Summary
Broadly neutralizing antibodies (bnAbs) recognize conserved features of rapidly mutating pathogens and confer universal protection, but they emerge rarely in natural infection. Increasing evidence indicates that seemingly passive antibodies may interfere with natural selection of B cells. Yet, how such interference modulates polyclonal responses is unknown. Here we provide a framework for understanding the role of antibody interference—mediated by multi-epitope antigens—in shaping B cell clonal makeup and the fate of bnAb lineages. We find that, under heterogeneous interference, clones with different intrinsic fitness can collectively persist. Furthermore, antagonism among fit clones (specific for variable epitopes) promotes expansion of unfit clones (targeting conserved epitopes), at the cost of repertoire potency. This trade-off, however, can be alleviated by synergy toward the unfit. Our results provide a physical basis for antigen-mediated clonal interactions, stress system-level impacts of molecular synergy and antagonism, and offer principles to amplify naturally rare clones.
Subject Areas: Biological Sciences, Immunology, Mathematical Biosciences
Graphical Abstract

Highlights
-
•
Multi-epitope antigens mediate antibody interference that couples B cell lineages
-
•
Trade-off exists between repertoire potency and persistence of broad lineages
-
•
Antigen-mediated synergy toward intrinsically unfit clones alleviates the trade-off
-
•
Amplifying rare clones by leveraging molecular interference structure
Biological Sciences; Immunology; Mathematical Biosciences
Introduction
Rapidly evolving viruses engage and enter host cells using highly variable surface proteins. The immune system counteracts with a diverse repertoire of antibodies (Abs) that bind to specific groups of residues (i.e., epitopes) on the viral envelope glycoprotein and interrupt viral life cycle. Most Abs can at best inhibit a narrow range of antigen (Ag) variants, whereas broadly neutralizing antibodies (bnAbs) can recognize epitope features common to almost all viable mutants, thus holding the promise for robust protection against highly mutable pathogens (Burton et al., 2012).
However, in the human immune system, bnAbs against rapidly adapting pathogens (e.g., HIV and influenza) only emerge rarely and last transiently (Liao et al., 2013; Corti et al., 2017). One main reason is that conserved epitopes—invariant targets of bnAbs—are often shielded by variable structures and hard to access; in contrast, highly mutable epitopes capable of evading Ab recognition are well exposed and greater in number (Zhou et al., 2010; West et al., 2014; Fera et al., 2014; Raymond et al., 2018; Angeletti et al., 2017). Consequently, bnAb-making B cell lineages that target most conserved yet poorly accessible epitopes would be outcompeted during affinity maturation Eisen and Siskind (1964); Victora and Nussenzweig (2012), a Darwinian process selecting for high-affinity clones, thereby contributing scarcely to polyclonal responses in an infected individual.
The reasoning above presumes that Abs targeting distinct epitopes are independent, so that native epitope accessibility determines competitive advantage of B cells expressing corresponding Abs as Ag receptors. If this were true, binding affinity of individual Abs should be predictive of the clonal makeup of a response, given that receptor affinity dictates cell proliferation (through the amount of Ag captured) (Gitlin et al., 2014). And yet, polyclonal responses cannot be fully explained by monoclonal binding, at least in part due to two factors. First, Abs present in a mixture do not behave additively (Zwick et al., 2001; Ndifon et al., 2009; Koefoed et al., 2011; Luo and Perelson, 2015; Derking et al., 2015; Howell et al., 2017; Caskey et al., 2019; Einav and Bloom, 2020; Greiff et al., 2012; Bachmann et al., 1997). Second, seemingly passive Abs can influence ongoing selection of B cells during affinity maturation (Ng et al., 2010; Zhang et al., 2013; Schoofs et al., 2016; Vono et al., 2019; Garg et al., 2019). Whether these observations are related and in what ways Ab interactions alter B cell clonal selection are important open questions.
We propose a mechanism—Ag-mediated Ab interference—by which interactions between Abs and B cell receptors, specific for the same or distinct epitopes on the same Ag, impact B cell clonal dynamics and the chance of bnAb expansion (Figure 1A). Specifically, existing Abs, either from prior and current affinity maturation or from passive immunization, may bind to a vacant epitope on a viral Ag and alter the accessibility of another epitope to B cell receptors, via steric blockade or allosteric coupling (Derking et al., 2015; Julien et al., 2013; Wang et al., 2016; Ozorowski et al., 2017; Vono et al., 2019). Steric blockade in general reduces epitope exposure, whereas allosteric coupling can make hidden epitopes more accessible through binding at a distal site.
Figure 1.
Antigen-Mediated Antibody Interference can Strongly Influence B Cell Clonal Composition
(A) A sketch of the mechanism. A viral particle (pink circle) presents copies of envelope spikes on its surface, each comprising multiple epitopes. Binding of an antibody j (blue Y-shaped molecule) to its target epitope (e.g., in the head domain) can interfere with binding of B cell receptor i (green Y-shaped molecule) targeting a different epitope (e.g., in the stem region) on the same antigen, via steric inhibition or allosteric coupling. Similarly, antibodies of type i may interfere with B cells of type j. This interference, mediated by viral antigen, can be synergistic, i.e., enhancing epitope accessibility, in one direction (lower blowup, cyan acute arrow, indicating αij < 0), and antagonistic, i.e., reducing epitope exposure, in the reverse direction (upper blowup, orange blunt arrow, indicating αji > 0). Such mutual influence via the antibody product alters the effective potency of B cell clones, thus affecting their rate of expansion (looping arrows).
(B–E) A specific example. (B) Interference matrix of bnAbs (PGT145, PGT121, PGT136, CH103, 35022, 3BC315) targeting six major epitopes (V1/V2 glycan, V3 glycan, OD-glycan, CD4bs, gp120-gp41, gp41) on the HIV envelope trimer (BG505 SOSIP.664), calculated from cross-competition data in Derking et al. (2015). (C) Predicted clonal dynamics with two experimental inputs: interference matrix in (B) and intrinsic reproduction rates (inversely related to 50% binding concentration, EC50). With this choice of competing clones, the broad CH103 lineage targeting poorly accessible conserved epitope CD4bs will go extinct (blue curve). (D) By replacing PGT121 with PGT122, strong mutual synergy between PGT122 and CH103 lineages (white stars) allows the latter to expand to considerable abundance (blue curve, panel E).
Such interference can be quantified using cross-competition assays (Koefoed et al., 2011; Laursen et al., 2018), in which binding signal of one Ab is measured with and without pre-incubation of Ag with the competitor Ab. Derking et al. have recently assembled an extensive dataset of known bnAbs targeting six major epitopes on the HIV envelope glycoprotein (Derking et al., 2015). The resulting interference matrix reveals a highly connected interaction network (Figure S3A): nearly full off-diagonal elements exhibit strong variation and considerable asymmetry. An intriguing consequence is that coupling between epitopes can mediate synergy (cyan) in one direction and incur antagonism (orange) in reverse direction, apparent from select submatrices (e.g., Figures 1B and 1D). Importantly, such mediated interactions can strongly impact clonal selection; synergy in particular may enable growth of intrinsically unfit clones: as shown in Figures 1C and 1E, changing one of six coexisting lineages (replacing PGT121 with PGT122) turns CH103, a desired broad lineage, from extinction to expansion (blue curve). Therefore, a quantitative understanding of how molecular interference influences B cell clonal makeup is a prerequisite of useful design for Ab therapy or vaccine Ags to enhance bnAb production Klein et al. (2012); Kong et al. (2015); Burton and Hangartner (2016); Caskey et al. (2019). Specifically, given an ensemble of interaction matrices, what are the survival probabilities of initially coexisting clones? Conversely, what structure of interference may ensure survival of the unfit? Last, can expansion of bnAb lineages and a potent repertoire be fulfilled simultaneously?
Here we use biophysically grounded models to study the role of Ag-mediated Ab interference in B cell clonal competition, and to predict under what conditions low-accessibility bnAb lineages can expand. We show that, without Ab interference, clones targeting poorly accessible conserved epitopes have little chance to survive. In contrast, provided a network of asymmetric interference, diverse lineages differing in inherent fitness can collectively persist and keep each other in balance. In particular, as intrinsically fitter clones engage in antagonism, unfit ones gain in abundance, although at the expense of diminishing neutralization potency. It is possible, however, to break this trade-off by choosing or designing viral particles in which synergy makes conserved epitopes more accessible.
Many works have studied lymphocyte clonal dynamics against evolving pathogens (De Boer and Perelson, 1991, 1994; De Boer et al., 2003; van Deutekom et al., 2013; Wang et al., 2015; Desponds et al., 2016; Amitai et al., 2017; Mayer et al., 2019), although these works do not consider epitope organization studied here. In cases where multiple epitopes are considered (Chaudhury et al., 2014; Childs et al., 2015; Luo and Perelson, 2015; Nourmohammad et al., 2016; Shaffer et al., 2016; Wang, 2017), they are treated as independent so that clones do not interact other than through a global resource limit. Here we emphasize mediated interactions between lineages that stem from spatial arrangement and physical coupling of epitopes via Ag. This scale-crossing mechanism suggests novel strategies to amplify naturally rare lineages; for instance, instead of fully masking variable epitopes with glycans, an attempt with little success in focusing response to conserved sites (Selvarajah et al., 2005), one can take advantage of potential synergy between conserved and variable targets to better promote bnAb expansion.
Results
Model
To describe polyclonal B cell response in a collection of germinal centers (Victora and Nussenzweig, 2012) (GCs, dynamic structures formed in lymph nodes upon infection or vaccination), we take a phenomenological approach that directly models the Ab interference aspect of competitive clonal expansion, without being concerned with full molecular details. This approach allows us to focus on a few ingredients essential for determining long-term clonal composition, which include (1) multiple coexisting B cell lineages each targeting a distinct epitope, (2) variation in accessibility among epitopes, (3) clonal competition for limited survival signal, and (4) interaction between B cell lineages due to Ag-mediated Ab interference. The last is the new and central ingredient in this study.
A minimal model that incorporates all these elements is an n-species Lotka-Volterra model for the multi-lineage B cell population, with viral Ag as a shared resource:
| (Equation 1a) |
| (Equation 1b) |
Here V is the concentration of viral particles and Bi is the concentration of B cells of specificity i (i.e., targeting epitope i), relative to that at half-maximum growth rate; i and j run over n specificities. The Lotka-Volterra models are long celebrated and widely used in ecology (Wangersky, 1978; Matsuda et al., 1992; Reichenbach et al., 2006; Bunin, 2017; Mickalide and Kuehn, 2019), but here we apply them in the immune context, viewing multi-epitope Ags as a resource. More importantly, Ab interference mediated by viral particles studied here has not been considered before. By linking molecular-scale physical coupling (Ab interference via Ag) to population-level clonal interactions (competition and cooperation of B cell lineages), our framework allows one to predict immunological consequences (survival of bnAb lineages) from biophysical measurements (Ab affinity and interference). We account for the key ingredients as follows:
-
(1)
As Abs targeting the same epitope directly compete (Derking et al., 2015; Koefoed et al., 2011), we only consider the B cell clone most effective at recognizing each epitope. Under this simplification, n B cell lineages {Bi(t)} are, respectively, associated with n distinct epitopes on a viral Ag.
-
(2)
Viral particles V are transported to the lymph nodes and presented at a steady rate Σ to GC B cells undergoing affinity maturation. We choose to focus on the late stage of this Darwinian process in which all B cell specificities of interest have developed, i.e., Bi(t = 0)>0 for i = 1, …,n, so that clonal selection dominates the dynamics; recent work indeed indicates that B cell mutation slows down in the chronic stage of HIV infection (Sheng et al., 2016). This allows us to decouple the ecological aspects explored here from evolution. We note that mutational effect would be important for clonal dynamics during acute infections and in earlier stages of chronic infections, which require future investigations with extended models.
The inherent reproductive potential of a B cell clone depends on both the accessibility of the target epitope and the binding affinity of B cell receptors (i.e., Abs in the membrane-anchored form). Assuming that affinity has saturated for mature B cells, one can regard the intrinsic reproduction rate ri as reflecting epitope accessibility and conservation level: Most unfit clones target least accessible yet most conserved epitopes, producing Abs of the largest breadth, whereas fitter clones binding more accessible and variable epitopes are of lower breadth.
-
(3)
The rate at which B cells bind and internalize Ags determines their promise in receiving T cell help (Gitlin et al., 2014). As the number of helper T cells is limited, B cells must compete for such limiting survival signal; this resource budget is characterized by a carrying capacity K. A small influx of naive B cells bi = b≪Σ/n is present for all specificities.
-
(4)
The extent to which the intrinsic reproduction rates govern clonal composition is strongly influenced by interaction between lineages. The actual reproduction rate of a B cell clone is a fraction of the intrinsic rate ri; this fraction depends on concentrations of existing Abs, assumed to be proportional to B cell concentrations. Therefore, the interaction matrix {αij} between B cell clones specifies the interference between binding of Abs produced by earlier generations of B cells and binding of receptors on contemporary B cells to the same Ag (Figure 1A). Interpreted as such, binding of Ab j is synergistic (antagonistic) with binding of B cell i if αij < 0 (αij > 0). Self-interactions are strongly antagonistic (Derking et al., 2015) (αii = 1); once self-exclusion by auto-Abs saturates (i.e., Bi = 1), clone i will have no net growth even in the absence of competing clones.
In essence, this model captures inter-generational feedback of clonal dynamics across GCs (Zhang et al., 2013), wherein the product of ancestral B cells can influence the effective fitness of contemporary clones. Such feedback, mediated by multi-epitope Ags, results in interactions between otherwise unrelated lineages via the Abs they produce. We present our results in terms of steady-state clonal composition and Ab quality.
To understand the physical requirements for expanding intrinsically unfit clones that target conserved epitopes of low accessibility, we need to consider how epitopes are spatially organized on the viral Ag (Murin et al., 2019). Many highly mutable viruses (especially HIV and influenza) use envelope glycoproteins on their surface to bind host cell receptors for entry. These protein spikes comprise a head and a stem. Some conserved epitopes are interspersed among variable epitopes in the head domain and partially shielded from Ab binding, whereas others are located in the stem or at the head-stem interface and thus are sterically difficult to access (Hedestam et al., 2008; Zhou et al., 2010; Burton and Hangartner, 2016; West et al., 2014).
We first consider conserved epitopes being spatially proximal to variable epitopes; here clones directed at different epitopes interfere with one another through random interactions, assuming no detailed knowledge of intermolecular interactions. We then consider a conserved epitope remotely coupled to a group of variable epitopes via, e.g., allosteric pathways. In fact, Abs were found to exert allosteric influence on viral glycoproteins (Julien et al., 2013; Wang et al., 2016), where binding to one location alters accessibility of a distal site. In this setting, fit clones specific for variable epitopes again interfere in a random manner, but interactions between this fit group and the unfit clone targeting a conserved site now become independent parameters, set free from the rest of the network.
In both cases, we study two quantities of interest: first, survival probability (ϕ0) of the unfit clone (i.e., persistence of a bnAb lineage) starting with B cells of all specificities at small equal concentrations and second, the overall quality of the polyclonal response, measured by the fitness (f) of the resulting Ab repertoire. An essential feature of our model is that clones of different intrinsic fitness (ri) can coexist at steady state, achieving equal productivity (collective fitness) by adjusting the relative abundance under cross-interference. This resembles clonal diversity enabled by resource dynamics (Posfai et al., 2017), but via a physical means instead of metabolic pathways. Therefore the repertoire fitness can be calculated by
| (Equation 2) |
Here the second equality is obtained at steady state and relates the viral load V inversely to repertoire fitness. This fitness function characterizes the neutralization efficiency of polyclonal Abs generated by a host. A comparative study of both scenarios of Ab interference, relevant for different pathogens with complex Ags (Murin et al., 2019), reveals the impact of how conserved and variable epitopes are organized on the Ab-binding surface of an Ag.
Spatially Proximal Conserved and Variable Epitopes
A major difficulty in evolving bnAbs is that conserved epitopes are often partially masked by variable epitopes (and glycans) from Ab access. To study interference due to spatial proximity, we assume that half of the epitopes are easy to access so that n/2 strain-specific B cell clones reproduce with a high intrinsic rate r1, whereas the other half are poorly accessible resulting in a low reproduction rate r0 for the remaining clones. As pairwise interactions between all specificities are typically not available, here they are drawn from distributions characterized by only a few model parameters. Considering random interference among n distinct clones, we draw each of the n(n−1) entries αij (note αii = 1) independently from the same distribution with mean μ/n and variance σ2/n; αij and αji are subject to possible correlations set by the asymmetry coefficient γ∈[−1,1], so that ⟨(αij−μ/n) (αji−μ/n)⟩ = γσ2/n; see Supplemental Information Section A for numerical implementation.
We use a mean-field method to solve self-consistently for the survival probability ϕ of all clones combined and the repertoire fitness f in the limit of many epitopes (large n; see Figure S1 in Supplemental Information Section B). We present in Figure 2 numerical results for modest n relevant for real viruses.
Figure 2.
Phase Diagrams for Spatially Proximal Conserved and Variable Epitopes
Survival probability of unfit broad clones, ϕ0 (color coded), as a function of the mean μ and standard deviation σ of random interactions. Left to right: γ = 1 (symmetric), γ = 0 (asymmetric), and γ = −1 (anti-symmetric).
(A–C) White lines in (A) and (B) indicate the predicted transition boundary between the multiple-attractor phase (above) and the unique-fixed-point phase (below). For γ = −1 (C), this boundary lies well above the plotted range of σ. Other parameters are n = 10, r0/r1 = 0.5, and K = Σ = 1.
See also Figure S2.
When epitopes interfere randomly regardless of accessibility, the survival probability ϕ0 of unfit clones vanishes as σ→0, starting from equal small concentrations of all clones. This indicates that, without variation in the interaction strength, unfit clones have no chance to survive, whether the average interference is synergistic or antagonistic. In other words, under uniform interactions, intrinsic fitness dominates selection; as a result, less fit clones are deemed to be outcompeted. As long as interactions are heterogeneous, ϕ0 is finite and increases monotonically with both an increasing mean μ and a growing variance σ of interference; this trend holds true for any level of asymmetry γ (Figure 2A–2C). Furthermore, survival is enhanced as interference becomes more anti-symmetric (γ being more negative). Hence, strong clonal competition (large positive μ), large variation of interaction strength (large σ), and anti-symmetric interference (negative γ) facilitate expansion of bnAb lineages.
The intuition, as will become more clear in the next section, is that when antagonistic interference is significant, the effective number of variable epitopes becomes small, so that unfit clones have fewer competitors and thus survive more readily. Meanwhile, when the variance of interference is large, it becomes more likely that an unfit clone receives synergistic influence from fit peers and persists. Notably, strong anti-symmetry (large negative γ) does both: it reduces the effective number of variable epitopes and at the same time enhances the chance of synergy toward unfit clones, even when interference is on average antagonistic. Importantly, synergy via indirect pathways can also support survival of the unfit, if, e.g., strong competitors of the bnAb lineage were suppressed by other fit clones (see Figure S4 for examples at various γ); and yet, repertoire fitness is likely degraded relative to direct synergy. This highlights the importance of considering specific interference structures when interpreting and predicting polyclonal responses.
Depending on the interference characteristics, our model exhibits distinct dynamical phases separated by sharp transitions at large n. Below the phase boundary (solid line in Figures 2A–2C), a unique fixed point of clonal coexistence is stable (see Supplemental Information Section C for analysis). Above the transition line, there appears a multiple-attractor phase in which a small subset of clones dominates the population at long times, and depending on initial composition, different subsets may prevail. This transition is primarily determined by the variance of interactions. Intuitively, when variation is strong, extreme values of interference overwhelms intrinsic fitness and drives alternative clonal fates: n clones segregate into low-diversity groups with heterogeneous compositions; within-group interactions are primarily synergistic, whereas between-group interactions tend to be antagonistic. We demonstrate this multiple-attractor phase numerically in Figure 3 (see Supplemental Information Section C for proof of its existence for large n). Given an identical interference matrix (Figure 3C), different founder clones lead to distinct steady-state compositions (Figure 3A vs Figure 3B).
Figure 3.
Numerical Examples of the Multiple-Attractor Phase
Given a common interference matrix (C), starting from different initial compositions, distinct clonal makeups result (A versus B).
(A) Predominant clones target epitopes 1 and 2 at t = 0; clones 2 and 4 prevail at steady state.
(B) Predominant clones target epitopes 1 and 3 at t = 0; clones 3 and 6 take over at steady state.
(C) Interference matrix drawn from a Gaussian distribution with mean μ = 0, width σ = 3, and symmetry γ = 0.
Simulations of full dynamics (Equation (Equation 1a), (Equation 1b)) reveal a pronounced trade-off between repertoire fitness f and survival probability ϕ0 of bnAb lineages (Figure 4): given asymmetry γ and randomness σ, stronger competition (larger μ; blue to red) not only promotes survival of the unfit but also curtails population fitness. In addition, under anti-symmetric interference (γ = −1), fitness is also traded for survival as σ increases (black arrow, Figure 4C), here due to predator-prey-like interactions: fit competitors are synergistic to unfit clones (ϕ0 rises) whereas the reverse interactions are antagonistic (f falls). Interestingly, when interference is relatively symmetric (γ≳0), both f and ϕ0 are enhanced by increasing σ (Figures 4A and 4B). That is, increasing variation of interference can potentially mitigate the survival-fitness trade-off: On one hand, strong mutual synergy due to large variation enhances collective fitness of all clones (most significant for γ = 1). On the other hand, random interactions overcome intrinsic fitness differences, stabilizing low-diversity multi-stable phases enriched in broad clones.
Figure 4.
Trade-off between Repertoire Fitness (f) and Survival Probability of bnAb Lineages (ϕ0)
(A–C) Color codes for μ; the black arrow points toward larger σ. The array of black circles in (A) and (C) indicates the frontier at γ = 0, σ = 8 in (B) as a reference. Every data point is an average over 1,000 realizations of the interference matrix for given (μ, σ, γ). Same parameters as in Figure 2.
Distantly Coupled Conserved and Variable Epitopes
We now consider the case where a conserved epitope is remotely coupled to a cluster of variable epitopes, like a stem site segregated from the head group in glycoproteins of many enveloped viruses (Murin et al., 2019). To single out this remote coupling, we consider n B cell clones reproducing at a high rate r1 and one clone at a lower rate r0; whereas clones within the fit group interact randomly (again characterized by μ, σ, and γ), they interfere with the unfit clone with a similar strength α1 and receive a reverse interaction of strength α0. Below we solve the model analytically by treating the fit group as a whole. This analysis not only sheds light on the origin of the survival-fitness trade-off but also reveals interference structures capable of resolving the trade-off thus yielding broad and potent responses.
By treating n fit clones as a whole, we reduce the problem to dynamics of a low-reproduction clone B0 and a high-reproduction group, , engaging in bidirectional interference:
| (Equation 3a) |
| (Equation 3b) |
| (Equation 3c) |
where
| (Equation 4) |
Here β is the effective self-competition strength of the fit group. Note that β depends linearly on the quantity Y. Known as participation ratio (Neher et al., 2013), represents the likelihood that two randomly picked surviving B cells belong to the same clone (see Supplemental Information Sections D and E). Y quantifies the degree of clonal condensation: Y = 1/n when all distinct clones coexist with similar size, whereas Y = 1 if the whole population coalesces into a single clone. Hence, β can be roughly interpreted as 1/neff, where neff is the effective number of independent high-accessibility clones.
Essentially, β is an emergent parameter that depends on all features of the random interactions within the fit group and characterizes its self-exclusion, thereby reducing the n+1 problem to a 1 + 1 problem. Typically β < 1; unless every cross-epitope interference is strongly competitive (i.e., αij = 1) then β = 1. Large β, and thus small neff, can result from strong antagonistic interactions (large μ), large variation of interference (large σ), and strong antisymmetry (large negative γ).
The simplified dynamics (Equation (Equation 3a), (Equation 3b), (Equation 3c)) yield three nontrivial steady-state solutions: survival of either fit or unfit clones and coexistence. Different phases are delineated by two stability boundaries in the interaction space (Figure 5, blue and black lines): α1∗ = β/ρ−(1−ρ)/(κρ) and α0∗ = ρ+(1−ρ)/κ. Here is the dimensionless capacity and ρ = r0/r1 the reproduction ratio. Four phases thus follow (Figure 5A):
-
•
High accessibility only: fit clones persist if asymmetric interference favors B cells targeting high-accessibility epitopes, manifested as α0<α0∗, whereas α1>α1∗;
-
•
Low accessibility only: the unfit clone expands if interference asymmetrically enhances growth of B cells targeting the conserved epitope, characterized by α1<α1∗, whereas α0>α0∗. However, antagonistic interaction from the unfit clone to the fit group results in a weak repertoire (blue region, Figure 5B);
-
•
Coexistence: both fit and unfit clones survive when α0<α0∗ and α1<α1∗. Meanwhile, mutual synergy yields a high collective fitness (yellow region, Figure 5B);
-
•
Multi-stability: either fit or unfit lineages eventually dominate, depending on initial composition. Although both single-survival solutions are stable when α0>α0∗ and α1>α1∗, fit clones will almost always overtake the population, due to faster expansion in approach to the carrying capacity.
Figure 5.
Clonal Composition and Repertoire Fitness for Distantly Coupled Conserved and Variable Epitopes
(A) Phase diagram of surviving lineages.
(B) Fitness (i.e., potency) of polyclonal responses. Blue and black lines are phase boundaries. The red symbol in (A) marks the point of no interference. The red dashed line in (B) indicates unbounded viral growth, hence vanishing repertoire fitness. Self-suppression of the fit group β = 0.17; accessibility ratio ρ = 0.5 and capacity κ = 2.4.
Without interference among epitopes, we have β = 1/n (Equation 4) and α0 = α1 = 0 (red symbol in Figure 5A), so the unfit clone survives only if α1∗≥0, i.e., 1−ρ≤βκ, or n(1−ρ)≤κ. It immediately follows that, for a given capacity κ, the range of accessibility ratio ρ in support of bnAb expansion significantly narrows as the number of variable epitopes grows. This potentially explains why pathogens often evolve diversity at multiple antigenic loci Georgieva et al. (2019). In addition, if variable epitopes are mutually cooperative (β < 0) and decoupled from the conserved site (α1 = 0), the broad lineage has no chance to survive. Also note that, α1∗ decreases with decreasing ), meaning that with a lower Ag supply (smaller Σ) or a smaller B cell capacity (smaller K), a stronger synergy is required to sustain bnAb lineages.
We can calculate an important quantity—the probability of bnAb persistence—as the fraction of unfit cells among all surviving ones at steady state, ϕ0/ϕ (see Equation S30 in Supplemental Information Section E). Curves of this probability at different numbers of variable epitopes collapse to a nonlinear function of (1−ρ)n, as shown in Figure 6, supporting the intuition that the chance of bnAb expansion falls as the conserved epitope gets harder to access or as the fit group enlarges. Furthermore, this suggests that partial masking of variable epitopes (reducing n) could help preserve unfit clones. However, a complete removal of the fit group is unfavorable, because it necessarily hurts the overall potency of polyclonal responses (see Equation (S31) in Supplemental Information Section E).
Figure 6.
Survival of the Unfit Depends on Antigenic Complexity (n) and Relative Epitope Accessibility (r_0/r_1)
The fraction of unfit cells among surviving ones shows a nonlinear dependence on (1−r0/r1)n. The black curve is a fit to Equation (S30) with c = 3.8 and σ = 2.6; Erf(⋅) denotes the error function.
In general, repertoire fitness declines with increasing self-exclusion of the fit group (larger β), but increases as the synergy between fit and unfit clones strengthens (smaller α0 and α1; see Figure 5B). Fierce competition (larger μ) not only results in effectively fewer competitors but also yields stronger antagonism between the fit group and the unfit clone. Hence, survival of the unfit is more likely as μ increases, but repertoire fitness inevitably drops. This is the origin of the survival-fitness trade-off observed in simulations (Figure 4); it is now captured analytically (Figure 7): the concentration B0 of the unfit clone increases with β (upper row), whereas the repertoire fitness f falls (lower row).
Figure 7.
Synergistic Interference can Resolve Survival-Fitness Trade-off
Top: concentration of unfit clones, B0, increases with β. Bottom: repertoire fitness f decreases with β.
(A–D) Synergistic (large negative) α1 both promotes expansion of broad clones (A) and enhances the quality of polyclonal responses (B).
Varying α0 cannot resolve the conflict (C and D). Here r1 = 1, r0 = 0.5, and κ = 1.
Is it possible to support survival of the unfit without compromising fitness? Intriguingly, synergy from the fit group to the unfit clone (α1 < 0) can break the trade-off (Figure 7 left column, purple to red): this synergy both boosts bnAb expansion (Figure 7A) and enhances repertoire potency (Figure 7B). In contrast, a reverse synergy (α0 < 0) is counterproductive (right column); it further diminishes bnAb survival (Figure 7C) with little fitness gain (Figure 7D).
Antigen Design Principles
Considering both the composition (hence breadth) and potency of polyclonal responses, we summarize graphically (Figure 8) key principles for choosing or engineering Ags based on measured {ri} and {αij}:
-
(1)
What kind of interactions between fit and unfit clones would enable the latter to expand?
Figure 8.
Structure of Antigen-Mediated Interference Strongly Impacts Composition and Potency of Polyclonal Response
(A) Without interference among epitopes, only B cell clones that target highly accessible epitopes (blue) can expand.
(B) Strong synergistic interaction (thick acute arrow) from fit clones (blue) to unfit ones (green) allows the latter to expand; fewer viral particles (spiky objects) indicate enhanced Ab potency compared with (A).
(C) Despite moderate synergy (thin arrows) from fit clones (dark and light blue), unfit clones (green) still perish if the number of high-accessibility epitopes/clones is large.
(D) If fit clones are mutually antagonistic (blunt arrows), unfit clones are more likely to expand, but at the cost of reduced repertoire potency and hence a higher viral load.
According to the phase diagram (Figure 5), an average interference of strength α1<α1∗ = −(1/κ−β)/ρ+1/κ from fit to unfit clones would suffice. Apparently, a small accessibility ratio ρ demands a strong synergy to support expansion of the unfit.
Then how does this interference affect the overall potency of Abs produced by surviving lineages? As sketched in Figure 8B, strong cooperative interactions (thick acute arrow) from the fit clone (blue) to the unfit (green) not only allow the latter to expand but also boost repertoire potency thus leaving fewer viral particles lingering (spiky objects near B cells) compared with without interference (Figure 8A).
-
(2)
What structure of interference among fit clones would promote persistence of unfit lineages?
Clearly, the key is to have effectively fewer fit competitors, i.e., a smaller neff∼1/β. Such clonal condensation can be achieved by increasing the participation ratio, which entails more intense competition among fit clones (larger μ), greater variation of interference strength (larger σ), or stronger antisymmetry in pairwise interactions (larger negative γ).
As illustrated in Figures 8C, if fit competitors (dark and light blue) do not interact, unfit clones (green) can hardly survive. In contrast, when fit clones interfere antagonistically (blunt arrows), unfit lineages are more likely to persist (Figure 8D). However, unfit clones grow at the expense of reduced repertoire potency, resulting in a higher viral load compared with the case of independent competitors (Figure 8C). Fortunately, this trade-off can be lifted via strong synergy toward the unfit, like in Figure 8B.
Interference Structure of HIV Env Glycoproteins
Are the principles developed above relevant for enhancing bnAb persistence against real viruses? We analyzed pairwise interactions between almost all known HIV bnAbs targeting six major epitopes on the Env glycoprotein, collected from extensive cross-competition experiments (Derking et al., 2015). We calculated an interference matrix from measured binding signal of each Ab in the presence or absence of a competitor, specific for the same or a different epitope (see details in Supplemental Information Section F). This matrix, shown in Figure S3A, directly quantifies Ag-mediated interactions between B cell clones. Note that each of the six epitope blocks along the diagonal contains multiple bnAbs known to target the same epitope but using different binding modes. The 6 × 6 submatrices in Figure 1 are constructed by choosing one bnAb for each epitope.
We find that interference between Abs targeting distinct epitopes (off-diagonal blocks of the full matrix in Figure S3A) follows a Gaussian distribution with mean μ = −0.09, width σ = 0.89, and asymmetry γ = 0.25; see Figure S3B. This simple analysis reveals surprising characteristics of the interference structure: First, interactions between epitopes are on average synergistic (μ < 0), in contrast to commonly considered mechanisms of Ab interference, such as steric inhibition or glycan reorientation, that are largely competitive. This observation hints at a potential role of allostery in mediating interference; indeed, physical communication between distant parts of HIV Env has been indicated in experimental (Sanders et al., 2013; Julien et al., 2013; Wang et al., 2016) and computational (Sethi et al., 2013) studies and is potentially generic to enveloped viruses (Dowd and Pierson, 2011; Fernando and Fernando, 2017). Furthermore, considerable variation in interaction strength (σ∼O(1)) indicates that some epitopes engage in strong synergy, whereas others are intensely antagonistic. Figures 1B–1E demonstrate dramatic impact of heterogeneous interactions on clonal fate: enhanced synergy, resulting from substitution of a single Ab, is sufficient to rescue an unfit lineage from extinction. Finally, interference is relatively asymmetric (γ being closer to 0 than to 1). This reflects that strong synergistic and antagonistic interactions are localized to a small subset of Abs, suggesting that it is both important and feasible to choose specific interference pathways for desired clonal composition.
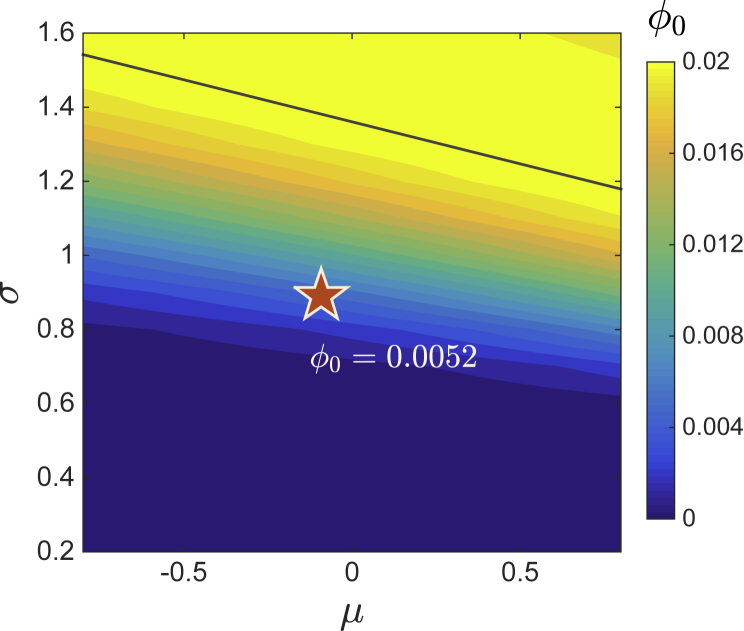
In sum, interference of the HIV Env glycoprotein exhibits appreciable synergy, heterogeneity, and asymmetry. Estimates from the cross-competition data (μ = −0.09, σ = 0.89, and γ = 0.25) allow us to place HIV Env on the theoretical phase diagram in the (σ,μ) plane with the corresponding γ; see Figure 9. Our model framework predicts a small but finite survival probability of the unfit broad lineages, ϕ0≃0.5%. This estimate indicates that natural production of HIV bnAbs of exceptional breadth is indeed rare, but not impossible, in line with the empirical view that bnAbs do emerge in a considerable fraction of patients with HIV but only at minute concentrations. Furthermore, HIV Env is located in the unique-fixed-point phase, below the transition line (black) at greater variation σ. This prediction suggests that design could help enhance bnAb persistence: When choosing Abs for combination therapy (Klein et al., 2012; Kong et al., 2015; Caskey et al., 2019), one should pick a small subset of favorably interfering Abs with strong variation of interactions, so that the subsystem is likely to reside in the multiple-attractor phase, allowing broad clones to prevail in some GCs. In the case of Ag design (Burton and Hangartner, 2016), this result recommends partial masking of variable epitopes (e.g., with glycans, Kulp et al., 2017) such that the exposed few act synergistically with the conserved epitope, overcoming the intrinsic fitness inferiority of bnAbs.
Figure 9.
Survival of HIV bnAbs
HIV Env glycoprotein (red star) is placed on the phase diagram using interference parameters, μ = −0.09, σ = 0.89, and γ = 0.25, estimated from HIV bnAb cross-competition data. Survival probability ϕ0 (color coded) of most broad clones is computed the same way as those in Figure 2, with r0 = 0.5 for two unfit clones and r1 = 1 for four fit clones. HIV Env is located in the unique-fixed-point phase, below the theoretical phase boundary (black line) given by Equation (S21); survival probability of very broad HIV bnAbs is around 0.5%.
Despite being inspired by the interference structure of HIV bnAbs, our framework applies generally to neutralizing Abs of highly mutable pathogens that present multiple epitopes with varying accessibility/conservation level. As non-neutralizing Abs do not target epitopes present in the native conformation of Env glycoproteins Burton and Mascola (2015), they are not included in the coexisting clones of the model.
Discussion
We have shown that mediated molecular interactions can strongly shape clonal fate: physical coupling between epitopes on the same Ag introduces interactions between otherwise unrelated lineages specific for distinct epitopes, because Ab binding to one epitope alters the accessibility of another hence changing reproductive fitness of the associated clone. Although competitive interference between fitter clones may encourage the expansion of unfit broad clones, it necessarily compromises the overall Ab potency. Importantly, this trade-off can be lifted if binding to accessible epitopes is synergistic to recognition of conserved sites otherwise hidden.
In contrast to clonal interference among multiple mutant lineages in a population that is primarily competitive, mediated interactions studied here can be synergistic. This Ag-mediated synergy is essential for expanding low-accessibility clones without sacrificing repertoire potency; it represents a physical mechanism of clonal cooperation, different from the evolutionary mechanism in which HIV escape mutations driven by one B cell lineage lead to recognition by a different lineage (Wibmer et al., 2013; Gao et al., 2014). Such molecular synergy could be provided by allostery, a ubiquitous process that mediates long-range coupling inside biomolecules (Süel et al., 2003; Eckmann et al., 2019) and is amenable to design for novel modes or channels (Yan et al., 2017; Rocks et al., 2017). There is indeed structural evidence that Abs exert allosteric influence on viral glycoproteins (Julien et al., 2013; Wang et al., 2016). Thus, one can exploit existing allosteric pathways in an Ag or search for or even create new ones that confer synergy to epitopes of interest. Our model can also be extended to account for homotypic interaction (Imkeller et al., 2018) between Abs that bind to repetitive epitopes on the same Ag. Synergistic binding of multiple receptors on a B cell may enhance reproductive fitness (increasing ri) due to increased avidity, whereas cooperativity between cells/Abs of the same clone, via binding different repeats of the target epitope, could mitigate self-exclusion of the clone (decreasing αii). Thus, homotypic interactions provide another means of Ag-mediated Ab synergy.
Given the complexity and nonlinear nature of mediated interactions between lineages, our quantitative framework can aid in the interpretation and prediction of polyclonal responses in diverse individuals. Model parameters pertinent to individual variations promoting bnAb generation (i.e., characteristics of “elite neutralizers”) include a higher intrinsic reproduction rate of poorly accessible broad clones (larger r0) and stronger heterogeneity of interference (larger σ) resulting from inherent repertoire composition shaped by infection histories. Our results offer novel principles to select or engineer Ags for preferred interference structure that suits an individual's Ab repertoire, or to inject synergistic Abs that promote bnAb expansion in the background of existing clones.
To predict the efficacy of injected Abs or vaccine Ag in an individual, one can obtain intrinsic fitness of coexisting clones (from EC50 of monoclonal Abs, Sanders et al., 2013) and the interference submatrix for targeted epitopes (from cross-competition assays, Derking et al., 2015). Using these as model inputs, survival probability of bnAb lineages and collective fitness can be calculated. Note that too short injections will be ineffective, because Ab concentrations are still low and interference effect is not yet significant; instead, sustained injections would allow interference to overcome intrinsic fitness and favorably shape the clonal makeup.
Our model predicts a trade-off in the immune repertoire between efficiency in suppressing viral populations (large f, by targeting easily accessible epitopes) and breadth in recognizing diverse mutants (large B0, by targeting poorly accessible yet highly conserved epitopes). Evolving pathogens seem to have exploited this trade-off and elicited diverse immune strategies. If accessible epitopes are only mildly competitive against each other while providing little synergy to the conserved epitope, immune responses would be specific and may clear infection quickly; but because bnAbs are unlikely to develop, hosts will be vulnerable to newly mutated strains, like for influenza. At the other extreme, bnAbs can be generated over the course of persistent infection; even though viral persistence slows host recovery, the broad memory acquired will provide a lasting protection against future variants, like for measles. In between, the trade-off between efficiency and breadth might leave room for the virus to both survive rapid immune attack and develop antigenic novelty before bnAbs can follow, which appears relevant for HIV and HCV.
Another interesting prediction is the existence of a phase with multiple attractors (above the transition line in Figure 2), where low-reproduction lineages have a finite probability to expand. This phase corresponds to the multi-stability region in the phase diagram (Figure 5A): depending on which clones initially dominate, founder effect will exclude others from outgrowing. More generally, this multistability stems from strong heterogeneity of interactions, so that interference dominates over intrinsic reproduction rates and determines clonal fitness. Consequently, only a few clones with strong synergy will persist, condensing the population. This multi-stable phase bears important biological implications: Within an organism, it may contribute to the strong variability in clonal diversity and condensation rate among GCs (Tas et al., 2016). On the population level, it implies that similar viruses can cause divergent immune outcomes, because individual infection histories result in distinct founder clones upon Ag encounter. Therefore, Ag-mediated interference represents a physical means by which Abs generated in the past may influence whether and which bnAbs can evolve in the future (Liao et al., 2013).
The simple models studied here ignore many ingredients present in B cell affinity maturation. For instance, stochasticity may be important when GCs are newly seeded or fluctuate to a small size; multiple GCs in the same lymph node can interact. Our results require all B cell specificities to be present initially and neglect the lag between B cell differentiation and Ab feedback. Nevertheless, our analysis has broad applicability, because it relies only on a simple phenomenological characterization of how conserved and variable epitopes are organized on the viral glycoprotein, depicted by statistical properties of pairwise interactions between coexisting clones. In particular, our results emphasize the influence of molecular interference on population structure, which is fundamentally linked to the fact that receptor-Ag binding dictates reproductive fitness of lymphocytes. In this sense, selection of specific clonal composition can potentially be accomplished by tuning intermolecular interactions. Quantitative principles developed here suggest new experiments to both understand and exploit this scale-crossing mechanism of clonal selection.
Limitations of the Study
In this work, we elucidate an important ecological drive of clonal turnover in the immune repertoire—clonal interactions mediated by molecular interference—via Ab binding to coupled Ag epitopes. We demonstrate how such ecological interactions of a physical origin lead to vastly different outcomes than one would expect from merely clonal competition due to a resource limit. To focus on ecology, we work in the regime where clonal selection dominates over mutation, and hence neglect mutation-induced changes in intrinsic fitness and interference. This decoupling of ecology from evolution is approximately valid when affinity maturation nearly saturates, such as in the late stage of chronic infections.
Clearly, an appropriate account for mutation is necessary for a faithful description of polyclonal responses to vaccine or virus. To this end, one needs to determine temporal changes in {ri} and {αij} from measured or modeled distributions of mutation effect. As a first step toward dynamic Ags, we consider a simplest scenario in which two distinct Ags (different {αij}, same statistics) are presented well apart in time, such that many rounds of selection could occur (see example clonal dynamics in Figure S5). We expect that the central finding of this study—a trade-off between bnAb expansion and repertoire fitness—still holds true. Indeed, as shown in Figure S6, over a wide range of interference structure, the trade-off remains, closely resembling that for a single fixed Ag (Figure 4). This simple setting can be tested in vitro: one can perform rounds of Ab selection between successive Ags until clonal makeup becomes steady, measure the composition and potency of the resulting Ab mixture, and compare with predictions based on monoclonal affinity and interference matrices. On the other hand, an extended model with B cell mutation (time-dependent {ri}) would allow one to estimate the minimum germline frequency required for a low-accessibility lineage to persist.
Resource Availability
Lead Contact
Further requests on data and code should be directed to and will be fulfilled by the Lead Contact, Shenshen Wang (shenshen@physics.ucla.edu).
Materials Availability
Not applicable.
Data and Code Availability
The simulation code and the data generated in this study are available at Github repository EpitopeInterference [https://github.com/lyyanlely/EpitopeInterference].
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported in part by the Chan Zuckerberg Biohub. S.W. gratefully acknowledges funding from UCLA and the National Science Foundation Grant No. NSF PHY-1748958. We are thankful for the hospitality of KITP at UCSB where this work was initiated.
Author Contributions
Conceptualization, S.W.; Methodology, L.Y. and S.W.; Software, L.Y.; Formal Analysis, L.Y. and S.W.; Investigation, L.Y. and S.W.; Data Curation, S.W.; Writing - Original Draft, L.Y. and S.W.; Writing – Review & Editing, L.Y. and S.W.; Visualization, L.Y. and S.W.; Supervision: S.W.; Funding Acquisition, S.W.
Declaration of Interests
The authors declare no competing interests.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101568.
Supplemental Information
References
- Amitai A., Mesin L., Victora G.D., Kardar M., Chakraborty A.K. A population dynamics model for clonal diversity in a germinal center. Front. Microbiol. 2017;8:1693. doi: 10.3389/fmicb.2017.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D., Gibbs J.S., Angel M., Kosik I., Hickman H.D., Frank G.M., Das S.R., Wheatley A.K., Prabhakaran M., Leggat D.J. Defining b cell immunodominance to viruses. Nat. Immunol. 2017;18:456–463. doi: 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M., Kalinke U., Althage A., Freer G., Burkhart C., Roost H.-P., Aguet M., Hengartner H., Zinkernagel R. The role of antibody concentration and avidity in antiviral protection. Science. 1997;276:2024–2027. doi: 10.1126/science.276.5321.2024. [DOI] [PubMed] [Google Scholar]
- Bunin G. Ecological communities with lotka-volterra dynamics. Phys. Rev. E. 2017;95:042414. doi: 10.1103/PhysRevE.95.042414. [DOI] [PubMed] [Google Scholar]
- Burton D.R., Hangartner L. Broadly neutralizing antibodies to hiv and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Mascola J.R. Antibody responses to envelope glycoproteins in hiv-1 infection. Nat. Immunol. 2015;16:571. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Poignard P., Stanfield R.L., Wilson I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M., Klein F., Nussenzweig M.C. Broadly neutralizing anti-hiv-1 monoclonal antibodies in the clinic. Nat. Med. 2019;25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S., Reifman J., Wallqvist A. Simulation of b cell affinity maturation explains enhanced antibody cross-reactivity induced by the polyvalent malaria vaccine ama1. J. Immunol. 2014;193:2073–2086. doi: 10.4049/jimmunol.1401054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs L.M., Baskerville E.B., Cobey S. Trade-offs in antibody repertoires to complex antigens. Philos. Trans. R Soc. B Biol. Sci. 2015;370:20140245. doi: 10.1098/rstb.2014.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Cameroni E., Guarino B., Kallewaard N.L., Zhu Q., Lanzavecchia A. Tackling influenza with broadly neutralizing antibodies. Curr. Opin. Virol. 2017;24:60–69. doi: 10.1016/j.coviro.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer R.J., Mohri H., Ho D.D., Perelson A.S. Turnover rates of b cells, t cells, and nk cells in simian immunodeficiency virus-infected and uninfected rhesus macaques. J. Immunol. 2003;170:2479–2487. doi: 10.4049/jimmunol.170.5.2479. [DOI] [PubMed] [Google Scholar]
- De Boer R.J., Perelson A.S. Size and connectivity as emergent properties of a developing immune network. J. Theor. Biol. 1991;149:381–424. doi: 10.1016/s0022-5193(05)80313-3. [DOI] [PubMed] [Google Scholar]
- De Boer R.J., Perelson A.S. T cell repertoires and competitive exclusion. J. Theor. Biol. 1994;169:375–390. doi: 10.1006/jtbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- Derking R., Ozorowski G., Sliepen K., Yasmeen A., Cupo A., Torres J.L., Julien J.-P., Lee J.H., van Montfort T., de Taeye S.W. Comprehensive antigenic map of a cleaved soluble hiv-1 envelope trimer. PLoS Pathog. 2015;11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desponds J., Mora T., Walczak A.M. Fluctuating fitness shapes the clone-size distribution of immune repertoires. Proc. Natl. Acad. Sci. 2016;113:274–279. doi: 10.1073/pnas.1512977112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd K.A., Pierson T.C. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann J.-P., Rougemont J., Tlusty T. Colloquium: proteins: the physics of amorphous evolving matter. Rev. Mod. Phys. 2019;91:031001. [Google Scholar]
- Einav T., Bloom J.D. When two are better than one: modeling the mechanisms of antibody mixtures. PLoS Comput. Biol. 2020;16:e1007830. doi: 10.1371/journal.pcbi.1007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen H.N., Siskind G.W. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- Fera D., Schmidt A.G., Haynes B.F., Gao F., Liao H.-X., Kepler T.B., Harrison S.C. Affinity maturation in an hiv broadly neutralizing b-cell lineage through reorientation of variable domains. Proc. Natl. Acad. Sci. 2014;111:10275–10280. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando S., Fernando T. Antivirals for allosteric inhibition of zika virus using a homology model and experimentally determined structure of envelope protein. BMC Res. Notes. 2017;10:354. doi: 10.1186/s13104-017-2685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Bonsignori M., Liao H.-X., Kumar A., Xia S.-M., Lu X., Cai F., Hwang K.-K., Song H., Zhou T. Cooperation of b cell lineages in induction of hiv-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.K., Desikan R., Dixit N.M. Preferential presentation of high-affinity immune complexes in germinal centers can explain how passive immunization improves the humoral response. Cell Rep. 2019;29:3946–3957. doi: 10.1016/j.celrep.2019.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva M., Buckee C.O., Lipsitch M. Models of immune selection for multi-locus antigenic diversity of pathogens. Nat. Rev. Immunol. 2019;19:55–62. doi: 10.1038/s41577-018-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., Shulman Z., Nussenzweig M.C. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiff V., Redestig H., Lück J., Bruni N., Valai A., Hartmann S., Rausch S., Schuchhardt J., Or-Guil M. A minimal model of peptide binding predicts ensemble properties of serum antibodies. BMC genomics. 2012;13:79. doi: 10.1186/1471-2164-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedestam G.B.K., Fouchier R.A., Phogat S., Burton D.R., Sodroski J., Wyatt R.T. The challenges of eliciting neutralizing antibodies to hiv-1 and to influenza virus. Nat. Rev. Microbiol. 2008;6:143. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- Howell K.A., Brannan J.M., Bryan C., McNeal A., Davidson E., Turner H.L., Vu H., Shulenin S., He S., Kuehne A. Cooperativity enables non-neutralizing antibodies to neutralize ebolavirus. Cell Rep. 2017;19:413–424. doi: 10.1016/j.celrep.2017.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkeller K., Scally S.W., Bosch A., Martí G.P., Costa G., Triller G., Murugan R., Renna V., Jumaa H., Kremsner P.G. Antihomotypic affinity maturation improves human b cell responses against a repetitive epitope. Science. 2018;360:1358–1362. doi: 10.1126/science.aar5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.-P., Sok D., Khayat R., Lee J.H., Doores K.J., Walker L.M., Ramos A., Diwanji D.C., Pejchal R., Cupo A. Broadly neutralizing antibody pgt121 allosterically modulates cd4 binding via recognition of the hiv-1 gp120 v3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Halper-Stromberg A., Horwitz J.A., Gruell H., Scheid J.F., Bournazos S., Mouquet H., Spatz L.A., Diskin R., Abadir A. Hiv therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koefoed K., Steinaa L., Søderberg J.N., Kjær I., Jacobsen H.J., Meijer P.-J., Haurum J.S., Jensen A., Kragh M., Andersen P.S. vol. 3. Taylor & Francis; 2011. Rational identification of an optimal antibody mixture for targeting the epidermal growth factor receptor; pp. 584–595. (MAbs). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R., Louder M.K., Wagh K., Bailer R.T., deCamp A., Greene K., Gao H., Taft J.D., Gazumyan A., Liu C. Improving neutralization potency and breadth by combining broadly reactive hiv-1 antibodies targeting major neutralization epitopes. J. Virol. 2015;89:2659–2671. doi: 10.1128/JVI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D., Steichen J., Pauthner M., Hu X., Schiffner T., Liguori A., Cottrell C., Havenar-Daughton C., Ozorowski G., Georgeson E. Structure-based design of native-like hiv-1 envelope trimers to silence non-neutralizing epitopes and eliminate cd4 binding. Nat. Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen N.S., Friesen R.H., Zhu X., Jongeneelen M., Blokland S., Vermond J., van Eijgen A., Tang C., van Diepen H., Obmolova G. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science. 2018;362:598–602. doi: 10.1126/science.aaq0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.-X., Lynch R., Zhou T., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z. Co-evolution of a broadly neutralizing hiv-1 antibody and founder virus. Nature. 2013;496:469. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Perelson A.S. Competitive exclusion by autologous antibodies can prevent broad hiv-1 antibodies from arising. Proc. Natl. Acad. Sci. 2015;112:11654–11659. doi: 10.1073/pnas.1505207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H., Ogita N., Sasaki A., Satō K. Statistical mechanics of population: the lattice lotka-volterra model. Prog. Theor. Phys. 1992;88:1035–1049. [Google Scholar]
- Mayer A., Zhang Y., Perelson A.S., Wingreen N.S. Regulation of t cell expansion by antigen presentation dynamics. Proc. Natl. Acad. Sci. 2019;116:5914–5919. doi: 10.1073/pnas.1812800116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickalide H., Kuehn S. Higher-order interaction between species inhibits bacterial invasion of a phototroph-predator microbial community. Cell Syst. 2019;9:521–533. doi: 10.1016/j.cels.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Murin C.D., Wilson I.A., Ward A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat. Microbiol. 2019;4:734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndifon W., Wingreen N.S., Levin S.A. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc. Natl. Acad. Sci. 2009;106:8701–8706. doi: 10.1073/pnas.0903427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R.A., Vucelja M., Mezard M., Shraiman B.I. Emergence of clones in sexual populations. J. Stat. Mech. Theor. Exp. 2013;2013:P01008. [Google Scholar]
- Ng C.T., Jaworski J.P., Jayaraman P., Sutton W.F., Delio P., Kuller L., Anderson D., Landucci G., Richardson B.A., Burton D.R. Passive neutralizing antibody controls shiv viremia and enhances b cell responses in infant macaques. Nat. Med. 2010;16:1117–1119. doi: 10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourmohammad A., Otwinowski J., Plotkin J.B. Host-pathogen coevolution and the emergence of broadly neutralizing antibodies in chronic infections. PLoS Genet. 2016;12:e1006171. doi: 10.1371/journal.pgen.1006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G., Pallesen J., de Val N., Lyumkis D., Cottrell C.A., Torres J.L., Copps J., Stanfield R.L., Cupo A., Pugach P. Open and closed structures reveal allostery and pliability in the hiv-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai A., Taillefumier T., Wingreen N.S. Metabolic trade-offs promote diversity in a model ecosystem. Phys. Rev. Lett. 2017;118:028103. doi: 10.1103/PhysRevLett.118.028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond D.D., Bajic G., Ferdman J., Suphaphiphat P., Settembre E.C., Moody M.A., Schmidt A.G., Harrison S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. 2018;115:168–173. doi: 10.1073/pnas.1715471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach T., Mobilia M., Frey E. Coexistence versus extinction in the stochastic cyclic lotka-volterra model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:051907. doi: 10.1103/PhysRevE.74.051907. [DOI] [PubMed] [Google Scholar]
- Rocks J.W., Pashine N., Bischofberger I., Goodrich C.P., Liu A.J., Nagel S.R. Designing allostery-inspired response in mechanical networks. Proc. Natl. Acad. Sci. 2017;114:2520–2525. doi: 10.1073/pnas.1612139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Derking R., Cupo A., Julien J.-P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J. A next-generation cleaved, soluble hiv-1 env trimer, bg505 sosip. 664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs T., Klein F., Braunschweig M., Kreider E.F., Feldmann A., Nogueira L., Oliveira T., Lorenzi J.C., Parrish E.H., Learn G.H. Hiv-1 therapy with monoclonal antibody 3bnc117 elicits host immune responses against hiv-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajah S., Puffer B., Pantophlet R., Law M., Doms R.W., Burton D.R. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 2005;79:12148–12163. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A., Tian J., Derdeyn C.A., Korber B., Gnanakaran S. A mechanistic understanding of allosteric immune escape pathways in the hiv-1 envelope glycoprotein. PLoS Comput. Biol. 2013;9:e1003046. doi: 10.1371/journal.pcbi.1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer J.S., Moore P.L., Kardar M., Chakraborty A.K. Optimal immunization cocktails can promote induction of broadly neutralizing abs against highly mutable pathogens. Proc. Natl. Acad. Sci. 2016;113:E7039–E7048. doi: 10.1073/pnas.1614940113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z., Schramm C.A., Connors M., Morris L., Mascola J.R., Kwong P.D., Shapiro L. Effects of darwinian selection and mutability on rate of broadly neutralizing antibody evolution during hiv-1 infection. PLoS Comput. Biol. 2016;12:e1004940. doi: 10.1371/journal.pcbi.1004940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel G.M., Lockless S.W., Wall M.A., Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Mol. Biol. 2003;10:59. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Tas J.M., Mesin L., Pasqual G., Targ S., Jacobsen J.T., Mano Y.M., Chen C.S., Weill J.-C., Reynaud C.-A., Browne E.P. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom H.W., Wijnker G., de Boer R.J. The rate of immune escape vanishes when multiple immune responses control an hiv infection. J. Immunol. 2013;191:3277–3286. doi: 10.4049/jimmunol.1300962. [DOI] [PubMed] [Google Scholar]
- Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Vono M., Eberhardt C.S., Auderset F., Mastelic-Gavillet B., Lemeille S., Christensen D., Andersen P., Lambert P.-H., Siegrist C.-A. Maternal antibodies inhibit neonatal and infant responses to vaccination by shaping the early-life b cell repertoire within germinal centers. Cell Rep. 2019;28:1773–1784. doi: 10.1016/j.celrep.2019.07.047. [DOI] [PubMed] [Google Scholar]
- Wang H., Cohen A.A., Galimidi R.P., Gristick H.B., Jensen G.J., Bjorkman P.J. Cryo-em structure of a cd4-bound open hiv-1 envelope trimer reveals structural rearrangements of the gp120 v1v2 loop. Proc. Natl. Acad. Sci. 2016;113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Optimal sequential immunization can focus antibody responses against diversity loss and distraction. PLoS Comput. Biol. 2017;13:e1005336. doi: 10.1371/journal.pcbi.1005336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Mata-Fink J., Kriegsman B., Hanson M., Irvine D.J., Eisen H.N., Burton D.R., Wittrup K.D., Kardar M., Chakraborty A.K. Manipulating the selection forces during affinity maturation to generate cross-reactive hiv antibodies. Cell. 2015;160:785–797. doi: 10.1016/j.cell.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangersky P.J. Lotka-volterra population models. Annu. Rev. Ecol. Syst. 1978;9:189–218. [Google Scholar]
- West A.P., Jr., Scharf L., Scheid J.F., Klein F., Bjorkman P.J., Nussenzweig M.C. Structural insights on the role of antibodies in hiv-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Bhiman J.N., Gray E.S., Tumba N., Karim S.S.A., Williamson C., Morris L., Moore P.L. Viral escape from hiv-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ravasio R., Brito C., Wyart M. Architecture and co-evolution of allosteric materials. PNAS. 2017;114:2526–2531. doi: 10.1073/pnas.1615536114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Meyer-Hermann M., George L.A., Figge M.T., Khan M., Goodall M., Young S.P., Reynolds A., Falciani F., Waisman A. Germinal center b cells govern their own fate via antibody feedback. J. Exp. Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.-Y., Dai K., Finzi A., Do Kwon Y., Scheid J.F., Shi W., Xu L. Structural basis for broad and potent neutralization of hiv-1 by antibody vrc01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M.B., Wang M., Poignard P., Stiegler G., Katinger H., Burton D.R., Parren P.W. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 2001;75:12198–12208. doi: 10.1128/JVI.75.24.12198-12208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The simulation code and the data generated in this study are available at Github repository EpitopeInterference [https://github.com/lyyanlely/EpitopeInterference].