Abstract
Minichromosome maintenance protein 2 (MCM2) is a highly conserved protein from the MCM protein family that plays an important role in eukaryotic DNA replication as well as in cell cycle progression. In addition, it maintains the ploidy level consistency in eukaryotic cells, hence, mutations or alteration of this protein could result in the disintegration of the fine-tuned molecular machinery that can lead to uncontrolled cell proliferation. Moreover, MCM2 has been found to be an important marker for progression and prognosis in different cancers. Therefore, we aimed to analyze the MCM2 expression and the associated outcome in breast cancer (BC) patients based on the publicly available online databases. In this study, server-based gene expression analyses indicate the upregulation of MCM2 (p < 10−6; fold change>2.0) in various BC subtypes as compared to the respective normal tissues. Besides, the evaluation of histological sections from healthy and cancer tissues showed strong staining signals indicating higher expression of MCM2 protein. The overexpression of MCM2 was significantly correlated to promoter methylation and was related to patients' clinical features. Further, mutation analysis suggested missense as the predominant type of mutation (71.4%) with 18 copy-number alterations and 0.2% mutation frequency in the MCM2 gene. This study revealed a significant correlation (Cox p ≤ 0.05) between the higher MCM2 expression and lower patient survival. Finally, we identified the co-expressed genes with gene ontological features and signaling pathways associated in BC development. We believe that this study will provide a basis for MCM2 to be a significant biomarker for human BC prognosis.
Keywords: Data analysis, Information retrieval, Computational chemistry, Computational biology, Bioinformatics, Biocomputational method, Gene expression, Gene mutation, Gene regulation, Microarray, Cancer research, MCM2, Breast cancer, Mutation analysis, Prognostic biomarker, Transcriptional expressions
Data analysis; Information retrieval; Computational chemistry; Computational biology; Bioinformatics; Biocomputational method; Gene expression; Gene mutation; Gene regulation; Microarray; Cancer research; MCM2; Breast cancer; Mutation analysis; Prognostic biomarker; Transcriptional expressions
1. Introduction
Cancer is a group of diseases that can develop anywhere in a body through abnormal cell proliferation. It is the second leading cause of death worldwide accounting for 1 out of every 6 deaths, hence, becomes one of the important concerns for human survival [1]. In 2018, about 18.1 million cancer cases were reported with an estimated 9.6 million deaths worldwide. Among them, about 2.1 million (11.6%) cases were BC in female patients with 0.63 million (6.6%) deaths [2]. At the early stages, BC shows no or a few symptoms. Due to the lack of awareness and difficulties in detection at an early stage, the death rate in BC patients is increasing gradually. Therefore, early diagnosis can potentially reduce the BC-associated death rate by providing time for the proper treatment plan. Furthermore, genetic or epigenetic alterations of genes can change their expression patterns which may facilitate the development of cancers. Differentially expressed genes (DEGs) can be the possible markers for the therapeutic target as it reflects cancer characteristics related to the patient's prognosis.
The cell cycle is the process of genome duplication and cell division which leads to the proliferation of cells [3]. It has four phases such as G1 (Gap 1), S (DNA synthesis), G2 (Gap 2), and M (mitosis and cytokinesis); and contains multiple checkpoints throughout the process to ensure the proper segregation and replication of chromosomes into daughter cells [4]. Each stage in the cell cycle undergoes a strict regulation by different factors [5]. For instance, proteins under the MCM family ensure that the chromosomal replication occurs only once per cell cycle [6]. MCM proteins 2–7 interact with origin recognition complex (ORC) and bind to replication origin, cell division cycle 6 (Cdc6), chromatin licensing, and DNA replication factor 1 in the early G1 phase resulting in the formation of the pre-replication complex which is essential for DNA replication, initiation, and elongation [7]. MCM proteins have also been found to play an interior role in genome stability [8]. Alteration in any of these factors can cause serious problems in cell cycles leading to uncontrolled cell growth followed by the development of neoplasm [4]. MCM proteins have been reported to be significant in cancer initiation and development. The gene expression pattern of these proteins has been found to be associated with a wide range of epithelial malignancies and indicates that the up-regulation of MCM proteins may occur at either genomic or transcriptional levels [9].
MCM2, also known as DNA replication licensing factor, is one of the members of the MCM protein family. It is an important element of the pre-replication complex (pre-RC) that regulates the helicase activity resulting in the formation of the replication forks. The helicase activity of the MCM2 protein plays a critical role in the initiation of DNA replication and unwinding of the DNA strands [10]. The overexpression of MCM2 protein correlates with cell proliferation and carcinogenesis [11]. Therefore, it can be utilized as the diagnostic and prognostic tool for human malignancy in the clinical setting [1]. The overexpression of MCM2 mRNA (messenger RNA) indicated that it can be a prospective biomarker for the diagnosis of human BC, colorectal cancer [12], anal neoplasia [13], esophageal [14], and bladder cancers [15]. In prostate, lung, ovary, and renal cancers, overexpression of MCM2 protein is related to regional recurrence, shorter survival of patients, and distant metastases [16, 17, 18]. Previously, two independent prognostic markers MCM5 and MCM6 have shown to overexpress in ovarian cancer and melanoma, respectively [17,19]. Likewise, MCM7 expression was demonstrated to be a remarkable biomarker for cervical cancer as well as a prognostic factor in colorectal, lung, and ovarian cancer [20,21].
Human BC is a heterogeneous disease with different neoplasms originating from the epithelial cells. The molecular underpinning of the BC cells led to the identification of prognostic tools to estimate therapeutic reaction and predict gene expression signatures which can lead to the long-term survival of the patients [22]. Traditional prognostic and predictive features including the status of lymph node, size of the tumor, histological grade, and types of the hormonal receptors (i.e., estrogen and progesterone receptors) are not sufficient for prognosticating and early-stage determination of the disease [23]. Hence, there is a need for potential biomarkers to detect primary and operable BC at early stages, which can reduce the burden of overtreatments [22]. In human BC, the MCM2 is thought to have a strong prognostic value since overexpression of MCM2 is correlated with patient survival, regional recurrence, and distant metastases [23]. Thus, the relevance of the distinct up-regulation feature of MCM2 during human BC led us to undertake a comprehensive analysis to further explore its expression pattern in human BC.
2. Materials and methods
2.1. Expression analysis of MCM2 in various cancers and normal tissues
The expression patterns of MCM2 transcript (mRNA) in various cancers and respective normal tissues were analyzed using the Oncomine (https://www.oncomine.org) [24,25], GEPIA2 (http://gepia2.cancer-pku.cn) [26,27], and GENT2 (http://gent2.appex.kr) databases [28,29]. The Oncomine database contains microarray data from 86,733 tumors and 12,764 normal tissues in which eight datasets were used for comparative analysis. The threshold value was set to fold change>2, p-value < 10−4, mRNA as data type, and 10% of gene ranking. The GEPIA2 database includes the expression data of 9,667 tumors and 602 healthy tissues from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) portal. On the other hand, the GENT2 contains normal and tumor tissue data from GPL570 platform (HG-U133_Plus_2). For GEPIA2 and GENT2 databases, all the MCM2 queries regarding breast cancer were performed with default settings.
2.2. Expression analysis of MCM2 in normal and cancerous human breast tissues
The mRNA expression levels of MCM2 gene in BC and normal tissue counterparts were evaluated with the Oncomine [24,25], UALCAN (http://ualcan.path.uab.edu/) [29,30], GEPIA2, and HPA (https://www.proteinatlas.org/) databases [31]. The GEPIA2 database contains data from 1,085 tumors and 291 normal tissues related to BC where the type of cancers can be predicted by the query sample based on the intensity of gene expression. The UALCAN is a web-resource to perform in-depth analysis of the cancer transcriptome such as TCGA expression data [29,30]. The UALCAN webserver contains data from 1097 cancers and 114 normal tissues where users can determine the potential biomarkers and validate the potential gene of interest. Using this server, we compared the relative patterns of MCM2 expression in cancers from the TCGA database. The analysis of MCM2 mRNA expression was performed with default settings and the cox p-value less than 0.05 was considered as statistically significant. Furthermore, the immunohistochemistry of MCM2 protein in BC and normal tissues was visualized using the HPA031496 dataset deposited in the HPA server [31].
2.3. Association of MCM2 expression in clinical features and promoter methylation
The mRNA expression of the MCM2 gene in BC patients was analyzed based on their clinicopathological characteristics in TCGA datasets with the UALCAN server [29,30]. The analysis was performed by comparing breast cancer tissues with healthy tissues where all the parameters were kept defaults. We used the UCSC Xena server (https://xenabrowser.net/) to visualize and analyze the functional genomics data, for example, DNA methylation. In doing so, we collected DNA methylation data of 1247 BC samples from TCGA datasets [32]. Herein, we considered cox p-value less than 0.05 as statistically significant.
2.4. Determination of mutations and copy number alterations of the MCM2 gene
The prevalence of mutations and copy number alterations of the MCM2 gene in human BC were examined using the cBioPortal server (https://www.cbioportal.org/) [33]. The cBioPortal is an interactive online platform designed for exploring, visualizing, and analyzing multidimensional cancer genomics datasets. Total 3834 samples were retrieved from the four TCGA database studies (Cell 2015, Firehose legacy, Nature 2016, and PanCancer Atlas), which were then analyzed by using the cBioPortal server. Mutation and somatic copy number alterations have become one of the major challenges for understanding the cancer genomics. In this study, the GISTIC algorithm (Genomic Identification of Significant Targets in Cancer) with default parameters was used to identify mutations and aberrant regions in our desired gene which may contribute to cancer pathogenesis.
2.5. Analyzing the relationship between MCM2 expression and survival of BC patients
The effect of MCM2 on BC patient's survival was analyzed with PrognoScan server (http://dna00.bio.kyutech.ac.jp/PrognoScan/) and the survival plots were generated using KM-Plotter [34,35]. The PrognoScan is a widely used server for survival analysis based on the genomics datasets from multiple cancers. It uses quickly confirmed disease prophecies including overall survival (OS), relapse-free survival (RFS), distant metastasis-free survival (DMFS), and post-progression survival (PPS) [36,37]. The cox p-value of less than 0.05 was considered as statistically significant.
2.6. Profiling of genes Co-expressed with MCM2 in human BC tissues
The co-expression profile of the MCM2 gene in BC was assessed and the corresponding heatmap was retrieved from the Oncomine server. In addition, co-expression profiling revealed that the MCM4 gene is highly correlated with the MCM2 gene in BC patients. To confirm their co-expression levels in BC, a heatmap relating to two genes was anticipated with the UCSC Xena web server based on the TCGA dataset [32]. The predicted co-expressed BC genes from TCGA were also analyzed with UCSC Xena server. Furthermore, a positive correlation between the levels of MCM2 and MCM4 transcripts in BC patients were confirmed by the GEPIA2 server.
2.7. Analysis of gene ontology and signaling pathways related to BC development
The gene ontology (GO) and signaling pathways of co-expressed genes and respective bar plots were retrieved from Enrichr server (http://amp.pharm.mssm.edu/Enrichr/) [38]. The Enrichr is a widely used comprehensive enrichment analysis server that works by comparing multiple genomics datasets. Based on the GO terms, the input genes were categorized into biological processes, molecular functions, and cellular components. Likewise, signaling pathways were determined using Bioplanet 2019, Reactome 2016, and KEGG 2019 databases via Enrichr server [38]. For both cases, we considered the cox p-value less than 0.05 as statistically significant.
3. Results
3.1. The mRNA expression of MCM2 in different types of cancer
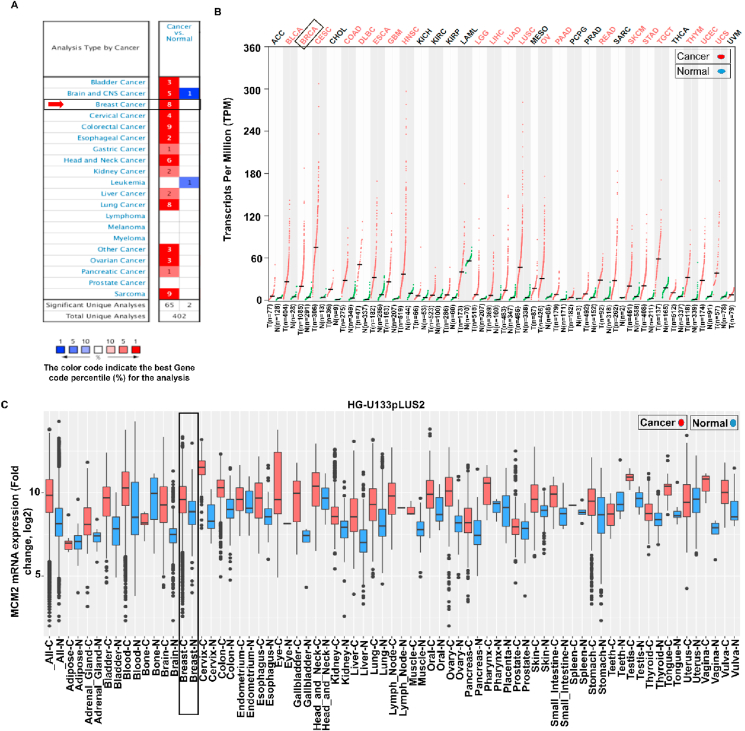
By utilizing the three-independent web-based platforms, we demonstrated the differential patterns of MCM2 transcript in different cancer tissues and their respective healthy counterparts. Among the 402 unique datasets in the Oncomine server, only 67 studies ranked in the top 10% showed statistically significant results. Interestingly, the upregulation of MCM2 was evident in different cancers including breast, bladder, brain, cervical, colorectal, esophageal, gastric, head and neck, kidney, liver, lung, other, ovarian, sarcoma, and pancreatic cancers (Figure 1A). Observation of the data also showed that MCM2 is highly expressed in BC patients while downregulated in leukemia only (Figure 1A). The expression of MCM2 in 33 different human cancers and their corresponding normal tissues were shown in Figure 1B. Further analysis of the data was performed using the Affymetrix HG-U133pLUS2 platform from the GENT2 database that reveals the overexpression of MCM2 in various cancers as depicted in Figure 1C. The results suggest strong and significant evidences relating the higher expression of MCM2 in BC tissues as compared to the normal tissues (Figure 1).
Figure 1.
Tissue-wide gene expression pattern of MCM2 in multiple human cancer, (A) comparison between cancer versus normal tissues in which high and low expression of mRNA has indicated by red and blue colors, respectively, (B) The dot plot shows the gene expression profile of the MCM2 gene in 33 types of human cancers including tumors and paired normal tissue samples. Herein, red and green dashed lines represent the average expression value in all tumor and normal tissues, respectively, and (C) box-plot showing the MCM2 mRNA expression in tumors and respective normal tissues based on the GENT2 database where boxes represent by the median, dots indicate outliers, red-boxes define the tumor tissues, and blue-boxes is normal tissues.
3.2. Expression of MCM2 transcript in human BC tissues
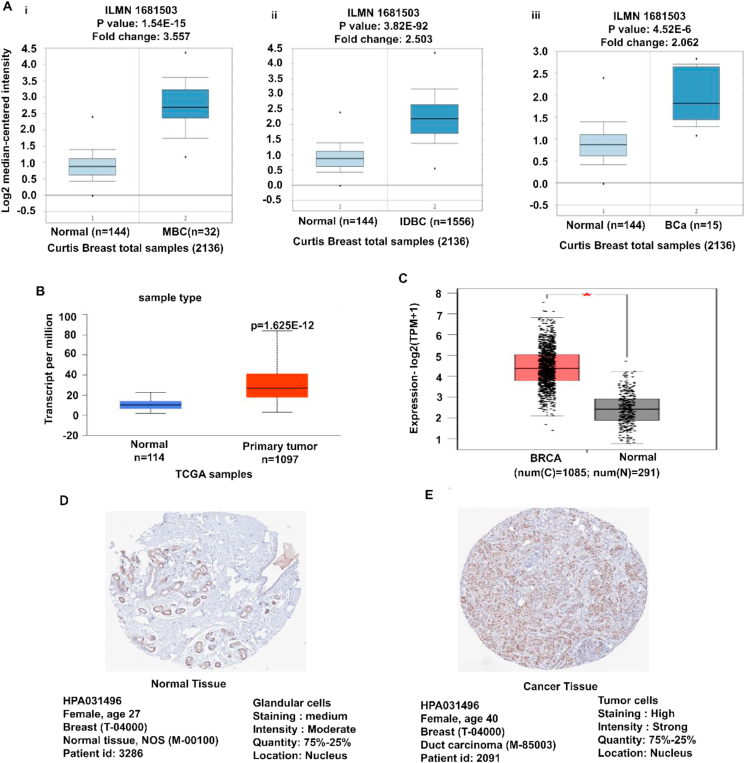
The Oncomine platform was used to assess the MCM2 gene expression for each subtype of BC compared to the normal tissues. The result revealed the overexpression of MCM2 in various BCs, such as medullary breast carcinoma, invasive ductal breast carcinoma, breast carcinoma, mixed lobular and ductal breast carcinoma, invasive lobular breast carcinoma, invasive breast carcinoma, and ductal breast carcinoma (Figure 2Ai-iii and Table 1). Further evaluation of TCGA datasets with UALCAN and GEPIA2 servers also exhibited the upregulation of MCM2 in BC (Figure 2B and 2C). Moreover, comparative immunohistochemistry analysis was performed between normal and cancer tissues using the HPA database. Among the 31 BC patients, sample 4 showed strong staining signals, while normal glandular cells showed moderate or weak staining signals indicating low MCM2 expression (Figure 2D and 2E). The results suggest that MCM2 is highly expressed in BC tissue compare to normal tissues (Figure 2 and Table 1).
Figure 2.
MCM2 expression analysis in various BC subtypes in which (A) box-plots showing comparative expression between normal (left) and cancer tissue (right) - MBC (i), IDBC (ii), and BCa (iii), (B–C) box-plots showing the expression of MCM2 mRNA in breast cancer and normal tissues using the UALCAN and GEPIA2 servers, respectively, and (D–E) the immunohistochemistry images of MCM2 in BC tissues and normal tissues retrieved from the HPA database. Abbreviation: MBC = Medullary breast carcinoma, IDBC = Invasive Ductal Breast Carcinoma, BCa = Breast carcinoma, n = number of samples, HPA = Human Protein Atlas.
Table 1.
The expression of MCM2 in the various subtype of BC from the Oncomine database.
| Datasets | Parameters | Samples | Cox p-value | Gene rank | Fold change |
|---|---|---|---|---|---|
| Curtis Breast (n = 2136) | |||||
| Normal | 144 | ||||
| Medullary breast carcinoma | 32 | 1.54E-15 | 112 | 3.557 | |
| Invasive Ductal Breast Carcinoma | 1556 | 3.82E-92 | 214 | 2.503 | |
| Breast carcinoma |
14 |
4.52E-6 |
371 |
2.062 |
|
| TCGA Breast (n = 553) | |||||
| Normal | 61 | ||||
| Mixed Lobule and Ductal Breast Carcinoma | 7 | 1.18E-6 | 183 | 2.200 | |
| Invasive Lobular Breast Carcinoma | 36 | 1.06E-12 | 506 | 2120 | |
| Invasive Ductal Breast Carcinoma | 389 | 5.24E-31 | 554 | 2.877 | |
| Invasive Breast Carcinoma |
76 |
1.30E-17 |
795 |
2.184 |
|
| Richardson Breast (n = 47) | |||||
| Normal | 7 | ||||
| Ductal Breast Carcinoma | 40 | 2.30E-5 | 1223 | 4.065 | |
3.3. Association of MCM2 expression with clinical characteristics of BC patients
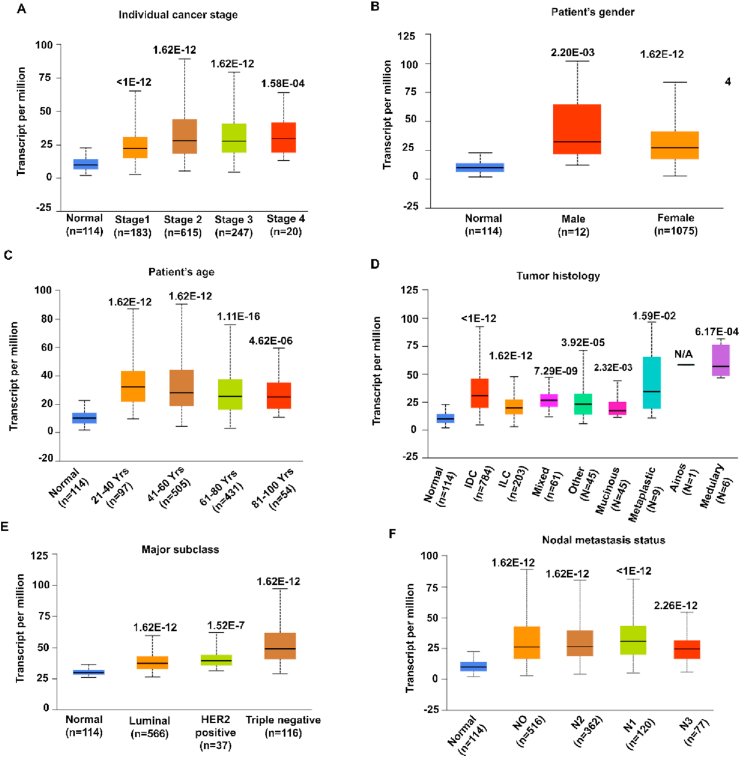
The relation between MCM2 expression and the clinical features of BC patients and healthy individuals (control) was performed using TCGA data via UALCAN database. The results exhibit enhanced expression of MCM2 irrespective of individual cancer stages, patient's race, gender, age, major subclass with and without different TNBC-type, menopause status, tumor histology, and nodal metastasis depicted in Figure 3 and listed in Table 2. The analysis asserted increased expression of MCM2 at later stages as compared to the early stages (Figure 3A). Besides, the expression level was enhanced in male patients compared to female patients. Also, Figure 3C shows higher expression at the early age group (21–40 years old). Upregulation of MCM2 expression can also be seen in terms of other clinicopathological parameters including patient's race (Table 2), tumor histology (Figure 3D), major subclasses (Figure 3E), major subclasses with TNBC-type (Table 2), nodal metastasis status (Figure 3F), and menopause status (Table 2). These results indicate that the expression of MCM2 and clinical characteristics is significantly higher in BC compared to healthy individuals.
Figure 3.
The expression analysis of MCM2 with clinical characteristics of BC patients. The MCM2 mRNA expression in BRCA showing (A) individual cancer stage, (B) patient's gender, (C) age group, (D) tumor histology, (E) major subclasses, and (F) nodal metastasis. These graphs were generated by comparing significant changes between normal variables and other variables. Abbreviation: HER2, human epidermal growth factor receptor 2; IDC, infiltrating ductal carcinoma; Mixed, mixed histology; Medullary, medullary carcinoma; INOS, infiltrating carcinoma NOS; ILC, infiltrating lobular carcinoma; Mucinous, mucinous carcinoma; Metaplastic, metaplastic carcinoma; N0, no regional lymph node metastasis; N1, metastases in 1–3 axillary lymph nodes; N2, metastases in 4–9 axillary lymph nodes; N3, metastases in 10 or more axillary lymph nodes.
Table 2.
The relationship between MCM2 and clinicopathological features of breast cancer.
| Type of samples | Expression of mRNA∗ | Number of samples | Cox P-value |
|---|---|---|---|
| Normal | ↓ | 114 | |
| Tumor | ↑ | 1097 | 1.62E-12 |
| Individual cancer stages | |||
| Normal | ↓ | 114 | |
| Stage 1 | ↑ | 183 | <1E-12 |
| Stage 2 | ↑ | 615 | 1.62E-12 |
| Stage 3 | ↑ | 247 | 1.62E-12 |
| Stage 4 |
↑ |
20 |
1.54E-04 |
| Patient's race | |||
| Normal | ↓ | 114 | |
| Caucasian | ↑ | 748 | 1.62E-12 |
| African American | ↑ | 179 | <1E-12 |
| Asian |
↑ |
61 |
6.65E-10 |
| Patient's gender | |||
| Normal | ↓ | 114 | |
| Male | ↑ | 12 | 2.20E-03 |
| Female |
↑ |
1075 |
1.60E-12 |
| Patient's age | |||
| Normal | ↓ | 114 | |
| 21-40 Yrs | ↑ | 97 | 1.62E-12 |
| 41-60 Yrs | ↑ | 505 | 1.62E-12 |
| 61-80 Yrs | ↑ | 431 | 1.11E-16 |
| 81-100 Yrs |
↑ |
54 |
4.62E-06 |
| Major subclasses | |||
| Normal | ↓ | 114 | |
| Luminal | ↑ | 566 | 1.62E-12 |
| HER2 positive | ↑ | 37 | 1.52E-07 |
| Triple-negative |
↑ |
116 |
1.62E-12 |
| Major subclasses (incl. TNBC type) | |||
| Normal | ↓ | 114 | |
| Luminal | ↑ | 566 | 1.62E-12 |
| HER2pos | ↑ | 37 | 1.52E-07 |
| TNBC-BL1 | ↑ | 13 | 2.74E-07 |
| TNBC-BL2 | ↑ | 1113 | 1.98E-04 |
| TNBC-IM | ↑ | 2011 | 1.25E-06 |
| TNBC-LAR | ↑ | 820 | 1.11E-01 |
| TNBC-MSL | ↑ | 8 | 4.70E-03 |
| TNBC-M | ↑ | 29 | 2.77E-06 |
| TNBC-UNS |
↑ |
27 |
3.90E-06 |
| Menopause Status | |||
| Normal | ↓ | 114 | |
| Pre-menopause | ↑ | 230 | 1.62E-12 |
| Perimenopause | ↑ | 37 | 5.18E-07 |
| Post-menopause |
↑ |
700 |
1.62E-12 |
| Tumor histology | |||
| Normal | ↓ | 114 | |
| IDC | ↑ | 784 | <1E-12 |
| ILC | ↑ | 203 | 1.62E-12 |
| Mixed | ↑ | 29 | 7.28E-09 |
| Other | ↑ | 45 | 3.92E-05 |
| Mucinous | ↑ | 17 | 2.32E-03 |
| Metaplastic | ↑ | 9 | 1.58E-02 |
| INOS | ↑ | 1 | N/A |
| Medullary |
↑ |
6 |
6.16E-04 |
| Nodal Metastasis status | |||
| Normal | ↓ | 114 | |
| NO | ↑ | 516 | 1.62E-12 |
| N1 | ↑ | 362 | 1.62E-12 |
| N2 | ↑ | 120 | <1E-12 |
| N3 | ↑ | 77 | 2.26E-12 |
∗Down-arrow (↓) indicates the underexpression while up-arrow (↑) indicates the overexpression.
3.4. Promoter methylation of BC from TCGA dataset
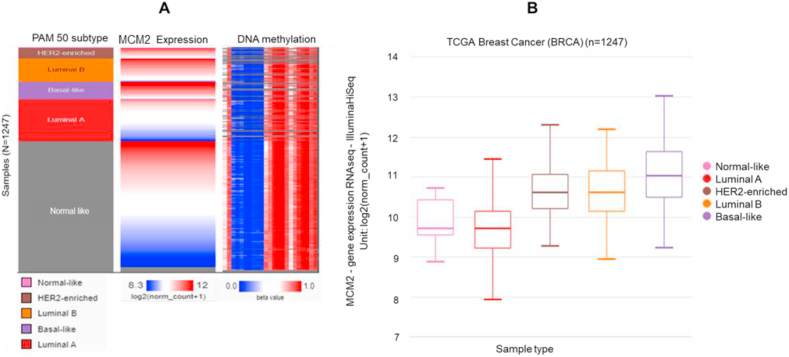
Promoter methylation is one of the essential epigenetic regulatory factors that plays a significant role in gene silencing, tissue differentiation, cellular development, and genetic imprinting. Aberrant hyper-methylation of high-density CpG regions, known as CpG Islands (CGIs), or genome-wide hypo-methylation have been found to be associated with cancers. Therefore, we examined the correlation between MCM2 expression and methylation in PAM50 BC subtypes. Comparing the MCM2 expression heatmap and DNA methylation status revealed that MCM2 expression might be negatively related with some CpG sites (blank frame) (Figure 4A). By comparing MCM2 expression in different DNA methylation clusters, we also confirmed that MCM2 expression gradually increased with decreasing DNA methylation in BC (Figure 4B).
Figure 4.
Promoter methylation of the MCM2 gene in BC tissue. (A) Heat map of MCM2 expression and DNA methylation status; (B) MCM2 expression in different breast cancer DNA methylation clusters. The results were generated using the UCSC Xena server based on TCGA dataset. Red color indicates higher expression while blue color indicates lower expression.
3.5. Mutations, copy number alterations, and expression of mutant MCM2 transcript
To determine the alterations of the MCM2 gene in BC, a total of 3834 samples from 4 BC studies were analyzed by utilizing the cBioPortal database (Table 3). The MCM2 were altered in 18 (<0.1%) of quarried samples having a somatic mutation frequency of 0.2%. A total of 7 mutations were detected in patients with multiple samples. Besides, there were 5 missenses and 2 non-sense mutations located within 1–904 residues of MCM2 and MCM domain. Furthermore, the highest mutation was reported as a missense among 1067 samples in breast invasive ductal carcinoma and fell in a hotspot of H287P (Figure 5A). Alteration frequency was found highest (0.91% of 1099 cases) for breast TCGA among the four categories of the studies (Figure 5B). Mutated MCM2 mRNA expression was profiled in the seven cases of analysis showing the highest five missense and 2 non-sense types of mutations in BC (Figure 5C). Cumulatively, these findings suggested that the up expression of MCM2 in BC might not be associated with mutations or copy number alterations even though these alterations were markedly found in MCM2 protein (Figure 5).
Table 3.
A list of MCM2 mutational positions and types in BC from the TCGA dataset.
| Sample ID | Cancer type | Protein change | Mutation type | Number of samples |
|---|---|---|---|---|
| TCGA-A2-A0T5-01 | Breast Invasive Ductal Carcinoma | H287P | Missense | 1067 |
| TCGA-BH-A2L8-01 | Breast Invasive Lobular Carcinoma | E254 | Nonsense | 279 |
| TCGA-BH-A0HF-01 | Breast Invasive Ductal Carcinoma | E184K | Missense | 551 |
| TCGA-BH-A2L8-01 | Breast Invasive Lobular Carcinoma | E254 | Nonsense | 423 |
| TCGA-A2-A0T5-01 | Breast Invasive Ductal Carcinoma | H287P | Missense | 1076 |
| TCGA-A2-A04Q-01 | Breast Invasive Ductal Carcinoma | M706I | Missense | 19 |
| TCGA-A8-A08R-01 | Invasive Breast Carcinoma | H287P | Missense | 128 |
Figure 5.
Genetic alteration and mutations of MCM2 in BC tissue. Herein (A) lollipop plot shows the type of alteration in seven mutation spots within the MCM2 peptide sequence (0–904 AA), (B) bar diagram shows the mutation frequencies and genome alteration in the MCM2 gene, and (C) the correlation between the expression and copy number alteration of MCM2 in TCGA dataset. Abbreviation: BC, breast cancer; TCGA, the cancer genome atlas; and CNA, copy number alteration.
3.6. MCM2 expression and clinical prognosis of BC patients
The relationship between the level of MCM2 expression and patient's survival in BC was analyzed with the PrognoScan database (significant at Cox P-value < 0.05). The analysis showed a negative correlation in all cases of overall survival, disease-free survival, disease-specific survival, relapse-free survival, and distant metastasis-free survival with a hazard ratio (HR) larger than 1 (Figure 6 and Table 4). For dataset GSE9893 and GSE7390, patients with low MCM2 expression (n = 120 and 100) had significantly higher overall survival whereas higher expression (n = 35 and 98) of MCM2 lead to lower overall survival as depicted in Figure 6A-B. Similarly, dataset GSE4922-GPL96 and GSE1456-GPL96 showed lower disease-free survival and disease-specific survival, respectively, with high MCM2 expression and vice-versa (Figure 6C-D). Furthermore, dataset GSE12276, GSE1456-GPL96, GSE11121, and GSE7390 indicate negative correlation since lower MCM2 expression (97, 94, 168, and 56) had significantly higher relapse-free survival and distant metastasis-free survival unlike its higher expression (107, 65, 32, and 142) as shown in Figure 6E-H. Therefore, these results confirmed that the increased expression of MCM2 could confer a poor prognosis in BC patients.
Figure 6.
The relationship between MCM2 gene expression and survival of BC patients. The survival curves demonstrate patients' survival with the high (red) and low (blue) expression of MCM2 in Kaplan-Meier plots where (A–B) showing overall survival, (C) diseases free survival, (E–F) relapse-free survival, and (G–H) distant metastasis-free survival. The analysis was focused on the MCM2 expression in BC patients. Abbreviation: HR, hazard ratio; CI, confidence interval; and BC, breast cancer.
Table 4.
The association of MCM2 expression and survival in human BC patients.
| Dataset | Endpoint | Probe ID | N | Cox P-value | HR∗ |
|---|---|---|---|---|---|
| GSE12276 | Relapse Free Survival | 202107_s_at | 204 | 0.002833 | 1.42 |
| GSE9195 | Distant Metastasis Free Survival | 202107_s_at | 77 | 0.001732 | 3.99 |
| GSE9195 | Relapse Free Survival | 202107_s_at | 77 | 0.003532 | 3.04 |
| GSE11121 | Distant Metastasis Free Survival | 202107_s_at | 200 | 0.005174 | 2 |
| GSE1378 | Relapse Free Survival | 8241 | 60 | 0.006214 | 1.15 |
| GSE9893 | Overall Survival | 3845 | 155 | 0.000003 | 1.69 |
| GSE2034 | Distant Metastasis Free Survival | 202107_s_at | 286 | 0.029374 | 1.43 |
| GSE1456-GPL96 | Relapse Free Survival | 202107_s_at | 159 | 0.000724 | 2.08 |
| GSE1456-GPL96 | Disease Specific Survival | 202107_s_at | 159 | 0.004401 | 2.07 |
| GSE1456-GPL96 | Overall Survival | 202107_s_at | 159 | 0.02056 | 1.7 |
| E-TABM-158 | Disease Specific Survival | 202107_s_at | 117 | 0.013239 | 0.59 |
| GSE3494-GPL96 | Disease Specific Survival | 202107_s_at | 236 | 0.011551 | 1.81 |
| GSE4922-GPL96 | Disease Free Survival | 202107_s_at | 249 | 0.001042 | 1.82 |
| GSE7390 | Distant Metastasis Free Survival | 202107_s_at | 198 | 0.034357 | 1.33 |
| GSE7390 | Overall Survival | 202107_s_at | 198 | 0.0388 | 1.34 |
| GSE7390 | Relapse Free Survival | 202107_s_at | 198 | 0.033839 | 1.27 |
∗HR denotes the hazard ratio and N is the number of samples.
3.7. Analysis of gene signatures linked to MCM2 and human BC
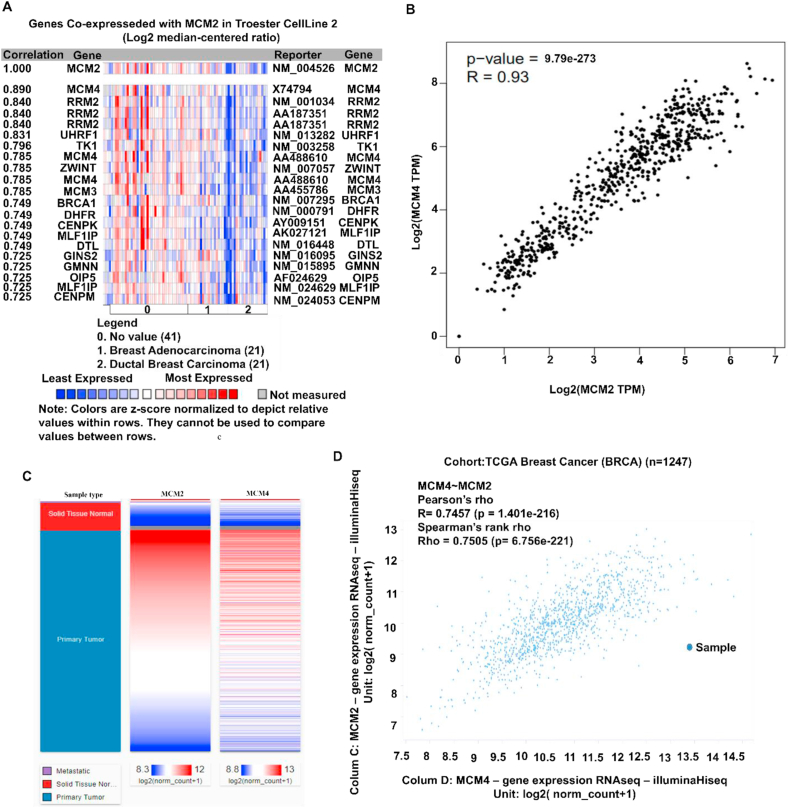
We explored the co-expression profile of MCM2 with 21 genes across 83 BC samples through the Oncomine database (Figure 7A). The result showing the expression of MCM4 (mini-chromosome maintenance 4) was mostly co-expressed (R = 0.890) among the total 21 genes. The positive correlation between MCM2 and MCM4 (R = 0.93) with the spearman coefficient was confirmed using the GEPIA2 server (Figure 7B). Further, the confirmation of positive correlation between MCM2 and MCM4 was done using Pearson (r = 0.7457) and Spearman (r = 0.7505) correlation analyses for BC patients using TCGA data through UCSCXena server (Figure 7C-D). These analyses suggest that MCM2 might be positively associated with the MCM4-mediated signaling pathway in BC progression.
Figure 7.
Co-expression profile of the MCM2 and co-expressed genes in human BC. The figure shows (A) co-expression profile of MCM2 derived from the Oncomine database, (B) correlation analysis between MCM2 and MCM4 obtained by GEPIA2 server, (C) heatmap of mRNA expression for MCM2 and MCM4 genes across BC in the TCGA database, and (D) co-expression analysis between MCM2 and MCM4 genes in BC using UCSC Xena server.
3.8. Gene ontologies and signaling pathways related to MCM2 and BC progression
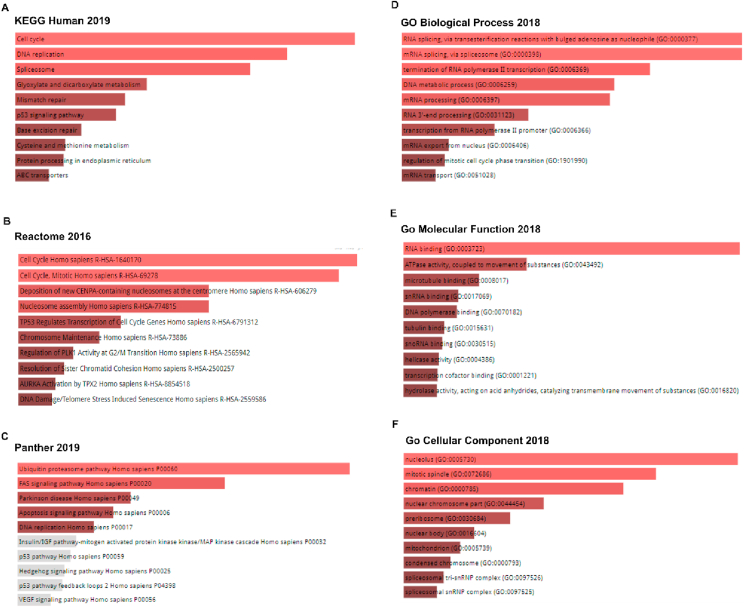
Based on the MCM2 and correlated genes, we identified signaling pathways and gene ontological features that lead to the progression of BC in humans. For pathway determination, we considered results from three databases depicted in Figure 8A-C. For KEGG human 2019 database, we found different significant pathways including cell cycle, DNA replication, spliceosome, mismatch repair, progesterone-mediated, p53 signaling pathway, base excision repair, protein processing, ABC transporters, etc. (Figure 8A). Likewise, analysis of Reactome 2016 showed pathways related to cell cycle, TP53 regulates transcription of cell cycle genes, resolution of sister chromatid cohesion, mitotic prometaphase, Rho-GTPase signaling, Rho-GTPases activate formin, deposition of new CENPA-containing nucleosomes at the centromere, nucleosome assembly, and separation of sister chromatids, etc. (Figure 8B). Finally, Panther 2019 suggested pathways such as beta1, beta2, and beta3adrenergic receptor signaling pathway, TGF-beta signaling pathway, ATP synthesis, nicotine pharmacodynamics pathway-amyloid secretase pathway, p53 pathway and its feedback loops 2, heme biosynthesis, etc. (Figure 8C). These pathways might be related to tumor development and involved in breast neoplasia. Furthermore, we computed the GO terms for MCM2 and positively correlated genes. The suggested GO features mainly include RNA splicing, via transesterification reactions with bulged adenosine as a nucleophile, transcription from RNA polymerase II promoter activity (Figure 8D), RNA binding, ATPase activity, coupled to the movement of substances (Figure 8E), nucleus and mitotic spindle activity (Figure 8F). Therefore, these pathways may also be related to cancer development and involved in BC tumorigenesis (Figure 8).
Figure 8.
Pathways and gene ontologies related to MCM2 and BC expression. Pathways are obtained from (A) KEGG human 2019, (B) Reactome 2016, (C) Panther 2019, (D) GO biological process 2018, (E) GO molecular function 2018, and (F) GO cellular component 2018. The length and the color gradient of the bar represent the level of significance (the brighter the color, the more significant the term is).
4. Discussion
Human BC is a heterogeneous disease and has different subtypes with a variety of biological behaviors and risk profiles that make the clinical management challenging [39]. Despite the declining mortality rate, BC is still one of the prominent causes of cancer deaths [40]. Therefore, the accurate prediction of the BC outcome is crucial to both patients and physicians as well as to researchers. Because, prognosis helps patients to know about their illness and to set up future courses [41,42]. For example, early prognosis by an Australian study ensured the opportunity of anticancer treatment for 85% of cancer patients in which 75% were incurable [41]. Besides, essential BC treatment relies on the precise prediction of the outcome [43]. Therefore, it is urgent to enroll a strong prognostic index to facilitate accurate predictions regarding the survival and/or response to the treatment of BC patients [44,45]. In this study, we evaluated the significance of MCM2 as a prognostic marker in BC prediction using bioinformatics [46].
Based on the multiple databases, we found that the expression level of MCM2 is positively correlated with the progression of BC. The analysis of MCM2 expression in BC exhibits a negative correlation with all cases of overall survival, diseases free survival, relapse-free survival, distant metastasis-free survival with the overall HR > 1. We have seen the noxious impact of high MCM2 levels on the survival rate than those with low MCM2 levels. A previous study suggested that the increased expression of MCM2 could confer a poor prognosis in BC patients [10]. The expression patterns of MCM2 in cancer tissues were significantly associated with different clinical characteristics of BC patients including, tumor histology, patient's race, gender, age, TNBC status, etc. Therefore, these results require further investigation since a high expression level of MCM2 may confer a risk of subsequent malignant transformation. The methodological aspects of modern immunohistochemistry that combines computer-aided systems and digital imaging will provide greater realization in immunohistochemical scoring [47,48]. The immunohistochemical data of MCM2 demonstrates strong nuclear immunoreactivity of MCM2 in each BC cell. Strong and highly intense staining of cancer cells compared to that of normal glandular cells ascertain the higher levels of MCM2 expression in BC tissues. Previous MCM2 expression was detected at higher levels than that of Ki-67 in normal breast tissues and breast cancers [49].
Most importantly, there are four facts, i.e., somatically acquired genetic, epigenetic, transcriptomic, and proteomic alterations which forms a series of histopathological process, thereby, causing cancer progression [47]. Any alteration in the genomic region either loss or gain can lead to either suppressive or oncogenic effects [50]. To explore the copy number alterations, mutations, and mutant mRNA expressions of MCM2, the cBioPortal webserver was utilized. Among the queried sample, 18 (<0.1%) found altered with the somatic mutation frequency of 0.2%. In total, 7 mutations were reported having the highest mutation to the H287P hotspot. Next, we compared MCM2 expression and DNA methylation status that reveals no significant association. The comparison between different DNA methylation clusters shows that MCM2 expression increases with the decrease in DNA methylation. It has also been shown that MCM2 DNA methylation status is decreased in human cancer [51].
Furthermore, co-expression and correlation analysis, we observed that 21 genes showed a positive correlation with the MCM2 gene and MCM4 was positively correlated with MCM2 expression (R = 0.89). Co-alteration of MCM2 and MCM4 was also confirmed by other analyses by other platforms as well. MCM4 regulates the initiation of DNA replication and may play an essential role in the proliferation of some NSCLC cells. Taken together with higher expression in NSCLCs and its correlation with clinicopathologic characteristics such as non-adenocarcinoma [52].
Finally, we analyzed the possible MCM2 related pathways in BC using the correlated genes. In the KEGG pathway analysis, the correlated genes were mostly related to the cell cycle. This is plausible since any disturbance in the cell cycle can lead to cancer progression by facilitating proliferation, genomic, and chromosomal instability [3]. In addition to the cell cycle, the correlated genes are found to be involved in DNA replication [53]. In GO analysis, the most enriched ontology terms were RNA splicing via trans-esterification reactions with bulged adenosine as the nucleophile that involves a bulge site during self-splicing [54]. Another mostly correlated molecular function was RNA binding in the cellular components. Collectively, the pathways and GO enrichment analysis imparts the importance of MCM2 and its correlated genes in different oncogenic processes.
5. Conclusion
In this study, we aimed to identify molecular signatures that play key roles in the development and progression of BC. In cancer, prognostic factors are important for efficient treatment, leaving patients with minimum risk profiles, and preventing side effects of overtreatment. To determine the candidacy of MCM2 as a potential prognostic marker in BC development, we analyzed the mRNA expression, DNA methylation, mutations and CNAs, correlated genes, and the prognostic features. Importantly, our evaluation exhibits marked upregulation and a positive correlation of MCM2 to the BC development. Furthermore, it revealed the possible signaling pathways and gene ontological features related to MCM2 and its expression in BC progression. These pathways could be the potential checkpoints to inhibit or reduce the development of cancer. In conclusion, MCM2 could be an effective biomarker and a potential therapeutic target to control BC in humans.
Declarations
Author contribution statement
A. Samad: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
F. Haque: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Z. Nain: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
R. Alam: Performed the experiments; Wrote the paper.
M. Al Noman: Performed the experiments; Contributed reagents, materials, analysis tools or data.
M. Molla and M. Khan: Contributed reagents, materials, analysis tools or data.
M. Hossen and M. Islam: Contributed reagents, materials, analysis tools or data; Wrote the paper.
F. Ahammad: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Special thanks to the Deanship of Scientific Research (DSR) at King Abdulaziz University and Biological Solution Centre (BioSol Centre) for their technical supports. A bunch of thanks to the anonymous reviewers for their thoughtful comments and suggestions which were truly helpful in revising the manuscript.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Canc. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Canc. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 4.Thu K., Soria-Bretones I., Mak T., Cescon D. Targeting the cell cycle in breast cancer: towards the next phase. Cell Cycle. 2018;17(15):1871–1885. doi: 10.1080/15384101.2018.1502567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez M.A., Pinder S.E., Callagy G., Vowler S.L., Morris L.S., Bird K., Bell J.A., Laskey R.A., Coleman N. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J. Clin. Oncol. 2003;21(23):4306–4313. doi: 10.1200/JCO.2003.04.121. [DOI] [PubMed] [Google Scholar]
- 6.Kearsey S.E., Maiorano D., Holmes E.C., Todorov I.T. The role of MCM proteins in the cell cycle control of genome duplication. Bioessays. 1996;18(3):183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- 7.Holmquist G.P. Role of replication time in the control of tissue-specific gene expression. Am. J. Hum. Genet. 1987;40(2):151. [PMC free article] [PubMed] [Google Scholar]
- 8.Forsburg S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68(1):109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tachibana K.e.K., Gonzalez M.A., Coleman N. “Cell-cycle-dependent regulation of DNA replication and its relevance to cancer pathology. J. Pathol. Soc. g. B. Irel. 2005;205(2):123–129. doi: 10.1002/path.1708. [DOI] [PubMed] [Google Scholar]
- 10.Issac M.S.M., Yousef E., Tahir M.R., Gaboury L.A. MCM2, MCM4, and MCM6 in breast cancer: clinical utility in diagnosis and prognosis. Neoplasia. 2019;21(10):1015–1035. doi: 10.1016/j.neo.2019.07.011. 2019/10/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu S., Driever W. Minichromosome maintenance proteins as markers for proliferation zones during embryogenesis. Cell Cycle. 2006;5(11):1140–1142. doi: 10.4161/cc.5.11.2779. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Li Y., Zhang W.-Y., Xia Q.-J., Li H.-G., Wang R., Yang L., Sun X.-F., Zhou Z.-G. mRNA expression of minichromosome maintenance 2 in colonic adenoma and adenocarcinoma. Eur. J. Canc. Prev. 2009;18(1):40–45. doi: 10.1097/CEJ.0b013e32830c8d5a. [DOI] [PubMed] [Google Scholar]
- 13.Scarpini C., White V., Muralidhar B., Patterson A., Hickey N., Singh N., Mullerat J., Winslet M., Davies R.J., Phillips M.-L. Improved screening for anal neoplasia by immunocytochemical detection of minichromosome maintenance proteins. Canc. Epidemiol. Prev. Biomakers. 2008;17(10):2855–2864. doi: 10.1158/1055-9965.EPI-08-0288. [DOI] [PubMed] [Google Scholar]
- 14.Sirieix P.S., O’Donovan M., Brown J., Save V., Coleman N., Fitzgerald R.C. “Surface expression of minichromosome maintenance proteins provides a novel method for detecting patients at risk for developing adenocarcinoma in barrett’s esophagus. Clin. Canc. Res. 2003;9(7):2560–2566. [PubMed] [Google Scholar]
- 15.Saeb-Parsy K., Wilson A., Scarpini C., Corcoran M., Chilcott S., McKean M., Thottakam B., Rai B., Nabi G., Rana D. Diagnosis of bladder cancer by immunocytochemical detection of minichromosome maintenance protein-2 in cells retrieved from urine. Br. J. Canc. 2012;107(8):1384–1391. doi: 10.1038/bjc.2012.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng M.V., Grossfeld G.D., Williams G.H., Dilworth S., Stoeber K., Mulley T.W., Weinberg V., Carroll P.R., Tlsty T.D. Minichromosome maintenance protein 2 expression in prostate: characterization and association with outcome after therapy for cancer. Clin. Canc. Res. 2001;7(9):2712–2718. [PubMed] [Google Scholar]
- 17.Gakiopoulou H., Korkolopoulou P., Levidou G., Thymara I., Saetta A., Piperi C., Givalos N., Vassilopoulos I., Ventouri K., Tsenga A. Minichromosome maintenance proteins 2 and 5 in non-benign epithelial ovarian tumours: relationship with cell cycle regulators and prognostic implications. Br. J. Canc. 2007;97(8):1124–1134. doi: 10.1038/sj.bjc.6603992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodins K., Cheale M., Coleman N., Fox S.B. Minichromosome maintenance protein 2 expression in normal kidney and renal cell carcinomas: relationship to tumor dormancy and potential clinical utility. Clin. Canc. Res. 2002;8(4):1075–1081. [PubMed] [Google Scholar]
- 19.Kwok H.F., Zhang S.-D., McCrudden C.M., Yuen H.-F., Ting K.-P., Wen Q., Khoo U.-S., Chan K.Y.-K. Prognostic significance of minichromosome maintenance proteins in breast cancer. Am. J. Canc. Res. 2015;5(1):52. [PMC free article] [PubMed] [Google Scholar]
- 20.Dumontet C., Jordan M.A. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010;9(10):790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ota T., Clayton A.C., Minot D.M., Shridhar V., Hartmann L.C., Gilks C.B., Chien J.R. Minichromosome maintenance protein 7 as a potential prognostic factor for progression-free survival in high-grade serous carcinomas of the ovary. Mod. Pathol. 2011;24(2):277–287. doi: 10.1038/modpathol.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi S., Watkins J., Gazinska P., Brown J.P., Gillett C.E., Grigoriadis A., Pinder S.E. Digital imaging in the immunohistochemical evaluation of the proliferation markers Ki67, MCM2 and Geminin, in early breast cancer, and their putative prognostic value. BMC Canc. 2015;15(1):546. doi: 10.1186/s12885-015-1531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shuch B., Bratslavsky G., Linehan W.M., Srinivasan R. Sarcomatoid renal cell carcinoma: a comprehensive review of the biology and current treatment strategies. Oncol. 2012;17(1):46. doi: 10.1634/theoncologist.2011-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha S.K., Kader M.A., Samad K.A., Biswas K.C., Rahman M.A., Parvez M.A.K., Rahman M.S. Prognostic and clinico-pathological significance of BIN1 in breast cancer. Inf. Med. Unlocked. 2020;19:100327. 2020/01/01/ [Google Scholar]
- 26.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samad A., Jafar T., Rafi J.H. Identification of angiotensin-converting enzyme 2 (ACE2) protein as the potential biomarker in SARS-CoV-2 infection-related lung cancer using computational analyses. Genomics. 2020;112(6):4912–4923. doi: 10.1016/j.ygeno.2020.09.002. 2020/11/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S.-J., Yoon B.-H., Kim S.-K., Kim S.-Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019;12(5):101. doi: 10.1186/s12920-019-0514-7. 2019/07/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karim M., Samad A., Adhikari U.K., Kader M., Kabir M., Islam M., Hasan M. A multi-omics analysis of bone morphogenetic protein 5 (BMP5) mRNA expression and clinical prognostic outcomes in different cancers using bioinformatics approaches. Biomedicines. 2020;8(2):19. doi: 10.3390/biomedicines8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V., Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A.-K., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 32.Goldman M., Craft B., Zhu J., Haussler D. The UCSC Xena system for cancer genomics data visualization and interpretation. AACR. 2017 [Google Scholar]
- 33.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. AACR. 2012 doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizuno H., Kitada K., Nakai K., Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics. 2009;2(1):18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barman U.D., Saha S.K., Kader M.A., Jamal M.A., Sharma S.P., Samad A., Rahman M.S. Clinicopathological and prognostic significance of GPC3 in human breast cancer and its 3D structure prediction. Netw. Model. Anal. Health Inform. Bioinform. 2020;9:1–18. [Google Scholar]
- 36.Wojnar A., Kobierzycki C., Krolicka A., Pula B., Podhorska-Okolow M., Dziegiel P. Correlation of Ki-67 and MCM-2 proliferative marker expression with grade of histological malignancy (G) in ductal breast cancers. Folia Histochem. Cytobiol. 2010;48(3):442–446. doi: 10.2478/v10042-010-0069-0. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Hu Z.-G., Li D., Xu J.-X., Zeng Z.-G. “Gene expression and prognosis of insulin-like growth factor-binding protein family members in non-small cell lung cancer. Oncol. Rep. 2019;42(5):1981–1995. doi: 10.3892/or.2019.7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A., Enrichr “. A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagerty R.G., Butow P.N., Ellis P.A., Lobb E.A., Pendlebury S., Leighl N., Goldstein D., Lo S.K., Tattersall M.H.N. Cancer patient preferences for communication of prognosis in the metastatic setting. 2004;22(9):1721–1730. doi: 10.1200/JCO.2004.04.095. [DOI] [PubMed] [Google Scholar]
- 40.Wolff A.C., Hammond M.E.H., Schwartz J.N., Hagerty K.L., Allred D.C., Cote R.J., Dowsett M., Fitzgibbons P.L., Hanna W.M., Langer A., McShane L.M., Paik S., Pegram M.D., Perez E.A., Press M.F., Rhodes A., Sturgeon C., Taube S.E., Tubbs R., Vance G.H., van de Vijver M., Wheeler T.M., Hayes D.F., P. American Society of Clinical Oncology/College of American American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch. Pathol. Lab Med. 2007;131(1):18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 41.Weigel M.T., Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr. Relat. Canc. 2010;17(4):R245–R262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 42.Moons K.G.M., Altman D.G., Vergouwe Y., Royston P. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ (Clinical research ed.) 2009;338:b606. doi: 10.1136/bmj.b606. b606. [DOI] [PubMed] [Google Scholar]
- 43.Jonkman N.H., Colpo M., Klenk J., Todd C., Hoekstra T., Del Panta V., Rapp K., van Schoor N.M., Bandinelli S., Heymans M.W., Mauger D., Cattelani L., Denkinger M.D., Rothenbacher D., Helbostad J.L., Vereijken B., Maier A.B., Pijnappels M. “Development of a clinical prediction model for the onset of functional decline in people aged 65–75 years: pooled analysis of four European cohort studies. BMC Geriatr. 2019;19(1):179. doi: 10.1186/s12877-019-1192-1. 2019/06/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rampaul R.S., Pinder S.E., Elston C.W., Ellis I.O., Nottingham Breast T. Prognostic and predictive factors in primary breast cancer and their role in patient management: the Nottingham Breast Team. Eur. J. Surg. Oncol. : J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2001;27(3):229–238. doi: 10.1053/ejso.2001.1114. [DOI] [PubMed] [Google Scholar]
- 45.Clark G.M. Do we really need prognostic factors for breast cancer? Breast Canc. Res. Treat. 1994;30(2):117–126. doi: 10.1007/BF00666054. 1994/01/01. [DOI] [PubMed] [Google Scholar]
- 46.Cui X., Yi Q., Jing X., Huang Y., Tian J., Long C., Xiang Z., Liu J., Zhang C., Tan B., Li Y., Zhu J. Mining prognostic significance of MEG3 in human breast cancer using bioinformatics analysis. Cell. Physiol. Biochem. : Int. J. Exp. Cell. Phys. Biochem. Pharm. 2018;50(1):41–51. doi: 10.1159/000493956. [DOI] [PubMed] [Google Scholar]
- 47.Laurinavicius A., Laurinaviciene A., Ostapenko V., Dasevicius D., Jarmalaite S., Lazutka J. Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagn. Pathol. 2012;7:27. doi: 10.1186/1746-1596-7-27. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu L., Xiao Z., Tu H., Tong B., Chen S. The expression and prognostic significance of Drp1 in lung cancer: a bioinformatics analysis and immunohistochemistry. Medicine. 2019;98(48):e18228. doi: 10.1097/MD.0000000000018228. e18228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yousef E.M., Furrer D., Laperriere D.L., Tahir M.R., Mader S., Diorio C., Gaboury L.A. MCM2: an alternative to Ki-67 for measuring breast cancer cell proliferation. Mod. Pathol. May, 2017;30(5):682–697. doi: 10.1038/modpathol.2016.231. [DOI] [PubMed] [Google Scholar]
- 50.Klonowska K., Czubak K., Wojciechowska M., Handschuh L., Zmienko A., Figlerowicz M., Dams-Kozlowska H., Kozlowski P. Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget. 2016;7(1):176–192. doi: 10.18632/oncotarget.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karpinski P., Pesz K., Sasiadek M.M. Pan-cancer analysis reveals presence of pronounced DNA methylation drift in CpG island methylator phenotype clusters. Epigenomics. 2017;9(11):1341–1352. doi: 10.2217/epi-2017-0070. [DOI] [PubMed] [Google Scholar]
- 52.Kikuchi J., Kinoshita I., Shimizu Y., Kikuchi E., Takeda K., Aburatani H., Oizumi S., Konishi J., Kaga K., Matsuno Y., Birrer M.J., Nishimura M., Dosaka-Akita H. Minichromosome maintenance (MCM) protein 4 as a marker for proliferation and its clinical and clinicopathological significance in non-small cell lung cancer. Lung Canc. 2011;72(2):229–237. doi: 10.1016/j.lungcan.2010.08.020. 2011/05/01/ [DOI] [PubMed] [Google Scholar]
- 53.Boyer A.-S., Walter D., Sørensen C.S. DNA replication and cancer: from dysfunctional replication origin activities to therapeutic opportunities. Semin. Canc. Biol. 2016;37–38:16–25. doi: 10.1016/j.semcancer.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Chu V.T., Liu Q., Podar M., Perlman P.S., Pyle A.M. More than one way to splice an RNA: branching without a bulge and splicing without branching in group II introns. RNA (New York, N.Y.) 1998;4(10):1186–1202. doi: 10.1017/s1355838298980724. [DOI] [PMC free article] [PubMed] [Google Scholar]