Abstract
Gram-negative phytopathogenic bacteria are a significant threat to food crops. These microbial invaders are responsible for a plethora of plant diseases and can be responsible for devastating losses in crops such as tomatoes, peppers, potatoes, olives, and rice. Current disease management strategies to mitigate yield losses involve the application of chemicals which are often harmful to both human health and the environment. Bacteriocins are small proteinaceous antibiotics produced by bacteria to kill closely related bacteria and thereby establish dominance within a niche. They potentially represent a safer alternative to chemicals when used in the field. Bacteriocins typically show a high degree of selectivity toward their targets with no off-target effects. This review outlines the current state of research on bacteriocins active against Gram-negative phytopathogenic bacteria. Furthermore, we will examine the feasibility of weaponizing bacteriocins for use as a treatment for bacterial plant diseases.
Keywords: bacteriocins, Gram-negative bacteria, phytopathogenic bacteria, plant disease, plant disease management, food security, crops
Introduction
By 2050 the global population is predicted to surpass 9 billion requiring food production to increase by 70%, equivalent to 127 × 1015 kcal (Cole et al., 2018). Major food crops suffer from a lack of genetic diversity allowing pathogens and pests to rapidly spread throughout fields and devastate crops, causing yield losses of up to 32% (Oerke and Dehne, 2004).
Gram-negative bacterial phytopathogens are an important contributor to crop losses due to plant disease (Mansfield et al., 2012). For example, Pseudomonas syringae pv. actinidiae, the causal agent of the kiwifruit canker pandemic, triggered enormous damage to the New Zealand economy (Vanneste et al., 2013) depreciating the land value of orchards growing the popular kiwifruit variety Hort16A from 300,000 to 46,000 USD per hectare (Vanneste, 2017). Enterobacterial soft rot phytopathogens such as Pectobacterium and Dickeya spp. are collectively responsible for diseases in potato like black leg and tuber soft rot pre- and post-harvest (Pérombelon, 2002; Toth et al., 2011). These diseases are responsible for losses of €30 m per annum in the Netherlands alone (Pérombelon, 2002; Toth et al., 2011).
Bacteriocins are proteinaceous antibiotics that are produced by both Gram-positive and Gram-negative bacteria (Cascales et al., 2007; Chavan and Riley, 2007). They target and kill related bacterial strains allowing producing strains to establish dominance within a niche (Chavan and Riley, 2007). Unlike conventional small molecule antibiotics, bacteriocins exhibit a narrow killing spectrum and cause minimal disruption to the commensal bacterial community (Chavan and Riley, 2007). A number of classification systems have been proposed to encompass the diversity of bacteriocins (Heng and Tagg, 2006; Chavan and Riley, 2007; Cotter et al., 2013). The classification of Chavan and Riley (2007) is based on size, splitting the bacteriocins into three groups; small peptide bacteriocins of <10 kDa, colicin-like bacteriocins (CLBs) which are multidomain proteins of 25–80 kDa and tailocins, which are large phage-like multimeric protein assemblies. This review focuses on the latter two of these groups as there is a dearth of information on small peptide bacteriocins active against phytopathogenic bacteria. In addition, in this review we cover an additional group, the lectin-like bacteriocins (LLBs), which although they fall within the size range of CBLs, are mechanistically distinct. We also provide examples of some orphan bacteriocins.
Bacteriocins have been identified in a number of important plant pathogenic bacterial genera including Xanthomonas, Pseudomonas, Pectobacterium, and Agrobacterium (Holtsmark et al., 2008; Grinter et al., 2012a). These include many important pathogens of crops such as rice, banana, potato, olives, peppers and tomatoes (Mansfield et al., 2012). In this review, we aim to outline the present landscape of research into bacteriocin plantibiotics (biological agents which selectively kill plant pathogenic bacteria) and discuss the practicalities of exploiting them to remedy plant disease.
Colicin-Like Bacteriocins
CLBs are multi-domain proteins that possess a modular domain structure usually consisting of translocation, receptor binding and cytotoxic domains. The translocation domain typically incorporates, or consists of an intrinsically disordered region (IDR) at the extreme N-terminus of the protein, which is first to cross the outer membrane during uptake (Behrens et al., 2020). To target a specific bacterial species, CLBs often parasitize existing nutrient uptake pathways involving TonB dependent transporters (TBDTs). These TBDTs are frequently involved with the uptake of iron siderophores and other metal chelate complexes, such as vitamin B12, from the environment (Michel-Briand and Baysse, 2002; Cascales et al., 2007). For most CLBs the IDR and translocation domains facilitate the import of bacteriocins across the outer membrane into the periplasmic space. Briefly, this is achieved by the IDR threading through the pore of an outer membrane transporter and interacting with components of the proton-motive force (PMF) responsive Ton or Tol complexes in the periplasm. Subsequently, the bacteriocin is actively pulled through the transporter in a PMF-dependent manner to cross the outer membrane (White et al., 2017). Methods of killing mediated by CLB cytotoxic domains include endonuclease activity (DNase, tRNase, and rRNase), depolarization of the inner-membrane, and inhibition of peptidoglycan synthesis (Michel-Briand and Baysse, 2002; Cascales et al., 2007).
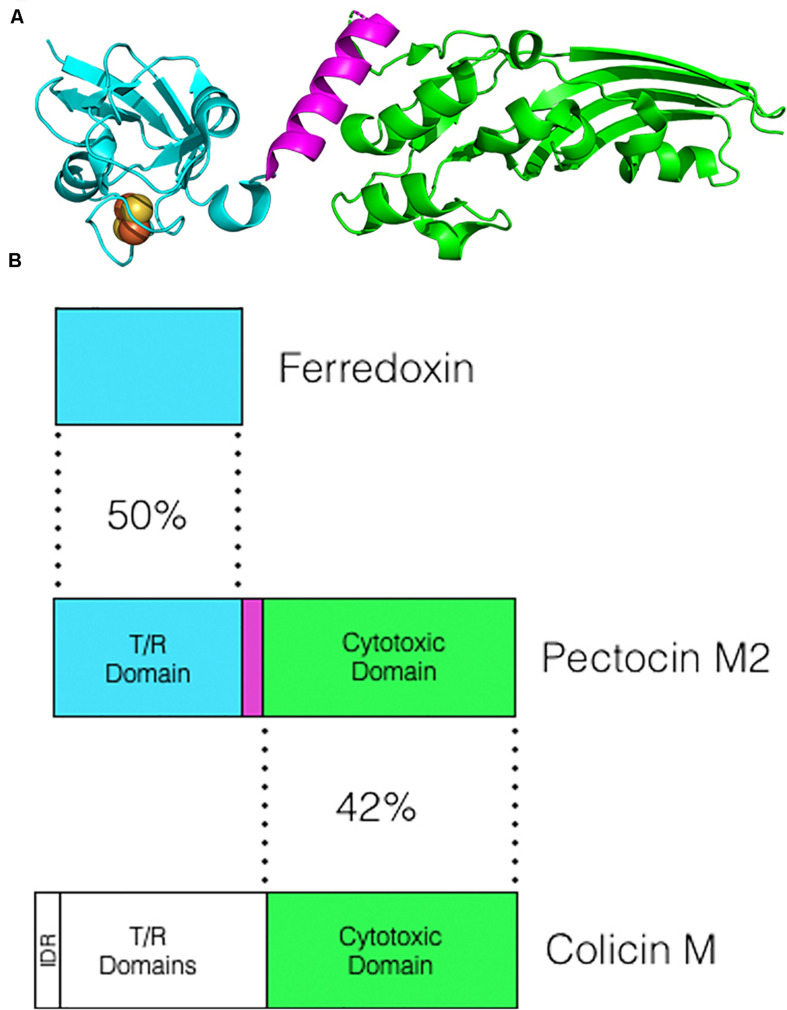
In Pectobacterium carotovorum, three CLB nucleases termed carocins have been characterized. Two of the carocins, S1K (40 kDa) and carocin D (91 kDa) are DNases while the third, S2 (85 kDa) is a tRNAse (Chuang et al., 2007; Chan et al., 2009, 2011; Roh et al., 2010; Atanaskovic et al., 2020). In addition, two pectobacterial CLBs, pectocins M1 and M2 (both 29 kDa) have been characterized that possess cytotoxic domains homologous to that of colicin M and have been shown to similarly target lipid II (Grinter et al., 2012b). Cleavage of lipid II by colicin M-like bacteriocins results in inhibition of peptidoglycan biosynthesis and cell death (Harkness and Ölschläger, 1991). Interestingly, pectocin M1 and M2 lack an IDR at their N-termini and instead contain a single globular domain N-terminal to the cytotoxic domain that is homologous in both sequence and structure to plant ferredoxin (Figure 1; Grinter et al., 2012b, 2014). Like plant ferredoxin, these CLBs also contain a 2Fe-2S iron sulfur cluster and as subsequent research has shown, have evolved to parasitize an existing ferredoxin uptake system utilized by Pectobacterium spp. to acquire iron from its plant hosts. Uptake of ferredoxin is mediated by the TBDT FusA and the TonB-like protein FusB which work in concert to translocate ferredoxin into the periplasm (Grinter et al., 2016; Wojnowska and Walker, 2020). FusB acts both in removal of the plug from the lumen of FusA and directly binding to ferredoxin mediating its active translocation across the outer membrane via the lumen of FusA (Wojnowska and Walker, 2020). Within the periplasm, the processing protease FusC cleaves ferredoxin in two specific locations releasing its iron into the periplasm (Mosbahi et al., 2018). Competition assays with spinach ferredoxin and killing assays under iron limiting conditions show that ferredoxin-containing bacteriocins are translocated using the same pathway (Grinter et al., 2012b). Bioinformatic analysis has revealed another putative pectobacterial bacteriocin, pectocin P (35 kDa), that also contains a ferredoxin domain (Grinter et al., 2012b). However, the cytotoxic domain of pectocin P shares structural homology to lysozyme implying that uptake using the ferredoxin domain can be utilized as a general translocation pathway to deliver cytotoxic proteins into the periplasm. Lastly, two CLBs from P. syringae have been reported, syringacin M (30 kDa), which shares homology with colicin M, and a nuclease bacteriocin, SE9a (64 kDa) related to pyocin S2 (Grinter et al., 2012c; Hockett et al., 2017). Unlike the colicin M-like pectocins M1 and M2, syringacin M does possess an N-terminal IDR and so likely has an uptake mechanism that is similar to the well-characterized colicins from E. coli (Grinter et al., 2012c).
FIGURE 1.
Structure and homology of the ferredoxin containing pectocin M2. (A) The crystal structure of pectocin M2 (PDB:4N58). The ferredoxin domain is in cyan, the linker region in purple and the colicin M-like cytotoxic domain in green. The iron sulfur cluster located in the ferredoxin domain is represented as yellow and orange spheres. (B) The N-terminal domain of pectocin M2 (ZP_03825528) shares homology with spinach ferredoxin (1704156A). The C-terminal domain which is separated from the ferredoxin domain by a short helical linker shares homology with the lipid II-cleaving cytotoxic domain of colicin M (WP_000449474). Numbers shown are percentage identities calculated using the Needleman-Wunsch algorithm (Needle program) from EBI.
Analysis of mutations in bacteria grown in the presence of bacteriocins suggest that resistance usually results from changes in the bacteriocin receptor (Cascales et al., 2007; Inglis et al., 2016). However, the development of resistance in the wild is still poorly understood and it may also depend on additional factors involving the receptor. For example, in iron limiting conditions, resistance to pyocin S2 is subject to negative selection as its receptor is required for the uptake of iron (Inglis et al., 2016). As these receptors are typically involved in processes that are important for competition and cell survival, resistant strains tend to be less fit and show reduced pathogenicity in some environments.
Lectin-Like Bacteriocins
LLBs are a distinct family of protein antibiotics found in Pseudomonas, Burkholderia, and Xanthomonas species (Ghequire et al., 2012a, 2013a). The hallmark of LLBs is the presence of monocot mannose-binding lectin (MMBLs) domains. MMBLs are expanded in both plants and animals and play a primitive defensive role against pests and pathogens (Ghequire et al., 2012b). LLBs possess at least 1 MMBL domain containing conserved QxDxNxVxYx sequences that constitute a carbohydrate-binding pocket. These MMBLs are instrumental in defining the selectivity of LLBs by enabling the docking onto D-rhamnose-containing lipopolysaccharide (LPS) on the cell surface (Ghequire et al., 2013b; McCaughey et al., 2014).
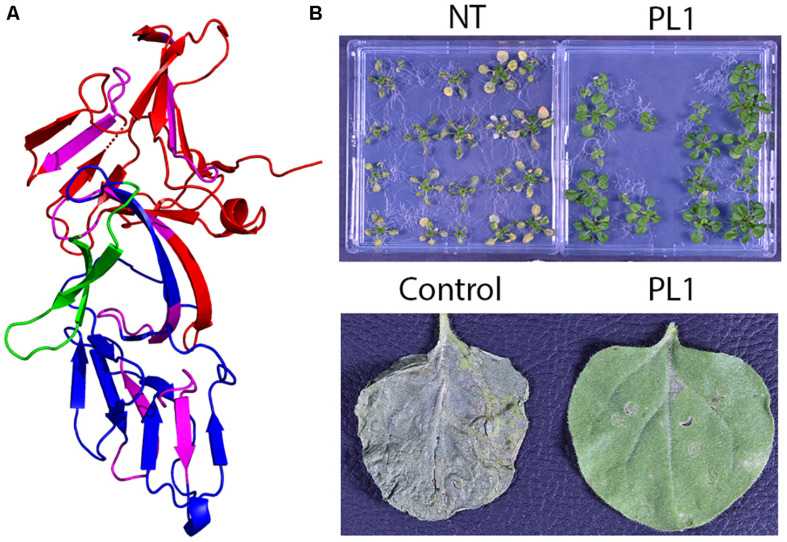
Our current understanding of LLBs arises predominantly from the study of pyocin L1 and putidacin L1 (PL1) isolated from P. aeruginosa and P. putida, respectively (Parret et al., 2003; McCaughey et al., 2014). PL1 (30 kDa) harbors 2 MMBL domains (Figure 2A) and phylogenetic analyses of the N- and C-terminal MMBL domains suggest distinct functions in LPS docking (Ghequire et al., 2013b). The N-terminal MMBL domains diverge substantially implying their importance in selectivity, whereas the C-terminal MMBL domains tightly cluster suggesting that their primary function are to bind to carbohydrates with high affinity (Ghequire et al., 2018b). Intriguingly, it was recently reported that LLBs containing 1 MMBL exhibit anti-microbial activity against Pseudomonads (Ghequire and De Mot, 2019). Although little is known about these bacteriocins isolated from soil- and plant-associated bacteria, their MMBLs share homology with the N-terminal MMBL domains of putidacin L1-type LLBs (Ghequire and De Mot, 2019).
FIGURE 2.
LLBs can be provide robust disease resistance against P. syringae. (A) Structure of the LLB PL1 (PDB:4GC2). The N- and C-terminal MMBL domains are shown in red and blue, respectively, and C-terminal extension is shown in green. Within these domains the QxDxNxVxYx sugar binding motifs are shown in purple. The C-terminal extension is predicted to play a role in the cytotoxic action of PL1 by disrupting the function of BamA. (B) Non-transgenic (NT) vs. transgenic expression of PL1 in both Arabidopsis seedlings (upper panel) and Nicotiniana benthamiana leaves (lower panel) provides robust resistance against strains of P. syringae that are susceptible to PL1. These images were adapted from Rooney et al. (2019).
Resistance to LLBs can arise from changes to LPS structure by susceptible bacteria (Ghequire et al., 2013b; McCaughey et al., 2014). However, LPS binding does not fully explain the selective nature of LLBs (Ghequire et al., 2013b). An exhaustive genetic study of resistant bacterial isolates identified novel changes in a surface-exposed extracellular loop of the outer membrane protein BamA (Ghequire et al., 2018a). BamA is a critical component of the β-barrel assembly machinery responsible for the chaperoning and insertion of β-barrel proteins into the outer membrane (Noinaj et al., 2017). Sequence alignments comparing PL1 sensitive and resistant strains identified the amino acid sequences of loop 6 of BamA as a genetic determinant of PL1 susceptibility. This was elegantly demonstrated when a “resistant” allele of BamA successfully rescued a PL1-sensitive strain from PL1-mediated killing in vitro (Ghequire et al., 2018a).
Tailocins
Tailocins are headless phage tail-like bacteriocins consisting of 8–14 individual components, including a sheath, core and baseplate (Ghequire et al., 2015b; Scholl, 2017). A producing cell releases 100s of particles and sometimes one particle is sufficient to eliminate a target cell (Scholl, 2017). Although tailocins from phytopathogenic bacteria share a high degree of similarity with contractile tail phages derived from Myoviridae, they have evolved independently, and represent an expansive group of protein complexes playing critical ecological roles like biofilm formation (Ghequire et al., 2015b; Turnbull et al., 2016). Tailocin-mediated killing occurs in two steps. Firstly, the tail fibers selectively bind to LPS of a target cell (Michel-Briand and Baysse, 2002). In turn, the sheath contracts and punctures the cell envelope, depolarizing the cell membrane and resulting in cell death (Scholl, 2017).
Tailocins are produced by a range of bacteria including Pseudomonads and Pectobacterium spp. (Nguyen et al., 2001; Hockett et al., 2015). Remarkably, tailocins from P. syringae have evolved independently of those of P. aeruginosa and do not share the same evolutionary ancestor (Ghequire and De Mot, 2015a; Hockett et al., 2015). This likely reflects the different environmental niches that P. aeruginosa and P. syringae occupy. Intriguingly, diversification and expansion of the tailocin family in P. syringae is driven by localized recombination of tailocin genes like those encoding the tail fibers (Baltrus et al., 2019).
Identification and genetic dissection of two distinct tailocins from P. chlororaphis has unmasked the robust competitive advantage tailocins provide in heterogeneous biofilms and the rhizosphere (Dorosky et al., 2017, 2018). Notably, P. fluorescens SF4c harbors a tailocin targeting X. vesicatoria, the causal agent of bacterial spot disease in tomatoes (Príncipe et al., 2018). In the case of Pectobacterium there are two highly similar tailocins, carotovoricin (Ctv)Er and CGE originating from P. carotovorum IAM 1068 and P. carotovorum CGE234-M403, respectively (Yamada et al., 2006). Indeed, both carotovoricins are identical apart from two 26 bp inverted repeats within and downstream of the tail fiber gene, which differentiates their killing spectrum (Nguyen et al., 2001). Notably, strains of Pectobacterium harboring CtvCGE are sold in Japan, under the name “Biokeeper” to manage bacterial soft rot infections in potatoes (Chuang et al., 1999).
Classically, tailocin tolerance arises from alterations in the LPS, enabling the targets to evade tailocin recognition (Scholl, 2017; Kandel et al., 2020). A recent study postulates that bacteria can persist in environments containing sub-lethal concentrations of tailocins (Kandel et al., 2020). Persistence is not a heritable genetic trait, and likely reflects heterogeneity of gene expression within a clonal population influenced by factors such as starvation and metabolic activity (Kandel et al., 2020). Persistence allows bacteria to bypass mutations which incur fitness costs providing the selective pressure is transient.
Orphan Bacteriocins and Plantibiotics
Several bacteriocins from phytopathogens do not display homology with other well-characterized classes. For example, X. campestris pv. glycines 8ra produces a bacteriocin called glycinecin A (55 kDa), which is unusually encoded by two genes (Heu et al., 2001). Interestingly, although glycinecin A can be produced recombinantly in E. coli, active bacteriocin is only obtained when glyA and glyB are co-expressed in the same cell; active bacteriocin cannot be reconstituted by combining the two separately expressed polypeptides in vitro (Heu et al., 2001). There are two bacteriocins identified from X. campestris and X. perforans, glycinecin R, and BCN-A, containing Rhs repeats (pfam05593) which share homology with the toxin complex of the insect pathogen Photorhabdus (Roh et al., 2008; Marutani-Hert et al., 2020). The mode of action for these Xanthomonas bacteriocins are unknown.
The production of a very narrow spectrum bacteriocin-like substance agrocin 84 (1.4 kDa) by some non-pathogenic strains of Agrobacterium tumefaciens has been characterized and exploited to control crown gall diseases caused by pathogenic strains of A. tumefaciens (Kerr and Htay, 1974; Ellis et al., 1979). This small nucleotide antibiotic represses leucyl-tRNA synthetase activity (Tate et al., 1979; Kim et al., 2006). Remarkably, A. tumefaciens strains that successfully evolve resistance against agrocin 84 become non-pathogenic (Kerr, 1980). The success of agrocin 84 as a strategy to control crown gall disease resulted in the development of an agrocin 84-producing A. tumefaciens strain which was successfully trademarked and sold under the name Nogall by Bio-care Technology (Jones et al., 1988).
Applications of Bacteriocins and Future Perspectives
The control of bacterial phytopathogens in agriculture often relies on the application of chemicals containing copper or antibiotics, notably streptomycin. These often have detrimental impacts on human health and the environment and their long term success as a control measure can be limited by the development of resistance (Sundin and Bender, 1993; Damalas and Eleftherohorinos, 2011). For example, streptomycin treatment has been used extensively in orchards to mitigate diseases like fire blight and citrus greening. However, widespread applications of antibiotics in a field context has the potential to create reservoirs of resistance that can potentially transfer from plant pathogenic bacteria into bacterial pathogens of clinical importance (Norelli et al., 2003; McKenna, 2019).
A major driver of the success of the insecticidal protein Bacillus thuringiensis (Bt) toxin has been its high degree of target selectivity and its ease of expression in planta. Bacteriocins share similar characteristics. In both clinical and agricultural contexts this is highly advantageous as their use would be expected to cause minimal disruption to the microbiome. Like Bt toxins, bacteriocins can be expressed in plants or directly applied to crops. Nomad Biosciences have neatly illustrated the feasibility of expressing bacteriocins (LLBs and CLBs) in several plant species (Schulz et al., 2015; Paškevičius et al., 2017; Schneider et al., 2018). Furthermore, there is little evidence of bacteriocin toxicity in various animal models (Behrens et al., 2017). Bacteriocins are naturally produced by environmental bacteria, it is thought that they have limited toxicity toward humans, animals or benign environmental bacterial species and some bacteriocins are already classified as generally regarded as safe for use in food preservation (Schulz et al., 2015). As we have recently shown, bacteriocins can be expressed transgenically in planta to provide resistance against P. syringae. Expression of PL1 in two model plant species provided a strong resistance phenotype in plants challenged with several unrelated PL1-sensitive P. syringae field isolates (Figure 2B; Rooney et al., 2019) with bacterial titres in PL1 transgenic lines 1.5 log-units lower than in non-transgenic controls (Rooney et al., 2019).
Non-GM-based protocols for bacteriocin-based control measures include examples where non-pathogenic but bacteriocin producing strains of bacteria have been directly applied to crops, for example, Nogall and Biokeeper (Jones et al., 1988; Chuang et al., 1999). Alternatively, treatments using bacteriocins as a direct application to crops have shown promise in laboratory conditions against olive knot disease and bacterial spot disease of tomato (Lavermicocca et al., 2002; Príncipe et al., 2018). One potential issue in utilizing bacteriocins as a direct treatment is the requirement for large scale bacteriocin production. This maybe technically difficult for multi-component bacteriocins (e.g., tailocins) but should not be a problem for LLBs and CLBs where successful production in planta has already been demonstrated (Schulz et al., 2015; Paškevičius et al., 2017; Rooney et al., 2019).
The organization of bacteriocins into functional domains enables them to be readily engineered, providing a potential route for producing further variants by domain swapping to create new chimeric bacteriocins with altered target activities and modes of killing (Lukacik et al., 2012). Similarly, for CLBs appropriate domain swapping could yield chimeric bacteriocins for which there is no immunity protein-based resistance in the targeted bacterial species (Akutsu et al., 1989). For tailocins, the exchange of tail fibers has already been shown to produce novel chimeras (Baltrus et al., 2019).
Despite the discovery and characterization of bacteriocins from phytopathogens, there is limited proof of a competitive advantage for the producing strain in vivo. Evidence suggests that soluble bacteriocins like CLBs function in the apoplastic space (endophytic fitness) whereas tailocins function in rhizosphere communities (epiphytic fitness) (Dorosky et al., 2018; Li et al., 2020). However, in vitro data suggests bacteriocins could work in concert in a conditionally redundant manner (Hockett et al., 2017).
Overall, bacteriocins represent an under-utilized resource of disease control. In the age of metagenomics, this can be easily remedied by the swift identification and characterization of new bacteriocins. This would allow bacteriocins to be rapidly deployed against current and emerging threats to important food crops.
Author Contributions
WR, RC, JM, and DW contributed to the original manuscript and the editorial changes. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The University of Glasgow has filed a patent on the transgenic expression of PL1 in plants with WR, JM, and DW listed as inventors (PCT/EP2018/057826). The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- CLB
Colicin-like bacteriocin
- GM
genetically modified
- IDR
intrinsically disordered region
- LLB
lectin-like bacteriocin
- LPS
lipopolysaccharide
- MMBL
monocot-mannose binding lectin
- PL1
putidacin L1
- TBDT
TonB-dependent transporter.
Footnotes
Funding. This work was funded by the BBSRC (Grant Refs: BB/T004207/1 and BB/L02022X/1).
References
- Akutsu A., Masaki H., Ohta T. (1989). Molecular structure and immunity specificity of colicin E6, an evolutionary intermediate between E-group colicins and cloacin DF13. J. Bacteriol. 171 6430–6436. 10.1128/jb.171.12.6430-6436.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanaskovic I., Mosbahi K., Sharp C., Housden N. G., Kaminska R., Walker D., et al. (2020). Targeted killing of Pseudomonas aeruginosa by Pyocin G occurs via the hemin transporter Hur. J. Mol. Biol. 432 3869–3880. 10.1016/j.jmb.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus D. A., Clark M., Smith C., Hockett K. L. (2019). Localized recombination drives diversification of killing spectra for phage-derived syringacins. ISME J. 13 237–249. 10.1038/s41396-018-0261-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens H. M., Lowe E. D., Gault J., Housden N. G., Kaminska R., Weber T. M., et al. (2020). Pyocin S5 import into Pseudomonas aeruginosa reveals a generic mode of bacteriocin transport. mBio 11:e3230-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens H. M., Six A., Walker D., Kleanthous C. (2017). The therapeutic potential of bacteriocins as protein antibiotics. Emerg. Top. Life Sci. 1 65–74. 10.1042/etls20160016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Buchanan S. K., Duche D., Kleanthous C., Lloubes R., Postle K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. C., Wu H. P., Chuang D. Y. (2009). Extracellular secretion of Carocin S1 in Pectobacterium carotovorum subsp. carotovorum occurs via the type III secretion system integral to the bacterial flagellum. BMC Microbiol. 9:181. 10.1186/1471-2180-9-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. C., Wu J. L., Wu H. P., Tzeng K. C., Chuang D. Y. (2011). Cloning, purification, and functional characterization of Carocin S2, a ribonuclease bacteriocin produced by Pectobacterium carotovorum. BMC Microbiol. 11:99. 10.1186/1471-2180-11-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan M. A., Riley M. A. (2007). Molecular Evolution of Bacteriocins in Gram-Negative Bacteria. In: Bacteriocins Ecology and Evolution. Berlin: Springer, 19–43. [Google Scholar]
- Chuang D. Y., Chien Y. C., Wu H. P. (2007). Cloning and expression of the Erwinia carotovora subsp. carotovora gene encoding the low-molecular-weight bacteriocin carocin S1. J. Bacteriol. 189 620–626. 10.1128/jb.01090-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang D. Y., Kyeremeh A. G., Gunji Y., Takahara Y., Ehara Y., Kikumoto T. (1999). Identification and cloning of an Erwinia carotovora subsp. carotovora bacteriocin regulator gene by insertional mutagenesis. J. Bacteriol. 181 1953–1957. 10.1128/jb.181.6.1953-1957.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. B., Augustin M. A., Robertson M. J., Manners J. M. (2018). The science of food security. NPJ Sci. Food 2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D., Ross R. P., Hill C. (2013). Bacteriocins-a viable alternative to antibiotics? Nat. Rev. Microbiol. 11 95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- Damalas C. A., Eleftherohorinos I. G. (2011). Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 8 1402–1419. 10.3390/ijerph8051402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorosky R. J., Pierson L. S., Pierson E. A. (2018). Pseudomonas chlororaphis produces multiple R-tailocin particles that broaden the killing spectrum and contribute to persistence in rhizosphere communities. Appl. Environ. Microbiol. 84:e01230-18. 10.1128/AEM.01230-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorosky R. J., Yu J. M., Pierson L. S., Pierson E. A. (2017). Pseudomonas chlororaphis produces two distinct R-tailocins that contribute to bacterial competition in biofilms and on roots. Appl. Environ. Microbiol. 83:e00706-17. 10.1128/AEM.00706-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. G., Kerr A., Van Montagu M., Schell J. (1979). Agrobacterium: genetic studies on agrocin 84 production and the biological control of crown gall. Physiol. Plant Pathol. 15 311–316. 10.1016/0048-4059(79)90082-1 [DOI] [Google Scholar]
- Ghequire M. G. K., De Canck E., Wattiau P., Van Winge I., Loris R., Coenye T., et al. (2013a). Antibacterial activity of a lectin-like Burkholderia cenocepacia protein. MicrobiologyOpen 2 566–575. 10.1002/mbo3.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G. K., De Mot R. (2015a). The tailocin tale: peeling off phage tails. Trends Microbiol. 23 587–590. 10.1016/j.tim.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Ghequire M. G. K., De Mot R. (2019). LlpB represents a second subclass of lectin-like bacteriocins. Microbial Biotechnol. 12 567–573. 10.1111/1751-7915.13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G. K., Dillen Y., Lambrichts I., Proost P., Wattiez R., De Mot R. (2015b). Different ancestries of R tailocins in rhizospheric Pseudomonas isolates. Genome Biol. Evol. 7 2810–2828. 10.1093/gbe/evv184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G. K., Garcia-Pino A., Lebbe E. K. M., Spaepen S., Loris R., de Mot R. (2013b). Structural determinants for activity and specificity of the bacterial toxin LlpA. PLoS Pathog. 9:e1003199. 10.1371/journal.ppat.1003199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G. K., Li W., Proost P., Loris R., De Mot R. (2012b). Plant lectin-like antibacterial proteins from phytopathogens Pseudomonas syringae and Xanthomonas citri. Environ. Microbiol. Rep. 4 373–380. 10.1111/j.1758-2229.2012.00331.x [DOI] [PubMed] [Google Scholar]
- Ghequire M. G. K., Loris R., De Mot R. (2012a). MMBL proteins: from lectin to bacteriocin. Biochem. Soc. Trans. 40 1553–1559. 10.1042/bst20120170 [DOI] [PubMed] [Google Scholar]
- Ghequire M. G. K., Öztürk B., De Mot R. (2018a). Lectin-like bacteriocins. Front. Microbiol. 9:2706. 10.3389/fmicb.2018.02706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G. K., Swings T., Michiels J., Buchanan S. K., De Mot R. (2018b). Hitting with a BAM: selective killing by lectin-like bacteriocins. mBio 9:e02138-17.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R., Josts I., Mosbahi K., Roszak A. W., Cogdell R. J., Bonvin A. M. J. J. (2016). Structure of the bacterial plant-ferredoxin receptor FusA. Nat. Commun. 7:133308. 10.1038/ncomms13308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R., Josts I., Zeth K., Roszak A. W., Mccaughey L. C., Cogdell R. J., et al. (2014). Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake. Mol. Microbiol. 93 234–246. 10.1111/mmi.12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R., Milner J., Walker D. (2012a). Bacteriocins active against plant pathogenic bacteria. Biochem. Soc. Trans. 40 1498–1502. 10.1042/bst20120206 [DOI] [PubMed] [Google Scholar]
- Grinter R., Milner J., Walker D. (2012b). Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS One 7:e33033. 10.1371/journal.pone.0033033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R., Roszak A. W., Cogdell R. J., Milner J. J., Walker D. (2012c). The crystal structure of the lipid II-degrading bacteriocin syringacin M suggests unexpected evolutionary relationships between colicin M-like bacteriocins. J. Biol. Chem. 287 38876–38888. 10.1074/jbc.m112.400150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness R. E., Ölschläger T. (1991). The biology of colicin M. FEMS Microbiol. Lett. 8 27–41. [DOI] [PubMed] [Google Scholar]
- Heng N. C. K., Tagg J. R. (2006). What’s in a name? Class distinction for bacteriocins. Nat. Rev. Microbiol. 4:160 10.1038/nrmicro1273-c2 [DOI] [Google Scholar]
- Heu S., Oh J., Kang Y., Ryu S., Cho S. K., Cho Y., et al. (2001). gly Gene cloning and expression and purification of glycinecin A, a bacteriocin produced by Xanthomonas campestris pv. glycines 8ra. Appl. Environ. Microbiol. 67 4105–4110. 10.1128/aem.67.9.4105-4110.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett K. L., Clark M., Scott S., Baltrus D. A. (2017). Conditionally redundant bacteriocin targeting by Pseudomonas syringae. bioRxiv [Preprint]. 10.1101/167593 [DOI] [Google Scholar]
- Hockett K. L., Renner T., Baltrus D. A. (2015). Independent co-option of a tailed bacteriophage into a killing complex in Pseudomonas. mBio 6:e00452-15. 10.1128/mBio.00452-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtsmark I., Eijsink V. G. H., Brurberg M. B. (2008). Bacteriocins from plant pathogenic bacteria. FEMS Microbiol. Lett. 280 1–7. 10.1111/j.1574-6968.2007.01010.x [DOI] [PubMed] [Google Scholar]
- Inglis R. F., Scanlan P., Buckling A. (2016). Iron availability shapes the evolution of bacteriocin resistance in Pseudomonas aeruginosa. ISME J. 10 2060–2066. 10.1038/ismej.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. A., Ryder M. H., Clare B. G., Farrand S. K., Kerr A. (1988). Construction of a Tra - deletion mutant of pAgK84 to safeguard the biological control of crown gall. MGG Mol. Gen. Genet. 212 207–214. 10.1007/bf00334686 [DOI] [Google Scholar]
- Kandel P. P., Baltrus D. A., Hockett K. L. (2020). Pseudomonas can survive tailocin killing via persistence-like and heterogenous resistance mechanisms. J. Bacteriol. 202:e00142-20. 10.1128/JB.00142-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr A. (1980). Biological control of crown gall through production of Agrocin 84. Plant Dis. 64 25–30. [Google Scholar]
- Kerr A., Htay K. (1974). Biological control of crown gall through bacteriocin production. Physiol. Plant Pathol. 4 37–40. 10.1016/0048-4059(74)90042-3 [DOI] [Google Scholar]
- Kim J. G., Byoung K. P., Sung-Uk K., Doil C., Baek H. N., Jae S. M., et al. (2006). Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan Horse antibiotic that controls crown gall. Proc. Natl. Acad. Sci. 103 8846–8851. 10.1073/pnas.0602965103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavermicocca P., Lisa Lonigro S., Valerio F., Evidente A., Visconti A. (2002). Reduction of olive knot disease by a bacteriocin from Pseudomonas syringae pv. ciccaronei. Appl. Environ. Microbiol. 68 1403–1407. 10.1128/aem.68.3.1403-1407.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Zhou L. Y., Peng Y. L., Fan J. (2020). Pseudomonas bacteriocin syringacin M released upon desiccation suppresses the growth of sensitive bacteria in plant necrotic lesions. Microbial Biotechnol. 13 134–147. 10.1111/1751-7915.13367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacik P., Barnard T. J., Keller P. W., Chaturvedi K. S., Seddiki N., Fairman J. W., et al. (2012). Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc. Natl. Acad. Sci. U.S.A. 109 9857–9862. 10.1073/pnas.1203472109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13 614–629. 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani-Hert M., Hert A. P., Tudor-Nelson S. M., Preston J. F., Minsavage G. V., Stall R. E., et al. (2020). Characterization of three novel genetic loci encoding bacteriocins associated with Xanthomonas perforans. PLoS One 15:e0233301. 10.1371/journal.pone.0233301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey L. C., Grinter R., Josts I., Roszak A. W., Waløen K. I., Cogdell R. J., et al. (2014). Lectin-like bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor. PLoS Pathog. 10:e1003898. 10.1371/journal.ppat.1003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M. (2019). Antibiotics set to flood Florida’s troubled orange orchards. Nature 567 302–303. 10.1038/d41586-019-00878-874 [DOI] [PubMed] [Google Scholar]
- Michel-Briand Y., Baysse C. (2002). The pyocins of Pseudomonas aeruginosa. Biochimie 26 329–336. [DOI] [PubMed] [Google Scholar]
- Mosbahi K., Wojnowska M., Albalat A., Walker D. (2018). Bacterial iron acquisition mediated by outer membrane translocation and cleavage of a host protein. Proc. Natl. Acad. Sci. U.S.A. 115 6840–6845. 10.1073/pnas.1800672115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. A., Tomita T., Hirota M., Kaneko J., Hayashi T., Kamio Y. (2001). DNA inversion in the tail fiber gene alters the host range specificity of carotovoricin Er, a phage-tail-like bacteriocin of phytopathogenic Erwinia carotovora subsp. carotovora Er. J. Bacteriol. 183 6274–6281. 10.1128/jb.183.21.6274-6281.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N., Gumbart J. C., Buchanan S. K. (2017). The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15 197–204. 10.1038/nrmicro.2016.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norelli J. L., Jones A. L., Aldwinckle H. S. (2003). Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis. 87 756–765. 10.1094/pdis.2003.87.7.756 [DOI] [PubMed] [Google Scholar]
- Oerke E. C., Dehne H. W. (2004). Safeguarding production—losses in major crops and the role of crop protection. Crop Protect. 23 275–285. 10.1016/j.cropro.2003.10.001 [DOI] [Google Scholar]
- Parret A. H. A., Schoofs G., Proost P., De Mot R. (2003). Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J. Bacteriol. 185 897–908. 10.1128/jb.185.3.897-908.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paškevičius Š, Starkevič U., Misiūnas A., Vitkauskienë A., Gleba Y., Ražanskienė A. (2017). Plant-expressed pyocins for control of Pseudomonas aeruginosa. PLoS One 12:e0185782. 10.1371/journal.pone.0185782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérombelon M. C. M. (2002). Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 51 1–12. 10.1046/j.0032-0862.2001.short [DOI] [Google Scholar]
- Príncipe A., Fernandez M., Torasso M., Godino A., Fischer S. (2018). Effectiveness of tailocins produced by prin in controlling the bacterial-spot disease in tomatoes caused by Xanthomonas vesicatoria. Microbiol. Res. 213 94–102. 10.1016/j.micres.2018.05.010 [DOI] [PubMed] [Google Scholar]
- Roh E., Heu S., Moon E. (2008). Genus-specific distribution and pathovar-specific variation of the glycinecin R gene homologs in Xanthomonas genomes. J. Microbiol. 46 681–686. 10.1007/s12275-008-0209-9 [DOI] [PubMed] [Google Scholar]
- Roh E., Park T. H., Lee S., Ryu S., Oh C. S., Rhee S., et al. (2010). Characterization of a new bacteriocin, carocin D, from Pectobacterium carotovorum subsp. carotovorum Pcc21. Appl. Environ. Microbiol. 76 7541–7549. 10.1128/aem.03103-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney W. M., Grinter R. W., Correia A., Parkhill J., Walker D. C., Milner J. J. (2019). Engineering bacteriocin-mediated resistance against the plant pathogen Pseudomonas syringae. Plant Biotechnol. J. 18 1296–1306. 10.1111/pbi.13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T., Hahn-Löbmann S., Stephan A., Schulz S., Giritch A., Naumann M., et al. (2018). Plant-made Salmonella bacteriocins salmocins for control of Salmonella pathovars. Sci. Rep. 8:4078. 10.1038/s41598-018-22465-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D. (2017). Phage tail–like bacteriocins. Ann. Rev. Virol. 4 453–467. 10.1146/annurev-virology-101416-041632 [DOI] [PubMed] [Google Scholar]
- Schulz S., Stephan A., Hahn S., Bortesi L., Jarczowski F., Bettmann U., et al. (2015). Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc. Natl. Acad. Sci. U.S.A. 112 E5454–E5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin G. W., Bender C. L. (1993). Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 59 1018–1024. 10.1128/aem.59.4.1018-1024.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M. E., Murphy P. J., Roberts W. P., Kerr A. (1979). Adenine N6-substituent of agrocin 84 determines its bacteriocin-like specificity [21]. Nature 280 697–699. 10.1038/280697a0 [DOI] [PubMed] [Google Scholar]
- Toth I. K., van der Wolf J. M., Saddler G., Lojkowska E., Hélias V., Pirhonen M., et al. (2011). Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 60 385–399. 10.1111/j.1365-3059.2011.02427.x [DOI] [Google Scholar]
- Turnbull L., Toyofuku M., Hynen A. L., Kurosawa M., Pessi G., Petty N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220. 10.1038/ncomms11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste J. L. (2017). The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of Kiwifruit (Pseudomonas syringae pv. actinidiae). Ann. Rev. Phytopathol. 55 377–399. 10.1146/annurev-phyto-080516-035530 [DOI] [PubMed] [Google Scholar]
- Vanneste J. L., Yu J., Cornish D. A., Tanner D. J., Windner R., Chapman J. R., et al. (2013). Identification, virulence, and distribution of two biovars of Pseudomonas syringae pv. actinidiae in New Zealand. Plant Dis. 97 708–719. 10.1094/pdis-07-12-0700-re [DOI] [PubMed] [Google Scholar]
- White P., Joshi A., Rassam P., Housden N. G., Kaminska R., Goult J. D., et al. (2017). Exploitation of an iron transporter for bacterial protein antibiotic import. Proc. Natl. Acad. Sci. U.S.A. 114 12051–12056. 10.1073/pnas.1713741114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnowska M., Walker D. (2020). FusB energises import across the outer membrane through direct interaction with its ferredoxin substrate. bioRxiv[Preprint]. 10.1101/749960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Hirota M., Niimi Y., Nguyen H. A., Takahara Y., Kamio Y., et al. (2006). Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci. Biotechnol. Biochem. 70 2236–2247. 10.1271/bbb.60177 [DOI] [PubMed] [Google Scholar]