Abstract
Tubular 3D liver tissue with enhanced capillary‐like structures branching from a large main channel is potentially useful for drug discovery because the perfusable main channel and capillary‐like structures enable mass transfer into and out from the tissue. Tubular liver tissue is comprised of the hepatocellular carcinoma cell line HepG2, human umbilical vein endothelial cells (HUVECs), and mesenchymal stem cells (MSCs), using a perfusion device functioning as the interface for an external pump. This study aimed to compare the expression of genes involved in drug metabolism between 2D‐cultured hepatocellular carcinoma cells and 3D‐cultured tubular liver tissue. Gene expression profiles of 2D‐cultured cells and tubular liver tissue were compared using RNA sequencing. Multidimensional scaling analysis revealed that culture dimensionality had a more prominent effect on gene expression profiles than perfusion conditions. More specifically, genes involved in drug metabolism such as CYP2D6, CYP2E1, NNMT, and SLC28A1 were slightly upregulated in the 3D cultures, while certain genes such as ALDH1B1, ALDH1A2, and SULT1E1 were downregulated. These results indicate that gene expression profiles are largely influenced by culture dimensionality and are potentially useful to researchers intending to switch from 2D culture to 3D culture of hepatocellular carcinoma or other tissue types.
Keywords: blood vessels, gene expression profile, hepatocellular carcinoma, liver, organoids, RNA‐seq, transcriptome
We compared drug‐metabolizing gene expression between 2D‐cultured hepatocellular carcinoma cells and 3D‐cultured perfusable liver tissue comprising hepatocellular carcinoma cells (HepG2), human umbilical vein endothelial cells, and mesenchymal stem cells by RNA sequencing. The results indicate that gene expression profiles are largely influenced by culture dimensionality and are potentially applicable to researchers intending to switch from 2D culturing to 3D culturing of hepatocellular carcinoma.
Abbreviations
- DEG
differentially expressed gene
- FC
fold change
- FDR
false discovery rate
- HUVEC
human umbilical vein endothelial cell
- MSC
mesenchymal stem cell
Various 3D liver‐like cultures comprising hepatocellular carcinoma cells, such as spheroids [1, 2], cell‐laden hydrogel [3, 4], and organoids [5, 6], have been developed. Drug discovery studies have increasingly focused on 3D liver‐like tissue cultures using cell culture plates owing to their various advantages over conventional 2D cultures of hepatocellular carcinoma cells, including increased cellular functions such as albumin secretion and xenobiotic metabolism and increased sensitivity to hepatotoxic compounds. To compensate for the lack of a perfusable vascular network in 3D liver‐like tissues, we previously developed a tubular 3D liver‐like culture, called tubular liver tissue, with enhanced capillary‐like structures branching from a large main channel [7], upon combining a perfusion device [8, 9] with a collagen gel populated with the hepatocellular carcinoma cell line HepG2, human umbilical vein endothelial cells (HUVECs), and mesenchymal stem cells (MSCs). The perfusable main channel and capillary‐like structures facilitate the mass transfer of not only oxygen and nutrients but also test substances and their metabolites both into and from the tissue, which can then be sampled, thereby facilitating their applications in drug discovery studies. However, differences in drug‐metabolizing gene expression between tubular liver tissue and the conventional 2D‐cultured HepG2 remain unknown. Herein, we constructed a tubular liver tissue and used RNA sequencing (RNA‐seq) to compare its gene expression profiles under perfused and nonperfused conditions, with that of 2D‐cultured HepG2 cells, HUVECs, and MSCs mixed at the same ratio as that used for generating the liver tissue (Fig. 1). We mainly focused on genes involved in drug metabolism, categorized as phase I, II, III, and nuclear receptor genes. Phase I enzymes are involved in oxidation, reduction, and hydrolysis of xenobiotics. Phase II enzymes are involved in conjugation reactions. Phase III enzymes are usually related to transportation of xenobiotics [10]. Since drug metabolism in vivo mainly involves these three phases, research in the field of drug discovery largely targets them. In addition, a set of nuclear receptors are also investigated because it is known that they control hepatic metabolism and hepatotoxicity [11]. Our results will potentially benefit researchers intending to switch from 2D‐cultured hepatocellular carcinoma cells or other 3D liver‐like tissues to tubular liver tissue.
Fig. 1.
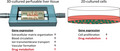
Schematic representation of the experimental design. (A) In the perfused group, the tubular liver tissue was cultured under perfusion with medium, using a tube pump. Total RNA was extracted from tissue detached from the device and used for RNA‐seq. (B) In the nonperfused group, the tubular liver tissue was submerged in the medium and statically cultured. Total RNA was extracted from the tissue detached from the device and subjected to RNA‐seq. (C) In the 2D‐cultured group, hepatocellular carcinoma cell line HepG2, HUVECs, and MSCs were cultured in cell culture dishes. The cells were dissociated, mixed at the same ratio as tubular liver tissues, and used for RNA‐seq after total RNA extraction.
Materials and methods
Construction of the tubular liver tissue
Tubular liver tissue was constructed using a perfusion device, as previously described (Fig. 1A,B) [7]. Briefly, the device was filled with a collagen matrix (IAC‐50; Koken Co., Tokyo, Japan) populated with HepG2 cells (3 × 107 cells/mL), HUVECs (3 × 107 cells/mL), and MSCs (0.5 × 107 cells/mL). The ratio of each cell type was determined based on the literature [5, 12]. The needle previously set in the device was extracted. Then, HUVECs were infused into the resultant tunnel to form the main channel. Subsequently, we connected the device to an external pump for perfusion (Fig. 1A) and immersed the device in culture medium for nonperfusion (Fig. 1B). The culture medium was composed of 1 : 1 mix of HepG2 and HUVEC media, which were individually used for the culture of cell populations in 2D conditions.
2D cell culture
HepG2 cells, HUVECs, and MSCs were independently cultured for 24 h. HepG2 cells were cultured in cell culture dishes (VTC‐D150; AS ONE Corporation, Osaka, Japan) with low glucose Dulbecco's modified Eagle's medium (DMEM; FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) containing 10% FBS (Thermo Fisher Scientific Inc., Waltham, MA, USA) and 1% penicillin/streptomycin solution (FUJIFILM Wako Pure Chemical). HUVECs (PromoCell GmbH, Heidelberg, Germany) were cultured in gelatin‐coated cell culture dishes (FUJIFILM Wako Pure Chemical Corporation) containing endothelial cell growth medium 2 (PromoCell) supplemented with 1% penicillin/streptomycin solution (×100). MSCs (SCRC‐4000; ATCC, Manassas, VA, USA) were cultured in gelatin‐coated cell culture dishes containing high‐glucose DMEM (FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% FBS, 1% nonessential amino acids solution (×100) (FUJIFILM Wako Pure Chemical), and 1% penicillin/streptomycin solution (×100). All cells were cultured at 37 °C in a 5% CO2 atmosphere. The cells were dissociated with TrypLE Express (Thermo Fisher Scientific) 1 day after plating and mixed at the same ratio as the tubular liver tissue (3 : 3 : 0.5) (Fig. 1C) for extraction of RNA.
RNA‐seq analysis
Total RNA was extracted from 3D‐ and 2D‐cultured perfused and nonperfused cells using NucleoSpin RNA (Macherey Nagel GmbH & Co. KG, Duren, Germany) and sequenced using NovaSeq 6000 (Illumina Inc., San Diego, CA, USA) with biological duplicates. Read data were processed and analyzed using STAR (2.7.1a) [13], RSEM (1.3.1) [14], and edgeR (3.28.1) [15, 16] with the hg38 reference genome and gene annotation Ensembl GRCh38. The read count data were normalized using the trimmed mean of M values. Gene ontology (GO) enrichment analysis was performed using the “database for annotation, visualisation and integrated discovery” (DAVID) [17, 18] via RDAVIDWebService (1.20.0) [19] and executed on R [20]. The enriched GO terms (adjusted P‐value < 0.01) obtained from a category GOTERM_BP_FAT of DAVID were classified into 4 clusters by clustering analysis based on semantic similarity computation [21]. The semantic similarities between all the pairs of the enriched GO terms were calculated using GOSemSim [22]. Then, hierarchical clustering according to the calculated similarities was performed using the hclust function of R.
Results and Discussion
Culture dimensionality had a prominent effect on gene expression
To assess overall differences among the three groups, the data were analyzed through multidimensional scaling (Fig. 2A). As expected, data points were clustered in accordance with the culture conditions in a plane of the first two dimensions, indicating that variations of the samples in each group were small enough to evaluate differences among groups and that these conditions had robust effects on gene expression profiles. Thereafter, pairwise group comparisons were carried out (Fig. 2B); 792 (upregulated: 381, downregulated: 411), 5974 (upregulated: 3437, downregulated: 2537), and 6016 (upregulated: 3598, downregulated: 2418) differentially expressed genes [DEGs; |log2 FC| ≥ 1 and false discovery rate (FDR) < 0.05] were identified between perfused and nonperfused cells, perfused and 2D‐cultured cells, and nonperfused and 2D‐cultured cells, respectively, suggesting that culture dimensionality (i.e., tubular liver tissue vs 2D) has a more prominent effect on gene expression than perfusion, even though previous studies have reported that perfusion retains tissue viability and functions. This finding was further confirmed through clustering analysis. As shown in the heat map (Fig. 2C), the perfused and nonperfused groups were closely clustered, compared to 2D‐cultured groups. GO enrichment analysis was also performed to investigate the differences between the perfused and 2D‐cultured groups in detail (Tables 1 and 2). The enriched GO terms were classified by clustering analysis based on semantic similarity computation. Consequently, it was revealed that genes involved in extracellular matrix organization, blood circulation, ion transmembrane transport, and vascular formation were enriched in the perfused condition (Table 1), whereas those involved in cell proliferation (Table 2) were enriched in 2D culture condition. This observation indicates that the perfused tubular tissue is physiologically more relevant than the 2D‐cultured cells.
Fig. 2.
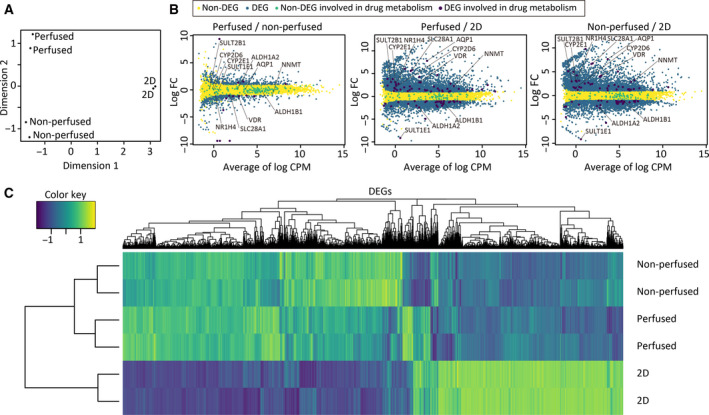
Analysis of gene expression profiles. (A) Multidimensional scaling plot of the perfused and nonperfused samples and 2D‐cultured samples. (B) Scatter plots of log2 FC values vs the average log2 CPM. CPM, counts per million. DEGs, |log2 FC| ≥ 1 and FDR < 0.05. (C) A heat map of the perfused and nonperfused samples, and the 2D‐cultured samples.
Table 1.
GO terms (biological process) enriched in the perfused tubular liver tissue compared with the 2D‐cultured cells. GO terms were classified into four clusters by clustering analysis based on semantic similarity computation, and the top three of each cluster are shown
| Cluster No. | GO ID | Description | Adjusted P‐value |
|---|---|---|---|
| 1 | GO:0007166 | Cell surface receptor signaling pathway | 1.63E‐16 |
| GO:0043062 | Extracellular structure organization | 8.05E‐14 | |
| GO:0030198 | Extracellular matrix organization | 8.05E‐14 | |
| 2 | GO:0003008 | System process | 3.68E‐16 |
| GO:0009605 | Response to external stimulus | 3.40E‐16 | |
| GO:0008015 | Blood circulation | 2.54E‐12 | |
| 3 | GO:0006811 | Ion transport | 4.55E‐14 |
| GO:0034220 | Ion transmembrane transport | 1.53E‐09 | |
| GO:0055085 | Transmembrane transport | 5.05E‐09 | |
| 4 | GO:0045595 | Regulation of cell differentiation | 3.49E‐13 |
| GO:2000026 | Regulation of multicellular organismal development | 1.13E‐11 | |
| GO:0001944 | Vasculature development | 6.01E‐09 |
Table 2.
GO terms (biological process) enriched in the 2D‐cultured cells compared with the perfused tubular liver tissue. GO terms were classified into four clusters by clustering analysis based on semantic similarity computation, and the top three of each cluster are shown
| Cluster No. | GO ID | Description | Adjusted P‐value |
|---|---|---|---|
| 1 | GO:1903047 | Mitotic cell cycle process | 1.43E‐39 |
| GO:0000278 | Mitotic cell cycle | 2.14E‐38 | |
| GO:0022402 | Cell cycle process | 1.01E‐35 | |
| 2 | GO:0006259 | DNA metabolic process | 5.12E‐29 |
| GO:0006260 | DNA replication | 3.20E‐28 | |
| GO:0034660 | ncRNA metabolic process | 8.64E‐28 | |
| 3 | GO:0000280 | Nuclear division | 5.51E‐25 |
| GO:0051276 | Chromosome organization | 2.32E‐22 | |
| GO:0048285 | Organelle fission | 2.37E‐22 | |
| 4 | GO:0032543 | Mitochondrial translation | 4.37E‐18 |
| GO:0070125 | Mitochondrial translational elongation | 5.78E‐15 | |
| GO:0006415 | Translational termination | 2.84E‐14 |
Drug‐metabolizing genes were basically upregulated in the tubular liver tissue
We evaluated the expression levels of drug‐metabolizing genes classified into phase I, phase II, and phase III, and nuclear receptor genes (Figs 2B and 3, Table 3, 4, 5, 6) as previously described [2, 10, 11]. For the phase I genes, mean log2 fold change (FC) values were 0.4 and 0.5 in the perfused and the nonperfused groups (Fig. 3A, Table 3), respectively, indicating that the phase I genes, particularly CYP2D6 and CYP2E1, in both groups were slightly upregulated on an average; CYP2D6 and CYP2E1 encoding cytochrome P450 members are involved in metabolism of 20% and 2% of known drugs, respectively [23]. Some phase I genes were downregulated, including aldehyde dehydrogenases such as ALDH1A2, which encodes a retinal dehydrogenase with retinaldehyde metabolizing potential but only very low activity with acetaldehyde and propanal, and ALDH1B1, which detoxifies alcohol‐derived acetaldehyde [24]. The downregulated genes might be related to cell proliferation since the 2D‐cultured cells were at a highly proliferative state as revealed by GO enrichment analysis (Table 4). For phase II genes, the trend was more neutral; mean log2 FC values were 0.0 for both perfused and the nonperfused groups (Fig. 3B, Table 4). These values may have been negatively biased owing to an extremely low log2 FC value (−9.1) of SULT1E1 which encodes a sulfotransferase catalyzing estradiol and estrone sulfation. Among the upregulated phase II genes, prominent drug‐metabolizing genes were identified, including SULT2B1 which catalyzes pregnenolone and dehydroepiandrosterone sulfation, and NNMT which encodes nicotinamide N‐methyltransferase involved in the biotransformation of numerous drugs and xenobiotic compounds [24]. Among phase III genes, mean log2 FC values were 0.5 both in perfused and in nonperfused groups (Fig. 3C, Table 5), wherein SLC28A1 and AQP1 were particularly upregulated. SLC28A1 encodes a sodium‐dependent and pyrimidine‐selective transporter involved in uridine, cytidine, thymidine, and nucleoside‐derived drug transport, whereas AQP1 encodes an aquaporin channel [24]. Among nuclear receptor genes, mean log2 FC values were 0.9 and 1.0 in the perfused and nonperfused groups, respectively, indicating higher expression levels than those of phase I–III genes (Fig. 3D, Table 6). In particular, VDR encoding the vitamin D receptor and NR1H4 encoding the bile acid receptor, both associated with hepatotoxicity and metabolism [11], were significantly upregulated. Together, drug‐metabolizing gene expression profiles were similar between perfused and nonperfused groups. Furthermore, these genes were basically upregulated in the tubular liver compared with 2D‐cultured cells, although it should be noted that some genes were downregulated and also that an interaction among different cell types was excluded in the 2D‐culture condition for simplicity. It is assumed that the upregulation of drug‐metabolizing genes was due to the physiologically relevant culture conditions, such as ECM, cell–cell interactions, and blood flow, of the tubular liver tissue (Fig. 4). The metabolism and hepatotoxicity of many drugs, such as midazolam, bufuralol, acetaminophen, and diclofenac, are assessed more accurately by using static 3D culture conditions (spheroid and cell‐laden ECM) rather than 2D conditions [2, 5]. Considering that the perfused tubular liver tissue is more viable (4.6‐fold RNA levels) and more feasible for drug injection into the tissue and metabolite collection than the nonperfused tissue (equivalent to static 3D culture), the perfusable tubular liver tissue might be a promising experimental model for drug discovery studies. Considering the utility of the perfusable tubular liver tissue, the difference with conventional 2D‐cultured cells should be considered.
Fig. 3.
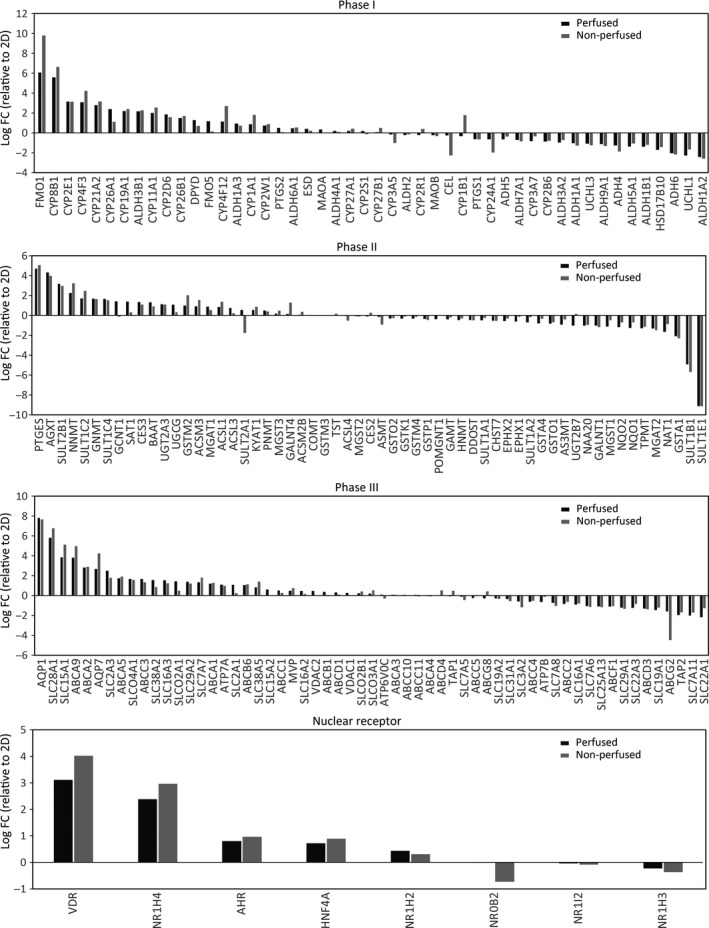
Graphs showing log2 FC values of genes associated with drug metabolism. (A) Phase I genes. (B) Phase II genes. (C) Phase III genes. (D) Nuclear receptor genes. The 2D‐cultured group; log2 FC = 0.
Table 3.
Phase I genes. Genes that were not detected by RNA‐seq or omitted due to extremely low counts during edgeR process are shown as blank.
| Ensembl ID | Symbol | Description | Log FC | Log CPM | FDR | ||
|---|---|---|---|---|---|---|---|
| Perfused/2D | Nonperfused/2D | Perfused/Nonperfused | |||||
| ENSG00000010932 | FMO1 | Flavin containing monooxygenase 1 | 6.07 | 9.79 | −3.73 | 1.10 | 2.87E‐05 |
| ENSG00000180432 | CYP8B1 | Cytochrome P450, family 8, subfamily B, polypeptide 1 | 5.58 | 6.63 | −1.05 | 1.77 | 3.59E‐06 |
| ENSG00000130649 | CYP2E1 | Cytochrome P450, family 2, subfamily E, polypeptide 1 | 3.15 | 3.13 | 0.02 | ‐0.02 | 6.09E‐04 |
| ENSG00000186529 | CYP4F3 | Cytochrome P450, family 4, subfamily F, polypeptide 3 | 3.08 | 4.23 | −1.15 | 2.92 | 4.67E‐06 |
| ENSG00000231852 | CYP21A2 | Cytochrome P450, family 21, subfamily A, polypeptide 2 | 2.78 | 3.16 | −0.38 | 0.36 | 1.96E‐04 |
| ENSG00000095596 | CYP26A1 | Cytochrome P450, family 26, subfamily A, polypeptide 1 | 2.38 | 1.12 | 1.26 | −0.89 | 2.14E‐02 |
| ENSG00000137869 | CYP19A1 | Cytochrome P450, family 19, subfamily A, polypeptide 1 | 2.20 | 2.38 | −0.18 | 1.67 | 3.37E‐05 |
| ENSG00000006534 | ALDH3B1 | Aldehyde dehydrogenase 3 family, member B1 | 2.16 | 2.26 | −0.10 | 4.45 | 6.43E‐07 |
| ENSG00000140459 | CYP11A1 | Cytochrome P450, family 11, subfamily A, polypeptide 1 | 1.99 | 2.54 | −0.55 | 1.01 | 2.56E‐04 |
| ENSG00000100197 | CYP2D6 | Cytochrome P450, family 2, subfamily D, polypeptide 6 | 1.86 | 1.57 | 0.29 | −0.12 | 1.67E‐02 |
| ENSG00000003137 | CYP26B1 | Cytochrome P450, family 26, subfamily B, polypeptide 1 | 1.50 | 1.70 | −0.21 | 0.59 | 5.72E‐03 |
| ENSG00000188641 | DPYD | Dihydropyrimidine dehydrogenase | 1.29 | 0.69 | 0.60 | 3.47 | 2.89E‐04 |
| ENSG00000131781 | FMO5 | Flavin containing monooxygenase 5 | 1.18 | 0.13 | 1.04 | −0.17 | 3.36E‐02 |
| ENSG00000186204 | CYP4F12 | Cytochrome P450, family 4, subfamily F, polypeptide 12 | 1.13 | 2.70 | −1.57 | 1.43 | 1.14E‐04 |
| ENSG00000184254 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 0.93 | 0.72 | 0.21 | 2.62 | 1.33E‐02 |
| ENSG00000140465 | CYP1A1 | Cytochrome P450, family 1, subfamily A, polypeptide 1 | 0.86 | 1.81 | −0.95 | 0.37 | 1.90E‐02 |
| ENSG00000073067 | CYP2W1 | Cytochrome P450, family 2, subfamily W, polypeptide 1 | 0.74 | 0.87 | −0.14 | 5.06 | 4.61E‐05 |
| ENSG00000073756 | PTGS2 | Prostaglandin‐endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 0.49 | 0.06 | 0.43 | 4.46 | 1.50E‐02 |
| ENSG00000119711 | ALDH6A1 | Aldehyde dehydrogenase 6 family, member A1 | 0.46 | 0.54 | −0.08 | 3.83 | 6.91E‐03 |
| ENSG00000139684 | ESD | Esterase D | 0.40 | 0.22 | 0.18 | 6.43 | 7.12E‐03 |
| ENSG00000189221 | MAOA | Monoamine oxidase A | 0.34 | 0.03 | 0.31 | 5.50 | 1.19E‐02 |
| ENSG00000159423 | ALDH4A1 | Aldehyde dehydrogenase 4 family, member A1 | 0.19 | 0.09 | 0.10 | 5.96 | 2.28E‐01 |
| ENSG00000135929 | CYP27A1 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | 0.19 | 0.42 | −0.23 | 5.10 | 1.45E‐02 |
| ENSG00000167600 | CYP2S1 | Cytochrome P450, family 2, subfamily S, polypeptide 1 | 0.18 | −0.13 | 0.31 | 4.62 | 2.59E‐01 |
| ENSG00000111012 | CYP27B1 | Cytochrome P450, family 27, subfamily B, polypeptide 1 | 0.07 | 0.51 | −0.44 | 0.24 | 4.59E‐01 |
| ENSG00000106258 | CYP3A5 | Cytochrome P450, family 3, subfamily A, polypeptide 5 | −0.16 | −1.03 | 0.87 | 2.92 | 6.08E‐04 |
| ENSG00000111275 | ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) | −0.22 | −0.14 | −0.08 | 5.98 | 6.16E‐02 |
| ENSG00000186104 | CYP2R1 | Cytochrome P450, family 2, subfamily R, polypeptide 1 | −0.22 | 0.39 | −0.61 | 0.43 | 3.86E‐01 |
| ENSG00000069535 | MAOB | Monoamine oxidase B | −0.24 | −0.33 | 0.10 | 4.23 | 6.00E‐02 |
| ENSG00000170835 | CEL | Carboxyl ester lipase (bile salt‐stimulated lipase) | −0.26 | −2.27 | 2.01 | −1.15 | 3.48E‐02 |
| ENSG00000138061 | CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | −0.32 | 1.78 | −2.10 | −0.28 | 2.64E‐02 |
| ENSG00000095303 | PTGS1 | Prostaglandin‐endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | −0.64 | −0.67 | 0.03 | 5.32 | 4.83E‐04 |
| ENSG00000019186 | CYP24A1 | Cytochrome P450, family 24, subfamily A, polypeptide 1 | −0.65 | −1.98 | 1.33 | 2.81 | 3.91E‐05 |
| ENSG00000197894 | ADH5 | Alcohol dehydrogenase 5 (class III), chi polypeptide | −0.65 | −0.37 | −0.28 | 6.86 | 9.12E‐05 |
| ENSG00000164904 | ALDH7A1 | Aldehyde dehydrogenase 7 family, member A1 | −0.71 | −0.86 | 0.15 | 5.99 | 3.09E‐05 |
| ENSG00000160870 | CYP3A7 | Cytochrome P450, family 3, subfamily A, polypeptide 7 | −0.83 | −0.34 | −0.49 | 0.12 | 2.38E‐01 |
| ENSG00000197408 | CYP2B6 | Cytochrome P450, family 2, subfamily B, polypeptide 6 | −0.87 | −0.79 | −0.08 | −0.77 | 2.42E‐01 |
| ENSG00000072210 | ALDH3A2 | Aldehyde dehydrogenase 3 family, member A2 | −0.97 | −0.72 | −0.25 | 5.54 | 8.87E‐06 |
| ENSG00000165092 | ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | −1.04 | −1.28 | 0.24 | 3.59 | 6.36E‐05 |
| ENSG00000118939 | UCHL3 | Ubiquitin carboxyl‐terminal esterase L3 (ubiquitin thiolesterase) | −1.08 | −1.25 | 0.17 | 4.84 | 8.72E‐07 |
| ENSG00000143149 | ALDH9A1 | Aldehyde dehydrogenase 9 family, member A1 | −1.12 | −1.31 | 0.19 | 5.06 | 8.58E‐07 |
| ENSG00000198099 | ADH4 | Alcohol dehydrogenase 4 (class II), pi polypeptide | −1.27 | −1.88 | 0.62 | 0.35 | 3.45E‐03 |
| ENSG00000112294 | ALDH5A1 | Aldehyde dehydrogenase 5 family, member A1 | −1.38 | −1.07 | −0.31 | 5.28 | 5.88E‐07 |
| ENSG00000137124 | ALDH1B1 | Aldehyde dehydrogenase 1 family, member B1 | −1.39 | −1.19 | −0.20 | 5.56 | 4.83E‐07 |
| ENSG00000072506 | HSD17B10 | Hydroxysteroid (17‐beta) dehydrogenase 10 | −1.71 | −1.44 | −0.27 | 5.50 | 7.48E‐08 |
| ENSG00000172955 | ADH6 | Alcohol dehydrogenase 6 (class V) | −2.04 | −2.18 | 0.14 | 2.41 | 2.59E‐05 |
| ENSG00000154277 | UCHL1 | Ubiquitin carboxyl‐terminal esterase L1 (ubiquitin thiolesterase) | −2.28 | −1.68 | −0.60 | 4.86 | 5.23E‐07 |
| ENSG00000128918 | ALDH1A2 | Aldehyde dehydrogenase 1 family, member A2 | −2.43 | −2.60 | 0.16 | 2.74 | 5.64E‐06 |
| ENSG00000114771 | AADAC | Arylacetamide deacetylase (esterase) | |||||
| ENSG00000187758 | ADH1A | Alcohol dehydrogenase 1A (class I), alpha polypeptide | |||||
| ENSG00000196616 | ADH1B | Alcohol dehydrogenase 1B (class I), beta polypeptide | |||||
| ENSG00000248144 | ADH1C | Alcohol dehydrogenase 1C (class I), gamma polypeptide | |||||
| ENSG00000196344 | ADH7 | Alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | |||||
| ENSG00000108602 | ALDH3A1 | Aldehyde dehydrogenase 3 family, member A1 | |||||
| ENSG00000132746 | ALDH3B2 | Aldehyde dehydrogenase 3 family, member B2 | |||||
| ENSG00000118514 | ALDH8A1 | Aldehyde dehydrogenase 8 family, member A1 | |||||
| ENSG00000160882 | CYP11B1 | Cytochrome P450, family 11, subfamily B, polypeptide 1 | |||||
| ENSG00000179142 | CYP11B2 | Cytochrome P450, family 11, subfamily B, polypeptide 2 | |||||
| ENSG00000148795 | CYP17A1 | Cytochrome P450, family 17, subfamily A, polypeptide 1 | |||||
| ENSG00000140505 | CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 | |||||
| ENSG00000187553 | CYP26C1 | Cytochrome P450, family 26, subfamily C, polypeptide 1 | |||||
| ENSG00000197838 | CYP2A13 | Cytochrome P450, family 2, subfamily A, polypeptide 13 | |||||
| ENSG00000108242 | CYP2C18 | Cytochrome P450, family 2, subfamily C, polypeptide 18 | |||||
| ENSG00000165841 | CYP2C19 | Cytochrome P450, family 2, subfamily C, polypeptide 19 | |||||
| ENSG00000138115 | CYP2C8 | Cytochrome P450, family 2, subfamily C, polypeptide 8 | |||||
| ENSG00000138109 | CYP2C9 | Cytochrome P450, family 2, subfamily C, polypeptide 9 | |||||
| ENSG00000197446 | CYP2F1 | Cytochrome P450, family 2, subfamily F, polypeptide 1 | |||||
| ENSG00000160868 | CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 | |||||
| ENSG00000021461 | CYP3A43 | Cytochrome P450, family 3, subfamily A, polypeptide 43 | |||||
| ENSG00000187048 | CYP4A11 | Cytochrome P450, family 4, subfamily A, polypeptide 11 | |||||
| ENSG00000162365 | CYP4A22 | Cytochrome P450, family 4, subfamily A, polypeptide 22 | |||||
| ENSG00000142973 | CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 | |||||
| ENSG00000171903 | CYP4F11 | Cytochrome P450, family 4, subfamily F, polypeptide 11 | |||||
| ENSG00000186115 | CYP4F2 | Cytochrome P450, family 4, subfamily F, polypeptide 2 | |||||
| ENSG00000186526 | CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | |||||
| ENSG00000167910 | CYP7A1 | Cytochrome P450, family 7, subfamily A, polypeptide 1 | |||||
| ENSG00000172817 | CYP7B1 | Cytochrome P450, family 7, subfamily B, polypeptide 1 | |||||
| ENSG00000100867 | DHRS2 | Dehydrogenase/reductase (SDR family) member 2 | |||||
| ENSG00000094963 | FMO2 | Flavin containing monooxygenase 2 (nonfunctional) | |||||
| ENSG00000007933 | FMO3 | Flavin containing monooxygenase 3 | |||||
| ENSG00000076258 | FMO4 | Flavin containing monooxygenase 4 | |||||
| ENSG00000145649 | GZMA | Granzyme A (granzyme 1, cytotoxic T lymphocyte‐associated serine esterase 3) | |||||
| ENSG00000100453 | GZMB | Granzyme B (granzyme 2, cytotoxic T lymphocyte‐associated serine esterase 1) | |||||
| ENSG00000158125 | XDH | Xanthine dehydrogenase | |||||
Table 4.
Phase II genes. Genes that were not detected by RNA‐seq or omitted due to extremely low counts during edgeR process are shown as blank.
| EnsemblID | Symbol | Description | Log FC | Log CPM | FDR | ||
|---|---|---|---|---|---|---|---|
| Perfused/2D | Nonperfused/2D | Perfused/Nonperfused | |||||
| ENSG00000148344 | PTGES | Prostaglandin E synthase | 4.70 | 5.06 | −0.37 | 4.83 | 2.99E‐07 |
| ENSG00000172482 | AGXT | Alanine‐glyoxylate aminotransferase | 4.32 | 3.98 | 0.34 | 2.76 | 2.16E‐06 |
| ENSG00000088002 | SULT2B1 | Sulfotransferase family, cytosolic, 2B, member 1 | 3.17 | 2.97 | 0.20 | −0.49 | 1.27E‐03 |
| ENSG00000166741 | NNMT | Nicotinamide N‐methyltransferase | 2.25 | 3.23 | −0.98 | 6.73 | 5.00E‐08 |
| ENSG00000198203 | SULT1C2 | Sulfotransferase family, cytosolic, 1C, member 2 | 1.71 | 2.47 | −0.76 | 1.88 | 4.77E‐05 |
| ENSG00000124713 | GNMT | Glycine‐N‐methyltransferase | 1.67 | 1.63 | 0.04 | −0.01 | 1.08E‐02 |
| ENSG00000198075 | SULT1C4 | Sulfotransferase family, cytosolic, 1C, member 4 | 1.65 | 1.53 | 0.12 | 2.75 | 9.40E‐05 |
| ENSG00000187210 | GCNT1 | Glucosaminyl (N‐acetyl) transferase 1, core 2 | 1.42 | −0.13 | 1.55 | 3.40 | 8.53E‐05 |
| ENSG00000130066 | SAT1 | Spermidine/spermine N1‐acetyltransferase 1 | 1.39 | 0.30 | 1.08 | 8.29 | 1.11E‐05 |
| ENSG00000172828 | CES3 | Carboxylesterase 3 | 1.35 | 1.10 | 0.25 | 3.66 | 4.72E‐05 |
| ENSG00000136881 | BAAT | Bile acid CoA: amino acid N‐acyltransferase (glycine‐N‐choloyltransferase) | 1.31 | 0.91 | 0.41 | 5.05 | 4.02E‐05 |
| ENSG00000135220 | UGT2A3 | UDP glucuronosyltransferase 2 family, polypeptide A3 | 1.12 | 1.09 | 0.02 | 0.79 | 8.30E‐02 |
| ENSG00000148154 | UGCG | UDP/glucose ceramide glucosyltransferase | 1.08 | 0.33 | 0.76 | 5.45 | 3.57E‐06 |
| ENSG00000213366 | GSTM2 | Glutathione S‐transferase mu 2 (muscle) | 0.99 | 2.03 | −1.04 | 3.76 | 6.29E‐07 |
| ENSG00000005187 | ACSM3 | Acyl‐CoA synthetase medium‐chain family member 3 | 0.91 | 1.55 | −0.64 | 3.61 | 4.27E‐05 |
| ENSG00000131446 | MGAT1 | Mannosyl (alpha‐1,3‐)‐glycoprotein beta‐1,2‐N‐acetylglucosaminyltransferase | 0.89 | 0.54 | 0.35 | 7.30 | 8.62E‐05 |
| ENSG00000151726 | ACSL1 | Acyl‐CoA synthetase long‐chain family member 1 | 0.85 | 1.37 | −0.53 | 6.10 | 1.08E‐06 |
| ENSG00000123983 | ACSL3 | Acyl‐CoA synthetase long‐chain family member 3 | 0.76 | 0.23 | 0.53 | 6.27 | 1.10E‐05 |
| ENSG00000105398 | SULT2A1 | Sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)‐preferring, member 1 | 0.54 | −1.76 | 2.31 | −0.81 | 9.08E‐02 |
| ENSG00000141744 | PNMT | Phenylethanolamine N‐methyltransferase | 0.48 | 0.39 | 0.09 | 0.42 | 4.40E‐01 |
| ENSG00000143198 | MGST3 | Microsomal glutathione S‐transferase 3 | 0.18 | 0.48 | −0.29 | 5.43 | 1.42E‐03 |
| ENSG00000257594 | GALNT4 | UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide N‐acetylgalactosaminyltransferase 4 (GalNAc‐T4) | 0.13 | 1.29 | −1.16 | 4.68 | 1.72E‐04 |
| ENSG00000066813 | ACSM2B | Acyl‐CoA synthetase medium‐chain family member 2B | 0.06 | 0.36 | −0.31 | −1.06 | 8.29E‐01 |
| ENSG00000093010 | COMT | Catechol‐O‐methyltransferase | 0.04 | 0.03 | 0.02 | 6.52 | 8.30E‐01 |
| ENSG00000134202 | GSTM3 | Glutathione S‐transferase mu 3 (brain) | −0.02 | 0.03 | −0.05 | 5.25 | 8.42E‐01 |
| ENSG00000128311 | TST | Thiosulfate sulfurtransferase (rhodanese) | −0.03 | 0.17 | −0.20 | 4.75 | 1.47E‐01 |
| ENSG00000068366 | ACSL4 | Acyl‐CoA synthetase long‐chain family member 4 | −0.04 | −0.51 | 0.47 | 7.15 | 1.03E‐04 |
| ENSG00000085871 | MGST2 | Microsomal glutathione S‐transferase 2 | −0.07 | −0.12 | 0.05 | 5.31 | 5.11E‐01 |
| ENSG00000172831 | CES2 | Carboxylesterase 2 | −0.10 | 0.27 | −0.37 | 4.85 | 3.06E‐02 |
| ENSG00000196433 | ASMT | Acetylserotonin O‐methyltransferase | −0.14 | −0.93 | 0.79 | −0.61 | 3.20E‐01 |
| ENSG00000065621 | GSTO2 | Glutathione S‐transferase omega 2 | −0.31 | −0.29 | −0.02 | 4.28 | 3.15E‐02 |
| ENSG00000197448 | GSTK1 | Glutathione S‐transferase kappa 1 | −0.33 | −0.11 | −0.21 | 5.83 | 6.86E‐03 |
| ENSG00000168765 | GSTM4 | Glutathione S‐transferase mu 4 | −0.33 | −0.14 | −0.19 | 3.96 | 1.21E‐01 |
| ENSG00000084207 | GSTP1 | Glutathione S‐transferase pi 1 | −0.35 | −0.45 | 0.09 | 6.00 | 5.48E‐03 |
| ENSG00000085998 | POMGNT1 | Protein O‐linked mannose beta1,2‐N‐acetylglucosaminyltransferase | −0.38 | −0.01 | −0.37 | 5.86 | 1.85E‐03 |
| ENSG00000130005 | GAMT | Guanidinoacetate N‐methyltransferase | −0.39 | −0.20 | −0.19 | 5.00 | 1.27E‐02 |
| ENSG00000150540 | HNMT | Histamine N‐methyltransferase | −0.46 | −0.30 | −0.17 | 5.56 | 2.70E‐02 |
| ENSG00000244038 | DDOST | Dolichyl‐diphosphooligosaccharide‐‐protein glycosyltransferase | −0.47 | −0.49 | 0.03 | 9.26 | 2.97E‐04 |
| ENSG00000196502 | SULT1A1 | Sulfotransferase family, cytosolic, 1A, phenol‐preferring, member 1 | −0.49 | −0.26 | −0.23 | 5.97 | 1.74E‐02 |
| ENSG00000147119 | CHST7 | Carbohydrate (N‐acetylglucosamine 6‐O) sulfotransferase 7 | −0.54 | −0.55 | 0.01 | 1.64 | 1.57E‐01 |
| ENSG00000120915 | EPHX2 | Epoxide hydrolase 2, cytoplasmic | −0.56 | −0.31 | −0.25 | 3.49 | 4.53E‐02 |
| ENSG00000143819 | EPHX1 | Epoxide hydrolase 1, microsomal (xenobiotic) | −0.62 | −0.14 | −0.47 | 5.03 | 1.12E‐03 |
| ENSG00000197165 | SULT1A2 | Sulfotransferase family, cytosolic, 1A, phenol‐preferring, member 2 | −0.71 | −0.17 | −0.53 | 3.00 | 7.07E‐02 |
| ENSG00000170899 | GSTA4 | Glutathione S‐transferase alpha 4 | −0.81 | −0.33 | −0.47 | 4.42 | 6.07E‐04 |
| ENSG00000148834 | GSTO1 | Glutathione S‐transferase omega 1 | −0.83 | −0.69 | −0.14 | 6.90 | 3.80E‐06 |
| ENSG00000214435 | AS3MT | Arsenic (+3 oxidation state) methyltransferase | −0.92 | −0.40 | −0.52 | 3.95 | 1.33E‐03 |
| ENSG00000171234 | UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | −1.02 | 0.15 | −1.17 | 2.23 | 2.21E‐03 |
| ENSG00000173418 | NAA20 | N(alpha)‐acetyltransferase 20, NatB catalytic subunit | −1.02 | −0.96 | −0.07 | 5.59 | 3.12E‐06 |
| ENSG00000141429 | GALNT1 | UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide N‐acetylgalactosaminyltransferase 1 (GalNAc‐T1) | −1.03 | −1.17 | 0.14 | 6.68 | 5.70E‐07 |
| ENSG00000008394 | MGST1 | Microsomal glutathione S‐transferase 1 | −1.13 | −0.49 | −0.64 | 6.33 | 1.33E‐06 |
| ENSG00000124588 | NQO2 | NAD(P)H dehydrogenase, quinone 2 | −1.17 | −0.70 | −0.47 | 4.87 | 3.92E‐06 |
| ENSG00000181019 | NQO1 | NAD(P)H dehydrogenase, quinone 1 | −1.27 | −0.70 | −0.56 | 7.46 | 2.89E‐05 |
| ENSG00000137364 | TPMT | Thiopurine S‐methyltransferase | −1.28 | −1.14 | −0.14 | 4.65 | 3.83E‐06 |
| ENSG00000168282 | MGAT2 | Mannosyl (alpha‐1,6‐)‐glycoprotein beta‐1,2‐N‐acetylglucosaminyltransferase | −1.31 | −1.49 | 0.18 | 4.68 | 8.21E‐07 |
| ENSG00000171428 | NAT1 | N‐acetyltransferase 1 (arylamine N‐acetyltransferase) | −1.65 | −0.87 | −0.78 | 1.49 | 8.77E‐03 |
| ENSG00000243955 | GSTA1 | Glutathione S‐transferase alpha 1 | −2.08 | −2.29 | 0.21 | −1.31 | 1.19E‐02 |
| ENSG00000173597 | SULT1B1 | Sulfotransferase family, cytosolic, 1B, member 1 | −4.93 | −5.69 | 0.76 | 3.47 | 1.05E‐08 |
| ENSG00000109193 | SULT1E1 | Sulfotransferase family 1E, estrogen‐preferring, member 1 | −9.13 | −9.13 | 0.00 | 0.57 | 2.27E‐05 |
| ENSG00000129673 | AANAT | Aralkylamine N‐acetyltransferase | |||||
| ENSG00000166743 | ACSM1 | Acyl‐CoA synthetase medium‐chain family member 1 | |||||
| ENSG00000171097 | CCBL1 | Cysteine conjugate‐beta lyase, cytoplasmic | |||||
| ENSG00000198848 | CES1 | Carboxylesterase 1 | |||||
| ENSG00000159398 | CES5A | Carboxylesterase 5A | |||||
| ENSG00000149124 | GLYAT | Glycine‐N‐acyltransferase | |||||
| ENSG00000174156 | GSTA3 | Glutathione S‐transferase alpha 3 | |||||
| ENSG00000182793 | GSTA5 | Glutathione S‐transferase alpha 5 | |||||
| ENSG00000134201 | GSTM5 | Glutathione S‐transferase mu 5 | |||||
| ENSG00000277656 | GSTT1 | Glutathione S‐transferase theta 1 | |||||
| ENSG00000241644 | INMT | Indolethylamine N‐methyltransferase | |||||
| ENSG00000156006 | NAT2 | N‐acetyltransferase 2 (arylamine N‐acetyltransferase) | |||||
| ENSG00000196228 | SULT1C3 | Sulfotransferase family, cytosolic, 1C, member 3 | |||||
| ENSG00000130540 | SULT4A1 | Sulfotransferase family 4A, member 1 | |||||
| ENSG00000138068 | SULT6B1 | Sulfotransferase family, cytosolic, 6B, member 1 | |||||
| ENSG00000241635 | UGT1A1 | UDP glucuronosyltransferase 1 family, polypeptide A1 | |||||
| ENSG00000244474 | UGT1A4 | UDP glucuronosyltransferase 1 family, polypeptide A4 | |||||
| ENSG00000241119 | UGT1A9 | UDP glucuronosyltransferase 1 family, polypeptide A9 | |||||
| ENSG00000173610 | UGT2A1 | UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus | |||||
| ENSG00000109181 | UGT2B10 | UDP glucuronosyltransferase 2 family, polypeptide B10 | |||||
| ENSG00000197888 | UGT2B17 | UDP glucuronosyltransferase 2 family, polypeptide B17 | |||||
| ENSG00000135226 | UGT2B28 | UDP glucuronosyltransferase 2 family, polypeptide B28 | |||||
| ENSG00000156096 | UGT2B4 | UDP glucuronosyltransferase 2 family, polypeptide B4 | |||||
| ENSG00000145626 | UGT3A1 | UDP glycosyltransferase 3 family, polypeptide A1 | |||||
| ENSG00000174607 | UGT8 | UDP glycosyltransferase 8 | |||||
Table 5.
Phase III genes. Genes that were not detected by RNA‐seq or omitted due to extremely low counts during edgeR process are shown as blank.
| EnsemblID | Symbol | Description | Log FC | Log CPM | FDR | ||
|---|---|---|---|---|---|---|---|
| Perfused/2D | Nonperfused/2D | Perfused/Nonperfused | |||||
| ENSG00000240583 | AQP1 | Aquaporin 1 (Colton blood group) | 7.81 | 7.65 | 0.16 | 3.65 | 1.45E‐06 |
| ENSG00000156222 | SLC28A1 | Solute carrier family 28 (sodium‐coupled nucleoside transporter), member 1 | 5.80 | 6.77 | −0.96 | 1.93 | 2.42E‐06 |
| ENSG00000088386 | SLC15A1 | Solute carrier family 15 (oligopeptide transporter), member 1 | 3.84 | 5.12 | −1.28 | −0.90 | 4.68E‐04 |
| ENSG00000154258 | ABCA9 | ATP‐binding cassette, subfamily A (ABC1), member 9 | 3.81 | 4.96 | −1.15 | −0.30 | 7.03E‐04 |
| ENSG00000107331 | ABCA2 | ATP‐binding cassette, subfamily A (ABC1), member 2 | 2.81 | 2.89 | −0.08 | 6.35 | 2.02E‐08 |
| ENSG00000165269 | AQP7 | Aquaporin 7 | 2.67 | 4.24 | −1.57 | 0.08 | 9.65E‐04 |
| ENSG00000059804 | SLC2A3 | Solute carrier family 2 (facilitated glucose transporter), member 3 | 2.49 | 1.77 | 0.72 | 10.64 | 6.39E‐08 |
| ENSG00000154265 | ABCA5 | ATP‐binding cassette, subfamily A (ABC1), member 5 | 1.74 | 1.89 | −0.16 | 4.89 | 1.71E‐06 |
| ENSG00000101187 | SLCO4A1 | Solute carrier organic anion transporter family, member 4A1 | 1.67 | 1.57 | 0.10 | 7.31 | 9.85E‐06 |
| ENSG00000108846 | ABCC3 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 3 | 1.66 | 1.32 | 0.33 | 6.00 | 8.97E‐07 |
| ENSG00000134294 | SLC38A2 | Solute carrier family 38, member 2 | 1.54 | 0.86 | 0.69 | 9.97 | 3.94E‐06 |
| ENSG00000141526 | SLC16A3 | Solute carrier family 16, member 3 (monocarboxylic acid transporter 4) | 1.53 | 1.23 | 0.30 | 8.84 | 1.53E‐07 |
| ENSG00000174640 | SLCO2A1 | Solute carrier organic anion transporter family, member 2A1 | 1.43 | 0.48 | 0.95 | 1.65 | 9.85E‐04 |
| ENSG00000174669 | SLC29A2 | Solute carrier family 29 (nucleoside transporters), member 2 | 1.39 | 1.20 | 0.18 | 0.20 | 2.40E‐02 |
| ENSG00000155465 | SLC7A7 | Solute carrier family 7 (amino acid transporter light chain, y + L system), member 7 | 1.32 | 1.81 | −0.48 | 5.40 | 8.39E‐07 |
| ENSG00000165029 | ABCA1 | ATP‐binding cassette, subfamily A (ABC1), member 1 | 1.18 | 1.26 | −0.08 | 6.33 | 9.56E‐05 |
| ENSG00000165240 | ATP7A | ATPase, Cu++ transporting, alpha polypeptide | 1.08 | 0.96 | 0.12 | 4.11 | 1.38E‐04 |
| ENSG00000117394 | SLC2A1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | 1.08 | 0.24 | 0.84 | 11.49 | 1.88E‐05 |
| ENSG00000115657 | ABCB6 | ATP‐binding cassette, subfamily B (MDR/TAP), member 6 | 1.05 | 1.13 | −0.08 | 6.17 | 1.07E‐06 |
| ENSG00000017483 | SLC38A5 | Solute carrier family 38, member 5 | 0.82 | 1.40 | −0.58 | 6.66 | 4.93E‐07 |
| ENSG00000163406 | SLC15A2 | Solute carrier family 15 (H+/peptide transporter), member 2 | 0.61 | 0.00 | 0.61 | −0.45 | 3.70E‐01 |
| ENSG00000103222 | ABCC1 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 1 | 0.50 | 0.26 | 0.24 | 7.39 | 3.48E‐04 |
| ENSG00000013364 | MVP | Major vault protein | 0.47 | 0.73 | −0.27 | 7.53 | 1.23E‐04 |
| ENSG00000147100 | SLC16A2 | Solute carrier family 16, member 2 (monocarboxylic acid transporter 8) | 0.46 | 0.15 | 0.31 | 2.16 | 2.28E‐01 |
| ENSG00000165637 | VDAC2 | Voltage‐dependent anion channel 2 | 0.46 | −0.01 | 0.46 | 8.19 | 1.51E‐03 |
| ENSG00000085563 | ABCB1 | ATP‐binding cassette, subfamily B (MDR/TAP), member 1 | 0.36 | −0.06 | 0.43 | 1.56 | 2.83E‐01 |
| ENSG00000101986 | ABCD1 | ATP‐binding cassette, subfamily D (ALD), member 1 | 0.31 | 0.09 | 0.22 | 5.17 | 1.33E‐02 |
| ENSG00000213585 | VDAC1 | Voltage‐dependent anion channel 1 | 0.26 | 0.03 | 0.23 | 8.31 | 1.24E‐02 |
| ENSG00000137491 | SLCO2B1 | Solute carrier organic anion transporter family, member 2B1 | 0.22 | 0.40 | −0.19 | 4.61 | 2.23E‐02 |
| ENSG00000176463 | SLCO3A1 | Solute carrier organic anion transporter family, member 3A1 | 0.19 | 0.53 | −0.33 | 0.28 | 4.53E‐01 |
| ENSG00000185883 | ATP6V0C | ATPase, H + transporting, lysosomal 16 kDa, V0 subunit c | 0.09 | −0.30 | 0.39 | 6.34 | 3.62E‐03 |
| ENSG00000167972 | ABCA3 | ATP‐binding cassette, subfamily A (ABC1), member 3 | 0.08 | 0.06 | 0.02 | 4.99 | 6.69E‐01 |
| ENSG00000124574 | ABCC10 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 10 | 0.06 | −0.02 | 0.08 | 5.09 | 6.46E‐01 |
| ENSG00000121270 | ABCC11 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 11 | 0.06 | −0.09 | 0.15 | 1.53 | 8.47E‐01 |
| ENSG00000198691 | ABCA4 | ATP‐binding cassette, subfamily A (ABC1), member 4 | 0.03 | −0.09 | 0.12 | 1.66 | 9.42E‐01 |
| ENSG00000119688 | ABCD4 | ATP‐binding cassette, subfamily D (ALD), member 4 | 0.00 | 0.52 | −0.52 | 5.08 | 4.08E‐04 |
| ENSG00000168394 | TAP1 | Transporter 1, ATP‐binding cassette, subfamily B (MDR/TAP) | −0.07 | 0.47 | −0.54 | 4.18 | 5.41E‐03 |
| ENSG00000103257 | SLC7A5 | Solute carrier family 7 (amino acid transporter light chain, L system), member 5 | −0.12 | −0.45 | 0.33 | 8.22 | 1.66E‐03 |
| ENSG00000114770 | ABCC5 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 5 | −0.25 | −0.06 | −0.19 | 5.58 | 1.61E‐01 |
| ENSG00000143921 | ABCG8 | ATP‐binding cassette, subfamily G (WHITE), member 8 | −0.29 | 0.41 | −0.70 | 0.50 | 2.40E‐01 |
| ENSG00000117479 | SLC19A2 | Solute carrier family 19 (thiamine transporter), member 2 | −0.29 | −0.34 | 0.05 | 4.68 | 2.23E‐02 |
| ENSG00000136868 | SLC31A1 | Solute carrier family 31 (copper transporters), member 1 | −0.35 | −0.56 | 0.21 | 5.77 | 1.84E‐03 |
| ENSG00000168003 | SLC3A2 | Solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | −0.59 | −1.18 | 0.58 | 7.14 | 3.69E‐06 |
| ENSG00000125257 | ABCC4 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 4 | −0.62 | −0.52 | −0.10 | 5.11 | 3.20E‐04 |
| ENSG00000123191 | ATP7B | ATPase, Cu++ transporting, beta polypeptide | −0.65 | −0.13 | −0.52 | 4.75 | 2.45E‐03 |
| ENSG00000092068 | SLC7A8 | Solute carrier family 7 (amino acid transporter light chain, L system), member 8 | −0.73 | −1.03 | 0.30 | 2.59 | 9.36E‐04 |
| ENSG00000023839 | ABCC2 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 2 | −0.84 | −0.63 | −0.22 | 8.62 | 1.38E‐04 |
| ENSG00000155380 | SLC16A1 | Solute carrier family 16, member 1 (monocarboxylic acid transporter 1) | −0.90 | −0.80 | −0.11 | 6.92 | 5.09E‐06 |
| ENSG00000103064 | SLC7A6 | Solute carrier family 7 (amino acid transporter light chain, y + L system), member 6 | −1.07 | −1.15 | 0.08 | 7.12 | 6.46E‐07 |
| ENSG00000004864 | SLC25A13 | Solute carrier family 25, member 13 (citrin) | −1.10 | −1.16 | 0.06 | 5.29 | 1.26E‐06 |
| ENSG00000204574 | ABCF1 | ATP‐binding cassette, subfamily F (GCN20), member 1 | −1.11 | −1.07 | −0.04 | 6.83 | 3.66E‐07 |
| ENSG00000112759 | SLC29A1 | Solute carrier family 29 (nucleoside transporters), member 1 | −1.23 | −1.33 | 0.11 | 6.80 | 2.45E‐07 |
| ENSG00000146477 | SLC22A3 | Solute carrier family 22 (extraneuronal monoamine transporter), member 3 | −1.24 | −0.85 | −0.39 | 3.45 | 8.53E‐05 |
| ENSG00000117528 | ABCD3 | ATP‐binding cassette, subfamily D (ALD), member 3 | −1.30 | −1.38 | 0.08 | 6.17 | 2.17E‐07 |
| ENSG00000173638 | SLC19A1 | Solute carrier family 19 (folate transporter), member 1 | −1.46 | −1.23 | −0.23 | 5.28 | 8.00E‐07 |
| ENSG00000118777 | ABCG2 | ATP‐binding cassette, subfamily G (WHITE), member 2 | −1.60 | −4.49 | 2.89 | −1.16 | 1.65E‐03 |
| ENSG00000204267 | TAP2 | Transporter 2, ATP‐binding cassette, subfamily B (MDR/TAP) | −1.96 | −1.70 | −0.27 | 4.74 | 1.17E‐06 |
| ENSG00000151012 | SLC7A11 | Solute carrier family 7 (anionic amino acid transporter light chain, xc‐ system), member 11 | −2.02 | −1.71 | −0.31 | 5.39 | 1.04E‐05 |
| ENSG00000175003 | SLC22A1 | Solute carrier family 22 (organic cation transporter), member 1 | −2.17 | −1.28 | −0.89 | −0.48 | 8.09E‐03 |
| ENSG00000144452 | ABCA12 | ATP‐binding cassette, subfamily A (ABC1), member 12 | |||||
| ENSG00000179869 | ABCA13 | ATP‐binding cassette, subfamily A (ABC1), member 13 | |||||
| ENSG00000073734 | ABCB11 | ATP‐binding cassette, subfamily B (MDR/TAP), member 11 | |||||
| ENSG00000005471 | ABCB4 | ATP‐binding cassette, subfamily B (MDR/TAP), member 4 | |||||
| ENSG00000004846 | ABCB5 | ATP‐binding cassette, subfamily B (MDR/TAP), member 5 | |||||
| ENSG00000140798 | ABCC12 | ATP‐binding cassette, subfamily C (CFTR/MRP), member 12 | |||||
| ENSG00000103569 | AQP9 | Aquaporin 9 | |||||
| ENSG00000100652 | SLC10A1 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | |||||
| ENSG00000125255 | SLC10A2 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 2 | |||||
| ENSG00000135917 | SLC19A3 | Solute carrier family 19, member 3 | |||||
| ENSG00000112499 | SLC22A2 | Solute carrier family 22 (organic cation transporter), member 2 | |||||
| ENSG00000197901 | SLC22A6 | Solute carrier family 22 (organic anion transporter), member 6 | |||||
| ENSG00000137204 | SLC22A7 | Solute carrier family 22 (organic anion transporter), member 7 | |||||
| ENSG00000149452 | SLC22A8 | Solute carrier family 22 (organic anion transporter), member 8 | |||||
| ENSG00000149742 | SLC22A9 | Solute carrier family 22 (organic anion transporter), member 9 | |||||
| ENSG00000137860 | SLC28A2 | Solute carrier family 28 (sodium‐coupled nucleoside transporter), member 2 | |||||
| ENSG00000197506 | SLC28A3 | Solute carrier family 28 (sodium‐coupled nucleoside transporter), member 3 | |||||
| ENSG00000163581 | SLC2A2 | Solute carrier family 2 (facilitated glucose transporter), member 2 | |||||
| ENSG00000138079 | SLC3A1 | Solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 | |||||
| ENSG00000100170 | SLC5A1 | Solute carrier family 5 (sodium/glucose cotransporter), member 1 | |||||
| ENSG00000100191 | SLC5A4 | Solute carrier family 5 (low affinity glucose cotransporter), member 4 | |||||
| ENSG00000021488 | SLC7A9 | Solute carrier family 7 (glycoprotein‐associated amino acid transporter light chain, bo,+ system), member 9 | |||||
| ENSG00000084453 | SLCO1A2 | Solute carrier organic anion transporter family, member 1A2 | |||||
| ENSG00000134538 | SLCO1B1 | Solute carrier organic anion transporter family, member 1B1 | |||||
| ENSG00000111700 | SLCO1B3 | Solute carrier organic anion transporter family, member 1B3 | |||||
Table 6.
Nuclear receptor genes. Genes that were not detected by RNA‐seq or omitted due to extremely low counts during edgeR process are shown as blank.
| EnsemblID | Symbol | Description | Log FC | Log CPM | FDR | ||
|---|---|---|---|---|---|---|---|
| Perfused/2D | Nonperfused/2D | Perfused/Nonperfused | |||||
| ENSG00000111424 | VDR | Vitamin D receptor | 3.11 | 4.02 | −0.91 | 2.76 | 3.96E‐06 |
| ENSG00000012504 | NR1H4 | Farnesoid X receptor; nuclear receptor subfamily 1 group H member 4 | 2.38 | 2.96 | −0.58 | 0.09 | 2.14E‐03 |
| ENSG00000106546 | AHR | Aryl hydrocarbon receptor | 0.80 | 0.97 | −0.17 | 4.06 | 5.54E‐04 |
| ENSG00000101076 | HNF4A | Hepatocyte nuclear factor 4 alpha | 0.72 | 0.89 | −0.18 | 6.26 | 1.32E‐05 |
| ENSG00000131408 | NR1H2 | Liver X receptor beta; nuclear receptor subfamily 1 group H member 2 | 0.43 | 0.31 | 0.12 | 5.01 | 2.81E‐03 |
| ENSG00000131910 | NR0B2 | Small heterodimer partner; nuclear receptor subfamily 0 group B member 2 | −0.01 | −0.73 | 0.72 | 4.34 | 4.39E‐04 |
| ENSG00000144852 | NR1I2 | Pregnane X receptor; nuclear receptor subfamily 1 group I member 2 | −0.04 | −0.08 | 0.05 | 3.48 | 8.72E‐01 |
| ENSG00000025434 | NR1H3 | Liver X receptor alpha; nuclear receptor subfamily 1 group H member 3 | −0.22 | −0.37 | 0.14 | 4.62 | 1.28E‐02 |
| ENSG00000143257 | NR1I3 | Constitutive androstane receptor; nuclear receptor subfamily 1 group I member 3 | |||||
Fig. 4.
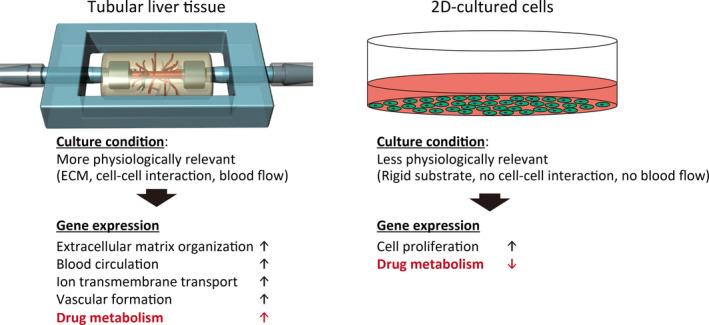
Schematic picture of the comparison between the tubular liver tissue and 2D‐cultured cells.
Conclusions
In conclusion, herein we compared the gene expression profiles of three groups: perfused and nonperfused tubular liver tissues and conventional 2D‐cultured hepatocellular carcinoma cells. Although all three groups displayed adequate differences among one another to be clearly clustered, differences resulting from culture dimensionality (i.e., tubular liver tissue or 2D‐cultured cells) were particularly significant. Furthermore, assessment of drug‐metabolizing gene expression revealed that expression patterns differed between tubular liver tissues and 2D‐cultured cells, being upregulated in the tubular liver tissue on average. The present results are potentially relevant to researchers using perfusable tubular liver tissues.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
NM and YSK conceived the study design, analyzed the data, and wrote the manuscript. YSK supervised the experimental design.
Acknowledgements
We thank Yasuko Ozaki and Tomoko Ataka for their administrative support. Computations were partially performed using the NIG supercomputer at ROIS National Institute of Genetics. We would like to thank Editage (www.editage.com) for English language editing. This study was partially supported by JSPS KAKENHI Grant Number JP18K14102 and JP18K19414, and AMED under Grant Number JP18be0304401j0002.
Data Accessibility
RNA‐seq data are available in the DNA Data Bank of Japan Sequence Read Archive under accession number DRA008972 and DRA010163 for the tubular liver tissues and 2D‐cultured cells, respectively. The raw data are available from the corresponding author upon reasonable request.
References
- 1. Takahashi Y, Mizukami Y, Tanaka Y, Nishikawa T, Ogino Y, Nishikawa M, Konishi S, Kusamori K, Mizuno N, Takakura Y et al (2017) Optimization of albumin secretion and metabolic activity of cytochrome P450 1A1 of human hepatoblastoma HepG2 cells in multicellular spheroids by controlling spheroid size. Biol Pharm Bull Pharm Bull 40, 334–338. [DOI] [PubMed] [Google Scholar]
- 2. Ramaiahgari SC, den Braver MW, Herpers B, Terpstra V and Commandeur JNM, van de Water B & Price LS (2014) A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver‐like properties for repeated dose high‐throughput toxicity studies. Arch Toxicol 88, 1083–1095. [DOI] [PubMed] [Google Scholar]
- 3. Luckert C, Schulz C, Lehmann N, Thomas M, Hofmann U, Hammad S, Hengstler JG, Braeuning A, Lampen A and Hessel S (2017) Comparative analysis of 3D culture methods on human HepG2 cells. Arch Toxicol 91, 393–406. [DOI] [PubMed] [Google Scholar]
- 4. Corstorphine L and Sefton MV (2011) Effectiveness factor and diffusion limitations in collagen gel modules containing HepG2 cells. J Tissue Eng Regen Med 5, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saheli M, Sepantafar M, Pournasr B, Farzaneh Z, Vosough M, Piryaei A and Baharvand H (2018) Three‐dimensional liver‐derived extracellular matrix hydrogel promotes liver organoids function. J Cell Biochem 119, 4320–4333. [DOI] [PubMed] [Google Scholar]
- 6. Au SH, Chamberlain MD, Mahesh S, Sefton MV and Wheeler AR (2014) Hepatic organoids for microfluidic drug screening. Lab Chip 14, 3290–3299. [DOI] [PubMed] [Google Scholar]
- 7. Mori N, Akagi Y, Imai Y, Takayama Y and Kida YS (2020) Fabrication of perfusable vascular channels and capillaries in 3D liver‐like tissue. Sci Rep 10, 5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mori N, Morimoto Y and Takeuchi S (2017) Skin integrated with perfusable vascular channels on a chip. Biomaterials 116, 48–56. [DOI] [PubMed] [Google Scholar]
- 9. Mori N, Morimoto Y and Takeuchi S (2018) Perfusable and stretchable 3D culture system for skin‐equivalent. Biofabrication 11, 011001. [DOI] [PubMed] [Google Scholar]
- 10. Guo L, Dial S, Shi L, Branham W, Liu J, Fang J‐L, Green B, Deng H, Kaput J and Ning B (2011) Similarities and differences in the expression of drug‐metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 39, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger J et al (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non‐parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87, 1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R‐R, Ueno Y, Zheng Y‐W, Koike N et al (2013) Vascularized and functional human liver from an iPSC‐derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- 13. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li B and Dewey CN (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robinson MD, McCarthy DJ and Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCarthy DJ, Chen Y and Smyth GK (2012) Differential expression analysis of multifactor RNA‐Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang DW, Sherman BT and Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang DW, Sherman BT and Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 19. Fresno C and Fernandez EA (2013) RDAVIDWebService: a versatile R interface to DAVID. Bioinformatics 29, 2810–2811. [DOI] [PubMed] [Google Scholar]
- 20. R Core Team (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- 21. Fedeli M, Riba M, Garcia Manteiga JM, Tian L, Viganò V, Rossetti G, Pagani M, Xiao C, Liston A, Stupka E et al (2016) miR‐17∼92 family clusters control iNKT cell ontogenesis via modulation of TGF‐β signaling. Proc Natl Acad Sci USA 113, E8286–E8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu G, Li F, Qin Y, Bo X, Wu Y and Wang S (2010) GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978. [DOI] [PubMed] [Google Scholar]
- 23. Zanger UM and Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138, 103–141. [DOI] [PubMed] [Google Scholar]
- 24. Bateman A (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA‐seq data are available in the DNA Data Bank of Japan Sequence Read Archive under accession number DRA008972 and DRA010163 for the tubular liver tissues and 2D‐cultured cells, respectively. The raw data are available from the corresponding author upon reasonable request.


