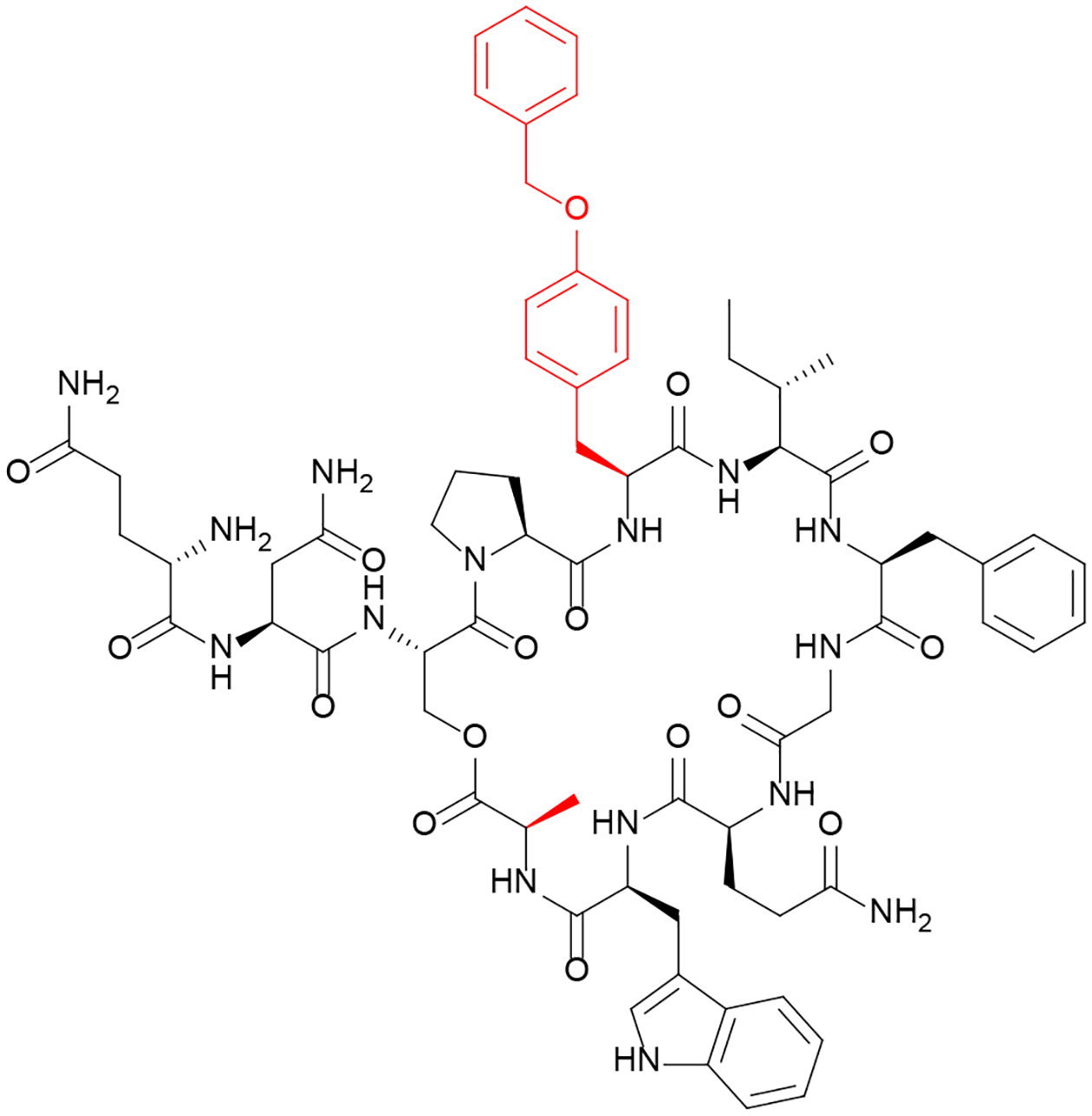
The designed analog GBAP-N5[YBzl]M11A was found to be a potent inhibitor of the Fsr QS circuitry.
2D. Progress in Identifying Streptococcus QS SARs and Lead Development
In order to gain insight into the effect of terminal residues on 21-CSP (competence stimulating peptide) activity in S. mutans, a library of peptides with truncations at both termini was synthesized.56 Results suggested that the C-terminal end of the peptide is more important for receptor binding than the N-terminus and that the peptide’s activity is not negatively impacted when cleaving the three N-terminal amino acids. These results are interesting since the protease SepM cleaves the three C-terminal residues of the 21 amino acid CSP (21-CSP) extracellularly into the final mature 18-CSP.57 It was also shown that modifications of the hydrophobic residues Phe-7, Phe-11, and Phe-15 resulted in loss of activity, suggesting that these hydrophobic residues play a key role in the activity of S. mutans CSP. Subsequent systematic SAR studies with S. mutans performed on both 21-CSP and 18-CSP against the ComD receptor confirmed the important hydrophobic groups and dispensable N-terminus residues.17 Additionally, 21-CSP alanine scan results revealed that G2A, S3A, and Q17A substitutions increase SepM recognition and processing of the peptide into 18-CSP. Contrary, Leu4, Asn12, and Arg13 were identified as important for SepM recognition and processing of 21-CSP. Interestingly, the identity of the three C-terminal residues are not important for SepM recognition and cleavage. An epimer scan revealed that changes to residues in the central and N-terminal regions are more tolerated than changes in the C-terminal region, suggesting that the C-terminal region is more important for recognition by SepM to facilitate processing into the active 18-CSP. Further investigation of the secondary structure of 21-CSP and 18-CSP by CD spectroscopy revealed that active analogs adopted an α-helix or a distorted α-helix conformation while inactive analogs adopted β-sheet conformations.17 These results suggest that α-helical secondary structure is important for CSP binding to ComD. N-methyl and reverse alanine scans were conducted on the important hydrophobic region of 18-CSP to differentiate between residues responsible for binding and those necessary for activation.58 In the N-methyl scan, terminal residues tolerated the changes in comparison to the modifications in the central region. These results align with the CD data and highlight the importance of hydrogen bonding of the peptide backbone in the central region. In the reverse alanine scan, all analogs were found to activate the pathway, indicating that the important hydrophobic residues in S. mutans CSP are involved in both receptor binding and activation, unlike the CSP of S. pneumoniae (see below).
In a less common approach, the SARs between CSP and ComD in S. mutans were investigated from the perspective of the receptor by using several topological prediction methods to construct a model of the ComD receptor. Reporter mutants were then used to assess the functionality of the predicted structural features.59 The model receptor consists of six transmembrane segments, three extracellular loops, and a C-terminal hydrophobic region in the cytoplasm. By using reporters with different in-frame insertion sites coding for partial ComD receptors, it was concluded that the three extracellular loops were responsible for CSP binding. Further study of loops A, B, and C, from N-terminus to C-terminus, concluded that deleting loop A did not affect CSP binding, suggesting that loop A does not participate in CSP recognition. Mutations of residues LDGT in loop B resulted in a reduction of reporter activity, suggesting that these residues participate in CSP recognition. In loop C, deletion of residues NVIP resulted in complete loss of activity, suggesting that these residues are essential for CSP recognition.
An SAR study that employed targeted mutations at conserved residues identified QS inhibitors for the two most common pherotype groups in S. pneumoniae.60 The first and third positions on the AIP for pherotype 1 (CSP1) were necessary for the peptide to induce competence as activity was lost when E1A and R3A substitutions were made. Substitutions and deletions were also conducted on N- and C- terminal residues. Synthetic peptides were tested for their ability to induce QS-dependent competence in a wildtype strain by monitoring transformation frequency and promoter activity of the gene responsible for QS-dependent competence. It was found that the two Lys residues on the C-terminus were unnecessary for competence induction. However, E1A and R3A substitutions significantly reduced CSP1’s ability to induce competence. Only the E1A analog could act as a competitive inhibitor of competence induction. This result suggests that Glu1 is important for activating the receptor while Arg3 is required for receptor binding. The E1A analog also delayed the onset of spontaneous competence development when used to pre-treat cells and inhibited virulence factor production in S. pneumoniae. These results strongly suggest that the E1A analog can act as an inhibitor of QS. It was also shown that an E1A substitution in the pherotype 2 (CSP2) AIP, resulted in an inhibitor that could reduce competence in pherotype 2 bacteria. Both pherotype E1A substituted analogs exhibited weak pan-group inhibitory effects (requiring higher concentrations to exert inhibitory activity), allowing the pherotype 1 analog to inhibit QS in type 2 bacteria and vice versa.
Systematic alanine and epimer scans were conducted on CSP1 and CSP2 of S. pneumoniae.8 Results revealed that the three N-terminal amino acids are vital for both CSP1 and CSP2 activity, while the C-terminal three amino acids are unnecessary for activity. CSP2 had a larger central hydrophobic region than CSP1 and tolerated single substitutions well in most positions. Interestingly, a d10 substitution increased CSP2 potency by 17-fold. In CSP1, the hydrophobic residues in positions 4, 7, 8, 11, 12, and 13 were found to be important for receptor binding. Cross-group experiments showed that CSP1 analogs were more potent against the ComD2 receptor than CSP2 analogs were against ComD1. This trend was found to be reversed when inhibitory potency was considered. CD experiments revealed that CSP1 analog potency increased with increased α-helical character. In a follow-up study, the important hydrophobic residues in CSP1 were evaluated by conducting conservative substitutions.61 Leu and Ile were replaced with Leu, Ile, Val, norleucine, or norvaline while Phe was replaced with phenylglycine, homophenylalanine, or Tyr. The results of this SAR study concluded that the naturally occurring aromatic residues are ideally occupying the binding pocket while Leu4 can accommodate all tested modifications. These results suggest that there is an important interaction between the δ carbon of Leu4 in CSP1 and the ComD1 binding pocket in S. pneumoniae. Further investigation of CSP1 provided insight into the optimal identities of important N-terminal residues to maximize activation of the ComD1 receptor.62 The N-terminal residues were further studied by replacing them with amino acids with similar size but different polarities. By replacing Glu1 with Ser, Asn, Dap, Asp, and Ala, it was discovered that Dap in the first position yielded an analog with the highest activity. Both the E1A (in agreement with earlier results) and E1S analogs exhibited inhibitory activity, suggesting that the N-terminal Glu side chain is necessary for activation, but not necessarily for binding. The 2-carbon linker in the Glu side chain was found to be optimal for activity. Capping experiments exhibited that an N-terminal positive charge is necessary for activity.
Structural analysis of AIP analogs using NMR for both pherotypes in S. pneumoniae identified generic structural features required for receptor recognition and activation.63 Analogs from earlier SAR studies (see above) were chosen to include inactive analogs, activators, inhibitors, and/or the native peptides derived from both pherotypes. The peptides were analyzed under membrane-mimicking conditions. CSP1 was found to adopt an amphiphilic α-helical conformation, confirming what had previously been observed,64 although with improved resolution. The hydrophobic “patch” of the α-helix represented many of the amino acids found to be important for receptor binding, strongly suggesting that this is the interacting part of the peptide. Furthermore, all the peptide analogs that were active against the type 1 receptor shared a similar “patch”. Interestingly, the E1A analog discussed above also retained the “patch”, emphasizing that the N-terminal glutamate is most likely responsible for receptor activation rather than binding. The inactive peptide analog containing an R3A substitution was shown to have a disrupted “patch”, explaining its inability to bind to the receptor. It was found that Phe11 is directly involved in receptor binding beyond its contribution to the formation of the hydrophobic “patch”, since the F11A analog lost activity despite the substitution retaining the rest of the “patch” intact, emphasizing the importance of the phenyl side chain for binding. Not too surprisingly, the incorporation of a D-amino acid at the 11 position was disruptive to helix formation. Like CSP1, CSP2-d10 also adopted an α-helical conformation resulting in a similar hydrophobic “patch”. These experiments emphasize the importance of α-helical character in the AIP for binding to and activating the associated receptors in S. pneumoniae.
A second generation library of CSP1 and CSP2 analogs in S. pneumoniae was constructed by combining changes informed by the earlier systematic studies.8 For both CSP1 and CSP2, truncation of the first two amino acids produced weak inhibitors. Three analogs were ComD2 inhibitors with one (CSP2-E1Ad10) being particularly potent and another (CSP2-E1Am2d10) exhibiting weak pan-group activity. Additional efforts were made to use these results to rationally design pan-group inhibitors in S. pneumoniae based on the CSP2 peptide sequence.67 In this design approach, an active truncated CSP2 sequence served as the starting scaffold and CSP1 sequence substitutions were introduced in an effort to create a “hybrid” AIP. While this approach has led to the identification of some additional pan-group activators, no improved pan-group inhibitors were obtained. Furthermore, it was found that individual modifications that resulted in the formation of an inhibitor when applied to the CSP2 sequence were incompatible when combined. Interestingly, although ComD2 inhibition activity was lost, ComD1 inhibition activity was gained. Additional efforts were made to develop a potent pan-group inhibitor.68 A thorough SAR combinatorial approach that sought to develop a potent pan-group activator and then convert it to an inhibitor by including the E1A and/or d10 substitutions failed to yield any pan-group inhibitors, but did yield additional data that were applied toward optimizing the most promising pan-group inhibitor scaffold, CSP2-E1AI4Nvad10 (Nva = norvaline). Eventually, the optimization resulted in the first pan-group inhibitor, CSP2-E1AI4Nvad10L14Q, with nanomolar potency against both S. pneumoniae pherotypes.
Previous NMR structural analysis was used to help direct the design of cyclic CSP1 AIPs.69 Since an α-helical conformation was found to be associated with interaction with the ComD1 receptor, stapling techniques were applied to design macrocyclic peptides with enhanced binding characteristics. Although all the initial ring-position modifications reduced the potency of the peptide, the most active peptide was selected to be further optimized. Ring-size optimization revealed that bridges of 18 to 19 atoms were optimal for peptide activity, leading to the identification of two potent pan-group activators. A data set was prepared including the two active peptides and two inactive peptides and additional NMR structural analysis was performed to compare the hydrophobic “patches” previously identified as being important for receptor binding. When SAR substitutions associated with the conversion of peptide agonists into antagonists were applied, several were able to convert the macrocyclic peptides from pan-group activators into potent (low nanomolar potency) pan-group inhibitors. The lead compound (Figure 7) to emerge from this design was CSP1-E1A-cyc(Dap6E10). NMR analysis of this peptide revealed that it had hydrophobic “patches” that were compatible for binding to both the ComD1 and ComD2 receptors.
Figure 7. Pan-Group Inhibitor of Streptococcus pneumoniae pherotype groups 1 and 2.
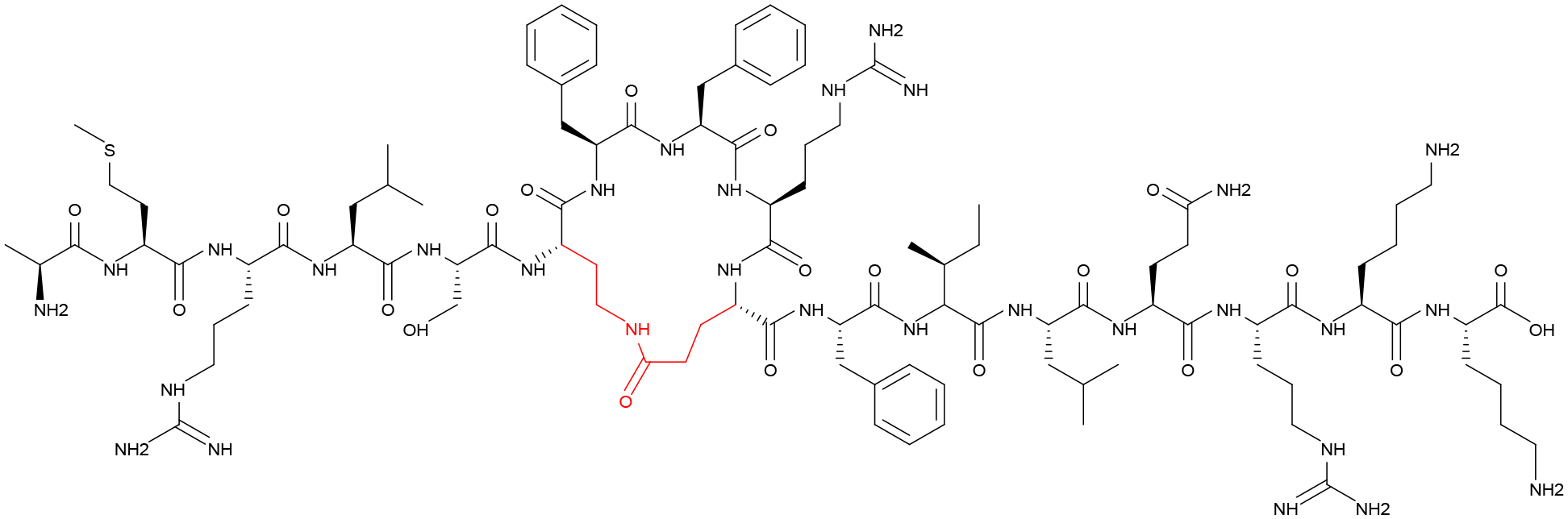
The designed analog CSP1-E1A-Cyc(Dap6E10) was found to be a potent inhibitor of the CSP QS circuitries in both CSP1 and CSP2.
The potential crosstalk between different streptococcal species and both S. pneumoniae pherotypes was investigated through synthesizing the putative AIPs (including the two most common pherotypes for S. pneumoniae and S. mitis) from Streptococci in the mitis group and a common AIP shared by Streptococci in the anginosis group and evaluating their ability to activate QS using a S. pneumoniae reporter assay.70 Out of all of the AIPs studied, only one, from S. mitis, was able to fully activate the receptors from both pherotypes. The sequence of this AIP, EIRQTHNIFFNFFKRR (S. mitis CSP2), was used as a template for the development of pan-group modulators of S. pneumoniae QS. Based on SAR results with the native S. pneumoniae AIPs, substitution efforts were focused on the N-terminal end of the template and key positions in the S. pneumoniae AIP were substituted into the S. mitis AIP. Although several substitutions resulted in improved potency, introduction of the conserved methionine from S. pneumoniae had the most significant effect, improving potency against both pherotypes by roughly 6-fold. Many of the substitutions in the hydrophobic “patch” region, tended to improve potency against ComD1 in particular. Analogs that combined modifications that individually gave improved potencies against both pherotypes in S. pneumoniae were prepared. One hydrophobic “patch” substitution, Q4L was found to be incompatible with other “patch” substitutions while one pairing of substitutions, N7I and I8F, resulted in the greatest improvement in pan-group potency relative to the original S. mitis AIP; roughly 7-fold enhancement for ComD1 and 26-fold enhancement for ComD2 interactions. Most triple modifications were poorly tolerated, however, several that included both the I2M and I8F substitutions resulted in pan-group nanomolar activators. In an effort to convert the most potent activator analogs into inhibitors, the E1A modification that worked for the native S. pneumoniae AIPs was applied. It was found that all analogs that also shared the N11F substitution were inactive as inhibitors, emphasizing that there are differences in the requirements for agonist and antagonist activity. This approach did yield three nanomolar potency pan-group inhibitors of S. pneumoniae QS: S. mitis-CSP-2-E1AN7II8F, S. mitis-CSP-2-E1AI2MN7II8F, and S. mitis-CSP-2-E1AI2MN7FI8F.
3. Identifying the Regulatory Role(s) and Therapeutic Potential of Analogs
The identification of regulatory roles of peptides often involves assessing the downstream effects of the AIP signal. As a result, reporter strains can once again be leveraged, and those results combined with phenotypic studies. There are two popular complementary approaches for conducting these studies. The first is through genetic manipulation. Mutant strains are developed that contain gene deletions or nonsense mutations to remove part of the downstream pathway and the bacteria are then tested with and without the native AIP to see if there is a loss of response due to the gene product and/or peptide being absent. Rescue experiments can then exogenously add (for instance via transformation with a plasmid containing the missing gene(s)) the missing component(s) to confirm that a wild type response can be restored. A second approach relies on the identification of potent activators or inhibitors of the QS system, usually peptides found from systematic SAR studies, rationally designed, or identified through screening experiments. With these tools, the ability to upregulate or downregulate phenotypes can be controlled through adjusting the concentration of the native peptide and/or the appropriate analog. Combining both approaches together provides the opportunity to clearly establish the regulatory role that the native peptide plays as part of the wider QS response. This combination of approaches is also necessary in order to evaluate how effective a given peptide analog might be as a therapeutic. Therapeutic assays can include both cell and/or animal infection models. Peptides that perform well in these may become candidates for clinical testing.
3A. Progress Understanding QS Regulation in Bacillus Species
The dependence on signaling by the AIP, ComX, on the expression of exoproteases was recently investigated.71 Experiments were conducted comparing AprE (a precursor for the dominant exoprotease produced by B. subtilis, subtilisin) reporter mutants containing a ComQ deletion (preventing formation of mature ComX) with wild type AprE reporter mutants. Results suggest that AprE production may be responsive to ComX. Furthermore, treating ComQ-deletion mutant cultures with spent media from an E. coli strain that produces mature ComX rescued the associated reporter activity. It was found that ComX induction was inhibited by elevated exoprotease concentrations. Doping in mature subtilisin into the spent media of the ComX-producing E. coli also inhibited the ability of the spent media to produce a signal in the ComX-responsive reporters. These results suggest that ComX may be degraded by the same exoproteases that it is responsible for inducing.
The interplay between the AIP (PapR) and a key transcription factor, CodY, in the RRNPP QS circuit in B. thuringiensis was further characeterized.72 The activity of CodY was detected through the expression of the phospholipase lecithinase in hemolysis and egg-agar assays. Rescue assays strongly suggested that CodY relies on PapR for its own upregulation. LC-MS/MS experiments suggested that CodY is needed to upregulate the production of protein permeases such as OppA and that several OppA-like proteins are needed for the effective import of mature PapR into the cell. Another recent study in B. thuringiensis looked more closely at the interactions between the mature heptapeptide PapR7 and its associated receptor complex, PlcR, and/or between PapRa7 and the PlcRa receptor.73,74 Deletion mutant and rescue experiments indicated that PapR7 is primarily responsible for the activation of PlcRa. However, results also indicated that PapRa7 is able to activate the PlcRa receptor in the absence of PapR7. PlcRa was shown to also be responsive to other PapR7 signals from different pherotypes, although the intensity of the response varied. Interestingly, although the strain tested was associated with pherotype I, the AIP signals from pherotypes II - IV all elicited significantly more intense responses, suggesting they are comparatively more potent. Peptide transporter deletion experiments with the two AIPs suggested that the peptide signals use different import transporters. In silico experiments suggested that binding between PapR7 and the PlcRa receptor is significantly more favorable than binding between the receptor and PapRa7. Hemolysis experiments with the PapR7-based inhibitors (see section 2A above) revealed that all four inhibitors with the following substitutions were capable of reducing hemolysis in both reporter strains and in wildtype B. cereus: P4A, E6A, F7A, e6, and f7.50 These inhibitors all exhibited half-minimal inhibitory concentrations (IC50 values) in the low micromolar range.
3B. Progress Understanding QS Regulation in Clostridium Species
Genetic analysis identified eight putative RRNPP QS circuits in C. acetobutylicum, and these were evaluated for their ability to influence the formation of industrially-relevant products such as butanol.40 The analysis indicated that these putative QS circuits were distinct from other previously identified RRNPP circuits. When evaluated for their ability to form butanol, insertion-inactivated receptor mutants distributed themselves into four categories: 1) similar to wildtype, 2) high early butanol producers, 3) low early butanol producers, and 4) low late butanol producers where early and late refer to the timing with respect to the initiation of solventogenesis. Acetone production was affected similarly to butanol production; however, some variation was seen in ethanol production with two strains showing very high concentrations in the late phase. A category 2 mutant was further evaluated. It was found that wildtype colony-forming behavior could be restored through genetic complementation with a plasmid harboring the native receptor promoter and gene. Insertion-inactivation mutants of the putative AIP propeptide gene within the chosen category 2 mutant were prepared and resulted in reduced butanol production while retaining wildtype growth and spore-forming behavior. Transformation with an overexpression plasmid containing the AIP propeptide sequence showed heightened butanol production although acetate/acetone formation was unaffected.
3C. Progress Understanding QS Regulation in Lactobacillus
Work was done to identify the PlnA (an AIP and bacteriocin)-dependent QS signaling response patterns adopted by a L. plantarum when co-cultured with other sourdough lactic acid bacteria.75 This analysis included identifying changes in protein expression as well as the production of excreted peptide signals. Several Lactobacilli species, including L. sanfranciscensis and L. pentosaceus experienced significant decreases in their growth and/or cell viability levels when co-cultured with L. plantarum. The levels of PlnA in the growth media were also elevated when L. plantarum was co-cultured with either of those two species. In the case of L. sanfranciscensis, mono-culture supplemented with exogenous PlnA was sufficient to replicate the observed outcome from co-culture with L. plantarum. Analysis with two-dimensional gel electrophoresis (2-DE) revealed that L. sanfranciscensis had specific changes in protein expression in response to the presence of L plantarum. These results demonstrate the largely selective bacteriocin effects of PlnA on L. sanfranciscensis as compared to most of the other Lactobacilli tested.
3D. Progress Understanding QS Regulation in Streptococcus Species
The minimal mature AIP (PhrA) identified in the TprA/PhrA QS circuit of S. pneumoniae (see Table 1) was also studied in phenotypic and reporter assays (evaluating its interaction with the receptor TprA).44 Experiments with phrA reporter strains including an amiC (gene that codes for the putative AIP importer) deletion mutant and an amiC+tprA double deletion mutant demonstrated that the addition of exogenous putative mature AIP did not lead to significant increase in reporter signal. This is consistent with reliance on AmiC for import of the extracellular peptide. Experiments with chemically defined media (CDM) containing either glucose or galactose and a reporter strain indicated that the TprA/PhrA circuit is inactive in the presence of glucose but is active in the presence of galactose. This activity was demonstrated to be dependent on excreted PhrA by treating reporter cells with culture supernatant from wildtype cells grown in the presence of galactose. RNA sequencing combined with mutant strains grown with or without exogenous putative AIP signal demonstrated that TprA and PhrA each regulate their own expression. Several genes that have been implicated in lantibioitic peptide resistance or production are likely repressed by TprA. Exogenously added putative AIP could also upregulate these genes.
The 21-mer AIP identified in S. gallolyticus (see Table 1) was used to identify phenotypes that it regulates.46 Although this AIP’s sequence aligned well with other competence-stimulating peptides (CSPs), no induction of competence was observed when trying to use the peptide to facilitate transformation with a plasmid containing an erythromycin-resistance gene under a variety of conditions that had been demonstrated to work with other Streptococci. Likewise, crystal violet assays for biofilm formation did not yield any measurable response to the peptide. However, assays for bacteriocin production (using co-cultures and spent media prepared in the presence or absence of exogenous AIP) exhibited AIP-dependent production of a substance that is toxic to the growth of other Streptococci species.
Recently, the importance of the fibrinogen binding-associated Rgf QS circuit in S. agalactiae was evaluated.76 An allelic variation analysis divided 40 clinical strains into 17 sequence types, largely grouped by whether strains possessed mutations in their QS circuitry and the nature of those mutations. An AIP propeptide truncation mutant was further investigated. Transcription levels for genes often associated with AIP expression, such as expression of fibrinogen, remained about the same between the wildtype and the deletion mutant. Experiments using decidualized telomerase-immortalized human endometrial stromal cells (T-HESCs) as a host cell in an infection model indicated that the deletion mutant was less effective at binding to the host cells compared to the wildtype and that the wildtype exhibited elevated levels of rgfC transcription in the presence of the host cells while the deletion mutant did not. Complementation with a strain harboring an intact rgfD gene rescued this response to the host cells.
Experiments with the ComS AIP identified in S. thermophilus (See Table 1), suggested that the AIP is not released to the supernatant in significant concentrations.16 Only the supernatant from a cultured overexpression mutant could cause significant response in the wildtype reporter strain. However, co-culture allowed for significant signal production, suggesting that the peptide is at least released to the cell surface. The signal was especially enhanced in an overexpression mutant containing a deletion of peptide importer. A mutant containing an Eep (a membrane protease) deletion was incapable of producing a signal without the addition of exogenous mature AIP. This strongly suggests that Eep is involved in the processing of ComS into its mature form. Experiments with defined growth media indicated that peptides such as those in tryptone can inhibit induction of the QS system. Bacteria cultured in milk (the presumed niche environment for S. thermophilus) did exhibit the ability to auto-induce competence. Growth experiments revealed that it takes cultures 80 to 100 minutes to produce a detectable reporter signal. This suggests that in the case of S. thermophilus, this putative QS system is used as a “timer” rather than as a measure of cell density.
3E. Progress in Streptococcus Species Therapeutic Development
The S. pneumoniae CSP1-E1A inhibitory analog (see section 2D above) was evaluated for its ability to attenuate pathogenicity in a mouse model of acute pneumonia caused by pherotype 1 S. pneumoniae infection.60 When administered intratracheally following establishment of the infection, the mortality rate dropped from 75% to 40% and the rapidity with which deaths began was attenuated. The CSP1-E1A analog could also significantly reduce horizontal gene transfer, preventing S. pneumoniae in the infection model from acquiring resistance to streptomycin. Later, a competitive peptide inhibitor based on the second pherotype AIP, CSP2-E1Ad10, was evaluated for its metabolic stability, ability to attenuate virulence factor production, and ability to attenuate pherotype 2 pneumonia infections in mice.67 Stability experiments were conducted on both CSP2-E1Ad10 and CSP1-E1A in vitro by evaluating their resistance to digestion by chymotrypsin/trypsin. CSP2-E1Ad10 exhibited an increase in half-life compared to the native peptide or single substitution analogs. It also protected against virulence factors as seen by Western blotting and a hemolysis assay. Mouse studies revealed that CSP2-E1Ad10 did not induce a pro-inflammatory response in healthy mice when administered intratracheally. Furthermore, when CSP2-E1Ad10 was administered in a model of acute infection, it increased the survival rate similarly to the CSP1-E1A analog. A third potent inhibitor, CSP1-E1A-cyc(Dap6E10) (see section 2D), exhibiting pan group reactivity, was also evaluated for resistance to proteases.69 It was found that the macrocyclic region was highly resistant to proteolytic cleavage, while linear portions remained susceptible. Serendipitously, it was found that the dominant cleavage product of CSP1-E1A-cyc(Dap6E10), CSP1-E1A-des-K16K17-cyc(Dap6E10), was a 10-fold more potent inhibitor, although the initial scaffold had better solubility. The “effective half-life” was estimated to be significantly greater than 4 hours, making this the most proteolytically stable analog of the three. CSP1-E1A-cyc(Dap6E10) also prevented hemolysis from both S. pneumoniae pherotypes and gave no contraindications in the lungs, heart, liver, kidneys, or spleen when given to healthy mice. When applied to acute pneumonia infection models using either group 1 or group 2 S. pneumoniae species, it was found that administration also substantially decreased death rates. These results suggest that CSP1-E1A-cyc(Dap6E10), with its potent pan-group activity, exhibits promise as potential lead compound for the development of treatment for S. pneumoniae infections.
Summary and Conclusions
Many of the most common approaches to studying QS in Gram-positive bacteria remain the “tried and true” methods of genetic analysis and manipulation combined with chemical synthesis of the peptides. The general workflow starting from the identification of the peptide and the related machinery followed by SAR and lead design, and ending in the assessment of triggered regulatory responses and therapeutic evaluation remain in place, with improvements made that accelerate the process in several places. Methods leveraging mass spectrometry are accelerating the confirmation of mature peptides from cell cultures, one of the more challenging steps. Furthermore, the growing body of QS knowledge has allowed in silico methods to become more viable, which may accelerate research efforts further as their predictions are tested and their models are refined. Potent modulators have been developed for several species, with therapeutically promising peptide analog modulators being identified that can attenuate infections by Streptococcus pneumoniae. There is an increasing number of studies looking at interspecies QS crosstalk, an effort that should aid in understanding the complexities of microbiomes and allow for the design of even more sophisticated peptide modulators of bacterial group behaviors.
Acknowledgements
Y.T acknowledges the National Institutes of Health (R35GM128651 and R01HL142626) for the generous support of research in his laboratory.
References
- 1.Fuqua WC, Winans SC and Greenberg EP, Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators., J. Bacteriol, 1994, 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutherford ST and Bassler BL, Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control, Cold Spring Harb. Perspect. Med, 2012, 2, a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook LC and Federle MJ, Peptide pheromone signaling in Streptococcus and Enterococcus, Fems Microbiol. Rev, 2014, 38, 473–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brackman G and Coenye T, Quorum Sensing Inhibitors as Anti-Biofilm Agents, Curr. Pharm. Des, 2015, 21, 5–11. [DOI] [PubMed] [Google Scholar]
- 5.Papenfort K and Bassler BL, Quorum sensing signal–response systems in Gram-negative bacteria, Nat. Rev. Microbiol, 2016, 14, 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monnet V, Juillard V and Gardan R, Peptide conversations in Gram-positive bacteria, Crit. Rev. Microbiol, 2016, 42, 339–351. [DOI] [PubMed] [Google Scholar]
- 7.Turan NB, Chormey DS, Büyükpınar Ç, Engin GO and Bakirdere S, Quorum sensing: Little talks for an effective bacterial coordination, TrAC Trends Anal. Chem, 2017, 91, 1–11. [Google Scholar]
- 8.Yang Y, Koirala B, Sanchez LA, Phillips NR, Hamry SR and Tal-Gan Y, Structure–Activity Relationships of the Competence Stimulating Peptides (CSPs) in Streptococcus pneumoniae Reveal Motifs Critical for Intra-group and Cross-group ComD Receptor Activation, ACS Chem. Biol, 2017, 12, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteley M, Diggle SP and Greenberg EP, Progress in and promise of bacterial quorum sensing research, Nature, 2017, 551, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abisado RG, Benomar S, Klaus JR, Dandekar AA and Chandler JR, Bacterial Quorum Sensing and Microbial Community Interactions, mBio, 2018, 9, e02331–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastings JW and Greenberg EP, Quorum Sensing: the Explanation of a Curious Phenomenon Reveals a Common Characteristic of Bacteria, J. Bacteriol, 1999, 181, 2667–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawver LA, Jung SA and Ng W-L, Specificity and complexity in bacterial quorum-sensing systems, FEMS Microbiol. Rev, 2016, 40, 738–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mull RW, Harrington A, Sanchez LA and Tal-Gan Y, Cyclic Peptides that Govern Signal Transduction Pathways: From Prokaryotes to Multi-Cellular Organisms, Curr. Top. Med. Chem, 2018, 18, 625–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reuter K, Steinbach A and Helms V, Interfering with Bacterial Quorum Sensing, Perspect. Med. Chem, 2016, 8, PMC.S13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbeke F, De Craemer S, Debunne N, Janssens Y, Wynendaele E, Van de Wiele C and De Spiegeleer B, Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In vitro and In vivo, Front. Neurosci, 2017, 11, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardan R, Besset C, Gitton C, Guillot A, Fontaine L, Hols P and Monnet V, Extracellular Life Cycle of ComS, the Competence-Stimulating Peptide of Streptococcus thermophilus, J. Bacteriol, 2013, 195, 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bikash CR, Hamry SR and Tal-Gan Y, Structure–Activity Relationships of the Competence Stimulating Peptide in Streptococcus mutans Reveal Motifs Critical for Membrane Protease SepM Recognition and ComD Receptor Activation, ACS Infect. Dis, 2018, 4, 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wynendaele E, Bronselaer A, Nielandt J, D’Hondt M, Stalmans S, Bracke N, Verbeke F, Van De Wiele C, De Tré G and De Spiegeleer B, Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides, Nucleic Acids Res, 2013, 41, D655–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM and Finlay B, The Intestinal Microbiome in Early Life: Health and Disease, Front. Immunol, 2014, 5, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, Sela DA, Muller AJ, Mullin JM, Albert K, Gilligan JP, DiGuilio K, Dilbarova R, Alexander W and Prendergast GC, The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists, Cancer Res, 2017, 77, 1783–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peñalver Bernabé B, Cralle L and Gilbert JA, Systems biology of the human microbiome, Curr. Opin. Biotechnol, 2018, 51, 146–153. [DOI] [PubMed] [Google Scholar]
- 22.Painter KL, Krishna A, Wigneshweraraj S and Edwards AM, What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia?, Trends Microbiol, 2014, 22, 676–685. [DOI] [PubMed] [Google Scholar]
- 23.Wang B and Muir TW, Regulation of Virulence in Staphylococcus aureus: Molecular Mechanisms and Remaining Puzzles, Cell Chem. Biol, 2016, 23, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balasubramanian D, Harper L, Shopsin B and Torres VJ, Staphylococcus aureus pathogenesis in diverse host environments, Pathog. Dis, 2017, 75, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salam AM and Quave CL, Targeting Virulence in Staphylococcus aureus by Chemical Inhibition of the Accessory Gene Regulator System In Vivo, mSphere, 2018, 3, e00500–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon CP, Synthetic strategies to access staphylococcus auto-inducing peptides as quorum sensing modulators, Org. Biomol. Chem, 2020, 18, 379–390. [DOI] [PubMed] [Google Scholar]
- 27.Horswill AR and Gordon CP, Structure–Activity Relationship Studies of Small Molecule Modulators of the Staphylococcal Accessory Gene Regulator, J. Med. Chem, 2020, 63, 2705–2730. [DOI] [PubMed] [Google Scholar]
- 28.Shpakov AO, Peptide autoinducers in bacteria, Microbiology, 2009, 78, 255–266. [Google Scholar]
- 29.Thoendel M, Kavanaugh JS, Flack CE and Horswill AR, Peptide Signaling in the Staphylococci, Chem. Rev, 2011, 111, 117–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haag AF and Bagnoli F, in Staphylococcus aureus: Microbiology, Pathology, Immunology, Therapy and Prophylaxis, eds. Bagnoli F, Rappuoli R and Grandi G, Springer International Publishing, Cham, 2017, pp. 145–198. [Google Scholar]
- 31.Rocha-Estrada J, Aceves-Diez AE, Guarneros G and de la Torre M, The RNPP family of quorum-sensing proteins in Gram-positive bacteria, Appl. Microbiol. Biotechnol, 2010, 87, 913–923. [DOI] [PubMed] [Google Scholar]
- 32.Do H and Kumaraswami M, Structural Mechanisms of Peptide Recognition and Allosteric Modulation of Gene Regulation by the RRNPP Family of Quorum-Sensing Regulators, J. Mol. Biol, 2016, 428, 2793–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neiditch MB, Capodagli GC, Prehna G and Federle MJ, Genetic and Structural Analyses of RRNPP Intercellular Peptide Signaling of Gram-Positive Bacteria, Annu. Rev. Genet. Vol 51, 2017, 51, 311–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perchat S, Dubois T, Zouhir S, Gominet M, Poncet S, Lemy C, Aumont‐Nicaise M, Deutscher J, Gohar M, Nessler S and Lereclus D, A cell–cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group, Mol. Microbiol, 2011, 82, 619–633. [DOI] [PubMed] [Google Scholar]
- 35.Wu L, Guo X, Liu X and Yang H, NprR-NprX Quorum-Sensing System Regulates the Algicidal Activity of Bacillus sp Strain S51107 against Bloom-Forming Cyanobacterium Microcystis aeruginosa, Front. Microbiol, 2017, 8, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esmaeilishirazifard E, De Vizio D, Moschos SA and Keshavarz T, Genomic and molecular characterization of a novel quorum sensing molecule in Bacillus licheniformis, Amb Express, 2017, 7, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha J, Flores V, Cabrera R, Soto-Guzman A, Granados G, Juaristi E, Guarneros G and de la Torre M, Evolution and some functions of the NprR-NprRB quorum-sensing system in the Bacillus cereus group, Appl. Microbiol. Biotechnol, 2012, 94, 1069–1078. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Mashooq M, Gandham RK, Alavandi SV and Nagaleekar VK, Characterization of quorum sensing system in Clostridium chauvoei, Anaerobe, 2018, 52, 92–99. [DOI] [PubMed] [Google Scholar]
- 39.Steiner E, Scott J, Minton NP and Winzer K, An agr Quorum Sensing System That Regulates Granulose Formation and Sporulation in Clostridium acetobutylicum, Appl. Environ. Microbiol, 2012, 78, 1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotte A-K, Severn O, Bean Z, Schwarz K, Minton NP and Winzer K, RRNPP-type quorum sensing affects solvent formation and sporulation in Clostridium acetobutylicum, Microbiol. Read. Engl, 2020, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal JE, Ma M, Saputo J, Garcia J, Uzal FA and McClane BA, Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity and pathogenicity of Clostridium perfringens type C isolate CN3685, Mol. Microbiol, 2012, 83, 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zetzmann M, Sánchez-Kopper A, Waidmann MS, Blombach B and Riedel CU, Identification of the agr Peptide of Listeria monocytogenes, Front. Microbiol, 2016, 7, 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gless BH, Bojer MS, Peng P, Baldry M, Ingmer H and Olsen CA, Identification of autoinducing thiodepsipeptides from staphylococci enabled by native chemical ligation, Nat. Chem, 2019, 11, 463–469. [DOI] [PubMed] [Google Scholar]
- 44.Hoover SE, Perez AJ, Tsui H-CT, Sinha D, Smiley DL, DiMarchi RD, Winkler ME and Lazazzera BA, A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniaeD39 that regulates a lantibiotic biosynthesis gene cluster, Mol. Microbiol, 2015, 97, 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aggarwal C, Jimenez JC, Nanavati D and Federle MJ, Multiple Length Peptide-Pheromone Variants Produced by Streptococcus pyogenes Directly Bind Rgg Proteins to Confer Transcriptional Regulation, J. Biol. Chem, 2014, 289, 22427–22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrington A and Tal-Gan Y, Identification of Streptococcus gallolyticus subsp. gallolyticus (Biotype I) Competence-Stimulating Peptide Pheromone, J. Bacteriol, 2018, 200, e00709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBrayer DN and Tal-Gan Y, Deciphering bacterial signalling, Nat. Chem, 2019, 11, 398–399. [DOI] [PubMed] [Google Scholar]
- 48.Dawson PE and Kent SBH, Synthesis of Native Proteins by Chemical Ligation, Annu. Rev. Biochem, 2000, 69, 923–960. [DOI] [PubMed] [Google Scholar]
- 49.Rajput A, Gupta AK and Kumar M, Prediction and Analysis of Quorum Sensing Peptides Based on Sequence Features, Plos One, 2015, 10, e0120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yehuda A, Slamti L, Bochnik-Tamir R, Malach E, Lereclus D and Hayouka Z, Turning off Bacillus cereus quorum sensing system with peptidic analogs, Chem. Commun, 2018, 54, 9777–9780. [DOI] [PubMed] [Google Scholar]
- 51.Zouhir S, Perchat S, Nicaise M, Perez J, Guimaraes B, Lereclus D and Nessler S, Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR, Nucleic Acids Res, 2013, 41, 7920–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh RP, Okubo K, Ohtani K, Adachi K, Sonomoto K and Nakayama J, Rationale design of quorum-quenching peptides that target the VirSR system of Clostridium perfringens, Fems Microbiol. Lett, 2015, 362, UNSP fnv188. [DOI] [PubMed] [Google Scholar]
- 53.McBrayer DN, Gantman BK, Cameron CD and Tal-Gan Y, An Entirely Solid Phase Peptide Synthesis-Based Strategy for Synthesis of Gelatinase Biosynthesis-Activating Pheromone (GBAP) Analogue Libraries: Investigating the Structure–Activity Relationships of the Enterococcus faecalis Quorum Sensing Signal, Org. Lett, 2017, 19, 3295–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McBrayer DN, Gantman BK and Tal-Gan Y, N-Methylation of Amino Acids in Gelatinase Biosynthesis-Activating Pheromone Identifies Key Site for Stability Enhancement with Retention of the Enterococcus faecalis fsr Quorum Sensing Circuit Response, ACS Infect. Dis, 2019, 5, 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang T, Tal-Gan Y, Paharik AE, Horswill AR and Blackwell HE, Structure–Function Analyses of a Staphylococcus epidermidis Autoinducing Peptide Reveals Motifs Critical for AgrC-type Receptor Modulation, ACS Chem. Biol, 2016, 11, 1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Syvitski RT, Tian X-L, Sampara K, Salman A, Lee SF, Jakeman DL and Li Y-H, Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans, J. Bacteriol, 2007, 189, 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biswas S, Cao L, Kim A and Biswas I, SepM, a Streptococcal Protease Involved in Quorum Sensing, Displays Strict Substrate Specificity, J. Bacteriol, 2016, 198, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bikash CR and Tal-Gan Y, Identification of highly potent competence stimulating peptide-based quorum sensing activators in Streptococcus mutans through the utilization of N-methyl and reverse alanine scanning, Bioorg. Med. Chem. Lett, 2019, 29, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong G, Tian X-L, Cyr K, Liu T, Lin W, Tziolas G and Li Y-H, Membrane Topology and Structural Insights into the Peptide Pheromone Receptor ComD, A Quorum-Sensing Histidine Protein Kinase of Streptococcus mutans, Sci. Rep, 2016, 6, 26502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu L and Lau GW, Inhibition of competence development, horizontal gene transfer and virulence in Streptococcus pneumoniae by a modified competence stimulating peptide, PLoS Pathog., 2011, 7, e1002241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koirala B, Hillman RA, Tiwold EK, Bertucci MA and Tal-Gan Y, Defining the hydrophobic interactions that drive competence stimulating peptide (CSP)-ComD binding in Streptococcus pneumoniae, Beilstein J. Org. Chem, 2018, 14, 1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y and Tal-Gan Y, Exploring the competence stimulating peptide (CSP) N-terminal requirements for effective ComD receptor activation in group1 Streptococcus pneumoniae, Bioorganic Chem., 2019, 89, 102987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y, Cornilescu G and Tal-Gan Y, Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae, Biochemistry, 2018, 57, 5359–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnsborg O, Kristiansen PE, Blomqvist T and Håvarstein LS, A Hydrophobic Patch in the Competence-Stimulating Peptide, a Pneumococcal Competence Pheromone, Is Essential for Specificity and Biological Activity, J. Bacteriol, 2006, 188, 1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McBrayer DN, Cameron CD, Gantman BK and Tal-Gan Y, Rational Design of Potent Activators and Inhibitors of the Enterococcus faecalis Fsr Quorum Sensing Circuit, ACS Chem. Biol, 2018, 13, 2673–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakayama J, Yokohata R, Sato M, Suzuki T, Matsufuji T, Nishiguchi K, Kawai T, Yamanaka Y, Nagata K, Tanokura M and Sonomoto K, Development of a Peptide Antagonist against fsr Quorum Sensing of Enterococcus faecalis, Acs Chem. Biol, 2013, 8, 804–811. [DOI] [PubMed] [Google Scholar]
- 67.Koirala B, Lin J, Lau GW and Tal-Gan Y, Development of a Dominant Negative Competence-Stimulating Peptide (dnCSP) that Attenuates Streptococcus pneumoniae Infectivity in a Mouse Model of Acute Pneumonia, ChemBioChem, 2018, 19, 2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koirala B and Tal-Gan Y, Development of Streptococcus pneumoniae Pan-Group Quorum-Sensing Modulators, ChemBioChem, 2020, 21, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Y, Lin J, Harrington A, Cornilescu G, Lau GW and Tal-Gan Y, Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity in Streptococcus pneumoniae, Proc. Natl. Acad. Sci, 2020, 117, 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milly TA and Tal-Gan Y, Biological evaluation of native streptococcal competence stimulating peptides reveals potential crosstalk between Streptococcus mitis and Streptococcus pneumoniae and a new scaffold for the development of S. pneumoniae quorum sensing modulators, RSC Chem. Biol, 2020, 1, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spacapan M, Danevcic T and Mandic-Mulec I, ComX-Induced Exoproteases Degrade ComX in Bacillus subtilis PS-216, Front. Microbiol, 2018, 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slamti L, Lemy C, Henry C, Guillot A, Huillet E and Lereclus D, CodY Regulates the Activity of the Virulence Quorum Sensor PlcR by Controlling the Import of the Signaling Peptide PapR in Bacillus thuringiensis, Front. Microbiol, 2016, 6, 1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huillet E, Tempelaars MH, André-Leroux G, Wanapaisan P, Bridoux L, Makhzami S, Panbangred W, Martin-Verstraete I, Abee T and Lereclus D, PlcRa, a New Quorum-Sensing Regulator from Bacillus cereus, Plays a Role in Oxidative Stress Responses and Cysteine Metabolism in Stationary Phase, PLoS ONE, 2012, 7, e51047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huillet E, Bridoux L, Barboza I, Lemy C, André-Leroux G and Lereclus D, The signaling peptide PapR is required for the activity of the quorum-sensor PlcRa in Bacillus thuringiensis, Microbiology, 2020, 166, 398–410. [DOI] [PubMed] [Google Scholar]
- 75.Di Cagno R, De Angelis M, Calasso M, Vincentini O, Vernocchi P, Ndagijimana M, De Vincenzi M, Dessi MR, Guerzoni ME and Gobbetti M, Quorum sensing in sourdough Lactobacillus plantarum DC400: Induction of plantaricin A (PlnA) under co-cultivation with other lactic acid bacteria and effect of PlnA on bacterial and Caco-2 cells, Proteomics, 2010, 10, 2175–2190. [DOI] [PubMed] [Google Scholar]
- 76.Parker RE, Knupp D, Al Safadi R, Rosenau A and Manning SD, Contribution of the RgfD Quorum Sensing Peptide to rgf Regulation and Host Cell Association in Group B Streptococcus, Genes, 2017, 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]


