Abstract
Dental caries (i.e., tooth decay), which is caused by biofilm formation on tooth surfaces, is the most prevalent oral disease worldwide. Unfortunately, many anti-biofilm drugs lack efficacy within the oral cavity due to poor solubility, retention, and penetration into biofilms. While drug delivery systems (DDS) have been developed to overcome these hurdles and improve traditional antimicrobial treatments, including farnesol, efficacy is still modest due to myriad resistance mechanisms employed by biofilms, suggesting that synergistic drug treatments may be more efficacious. Streptococcus mutans (S. mutans), a cariogenic pathogen and biofilm forming model organism, has several key virulence factors including acidogenicity and exopolysaccharide (EPS) matrix synthesis. Flavonoids, such as myricetin, can reduce both biofilm acidogenicity and EPS synthesis. Therefore, a nanoparticle carrier (NPC) DDS with flexibility to co-load farnesol in the hydrophobic core and myricetin within the cationic corona, was tested in vitro using established and developing S. mutans biofilms. Co-loaded NPC treatments effectively disrupted biofilm biomass (i.e., dry weight) and reduced biofilm viability by ~3 log CFU/mL versus single drug-only controls in developing biofilms, suggesting dual-drug delivery exhibits synergistic anti-biofilm effects. Mechanistic studies revealed that co-loaded NPCs synergistically inhibited planktonic bacterial growth compared to controls and reduced S. mutans acidogenicity due to decreased atpD gene expression associated with acid tolerance. Moreover, the myricetin-loaded NPC corona enhanced NPC binding to tooth-mimetic surfaces, which can increase drug efficacy through improved retention at the biofilm-apatite interface. Altogether, these findings suggest promise for co-delivery of myricetin and farnesol DDS as an alternative anti-biofilm treatment to prevent dental caries.
Keywords: Biofilm, nanoparticle, co-loading, myricetin, farnesol, drug delivery, synergy
Graphical Abstract
Introduction
Biofilms are implicated in 65% of infections and 80% of chronic infections in humans [1–3]. Biofilms are particularly difficult to treat due to their physical and chemical complexity and the presence of persister cells, which together contribute to antibiotic resistance and cost the United States more than $55 billion per year [2, 4–7]. One of the most common biofilm-associated infections globally is dental caries (i.e., tooth decay) [8, 9], which affects >60% of children and nearly all adults worldwide [10, 11]. Tooth decay is caused by oral biofilms (i.e., dental plaque) on the tooth surface, which elicit localized acidification due to sugar metabolism and lead to an imbalance in the enamel demineralization and remineralization processes, ultimately resulting in carious lesion formation [9, 11]. Although true for all biofilms, oral biofilms are particularly challenging to treat or prevent with small molecule drugs due to poor drug residence time caused by low drug solubility, brief topical-exposure regimens, salivary clearance, localized acid production (i.e., acidogenicity) and tolerance (i.e., aciduricity), and limited drug diffusion into the exopolysaccharide (EPS) biofilm matrix [12–14].
Recent efforts have utilized nanotechnology in drug delivery applications to overcome these hurdles and inhibit oral biofilms responsible for tooth decay [15–37]. Drug delivery systems (DDS) have been developed using myriad nanomaterial platforms, including natural [31, 36] and synthetic polymers [21, 23, 26, 27, 32–35] as well as inorganic materials [20, 22, 24, 25, 28, 30, 37] to inhibit the cariogenic pathogen and biofilm-forming model organism, Streptococcus mutans [38–40]. DDS have largely focused on delivery of singular antimicrobial compounds, such as the membrane-disrupting agents farnesol [21, 26, 27, 33, 34] and chlorhexidine [24, 28, 41], to disrupt S. mutans biofilm formation. These DDS approaches improve drug solubility and residence time, and some also exploit changes in pH associated with the biofilm microenvironment to trigger drug release and reduce S. mutans burden [21, 24, 26, 27, 31–35], yielding modest-to-moderate ~2-4 log and, for one, up to 6-log reductions in CFU/mL. However, to overcome antimicrobial treatments, biofilms employ many antimicrobial resistance mechanisms, including EPS matrix formation to inhibit drug penetration, altered growth and adaptive stress responses to inhibit drug uptake and metabolism, and efflux pumps to actively recognize and transport drugs out of cells [42, 43]. Therefore, more clinically relevant approaches that are more effective and efficient against S. mutans biofilms may result by exploiting multi-drug co-delivery strategies that combine antimicrobial agents with synergistic mechanisms of action against one or more of these antimicrobial resistance mechanisms.
Synergistic targets for enhancing the effects of antimicrobial agents include virulence factors, such as acid production and acid-stress adaptive responses (i.e., acidogenicity and aciduricity for S. mutans) as well as the EPS [15, 18, 44]. S. mutans’ acidogenicity and aciduricity rely on proton pumps, such as the F1F0-H/ATP synthase [38, 45–50]. In particular, the beta subunit of this ATP synthase, atpD, supports the assembly of the F1F0-H/ATPase complex, which maintains the pH gradient across the bacterial membrane by pumping protons out of the cell [46, 51]. Glucans within the EPS, which serve as the architectural scaffold, protective barrier, and prime nutrient source for bacteria within dental plaque [13, 14, 52, 53], also appear to increase retention of protons in close proximity to S. mutans cell-surface, helping to trigger the acid-adaptive response [54]. This unique association between the acid tolerance response and the EPS matrix suggests that the atp operon, which encodes the F1F0-H/ATPase complex [46, 55, 56], and glucosyltransferases (Gtf), which synthesize EPS glucans [13, 57–67], may contain promising targets. Indeed, natural products, such as catechin-based polyphenols, flavonoids, and polymeric polyphenols, have been shown to be potent inhibitors of F-ATPases [64, 68, 69] and Gtfs [13, 57–67]. The generally recognized as safe (GRAS) flavonoid myricetin inhibits both F-ATPase and Gtf activity [63–65], thereby making myricetin a key anti-biofilm drug of interest. Furthermore, treatments of farnesol, which is also considered GRAS by the US Food and Drug Administration, and flavonoids show decreased mRNA levels for both acid tolerance and Gtf genes [67] and reductions of up to ~50% in oral biofilm EPS via reduced Gtf activity [61–63, 65, 70]. These results are particularly striking considering the primary limitations of farnesol and myricetin: 1) poor solubility and biofilm retention [71, 72], which can lead to premature salivary drug clearance in the oral cavity, and 2) myricetin’s sensitivity to temperature, pH, and light [19, 72, 73]. However, the lack of a suitable delivery method for farnesol and myricetin has stymied the development of these compounds for clinical anti-biofilm use to date.
Therefore, the goal of this work is to use a polymer nanoparticle DDS to control delivery of farnesol and myricetin to a representative model oral biofilm pathogen (i.e., S. mutans), thereby enhancing their anti-biofilm efficacy. In particular, the complementary and potentially synergistic mechanisms of drug action by farnesol and myricetin against S. mutans oral biofilms when administered using controlled drug delivery was hypothesized to enhance anti-biofilm efficacy. This work tested this hypothesis on established and developing S. mutans biofilms grown on tooth-mimetic surfaces. Specifically, nanoparticle carriers (NPCs) uniquely designed to enable farnesol loading into hydrophobic cores and myricetin binding to cationic coronas were used. The pH-responsive nature of these NPCs facilitates controlled drug release of farnesol and myricetin, as previously described [27, 34, 74]. The effect of co-loaded NPC treatments on biofilm bacteria survival, biomass (i.e., dry weight), pH, EPS, and acidogenicity-related factors were explored. Furthermore, the impact of myricetin loading on the electrostatic interactions of NPC-tooth-mimetic binding, which enhances drug efficacy through improved retention on apatitic surfaces, was tested. Overall, this work suggests promise for co-delivery of myricetin and farnesol DDS as an anti-biofilm treatment for dental caries and demonstrates the feasibility of dual-drug delivery that may be applicable to other biofilm systems.
Materials and Methods
Materials
All materials were supplied by Sigma-Aldrich unless otherwise specified. Dimethylaminoethyl methacrylate (DMAEMA) and butyl methacrylate (BMA) were purified by distillation prior to use. The reversible addition-fragmentation chain transfer (RAFT) polymerization chain transfer agent (CTA), 4-cyano-4-[(ethylsulfanylthiocarbonyl)sulfanyl]pentanoic acid (ECT), and propylacrylic acid (PAA) were synthesized as described previously [75–77]. The RAFT radical initiator, 2,2-Azobisisobutyronitrile (AIBN), was recrystallized from methanol. All phosphate-buffered saline (PBS) used in this study was 1x Dulbecco’s phosphate-buffered saline (1x DPBS) unless otherwise specified. All water used was deionized and distilled with resistivity of at least 18 MΩ and is referred to as ddH2O.
Polymer Synthesis and Characterization
Cationic corona block (Block 1) synthesis
Poly(dimethylaminoethyl methacrylate), or p(DMAEMA), was synthesized via RAFT polymerization using the CTA ECT and the radical initiator AIBN. Distilled DMAEMA was mixed with specific molar ratios of ECT and AIBN so the [monomer]/[CTA]/[Initiator] = 1325/5/1 in dimethylformamide (DMF) at 40 wt% to obtain the Block 1 molecular weights shown in Table 1. Nitrogen was used with a Schlenk line to purge the reaction vessel for 45 minutes prior to submerging the vessel in an oil bath at 60 °C for 6 hours. The polymerization reaction was terminated by exposing the vessel contents to atmospheric oxygen. The product was precipitated and washed 4 times in 80:20 (v/v) pentane:diethyl ether with centrifugation and dried under vacuum overnight.
Table 1.
Characterization summary of polymers and NPCs used in this studyΔ
| Polymers |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Corona Block |
Core Block |
Diblock Copolymers |
NPC |
|||||||||||
| Polymer | DP | Block 1 Mn1 (kDa) | PDI1 | DP | Block 2 Mn1 (kDa) | %DMAEMA2 | %BMA2 | %PAA2 | Overall Mn1 (kDa) | PDI1 | CCR | Size3 (d.nm) | Size PDI3 | Z4 (mV) |
| NP25/8 | 265 | 24.5 | 1.04 | 310 | 8.0 | 12 | 81 | 7 | 32.5 | 1.02 | 3.1 | 34.3 ± 8.8 | 0.08 | 21.0 ± 5.1 |
| NP25/10 | 265 | 24.5 | 1.04 | 310 | 9.8 | 15 | 57 | 29 | 34.3 | 1.04 | 2.5 | 30.6 ± 8.2 | 0.09 | 16.8 ± 4.7 |
As characterized by
gel permeation chromatography,
1H nuclear magnetic resonance spectroscopy,
dynamic light scattering, and
electrophoretic light scattering. Abbreviations: CCR, corona-to-core molecular weight ratio; DP, target degree of polymerization; PDI, polydispersity index (Mw/Mn); ζ, zeta potential.
Hydrophobic core block (Block 2) synthesis
Poly(dimethylaminoethyl methacrylate)-b-poly(dimethylaminoethyl methacrylate-co-butyl methacrylate-co-propylacrylic acid), or p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA), was synthesized using RAFT polymerization with p(DMAEMA) as the macroCTA, AIBN as the radical initiator, and 15-25 wt% DMAEMA, 50-55 wt% BMA, and 25-30 wt% PAA dissolved in DMF (40 wt% monomers, initiator, and macroCTA to solvent volume) as done previously [78]. The target degree of polymerization (DP), or [monomer]/[CTA], was varied according to Table 1 to control block Mn, and [ECT]/[AIBN] = 5 for all polymerizations based on established rigor and reproducibility parameters [78]. After purging the reaction vessel with nitrogen for 45 minutes, the reaction proceeded at 60 °C for 24 hours. The polymerization reaction was terminated by exposing the vessel contents to atmospheric oxygen. The product was precipitated 4 times using 80:20 (v/v) pentane:diethyl ether and centrifugation before being dried overnight under vacuum.
Polymer purification and storage
Dried diblock copolymer was removed from vacuum and dissolved in ~5 mL 100% ethanol. After dissolution, ~25 mL PBS was added. The combined solution was dialyzed using 6-8 kDa dialysis membrane tubing (Spectrum Laboratories) for at least 4 days with ~2-3 water changes each day. The dialyzed solution was frozen at −80 °C and lyophilized for at least 4 days using a Labconco FreeZone 2.5 freeze dryer. The lyophilized polymer was stored in closed containers at room temperature prior to use.
Polymer Characterization
First block and diblock copolymer molecular weights and polydispersities (PDI, Mw/Mn) were determined using gel permeation chromatography (GPC, Shimadzu Technologies) with a miniDAWN TREOS multi-angle light scattering detector (Wyatt Technology) in line with an Optilab T-rEX refractive index detector (Wyatt Technology). High Performance Liquid Chromatography (HPLC) grade DMF + 0.05 mM LiCl (0.2 μm filtered) was used as the mobile phase at a flow rate of 0.35 mL/min through a TSKgel SuperH-H guard column and TSKgel SuperHM-N column (Tosoh Biosciences) at 60 °C. ASTRA® 6.1 light scattering software (Wyatt Technology) and a previously reported dn/dc value of 0.06 [79, 80] were used to calculate molecular weight. Diblock copolymer percent composition was characterized using proton nuclear magnetic resonance (1H NMR) spectroscopy (Bruker Avance 400), as described previously [75].
Nanoparticle Carrier (NPC) Self-Assembly Characterization
NPC size and zeta potential were measured using a Zetasizer Nano ZS (Malvern Panalytical). NPC size measurements were performed via dynamic light scattering (DLS) analysis using lyophilized polymer concentrations of ~0.2-0.3 mg/mL in PBS that were self-assembled into NPCs via sonication prior to filtering using a 0.45 μm PVDF aqueous syringe filter into disposable cuvettes. Zeta potential was determined using polymer concentrations of ~0.2-0.5 mg/mL in 90:10 (v/v) water:PBS solutions and filtered using 0.45 μm PVDF aqueous syringe filters into disposable p1070 capillary cells to ensure sample conductivity values permitted analysis via General Purpose analysis mode in the Malvern Zetasizer software.
Drug Loading
NPCs were loaded with farnesol and/or myricetin using well-established standard operating procedures, as described previously [21, 27, 34, 74]. Briefly, trans,trans-farnesol was obtained from Krackeler Scientific (CAS Number: 106-28-5 and Product Number: 277541), and myricetin was obtained from TCI America (CAS Number: 529-44-2 and Product Number: M2131). Emulsions of farnesol/PBS at 1.0 mg/mL (4.5 mM) were prepared by tip sonication (Fisher Scientific Sonic Dismembrator Model 100 at 4 W power setting) for ~30 seconds and were immediately added to pre-weighed lyophilized diblock copolymers in 20 mL glass scintillation vials to achieve desired polymer NPC concentrations (e.g., 2.7 mg/mL). Myricetin was prepared at 25 mM in dimethylsulfoxide (DMSO) and diluted further using PBS to achieve final sample concentrations. NPCs were loaded with myricetin by adding calculated volumes of myricetin dissolved in DMSO (from either bulk or experimental stock solutions) to NPC and/or farnesol solutions as necessary. Myricetin loading within the NPC shell was confirmed by absorbance changes in two main absorption bands known to be characteristic of flavonoids. These changes occurred for Band I (from 330 nm to 380 nm) and Band II (230 nm to 240 nm), as described in a recently published paper [74]. Moreover, these absorbance shifts are associated with copigmentation phenomena caused by the electrostatic interactions between myricetin and the NPC corona, which also results in a distinctive visual color change from yellow to dark red that added a real-time visual confirmation of myricetin loading within NPCs. Every sample solution contained a variation of nanoparticle concentration, drug concentration, and/or buffer for a total volume necessary for each study as shown in Supplemental Table S1. All of these solutions were bath sonicated using VWR® Ultrasonic Cleaner (Model 50HT Bath Sonicator) for 15 minutes to enable drug loading with similar drug loading efficiencies to previously reported values (>90% for farnesol [21] and ~10-20% for myricetin [74] prior to experimental use.
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was used to characterize the size and morphology of unloaded, single drug-loaded, and dual drug-loaded NPCs. Samples were prepared using lyophilized polymer at concentrations of 2.7 mg/mL (0.08 mM) in ddH2O that were self-assembled into NPCs and loaded with drugs (4.5 mM farnesol and/or 1.0 mM myricetin) via sonication prior to passing through a 0.45 μm PVDF filter. These samples were diluted 1:20 (v/v) with ddH2O and submitted to the University of Rochester Medical Center’s Electron Microscopy Core for standard processing (i.e., staining, drying, mounting, and imaging). The images of NPCs were taken at magnifications of 200,000x using a Hitachi 7650 transmission electron microscope (Hitachi, Schaumburg, IL, USA) and a Gatan 11 megapixel Erlangshen digital camera system (Gatan, Pleasanton, CA, USA).
Characterization of NPC Binding to Plain (HA) and Saliva-Coated Hydroxyapatite (sHA) Beads
NPC solutions (1.35 mg/mL) were prepared by dissolving nanoparticles in 5 mM phosphate buffer at pH 7.0. NPC solutions were sonicated (VWR Model 50HT Bath Sonicator) at room temperature for approximately 15 minutes or until the NPCs were fully dispersed. Hydroxyapatite (HA) beads (i.e., (Ca5((PO4)3)OH)2) with diameters of 80 μm were purchased from Bio-Rad (catalog #1570080), rinsed with ddH2O, dried overnight, and massed into Eppendorf tubes using 10 ±0.1 mg beads per tube. These tubes were either used without further processing (i.e., plain HA beads) or were coated with clarified whole saliva (i.e., saliva-coated HA beads) prior to use.
Clarified whole saliva was prepared using an established protocol [81] with a few modifications. First, ~15 mL of fresh saliva was collected after rinsing the mouth with water and discarding the initial 5 mL of saliva produced. This fresh saliva was centrifuged at 3,300 g for 10 minutes, and the supernatant was collected. Adsorption buffer (50 mM KCl, 1.0 mM KPO4; 1.0 mM CaCl2; 0.1 mM MgCl2; pH 6.5) was added at a 1:1 ratio (v/v), and the solution was centrifuged at 4,000 g for 10 minutes. The volume needed per experiment was taken from this supernatant and transferred to 1.5 mL Eppendorf tubes for a final round of centrifugation at 16,000 g for 10 minutes. The supernatant from these samples was combined into a 20 mL vial with 0.1 M phenylmethylsulfonyl fluoride in isopropyl alcohol, and the resulting solution was heated at 60 °C for 30 minutes. Once cooled to room temperature, 1 mL of this clarified saliva was added to each tube containing 10 mg HA beads and incubated at 37 °C for 2 hours. Following incubation, these tubes were centrifuged at 1,000 g for 2 minutes, and the clarified whole saliva supernatant was discarded, leaving only the saliva-coated HA (sHA) beads for further experimental use.
For both plain and saliva-coated bead experiments, the following groups were prepared in triplicate: Bead control (10 mg bead + 1 mL phosphate buffer), Bead-NPC (10 mg bead + 1 mL of 1.35 mg/mL NPC in phosphate buffer), NPC alone (1 mL of 1.35 mg/mL NPC in phosphate buffer), and phosphate buffer (1 mL). Bead-NPC and Bead control groups were gently hand shaken for 2-3 seconds to allow beads to spread throughout the Eppendorf tube. The tubes were positioned on their sides at 37 °C for 3 hours on an orbital shaker to maximize the potential for contact between beads and NPCs. After incubation, all samples were centrifuged for 10 minutes at ~16,000 g. After centrifugation, samples were plated on 96 well Flat Bottomed Black plates with 100 μL in each well. The sample wells were analyzed using an Infinite M200 PRO microplate reader (Tecan, Switzerland) at an Excitation of 265 nm and Emission of 440 nm, with a gain of 255. Additional sample volumes were taken from the Eppendorf tubes and diluted with phosphate buffer at pH 7.0 either 10 or 100-fold for use in particle count analysis. Specifically, a Nanosight NS300 (Malvern Panalytical) was used according to the manufacturer’s instructions using camera level of 13 and detection threshold of 2 to determine average particle count per mL. A fold-change of NPC-bead binding was calculated by converting particles per mL to NPC concentration using a standard curve of measured particle counts corresponding to known NPC concentrations and comparing the average measured value per group to the NPC alone control for each experiment. Three separate experiments with n=3-6 independent measurements per experiment were completed.
Characterization of Myricetin Loaded NPC Binding to Plain and Saliva-Coated HA Beads
This binding assay was completed as described above with a few differences due to the presence of myricetin. Myricetin dilutions of 0.01, 0.1, and 1.0 mM were prepared by adding myricetin dissolved in DMSO to various amounts of 1.35 mg/mL nanoparticle solutions. The drug-NPC mixture was sonicated for approximately 5 minutes and then stored in the dark, as myricetin is light sensitive. NPC + Myricetin, NPC + Myricetin + Bead, and phosphate buffer control groups of 1 mL each were prepared in 1.5 mL Eppendorf tubes in triplicate. NPC, HA, and sHA bead control samples were prepared as discussed above.
After incubation at 37 °C for 3 hours, samples were initially centrifuged for 5 minutes at ~16,000 g, and then 500 μL sample volumes were transferred into 3,000 kDa Eppendorf filters to be centrifuged at 14,000 g for 10 minutes. Following the second centrifugation step, filters were inverted into new Eppendorf tubes and centrifuged again for 2 minutes at 1,000 g. The concentrate was reconstituted to 500 μL, and sample volumes were taken from the Eppendorf tubes and diluted with phosphate buffer at pH 7.0 either 10- or 100-fold for use in particle count analysis, as described above.
Saliva-coated Hydroxyapatite (sHA) Developing Biofilm Assay
Anti-biofilm effects of NPC drugs on developing S. mutans biofilm formation were assessed following a modified topical treatment regimen [27], S. mutans biofilms were formed on saliva-coated hydroxyapatite (sHA) discs (surface area, 2.7 ± 0.2 cm2, Clarkson Chromatography Products Inc., South Williamsport, PA, USA) vertically suspended in 24-well plates using a custom-made wire disk holder, mimicking the smooth surfaces of the pellicle-coated tooth [21, 27, 52]. Each sHA disk was inoculated with ~2 × 105 CFU of S. mutans per mL in ultra-filtered (10 kDa molecular-weight cut-off membrane; Prep/Scale, Millipore, MA) buffered tryptone-yeast extract (UFTYE) broth containing 1% (w/v) sucrose at 37 °C with 5% CO2. The sHA discs and biofilms were treated with the NPC drug solutions for 10 minutes and transferred to culture media. Specifically, 25 μL NPC and drug solutions were applied to each side of each disk at 0, 2.5, 5, and 7.5 minutes with a total application volume of 400 μL for each pair of discs. The first treatment was applied directly after salivary pellicle formation and the treated discs were transferred to culture media. Biofilms were allowed to form on the discs without interruption for 6 hours at which point a second treatment was applied. The next day, biofilms were treated three times (at 20, 26, and 32 hours) and the culture media was changed twice (at 20 hours and 30 hours). After 44 hours, biofilms were removed and homogenized by sonication in 0.89% NaCl (30 seconds pulse; 7 W). CFU and the dry weight of biofilm were determined as described elsewhere [21, 27, 52]. The bacteria viability limit of detection for this assay was 1 × 103 CFU/mL.
Saliva-coated Hydroxyapatite (sHA) Established Biofilm Assay
In addition to the developing biofilm assay described above, anti-biofilm effect of NPC drugs on S. mutans biofilms established for 20 hours was assessed following a modified topical treatment regimen [27]. Briefly, S. mutans biofilms were formed on sHA discs as described above but for the 20-hour incubation period. Each sHA disk was inoculated with ~2 × 105 CFU of S. mutans per mL in UFTYE containing 1% (w/v) sucrose at 37 °C with 5% CO2. These discs were incubated at static conditions for 20 hours, and baseline (i.e. control) samples were collected and tested for bacterial viability (CFU/mL) and biomass (dry weight) after 20 hours. Then, the biofilms were treated three times (at 20, 26, and 32 hours) as described above. Culture media was changed at 20 hours and 30 hours. After 44 hours, biofilms were removed and homogenized by sonication in 0.89% NaCl (30 seconds pulse; 7 W). CFU and dry weight of biofilm were determined as described elsewhere [21, 27, 52], and the baseline data was subtracted from the final samples. The bacteria viability limit of detection for this assay was 1 × 103 CFU/mL.
Glucosyltransferase Activity and Inhibition of Insoluble Glucan Formation Assays
The inhibition of GtfB enzyme activity was examined using methods detailed previously [82]. Briefly, 200 μL of each sample was mixed with 200 μL of ([14C]glucosyl)-sucrose substrate (0.2 μCi/ml; 200.0 mmol/L sucrose, 40 μmol/L dextran 9000, 0.02% sodium azide in adsorption buffer, pH6.5) with 2 units of GtfB enzyme. The mixed solution was incubated at 37 °C with rocking for 2 h to allow glucan synthesis. Subsequently, insoluble glucans were determined by scintillation counting to assess GtfB activity.
Gene Expression Assay
Quantitative reverse transcription-PCR (qRT-PCR) was performed to measure the expression profile of S. mutans specific target genes (gtfB, gtfC. gtfD, atpD). S. mutans biofilms were formed on sHA discs vertically suspended in 24-well plates using a custom-made wire disk holder as described above but for a 19 hour incubation period. Each sHA disk was inoculated with ~2 × 105 CFU of S. mutans per mL in UFTYE containing 1% (w/v) sucrose at 37 °C with 5% CO2. These discs were incubated at static conditions for 20 hours, and treatments were applied at 20 hours. The treatment consisted of removing the sHA discs and biofilms from culture media, washing them 3 times in 0.89% NaCl, and the applying 25 μL NPC and drug solutions to each side of each disk at 0, 2.5, 5, and 7.5 minutes (total application volume of 400 μL) for each pair of discs. After 10 minutes of treatment, the biofilms were transferred back to culture media and continued to grow without interruption for 4 hours. Then, biofilms were harvested and RNA was extracted and purified using standard protocols optimized for biofilms [51, 83]. The RNA integrity numbers (RIN) of purified samples used for qRT-PCR were determined by microcapillary electrophoresis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The cDNA was synthesized from 1 μg of purified RNA (RIN, ≥9) with the Bio-Rad iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA), and quantitative amplification conditions using Bio-Rad iTaq Universal SYBR green Supermix and Bio-Rad CFX96 system (Bio-Rad Laboratories, Inc.). The primers used in present study are listed in Supplemental Table S2 [51, 84]. Standard curves for each primer were used to determine the relative number of cDNA molecules, and relative expression was calculated by normalizing to the 16S rRNA gene transcripts [51, 84].
Minimum Inhibition Concentration and Minimum Bactericidal Concentration Testing
Minimum inhibition concentration (MIC) and minimum bactericidal concentration (MBC) values for unloaded NPCs, farnesol loaded NPCs, myricetin loaded NPCs, and farnesol + myricetin co-loaded NPCs were measured using planktonic Streptococcus mutans UA159 cells using a microdilution (microplate) assay, as previously described [21, 60]. Briefly, S. mutans UA159 cells were grown in ultra-filtered tryptone-yeast extract (UFTYE) broth containing 1% glucose (w/v) at 37 °C and 5% CO2 to mid-exponential phase. Serially diluted treatments of farnesol, myricetin, NPCs, or farnesol + myricetin co-loaded NPCs were added to wells of 96 well plates. The highest concentrations were 4.5 mM farnesol, 1.0 mM myricetin, and 2.7 mg/mL (0.08 mM) NPC. UFTYE was added up to 90 μL per well. A 10 μL suspension of S. mutans (106 colony forming units (CFU) per mL) was added to each well to yield a final S. mutans concentration of 105 CFU/mL per well except for media only controls. After incubating at 37 °C and 5% CO2 for 24 hours, the MIC was visually determined to be the lowest treatment concentration that inhibited bacterial growth, as previously described [21, 60]. To quantify the antibacterial effectiveness in terms of CFU/mL, samples from each of testing group were plated on blood agar using serial dilutions and the CFUs were counted. MBC was defined as the lowest treatment concentration that demonstrated ≥ 3 log reduction (99.9% killing efficacy) from the initial colony count in the S. mutans suspension, as previously described [61, 85].
Statistical Analysis
GraphPad Prism® 6 software was used to perform statistical analyses as indicated in the figure captions. Multiple statistical tests including One-way ANOVA with Tukey’s or Dunnett’s correction for multiple comparisons or Two-way ANOVA with Sidak’s multiple comparisons and p-values of p < 0.05 were used to assess statistical differences. Specific tests are denoted in figure legends. All data is shown as the mean plus or minus the standard deviation of the data presented. Therefore, all error bars represent the standard deviation.
Results
Here, a NPC drug delivery system was evaluated for anti-biofilm efficacy of NPCs co-loaded with farnesol and myricetin, as illustrated in Figure 1. Table 1 summarizes the characterization of the two poly(dimethylaminoethyl methacrylate)-b-poly(dimethylaminoethyl methacrylate-co-butyl methacrylate-co-propylacrylic acid) (i.e., p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA)) NPCs (NP25/8 and NP25/10) used in this study. In particular, diblock copolymers were composed of a cationic corona (i.e., Block 1; molecular weight ~25 kDa) and a hydrophobic core (i.e., Block 2; molecular weight ~8 and ~10 kDa, respectively) that facilitated self-assembly into spherical nanoparticle carriers (i.e., NPCs) with diameters of ~30-34 nm (per DLS) and zeta potentials of ~17-21 mV (Table 1). Transmission electron microscopy images in Figure 1B demonstrated a ~2-fold increase in nanoparticle size from ~20-25 nm to ~40-50 nm upon loading with farnesol (ii and iv), which corroborated previous reports [27, 34], but no change in size was observed upon loading with myricetin (iii) compared to the unloaded NPC control (i). Additional details pertaining to the NPC drug loading and release mechanisms of farnesol and myricetin as well as transmission electron microscopy analyses of these NPCs have been presented previously [27, 34, 74].
Figure 1. Proposed anti-biofilm approach using cationic NPCs co-loaded with farnesol and myricetin.
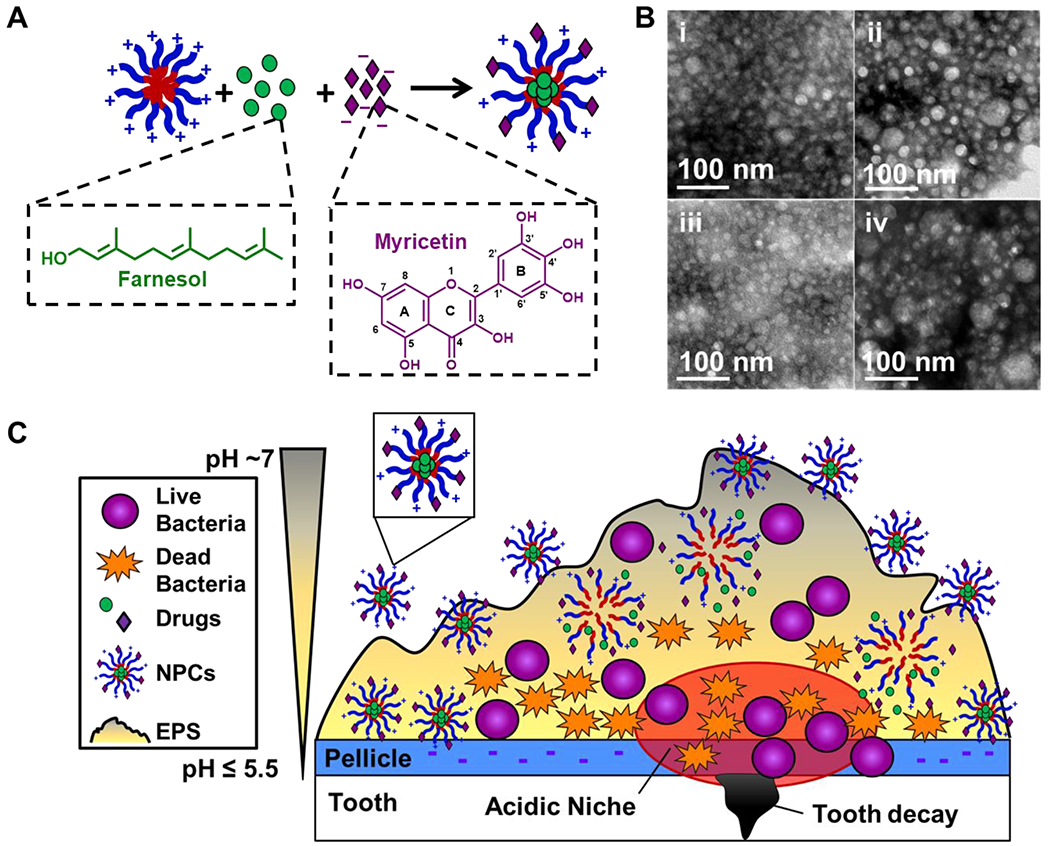
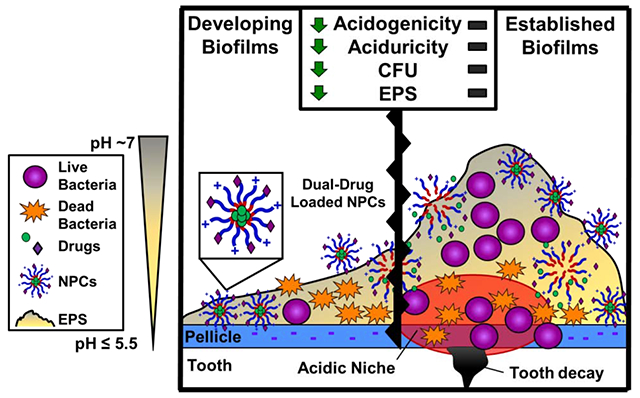
A) Cartoon illustrating the known mechanisms of farnesol loading within NPC hydrophobic core (i.e., hydrophobic drug farnesol, which is represented by green circles, partitioning into hydrophobic NPC core [21, 27, 34]) and myricetin loading via electrostatic interactions with cationic NPCs (i.e., anionic flavonoid myricetin, which is represented by purple diamonds, interacts electrostatically with NPC cationic corona). B) Transmission electron microscopy (TEM) images of (i) unloaded NPCs, (ii) farnesol-loaded NPCs, (iii) myricetin-loaded NPCs, and (iv) NPCs co-loaded with farnesol and myrictin. C) Cartoon illustrating the hypothesized interaction and anti-biofilm efficacy of dual drug-loaded NPCs on S. mutans oral biofilms. The red encircled area illustrates the acidic microenvironment commonly found within cariogenic biofilms, which is promoted by S. mutans’ key virulence factors (i.e., EPS synthesis, acidogenicity and aciduricity) that are responsible for hydroxyapatite demineralization leading to tooth decay.
The antibacterial efficacy of drug-loaded NPCs was assessed in developing S. mutans biofilms using the experimental scheme shown in Figure 2A. Synergistic improvements in anti-biofilm efficacy were observed for NP25/10 co-loaded with farnesol and myricetin when treating developing biofilms. As shown in Figure 2B, the biofilm dry weights were significantly reduced (~97-99%, p ≤ 0.001) for NPCs co-loaded with farnesol and myricetin compared to controls. NPCs loaded with only farnesol also showed a significant (p ≤ 0.0001) yet more modest dry weight reduction of ~62%. However, NPCs loaded only with myricetin showed no effect on biofilm dry weight (Figure 2B). Furthermore, co-loaded NPCs significantly (p ≤ 0.0001) inhibited developing biofilm viability by ~2-3 log CFU per mL while farnesol-only loaded NPCs resulted in only a ~1-log CFU/mL reduction (Figure 2C). Interestingly, co-loaded NPCs using the lower myricetin concentration (0.1 mM) reduced developing biofilm dry weight and inhibited biofilm viability to a greater extent compared to the higher myricetin concentration (1.0 mM); however, these differences were not statistically significant. These nonsignificant results are likely due to variability in the data, low replicate numbers, as well as the high level of degrees of freedom among the groups.
Figure 2. In vitro biofilm studies show co-delivery of farnesol and myricetin synergistically improved anti-biofilm efficacy and maintained neutral pH in developing biofilms.
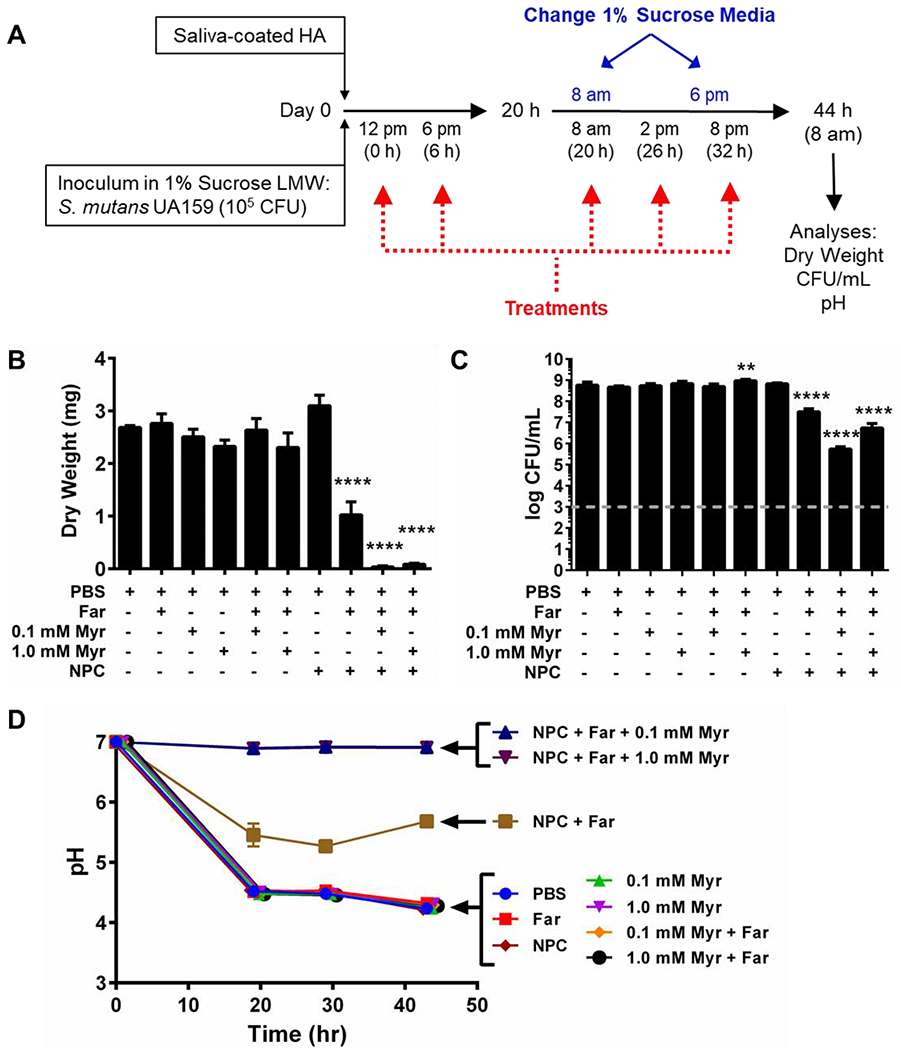
A) Scheme showing the treatment regimen used with developing S. mutans biofilms grown in vitro on saliva-coated hydroxyapatite (sHA) disks.B) Biofilm dry weights obtained for biofilms formed on sHA disks for each treatment show significant reduction (~97-99%) for NPCs co-loaded with farnesol and myricetin. C) Log colony forming unit per mL data for NPCs co-loaded with farnesol and myricetin show ~2-3 log reduction compared to controls. Grey dashed line indicates the log CFU/mL limit of detection for this assay. Dry weight and CFU/mL data shown as mean ± standard deviation from n=2-4 independent measurements. **p<0.01 and ****p<0.001 from One-way ANOVA with Dunnett’s multiple comparisons test (versus PBS control). D) Plots showing changes in pH over time for developing S. mutans biofilms treated with control or experimental groups listed. pH data shown as mean ± standard deviation from n = 2 measurements per group at each experimental time point (i.e., t = 0, 20, 30, and 44 hours).
Based on the synergistic results of NPCs co-loaded with farnesol and myricetin in the developing biofilm model, further studies were performed. Changes in the pH of the culture media surrounding the S. mutans biofilms grown on sHA disks were evaluated. As shown in Figure 2D, media pH for biofilms treated with controls of PBS or farnesol, myricetin, NP25/10, or the combination of farnesol and myricetin without NPCs dropped below pH 5 within the first 20 hours and remained at pH ~4.5 at each time check point; such low pH values can lead to enamel demineralization and development of dental caries. However, media from treatments using NP25/10 co-loaded with farnesol and myricetin (regardless of myricetin concentration) did not vary from neutral pH values (i.e., pH ~7) throughout the 44-hour study period (Figure 2D). Media pH values for biofilms treated with NPCs loaded with farnesol alone were intermediary compared to the other controls, remaining between pH 5.3 – 5.7 after 20 hours of treatment.
Dual drug-loaded NPCs were also used to treat established S. mutans biofilms grown using the saliva-coated hydroxyapatite (sHA) disk model [86] (see experimental scheme in Figure 3A). The combination of 0.08 mM NP25/8 or NP25/10 with 4.5 mM farnesol and either 0.1 mM or 1.0 mM myricetin was tested using 20-hour established S. mutans biofilms. As shown in Figure 3B, biofilm dry weights were significantly (p ≤ 0.01) reduced by ~15-17% for NPCs co-loaded with farnesol and myricetin compared to PBS controls. Moreover, NPCs co-loaded with 4.5 mM farnesol and 1.0 mM myricetin significantly (p ≤ 0.0001) inhibited established biofilm viability by ~0.8 log colony forming units (CFU) per mL while NPCs loaded with farnesol alone or farnesol and 0.1 mM myricetin resulted in ~0.4 log and ~0.3 log CFU per mL statistically significant reductions (p ≤ 0.001 and p ≤ 0.01), respectively (Figure 3C).
Figure 3. In vitro 20-hour biofilm studies show co-delivery of farnesol and myricetin had limited anti-biofilm efficacy against established biofilms.
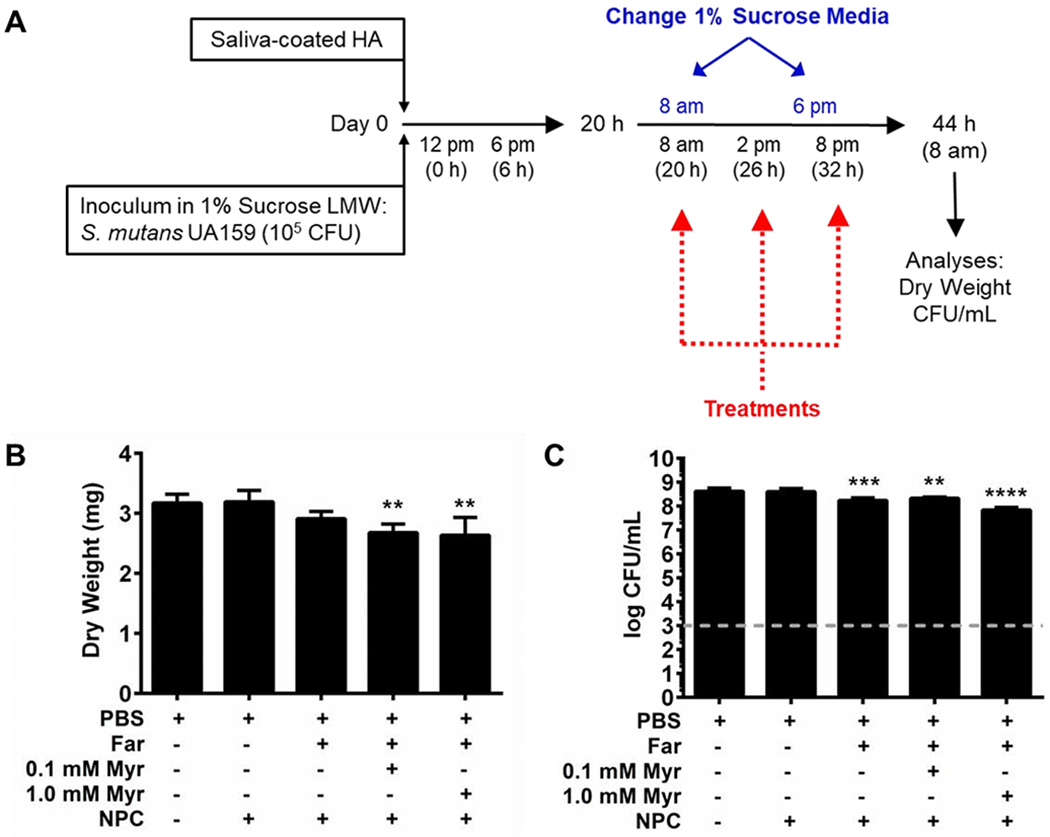
A) Schematic showing in vitro 20-hour established biofilm procedure. B) Biofilm dry weights obtained for biofilms formed on sHA disks for each treatment show modest reduction (~8-17% decrease) for NPCs (0.08 mM) co-loaded with farnesol (4.5 mM) and myricetin (0.1 mM or 1.0 mM). C) Colony forming unit per mL (CFU/mL) data for NPCs co-loaded with farnesol and myricetin show ≤1 log reduction compared to controls. Grey dashed line indicates the log CFU/mL limit of detection for this assay. Data shown as mean ± standard deviation from n=4-8 independent experiments. **p < 0.01, *** p < 0.001, and **** p < 0.0001 from One-way ANOVA with Dunnett’s multiple comparisons test (versus PBS control).
Given the enhanced inhibition efficacy on developing versus established biofilms, additional studies were conducted to better understand the effects of dual-drug loaded NPC delivery on biological actions related to binding to the apatitic surface, EPS synthesis, and acidogenicity. First, drug-loaded NPC interactions with the tooth-mimetic surface were investigated using a hydroxyapatite (HA) bead binding assay (Figure 4A). As summarized in Figure 4A, this assay indirectly calculates NPC binding to tooth-mimetic surfaces by measuring average particle concentration of supernatant solutions after NPC incubation at 37 °C for 3 hours with or without HA or saliva-coated HA (sHA) beads. Results from this study revealed slight, yet significant increases in fold-change of NP25/8 binding to HA bead surfaces versus the unloaded NPC control regardless of myricetin concentration (p ≤ 0.01, p ≤ 0.001, and p ≤ 0.0001 for 1.0 mM, 0.1 mM, and 0.01 mM myricetin, respectively; Figure 4B, black bars). In addition, significant increases (p ≤ 0.0001) of approximately three times as much NPC binding to sHA bead surfaces occurred when NPCs were loaded with myricetin compared to unloaded NPCs (Figure 4B, grey bars). No statistically significant differences in NPC binding were observed among the three myricetin concentrations tested for the HA assay; however, NPC binding for NPCs loaded with 0.1 mM myricetin was significantly less (p ≥ 0.5) than either of the other two drug concentrations tested in the sHA assay.
Figure 4. Effects of NPC-myricetin electrostatic interactions on NPC-hydroxyapatite binding.
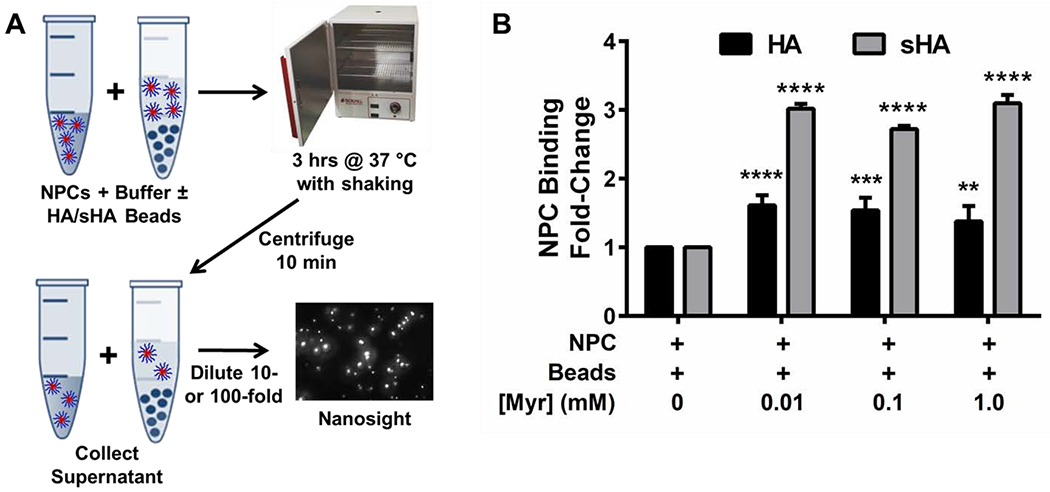
A) Schematic overview of NPC binding assay using hydroxyapatite (HA) and saliva-coated hydroxyapatite (sHA) beads and Nanosight particle count analysis. NPC solution and NPC + HA or NPC + sHA bead solution were incubated at 37 °C for three hours, centrifuged, diluted, and assessed via Nanosight analysis.B) Fold-change of NPC binding to HA (black bars) and sHA (grey bars) beads as a function of myricetin concentration, including unloaded NPCs and NPCs loaded with 0.01 mM, 0.1 mM, or 1.0 mM myricetin. Results are shown as mean ± standard deviation from three independent experiments with each containing three replicates and five measurements per replicate, ns = no significant difference among groups, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 from Two-way ANOVA with Sidak’s correction for multiple comparisons compared to NPC + Beads controls without myricetin.
Effects on gtfBCD (associated with glucan synthesis) and atpD expression (associated with aciduric and acidogenic properties of S. mutans) caused by the dual-drug delivery approach were examined. As shown in Figure 5A, the sHA disk model [86] was used to generate biofilms for gene expression analysis. Biofilm gene expression results revealed that NP25/10 co-loaded with 4.5 mM farnesol and 1.0 mM myricetin reduced expression of the metabolism and acid tolerance gene atpD [87, 88] by ~43% compared to the PBS control in the developing biofilm model (Figure 5B). A significant decrease in atpD gene expression (~33%) was also observed for farnesol-loaded NPCs (Figure 4B). Expression of three additional genes involved in S. mutans Gtf production (i.e., gtfB, gtfC, and gtfD) was also evaluated (Supplemental Figure S2). Despite similar trends in reducing gene expression to those seen for atpD in Figure 5, no other significant decreases were observed (Supplemental Figure S2).
Figure 5. Effects of NPC co-delivery of farnesol and myricetin on atpD gene expression, GtfB activity, and MIC/MBC.
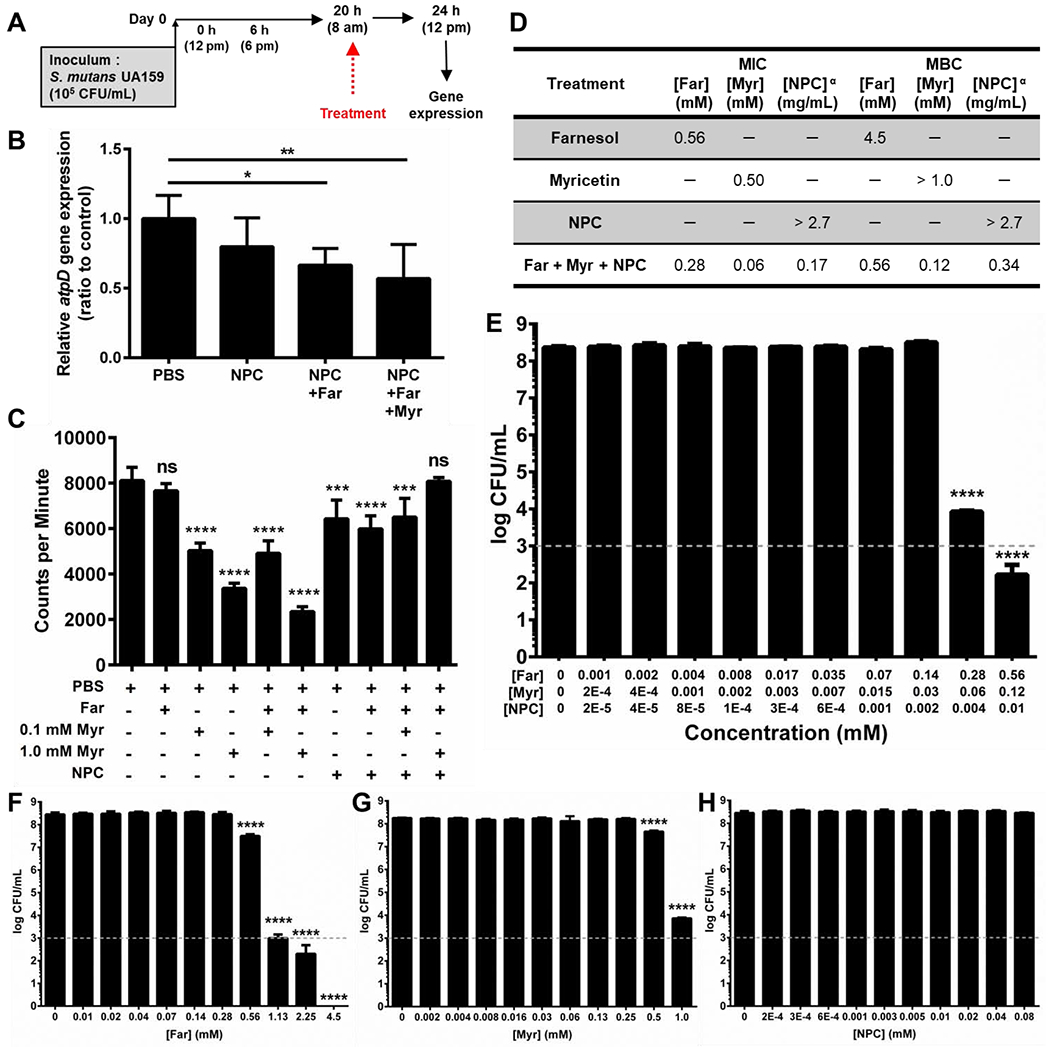
A) Scheme showing the treatment regimen used with developing S. mutans biofilms grown in vitro on saliva-coated hydroxyapatite (sHA) disks for gene expression studies. B) After completion of the biofilm growth and treatment procedure shown in (A), the biofilms were returned to fresh medium to grow for 4 hours. Then, gene expression analysis was completed for the atpD gene that encodes proteins associated with S. mutans biofilm metabolism and acid tolerance. These data revealed a trend indicating the co-loaded NPCs reduced expression of atpD in the developing biofilm model. Data shown as mean ± standard deviation from n = 6 measurements. * p ≤ 0.05 and ** p ≤ 0.01 from One-way ANOVA with Tukey’s multiple comparisons test (versus PBS control). C) In vitro GtfB activity analysis. Data shown as mean ± standard deviation from n = 4 measurements. *** p ≤ 0.001, **** p ≤ 0.0001, and ns = not significant from One-way ANOVA with Dunnett’s multiple comparisons test (versus PBS control). D) Table showing minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values for farnesol, myricetin, NPCs, and NPCs co-loaded with farnesol and myricetin versus planktonic S. mutans UA159. α Due to the high molecular weight of the NPCs compared to the small molecule drugs, the MIC and MBC values for NPCs are recorded in mg/mL to avoid confusion as the NPC concentrations in mM are not directly comparable to the small molecule concentrations shown. E) Log colony forming unit per mL (CFU/mL) data for NPCs co-loaded with farnesol and myricetin show ~4 and ~6 log CFU/mL reductions in planktonic S. mutans UA159 for dual drug-loaded NPCs at MIC and 2x MIC concentrations, respectively. Data shown as mean ± standard deviation from n = 9 measurements. F) Log CFU/mL data for farnesol alone show ~1, ~5, and ~6 log CFU/mL reductions in planktonic S. mutans UA159 for farnesol MIC, 2x MIC, and 4x MIC concentrations, respectively. Complete eradication of bacterial growth was observed at farnesol MBC. G) Log CFU/mL reduction data for myricetin alone show ~1 and ~4 log CFU/mL reductions in planktonic S. mutans UA159 for myricetin MIC and 2x MIC, respectively. H) Log CFU/mL data for unloaded NPCs show no change in planktonic S. mutans UA159 bacterial concentration for any of the NPC concentrations tested. F-H) Data shown as mean ± standard deviation from n = 4 measurements. E-H) Grey dashed line indicates the log CFU/mL limit of detection for this assay. **** p ≤ 0.0001 from One-way ANOVA with Dunnett’s multiple comparisons test versus PBS control (0 mM drug or NPC).
GtfB activity was measured in vitro for NP25/10 co-loaded with farnesol and for both high and low myricetin concentrations (Figure 5C). Unexpectedly, the results showed that myricetin alone or in combination with farnesol without NPCs exhibited the highest inhibition of GtfB activity (~37-71%). A ~20% reduction in GtfB activity was observed for NP25/10 alone, NP25/10 loaded with farnesol, and NP25/10 co-loaded with 4.5 mM farnesol and 0.1 mM myricetin. However, GtfB activity did not vary from PBS when treated with NP25/10 co-loaded with farnesol and 1.0 mM myricetin (Figure 5C).
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were identified for farnesol alone, myricetin alone, NPC alone, and dual drug-loaded NPCs using planktonic S. mutans UA159. The MIC values for farnesol alone, myricetin alone, and NPC alone were 0.56 mM, 0.50 mM, and >2.7 mg/mL (>0.08 mM), respectively (Figure 5D). Moreover, the MIC values for the dual drug-loaded NPC treatments were 0.28 mM farnesol, 0.06 mM myricetin, and 0.17 mg/mL (0.004 mM) NPC (Figure 5D). The MBC values for farnesol alone, myricetin alone, and NPC alone were 4.5 mM, >1.0 mM, and >2.7 mg/mL (>0.08 mM), respectively, while the MBC values for the combination treatment were >0.56 mM farnesol, >0.12 mM myricetin, and >0.34 mg/mL (>0.01 mM) NPC. It should be noted that due to the high molecular weight of the NPCs compared to the small molecule drugs, the MIC and MBC values for NPCs are recorded in Figure 5D using mg/mL rather than mM to avoid confusion as the NPC concentrations in mM are not directly comparable to the small molecule concentrations shown. Samples from MIC tests were used to quantify CFUs, and log colony forming unit per mL (CFU/mL) data for NPCs co-loaded with farnesol and myricetin show ~4 and ~6 log CFU/mL reductions in planktonic S. mutans UA159 for dual drug-loaded NPCs at MIC and 2x MIC concentrations, respectively (Figure 5E). However, no change in planktonic S. mutans UA159 bacterial concentration was observed for any of the NPC concentrations tested (Figure 5H). Interestingly, the log CFU/mL data corresponding to the MIC values for farnesol (0.56 mM, Figure 5F) and myricetin (0.50 mM, Figure 5G) only showed ~1 log CFU/mL reduction for both drugs. Furthermore, log CFU/mL data for 2x and 4x the farnesol MIC showed ~5 and ~6 log CFU/mL reductions, respectively (Figure 5F), while the farnesol MBC (4.5 mM), which was 8 times higher than the farnesol MIC (Figure 5D), showed eradication of bacterial growth. Myricetin at 2x MIC showed a ~4 log CFU/mL reduction (Figure 5G).
1. Discussion
Here, a synergistic dual-drug delivery approach using NPCs co-loaded with farnesol and myricetin (Figure 1) was assessed for anti-biofilm efficacy using developing and established S. mutans biofilms. Coloaded NPC treatments showed drug synergism in developing biofilms with substantial disruption of biofilm dry weight, reduction of S. mutans viability by ~3 log CFU/mL, and prevention of pH drops associated with biofilm virulence compared to controls (Figure 2). However, limited efficacy was observed against established biofilms (Figure 3). In developing biofilms, NPCs co-loaded with farnesol and myricetin showed ~97% reduction in biomass (i.e., dry weight) compared to controls. Interestingly, this reduction was not strongly linked with inhibition of gtf gene expression, which was either moderate or negligible, thus indicating bacterial growth inhibition or disruption in bacterial metabolism was responsible for anti-biofilm efficacy. Indeed, the most striking result was arrested acidogenicity and aciduricity of the biofilm when treated with NPCs loaded with farnesol or co-loaded with farnesol and myricetin, indicated by minimal changes in the biofilm media pH (Figure 2C) and associated decreases in gene expression of the acid tolerance gene, atpD (Figure 5B). While the MBC results in Figure 5D–H suggest that bacterial eradication was elusive, topical treatments of drug-loaded NPCs were able to reduce S. mutans acidogenic and aciduric properties, which are key virulence attributes associated with dental caries. In addition, an in vitro sHA binding assay showed myricetin loading improved NPC binding to tooth-mimetic surfaces (Figure 4), suggesting that myricetin-bound NPC could enhance efficacy by improving drug retention at the biofilm-apatite interface. Finally, a reduction of up to 20% in GtfB activity was observed for farnesol and myricetin co-loaded NPCs compared to PBS controls (Figure 5C). Based on the data in Figure 5C, inclusion of myricetin was necessary to inhibit GtfB activity, likely due to the structural resemblance of myricetin to sucrose and similar locations of putative Gtf binding sites within these molecules (Supplemental Figure S1) [66, 89, 90]. Thus, the data suggest co-delivery of farnesol and myricetin via NPC enhances anti-biofilm efficacy through synergistic drug activity but only for developing biofilms.
Although other reports have demonstrated upwards of 4 to 6 log reductions in S. mutans biofilm viability following treatment with drug-loaded nanoparticles [20, 22, 25, 32, 33], few nanoparticle studies have investigated the impact on S. mutans’ key adaptive stress responses, such as acidogenicity and aciduricity [91]. Rather, these studies were focused on assessing delivery and effectiveness of polymer nanoparticles alone or singular drugs in nanoparticle formulations, and collectively lacked any substantial improvements regarding biofilm pH while biomass reductions ranged from ~50-80% [20, 22, 23, 25] when tested on similar developing biofilm models or lacked quantification altogether [91]. These modest results likely stem from the resilient effects of multiple antimicrobial resistance mechanisms present in S. mutans biofilms, including EPS matrix formation to limit drug diffusion and establish localized acidic microenvironments, altered adaptive stress responses to inhibit drug uptake and metabolism, natural acid competence and increasing aciduricity, and efflux pumps to remove drugs from cells [42, 43]. While previous studies have reported nanoparticles designed to act synergistically to reduce medically relevant biofilms, such studies have mainly focused on the use of metal nanoparticles [92–94], nitric oxide releasing polymers [95], or nanoparticles requiring stimulus by outside systems, such as ultrasound [96].
Here, the unique combination of farnesol, myricetin, and NPCs yielded synergistic results when tested for bacterial viability (i.e., planktonic and biofilm), biofilm biomass (dry weight), and resistance to pH change using developing biofilms (Figure 2). As expected, NPC alone did not exhibit any antibacterial activity versus S. mutans UA159 (Figure 5H), and the MIC for farnesol alone (Figure 5D) matched previous findings [21]. The MIC for myricetin alone (0.5 mM) was ~8-fold higher than the MIC for myricetin when combined with farnesol and NPCs (0.06 mM), suggesting that myricetin is primarily responsible for the synergistic results observed versus planktonic S. mutans and developing biofilms herein. Moreover, the combined results of individual farnesol, myricetin, and NPC MIC testing (Figures 5F–H) only accounted for ~1 log CFU/mL reduction, while the dual drug-loaded NPC treatment using the same component concentrations in Figure 5E resulted in ~6 log CFU/mL reduction, thereby indicating a synergistic relationship for this treatment combination. Altogether, these data suggest that the mechanisms of action were inhibition of planktonic bacterial growth and reduction in S. mutans acidogenicity and aciduricity. Indeed, this combination resulted in ~100-150-fold higher CFU/mL inhibition, ~500-fold decrease in proton concentration (i.e., acid levels), and ~60-fold lower biofilm dry weight (i.e., substantial disruption of biomass) compared to the effects of individual components of the treatment regimen.
S. mutans’ acidogenicity and aciduricity rely on several factors, including atpD expression. The atp operon encodes the F1F0-H/ATPase enzyme responsible for pumping protons out of cells to maintain intracellular pH [46, 48, 54, 97]. Through F-ATPase activity, S. mutans withstands changes in extracellular acid levels and thus, outcompetes other bacterial species within the oral cavity [48, 54]. In contrast to other studies showing significant gene expression differences in gtfB and gtfC but not in atpD following treatment with the antioxidant ethyl gallate [98], the work presented here shows a statistically significant decrease in gene expression only for atpD from biofilms treated with NPCs loaded with farnesol or co-loaded with farnesol and 1 mM myricetin (Figure 5B; Supplemental Figure S2). Moreover, previous studies have shown treatment using multiple combinations of farnesol, myricetin, and fluoride alone had no effect on atpD expression [67]. These results suggest that co-loaded NPC treatments disrupted acid tolerance processes associated with F1F0-H/ATPase, which can inhibit bacterial metabolism and lower pH for developing biofilms in Figure 2C, ultimately affecting bacterial growth and biomass accumulation. These findings also help to explain the previously-described effect of farnesol on S. mutans acid production, acid tolerance, and biofilm accumulation [86].
While NPC-mediated co-delivery of myricetin and farnesol resulted in multi-log antibacterial efficacy versus planktonic bacteria and multi-log anti-biofilm efficacy in developing biofilms, treatment of established biofilms showed little anti-biofilm effects. In fact, the combination proved ineffective against established biofilms, whereby only ~1-2 fold differences in either CFU/mL or dry weight and a modest ~13-fold decrease in proton concentrations after 24 hours were observed (Supplemental Figure S3). These results may be due to poor drug accessibility, penetration, or retention in the established biofilm even when NPC was used for drug delivery. In the process of biofilm formation, Gtfs quickly adsorb to the salivary pellicle and cleave dietary sucrose to synthesize homopolymers of glucose, termed glucans, and initiate the formation of the EPS matrix [13, 57, 58]. Approximately 10-40% of dental plaque dry weight is due to glucans [13, 99]. The glucans-rich EPS matrix has multiple functions including being an architectural scaffold, aiding in surface adhesion and cohesion of bacterial cells, serving as a nutrient source for bacteria, scavenging biomolecules, and preventing penetration of antimicrobial drugs into biofilms, thereby reducing drug efficacy [9, 14, 53, 100]. During the 20-hour duration for biofilm growth in the established biofilm model, significant biofilm formation occurred, including Gtf production and abundant glucan-matrix synthesis within the biofilm as well as upregulation of other S. mutans EPS and co-adhesion factors [13, 14, 48, 101–103], which could have hindered penetration of NPC-loaded drugs. Therefore, the complexity and protective characteristics of the EPS matrix combined with S. mutans phenotype in the established biofilms likely impeded drug access and efficacy (Figure 3), resulting in limited effects on bacterial viability and biofilm dry weight.
Inhibiting acidogenicity and aciduricity directly via farnesol and myricetin co-delivery underpins the dominant aspect of anti-biofilm efficacy, yet neither farnesol nor myricetin exhibited anti-biofilm efficacy in the absence of NPC. The use of NPCs is necessary for enhanced drug solubility, drug retention at the pellicle-coated apatitic surface through electrostatic interactions, and pH-responsive drug release as a consequence of developing biofilm virulence, as previously described [27, 34]. However, NPC co-delivered drugs were less efficacious at inhibiting Gtf, a common mechanism of action of flavonoids, versus free drug treatments (Figure 5C), and NPC alone reduced Gtf activity to a similar extent as drug-loaded NPC, though no impact to biofilm biomass (dry weight) or bacterial viability was observed for NPC alone. Cationic moieties, such as those within the NPC corona, can interact directly with enzymes, including Gtfs, and cause changes, such as enzyme activation or destabilization [104, 105]. However, myricetin, which is anionic, can neutralize the cationic NPC corona in a dose-dependent fashion, thereby preventing interactions between the NPCs and Gtf. Others have reported that the attenuation or neutralization of nanoparticle surface charge can inhibit nanoparticle-enzyme interactions and displace nanoparticles from enzyme binding sites [105, 106]. Therefore, reductions in NPC-enzyme affinity for NPCs loaded with 1.0 M myricetin may elucidate lackluster GtfB inhibition shown in Figure 5C. While dichotomous effects on Gtf inhibition were observed, robust anti-biofilm efficacy through synergistic targeting of F1F0-H/ATPase-mediated S. mutans acidogenicidy and aciduricity suggest a promising approach to treat oral biofilms and show the feasibility of co-loading different drugs to enhance overall bioactivity.
Cationic moieties are also thought to aid in nanoparticle binding to tooth enamel [16], yet myricetin loading via electrostatic charge neutralization with NPCs [74] appeared to improve NPC binding to tooth-mimetic surfaces in vitro (Figure 4), thus likely increasing drug retention at the biofilm-apatite interface. Although farnesol was not included in this testing for simplicity, the inclusion of farnesol is not expected to affect the electrostatic nature of NPC binding to HA or sHA surfaces since farnesol loads within the NPC hydrophobic core [27]. However, loading NPCs with farnesol is known to increase the NPC diameter (Figure 1B), which has been previously shown to generally reduce the quantity of NPCs bound to a fixed surface area since NPC surface area will be larger [34]. Therefore, it is expected that the presence of farnesol would slightly reduce the NPC binding fold-change observed for myricetin-loaded NPCs on HA and sHA surfaces, but not to NPC alone levels, given the added benefit of myricetin electrostatic interactions between NPCs and apatitic surfaces.
Lastly, it is important to note that dental caries is a multifactorial disease associated with multi-species oral biofilms that are more complex than the S. mutans biofilms used for this proof-of-principle study. However, S. mutans is known to be a major pathogen responsible for dental caries due to its ability to outcompete other bacterial species under sugar-rich, cariogenic conditions in mixed-species biofilm models [52, 107], thereby promoting localized acidic microenvironments on tooth surfaces [38–40]. Thus, S. mutans is a suitable simple model system with minimal experimental variables to consider for initial interrogation of new anti-biofilm treatment approaches. Importantly, the ~3 log reduction in biofilm CFU/mL observed here is significant since this level of cariogenic pathogen reduction has been shown to reduce dental caries in vivo [61, 70]. Furthermore, the goal of this type of treatment is to adequately keep S. mutans and its virulence attributes (e.g., acidogenicity and aciduricity) at bay rather than complete eradication of all bacterial species, as may be the case for wound healing or sepsis therapies. Complete eradication is not desirable given that the complex oral microbiota also harbor commensals. However, future work investigating how dual drug-loaded NPCs affect other bacteria, including anaerobes such as fusobacterium and actinobacteria, via bacteriostatic and bactericidal evaluations is warranted. In sum, the single species S. mutans biofilms used in this work served as a first and very necessary step on the journey to developing a clinically-relevant anti-biofilm treatment using dual-drug loaded nanoparticles.
2. Conclusions
This study showed that a nanoparticle carrier drug delivery system with capabilities to co-load myricetin and farnesol synergistically improves S. mutans anti-biofilm efficacy in vitro. Evaluations of co-loaded NPC treatments using both developing and established S. mutans biofilms yielded contrasting results. Although treatments of established biofilm testing showed limited efficacy presumably due to high biomass accumulation covering the apatite surface, tests using developing biofilms showed substantial disruption of biofilm biomass (dry weight) and a ~3 log reduction in CFU/mL for the co-loaded NPC treatments compared to controls. Mechanistic studies revealed that NPCs co-loaded with farnesol and myricetin (regardless of myricetin concentration) prevented virulence-related pH drop likely due to decreased atpD expression and disrupted acid production (i.e., acidogenicidy) and acid tolerance (i.e., aciduricity) associated with F1F0-H/ATPase levels. Moreover, MIC testing using planktonic bacteria showed an ~8-fold difference in myricetin concentration necessary to inhibit bacterial growth when comparing myricetin to NPCs co-loaded with myricetin and farnesol. Additionally, dual drug-loaded NPC treatment demonstrated a synergistic relationship with a ~6 log CFU/mL reduction versus planktonic bacteria compared to the combined ~1 log CFU/mL reduction observed for the individual components alone. Finally, myricetin loading resulted in concentration-dependent effects on NPC binding to saliva-coated hydroxyapatite (sHA) surfaces, which can increase drug efficacy through improved retention at the biofilm-apatite interface. This effect can create an in situ reservoir at the apatite surface, thereby disrupting biofilm initiation and development consistent with the efficacy shown against developing biofilms. Altogether, this work suggests promise for co-delivery of myricetin and farnesol DDS as an alternative anti-biofilm treatment to prevent dental caries.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the National Institutes of Health (R01 DE018023 to HK and DB, and F31 DE026944 to KS), the National Science Foundation (DMR 1206219 to DB), and the Sao Paulo Research Foundation (FAPESP 2019/01429-4 to GR) for funding that supported this work. Research reported in this publication was supported by the National Institute of Dental & Craniofacial Research of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank James L. McGrath for access to the Malvern Zetasizer and Karen Bentley for assistance acquiring the TEM images within the University of Rochester Medical Center’s Electron Microscopy Core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors have no interests to declare.
References
- [1].Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, Hussain T, Ali M, Rafiq M, Kamil MA, Bacterial biofilm and associated infections, Journal of the Chinese Medical Association : JCMA 81(1) (2018) 7–11. [DOI] [PubMed] [Google Scholar]
- [2].Lewis K, Persister cells, dormancy and infectious disease, Nature reviews. Microbiology 5(1) (2007) 48–56. [DOI] [PubMed] [Google Scholar]
- [3].Wolcott RD, Ehrlich GD, Biofilms and chronic infections, Jama 299(22) (2008) 2682–4. [DOI] [PubMed] [Google Scholar]
- [4].Hall-Stoodley L, Costerton JW, Stoodley P, Bacterial biofilms: from the natural environment to infectious diseases, Nature reviews. Microbiology 2(2) (2004) 95–108. [DOI] [PubMed] [Google Scholar]
- [5].Mah TF, O’Toole GA, Mechanisms of biofilm resistance to antimicrobial agents, Trends Microbiol 9(1) (2001) 34–9. [DOI] [PubMed] [Google Scholar]
- [6].Smith R, Coast J, The true cost of antimicrobial resistance, BMJ (Clinical research ed.) 346 (2013) f1493. [DOI] [PubMed] [Google Scholar]
- [7].Donlan RM, Costerton JW, Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms, Clinical microbiology reviews 15(2) (2002) 167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ozdemir D, Dental Caries : The Most Common Disease Worldwide and Preventive Strategies, 2013.
- [9].Bowen WH, Burne RA, Wu H, Koo H, Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments, Trends in Microbiology (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C, The global burden of oral diseases and risks to oral health, Bulletin of the World Health Organization 83(9) (2005) 661–9. [PMC free article] [PubMed] [Google Scholar]
- [11].Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, Tagami J, Twetman S, Tsakos G, Ismail A, Dental caries, Nature Reviews Disease Primers 3 (2017) 17030. [DOI] [PubMed] [Google Scholar]
- [12].Marsh PD, Dental plaque as a microbial biofilm, Caries research 38(3) (2004) 204–11. [DOI] [PubMed] [Google Scholar]
- [13].Bowen WH, Koo H, Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms, Caries research 45(1) (2011) 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Flemming H-C, Neu TR, Wozniak DJ, The EPS Matrix: The “House of Biofilm Cells”, Journal of bacteriology 189(22) (2007) 7945–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Priyadarsini S, Mukherjee S, Mishra M, Nanoparticles used in dentistry: A review, Journal of oral biology and craniofacial research 8(1) (2018) 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Besinis A, De Peralta T, Tredwin CJ, Handy RD, Review of Nanomaterials in Dentistry: Interactions with the Oral Microenvironment, Clinical Applications, Hazards, and Benefits, ACS nano 9(3) (2015) 2255–2289. [DOI] [PubMed] [Google Scholar]
- [17].Allaker RP, Memarzadeh K, Nanoparticles and the control of oral infections, International Journal of Antimicrobial Agents 43(2) (2014) 95–104. [DOI] [PubMed] [Google Scholar]
- [18].Dos Santos Ramos MA, Da Silva PB, Spósito L, De Toledo LG, Bonifácio BV, Rodero CF, Dos Santos KC, Chorilli M, Bauab TM, Nanotechnology-based drug delivery systems for control of microbial biofilms: a review, International journal of nanomedicine 13 (2018) 1179–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bilia AR, Isacchi B, Righeschi C, Guccione C, Bergonzi MC, Flavonoids Loaded in Nanocarriers: An Opportunity to Increase Oral Bioavailability and Bioefficacy, Food and Nutrition Sciences Vol.05No. 13 (2014) 16. [Google Scholar]
- [20].Naha PC, Liu Y, Hwang G, Huang Y, Gubara S, Jonnakuti V, Simon-Soro A, Kim D, Gao L, Koo H, Cormode DP, Dextran-Coated Iron Oxide Nanoparticles as Biomimetic Catalysts for Localized and pH-Activated Biofilm Disruption, ACS nano (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sims KR, Liu Y, Hwang G, Jung HI, Koo H, Benoit DSW, Enhanced design and formulation of nanoparticles for anti-biofilm drug delivery, Nanoscale 11(1) (2018)219–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu Y, Naha PC, Hwang G, Kim D, Huang Y, Simon-Soro A, Jung H-I, Ren Z, Li Y, Gubara S, Alawi F, Zero D, Hara AT, Cormode DP, Koo H, Topical ferumoxytol nanoparticles disrupt biofilms and prevent tooth decay in vivo via intrinsic catalytic activity, Nature Communications 9(1) (2018) 2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Takahashi H, Nadres ET, Kuroda K, Cationic Amphiphilic Polymers with Antimicrobial Activity for Oral Care Applications: Eradication of S. mutans Biofilm, Biomacromolecules 18(1) (2017) 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li X, Wong CH, Ng TW, Zhang CF, Leung KC, Jin L, The spherical nanoparticle-encapsulated chlorhexidine enhances anti-biofilm efficiency through an effective releasing mode and close microbial interactions, International journal of nanomedicine 11 (2016)2471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gao L, Liu Y, Kim D, Li Y, Hwang G, Naha PC, Cormode DP, Koo H, Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo, Biomaterials 101 (2016) 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mogen AB, Chen F, Ahn SJ, Burne RA, Wang D, Rice KC, Pluronics-Formulated Farnesol Promotes Efficient Killing and Demonstrates Novel Interactions with Streptococcus mutans Biofilms, PloS one 10(7) (2015) e0133886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Horev B, Klein MI, Hwang G, Li Y, Kim D, Koo H, Benoit DS, pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence, ACS nano 9(3) (2015) 2390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Seneviratne CJ, Leung KC, Wong CH, Lee SF, Li X, Leung PC, Lau CB, Wat E, Jin L, Nanoparticle-encapsulated chlorhexidine against oral bacterial biofilms, PloS one 9(8) (2014) e103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yamakami K, Tsumori H, Sakurai Y, Shimizu Y, Nagatoshi K, Sonomoto K, Sustainable inhibition efficacy of liposome-encapsulated nisin on insoluble glucan-biofilm synthesis by Streptococcus mutans, Pharmaceutical biology 51(2) (2013) 267–70. [DOI] [PubMed] [Google Scholar]
- [30].Hernandez-Delgadillo R, Velasco-Arias D, Diaz D, Arevalo-Ni ntilde, K. o M, Garza-Enriquez M, De la Garza-Ramos MA, Cabral-Romero C, Zerovalent bismuth nanoparticles inhibit Streptococcus mutans growth and formation of biofilm, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen C-P, Chen C-T, Tsai T, Chitosan Nanoparticles for Antimicrobial Photodynamic Inactivation: Characterization and In Vitro Investigation†, Photochemistry and Photobiology 88(3) (2012) 570–576. [DOI] [PubMed] [Google Scholar]
- [32].Chen F, Rice KC, Liu XM, Reinhardt RA, Bayles KW, Wang D, Triclosan-loaded tooth-binding micelles for prevention and treatment of dental biofilm, Pharmaceutical research 27(11) (2010) 2356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen F, Liu XM, Rice KC, Li X, Yu F, Reinhardt RA, Bayles KW, Wang D, Tooth-binding micelles for dental caries prevention, Antimicrob Agents Chemother 53(11) (2009) 4898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou J, Horev B, Hwang G, Klein MI, Koo H, Benoit DSW, Characterization and optimization of pH-responsive polymer nanoparticles for drug delivery to oral biofilms, Journal of Materials Chemistry B 4(18) (2016) 3075–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen F, Jia Z, Rice KC, Reinhardt RA, Bayles KW, Wang D, The development of dentotropic micelles with biodegradable tooth-binding moieties, Pharmaceutical research 30(11) (2013) 2808–17. [DOI] [PubMed] [Google Scholar]
- [36].Chávez de Paz LE, Resin A, Howard KA, Sutherland DS, Wejse PL, Antimicrobial effect of chitosan nanoparticles on streptococcus mutans biofilms, Applied and environmental microbiology 77(11) (2011) 3892–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leung KC-F, Seneviratne CJ, Li X, Leung PC, Lau CBS, Wong C-H, Pang KY, Wong CW, Wat E, Jin L, Synergistic Antibacterial Effects of Nanoparticles Encapsulated with Scutellaria baicalensis and Pure Chlorhexidine on Oral Bacterial Biofilms, Nanomaterials (Basel, Switzerland) 6(4) (2016) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Krzysciak W, Jurczak A, Koscielniak D, Bystrowska B, Skalniak A, The virulence of Streptococcus mutans and the ability to form biofilms, European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 33(4) (2014) 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lemos JA, Quivey RG Jr., Koo H, Abranches J, Streptococcus mutans: a new Gram-positive paradigm?, Microbiology (Reading, England) 159(Pt 3) (2013) 436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Loesche WJ, Role of Streptococcus mutans in human dental decay, Microbiological reviews 50(4) (1986) 353–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Priyadarshini BM, Mitali K, Lu TB, Handral HK, Dubey N, Fawzy AS, PLGA nanoparticles as chlorhexidine-delivery carrier to resin-dentin adhesive interface, Dental materials : official publication of the Academy of Dental Materials 33(7) (2017) 830–846. [DOI] [PubMed] [Google Scholar]
- [42].Stewart PS, Mechanisms of antibiotic resistance in bacterial biofilms, International Journal of Medical Microbiology 292(2) (2002) 107–113. [DOI] [PubMed] [Google Scholar]
- [43].Roberts AP, Mullany P, Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance, Expert review of anti-infective therapy 8(12) (2010) 1441–50. [DOI] [PubMed] [Google Scholar]
- [44].Benoit DSW, Sims KR, Fraser D, Nanoparticles for Oral Biofilm Treatments, ACS nano 13(5) (2019) 4869–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nakanishi-Matsui M, Sekiya M, Nakamoto RK, Futai M, The mechanism of rotating proton pumping ATPases, Biochimica et biophysica acta 1797(8) (2010) 1343–52. [DOI] [PubMed] [Google Scholar]
- [46].Kuhnert WL, Zheng G, Faustoferri RC, Quivey RG Jr., The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH, Journal of bacteriology 186(24) (2004) 8524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Quivey RG Jr., Kuhnert WL, Hahn K, Adaptation of oral streptococci to low pH, Adv Microb Physiol 42 (2000) 239–74. [DOI] [PubMed] [Google Scholar]
- [48].Matsui R, Cvitkovitch D, Acid tolerance mechanisms utilized by Streptococcus mutans, Future Microbiol 5(3) (2010) 403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Colby SM, Russell RR, Sugar metabolism by mutans streptococci, Society for Applied Bacteriology symposium series 26 (1997) 80s–88s. [PubMed] [Google Scholar]
- [50].Takahashi N, Yamada T, Acid-induced acid tolerance and acidogenicity of non-mutans streptococci, Oral microbiology and immunology 14(1) (1999) 43–8. [DOI] [PubMed] [Google Scholar]
- [51].Klein MI, Xiao J, Lu B, Delahunty CM, Yates JR 3rd, Koo H, Streptococcus mutans protein synthesis during mixed-species biofilm development by high-throughput quantitative proteomics, PloS one 7(9) (2012) e45795–e45795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR III, Heydorn A, Koo H, The Exopolysaccharide Matrix Modulates the Interaction between 3D Architecture and Virulence of a Mixed-Species Oral Biofilm, PLOS Pathogens 8(4) (2012) e1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Flemming HC, Wingender J, The biofilm matrix, Nature reviews. Microbiology 8(9) (2010) 623–33. [DOI] [PubMed] [Google Scholar]
- [54].Guo L, McLean JS, Lux R, He X, Shi W, The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of Streptococcus mutans, Scientific Reports 5 (2015) 18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Santana M, Ionescu MS, Vertes A, Longin R, Kunst F, Danchin A, Glaser P, Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants, Journal of bacteriology 176(22) (1994) 6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Smith AJ, Quivey RG, Faustoferri RC, Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase operon, Gene 183(1) (1996) 87–96. [DOI] [PubMed] [Google Scholar]
- [57].Klein MI, Hwang G, Santos PH, Campanella OH, Koo H, Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms, Frontiers in cellular and infection microbiology 5 (2015) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schilling KM, Bowen WH, Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans, Infection and immunity 60(1) (1992) 284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ren Z, Chen L, Li J, Li Y, Inhibition of Streptococcus mutans polysaccharide synthesis by molecules targeting glycosyltransferase activity, Journal of Oral Microbiology 8(1) (2016) 31095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Koo H, Rosalen PL, Cury JA, Park YK, Bowen WH, Effects of Compounds Found in Propolis on Streptococcus mutans Growth and on Glucosyltransferase Activity, Antimicrobial Agents and Chemotherapy 46(5) (2002) 1302–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Koo H, Pearson SK, Scott-Anne K, Abranches J, Cury JA, Rosalen PL, Park YK, Marquis RE, Bowen WH, Effects of apigenin and tt-farnesol on glucosyltransferase activity, biofilm viability and caries development in rats, Oral microbiology and immunology 17(6) (2002) 337–43. [DOI] [PubMed] [Google Scholar]
- [62].Koo H, Hayacibara MF, Schobel BD, Cury JA, Rosalen PL, Park YK, Vacca-Smith AM, Bowen WH, Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol, Journal of Antimicrobial Chemotherapy 52(5) (2003) 782–789. [DOI] [PubMed] [Google Scholar]
- [63].Rocha GR, Florez Salamanca EJ, de Barros AL, Lobo CIV, Klein MI, Effect of tt-farnesol and myricetin on in vitro biofilm formed by Streptococcus mutans and Candida albicans, BMC complementary and alternative medicine 18(1) (2018) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gregoire S, Singh AP, Vorsa N, Koo H, Influence of cranberry phenolics on glucan synthesis by glucosyltransferases and Streptococcus mutans acidogenicity, Journal of applied microbiology 103(5) (2007) 1960–8. [DOI] [PubMed] [Google Scholar]
- [65].Falsetta ML, Klein MI, Lemos JA, Silva BB, Agidi S, Scott-Anne KK, Koo H, Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in Streptococcus mutans to modulate virulence expression in vivo, Antimicrob Agents Chemother 56(12) (2012) 6201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jeon JG, Rosalen PL, Falsetta ML, Koo H, Natural products in caries research: current (limited) knowledge, challenges and future perspective, Caries research 45(3) (2011) 243–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jeon J-G, Klein MI, Xiao J, Gregoire S, Rosalen PL, Koo H, Influences of naturally occurring agents in combination with fluoride on gene expression and structural organization of Streptococcus mutans in biofilms, BMC Microbiology 9(1) (2009) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hong S, Pedersen PL, ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas, Microbiology and molecular biology reviews : MMBR 72(4) (2008) 590–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Thimothe J, Bonsi IA, Padilla-Zakour OI, Koo H, Chemical characterization of red wine grape (Vitis vinifera and Vitis interspecific hybrids) and pomace phenolic extracts and their biological activity against Streptococcus mutans, Journal of agricultural and food chemistry 55(25) (2007) 10200–7. [DOI] [PubMed] [Google Scholar]
- [70].Koo H, Schobel B, Scott-Anne K, Watson G, Bowen WH, Cury JA, Rosalen PL, Park YK, Apigenin and tt-farnesol with fluoride effects on S. mutans biofilms and dental caries, Journal of dental research 84(11) (2005) 1016–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Shchepin R, Hornby JM, Burger E, Niessen T, Dussault P, Nickerson KW, Quorum Sensing in Candida albicans: Probing Farnesol’s Mode of Action with 40 Natural and Synthetic Farnesol Analogs, Chemistry & biology 10(8) (2003) 743–750. [DOI] [PubMed] [Google Scholar]
- [72].Yao Y, Lin G, Xie Y, Ma P, Li G, Meng Q, Wu T, Preformulation studies of myricetin: a natural antioxidant flavonoid, Die Pharmazie 69(1) (2014) 19–26. [PubMed] [Google Scholar]
- [73].Semwal DK, Semwal RB, Combrinck S, Viljoen A, Myricetin: A Dietary Molecule with Diverse Biological Activities, Nutrients 8(2) (2016) 90–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sims KR, He B, Koo H, Benoit DSW, Electrostatic Interactions Enable Nanoparticle Delivery of the Flavonoid Myricetin, ACS Omega 5(22) (2020) 12649–12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS, Development of a novel endosomolytic diblock copolymer for siRNA delivery, Journal of controlled release : official journal of the Controlled Release Society 133(3) (2009) 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Moad G, Chong YK, Postma A, Rizzardo E, Thang SH, Advances in RAFT polymerization: the synthesis of polymers with defined end-groups, Polymer 46(19) (2005) 8458–8468. [Google Scholar]
- [77].Murthy N, Robichaud JR, Tirrell DA, Stayton PS, Hoffman AS, The design and synthesis of polymers for eukaryotic membrane disruption, Journal of Controlled Release 61(1–2) (1999) 137–143. [DOI] [PubMed] [Google Scholar]
- [78].Sims KR, Maceren JP, Strand AI, He B, Overby C, Benoit DSW, Rigor and reproducibility in polymer nanoparticle synthesis and characterization, RSC Advances 10(5) (2020) 2513–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gallow KC, Jhon YK, Genzer J, Loo Y-L, Influence of gradient strength and composition profile on the onset of the cloud point transition in hydroxyethyl methacrylate/dimethylaminoethyl methacrylate gradient copolymers, Polymer 53(5) (2012) 1131–1137. [Google Scholar]
- [80].Vesterinen A, Lipponen S, Rich J, Seppälä J, Effect of block composition on thermal properties and melt viscosity of poly(2-(dimethylamino)ethyl methacrylate), poly(ethylenen oxide) and poly(propylene oxide) block co-polymers, EXPRESS POLYMER LETTERS 5(9) (2011) 754–765. [Google Scholar]
- [81].Koo H, Vacca Smith AM, Bowen WH, Rosalen PL, Cury JA, Park YK, Effects of Apis mellifera Propolis on the Activities of Streptococcal Glucosyltransferases in Solution and Adsorbed onto Saliva–Coated Hydroxyapatite, Caries research 34(5) (2000) 418–426. [DOI] [PubMed] [Google Scholar]
- [82].Hayacibara MF, Koo H, Vacca-Smith AM, Kopec LK, Scott-Anne K, Cury JA, Bowen WH, The influence of mutanase and dextranase on the production and structure of glucans synthesized by streptococcal glucosyltransferases, Carbohydr Res 339(12) (2004) 2127–37. [DOI] [PubMed] [Google Scholar]
- [83].Cury JA, Koo H, Extraction and purification of total RNA from Streptococcus mutans biofilms, Analytical biochemistry 365(2) (2007) 208–214. [DOI] [PubMed] [Google Scholar]
- [84].He J, Hwang G, Liu Y, Gao L, Kilpatrick-Liverman L, Santarpia P, Zhou X, Koo H, l-Arginine Modifies the Exopolysaccharide Matrix and Thwarts Streptococcus mutans Outgrowth within Mixed-Species Oral Biofilms, Journal of bacteriology 198(19) (2016) 2651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Koo H, Rosalen PL, Cury JA, Ambrosano GM, Murata RM, Yatsuda R, Ikegaki M, Alencar SM, Park YK, Effect of a new variety of Apis mellifera propolis on mutans Streptococci, Current microbiology 41 (3) (2000) 192–6. [DOI] [PubMed] [Google Scholar]
- [86].Jeon JG, Pandit S, Xiao J, Gregoire S, Falsetta ML, Klein MI, Koo H, Influences of trans-trans farnesol, a membrane-targeting sesquiterpenoid, on Streptococcus mutans physiology and survival within mixed-species oral biofilms, International journal of oral science 3(2) (2011) 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kuhnert WL, Quivey J, Robert G, Genetic and Biochemical Characterization of the F-ATPase Operon from Streptococcus sanguis 10904, Journal of bacteriology 185(5) (2003) 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gong Y, Tian X-L, Sutherland T, Sisson G, Mai J, Ling J, Li Y-H, Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation, Microbiology (Reading, England) 155(10) (2009) 3322–3332. [DOI] [PubMed] [Google Scholar]
- [89].Sakanaka S, Kim M, Taniguchi M, Yamamoto T, Antibacterial Substances in Japanese Green Tea Extract against Streptococcus mutans, a Cariogenic Bacterium, Agricultural and Biological Chemistry 53(9) (1989) 2307–2311. [Google Scholar]
- [90].Ito K, Ito S, Shimamura T, Weyand S, Kawarasaki Y, Misaka T, Abe K, Kobayashi T, Cameron AD, Iwata S, Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans, Journal of molecular biology 408(2) (2011) 177–86. [DOI] [PubMed] [Google Scholar]
- [91].Zhang K, Cheng L, Weir MD, Bai Y-X, Xu HHK, Effects of quaternary ammonium chain length on the antibacterial and remineralizing effects of a calcium phosphate nanocomposite, International journal of oral science 8(1) (2016) 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Franci G, Falanga A, Galdiero S, Palomba L, Rai M, Morelli G, Galdiero M, Silver Nanoparticles as Potential Antibacterial Agents, Molecules 20(5) (2015) 8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ping L, Juan L, Changzhu W, Qingsheng W, Jian L, Synergistic antibacterial effects of β-lactam antibiotic combined with silver nanoparticles, Nanotechnology 16(9) (2005) 1912. [Google Scholar]
- [94].Yeh Y-C, Huang T-H, Yang S-C, Chen C-C, Fang J-Y, Nano-Based Drug Delivery or Targeting to Eradicate Bacteria for Infection Mitigation: A Review of Recent Advances, Front Chem 8(286) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Namivandi-Zangeneh R, Sadrearhami Z, Bagheri A, Sauvage-Nguyen M, Ho KKK, Kumar N, Wong EHH, Boyer C, Nitric Oxide-Loaded Antimicrobial Polymer for the Synergistic Eradication of Bacterial Biofilm, ACS Macro Letters 7(5) (2018) 592–597. [DOI] [PubMed] [Google Scholar]
- [96].Yang M, Du K, Hou Y, Xie S, Dong Y, Li D, Du Y, Synergistic Antifungal Effect of Amphotericin B-Loaded Poly(Lactic-Co-Glycolic Acid) Nanoparticles and Ultrasound against <span class="named-content genus-species" id="named-content-1">Candida albicans<span> Biofilms, Antimicrobial Agents and Chemotherapy 63(4) (2019) e02022–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Quivey RG, Kuhnert WL, Hahn K, Genetics of acid adaptation in oral streptococci, Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists 12(4) (2001) 301–14. [DOI] [PubMed] [Google Scholar]
- [98].Gabe V, Kacergius T, Abu-Lafi S, Kalesinskas P, Masalha M, Falah M, Abu-Farich B, Melninkaitis A, Zeidan M, Rayan A, Inhibitory Effects of Ethyl Gallate on Streptococcus mutans Biofilm Formation by Optical Profilometry and Gene Expression Analysis, Molecules (Basel, Switzerland) 24(3) (2019) 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Koo H, Xiao J, Klein MI, Jeon JG, Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms, Journal of bacteriology 192(12) (2010) 3024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang Q, Kang F, Gao Y, Mao X, Hu X, Sequestration of nanoparticles by an EPS matrix reduces the particle-specific bactericidal activity, Scientific Reports 6 (2016) 21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lemos JA, Palmer SR, Zeng L, Wen ZT, Kajfasz JK, Freires IA, Abranches J, Brady LJ, The Biology of Streptococcus mutans, Microbiol Spectr 7(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AHM, Hamaker BR, Lemos JA, Koo H, Dynamics of Streptococcus mutans Transcriptome in Response to Starch and Sucrose during Biofilm Development, PloS one 5(10) (2010) e13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Takahashi N, Nyvad B, Caries ecology revisited: microbial dynamics and the caries process, Caries research 42(6) (2008) 409–18. [DOI] [PubMed] [Google Scholar]
- [104].Fears KP, Gonzalez-Begne M, Love CT, Day DE, Koo H, Surface-Induced Changes in the Conformation and Glucan Production of Glucosyltransferase Adsorbed on Saliva-Coated Hydroxyapatite, Langmuir 31 (16) (2015) 4654–4662. [DOI] [PubMed] [Google Scholar]
- [105].Wu Z, Zhang B, Yan B, Regulation of Enzyme Activity through Interactions with Nanoparticles, International Journal of Molecular Sciences 10(10) (2009) 4198–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM, Control of protein structure and function through surface recognition by tailored nanoparticle scaffolds, J Am Chem Soc 126(3) (2004) 739–43. [DOI] [PubMed] [Google Scholar]
- [107].Liu Y, Palmer SR, Chang H, Combs AN, Burne RA, Koo H, Differential oxidative stress tolerance of Streptococcus mutans isolates affects competition in an ecological mixed-species biofilm model, Environmental microbiology reports 10(1) (2018) 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


