Abstract
The purpose of this study was to characterize the complete sequence of a novel plasmid carrying tigecycline resistance gene tet(X) and carbapenemase gene blaOXA-58 from a swine Acinetobacter sp. strain SH19PTT10. Minimal inhibitory concentration (MIC) was performed using microbroth dilution method. The isolate SH19PTT10 was highly resistant (16 mg/L) to tigecycline, and also exhibited resistance to ampicillin, streptomycin, tetracycline, chloramphenicol, florfenicol, ciprofloxacin, and sulfamethoxazole/trimethoprim. Although SH19PTT10 harbored blaOXA-58, it was susceptible to cefotaxime and meropenem. The genome sequence of SH19PTT10 was determined using PacBio single-molecule real-time sequencing. Plasmid pYUSHP10-1 had a size of 174,032 bp and showed partial homology to several plasmids found in Acinetobacter isolates. It contained two repA genes, putative toxin-antitoxin systems (HipA/HipB, RelE/RelB, and BrnT/BrnA), partitioning genes (parA and parB), and heavy metal resistance-associated genes (copA/copB, nrp, and czcA/czcD) but the transfer region or proteins was not found. pYUSHP10-1 carried 16 resistance genes, mainly clustered in two mosaic multiresistance regions (MRRs). The first MRR contained sul3, qacI-aadA1-clmA1-aadA2-blaCARB-2-dfrA16 cassette, aac(3)-IId, and blaOXA-58. The blaOXA-58 gene was associated with ISAba3, as previously described. The second MRR is the tet(X) region (ISAcsp12-aph(3')-Ia-IS26-ΔxerD-tet(X)-res-ISCR2-sul2) related to the corresponding region in other tet(X)-bearing plasmids. The pdif sites, as well as mobile elements, play an important role in mobilization of DNA modules and plasmid evolution. Coexistence of numerous resistance genes on a single plasmid may contribute to the dissemination of these genes under pressure posed by different agents, which may explain the presence of clinically crucial resistance genes tet(X) and blaOXA-58 in livestock. Thus, rational drug use and continued surveillance of tet(X) and blaOXA-58 in livestock are warranted.
Keywords: Acinetobacter, blaOXA-58, plasmid, tet(X), tigecycline resistance
Introduction
The genus Acinetobacter currently includes more than 60 species with valid species names1, and most of them are important nosocomial pathogens. Carbapenems are clinically crucial antimicrobial agents for treating multidrug-resistant Gram-negative pathogens, including Acinetobacter isolates (Asif et al., 2018; Rodríguez-Baño et al., 2018). The rapid increase in the prevalence of carbapenem-resistant Acinetobacter is mainly attributed to the acquisition of carbapenem-hydrolyzing class D β-lactamases (e.g., OXA-23, -40, -51, -58, and -143; Evans and Ameyes, 2014). OXA-58 has been detected in Acinetobacter isolates from patients, animals, and the environment from distinct geographical areas, particularly in clinics (Poirel et al., 2005; Fu et al., 2014; Feng et al., 2016; Klotz et al., 2017; Narciso et al., 2017; Chen et al., 2019; Matos et al., 2019; Suzuki et al., 2019). Though OXA-58 shows weak carbapenem-hydrolyzing activity, the insertion of other insertion sequence (IS) elements into the upstream ISAba3 of blaOXA-58 may provide an alternative promoter that enhances its transcription and the level of carbapenem resistance (Poirel et al., 2005; Narciso et al., 2017; Chen et al., 2019; Matos et al., 2019).
Tigecycline, belonging to the novel glycylcycline class, has been a last-resort antibiotic to treat serious infections caused by extensively drug-resistant Gram-negative bacteria, including Acinetobacter (Noskin, 2005; Asif et al., 2018). However, novel plasmid-mediated high-level tigecycline resistance genes tet(X) [former name tet(X3)~tet(X5)]2 have been identified in Acinetobacter isolates from animals and humans in China in 2019 (He et al., 2019; Wang et al., 2019). The emergence and dissemination of tet(X) will impair the efficacy of tigecycline in clinical treatment, thus would pose a significant threat to public health. The co-location of tet(X) and carbapenem resistance gene blaNDM-1 was previously described in Acinetobacter isolates from animals (duck, goose, and cow) and the environment (soil and sewage; Cui et al., 2020; He et al., 2020). In this study, we aimed to determine and analyze the complete sequence of a single plasmid bearing tet(X) [formerly designated as tet(X3), GenBank accession no. MK134375] and blaOXA-58 obtained from a swine Acinetobacter sp. strain in Shanghai, China, providing insights into the genetic structures of the plasmid and these genes.
Materials and Methods
Bacterial Strain and tet(X) Detection
In September 2019, one strain SH19PTT10 was isolated from the feces sample of a pig by Tryptic Soy Agar plate containing tigecycline (2 mg/L) from a pig farm located in Shanghai, China and was identified using 16S ribosomal RNA (rRNA) gene sequencing (Kim et al., 2010). The presence of tet(X) [former name tet(X3), tet(X4), and tet(X5)] was detected by PCR and sequencing (He et al., 2019; Wang et al., 2019).
Antimicrobial Susceptibility Testing
The isolate SH19PTT10 was tested for minimal inhibitory concentrations (MICs) of ampicillin, cefotaxime, meropenem, amikacin, streptomycin, tetracycline, minocycline, tigecycline, chloramphenicol, florfenicol, ciprofloxacin, colistin, and sulfamethoxazole/trimethoprim using microbroth dilution method recommended by the guidelines of the Clinical and Laboratory Standards Institute [CLSI, Wayne, PA, United States; Clinical and Laboratory Standards Institute (CLSI), 2012]. The results were interpreted according to CLSI M100, 28th edition [Clinical and Laboratory Standards Institute (CLSI), 2018]. Tigecycline (≧1 mg/L), streptomycin (≧32 mg/L), and florfenicol (≧32 mg/L) were interpreted according to the clinical breakpoint or epidemiological cutoff values for Escherichia coli set by EUCAST3. The E. coli strain ATCC 25922 was used for quality control.
Conjugation/Transformation Experiments
Conjugation experiments were performed using streptomycin-resistant E. coli C600 as the recipient strain, as previously described (Chen et al., 2007). Transconjugants were selected using 2 mg/L tigecycline and 3,000 mg/L streptomycin. Transformation was carried out by heat-shock and electroporation using E. coli DH5α and Acinetobacter baumannii ATCC 19606. Transformants were selected by 2 mg/L tigecycline.
Whole Genome Sequencing and Analysis
The whole genome of SH19PTT10 was extracted and sequenced using PacBio single-molecule real-time sequencing (RSII platform, Pacific Biosciences, Menlo Park, CA, United States). Raw sequence data were introduced into the non-hybrid hierarchical genome assembly process (HGAP version 4). The 16S rRNA gene sequences of SH19PTT10 and other representatives across the genus Acinetobacter were aligned using ClustalW, and a phylogenetic tree was constructed by the neighbor joining algorithm using MEGA 7.0 (Kumar et al., 2008). The plasmid sequence was analyzed and annotated using the RAST server (Aziz et al., 2008), ResFinder 3.2 (Zankari et al., 2012), ISfinder (Siguier et al., 2006), PlasmidFinder (Carattoli et al., 2014), BLAST4, and Gene Construction Kit 4.5 (Textco BioSoftware, Inc., Raleigh, NC, United States). The replicase genes (rep) of plasmids from SH19PTT10 were assigned to a group according to the typing scheme for plasmids in A. baumannii (Bertini et al., 2010). Plasmid pYUSHP10-1 was compared with other plasmids using BLASTn and BRIG (Alikhan et al., 2011).
Nucleotide Sequence Accession Number
The complete sequence of pYUSHP10-1 was deposited in GenBank under the accession number MT107270.
Results and Discussion
tet(X)-Encoding Acinetobacter sp. Strain SH19PTT10
The strain SH19PTT10 was positive for tet(X) [formerly designated as tet(X3), MK134375] and was highly resistant (16 mg/L) to tigecycline. SH19PTT10 also showed resistance to ampicillin, streptomycin, tetracycline, chloramphenicol, florfenicol, ciprofloxacin, and sulfamethoxazole/trimethoprim but was susceptible to cefotaxime, meropenem, amikacin, minocycline, and colistin (Table 1). The 16S rRNA gene sequencing showed SH19PTT10 belonging to Acinetobacter but not to any described species, thus whole genome sequencing (WGS) was further performed.
Table 1.
Results of antimicrobial susceptibility test.
| Antimicrobial agents | MIC (mg/L) | Interpretation |
|---|---|---|
| ampicillin | >128 | R |
| cefotaxime | 4 | S |
| meropenem | 0.5 | S |
| amikacin | 1 | S |
| streptomycin | >128 | R |
| tetracycline | >128 | R |
| minocycline | ≤2 | S |
| tigecycline | 16 | R |
| chloramphenicol | 128 | R |
| florfenicol | 128 | R |
| ciprofloxacin | >8 | R |
| colistin | 0.25 | S |
| sulfamethoxazole/trimethoprim | >32 | R |
SH19PTT10 consisted of a 3,440,498-bp chromosome and five plasmids designated as pYUSHP10-1 to pYUSHP10-5, ranging from 5.7 to 174 kb (Supplementary Table S1). Among them, pYUSHP10-1, the largest plasmid found in SH19PTT10, carried numerous resistance genes, particularly tigecycline resistance gene tet(X) and carbapenemase gene blaOXA-58; pYUSHP10-3 also carried resistance gene tet(39) (Supplementary Table S1). The tet(X)-carrying plasmid pYUSHP10-1 was failed to transfer to E. coli C600/DH5α by conjugation/transformation or A. baumannii ATCC 19606 by transformation. The complete sequence of the 16S rRNA gene exhibited 98.76% identity (1515/1534) with A. cumulans strain WCHAc060092 (CP035934) and 98.37% identity (1509/1534) with Acinetobacter haemolyticus strains AN54 (CP041224). The neighbor-joining tree based on partial 16S rRNA gene sequences within Acinetobacter suggested that the isolate SH19PTT10 may take a distinct position within the genus; however, the branch support for SH19PTT10 was low (bootstrap value 36%; Supplementary Figure S1).
The Backbone of tet(X)‐ and blaOXA-58-Bearing Plasmid pYUSHP10-1
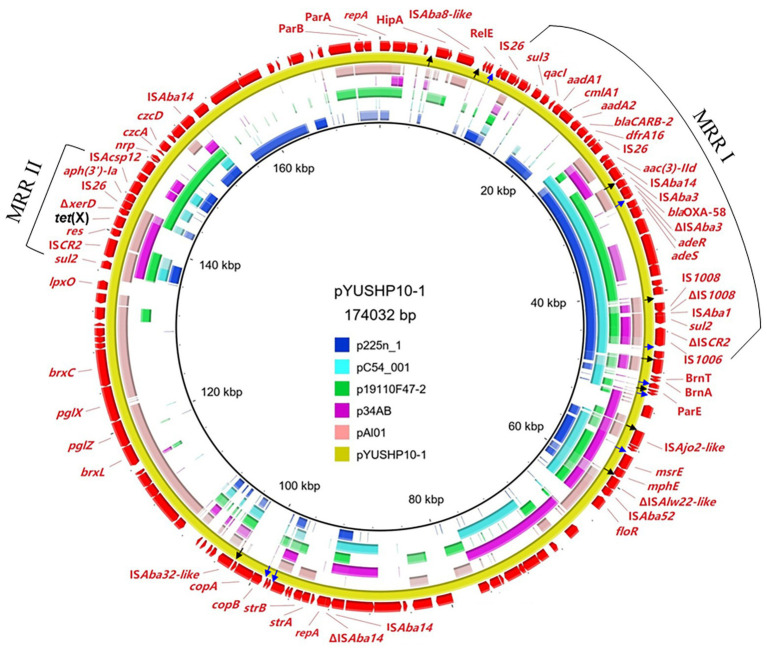
The plasmid pYUSHP10-1 had a size of 174,032 bp with a 41.32% GC content; it could not be assigned to any previously known incompatibility group. pYUSHP10-1 showed partial homology to several plasmids obtained from Acinetobacter isolates, such as OXA-58-encoding plasmids p225n_1 (KT852971, A. baumannii, patient, Vietnam), pC54_001 (CP042365, Acinetobacter pittii, patient, Australia), and p19110F47-2 (CP046044, Acinetobacter towneri, pig, China), as well as tet(X)-carrying plasmids p34AB (MK134375, A. baumannii, swine, China) and pAl01 (CP044019, Acinetobacter indicus, feces, China; 29–41% coverage, 97.3–99.9% nucleotide identity; Figure 1).
Figure 1.
Sequence comparison of plasmids pYUSHP10-1 with plasmids p225n_1 (GenBank accession number KT852971), pC54_001 (CP042365), p19110F47-2 (CP046044), p34AB (MK134375), and pAl01 (CP044019) using BRIG. The reference sequence pYUSHP10-1 is indicated in red in the outer circle. The arrows in black color indicate pdif sites (XerD-XerC) and arrows in blue color indicate pdif sites (XerD-XerC).
pYUSHP10-1 contained two repA genes, both of them belonged to the Rep_3 type family (pfam01051). The first repA (positions 1–1068) shared highly similarity (>99.9%) with those of tigecycline-resistant plasmids p18TQ-X3 (CP045132, 1067/1068) and pAB17H194-1 (CP040912, 1067/1068) obtained from Acinetobacter strains in China. The second repA (positions 92451–93302) was identical to those of plasmids pAHTJR1 (CP038010, A. haemolyticus, patient) and pBXX1-9 (CP010351, Acinetobacter johnsonii, hospital sewage; Feng et al., 2016) in China. In addition, pYUSHP10-1 harbored multiple putative toxin-antitoxin systems, such as HipA/HipB, RelE/RelB, and BrnT/BrnA, and the partitioning genes (parA and parB), which may ensure plasmid maintenance at cell division (Zielenkiewicz and Ceglowski, 2001). However, the transfer region or proteins for plasmid mobilization was not found in pYUSHP10-1. Putative part of the BREX system (brxC-pglX-pglZ-brxL) was identified in plasmid pYUSHP10-1 (Figure 1), which has been reported as a novel phage defense system which confers resistance to a broad range of phages (Goldfarb et al., 2015). Similar structure was also found in plasmids from Acinetobacter, such as pOXA58_010062 (CP033131, Acinetobacter wuhouensis, sewage, China) with 91% coverage and 96.21% identity and pAl01 shared 93% coverage and 93.93% identity. Additionally, heavy metal resistance-associated genes were also detected in pYUSHP10-1, such as copA/copB (copper resistance), nrp (nickel resistance), and czcA/czcD (cobalt-zinc-cadmium resistance; Figure 1).
The pdif Sites of pYUSHP10-1
Recently, numerous plasmids from various Acinetobacter species have been identified to contain 28-bp pdif site, consisting of 11-bp inversely-oriented binding sites for the XerC and XerD recombinases separated by a spacer of 6 bp (Supplementary Table S2; D’Andrea et al., 2009; Merino et al., 2010; Blackwell and Hall, 2017; Cameranesi et al., 2018). We also found 17 pdif sites on pYUSHP10-1 (Figure 1; Supplementary Tables S2 and S3). The modules flanked by inversely-oriented pdif sites either with the XerC sites internal (D/C and C/D) or the XerD sites internal (C/D and D/C) are identified as dif module and may be able to mobilize mediated by pdif sites using XerC-XerD recombination system (D’Andrea et al., 2009; Merino et al., 2010; Blackwell and Hall, 2017; Cameranesi et al., 2018). For example, one module in pYUSHP10-1 consisting of ISAjo2-like and putative toxin-antitoxin system BrnT/BrnA, was surrounded by inversely-oriented pdif sites (XerD/C and XerC/D; Supplementary Figure S2). This dif module was adjacent to a hypothetical protein and a further dif module carrying another putative toxin-antitoxin system encoding ParE toxin and helix-turn-helix family protein (Supplementary Figure S2). Interestingly, the pdif sites flanking two dif modules were also surrounding the hypothetical protein in the opposite orientation, making it being a putative dif module that may be mobile (Supplementary Figure S2). Different dif module combinations were found in some Acinetobacter plasmids such as pABIR (EU294228), pJ9-3 (CP041590), and pM131-2 (JX101647), indicating that the presence of the pdif sites facilitated the mobilization of discrete DNA segment through multiple events (Supplementary Figure S2). Additional dif modules identified in pYUSHP10-1 mostly include toxin-antitoxin systems, and copA/copB were also found in dif module (Figure 1).
pYUSHP10-1 Carrying Resistance Genes and dif Modules
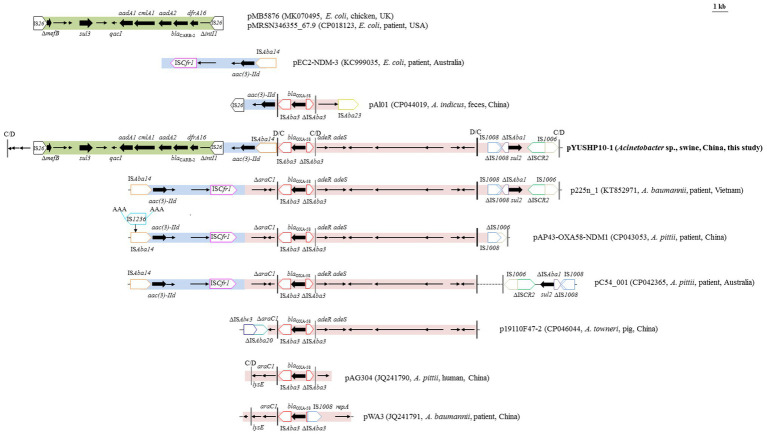
pYUSHP10-1 contained 16 resistance genes, which were mainly clustered in two mosaic multiresistance regions (MRRs; Figure 1). The first MRR carried an approximately 32.2-kb segment consisting of three parts. The first of these (~11.6 kb) was bounded at both ends by IS26 and comprised a truncated mefB (encoding macrolide efflux protein), sul3 (sulfonamide resistance), and an incomplete class 1 integron with ΔintI1 and the qacI-aadA1-clmA1-aadA2-blaCARB-2-dfrA16 cassette array (Figure 2). This fragment showed 99.9% identity to those of plasmids from E. coli such as pMB5876 (MK070495) and pMRSN346355_67.9 (CP018123), suggesting that pYUSHP10-1 may capture this segment from E. coli plasmids by IS26-mediated transposition or homologous recombination (Figure 2).
Figure 2.
Genetic organization of the multiresistance region I of plasmid pYUSHP10-1, and structural comparison with other plasmids. The extents and directions of antibiotic resistance (thick arrows) and other genes are indicated. Δ indicates a truncated gene or mobile element. Insertion sequences (ISs) are shown as boxes labeled with their name. Labeled vertical arrows with IS box indicate the insertion site of IS element. Direct repeats are indicated by arrows and sequences. Vertical bars in black color indicate pdif sites (XerD-XerC) and vertical bars in gray color indicate pdif sites (XerD-XerC).
The second part (3,296 bp) contained an open reading frame (ORF) encoding AAA family ATPase, aminoglycoside resistance gene aac(3)-IId, and mobile element ISAba14 (Figure 2). It was highly similar (>99%) to the corresponding regions of multiple plasmids found in Acinetobacter and Enterobacteriaceae isolates, such as Acinetobacter plasmids p225n_1 and pC54_001 and E. coli plasmid pEC2-NDM-3 (KC999035). Interestingly, the pdif site adjacent to ISAba14 and the pdif site upstream of IS26 were in inverse orientation, suggesting that the acquisition of ~16.5 kb segment including two hypothetical proteins and the first and second parts of MRR I in pYUSHP10-1 was possibly mediated by the pdif sites via site-specific recombination (Figure 2).
The last part of MRR I (17,301 bp) was highly similar to those of Acinetobacter plasmids p225n_1 (one single nucleotide polymorphism), p19110F47-2, pC54_001, and pAP43-OXA58-NDM1 with additional deletions (Figure 2). The primary component of this fragment is the blaOXA-58 region. As previously described (Fu et al., 2014; Feng et al., 2016; Klotz et al., 2017), blaOXA-58 was flanked by two copies of ISAba3 with opposite orientation, although the upstream ISAba3 was incomplete in pYUSHP10-1 (Figure 2). OXA-58 shows weak activity against the carbapenems and is unable to hydrolyze some cephalosporin such as ceftazidime and cefotaxime (Poirel et al., 2005). The insertion of other IS elements, such as ISAba825, ISOur1, IS1008, and IS1006 into the upstream ISAba3 may provide a promoter to enhance the expression of OXA-58 and leading to the carbapenem resistance (Fu et al., 2014; Narciso et al., 2017; Chen et al., 2019; Matos et al., 2019). In this study, we did not observe insertion of IS element into ISAba3, suggesting the lacking of a putative promoter for blaOXA-58 overexpression, thus the strain SH19PTT10 showed susceptibility to cefotaxime (4 mg/L) and meropenem (0.5 mg/L).
The blaOXA-58 region has been frequently bracketed by two Re27 sequences, recognized as pdif sites later (D’Andrea et al., 2009), which have been shown to mediate the mobilization of blaOXA-58 (Poirel and Nordmann, 2006). In this study, two pdif sites in inverse orientation (XerD-XerC and XerC-XerD) were identified flanking the 2,257-bp segment containing ISAba3-blaOXA-58-ΔISAba3 arrangement in pYUSHP10-1, as observed in numerous Acinetobacter plasmids (Figure 2). The downstream ISAba3 was commonly followed by the putative transcriptional regulator gene araC1 and threonine efflux protein gene lysE in Acinetobacter isolates (Figure 2; Fu et al., 2014; Feng et al., 2016; Matos et al., 2019). An additional pdif site was found adjacent to lysE (Figure 2), which may account for the mobilization of this segment together with the blaOXA-58 dif module. However, the conserved structure araC1-lysE was not found in pYUSHP10-1. A truncated araC1 was identified in p225n_1, pAP43-OXA58-NDM1, and pC54_001, possibly due to homologous recombination between araC1 and the downstream putative gene encoding site-specific integrase, which shared a sequence of 6 bp (TAAGTT) that might be the site of recombination (Figure 2). The same ΔaraC1 was also observed in p19110F47-2, which truncated by an incomplete ISAba20 (Figure 2).
The genetic contexts downstream of ISAba3-blaOXA-58-ΔISAba3 is diverse in Acinetobacter (Figure 2; Fu et al., 2014; Feng et al., 2016; Klotz et al., 2017; Matos et al., 2019). In pYUSHP10-1, several putative ORFs encoding two-component regulatory system AdeR/AdeS, RND family efflux transporters, LysE family protein, and AraC family transcriptional regulator were identified (Figure 2). In addition, a ~5.1 kb segment was further present downstream consisting of sulfonamide resistance gene sul2 and several intact or truncated insertion sequences, including IS1008, ISAba1, ISCR2, and IS1006 (Figure 2). Two pdif sites in inverse orientation (XerD-XerC and XerC-XerD) were found within this segment to create a dif module (Figure 2). The first one disclosed an additional adeR/adeS dif module with the pdif site downstream of ΔISAba3, which may readily explain its co-transfer with the blaOXA-58 dif module in several plasmids, e.g., p19119F47-2 (Figure 2). The structure ISAba1-sul2-ΔISCR2-IS1006 was also observed in many plasmids found in Acinetobacter and Enterobacteriaceae isolates, such as pMCR_WCHEC050613 (CP019214, E. coli, sewage, China), pOXA58_005069 (CP026086, A. pittii, patient, China), and 1205p1 (CP012141, Shigella flexneri, China), although ISAba1 was truncated by IS1008 at 3' end in pYUSHP10-1.
The second MRR module (~9.2 kb) is the tet(X) region. In pYUSHP10-1, tet(X) had a single nucleotide substitution at position 21 (A21) compared with that of the first described tet(X)-bearing plasmid p34AB from swine A. baumanii (He et al., 2019) but it did not result in any amino acid change. In p34AB, tet(X) was located within the arrangement ISCR2-xerD-tet(X)-res-ISCR2 with three copies (He et al., 2019). pYUSHP10-1 possessed a structure [ISAcsp12-aph(3')-Ia-IS26-ΔxerD-tet(X)-res-ISCR2-sul2] closely related to those of p34AB, pHH1107 (FJ012881, soil), and pAl01 (Figure 3). A new IS element was identified adjacent to the aminoglycoside resistance gene aph(3')-Ia and showed 87% identity to ISAba21. It was 1,274 bp and had 36-bp imperfect IR, and it was designated as ISAcsp12 in ISFinder database5. The fragment (res-ISCR2-sul2) located downstream of tet(X) was similar to the corresponding region in pHH1107, differed by only one nucleotide change within ISCR2. In pYUSHP10-1, xerD which encoded a recombinase was truncated by IS26 element at the 5' end; it may explain the absence of the upstream ISCR2 observed in p34AB. A similar segment was found in pAl01 except that aph(3')-Ia was present upstream of IS26. In pHH1107, the upstream ISCR2 was truncated by a complete transposon Tn5393 at the 5′ end, which contained streptomycin resistance genes strA/strB and was interrupted by IS26 at tnpA of Tn5393, generated 8-bp direct repeats (CTCGCGAT; Figure 3).
Figure 3.
Genetic organization of the multiresistance region II of plasmid pYUSHP10-1, and structural comparison with other plasmids. Arrows indicate the position of the genes and the direction. Regions with >99% homology are shaded in gray. Δ indicates a truncated gene or mobile element. ISs are shown as boxes labeled with their name. Labeled vertical arrows with IS box indicate the insertion site of IS element. Direct repeats are indicated by arrows and sequences. Tall bars represent the 81 bp inverted repeats (IRs) of Tn5393.
The horizontal transfer of tet(X) is likely to be associated with ISCR2 (He et al., 2019). The generation of a circular molecule [ISCR2-xerD-tet(X)-res] by recombination between the two copies of ISCR2 in the same orientation could lead to the insertion of the tet(X) module (He et al., 2019). Although two copies of ISCR2 were only intact in p34AB, the formation of different but related tet(X) structures may have resulted from additional molecular events mediated by mobile elements (e.g., IS26 and Tn5393) via transposition or homologous recombination and have the potential to evolve diverse tet(X) genetic contexts.
pYUSHP10-1 also carried other resistance genes, including macrolide resistance genes mph(E)/msr(E), florfenicol resistance gene floR, and streptomycin resistance genes strA/strB (Figure 1). As previously described (Blackwell and Hall, 2017), a 2,950-bp segment including the macrolide resistance genes mph(E)/msr(E) was surrounded by two pdif sites in inverse orientation (Figure 1), further suggesting that the pdif sites may mediate mph(E)/msr(E) mobilization in Acinetobacter plasmids. Furthermore, many intact or truncated insertion sequences were present in the backbone of pYUSHP10-1, such as ISAba14 (Figure 1). These IS elements may enable this plasmid to capture more genes and evolve through IS-mediated recombination events.
Conclusion
We report the isolation and genetic characterization of an Acinetobacter sp. strain exhibited multiresistance phenotype, due to the acquisition of a novel plasmid carrying 16 resistance genes, including tigecycline resistance gene tet(X) and carbepenemase gene blaOXA-58. The Acinetobacter sp. may serve as an important reservoir of antimicrobial resistance genes. Coexistence of numerous resistance genes on a single plasmid may facilitate its dissemination and persistence under different selection pressure, which may explain the presence of clinically crucial antibiotic resistance genes tet(X) and blaOXA-58 in livestock. Additionally, at least two different mechanisms, site-specific recombination via the pdif sites and transposition of mobile elements (e.g., ISCR2 and IS26) could account for the acquisition of DNA modules containing resistance structures and/or other genes in pYUSHP10-1. It highlights the ability of resistance structures to be captured by multiple events and their capacity to evolve during horizontal transfer.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Z-MP, XJ, and JW conceived the study. YW, HW, P-CS, JW, Y-QT, and FS carried out the experiments. JW and Z-YW analyzed the data. JW wrote the manuscript. Z-MP and XJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by National Natural Science Foundation of China (grant number 31902319), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and grant supported by Ministry of Agriculture and Rural Affairs, China (no. 14162130110239004).
2According to the standards of the nomenclature center (http://faculty.washington.edu/marilynr/), all tet(X) genes [former name tet(X1)-tet(X5)] at present can only be designated as tet(X).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmicb.2020.578020/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Alvi I. A., Rehman S. U. (2018). Insight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect. Drug Resist. 11, 1249–1260. 10.2147/IDR.S166750, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini A., Poirel L., Mugnier P. D., Villa L., Nordmann P., Carattoli A. (2010). Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4168–4177. 10.1128/AAC.00542-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell G. A., Hall R. M. (2017). The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely-oriented pdif (XerC-XerD) sites. Antimicrob. Agents Chemother. 61:e00780-17. 10.1128/AAC.00780-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameranesi M. M., Morán-Barrio J., Limansky A. S., Repizo G. D., Viale A. M. (2018). Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a blaOXA-58‐ and TnaphA6-resistance module in Acinetobacter baumannii. Front. Microbiol. 9:66. 10.3389/fmicb.2018.00066, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garcìa-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. 10.1128/AAC.02412-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chen Z. L., Liu J. H., Zeng Z. L., Ma J. Y., Jiang H. X. (2007). Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. 10.1093/jac/dkm065, PMID: [DOI] [PubMed] [Google Scholar]
- Chen F. J., Huang W. C., Liao Y. C., Wang H. Y., Lai J. F., Kuo S. C., et al. (2019). Molecular epidemiology of emerging carbapenem resistance in Acinetobacter nosocomialis and Acinetobacter pittii in Taiwan, 2010 to 2014. Antimicrob. Agents Chemother. 63:e02007-18. 10.1128/AAC.02007-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2012). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard–ninth edition. CLSI document M07-A9 Wayne, PA, USA: CLSI. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2018). Performance standards for antimicrobial susceptibility testing. 28th Edn. CLSI supplement M100 Wayne, PA, USA: CLSI. [Google Scholar]
- Cui C. Y., Chen C., Liu B. T., He Q., Wu X. T., Sun R. Y., et al. (2020). Co-occurrence of plasmid-mediated tigecycline and carbapenem resistance in Acinetobacter spp. from waterfowls and their neighboring environment. Antimicrob. Agents Chemother. 64:e02502-19. 10.1128/AAC.02502-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea M. M., Giani T., D’Arezzo S., Capone A., Petrosillo N., Visca P., et al. (2009). Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 3528–3533. 10.1128/AAC.00178-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. A., Ameyes S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. 10.1128/CMR.00117-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yang P., Wang X., Zong Z. (2016). Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J. Antimicrob. Chemother. 71, 71–75. 10.1093/jac/dkv324, PMID: [DOI] [PubMed] [Google Scholar]
- Fu Y., Jiang J., Zhou H., Jiang Y., Fu Y., Yu Y., et al. (2014). Characterization of a novel plasmid type and various genetic contexts of blaOXA-58 in Acinetobacter spp. from multiple cities in China. PLoS One 9:e84680. 10.1371/journal.pone.0084680, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T., Sberro H., Weinstock E., Cohen O., Doron S., Charpak-Amikam Y., et al. (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183. 10.15252/embj.201489455, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T., Li R., Wei R., Liu D., Bai L., Zhang L., et al. (2020). Characterization of Acinetobacter indicus co-harbouring tet(X3) and blaNDM-1 of dairy cow origin. J. Antimicrob. Chemother. 10.1093/jac/dkaa182, PMID: [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- He T., Wang R., Liu D., Walsh T. R., Zhang R., Lv Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. 10.1038/s41564-019-0445-2, PMID: [DOI] [PubMed] [Google Scholar]
- Kim T. W., Kim Y. H., Kim S. E., Lee J. H., Park C. S., Kim H. Y. (2010). Identification and distribution of Bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214. 10.4014/jmb.1002.02008, PMID: [DOI] [PubMed] [Google Scholar]
- Klotz P., Jacobmeyer L., Leidner U., Stamm I., Semmler T., Ewers C. (2017). Acinetobacter pittii from companion animals coharboring blaOXA-58, the tet(39) region, and other resistance genes on a single plasmid. Antimicrob. Agents Chemother. 62:e01993-17. 10.1128/AAC.01993-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306. 10.1093/bib/bbn017, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos A. P., Cayô R., Almeida L. G. P., Streling A. P., Nodari C. S., Martins W. M. B. S., et al. (2019). Genetic characterization of plasmid-borne blaOXA-58 in distinct Acinetobacter species. mSphere 4:e00376-19. 10.1128/mSphere.00376-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino M., Acosta J., Poza M., Sanz F., Beceiro A., Chaves F., et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54, 2724–2727. 10.1128/AAC.01674-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso A. C., Martins W. M. B. S., Cayô R., Pereira de Matos A., Santos S. V., Ramos P. L., et al. (2017). Detection of OXA-58-producing Acinetobacter seifertii recovered from a black-necked swan at a zoo lake. Antimicrob. Agents Chemother. 61:e01360-17. 10.1128/AAC.01360-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskin G. A. (2005). Tigecycline: a new glycylcycline for treatment of serious infections. Clin. Infect. Dis. 41(Suppl. 5), S303–S314. 10.1086/431672, PMID: [DOI] [PubMed] [Google Scholar]
- Poirel L., Marqué S., Héritier C., Segonds C., Chabanon G., Nordmann P. (2005). OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 202–208. 10.1128/AAC.49.1.202-208.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Nordmann P. (2006). Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 1442–1448. 10.1128/AAC.50.4.1442-1448.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Baño J., Gutiérrez-Gutiérrez B., Machuca I., Pascual A. (2018). Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 31:e00079-17. 10.1128/CMR.00079-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. 10.1093/nar/gkj014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Endo S., Nakano R., Nakano A., Saito K., Kakuta R., et al. (2019). Emergence of IMP-34‐ and OXA-58-producing carbapenem-resistant Acinetobacter colistiniresistens. Antimicrob. Agents Chemother. 63:e02633-18. 10.1128/AAC.02633-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu D., Lv Y., Cui L., Li Y., Li T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline and omadacycline in clinical Acinetobacter baumannii. Antimicrob. Agents Chemother. 64:e01326-19. 10.1128/AAC.01326-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. 10.1093/jac/dks261, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenkiewicz U., Ceglowski P. (2001). Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48, 1003–1023. 10.18388/abp.2001_3863, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.