Abstract
Keratin 8 (K8) expressed at the surface of cancer cells, referred as externalized K8 (eK8), has been observed in a variety of carcinoma cell lines. K8 has been previously reported to be expressed in poorly differentiated head and neck squamous cell carcinoma (HNSCC); however, its role during the invasive phase of upper aerodigestive tract tumorigenesis is unknown. Cohorts of HNSCC tumors for protein and mRNA expression and panel of cell lines were used for investigation. K8 was found to be externalized in a majority of HNSCC cell lines. Among the two main K8 protein isoforms only the 54 kDa was found to be present at the plasma membrane of HNSCC cells. The plasminogen-induced invasion of HNSCC cells was inhibited by the anti-eK8 D-A10 antagonist monoclonal antibody. Overexpression of K8 mRNA and protein were both correlated with tumor aggressive features and poor outcome. The effect of eK8 neutralization on invasion, its presence exclusively in cancer cells and the association of K8 expression with aggressive features and poor clinical outcome in HNSCC unravel eK8 as key player in invasion and a promising therapeutic target in HNSCC.
Introduction
Head and neck cancers represent 5% of all malignancies [1,2]. The vast majority of head and neck cancers are squamous cell carcinoma (SCC). Multimodal strategies including surgery, radiation therapy, and platinum-based chemotherapy are offered to patients with head and neck SCC (HNSCC) depending on the stage of the disease. Only half of patients diagnosed with early stage disease are alive at 5 years. Moreover, these poor oncological outcomes reflect advanced stages at the time of diagnosis, as well as a high rate of recurrence and frequent second primary tumors of the upper aerodigestive tract [[3], [4], [5]]. Despite observations of some notable efficiency of cetuximab, a monoclonal antibody (mAb) targeting epidermal growth factor receptor (EGFR), in combination with radiation therapy for locally advanced disease, and with chemotherapy for distant metastatic disease, its benefits remain limited. Other targeting agents have also been evaluated in HNSCC patients but have yielded low efficacy rates. Although targeting PD-1/PD-L1 immune checkpoints has recently been approved, only 20% of the patients may benefit from this approach. Therefore, new strategies are urgently needed to identify promising targets.
Keratin 8 (K8) is present in the cytoplasm of healthy cells; it is a well-known constitutive element of the intermediate filament network in normal glandular epithelial cells. In adenocarcinomas, K8 has been reported to show ectopic localization to the plasma membrane despite lack of transmembrane domains and signal peptide to facilitate its delivery [[6], [7], [8]]. K8 localized on the external leaflet of the plasma membrane, subsequently referred as externalized K8 (eK8), was initially described in breast adenocarcinomas [9]. More recently, we have reported that eK8 inhibition in colorectal adenocarcinomas was associated with caspase-mediated apoptosis, decrease of plasminogen activation-induced invasion, and anti-tumor activity in vivo. We demonstrated that this dual function was supported by anchorage to the plasma membrane [8].
As opposed to normal glandular epithelium that expresses K8 in the cytoplasm, normal squamous stratified epithelium of the head and neck [10,11] is typically devoid of it [[10], [11], [12]]. Interestingly, K8 protein expression measured by immunohistochemistry has been reported in various carcinomas including HNSCC [9,[12], [13], [14], [15]], where it was preferentially observed at the tumor invasion front and was associated with morphological dedifferentiation [16,17]. However, the mechanism underlying these observations remains to be elucidated. These intriguing observations led us to hypothesize that K8 is externalized in HNSCC, and to examine the underlying mechanisms associated with tumor progression.
Materials and methods
Cell line culture
In total, 7 HNSCC cell lines were included in the study: SCC4, SCC9, SCC15, SCC61, Tr146, and PCI-24, derived from oral SCC (OSCC), along with BICR 18 cells originating from the larynx. All cell lines were obtained from JM Myers (The University of Texas MD Anderson Cancer Center) and are well characterized [18]. The cells were cultured in DMEM F12 (Gibco, Waltham, USA) supplemented with 10% fetal bovine serum (FBS), prophylactic antibiotics (penicillin/streptomycin 50 U/mL, ciflox 25 μg/mL) and hydrocortisone (0.4 μg/mL). PCI-24 and Tr146 were cultured in DMEM high glucose (Gibco, Waltham, USA) media, supplemented with 10% FBS and prophylactic antibiotics (penicillin/streptomycin 50 U/mL). All cell lines were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
Immunohistochemistry
Tumor specimens including 64 HNSCC were collected retrospectively from the Centre Léon Bérard (Lyon, France), and Trousseau Hospital (University Hospital, Tours, France). Tumor specimens included 36 oral SCC, 24 larynx SCC, and 4 oropharynx SCC, 20 presenting lymph node involvement, and 4 oral leukoplakia. Written informed consent was obtained from all patients.
Four-micron sections were cut from formalin-fixed paraffin-embedded blocks. One section was stained using standard hematoxylin, eosin, and saffron. Another section was stained according to the Ventana automat protocols (Ventana Medical System, Arizona, USA) using a mouse anti-K8 M20 monoclonal antibody (dilution 1:600, Sigma-Aldrich, Darmstadt, Germany). In the same experiment, an identical condition without the primary antibody was used as the negative control. Normal glandular epithelium from salivary glands and normal mucosa were used as positive and negative controls respectively.
K8 immunostaining was scored according to the intensity of staining in the membrane and in the cytosol (for each, 0: no staining, 1: weak staining, 2: moderate staining, 3: strong staining) and the percentage of stained cells in the tumor (0: no stained cells, 1: staining in less than 25% of malignant cells, 2: staining between 26 and 50% of malignant cells, 3: staining in more than 50% of malignant cells). The final score was calculated as the sum of the intensity and percentage scores [19]. For the correlations and survival analyses, tumors were considered to have low expression of K8 if they had a score ≤ 5, and a high expression if they had a score > 5. Because of our interest in the externalized form of K8, a membrane staining analysis was performed. Tumors with more than 10% malignant cells with positive membrane staining were considered to have high membrane expression of K8. The choice of cut-off for high versus low K8 expression was based on the most discriminative value in terms of survival analysis.
Western blot analysis and subcellular fractionation
When cells reached subconfluence, they were washed with phosphate buffered saline (PBS) and lysed using the Laemmli lysis buffer [20], solubilized at 95 °C during 10 min, and then stored at −20 °C. Five micrograms of protein were separated on a 12.5% acrylamide gel by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in Tris-buffered saline with Tween 20 (TBST 1×) containing 5% milk for 90 min and then incubated with the primary antibody in TBST 1× containing 2.5% milk during 60 min at room temperature. Membranes were then washed three times with TBST 1× and incubated at room temperature with TBST 1× containing HRP-secondary antibodies at a dilution of 1:3000. Immunostaining was revealed using ECL clarity (Amersham, Little Chalfont, UK). Protein quantification was performed using densitometric analysis with ImageLab software version 4.1 (ChemiDoc™ MP Imaging System - Bio-Rad). Subcellular fractionation was performed by successive centrifugation using the Qproteome cell compartment kit, (Qiagen®, Hilden, Germany). In brief, cytosolic proteins (fraction 1) were obtained in the first supernatant (1000 × g), membrane proteins in the second supernatant (fraction 2) (6000 × g) and nuclear proteins (fraction 3) in the third supernatant (6800 × g). Cytoskeletal proteins (fraction 4) were obtained from the last pellet. All protein subcellular fractions were precipitated by adding ice-cold acetone and centrifugation at 12,000 ×g. All lysates were prepared using the Laemmli lysis buffer. Each protein fraction was then analyzed by western blot as mentioned above (10% acrylamide gel). The primary antibodies used for western blot analysis included the following: mouse anti-K8 M20 (dilution 1 μg/ml, Sigma-Aldrich, Darmstadt, Germany), mouse monoclonal anti-K8 D-A10 [8] (dilution 1 μg/mL), rabbit monoclonal anti-K8 phospho S73 (dilution 1:750, Abcam, Cambridge, UK), rabbit monoclonal anti-K8 phospho S431 (dilution 1:750, Abcam, Cambridge, UK), mouse monoclonal anti-Ku80 (dilution 1:2000, Abcam, Cambridge, UK), and rabbit monoclonal anti-histone H3 (dilution 1:2000, Abcam, Cambridge, UK) antibodies.
Immunofluorescence
Cells were grown on glass slides. To preserve the membrane-associated components, non-permeabilized cells were fixed in 4% paraformaldehyde (PFA) at room temperature (RT) for 5 min. In contrast, permeabilized cells were fixed in 4% PFA at RT for 30 min. Membrane permeabilization was performed by incubation with Triton X-100 at the concentration of 1% (Sigma-Aldrich, Darmstadt, Germany) for 5 min. For protein expression analysis at the plasma membrane, cells on glass coverslips were incubated with a solution of PBS containing primary antibodies for 2 h at RT. For intracellular protein expression analysis, primary antibodies were added to PBS containing 0.1 M bovine serum albumin (BSA), 0.3 M NaCl, 0.5% Tween-20, and 1% FBS, and then added to cells on glass coverslips for 1 h at RT.
Primary antibodies used for immunofluorescence analysis were as follows: mouse anti-K8 M20 antibody (dilution 20 μg/mL, Sigma-Aldrich, Darmstadt, Germany), mouse monoclonal anti-K8 antibody D-A10 [8] (dilution 20 μg/mL), rabbit polyclonal anti-N terminal K8 antibody (1:250, Abcam, Cambridge, UK), mouse monoclonal anti-plasminogen antibody (1:100, Abcam, Cambridge, UK), and mouse monoclonal anti-uPA antibody (1:100 Abcam, Cambridge, UK). Hoechst dye was used to counterstain the nucleus (blue signal). A negative control was performed by staining with secondary antibodies only. Images were obtained by confocal microscopy (Zeiss LSM 780 confocal microscope).
Real-time analysis of invasion by x-CELLigence technology
Cell invasion was assessed using the x-CELLigence Real-Time Cell Analysis (RTCA) system (ACEA Biosciences, San Diego, CA, USA) using 8 μm pore migration/invasion plates containing 16 wells (CIM-plate 16). Wells are formed by a lower and an upper chamber placed at 37 °C and 5% CO2. The lower chamber contained culture medium supplemented with 10% FBS and the upper chamber was deprived of FBS (SF medium). The bottom of the upper chamber was previously covered with a layer of matrigel diluted 1/20 (Matrigel™, BD biosciences, San Jose, CA, USA). This technology allows continuous and real-time measurement of electrical impedance, reflecting cell-microelectrode contact, further interpreted as a cell index. The impedance generated by cell contact is measured every 15 min and is recorded by a computer linked to the RTCA DP system. This process was evaluated over 60 h and the cell index was calculated by integrating the slopes of the mean curves from triplicate experiments +/− SEM. SCC4 and/or BICR 18 cells (2 × 104 cells) were placed in the upper chamber with or without the D-A10 anti-K8 mAb at 50 μg/mL for testing the effects of this antibody; with or without purified plasminogen (Abcam, Cambridge, UK) at different concentrations (500, 50, or 5 nM) or with purified plasminogen at 50 nM and D-A10 Fab mAb or isotypic control Fab mAb at 160 nM (25 μg/mL) when competition for eK8 binding was analyzed. For experiments with purified plasminogen, cells were previously grown for 24 h in SF medium. Integration of the mean invasion curve slope (triplicate of three wells per condition ± SEM) over 45 h compared to the control for the cell line invasive properties analysis, over 22 h compared to the control for the effect of D-A10 mAb and over 30 h compared to the control for eK8/plasminogen competition analysis, were represent as an histogram. Negative controls included non-treated cells and/or isotype controls.
In silico gene expression analysis
KRT8 gene and transcript isoform [UCSC annotation: uc009zmk.1, uc001sbd.2, uc009zml.1, uc009zmm.1, uc009zmj.2, uc003xuk.2] expression levels (level 3, RNAseqV2 dataset) from 502 HNSCC samples and associated clinical data were obtained from The Cancer Genome Atlas (TCGA) [21]. Paired normal mucosa were also available in 11 cases.
Nucleotide sequences of K8 mRNA and its isoforms were obtained from Expasy portal [22], the UCSC genome browser [23], and from the National Center for Biotechnology Information [24]. Sequence similarity between the different coding RNA isoforms was performed using Basic Local Alignment Search tool (BLAST) from NCBI.
Statistical analysis
The association between clinical variables and the KRT8 gene or K8 transcript isoform expression levels, as well as semi-quantitative analysis of K8 protein staining in tumor samples was analyzed using t-tests and ANOVA tests (continuous variables). Fisher's exact test was used for categorical variables. Disease-free survival (DFS) was defined as the time from the date of HNSCC diagnosis to the date of cancer recurrence (local, regional or distant) or death. Overall survival (OS) was defined as the time from the date of HNSCC diagnosis to the date of death. Groups with high and low K8 mRNA levels were defined by the median expression level from 502 patients. Survival curves were constructed according to the Kaplan Meier method and were compared using the log-rank test. Hazard ratios and 95% CIs were calculated using Cox regression model. Univariate analysis was performed using Cox regression model to determine whether a factor is an independent predictor of DFS or OS. A P-value <0.05 was considered significant. Analysis was performed using GraphPad prism 6 software for transcriptomic data. SAS software, version 9.3 (SAS Institute Inc. Cary NC, USA) was used for clinical data from tumor specimens. K8 expression was then correlated with clinical data: age, sex, tumor anatomical site, pathological TNM status (pTNM), stage of the disease, squamous differentiation according to the neoplasm histologic grade, lymph node extra capsular spread, lymphovascular invasion, perineural invasion, overall survival (OS), and disease-free survival (DFS).
Results
eK8 is expressed at the surface of HNSCC cells and its neutralization decreases plasminogen-induced cell invasion
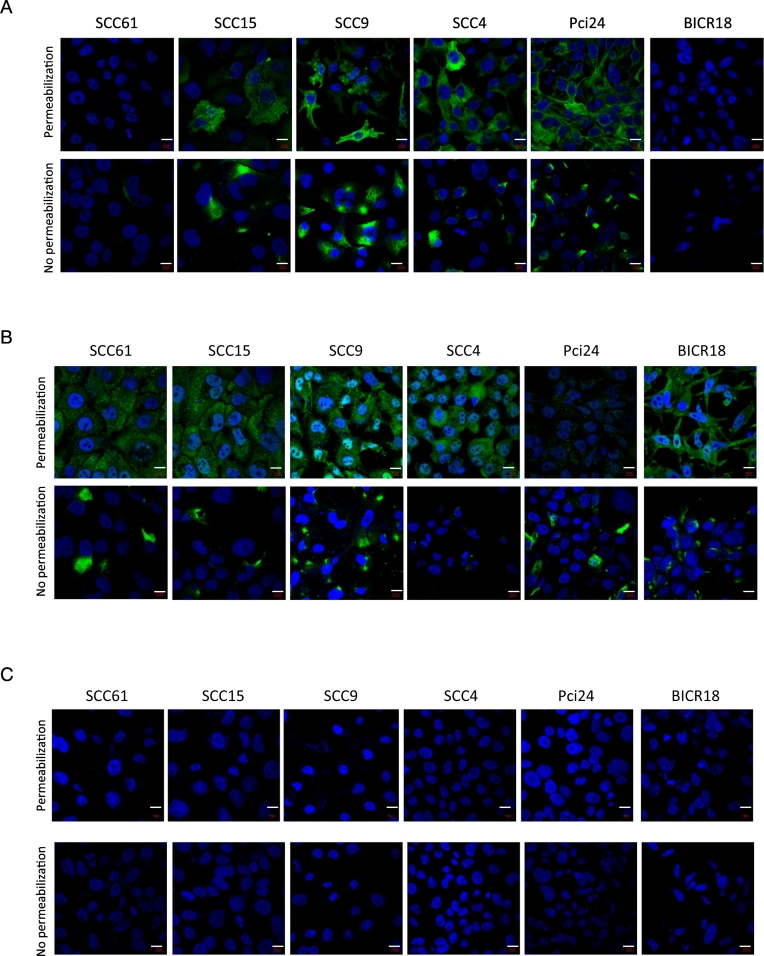
Externalization of K8 was investigated by immunofluorescence (IF) analyses performed on a panel of HNSCC cell lines using two different mAbs: M20 (commercially available) and D-A10, an anti-K8 antagonist mAb that we have reported previously [8] to target eK8 (Fig. 1A–B). Staining of both non permeabilized and permeabilized fixed cells was performed to study the presence of K8 at the external leaflet of the plasma membrane (eK8) or its intracellular localization. A Negative control involving staining with secondary antibodies only was realized (Fig. 1C). Hoechst dye was used to reveal the nucleus (blue). In permeabilized cells, K8 was observed in most cell lines with M20 (Fig. 1A), and in all cell lines with D-A10 (Fig. 1B). Using M20 antibody, the cytoplasmic accumulation of K8 with a polymerized form and fibrous organization was observed, as expected for intermediate filament proteins. Interestingly, when cells were not permeabilized, accumulation of eK8 at the external surface of the plasma membrane was observed with both anti-K8 mAbs, with cell surface expression as spots of different sizes and localization in some areas of the cell surface.
Fig. 1.
An externalized form of K8 (e-K8) is present at the cell surface of HNSCC cell lines.
(A–B) Confocal fluorescence micrographs of K8 staining (green signal) in SCC61, SCC15, SCC9, SCC4, Pci24 and BICR18 HNSCC cell lines with mouse anti-K8 M20 antibody (A) and the mouse monoclonal anti-K8 antibody D-A10 (B) on permeabilized cells (top panels) allowing the intracellular K8 visualization and on non permeabilized cells (bottom panels) allowing externalized form of K8 visualization at cell surface. Hoechst dye was used to reveal the nucleus (blue) (scale bar is 10 μm).
(C) Negative control: confocal fluorescence micrographs of staining with secondary antibodies only. Hoechst dye was used to reveal the nucleus (blue) (scale bar is 10 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
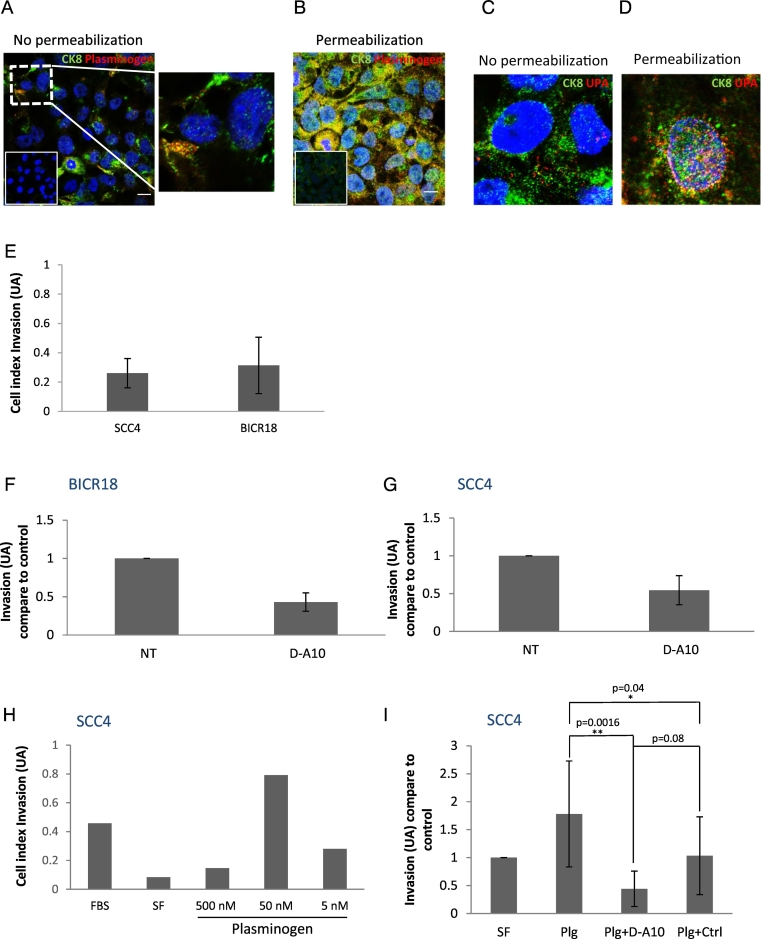
The colocalization of eK8, plasminogen (Plg), and urokinase-type plasminogen activator (uPA) was then investigated by confocal microscopy using the N-terminal anti-K8 antibody, anti-Plg, and anti-uPA mAbs in non-permeabilized and permeabilized SCC4 cells (Fig. 2A–D). Colocalization of eK8 and Plg as well as eK8 and uPA was observed at the cell surface of non-permeabilized SCC4 cells supporting the previous observations that eK8 may function as a Plg activator [8].
Fig. 2.
eK8 co-localizes with plasminogen and uPA and neutralization of eK8 limits the plasminogen-induced invasion.
(A–D) Sub-cellular localization of K8 and plasminogen or urokinase plasminogen activator (uPA) by immunofluorescence (IF) analysis and confocal microscopy in SCC4 cells previously grown on matrigel coated slides. (A–B) Fluorescence micrographs of K8 staining (N-ter antibody, green signal) and plasminogen (anti-plasminogen antibody, red signal) on non permeabilized cells (A) or on permeabilized cells (B). The inserts represent IF controls with omission of primary antibodies. Hoechst dye was used to reveal the nucleus (blue) (scale bar is 10 μm). Superposition or not of green and red signals is presented on enlarged views of the merged images (right panel Fig. 2A). (C–D) Fluorescence micrographs of K8 staining (N-ter antibody, green signal) and uPA (anti-uPA antibody, red signal) on non permeabilized cells (C) or on permeabilized cells (D). Hoechst dye was used to reveal the nucleus (blue).
(E–G) Real-time analysis of SCC4 and BICR18 cell lines invasion-kinetics with or without addition of D-A10 mAb using the xCELLigence system. (E) Comparison of invasive properties of SCC4 and BICR18 cell lines. Histogram representing the integration of the mean invasion curve slopes (triplicate of three wells per condition ± SEM) over 45 h calculated using the RTCA software®. AU: Arbitrary Unit. (n = 2 ± SEM). (F–G) D-A10 mAb effect on invasive properties of BICR18 (F) and SCC4 (G) cell lines at concentration of 50 μg/mL. Histogram representing the integration of the mean invasion curve slopes (triplicate of three wells per condition ± SEM) over 22 h compare to control. AU: Arbitrary Unit, NT: Not treated (n = 2 ± SEM).
(H–I) Real-time analysis of plasminogen effect on SCC4 cell line with or without addition of D-A10 mAb using the xCELLigence system. (H) Dose-dependent effect of plasminogen on SCC4 cell invasion. Cells were placed for 24 h either in 10% FBS or in SF medium and resuspended in SF medium containing or not increasing concentrations of plasminogen (5, 50 or 500 nM). Histogram representing the integration of the mean invasion curve slopes (triplicate of three wells per condition ± SEM) over 30 h. AU: Arbitrary Unit, (n = 1). (I) Effect of plasminogen in the presence of D-A10 Fab mAb or isotypic control Fab mAb on SCC4 cell invasion. Histogram representing the integration of the mean invasion curve slopes (triplicate of three wells per condition ± SEM) over 30 h compare to control (SF condition). AU: Arbitrary Unit, FBS: fetal bovine serum, SF: serum free. Student's t-test: **p < 0.01.(n = 6 ± SEM). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The effect of K8 inhibition on cell invasion was then evaluated in 2 HNSCC cell lines using the xCELLigence RTCA system: SCC4 OSCC cell line, as well as the larynx SCC cell line BICR 18. We first confirmed the invasive capacity of the two cell lines stimulated by an FBS gradient (Fig. 2E). Cell invasion was then studied with anti eK8 D-A10 mAb at 50 μg/mL and showed a decrease in BICR18 and SCC4 invasive properties (Fig. 2F–G). To define the increased invasive ability of SCC4 cells though the Plg pathway, invasion was evaluated by adding increasing concentrations of Plg. Fig. 2H shows increased invasion of SCC4 cells with Plg at 50 nM. To determine if the Plg-induced invasion could be inhibited by D-A10 mAb, we used a strategy used previously [8], that prevents binding of Plg to the lysine C terminal of D-A10 mAb. The Fab fragment of D-A10 lacking the Fc domain that contains a C-terminal lysine (D-A10 Fab mAb) was thus used. K8 inhibition, using mAb D-A10 at 25 μg/mL and Plg at 50 nM (Fig. 2I), showed a significant decrease in cell invasion.
These results showed the presence of K8 at the surface of HNSCC cells where eK8 plays a role in their plasminogen induced-invasion.
Among the two main isoforms of K8, the 54 kDa isoform phosphorylated on serine 431 is present in the cell membranes
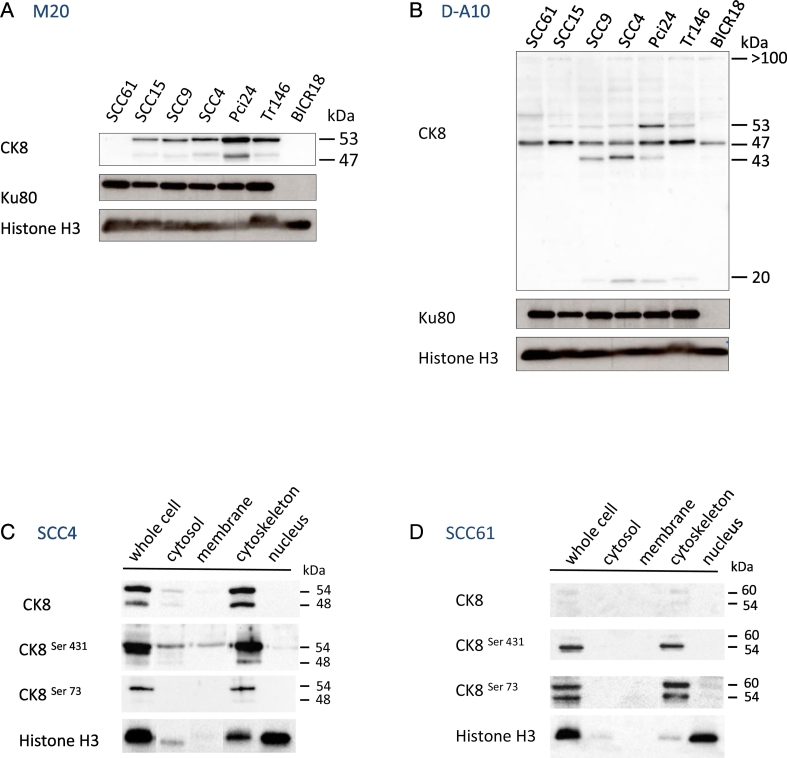
To better characterize K8 expression and its protein isoforms in HNSCC, we first performed western-blot analysis in HNSCC cells with M20 antibody and D-A10 mAb. Except for two cell lines (SCC61 and BICR18), M20 antibody and D-A10 mAb revealed the accumulation of two K8 isoforms at 54 and 47 kDa that could include the eK8 protein (Fig. 3A–B). In SCC61 and BICR18 cells, D-A10 mAb detected the 47 kDa isoform and slightly detected the 54 kDa isoform of K8, whereas the M20 antibody did not detect the K8 isoforms.
Fig. 3.
Proteomic analysis identifies the 54 kDa isoform as the main membrane K8 form and phosphorylation of Serine 431 in this 54 kDa isoform.
(A–B) 5 μg of proteins from SCC61, SCC15, SCC9, SCC4, Pci24, Tr146 and BICR18 whole cell lysates were separated by SDS-PAGE electrophoresis and transferred onto nitrocellulose membrane. Detection of isoforms of K8 was performed by anti-K8 M20 antibody (A) and anti-K8 D-A10 mAb (B). Ku80 and Histone H3 were used as loading control.
(C–D) 5 μg of proteins from SCC4 (C) and SCC 61 (D) whole cell lysates or from subcellular fractionation lysate, (cytosol, membrane, cytoskeleton and nucleus fractions) were separated by SDS-PAGE electrophoresis and transferred onto nitrocellulose membrane. Detection of all forms or phosphorylated forms of K8 respectively were performed by anti-K8 M20 antibody, and anti-K8 phospho serine 431 (S431) or serine 73 (S73) mAbs. Histone H3 was used as internal reference for subcellular fractionation.
To study the association of K8 with the plasma membrane, we then performed subcellular fractionation of SCC4 cells expressing the 2 isoforms and of SCC61 with lower expression as the negative control. Thus, we obtained enrichment of cytosolic proteins (cytosol fraction 1), membrane proteins (membrane fraction 2), cytoskeletal proteins (cytoskeleton fraction 4), and nuclear proteins (nucleus fraction 3). Then, by western-blot analysis, we characterized the accumulation of all forms of K8 (with M20 antibody), or the phosphorylated forms of K8 at serine 431 or 73 in the whole cell lysates and in different subcellular fractions of SCC4 (Fig. 3C) and SCC61 cells (Fig. 3D). As shown previously by western-blot analysis with the M20 antibody, accumulation of all K8 isoforms was largely increased in SCC4 cells versus SCC61 cells. Moreover, we showed that the distribution of K8 is quite different in SCC4 cells versus SCC61 cells. Quantification of western-blot signals indicated preferential accumulation of K8 in the cytoskeletal fraction (89.6%), and a weak presence in the cytosol (9%) and membrane (1.4%) fractions of SCC4 cells, whereas K8 was exclusively accumulated in the cytoskeleton of the SCC61 cells (100%).
These results demonstrated the presence of the 54 kDa isoform of K8 in the membrane of SCC4 cells suggesting that this isoform could be an externalized form of K8. This isoform is phosphorylated on serine 431. This result suggests the potential role of phosphorylation as a post-translational mechanism in the externalization of K8.
Two K8 mRNA variants are overexpressed in HNSCC, associated with poor prognosis, and potentially code for the two main K8 protein isoforms
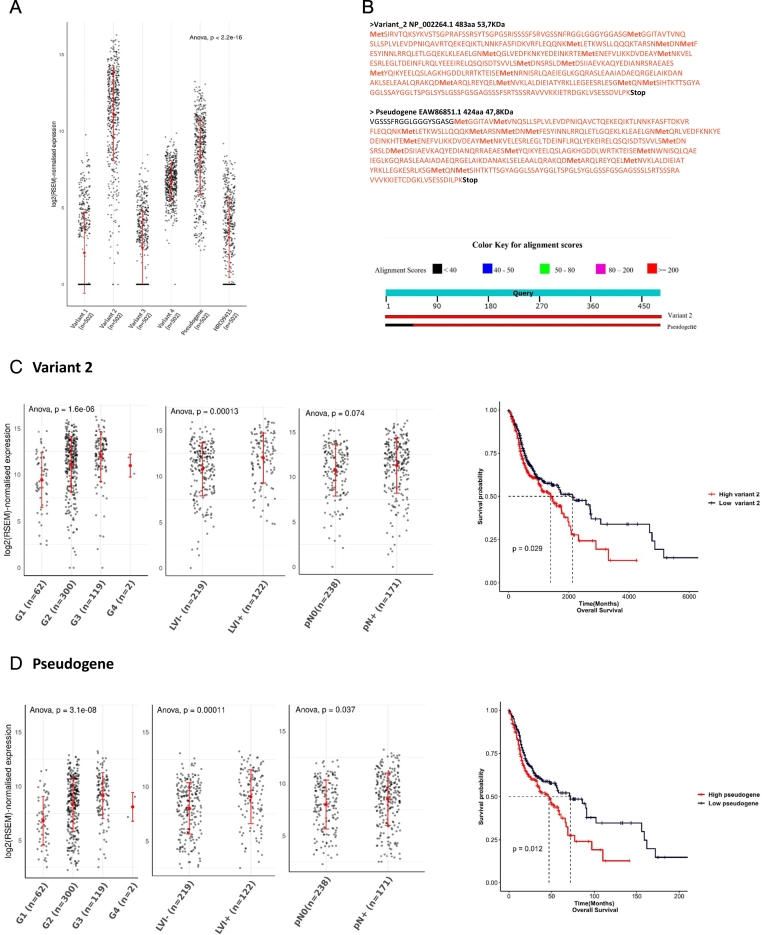
We then investigated the RNA transcripts expression levels of K8 to identify the potential mRNA variants coding for the two main isoforms observed in western blot analyses and to analyze their impact on the prognosis of HNSCC. The KRT8 gene is localized in chromosome 12 (12q13) and is composed of 9 coding exons. In UCSC, six transcripts were described named variants 1 to 4, the clone HRC09415 and one derived from a sequence annotated as a pseudogene. Variant 1 (UCSC annotation: uc009zmk.1) was described as the canonical variant with 9 exons encoding a 511 amino acids (AA) long protein (56.6 kDa); variants 2 (uc001sbd.2) and 3 (uc009zml.1) encode for proteins of 483 amino acid long (53.7 kDa) truncated at their N-terminus due to the loss of exon 1 in these two variants; variant 4 (uc009zmm.1) was described as a non-coding RNA because of the presence of an upstream Open Reading Frame (ORF) sequence. The clone HRC09415 (uc009zmj.2) comprised 6 coding exons for a sequence of 329 AA (36.2 kDa). Finally, an additional transcript derived from a K8 sequence annotated as a pseudogene (uc003xuk.2) was also described in the intron site of chromosome 8 and comprised one potential coding exon for a sequence of 424 AA (46.6 kDa). We named this transcript: K8-psd.
In 502 cases of HNSCC from TCGA, among the polyadenylated transcript characterized, variant 2, the K8-psd and the non-coding variant 4 were the three isoforms significantly overexpressed (P < 0.001) in tumor tissues (Fig. 4A) and also in paired or not normal mucosa samples (Suppl Fig. 1A–B). To confirm the homology between K8-psd and K8, we translated the mRNA coding sequence in silico to compare between the protein sequences of K8-psd versus variant 2 using basic local alignment search tool (BLAST) from the NCBI (Fig. 4B). We found an important homology between the two protein isoforms. The main difference was the occurrence of a stop codon at the N terminal extremity in K8-psd protein isoform, leading to a loss of 59 AA and a shorter protein sequence of 424 AA. The query cover and homology between variant 2 and K8-psd translation was 95% and 96%, respectively. Indeed, we revealed that these two variants that were highly expressed in HNSCC tumors and that were polyadenylated could potentially encode the two main protein isoforms of 54 and 47 kDa observed in western blot analysis of HNSCC cells. Association between variant 2 and K8-psd mRNA expression with histological prognostic factors and OS was then investigated (Fig. 4C–D). We obtained the normalized expression data from TCGA database and observed a higher level of Variant 2 and K8-psd in dedifferentiated tumors (P < 0.001 for both), positive cervical lymph nodes (P = 0.074 and P = 0.037), and lymphovascular invasion (P < 0.001 for both). Furthermore, high expression levels of both variants were associated with a shorter OS (P = 0.029 and P = 0.012).
Fig. 4.
Overexpression of K8 transcript isoforms (variants 2 and K8 pseudogene) is associated with poor pathological prognostic factors and could code for 47 and 54 kDa protein isoforms.
The mRNA level of the K8 variants was extracted from 502 cases of HNSCC (TCGA data portal). The variant 2, the K8 pseudogene and the non-coding variant 4 were the three isoforms significantly overexpressed (P < 0.001) (A). The comparison of the variant 2 and the K8 pseudogene (K8 psd) using of basic local alignment search tool (BLAST) from the NCBI found 2 potential isoforms of 53.7 and 47.8.6 kDa with a query cover of 95% and sequence homology of 96% (B). mRNA level from HNSCC cases was analyzed for the variant 2 (C) and the K8 psd (D): expression was correlated and was associated with poor prognostic factors as dedifferentiated grade (n = 483) (P < 0.001 for the K8 psd and P < 0.001for the variant 2), pathological lymph node involvement (n = 409) (P = 0.037 for the K8 psd and P = 0.074 for the variant 2) and lymphovascular invasion (n = 341) (P < 0.001for the K8 psd and P < 0.001for the variant 2). Overall survival was decreased in high expression group (P = 0.012 for the K8 psd and P = 0.029 for the variant 2).
These results demonstrate the association of the mainly expressed K8 mRNA variants (variant 2 and K8-psd) with invasiveness and the poor prognostic factors of HNSCC tumors. They suggest the role of K8 and eK8 through the expression of 54 and 47 kDa isoforms, in HNSCC tumor progression and invasion.
KRT8 gene and K8 protein overexpression are associated with invasiveness features and poor prognosis in HNSCC
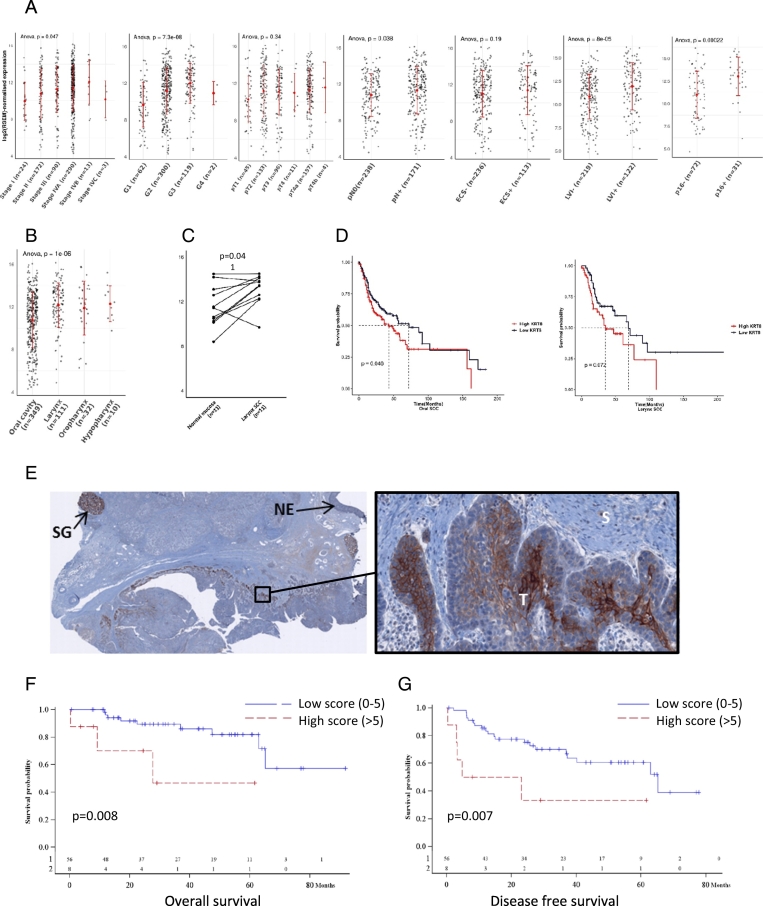
We then expanded the statistical analysis to KRT8 gene expression and its association with clinical prognostic factors in the TCGA data set (n = 502 patients with HNSCC). The group with higher K8 mRNA expression was associated with advanced pathological stages of the disease (P = 0.047), poor differentiation (P < 0.001), and p16 positive tumors (P < 0.001). Histological analyses reflecting the invasiveness of the disease such as the lymph node involvement (P = 0.038) or the lymphovascular invasion (P < 0.001) were also significantly associated with higher expression (Fig. 5A).
Fig. 5.
Overexpression of KRT8 gene and K8 protein is associated with poor histological prognostic factors, decreased survival and K8 is accumulated at the tumor invasion front of HNSCC.
KRT8 gene expression level was obtained for 502 HNSCC tumor samples from TCGA (A). Higher expression was observed for greater pathological stage (P = 0.047), squamous dedifferentiation (P<0,001), greater pathological primary tumor (P = 0.336), pathological lymph node involvement (p = 0.038), lymph node extra capsular spread (P = 0.201), lymphovascular invasion (P < 0,001) and for positive P16 status, (P < 0,001). Various expression pattern of KRT8 gene was observed according to anatomical subsite of HNSCC (P < 0.001) (B). Laryngeal tumor tissue had higher level of mRNA compared to normal laryngeal mucosa (P = 0.041) (C). Overall survival comparison in oral (P = 0.046) and larynx SCC (P = 0.072) underscored the altered OS in group with higher level of mRNA (D).
IHC was performed using M20 K8 antibody staining in 64 cases of HNSCC. A K8 protein higher staining in invasion front of the tumor was observed (E). Survival curve comparison between high and low K8 staining from the 64 cases of HNSCC showed altered OS (P = 0.008) (F) and DFS (P = 0.007) (G) in group with higher expression level (score > 5).
Legends: Grade I: well differentiated; Grade II: moderately differentiated; Grade III: poorly differentiated; Grade IV: anaplastic tumors pT, primary tumor; pN0, negative lymph node involvement; pN+, positive lymph node involvement; ECS, lymph node extracapsular spread; LVI, lymphovascular invasion; SCC, squamous cell carcinoma; SG: salivary gland; NE: normal epithelia; S: Stroma; T: Tumor.
We then focused on the anatomical subsites of the head and neck area and observed that K8 mRNA expression was heterogeneous, with a higher level in larynx and oropharyngeal SCC (P < 0.001) (Fig. 5B). Oral and larynx SCC with metastatic lymph node involvement and lymphovascular invasion had higher K8 mRNA expression levels. While KRT8 gene expression was significantly associated with these two histological criteria, no difference was observed in terms of extra capsular spread (Suppl Table 1). We notably observed a significant increase of K8 mRNA in tumors compared to the normal mucosa in larynx SCC (P = 0.041) (Fig. 5C). We also observed a decreased OS in the group with the highest level of K8 mRNA in oral (P = 0.046) and larynx SCC (P = 0.072) (Fig. 5D). Indeed, high KRT8 gene expression was associated with poor survival in patients with oral SCC, while no significant association was observed in patients with laryngeal cancer. Therefore, KRT8 gene expression was significantly associated with the local and the regional spread of the disease in both oral and larynx SCC with poor survival outcomes in oral SCC particularly.
To reinforce these findings, we then investigated the level of K8 protein in tumor samples and its potential association with similar histological factors and patient survival. IHC was performed to determine the accumulation of K8 in tumor specimens as well as its subcellular localization. Using M20 antibody staining in 64 cases of HNSCC, K8 protein was observed in all subsites of HNSCC. Positive staining was observed with a higher proportion for larynx and oropharyngeal SCC with 54% (13 out of 24) and 75% (3 out of 4) of the cases, respectively. A total of 19.4% oral SCC (7 out of 36) showed positive staining. Eight of 20 cases (40%) with metastatic lymph nodes showed positive staining. As expected, normal stratified epithelium did not express K8 as oral leukoplakia contrary to the normal salivary gland (SG) expressing K8 with intense staining (glandular epithelium) and considered as a positive control.
Various patterns of expression were observed in tumor specimens: oral SCC had low membrane and cytosolic K8 accumulation whereas cytosol and membrane staining was higher in laryngeal tumors or involved lymph nodes. In case of positive staining, K8 protein was preferentially observed in the deep part of the HNSCC tumor invasive front as presented in Fig. 5E. Higher magnification showed heterogeneous expression of K8 in cells present in the front of invasion with staining in membrane or cytosol compartment or both.
We performed a statistical analysis in order to evaluate the association between the high expression of K8 in the different subcellular compartment and the clinicopathological factors as well as the survival rates. The isolated membrane staining (>10%) and the overall K8 expression (membrane and cytosol) (score > 5) were both studied. The results are detailed in Table 1. We observed similar results for the two expression patterns with significant and higher expression in tumors associated with: age higher than 60 years, advanced stages III and IV, site of origin with higher expression for positive staining laryngeal SCC, pathological primary tumor with higher staining in pT3 and pT4 tumors, positive pathological lymph node involvement, and a trend for differentiation grade with a higher level in moderately differentiated tumors. In addition, OS (P = 0.008) (Fig. 5F) and progression free survival (PFS) (P = 0.007) (Fig. 5G) were both associated with higher overall K8 expression in HNSCC tumors. Decreased OS was also observed in groups with stronger membrane staining (P = 0.033).
Table 1.
Overexpression of K8 protein is associated with clinico-pathological prognostic factors in HNSCC.
| Clinico-pathological data | Overall K8 expression(Score > 5) P-value | Membrane K8 expression(> 10%) P-value |
|---|---|---|
| Age > 60 Y.O | 0.017 | 0.035 |
| Sex | 0.417 | 0.152 |
| Larynx vs other locations | <0.001 | <0.001 |
| Moderately vs Well differentiated | 0.069 | 0.039 |
| p stage III/IV vs I/II | 0.026 | 0.043 |
| pT3/T4 vs pT1/T2 | 0.005 | 0.006 |
| pN + | 0.009 | 0.008 |
| LN extracapsular spread | 0.613 | 1.000 |
Abbreviation: HNSCC, head and neck squamous cell carcinoma; SCC, squamous cell carcinoma; p stage, pathological stage; pT, pathological primary tumor; pN+, positive pathological lymph node involvement; LN, lymph node. Significative P-value is indicated in bold.
Altogether these results show that mRNA and IHC analyses underscored the prognostic impact of K8 expression in HNSCC, particularly in oral and laryngeal SCC. Overexpression of K8 mRNA and protein (membrane and overall K8) were both significantly associated with poor prognostic histological factors like invasiveness and decreased survival. We also observed an incremental expression in tumor samples with higher accumulation of K8 protein at the invasion front of the tumor. All these results reinforce the role of K8 and its membrane form in tumor invasion.
Discussion
Herein we report the presence of K8 at the external surface of the cell membrane of HNSCC cells, and the accumulation of a 54 kDa phosphorylated form of K8 associated to membranes. We show the preferential accumulation of two coding transcripts of K8, named variant 2 and K8-psd, in HNSCC tumors, and suggest that variant 2 could encode for the phosphorylated form of K8 associated to membranes. We then demonstrate that eK8, that was previously described by others and us in other malignancies [9,25,26] plays an important role in plasminogen-induced invasion in HNSCC cells. Finally, we describe the association between K8 and negative prognostic variables, which provides key insights on the role of K8 in tumor invasion. All together, these results suggest that eK8 may represent a relevant target in patients with HNSCC.
In order to evaluate the association of K8 expression with disease aggressiveness, we used a comprehensive approach including the analysis of both mRNA expression at the gene and isoforms levels (TCGA) and protein expression using IHC. For this, we determined the association between KRT8 gene and mRNA isoforms expression and clinical and pathological factors which have not been investigated so far in the literature. This analysis revealed an association between KRT8 gene expression, along with the two most expressed K8 transcripts, variant 2 and K8-psd, and disease stage. Consistently, tumors with lymphovascular invasion and lymph node involvement had higher levels of K8 transcripts, variant 2 and K8-psd expression levels. These observations and the impact of KRT8 gene expression on survival, support the involvement of K8 in tumor aggressiveness. In addition, these observations strongly support the existence of multiple mRNA variants coding for different K8 isofoms that play crucial roles in HNSCC cancer biology.
Impact of KRT8 gene expression on survival was different depending on the anatomical subsites considered and was significantly associated with oral cavity but not with larynx. While SCC from different anatomical subsites of the head and neck share similar pathological and molecular characteristics, they differ on several aspects. Larynx SCC is strongly associated with smoking history, while alcohol and smoking histories are frequently associated in patients with oral SCC [27,28] Therapeutically, oral cancer is commonly considered to be resistant to chemotherapy while neoadjuvant chemotherapy is often offered to patients with laryngeal SCC for organ-preservation strategies [29,30] Using gene expression profiles, TCGA has validated 4 subtypes: atypical, classical, mesenchymal and basal. Larynx SCC were more frequently classified as atypical and classical, while oral cavity SCC were more frequently classified as mesenchymal and basal [31] These subtle differences outline that the underlying biology of SCC arising in larynx and oral cavity might be difference, and explain why the impact of KRT8 gene expression was observed in SCC developing in the oral cavity.
These results obtained at the RNA level were confirmed at the protein level by IHC. In line with our transcriptomic analysis, K8 protein overexpression was also associated with poor prognostic factors and outcomes. High K8 protein expression, both in total cell and membrane fractions, were associated with advanced disease stage, tumor size, lymph node involvement, and decreased OS and DFS. Together, these results confirmed K8 and its membrane form as biomarkers for poor prognosis of HNSCC tumors. While few laboratories had previously characterized the overexpression of K8 protein as a marker of poor prognostic, they did not report on the specific pattern of expression at the plasma membrane [15,32]. Interestingly, the expression of K8 protein has been reported in oral cavity samples during embryogenesis [33,34], and in immature basal cells of normal head and neck epithelium [11,12]. Consequently, K8 seems to be a relevant marker of epithelial cell immaturity and a tumor-associated antigen, consistent with an increased expression we observed in poorly differentiated HNSCC. Andratschke et al. [16] detected high expression of K8 and its membrane form in the dedifferentiated larynx tissues of HNSCC. Here, IHC analysis mainly revealed membrane and cytosol staining in the deep part of the HNSCC tumor, which is in contact with the extra cellular matrix (ECM). Indeed, a gradient of K8 staining with higher expression at the invasive front of the tumor was observed in the majority of tumor samples. These observations strongly support the involvement of K8 in the late stage of invasion of established HNSCC, as well as a potential implication of its membrane form, eK8. Conversely, K8 expression was not observed in normal mucosa [11,12] or in oral leukoplakia, a premalignant condition. Despite a small set of leukoplakia data, this suggests that K8 is likely not involved in early stage of tumorigenesis.
Our work also sheds some light on the complex expression pattern of the KRT8 gene that takes place in HNSCC and that could lead to the generation of eK8 in this particular cancer. Among the different hypothesis that could explain the ectopic localization of eK8 to the plasma membrane, we considered the possible involvement of the specific alternative splicing transcripts expression. Through TCGA data mining, we identified two transcripts overexpressed in HNSCC tumors, variant 2 and K8-psd. We demonstrated by in silico translation that one of them, variant 2 could encode for the 54 kDa K8 protein isoform. Furthermore, we characterized for the first time the subcellular accumulation of K8 protein isoforms in HNSCC cells by western-blot analysis. We demonstrated that 54 kDa K8 isoform, in addition to its expected localization in cytoskeleton and cytosol fractions, was accumulated in membrane fraction suggesting that it could correspond to eK8. Confirmation of the relationship between K8 mRNA variant 2 and membrane associated K8 protein isoform would require additional investigations.
Post-translational modifications such as phosphorylation could also be at stake in the externalization of K8. The finding of the presence of K8 phosphorylated on serine 73 at cell surface of HNSCC cells sustains this hypothesis [26]. Here, we demonstrated that the 54 kDa membrane form of K8 is phosphorylated on serine 431. Indeed, phosphorylation/de-phosphorylation of cytokeratin proteins is responsible for an increase in their soluble forms by triggering a constant turnover between polymerized/depolymerized forms. The p38 mitogen-activated protein kinases (p38MAPK) and protein kinase C epsilon type (PRKCE) are well-characterized enzymes [35,36], responsible for the phosphorylation of K8 on Serine 73 or Serine 431 [14,15,35,37]. A modulation of phosphorylation of these two particular residues has been shown to lead to poor prognosis in HNSCC and colorectal cancer [14,15,37]. In OSCC particularly, an association between the dephosphorylation of K8 and advanced stages, or tumor progression has been reported [15]. However, in their study, the authors do not evaluate eK8 form and concentrate their analysis to the cytoplasmic form of K8. Consequently, post-translational modifications like phosphorylation probably play a key role in the K8 externalization process but this remains to be determined.
After confirming the presence of eK8 at the cell surface of HNSCC cell lines, we tested its role in tumor invasion modulated by Plg. We performed an experiment based on the competing binding of Plg between eK8 and the Fab fragment of D-A10 mAb. By removing the Fc domain of the antibody that contains a C-terminal lysine, illegitimate fixation of Plg on this residue is avoided allowing the potential fixation of Plg on the C-terminal lysine of eK8. In these conditions, we demonstrated inhibition of Plg-induced invasion when the Fab fragment of D-A10 mAb was added. In addition, we used IF on non-permeabilized cells to demonstrate the co-localization of Plg with K8, and also observed Plg and uPA at the cell surface. Together, these results strongly suggest that eK8 modulates the invasion ability of HNSCC cells via the Plg-uPA system activated through interaction with eK8.
These results strengthen the role of eK8 as a membrane plasminogen system activator [7,9,25,38] and demonstrate its involvement in the tumor invasion process, making eK8 an important element in HNSCC tumor progression.
Conclusion
In conclusion, externalization of eK8 in HNSCC cell lines, its involvement in plasminogen-dependent invasion, and the association of K8 overexpression with tumor invasion and poor prognostic factors make eK8 a marker of tumor progression and also a promising therapeutic target deserving future development of specific eK8 targeting-antibodies. Further investigations including a larger cohort as well as in vivo experiments will be required to validate these encouraging results. Indeed, if validated, the ability to modulate the invasion behavior of cells expressing eK8 will be a hopeful and promising finding that may provide new therapeutic options for patients with HNSCC.
The following are the supplementary data related to this article.
Expression of KRT8 gene is associated with clinico-pathological prognostic factors in oral and larynx SCC. Significative P-value is indicated in bold.
Variant 2, the K8-psd and the non-coding variant 4 K8 mRNA are the three isoforms overexpressed in HNSCC tumor and healthy tissues.
The mRNA level of the K8 variants was extracted from 502 cases of HNSCC and 44 available normal tissues (TCGA data portal). Despite no significative difference in mean expression of each K8 mRNA isoform, comparison between tumor and paired (B) or not (A) healthy tissues highlight a higher expression of variant 2, the K8 pseudogene and the non-coding variant 4 in these two tissues.
Grant support
This work was supported by the Cancéropôle Lyon Auvergne Rhône-Alpes (CLARA) 2014–2016 – Programme structurant (N°CVPPRCAN000153-International Head and Neck Prevention Act-IHNPACT), France, the Integrated Cancer Research Site LYriCAN (INCa-DGOS-Inserm_12563), France and a 2014–2015 clinical fellowship award from CHRU of Tours, France.
CRediT authorship contribution statement
Marie Alexandra Albaret: Conceptualization, Methodology, Formal analysis, Writing-Original Draft Preparation, Writing-Review & Editing, Visualization, Supervision, Project Administration Arnaud Paré: Investigation, Validation, Writing-Original Draft Preparation Lucie Malet: Investigation, Validation, Geneviève De Souza: Investigation, Validation Emilie Lavergne: Investigation, Validation, Formal analysis Dominique Goga: Investigation, Validation Gonzague De Pinieux: Investigation, Validation Claire Castellier: Investigation, Validation Aurélie Swalduz: Investigation, Validation Vivian Robin: Investigation, Validation, Formal analysis Vincent Lavergne: Investigation, Validation, Formal analysis, software Hichem-Claude Mertani: Investigation, Validation, Formal analysis Isabelle Treilleux: Investigation, Validation Claudine Vermot-Desroches: Formal analysis Jean Jacques Diaz: Conceptualization, Methodology, Formal analysis, Resources, Writing-Review & Editing, Visualization, Supervision, Funding Acquisition Pierre Saintigny: Conceptualization, Methodology, Formal analysis, Resources, Writing-Review & Editing, Visualization, Supervision, Funding Acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Jean-Jacques Diaz, Email: jean-jacques.diaz@lyon.unicancer.fr.
Pierre Saintigny, Email: pierre.saintigny@lyon.unicancer.fr.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Marur S., Forastiere A.A. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2008;83:489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]
- 3.Fitzmaurice C., Dicker D., Pain A., Hamavid H., Moradi-Lakeh M., MacIntyre M.F. The global burden of cancer 2013. JAMA Oncol. 2015;1:505. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrino F., Gatta G. Variation in survival of patients with head and neck cancer in Europe by the site of origin of the tumours. EUROCARE Working Group. Eur J Cancer Oxf Engl 1990. 1998;34:2154–2161. doi: 10.1016/s0959-8049(98)00328-1. [DOI] [PubMed] [Google Scholar]
- 5.Lin S.S., Massa S.T., Varvares M.A. Improved overall survival and mortality in head and neck cancer with adjuvant concurrent chemoradiotherapy in national databases: improved overall survival with adjuvant chemoradiotherapy. Head Neck. 2016;38:208–215. doi: 10.1002/hed.23869. [DOI] [PubMed] [Google Scholar]
- 6.Weidle U.H., Maisel D., Klostermann S., Schiller C., Weiss E.H. Intracellular proteins displayed on the surface of tumor cells as targets for therapeutic intervention with antibody-related agents. Cancer Genomics Proteomics. 2011;8:49–63. [PubMed] [Google Scholar]
- 7.Plow E.F., Das R. Enolase-1 as a plasminogen receptor. Blood. 2009;113:5371–5372. doi: 10.1182/blood-2009-03-208546. [DOI] [PubMed] [Google Scholar]
- 8.Albaret M.A., Vermot-Desroches C., Paré A., Roca-Martinez J.-X., Malet L., Esseily J. Externalized keratin 8: a target at the interface of microenvironment and intracellular signaling in colorectal cancer cells. Cancers. 2018;10 doi: 10.3390/cancers10110452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hembrough T.A., Li L., Gonias S.L. Cell-surface cytokeratin 8 is the major plasminogen receptor on breast cancer cells and is required for the accelerated activation of cell-associated plasminogen by tissue-type plasminogen activator. J. Biol. Chem. 1996;271:25684–25691. doi: 10.1074/jbc.271.41.25684. [DOI] [PubMed] [Google Scholar]
- 10.Barrett A.W., Selvarajah S., Franey S., Wills K.A., Berkovitz B.K. Interspecies variations in oral epithelial cytokeratin expression. J. Anat. 1998;193(Pt 2):185–193. doi: 10.1046/j.1469-7580.1998.19320185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gires O., Mack B., Rauch J., Matthias C. CK8 correlates with malignancy in leukoplakia and carcinomas of the head and neck. Biochem. Biophys. Res. Commun. 2006;343:252–259. doi: 10.1016/j.bbrc.2006.02.139. [DOI] [PubMed] [Google Scholar]
- 12.Matthias C., Mack B., Berghaus A., Gires O. Keratin 8 expression in head and neck epithelia. BMC Cancer. 2008;8:267. doi: 10.1186/1471-2407-8-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukunaga Y., Bandoh S., Fujita J., Yang Y., Ueda Y., Hojo S. Expression of cytokeratin 8 in lung cancer cell lines and measurement of serum cytokeratin 8 in lung cancer patients. Lung Cancer Amst Neth. 2002;38:31–38. doi: 10.1016/s0169-5002(02)00109-5. [DOI] [PubMed] [Google Scholar]
- 14.Mizuuchi E., Semba S., Kodama Y., Yokozaki H. Down-modulation of keratin 8 phosphorylation levels by PRL-3 contributes to colorectal carcinoma progression. Int. J. Cancer. 2009;124:1802–1810. doi: 10.1002/ijc.24111. [DOI] [PubMed] [Google Scholar]
- 15.Alam H., Gangadaran P., Bhate A.V., Chaukar D.A., Sawant S.S., Tiwari R. Loss of keratin 8 phosphorylation leads to increased tumor progression and correlates with clinico-pathological parameters of OSCC patients. Oshima R, editor. PLoS ONE. 2011;6:e27767. doi: 10.1371/journal.pone.0027767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andratschke M., Hagedorn H., Nerlich A. Expression of the epithelial cell adhesion molecule and cytokeratin 8 in head and neck squamous cell cancer: a comparative study. Anticancer Res. 2015;35:3953–3960. [PubMed] [Google Scholar]
- 17.van der Velden L.A., Schaafsma H.E., Manni J.J., Ruiter D.J., Ramaekers F.C., Kuijpers W. Cytokeratin and vimentin expression in normal epithelium and squamous cell carcinomas of the larynx. Eur Arch Oto-Rhino-Laryngol Off J Eur Fed Oto-Rhino-Laryngol Soc EUFOS Affil Ger Soc Oto-Rhino-Laryngol - Head Neck Surg. 1997;254:376–383. doi: 10.1007/BF01642554. [DOI] [PubMed] [Google Scholar]
- 18.Zhao M., Sano D., Pickering C.R., Jasser S.A., Henderson Y.C., Clayman G.L. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin. Cancer Res. 2011;17:7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allred D.C., Bustamante M.A., Daniel C.O., Gaskill H.V., Cruz A.B. Immunocytochemical analysis of estrogen receptors in human breast carcinomas. Evaluation of 130 cases and review of the literature regarding concordance with biochemical assay and clinical relevance. Arch Surg Chic Ill 1960. 1990;125:107–113. doi: 10.1001/archsurg.1990.01410130113018. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.The results shown here are in whole or part based upon data generated by the TCGA Research Network. https://www.cancer.gov/tcga
- 22.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hembrough T.A., Vasudevan J., Allietta M.M., Glass W.F., Gonias S.L. A cytokeratin 8-like protein with plasminogen-binding activity is present on the external surfaces of hepatocytes, HepG2 cells and breast carcinoma cell lines. J. Cell Sci. 1995;108(Pt 3):1071–1082. doi: 10.1242/jcs.108.3.1071. [DOI] [PubMed] [Google Scholar]
- 26.Gires O., Andratschke M., Schmitt B., Mack B., Schaffrik M. Cytokeratin 8 associates with the external leaflet of plasma membranes in tumour cells. Biochem. Biophys. Res. Commun. 2005;328:1154–1162. doi: 10.1016/j.bbrc.2005.01.074. [DOI] [PubMed] [Google Scholar]
- 27.Turati F., Garavello W., Tramacere I., Bagnardi V., Rota M., Scotti L. A meta-analysis of alcohol drinking and oral and pharyngeal cancers. Part 2: results by subsites. Oral Oncol. 2010;46:720–726. doi: 10.1016/j.oraloncology.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Chang C.-P., Chang S.-C., Chuang S.-C., Berthiller J., Ferro G., Matsuo K. Age at start of using tobacco on the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium (INHANCE) Cancer Epidemiol. 2019;63:101615. doi: 10.1016/j.canep.2019.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonomi M.R., Blakaj A., Blakaj D. Organ preservation for advanced larynx cancer: a review of chemotherapy and radiation combination strategies. Oral Oncol. 2018;86:301–306. doi: 10.1016/j.oraloncology.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Dhar H., Vaish R., D'Cruz A.K. Management of locally advanced oral cancers. Oral Oncol. 2020;105:104662. doi: 10.1016/j.oraloncology.2020.104662. [DOI] [PubMed] [Google Scholar]
- 31.The Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillies T., Werkmeister R., Packeisen J., Brandt B., Morin P., Weingart D. Cytokeratin 8/18 expression indicates a poor prognosis in squamous cell carcinomas of the oral cavity. BMC Cancer. 2006;6:10. doi: 10.1186/1471-2407-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krauss S., Franke W.W. Organization and sequence of the human gene encoding cytokeratin 8. Gene. 1990;86:241–249. doi: 10.1016/0378-1119(90)90285-y. [DOI] [PubMed] [Google Scholar]
- 34.Mbiene J.-P., Roberts J.D. Distribution of keratin 8-containing cell clusters in mouse embryonic tongue: Evidence for a prepattern for taste bud development. J. Comp. Neurol. 2003;457:111–122. doi: 10.1002/cne.10551. [DOI] [PubMed] [Google Scholar]
- 35.Ku N.-O., Azhar S., Omary M.B. Keratin 8 phosphorylation by p38 kinase regulates cellular keratin filament reorganization: modulation by a keratin 1-like disease-causing mutation. J. Biol. Chem. 2002;277:10775–10782. doi: 10.1074/jbc.M107623200. [DOI] [PubMed] [Google Scholar]
- 36.Windoffer R., Beil M., Magin T.M., Leube R.E. Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J. Cell Biol. 2011;194:669–678. doi: 10.1083/jcb.201008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khapare N., Kundu S.T., Sehgal L., Sawant M., Priya R., Gosavi P. Plakophilin3 loss leads to an increase in PRL3 levels promoting K8 dephosphorylation, which is required for transformation and metastasis. Gottardi C, editor. PLoS ONE. 2012;7:e38561. doi: 10.1371/journal.pone.0038561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ceruti P., Principe M., Capello M., Cappello P., Novelli F. Three are better than one: plasminogen receptors as cancer theranostic targets. Exp Hematol Oncol. 2013;2:12. doi: 10.1186/2162-3619-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of KRT8 gene is associated with clinico-pathological prognostic factors in oral and larynx SCC. Significative P-value is indicated in bold.
Variant 2, the K8-psd and the non-coding variant 4 K8 mRNA are the three isoforms overexpressed in HNSCC tumor and healthy tissues.
The mRNA level of the K8 variants was extracted from 502 cases of HNSCC and 44 available normal tissues (TCGA data portal). Despite no significative difference in mean expression of each K8 mRNA isoform, comparison between tumor and paired (B) or not (A) healthy tissues highlight a higher expression of variant 2, the K8 pseudogene and the non-coding variant 4 in these two tissues.